The presence of antibiotic-resistant bacteria at four Norwegian wastewater treatment plants: seasonal and wastewater-source effects
- Department of Chemistry, Bioscience and Environmental Engineering, Faculty of Science and Technology, University of Stavanger, Stavanger, Norway
Wastewater treatment plants receive low concentrations of antibiotics. Residual concentrations of antibiotics in the effluent may accelerate the development of antibiotic resistance in the receiving environments. Monitoring of antimicrobial resistance genes (ARGs) in countries with strict regulation of antibiotic use is important in gaining knowledge of how effective these policies are in preventing the emergence of ARGs or whether other strategies are required, for example, at-source treatment of hospital effluents. This study evaluates the presence of certain common resistance genes (blaSHV-1, blaTEM-1, msrA, ermA, ermC, tetM, tetL, tetA, vanA, and vanC) in the influent, sludge, and effluent of four wastewater treatment plants (WWTPs) in the North Jæren region of Norway at two different sampling times (January and May). These WWTPs vary in drainage area and wastewater composition and were selected based on their differing wastewater characteristics. Randomly selected colonies from the activated sludge samples were used to determine the minimum inhibitory concentration (MIC) for ampicillin, vancomycin, and tetracycline. In addition, variations in the bacterial composition of the wastewater were characterized via 16S rRNA sequencing and were analyzed in terms of bacterial host taxa that explain the presence of the ARGs in wastewater. The MIC tests revealed MIC90 values of >128 µg/mL for ampicillin, ≥128 µg/mL for vancomycin, and 32 µg/mL for tetracycline. In addition, the three resistance genes, ermB, tetA, and tetM, that were present in the influent and activated sludge were still present in the effluent. These results indicate that WWTPs represent a direct route into the environment for resistance genes and do not significantly reduce their abundance. Hence, the development of treatment methods for the removal of these genes from WWTPs in the future is of utmost importance.
1 Introduction
The rapidly increasing global development of antimicrobial resistance (AMR) represents a threat to health worldwide. It is predicted that antibiotic-resistant bacteria (ARBs) will become one of the leading causes of death and their development will lead to major financial losses, especially in low- to middle-income countries (Morrison and Zembower, 2014; Durand et al., 2019). Identifying and limiting probable locations for the development and distribution of ARBs is pivotal in gaining understanding of them and in the development of new strategies and policies to help prevent further resistance development. Wastewater treatment plants (WWTPs) have been identified as hotspots for and distributors into the environment of antibiotic resistance genes (ARGs) (Karkman et al., 2018; Pazda et al., 2019). When humans or animals ingest antibiotics, large amounts are excreted intact via urine and can enter the sewer system (Kümmerer, 2009). Gut bacteria from humans and animals are a major source of ARGs (Kent et al., 2020). Hence, it is very likely that WWTPs receive a constant stream of low levels of both antibiotics and ARBs. WWTP design has focused on the removal of organic matter, nitrogen, and phosphorous from the influent, with the removal of ARGs and ARBs as a lower priority (Pruden, 2014).
Wastewater is rich in nutrients (Morée et al., 2013), contains a diverse and dense microbial community (Qin et al., 2018), and often contains low, sublethal doses of antibiotics (Chow et al., 2021), rendering it a good environment for the transfer of ARGs. Antibiotic concentrations below the minimum inhibitory concentration (MIC) are known to lead to selectivity for ARGs. The minimal selective concentration (MSC) can be much lower than the MIC. Gullberg et al. (2011) found that the MSC values of various antibiotics could be anywhere from 1/4 to 1/230 of the MIC. In addition, exposure to antibiotics can result in increased mutation rates due to the SOS response (Chow et al., 2021). ARGs can be transferred between species by horizontal gene transfer (HGT) mechanisms such as integrons, plasmids, and transposons (Abby et al., 2012; Redondo-Salvo et al., 2020). It has been shown in the case of the human gut microbiota that HGT can play a major role in the spread of ARGs (Kent et al., 2020; Li et al., 2020).
Recently, the use of sewage systems for monitoring of ARGs has garnered interest (Kwak et al., 2015; Hutinel et al., 2019; Huijbers et al., 2020). By investigating the microbial communities and the distribution of ARGs present in wastewater, the probable abundance and types of ARBs that may be circulating in that area may be determined. Norway has relatively low antibiotic consumption and abundance of ARBs (World Health Organization, 2018), and has decreased its consumption of antibiotics further below the European average in 2021 (European Centre for Disease Prevention and Control (ECDC), 2021). Norway therefore represents a good example of a country on the lower end of antibiotic consumption (World Health Organization, 2018). To determine whether decreased antibiotic consumption affects the ARBs present in WWTPs, it is important to investigate the antimicrobial resistance of genes and bacteria circulating in Norwegian WWTPs. This study aimed to evaluate the presence of ARGs of concern (blaSHV-1, blaTEM-1, msrA, ermA, ermC, tetA, tetL, tetM, vanA, and vanC) in the inlet, the sludge produced, and the effluent of four Norwegian WWTPs with differing wastewater characteristics, catchment area activities, and process configurations. To evaluate seasonal changes in the abundance of ARGs and ARBs, two contrasting months, one in the winter and one in the spring, were selected for sampling. In addition, the bacterial community composition was sequenced for detailed mapping of the microorganisms present and to assess changes in the microbial communities. Finally, random colonies were isolated, and minimum inhibitory concentration tests for ampicillin, tetracycline, and vancomycin were conducted to understand the dynamics of resistance to popular antibiotics.
2 Methods
2.1 Study sites and sample collection
Grab samples were obtained from four different WWTPs [Vik RA, Bore, Grødaland, and Mekjarvik (Sentralrenseanlegg Nord-Jæren [SNJ])] in the North Jæren region of southwest Norway. One-liter samples were taken from the wastewater inlet and outlet and from the produced sludge at two separate sampling times in January and May 2022. These two months were selected to represent the beginning and end of the winter flu season. The WWTPs were selected to reflect variability in wastewater composition, mass loadings, industrial to municipal wastewater fraction, and treatment process configuration. Of particular interest was the ratio of agro-industrial to municipal household loading. Mekjarvik (SNJ) represents typical urban municipality wastewater; Vik RA, mixed municipal and food industry wastewater; Grødaland, agro- and food industry-dominated wastewater; and Bore, a typical rural municipal sewage treatment plant. Refer to Supplementary Table S1 for details and Supplementary Figure S1 for the locations. Samples were stored in cool bags and transported by car to the laboratory immediately after sampling; biological and analytical tests were conducted immediately upon arrival at the laboratory.
2.2 Wastewater compositional analysis
Total suspended solids (TSS) was measured following the standard method 2540D (Lipps et al., 2022) using duplicate filtered samples (preweighed glass fiber filter, pore size 0.45 μm, diameter 47 mm, 934-AH RTU; Whatman, Kent, UK), which were dried at 105°C for 1 h and weighed analytically (XP205, Mettler Toledo, Switzerland). Volatile suspended solids (VSS) was determined following the standard method 2540E using the same filters as for the TSS analysis, with the retained particles combusted at 550°C for 30 minutes (Rice et al., 2013).
2.3 Isolation of strains
Strains were isolated for the MIC tests by diluting 1 g of centrifuged activated sludge in 99 mL Mueller–Hinton broth. A 1:10 serial dilution series was then spread on Mueller–Hinton agar plates and incubated overnight at room temperature (20°C–25°C) (as the WWTPs were not operated at 37°C). The procedure was adapted with modifications from Reddy et al. (1986). As not all colonies could be tested, the next morning, 24 colonies were randomly picked and grown in liquid Mueller–Hinton broth, and a cryostock for each of the isolated strains was stored in a freezer at −80°C.
2.4 Antibiotic susceptibility testing
The MICs of antibiotics for the 24 bacterial isolates from each sludge sample were determined by a standard twofold serial broth dilution method using Mueller–Hinton broth in 96-well plates (Greiner Bio-One, Kremsmünster, Austria). This procedure was conducted according to the international standard ISO 20776-1:2019 (212, T. C. I, 2019) and Wiegand et al. (2008) using antibiotic concentrations ranging from 0.25 to 128 mg/L. The inoculum was grown to a turbidity of 0.5 McFarland standard, measured using a photometer (Genesys 50; Thermo Fisher Scientific, Waltham, MA, USA). The 96-well plates were then inoculated and incubated for 20 h at 25°C. In this study, three antibiotics were tested: ampicillin (AMP) (Sigma-Aldrich, St. Louis, MO, USA), tetracycline (TET) (Sigma-Aldrich), and vancomycin (VAN) (Millipore, Billerica, MA, USA). Ampicillin was included as it is extensively used in human and veterinary medicine (Klein et al., 2021); tetracycline, on the basis that it is one of the major antimicrobials used in animal husbandry (Rushton et al., 2014); and vancomycin, as it is one important treatment for methicillin-resistant Staphylococcus aureus (Shariati et al., 2020). In addition to the varying antibiotic concentrations, growth was verified via positive and negative wells, which contained no antibiotic and no bacterial isolate, respectively.
2.5 DNA extraction and PCR
DNA was extracted from activated sludge and water samples upon arrival at the laboratory. For each sample, 50 mL activated sludge was taken and centrifuged at 5,000 g for 10 minutes (Falcon tube), and 0.3 mg of the pellet was used for DNA extraction. Subsequently, 50 mL water samples were filtered through autoclaved OE66 membrane filters (cellulose acetate, pore size 0.2 μm, diameter 47 mm; Whatman, UK) cut into small pieces. DNA extraction was then conducted using the DNeasy PowerSoil Pro Kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. A NanoVue™ Plus Spectrophotometer (GE Healthcare, Chicago, IL, USA) was used to quantify the extracted DNA and ensure the quality of DNA using an A260/280 measurement of approximately 1.8.
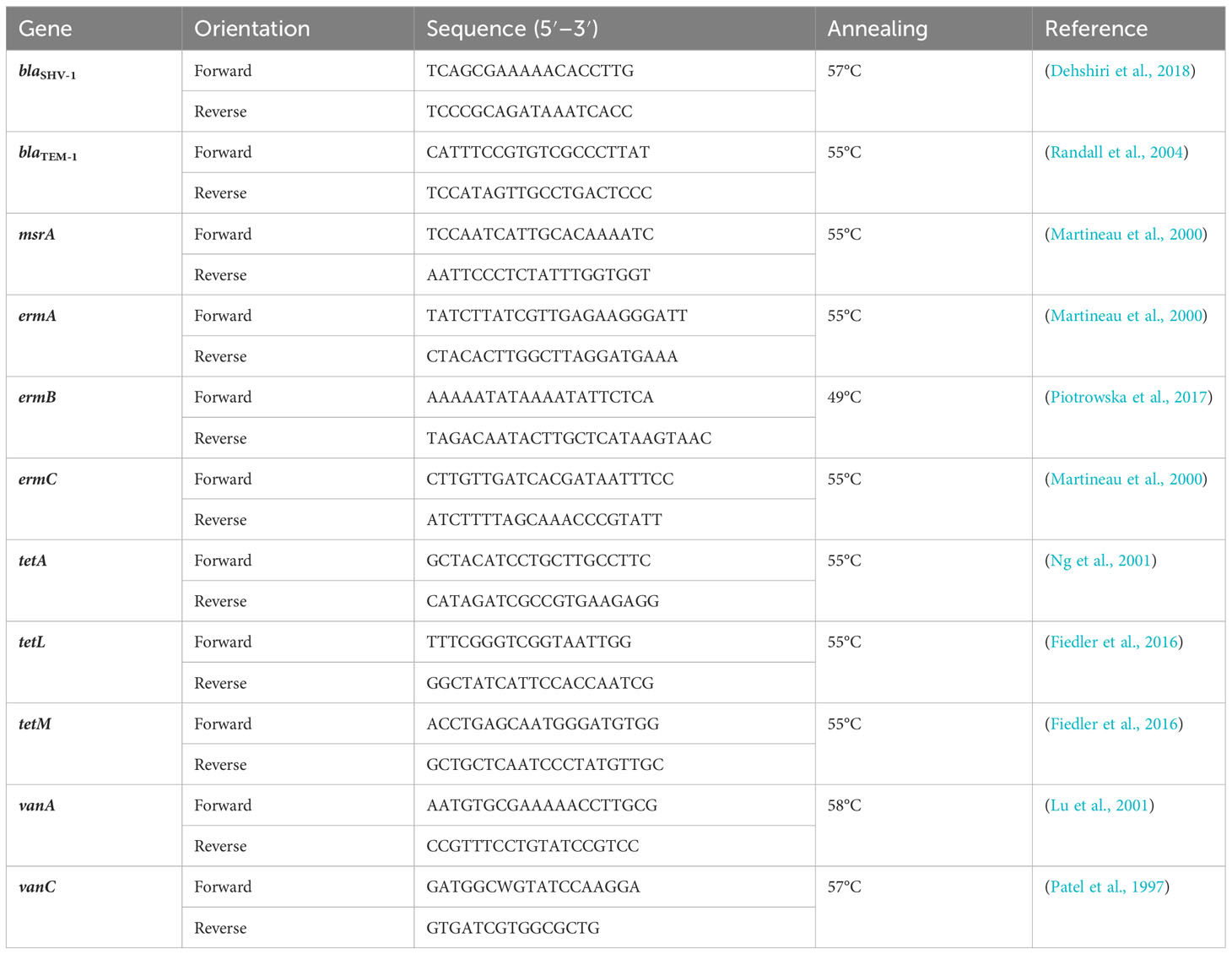
A 20-µL final volume was used for polymerase chain reaction (PCR) amplification. The PCR mix contained 2 µL 10x Key Buffer Mg2+ free (VWR, Radnor, PA, USA), 1 µL dNTP, 0.3 µL of each primer (20 µM), Taq DNA Polymerase (Invitrogen, Carlsbad, CA, USA), 1 µL template DNA, and 15.7 µL molecular-grade water (Mediatech, Inc., Manassas, VA, USA). A 2720 thermal cycler (Applied Biosystems, Foster City, CA, USA) was used to amplify the targeted sequences. The sequences of the PCR primers and the amplification cycles used are presented in Table 1. All samples from the four WWTPs were tested for resistance genes with positive and negative controls.
The genes were selected based on common resistance genes present in WWTPs and after reviewing the literature (Özkök, 2012; Jang et al., 2018; Hubeny et al., 2019; Pallares-Vega et al., 2019; Peirano and Pitout, 2019; Mirkalantari et al., 2020). Table 2 shows the corresponding gene family and the drugs they work against.
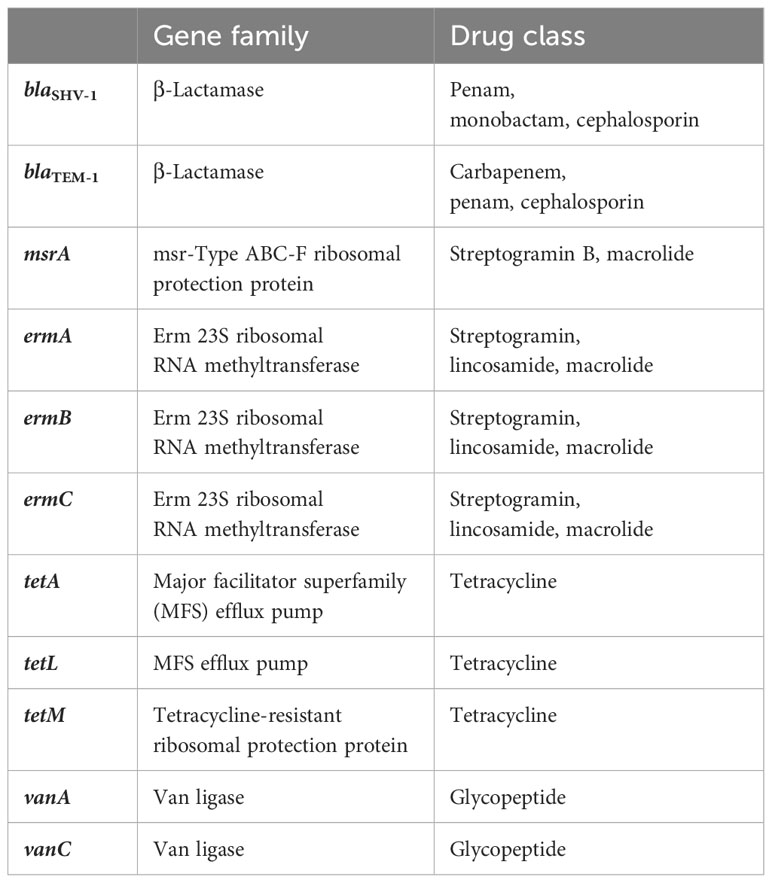
Table 2 Antibiotic resistance genes, their gene families, and the target antibiotic classes against which resistance is conveyed.
2.6 Sequencing
Extracted DNA from the inlet, activated sludge, and effluent was normalized to ~100 ng/µL, frozen at −20°C until all samples had been collected, and subsequently submitted to Novogene (UK) Company Limited (Cambridge, UK) for external sequencing.
The V3–V4 region of the 16S rRNA gene for bacterial DNA was targeted using the B-341F (5′-CCTAYGGGRBGCASCAG) and B-806R (5′-GGACTACNNGGGTATCTAAT) primers amplifying 470 base pairs (bp). The library was created by “sequencing by synthesis” using the Illumina NovaSeq 6000 platform, and 250-bp conducted paired-end reads were then created by Novogene. Raw reads were assigned to samples and merged using the FLASH (fast length adjustment of short reads) software. The clean tags were clustered into operational taxonomic units (OTUs) with 97% similarity (Youssef et al., 2009; Caporaso et al., 2011; Hess et al., 2011). The sequencing data can also be found in the European Nucleotide Archive (accession number PRJEB70859).
3 Results
3.1 Minimum inhibitory concentration of strains
The antibiotic resistance levels for tested bacteria obtained from randomly selected colonies from the 16 sludge samples, displayed as MIC50 and MIC90, are presented in Table 3. MIC50 and MIC90 respectively represent the MIC values at which 50% and 90% of the isolates were inhibited by the administered antibiotic concentrations. The individual values for each isolate can be found in Table S3.
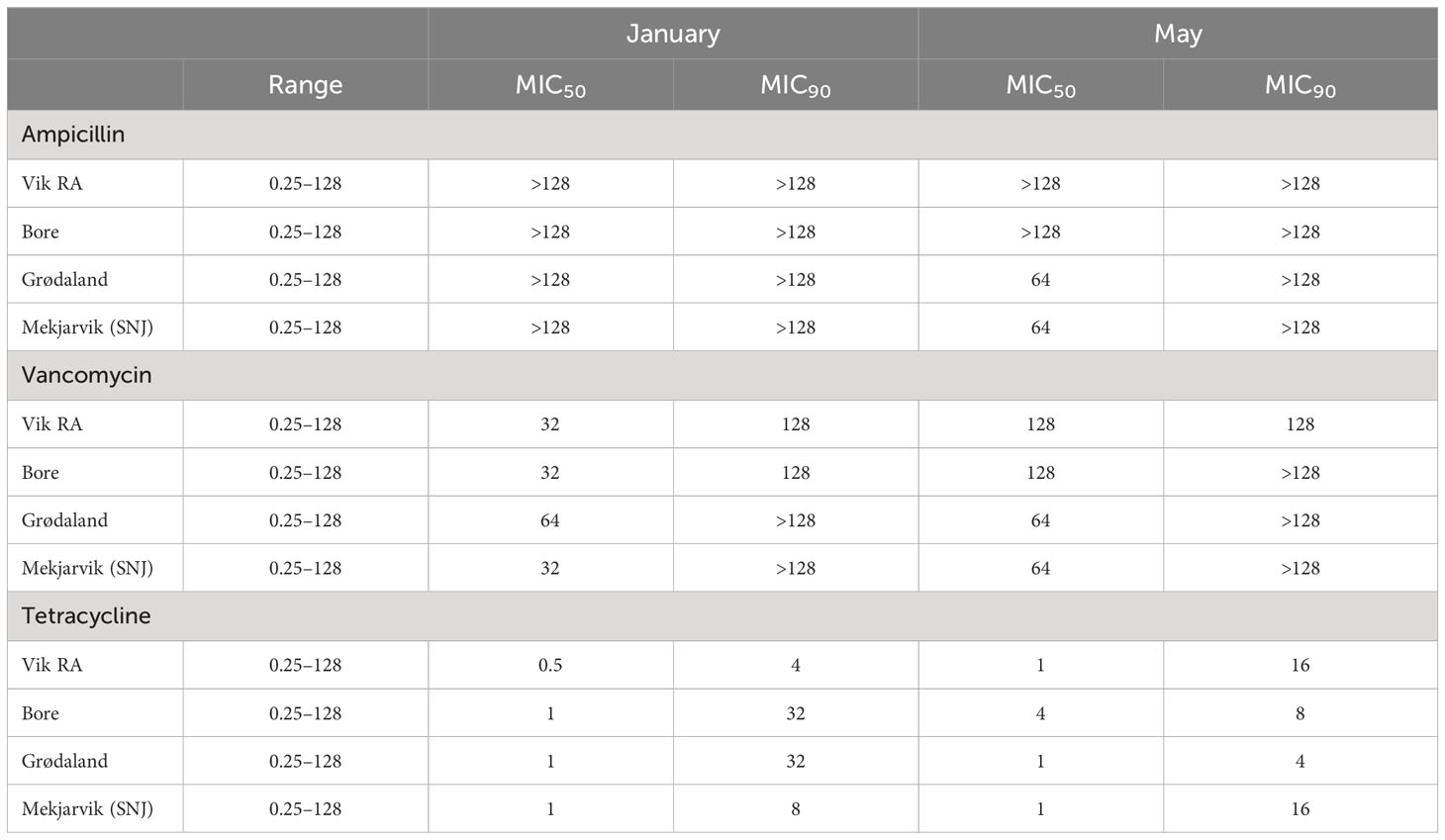
Table 3 MIC50 and MIC90 values for the antibiotics ampicillin, vancomycin, and tetracycline in mg/L, representing the MIC value that includes 50% or 90% of the samples, respectively.
The MIC50 and MIC90 values for ampicillin were above the measured concentration range of 128 µg/mL in all WWTPs in January. The same result was observed in May for the Vik RA and Bore WWPTs. For the other two WWTPs, the MIC50 values decreased to 64 µg/mL in May, exhibiting a reduction in overall ampicillin resistance.
In general, an increase in vancomycin susceptibility could be observed from January to May. The MIC90 for vancomycin remained the same from January to May except in Bore, where it increased from 128 µg/mL to above the tested values. The MIC50 values increased in Vik RA and Bore to 128 µg/mL in May, and to 64 µg/mL in Mekjarvik (SNJ). The WWTPs in Vik RA and Mekjarvik (SNJ) had higher MIC90 values in May. In Bore and Grødaland, a reduction in the top 10% occurred.
The MIC50 and MIC90 values for tetracycline were far lower than those for ampicillin and vancomycin at both sampling times and at all four WWTPs: the MIC50 values were 1 µg/mL in January and May, with the exception of Vik RA in January, where the value was 0.5 µg/mL, and Bore in May, with 4 µg/mL. There was more variability in values of MIC90. In January, Bore and Grødaland had the highest values at 32 µg/mL, whereas Vik RA had the lowest at 4 µg/mL. In May, the highest values were in Vik RA and Mekjarvik (SNJ) at 16 µg/mL, whereas the lowest was in Grødaland at 4 µg/mL.
3.2 Antibiotic resistance genes
The presence or absence of the resistance genes blaSHV-1, blaTEM-1, msrA, ermA, ermC, tetA, tetL, tetM, vanA, and vanC was examined via PCR testing; the results are presented in Table 4. The majority of the resistance genes were absent from all samples tested. However, ermB, tetA, and tetM were found in all samples, with the exceptions of the outlet samples taken from Vik RA and Mekjarvik (SNJ) in May. In Vik RA, none of the three resistance genes (ermB, tetA, and tetM) were found. In Mekjarvik (SNJ), only ermB was present. The msrA resistance gene was found in the samples taken from Bore in May and from Grødaland in both January and May, except in the case of the sludge sample taken in January, where the msrA gene was not found. Common resistance genes such as blaTEM-1 were not found in any of the samples.
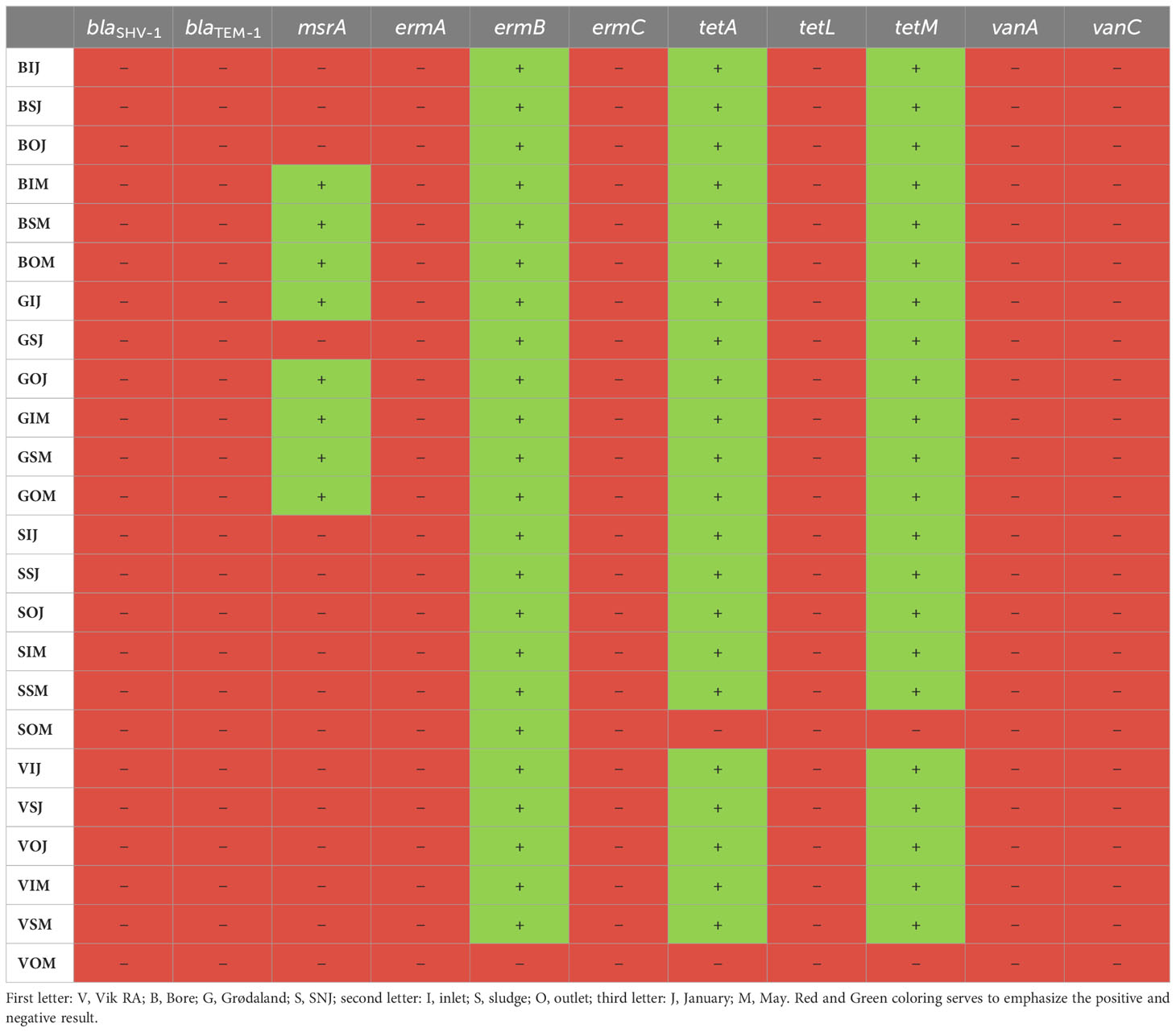
Table 4 The presence of different antibiotic resistance genes at the WWTPs in January and May, as tested via PCR.
3.3 Microbial community
The sequencing data from the four WWTPs exhibited a Good’s coverage of at least 0.95 for all samples. The Shannon index was slightly higher for the May samples (5.9–9.8) than for the January samples (5.5–7.5). The number of OTUs over all samples ranged from 750 to 2,700. In general, the number of OTUs was higher in the sludge samples than in the inlet and outlet samples. The exceptions to this were the May samples from Vik RA and Mekjarvik (SNJ). The inlet and outlet samples for both WWTPs had numbers of OTUs at least 45% higher than those of the sludge samples. The precise Shannon and Chao1 values and the coverage can be found in Supplementary Table S4.
Figure 1 presents the most abundant phyla from the four WWTPs, Vik RA, Bore, Grødaland, and Mekjarvik, in January and May, with at least 1% relative abundance of the total population. The five most abundant phyla were Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Campylobacterota, with varying relative abundance. In addition to the five most common phyla, Acidobacteria could be detected in all the sludge samples, with an average abundance of 2.8%. This phylum was also detected in the May inlet sample from Mekjarvik and in the outlet sample from Vik RA. The phylum also Fusobacteriota was present, with higher relative abundance in the inlet samples from Vik RA (4% and 3%) and Bore (2% and 2%) in January and May and from Mekjarvik (2%) in January. Fusobacteriota was also detected in the outlet samples with lower than 1% abundance, with the exception of Bore in January, with an abundance of 3%. The noticeable outlier was the January sample from Mekjarvik, where the most dominant phylum was Archaea Halobacterota, with 28% abundance. In addition, the phyla Synergistota, Cloacimonadota, Desulfobacterota, Chloroflexi, Chermotogota, and Fermentibacterota, were also present, with very low relative abundance in most samples. The aforementioned phyla appeared in proportions between 1% and 4% in the same sample from Mekjarvik.
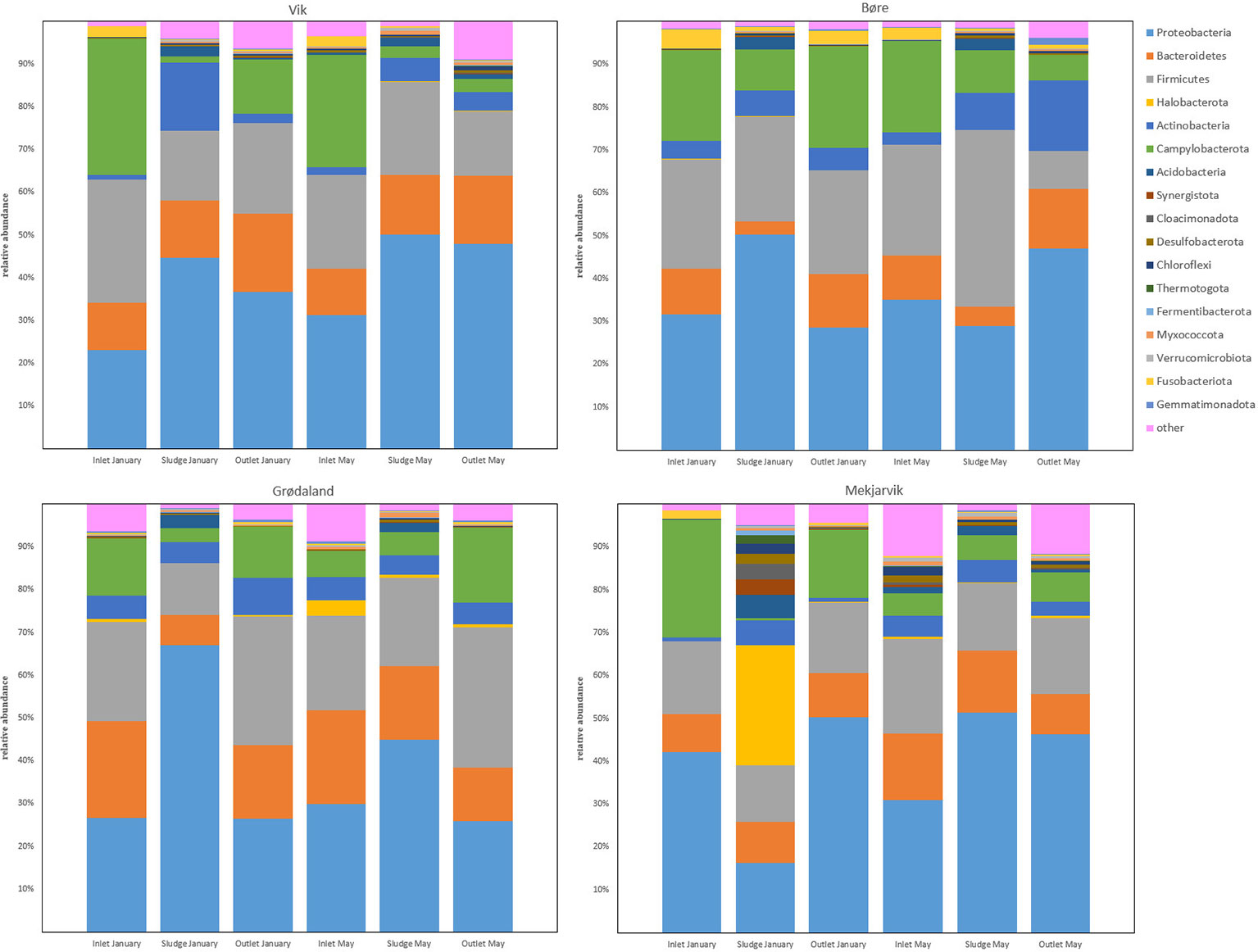
Figure 1 The common bacterial phyla with more than 1% relative abundance in at least one sample obtained. The legend indicates the color corresponding to each phylum.
The most common genera at Vik RA were Acidovorax, Acinetobacter, Agitococcus, Arcobacter, Bacteroides, Flavobacterium, Lactococcus, Hydrogenophaga, Paludibacter, Pseudarcobacter, Rhodoferax, and Trichococcus. The 13% proportion of Acinetobacter in the inlet samples (January and May) was relatively high, but its relative abundance fell in the sludge and the outlet water samples to 1% and 3% in January and 2% and 4% in May, respectively. Agitococcus was only detected at low levels (<0.2%) in the inlet and had a higher relative abundance in the outlet samples, at 2% in January and 6% in May. Lactococcus was present at 10% and 8% in the inlet in January and May, respectively, and its relative abundance was reduced to 2% and 1% in the outlet samples, respectively. Rhodoferax was present at 1% in the inlet, which increased to 10% and 8% in the sludge and 13% and 9% in the outlet in January and May, respectively.
At Bore, the most common genera were Acidovorax, Acinetobacter, Aeromonas, Arcobacter, Lactococcus, Pseudarcobacter, and Trichococcus. In January, Acinetobacter was present with a relative abundance of 18% in the inlet and 26% in the sludge, which decreased to 10% in the outlet. In May, the relative abundances were 23%, 6%, and 5% in the inlet, sludge, and outlet samples, respectively. These results indicate a large difference in the relative abundance of Acinetobacter in the sludge in January compared with May. Lactococcus exhibited a drastic reduction in the relative abundance between the sludge and outlet samples in May, in which the overall abundance fell from 13% in the sludge to 0.7% in the outlet. This decrease was not observed in January samples, where Lactococcus fell from 11% to 8%. Leucobacter, which was only present in the inlet samples at <2.2%, was present in the May outlet at 10%. Although the relative abundances of Pseudarcobacter in the inlet and sludge were comparable in the January and May samples, in the outlet samples, more variation was observed, with the abundance of Pseudarcobacter at 14% in January, decreasing to 3% in May.
Figure 2 shows the most abundant bacterial genera from the four WWTPs Vik RA, Bore, Grødaland and Mekjarvik in January and May with at least 1% relative abundance of the total population. The most common genera found at Grødaland were Acinetobacter, Aeromonas, Arcobacter, Brachymonas, Chryseobacterium, Lactococcus, Leucobacter, Prevotella, Pseudarcobacter, Trichococcus, and Zoogloea. Compared with the other WWTPs, Acinetobacter comprised a lower proportion of all samples, at approximately 2% of the community. Another outlier was Brachymonas, which was present with an abundance >2.4% only in the Grødaland samples. The abundance of Brachymonas as a percentage of the total community decreased from 8% and 11% in January to 7% and 6% in May at Grødaland. Its proportion in the sludge was 40% in January and 21% in May. Trichococcus was also present in higher proportions at Grødaland compared with the other WWTPs, at 13% and 8% in January and May in the inlet, and exhibited a further increased in the outlet at 21% and 20%.
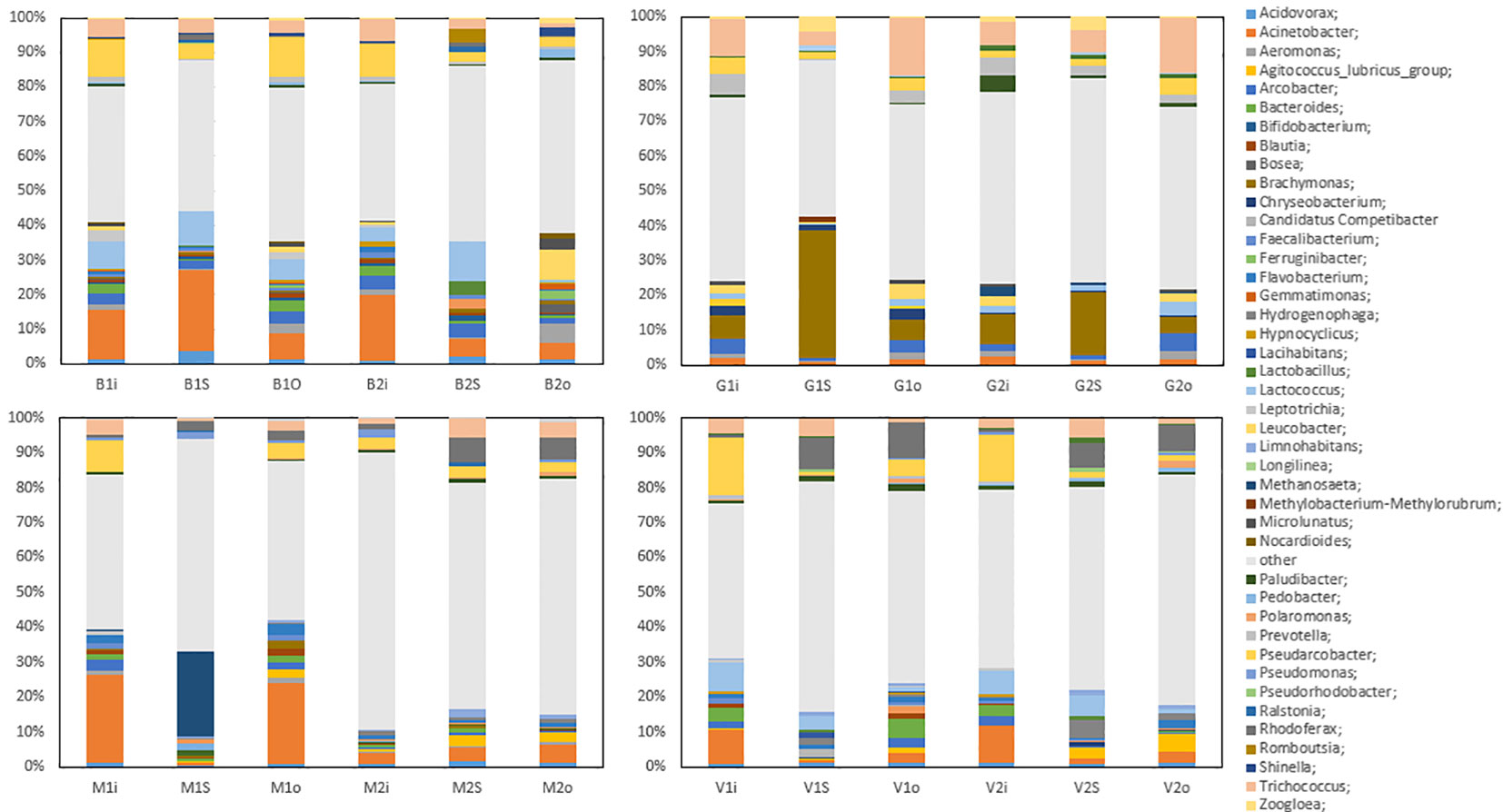
Figure 2 The common bacterial genera with more than 1% relative abundance in at least one sample obtained. The abbreviations B, G, S, and V represent the four WWTPs: Bore, Grødaland, SNJ, and Vik RA, respectively. The legend indicates the color corresponding to each genus.
At Mekjarvik (SNJ), the most common genera were Acinetobacter, Methanosaeta, Pseudarcobacter, Rhodoferax, and Trichococcus. Acinetobacter was found in high proportions in the January samples, at 29% in the inlet and 26% in the outlet. The main outlier was the Archaea genus Methanosaeta, at 25% in the January sludge sample, corresponding to the Halobacterota phylum.
Of the pathogens from the ESKAPEE group (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli), only Enterococcus faecium and E. coli were detected. E. faecium was at less than 1% in all inlet and outlet samples (both months), and E. coli was at less than 1% in all inlet, outlet, and sludge samples (both months).
4 Discussion
To determine whether wastewater influents have an effect on the microbial community and resistance pattern, samples were collected from four WWTPs in Rogaland County, Norway, which received wastewater from areas with varying population densities and industries.
The OTU values (750–2,700) and the Shannon index (5.5–9.8) (Supplementary Table S4) indicated that the WWTPs investigated in this study contained a rich and diverse bacterial community. The literature on WWTP microbial communities reports OTU counts in the range of 750–970 and Shannon index values of 3.7–5.7, which are lower than those observed for the microbial communities in this study, demonstrating that the microbial communities of the WWTPs in this study were very diverse (Wolff et al., 2018; Do et al., 2019; Yang et al., 2020). The WWTPs at Mekjarvik (SNJ) and Vik RA, receiving municipal wastewater with higher population equivalents, had the highest OTUs (particularly in May).
Many of the ARGs that are commonly present, such as the β-lactamases blaSHV-1 and blaTEM-1 (Peirano and Pitout, 2019; Mirkalantari et al., 2020), were not detected at the four treatment plants in this study. Only the msrA, ermB, tetA, and tetM genes were detected via PCR assay. Exceptions to this were the outlet samples from Vik RA and Mekjarvik (SNJ) in May. In the Vik RA sample, no resistance genes were detected, and in the case of Mekjarvik (SNJ), only ermB was detected. For the most part, when these genes were present in the influent, they were not removed during the treatment process and were still present in the effluent, and were thus released into the surrounding environments. This was expected, as none of the four treatment plants was designed with the removal of ARGs in mind. Putting this into perspective, other Norwegian WWTPs have been found to have a higher prevalence of ampicillin resistance than WWTPs in the Rogaland region. In a study conducted in the Oslo region, blaTEM-1 and tetM were the only resistance genes found; this finding concurs with the low number of resistance genes found in this study, and both genes could be found in the influent and effluent of one Oslo-based WWTP (Cacace et al., 2019). Cacace et al. (2019) also investigated six additional ampicillin resistance genes, which were not tested in our study, and only blaOXA-58 was found to be present in higher numbers than blaTEM-1, with tetM numbers being comparable to those observed for blaOXA-58 in Oslo. This observation also applied to other European WWTPs tested in the study, such as in Germany and the Netherlands (Cacace et al., 2019). In Bergen, Radisic et al. (2023) examined K. pneumoniae at five different WWTPs and found that the most common forms of resistance among K. pneumoniae were against ampicillin and azithromycin. The most common ampicillin resistance genes originated from the extended-spectrum β-lactamase (ESBL) family CTX-M with several SHV and TEM representatives (Radisic et al., 2023), with the latter not being detected in Rogaland. Alexander et al. (2020) studied different WWTP clusters (communal, food industry, and hospital) and found that more than 99% of ARGs consisted of ermB, tetM, blaTEM, and sul1, with slightly higher abundances observed in hospital wastewater. The wastewater was also tested for vanA, which was categorized as rarely occurring (Alexander et al., 2020). These results correspond to our findings in terms of the presence of ermB and tetM in nearly all samples and the absence of vanA.
The MIC50 and MIC90 values indicated higher resistance to ampicillin and vancomycin. The ampicillin resistance observed in this study is very common, as ampicillin resistance occurs naturally in soil bacteria, and the number of variants for blaSHV and blaTEM combined exceeds 450 (NCBI, 2023a; NCBI, 2023b). As these were not found in our study, it can be assumed that the ampicillin resistance was conferred by a different resistance gene, for example, from the CTX-M gene family (Peirano and Pitout, 2019). As the two tested vancomycin resistance genes were not found in the samples, the high MIC values can be explained by either the presence of untested resistance genes or the presence of mostly gram-negative bacteria. Vancomycin’s main targets are gram-positive bacteria, and it has nearly no effect on gram-negative bacteria (Mühlberg et al., 2020). In Buenos Aires, Argentina, the MIC50 and MIC90 values were compared for ampicillin and vancomycin in hospital and municipal wastewater. For vancomycin, both values for both sampling sites exceeded 960 mg/L. Ampicillin in the municipality wastewater was also >960 mg/L, whereas in the hospital wastewater, the MIC50 value was 240 and the MIC90 value was 480 mg/L (Nuñez et al., 2016). Li et al. (2009) studied bacterial strains from a WWTP treating penicillin production wastewater. For ampicillin, the MIC50 and MIC90 values were >1,024 mg/L. For tetracycline, the MIC50 value was 8 mg/L and the MIC90 value was 512 mg/L (Li et al., 2009). MIC values for ampicillin are challenging to use as a point of comparison, as the values exceeded the established scale set by the experiment. In Norway, the measured values were lower for vancomycin at MIC50 and tetracycline at MIC50 and MIC90.
The low MIC values and the detection of only a small number of resistance genes indicate that the maintenance of human consumption of antibiotics below the European average, along with Norway being a country with one of the lowest levels in Europe of antibiotic consumption by food-producing animals, is a successful strategy for reducing the presence of antibiotic resistance genes in wastewater (European Centre for Disease Prevention and Control (ECDC), EFSA and EMA, 2017; European Centre for Disease Prevention and Control (ECDC), 2021). Comparing these results with those obtained in Germany, which has a comparable human development index and a lower human community use of antibiotics, but has considerably higher antibiotic use in food-producing animals (European Centre for Disease Prevention and Control (ECDC), EFSA and EMA, 2017; European Centre for Disease Prevention and Control (ECDC), 2021), certain similarities and differences are recognizable. Alexander et al. (2020) studied different-sized WWTPs in Germany with municipal, food, and hospital wastewater. They demonstrated that, independently of the catchment area, all WWTPs were contributing to the release of ARGs into the environment. The greatest difference was detected in hospital wastewater, where a stronger correlation between ARGs and facultative pathogenic bacteria was observed (Alexander et al., 2020). Pärnänen et al. (2019) examined ARGs in urban wastewater treatment plants (UWTPs) in different European countries via qPCR. The Norwegian UWTP (eastern Norway) exhibited very low numbers of AMP and VAN resistance genes in the inlet and outlet. This is consistent with our detection of no AMP or VAN resistance genes via PCR. They also concluded that antibiotic consumption had a lower impact than external environmental factors (e.g., temperature) (Pärnänen et al., 2019). This is also concordant with the findings of this study and the study by Alexander et al. (2020), with results indicating that the source of the wastewater did not influence the resistome strongly but rather that temperature is a controlling factor. MacFadden et al. (2018) demonstrated by studying three common bacterial pathogens (E. coli, K. pneumoniae, and S. aureus) in the USA that an average increase of 10°C can lead to an increase in ARGs in this population by up to 10%. McGough et al. (2020) demonstrated using the same bacterial pathogens (E. coli, K. pneumoniae, and S. aureus) that higher ambient temperatures favor antibiotic resistance in E. coli and K. pneumoniae (McGough et al., 2020).
4.1 Microbial analysis
Typical phylum compositions observed at WWTPs with different catchment areas in China, Canada, the USA, and Germany (Hu et al., 2012; Zhang et al., 2012; Ju et al., 2014; Wolff et al., 2018; Ma et al., 2023) are similar to the those observed for all four WWTPs in this work. This includes the five most common phyla, Proteobacteria, Firmicutes, Bacteroidetes, Campylobacter, and Actinobacteria, along with phyla such as Acidobacteria and Verrucomicrobia.
Fusobacteriota was found to comprise more than 1% of the OTUs detected in the inlet samples from the Bore, SNJ, and Vik RA WWTPs in January and May and was present in the outlet sample from Bore in May. The Fusobacteriota phylum has recently been found to be statistically associated with carbapenemase-encoding genes in hospital wastewater and its members have been identified as potential ARG reservoirs (Hubeny et al., 2021). Most of the fusobacteria detected in this study were determined to be composed of the pathogen Leptotrichia, which occurs in the human oral cavity, among other biomes (Eribe and Olsen, 2017). Quite recently, Leptotrichia has also been found in WWTPs in Finland and South Africa (Leiviskä and Risteelä, 2022; Poopedi et al., 2023). As the Leptotrichia genus can be involved in opportunistic infections through blood or oral transmission (Eribe and Olsen, 2017) and may be a reservoir for ARGs, it is therefore important for public health. Leptotrichia was reduced in the outlet on average from 1.2%–2.2% to <0.6% in SNJ and Vik RA. The percentage of Leptotrichia at the Bore WWTP was reduced from 3.7% to 1.8%. Bore, unlike the other WWTPs included in this study, does not include secondary treatment (Supplementary Table S1). Although Fusobacteriota has previously been found to be associated with common carbapenemases, such as blaTEM, this gene was not found at any of the WWTPs in this study.
One exception observed in the sequencing results when compared with the other three WWTPs in this study was the high number of the Archaea phylum Halobacterota found at the SNJ WWTP in January, comprising 28% of the total population. The largest part of the Halobacterota community was found to be composed of the genus Methanosaeta, comprising 25% of the total microbial community. Methanosaeta is normally found in anaerobic digestion reactors and in upflow anaerobic sludge blanket (UASB) reactors, as it is rather effective in adhesion and granulation (Liu and Whitman, 2008). We assume the reason that Methanosaeta represents such a high proportion of the microbial community at SNJ is the anaerobic digester return flow following sludge dewatering of digested sludge, hence leading to the presence of the Halobacterota phylum in the activated sludge despite the fact that the aerobic environment is not suited to this phylum.
The genus Acinetobacter has often been found to be associated with the removal of phosphate from wastewater and activated sludge (Oerther et al., 2002). At the same time, it includes species such as A. baumannii, Acinetobacter calcoaceticus, and Acinetobacter lwoffii, which have been described as highly virulent pathogens (Rosalino et al., 2020). These three species were not detected in the samples in this study, and Acinetobacter johnsonii comprised the majority of the Acinetobacter genus detected. A. johnsonii has been described as a bioaccumulator of phosphate (Boswell et al., 2001).
Detected ESKAPEE pathogens were limited to E. faecium and E. coli. Although many of these pathogens can grow in lower temperatures, the optimal growth temperatures are >30°C. Specifically, the optimal growth temperatures of these pathogens are as follows: E. faecium, 42°C (Fisher and Phillips, 2009); S. aureus, 30°C–37°C (Kadariya et al., 2014); K. pneumoniae, 30°C–35°C; A. baumannii, 33°C–35°C; P. aeruginosa, 37°C; and E. coli, 37°C (Garrity, 2007). As Stavanger (the administrative center of Rogaland County) had an average temperature of 4.3°C and 10°C in January and May, respectively (Norwegian Meteorological Institute, 2022), the low number of ESKAPEE microbes is to be expected. This ties in with the previously presented argument that higher temperatures are a major factor in high ARG and pathogen presence. A putative hypothetical explanation for this effect may be that cellular growth, rather than horizontal gene transfer, is the dominant mechanism in ARG spread. As pathogens, especially colon bacteria, are expected to have a stricter growth temperature range, low mesophilic temperate conditions may impose a strong selective pressure for non-pathogens, and thus, vertical spreading becomes limited. Theoretical modeling conducted in our group indicates that vertical ARG transfer is a determinative mechanism in high-growth environments such as nutrient-rich WWTPs (Stokka, 2024). Temperature may also affect HGT directly (e.g., via reduced competence or transfer dynamics), which is also a possible explanation. This aspect of how temperature affects the spread of ARGs should be further investigated in controlled growth and gene transfer studies.
5 Conclusion and outlook
The results presented here demonstrate that, overall, differences in the presence of ARGs and in MIC values are only slightly influenced by catchment area activities, wastewater sources, and/or treatment process configuration. The same is true for the different microbial community compositions, which do not deviate significantly in Norway compared with communities observed in studies from other countries around the world. It can be observed that the resistance genes that were present were not removed from the wastewater, meaning that additional unit treatment processes are required for active removal. On the basis of the same comparative observations, we hypothesize that environmental factors (e.g., temperature) influence the resistome by vertical rather than horizontal gene transfer to a larger degree than antibiotic concentrations or wastewater origin.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena/browser/view/PRJEB70859.
Author contributions
DB: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. RK: Data curation, Funding acquisition, Methodology, Writing – review & editing. KK: Conceptualization, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the European Union JPI-AMR grant “Wastewater treatment plants as critical reservoirs for resistance genes (Gene-gas)” through the Norwegian Research Council grant NFR 271177/H10. Open access publication funds were received from the University of Stavanger.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frabi.2024.1351999/full#supplementary-material
Abbreviations
AMP, ampicillin; AMR, antimicrobial resistance; ARBs, antibiotic-resistant bacteria; ARG, antibiotic resistance gene; bla, β-lactamase; DNA, deoxyribonucleic acid; FLASH, fast length adjustment of short reads; HGT, horizontal gene transfer; MIC, minimum inhibitory concentration; MSC, minimal selective concentration; OTU, operational taxonomic unit; PCR, polymerase chain reaction; rRNA, ribosomal ribonucleic acid; SNJ, Sentralrenseanlegg Nord-Jæren; TET, tetracycline; TSS, total suspended solids; UWTP, urban wastewater treatment plant; VAN, vancomycin; VSS, volatile suspended solids; WWTP, wastewater treatment plant.
References
212, T. C. I (2019). ISO 20776-1:2019. Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices — Part 1: Broth micro-dilution reference method for testing the in vitro activity of antimicrobial agents against rapi. Geneva, Switzerland: ISO.
Abby S. S., Tannier E., Gouy M., Daubin V. (2012). Lateral gene transfer as a support for the tree of life. Proc. Natl. Acad. Sci. United States America 109 (13), 4962–4967. doi: 10.1073/pnas.1116871109
Alexander J., Hembach N., Schwartz T. (2020). Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 10 (1), 1–9. doi: 10.1038/s41598-020-65635-4
Boswell C. D., Dick R. E., Eccles H., Macaskie L. E. (2001). Phosphate uptake and release by acinetobacter johnsonii in continuous culture and coupling of phosphate release to heavy metal accumulation. J. Ind. Microbiol. Biotechnol. 26 (6), 333–340. doi: 10.1038/sj.jim.7000139
Cacace D., Fatta-Kassinos D., Manaia C. M., Cytryn E., Kreuzinger N., Rizzo L., et al. (2019). Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 162, 320–330. doi: 10.1016/j.watres.2019.06.039
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., et al. (2011). ‘Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. United States America 108 (SUPPL. 1), 4516–4522. doi: 10.1073/pnas.1000080107
Chow L. K. M., Ghaly T. M., Gillings M. R. (2021). A survey of sub-inhibitory concentrations of antibiotics in the environment. J. Environ. Sci. (China) 99, 21–27. doi: 10.1016/j.jes.2020.05.030
Dehshiri M., Khoramrooz S. S., Zoladl M., Khosravani S. A., Parhizgari N., Motazedian M. H., et al. (2018). The frequency of Klebsiella pneumonia encoding genes for CTX-M, TEM-1 and SHV-1 extended-spectrum beta lactamases enzymes isolated from urinary tract infection. Ann. Clin. Microbiol. Antimicrobials 17 (1), 1–7. doi: 10.1186/s12941-018-0256-y
Do T. T., Delaney S., Walsh F. (2019). 16S rRNA gene based bacterial community structure of wastewater treatment plant effluents. FEMS Microbiol. Lett. 366 (3), 1–10. doi: 10.1093/femsle/fnz017
Durand G. A., Raoult D., Dubourg G. (2019). Antibiotic discovery: history, methods and perspectives. Int. J. Antimicrobial Agents 53 (4), 371–382. doi: 10.1016/j.ijantimicag.2018.11.010
Eribe E. R. K., Olsen I. (2017). Leptotrichia species in human infections II. J. Oral. Microbiol. 9 (1). doi: 10.1080/20002297.2017.1368848
European Centre for Disease Prevention and Control (ECDC) (2021). Antimicrobial consumption in the EU/EEA (ESAC-net)’, surveillance report, (November). Stockholm, Sweden: ECDC.
European Centre for Disease Prevention and Control (ECDC), EFSA and EMA (2017). ECDC, EFSA and EMA Joint Scientific Opinion on a list of outcome indicators as regards surveillance of antimicrobial resistance and antimicrobial consumption in humans and food-producing animals. EFSA J. 15 (10). doi: 10.2903/j.efsa.2017.5017
Fiedler S., Bender J. K., Klare I., Halbedel S., Grohmann E., Szewzyk U. (2016). Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrobial Chemotherapy 71 (4), 871–881. doi: 10.1093/jac/dkv420
Fisher K., Phillips C. (2009). The ecology, epidemiology and virulence of Enterococcus. Microbiology 155 (6), 1749–1757. doi: 10.1099/mic.0.026385-0
Garrity G. M. (2007). Bergey’s manual of systematic bacteriology volume two the proteobacteria, part C-the alpha-, beta-, delta- and epsilonproteobacteria, ed. Brenner D. J., Krieg N. R., Staley J. T.. (Springer New York, NY: Springer Verlag). doi: 10.1007/0-387-29298-5
Gullberg E., Cao S., Berg O. G., Ilbäck C., Sandegren L., Hughes D., et al. (2011). Selection of resistant bacteria at very low antibiotic concentrations. PloS Pathog. 7 (7), 1–9. doi: 10.1371/journal.ppat.1002158
Hess M., Sczyrba A., Egan R., Kim T.-W., Chokhawala H., Schroth G., et al. (2011). Metagenomic Discovery of Biomass-degrading genes and genomes from Cow rumen. Science 463 (6016), 463–467. doi: 10.1126/science.1200387
Hu M., Wang X., Wen X., Xia Y. (2012). Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresource Technol. 117, 72–79. doi: 10.1016/j.biortech.2012.04.061
Hubeny J., Buta M., Zieliński W., Harnisz M., Korzeniewska E., Nowrotek M., et al. (2019). The prevalence of tet(A) and tet(M) tetracycline resistance genes in municipal wastewater. J. Ecol. Eng. 20 (10), 1–6. doi: 10.12911/22998993/112714
Hubeny J., Ciesielski S., Harnisz M., Korzeniewska E., Dulski T., Jałowiecki Ł., et al. (2021). ‘Impact of hospital wastewater on the occurrence and diversity of beta-lactamase genes during wastewater treatment with an emphasis on carbapenemase genes: A metagenomic approach. Front. Environ. Sci. 9 (October). doi: 10.3389/fenvs.2021.738158
Huijbers P. M. C., Larsson D. G. J., Flach C. F. (2020). Surveillance of antibiotic resistant Escherichia coli in human populations through urban wastewater in ten European countries. Environ. pollut. 261, 114200. doi: 10.1016/j.envpol.2020.114200
Hutinel M., Huijbers P. M. C., Fick J., Åhrén C., Larsson D. G. J., Flach C. F., et al. (2019). Population-level surveillance of antibiotic resistance in Escherichia coli through sewage analysis. Eurosurveillance 24 (37), 1–11. doi: 10.2807/1560-7917.ES.2019.24.37.1800497
Jang H. M., Kim Y. B., Choi S., Lee Y., Shin S. G., Unno T., et al. (2018). Prevalence of antibiotic resistance genes from effluent of coastal aquaculture, South Korea. Environ. pollut. 233, 1049–1057. doi: 10.1016/j.envpol.2017.10.006
Ju F., Guo F., Ye L., Xia Y., Zhang T. (2014). Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 6 (1), 80–89. doi: 10.1111/1758-2229.12110
Kadariya J., Smith T. C., Thapaliya D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed. Res. Int. 150, 528–540. doi: 10.1155/2014/827965
Karkman A., Do T. T., Walsh F., Virta M. P. J. (2018). Antibiotic-resistance genes in waste water. Trends Microbiol. 26 (3), 220–228. doi: 10.1016/j.tim.2017.09.005
Kent A. G., Vill A. C., Shi Q., Satlin M. J., Brito I. L. (2020). Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11 (1), 1–9. doi: 10.1038/s41467-020-18164-7
Klein A. R., Sarri E., Kelch S. E., Basinski J. J., Vaidya S., Aristilde L., et al. (2021). Probing the fate of different structures of beta-lactam antibiotics: hydrolysis, mineral capture, and influence of organic matter. ACS Earth Space Chem. 5 (6), 1511–1524. doi: 10.1021/acsearthspacechem.1c00064
Kümmerer K. (2009). Antibiotics in the aquatic environment - A review - Part I. Chemosphere 75 (4), 417–434. doi: 10.1016/j.chemosphere.2008.11.086
Kwak Y. K., Colque P., Byfors S., Giske C. G., Möllby R., Kühn I., et al. (2015). Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: Does it reflect the resistance trends in the society? Int. J. Antimicrobial Agents 45 (1), 25–32. doi: 10.1016/j.ijantimicag.2014.09.016
Leiviskä T., Risteelä S. (2022). Analysis of pharmaceuticals, hormones and bacterial communities in a municipal wastewater treatment plant – Comparison of parallel full-scale membrane bioreactor and activated sludge systems. Environ. pollut. 292 (July 2021). doi: 10.1016/j.envpol.2021.118433
Li C., Chen J., Li S. C. (2020). Understanding Horizontal Gene Transfer network in human gut microbiota. Gut Pathog. 12 (1), 1–20. doi: 10.1186/s13099-020-00370-9
Li D., Yang M., Hu J., Zhang J., Liu R., Gu X., et al. (2009). Antibiotic-resistance profile in environmental bacteria isolated from penicillin production wastewater treatment plant and the receiving river. Environ. Microbiol. 11 (6), 1506–1517. doi: 10.1111/j.1462-2920.2009.01878.x
Lipps W., Baxter T., Braun-Howland E. (2022). “Standard methods committee of the american public health association, american water works association, and water environment federation,” in Standard methods for the examination of water and wastewater, 24th edn. Eds. Lipps W., Baxter T., Braun-Howland E. (Washington DC: APHA Press). doi: 10.2105/SMWW.2882.030
Liu Y., Whitman W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. New York Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Lu J. J., Perng C. L., Chiueh T. S., Lee S. Y., Chen C. H., Chang F. Y., et al. (2001). Detection and typing of vancomycin-resistance genes of enterococci from clinical and nosocomial surveillance specimens by multiplex PCR. Epidemiol. Infection 126 (3), 357–363. doi: 10.1017/S0950268801005453
Ma J., Perng C. L., Chiueh T. S., Lee S. Y., Chen C. H., Chang F. Y., et al. (2023). Microbial source tracking identifies sources of contamination for a river flowing into the Yellow Sea. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1111297
MacFadden D. R., McGough S. F., Fisman D., Santillana M., Brownstein J. S. (2018). Antibiotic resistance increases with local temperature. Nat. Climate Change 8 (6), 510–514. doi: 10.1038/s41558-018-0161-6
Martineau F., Picarda F. J., Greniera L., Roya P. H., Ouellettea M., Bergerona M. G., et al. (2000). Multiplex PCR assays for the detection of clinically relevant antibiotic resistance genes in staphylococci isolated from patients infected after cardiac surgery. J. Antimicrobial Chemotherapy 46 (6), 527–533. doi: 10.1093/jac/46.4.527
McGough S. F., MacFadden D. R., Hattab M. W., Mølbak K., Santillana M. (2020). Rates of increase of antibiotic resistance and ambient temperature in Europe: A cross-national analysis of 28 countries between 2000 and 2016. Eurosurveillance 25 (45), 1–12. doi: 10.2807/1560-7917.ES.2020.25.45.1900414
Mirkalantari S., Masjedian F., Irajian G., Siddig E. E., Fattahi A. (2020). Determination of the frequency of β-lactamase genes (bla SHV, bla TEM, bla CTX-M) and phylogenetic groups among ESBL-producing uropathogenic Escherichia coli isolated from outpatients. J. Lab. Med. 44 (1), 27–33. doi: 10.1515/labmed-2018-0136
Morée A. L., Beusen A. H. W., Bouwman A. F., Willems W. J. (2013). Exploring global nitrogen and phosphorus flows in urban wastes during the twentieth century. Global Biogeochemical Cycles 27 (3), 836–846. doi: 10.1002/gbc.20072
Morrison L., Zembower T. R. (2014). Antimicrobial resistance - global report on surveillance. WHO 30 (4), 619–635. doi: 10.1016/j.giec.2020.06.004
Mühlberg E., Umstätter F., Kleist C., Domhan C., Mier W., Uhl P., et al. (2020). Renaissance of vancomycin: Approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 66 (1), 11–16. doi: 10.1139/cjm-2019-0309
NCBI (2023a) SHV variant list. Available at: https://www.ncbi.nlm.nih.gov/pathogens/refgene/SHV.
NCBI (2023b) TEM variant list. Available at: https://www.ncbi.nlm.nih.gov/pathogens/refgene/#TEM.
Ng L. K., Martin I., Alfa M., Mulvey M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15 (4), 209–215. doi: 10.1006/mcpr.2001.0363
Norwegian Meteorological Institute. (2022). Temperature in stavanger. Available at: https://www.yr.no/en/statistics/graph/1-15183/Norway/Rogaland/Stavanger/Stavanger?q=2022 (Accessed 2 November 2023).
Nuñez L., Tornello C., Puentes N., Espigares E., Moreno E., Espigares M., et al. (2016). Hospital effluent constitutes a source of vancomycin-resistant enterococci. Ars Pharm. 57 (3), 121–126. doi: 10.4321/S2340-98942016000300003
Oerther D. B., Van Loosdrecht M. C. M., Raskin L. (2002). Quantifying the impact of wastewater micronutrient composition on in situ growth activity of Acinetobacter spp. Water Sci. Technol. 46 (1–2), 443–447. doi: 10.2166/wst.2002.0514
Özkök I. (2012). Inhibitory impact of selected antibiotics on biodegradation characteristic and microbial population under aerobic conditions, Экономика Региона. Istanbul, Turkey: Istanbul Technical University.
Pallares-Vega R., Blaak H., van der Plaats R., de Roda Husman A. M., Hernandez Leal L., van Loosdrecht M. C. M., et al. (2019). Determinants of presence and removal of antibiotic resistance genes during WWTP treatment: A cross-sectional study. Water Res. 161, 319–328. doi: 10.1016/j.watres.2019.05.100
Pärnänen K. M. M., Narciso-Da-Rocha C., Kneis D., Berendonk T. U., Cacace D., Do T. T., et al. (2019). Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5 (3). doi: 10.1126/sciadv.aau9124
Patel R., Uhl J. R., Kohner P., Hopkins M. K., Ill Cockerill F. R., et al. (1997). Multiplex PCR Detection of vanA, vanB, vanC-1, and vanC-2/3 vanC-2/3 Genes in Enterococci. J. Clin. Microbiol. 35 (3), 703–707. doi: 10.1128/jcm.35.3.703-707.1997
Pazda M., Kumirska J., Stepnowski P., Mulkiewicz E. (2019). ‘Antibiotic resistance genes identified in wastewater treatment plant systems – A review. Sci. Total Environ. 697, 134023. doi: 10.1016/j.scitotenv.2019.134023
Peirano G., Pitout J. D. D. (2019). Extended-spectrum β-lactamase-producing enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79 (14), 1529–1541. doi: 10.1007/s40265-019-01180-3
Piotrowska M., Rzeczycka M., Ostrowski R., Popowska M. (2017). Diversity of antibiotic resistance among bacteria isolated from sediments and water of carp farms located in a Polish nature reserve. Polish J. Environ. Stud. 26 (1), 239–252. doi: 10.15244/pjoes/64910
Poopedi E., Singh T., Gomba A. (2023). Potential exposure to respiratory and enteric bacterial pathogens among wastewater treatment plant workers, South Africa. Int. J. Environ. Res. Public Health 20 (5). doi: 10.3390/ijerph20054338
Pruden A. (2014). Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 48 (1), 5–14. doi: 10.1021/es403883p
Qin H., Ji B., Zhang S., Kong Z. (2018). Study on the bacterial and archaeal community structure and diversity of activated sludge from three wastewater treatment plants. Mar. pollut. Bull. 135 (August), 801–807. doi: 10.1016/j.marpolbul.2018.08.010
Radisic V., Grevskott D. H., Lunestad B. T., Øvreås L., Marathe N. P. (2023). Sewage-based surveillance shows presence of Klebsiella pneumoniae resistant against last resort antibiotics in the population in Bergen, Norway’. Int. J. Hygiene Environ. Health 248 (September 2022). doi: 10.1016/j.ijheh.2022.114075
Randall L. P., Cooles S. W., Osborn M. K., Piddock L. J. V., Woodward M. J. (2004). Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrobial Chemotherapy 53 (2), 208–216. doi: 10.1093/jac/dkh070
Reddy G. B., Ford E., Aldridge D. (1986). Seasonal changes in bacterial numbers and plant nutrients in point and non-point source ponds. Environ. Pollution. Ser. A Ecol. Biol. 40 (4), 359–367. doi: 10.1016/0143-1471(86)90067-X
Redondo-Salvo S., Fernández-López R., Ruiz R., Vielva L., de Toro M., Rocha E. P.C., et al. (2020). Pathways for horizontal gene transfer in bacteria revealed by a global map of their plasmids. Nat. Commun. 11 (1). doi: 10.1038/s41467-020-17278-2
Rice E. W., Baird R. B., Eaton A. D., Clesceri L. S. (2013). Standard methods for the examination of water and wastewater (23rd ed.). APHA, AWWA, WEF 20th edn.
Rosalino V., Georgina S., Andr L. Á., Muleiro Nabil M. E., Vega L., et al. (2020). Acinetobacter baumannii resistance : A real challenge for clinicians. Antibiotics 9 (205), 1–22. doi: 10.3390/antibiotics9040205
Rushton J., Ferreira J. P., Stärk K. D. C. (2014). Antimicrobial resistance: the use of antimicrobials in the mouth. OECD Food Agric. Fisheries 68), 1–43. doi: 10.1787/18156797
Shariati A., Dadashi M., Moghadam M. T., van Belkum A., Yaslianifard S., Darban-Sarokhalil D., et al. (2020). Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci. Rep. 10 (1), 1–16. doi: 10.1038/s41598-020-69058-z
Stokka S. H. (2024). Spread of antibiotic resistance in wastewater treatment plants: mechanisms and a mathematical model compatible with ASM1. in preperation.
Wiegand I., Hilpert K., Hancock R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3 (2), 163–175. doi: 10.1038/nprot.2007.521
Wolff D., Krah D., Dötsch A., Ghattas A. K., Wick A., Ternes T. A., et al. (2018). Insights into the variability of microbial community composition and micropollutant degradation in diverse biological wastewater treatment systems. Water Res. 143, 313–324. doi: 10.1016/j.watres.2018.06.033
World Health Organization (2018). WHO report on surveillance of antibiotic consumption (Geneva, Switzerland: Who). Available at: http://apps.who.int/iris%0Ahttps://apps.who.int/iris/bitstream/handle/10665/277359/9789241514880-eng.pdf.
Yang Y., Wang L., Xiang F., Zhao L., Qiao Z. (2020). Activated sludge microbial community and treatment performance of wastewater treatment plants in industrial and municipal zones. Int. J. Environ. Res. Public Health 17 (2). doi: 10.3390/ijerph17020436
Youssef N., Sheik C. S., Krumholz L. R., Najar F. Z., Roe B. A., Elshahed M. S., et al. (2009). Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl. Environ. Microbiol. 75 (16), 5227–5236. doi: 10.1128/AEM.00592-09
Keywords: wastewater treatment plant, sequencing, antibiotic, antibiotic resistance genes, seasonal variation, Norway, microbial community effects
Citation: Basiry D, Kommedal R and Kaster KM (2024) The presence of antibiotic-resistant bacteria at four Norwegian wastewater treatment plants: seasonal and wastewater-source effects. Front. Antibiot. 3:1351999. doi: 10.3389/frabi.2024.1351999
Received: 07 December 2023; Accepted: 17 January 2024;
Published: 07 February 2024.
Edited by:
Tugce Katipoglu-Yazan, Istanbul Technical University, TürkiyeReviewed by:
Mohammad Tahir Siddiqui, Indian Institute of Technology Delhi, IndiaSamy Selim, Al Jouf University, Saudi Arabia
Copyright © 2024 Basiry, Kommedal and Kaster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Krista Michelle Kaster, krista.kaster@uis.no
 Daniel Basiry
Daniel Basiry Roald Kommedal
Roald Kommedal Krista Michelle Kaster
Krista Michelle Kaster