Simultaneous determination of small molecules and proteins in wastewater-based epidemiology
- 1Food and Environmental Research Group of the University of Valencia (SAMA-UV), Research Desertification Centre (CIDE) (CSIC-GV-UV), Moncada, Spain
- 2Department of Environmental Chemistry, Institute of Environmental Assessment and Water Studies, Spanish National Research Council (IDAEA-CSIC), Barcelona, Spain
- 3Proteomics Laboratory CSIC/UAB, Institute of Biomedical Research of Barcelona, Spanish National Research Council (IIBB-CSIC/IDIBAPS), Barcelona, Spain
Wastewater-based epidemiology (WBE) aims to understand a population’s consumption habits, exposure to chemicals, and the prevalence of specific diseases or pathogens. This is achieved by the chemical or biological/genomic determination of biomarkers (e.g., excreted metabolic products), which are in urban wastewater generated by that population. WBE has been mostly linked to the determination of small molecules of human origin using liquid-chromatography mass spectrometry (LC-MS). In this Perspective, we provide a state-of-the-art and critical evaluation of further developments in the information achieved by determining small molecules as well as the most promising analytical techniques to enlarge the information obtained. By simultaneously monitoring small and large molecules we can comprehensively trace the population’s health by their consumption of prescribed pharmaceuticals and illegal drugs, as well as by the amount of excreted macromolecule biomarkers such as peptides and proteins. Moreover, species-specific protein sequences allow us to monitor animal populations reflecting farming and slaughterhouse activities (poultry, pigs…) or pest occurrences (rats). To this end, the capability of proteomic studies using high-resolution tandem mass spectrometry is highlighted and compared in the context of other advances in the broader field of high-resolution mass spectrometry (HRMS).
1 Introduction
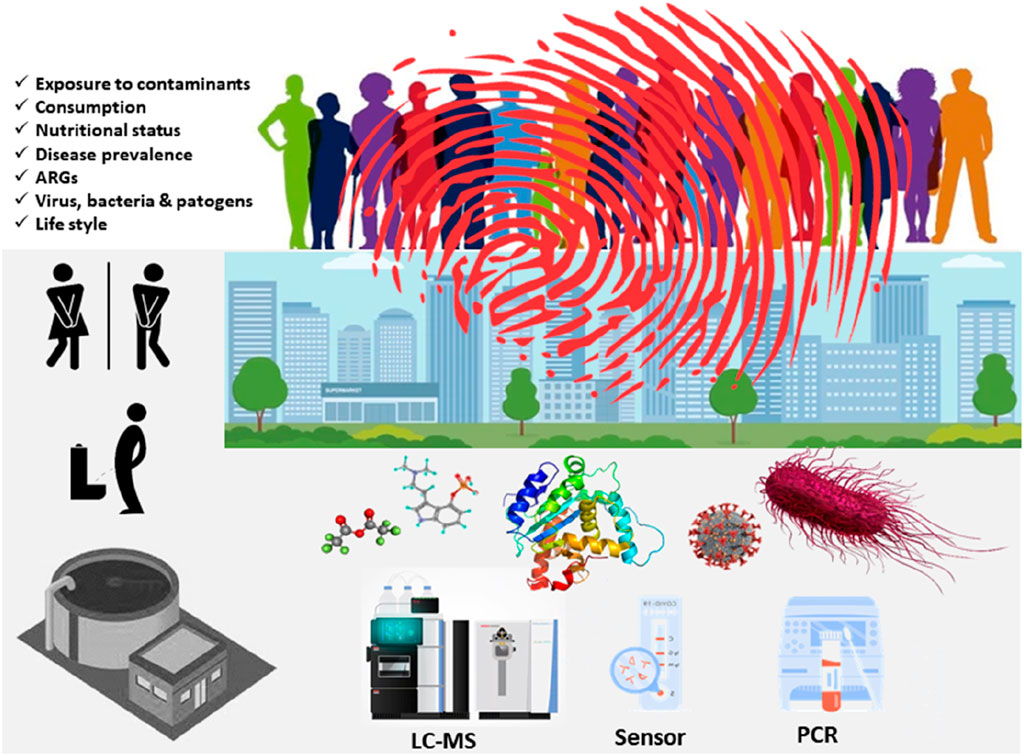
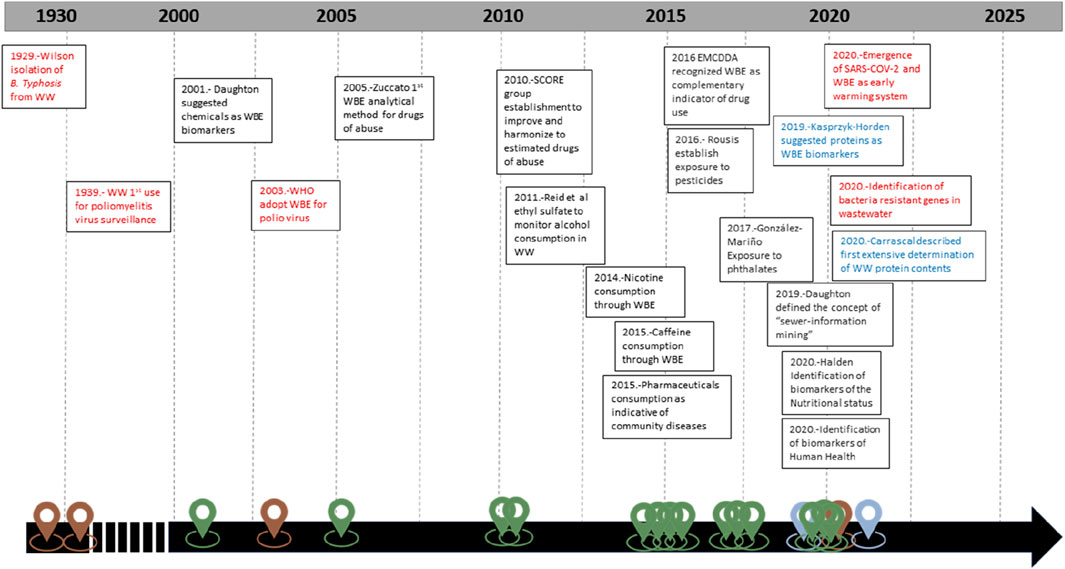
Wastewater-based epidemiology (WBE), also known as sewage epidemiology or wastewater epidemiology, is an approach to monitoring public health by analyzing the presence of various substances, including pathogens, chemicals, and drugs, in municipal wastewater (Lorenzo and Picó, 2019; Singer et al., 2023). Figure 1 illustrates the rationale of WBE. Wastewater can be considered as an anonymous pool of feces and urine and in a lesser extent respiratory discharges of the population that is served by the wastewater treatment plant (WWTP) (García-Encina, 2021). Figure 2 summarizes chronologically the main steps in the evolution of WBE. Traditionally, this approach involves identifying microorganisms and analyzing small molecules. Advancements in molecular techniques, such as qPCR and sequencing, have enabled the detection of viruses, bacteria, and other potential pathogens. Since 2020, these techniques have become increasingly important due to the need to control the spread of SARS-CoV-2 (Barcellos et al., 2023). As with any research related to SARS-CoV-2, there is an extensive literature on its surveillance in wastewater (Amman and Bergthaler, 2022; Dutta et al., 2022; Gonçalves et al., 2022; Greenbaum et al., 2022; Kumar et al., 2022; Burdorf and Rugulies, 2023; Du Toit, 2023; Gahlot et al., 2023; Hopkins et al., 2023; Oloye et al., 2023; Sodhi and Singh, 2023; Tavazzi et al., 2023). This has resulted in a renaissance of WBE and early warning systems based on pathogen surveillance (O’Brien and Xagoraraki, 2019; Guo et al., 2022a; Guo et al., 2022b; Fitzmorris-Brisolara et al., 2022; Demeter et al., 2023; Hassard et al., 2023; Shaw et al., 2023; Wolfe et al., 2023)
On the other hand, WBE also place significant emphasis on detecting small molecules and/or metabolites excreted by humans. These substances can offer valuable insights into population habits, particularities, and health status serving as biomarkers (Vitale et al., 2021). In this aspect, WBE has been closely linked to the development of liquid-chromatography mass spectrometry (LC-MS) protocols. The recent advent of high-resolution mass spectrometry (HRMS) has opened new insights not only for the determination of target small molecules but also for the application to larger ones (e.g., proteins) as well as for expanding non-targeted “omic” approaches in this field (Rice and Kasprzyk-Hordern, 2019).
In this Perspective article, we will discuss the recent advances of MS-based WBE in the detection and monitoring of the metabolites excreted by living beings in wastewater (including those from licit and illicit drugs consumption), based on the analysis of small molecules and the complementarity and role of proteomics, which is still in a very early stage of development and exploitation.
2 State-of-art of WBE
Firstly, on the detection of small molecules, WBE first gained recognition for monitoring the consumption of illegal drugs within the population (Gao et al., 2023). A scheme of how this consumption is estimated from wastewater analysis is shown in Supplementary Figure S1. Many studies have addressed sampling, sample stability, analytical protocol validation, and back-calculation of drug consumption (Huizer et al., 2021). The examination of these drugs primarily relied on solid-phase extraction (SPE) from wastewater, coupled with subsequent targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS). Passive samplers that provide a more representative sample in time are also applied. Traditionally, the MS/MS analysis involved triple quadrupoles or linear ion trap configurations; however, contemporary practices have incorporated HRMS systems. Furthermore, enantiomeric separation is particularly crucial in WBE, as metabolism is often enantioselective and racemates could be transformed in one or other enantiomers in the human body (Langa et al., 2021). In any case, the analysis always follows a targeted strategy as needed for the identification of minority compounds. (Rousis et al., 2023). (Bio)sensors were developed to enhance the speed and efficiency of WBE. These sensors play a crucial role in rapidly detecting and measuring specific substances in wastewater, contributing to the overall effectiveness and velocity of WBE processes (Bilge et al., 2022; Kim et al., 2023). Their use enabled quicker and more precise analysis of key biomarkers, enhancing the capabilities of monitoring systems for various substances in wastewater (Pan et al., 2022).
Most studies primarily sample from WWTPs to estimate drug consumption trends in various regions (Shimko et al., 2021; Asadi et al., 2023; Laimou-Geraniou et al., 2023). However, a growing number of studies now employ WBE to monitor substance use in specific communities, focusing on locations such as educational institutions, prisons, music festivals, and sporting events, as well as on special dates, such as, holidays (Verovšek et al., 2020). Advancements in technology and theory have expanded the application of WBE to include surveys on the consumption of legally sold addictive substances like alcohol, nicotine, and caffeine (Gao et al., 2023).
In addition, fewer WBE studies explored other health biomarkers like pharmaceuticals. The significant medical and social implications of pharmaceutical misuse underscore the necessity for comprehensive drug utilization research (Jaunay et al., 2023; Massano et al., 2023). WBE has also been applied to monitor temporal patterns of pharmaceutical use or document interventions such as rescheduling (e.g., changing from over-the-counter to prescription only), sales-restrictions, and educational programs to influence prescription behaviour or consumer choice. WBE can reveal shifts in pharmaceutical consumption patterns during public health crises, like the COVID-19 pandemic (Picó and Barceló, 2023). Although interest in WBE for pharmaceutical monitoring is growing, further background research, including addressing compound-specific uncertainties, is essential to connect WBE data with routine pharmacoepidemiological information and workflows. WBE presents the opportunity to 1) estimate pharmaceutical consumption by analyzing metabolic excretion products in wastewater; 2) continuously and near real-time monitor spatial and temporal consumption patterns of pharmaceuticals; and 3) cross-reference data with other drug utilization research sources to evaluate the impact of strategies or interventions aimed at reducing inappropriate pharmaceutical use (Boogaerts et al., 2021)
Recently, WBE has evolved into a method for creating a city’s fingerprint, indicating health, lifestyle habits, and exposure to contaminants in comparison to other cities (Picó and Barceló, 2021a; Singer et al., 2023). This expansion is attributed to identifying human biomarkers and profiling wastewater catchment characteristics that define these conditions.
3 Perspectives and future applications
3.1 Within small molecule biomarkers
WBE shows potential for swiftly assessing the impact of chemical contaminants and dietary nutrients on public health. Over the last decade, WBE studies have broadened to monitor a wide range of biomarkers and health determinants (HDs), including pharmaceuticals (e.g., antibiotics, benzodiazepines), pesticides, plasticizers, alcohol, tobacco, indicators of oxidative stress (e.g., isoprostanes), and so on (detailed list is outlined in the Supplementary Table S1). WBE has also been proposed for assessing nutritional status in human populations using reported dietary metabolites and known linkages to health effects in human populations (Bowes and Halden, 2019). Analytes examined included 1-methylhistidine (meat intake), isoflavones and lignans (phytoestrogens), allyl isothiocyanates (cruciferous vegetables), and alkyl resorcinols (whole-wheat intake). Results obtained from theory identify WBE as a promising tool for tracking dietary trends in human populations, since it can complement existing tools by providing aggregated quantitative information regarding average consumption.
One key challenge in Public Health is to understand substances which individuals are exposed to in their work places and daily routines. In this sense, Dr. Christopher Wild introduced the term “exposome” in 2005, defining it as “every exposure to which an individual is subjected from conception to death” (Wild, 2005; Wild, 2012). The exposome exhibits significant variability and undergoes evolution throughout one’s lifespan. However, comprehending the intricate interplay between environmental exposures such as air pollutants, radiation, chemicals in consumer goods, extreme weather conditions, pathogens, toxic substances, pesticides, heavy metals, dietary factors, physical activity, medications, genetics, epigenetics, and physiology remains a challenge. WBE is well-suited for studying the exposome and its effects. However, discovering biomarkers that reflect different population influences has been challenging and has entailed significant analytical effort. Picó and Barceló (2021a) classified the different methodological schemes into 1) top-down and 2) bottom-up approaches. The top-down approach, the most used in WBE, examines substance metabolism in humans using human biomonitoring (HBM) data, identifying suitable HDs and biomarkers for detection in wastewater. This method involves studying human metabolism, identifying urine-excreted substances, selecting highly representative ones, developing analytical methods for their determination, assessing their stability in wastewater, and ultimately analyzing them in wastewater. The alternative bottom-up approach involves non-targeted analysis of wastewater to identify substances from human sources and evaluate their potential as biomarkers. This method identifies various substances, including biomarkers, transformation products, and other compounds, through the application of non-targeted approaches involving HRMS. The introduction of HRMS has created new avenues for the identification of previously undiscovered chemicals of concern since it can provide the more probable empirical formula and the MS/MS spectrum to aid with structural elucidation (Gil-Solsona et al., 2021). Despite being less utilized than the top-down approach due to wastewater complexity, it holds promise for discovering unknown substances critical to advancing WBE (Henriot et al., 2024). Furthermore, advances in the most recent HRMS instruments equipped with operational commercial or free tools for wide-scope suspect screening and/or automatic annotation of unknowns, have offered an unparalleled opportunity to decipher those molecules that indicate exposure to pollutants (Picó and Barceló, 2021b). These features permit to perform simultaneous wide screening against a target list of compounds and non-target with extensive searching in on-line databases and chemical repositories housing MS/HRMS (such as ChemSpider, Human Metabolism Database (HMDB), Massbank, Metlin, mzCloud, NIST) and are highly promising to discover new biomarkers (Gil-Solsona et al., 2021; Perez-Lopez et al., 2024).
Connecting marker concentrations in wastewater to human exposure levels is challenging due to uncertainties in real sewer system stability and marker excretion rates. Overcoming this challenge involves conducting intensive human biomonitoring using representative specimens alongside well-defined WBE studies in specific catchments. Establishing correlations between concentrations in wastewater and human biological samples can help to forge these crucial links. Notably, Eaton et al. (2022) developed a framework to help public health authorities decide which HDs may be appropriate for WBE and which biomarkers could be used. This framework consists of an assessment tree that summarizes 1) the requirement for individual- or population-level information, 2) alternative methodologies for monitoring the HD, 3) the availability of a suitable biomarker, and 4) the requirement for changes in biomarker levels to be reflective of changes in the prevalence of the HD. This could through light to solve challenges in the future.
WBE has the potential for real-time, tracking of progress in attaining United Nations Sustainable Development Goals (SDGs) globally as a non-expensive method using existing infrastructure. As an example, Rousis et al. (2023) described that WBE approach can be used to monitor the achievement of the SDGs, such as reducing alcohol consumption and reducing the use of tobacco. In addition, it can be applied to monitor total caffeine consumption and assess levels according to the recommended daily safety limit as suggested by the European Food Safety Authority. Furthermore, in a promising effort, Adhikari and Halden (2022) compiled and analyzed existing data to 1) inventory the totality of centralized wastewater infrastructure globally, 2) identify countries featuring and lacking such infrastructure, 3) determine the fraction of the global population that is readily accessible to conventional WBE that leverages centralized sewerage infrastructure, 4) rank countries based on income level (as per UN“s” classification system) and on other factors to identify geographic regions which could benefit most from infrastructure improvements, and 5) compile an initial list of wastewater-borne markers that hold promise for tracking attainment of UN SDGs. Reviewing literature from 2005 to 2021, the authors identified 25 classes of biomarkers, both endogenous and exogenous, that can aid in tracking progress towards achieving the SDGs. These biomarkers encompass hunger and stress hormones, indicators for cardiovascular and pulmonary diseases, cancer, illicit drugs, personal care products, surfactants, hazardous chemicals, drug-resistant pathogens, antimicrobial-resistant genes (ARGs), and psychotropic drugs. These biosignatures offer valuable insights for monitoring SDG attainment.
WBE shows prospects in the growing field of the “One-Health” approach. This approach integrates human, animal, and environmental health efforts, offering a potential avenue to predict and control diseases at the interface of human, animal, and ecosystem interactions. The clearest example of this application within the field of small chemical molecules analysis is to help in the study of antibiotic resistance spread in the environment. This is, according to the WHO, a forefront challenge for the future of the human being. Antibiotic residues, antibiotic-resistant bacteria, or ARGs are detectable in wastewater and constitute a study system that provides a wealth of information. In addition to the identification of antimicrobial resistance (AMR) by several microbiological techniques (Sims and Kasprzyk-Hordern, 2020; Chau et al., 2022; Foyle et al., 2023; Larsson et al., 2023; Pandey et al., 2023; Sharma et al., 2023), it is crucial to monitor and assess antibiotic presence in wastewater, and other environmental matrices to address the problem of AMR. WBE is potentially the most reliable approach to estimate antibiotics use (Henriot et al., 2024).
WBE has an environmental safety aspect since many of these small molecules of chemical compounds that humans use or are exposed to directly or indirectly (including potential metabolites and degradation products) are currently considered emerging contaminants of concern (CECs). These molecules are initially collected in sewer systems and, if the treatment in WWTPs is ineffective in removing them, they are subsequently released into the environment, posing associated problems. Interestingly, Zillien et al. (2022) collated the fragmented knowledge and data on in-sewer fate of CECs to develop practical guidelines for water managers on how to deal with in-sewer fate of CECs. To this end, these authors collected experimental half-lives of 96 organic CECs from literature to support environmental modelling efforts and to optimize monitoring campaigns, including field studies in the context of WBE. Furthermore, a pending issue is also hospital wastewater, which is a complex mixture of pharmaceuticals, drugs, and their metabolites as well as different susceptible and antibiotic-resistant microorganisms, including viruses. Many studies pointed out that wastewater from healthcare facilities (including hospital wastewater), significantly contributes to higher loads of CECs in municipal wastewater. Because many pharmaceuticals, drugs, and microorganisms can pass through wastewater treatment plants without any significant change in their structure and toxicity and enter surface waters, treatment technologies need to be improved in terms of efficiency as well as economy (Mackull’ak et al., 2021).
3.2 Within proteomics field
Since more than 1 decade ago, proteomic studies can be found in the literature as useful tools for the monitoring of changes occurring in the complex microbiomes associated with biotechnological processes such as those used in the treatment of wastewater and sludge (Park et al., 2008; Westgate and Park, 2010; Kuhn et al., 2011; Zhang et al., 2019). This research area is currently active and has found recently specific applications, such as the effects caused by the presence of toxicants in the proteome of the bacterial consortia associated with the anammox process (Zhang et al., 2013; Wang et al., 2021; Kennes-Veiga et al., 2022; Guzmán-Fierro et al., 2024). Owing to these reasons, these proteomic studies are exclusively focused on the prokaryote (i.e., microbial) fraction of the whole wastewater proteome as ultimately responsible for the biological treatment processes. In contrast, the eukaryote higher organisms’ proteome (i.e., plants, animals, and humans) has been systematically disregarded, despite being a significant portion of it (the largest in the water phase) (Figure 3).
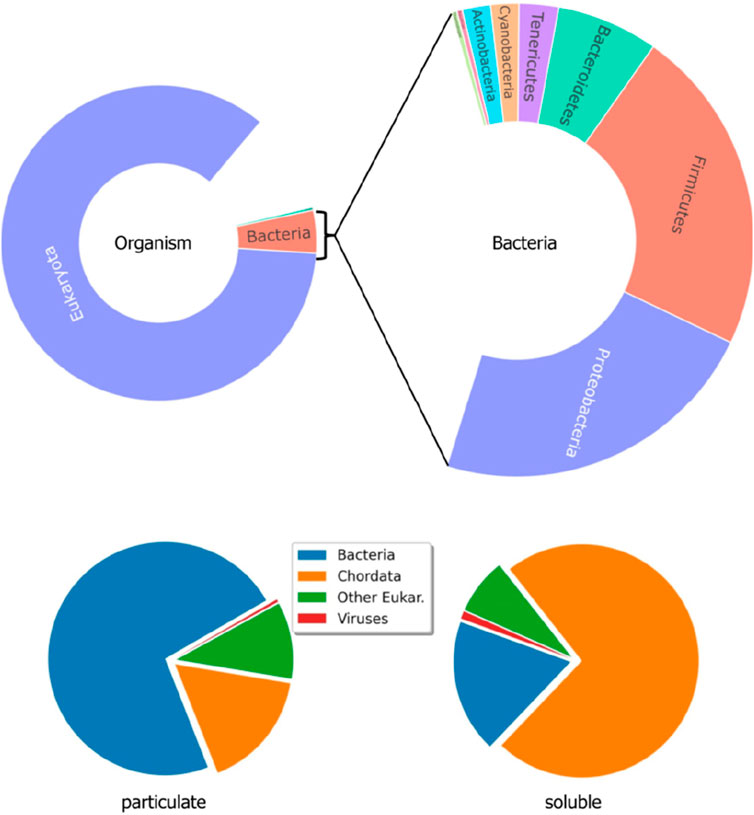
Figure 3. Distribution of Bacteria and Eukaryota proteins in the soluble fraction of wastewater and comparison with the particulate fraction (adapted from Carrascal et al., 2023).
The use of human proteins occurring in wastewater as potential biomarkers in the assessment of community health was conceptually suggested for the first time by Rice and Kasprzyk-Horden (2019). Later on, Devianto and Sano (2023), reviewed the feasibility of biosensor technology for the real-time analysis of protein-based biomarkers to be used in early warning WBE surveys of human disease. In their study, though a meta-analysis based on 231 articles, they assessed the possible candidate proteins taking into consideration their respective concentration levels in urine and feces, their dilution in wastewater, and available biosensors’ detection/quantification limits AMR. These authors suggested up to 64 proteins present either in feces or urine (including, among others, calprotectin and uromodulin), as feasible biomarkers. Altogether, there is a consensus on the requirements for a protein to be utilized as a biomarker in WBE, namely, (a) a well-defined disease−biomarker correlation, (b) its excretion must take place in sufficient amount to allow for its analytical detection and quantification, once it is diluted in wastewater, and (c) its stability both in vivo and in the wastewater media must be ensured so that the integrity and representativity of the sample are guaranteed. As for now, the number of potential candidates is scarce and they have not been proven yet.
Despite the interest of proteins for WBE manifested by the above-referred authors, the need (and lack) for experimental research studies addressed to this end is recognized. Advances in liquid chromatography coupled with HR MS non-targeted shotgun proteomic methods have enabled substantial progress in the profiling of wastewater proteome (Picó and Barceló, 2021b) thus allowing to achieve for the first time its comprehensive characterization, encompassing both prokaryotes and eukaryotes (Carrascal et al., 2020; Pérez-López et al., 2021) using passive polymeric devices.
This seminal work paved the way for further research (Carrascal et al., 2023) carried out at the influent collection points of 10 WWTPs using optimized analytical methods for both the soluble and particulate fractions that have been fully described elsewhere (Sánchez-Jiménez et al., 2023). This improved analyses of the wastewater proteome profile and distinguished signals from different groups of organisms. Overall, eukaryotic proteins, primarily from Chordata (i.e., mammals and birds) but also from non-Chordata (plants), were the major proteome components of the liquid phase, followed by bacterial proteins, while the particulate phase shows the opposite trend. Viral proteins were detected in small quantities as well (Figure 3). Over 4,000 peptides, associated with ca. 800 proteins (243 human), were identified through the semiquantitative analysis of the main constituents (Table 1). Despite not being a focused study on human epidemiology, it provided valuable information regarding the presence of endogenous human molecules potentially relevant to WBE. Pancreatic enzymes, dominated by α-amylases, were the most abundant human proteins identified in wastewater, making them the primary markers of human presence. Blood proteins, including albumin, Igs, and complement proteins, and skin-derived proteins, such as keratins, were found in significant amounts as well. A gene ontology analysis (DAVID) (Sherman et al., 2022), highlighted several functional terms that were enriched, such as those associated with the immune response (Igs, calprotectin, lactoferrin, lipocain, and dermcidin) or the anti-inflammatory response (meprin A, orosomucoid, and the serpin family).
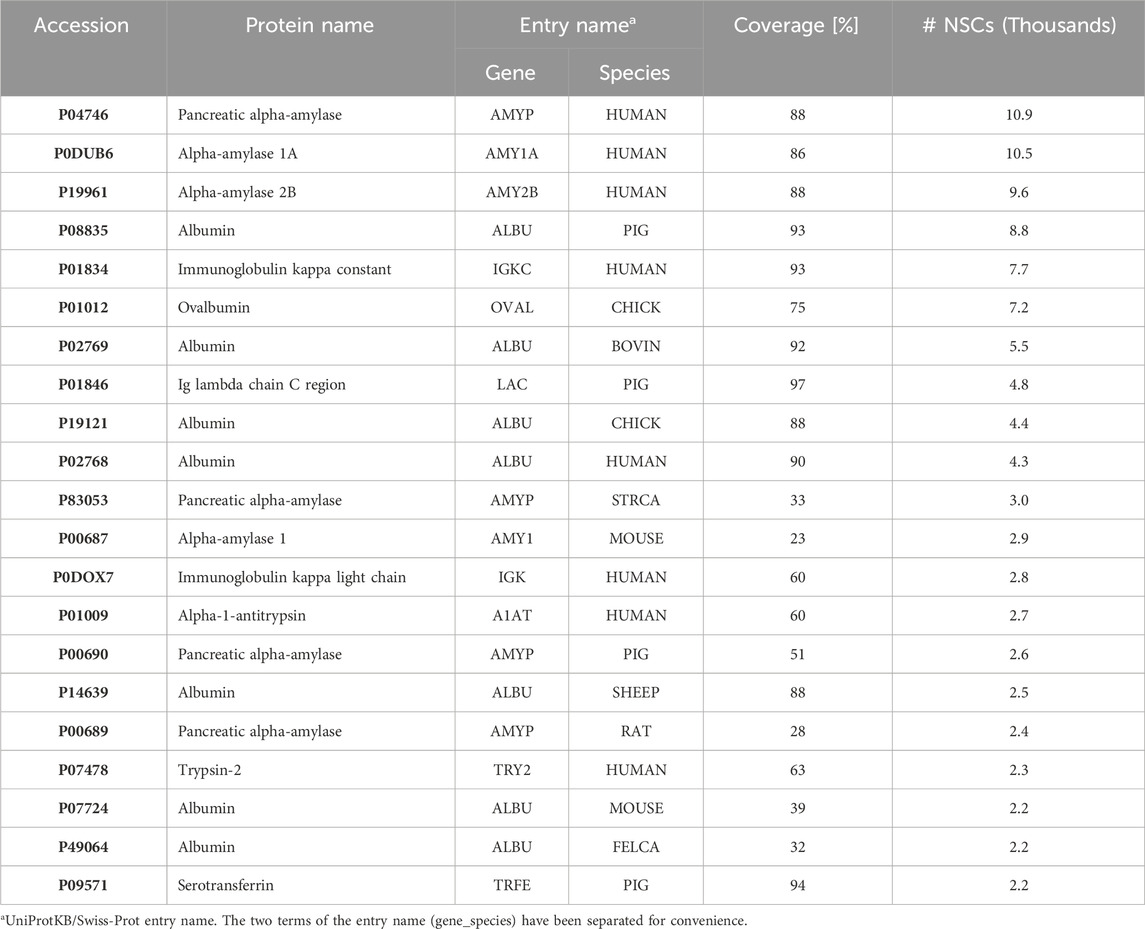
Table 1. The 20 most abundant proteins in the wastewater samples based on the normalized number of MS/MS spectra identified (NSCs) (Carrascal et al., 2023). STRCA: Ostrich; FELCA: Cat.
A potential application of protein biomarkers of great interest would be the use of rat/mouse pancreatic amylases and immunoglobulins to monitor rodent populations in urban areas (Carrascal et al., 2023). Rat pests not only constitute a human health hazard as potential disease transmitters but also a threat to the integrity of infested infrastructures. Moreover, available rat pest surveillance methods (i.e., live or photo trapping) are costly and typically limited to point (local) monitoring.
The study of the wastewater proteome is currently in its infancy, but their preliminary findings have prompted new and unanticipated scientific inquiries. This is an outcome of our limited understanding of the many aspects that influence protein dynamics and fate during their journey from the emission source to the sampling site, including the actual discharge rates of these proteins over time, hydraulic retention times, stability, and other uncertainties and confounding factors. Still, the exploitation of the potential of proteins as health and environmental biomarkers is deeply recognized as an invaluable source of information about the population’s health and lifestyle status.
4 Conclusion
WBE has expanded its potential to become a benchmark for monitoring a population’s health and providing information of the population discharges in wastewater. In order to expand the scope of WBE, the effort made in the identification of small molecules is crucial. WBE also has an important future in monitoring the scope of the SDGs and to become a tool within the global “One Health” system bridging the gap between human and veterinary health. The development of faster monitoring systems and advancements in mass spectrometry techniques will make these achievements possible. However, we strongly believe that the simultaneous use of small molecule chemical analysis and proteomic identification is a powerful chemical analytical tool in WBE. This methodology can be even more sophisticated when combined with microbiological and genetic information providing a comprehensive picture of the powerful information that can be extracted from a given wastewater sample.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
YP: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing. AG: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing. MC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing. JA: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing. DB: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work has been supported by Grants PID 2020-114065RB-C22, PID 2020-114065RB-C21), and PID 2022-138556OB-C22 funded by MCIN/AEI/10.13039/501100011033, by “ERDF A way of making Europe” and by the Grant CIPROM/2021/032 funded by the Conselleria d'Educació, Universitats i Ocupació (Generalitat Valenciana).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frans.2024.1367448/full#supplementary-material
References
Adhikari, S., and Halden, R. U. (2022). Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environ. Int. 163, 107217. doi:10.1016/j.envint.2022.107217
Amman, F., and Bergthaler, A. (2022). Wastewater is a robust proxy for monitoring circulating SARS-CoV-2 variants. Nat. Biotechnol. 40, 1768–1769. doi:10.1038/s41587-022-01388-x
Asadi, A., Fakhri, Y., Salimi, Y., Daglioglu, N., Tahmasebifard, M., and Aghajarinezhad, M. (2023). Nicotine consumption rate through wastewater-based epidemiology: a systematic review, meta-analysis and probabilistic risk assessment. Environ. Sci. Pollut. Res. 30, 63416–63426. doi:10.1007/s11356-023-27017-x
Barcellos, D. S., Barquilha, C. E. R., Oliveira, P. E., Prokopiuk, M., and Etchepare, R. G. (2023). How has the COVID-19 pandemic impacted wastewater-based epidemiology? Sci. Total Environ. 892, 164561. doi:10.1016/j.scitotenv.2023.164561
Bilge, S., Dogan-Topal, B., Gürbüz, M. M., Yücel, A., Sınağ, A., and Ozkan, S. A. (2022). Recent advances in electrochemical sensing of cocaine: a review. TrAC - Trends Anal. Chem. 157, 116768. doi:10.1016/j.trac.2022.116768
Boogaerts, T., Ahmed, F., Choi, P. M., Tscharke, B., O'brien, J., De Loof, H., et al. (2021). Current and future perspectives for wastewater-based epidemiology as a monitoring tool for pharmaceutical use. Sci. Total Environ. 789, 148047. doi:10.1016/j.scitotenv.2021.148047
Bowes, D. A., and Halden, R. U. (2019). Theoretical evaluation of using wastewater-based epidemiology to assess the nutritional status of human populations. Curr. Opin. Environ. Sci. Health 9, 58–63. doi:10.1016/j.coesh.2019.05.001
Burdorf, A., and Rugulies, R. (2023). The importance of occupation in the development of the COVID-19 pandemic. Scand. J. Work, Environ. Health 49, 231–233. doi:10.5271/sjweh.4094
Carrascal, M., Abian, J., Ginebreda, A., and Barceló, D. (2020). Discovery of large molecules as new biomarkers in wastewater using environmental proteomics and suitable polymer probes. Sci. Total Environ. 747, 141145. doi:10.1016/j.scitotenv.2020.141145
Carrascal, M., Sánchez-Jiménez, E., Fang, J., Pérez-López, C., Ginebreda, A., Barceló, D., et al. (2023). Sewage protein information mining: discovery of large biomolecules as biomarkers of population and industrial activities. Environ. Sci. Technol. 57, 10929–10939. doi:10.1021/acs.est.3c00535
Chau, K. K., Barker, L., Budgell, E. P., Vihta, K. D., Sims, N., Kasprzyk-Hordern, B., et al. (2022). Systematic review of wastewater surveillance of antimicrobial resistance in human populations. Environ. Int. 162, 107171. doi:10.1016/j.envint.2022.107171
Demeter, K., Linke, R., Balleste, E., Reischer, G., Mayer, R. E., Vierheilig, J., et al. (2023). Have genetic targets for faecal pollution diagnostics and source tracking revolutionized water quality analysis yet? FEMS Microbiol. Rev. 47, fuad028. doi:10.1093/femsre/fuad028
Devianto, L. A., and Sano, D. (2023). Systematic review and meta-analysis of human health-related protein markers for realizing real-time wastewater-based epidemiology. Sci. Total Environ. 897, 165304. doi:10.1016/j.scitotenv.2023.165304
Du Toit, A. (2023). Sewer biofilms and SARS-CoV-2. Nat. Rev. Microbiol. 21, 277. doi:10.1038/s41579-023-00882-6
Dutta, H., Kaushik, G., and Dutta, V. (2022). Wastewater-based epidemiology: a new frontier for tracking environmental persistence and community transmission of COVID-19. Environ. Sci. Pollut. Res. 29, 85688–85699. doi:10.1007/s11356-021-17419-0
Eaton, C. J., Coxon, S., Pattis, I., Chappell, A., Hewitt, J., and Gilpin, B. J. (2022). A framework for public health authorities to evaluate health determinants for wastewater-based epidemiology. Environ. Health Perspect. 130, 125001. doi:10.1289/EHP11115
Fitzmorris-Brisolara, K., Maal-Bared, R., Worley-Morse, T., Danley-Thomson, A., and Sobsey, M. (2022). Monitoring coliphages to reduce waterborne infectious disease transmission in the One Water framework. Int. J. Hyg. Environ. Health 240, 113921. doi:10.1016/j.ijheh.2022.113921
Foyle, L., Burnett, M., Creaser, A., Hens, R., Keough, J., Madin, L., et al. (2023). Prevalence and distribution of antimicrobial resistance in effluent wastewater from animal slaughter facilities: a systematic review. Environ. Pollut. 318, 120848. doi:10.1016/j.envpol.2022.120848
Gahlot, P., Alley, K. D., Arora, S., Das, S., Nag, A., and Tyagi, V. K. (2023). Wastewater surveillance could serve as a pandemic early warning system for COVID-19 and beyond. Wiley Interdiscip. Rev. Water 10, e1650. doi:10.1002/wat2.1650
Gao, Z., Gao, M., Chen, C. H., Zhou, Y., Zhan, Z. H., and Ren, Y. (2023). Knowledge graph of wastewater-based epidemiology development: a data-driven analysis based on research topics and trends. Environ. Sci. Pollut. Res. 30, 28373–28382. doi:10.1007/s11356-023-25237-9
García-Encina, P. A. (2021). Wastewater-based epidemiology (WBE). Water Environ. J. 35, 1162–1163. doi:10.1111/wej.12761
Gil-Solsona, R., Nika, M.-C., Bustamante, M., Villanueva, C. M., Foraster, M., Cosin-Tomás, M., et al. (2021). The potential of sewage sludge to predict and evaluate the human chemical exposome. Environ. Sci. Technol. Lett. 8, 1077–1084. doi:10.1021/acs.estlett.1c00848
Gonçalves, J., Torres-Franco, A., Rodriguéz, E., Diaz, I., Koritnik, T., Silva, P. G. D., et al. (2022). Centralized and decentralized wastewater-based epidemiology to infer COVID-19 transmission – a brief review. One Health 15, 100405. doi:10.1016/j.onehlt.2022.100405
Greenbaum, D., Gurwitz, D., and Joly, Y. (2022). Editorial: COVID-19 pandemics: ethical, legal and social issues. Front. Genet. 13, 1021865. doi:10.3389/fgene.2022.1021865
Guo, Y., Li, J., O'Brien, J., Sivakumar, M., and Jiang, G. (2022a). Back-estimation of norovirus infections through wastewater-based epidemiology: a systematic review and parameter sensitivity. Water Res. 219, 118610. doi:10.1016/j.watres.2022.118610
Guo, Y., Sivakumar, M., and Jiang, G. (2022b). Decay of four enteric pathogens and implications to wastewater-based epidemiology: effects of temperature and wastewater dilutions. Sci. Total Environ. 819, 152000. doi:10.1016/j.scitotenv.2021.152000
Guzmán-Fierro, V., Dieguez-Seoane, A., Roeckel, M., Lema, J. M., and Trueba-Santiso, A. (2024). Environmental proteomics as a useful methodology for early-stage detection of stress in anammox engineered systems. Sci. Total Environ. 912, 169349. doi:10.1016/j.scitotenv.2023.169349
Hassard, F., Bajón-Fernández, Y., and Castro-Gutierrez, V. (2023). Wastewater-based epidemiology for surveillance of infectious diseases in healthcare settings. Curr. Opin. Infect. Dis. 36, 288–295. doi:10.1097/QCO.0000000000000929
Henriot, P., Buelow, E., Petit, F., Ploy, M. C., Dagot, C., and Opatowski, L. (2024). Modeling the impact of urban and hospital eco-exposomes on antibiotic-resistance dynamics in wastewaters. Sci. Total Environ. 924, 171643. doi:10.1016/j.scitotenv.2024.171643
Hopkins, L., Ensor, K. B., Stadler, L., Johnson, C. D., Schneider, R., Domakonda, K., et al. (2023). Public health interventions guided by houston’s wastewater surveillance program during the COVID-19 pandemic. Public Health Rep. 138, 856–861. doi:10.1177/00333549231185625
Huizer, M., Ter Laak, T. L., De Voogt, P., and Van Wezel, A. P. (2021). Wastewater-based epidemiology for illicit drugs: a critical review on global data. Water Res. 207, 117789. doi:10.1016/j.watres.2021.117789
Jaunay, E. L., Simpson, B. S., White, J. M., and Gerber, C. (2023). Using wastewater-based epidemiology to evaluate the relative scale of use of opioids. Sci. Total Environ. 897, 165148. doi:10.1016/j.scitotenv.2023.165148
Kennes-Veiga, D. M., Trueba-Santiso, A., Gallardo-Garay, V., Balboa, S., Carballa, M., and Lema, J. M. (2022). Sulfamethoxazole enhances specific enzymatic activities under aerobic heterotrophic conditions: a metaproteomic approach. Environ. Sci. Technol. 56, 13152–13159. doi:10.1021/acs.est.2c05001
Kim, K., Stoll, S., Singh, R., Lee, W. H., and Hwang, J. H. (2023). Recent advances in illicit drug detection sensor technology in water. TrAC - Trends Anal. Chem. 168, 117295. doi:10.1016/j.trac.2023.117295
Kuhn, R., Benndorf, D., Rapp, E., Reichl, U., Palese, L. L., and Pollice, A. (2011). Metaproteome analysis of sewage sludge from membrane bioreactors. Proteomics 11, 2738–2744. doi:10.1002/pmic.201000590
Kumar, M., Vithanage, M., An, A. K., Weber, K. A., and Bhattacharya, P. (2022). Geogenic and pathogenic contamination: issues and innovation in the post-COVID-Anthropocene. J. Hazard. Mater. 429, 127501. doi:10.1016/j.jhazmat.2021.127501
Laimou-Geraniou, M., Heath, D., and Heath, E. (2023). Analytical methods for the determination of antidepressants, antipsychotics, benzodiazepines and their metabolites through wastewater-based epidemiology. Trends Environ. Anal. Chem. 37, 200192. doi:10.1016/j.teac.2022.e00192
Langa, I., Gonçalves, R., Tiritan, M. E., and Ribeiro, C. (2021). Wastewater analysis of psychoactive drugs: non-enantioselective vs enantioselective methods for estimation of consumption. Forensic Sci. Int. 325, 110873. doi:10.1016/j.forsciint.2021.110873
Larsson, D. G. J., Flach, C. F., and Laxminarayan, R. (2023). Sewage surveillance of antibiotic resistance holds both opportunities and challenges. Nat. Rev. Microbiol. 21, 213–214. doi:10.1038/s41579-022-00835-5
Lorenzo, M., and Picó, Y. (2019). Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health 9, 77–84. doi:10.1016/j.coesh.2019.05.007
Mackull’ak, T., Cverenkárová, K., Staňová, A. V., Fehér, M., Tamáš, M., Škulcová, A. B., et al. (2021). Hospital wastewater—source of specific micropollutants, antibiotic-resistant microorganisms, viruses, and their elimination. Antibiotics 10, 1070. doi:10.3390/antibiotics10091070
Massano, M., Salomone, A., Gerace, E., Alladio, E., Vincenti, M., and Minella, M. (2023). Wastewater surveillance of 105 pharmaceutical drugs and metabolites by means of ultra-high-performance liquid-chromatography-tandem high resolution mass spectrometry. J. Chromatogr. A 1693, 463896. doi:10.1016/j.chroma.2023.463896
O'Brien, E., and Xagoraraki, I. (2019). A water-focused one-health approach for early detection and prevention of viral outbreaks. One Health 7, 100094. doi:10.1016/j.onehlt.2019.100094
Oloye, F. F., Xie, Y., Challis, J. K., Femi-Oloye, O. P., Brinkmann, M., Mcphedran, K. N., et al. (2023). Understanding common population markers for SARS-CoV-2 RNA normalization in wastewater – a review. Chemosphere 333, 138682. doi:10.1016/j.chemosphere.2023.138682
Pan, Y., Mao, K., Hui, Q., Wang, B., Cooper, J., and Yang, Z. (2022). Paper-based devices for rapid diagnosis and wastewater surveillance. TrAC - Trends Anal. Chem. 157, 116760. doi:10.1016/j.trac.2022.116760
Pandey, R. P., Yousef, A. F., Alsafar, H., and Hasan, S. W. (2023). Surveillance, distribution, and treatment methods of antimicrobial resistance in water: a review. Sci. Total Environ. 890, 164360. doi:10.1016/j.scitotenv.2023.164360
Park, C., Novak, J. T., Helm, R. F., Ahn, Y. O., and Esen, A. (2008). Evaluation of the extracellular proteins in full-scale activated sludges. Water Res. 42, 3879–3889. doi:10.1016/j.watres.2008.05.014
Perez-Lopez, C., Ginebreda, A., Carrascal, M., Barceló, D., Abian, J., and Tauler, R. (2021). Non-target protein analysis of samples from wastewater treatment plants using the regions of interest-multivariate curve resolution (ROIMCR) chemometrics method. J. Environ. Chem. Eng. 9, 105752. doi:10.1016/j.jece.2021.105752
Perez-Lopez, C., Ginebreda, A., Jaumot, J., Yamamoto, F. Y., Barceló, D., and Tauler, R. (2024). MSident: straightforward identification of chemical compounds from MS-resolved spectra. Chemom. Intelligent Laboratory Syst. 245, 105063. doi:10.1016/j.chemolab.2024.105063
Picó, Y., and Barceló, D. (2021a). Identification of biomarkers in wastewater-based epidemiology: main approaches and analytical methods. TrAC - Trends Anal. Chem. 145, 116465. doi:10.1016/j.trac.2021.116465
Picó, Y., and Barceló, D. (2021b). Mass spectrometry in wastewater-based epidemiology for the determination of small and large molecules as biomarkers of exposure: toward a global view of environment and human health under the COVID-19 outbreak. ACS Omega 6, 30865–30872. doi:10.1021/acsomega.1c04362
Picó, Y., and Barceló, D. (2023). Microplastics and other emerging contaminants in the environment after COVID-19 pandemic: the need of global reconnaissance studies. Curr. Opin. Environ. Sci. Health 33, 100468. doi:10.1016/j.coesh.2023.100468
Rice, J., and Kasprzyk-Hordern, B. (2019). A new paradigm in public health assessment: water fingerprinting for protein markers of public health using mass spectrometry. TrAC - Trends Anal. Chem. 119, 115621. doi:10.1016/j.trac.2019.115621
Rousis, N., Bade, R., and Gracia-Lor, E. (2023). Wastewater-based epidemiology as a surveillance tool to assess human consumption of psychotropic substances: alcohol, nicotine and caffeine as case studies. TrAC - Trends Anal. Chem. 167, 117230. doi:10.1016/j.trac.2023.117230
Sánchez-Jiménez, E., Abian, J., Ginebreda, A., Barceló, D., and Carrascal, M. (2023). Shotgun proteomics to characterize wastewater proteins. MethodsX 11, 102403. doi:10.1016/j.mex.2023.102403
Sharma, A., Karande, M., and Zumla, A. (2023). Advancing sewage surveillance at mass gathering events for reducing transmission of antimicrobial resistant bacterial pathogens. Travel Med. Infect. Dis. 54, 102619. doi:10.1016/j.tmaid.2023.102619
Shaw, A. G., Troman, C., Akello, J. O., O’Reilly, K. M., Gauld, J., Grow, S., et al. (2023). Defining a research agenda for environmental wastewater surveillance of pathogens. Nat. Med. 29, 2155–2157. doi:10.1038/s41591-023-02457-7
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi:10.1093/nar/gkac194
Shimko, K. M., Piatkowski, T., Thomas, K. V., Speers, N., Brooker, L., Tscharke, B. J., et al. (2021). Performance- and image-enhancing drug use in the community: use prevalence, user demographics and the potential role of wastewater-based epidemiology. J. Hazard. Mater. 419, 126340. doi:10.1016/j.jhazmat.2021.126340
Sims, N., and Kasprzyk-Hordern, B. (2020). Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 139, 105689. doi:10.1016/j.envint.2020.105689
Singer, A. C., Thompson, J. R., Filho, C. R. M., Street, R., Li, X., Castiglioni, S., et al. (2023). A world of wastewater-based epidemiology. Nat. Water 1, 408–415. doi:10.1038/s44221-023-00083-8
Sodhi, K. K., and Singh, C. K. (2023). A systematic review on the occurrence, fate, and remediation of SARS-CoV-2 in wastewater. Int. J. Environ. Sci. Technol. 20, 8073–8086. doi:10.1007/s13762-022-04326-1
Tavazzi, S., Cacciatori, C., Comero, S., Fatta-Kassinos, D., Karaolia, P., Iakovides, I. C., et al. (2023). Short-term stability of wastewater samples for storage and shipment in the context of the EU Sewage Sentinel System for SARS-CoV-2. J. Environ. Chem. Eng. 11, 109623. doi:10.1016/j.jece.2023.109623
Verovšek, T., Krizman-Matasic, I., Heath, D., and Heath, E. (2020). Site- and event-specific wastewater-based epidemiology: current status and future perspectives. Trends Environ. Anal. Chem. 28, e00105. doi:10.1016/j.teac.2020.e00105
Vitale, D., Morales Suárez-Varela, M., and Picó, Y. (2021). Wastewater-based epidemiology, a tool to bridge biomarkers of exposure, contaminants, and human health. Curr. Opin. Environ. Sci. Health 20, 100229. doi:10.1016/j.coesh.2021.100229
Wang, S., Ishii, K., Yu, H., Shi, X., Smets, B. F., Palomo, A., et al. (2021). Stable nitrogen removal by anammox process after rapid temperature drops: insights from metagenomics and metaproteomics. Bioresour. Technol. 320, 124231. doi:10.1016/j.biortech.2020.124231
Westgate, P. J., and Park, C. (2010). Evaluation of proteins and organic nitrogen in wastewater treatment effluents. Environ. Sci. Technol. 44, 5352–5357. doi:10.1021/es100244s
Wild, C. P. (2005). Complementing the genome with an "exposome": the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14, 1847–1850. doi:10.1158/1055-9965.EPI-05-0456
Wolfe, M. K., Yu, A. T., Duong, D., Rane, M. S., Hughes, B., Chan-Herur, V., et al. (2023). Use of wastewater for mpox outbreak surveillance in California. N. Engl. J. Med. 388, 570–572. doi:10.1056/NEJMc2213882
Zhang, P., Zhu, J., Xu, X. Y., Qing, T. P., Dai, Y. Z., and Feng, B. (2019). Identification and function of extracellular protein in wastewater treatment using proteomic approaches: a minireview. J. Environ. Manage. 233, 24–29. doi:10.1016/j.jenvman.2018.12.028
Zhang, Y., Fonslow, B. R., Shan, B., Baek, M. C., and Yates, J. R. (2013). Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 113, 2343–2394. doi:10.1021/cr3003533
Keywords: biomarkers, proteomics, wastewater, high-resolution mass spectrometry (HRMS), metabolites
Citation: Picó Y, Ginebreda A, Carrascal M, Abian J and Barceló D (2024) Simultaneous determination of small molecules and proteins in wastewater-based epidemiology. Front. Anal. Sci. 4:1367448. doi: 10.3389/frans.2024.1367448
Received: 08 January 2024; Accepted: 21 May 2024;
Published: 12 June 2024.
Edited by:
Ernesto Satoshi Nakayasu, Pacific Northwest National Laboratory (DOE), United StatesReviewed by:
Judith Wong, National Environment Agency, SingaporeCopyright © 2024 Picó, Ginebreda, Carrascal, Abian and Barceló. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yolanda Picó, Yolanda.Pico@uv.es
 Yolanda Picó
Yolanda Picó Antoni Ginebreda2
Antoni Ginebreda2  Montserrat Carrascal
Montserrat Carrascal Damià Barceló
Damià Barceló