Disrupting the SKN-1 homeostat: mechanistic insights and phenotypic outcomes
- 1Leonard Davis School of Gerontology, University of Southern California, Los Angeles, CA, United States
- 2Dornsife College of Letters, Arts, and Sciences, Department of Molecular and Computational Biology, University of Southern California, Los Angeles, CA, United States
The mechanisms that govern maintenance of cellular homeostasis are crucial to the lifespan and healthspan of all living systems. As an organism ages, there is a gradual decline in cellular homeostasis that leads to senescence and death. As an organism lives into advanced age, the cells within will attempt to abate age-related decline by enhancing the activity of cellular stress pathways. The regulation of cellular stress responses by transcription factors SKN-1/Nrf2 is a well characterized pathway in which cellular stress, particularly xenobiotic stress, is abated by SKN-1/Nrf2-mediated transcriptional activation of the Phase II detoxification pathway. However, SKN-1/Nrf2 also regulates a multitude of other processes including development, pathogenic stress responses, proteostasis, and lipid metabolism. While this process is typically tightly regulated, constitutive activation of SKN-1/Nrf2 is detrimental to organismal health, this raises interesting questions surrounding the tradeoff between SKN-1/Nrf2 cryoprotection and cellular health and the ability of cells to deactivate stress response pathways post stress. Recent work has determined that transcriptional programs of SKN-1 can be redirected or suppressed to abate negative health outcomes of constitutive activation. Here we will detail the mechanisms by which SKN-1 is controlled, which are important for our understanding of SKN-1/Nrf2 cytoprotection across the lifespan.
1 SKN-1 is a multifunctional cytoprotective transcriptional regulator
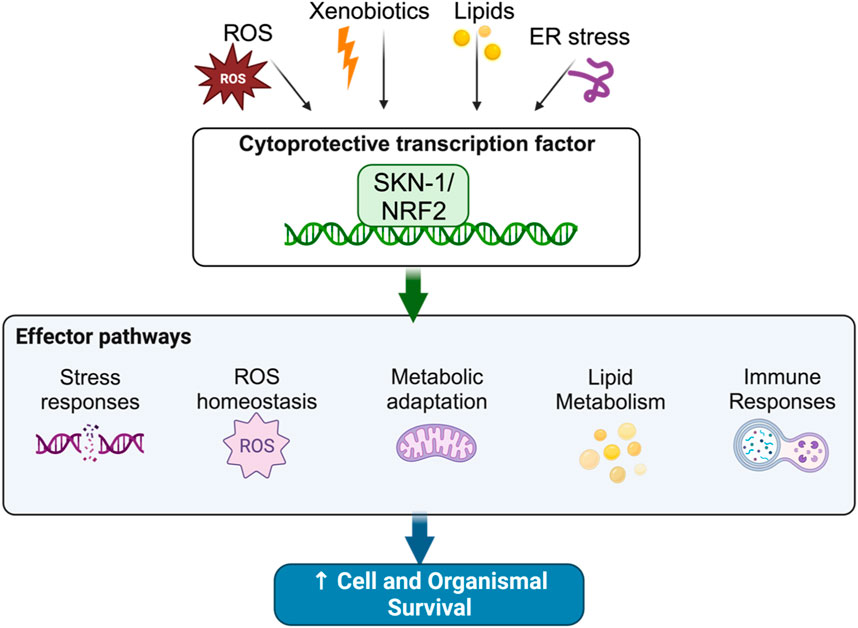
Homeostatic control mechanisms are crucial to the maintenance of cellular health and organismal longevity (Sykiotis and Bohmann, 2010; López-Otín et al., 2013; Blackwell et al., 2015; Torday, 2015; Cohen, 2016; Röder et al., 2016; Lemberg and Strisovsky, 2021; Ni and Buszczak, 2023). Indeed, several of the hallmarks of aging including genomic instability, mitochondrial dysfunction, and proteostatic disruption are all caused by the inability to maintain cellular homeostasis (Sykiotis and Bohmann, 2010; López-Otín et al., 2013; Blackwell et al., 2015; Torday, 2015; Cohen, 2016; Röder et al., 2016; Lemberg and Strisovsky, 2021; Ni and Buszczak, 2023). Xenobiotics and Reactive Oxygen Species (ROS) drive cellular instability by damaging cellular components; to respond to this, organisms have evolved stress tolerance pathways that can abate damage caused by xenotoxic stress (Sykiotis and Bohmann, 2008; Sykiotis and Bohmann, 2010; Kansanen et al., 2013; López-Otín et al., 2013; Blackwell et al., 2015; Torday, 2015; Cohen, 2016; Röder et al., 2016; Lemberg and Strisovsky, 2021; Ni and Buszczak, 2023). At the center of this homeostat, in worms and mammals, are the “Cap’n’Collar” (CnC) transcription factors skinhead-1 (SKN-1) and NF E2-Related Factor (Nrf2), respectively (Figure 1).
1.1 Structural comparisons of Nrf2 and SKN-1
Nrf2 is comprised of four major domains (Itoh et al., 1995). The DNA binding domain, which contains the CnC domain and Basic Region domain, followed by a basic leucine zipper domain (bZIP) that facilitates heterodimerization with Maf proteins to promote DNA binding (Itoh et al., 1995). Upstream of the DNA binding domain and ZIP region is the transcription activation DIDLID domain (Katoh et al., 2005). While SKN-1 is generally structurally diverged from its mammalian orthologue Nrf2, the CnC and Basic Region domains remain conserved (Blackwell et al., 1994; Carroll et al., 1997). Interestingly, SKN-1 lacks the leucine zipper domain that characterizes Nrf2 and other species of CnC transcription factors (Blackwell et al., 1994; Carroll et al., 1997). In vitro studies suggest that SKN-1 has adapted to bind DNA as a monomer at similar bZIP recognition sequences to Nrf2, via an enhanced Homeodomain Arm that interacts with the minor groove of the DNA helix where Maf proteins would bind in mammals (Blackwell et al., 1994; Carroll et al., 1997; Lo et al., 1998; Rupert et al., 1998; Kophengnavong et al., 1999). It is currently unclear if SKN-1 forms any dimers in vivo.
Nrf2 is part of a family that comprises three proteins each with distinct expression patterns, regulation, and gene targets (Sykiotis and Bohmann, 2010). Nrf2 is the most well characterized of these three proteins, it controls the expression of Phase II Detoxification genes that remove ROS-damaged molecules from the cell (Motohashi and Yamamoto, 2004). Nrf2 is negatively regulated by the cytoplasmic protein Kelch-like ECH associated protein 1 (Keap1) (Motohashi and Yamamoto, 2004). Keap1 binds to Nrf2 and promotes proteasomal degradation (Motohashi and Yamamoto, 2004). Upon electrophilic stress, cystine residues on Keap1 are reduced, weakening association with Nrf2 (Itoh et al., 2004; Motohashi and Yamamoto, 2004; Uruno and Motohashi, 2011). Once free of this negative regulation, Nrf2 localizes to the nucleus and binds at the Antioxidant Response Element (ARE), a recognition sequence in the promoters of detoxification response genes, which upregulates transcription at these loci (Itoh et al., 2004; Motohashi and Yamamoto, 2004; Uruno and Motohashi, 2011). Nrf1 is embedded into the membrane of the Endoplasmic Reticulum (ER) via an N-terminal transmembrane domain (Chan et al., 1993; Ruvkun and Lehrbach, 2023). During proteasome stress, Nrf1 is cleaved from the ER membrane, translocates into the nucleus, and upregulates proteasome subunits (Ruvkun and Lehrbach, 2023). The least well characterized Nrf family member is Nrf3 (Chevillard and Blank, 2011). Like Nrf1, Nrf3 also associates with the ER membrane, however, its methods of regulation are more complex, and its downstream targets are not well characterized (Chevillard and Blank, 2011).
Unlike mammals, C. elegans only have one orthologue to the Nrf family, SKN-1 (Blackwell et al., 2015). SKN-1 has three functional isoforms SKN-1A, SKN-1B, and SKN-1C (Blackwell et al., 2015). Remarkably the isoforms of SKN-1 have functional overlap with the different members of the Nrf family (Blackwell et al., 2015). SKN-1A is the only isoform with a transmembrane domain whereby it associates with the ER membrane (Ruvkun and Lehrbach, 2023). During proteasome stress, SKN-1 is cleaved from the ER and upregulates proteasome subunits (Ruvkun and Lehrbach, 2023). SKN-1C, like Nrf2, is thought to be primarily involved in the cytoprotective response against ROS (Blackwell et al., 2015). Lastly, SKN-1B is the less well understood SKN-1 isoform (Blackwell et al., 2015). GFP tagging of SKN-1 suggests that SKN-1B remains entirely localized to a pair of sensory neurons called ASI and contributes to longevity induced by caloric restriction (Bishop and Guarente, 2007). While SKN-1 has significantly diverged from the Nrf proteins in mammals in both expansion and DNA binding mode, SKN-1 still shares functional overlap with the mammalian Nrf transcription factors. The functional similarities between each isoform of SKN-1 and the different members of the Nrf family indicate that there is evolutionary conservation of CnC transcription factors and that C. elegans could be used as a model to understand Nrf biology.
1.2 SKN-1 roles in embryonic development
SKN-1 was initially described as a maternally deposited mRNA for a tissue specification factor (Bowerman et al., 1992; Bowerman et al., 1993). SKN-1 is required for the differentiation of the EMS blastomere into the MS and E cells, which will further differentiate into the pharyngeal and intestinal tissues, respectively (Bowerman et al., 1992; Bowerman et al., 1993). Genetic analysis of loss-of-function alleles of skn-1 (zu67 and zu138) demonstrated a failure of embryos to differentiate the EMS blastomere into the proper tissue types, instead differentiating into additional hypodermis and body wall muscle (Bowerman et al., 1992; Bowerman et al., 1993). SKN-1 is positioned at the top of the specification/differentiation cascade of the EMS blastomere (Maduro et al., 2001a). SKN-1 initiates this by binding to the genes med-1 and med-2, which will bind other differentiation factors, determining which daughter cell will become the MS or E blastomere (Maduro et al., 2001a). Only Nrf1 is required for development in mice, with Nrf1−/− mice having liver development abnormalities (Chen et al., 2003). However, Nrf1 in mice contributes to hepatocyte maturation by protecting from Tumor Necrosis Factor mediated apoptosis and maintaining redox balance and does not act as a specification factor in early embryogenesis like SKN-1 (Chen et al., 2003). While the requirement for SKN-1 in the development of the intestine is clear, what remains unknown is which isoforms of SKN-1 contribute to development and how. The skn-1 loss of function alleles zu67 and zu138 ablate the function of all SKN-1 isoforms, making it difficult to determine which isoform(s) contribute to development. If we look to mammals as an example where Nrf1 is involved in development, it may mean that SKN-1A is responsible for development in the worm. Using genetic nulls for each isoform will inform which isoform is responsible for intestinal specification.
1.3 SKN-1 roles in oxidative stress response
Many of the postembryonic investigations done on SKN-1 have focused on its role in upregulating detoxifying pathways. Like mammals, C. elegans requires a robust mechanism for detoxifying xenobiotics and endogenous toxins. This process is broken into three phases (Xu et al., 2005; Lindblom and Dodd, 2006). Phase I detoxification occurs as cytochrome P450s and short chain dehydrogenases conjugate reactive groups onto toxic molecules increasing solubility and allowing further detoxifying metabolism to occur (Xu et al., 2005; Lindblom and Dodd, 2006). Phase II detoxification is comprised of enzymes that conjugate glutathione, a reducing agent, onto reactive groups (Xu et al., 2005; Lindblom and Dodd, 2006). These enzymes include Glutathione-S Transferases (GSTs) and UDP-glucosyl Transferases (UGTs) (Xu et al., 2005; Lindblom and Dodd, 2006). Finally, Phase III detoxification is comprised of transporters, including ATP-Binding Cassette (ABC) transporters (Xu et al., 2005; Lindblom and Dodd, 2006). These will move the fully solubilized and neutralized toxins out from the cytoplasm (Xu et al., 2005; Lindblom and Dodd, 2006). Microarray, RNA-seq, and ChIP-seq studies have determined that SKN-1 upregulates a bevy of Phase I, II, and III components as direct targets (Oliveira et al., 2009; Staab et al., 2013; Staab et al., 2014; Steinbaugh et al., 2015; Ramos and Curran, 2023; Turner et al., 2023).
Early work on the role of SKN-1 in the detoxifying response to ROS relied on GFP fusion expression arrays that tagged SKN-1 at the C-terminus, tagging each isoform simultaneously (An and Blackwell, 2003). These visualization experiments determined that under normal conditions SKN-1GFP accumulates within two sensory neurons at the head of the animals called ASI (An and Blackwell, 2003). Furthermore, when acutely exposed to sodium arsenite, SKN-1GFP accumulated in the intestinal cell nuclei, marking the intestine as a major site for SKN-1-mediated detoxification (An and Blackwell, 2003). In addition to visualization experiments, genetic analysis using the skn-1 loss-of-function alleles zu67 and zu129 determined that without functional SKN-1, animals had diminished survival when exposed to acute oxidative stress (An and Blackwell, 2003). Furthermore, SKN-1 loss-of-function alleles display shortened lifespans in comparison to wildtype worms, establishing SKN-1 as both a stress response factor and a longevity maintenance factor (An and Blackwell, 2003). While SKN-1 is implicated in both stress adaptation and longevity, it is likely that these roles are deeply interconnected, given that in situations where SKN-1 is required for longevity, loss of SKN-1 not only abates longevity but also abates stress adaptation. Furthermore, in constitutively active skn-1 mutants, where longevity is decreased, stress adaptation is improved (Paek et al., 2012; Blackwell et al., 2015).
It has long been appreciated that SKN-1 mediates the oxidative stress response via the intestine. However, it remains less clear what SKN-1 does in the ASI neuron pair. Bishop and Guarente used isoform specific RNAi to knockdown skn-1b gene products, and found that RNAi to skn-1b diminished the brightness of a SKN-1 activity reporter in the ASI neurons (Bishop and Guarente, 2007). Additionally, they found that SKN-1B was required for lifespan extension caused by caloric restriction, while SKN-1A/C had no effect on this phenotype (Bishop and Guarente, 2007). A recent investigation into cell non-autonomous roles of SKN-1 has given better insight into SKN-1 activity in the ASI neurons (Turner et al., 2023). Cell type-specific RNA-seq on the ASI neurons, in an activated SKN-1 background, showed an increase in typical SKN-1 targets including Phase II detoxification, components of the ubiquitin proteasome, and pathogen response genes (Turner et al., 2023). This introduces new hypotheses about SKN-1 in the ASI neurons. Either SKN-1B has remarkable overlap with SKN-1A/C targets and regulates all SKN-1 targets in the ASI neurons, or the other isoforms of SKN-1 are active in the ASI neurons. Supporting the hypothesis that other SKN-1 isoforms are active in the ASI neurons, a cell type expression construct expressing an activated allele of skn-1c in the ASI neurons resulted in the activation of the gst-4p:GFP SKN-1 activity reporter across multiple tissues (Turner et al., 2023). However, when activated, skn-1c was expressed in singular tissues and animals were assessed for survival from acute oxidative stress, none could achieve a robust level of survival associated with activated SKN-1 (Turner et al., 2023). Taken together, these data suggest that neuronal SKN-1 controls more than the longevity response to caloric restriction. The activation of SKN-1 transcriptional targets in the neurons can initiate a multi-tissue stress response (Turner et al., 2023). The initiation of the stress response may require one or more tissues or the activity of more than one SKN-1 isoform to produce a fully effective response to oxidative stress (Turner et al., 2023); delineation of the tissue-specific roles of each isoform is of critical importance.
1.4 SKN-1 roles in proteasome stress
While SKN-1C/Nrf2 manages the response to oxidative stress, the membrane-bound SKN-1A/Nrf1 contributes to the response to proteasomal stress (Glover-Cutter et al., 2013). Under conditions where the proteasome is functional, SKN-1A is typically stored within the ER lumen where it is N-linked Glycosylated at several sites (Glover-Cutter et al., 2013; Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). SKN-1A is extruded into the cytoplasm via the ER Associated Degradation pathway and degraded by the proteasome. During conditions where the proteasome is inhibited, SKN-1A is extruded into the cytoplasm but is not degraded by the proteasome (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). Instead SKN-1A undergoes significant post-translational modification (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). The protease DDI-1 interacts with SKN-1A and cleaves the transmembrane domain from SKN-1A (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). Further modification is performed by the peptide N-glycanase PNG-1. PNG-1 deglycosylates SKN-1A at the residues that were glycosylated inside the ER lumen (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). Interestingly, the N-glycosylation within the ER lumen and subsequent deglycosylation by PNG-1 catalyzes sequence editing on SKN-1A, whereby the N-glycosylated asparagine residues are converted to aspartic acid residues (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). Both mutation analysis, mutating the asparagine residues to aspartic acid, and assays reducing the function of PNG-1 have demonstrated that the sequence editing of SKN-1A is required for it to upregulate target genes (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). SKN-1A will upregulate proteasome subunits thus increasing proteasome function and alleviating acute proteasome stress (Lehrbach and Ruvkun, 2016; Lehrbach et al., 2019; Yoshida et al., 2021). Remarkably SKN-1C and SKN-1A share many regulators such as, ERK1/2, mTORC1, and OGT, and while evidence suggests that sequence editing on SKN-1A fine tunes it to proteasome subunit targets, it is unclear if SKN-1A and SKN-1C synergize or overlap with each other. While large strides have been made in defining the post-translational modification and subsequent activation of SKN-1A from the ER, further studies should focus on the role SKN-1A might play in metabolism and nutrient sensing given its regulation by these pathways, along with potential synergies with SKN-1C that is also implicated in metabolism regulation. Furthermore, advancements in tissue-specific expression will allow for tissue and cell type investigation into SKN-1A function.
1.5 SKN-1 roles in mitochondrial homeostasis
The impairment of mitochondrial homeostasis is a hallmark of aging and age-related pathologies. Healthy cells utilize autophagy of mitochondria organelles (mitophagy), that selectively degrade damaged mitochondria (Youle and Narendra, 2011). C. elegans requires a robust mitophagy pathway to promote longevity in canonically long-lived mutants, such as daf-2 mutants (Palikaras et al., 2015), that disrupt insulin signaling. In C. elegans, mitophagy is primarily controlled by DCT-1 (Palikaras et al., 2015). DCT-1 is localized to the outer mitochondrial membrane where it co-localizes with autophagosomal protein LGG-1, inducing mitophagy (Palikaras et al., 2015). Loss of DCT-1 via RNAi knockdown causes a loss of mitochondrial network integrity and under nominal conditions, loss of DCT-1 does not shorten lifespan (Palikaras et al., 2015). Loss of DCT-1 activity results in SKN-1 activation, SKN-1 in turn upregulates mitophagy factors including DCT-1 (Palikaras et al., 2015). During dietary restriction, respiration rates increase, and ablation of mitochondrial electron transport chain suppresses the longevity increase in dietarily restricted worms (Bishop and Guarente, 2007). Correspondingly, loss-of-function alleles of skn-1 subjected to dietary restriction, fail to increase mitochondrial respiration, and rescue of the skn-1b isoform in the ASI neurons facilitates increased respiration and longevity under dietary restriction (Bishop and Guarente, 2007). Recently, the natural product tomatidine was shown to upregulate mitophagy in both worms and human cell culture by inducing SKN-1/Nrf2 activity, improving healthspan in C. elegans (Fang et al., 2017). In addition, SKN-1 was previously shown to interact with the mitochondrial surface protein Phosphoglycerate Mutase 5 (PGAM-5) via in vitro co-immunoprecipitation assays (Paek et al., 2012). Activating mutations in skn-1 weaken the interaction between PGAM-5 and SKN-1, suggesting that SKN-1 activation may release SKN-1 from associations with PGAM-5 at the mitochondrial outer membrane (Paek et al., 2012). These studies establish SKN-1 as a mediator of mitochondrial homeostasis. Additional work should focus on establishing a mechanism for SKN-1 association at the outer mitochondrial membrane and possible interaction with mitophagy factors as a sensor for mitochondrial network integrity. Further exploration of natural products and derivatives is warranted as enhancing mitophagy at advanced age may be a beneficial treatment for age related disease.
2 SKN-1 responses to diet and metabolism
2.1 Dietary impact on physiology
The influence of bacterial diets on the physiology of C. elegans extends across various life history traits, encompassing development, reproduction, healthspan, and longevity (Soukas et al., 2009; Hansen et al., 2013; Bansal et al., 2015; Stuhr and Curran, 2020). Numerous studies have elucidated diet-induced effects, correlating with observable differences in metabolic profiles, lipid homeostasis, and feeding behavior (Soukas et al., 2009; Xiao et al., 2015). In a study from Stuhr and Curran (2020), they demonstrated that distinct bacterial species isolated from the laboratory environment, which mirror those found in C. elegans’ natural habitats, can selectively alter development, reproduction, and metabolism (Stuhr and Curran, 2020). Their analysis of Gene Ontology (GO) terms associated with unique genes differentially expressed in animals fed Methylobacterium, Xanthomonas, or Sphingomonas revealed distinct molecular signatures for each diet. When compared to age-matched animals fed the standard laboratory diet E. coli/OP50 B, significant alterations were identified, among the identified genes, many played roles in development (sqt-2, mlt-7, mlt-10), metabolism (fat-5, fat-7), reproduction (col-71, ptr-1, lon-1), and aging (downregulation of daf-2 and age-1, and upregulation of DAF-16/FoxO target genes). Notably, each bacterial diet corresponded to a specific metabolic profile, and C. elegans reared on these bacteria exhibited variations in intracellular lipid distribution. Examination of gene expression related to lipid metabolism revealed that approximately one-third of the differentially expressed genes on each bacterial food source has the potential to impact a pleiotropic physiological response, highlighting the intricate interplay between diet and regulation of fundamental biological processes in C. elegans. The understanding of how specific dietary components affect both health and lifespan in C. elegans can contribute to explaining variations in aging rates and the severity of age-related diseases.
It is notable that SKN-1 serves as a mediator in translating dietary influences into physiological outcomes. While live bacteria are conventionally utilized as the primary source in C. elegans studies, studies have indicated host-pathogen defenses stemming from the pathogenic nature of E. coli (Garigan et al., 2002; Irazoqui et al., 2010; Kumar et al., 2019). Studies employing dead bacteria, either through UV-killing or bactericidal antibiotics, have demonstrated an extension in lifespan by mitigating bacterial proliferation within the pharynx and intestine (Garigan et al., 2002), along with distinct impacts on physiological metrics such as growth, lipid metabolism, and movement (Stuhr and Curran, 2023). Nhan et al (2019) examined that feeding skn-1 gain-of-function mutant animals with dead bacteria resulted in healthier lipid metabolic outcomes compared to their counterparts fed live bacteria (Nhan et al., 2019). This intricate relationship between diet and physiological outcomes, influenced by specific bacterial species, underscores the relevance of SKN-1 in orchestrating diverse responses, providing a potential link to the regulation of fundamental biological processes across organisms.
2.2 Gene-diet interactions and SKN-1 coordination
Diet and genetics are pivotal in shaping the intricate web of metabolic regulation, healthspan, and lifespan. Studies have identified neuroendocrine signaling pathways, such as the neuromedin U receptor (NMUR), are important for sensing dietary cues from different food types and are involved in regulation of lifespan in a diet-dependent manner (Maier et al., 2010). Specifically, nmur-1 mutants lived longer on the standard E. coli/OP50 B diet than on the E. coli/HT115 K-12 diet, illustrating a notable gene-diet interaction (Maier et al., 2010). Recent work has revealed insight into adaptive response mechanisms that organisms utilize to maintain physiological homeostasis when exposed to different diets (Pang and Curran, 2012; Pang and Curran, 2014). Work by Pang and Curran (2014) on mutants of alh-6, a proline metabolism gene, displayed a shortened lifespan on the standard OP50 diet, but not on the HT115 diet, their findings further revealed a mechanism by which diets will signal a mitochondrial adaptation through coordination of neuronal and metabolic tissues upon exposure to the E. coli/HT115 K-12 diet (Pang and Curran, 2014). Within this context, the transcription factor SKN-1 has emerged as a key player; from its activation in long-lived mutants to its intricate involvement in oxidative stress and detoxification pathways, offering potential avenues for further exploration in this intricate landscape of gene-diet interactions. While significant strides have been made in deciphering gene-diet interactions and understanding the impact of diverse bacterial diets on life history traits in C. elegans, the molecular underpinnings of these dietary responses remain a subject of ongoing investigation (Lynn and Curran, 2015).
2.3 Nutrient sensing and SKN-1 activation
SKN-1 activity and regulation extend to changes in nutrient availability, redox status, and other environmental signals, highlighting its dynamic engagement with the cellular environment (Pang et al., 2014). Caloric restriction (CR) or dietary restriction (DR), characterized by a reduction in nutrient availability and/or metabolic energy without malnutrition, serves as a potent activator of SKN-1 (Onken and Driscoll, 2010). Dietary restriction has been established as a mechanism that extends the lifespan of C. elegans, conferring diverse health benefits. Notably, SKN-1 is one of the transcription factors involved in mediating this effect. In a study by Bishop and Guarente (2007), they showed that lifespan extension was nullified in skn-1 mutants subjected to DR, and this attenuation was restored by introduction of a skn-1 transgene (Cypser et al., 2013). Interestingly, the activation of skn-1 proved necessary exclusively in the ASI neurons for DR-induced longevity. Expression of a skn-1 transgene solely in the ASI neurons rescued the loss of DR-induced lifespan extension caused by skn-1 deletion (Bishop and Guarente, 2007). This study suggests a neuroendocrine function for SKN-1, particularly in the ASI neurons, and points to a cell non-autonomous effect involving a systemic longevity signal downstream of SKN-1 (Bishop and Guarente, 2007; Sykiotis et al., 2011).
Further recognized as a longevity factor, SKN-1 is activated in various long-lived mutant backgrounds exhibiting altered metabolic homeostasis. SKN-1 stands as an indispensable contributor to the observed lifespan extension in these mutants, underscoring its significance in the regulation of aging processes (Bishop and Guarente, 2007; Tullet et al., 2008; Wang et al., 2010). SKN-1 is integrated into the insulin/IGF-1 signaling pathway, which plays a central role in nutrient sensing and metabolism. Specifically, the insulin/IGF-1 signaling (IIS) receptor DAF-2 plays a role in this regulatory cascade. Activation of DAF-2 triggers a downstream modulation of transcription factors, including the inhibition of SKN-1. Consequently, SKN-1 is hindered from regulating its target gene networks (Tullet et al., 2008; Onken and Driscoll, 2010). Experimental manipulations such as knockdown of DAF-2, either through mutation or RNA interference (RNAi), have been shown to activate SKN-1, resulting in long-lived and stress-resistant phenotypes in worms (Carroll et al., 1997). Importantly, SKN-1 is needed during adulthood to confer these beneficial effects (Onken and Driscoll, 2010; Grushko et al., 2021). The observed benefits of DR and decreased insulin signaling may be explained by a metabolic shift that optimally allocates available resources for the maintenance of overall fitness and viability (Sykiotis et al., 2011). Drawing parallels with worms, several studies reveal that reduced IGF-1 signaling exerts a comparable influence on lifespan extension in mice (Cohen et al., 2009; Vitale et al., 2019). Intriguingly, mutations in components of the IGF-1 pathway also correlate with longevity observed in humans (Vitale et al., 2019). Together, these findings indicate that the aging-regulating roles attributed to the IIS pathway are conserved from worms to mammals (Holzenberger et al., 2003; Willcox et al., 2008).
In recent investigations, microRNAs (miRNAs) have been identified to promote longevity, particularly under conditions of DR and reduced insulin signaling (Matai et al., 2023). miRNAs are short, non-coding RNAs that bind to complementary sequences in mRNA molecules, orchestrating the inhibition of protein translation or mRNA degradation, thereby crucially contributing to post-transcriptional regulation. A study by Matai et al. (2023) has shed light on the involvement of the mir-229,-64,-65,-66 cluster of miRNAs in the context of DR and low-IIS scenarios. Intriguingly, this miRNA cluster is upregulated under DR and low-IIS scenarios, and in turn this cluster upregulates skn-1 mRNA levels, revealing an interplay between miRNAs and SKN-1 (Matai et al., 2023). The interaction of the miRNA cluster (miR-229–66) together with SKN-1 introduces an indirect regulatory mechanism that transduces the effects of DR and low-IIS into lifespan extension. This regulatory cascade involves the modulation of several ubiquitin-mediated proteolysis and xenobiotic detoxification pathway genes (Matai et al., 2023). Understanding the molecular mechanisms by which SKN-1 responds to diet and metabolism in C. elegans can provide insights into the broader connections between nutrient sensing, oxidative stress response, and lifespan regulation in other organisms, including mammals.
2.4 Role of SKN-1 in lipid metabolism
Maintaining energy homeostasis is a fundament aspect of animal physiology, requiring intricate coordination in the metabolism of intracellular nutrients such as glucose, lipids, and amino acids (Pang et al., 2014). Animals employ sophisticated molecular mechanisms to ensure the continual fulfillment of organismal demands for growth, cellular maintenance, and reproduction (Efeyan et al., 2015; Lynn et al., 2015). SKN-1 has been implicated in the regulation of lipid metabolism, impacting both lipid storage and utilization in response to fluctuating nutrient availability (Pang et al., 2014; Lynn and Curran, 2015; Lynn et al., 2015). SKN-1 responds dynamically to changes in nutrient availability and can modulate its activity in response to shifting nutritional landscapes through interactions with signaling pathways, including the mitochondrial amino acid catabolism pathway, and insulin and IGF-1 signaling pathways (Tullet et al., 2008; Zarse et al., 2012; Pang et al., 2014). Recent studies propose that SKN-1-mediated transcriptional programs mechanistically link proline and fatty acid metabolism, providing a deeper understanding of the crosstalk between these pathways (Pang et al., 2014; Yen et al., 2020). Mutations in mitochondrial proline catabolism (alh-6) lead to mobilization of intestinal lipids during starvation, indicating a coupling with lipid metabolism in response to nutrient depletion (Pang and Curran, 2014). In these mutants, several lipid metabolism and fatty acid synthesis genes were upregulated during starvation, such as the fasting-induced lipase-1 (flp-1) and the fatty acid oxidation enzyme (cpt-5) (Pang and Curran, 2014). Notably, a loss-of-function mutation in skn-1 abolishes the enhanced depletion of somatic lipid stores observed in alh-6 mutants after fasting, indicating SKN-1’s role in mediating fasting metabolic responses (Pang and Curran, 2014).
Subsequently, gain-of-function mutations in skn-1 (lax188) lead to a starvation-like status and can induce the expression of several metabolic genes; however, the full extent to which SKN-1 participates in organismal physiology and metabolism remains unknown (Paek et al., 2012). Work on skn-1 gain-of-function mutant animals show these animals have an upregulation of FAO genes, such as acs-1, cpt-4, cpt-6 (Pang et al., 2014), as well as metabolism and starvation adaptation genes, such as lips-3, atgp-2, acdh-6 (Paek et al., 2012). Interestingly, these skn-1gf mutants exhibit the age-dependent somatic depletion of fat (Asdf) phenotype, characterized by the depletion of intestinal lipid stores without affecting germline lipid stores. This phenotype, associated with a starvation response dependent on vitellogenesis, which facilitates transport of stored lipids from the intestine to developing oocytes, emphasized the role of SKN-1 in the reallocation of lipids from soma to germline, potentially ensuring survival and fitness under challenging conditions (Lynn et al., 2015). This Asdf phenotype exhibits specificity to oxidative stress, as wildtype worms subjected to heat or osmotic stress failed to induce the observed lipid shift to the germline. Notably, treatment with N-acetylcysteine (NAC), a potent antioxidant, effectively suppressed the Asdf phenotype in skn-1gf mutants. This highlights the oxidative stress-dependent nature of the observed lipid distribution, emphasizing the central role of SKN-1 in coordinating the cellular response to oxidative challenges. Given its high expression in the intestine, SKN-1’s crucial role in responding to dietary changes becomes evident. The intestine, as a central tissue for nutrient absorption and metabolism, positions SKN-1 as a contributor to the overall metabolic homeostasis of C. elegans, underscoring SKN-1’s role in orchestrating lipid homeostasis and metabolic responses essential for the organism’s adaptation and survival.
Dietary lipids and lipid metabolism are critical for organismal adaptation to external conditions, lifespan regulation, and reproduction (Ma et al., 2020; Mejhert et al., 2022; Papsdorf et al., 2023). Lipids encompass a diverse array of hydrophobic molecules, including triglycerides, phospholipids, and steroids. The impact of lipids on longevity and the regulatory mechanisms of lipid metabolism during aging are subjects of ongoing investigation. Specific lipid profiles are associated with survival, with increased fat storage linked to aging, while certain lipids, such as mono-unsaturated fatty acids (MUFAs) like oleic acid and polyunsaturated fatty acids (PUFAs) like eicosapentaenoic acid (EPA), alpha-linolenic acid (ALA), and arachidonic acid (ARA), are implicated in regulating transcription factors like SKN-1 (Papsdorf and Brunet, 2019). During nutrient deprivation, stored lipids and amino acids are used instead of dietary glucose to satisfy organismal energy requirements. Lipids are mobilized as an energy resource through lipolysis and fatty acid oxidation (FAO) (Pang et al., 2014). Analysis of the skn-1gf animals revealed a reduction in oleic acid (C18:1), suggesting its deficiency as causative for the Asdf phenotype (Lynn et al., 2015). Knockdown of fat-6 and fat-7, which encode enzymes converting stearic acid to oleic acid, phenocopied the Asdf phenotype (Lynn et al., 2015). Supplementation with biosynthetic precursors to oleic acid, including stearic acid, lauric acid, and alpha-linolenic acid, did not suppress Asdf in skn-1gf mutants. Investigations into lipid biosynthesis pathways in Lynn et al. (2015) indicated that under nutrient-poor and oxidative stress conditions, omega-3 and omega-6 PUFAs mediate the balance of lipid stores between the soma and germline, impacting reproduction and somatic aging (Lynn et al., 2015). Mutant animals unable to generate EPA or ARA exhibited the Asdf phenotype at a later time point than skn-1gf animals, highlighting the role of specific PUFAs in regulating this metabolic shift. The complexity of the relationship between lipid metabolism and SKN-1-mediated responses in maintaining cellular homeostasis is underscored by these studies. In a study by Qi et al (2017), the supplementation of ALA was found to extend lifespan in C. elegans through the regulation of the NHR-49 and SKN-1 transcription factors. Notably, ALA indirectly activated SKN-1 activation via oxidation of ALA to produce oxylipins, ultimately leading to the activation of SKN-1 and conferred longevity (Qi et al., 2017). These studies emphasize the complexity between lipid metabolism and SKN-1-mediated responses in maintaining cellular homeostasis, with an emphasis on how specific lipids can influence and modulate the organism’s lifespan.
A study by Cedillo et al. (2023), elucidated the connection between SKN-1, biguanides, and ether lipid metabolism, highlighting their collective role in coordinating biguanide-mediated responses to dietary conditions and innate immune responses to promote longevity (Cedillo et al., 2023). Ether lipids, a diverse class of membrane lipids integral to cellular functions and homeostasis, contribute to membrane structure, stability, and fluidity, impacting crucial cellular processes such as membrane fusion and cellular signaling (Mejhert et al., 2022). Biguanides, including the widely prescribed metformin, are drugs commonly used to lower blood sugar levels by reducing glucose production in the liver. Metformin is often used to manage and treat type 2 diabetes, and is associated with various metabolic benefits, including weight control and improved insulin sensitivity (Martin-Montalvo et al., 2013). SKN-1 has been previously established as necessary for metformin-induced lifespan extension (Onken and Driscoll, 2010; Cedillo et al., 2023). This study further demonstrated that treatment with phenformin, a biguanide akin to metformin, induces the Asdf phenotype at day 3 of adulthood, similar to skn-1gf mutants (Lynn et al., 2015; Nhan et al., 2019). Notably, loss-of-function mutations in ether lipid biosynthetic genes thwarted the phenformin-induced Asdf phenotype, suggesting a mechanistic link between ether lipids and skn-1-dependent metabolic defenses (Cedillo et al., 2023). Moreover, loss-of-function skn-1 animals nullified the Asdf phenotype associated with phenformin treatment, indicating the essential role of SKN-1 in this biguanide-mediated Asdf lipid shift. Phenformin treatment induced expression of the innate immune response gene dod-24 in a SKN-1-dependent manner. Collectively, this study indicates that biguanides, specifically phenformin, can extend the lifespan by activating metabolic stress defenses through ether lipids, with SKN-1 playing a pivotal role. Although the precise molecular mechanisms underlying metformin’s interaction with SKN-1 are an area of ongoing research, these studies offer valuable insights into the potential involvement of Nrf2 in mediating the health-promoting effects of biguanides, with implications for understanding the broader impacts of metformin on cellular health and longevity in higher organisms, including humans.
3 SKN-1 responses to pathogens
Transitioning from the influence of specific lipids on SKN-1-mediated responses, an exploration into the transcripts regulated by SKN-1 that contribute to the Asdf phenotype unveils a deeper understanding of the molecular mechanisms involved. Previous findings revealed that SKN-1 triggers a metabolic stress defense response, facilitating somatic lipid depletion by enhancing lipid utilization and promoting innate immunity gene expression, counteracting canonical oxidative stress responses (Nhan et al., 2019). Lynn et al. (2019) demonstrated that manipulating methylation states, specifically H3K4me3, Histone H3 at lysine 4, through knockdown of wdr-5 or rbbp-5, or with studies on wdr-5lf;skn-1gf double mutants, reversed the Asdf phenotype in skn-1(lax188) gain-of-function mutant animals, restoring somatic lipids and extending lifespan (Nhan et al., 2019). This study highlights the role of H3K4me3 trimethylation in fine-tuning deregulated SKN-1 transcriptional activity away from innate immunity targets, mitigating the negative metabolic outcomes. Furthermore, exposure to subliminal paraquat-induced oxidative stress redirected skn-1gf activity from pathogen response genes, simultaneously restoring somatic lipid distribution. Transcriptional analyses of this paraquat exposure unveiled reduced skn-1gf association with the promoter regions of innate immunity genes, dod-24 and endu-2, and enhanced association with the oxidative stress gene, gst-4. This study opened a novel avenue for activated SKN-1 transcription factor studies, emphasizing the critical role of immune activation in coordinating pathogen responses and adaptive adjustments.
The use of bacteria as the primary nutritional source for C. elegans serves as a valuable approach to investigate host-pathogen interactions. The immune defense of C. elegans against pathogenic stress involves a sophisticated network of molecular pathways, with SKN-1 emerging as a master regulator (Papp et al., 2012a; Liu et al., 2022). In response to pathogenic encounters, SKN-1 orchestrates a range of responses to maintain host homeostasis, ensuring survival and contribute to longevity. In adult skn-1gf mutants exhibiting the Asdf phenotype, transcriptional and Gene Ontology analyses revealed a strong enrichment of immune and pathogen response genes, including dod-24, endu-2, and clec-66, suggesting that SKN-1 plays a crucial role in regulating innate immune response genes during later stages of development (Nhan et al., 2019). Interestingly, a nsy-1gf mutant, which constitutively activates the immune response via the p38 mitogen-activated protein kinase (MAPK) PMK-1 pathway, revealed a similarly Asdf phenotype. The suppression of this phenotype in the skn-1 (zu135) loss-of-function mutant points to the essential role of SKN-1 activity for the NSY-1/PMK-1 pathways and the subsequent transcription of innate immunity target genes (Nhan et al., 2019). While these studies were conducted on the non-pathogenic OP50 diet, other studies utilized the opportunistic human pathogen Pseudomonas aeruginosa (PA14) strain, known to infect and ultimately kill nematodes (Cezairliyan et al., 2013; Pukkila-Worley, 2016). In the context of PA14 infection, SKN-1 was identified as a regulator of pathogen resistance, and this activation was dependent on the TIR-1/PMK-1 signaling pathway (Papp et al., 2012b). By dissecting these pathogen-specific interactions, a deeper understanding of the adaptability and specificity of SKN-1 in response to diverse metabolic threats is achieved. These studies, ranging from SKN-1 activation pathways to its contributions to immune effectors and stress resilience, can unravel additional layers of complexity in host-pathogen dynamics.
4 Mechanisms of molecular regulation of SKN-1
4.1 Inhibitory regulation of SKN-1 via the ubiquitin proteasome
Like Nrf2, proteostatic turnover is the primary way SKN-1 is regulated in C. elegans (Figure 2) (Choe et al., 2009; Taguchi et al., 2011; Tang and Choe, 2015). Worms do not have a Keap1 orthologue, however SKN-1 associates with the WD Repeat Protein 23 (WDR-23) which is an E3 ubiquitin ligase substrate adaptor (Choe et al., 2009). WDR-23 will associate with CULlin-4 (CUL-4) allowing for the polyubiquitylation of SKN-1 and subsequent degradation by the proteasome (Choe et al., 2009). RNAi knockdown of wdr-23 will result in the accumulation of SKN-1 in the intestinal nuclei (Choe et al., 2009). However, wdr-23 has two isoforms, wdr-23a and wdr-23b, each having a distinct subcellular localization, WDR-23A localizing to mitochondria and WDR-23B localized to the nucleus (Staab et al., 2013). Rescue constructs that restore only WDR-23b in wdr-23 loss-of-function mutants display normal SKN-1 activity in SKN-1 activity reporters, while rescues restoring only WDR-23a do not (Spatola et al., 2019). Furthermore, knockdown of wdr-23 resulted in a synergistic activation of SKN-1 which indicate the mechanisms that control SKN-1 functions expand beyond proteostatic levels of SKN-1 abundance (Paek et al., 2012). This presents an interesting model for WDR-23 regulation of SKN-1 where SKN-1 is turned over by nuclear WDR-23b while cytoplasmic WDR-23 associates at the mitochondria and binds mitochondrial SKN-1 and acts a sensor for mitochondrial stress and ROS accumulation (Spatola et al., 2019). This model would perfectly mirror the mammalian model for Nrf2 regulation via Keap1; however, to date there is no evidence that the cystine residues on WDR-23 are oxidized by electrophilic compounds. Furthermore, mammals have a conserved orthologue of WDR23 which has been shown to regulate Nrf2 independently of KEAP1 (Lo et al., 2017), this suggests that mammalian regulation is diverged from C. elegans and that insights into WDR-23 in the worm may yield information on how WDR23 functions in mammals.
Regulation of SKN-1 by WDR-23/CUL-4 and the canonical ubiquitin proteasome has been established, however proteostasis is also mediated via two other ubiquitin-like modifying pathways (Enchev et al., 2015; Vertegaal, 2022). These include the SUMOylation pathway and NEDDylation pathway (Enchev et al., 2015; Vertegaal, 2022). Both pathways mediate the addition of small ubiquitin like moieties to proteins, chiefly to signal degradation; however, both pathways are involved in other processes such as signaling, cell cycle, DNA damage response, and immunity (Enchev et al., 2015; Vertegaal, 2022). The constitutively active skn-1 allele, lax188, contains a glutamic acid to lysine substitution (Paek et al., 2012). This mutant displays a trade-off between increased stress tolerance and decreased longevity (Paek et al., 2012; Nhan et al., 2019; Turner et al., 2023). Using RNAi to knockdown both the SUMOylation and NEDDylation enzymes in the worm, the activated SKN-1 allele was shown to be more sensitive to loss of NEDDylation, accumulating in the intestinal nuclei, while wildtype SKN-1 remained excluded (Turner et al., 2023). Loss of SUMOylation had no effect on the wildtype or constitutively active SKN-1 (Turner et al., 2023). These results have given new insight into the regulation of SKN-1. Given that SUMOylation did not have an effect on the localization of SKN-1, it is likely that SKN-1 is a specific substrate of NED8. There is much left to understand about NEDDylation on SKN-1, whether it is mono- or poly-NEDDylated and if it has chains of NEDDylation. It is also unknown if SKN-1 is broadly NEDDylated or if specific conditions are required for it to be NEDDylated. Investigations into the proteostatic turnover of SKN-1 have yielded insights into the intricate nature of SKN-1 regulation via the ubiquitin proteasome. Additional studies are needed to determine the sites of ubiquitylation and NEDDylation on SKN-1, these may also reveal if SKN-1 is poly- or mono-ubiquitinylated/NEDDylated. In vivo identification of the subcellular localization of SKN-1C and SKN-1B isoforms will be crucial to understand where SKN-1 is sequestered in the cell and how it may evade degradation as a form of signaling.
4.2 Inhibitory regulation by kinases
SKN-1 regulation outside of proteasomal degradation is mediated by post-translational phosphorylation modifications (Blackwell et al., 2015). Among the various kinases that regulate SKN-1, one of the more defined is GSK-3, a kinase first discovered to control the synthesis of glycogen (An et al., 2005). Interestingly, GSK-3 is another developmentally required protein (Maduro et al., 2001a). In the embryo, it controls the establishment of C blastomere fate that will develop into the epidermis, it does so by inhibiting the transcriptional activity of SKN-1 in the P2 blastomere (Maduro et al., 2001b). Post-embryonically, GSK-3 phosphorylates SKN-1 at serine residues S393 and S432 (An et al., 2005). This phosphorylation prevents the intestinal accumulation of SKN-1, with both post-embryonic RNAi to gsk-3 as well as serine to alanine mutations at GSK-3 phosphorylation sites, resulting in the nuclear accumulation of SKN-1 in intestinal nuclei (An et al., 2005). Another inhibitory phosphorylation event is mediated via AKT kinase. Insulin-like signaling in C. elegans is mediated through DAF-2 activation of AKT (Tullet et al., 2008; Ewald et al., 2015). AKT inhibits SKN-1 by phosphorylating serine residue S12, preventing intestinal accumulation (Tullet et al., 2008; Ewald et al., 2015). Reduction of Insulin-like signaling results in the accumulation of SKN-1 (Tullet et al., 2008; Blackwell et al., 2015). Regulation of SKN-1 via insulin-like signaling places SKN-1 in longevity promoting pathways in parallel to those mediated via DAF-16 (Tullet et al., 2008; Ewald et al., 2015), further connecting SKN-1 to mechanisms controlling metabolic homeostasis.
SKN-1 is also regulated by the mTORC-2 pathway (Jones et al., 2009; Robida-Stubbs et al., 2012; Ruf et al., 2013; Mizunuma et al., 2014). mTORC-2 subunit RICT-1 is responsible for phosphorylating and activating kinase SGK-1 (Jones et al., 2009). A particular serine residue has yet to be determined for SGK-1, however western blots for phosphorylated SKN-1 show that SGK-1 phosphorylates SKN-1 in the C-terminal region 300-623aa (Jones et al., 2009). Like other inhibitory kinase systems, loss of either rict-1 or sgk-1 results in the nuclear accumulation of SKN-1 in the intestinal nuclei (Jones et al., 2009; Robida-Stubbs et al., 2012; Ruf et al., 2013; Mizunuma et al., 2014). Phosphorylation of SKN-1 by the mTORC-2 pathway implicates SKN-1 in nutrient sensing and growth regulation. Previously it was shown that feeding two different bacterial diets had opposing effects on lifespan under inhibited mTORC-2 conditions (Mizunuma et al., 2014). Determining the bacterial or metabolic signals that cause differential effects on lifespan as well as the tissue-specificity will be key in understanding mTORC-2-mediated SKN-1 longevity.
4.3 Positive regulation by the MAPK pathway
Kinases can also positively regulate SKN-1. The p38 mitogen-activated protein kinase (MAPK) pathways are evolutionarily conserved kinase cascades that are required for responses to various stressors including oxidative stress and pathogenic infection (Kurz and Tan, 2004; Inoue et al., 2005; Hoeven et al., 2011). Canonical MAPK members in C. elegans include NSY-1,SEK-1, and PMK-1 (Kurz and Tan, 2004; Inoue et al., 2005; Hoeven et al., 2011). Genetic ablation of any of these results in an inability of intestinal SKN-1 to accumulate and respond to stress (Inoue et al., 2005). The terminal kinase PMK-1 phosphorylates SKN-1 at two known serine residues, S164 and S430, accordingly genetic ablation of these serine residues to alanine ablates the ability of SKN-1 to accumulate into the intestinal nuclei (Inoue et al., 2005). The p38 MAPK network is nearly universally required for stress adaptation in the intestine of the worm, however the importance of the p38 MAPK pathway and Nrf2 activation is less understood (Kurz and Tan, 2004; Inoue et al., 2005; Sun et al., 2009; Hoeven et al., 2011). While phosphorylation via the MAPK pathway is required for response to most biotic stimuli, it is unclear if constitutively active SKN-1 is continually phosphorylated by the MAPK pathway or if it circumvents it entirely. Studies on the phosphorylation of activated SKN-1 mutants will provide insight into the regulation of activated SKN-1. Furthermore, PMK-1 phosphorylation sites are shared between all SKN-1 isoforms, and it is currently unknown if there is any selectivity of the MAPK pathway to phosphorylate certain SKN-1 isoforms over others, and given the proposed specific roles each isoform plays, investigating this may provide insight into SKN-1 target selectivity.
4.4 Additional regulation by post-translational modifications
A bevy of other post-translational modifications regulate SKN-1 activity (Figure 3). Recently, SKN-1 was shown to be regulated via O-linked N-acetylglucosamine (O-GlcNAc) cycling. Mutants for ogt-1, the transferase responsible for the addition of O-GlcNAc show impaired response to oxidative stress and a reduced SKN-1 accumulation in the intestinal nuclei (Li et al., 2017). Accordingly, mutants for oga-1, the acetylase responsible for removing the modification, have elevated SKN-1 target gene expression and increased SKN-1 intestinal accumulation (Li et al., 2017). Further biochemical analysis demonstrated that SKN-1 is O-GlcNAcylated at serine residue S470 and threonine residue T493 which neighbor the GSK-3 phosphorylation site and that during stress conditions, the GSK-3 site was less phosphorylated and the OGT-1-targeted residues were O-GlcNAcylated (Li et al., 2017). This analysis proposes an interesting competition dynamic between GSK-3 inhibitory phosphorylation and activating O-GlcNAcylation (Li et al., 2017). O-GlcNAcylation on mammalian Nrf2 has yet to be deeply understood, however genetic analysis of Ogt in mammals is difficult since genetic ablations of O-GlcNAc cycling enzymes is lethal (Li et al., 2017; Levine et al., 2021).
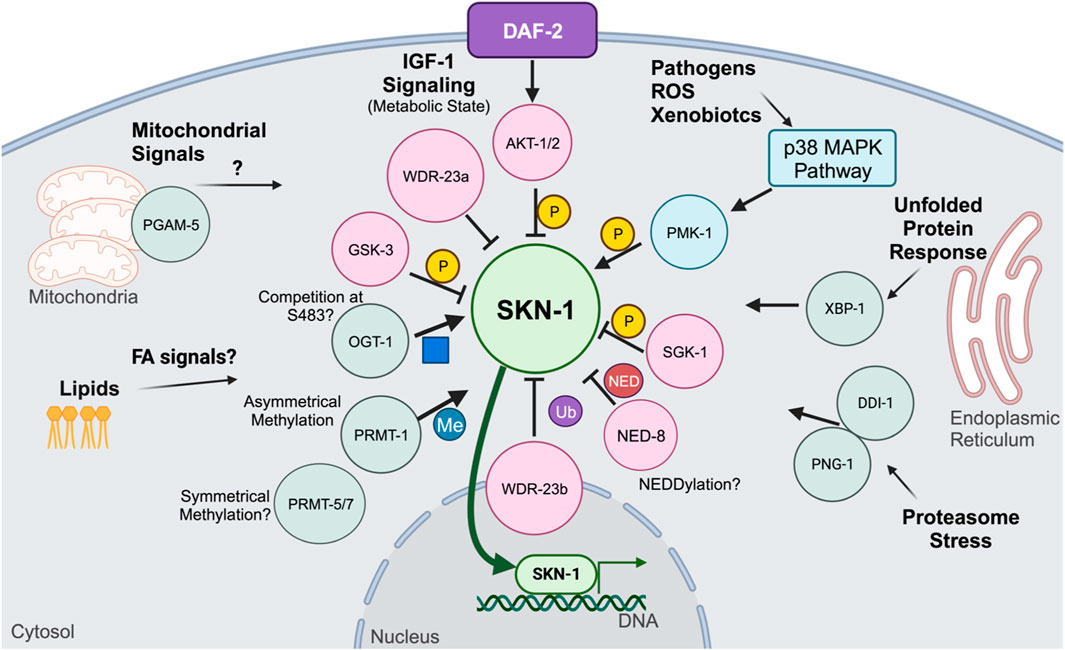
FIGURE 3. Post-translation modifications and signaling pathways that mechanistically affect SKN-1 activity.
Another post-translational modification shown to regulate SKN-1 activity is arginine methylation (Li et al., 2019). SKN-1 has been shown to be asymmetrically methylated by PRMT-1 at arginine residues 484 and 516 (Li et al., 2019). During oxidative stress, PRMT-1 binds to SKN-1 and methylates its target arginine residues, which results in SKN-1 activation (Li et al., 2019). Loss of PRMT-1, either through RNAi knockdown or genetic mutation, results in the inability for SKN-1 to accumulate in the intestinal nuclei and a reduction in the expression of phase II detoxification genes (Li et al., 2019). Loss of the Arginine residues where SKN-1 is methylated did not alter its nuclear accumulation like total loss of PRMT-1, suggesting that phosphorylation contributes significantly to the nuclear accumulation and arginine methylation may play a role in DNA binding enhancement (Li et al., 2019). SKN-1 contains several other Arginine residues that may be methylated, further proteomic analysis may reveal additional methylation on SKN-1 (Li et al., 2019). Furthermore, while PRMT-1 catalyzes asymmetrical methylation, it is unknown if any symmetrical methylation occurs on SKN-1 and how it may affect DNA binding dynamics. Further analysis of arginine methylation may yield new insights into how SKN-1 targets are specified and selected, and future work should focus on the methylation status of different SKN-1 isoforms as well as the methylation status under various stressors, such as pathogenic and metabolic.
Additional RNAi-based screens of kinases in C. elegans have revealed additional regulatory kinases that function to inhibit SKN-1 activity. Kinases from this screen include nekl-2, ikke-1, and mkk-4 (Kell et al., 2007). Each of these kinases are from disparate biological processes which include the cell cycle and glucose metabolism, however no biochemical analyses have been done to determine if such kinases interact directly with SKN-1 or if these kinases regulate processes that modulate SKN-1 downstream (Kell et al., 2007). A determination of whether SKN-1 is a direct target of each of these kinases and comprehensive identification of the residues modified is critical to understanding the mechanistic roles these PTMs play in controlling SKN-1 activity.
5 Modulators of SKN-1 activity
5.1 Molecular triggers of SKN-1 activity
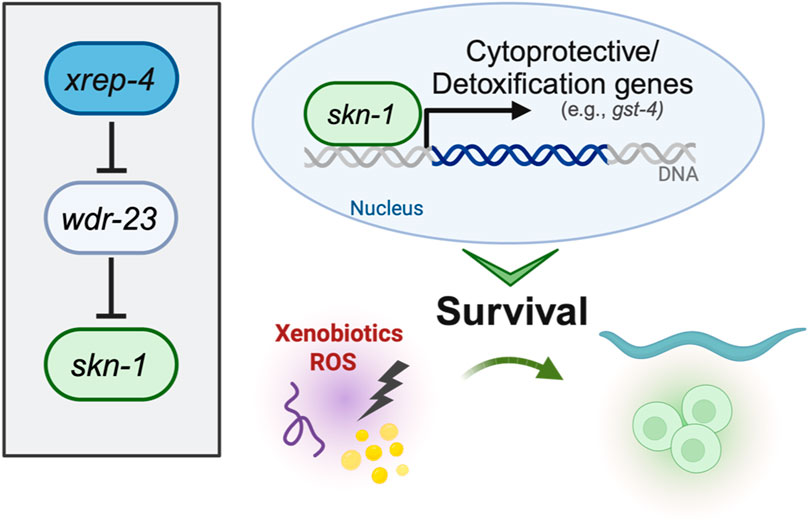
The activation of SKN-1 in C. elegans represents a complex regulatory network crucial for its adaptive responses to environmental cues. SKN-1 activity is finely tuned by a spectrum of stimuli, with oxidative stress serving as a primary inducer. Positive regulators, including phosphorylation by p38 MAPK PMK-1, and the ubiquitin ligase adaptor protein SKR-1, activate SKN-1 in response to ROS (Troemel et al., 2006; Wu et al., 2016). Additionally, the acetyltransferase enzyme CBP-1, a transcriptional regulator in cellular processes such as oxidative stress resistance and aging, has been shown to directly modulate SKN-1 translocation to the nucleus. In response to stress, CBP-1 promotes SKN-1-dependent transcription of protective genes (Ganner et al., 2019). Knockdown of cbp-1 suppressed the expression of said SKN-1 target genes, indicating that CBP-1 drives SKN-1 transcription. Conversely, negative regulators such as serum and glucocorticoid-inducible kinase-1 (SGK-1) and WDR-23 inhibit SKN-1 nuclear accumulation and target it for degradation, respectively (Tullet et al., 2008; Choe et al., 2009). Additional positive regulators have been studied, including the F-box protein XREP-4, which functions with SKR-1 to negatively inhibit the activity of WDR-23 (Fukushige et al., 2017; Wu et al., 2017). Recently, we have defined a novel gain-of-function mutation in the xrep-4 locus, which contributes to constitutive activation of wildtype SKN-1 (Ramos and Curran, 2023). In established genetic models of constitutive SKN-1 activation, two paradigms include loss-of-function alleles of wdr-23 and gain-of-function alleles of skn-1. Both hinder the ubiquitin proteasome-mediated turnover of SKN-1, leading the sustained activation (Paek et al., 2012; Spatola et al., 2019). wdr-23lf mutants exhibit enhanced resistance to oxidative stress, accompanied by an upregulation of stress response, metabolic, and signaling genes (Staab et al., 2013; Tang and Choe, 2015). skn-1gf mutants, as discussed earlier, undergo a transcriptional redirection that influences altered survival responses (Nhan et al., 2019). Extending this paradigm, xrep-4gf mutants also show increased expression of genes associated with glutathione transferase activity, immune system responses, and xenobiotic responses. Intriguingly, there is an overlap of 1796 genes with skn-1gf mutants, indicating shared molecular signatures. Despite exhibiting a similar premature age-related decline as skn-1gf animals, the phenotypic impact of xrep-4gf animals is comparatively less severe in terms of both healthspan and lifespan, emphasizing nuances in the outcomes of SKN-1 activation through distinct genetic models (Ramos and Curran, 2023).
In addition to genetic regulators, SKN-1-mediated transcriptional activation through disruption of amino acid catabolism or exogenous amino acid supplementation have also been elucidated. Perturbation of amino acid catabolic pathways, such as proline catabolism, leads to a buildup of toxic catabolites, triggering SKN-1-related transcription through ROS accumulation (Pang et al., 2014; Yen et al., 2020). Similar SKN-1 transcriptional responses have been reported with perturbations of tryptophan, threonine, and most recently, histidine catabolism (Frankino et al., 2022). In a study by Frankino et al. (2022), knockdown of the conserved amidohydrolase T12A2.1/amdh-1 results in the accumulation of the catabolite 4-imidazolone-5-propanoate, serving as a mediator for the activation of a specific subset of SKN-1-regulated genes. Interestingly, this activation is independent of the P38-MAPK pathway but involves the nuclear factor ELT-3, nuclear hormone receptor NHR-49, and mediator complex subunit MDT15. Most recently, Wu et al. (2023) proposed a cascade of events activating SKN-1 initiated by alpha-ketobutyrate (α-KB) supplementation (Wu et al., 2023). Addition of α-KB leads to increased expression of the enzyme ACOX1, which is involved in the peroxisomal fatty acid β-oxidation pathway. This process generates hydrogen peroxide (H2O2), activating SKN-1, and subsequently promoting the translocation of SKN-1 to the nucleus to upregulate genes involved in cellular defense against oxidative stress (Wu et al., 2023). These findings highlight the multifaceted regulator mechanisms modulating SKN-1 activity, in addition to the intersection between metabolism and stress response pathways in orchestrating cellular homeostasis.
The intricate regulatory mechanisms uncovered in C. elegans’ SKN-1 have parallels in mammalian systems, particularly Nrf2, where constitutive activation is implicated in cancer progression and chemoresistance (Sykiotis and Bohmann, 2010; Wu et al., 2019). Understanding these regulatory networks not only sheds light on fundamental cell biology questions but also holds clinical relevance, offering insights into diseases characterized by dysregulated transcription factors like Nrf2. Such findings could have implications in various fields, including pharmacology, as they suggest potential therapeutic avenues for modulating oxidative stress-related pathways. Understanding these molecular triggers not only provides insights into the nuanced regulatory mechanisms governing SKN-1 but also unravels the interconnected signaling pathways that contribute to the adaptive responses to changing environment conditions.
5.2 Insights into SKN-1 regulatory control
The importance of precisely regulating SKN-1/NRF2 activity becomes evident when considering the repercussions of chronic activation. Despite the crucial role of SKN-1 in mounting a protective response to various stressors, animals with sustained or constitutive activation of SKN-1 experience detrimental outcomes, highlighting the essentiality of regulating this cytoprotective pathway (Pang et al., 2014; Blackwell et al., 2015; Nhan et al., 2019). Studies on constitutively active SKN-1 mutants, such as skn-1gf and xrep-4gf, reveal age-dependent phenotypes, initially presenting enhanced stress resistance early in life but eventually exhibiting the Asdf phenotype, prioritizing reproductive fitness over somatic health (Lynn et al., 2015). This dysregulation of cellular lipid homeostasis contributes to diminished health and alters the organism’s overall lifespan (Pang et al., 2014; Lynn et al., 2015; Nhan et al., 2019). The capacity of epigenetic modifications to reshape the regulation of crucial physiological processes by influencing chromatin association underscores an exciting avenue for research. Chromatin modifications, including alterations in DNA packaging, emerge as pivotal, long-lasting mechanisms governing gene expression, particularly those implicated in cellular maintenance and longevity (Papsdorf and Brunet, 2019). Studies of the transcriptional redirection of constitutively active SKN-1 in skn-1gf mutants unveil the remarkable ability to fine-tune protective responses, adapting them to the specific requirements for reinstating homeostatic balance (Nhan et al., 2019). Of significance is the finding that activated SKN-1 transcription can be intentionally redirected, offering a potentially adaptable strategy to address diseases marked by aberrant transcriptional regulation. In the context of skn-1gf mutants, an RNAi screen targeting transcriptional regulators identified mediator subunit MDT-15, a lipid metabolism-regulating transcription factor, as a suppressor of constitutive SKN-1 activation (Pang et al., 2014). This study suggests that MDT-15 interacts with SKN-1 in the intestine, coordinating responses to different dietary conditions, metabolic regulation, and oxidative stress (Pang et al., 2014; Hu et al., 2018). Recent findings have identified another suppressor, a gain-of-function mutation in the dicer-related helicase gene, drh-1, which temporarily delays the adverse effects of SKN-1 activation (Turner et al., 2023). DRH-1, known for detecting viral double-stranded RNA (dsRNA) and regulating transcription of immune response genes upon viral infection, works to oppose the increased transcriptional load resulting from skn-1gf, highlighting the intricate balance required for maintaining cellular homeostasis (Sowa et al., 2020; Turner et al., 2023). However, the temporary relief provided by DRH-1 activation (drh-1gf) emphasized the critical importance of turning off cytoprotection to ensure proper physiological outcomes and prevent the adverse consequences stemming from chronic SKN-1 activation. Together, these findings hold promise for innovative therapeutic strategies aimed at correcting dysregulated molecular pathways to improve overall health.
6 Concluding remarks and perspectives
6.1 Mechanistic questions about SKN-1 biology and implications for human aging
Regulation of SKN-1 activity is incredibly complex. Foundational studies have given solid framework to our understanding of the mechanisms that govern transcriptional activation. However, to achieve a deeper understanding of SKN-1 biology, tissue, cell, and even sub-cellular investigations on the function of SKN-1 will need to be done. Understanding how SKN-1 interacts in each tissue will be paramount to further understanding tissue-specific roles that can be paralleled to mammalian tissues. Additionally, we do not understand what signaling molecules are used to communicate between tissues. While specific targets of each isoform of SKN-1 have been proposed, understanding how each SKN-1 isoform synergizes with the others is a key gap in knowledge. Continuing to study SKN-1 and metabolism will be paramount as healthy diets become therapeutic strategies for aging human populations. Using constitutively active SKN-1/Nrf models will also help fill the major knowledge gap about the deactivation of SKN-1/Nrf. Considering that hyperactive Nrf2 is a cancer hallmark in some tissues (Kansanen et al., 2013), it is paramount to understand how SKN-1/Nrf2 can be turned off or redirected once activated. The finding that activated SKN-1 can have its transcriptional profile redirected by exposure to different acute stresses has implications for the control of SKN-1 in constitutively active pathologies, such as cancer. SKN-1 is regulated by many post-translational modifications where some, like arginine methylation, contribute to target specificity and not SKN-1 function as a whole. Thus, it is possible that pharmacological targeting of SKN-1 post-translational modification could fine tune SKN-1/Nrf2 activity. Continual study of the intricate regulatory networks of SKN-1/Nrf2 may help identify future therapeutics to allow for beneficial activation of SKN-1/Nrf2 and the suppression of runaway activation in human pathologies.
Author contributions
CT: Writing–review and editing, Writing–original draft. CR: Writing–review and editing, Writing–original draft. SC: Writing–review and editing, Writing–original draft, Supervision, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH R01AG058610 and Hevolution Foundation award HF AGE-004 to SC and T32AG052374 to CT.
Acknowledgments
We thank N. Stuhr for thoughtful discussion and edits to the manuscript. Figures created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
References
An, J. H., and Blackwell, T. K. (2003). SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes and Dev. 17 (15), 1882–1893. doi:10.1101/gad.1107803
An, J. H., Vranas, K., Lucke, M., Inoue, H., Hisamoto, N., Matsumoto, K., et al. (2005). Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc. Natl. Acad. Sci. U. S. A. 102 (45), 16275–16280. doi:10.1073/pnas.0508105102
Bansal, A., Zhu, L. J., Yen, K., and Tissenbaum, H. A. (2015). Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc. Natl. Acad. Sci. U. S. A. 112 (3), E277–E286. doi:10.1073/pnas.1412192112
Bishop, N. A., and Guarente, L. (2007). Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447 (7144), 545–549. doi:10.1038/nature05904
Blackwell, T. K., Bowerman, B., Priess, J. R., and Weintraub, H. (1994). Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science 266 (5185), 621–628. doi:10.1126/science.7939715
Blackwell, T. K., Steinbaugh, M. J., Hourihan, J. M., Ewald, C. Y., and Isik, M. (2015). SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 88, 290–301. doi:10.1016/j.freeradbiomed.2015.06.008
Bowerman, B., Draper, B. W., Mello, C. C., and Priess, J. R. (1993). The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74 (3), 443–452. doi:10.1016/0092-8674(93)80046-h
Bowerman, B., Eaton, B. A., and Priess, J. R. (1992). skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 68 (6), 1061–1075. doi:10.1016/0092-8674(92)90078-q
Carroll, A. S., Gilbert, D. E., Liu, X., Cheung, J. W., Michnowicz, J. E., Wagner, G., et al. (1997). SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes Dev. 11 (17), 2227–2238. doi:10.1101/gad.11.17.2227
Cedillo, L., Ahsan, F. M., Li, S., Stuhr, N. L., Zhou, Y., Zhang, Y., et al. (2023). Ether lipid biosynthesis promotes lifespan extension and enables diverse pro-longevity paradigms in Caenorhabditis elegans. Elife 12, 12. doi:10.7554/eLife.82210
Cezairliyan, B., Vinayavekhin, N., Grenfell-Lee, D., Yuen, G. J., Saghatelian, A., and Ausubel, F. M. (2013). Identification of Pseudomonas aeruginosa phenazines that kill Caenorhabditis elegans. PLoS Pathog. 9 (1), e1003101. doi:10.1371/journal.ppat.1003101
Chan, J. Y., Han, X. L., and Kan, Y. W. (1993). Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. U. S. A. 90 (23), 11371–11375. doi:10.1073/pnas.90.23.11371
Chen, L., Kwong, M., Lu, R., Ginzinger, D., Lee, C., Leung, L., et al. (2003). Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell Biol. 23 (13), 4673–4686. doi:10.1128/MCB.23.13.4673-4686.2003
Chevillard, G., and Blank, V. (2011). NFE2L3 (NRF3): the Cinderella of the Cap'n'Collar transcription factors. Cell Mol. Life Sci. 68 (20), 3337–3348. doi:10.1007/s00018-011-0747-x
Choe, K. P., Przybysz, A. J., and Strange, K. (2009). The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol. Cell Biol. 29 (10), 2704–2715. doi:10.1128/MCB.01811-08
Cohen, A. A. (2016). Complex systems dynamics in aging: new evidence, continuing questions. Biogerontology. 17 (1), 205–220. doi:10.1007/s10522-015-9584-x
Cohen, E., Paulsson, J. F., Blinder, P., Burstyn-Cohen, T., Du, D., Estepa, G., et al. (2009). Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139 (6), 1157–1169. doi:10.1016/j.cell.2009.11.014
Cypser, J. R., Kitzenberg, D., and Park, S. K. (2013). Dietary restriction in C. elegans: recent advances. Exp. Gerontol. 48 (10), 1014–1017. doi:10.1016/j.exger.2013.02.018
Efeyan, A., Comb, W. C., and Sabatini, D. M. (2015). Nutrient-sensing mechanisms and pathways. Nature 517 (7534), 302–310. doi:10.1038/nature14190
Enchev, R. I., Schulman, B. A., and Peter, M. (2015). Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16 (1), 30–44. doi:10.1038/nrm3919
Ewald, C. Y., Landis, J. N., Porter Abate, J., Murphy, C. T., and Blackwell, T. K. (2015). Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519 (7541), 97–101. doi:10.1038/nature14021
Fang, E. F., Waltz, T. B., Kassahun, H., Lu, Q., Kerr, J. S., Morevati, M., et al. (2017). Tomatidine enhances lifespan and healthspan in C. elegans through mitophagy induction via the SKN-1/Nrf2 pathway. Sci. Rep. 7, 46208. doi:10.1038/srep46208
Frankino, P. A., Siddiqi, T. F., Bolas, T., Bar-Ziv, R., Gildea, H. K., Zhang, H., et al. (2022). SKN-1 regulates stress resistance downstream of amino catabolism pathways. iScience 25 (7), 104571. doi:10.1016/j.isci.2022.104571
Fukushige, T., Smith, H. E., Miwa, J., Krause, M. W., and Hanover, J. A. (2017). A genetic analysis of the Caenorhabditis elegans detoxification response. Genetics 206 (2), 939–952. doi:10.1534/genetics.117.202515
Ganner, A., Gerber, J., Ziegler, A. K., Li, Y., Kandzia, J., Matulenski, T., et al. (2019). CBP-1/p300 acetyltransferase regulates SKN-1/Nrf cellular levels, nuclear localization, and activity in C. elegans. Exp. Gerontol. 126, 110690. doi:10.1016/j.exger.2019.110690
Garigan, D., Hsu, A. L., Fraser, A. G., Kamath, R. S., Ahringer, J., and Kenyon, C. (2002). Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 161 (3), 1101–1112. doi:10.1093/genetics/161.3.1101
Glover-Cutter, K. M., Lin, S., and Blackwell, T. K. (2013). Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 9 (9), e1003701. doi:10.1371/journal.pgen.1003701
Grushko, D., Boocholez, H., Levine, A., and Cohen, E. (2021). Temporal requirements of SKN-1/NRF as a regulator of lifespan and proteostasis in Caenorhabditis elegans. PLoS One 16 (7), e0243522. doi:10.1371/journal.pone.0243522
Hansen, M., Flatt, T., and Aguilaniu, H. (2013). Reproduction, fat metabolism, and life span: what is the connection? Cell Metab. 17 (1), 10–19. doi:10.1016/j.cmet.2012.12.003
Hoeven, R., McCallum, K. C., Cruz, M. R., and Garsin, D. A. (2011). Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 7 (12), e1002453. doi:10.1371/journal.ppat.1002453
Holzenberger, M., Dupont, J., Ducos, B., Leneuve, P., Géloën, A., Even, P. C., et al. (2003). IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421 (6919), 182–187. doi:10.1038/nature01298
Hu, Q., D'Amora, D. R., MacNeil, L. T., Walhout, A. J. M., and Kubiseski, T. J. (2018). The Caenorhabditis elegans oxidative stress response requires the NHR-49 transcription factor. G3 (Bethesda) 8 (12), 3857–3863. doi:10.1534/g3.118.200727
Inoue, H., Hisamoto, N., An, J. H., Oliveira, R. P., Nishida, E., Blackwell, T. K., et al. (2005). The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 19 (19), 2278–2283. doi:10.1101/gad.1324805
Irazoqui, J. E., Troemel, E. R., Feinbaum, R. L., Luhachack, L. G., Cezairliyan, B. O., and Ausubel, F. M. (2010). Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6, e1000982. doi:10.1371/journal.ppat.1000982
Itoh, K., Igarashi, K., Hayashi, N., Nishizawa, M., and Yamamoto, M. (1995). Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell Biol. 15 (8), 4184–4193. doi:10.1128/MCB.15.8.4184
Itoh, K., Tong, K. I., and Yamamoto, M. (2004). Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 36 (10), 1208–1213. doi:10.1016/j.freeradbiomed.2004.02.075
Jones, K. T., Greer, E. R., Pearce, D., and Ashrafi, K. (2009). Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7 (3), e60. doi:10.1371/journal.pbio.1000060
Kansanen, E., Kuosmanen, S. M., Leinonen, H., and Levonen, A. L. (2013). The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 1 (1), 45–49. doi:10.1016/j.redox.2012.10.001
Katoh, Y., Iida, K., Kang, M. I., Kobayashi, A., Mizukami, M., Tong, K. I., et al. (2005). Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 433 (2), 342–350. doi:10.1016/j.abb.2004.10.012
Kell, A., Ventura, N., Kahn, N., and Johnson, T. E. (2007). Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic. Biol. Med. 43 (11), 1560–1566. doi:10.1016/j.freeradbiomed.2007.08.025
Kophengnavong, T., Carroll, A. S., and Blackwell, T. K. (1999). The SKN-1 amino-terminal arm is a DNA specificity segment. Mol. Cell Biol. 19 (4), 3039–3050. doi:10.1128/MCB.19.4.3039
Kumar, S., Egan, B. M., Kocsisova, Z., Schneider, D. L., Murphy, J. T., Diwan, A., et al. (2019). Lifespan extension in C. elegans caused by bacterial colonization of the intestine and subsequent activation of an innate immune response. Dev. Cell. 49 (1), 100–117. doi:10.1016/j.devcel.2019.03.010
Kurz, C. L., and Tan, M. W. (2004). Regulation of aging and innate immunity in C. elegans. Aging Cell. 3 (4), 185–193. doi:10.1111/j.1474-9728.2004.00108.x
Lehrbach, N. J., Breen, P. C., and Ruvkun, G. (2019). Protein sequence editing of SKN-1A/Nrf1 by peptide:N-glycanase controls proteasome gene expression. Cell 177 (3), 737–750. doi:10.1016/j.cell.2019.03.035
Lehrbach, N. J., and Ruvkun, G. (2016). Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. Elife 5, e17721. doi:10.7554/eLife.17721
Lemberg, M. K., and Strisovsky, K. (2021). Maintenance of organellar protein homeostasis by ER-associated degradation and related mechanisms. Mol. Cell 81 (12), 2507–2519. doi:10.1016/j.molcel.2021.05.004
Levine, Z. G., Potter, S. C., Joiner, C. M., Fei, G. Q., Nabet, B., Sonnett, M., et al. (2021). Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. Proc. Natl. Acad. Sci. U. S. A. 118 (4), e2016778118. doi:10.1073/pnas.2016778118
Li, H., Liu, X., Wang, D., Su, L., Zhao, T., Li, Z., et al. (2017). O-GlcNAcylation of SKN-1 modulates the lifespan and oxidative stress resistance in Caenorhabditis elegans. Sci. Rep. 7, 43601. doi:10.1038/srep43601
Li, H., Su, L., Su, X., Liu, X., Wang, D., Li, H., et al. (2019). Arginine methylation of SKN-1 promotes oxidative stress resistance in Caenorhabditis elegans. Redox Biol. 21, 101111. doi:10.1016/j.redox.2019.101111
Lindblom, T. H., and Dodd, A. K. (2006). Xenobiotic detoxification in the nematode Caenorhabditis elegans. J. Exp. Zool. A Comp. Exp. Biol. 305 (9), 720–730. doi:10.1002/jez.a.324
Liu, F., Wang, H., Zhu, X., Jiang, N., Pan, F., Song, C., et al. (2022). Sanguinarine promotes healthspan and innate immunity through a conserved mechanism of ROS-mediated PMK-1/SKN-1 activation. iScience 25 (3), 103874. doi:10.1016/j.isci.2022.103874
Lo, J. Y., Spatola, B. N., and Curran, S. P. (2017). WDR23 regulates NRF2 independently of KEAP1. PLoS Genet. 13 (4), e1006762. doi:10.1371/journal.pgen.1006762
Lo, M. C., Ha, S., Pelczer, I., Pal, S., and Walker, S. (1998). The solution structure of the DNA-binding domain of Skn-1. Proc. Natl. Acad. Sci. U. S. A. 95 (15), 8455–8460. doi:10.1073/pnas.95.15.8455
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cell 153 (6), 1194–1217. doi:10.1016/j.cell.2013.05.039
Lynn, D. A., and Curran, S. P. (2015). The SKN-1 hunger games: may the odds be ever in your favor. Worm 4 (3), e1078959. doi:10.1080/21624054.2015.1078959
Lynn, D. A., Dalton, H. M., Sowa, J. N., Wang, M. C., Soukas, A. A., and Curran, S. P. (2015). Omega-3 and -6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 112 (50), 15378–15383. doi:10.1073/pnas.1514012112
Ma, Q. L., Zhu, C., Morselli, M., Su, T., Pelligrini, M., Lu, Z., et al. (2020). The novel omega-6 fatty acid docosapentaenoic acid positively modulates brain innate immune response for resolving neuroinflammation at early and late stages of humanized APOE-based alzheimer's disease models. Front. Immunol. 11, 558036. doi:10.3389/fimmu.2020.558036
Maduro, M. F., Meneghini, M. D., Bowerman, B., Broitman-Maduro, G., and Rothman, J. H. (2001a). Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7 (3), 475–485. doi:10.1016/s1097-2765(01)00195-2
Maduro, M. F., Meneghini, M. D., Bowerman, B., Broitman-Maduro, G., and Rothman, J. H. (2001b). Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7 (3), 475–485. doi:10.1016/s1097-2765(01)00195-2
Maier, W., Adilov, B., Regenass, M., and Alcedo, J. (2010). A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 8 (5), e1000376. doi:10.1371/journal.pbio.1000376
Martin-Montalvo, A., Mercken, E. M., Mitchell, S. J., Palacios, H. H., Mote, P. L., Scheibye-Knudsen, M., et al. (2013). Metformin improves healthspan and lifespan in mice. Nat. Commun. 4, 2192. doi:10.1038/ncomms3192
Matai, L., Stathis, T., Lee, J. D., Parsons, C., Saxena, T., Shlomchik, K., et al. (2023). The conserved microRNA-229 family controls low-insulin signaling and dietary restriction induced longevity through interactions with SKN-1/NRF2. Aging Cell 22 (4), e13785. doi:10.1111/acel.13785
Mejhert, N., Gabriel, K. R., Frendo-Cumbo, S., Krahmer, N., Song, J., Kuruvilla, L., et al. (2022). The Lipid Droplet Knowledge Portal: a resource for systematic analyses of lipid droplet biology. Dev. Cell. 57 (3), 387–397.e4. doi:10.1016/j.devcel.2022.01.003
Mizunuma, M., Neumann-Haefelin, E., Moroz, N., Li, Y., and Blackwell, T. K. (2014). mTORC2-SGK-1 acts in two environmentally responsive pathways with opposing effects on longevity. Aging Cell 13 (5), 869–878. doi:10.1111/acel.12248
Motohashi, H., and Yamamoto, M. (2004). Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10 (11), 549–557. doi:10.1016/j.molmed.2004.09.003
Nhan, J. D., Turner, C. D., Anderson, S. M., Yen, C. A., Dalton, H. M., Cheesman, H. K., et al. (2019). Redirection of SKN-1 abates the negative metabolic outcomes of a perceived pathogen infection. Proc. Natl. Acad. Sci. U. S. A. 116 (44), 22322–22330. doi:10.1073/pnas.1909666116
Ni, C., and Buszczak, M. (2023). The homeostatic regulation of ribosome biogenesis. Semin. Cell Dev. Biol. 136, 13–26. doi:10.1016/j.semcdb.2022.03.043
Oliveira, R. P., Porter Abate, J., Dilks, K., Landis, J., Ashraf, J., Murphy, C. T., et al. (2009). Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. Sep. 8 (5), 524–541. doi:10.1111/j.1474-9726.2009.00501.x
Onken, B., and Driscoll, M. (2010). Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS One 5 (1), e8758. doi:10.1371/journal.pone.0008758
Paek, J., Lo, J. Y., Narasimhan, S. D., Nguyen, T. N., Glover-Cutter, K., Robida-Stubbs, S., et al. (2012). Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 16 (4), 526–537. doi:10.1016/j.cmet.2012.09.007
Palikaras, K., Lionaki, E., and Tavernarakis, N. (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521 (7553), 525–528. doi:10.1038/nature14300
Pang, S., and Curran, S. P. (2012). Longevity and the long arm of epigenetics: acquired parental marks influence lifespan across several generations. Bioessays. 34 (8), 652–654. doi:10.1002/bies.201200046
Pang, S., and Curran, S. P. (2014). Adaptive capacity to bacterial diet modulates aging in C. elegans. Cell Metab. 19 (2), 221–231. doi:10.1016/j.cmet.2013.12.005
Pang, S., Lynn, D. A., Lo, J. Y., Paek, J., and Curran, S. P. (2014). SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 5, 5048. doi:10.1038/ncomms6048
Papp, D., Csermely, P., and Sőti, C. (2012a). A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. Plos Pathog. 8, e1002673. doi:10.1371/journal.ppat.1002673
Papp, D., Csermely, P., and Soti, C. (2012b). A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog. 8 (4), e1002673. doi:10.1371/journal.ppat.1002673
Papsdorf, K., and Brunet, A. (2019). Linking lipid metabolism to chromatin regulation in aging. Trends Cell Biol. 29 (2), 97–116. doi:10.1016/j.tcb.2018.09.004
Papsdorf, K., Miklas, J. W., Hosseini, A., Cabruja, M., Morrow, C. S., Savini, M., et al. (2023). Lipid droplets and peroxisomes are co-regulated to drive lifespan extension in response to mono-unsaturated fatty acids. Nat. Cell Biol. 25 (5), 672–684. doi:10.1038/s41556-023-01136-6
Pukkila-Worley, R. (2016). Surveillance immunity: an emerging paradigm of innate defense activation in Caenorhabditis elegans. PLoS Pathog. 12 (9), e1005795. doi:10.1371/journal.ppat.1005795
Qi, W., Gutierrez, G. E., Gao, X., Dixon, H., McDonough, J. A., Marini, A. M., et al. (2017). The ω-3 fatty acid α-linolenic acid extends Caenorhabditis elegans lifespan via NHR-49/PPARα and oxidation to oxylipins. Aging Cell. 16 (5), 1125–1135. doi:10.1111/acel.12651
Ramos, C. M., and Curran, S. P. (2023). Comparative analysis of the molecular and physiological consequences of constitutive SKN-1 activation. Geroscience 45 (6), 3359–3370. doi:10.1007/s11357-023-00937-9
Robida-Stubbs, S., Glover-Cutter, K., Lamming, D. W., Mizunuma, M., Narasimhan, S. D., Neumann-Haefelin, E., et al. (2012). TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15 (5), 713–724. doi:10.1016/j.cmet.2012.04.007
Röder, P. V., Wu, B., Liu, Y., and Han, W. (2016). Pancreatic regulation of glucose homeostasis. Exp. Mol. Med. 48 (3), e219. doi:10.1038/emm.2016.6
Ruf, V., Holzem, C., Peyman, T., Walz, G., Blackwell, T. K., and Neumann-Haefelin, E. (2013). TORC2 signaling antagonizes SKN-1 to induce C. elegans mesendodermal embryonic development. Dev. Biol. 384 (2), 214–227. doi:10.1016/j.ydbio.2013.08.011
Rupert, P. B., Daughdrill, G. W., Bowerman, B., and Matthews, B. W. (1998). A new DNA-binding motif in the Skn-1 binding domain-DNA complex. Nat. Struct. Biol. 5 (6), 484–491. doi:10.1038/nsb0698-484
Ruvkun, G., and Lehrbach, N. (2023). Regulation and functions of the ER-associated Nrf1 transcription factor. Cold Spring Harb. Perspect. Biol. 15 (1), a041266. doi:10.1101/cshperspect.a041266
Soukas, A. A., Kane, E. A., Carr, C. E., Melo, J. A., and Ruvkun, G. (2009). Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes and Dev. 23 (4), 496–511. doi:10.1101/gad.1775409
Sowa, J. N., Jiang, H., Somasundaram, L., Tecle, E., Xu, G., Wang, D., et al. (2020). The Caenorhabditis elegans RIG-I homolog DRH-1 mediates the intracellular pathogen response upon viral infection. J. Virol. 94 (2), e01173-19. doi:10.1128/JVI.01173-19
Spatola, B. N., Lo, J. Y., Wang, B., and Curran, S. P. (2019). Nuclear and cytoplasmic WDR-23 isoforms mediate differential effects on GEN-1 and SKN-1 substrates. Sci. Rep. 9 (1), 11783. doi:10.1038/s41598-019-48286-y
Staab, T. A., Evgrafov, O., Knowles, J. A., and Sieburth, D. (2014). Regulation of synaptic nlg-1/neuroligin abundance by the skn-1/Nrf stress response pathway protects against oxidative stress. PLoS Genet. 10 (1), e1004100. doi:10.1371/journal.pgen.1004100
Staab, T. A., Griffen, T. C., Corcoran, C., Evgrafov, O., Knowles, J. A., and Sieburth, D. (2013). The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans. PLoS Genet. 9 (3), e1003354. doi:10.1371/journal.pgen.1003354
Steinbaugh, M. J., Narasimhan, S. D., Robida-Stubbs, S., Moronetti Mazzeo, L. E., Dreyfuss, J. M., Hourihan, J. M., et al. (2015). Lipid-mediated regulation of SKN-1/Nrf in response to germ cell absence. Elife 4, e07836. doi:10.7554/eLife.07836
Stuhr, N., and Curran, S. (2023). Different methods of killing bacteria diets differentially influence Caenorhabditis elegans physiology. Micropubl. Biol. 2023. doi:10.17912/micropub.biology.000902
Stuhr, N. L., and Curran, S. P. (2020). Bacterial diets differentially alter lifespan and healthspan trajectories in C. elegans. Commun. Biol. 3 (1), 653. doi:10.1038/s42003-020-01379-1
Sun, Z., Huang, Z., and Zhang, D. D. (2009). Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One 4 (8), e6588. doi:10.1371/journal.pone.0006588
Sykiotis, G. P., and Bohmann, D. (2008). Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell. 14 (1), 76–85. doi:10.1016/j.devcel.2007.12.002
Sykiotis, G. P., and Bohmann, D. (2010). Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 3 (112), re3. doi:10.1126/scisignal.3112re3
Sykiotis, G. P., Habeos, I. G., Samuelson, A. V., and Bohmann, D. (2011). The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care 14 (1), 41–48. doi:10.1097/MCO.0b013e32834136f2
Taguchi, K., Motohashi, H., and Yamamoto, M. (2011). Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes cells. 16 (2), 123–140. doi:10.1111/j.1365-2443.2010.01473.x
Tang, L., and Choe, K. P. (2015). Characterization of skn-1/wdr-23 phenotypes in Caenorhabditis elegans; pleiotrophy, aging, glutathione, and interactions with other longevity pathways. Mech. Ageing Dev. 149, 88–98. doi:10.1016/j.mad.2015.06.001
Torday, J. S. (2015). Homeostasis as the mechanism of evolution. Biol. (Basel) 4 (3), 573–590. doi:10.3390/biology4030573
Troemel, E. R., Chu, S. W., Reinke, V., Lee, S. S., Ausubel, F. M., and Kim, D. H. (2006). p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2 (11), e183. doi:10.1371/journal.pgen.0020183
Tullet, J. M., Hertweck, M., An, J. H., Baker, J., Hwang, J. Y., Liu, S., et al. (2008). Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132 (6), 1025–1038. doi:10.1016/j.cell.2008.01.030
Turner, C. D., Stuhr, N. L., Ramos, C. M., Van Camp, B. T., and Curran, S. P. (2023). A dicer-related helicase opposes the age-related pathology from SKN-1 activation in ASI neurons. Proc. Natl. Acad. Sci. U. S. A. 120 (52), e2308565120. doi:10.1073/pnas.2308565120
Uruno, A., and Motohashi, H. (2011). The Keap1-Nrf2 system as an in vivo sensor for electrophiles. Nitric Oxide 25 (2), 153–160. doi:10.1016/j.niox.2011.02.007
Vertegaal, A. C. O. (2022). Signalling mechanisms and cellular functions of SUMO. Nat. Rev. Mol. Cell Biol. 23 (11), 715–731. doi:10.1038/s41580-022-00500-y
Vitale, G., Pellegrino, G., Vollery, M., and Hofland, L. J. (2019). ROLE of IGF-1 system in the modulation of longevity: controversies and new insights from a centenarians' perspective. Front. Endocrinol. (Lausanne) 10, 27. doi:10.3389/fendo.2019.00027
Wang, J., Robida-Stubbs, S., Tullet, J. M., Rual, J. F., Vidal, M., and Blackwell, T. K. (2010). RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. Aug 6 (8), e1001048. doi:10.1371/journal.pgen.1001048
Willcox, B. J., Donlon, T. A., He, Q., Chen, R., Grove, J. S., Yano, K., et al. (2008). FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U. S. A. 105 (37), 13987–13992. doi:10.1073/pnas.0801030105
Wu, C. W., Deonarine, A., Przybysz, A., Strange, K., and Choe, K. P. (2016). The Skp1 homologs SKR-1/2 are required for the Caenorhabditis elegans SKN-1 antioxidant/detoxification response independently of p38 MAPK. PLoS Genet. 12 (10), e1006361. doi:10.1371/journal.pgen.1006361
Wu, C. W., Wang, Y., and Choe, K. P. (2017). F-box protein XREP-4 is a new regulator of the oxidative stress response in Caenorhabditis elegans. Genetics 206 (2), 859–871. doi:10.1534/genetics.117.200592
Wu, N., Ma, Y. C., Gong, X. Q., Zhao, P. J., Jia, Y. J., Zhao, Q., et al. (2023). The metabolite alpha-ketobutyrate extends lifespan by promoting peroxisomal function in C. elegans. Nat. Commun. 14 (1), 240. doi:10.1038/s41467-023-35899-1
Wu, S., Lu, H., and Bai, Y. (2019). Nrf2 in cancers: a double-edged sword. Cancer Med. 8 (5), 2252–2267. doi:10.1002/cam4.2101
Xiao, R., Chun, L., Ronan, E. A., Friedman, D. I., Liu, J., and Xu, X. Z. (2015). RNAi interrogation of dietary modulation of development, metabolism, behavior, and aging in C. elegans. Cell Rep. 11 (7), 1123–1133. doi:10.1016/j.celrep.2015.04.024
Xu, C., Li, C. Y., and Kong, A. N. (2005). Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch. Pharm. Res. 28 (3), 249–268. doi:10.1007/BF02977789
Yen, C.-A., Ruter, D. L., Turner, C. D., Pang, S., and Curran, S. P. (2020). Loss of flavin adenine dinucleotide (FAD) impairs sperm function and male reproductive advantage in C. elegans. Elife 9, e52899. doi:10.7554/eLife.52899
Yoshida, Y., Asahina, M., Murakami, A., Kawawaki, J., Yoshida, M., Fujinawa, R., et al. (2021). Loss of peptide: N -glycanase causes proteasome dysfunction mediated by a sugar-recognizing ubiquitin ligase. Proc. Natl. Acad. Sci. U. S. A. 118 (27), 118. doi:10.1073/pnas.2102902118
Youle, R. J., and Narendra, D. P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12 (1), 9–14. doi:10.1038/nrm3028
Keywords: SKN-1, C. elegans, Nrf2, aging, cytoprotection, genetics, physiology, metabolism
Citation: Turner CD, Ramos CM and Curran SP (2024) Disrupting the SKN-1 homeostat: mechanistic insights and phenotypic outcomes. Front. Aging 5:1369740. doi: 10.3389/fragi.2024.1369740
Received: 12 January 2024; Accepted: 15 February 2024;
Published: 04 March 2024.
Edited by:
Robert Joseph Shmookler Reis, United States Department of Veterans Affairs, United StatesReviewed by:
Mark A. McCormick, University of New Mexico, United StatesIlke Sen, INSERM U955 Institut Mondor de Recherche Biomédicale (IMRB), France
Copyright © 2024 Turner, Ramos and Curran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sean P. Curran, spcurran@usc.edu
†These authors have contributed equally to this work
 Chris D. Turner
Chris D. Turner Carmen M. Ramos
Carmen M. Ramos Sean P. Curran
Sean P. Curran