Activity assay based on the immobilized enzyme kallikrein and mass spectrometry
- 1Departamento de Química, Grupo de Cromatografia de Bioafinidade e Produtos Naturais, Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, Brazil
- 2Division of BioAnalytical Chemistry, Department of Chemistry and Pharmaceutical Sciences, Amsterdam Institute of Molecules, Medicine, and Systems, Vrije Universiteit Amsterdam, Amsterdam, Netherlands
Deregulated activity and expression of human kallikreins (KLKs) may be involved in various pathologies, so these enzymes are an attractive biological target for identifying molecules that can modulate KLK activity. This identification involves applying fast and efficient screening methods. This work describes an off-line assay with mass spectrometry (MS) detection that uses KLK immobilized on Sepharose-NHS as a micro-column configuration (IMER-KLK-Sepharose-NHS). The mass spectrometry used has an ion trap analyzer and electrospray ionization (EIS). The HPLC-MS method for quantifying KLK activity was developed. The enzymatic assay conditions were optimized, and the IMER-KLK-Sepharose-NHS kinetic parameter (KMapp = 15.48 ± 3 μmol L−1) was evaluated. Finally, the method was validated by using leupeptin as a reference inhibitor (IC50 = 0.85 ± 0.10 μmol L−1). The developed method was able to identify the reference inhibitor and can be an alternative for screening KLK inhibitors.
Introduction
Kallikreins (KLKs) are a group of serine protease enzymes that play an important role in biological fluids and tissues. KLKs can be divided into two groups: plasma kallikrein (KLK 1B) and tissue KLKs (KLK1–15) (Prassas et al., 2015). KLK 1B is exclusively expressed by hepatic cells and is part of the plasma kallikrein-kinin system, which, among other things, participates in the processes of fibrinolysis, blood coagulation, inflammation, cancer, and pathologies related to the cardiovascular system (Schmaier and Mccrae, 2007; Costa-Neto et al., 2008). Under physiological conditions, plasma kallikrein serves as a cardioprotective enzyme. However, its increased plasma concentration or hyperactivity perpetuates cardiovascular disease (CVD) (Kolte and Shariat-Madar, 2016). Tissue KLKs comprise human tissue kallikrein (KLK1) and 14 kallikrein-related peptidases (KLK2-15) (Prassas et al., 2015) that share genetic and protein structural similarities. Amino acid identity among human tissue KLKs ranges from 40 to 80% (Yousef and Diamandis, 2001).
Tissue KLKs are present in several tissues of the human organism, and their unregulated expression may be associated with various diseases (Costa-Neto et al., 2008). For example, in patients with Alzheimer’s and Parkinson’s disease, KLK6 has decreased expression in brain tissue (Diamandis et al., 2000; Bayani and Diamandis, 2012). The human kallikrein-related peptidases 5 (hKLK5), 6 (hKLK), 7 (hKLK7), 8 (hKLK8), and 14 (hKLK14) are also involved in inflammatory skin disorders (Di Paolo, Diamandis and Prassas, 2020) with abnormal desquamation and inflammation, such as atopic dermatitis (AD), psoriasis, and the rare disease Netherton syndrome (NS) (Zani et al., 2022). In addition, KLKs are dysregulated in most solid tumors, especially in hormone-dependent tumors (Kryza et al., 2016). KLK3 also known as prostate specific antigen (PSA) is used as a biomarker for prostate cancer because this enzyme is overexpressed in these types of cancers and KLK3 presents dysregulated expression in breast tumors (Yu et al., 1995). KLK2 is overexpressed in prostate tumors (Darson et al., 1997). KLK7 overexpression in colon cancer tissues is involved in cancer cell proliferation (Walker et al., 2014).
Given that some enzymes underlie pathologies that affect humanity, searching for molecules that can inhibit them is a relevant topic when searching for a treatment or cure for these pathologies (Copeland, 2013). In this scenario, KLKs are important targets when it comes to developing drugs that can inhibit their activities or restore their expression (Kolte and Shariat-Madar, 2016; Xie et al., 2020). This is particularly relevant in the context of anticancer studies because KLKs have been shown to modulate cancer cell multiplication (Shang et al., 2014; Walker et al., 2014). The PROSTVAC vaccine, developed for prostate cancer treatment, is an example of a therapeutic resource that targets KLK3 and multiple T cell co-stimulatory molecules (TRICOM) (Madan et al., 2009).
Developing tools that allow enzyme inhibitors to be discovered and developed is therefore essential. In this sense, enzyme immobilization is an important tool for screening ligands that can potentially inhibit enzymes. Assays with immobilized enzymes offer advantages over the use of enzymes in solution—they allow enzymes to be better handled and reused, and they increase enzyme stability under varying conditions (Brena, González-Pombo and Batista-Viera, 2013). Additionally, enzyme immobilization allows enzymatic activity modulation to be monitored in the presence of libraries of molecules (synthetic or natural) and complex matrixes, like crude extracts (Singh et al., 2014). Moreover, enzyme immobilization enables high-throughput screening (HTS) of ligands from such matrixes (De Simone et al., 2019). Indeed, different enzyme immobilization protocols (Rodrigues et al., 2021) and ligand screening assays, ranging from online (flow) to offline assays, have been developed (Wubshet et al., 2019; Cieśla and Moaddel, 2016; Zhuo et al., 2016; Cheng and Chen, 2018; de Moraes, Cardoso and Cass, 2019; Q. Chen et al., 2021; Trindade Ximenes et al., 2021; de Oliveira et al., 2022).
In online assays, ligands are analyzed and screened simultaneously (Zhuo et al., 2016). These assays basically consist of an immobilized enzyme reactor (IMER) coupled to an automated system, such as high-performance liqbib_Chen_et_al_2021buid chromatography (HPLC) or capillary electrophoresis (Cheng and Chen, 2018; G. Y. Chen et al., 2021), which can be coupled with different detection techniques, including ultraviolet (UV) (da Silva et al., 2013), fluorescence (Hai et al., 2011), electrochemical detection (Sun, Gao and Jin, 2006), and mass spectrometry (MS) (Vilela and Cardoso, 2017).
HPLC-MS is considered the most powerful analytical tool for analyzing polar compounds, including peptides, phenols, polymers, and biomolecules (Loos, Van Schepdael and Cabooter, 2016). HPLC-MS combines separation by HPLC with the broad analysis capability of MS. The advantages of HPLC-MS include the need for a small amount of sample, high resolution power, high molecular specificity, and high detection sensitivity (Beccaria and Cabooter, 2020). The ionization mode is crucial for the performance of HPLC-MS method depending on the analytes targets, between others the electrospray ionization source (ESI) is the main ionization mode analysis for thermally labile, nonvolatile, and polar compounds. (Beccaria and Cabooter, 2020). Specifically, ESI ionization source and ion trap mass analyzer (IT) has been successfully used in IMER coupled with HPLC-MS. For example, Beta secretase1 and acetylcholinesterase (AChE) were co-immobilized and used to dual-ligand screening (Vilela, Narciso dos Reis, and Cardoso 2021) AChE-IMER (Vanzolini et al., 2013) used to screening coumarin derivatives and butyrylcholinesterase (BChE)-IMER and AChE-IMER used in on-flow mass spectrometry dual enzyme assay (Seidl et al., 2019).
On the other hand, off-line assays entail independent screening and analysis steps. However, the detection techniques mentioned above can be used in offline assays, as well (Zhuo et al., 2016; Zhang et al., 2018).
Numerous supports, such as magnetic particles (Wang et al., 2018; Q. Chen et al., 2021), silica gel microspheres (Shi et al., 2015), and Sepharose (Zofair et al., 2020), can be used to construct bioreactors that can be applied in off-line assays. Previously, we immobilized the KLK enzyme on Sepharose-NHS solid support and employed it in ligand screening assays during which we used a 96-well flat-bottom microplate to measure fluorescence (Carvalho et al., 2021). The immobilized KLK showed excellent operational and storage stability and proved an efficient tool for identifying KLK inhibitors (Carvalho et al., 2021).
Bearing the advantages of enzyme immobilization and the growing search for KLK inhibitors in mind, we present an off-line assay based on KLK immobilized on Sepharose-NHS as a micro-column configuration and MS detection. We will show that this bioreactor, IMER-KLK-Sepharose-NHS, exhibits excellent stability, can be reused, is a useful tool in off-line ligand screening, and can be combined with an HPLC-MS method.
Materials and methods
Chemical and reagents
Porcine pancreas kallikrein (KLK, 250 units), N-hydroxysuccinimidyl-Sepharose® 4 Fast Flow, CBZ-Phe-Arg-7-amido-4-methylcoumarin hydrochloride (Z-Phe-Arg-AMC), leupeptin hemisulfate salt (Acetyl-Leu-Leu-Arg-al), and 7-amino-4-methylcoumarin (AMC) were purchased from Sigma-Aldrich (St. Louis, MO, United States). Buffer components and other chemicals were acquired from Sigma-Aldrich, Merck (Darmstadt, Germany), Synth (São Paulo, Brazil), or Acros (Geel, Belgium). The water that was used in all the preparations was obtained from a MILLI-Q® system (Millipore®, São Paulo, Brazil). All the chemicals and solvents used here were analytical or HPLC grade and were employed without any further purification.
Instrumentation and system configuration for analyses
HPLC-MS analyses were carried out on a NexeraXR high-performance liquid chromatography system (Shimadzu, Kyoto, Japan) equipped with three LC 20ADXR pumps, an SIL-20A automatic injector, a DGU-20A degasser, a 10-port two-position high-pressure switching valve (Valco Instruments Co. Inc., Houston, United States), and a CBM-20A system controller. The HPLC system was coupled to ion trap mass spectrometer, model AmaZon speed dual ion funnel, QIT technology (IT-MS) (Bruker Daltonics, Bremen, Germany) equipped with an electrospray source (ESI). The system was controlled by Bruker Compass Hystar software (version 4.5, Bruker Daltonics Inc., Billerica, United States). Data were acquired and analyzed by using the Compass DataAnalysis software (version 4.3, Bruker Daltonics Inc., Billerica, United States).
Preparation of IMER-KLK-sepharose-NHS micro-column
KLK from porcine pancreas (100 μg ml−1) was covalently immobilized on N-hydroxysuccinimidyl-Sepharose® 4 Fast Flow (Sepharose-NHS, solid support) as previously described (Carvalho et al., 2021). After immobilization, 80 mg of the immobilized enzyme was placed in a micro-column device (Mobitec, Göttingen, Germany), to give IMER-KLK-Sepharose-NHS. The micro-column was washed with 50 mmol L−1 Tris-HCl buffer (pH 8.0), and stored in the same buffer solution at 4 C. Control columns containing 80 mg of Sepharose-NHS (without enzyme) were prepared as described above.
IMER-KLK-sepharose-NHS micro-column off-line assay
Aliquots of 120 µl of 10 mmol L−1 ammonium acetate solution (pH 8.0), and 50 µl of 200 μmol L−1 Z-Phe-Arg-AMC substrate were added to the micro-column and incubated under gentle agitation at room temperature for 5 min. After incubation, the reaction medium was filtered and collected. Then, 20-µL aliquots of the reaction mixture (filtered) were transferred to vials, diluted with 80 µl of 10 mmol L−1 ammonium acetate solution (pH 8.0), and analyzed by HPLC-MS. At the end of each reaction, the IMER-KLK-Sepharose-NHS micro-column was washed with 5 ml of Milli-Q water (three times), followed by 5 ml of 5 mmol L−1 ammonium acetate solution (pH 8.0) three times. At the end of each reaction, the immobilized KLK micro-column was washed with 5 ml of Milli-Q water (three times), followed by 5 ml of 5 mmol L−1 ammonium acetate solution pH 8.0 (for 3 times). Blank samples were prepared under the same reaction conditions by using control micro-columns (without enzyme). All the experiments were performed in triplicate.
The optimization of IMER-KLK-Sepharose-NHS off-line assay conditions included the effect of the reaction time and ammonium acetate solution concentration on the enzymatic activity. The effect of ammonium acetate solution concentration was evaluated at concentrations ranging from 2 to 15 mmol L−1 (pH 8.0). For this, the immobilized KLK micro-column was previously washed with 5 ml of each concentration of ammonium acetate pH 8.0 (three times) and left under conditioning for 10 min at each concentration. The reaction time was evaluated for incubation times ranging from 5 to 40 min. The enzymatic reactions were carried out under the same conditions described above, and Z-Phe-Arg-AMC was employed as substrate. The reproducibility of the IMER-KLK-Sepharose-NHS activity assay was verified by carrying out five consecutive reaction cycles.
Analysis of the IMER-KLK-sepharose-NHS activity by HPLC-MS
HPLC analyses were carried out by using an Ascentis® Express C18 micron guard cartridge (5 mm × 2.1 mm x 2.7 µm, Supelco, Bellefonte, PA, United States) connected to a 10-port/two-position valve in the HPLC-MS system (Figure 1); the mobile phase consisted of water (solvent A) and acetonitrile (solvent B). Initially, with the valve in position 1, the enzymatic reaction products were eluted from the column with an initial mobile phase composed of 30% solvent B at a flow rate of 0.2 ml min−1 for analysis in the mass spectrometer. After 2.0 min, the valve was switched to position 2; solvent B was increased to 90% at a flow rate of 0.5 ml min−1, which was maintained until 5 min, to wash the column and to remove excess substrate. At 5.1 min, the valve was switched to position 1 again, and the mobile phase was returned to the initial condition. The total run time was 6 min. Pump C (solvent water) at a flow rate of 0.2 ml min−1 was used to deliver the mobile phase to the MS during the column washing step. All the analyses were performed at 21 C (controlled room temperature), and the injection volume was 5 µl. The ESI ionization parameters were as follows: positive ionization mode, capillary voltage = 4500 V, end plate voltage = 550 V, drying gas flow = 7 L min−1, drying temperature = 270 C, and nebulizer pressure = 27 psi. The product of the enzymatic reaction, AMC, was monitored at m/z 176 [M + H]+.
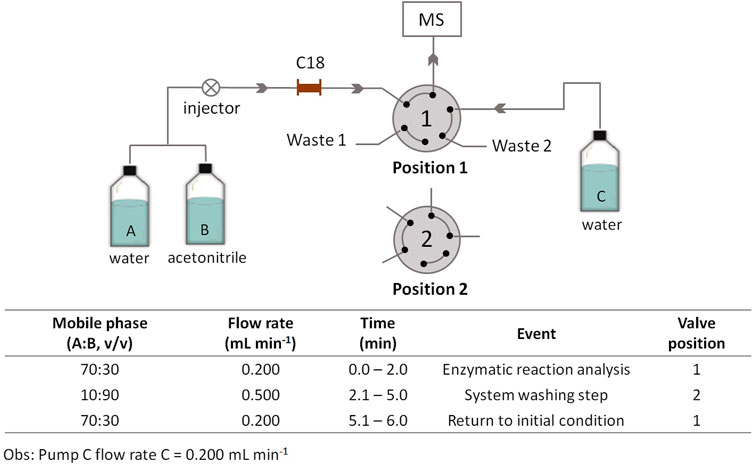
FIGURE 1. Schematic representation of the HPLC–MS system for monitoring the IMER-KLK-Sepharose-NHS activity. Source: Designed by the authors.
Method qualification
Method qualification was performed on the basis of literature criteria (FDA 2011). Method linearity was evaluated by constructing a standard curve; AMC at the following concentrations was used: 50, 70, 100, 200, 300, 400, 500, 600, 800, and 1,000 nmol L−1. For this purpose, from a standard 10 mmol L−1 AMC solution (in DMSO), the AMC solutions at concentrations ranging from 50 to 1,000 nmol L−1 were prepared in 10 mmol L−1 ammonium acetate solution. For the analytical curve, the solutions were prepared in triplicate from the concentrated stock solutions, and 5-μL aliquots were injected into the HPLC-MS system as described above. The analytical curve was constructed by linear regression of the plot of the chromatogram area of the extracted ions with m/z 176 [M + H]+ as a function of the concentration. Method selectivity was evaluated by buffer analysis under the same conditions described above, but without adding AMC. The limit of quantification and the limit of detection were evaluated from the injection of AMC solutions at concentrations of 50, 30, 10, 5, and 2.5 nmol L−1. The -limit of quantification was defined as the lowest concentration that produced a signal-to-noise value higher than ten times that response obtained by analyzing the blank (buffer without AMC), while the limit of detection was defined as the lowest concentration of an analyte in a sample that can be consistently detected with a stated probability and could be reliably differentiated from the baseline. When based on signal-to-noise establishing the minimum concentration at which the analyte can reliably be detected. Typically, acceptable signal-to-noise ratios are 2:1 or 3:1. (FDA 2011). The intra- and inter-day precision and accuracy were evaluated by analyzing samples at three different concentration levels (50, 500, and 1,000 nmol L−1); each concentration level was prepared and analyzed in quintuplicate.
IMER-KLK-sepharose-NHS kinetic constant (KMapp)
The IMER-KLK-Sepharose-NHS kinetic constant (KMapp) was determined by using different Z-Phe-Arg-AMC concentrations (4–1,200 μmol L−1) in 10 mmol L−1 ammonium acetate solution, pH 8.0. After incubation for 5 min, the reaction medium was filtered and collected, and 20-µL aliquots of the solution were transferred to vials, diluted with 80 µl of 10 mmol L−1 ammonium acetate solution (pH 8.0), and analyzed by HPLC-MS as describe above. The enzymatic activity was monitored by quantifying the AMC that was formed (m/z 176 [M + H]+). Data were fitted by using nonlinear regression into a Michaelis-Menten plot, and KMapp values were obtained with the GraphPad Prism 8.0 software.
Inhibition studies
Inhibition studies were carried out by using leupeptin as standard inhibitor. To determine their half maximum inhibitory concentration (IC50), 110-µL aliquots of 10 mmol L−1 ammonium acetate solution (pH 8.0), 10 µl of leupeptin (1–2,500 μmol L−1), and 50 µl of the substrate Z-Phe-Arg-AMC at 200 μmol L−1 were added to the micro-column and incubated under gentle agitation at room temperature for 5 min. After incubation, the reaction medium was filtered and collected, and 20-µl aliquots of the solution were transferred to vials, diluted with 80 µl of 10 mmol L−1 ammonium acetate solution (pH 8.0), and analyzed by HPLC-MS as describe above. The enzymatic activity was monitored by quantifying the AMC that was formed (m/z 176 [M + H]+). The activities in the presence (Ai) and absence (A0) of the inhibitor were compared, and the percentage of inhibition was calculated by the expression: %I = 100—[(Ai/A0) × 100]. The inhibition curve was constructed by plotting the percentage inhibition versus the corresponding leupeptin concentration, and the IC50 value was determined by plot sigmoidal curve build by log [leupeptin] versus % inhibition with the GraphPad Prism 5.0 software.
Results and discussion
Assays based on immobilized enzyme coupled to HPLC–MS systems have emerged as a promising alternative to colorimetric screening assays in microplates. Here, we developed an off-line assay with MS detection by using KLK immobilized on Sepharose-NHS as a micro-column configuration. We prepared the immobilized enzyme by a batch approach. After we covalently immobilized KLK onto Sepharose-NHS, we packed it into micro-column devices, to obtain IMER-KLK-Sepharose-NHS.
HPLC-MS analysis method development
We performed chromatographic analysis in the isocratic elution mode with a mobile phase composed of water/acetonitrile; a C18 micron guard cartridge (5 mm × 2.1 mm × 2.7 µm) was used. To eliminate the substrate excess and to avoid possible problems caused by ionization suppression and/or contamination in the MS ionization source, we added a clean-up step, as described in Figure 1.
Figure 2 shows the chromatogram corresponding to the analysis of the IMER-KLK-Sepharose-NHS enzymatic reaction by the proposed method. Under the employed conditions, we eliminated the interference caused by excess substrate, but the products AMC and Z-Phe-Arg-OH co-eluted. In this case, there was no need for high-resolution chromatographic separation even though the analytes co-eluted because we later identified them according to their respective m/z ratio. Therefore, this method allowed us to monitor the KLK activity by fast analysis, which lasted only 6 min. In routine analyses, we evaluated the IMER-Sepharose-NHS enzymatic activity by monitoring the ion with m/z 176 [M + H]+, which refers to the AMC product.
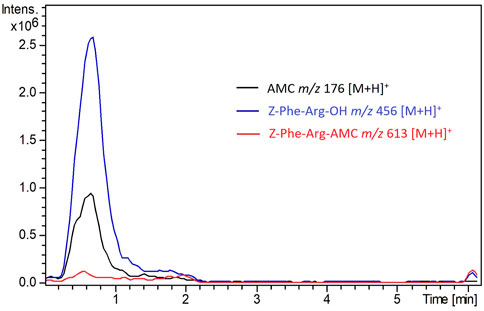
FIGURE 2. Overlap of peaks in the extracted ion chromatograms recorded for the IMER-KLK-Sepharose-NHS activity. See HPLC-MS conditions described in the experimental section Source: Designed by the authors.
Method qualification
We evaluated method linearity by constructing a calibration curve for the AMC product. The curve showed a linear response and R2 = 0.99 (n = 3) for AMC concentrations ranging from 50 to 1,000 nmol L−1 (y = 5.07 × 107 x + 8.85 × 105); the relative standard deviation (RSD) values were below 15% for the triplicates. When we injected only the buffer under the same assay conditions, no signal due to AMC emerged, which demonstrated method selectivity. The limits of detection and quantification for AMC were 5 and 30 nmol L−1, respectively. The intra- and inter-day precision values were expressed in RSD and ranged from 5 to 15%. The intra-day accuracy of the method ranged from 85 to 110%. The inter-day accuracy lay between 87 and 101%. Thus, the developed HPLC–MS method can be applied to monitor the immobilized KLK activity.
IMER-KLK-sepharose-NHS off-line assay conditions
We evaluated the reaction conditions for the IMER-KLK-Sepharose-NHS activity assay, namely reaction time, ammonium acetate solution concentration, and immobilized KLK micro-column washing steps. We evaluated the IMER-KLK-Sepharose-NHS activity at different incubation times (Figure 3A). We obtained a linear response for AMC formation as a function of reaction time, which showed that the proposed assay model can be used to monitor the IMER-KLK-Sepharose-NHS activity. Among the analyzed conditions, incubation for 5 min was sufficient to measure the enzymatic activity, so we chose this time for further analyses.
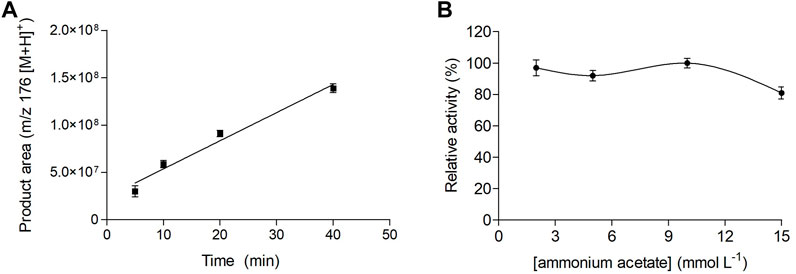
FIGURE 3. Effect of reaction time (A) and ammonium acetate solution concentration (B) on the IMER-KLK-Sepharose-NHS activity. Source: Designed by the authors.
Figure 3B shows how the ammonium acetate solution concentration, pH 8.0, affected the immobilized KLK activity. The immobilized enzyme remained active between 2 and 10 mmol L−1 ammonium acetate. In contrast, IMER-KLK-Sepharose-NHS showed lower activity at higher ammonium acetate concentration (15 mmol L−1).
Hence, the optimized assay conditions for the IMER-KLK-Sepharose-NHS activity off-line assay were incubation time of 5 min and 10 mmol L−1 ammonium acetate solution (pH 8.0).
Another important parameter to evaluate is the clean-up step of the immobilized KLK micro-column, to ensure that it can be reused and that results are reproducible. KLK immobilized on Sepharose-NHS had previously been shown to have excellent reusability after simple washing steps with washing buffer (10 mmol L−1 Tris-HCl, pH 8.0) (Carvalho et al., 2021). Here, we adjusted the clean-up step to the conditions used during MS detection, and we replaced the washing buffer with 5 mmol L−1 ammonium acetate (pH 8.0) 0. The final clean-up conditions of the IMER-KLK-Sepharose-NHS were 5 ml of Milli-Q water followed by 5 ml of 5 mmol L−1 ammonium acetate solution (pH 8.0). The washing procedure proved adequate for cleaning the immobilized KLK micro-column. In routine analyses, we repeated this washing procedure three times.
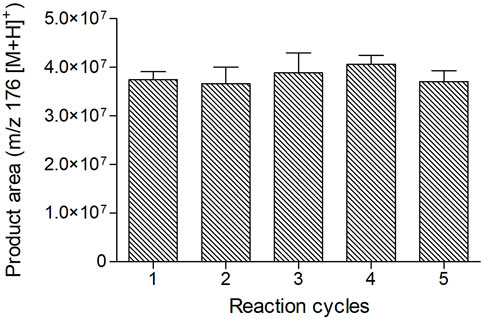
In addition, we tested IMER-KLK-Sepharose-NHS in repeated reaction cycles (Figure 4), which showed excellent reproducibility in five consecutive reactions (CV = 4.3%, n = 5) and indicated that the proposed model assay was suitable for monitoring the IMER-KLK-Sepharose-NHS activity. The immobilized enzyme showed good stability and remained active throughout the assay.
IMER-KLK-sepharose-NHS kinetic constant (KMapp)
We determined the kinetic parameter KMapp for Z-Phe-Arg-AMC hydrolysis by IMER-KLK-Sepharose-NHS under the conditions established for the enzymatic reaction and for the HPLC–MS analysis method. The KMapp value was 15.48 ± 3 μmol L−1 (Figure 5), corroborating the value previously obtained by the fluorescence assay on microplates of 10.3 ± 0.9 μmol L−1 (Carvalho et al., 2021). Thus, we selected 40 μmol L−1 substrate (corresponding to more than twice the KMapp) for the inhibition studies.
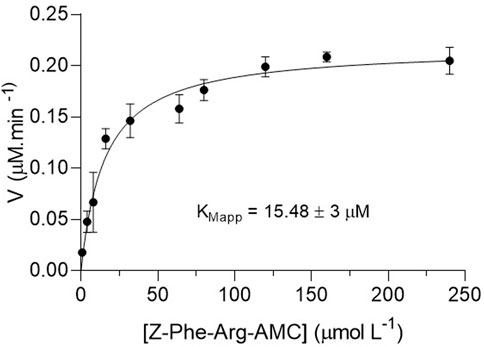
FIGURE 5. Michaelis-Menten plots of the product area obtained for IMER-KLK-Sepharose-NHS by using Z-Phe-Arg-AMC as substrate. Source: Elaborated by the authors.
Inhibition studies
As a proof of concept, we used the peptide leupeptin as a reference inhibitor to validate the off-line assay employing IMER-KLK-Sepharose-NHS as a tool for screening ligands. To determine the inhibitory potency, we evaluated the IMER-KLK-Sepharose-NHS activity in the presence of increasing leupeptin concentrations and obtained IC50 of 0.85 ± 0.10 μmol L−1 (Figure 6), which is in the same order of magnitude as the value obtained by the microplate assay and fluorescence detection IC50 of 0.13 ± 0.01 μmol L−1 and to IC50 obtained for the free enzyme in solution (IC50 = 1.62 ± 0.18 μmol L−1 (Carvalho et al., 2021). The IC50 values vary when the test conditions are modified, so it is a relative comparison parameter (Holdgate, Meek and Grimley, 2018). In general, IMER-KLK-Sepharose-NHS was able to identify the reference inhibitor and proved to be effective in determining the IC50 parameter.
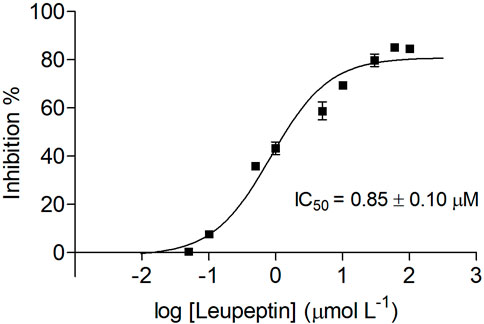
FIGURE 6. Dose-response curve plot of inhibition percentage for IMER-KLK-Sepharose-NHS in the presence of leupeptin. Source: Designed by the authors.
Conclusion
KLK immobilized on Sepharose-NHS as a micro-column configuration is a useful approach for measuring KLK activity; exhibits satisfactory stability; allows the enzyme to be reused; and can be combined with an HPLC–MS method off-line. We determined the KLK activity by quantifying the AMC product through the proposed HPLC–MS method. The developed assay was also able to identify the known KLK inhibitor leupeptin, demonstrating that it can be used for screening inhibitors. Thus, the off-line assay with MS detection reported here represents a good alternative to the fluorescence microplate assays.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
DC: methodology, validation, formal analysis, investigation, writing—original draft, BL: formal analysis, validation. CR: writing—original draft formal analysis. EA: software, writing—review GS: writing—review. JK: writing—review. CC: conceptualization, funding acquisition, project administration, resources, visualization, supervision, writing—review and editing.
Funding
This research was supported by the São Paulo State Foundation (FAPESP grants 2019/05363-0 and 2014/50249-8, 2014/50299-5 and GSK), and by the National Council for Scientific and Technological Development (CNPq) DCR and CLR acknowledge FAPESP scholarship (grand numbers 2016/14482-5, CNPq—Grants 141748/2017-6, 303723/2018-1, and 311969/2019-4). This study was partially financed by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (CAPES), Finance Code 001. The opinions, hypotheses and criteria or recommendations expressed in this material are the responsibility of the author(s) and do not necessarily reflect the views of FAPESP.
Acknowledgments
CC and DC acknowledge the Sao Paulo State Research Foundation (FAPESP) 2016-14482-5 and CNPq (307108-2021-0; 141748/2017-6).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frans.2022.1018115/full#supplementary-material
References
Bayani, J., and Diamandis, E. P. (2012). The physiology and pathobiology of human kallikrein-related peptidase 6 (KLK6). Clin. Chem. Lab. Med. 50 (2), 211–233. doi:10.1515/cclm.2011.750
Beccaria, M., and Cabooter, D. (2020). Current developments in LC-MS for pharmaceutical analysis. Analyst 145 (4), 1129–1157. doi:10.1039/C9AN02145K
Brena, B., González-Pombo, P., and Batista-Viera, F. (2013). “Immobilization of enzymes: A literature survey,” in Immobilization of enzymes and cells. Editor J. Guisan. 2nd edn. (Totowa: Humana Press), 15–31. doi:10.1007/978-1-62703-550-7_2
Chen, G. Y., Zhang, H., Zhang, C. Y., Wang, Y., Zhao, C. P., Chen, H., et al. (2021). Immobilized kallikrein microreactor based on capillary electrophoresis for online enzyme kinetics analysis and inhibitor screening. Chromatographia 84 (12), 1141–1150. doi:10.1007/s10337-021-04098-9
Chen, Q., Zhu, L., Yip, K. M., Tang, Y., Liu, Y., Jiang, T., et al. (2021). A hybrid platform featuring nanomagnetic ligand fishing for discovering COX-2 selective inhibitors from aerial part of Saussurea laniceps Hand.-Mazz. J. Ethnopharmacol. 271, 113849. doi:10.1016/j.jep.2021.113849
Cheng, M., and Chen, Z. (2018). Recent advances in screening of enzymes inhibitors based on capillary electrophoresis. J. Pharm. Anal. 8 (4), 226–233. doi:10.1016/j.jpha.2018.05.002
Cieśla, Ł., and Moaddel, R. (2016). Comparison of analytical techniques for the identification of bioactive compounds from natural products. Nat. Prod. Rep. 33 (10), 1131–1145. doi:10.1039/c6np00016a
Copeland, R. A. (2013). Evaluation of enzyme inhibitors in drug discovery. Hoboken: Wiley. doi:10.1002/9781118540398
Costa-Neto, C. M., Dillenburg-Pilla, P., Heinrich, T. A., Parreiras-e-Silva, L. T., Pereira, M. G. A. G., Reis, R. I., et al. (2008). Participation of kallikrein–kinin system in different pathologies. Int. Immunopharmacol. 8 (2), 135–142. doi:10.1016/j.intimp.2007.08.003
da Silva, J. I., de Moraes, M. C., Vieira, L. C. C., Correa, A. G., Cass, Q. B., and Cardoso, C. L. (2013). Acetylcholinesterase capillary enzyme reactor for screening and characterization of selective inhibitors. J. Pharm. Biomed. Anal. 73, 44–52. doi:10.1016/j.jpba.2012.01.026
Darson, M. F., Pacelli, A., Roche, P., Rittenhouse, H. G., Wolfert, R. L., Young, C. Y., et al. (1997). Human glandular kallikrein 2 (hK2) expression in prostatic intraepithelial neoplasia and adenocarcinoma: A novel prostate cancer marker. Urology 49 (6), 857–862. doi:10.1016/S0090-4295(97)00108-8
de Moraes, M. C., Cardoso, C. L., and Cass, Q. B. (2019). Solid-supported proteins in the liquid chromatography domain to probe ligand-target interactions. Front. Media S.A.. doi:10.3389/fchem.2019.00752
De Simone, A., Naldi, M., Bartolini, M., Davani, L., and Andrisano, V. (2019). Immobilized enzyme reactors: An overview of applications in drug discovery from 2008 to 2018. Chromatographia 82 (1), 425–441. doi:10.1007/s10337-018-3663-5
Di Paolo, C. T., Diamandis, E. P., and Prassas, I. (2020). The role of kallikreins in inflammatory skin disorders and their potential as therapeutic targets. Crit. Rev. Clin. Lab. Sci. 58 (1), 1–16. doi:10.1080/10408363.2020.177517110.1080/10408363.2020.1775171
Diamandis, E. P., Yousef, G. M., Petraki, C., and Soosaipillai, A. R. (2000). Human kallikrein 6 as a biomarker of Alzheimer’s disease. Clin. Biochem. 33 (8), 663–667. doi:10.1016/S0009-9120(00)00185-5
Vilela, A. F. L., and Cardoso, C. L. (2017). An on-flow assay for screening of β-secretase ligands by immobilised capillary reactor-mass spectrometry. Anal. Methods 9 (14), 2189–2196. doi:10.1039/C7AY00284J
Hai, X., Wang, X., El-Attug, M., Adams, E., Hoogmartens, J., and Van Schepdael, A. (2011). In-capillary screening of matrix metalloproteinase inhibitors by electrophoretically mediated microanalysis with fluorescence detection. Anal. Chem. 83 (1), 425–430. doi:10.1021/ac1027098
Holdgate, G. A., Meek, T. D., and Grimley, R. L. (2018). Mechanistic enzymology in drug discovery: A fresh perspective. Nat. Rev. Drug Discov. 17, 115–132. doi:10.1038/nrd.2017.219
Kolte, D., and Shariat-Madar, Z. (2016). Plasma Kallikrein inhibitors in cardiovascular disease an innovative therapeutic approach. Cardiol. Rev. 24 (3), 99–109. doi:10.1097/CRD.0000000000000069
Kryza, T., Silva, M. L., Loessner, D., Heuze-Vourc'hN., , and Clements, J. A. (2016). The kallikrein-related peptidase family: Dysregulation and functions during cancer progression. Biochimie 122, 283–299. doi:10.1016/j.biochi.2015.09.002
Loos, G., Van Schepdael, A., and Cabooter, D. (2016). Quantitative mass spectrometry methods for pharmaceutical analysis. Philos. Trans. A Math. Phys. Eng. Sci. 374, 20150366. doi:10.1098/rsta.2015.0366
Madan, R. A., Arlen, P. M., Mohebtash, M., Hodge, J. W., and Gulley, J. L. (2009). Prostvac-VF: A vector-based vaccine targeting PSA in prostate cancer. Expert Opin. Investig. Drugs 18 (7), 1001–1011. doi:10.1517/13543780902997928
Prassas, I., Eissa, A., Poda, G., and Diamandis, E. P. (2015). Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat. Rev. Drug Discov. 14 (3), 183–202. doi:10.1038/nrd4534
Rodrigues, R. C., Berenguer-Murcia, A., Carballares, D., Morellon-Sterling, R., and Fernandez-Lafuente, R. (2021). Stabilization of enzymes via immobilization: Multipoint covalent attachment and other stabilization strategies. Biotechnol. Adv. 52, 107821. doi:10.1016/j.biotechadv.2021.107821
Carvalho, R. de. D., Farias Ximenes, V., Groppo, M., and Cardoso, C. L. (2021). Ligand screening assay for the enzyme kallikrein immobilized on NHS-activated Sepharose. J. Pharm. Biomed. Anal. 199, 114026. doi:10.1016/j.jpba.2021.114026
Schmaier, A. H., and Mccrae, K. R. (2007). The plasma kallikrein-kinin system: Its evolution from contact activation. J. Thromb. Haemost. 5 (12), 2323–2329. doi:10.1111/j.1538-7836.2007.02770.x
Shang, Z., Niu, Y., Cai, Q., Chen, J., Tian, J., Yeh, S., et al. (2014). Human kallikrein 2 (KLK2) promotes prostate cancer cell growth via function as a modulator to promote the ARA70-enhanced androgen receptor transactivation. Tumour Biol. 35 (3), 1881–1890. doi:10.1007/s13277-013-1253-6
Shi, Q., Chen, J., Wang, Y., Li, Z., Li, X., Sun, C., et al. (2015). Immobilization of cyclooxygenase-2 on silica gel microspheres: Optimization and characterization. Molecules 20 (11), 19971–19983. doi:10.3390/molecules201119670
Singh, N., Ravichandran, S., Spelman, K., Fugmann, S. D., and Moaddel, R. (2014). The identification of a novel SIRT6 modulator from Trigonella foenum-graecum using ligand fishing with protein coated magnetic beads. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 968, 105–111. doi:10.1016/j.jchromb.2014.03.016
Sun, X., Gao, N., and Jin, W. (2006). Monitoring yoctomole alkaline phosphatase by capillary electrophoresis with on-capillary catalysis-electrochemical detection. Anal. Chim. Acta 571 (1), 30–33. doi:10.1016/j.aca.2006.04.063
Trindade Ximenes, I. A., de Oliveira, P. C. O., Wegermann, C. A., and de Moraes, M. C. (2021). Magnetic particles for enzyme immobilization: A versatile support for ligand screening. J. Pharm. Biomed. Anal. 204, 114286. doi:10.1016/j.jpba.2021.114286
Walker, F., Nicole, P., Jallane, A., Soosaipillai, A., Mosbach, V., Oikonomopoulou, K., et al. (2014). Kallikrein-related peptidase 7 (KLK7) is a proliferative factor that is aberrantly expressed in human colon cancer. Biol. Chem. 395 (9), 1075–1086. doi:10.1515/hsz-2014-0142
Wang, Z., Li, X., Chen, M., Liu, F., Han, C., Kong, L., et al. (2018). A strategy for screening of α-glucosidase inhibitors from Morus alba root bark based on the ligand fishing combined with high-performance liquid chromatography mass spectrometer and molecular docking. Talanta 180, 337–345. doi:10.1016/j.talanta.2017.12.065
Wubshet, S. G., Liu, B., Kongstad, K. T., Böcker, U., Petersen, M. J., Li, T., et al. (2019). Combined magnetic ligand fishing and high-resolution inhibition profiling for identification of α-glucosidase inhibitory ligands: A new screening approach based on complementary inhibition and affinity profiles. Talanta 200, 279–287. doi:10.1016/j.talanta.2019.03.047
Xie, Z., Li, Z., Shao, Y., and Liao, C. (2020). Discovery and development of plasma kallikrein inhibitors for multiple diseases. Eur. J. Med. Chem. 190, 112137. doi:10.1016/j.ejmech.2020.112137
Yousef, G. M., and Diamandis, E. P. (2001). The new human tissue kallikrein gene family: Structure, function, and association to disease. Endocr. Rev. 22 (2), 184–204. doi:10.1210/edrv.22.2.0424
Yu, H., GiaiM., , Diamandis, E. P., Katsaros, D., Sutherland, D. J., Levesque, M. A., et al. (1995). Prostate-specific antigen is a new favorable prognostic indicator for women with breast cancer. Cancer Res. 55 (10), 2104–2110.
Zani, M. B., Sant'Ana, A. M., Tognato, R. C., Chagas, J. R., and Puzer, L. (2022). Human tissue kallikreins-related peptidases are targets for the treatment of skin desquamation diseases. Front. Med. 8, 777619. doi:10.3389/fmed.2021.777619
Zhuo, R., Liu, H., Liu, N., and Wang, Y. (2016). Ligand fishing: A remarkable strategy for discovering bioactive compounds from complex mixture of natural products. Molecules 21 (11), 1516. doi:10.3390/molecules21111516
Keywords: immobilized enzyme, HPLC-MS, off-line assay, kallikerin enzyme, ligand screening assay
Citation: Carvalho DRD, Laurentino BB, Rocha CL, Kool J, Somsen G, Amstalden van Hove E and Cardoso CL (2022) Activity assay based on the immobilized enzyme kallikrein and mass spectrometry. Front. Anal. Sci. 2:1018115. doi: 10.3389/frans.2022.1018115
Received: 12 August 2022; Accepted: 05 October 2022;
Published: 19 October 2022.
Edited by:
Luzineide Wanderley Tinoco, Federal University of Rio de Janeiro, BrazilReviewed by:
Felipe Moura A. da Silva, Federal University of Amazonas, BrazilAngela De Simone, University of Turin, Italy
Copyright © 2022 Carvalho, Laurentino, Rocha, Kool, Somsen, Amstalden van Hove and Cardoso. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carmen Lúcia Cardoso, cccardoso@ffclrp.usp.br
†ORCID ID:Erika Amstalden van Hoveorcid.org/0000-0002-1150-3805
 Daniella Romano De Carvalho1
Daniella Romano De Carvalho1  Bruna Barbosa Laurentino
Bruna Barbosa Laurentino Camila Loreta Rocha
Camila Loreta Rocha Jeroen Kool
Jeroen Kool Carmen Lúcia Cardoso
Carmen Lúcia Cardoso