Reliable particle sizing in vaccine formulations using advanced dynamic light scattering
- 1LS Instruments AG, Fribourg, Switzerland
- 2Solvias AG, Large Molecule & ATMP Services, Kaiseraugst, Switzerland
Understanding the impact of lipid nanoparticle size on immunogenicity represents an important step for enabling the rapid development of novel vaccines against known or emergent diseases. Dynamic light scattering, also known as quasi-elastic light scattering or photon correlation spectroscopy, has established itself as an optimal analytical method to determine particle size due to its in-situ approach and fast measurements. However, its application to many systems of industrial relevance has been hindered due to artifacts arising from multiple scattering. Result interpretation becomes severely compromised depending on the concentration of the system and the size of the particles. In this context, strong sample dilution is often required, bringing additional uncertainties to the formulation development process. Here, we show how advanced dynamic light scattering technology can filter out multiple scattering from the signal and yield fully accurate sizing measurements regardless of the sample concentration. We illustrate this in a comparative study with standard dynamic light scattering using polystyrene beads as model suspension as well as a concentrated commercial lipid nanoparticle adjuvant (AddaVax™).
Introduction
Vaccines are among the most remarkable achievements of mankind and continue to be under the spotlight of currently ongoing research (Liu, 2003; Delany, et al., 2014; Pardi, et al., 2018). Despite the successful eradication of several critical diseases (Pollard and Bijker, 2021) and immense contribution to global public health, there is still a great need for innovative vaccine formulations. Common approaches, such as the use of aluminum adjuvants or direct injection, have proven inadequate for several important pathogens (Pollard and Bijker, 2021) and challenges inherent to such complex formulations are gradually being overcome as new vaccines continue to appear on the market (Beg et al., 2020; Wang, et al., 2020; Yenkoidiok-Douti and Jewell, 2020). However, the scientific developments remaining to be done are considerable (Beg et al., 2020; Crommelin, et al., 2021; Hou, et al., 2021; Pulendran, et al., 2021).
The COVID-19 pandemic has brought mRNA technology to the spotlight, and in particular, the use of lipid nanoparticles (LNPs) as a delivery system for mRNA vaccines (Kauffman, et al., 2015; Oberli, et al., 2017; Kulkarni, et al., 2018; Hou, et al., 2021). The advantages of using LNPs for vaccines are numerous, as they are highly customizable, can be manufactured on a large scale, and constitute a safer alternative to viral vectors (Hou, et al., 2021; Aldosari, et al., 2021; Reichmuth, et al., 2016; Zhao and Huang, 2014; Allen and Cullis, 2013; Behzadi, et al., 2017; zur Mühlen, et al., 1998). However, as with any new approach, there remain unknowns and caveats to be exploited. Therefore, accurate characterization is crucial for the complex systems used in human vaccination. Vaccine performance is affected by biophysical properties, and among these properties, LNP size provides a significant contribution to many functional parameters such as biodistribution and cellular uptake. In particular, it determines the entry route of particles into the cells (Reichmuth, et al., 2016; Hou, et al., 2021). Typical particle diameters of LNPs are in the order of 10s–100s of nanometers, can vary widely, and have even been optimized to target certain organs specifically (Pardeike, et al., 2009; Iyer, et al., 2015; Pardi, et al., 2018; Hou, et al., 2021; Tenchov, et al., 2021). Furthermore, recent studies have evidenced that high antibody titers are generated for an average LNP size of 100 nm in an RNA LNP vaccine in mice (Fortpied, et al., 2020; Hassett, et al., 2021).
While aluminum salts have long been the only adjuvants included in licensed human vaccines (Mold, et al., 2016; Shardlow, et al., 2017; Shardlow, et al., 2018; Orr, et al., 2019), engineered LNPs have also emerged as an excellent platform for the delivery of adjuvants (Swaminathan, et al., 2016; Pulendran, et al., 2021). As for their role as nanocarriers, also here the size is a decisive parameter in the adjuvating properties of LNPs. For example, particle size has been shown to impact cell recruitment in a commercial self-emulsifying vaccine adjuvant. Formulations with droplet sizes of 160 nm and 20 nm were compared to a commercial benchmark influenza A adjuvant, MF59. The results showed larger recruitment of immune cells to the injection site for emulsion droplets of 160 nm as compared to smaller particles of 20 and 90 nm size with identical composition (Shah, et al., 2019). Other adjuvant nanocarriers comprise the AS0 family (Pollard and Bijker, 2021) such as AS03 used in a pandemic influenza vaccine (ca. 150–180 nm emulsion droplets) (Iyer, et al., 2015; Shah, et al., 2019; Garçon and di Pasquale, 2017) or AS01 used in Malaria (Olotu, et al., 2016) and Varicella Zoster (Lal, et al., 2015) vaccine (ca. 105 nm liposomes) (AboulFotouh, et al., 2022a).
Knowledge of particle size distribution is not only important for vaccine formulations but also of interest for the characterization of aggregation in biologics, such as for monoclonal antibody (mAb) drugs (le Basle, et al., 2020). While dimer or trimer aggregate formation from mAb monomers has to be strongly controlled, also higher molecular weight (HMW) aggregates can form in the size range of 10s–100s of nanometers or even micrometers (den Engelsman, et al., 2011). These issues gain even more importance nowadays as biologics are manufactured at increasing concentrations in order to administer higher doses (le Basle, et al., 2020; Kollár, et al., 2020; Garidel, et al., 2017). With increasing concentration, stability and processibility of the formulations become a major concern due to the higher probability of aggregate formation (Mahler, et al., 2009) that is suspected to increase the immunogenicity potential depending on their size (le Basle, et al., 2020; den Engelsman, et al., 2011).
As particle size is one of the key parameters for vaccine efficacy and an important indicator of the quality of biologics, it is crucial to be able to accurately determine the sizes, at best within the original formulation.
Various analytical methods for size determination are available nowadays and summarized in the literature (Shekunov, et al., 2007; Szebeni, et al., 2023). One widely used technique for this purpose is Dynamic Light Scattering (DLS)1, due to its rapidity, simplicity, low sample volume requirement, high throughput capability, and ability to measure the original formulation in a non-invasive manner, i.e., no requirement of column material, vacuum injection, staining, etc. (Brito, et al., 2014; Till, et al., 2014; Stetefeld, et al., 2016; Levin, et al., 2020). Nevertheless, in the case of concentrated systems, due to the likely presence of multiple scattering effects, DLS can lead to undetected erroneous results (Urban and Schurtenberger, 1999; Alexander and Dalgleish, 2006; Ragheb and Nobbmann, 2020). DLS tools are ill-equipped to provide an accurate size determination in complex, concentrated formulations, such as for vaccines, without substantial sample dilution. However, dilution can be a time-consuming step that may also alter the properties of the sample and therefore lead to erroneous interpretation. In the case of the traditionally used aluminum adjuvants, it has already been shown that dilution severely impacts nanoparticle size (Shardlow, et al., 2017). In such cases, reliability in the investigation of native formulations is simply impossible and can substantially impact the quality of the product.
More specifically, the limitations of DLS arise from multiple scattering of photons. In simple terms, this means that photons are scattered by multiple particles before reaching the detection unit. This means that the experimental conditions for the model, that is used to calculate the particle size, cannot be met. One way to overcome this problem is to dilute the sample, which reduces the likelihood of photons being scattered by multiple particles before reaching the detector. If dilution of the sample is undesirable or the sample tends to multiple scattering even at low concentrations (as, for example, in case of LNP due to their relatively larger size), another option is to reduce the light path within the sample volume. One available and widely used solution for this issue is called backscattering DLS. Both approaches, individually or in combination, offer the possibility of minimizing but not eliminating multiple scattering. If the probability of multiple scattering is not only to be reduced but fully eliminated, Modulated 3D cross-correlation DLS technology can be used together with the light path reduction. In this work, we detail the challenge of correctly determining the particle size using DLS, especially for sizes in the range of 100s–200s of nanometers. For this purpose, we experimentally illustrate result differences between Modulated 3D cross-correlation, and the commonly used backscattering DLS approach. The results are obtained from suspensions of polystyrene beads with a diameter of around 132 nm as a model system, as well as the commercial LNP system Addavax™ (approx. 146 nm), similar to currently used vaccine adjuvants (Iyer, et al., 2015; Shah, et al., 2019; Garçon and di Pasquale, 2017).
Dynamic light scattering and the multiple scattering limitation
In a DLS experiment, a laser beam is shone onto a sample loaded in a cuvette and the light flux is measured that results from the interference of the photons which are scattered by each particle (Berne and Pecora, 2020). The measurement principle is schematized in Figure 1. Due to Brownian motion, the nanoparticles in suspension undergo constant movement. The ensuing continuous rearrangement of the particle positions results in a fluctuation of the measured intensity which allows for the straightforward determination of the diffusion coefficient of the particles under investigation. Small particles diffuse relatively rapidly in the medium, resulting in a rapidly fluctuating intensity signal as compared to larger particles, which diffuse more slowly. Quantitative information regarding the time scale of these fluctuations in the scattered intensity is obtained by the time correlation of the raw signal trace. From there, the diffusion coefficient(s) of the sample is obtained and related to the hydrodynamic diameters of the particles in motion via the Stokes-Einstein equation (Berne and Pecora, 2020).
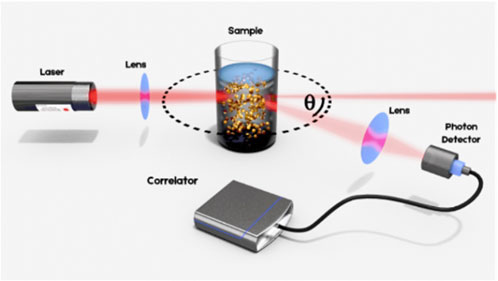
FIGURE 1. Illustration of the Dynamic Light Scattering principle. The laser beam is focused into the sample suspension containing particles that undergo Brownian motion. The resulting intensity fluctuations in the scattered light are detected at an angle Ѳ for analysis.
This principle, however, requires the measurement of photons scattered only once by the particles: the so-called single scattering events. Apart from highly dilute conditions, multiple scattering may be, in general, present in the signal. In such cases, the sizing results will contain significant errors that are undetectable when analyzing a standard DLS measurement. This undetectability is particularly problematic since such an erroneous result can still provide a good signal-to-noise ratio and will thus appear reliable to the analyst.
Mitigation of multiple scattering has been addressed in several DLS instruments through the backscattering detection scheme consisting of measuring the signal scattered at large angles, typically 173°. This scheme allows for a reduction of the path length of the light within the sample, hence reducing the number of multiple scattering events recorded. However, it does not provide a guarantee of avoiding multiple scattering entirely, especially with highly concentrated formulations and larger species due to their increased scattering cross-section. Additionally, one does not know when the sample is dilute enough such that there are no errors present in the scattering signal and therefore additional pre-dilution testing is required.
Advanced light scattering techniques developed within the last 15 years have provided a robust solution to this limitation (Urban and Schurtenberger, 1998; Urban and Schurtenberger, 1999). The general idea is to isolate singly scattered light and suppress undesired contributions from multiple scattering in the experiment. This can be achieved by performing two scattering experiments simultaneously on the same scattering volume, in the so-called 3D cross-correlation scheme. A schematic of the principle of this technique is provided in Figure 2.
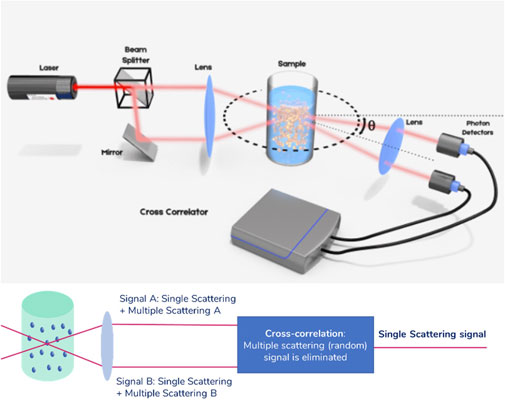
FIGURE 2. Principle of the 3D cross-correlation technique. Before being focused into the sample solution, the laser beam is split in two parallel beams. The resulting intensity fluctuations in the scattering light are detected similarly as explained in Figure 1, but using two photon detectors. The analysis is done using a cross correlator. The extension to a scheme in which the illumination beams are alternated and the detection electronics are gated is called Modulated 3D cross-correlation.
In this scheme, the sample is illuminated by two laser beams and the scattered light is collected by two detectors placed at the same observation angle, above and below the symmetry plane normal to the cuvette axis. Singly scattered photons follow a deterministic path before being detected, while multiply scattered photons follow a completely random path before reaching the detector. As a result, the two measured multiply scattered photon count signals are temporally uncorrelated. The cross-correlation of the two total count rate signals, within this detection scheme, effectively filters out multiple scattering. This technology offers a fully reliable way of performing DLS experiments, no matter the concentration of the sample. However, the implementation suffers from a reduction in signal quality due to cross-talk between detectors. This scheme has been further improved by temporally separating the two experiments in the Modulated 3D cross-correlation scheme (Block and Scheffold, 2010a; Block and Scheffold, 2010b). In this scheme, the illumination beams are alternately activated with high-speed intensity modulators while the detection electronics are synchronously gated. In this implementation, the quality of the signal for dilute samples is comparable to that of a standard auto-correlation experiment as in standard DLS. This technology effectively removes the upper limit to the concentration of the formulations measured in DLS, allowing for error-free sizing for all kinds of samples, including those showing substantial turbidity. In the following sections, this is illustrated using two types of nanosuspensions.
Materials and methods
Measurements in the Modulated 3D cross-correlation scheme were performed using a NanoLab 3D (LS Instruments AG). This device is equipped with a 120 mW laser operating at a 638 nm wavelength. The detection optics are located at a measurement angle of 90° relative to the incident beam. The particle size was obtained by means of the cumulant fitting method (Koppel, 1972). The results obtained with this instrument are referred to as „Modulated 3D DLS” in the following sections.
To compare the results with those obtained by means of a standard backscattering DLS technology, a Malvern ZetaSizer Nano ZS was used as a benchmarking tool. The device is equipped with a 4 mW laser operating at a 633 nm wavelength. The detection electronics are located at a measurement angle of 173° relative to the incident beam. This backscattering mode minimizes artifacts introduced by multiple scattering. The particle size was obtained by means of the cumulant fitting method. The results obtained with this instrument are referred to as „Standard Backscattering DLS” in the following sections.
12 measurements of 90 s were carried out in the Modulated 3D DLS scheme for each data point to maximize results precision (DLS measurements may be as short as 10–15 s for such samples, which yield a high scattering signal). In the case of the Standard Backscattering DLS, 8 measurements of 10 s per repetition were set by the software of the instrument for each data point.
For both instruments, measurement repeatability was well below 2%, which is the limit described in the ISO 22412 standard (22412-2017, 2017).
Owing to the high particle concentration studied here the classical DLS data treatment was extended to take into consideration the effects of the volume fraction of the particles’ diffusion process. Details are given in the Supplementary Material. The temperature was set to 25°C in both instruments.
Suspensions of polystyrene beads in water were obtained from Bangs Laboratories Inc. at a concentration of 10.06% (solids) and a nominal diameter of 132 nm.
A squalene-based oil-in-water emulsion of AddaVax™ adjuvant was obtained from InvivoGen, with a nominal particle size of around 146 nm and a concentration of 38.7 mg/mL. The concentration was converted to a volume fraction of 3.9% using the density value of MF59 (0.9963 g/mL) which has the same composition as AddaVax™ (O'Hagan, 2007; AboulFotouh, et al., 2022b).
Dilutions were prepared using phosphate-buffered saline (PBS) from Sigma Aldrich. The buffer was prepared by dissolving a PBS tablet (2 g) in 200 mL purified water yielding a solution of pH 7.5.
Results and discussion
Measurements on a model system
In order to conduct experiments on suspensions of polystyrene beads with a nominal diameter of 132 nm, a concentration series between 0.001% v/v and 10% v/v was analyzed. Given the fact that the particles are solid at room temperature and given their high bound surface charge, dilution is expected not to affect the particle size, and hence can be considered a model system.
Figure 3 shows the sizing results obtained using both DLS measurement methods, together with the error relative to the nominal size. Both instruments produce the same sizing results under highly dilute conditions2. Once the concentration increases, the results start to differ substantially. While the measured size does not vary significantly for concentrations lower than 0.1% v/v, they deviate from the initial size measurement at higher concentrations in the Standard Backscattering DLS scheme. This deviation continues to increase with increasing concentration, to reach a situation whereby the measured size is less than half the value of a dilute sample. For example, the measurement error is more than 60% for samples at volume fractions of 10% v/v (Figure 3 bottom graph).
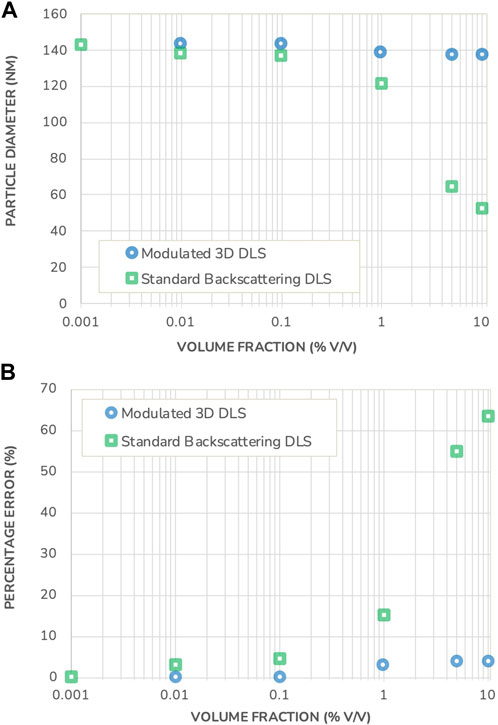
FIGURE 3. (A) Particle diameter measured in a polystyrene suspension using Standard Backscattering DLS and Modulated 3D DLS as a function of the volume fraction. (B) Percentage error on the DLS sizing results in both measurement modes, as a function of the volume fraction. Note that the error bars for measurement repeatability are substantially smaller than the data points and therefore not shown.
This observed size reduction is typical for the presence of multiple scattering events and, given the attributes of the model system under investigation, cannot be ascribed to an actual particle size change. Furthermore, the onset of multiple scattering is already visible at 0.1% v/v volume fraction where the error in the measured particle size by Standard Backscattering DLS exceeds the typical DLS reproducibility. Generally, the onset of multiple scattering is difficult, if not impossible, to predict as it requires advanced modeling and a solid knowledge of the optical properties of the suspension. These demands would however put a tremendous barrier to quality control and routine measurements, especially when dealing with multiple complex formulations of differing compositions and properties. As a result, to guarantee trustable measurements when working with Standard Backscattering DLS, there is no reliable alternative to dilution with its accompanying uncertainties of sample alteration.
On the other hand, the size of the polystyrene beads measured in the Modulated 3D DLS mode remains constant, as expected for the polystyrene beads sample under investigation with less than a 4% variation relative to the nominal value. The Modulated 3D DLS technology thus demonstrates its ability to reliably filter out multiple scattering and to deliver correct and trustable results under any concentration considered.
Measurements on a commercial system: AddaVax™
In the following, a comparison between Standard Backscattering DLS and Modulated 3D DLS is made on a commercial LNP system: AddaVax™, which is a squalene-based oil-in-water emulsion adjuvant with a nominal particle size of around 146 nm, similar to commonly used vaccine adjuvants (Iyer, et al., 2015; Shah, et al., 2019; Garçon and di Pasquale, 2017). The adjuvant was supplied at an initial concentration of 38.7 mg/mL, from which a dilution series was created. A picture of the formulation is displayed in Figure 4 and shows the turbid appearance of the sample. The sizing results, corrected for volume effects following the procedure as detailed in the Supplementary Material, are presented in Figure 5 together with the measurement error relative to the size measured at the lowest concentration.
FIGURE 4. Photograph of AddaVax™ in a measurement cuvette at original formulation concentration (3.9% volume fraction).
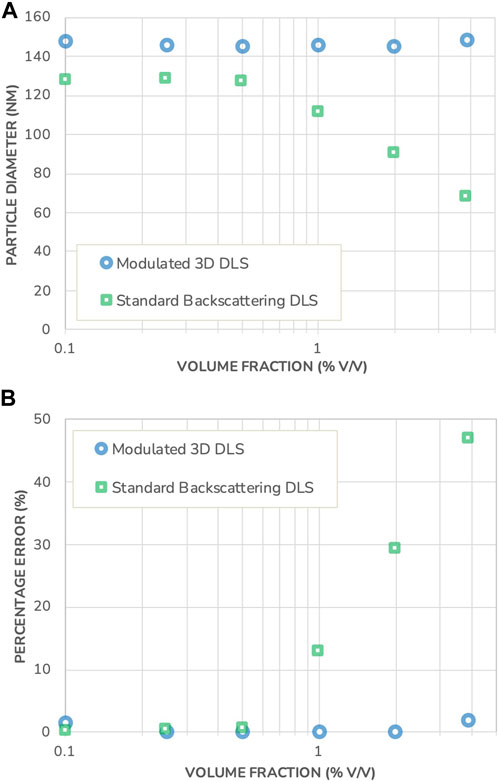
FIGURE 5. (A) Particle diameter measured in AddaVax™ using Standard Backscattering DLS and Modulated 3D DLS, as a function of the volume fraction. (B) Percentage error on the DLS sizing results in both instruments, as a function of the volume fraction. Note that the error bars for measurement repeatability are substantially smaller than the data points and therefore not shown.
Comparing the results, a size difference of 14% is observed between the two different measuring techniques at the highest dilution. This apparent discrepancy is typical of DLS when applied to polydisperse samples and is not related to multiple-scattering events (Jin, et al., 2022). In DLS, to obtain the hydrodynamic size, one measures the intensity-weighted diffusion coefficient. The active intensity weighting scheme for polydisperse samples changes with the measurement angle. When the measurement angle is decreased, the signal originating from large particles becomes dominant and they contribute more to the result thus leading to an increase in the measured hydrodynamic size. On the contrary, an increase in the angle, as in the backscattering scheme, gives more weight to the smaller particles of the polydisperse sample.
The difference is observed in Figure 5 already for the highest dilution since the Modulated 3D scheme collects intensity at 90° while the Standard Backscattering scheme uses 173°. In comparison, this was not observed in Figure 3 with a model monodisperse system owing to its very low polydispersity. This emphasizes the importance of the scattering angle chosen, as the Standard Backscattering DLS scheme yields measurement results biased toward lower particle sizes and does not provide an accurate representation of the particle size distribution in the case of polydisperse systems. On the other hand, a measurement angle of 90° provides a more “neutral” characterization of such systems, with less bias towards smaller particles.
Similarly, as with the polystyrene beads model system, the measurement results obtained with the Modulated 3D DLS are constant across all concentrations evaluated, indicating that the AddaVax™ suspension is stable against dilution. However, the results obtained with the Standard Backscattering DLS technique show a sharp decrease in size with an onset that begins at a volume fraction just below 1% v/v. As mentioned previously, this onset is not predictable unless resorting to advanced modeling techniques that require precise knowledge of the optical properties of the sample. The measurement error reaches up to 47% in this set of experiments. This illustrates the ability of the Modulated 3D DLS to accurately determine particle size for opalescent industrial formulations, while Standard Backscattering DLS fails to do so.
Conclusion
The field of vaccine formulation is growing and new mRNA technologies are being developed at an ever increasingly fast pace. LNP size is among the most crucial parameters to provide an understanding of improved biodistribution, cellular uptake, and adjuvating potency. Unlike proteins, for which Standard Backscattering DLS is widely used and marketed with the claim to limit the influence from multiple scattering to allow for measurements with more concentrated and/or turbid samples, LNP systems such as AddaVax™ are much larger in size and introduce significant measurement errors caused by multiple scattering already at low concentrations. Furthermore, the user cannot determine from the generated data whether the results can be trusted. Proper multiple scattering filtering, as introduced with Modulated 3D DLS, opens the possibility of fail-safe and trustable measurements. As an illustration, a polystyrene bead model suspension and a technologically relevant emulsion system (AddaVax™) formulated at high concentrations have been investigated. In this work, we demonstrated that characterization of AddaVax™ yielded measurement errors of up to 47% at concentrations below 4% v/v when using Standard Backscattering DLS, while Modulated 3D cross-correlation DLS provided error-free measurements. These findings are generally applicable to LNP systems of similar size ranges. The knowledge translates similarly to high-concentrated biologics formulations with their increased possibility of forming larger aggregates.
With the help of dilution-free measurements of samples, experiment times are significantly shortened and human- or dilution-introduced sample alteration is prevented. The reuse of highly concentrated samples furthermore saves sample material and opens roads for particle size determination of unopened containers as it might be of value for stability studies and quality control of pharmaceuticals. The inevitable presence of multiple scattering events for such a situation, with its erroneous results, can be properly taken care of by using Modulated 3D DLS.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
CB: Investigation, Project administration, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. AJ: Investigation, Writing–review and editing. DL: Funding acquisition, Project administration, Resources, Writing–review and editing. MK: Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. AV: Conceptualization, Project administration, Supervision, Validation, Investigation, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was funded by LS Instruments AG and Solvias AG.
Acknowledgments
AJ and MK would like to thank the Solvias AG for the support in carrying out the study.
Conflict of interest
Authors CB, DL, and AV were employed by LS Instruments AG. Authors AJ and MK were employed by Solvias AG.
The authors declare that this study received funding from LS Instruments AG and Solvias AG. The funders LS Instruments AG and Solvias AG had the following involvement in the study: study design, collection, analysis, interpretation of data, the writing of this article, and the decision to submit it for publication.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frans.2024.1358893/full#supplementary-material
Footnotes
1Depending on the context, DLS is also referred to as quasi-elastic light scattering, QELS, or photon correlation spectroscopy, PCS.
2The nominal size of 132 nm reported by the particle manufacturer is obtained by means of DLS and by strongly diluting the sample with a 10 mM electrolyte solution. This compresses the electrical double layer around the particle thus explaining the difference between the nominal size and the size measured in this work in both DLS instruments in dilute conditions.
References
22412-2017 ISO, 2017 22412-2017 ISO (2017). Particle size analysis – dynamic light scattering (DLS). ISO Geneva, Switzerland, s.l.:s.n.
AboulFotouh, K., Uno, N., Xu, H., Moon, C., Sahakijpijarn, S., Christensen, D. J., et al. (2022a). Formulation of dry powders of vaccines containing MF59 or AddaVax by Thin-Film Freeze-Drying: towards a dry powder universal flu vaccine. Int. J. Pharm. 624, 1–13. doi:10.1016/j.ijpharm.2022.122021
AboulFotouh, K., Xu, H., Moon, C., Williams, R. O., and Cui, Z. (2022b). Development of (inhalable) dry powder formulations of AS01B-containing vaccines using thin-film freeze-drying. Int. J. Pharm. 622 (121825), 1–12. doi:10.1016/j.ijpharm.2022.121825
Aldosari, B. N., Alfagih, I. M., and Almurshed, A. S. (2021). Lipid nanoparticles as delivery systems for RNA-based vaccines. Pharmaceutics 13 (2), 1–26. doi:10.3390/pharmaceutics13020206
Alexander, M., and Dalgleish, D. G. (2006). Dynamic light scattering techniques and their applications in food science. Food Biophys. 1, 2–13. doi:10.1007/s11483-005-9000-1
Allen, T. M., and Cullis, P. R. (2013). Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 65 (1), 36–48. doi:10.1016/j.addr.2012.09.037
Beg, S., Alharbi, K. S., Alruwaili, N. K., Alotaibi, N. H., Almalki, W. H., Alenezi, S. K., et al. (2020). Nanotherapeutic systems for delivering cancer vaccines: recent advances. Nanomedicine 15 (15), 1527–1537. doi:10.2217/nnm-2020-0046
Behzadi, S., Serpooshan, V., Tao, W., Hamaly, M. A., Alkawareek, M. Y., Dreaden, E. C., et al. (2017). Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 46 (14), 4218–4244. doi:10.1039/c6cs00636a
Berne, B. J., and Pecora, R. (2020). Dynamic light scattering: with applications to chemistry, biology, and physics. Dover: Dover Books on Physics.
Block, I. D., and Scheffold, F. (2010a). Cross-correlation dynamic light scattering (DLS) method and system. Munich, Germany: European Patent Office. No. EP2365313A1.
Block, I. D., and Scheffold, F. (2010b). Modulated 3D cross-correlation light scattering: improving turbid sample characterization. Rev. Sci. Instrum. 81, 123107. doi:10.1063/1.3518961
Brito, L. A., Chan, M., Shaw, C. A., Hekele, A., Carsillo, T., Schaefer, M., et al. (2014). A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 22 (12), 2118–2129. doi:10.1038/mt.2014.133
Crommelin, D. J. A., Anchordoquy, T. J., Volkin, D. B., Jiskoot, W., and Mastrobattista, E. (2021). Addressing the cold reality of mRNA vaccine stability. J. Pharmceutical Sci. 110 (3), 997–1001. doi:10.1016/j.xphs.2020.12.006
Delany, I., Rappuoli, R., and de Gregorio, E. (2014). Vaccines for the 21st century. EMBO Mol. Med. 6 (6), 708–720. doi:10.1002/emmm.201403876
den Engelsman, J., Garidel, P., Smulders, R., Koll, H., Smith, B., Bassarab, S., et al. (2011). Strategies for the assessment of protein aggregates in pharmaceutical biotech product development. Pharm. Res. 28 (4), 920–933. doi:10.1007/s11095-010-0297-1
Fortpied, J., Collignon, S., Moniotte, N., Renaud, F., Bayat, B., and Lemoine, D. (2020). The thermostability of the RTS,S/AS01 malaria vaccine can be increased by co-lyophilizing RTS,S and AS01. Malar. J. 19 (202), 202–215. doi:10.1186/s12936-020-03253-1
Garçon, N., and di Pasquale, A. (2017). From discovery to licensure, the Adjuvant System story. Hum. Vaccines Immunother. 13 (1), 19–33. doi:10.1080/21645515.2016.1225635
Garidel, P., Kuhn, A. B., Schäfer, L. V., Karow-Zwick, A. R., and Blech, M. (2017). High-concentration protein formulations: how high is high? Eur. J. Pharm. Biopharm. 119, 353–360. doi:10.1016/j.ejpb.2017.06.029
Hassett, K. J., Higgins, J., Woods, A., Levy, B., Xia, Y., Hsiao, C. J., et al. (2021). Impact of lipid nanoparticle size on mRNA vaccine immunogenicity. J. Control. Release 335, 237–246. doi:10.1016/j.jconrel.2021.05.021
Hou, X., Zaks, T., Langer, R., and Dong, Y. (2021). Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6 (12), 1078–1094. doi:10.1038/s41578-021-00358-0
Iyer, V., Cayatte, C., Guzman, B., Schneider-Ohrum, K., Matuszak, R., Snell, A., et al. (2015). Impact of formulation and particle size on stability and immunogenicity of oil-in-water emulsion adjuvants. Hum. Vaccines Immunother. 11 (7), 1853–1864. doi:10.1080/21645515.2015.1046660
Jin, L., Jarand, C. W., Brader, M. L., and Reed, W. F. (2022). Angle-dependent effects in DLS measurements of polydisperse particles. Meas. Sci. Technol. 33 (4), 045202–045214. doi:10.1088/1361-6501/ac42b2
Kauffman, K. J., Dorkin, J. R., Yang, J. H., Heartlein, M. W., DeRosa, F., Mir, F. F., et al. (2015). Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 15 (11), 7300–7306. doi:10.1021/acs.nanolett.5b02497
Kollár, É., Balázs, B., Tari, T., and Siró, I. (2020). Development challenges of high concentration monoclonal antibody formulations. Drug Discov. Today Technol. 37, 31–40. doi:10.1016/j.ddtec.2020.08.005
Koppel, D. E. (1972). Analysis of macromolecular polydispersity in intensity correlation spectroscopy: the method of cumulants. J. Chem. Phys. 57 (11), 4814–4820. doi:10.1063/1.1678153
Kulkarni, J. A., Cullis, P. R., and van der Meel, R. (2018). Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid. Ther. 28 (3), 146–157. doi:10.1089/nat.2018.0721
Lal, H., Cunningham, A. L., Godeaux, O., Chlibek, R., Diez-Domingo, J., Hwang, S. J., et al. (2015). Efficacy of an adjuvanted herpes zoster subunit vaccine in older adults. N. Engl. J. Med. 372 (22), 2087–2096. doi:10.1056/NEJMoa1501184
le Basle, Y., Chennell, P., Tokhadze, N., Astier, A., and Sautou, V. (2020). Physicochemical stability of monoclonal antibodies: a review. J. Pharm. Sci. 109 (1), 169–190. doi:10.1016/j.xphs.2019.08.009
Levin, A. D., Ringaci, A., Alenichev, M. K., Drozhzhennikova, E. B., Shevchenko, K. G., Cherkasov, V. R., et al. (2020). Dynamic light scattering biosensing based on analyte-induced inhibition of nanoparticle aggregation. Anal. Bioanal. Chem. 412 (14), 3423–3431. doi:10.1007/s00216-020-02605-9
Liu, M. A. (2003). DNA vaccines: a review. J. Intern. Med. 253 (4), 402–410. doi:10.1046/j.1365-2796.2003.01140.x
Mahler, H. C., Friess, W., Grauschopf, U., and Kies, S. (2009). Protein aggregation: pathways, induction factors and analysis. J. Pharm. Sci. 98 (9), 2909–2934. doi:10.1002/jps.21566
Mold, M., Shardlow, E., and Exley, C. (2016). Insight into the cellular fate and toxicity of aluminium adjuvants used in clinically approved human vaccinations. Sci. Rep. 6 (31578), 1–13. doi:10.1038/srep31578
Oberli, M. A., Reichmuth, A. M., Dorkin, J. R., Mitchell, M. J., Fenton, O. S., Jaklenec, A., et al. (2017). Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 17 (3), 1326–1335. doi:10.1021/acs.nanolett.6b03329
O'Hagan, D. T. (2007). MF59 is a safe and potent vaccine adjuvant that enhances protection against influenza virus infection. Expert Rev. Vaccines 6 (5), 699–710. doi:10.1586/14760584.6.5.699
Olotu, A., Fegan, G., Wambua, J., Nyangweso, G., Leach, A., Lievens, M., et al. (2016). Seven-year efficacy of RTS,S/AS01 malaria vaccine among young african children. N. Engl. J. Med. 374 (26), 2519–2529. doi:10.1056/NEJMoa1515257
Orr, M. T., Khandhar, A. P., Seydoux, E., Liang, H., Gage, E., Mikasa, T., et al. (2019). Reprogramming the adjuvant properties of aluminum oxyhydroxide with nanoparticle technology. npi Vaccines 4 (1), 1–10. doi:10.1038/s41541-018-0094-0
Pardeike, J., Hommoss, A., and Müller, R. H. (2009). Lipid nanoparticles (SLN, NLC) in cosmetic and pharmaceutical dermal products. Int. J. Pharm. 366 (1-2), 170–184. doi:10.1016/j.ijpharm.2008.10.003
Pardi, N., Hogan, M. J., Porter, F. W., and Weissman, D. (2018). mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 17 (4), 261–279. doi:10.1038/nrd.2017.243
Pollard, A. J., and Bijker, E. M. (2021). A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 21 (2), 83–100. doi:10.1038/s41577-020-00479-7
Pulendran, B., Arunachalam, P. S., and O’Hagan, D. T. (2021). Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 20 (6), 454–475. doi:10.1038/s41573-021-00163-y
Ragheb, R., and Nobbmann, U. (2020). Multiple scattering effects on intercept, size, polydispersity index, and intensity for parallel (VV) and perpendicular (VH) polarization detection in photon correlation spectroscopy. Sci. Rep. 10 (21768), 21768–21769. doi:10.1038/s41598-020-78872-4
Reichmuth, A. M., Oberli, M. A., Jaklenec, A., Langer, R., and Blankschtein, D. (2016). mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 7 (5), 319–334. doi:10.4155/tde-2016-0006
Shah, R. R., Taccone, M., Monaci, E., Brito, L. A., Bonci, A., O'Hagan, D. T., et al. (2019). The droplet size of emulsion adjuvants has significant impact on their potency, due to differences in immune cell-recruitment and -activation. Sci. Rep. 9 (11520), 11520–11529. doi:10.1038/s41598-019-47885-z
Shardlow, E., Mold, M., and Exley, C. (2017). From stock bottle to vaccine: elucidating the particle size distributions of aluminum adjuvants using dynamic light scattering. Front. Chem. 4 (48), 48–10. doi:10.3389/fchem.2016.00048
Shardlow, E., Mold, M., and Exley, C. (2018). Unraveling the enigma: elucidating the relationship between the physicochemical properties of aluminium-based adjuvants and their immunological mechanisms of action. Allergy, Asthma and Clin. Immunol. 14 (8), 80–19. doi:10.1186/s13223-018-0305-2
Shekunov, B. Y., Chattopadhyay, P., Tong, H. H. Y., and Chow, A. H. L. (2007). Particle size analysis in pharmaceutics: principles, methods and applications. Pharm. Res. 24 (2), 203–227. doi:10.1007/s11095-006-9146-7
Stetefeld, J., McKenna, S. A., and Patel, T. R. (2016). Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 8 (4), 409–427. doi:10.1007/s12551-016-0218-6
Swaminathan, G., Thoryk, E. A., Cox, K. S., Meschino, S., Dubey, S. A., Vora, K. A., et al. (2016). A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccines 34 (1), 110–119. doi:10.1016/j.vaccine.2015.10.132
Szebeni, J., Kiss, B., Bozó, T., Turjeman, K., Levi-Kalisman, Y., Barenholz, Y., et al. (2023). Insights into the structure of comirnaty covid-19 vaccine: a theory on soft, partially bilayer-covered nanoparticles with hydrogen bond-stabilized mRNA-lipid complexes. ACS Nano 17, 13147–13157. doi:10.1021/acsnano.2c11904
Tenchov, R., Bird, R., Curtze, A. E., and Zhou, Q. (2021). Lipid nanoparticles from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano 15 (11), 16982–17015. doi:10.1021/acsnano.1c04996
Till, U., Gaucher-Delmas, M., Saint-Aguet, P., Hamon, G., Marty, J. D., Chassenieux, C., et al. (2014). Asymmetrical flow field-flow fractionation with multi-angle light scattering and quasi-elastic light scattering for characterization of polymersomes: comparison with classical techniques. Anal. Bioanal. Chem. 406 (30), 7841–7853. doi:10.1007/s00216-014-7891-8
Urban, C., and Schurtenberger, P. (1998). Characterization of turbid colloidal suspensions using light scattering techniques combined with cross-correlation methods. J. Colloid Interface Sci. 207 (1), 150–158. doi:10.1006/jcis.1998.5769
Urban, C., and Schurtenberger, P. (1999). Application of a new light scattering technique to avoid the influence of dilution in light scattering experiments with milk. Phys. Chem. Chem. Phys. 1 (17), 3911–3915. doi:10.1039/a903906f
Wang, J., Peng, Y., Xu, H., Cui, Z., and Williams, R. O. (2020). The COVID-19 vaccine race: challenges and opportunities in vaccine formulation. AAPS PharmSciTech 21 (225), 1–12. doi:10.1208/s12249-020-01744-7
Yenkoidiok-Douti, L., and Jewell, C. M. (2020). Integrating biomaterials and immunology to improve vaccines against infectious diseases. ACS Biomaterials Sci. Eng. 6 (2), 759–778. doi:10.1021/acsbiomaterials.9b01255
Zhao, Y., and Huang, L. (2014). Lipid nanoparticles for gene delivery. Adv. Genet. 88, 13–36. doi:10.1016/B978-0-12-800148-6.00002-X
Keywords: bioanalytical methods, biopolymers/lipids, nanoparticles/nanotechnology, multiple scattering, dynamic light scattering, cross-correlation
Citation: Bretz C, Jauslin A, Leumann D, Koch M and Vaccaro A (2024) Reliable particle sizing in vaccine formulations using advanced dynamic light scattering. Front. Anal. Sci. 4:1358893. doi: 10.3389/frans.2024.1358893
Received: 20 December 2023; Accepted: 12 February 2024;
Published: 29 February 2024.
Edited by:
Quezia B. Cass, Federal University of São Carlos, BrazilReviewed by:
Janos Szebeni, Semmelweis University, HungaryLuiz Cláudio Rodrigues Pereira da Silva, Federal University of Rio de Janeiro, Brazil
Copyright © 2024 Bretz, Jauslin, Leumann, Koch and Vaccaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marius Koch, marius.koch@solvias.com
 Coline Bretz1
Coline Bretz1  Marius Koch
Marius Koch Andrea Vaccaro
Andrea Vaccaro