Protein arginine methylation in transcription and epigenetic regulation
- 1Department of Cancer Genetics and Epigenetics, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA, United States
- 2Irell and Manella Graduate School of Biological Sciences, Beckman Research Institute, City of Hope National Cancer Center, Duarte, CA, United States
Arginine methylation is a prevalent post-translational modification found in all eukaryotic systems. It involves the addition of a methyl group to the guanidino nitrogen atoms of arginine residues within proteins, and this process is catalyzed by a family of enzymes called protein arginine methyltransferases (PRMTs). In mammals, there exist nine PRMTs (PRMT1–9) that catalyze three distinct types of arginine methylation: monomethylarginine, asymmetric dimethylarginine, and symmetric dimethylarginine. These modifications play critical roles in numerous fundamental cellular processes, including transcription, RNA metabolism, genome maintenance, and signaling transduction. Aberrations in protein arginine methylation have been implicated in various human diseases, such as neurodevelopmental disorders and cancer. This review offers a general overview of arginine methylation, covering its deposition, its impact on protein function, and the diverse regulatory mechanisms involved. We specifically focus on an in-depth view of the role of arginine methylation in transcription and the epigenetic regulation of gene expression. Readers are directed towards additional reviews that encompass other aspects of arginine methylation biology.
Introduction
Among the twenty amino acids, arginine has a distinguishing feature for its positively charged guanidino group. This unique moiety allows arginine to engage in molecular interactions by forming up to six hydrogen bonds, and its chemical reactivity enables a wide array of chemical modifications (Lassak et al., 2019). These modifications, including methylation, phosphorylation, and ADP-ribosylation, in turn, play a crucial role in regulating the physiological properties of arginine-mediated molecular interactions. In eukaryotic cells, methylation is the most abundant modification found on arginine residues. It is estimated that about 2% of arginine residues are methylated in rat liver nuclei (Boffa et al., 1977), while mammalian tissues contain approximately 0.5% of arginine residues in a methylated state (Matsuoka, 1972). Consequently, arginine methylation is as abundant as other widely known modifications, such as phosphorylation and ubiquitination (Khoury et al., 2011; Zhang et al., 2021).
There are three types of methylated arginine resides, namely, ω-NG-monomethylarginine (MMA), ω-NG,NG-asymmetric dimethylarginine (ADMA) and ω-NG,N'G-symmetric dimethylarginine (SDMA). MMA is produced in the initial reaction, followed by a sequential catalytic reaction that drives further methylation resulting in dimethylarginines (Figure 1A). In mammalian cells, the abundance of ADMA modification surpasses that of MMA and SDMA combined, with an estimated ratio of 9:1 (Maron et al., 2021; Zhang et al., 2021). However, this ratio can vary depending on the specific cell types being studied and the method being used. Mounting evidence emerged from the past two decades has clearly established arginine methylation as a key regulator of protein function in various cellular processes, including transcription, RNA metabolism, genome maintenance, and signaling transduction (Bedford and Clarke, 2009; Blanc and Richard, 2017), and dysregulation of arginine methylation has been implicated in various human diseases, such as neurological disorders (Quan et al., 2015; Hofweber et al., 2018; Qamar et al., 2018; Ryan et al., 2018; Yoshizawa et al., 2018) and cancers (Yang and Bedford, 2013; Jarrold and Davies, 2019; Hwang et al., 2021). Additionally, the rapid development of small molecule inhibitors of protein arginine methylation has greatly boosted the interest of clinical and translational research to tackle arginine methylation in diseases (Kaniskan et al., 2018). In more recent years, the study of protein arginine methylation has entered a new era. This is largely benefited from the development of new tools and technologies that have provided unprecedented opportunities for detecting, monitoring, and manipulating cellular arginine methylation levels. Specifically, the combination of pan-methylarginine antibodies raised against short methylated-peptides with the advanced proteomic techniques, such as stable isotope labeling by amino acids in cell culture (SILAC) and strong cation exchange (SCX) chromatography, as well as nuclear magnetic resonance (NMR), have enabled the identification of arginine-methylated protein substrates and quantification of arginine methylation levels in cell lines and mouse tissues to a prodigious scale and depth (Boisvert et al., 2003; Ong et al., 2004; Hung et al., 2009; Uhlmann et al., 2012; Guo et al., 2014; Hartel et al., 2019; Maron et al., 2021; Zhang et al., 2021). These new discoveries have provided us with an ever-evolving comprehensive view of this important PTM in cellular function. Particularly relevant to this review, proteins involved in transcription and epigenetic regulation have been identified as the most significantly enriched category among all the arginine-methylated substrates (Boisvert et al., 2003; Guo et al., 2014; Hartel et al., 2019). Thus, we focus our review on the role of arginine methylation in this area. Notably, aberrant protein arginine methylation underlies many human diseases, and significant progress has been made in the development of small molecule inhibitors of arginine methylation. We direct readers that are more interested in these topics to these recent reviews (Fuhrmann and Thompson, 2016; Blanc and Richard, 2017; Guccione and Richard, 2019; Lorton and Shechter, 2019; Tewary et al., 2019; Couto e Silva et al., 2020; Qin and Xu, 2021; Wu et al., 2021).
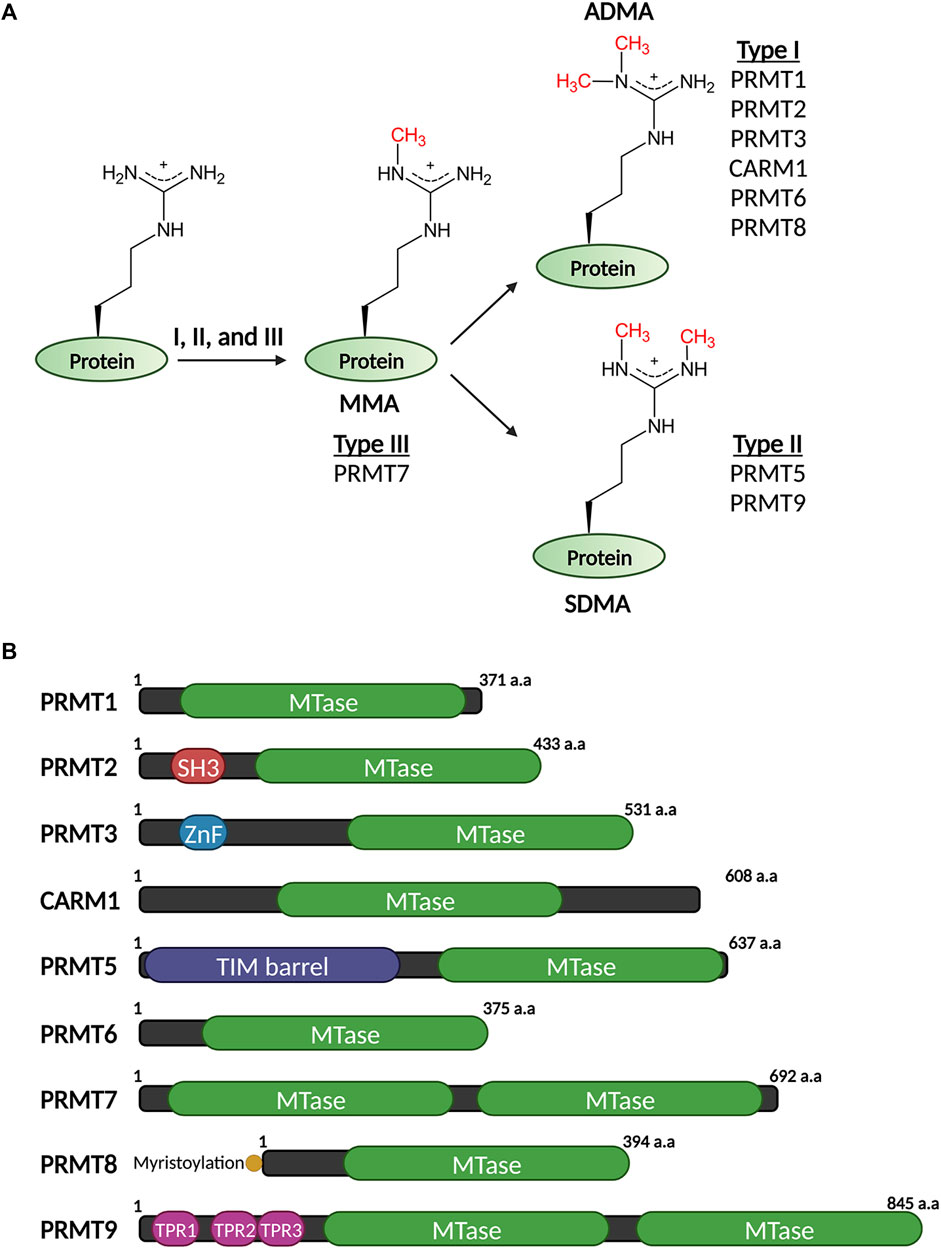
FIGURE 1. Three types of methylation on arginine residues (A) and domain structures of mammalian PRMTs that catalyze these modifications (B).
Writers, readers, and erasers of protein arginine methylation
Protein arginine methyltransferases (PRMTs)
Arginine methylation is catalyzed by a family of protein arginine methyltransferases (PRMTs), also known as writers of arginine methylation. They belong to the seven-β-strand methyltransferases characterized by the twisted beta-sheet structures in their catalytic domains that contain the signature methyltransferase motifs I, post-I, II, and III. They also harbor additional “double E” (two glutamate residues) and “THW” (threonine–histidine–tryptophan) sequence motifs, which are unique to the PRMT subfamily methyltransferases (Bedford and Clarke, 2009). Like all other methyltransferases, PRMTs use S-adenosyl-l-methionine (SAM) as a methyl-donating cofactor, which is subsequently converted into S-adenosyl-l-homocysteine (SAH) after the methyl-group being transferred to the guanidino group of a peptidyl arginine residue. The X-ray crystallography of the PRMT catalytic domain revealed a doughnut-shaped homodimer structure arranged in a head-to-tail pattern (Zhang and Cheng, 2003). Human genome encodes nine PRMTs (PRMT1–9) (Figure 1B), with most of them, except PRMT7 and PRMT9 that themselves form pseudo-dimers, require the dimerization state to be functionally active (Weiss et al., 2000; Zhang and Cheng, 2003). In addition to the highly similar organization in their catalytic domains, PRMTs harbor divergent amino (N)-terminal protein–protein interaction domains and/or signaling peptides that are critical for their substrate specificities and distinct subcellular localization. For example, the N-terminus of PRMT2 contains a SRC homology 3 domain (SH3 domain), which in general mediates protein–protein interactions by recognizing the proline-rich sequence motifs on cellular proteins (Weng et al., 1995). Without the SH3 domain, truncated PRMT2 exhibits seven-fold reduction of the methyltransferase activity in comparison to the full-length enzyme (Cura et al., 2017). The N-terminus of PRMT9 contains three tetratricopeptide repeats (TPRs) in tandem that are essential for PRMT9 to interact with and methylate its protein substrate, SF3B2 (splicing factor 3B subunit 2) (Hadjikyriacou et al., 2015; Yang et al., 2015). Furthermore, PRMT8, a neuronal specific PRMT, harbors a unique myristoylation motif at its N-terminus. This short motif distinguishes PRMT8 from its closest paralog PRMT1 and targets PRMT8 to the plasma membrane, where it likely methylates a unique set of substrates (Lee J. et al., 2005; Lin et al., 2013; Penney et al., 2017).
Mammalian PRMTs can be classified into three catalytic groups based on the methylation products that they produce: type I PRMTs that produce MMA and ADMA include PRMT1, PRMT2, PRMT3, PRMT4/CARM1, PRMT6 and PRMT8; type II PRMTs that produce MMA and SDMA include PRMT5 and PRMT9, and type III PRMT PRMT7, which only produces MMA (Figure 1A). Although some PRMTs share overlapping methylation substrates and exhibit marginal specificity in vitro, individual PRMT can demonstrate strong substrate preference and fulfill distinct cellular functions in vivo. For example, PRMT1, PRMT3, PRMT6, and PRMT8 have similar substrate preferences of Arg-Gly-Gly (RGG) motif for methylation in vitro, but PRMT1 is the predominant PRMT that catalyzes majority (>85%) of the cellular methylation, whereas PRMT3 has a very focused methylation substrate–the ribosomal protein S2 (RPS2) (Bachand and Silver, 2004; Swiercz et al., 2005; Swiercz et al., 2007). Similarly, although PRMT5 is the major SDMA enzyme that is responsible for almost all cellular SDMA deposition, it does not methylate PRMT9’s only known substrate–the R508 of SF3B2 (Hadjikyriacou et al., 2015; Yang et al., 2015). Notably, research in the past decade using loss-of-function cell lines and genetic mouse models, as well as small molecule PRMT inhibitors has also revealed significant functional redundancies among PRMT family members, many of which compete for the same substrate/site for methylation. For example, the asymmetrical dimethylation of histone 3 arginine 17 site (H3R17me2a), an active mark associated with gene promoters and enhancers, is deposited by the coactivator associated arginine methyltransferase CARM1 (Bauer et al., 2002). However, knockout of CARM1 does not lead to significant loss of global H3R17me2a. It was recently showed that PRMT6 can also catalyze H3R17me2a, and this mark only diminishes upon CARM1 and PRMT6 double knockout (Cheng et al., 2020). Another pronounced example of the redundancy among different PRMTs is the observation that loss of or inhibiting the major ADMA methyltransferase PRMT1 leads to methylation substrate scavenging by the SDMA enzyme PRMT5 (Dhar et al., 2013; Zhang et al., 2021), further suggesting that different types of methylation could compete for the same substrates. Further supporting this principle, knockdown PRMT1 expression or inhibiting PRMT1 activity using small molecule inhibitors sensitizes cancer cells to PRMT5 inhibition or knockdown (Gao et al., 2019). Further exploration of this redundancy is likely to reveal cancer cell vulnerabilities that could be harnessed for therapeutic interventions.
Methylarginine reader proteins
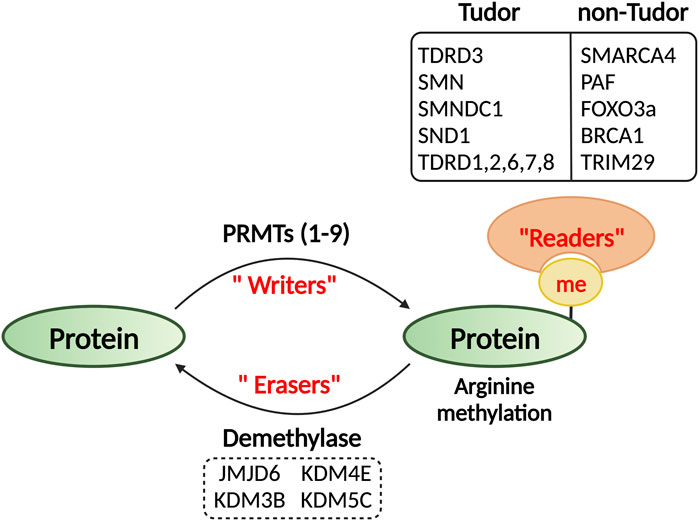
Arginine methylation could potentially alter protein structures, impact protein–DNA/RNA integrations, and generate docking sites for effector proteins (Figure 2). The Tudor domain-containing proteins are the “primary” readers of methylarginine modifications (Chen et al., 2011; Pek et al., 2012). Tudor domains are ∼60 amino acids in size and use conserved aromatic residues to build an “aromatic cage” and bind methylated-arginine through cation–π and π–π stacking interactions (Selenko et al., 2001; Sprangers et al., 2003; Friberg et al., 2009; Liu et al., 2010). In humans, there are at least thirty-six Tudor domain containing proteins that can be classified into methylarginine-binding and methyllysine-binding groups (Chen et al., 2011). Although it is not possible to predict the binding specificity based on their primary amino acid sequences, structure analyses suggest that the methylarginine binding Tudor domains form a relatively narrower aromatic cage than the methyllysine Tudor domains, thus, favoring the docking of the planar methyl-guanidinium group of the arginine (Liu et al., 2012). A “common” mechanism of action for the Tudor domain containing proteins is to function as a scaffold that links methylated arginine or lysine marks to downstream effectors with specific catalytic activities, so that the methylation signal can be interpretated or transduced in support of cellular function (Pek et al., 2012). This working model is well exemplified by the Tudor domain containing protein 3 (TDRD3), which harbors a Tudor domain at C-terminus that reads active methylarginine histone marks and an oligonucleotide/oligosaccharide binding (OB) fold at N-terminus that interacts with the DNA topoisomerase 3B (TOP3B) (Yang et al., 2010; Siaw et al., 2016). TDRD3 recruits TOP3B to chromatin regions enriched for active methylarginine histone marks to resolve the DNA negative supercoiling in the wake of transcribing RNA polymerase II (RNAPII) to facilitate transcription elongation (Yang et al., 2014; Zhang et al., 2019). Similarly, the Tudor domain of another methylarginine reader protein SMN (survival of motor neuron) can recognize SDMA mark on the C-terminal domain (CTD) of RNAPII deposited by PRMT5, and concomitantly, recruits RNA/DNA helicase Senataxin to promote transcription termination (Yanling Zhao et al., 2016).
In addition to these structurally defined Tudor domain containing reader proteins, several other domains and protein folds from specific proteins also exhibited enhanced affinity towards methylated arginine residues, such as the BRCT domain of BRCA1 (Lee et al., 2011), the HELICc domain of SMARCA4 (Yao et al., 2021), the CR3 domain of FOXO3a (Yu et al., 2020), and the C-terminal domain of TRIM29 (Gao et al., 2023). Although the structural basis underlying their interactions with methylated arginine has yet been defined, the involvement of aromatic residues strongly indicates the putative aromatic cage formation for methyl-binding.
Demethylases/erasers of arginine methylation
Once considered a stable mark, protein arginine methylation on both histone and non-histone protein substrates was recently found to be dynamic (Sarmento et al., 2004; Le Romancer et al., 2008; Litt et al., 2009), indicating the existence of an arginine demethylase or demethylases (Figure 2). However, the identification of a bona fide arginine demethylase is still under debate. The Jumonji domain containing 6 (JMJD6) was identified as the first candidate arginine demethylase for histone methylarginine marks (Chang et al., 2007). However, it was later characterized as a lysine hydroxylase that catalyzes lysyl-5-hydroxylation of the splicing factor U2 small nuclear ribonucleoprotein auxiliary factor 65-kDa subunit (U2AF65) (Webby et al., 2009). Nevertheless, recent biochemical studies using recombinant enzymes and methylated peptides suggested that the enzymes that remove methyl groups from arginine residues belong to the oxygen-sensitive dioxygenase family (Herr and Hausinger, 2018; Islam et al., 2018). For instance, biochemical analyses of a few JmjC domain-containing demethylases, including the well-defined histone lysine demethylases KDM4E and KDM5C, demonstrated that they are also capable of removing methyl groups from arginine-methylated histone peptides (Walport et al., 2016). The first evidence of arginine demethylase activity in vivo was reported on the H3K9me2 demethylase, JMJD1B (KDM3B), which can also catalyze demethylation of symmetrical dimethylation at histone 4 arginine 3 (H4R3me2s) (Li et al., 2018). More recently, KDM5C was reported to demethylate the autophagy activating kinase ULK1 at R170me2s deposited by PRMT5 (Li et al., 2022). It is likely that more evidence supporting arginine demethylation will emerge, whereas distinguishing the individual roles of these dual lysine and arginine demethylases remains a significant challenge.
How arginine methylation regulates protein function?
At physiological pH, arginine is positively charged and hydrophilic. It has the potential to form up to six hydrogen bonds (Bedford and Clarke, 2009). Thus, arginine residue per se has great capability for mediating molecular interactions with either nucleic acids or proteins. Arginine methylation does not alter the cationic charge of the arginine residue but removes its potential hydrogen bond donors and imparts hydrophobicity of the protein substrates (Tripsianes et al., 2011), thus, could potentially alter protein structures, affect protein–protein and protein–DNA/RNA interactions.
Protein–protein interaction
The best illustration of how arginine methylation affects protein–protein interaction is demonstrated by the interaction with Tudor domains, as well as protein sequences that have the potential to form aromatic cages (Chen et al., 2011; Wang and Bedford, 2023). This has been discussed in the previous section. Beyond the reader mediated interactions, arginine methylation can both negatively and positively regulate protein-protein interactions. For example, PRMT4/CARM1 catalyzed arginine methylation of pyruvate kinase M2 (PKM2) at R445 and 447 sites promotes the tetramerization of this key glycolytic enzyme, which greatly enhances its kinase activity (Abeywardana et al., 2018). The methylation of arginine residues neighboring proline-rich motifs in Sam68 protein prevents its association with SH3 (Src homology 3) domains, but not with WW domains (Bedford et al., 2000). Additionally, histone H3R2 is critically involved in interactions with several H3 N-terminal tail binding domains, such as PHD and WD40 (Couture et al., 2006; Taverna et al., 2007), and PRMT6 catalyzed H3R2 methylation (H3R2me2a) blocks the binding of these H3 N-terminal binders (Iberg et al., 2008).
Protein–DNA/RNA interaction
Proteomic studies have revealed that arginine methylated proteins are highly enriched in the nucleus and RNA-binding proteins (RBPs) form the largest group of arginine-methylated substrates (Boisvert et al., 2003; Guo et al., 2014; Geoghegan et al., 2015). In principle, the positive charge of an arginine residue promotes its interaction with negatively charged nucleic acids via electrostatic interactions. Addition of a methyl group, although does not change the positive charge, could impact the interactions between protein and DNA/RNA either positively or negatively. For example, PRMT1 catalyzed arginine methylation of FMRP (Fragile X Messenger Ribonucleoprotein) at its RGG motif region reduces its RNA binding capacity (Denman, 2002; Stetler et al., 2006), whereas arginine methylation of the RNA m6A methyltransferase METTL14 at its C-terminal RGG motif enhances its interaction with RNA substrates (Wang et al., 2021). The molecular basic underlying this differential regulation remains unclear, but some evidence suggests that the formation of RNA secondary structures might be a consideration (Vasilyev et al., 2015; Yoshida et al., 2022).
Arginine methylation crosstalk with other PTMs
The best characterized crosstalk between arginine methylation and other types of PTM is with phosphorylation at the consensus Akt substrate motif, RXRXXS/T (Obata et al., 2000). Demonstrated both in a nuclear transcription factor FOXO1 and a cytosolic BCL-2 antagonist of cell death (BAD), PRMT1-catalyzed methylation of arginine residues in this motif blocks Akt-mediated phosphorylation of downstream Serine (Yamagata et al., 2008; Sakamaki et al., 2011). Furthermore, the SRGG motif seems to be another hotspot of methylation–phosphorylation crosstalk (Lenard et al., 2021). Yeast Npl3p has six repeats of SRGG motif in its disordered region, which can be arginine methylated and phosphorylated (Gilbert et al., 2001; Hart-Smith et al., 2012). In this scenario, phosphorylation of Serine by the yeast kinase Sky1p inhibits the arginine methylation by the only yeast PRMT, Hmt1p (Smith et al., 2020). It is likely that this inhibitory effect is mutual, as arginine methylation of CIRBP-RGG could suppress its phosphorylation at neighboring serine by the serine-arginine protein kinase 1 (SRPK1) (Lenard et al., 2021). As these modifications are often located at the intrinsically disordered regions, it is likely that they could involve in the regulation of the biophysical property of the protein, such as phase separation. Additionally, arginine methylation could also crosstalk with other PTMs, such as acetylation (Yue et al., 2007; Yi et al., 2017) and lysine methylation (Kirmizis et al., 2007; Casadio et al., 2013).
Arginine methylation and phase separation
Most PRMTs prefer RG/RGG motif as methylation substrates (Boisvert et al., 2003; Guo et al., 2014; Geoghegan et al., 2015). Importantly, this motif is often found at the low complexity domain or disordered region of a protein, which are critically involved in liquid-liquid phase separation (LLPS) (Rajyaguru and Parker, 2012; Thandapani et al., 2013; Chong et al., 2018; Chowdhury and Jin, 2023), a fundamental process involved in the organization of many cellular biomolecular condensates, such as nuclear speckles and cytoplasmic stress granules (Banani et al., 2017; Lyon et al., 2021). Thus, it is not surprising that arginine methylation has emerged as an important regulatory mechanism for LLPS (Wang et al., 2022). The first evidence demonstrating this type of regulation comes from the study of a protein called fused in sarcoma (FUS), a multifunctional RNA-binding protein involved in transcription, splicing, transport, and translation (Hofweber et al., 2018; Qamar et al., 2018). FUS undergoes physiologically reversible phase separation; however, pathogenic missense mutations in FUS disrupt this balance and could trigger disease, such as familial amyotrophic lateral sclerosis (fALS) and frontotemporal lobar degeneration (FTLD) (Hofweber et al., 2018; Qamar et al., 2018). As FUS phase separation is driven by Cation-π interaction between arginines and the tyrosines, arginine methylation was shown to suppress phase separation. Hypomethylation, which often occurs in FUS-associated FTLD, induces FUS condensation into stable hydrogels that disrupt RNP function in neurons. This negative regulation of LLPS by arginine methylation is also exemplified by a few stress granule proteins, including G3BP1, hnRNPA1, and KHDRBS1 (Gill et al., 2021; Wang et al., 2022). Interestingly, many of the Tudor domain containing proteins undergo LLPS in a dimethylarginine dependent manner (Goulet et al., 2008; Linder et al., 2008; Narayanan et al., 2017; Huang et al., 2018; Courchaine et al., 2021; Simcikova et al., 2023), suggesting that Tudor–methylarginine interaction could promote phase separation or modulate the dynamics of the condensates. Thus, while methylation itself may have a negative impact on the biophysical characteristics of arginine regarding phase separation in vitro, it can still exert a positive effect on LLPS by enhancing protein–protein interactions, and possibly also protein–RNA interactions, within a more complex in vivo environment. We suggest that the biological role of arginine methylation in cellular biomolecular condensates should be determined in a case-by-case basis.
Arginine methylation in transcription
Arginine methylation of transcription factors
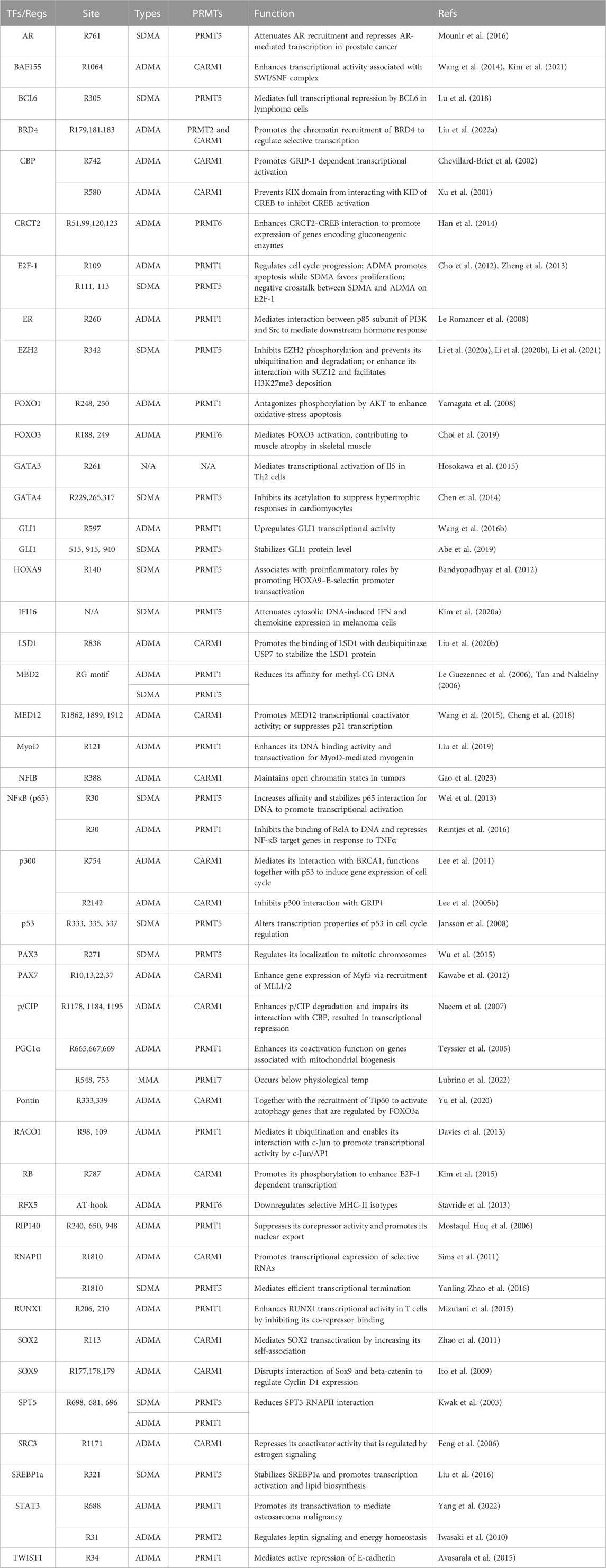
Transcription factors (TFs) bind sequence-specific DNA elements to control gene expression in various cellular processes, including development, differentiation, and response to environment cues (Spitz and Furlong, 2012; Lambert et al., 2018). Arginine methylation of TFs can have significant effects on their activity, DNA binding affinity, and interaction with other co-regulators. For example, tumor suppressor p53 is arginine methylated at R333, R335, and R337 by PRMT5 (Jansson et al., 2008). Although these sites reside within p53 oligomerization domain, methylation seems to have more specific impact on p53’s function in cell cycle regulation rather than having a global “on” or “off” effect. In lymphomagenesis, this mechanism is harnessed by cancer cells to selectively suppresses expression of crucial proapoptotic and antiproliferative target genes, thereby sustaining tumor cell self-renewal and proliferation (Li et al., 2015). Another well-characterized arginine methylated TF is E2F Transcription Factor 1 (E2F1), which is subjected to ADMA and SDMA modifications by PRMT1 and PRMT5, respectively (Cho et al., 2012; Choi et al., 2019). The methylation sites, R109 by PRMT1 and R111/113 by PRMT5 are located in the RGRGR motif but were not present in other E2F family proteins. Interestingly, R109me2a and R111/113me2s impose opposing effects on E2F1 function in cell fate decision. R109me2a is induced upon DNA damage and associated with transcription activation of apoptotic gene expression, whereas R111/113me2s is associated with the activation of growth promoting genes (Cho et al., 2012; Choi et al., 2019). A more comprehensive list of TFs regulated by arginine methylation is summarized in Table 1.
Arginine methylation of RNAPII
The C-terminal domain (CTD) of mammalian RNAPII contains 52 heptapeptide repeats (YSPTSPS) (Hsin and Manley, 2012) and the R1810 located in repeat 31 is methylated by PRMT4/CARM1 and PRMT5, generating ADMA and SDMA marks, respectively (Mizutani et al., 2015; Yanling Zhao et al., 2016). Importantly, these two types of methylation recruit different reader proteins that transduce the methylation signal to distinct downstream effects. Reading of R1810me2a by TDRD3 is proposed to recruit the DNA topoisomerase 3B (TOP3B) to resolve negative supercoiling DNA generated in the wake of transcribing RNAPII (Mizutani et al., 2015), whereas reading of R1810me2s by SMN enables the association of Senataxin helicase with RNAPII for R-loop resolution and transcription termination (Yanling Zhao et al., 2016). Interestingly, R1810 could also be deiminated to citrulline by peptidyl arginine deiminase 2 (PADI2), which is important for RNAPII pause release and efficient transcription (Sharma et al., 2019). Thus, proper spatial and temporal control of RNAPII R1810 modification by methylation and citrullination could be essential for RNAPII function.
Arginine methylation of transcription elongation factors
The initial observation of direct involvement of arginine methylation in transcription elongation is from yeast Hmt1p (the only PRMT in yeast) mutants, which exhibited reduced elongation rate and increased transcription termination at cryptic terminators (Wong et al., 2010). Mechanistically, arginine methylation of Npl3p at its RGG motif is essential for the recruitment of elongation factors, such as Tho2p, for antitermination (Wong et al., 2010). In mammals, transcription elongation factor SPT5 is methylated by PRMT1 and PRMT5, and methylation regulates its interaction with RNAPII (Kwak et al., 2003). In this case, it seems that ADMA and SDMA exhibit similar effects in suppressing SPT5–RNAPII interaction and transcription elongation. Interestingly, methylation specifically affect SPT5 interaction with serine-2 phosphorylated, but not with serine 5 phosphorylated RNAPII, which mainly enriched at gene promoters (Kwak et al., 2003). On the other hand, the activity of transcription elongation can, in turn, impact arginine methylation levels of several elongation regulators, including SPT5. Upon transcription arrest by Actinomycin D, the levels of SPT5 monomethylation decreases (Sylvestersen et al., 2014); however, the functional significance of this regulation remains unknown.
Arginine methylation of mediators and transcription co-regulators
Mediator is a large, conformationally flexible protein complex with a variable subunit composition (Allen and Taatjes, 2015). It functions by communicating regulatory signals from DNA-binding TFs directly to RNAPII. The mediator subunit 12 (MED12) is a major substrate of PRMT4/CARM1 in transcription regulation. Multiple arginine residues of MED12 are methylated, including R1862, R1912, and R1899 (Wang et al., 2015; Cheng et al., 2018). Although it is unclear how methylation affects mediator function, the methylarginine mark did promote its interaction with reader protein TDRD3, which functions in resolving co-translational R-loops to facilitate transcription (Yang et al., 2010; Yang et al., 2014). It is worth noting that PRMT5 was also identified to associate with mediator complex using a Flag-tag affinity purification of CDK9 and CDK19 interaction proteins (Tsutsui et al., 2013). However, PRMT5 is a known common contaminant of Flag-tag based affinity purification (Nishioka and Reinberg, 2003; Mellacheruvu et al., 2013), further validation is necessary to confirm this observation. Additionally, the histone acetyltransferases CREB binding protein (CBP) and the related p300 protein, which are key transcriptional co-activators, are methylated by PRMT4/CARM1 (Chen et al., 1999; Xu et al., 2001; Chevillard-Briet et al., 2002; Lee Y. H. et al., 2005). The methylation sites are located both at the N-terminal KIX domain (Chen et al., 1999; Xu et al., 2001; Chevillard-Briet et al., 2002) and C-terminal GRIP1 binding domain (GBD) (Lee Y. H. et al., 2005). The N-terminal methylation is important for hormone-dependent transcriptional activation, whereas the methylation of GBD inhibits the interaction of GRIP1 with p300. In Testis, GBD methylation of p300 inhibits its interaction with the haploid spermatid specific coactivator, activator of CREMτ in the testis (ACT), to suppress CREMτ target gene expression (Bao et al., 2018). These studies demonstrated that methylation at different sites of a protein could have opposing effects on co-regulator function.
Arginine methylation in epigenetic regulation
Arginine methylation of Histones
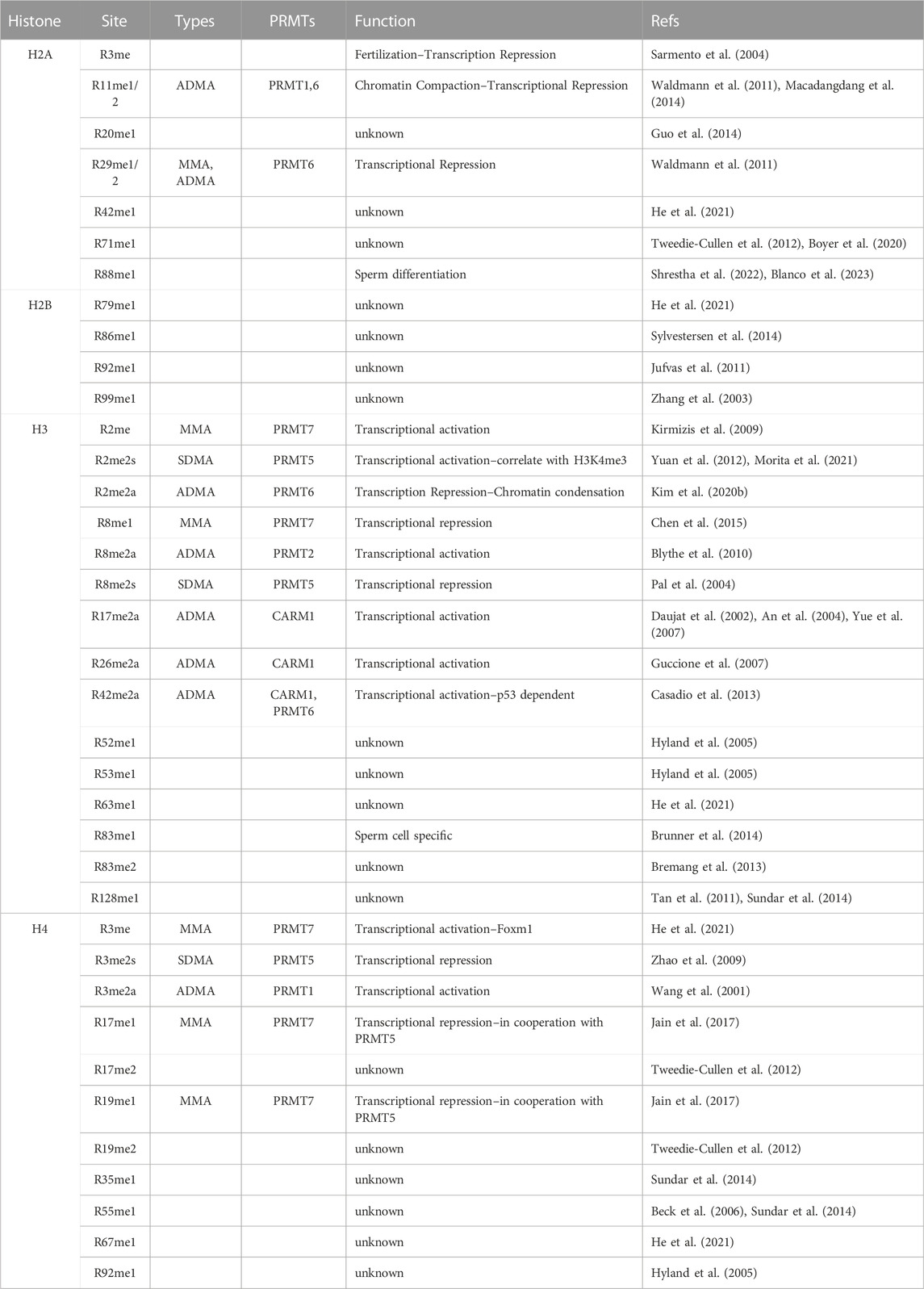
Histones are especially enriched for arginine residues and are subjected to extensive arginine methylation (Wysocka et al., 2006; Di Lorenzo and Bedford, 2011; Huang et al., 2015; Fuhrmann and Thompson, 2016). Most of the methylation sites are located in histone tails due to their accessibility, but methylarginine can also be found in the globular domain (Table 2). Both the location and type of methylation are critical for determining how arginine methylation might impact chromatin biology, including transcription and genome stability. For example, R3 site of Histone H4 can be modified by ADMA (H4R3me2a) or SDMA (H4R3me2s) marks by PRMT1 and PRMT5, respectively (Di Lorenzo and Bedford, 2011). These two distinct marks generate opposite effects in transcription regulation: H4R3me2a is generally considered as an active histone mark that recruits at least two effector complexes, TDRD3/TOP3B and SMARCA4 SWI/SNF chromatin remodeling complex, to promote transcription activation (Wang et al., 2001; Yang et al., 2010; Yang et al., 2014; Yao et al., 2021); whereas H4R3me2s is often associated with a repressive chromatin status (Xu et al., 2010), possibly by recruiting DNMT3A for de novo DNA methylation (Zhao et al., 2009). Although the methylation levels of individual sites vary between tissues, cell types, developmental stages, as well as physiological vs. pathological conditions, H4R3me2a seems to be the most abundant methylarginine histone mark with ∼10% stoichiometry in adult mouse brain (Tweedie-Cullen et al., 2012). As technology continues to advance in mass spectrometry, an ever-expanding repertoire of arginine methylation sites is being discovered (Huang et al., 2015), presenting opportunities for further characterization and investigation. For example, mono- and di-methylation of H4R17 and R19 were identified at considerable level (2–8%) in mouse brain (Tweedie-Cullen et al., 2012), but their biological function is still unknown. PRMT7 has been shown to methylate both sites on a peptide substrate (Feng et al., 2013), and stimulate PRMT1 and PRMT5 activity in methylating H4R3 (Beck et al., 2006) in vitro. Interestingly, R17 and R19 are located within what is known as the “basic patch” of histone H4 and have been shown to be critical in regulating chromatin dynamics and the binding of multiple chromatin-modifying enzymes (Liu et al., 2017; Meriesh et al., 2020; Zhang X. et al., 2022). It is reasonable to speculate that methylation at these two sites would have a significant impact on chromatin remodeling. A more comprehensive list of histone arginine methylation sites is summarized in Table 2.
Arginine methylation of epigenetic modifiers
In addition to arginine methylation, histones are also subjected to other types of PTMs, such as acetylation and lysine methylation (Huang et al., 2014; Millan-Zambrano et al., 2022). Many of the epigenetic modifiers that deposit these modifications are regulated by arginine methylation, which in turn impacts histone codes. For example, the histone methyltransferase EZH2 is methylated by PRMT1 at R342 (Li et al., 2020a; Li et al., 2020b; Li et al., 2021). R342me2a inhibits the CDK1-mediated EZH2 phosphorylation at T345 and T487, which otherwise would promote EZH2 ubiquitination and degradation (Li et al., 2020b). On the other hand, the same R342me2a was shown to prevent AMPK-mediated EZH2 phosphorylation at T311 to enhance EZH2 interaction with SUZ12 and to promote H3K27me3 deposition (Li et al., 2021). Interestingly, PRMT5 has also been shown to either associated with EZH2 or impact H3K27me3, although whether there is a direct methylation of EZH2 by PRMT5 is not clear (Liu F. et al., 2020; Yang et al., 2021). Ash2L is a shared component of mammalian histone H3K4 methyltransferase complexes, which include the mixed lineage leukemia (MLL) family members 1–4, Setd1A, and Setd1B, and is essential to maintain global H3K4me3 levels (Milne et al., 2002). Both PRMT1 and PRMT5 can methylate Ash2L at R296. Although R296 methylation is not required for Ash2L nuclear localization or global H3K4me3 levels, it remains unchecked if this modification affects locus-specific H3K4me3 deposition. Additionally, lysine demethylase LSD1 is also a substrate of arginine methylation (Liu et al., 2020b). CARM1 catalyzes LSD1 methylation at R838, which promotes the binding of the deubiquitinase USP7, resulting in the deubiquitination and stabilization of LSD1. Consequently, two of LSD1 substrates, H3K4me2 and H3K9me2, were affected (Liu et al., 2020b). Interestingly, PRMT5 was shown to cooperate with LSD1 in Slug-mediated transcription activation through histone arginine methylation (Zhang J. et al., 2022), whether PRMT5 can methylate LSD1 has not been tested. Altogether, these studies highlighted a broad impact of PRMTs in histone modifications and epigenetic regulation beyond its direct activity on histone arginine methylation.
Arginine methylation of chromatin remodeling factors
SWI/SNF is a multisubunit chromatin-remodeling complex that utilizes ATP to alter high-order chromatin structures for transcription regulation (Mathur and Roberts, 2018; Centore et al., 2020). One of the core subunits BAF155 is methylated by PRMT4/CARM1 (Wang et al., 2014; Kim et al., 2021). Significantly, CARM1 methylates BAF155 at a single site R1064, and majority of endogenous BAF155 carries R1064me2a, indicating a critical function of this methylation site in BAF155 chromatin remodeling activity (Wang et al., 2014; Kim et al., 2021). The nucleosome remodeling and deacetylase (NuRD; also known as Mi-2) complex is another group of multisubunit ATP-dependent chromatin remodeling complexes with histone deacetylase activity (Lai and Wade, 2011). The non-enzymatic subunits, including methyl-CpG-binding domain 2 (MBD2) and MDB3, are important for targeting NuRD complex to regions with methylated DNA (Lai and Wade, 2011). MBD2/NuRD and MBD3/NuRD were shown to have different biochemical and functional properties–PRMT5 and its cofactor MEP50 were identified as specific MBD2/NuRD components (Le Guezennec et al., 2006). PRMT5 methylates the RG rich region of MBD2, which reduces its interaction with methylated DNA and subsequently the recruitment of histone deacetylases (Tan and Nakielny, 2006). Thus, MBD2 methylation by PRMT5 inhibits the repressive function of the NuRD complex. Additionally, the Bromodomain-containing protein 4 (BRD4), a chromatin-binding protein that functions as a scaffold for transcription factors at promoters and super-enhancers, is methylated by PRMT2 and PRMT4/CARM1 at R179, 181, and 183 upon DNA damage (Liu L. et al., 2022). Methylation selectively controls a transcriptional program by promoting BRD4 recruitment to acetylated histones/chromatin. BRD4 has been shown to harbor histone acetyltransferase activity and can evicts nucleosomes from chromatin (Devaiah et al., 2016). Thus, arginine methylation might impact its function in chromatin remodeling.
Arginine methylation in the regulation of epitranscriptomics
Another research area of gene regulation that arises in recent years is the chemical modification of RNA molecules (Song and Yi, 2017). In the last two decades, with the advance of technologies in deep sequencing and mass spectrometry, hundreds of modifications have been identified across all RNA species and domains of life (Frye et al., 2016). These RNA post-transcriptional modifications are collectively called epitranscriptomics (Meyer and Jaffrey, 2014; Wiener and Schwartz, 2021). Among them, N6-Methyladenosine (m6A) is the most abundant internal modification of mRNA and involves in every step of mRNA life cycle, including splicing, translation, and stability (He and He, 2021). Additionally, m6A modification of chromosome-associated regulatory RNA (carRNA), such as promoter-associated RNAs, enhancer RNAs, and repeat RNAs, play critical roles in controlling their expression levels, and consequently the chromatin state and transcription (Liu et al., 2020c; Huang et al., 2020). Majority of mRNA m6A is catalyzed by a multicomponent methyltransferase complex containing the methyltransferase-like 3 (METTL3)/methyltransferase-like 14 (METTL14) heterodimer, and other regulatory factors (Wang P. et al., 2016). Three studies have independently reported arginine methylation of METTL14 by PRMT1 (Liu et al., 2021; Wang et al., 2021; Wang et al., 2023), all of which revealed a positive regulation of m6A methyltransferase activity by arginine methylation. Although these studies focused on m6A on mRNA, reduced methyltransferase activity could also cause decrease of m6A levels in carRNA, which might provide a novel functional role of arginine methylation in chromatin biology.
Regulation of arginine methylation
Recombinant PRMTs purified from bacteria are biochemically active and share many common substrates in vitro. Thus, to ensure their biological function in vivo, proper regulatory mechanisms are imperative for these otherwise promiscuous enzymes. PRMTs undergo regulation at various levels, including isoform-specific gene expression, subcellular compartmentalization, substrate recognition, and modulation of catalytic activity, among others. These multifaceted regulatory processes collectively contribute to the precise control and effective functioning of PRMTs within living organisms.
Regulation of PRMT expression
At the RNA level, the expression of PRMTs can be regulated through alternative splicing. Although the functional significance of many isoforms has yet to be determined due to their low abundance, a few do exhibit distinct biological characteristics. For example, next-generation sequencing (NGS) identified more than 30 protein coding isoforms of PRMT1 (Adamopoulos et al., 2019), some of which are associated with specific substrates and unique nucleocytoplasmic patterns (Goulet et al., 2007). The majority of studies have focused on the most abundant isoform, PRMT1v1, which has a broad substrate spectrum in both nuclear and cytoplasm. The v2 isoform differs from v1 by harboring an additional 18-amino acid sequence of a nuclear export sequence and thus is exclusively localized in the cytoplasm. PRMT1v2 exhibits oncogenic function by promoting the survival and invasiveness of breast cancer cells (Baldwin et al., 2012), suggesting that cytoplasmic PRMT1 is sufficient for promoting tumorigenesis. The splicing of PRMT5 is uniquely regulated by a novel class of splicing regulatory elements located between the polypyrimidine tract (Py) and 3′AG (REPA) at intron ends (Sohail et al., 2014). This cis-regulatory element functions as a silencer by binding to hnRNPH and competing off the interaction of U2AF65 with the Py, thus leading to exon skipping. The product of this alternative splicing, PRMT5s, misses 44 amino acids located at the N-terminus of the full-length protein without affecting overall PRMT5 enzyme activity (Sohail and Xie, 2015). However, the PRMT5s exhibits drastic differences in subcellular localization, indicating that it might methylate different set of substrates (Sohail et al., 2015; Sohail and Xie, 2015; Wen et al., 2022). Additionally, knockdown the key splicing factor SF3B1 leads to the exon 5 skipping of PRMT9 (Choi et al., 2020), which could lead to translation frame shift and reduce PRMT9 expression through nonsense-mediated RNA decay.
Subcellular compartmentalization
The best example for this type of regulation is demonstrated on PRMT8, which harbors an N-terminal myristoylation site that directs PRMT8 to the plasma membrane. The expression of PRMT8 is mainly found in the brain (Lee J. et al., 2005; Kousaka et al., 2009), suggesting its function in methylating membrane associated proteins and involving in signaling transduction (Kim et al., 2008; Pahlich et al., 2008). Interestingly, the enzymatic activity and the interaction with its methylation substrates are necessary for PRMT1 and PRMT3’s subcellular localization. Catalytic inactive PRMT1 accumulates in the nucleus (Herrmann and Fackelmayer, 2009), and treatment of cells with a global methyltrasferase inhibitor, AdOx, causes PRMT3 (a cytoplasmic protein) to be nuclear (Xie and Denman, 2011). Consistent with these earlier reports, a recent study revealed that phosphorylation of PRMT1 at S307 by the cyclin-dependent kinase 5 (CDK5) promotes its translocation from nucleus to cytoplasm, which is associated with an enhanced methyltransferase activity (Yin et al., 2023). Furthermore, during the specification of primordial germ cells (PGCs) in mice, PRMT5 is in complex with the SET–PR domain protein Blimp1, also known as PRDM1, in the nucleus at the embryonic day 8.5 (E8.5) and subsequently translocated to the cytoplasm at E11.5. Although the molecular mechanism underlying this nuclear-cytoplasm translocation is unknown, this process could be important for the extensive epigenetic programming during germ cell development (Ancelin et al., 2006). Notably, specific subcellular compartmentalization of PRMTs has been associated with the specific subtypes of cancer and the progression of the malignancy (Davis et al., 2013; Shilo et al., 2013), highlighting the prognostic value for understanding this type of regulation in cancer treatment.
PTMs of PRMTs
PTMs are extensively identified on PRMTs and have emerged as another key layer of PRMTs’ regulation. The impacts of these modifications, including phosphorylation (auto)methylation, OGT-glycosylation, and ubiquitination, on various aspects of PRMTs’ function were recently reviewed (Hartley and Lu, 2020). Here, we added a few examples that were not previously covered. Whereas polyubiquitination of PRMT5 by the E3 ligase CHIP was found to negatively regulate PRMT5 protein expression through K48-linked ubiquitin-dependent proteasomal degradation (Zhang et al., 2016), PRMT5 is also a substrate of TRAF6, which deposits K63-linked ubiquitin chain (Liu et al., 2023). Multiple K63 ubiquitination sites were identified at the N-terminus of PRMT5, which enhances its interaction with co-factor MEP50 and its enzymatic activity (Liu et al., 2023). Note that the K48 ubiquitination sites are located in the central region of PRMT5, away from the K63 ubiquitination sites (Zhang et al., 2016).
Substrate recognition of PRMTs
PRMTs do not contain any specific domains for DNA or histone interaction. Their functions in transcription and epigenetic regulation are largely achieved by the recruitment through protein–protein interactions with transcription factors and co-regulators. These interactions significantly contribute to their substrate specificity. For example, PRMT5 interacts with the hSWI/SNF chromatin remodeler complex, which in turn enhances its methyltransferase activity towards histone substrates (An et al., 2004). The histone-binding protein COPR5 specifically interacts with the N-terminus of histone H4 and guides PRMT5 to methylate H4R3 rather than H3R8 (Lacroix et al., 2008). Recombinant PRMT4/CARM1 preferentially methylates free histone 3, whereas upon association with the nucleosomal methylation activator complex (NUMAC), PRMT4/CARM1 obtains the activity to methylate nucleosome histone 3 (Xu et al., 2004). Another important layer of regulation on PRMT’s substrate recognition resides the sequence context of the substrates, specifically methylation site could be influenced by other PTMs in close proximity. Such regulations are prevalent on histone substrates because of their extensive PTMs. For example, PRMT4/CARM1 prefers Histone H3 lysine 18 acetylation (H3K18ac) modified histone for depositing methylation mark at arginine 17 residue (H3R17me2a) (Daujat et al., 2002; Guccione et al., 2007; Yue et al., 2007). H3K9ac impedes PRMT5 for the deposition of H3R8me2s mark (An et al., 2004). Histone H4 lysine 5 acetylation (H4K5ac) primes the H4R3 site for methylation by PRMT5, but not PRMT1, and consequently switching an active mark to a repressive mark (H4R3me2s) (Feng et al., 2011). At the CTD of RNAPII, phosphorylation of either Serine 2 or Serine 5 within the heptameric repeats could block the methylation of R1810 by CARM1 (Mizutani et al., 2015). Furthermore, in glioblastoma, 5-hydroxymethylcytosine (5 hmC) mark on DNA can recruit PRMT1 to chromatin by interacting with an accessory protein of PRMT1, chromatin target of PRMT1 (CHTOP), linking the modification of DNA with histone methylation (Takai et al., 2014). Additional examples of this regulation can be found in the “Arginine methylation crosstalk with other PTMs” section.
Regulation of PRMT activity by cellular metabolites
S-adenosylmethionine (SAM) is the universal donor for all methylation reactions, including DNA, RNA, and protein methylation (Mentch et al., 2015). It is catalyzed from amino acid methionine and ATP by methionine adenosyl transferase enzymes (MATs), which are encoded by the MAT1A and MAT2A genes (Cantoni, 1953). In the methionine salvage pathway, the methylthioadenosine phosphorylase (MTAP) cleaves the byproduct of polyamine biosynthesis, methylthioadenosine (MTA), into adenine and MTR-1-P (5-methythioribose-1-phosphate), which eventually leads to the regeneration of methionine (Zappia et al., 1988). Additionally, methylation reaction generates S-adenosylhomocysteine (SAH) as a byproduct that needs to be hydrolyzed to homocysteine (Hcy) and adenine by S-adenosylhomocysteine hydrolase (AHCY) (Obeid and Herrmann, 2009). Cellular accumulation of SAH and MTA, in principle, could suppress all methylation reactions. For example, deletion of MTAP, which occurs in approximately 15% of human cancers causes the accumulation of MTA that imposes the strongest inhibitory effect on PRMT5. Thus, MTAP deleted tumors are highly sensitive to further inhibition of PRMT5 activity (Kryukov et al., 2016; Mavrakis et al., 2016). On top of MTAP deletion, inhibition of MAT2A, which reduces available SAM, further strengthens PRMT5 dependency (Marjon et al., 2016). Thus, PRMT5 inhibitor exhibited highest efficacy in tumor suppression under this context (Fedoriw et al., 2019; Marjon et al., 2021). Mechanistically, PRMT5 seems to be the most sensitive one among many protein methyltransferases upon MTA inhibition (Kryukov et al., 2016). On the other hand, fluctuations in SAM level could also differentially impact PRMTs activity due to their different apparent Km for SAM. Among all PRMTs, PRMT9 has the highest SAM Km (40.5 ± 1 μM), which is 10-20 fold higher than other PRMTs (Li A. S. M. et al., 2020), suggesting that PRMT9 might be a frontline sensor for SAM alternation in response to methionine metabolism.
Future perspectives
Arginine demethylases
Dynamic arginine methylation and demethylation play crucial roles in the regulation of protein function. In comparison to the well-characterized writers and readers of arginine methylation, the biochemical properties and biological functions of erasers or demethylases that reverse the methylation reaction remain elusive. However, it becomes clear that lysine demethylases are capable of catalyzing arginine demethylation in vitro (Walport et al., 2016). These enzymes belong to 2-Oxoglutarate-dependent dioxygenases, which require oxygen, reduced iron and 2-oxoglutarate (also known as α-ketoglutarate) as cofactors to function (Herr and Hausinger, 2018; Islam et al., 2018; Losman et al., 2020). Thus, when these metabolites become limited, their enzymatic activities are often inhibited. Although this connection has been well documented with histone lysine methylation marks, such as H3K4me3 and H3K27me3 (Mentch et al., 2015; Pan et al., 2016; Batie et al., 2019; Chakraborty et al., 2019), whether and how metabolic alterations impact histone arginine methylation is unknown. Furthermore, if demethylation of lysine and arginine share the same family of demethylases, what are the factors that determine their substrate selectivity and specificity, if any? As the levels of histone lysine methylation are in general higher than that of arginine methylation (Huang et al., 2015), the availability and concentrations of specific co-factors might differentially influence the enzymatic activity and substrate specificity. The surrounding amino acid sequence and structural context of the target residue could also differentially influence the substrate recognition by demethylases. Understanding the precise mechanisms of substrate selectivity and specificity of lysine and arginine demethylases will provide more comprehensive understanding of epigenetic regulation through these two types of histone marks.
Tudor domain inhibitors
Targeting protein–protein interactions (PPIs) for drug discovery are considered challenging because PPIs are often characterized by large and flat protein surfaces, making it difficult to design small molecules that can effectively bind and disrupt the interaction (Scott et al., 2016). Recently, the development of chemical inhibitors that target bromodomain and extra-terminal domain (BET) containing proteins has gained significant attention in the field of cancer research. These inhibitors, represented by JQ1, competes with acetylated lysine residues for binding to the bromodomain pockets of BET proteins, thus inhibiting their transcriptional activity (Filippakopoulos et al., 2010; Dawson et al., 2011; Delmore et al., 2011; Mertz et al., 2011). A few inhibitors have been developed to target methyl lysine binding Tudor domains, such as those of UHRF1 and SETDB1 (Senisterra et al., 2018; Chang et al., 2021; Guo et al., 2021), whereas the development of methylarginine Tudor domain inhibitors is lagging. The SMN Tudor-selective antagonist was recently discovered by serendipity while searching for inhibitors against the histone H3K9me3 binding tandem Tudor domain (TTD) of UHRF1 (Liu Y. L. et al., 2022). This inhibitor binds to Tudor domain of SMN, SMNDC1, and TDRD3 with slightly higher affinity than to UHRF1 (Liu Y. L. et al., 2022). In cells, it inhibits SMN interaction with RNAPII and causes transcription termination, mirroring the effect of SMN knockdown (Liu Y. L. et al., 2022). However, its selectivity for SMN is only ∼4-fold over TDRD3. Thus, the observed in vivo effect could partially contribute from TDRD3 inhibition. Using nuclear magnetic resonance (NMR) based fragment screening, Liu et al. identified several small molecule inhibitors of TDRD3 Tudor domain with a Kd of ∼50 uM. These compounds show reduced binding to SMN Tudor domain and no detectable binding with the methyl lysine binding Tudor domain of 53BP1 (Liu et al., 2018). Although the in vivo efficacy of these inhibitors has yet been determined, this study provided useful information for future structure-guided discovery of Tudor domain inhibitors.
Author contributions
HP, XT, and YY structured and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research in YY laboratory is funded by an R01 grant (GM133850) from the National Institutes of Health.
Acknowledgments
We apologize to the colleagues whose work we are unable to cite due to space constraints.
Conflict of interest
The authors YY declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, Y., Suzuki, Y., Kawamura, K., and Tanaka, N. (2019). MEP50/PRMT5-mediated methylation activates GLI1 in Hedgehog signalling through inhibition of ubiquitination by the ITCH/NUMB complex. Commun. Biol. 2, 23. doi:10.1038/s42003-018-0275-4
Abeywardana, T., Oh, M., Jiang, L., Yang, Y., Kong, M., Song, J., et al. (2018). CARM1 suppresses de novo serine synthesis by promoting PKM2 activity. J. Biol. Chem. 293, 15290–15303. doi:10.1074/jbc.ra118.004512
Adamopoulos, P. G., Mavrogiannis, A. V., Kontos, C. K., and Scorilas, A. (2019). Novel alternative splice variants of the human protein arginine methyltransferase 1 (PRMT1) gene, discovered using next-generation sequencing. Gene 699, 135–144. doi:10.1016/j.gene.2019.02.072
Allen, B. L., and Taatjes, D. J. (2015). The mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166. doi:10.1038/nrm3951
An, W., Kim, J., and Roeder, R. G. J. C. (2004). Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748. doi:10.1016/j.cell.2004.05.009
Ancelin, K., Lange, U. C., Hajkova, P., Schneider, R., Bannister, A. J., Kouzarides, T., et al. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8, 623–630. doi:10.1038/ncb1413
Avasarala, S., Van Scoyk, M., Karuppusamy Rathinam, M. K., Zerayesus, S., Zhao, X., Zhang, W., et al. (2015). PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J. Biol. Chem. 290, 13479–13489. doi:10.1074/jbc.m114.636050
Bachand, F., and Silver, P. A. (2004). PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23, 2641–2650. doi:10.1038/sj.emboj.7600265
Baldwin, R. M., Morettin, A., Paris, G., Goulet, I., and Cote, J. (2012). Alternatively spliced protein arginine methyltransferase 1 isoform PRMT1v2 promotes the survival and invasiveness of breast cancer cells. Cell Cycle 11, 4597–4612. doi:10.4161/cc.22871
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298. doi:10.1038/nrm.2017.7
Bandyopadhyay, S., Harris, D. P., Adams, G. N., Lause, G. E., McHugh, A., Tillmaand, E. G., et al. (2012). HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol. Cell. Biol. 32, 1202–1213. doi:10.1128/mcb.05977-11
Bao, J., Rousseaux, S., Shen, J., Lin, K., Lu, Y., and Bedford, M. T. (2018). The arginine methyltransferase CARM1 represses p300•ACT•CREMτ activity and is required for spermiogenesis. Nucleic acids Res. 46, 4327–4343. doi:10.1093/nar/gky240
Batie, M., Frost, J., Frost, M., Wilson, J. W., Schofield, P., and Rocha, S. (2019). Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363, 1222–1226. doi:10.1126/science.aau5870
Bauer, U. M., Daujat, S., Nielsen, S. J., Nightingale, K., and Kouzarides, T. (2002). Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3, 39–44. doi:10.1093/embo-reports/kvf013
Beck, H. C., Nielsen, E. C., Matthiesen, R., Jensen, L. H., Sehested, M., Finn, P., et al. (2006). Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics 5, 1314–1325. doi:10.1074/mcp.m600007-mcp200
Bedford, M. T., and Clarke, S. G. (2009). Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 33, 1–13. doi:10.1016/j.molcel.2008.12.013
Bedford, M. T., Frankel, A., Yaffe, M. B., Clarke, S., Leder, P., and Richard, S. (2000). Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275, 16030–16036. doi:10.1074/jbc.m909368199
Blanc, R. S., and Richard, S. (2017). Arginine methylation: The coming of age. Mol. Cell 65, 8–24. doi:10.1016/j.molcel.2016.11.003
Blanco, M., El Khattabi, L., Gobé, C., Crespo, M., Coulée, M., de la Iglesia, A., et al. (2023). DOT1L regulates chromatin reorganization and gene expression during sperm differentiation. EMBO Rep. 24, e56316. doi:10.15252/embr.202256316
Blythe, S. A., Cha, S.-W., Tadjuidje, E., Heasman, J., and Klein, P. S. (2010). β-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev. Cell 19, 220–231. doi:10.1016/j.devcel.2010.07.007
Boffa, L. C., Karn, J., Vidali, G., and Allfrey, V. G. (1977). Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem. Biophysical Res. Commun. 74, 969–976. doi:10.1016/0006-291x(77)91613-8
Boisvert, F. M., Cote, J., Boulanger, M. C., and Richard, S. (2003). A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2, 1319–1330. doi:10.1074/mcp.m300088-mcp200
Boyer, J. A., Spangler, C. J., Strauss, J. D., Cesmat, A. P., Liu, P., McGinty, R. K., et al. (2020). Structural basis of nucleosome-dependent cGAS inhibition. Science 370, 450–454. doi:10.1126/science.abd0609
Bremang, M., Cuomo, A., Agresta, A. M., Stugiewicz, M., Spadotto, V., and Bonaldi, T. (2013). Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. Biosyst. 9, 2231–2247. doi:10.1039/c3mb00009e
Brunner, A. M., Nanni, P., and Mansuy, I. M. (2014). Epigenetic marking of sperm by post-translational modification of histones and protamines. Epigenetics Chromatin 7, 2. doi:10.1186/1756-8935-7-2
Cantoni, G. L. (1953). S-ADENOSYLMETHIONINE; A NEW INTERMEDIATE FORMED ENZYMATICALLY FROM l-METHIONINE AND ADENOSINETRIPHOSPHATE. J. Biol. Chem. 204, 403–416. doi:10.1016/s0021-9258(18)66148-4
Casadio, F., Lu, X., Pollock, S. B., LeRoy, G., Garcia, B. A., Muir, T. W., et al. (2013). H3R42me2a is a histone modification with positive transcriptional effects. Proc. Natl. Acad. Sci. 110, 14894–14899. doi:10.1073/pnas.1312925110
Centore, R. C., Sandoval, G. J., Soares, L. M. M., Kadoch, C., and Chan, H. M. (2020). Mammalian SWI/SNF chromatin remodeling complexes: Emerging mechanisms and therapeutic strategies. Trends Genet. 36, 936–950. doi:10.1016/j.tig.2020.07.011
Chakraborty, A. A., Laukka, T., Myllykoski, M., Ringel, A. E., Booker, M. A., Tolstorukov, M. Y., et al. (2019). Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222. doi:10.1126/science.aaw1026
Chang, B., Chen, Y., Zhao, Y., and Bruick, R. K. (2007). JMJD6 is a histone arginine demethylase. Science 318, 444–447. doi:10.1126/science.1145801
Chang, L., Campbell, J., Raji, I. O., Guduru, S. K. R., Kandel, P., Nguyen, M., et al. (2021). Discovery of small molecules targeting the tandem tudor domain of the epigenetic factor UHRF1 using fragment-based ligand discovery. Sci. Rep. 11, 1121. doi:10.1038/s41598-020-80588-4
Chen, C., Nott, T. J., Jin, J., and Pawson, T. (2011). Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 12, 629–642. doi:10.1038/nrm3185
Chen, D., Ma, H., Hong, H., Koh, S. S., Huang, S. M., Schurter, B. T., et al. (1999). Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177. doi:10.1126/science.284.5423.2174
Chen, H., Zhang, C., Sheng, Y., Yao, S., Liu, Z., Zhang, C., et al. (2015). Frequent SOCS3 and 3OST2 promoter methylation and their epigenetic regulation in endometrial carcinoma. Am. J. cancer Res. 5, 180–190.
Chen, M., Yi, B., and Sun, J. (2014). Inhibition of cardiomyocyte hypertrophy by protein arginine methyltransferase 5. J. Biol. Chem. 289, 24325–24335. doi:10.1074/jbc.m114.577494
Cheng, D., Gao, G., Di Lorenzo, A., Jayne, S., Hottiger, M. O., Richard, S., et al. (2020). Genetic evidence for partial redundancy between the arginine methyltransferases CARM1 and PRMT6. J. Biol. Chem. 295, 17060–17070. doi:10.1074/jbc.ra120.014704
Cheng, D., Vemulapalli, V., Lu, Y., Shen, J., Aoyagi, S., Fry, C. J., et al. (2018). CARM1 methylates MED12 to regulate its RNA-binding ability. Life Sci. Alliance 1, e201800117. doi:10.26508/lsa.201800117
Chevillard-Briet, M., Trouche, D., and Vandel, L. (2002). Control of CBP co-activating activity by arginine methylation. EMBO J. 21, 5457–5466. doi:10.1093/emboj/cdf548
Cho, E. C., Zheng, S., Munro, S., Liu, G., Carr, S. M., Moehlenbrink, J., et al. (2012). Arginine methylation controls growth regulation by E2F-1. EMBO J. 31, 1785–1797. doi:10.1038/emboj.2012.17
Choi, N., Liu, Y., Oh, J., Ha, J., Zheng, X., and Shen, H. (2020). U2AF65-Dependent SF3B1 function in SMN alternative splicing. Cells 9, 2647. doi:10.3390/cells9122647
Choi, S., Jeong, H. J., Kim, H., Choi, D., Cho, S. C., Seong, J. K., et al. (2019). Skeletal muscle-specific Prmt1 deletion causes muscle atrophy via deregulation of the PRMT6-FOXO3 axis. Autophagy 15, 1069–1081. doi:10.1080/15548627.2019.1569931
Chong, P. A., Vernon, R. M., and Forman-Kay, J. D. (2018). RGG/RG motif regions in RNA binding and phase separation. J. Mol. Biol. 430, 4650–4665. doi:10.1016/j.jmb.2018.06.014
Chowdhury, M. N., and Jin, H. (2023). The RGG motif proteins: Interactions, functions, and regulations. Wiley Interdiscip. Rev. RNA 14, e1748. doi:10.1002/wrna.1748
Courchaine, E. M., Barentine, A. E. S., Straube, K., Lee, D. R., Bewersdorf, J., and Neugebauer, K. M. (2021). DMA-tudor interaction modules control the specificity of in vivo condensates. Cell 184, 3612–3625 e17. doi:10.1016/j.cell.2021.05.008
Couto e Silva, A., Wu, C. Y., Citadin, C. T., Clemons, G. A., Possoit, H. E., Grames, M. S., et al. (2020). Protein arginine methyltransferases in cardiovascular and neuronal function. Mol. Neurobiol. 57, 1716–1732. doi:10.1007/s12035-019-01850-z
Couture, J. F., Collazo, E., and Trievel, R. C. (2006). Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 13, 698–703. doi:10.1038/nsmb1116
Cura, V., Marechal, N., Troffer-Charlier, N., Strub, J. M., Haren, M. J., Martin, N. I., et al. (2017). Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors. FEBS J. 284, 77–96. doi:10.1111/febs.13953
Daujat, S., Bauer, U.-M., Shah, V., Turner, B., Berger, S., and Kouzarides, T. J. C. b. (2002). Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 12, 2090–2097. doi:10.1016/s0960-9822(02)01387-8
Davies, C. C., Chakraborty, A., Diefenbacher, M. E., Skehel, M., and Behrens, A. (2013). Arginine methylation of the c-Jun coactivator RACO-1 is required for c-Jun/AP-1 activation. Embo J. 32, 1556–1567. doi:10.1038/emboj.2013.98
Davis, M. B., Liu, X., Wang, S., Reeves, J., Khramtsov, A., Huo, D., et al. (2013). Expression and sub-cellular localization of an epigenetic regulator, co-activator arginine methyltransferase 1 (CARM1), is associated with specific breast cancer subtypes and ethnicity. Mol. Cancer 12, 40. doi:10.1186/1476-4598-12-40
Dawson, M. A., Prinjha, R. K., Dittmann, A., Giotopoulos, G., Bantscheff, M., Chan, W. I., et al. (2011). Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. doi:10.1038/nature10509
Delmore, J. E., Issa, G. C., Lemieux, M. E., Rahl, P. B., Shi, J., Jacobs, H. M., et al. (2011). BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917. doi:10.1016/j.cell.2011.08.017
Denman, R. B. (2002). Methylation of the arginine-glycine-rich region in the fragile X mental retardation protein FMRP differentially affects RNA binding. Cell Mol. Biol. Lett. 7, 877–883.
Devaiah, B. N., Case-Borden, C., Gegonne, A., Hsu, C. H., Chen, Q. R., Meerzaman, D., et al. (2016). BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 23, 540–548. doi:10.1038/nsmb.3228
Dhar, S., Vemulapalli, V., Patananan, A. N., Huang, G. L., Di Lorenzo, A., Richard, S., et al. (2013). Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 3, 1311. doi:10.1038/srep01311
Di Lorenzo, A., and Bedford, M. T. (2011). Histone arginine methylation. FEBS Lett. 585, 2024–2031. doi:10.1016/j.febslet.2010.11.010
Fedoriw, A., Rajapurkar, S. R., O'Brien, S., Gerhart, S. V., Mitchell, L. H., Adams, N. D., et al. (2019). Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell 36, 100–114.e25. doi:10.1016/j.ccell.2019.05.014
Feng, Q., Yi, P., Wong, J., and O'Malley, B. W. (2006). Signaling within a coactivator complex: Methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell. Biol. 26, 7846–7857. doi:10.1128/mcb.00568-06
Feng, Y., Maity, R., Whitelegge, J. P., Hadjikyriacou, A., Li, Z., Zurita-Lopez, C., et al. (2013). Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J. Biol. Chem. 288, 37010–37025. doi:10.1074/jbc.m113.525345
Feng, Y., Wang, J., Asher, S., Hoang, L., Guardiani, C., Ivanov, I., et al. (2011). Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism. J. Biol. Chem. 286, 20323–20334. doi:10.1074/jbc.m110.207258
Filippakopoulos, P., Qi, J., Picaud, S., Shen, Y., Smith, W. B., Fedorov, O., et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. doi:10.1038/nature09504
Friberg, A., Corsini, L., Mourao, A., and Sattler, M. (2009). Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J. Mol. Biol. 387, 921–934. doi:10.1016/j.jmb.2009.02.018
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G., and Suzuki, T. (2016). RNA modifications: What have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372. doi:10.1038/nrg.2016.47
Fuhrmann, J., and Thompson, P. R. (2016). Protein arginine methylation and citrullination in epigenetic regulation. ACS Chem. Biol. 11, 654–668. doi:10.1021/acschembio.5b00942
Gao, G., Hausmann, S., Flores, N. M., Benitez, A. M., Shen, J., Yang, X., et al. (2023). The NFIB/CARM1 partnership is a driver in preclinical models of small cell lung cancer. Nat. Commun. 14, 363. doi:10.1038/s41467-023-35864-y
Gao, G., Zhang, L., Villarreal, O. D., He, W., Su, D., Bedford, E., et al. (2019). PRMT1 loss sensitizes cells to PRMT5 inhibition. Nucleic acids Res. 47, 5038–5048. doi:10.1093/nar/gkz200
Geoghegan, V., Guo, A., Trudgian, D., Thomas, B., and Acuto, O. (2015). Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat. Commun. 6, 6758. doi:10.1038/ncomms7758
Gilbert, W., Siebel, C. W., and Guthrie, C. (2001). Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7, 302–313. doi:10.1017/s1355838201002369
Gill, A. L., Premasiri, A. S., and Vieira, F. G. (2021). Hypothesis and theory: Roles of arginine methylation in C9orf72-mediated ALS and FTD. Front. Cell Neurosci. 15, 633668. doi:10.3389/fncel.2021.633668
Goulet, I., Boisvenue, S., Mokas, S., Mazroui, R., and Cote, J. (2008). TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 17, 3055–3074. doi:10.1093/hmg/ddn203
Goulet, I., Gauvin, G., Boisvenue, S., and Cote, J. (2007). Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 282, 33009–33021. doi:10.1074/jbc.m704349200
Guccione, E., Bassi, C., Casadio, F., Martinato, F., Cesaroni, M., Schuchlautz, H., et al. (2007). Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937. doi:10.1038/nature06166
Guccione, E., and Richard, S. (2019). The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657. doi:10.1038/s41580-019-0155-x
Guo, A., Gu, H., Zhou, J., Mulhern, D., Wang, Y., Lee, K. A., et al. (2014). Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 13, 372–387. doi:10.1074/mcp.o113.027870
Guo, Y., Mao, X., Xiong, L., Xia, A., You, J., Lin, G., et al. (2021). Structure-guided discovery of a potent and selective cell-active inhibitor of SETDB1 tudor domain. Angew. Chem. 133, 8842–8847. doi:10.1002/ange.202017200
Hadjikyriacou, A., Yang, Y., Espejo, A., Bedford, M. T., and Clarke, S. G. (2015). Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. J. Biol. Chem. 290, 16723–16743. doi:10.1074/jbc.m115.659433
Han, H.-S., Jung, C.-Y., Yoon, Y.-S., Choi, S., Choi, D., Kang, G., et al. (2014). Arginine methylation of CRTC2 is critical in the transcriptional control of hepatic glucose metabolism. Sci. Signal. 7, ra19. doi:10.1126/scisignal.2004479
Hart-Smith, G., Low, J. K., Erce, M. A., and Wilkins, M. R. (2012). Enhanced methylarginine characterization by post-translational modification-specific targeted data acquisition and electron-transfer dissociation mass spectrometry. J. Am. Soc. Mass Spectrom. 23, 1376–1389. doi:10.1007/s13361-012-0417-8
Hartel, N. G., Chew, B., Qin, J., Xu, J., and Graham, N. A. (2019). Deep protein methylation profiling by combined chemical and immunoaffinity approaches reveals novel PRMT1 targets. Mol. Cell. Proteomics 18, 2149–2164. doi:10.1074/mcp.ra119.001625
Hartley, A. V., and Lu, T. (2020). Modulating the modulators: Regulation of protein arginine methyltransferases by post-translational modifications. Drug Discov. Today 25, 1735–1743. doi:10.1016/j.drudis.2020.06.031
He, H., Chen, J., Zhao, J., Zhang, P., Qiao, Y., Wan, H., et al. (2021). PRMT7 targets of Foxm1 controls alveolar myofibroblast proliferation and differentiation during alveologenesis. Cell Death Dis. 12, 841. doi:10.1038/s41419-021-04129-1
He, P. C., and He, C. (2021). m(6 A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40, e105977. doi:10.15252/embj.2020105977
Herr, C. Q., and Hausinger, R. P. (2018). Amazing diversity in biochemical roles of Fe(II)/2-Oxoglutarate oxygenases. Trends Biochem. Sci. 43, 517–532. doi:10.1016/j.tibs.2018.04.002
Herrmann, F., and Fackelmayer, F. O. (2009). Nucleo-cytoplasmic shuttling of protein arginine methyltransferase 1 (PRMT1) requires enzymatic activity. Genes Cells 14, 309–317. doi:10.1111/j.1365-2443.2008.01266.x
Hofweber, M., Hutten, S., Bourgeois, B., Spreitzer, E., Niedner-Boblenz, A., Schifferer, M., et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719 e13. doi:10.1016/j.cell.2018.03.004
Hosokawa, H., Kato, M., Tohyama, H., Tamaki, Y., Endo, Y., Kimura, M. Y., et al. (2015). Methylation of Gata3 protein at Arg-261 regulates transactivation of the Il5 gene in T helper 2 cells. J. Biol. Chem. 290, 13095–13103. doi:10.1074/jbc.m114.621524
Hsin, J. P., and Manley, J. L. (2012). The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & Dev. 26, 2119–2137. doi:10.1101/gad.200303.112
Huang, H., Lin, S., Garcia, B. A., and Zhao, Y. (2015). Quantitative proteomic analysis of histone modifications. Chem. Rev. 115, 2376–2418. doi:10.1021/cr500491u
Huang, H., Sabari, B. R., Garcia, B. A., Allis, C. D., and Zhao, Y. (2014). SnapShot: Histone modifications. Cell 159, 458–458 e1. doi:10.1016/j.cell.2014.09.037
Huang, H., Weng, H., and Chen, J. (2020). m(6 A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 37, 270–288. doi:10.1016/j.ccell.2020.02.004
Huang, L., Wang, Z., Narayanan, N., and Yang, Y. (2018). Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic acids Res. 46, 3061–3074. doi:10.1093/nar/gky103
Hung, C. J., Lee, Y. J., Chen, D. H., and Li, C. (2009). Proteomic analysis of methylarginine-containing proteins in HeLa cells by two-dimensional gel electrophoresis and immunoblotting with a methylarginine-specific antibody. Protein J. 28, 139–147. doi:10.1007/s10930-009-9174-3
Hwang, J. W., Cho, Y., Bae, G. U., Kim, S. N., and Kim, Y. K. (2021). Protein arginine methyltransferases: Promising targets for cancer therapy. Exp. Mol. Med. 53, 788–808. doi:10.1038/s12276-021-00613-y
Hyland, E. M., Cosgrove, M. S., Molina, H., Wang, D., Pandey, A., Cottee, R. J., et al. (2005). Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 10060–10070. doi:10.1128/mcb.25.22.10060-10070.2005
Iberg, A. N., Espejo, A., Cheng, D., Kim, D., Michaud-Levesque, J., Richard, S., et al. (2008). Arginine methylation of the histone H3 tail impedes effector binding. J. Biol. Chem. 283, 3006–3010. doi:10.1074/jbc.c700192200
Islam, M. S., Leissing, T. M., Chowdhury, R., Hopkinson, R. J., and Schofield, C. J. (2018). 2-Oxoglutarate-Dependent oxygenases. Annu. Rev. Biochem. 87, 585–620. doi:10.1146/annurev-biochem-061516-044724
Ito, T., Yadav, N., Lee, J., Furumatsu, T., Yamashita, S., Yoshida, K., et al. (2009). Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev. Biol. 9, 47. doi:10.1186/1471-213x-9-47
Iwasaki, H., Kovacic, J. C., Olive, M., Beers, J. K., Yoshimoto, T., Crook, M. F., et al. (2010). Disruption of protein arginine N-methyltransferase 2 regulates leptin signaling and produces leanness in vivo through loss of STAT3 methylation. Circ. Res. 107, 992–1001. doi:10.1161/circresaha.110.225326
Jain, K., Jin, C. Y., and Clarke, S. G. (2017). Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc. Natl. Acad. Sci. 114, 10101–10106. doi:10.1073/pnas.1706978114
Jansson, M., Durant, S. T., Cho, E. C., Sheahan, S., Edelmann, M., Kessler, B., et al. (2008). Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439. doi:10.1038/ncb1802
Jarrold, J., and Davies, C. C. (2019). PRMTs and arginine methylation: Cancer's best-kept secret? Trends Mol. Med. 25, 993–1009. doi:10.1016/j.molmed.2019.05.007
Jufvas, Å., Strålfors, P., and Vener, A. V. (2011). Histone variants and their post-translational modifications in primary human fat cells. PloS one 6, e15960. doi:10.1371/journal.pone.0015960
Kaniskan, H. U., Martini, M. L., and Jin, J. (2018). Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 118, 989–1068. doi:10.1021/acs.chemrev.6b00801
Kawabe, Y.-i., Wang, Yu X., McKinnell, Iain W., Bedford, Mark T., and Rudnicki, Michael A. (2012). Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11, 333–345. doi:10.1016/j.stem.2012.07.001
Khoury, G. A., Baliban, R. C., and Floudas, C. A. (2011). Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90. doi:10.1038/srep00090
Kim, E. J., Liu, P., Zhang, S., Donahue, K., Wang, Y., Schehr, J. L., et al. (2021). BAF155 methylation drives metastasis by hijacking super-enhancers and subverting anti-tumor immunity. Nucleic acids Res. 49, 12211–12233. doi:10.1093/nar/gkab1122
Kim, H., Kim, H., Feng, Y., Li, Y., Tamiya, H., Tocci, S., et al. (2020a). PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 12, eaaz5683. doi:10.1126/scitranslmed.aaz5683
Kim, J. D., Kako, K., Kakiuchi, M., Park, G. G., and Fukamizu, A. (2008). EWS is a substrate of type I protein arginine methyltransferase, PRMT8. Int. J. Mol. Med. 22, 309–315.
Kim, K. Y., Wang, D. H., Campbell, M., Huerta, S. B., Shevchenko, B., Izumiya, C., et al. (2015). PRMT4-mediated arginine methylation negatively regulates retinoblastoma tumor suppressor protein and promotes E2F-1 dissociation. Mol. Cell. Biol. 35, 238–248. doi:10.1128/mcb.00945-14
Kim, S., Kim, N. H., Park, J. E., Hwang, J. W., Myung, N., Hwang, K.-T., et al. (2020b). PRMT6-mediated H3R2me2a guides Aurora B to chromosome arms for proper chromosome segregation. Nat. Commun. 11, 612. doi:10.1038/s41467-020-14511-w
Kirmizis, A., Santos-Rosa, H., Penkett, C. J., Singer, M. A., Green, R. D., and Kouzarides, T. (2009). Distinct transcriptional outputs associated with mono- and dimethylated histone H3 arginine 2. Nat. Struct. Mol. Biol. 16, 449–451. doi:10.1038/nsmb.1569
Kirmizis, A., Santos-Rosa, H., Penkett, C. J., Singer, M. A., Vermeulen, M., Mann, M., et al. (2007). Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932. doi:10.1038/nature06160
Kousaka, A., Mori, Y., Koyama, Y., Taneda, T., Miyata, S., and Tohyama, M. (2009). The distribution and characterization of endogenous protein arginine N-methyltransferase 8 in mouse CNS. Neuroscience 163, 1146–1157. doi:10.1016/j.neuroscience.2009.06.061
Kryukov, G. V., Wilson, F. H., Ruth, J. R., Paulk, J., Tsherniak, A., Marlow, S. E., et al. (2016). MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science 351, 1214–1218. doi:10.1126/science.aad5214
Kwak, Y. T., Guo, J., Prajapati, S., Park, K. J., Surabhi, R. M., Miller, B., et al. (2003). Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11, 1055–1066. doi:10.1016/s1097-2765(03)00101-1
Lacroix, M., Messaoudi, S. E., Rodier, G., Le Cam, A., Sardet, C., and Fabbrizio, E. (2008). The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 9, 452–458. doi:10.1038/embor.2008.45
Lai, A. Y., and Wade, P. A. (2011). Cancer biology and NuRD: A multifaceted chromatin remodelling complex. Nat. Rev. Cancer 11, 588–596. doi:10.1038/nrc3091
Lambert, S. A., Jolma, A., Campitelli, L. F., Das, P. K., Yin, Y., Albu, M., et al. (2018). The human transcription factors. Cell 172, 650–665. doi:10.1016/j.cell.2018.01.029
Lassak, J., Koller, F., Krafczyk, R., and Volkwein, W. (2019). Exceptionally versatile - arginine in bacterial post-translational protein modifications. Biol. Chem. 400, 1397–1427. doi:10.1515/hsz-2019-0182
Le Guezennec, X., Vermeulen, M., Brinkman, A. B., Hoeijmakers, W. A., Cohen, A., Lasonder, E., et al. (2006). MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26, 843–851. doi:10.1128/mcb.26.3.843-851.2006
Le Romancer, M., Treilleux, I., Leconte, N., Robin-Lespinasse, Y., Sentis, S., Bouchekioua-Bouzaghou, K., et al. (2008). Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol. Cell 31, 212–221. doi:10.1016/j.molcel.2008.05.025
Lee, J., Sayegh, J., Daniel, J., Clarke, S., and Bedford, M. T. (2005a). PRMT8, a new membrane-bound tissue-specific member of the protein arginine methyltransferase family. J. Biol. Chem. 280, 32890–32896. doi:10.1074/jbc.m506944200
Lee, Y. H., Bedford, M. T., and Stallcup, M. R. (2011). Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes & Dev. 25, 176–188. doi:10.1101/gad.1975811
Lee, Y. H., Coonrod, S. A., Kraus, W. L., Jelinek, M. A., and Stallcup, M. R. (2005b). Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U. S. A. 102, 3611–3616. doi:10.1073/pnas.0407159102
Lenard, A. J., Hutten, S., Zhou, Q., Usluer, S., Zhang, F., Bourgeois, B. M. R., et al. (2021). Phosphorylation regulates CIRBP arginine methylation, transportin-1 binding and liquid-liquid phase separation. Front. Mol. Biosci. 8, 689687. doi:10.3389/fmolb.2021.689687
Li, A. S. M., Li, F. L., Eram, M. S., Bolotokova, A., dela Sena, C. C., and Vedadi, M. (2020c). Chemical probes for protein arginine methyltransferases. Methods 175, 30–43. doi:10.1016/j.ymeth.2019.11.017
Li, J., Zhang, T., Ren, T., Liao, X., Hao, Y., Lim, J. S., et al. (2022). Oxygen-sensitive methylation of ULK1 is required for hypoxia-induced autophagy. Nat. Commun. 13, 1172. doi:10.1038/s41467-022-28831-6
Li, S., Ali, S., Duan, X., Liu, S., Du, J., Liu, C., et al. (2018). JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 23, 389–403. doi:10.1016/j.celrep.2018.03.051
Li, Y., Chitnis, N., Nakagawa, H., Kita, Y., Natsugoe, S., Yang, Y., et al. (2015). PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 5, 288–303. doi:10.1158/2159-8290.cd-14-0625
Li, Z., Wang, D., Chen, X., Wang, W., Wang, P., Hou, P., et al. (2021). PRMT1-mediated EZH2 methylation promotes breast cancer cell proliferation and tumorigenesis. Cell Death Dis. 12, 1080. doi:10.1038/s41419-021-04381-5
Li, Z., Wang, D., Lu, J., Huang, B., Wang, Y., Dong, M., et al. (2020b). Methylation of EZH2 by PRMT1 regulates its stability and promotes breast cancer metastasis. Cell Death Differ. 27, 3226–3242. doi:10.1038/s41418-020-00615-9
Li, Z., Wang, D., Wang, W., Chen, X., Tang, A., Hou, P., et al. (2020a). Macrophages-stimulated PRMT1-mediated EZH2 methylation promotes breast cancer metastasis. Biochem. Biophysical Res. Commun. 533, 679–684. doi:10.1016/j.bbrc.2020.10.037
Lin, Y. L., Tsai, Y. J., Liu, Y. F., Cheng, Y. C., Hung, C. M., Lee, Y. J., et al. (2013). The critical role of protein arginine methyltransferase prmt8 in zebrafish embryonic and neural development is non-redundant with its paralogue prmt1. PloS one 8, e55221. doi:10.1371/journal.pone.0055221
Linder, B., Plottner, O., Kroiss, M., Hartmann, E., Laggerbauer, B., Meister, G., et al. (2008). Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 17, 3236–3246. doi:10.1093/hmg/ddn219
Litt, M., Qiu, Y., and Huang, S. (2009). Histone arginine methylations: Their roles in chromatin dynamics and transcriptional regulation. Biosci. Rep. 29, 131–141. doi:10.1042/bsr20080176
Liu, F., Xu, Y., Lu, X., Hamard, P. J., Karl, D. L., Man, N., et al. (2020a). PRMT5-mediated histone arginine methylation antagonizes transcriptional repression by polycomb complex PRC2. Nucleic acids Res. 48, 2956–2968. doi:10.1093/nar/gkaa065
Liu, J., Dou, X., Chen, C., Chen, C., Liu, C., Xu, M. M., et al. (2020c). N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586. doi:10.1126/science.aay6018
Liu, J., Feng, J., Li, L., Lin, L., Ji, J., Lin, C., et al. (2020b). Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 21, e48597. doi:10.15252/embr.201948597
Liu, J., Zhang, S., Liu, M., Liu, Y., Nshogoza, G., Gao, J., et al. (2018). Structural plasticity of the TDRD3 Tudor domain probed by a fragment screening hit. FEBS J. 285, 2091–2103. doi:10.1111/febs.14469
Liu, K., Chen, C., Guo, Y., Lam, R., Bian, C., Xu, C., et al. (2010). Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. U. S. A. 107, 18398–18403. doi:10.1073/pnas.1013106107
Liu, K., Guo, Y., Liu, H., Bian, C., Lam, R., Liu, Y., et al. (2012). Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PloS one 7, e30375. doi:10.1371/journal.pone.0030375
Liu, L., Lin, B., Yin, S., Ball, L. E., Delaney, J. R., Long, D. T., et al. (2022a). Arginine methylation of BRD4 by PRMT2/4 governs transcription and DNA repair. Sci. Adv. 8, eadd8928. doi:10.1126/sciadv.add8928
Liu, L., Yin, S., and Gan, W. (2023). TRAF6 promotes PRMT5 activity in a ubiquitination-dependent manner. Cancers (Basel) 15, 2501. doi:10.3390/cancers15092501
Liu, L., Zhao, X., Zhao, L., Li, J., Yang, H., Zhu, Z., et al. (2016). Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Res. 76, 1260–1272. doi:10.1158/0008-5472.can-15-1766
Liu, Q., Zhang, X.-l., Cheng, M.-b., and Zhang, Y. (2019). PRMT1 activates myogenin transcription via MyoD arginine methylation at R121. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1862, 194442. doi:10.1016/j.bbagrm.2019.194442
Liu, X., Li, M., Xia, X., Li, X., and Chen, Z. (2017). Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445. doi:10.1038/nature22036
Liu, X. N., Wang, H. L., Zhao, X. Y., Luo, Q. Z., Wang, Q. W., Tan, K. F., et al. (2021). Arginine methylation of METTL14 promotes RNA N-6-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat. Commun. 12, 3780. doi:10.1038/s41467-021-24035-6
Liu, Y. L., Iqbal, A., Li, W. G., Ni, Z. Y., Wang, Y. L., Ramprasad, J., et al. (2022b). A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II. Nat. Commun. 13, 5453. doi:10.1038/s41467-022-33229-5
Lorton, B. M., and Shechter, D. (2019). Cellular consequences of arginine methylation. Cell Mol. Life Sci. 76, 2933–2956. doi:10.1007/s00018-019-03140-2
Losman, J. A., Koivunen, P., and Kaelin, W. G. (2020). 2-Oxoglutarate-dependent dioxygenases in cancer. Nat. Rev. Cancer 20, 710–726. doi:10.1038/s41568-020-00303-3
Lu, X., Fernando, T. M., Lossos, C., Yusufova, N., Liu, F., Fontán, L., et al. (2018). PRMT5 interacts with the BCL6 oncoprotein and is required for germinal center formation and lymphoma cell survival. Blood 132, 2026–2039. doi:10.1182/blood-2018-02-831438
Lubrino, T., Mendoza, M., Mendoza, M., Briski, S., Osuji, I., Cuala, J., et al. (2022). Arginine methylation of the PGC-1 alpha C-terminus is temperature- dependent. Biochemistry 62 (1), 22–34. doi:10.1021/acs.biochem.2c00363
Lyon, A. S., Peeples, W. B., and Rosen, M. K. (2021). A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235. doi:10.1038/s41580-020-00303-z
Macadangdang, B. R., Oberai, A., Spektor, T., Campos, O. A., Sheng, F., Carey, M. F., et al. (2014). Evolution of histone 2A for chromatin compaction in eukaryotes. Elife 3, e02792. doi:10.7554/elife.02792
Marjon, K., Cameron, M. J., Quang, P., Clasquin, M. F., Mandley, E., Kunii, K., et al. (2016). MTAP deletions in cancer create vulnerability to targeting of the mat2a/PRMT5/RIOK1 Axis. Cell Rep. 15, 574–587. doi:10.1016/j.celrep.2016.03.043
Marjon, K., Kalev, P., and Marks, K. (2021). Cancer dependencies: PRMT5 and MAT2A in MTAP/p16-Deleted cancers. Annu. Rev. Cancer Biol. 5, 371–390. doi:10.1146/annurev-cancerbio-030419-033444
Maron, M. I., Lehman, S. M., Gayatri, S., DeAngelo, J. D., Hegde, S., Lorton, B. M., et al. (2021). Independent transcriptomic and proteomic regulation by type I and II protein arginine methyltransferases. iScience 24, 102971. doi:10.1016/j.isci.2021.102971
Mathur, R., and Roberts, C. W. M. (2018). SWI/SNF (BAF) complexes: Guardians of the epigenome. Annu. Rev. Cancer Biol. 2, 413–427. doi:10.1146/annurev-cancerbio-030617-050151
Matsuoka, M. (1972). Epsilon-N-methylated lysine and guanidine-N-methylated arginine of proteins. 3. Presence and distribution in nature and mammals. Seikagaku 44, 364–370.
Mavrakis, K. J., McDonald, E. R., Schlabach, M. R., Billy, E., Hoffman, G. R., deWeck, A., et al. (2016). Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351, 1208–1213. doi:10.1126/science.aad5944
Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J. P., St-Denis, N. A., Li, T., et al. (2013). The CRAPome: A contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736. doi:10.1038/nmeth.2557
Mentch, S. J., Mehrmohamadi, M., Huang, L., Liu, X., Gupta, D., Mattocks, D., et al. (2015). Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab. 22, 861–873. doi:10.1016/j.cmet.2015.08.024
Meriesh, H. A., Lerner, A. M., Chandrasekharan, M. B., and Strahl, B. D. (2020). The histone H4 basic patch regulates SAGA-mediated H2B deubiquitination and histone acetylation. J. Biol. Chem. 295, 6561–6569. doi:10.1074/jbc.ra120.013196
Mertz, J. A., Conery, A. R., Bryant, B. M., Sandy, P., Balasubramanian, S., Mele, D. A., et al. (2011). Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U. S. A. 108, 16669–16674. doi:10.1073/pnas.1108190108
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. doi:10.1038/nrm3785
Millan-Zambrano, G., Burton, A., Bannister, A. J., and Schneider, R. (2022). Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 23, 563–580. doi:10.1038/s41576-022-00468-7
Milne, T. A., Briggs, S. D., Brock, H. W., Martin, M. E., Gibbs, D., Allis, C. D., et al. (2002). MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117. doi:10.1016/s1097-2765(02)00741-4
Mizutani, S., Yoshida, T., Zhao, X., Nimer, S. D., Taniwaki, M., and Okuda, T. (2015). Loss of RUNX1/AML1 arginine-methylation impairs peripheral T cell homeostasis. Br. J. Haematol. 170, 859–873. doi:10.1111/bjh.13499
Morita, K., Hatanaka, Y., Ihashi, S., Asano, M., Miyamoto, K., and Matsumoto, K. (2021). Symmetrically dimethylated histone H3R2 promotes global transcription during minor zygotic genome activation in mouse pronuclei. Sci. Rep. 11, 10146. doi:10.1038/s41598-021-89334-w
Mostaqul Huq, M. D., Gupta, P., Tsai, N. P., White, R., Parker, M. G., and Wei, L. N. (2006). Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 25, 5094–5104. doi:10.1038/sj.emboj.7601389
Mounir, Z., Korn, J. M., Westerling, T., Lin, F., Kirby, C. A., Schirle, M., et al. (2016). ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the Androgen Receptor. Elife 5, e13964. doi:10.7554/elife.13964
Naeem, H., Cheng, D., Zhao, Q., Underhill, C., Tini, M., Bedford, M. T., et al. (2007). The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 27, 120–134. doi:10.1128/mcb.00815-06
Narayanan, N., Wang, Z., Li, L., and Yang, Y. (2017). Arginine methylation of USP9X promotes its interaction with TDRD3 and its anti-apoptotic activities in breast cancer cells. Cell Discov. 3, 16048. doi:10.1038/celldisc.2016.48
Nishioka, K., and Reinberg, D. (2003). Methods and tips for the purification of human histone methyltransferases. Methods 31, 49–58. doi:10.1016/s1046-2023(03)00087-2
Obata, T., Yaffe, M. B., Leparc, G. G., Piro, E. T., Maegawa, H., Kashiwagi, A., et al. (2000). Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J. Biol. Chem. 275, 36108–36115. doi:10.1074/jbc.m005497200
Obeid, R., and Herrmann, W. (2009). Homocysteine and lipids: S-adenosyl methionine as a key intermediate. FEBS Lett. 583, 1215–1225. doi:10.1016/j.febslet.2009.03.038
Ong, S. E., Mittler, G., and Mann, M. (2004). Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126. doi:10.1038/nmeth715
Pahlich, S., Zakaryan, R. P., and Gehring, H. (2008). Identification of proteins interacting with protein arginine methyltransferase 8: The ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins 72, 1125–1137. doi:10.1002/prot.22004
Pal, S., Vishwanath, S. N., Erdjument-Bromage, H., Tempst, P., and Sif, S. (2004). Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645. doi:10.1128/mcb.24.21.9630-9645.2004
Pan, M., Reid, M. A., Lowman, X. H., Kulkarni, R. P., Tran, T. Q., Liu, X., et al. (2016). Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 18, 1090–1101. doi:10.1038/ncb3410
Pek, J. W., Anand, A., and Kai, T. (2012). Tudor domain proteins in development. Development 139, 2255–2266. doi:10.1242/dev.073304
Penney, J., Seo, J., Kritskiy, O., Elmsaouri, S., Gao, F., Pao, P. C., et al. (2017). Loss of protein arginine methyltransferase 8 alters synapse composition and function, resulting in behavioral defects. J. Neurosci. 37, 8655–8666. doi:10.1523/jneurosci.0591-17.2017
Qamar, S., Wang, G., Randle, S. J., Ruggeri, F. S., Varela, J. A., Lin, J. Q., et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 173, 720–734 e15. doi:10.1016/j.cell.2018.03.056
Qin, J., and Xu, J. (2021). Arginine methylation in the epithelial-to-mesenchymal transition. FEBS J. 289, 7292–7303. doi:10.1111/febs.16152
Quan, X., Yue, W., Luo, Y., Cao, J., Wang, H., Wang, Y., et al. (2015). The protein arginine methyltransferase PRMT5 regulates Aβ-induced toxicity in human cells and Caenorhabditis elegans models of Alzheimer's disease. J. Neurochem. 134, 969–977. doi:10.1111/jnc.13191
Rajyaguru, P., and Parker, R. (2012). RGG motif proteins: Modulators of mRNA functional states. Cell Cycle 11, 2594–2599. doi:10.4161/cc.20716
Reintjes, A., Fuchs, J. E., Kremser, L., Lindner, H. H., Liedl, K. R., Huber, L. A., et al. (2016). Asymmetric arginine dimethylation of RelA provides a repressive mark to modulate TNFα/NF-κB response. Proc. Natl. Acad. Sci. U. S. A. 113, 4326–4331. doi:10.1073/pnas.1522372113
Ryan, V. H., Dignon, G. L., Zerze, G. H., Chabata, C. V., Silva, R., Conicella, A. E., et al. (2018). Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell 69, 465–479 e7. doi:10.1016/j.molcel.2017.12.022
Sakamaki, J., Daitoku, H., Ueno, K., Hagiwara, A., Yamagata, K., and Fukamizu, A. (2011). Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc. Natl. Acad. Sci. U. S. A. 108, 6085–6090. doi:10.1073/pnas.1015328108
Sarmento, O. F., Digilio, L. C., Wang, Y., Perlin, J., Herr, J. C., Allis, C. D., et al. (2004). Dynamic alterations of specific histone modifications during early murine development. J. Cell Sci. 117, 4449–4459. doi:10.1242/jcs.01328
Scott, D. E., Bayly, A. R., Abell, C., and Skidmore, J. (2016). Small molecules, big targets: Drug discovery faces the protein-protein interaction challenge. Nat. Rev. Drug Discov. 15, 533–550. doi:10.1038/nrd.2016.29
Selenko, P., Sprangers, R., Stier, G., Bühler, D., Fischer, U., and Sattler, M. (2001). SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8, 27–31. doi:10.1038/83014
Senisterra, G., Zhu, H. Y., Luo, X., Zhang, H., Xun, G., Lu, C., et al. (2018). Discovery of small-molecule antagonists of the H3K9me3 binding to UHRF1 tandem tudor domain. SLAS Discov. 23, 930–940. doi:10.1177/2472555218766278
Sharma, P., Lioutas, A., Fernandez-Fuentes, N., Quilez, J., Carbonell-Caballero, J., Wright, R. H. G., et al. (2019). Arginine citrullination at the C-terminal domain controls RNA polymerase II transcription. Mol. Cell 73, 84–96.e7. doi:10.1016/j.molcel.2018.10.016
Shilo, K., Wu, X., Sharma, S., Welliver, M., Duan, W., Villalona-Calero, M., et al. (2013). Cellular localization of protein arginine methyltransferase-5 correlates with grade of lung tumors. Diagn Pathol. 8, 201. doi:10.1186/1746-1596-8-201
Shrestha, S., Lucky, A. B., Brashear, A. M., Li, X., Cui, L., and Miao, J. (2022). Distinct histone post-translational modifications during plasmodium falciparum gametocyte development. J. Proteome Res. 21, 1857–1867. doi:10.1021/acs.jproteome.2c00108
Siaw, G. E., Liu, I. F., Lin, P. Y., Been, M. D., and Hsieh, T. S. (2016). DNA and RNA topoisomerase activities of Top3β are promoted by mediator protein Tudor domain-containing protein 3. Proc. Natl. Acad. Sci. U. S. A. 113, E5544–E5551. doi:10.1073/pnas.1605517113
Simcikova, D., Gelles-Watnick, S., and Neugebauer, K. M. (2023). Tudor-dimethylarginine interactions: The condensed version. Trends Biochem. Sci. 48, 689–698. doi:10.1016/j.tibs.2023.04.003
Sims, R. J., Rojas, L. A., Beck, D. B., Bonasio, R., Schüller, R., Drury, W. J., et al. (2011). The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332, 99–103. doi:10.1126/science.1202663
Smith, D. L., Erce, M. A., Lai, Y. W., Tomasetig, F., Hart-Smith, G., Hamey, J. J., et al. (2020). Crosstalk of phosphorylation and arginine methylation in disordered SRGG repeats of saccharomycescerevisiae fibrillarin and its association with nucleolar localization. J. Mol. Biol. 432, 448–466. doi:10.1016/j.jmb.2019.11.006
Sohail, M., Cao, W., Mahmood, N., Myschyshyn, M., Hong, S. P., and Xie, J. (2014). Evolutionarily emerged G tracts between the polypyrimidine tract and 3' AG are splicing silencers enriched in genes involved in cancer. BMC Genomics 15, 1143. doi:10.1186/1471-2164-15-1143
Sohail, M., and Xie, J. (2015). Evolutionary emergence of a novel splice variant with an opposite effect on the cell cycle. Mol. Cell. Biol. 35, 2203–2214. doi:10.1128/mcb.00190-15
Sohail, M., Zhang, M., Litchfield, D., Wang, L., Kung, S., and Xie, J. (2015). Differential expression, distinct localization and opposite effect on Golgi structure and cell differentiation by a novel splice variant of human PRMT5. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1853, 2444–2452. doi:10.1016/j.bbamcr.2015.07.003
Song, J., and Yi, C. (2017). Chemical modifications to RNA: A new layer of gene expression regulation. ACS Chem. Biol. 12, 316–325. doi:10.1021/acschembio.6b00960
Spitz, F., and Furlong, E. E. (2012). Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626. doi:10.1038/nrg3207
Sprangers, R., Groves, M. R., Sinning, I., and Sattler, M. (2003). High-resolution X-ray and NMR structures of the SMN tudor domain: Conformational variation in the binding site for symmetrically dimethylated arginine residues. J. Mol. Biol. 327, 507–520. doi:10.1016/s0022-2836(03)00148-7
Stavride, P., Arampatzi, P., and Papamatheakis, J. (2013). Differential regulation of MHCII genes by PRMT6, via an AT-hook motif of RFX5. Mol. Immunol. 56, 390–398. doi:10.1016/j.molimm.2013.05.235
Stetler, A., Winograd, C., Sayegh, J., Cheever, A., Patton, E., Zhang, X., et al. (2006). Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 15, 87–96. doi:10.1093/hmg/ddi429
Sundar, I. K., Nevid, M. Z., Friedman, A. E., and Rahman, I. (2014). Cigarette smoke induces distinct histone modifications in lung cells: Implications for the pathogenesis of COPD and lung cancer. J. Proteome Res. 13, 982–996. doi:10.1021/pr400998n
Swiercz, R., Cheng, D., Kim, D., and Bedford, M. T. (2007). Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 282, 16917–16923. doi:10.1074/jbc.m609778200
Swiercz, R., Person, M. D., and Bedford, M. T. (2005). Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 386, 85–91. doi:10.1042/bj20041466
Sylvestersen, K. B., Horn, H., Jungmichel, S., Jensen, L. J., and Nielsen, M. L. (2014). Proteomic analysis of arginine methylation sites in human cells reveals dynamic regulation during transcriptional arrest. Mol. Cell. Proteomics 13, 2072–2088. doi:10.1074/mcp.o113.032748
Takai, H., Masuda, K., Sato, T., Sakaguchi, Y., Suzuki, T., Suzuki, T., et al. (2014). 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 9, 48–60. doi:10.1016/j.celrep.2014.08.071
Tan, C. P., and Nakielny, S. (2006). Control of the DNA methylation system component MBD2 by protein arginine methylation. Mol. Cell. Biol. 26, 7224–7235. doi:10.1128/mcb.00473-06
Tan, M., Luo, H., Lee, S., Jin, F., Yang, J. S., Montellier, E., et al. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028. doi:10.1016/j.cell.2011.08.008
Taverna, S. D., Li, H., Ruthenburg, A. J., Allis, C. D., and Patel, D. J. (2007). How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040. doi:10.1038/nsmb1338
Tewary, S. K., Zheng, Y. G., and Ho, M. C. (2019). Protein arginine methyltransferases: Insights into the enzyme structure and mechanism at the atomic level. Cell Mol. Life Sci. 76, 2917–2932. doi:10.1007/s00018-019-03145-x
Teyssier, C., Ma, H., Emter, R., Kralli, A., and Stallcup, M. R. (2005). Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes & Dev. 19, 1466–1473. doi:10.1101/gad.1295005
Thandapani, P., O'Connor, T. R., Bailey, T. L., and Richard, S. (2013). Defining the RGG/RG motif. Mol. Cell 50, 613–623. doi:10.1016/j.molcel.2013.05.021
Tripsianes, K., Madl, T., Machyna, M., Fessas, D., Englbrecht, C., Fischer, U., et al. (2011). Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18, 1414–1420. doi:10.1038/nsmb.2185
Tsutsui, T., Fukasawa, R., Shinmyouzu, K., Nakagawa, R., Tobe, K., Tanaka, A., et al. (2013). Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J. Biol. Chem. 288, 20955–20965. doi:10.1074/jbc.m113.486746
Tweedie-Cullen, R. Y., Brunner, A. M., Grossmann, J., Mohanna, S., Sichau, D., Nanni, P., et al. (2012). Identification of combinatorial patterns of post-translational modifications on individual histones in the mouse brain. PloS one 7, e36980. doi:10.1371/journal.pone.0036980
Uhlmann, T., Geoghegan, V. L., Thomas, B., Ridlova, G., Trudgian, D. C., and Acuto, O. (2012). A method for large-scale identification of protein arginine methylation. Mol. Cell. Proteomics 11, 1489–1499. doi:10.1074/mcp.m112.020743
Vasilyev, N., Polonskaia, A., Darnell, J. C., Darnell, R. B., Patel, D. J., and Serganov, A. (2015). Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U. S. A. 112, E5391–E5400. doi:10.1073/pnas.1515737112
Waldmann, T., Izzo, A., Kamieniarz, K., Richter, F., Vogler, C., Sarg, B., et al. (2011). Methylation of H2AR29 is a novel repressive PRMT6 target. Epigenetics Chromatin 4, 11. doi:10.1186/1756-8935-4-11
Walport, L. J., Hopkinson, R. J., Chowdhury, R., Schiller, R., Ge, W., Kawamura, A., et al. (2016). Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974. doi:10.1038/ncomms11974
Wang, H., Huang, Z. Q., Xia, L., Feng, Q., Erdjument-Bromage, H., Strahl, B. D., et al. (2001). Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857. doi:10.1126/science.1060781
Wang, J., Wang, Z., Inuzuka, H., Wei, W., and Liu, J. (2023). PRMT1 methylates METTL14 to modulate its oncogenic function. Neoplasia 42, 100912. doi:10.1016/j.neo.2023.100912
Wang, L., Zeng, H., Wang, Q., Zhao, Z., Boyer, T. G., Bian, X., et al. (2015). MED12 methylation by CARM1 sensitizes human breast cancer cells to chemotherapy drugs. Sci. Adv. 1, e1500463. doi:10.1126/sciadv.1500463
Wang, L., Zhao, Z., Meyer, M. B., Saha, S., Yu, M., Guo, A., et al. (2014). CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell 25, 21–36. doi:10.1016/j.ccr.2013.12.007
Wang, P., Doxtader, K. A., and Nam, Y. (2016a). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317. doi:10.1016/j.molcel.2016.05.041
Wang, Q., Li, Z., Zhang, S., Li, Y., Wang, Y., Fang, Z., et al. (2022). Global profiling of arginine dimethylation in regulating protein phase separation by a steric effect-based chemical-enrichment method. Proc. Natl. Acad. Sci. U. S. A. 119, e2205255119. doi:10.1073/pnas.2205255119
Wang, Y., and Bedford, M. T. (2023). Effectors and effects of arginine methylation. Biochem. Soc. Trans. 51, 725–734. doi:10.1042/bst20221147
Wang, Y., Hsu, J. M., Kang, Y., Wei, Y., Lee, P. C., Chang, S. J., et al. (2016b). Oncogenic functions of Gli1 in pancreatic adenocarcinoma are supported by its PRMT1-mediated methylation. Cancer Res. 76, 7049–7058. doi:10.1158/0008-5472.can-16-0715
Wang, Z., Pan, Z., Adhikari, S., Harada, B. T., Shen, L., Yuan, W., et al. (2021). m(6 A deposition is regulated by PRMT1-mediated arginine methylation of METTL14 in its disordered C-terminal region. EMBO J. 40, e106309. doi:10.15252/embj.2020106309
Webby, C. J., Wolf, A., Gromak, N., Dreger, M., Kramer, H., Kessler, B., et al. (2009). Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93. doi:10.1126/science.1175865
Wei, H., Wang, B., Miyagi, M., She, Y., Gopalan, B., Huang, D.-B., et al. (2013). PRMT5 dimethylates R30 of the p65 subunit to activate NF-κB. Proc. Natl. Acad. Sci. 110, 13516–13521. doi:10.1073/pnas.1311784110
Weiss, V. H., McBride, A. E., Soriano, M. A., Filman, D. J., Silver, P. A., and Hogle, J. M. (2000). The structure and oligomerization of the yeast arginine methyltransferase, Hmt1. Nat. Struct. Biol. 7, 1165–1171. doi:10.1038/82028
Wen, C. W., Tian, Z. J., Li, L., Chen, T. K., Chen, H. J., Dai, J. C., et al. (2022). SRSF3 and HNRNPH1 regulate radiation-induced alternative splicing of protein arginine methyltransferase 5 in hepatocellular carcinoma. Int. J. Mol. Sci. 23, 14832. doi:10.3390/ijms232314832
Weng, Z., Rickles, R. J., Feng, S., Richard, S., Shaw, A. S., Schreiber, S. L., et al. (1995). Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol. Cell. Biol. 15, 5627–5634. doi:10.1128/mcb.15.10.5627
Wiener, D., and Schwartz, S. (2021). The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 22, 119–131. doi:10.1038/s41576-020-00295-8
Wong, C. M., Tang, H. M., Kong, K. Y., Wong, G. W., Qiu, H., Jin, D. Y., et al. (2010). Yeast arginine methyltransferase Hmt1p regulates transcription elongation and termination by methylating Npl3p. Nucleic acids Res. 38, 2217–2228. doi:10.1093/nar/gkp1133
Wu, Q., Schapira, M., Arrowsmith, C. H., and Barsyte-Lovejoy, D. (2021). Protein arginine methylation: From enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 20, 509–530. doi:10.1038/s41573-021-00159-8
Wu, T. F., Yao, Y. L., Lai, I. L., Lai, C. C., Lin, P. L., and Yang, W. M. (2015). Loading of PAX3 to mitotic chromosomes is mediated by arginine methylation and associated with waardenburg syndrome. J. Biol. Chem. 290, 20556–20564. doi:10.1074/jbc.m114.607713
Wysocka, J., Allis, C. D., and Coonrod, S. (2006). Histone arginine methylation and its dynamic regulation. Front. Biosci. 11, 344–355. doi:10.2741/1802
Xie, W., and Denman, R. B. (2011). Protein methylation and stress granules: Posttranslational remodeler or innocent bystander? Mol. Biol. Int. 2011, 1–14. doi:10.4061/2011/137459
Xu, W., Chen, H., Du, K., Asahara, H., Tini, M., Emerson, B. M., et al. (2001). A transcriptional switch mediated by cofactor methylation. Science 294, 2507–2511. doi:10.1126/science.1065961
Xu, W., Cho, H., Kadam, S., Banayo, E. M., Anderson, S., Yates, J. R., et al. (2004). A methylation-mediator complex in hormone signaling. Genes & Dev. 18, 144–156. doi:10.1101/gad.1141704
Xu, X., Hoang, S., Mayo, M. W., and Bekiranov, S. (2010). Application of machine learning methods to histone methylation ChIP-Seq data reveals H4R3me2 globally represses gene expression. BMC Bioinforma. 11, 396. doi:10.1186/1471-2105-11-396
Yamagata, K., Daitoku, H., Takahashi, Y., Namiki, K., Hisatake, K., Kako, K., et al. (2008). Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231. doi:10.1016/j.molcel.2008.09.013
Yang, L., Ma, D. W., Cao, Y. P., Li, D. Z., Zhou, X., Feng, J. F., et al. (2021). PRMT5 functionally associates with EZH2 to promote colorectal cancer progression through epigenetically repressing CDKN2B expression. Theranostics 11, 3742–3759. doi:10.7150/thno.53023
Yang, M., Zhang, Y., Liu, G., Zhao, Z., Li, J., Yang, L., et al. (2022). TIPE1 inhibits osteosarcoma tumorigenesis and progression by regulating PRMT1 mediated STAT3 arginine methylation. Cell Death Dis. 13, 815. doi:10.1038/s41419-022-05273-y
Yang, Y., and Bedford, M. T. (2013). Protein arginine methyltransferases and cancer. Nat. Rev. Cancer 13, 37–50. doi:10.1038/nrc3409
Yang, Y., Hadjikyriacou, A., Xia, Z., Gayatri, S., Kim, D., Zurita-Lopez, C., et al. (2015). PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. 6, 6428. doi:10.1038/ncomms7428
Yang, Y., Lu, Y., Espejo, A., Wu, J., Xu, W., Liang, S., et al. (2010). TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell 40, 1016–1023. doi:10.1016/j.molcel.2010.11.024
Yang, Y., McBride, K. M., Hensley, S., Lu, Y., Chedin, F., and Bedford, M. T. (2014). Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497. doi:10.1016/j.molcel.2014.01.011
Yanling Zhao, D., Gish, G., Braunschweig, U., Li, Y., Ni, Z., Schmitges, F. W., et al. (2016). SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53. doi:10.1038/nature16469
Yao, B., Gui, T., Zeng, X., Deng, Y., Wang, Z., Wang, Y., et al. (2021). PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 13, 58. doi:10.1186/s13073-021-00871-5
Yi, P., Wang, Z., Feng, Q., Chou, C. K., Pintilie, G. D., Shen, H., et al. (2017). Structural and functional impacts of ER coactivator sequential recruitment. Mol. Cell 67, 733–743 e4. doi:10.1016/j.molcel.2017.07.026
Yin, S., Liu, L., Ball, L. E., Wang, Y., Bedford, M. T., Duncan, S. A., et al. (2023). CDK5-PRMT1-WDR24 signaling cascade promotes mTORC1 signaling and tumor growth. Cell Rep. 42, 112316. doi:10.1016/j.celrep.2023.112316
Yoshida, A., Oyoshi, T., Suda, A., Futaki, S., and Imanishi, M. (2022). Recognition of G-quadruplex RNA by a crucial RNA methyltransferase component, METTL14. Nucleic acids Res. 50, 449–457. doi:10.1093/nar/gkab1211
Yoshizawa, T., Ali, R., Jiou, J., Fung, H. Y. J., Burke, K. A., Kim, S. J., et al. (2018). Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173, 693–705 e22. doi:10.1016/j.cell.2018.03.003
Yu, Y. S., Shin, H. R., Kim, D., Baek, S. A., Choi, S. A., Ahn, H., et al. (2020). Pontin arginine methylation by CARM1 is crucial for epigenetic regulation of autophagy. Nat. Commun. 11, 6297. doi:10.1038/s41467-020-20080-9
Yuan, C.-C., Matthews, A. G. W., Jin, Y., Chen, C. F., Chapman, B. A., Ohsumi, T. K., et al. (2012). Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell Rep. 1, 83–90. doi:10.1016/j.celrep.2011.12.008
Yue, W. W., Hassler, M., Roe, S. M., Thompson Vale, V., and Pearl, L. H. (2007). Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 26, 4402–4412. doi:10.1038/sj.emboj.7601856
Zappia, V., Della Ragione, F., Pontoni, G., Gragnaniello, V., and Carteni-Farina, M. (1988). Human 5'-deoxy-5'-methylthioadenosine phosphorylase: Kinetic studies and catalytic mechanism. Adv. Exp. Med. Biol. 250, 165–177. doi:10.1007/978-1-4684-5637-0_15
Zhang, F., Kerbl-Knapp, J., Rodriguez Colman, M. J., Meinitzer, A., Macher, T., Vujic, N., et al. (2021). Global analysis of protein arginine methylation. Cell Rep. Methods 1, 100016. doi:10.1016/j.crmeth.2021.100016
Zhang, H. T., Zeng, L. F., He, Q. Y., Tao, W. A., Zha, Z. G., and Hu, C. D. (2016). The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1863, 335–346. doi:10.1016/j.bbamcr.2015.12.001
Zhang, J., Fan, X., Zhou, Y., Chen, L., and Rao, H. (2022b). The PRMT5-LSD1 axis confers Slug dual transcriptional activities and promotes breast cancer progression. J. Exp. Clin. Cancer Res. 41, 191. doi:10.1186/s13046-022-02400-7
Zhang, L., Eugeni, E. E., Parthun, M. R., and Freitas, M. A. (2003). Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112, 77–86. doi:10.1007/s00412-003-0244-6
Zhang, T., Wallis, M., Petrovic, V., Challis, J., Kalitsis, P., and Hudson, D. F. (2019). Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 9, 190222. doi:10.1098/rsob.190222
Zhang, X., and Cheng, X. (2003). Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11, 509–520. doi:10.1016/s0969-2126(03)00071-6
Zhang, X., Wu, X., Peng, J., Sun, A., Guo, Y., Fu, P., et al. (2022a). Cis- and trans-regulation by histone H4 basic patch R17/R19 in metazoan development. Open Biol. 12, 220066. doi:10.1098/rsob.220066
Zhao, H.-y., Zhang, Y.-j., Dai, H., Zhang, Y., and Shen, Y.-f. (2011). CARM1 mediates modulation of Sox2. PloS one 6, e27026. doi:10.1371/journal.pone.0027026
Zhao, Q., Rank, G., Tan, Y. T., Li, H., Moritz, R. L., Simpson, R. J., et al. (2009). PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311. doi:10.1038/nsmb.1568
Keywords: post-translational modification, arginine methylation, transcription, epigenetics, histone modification
Citation: Pham HQH, Tao X and Yang Y (2023) Protein arginine methylation in transcription and epigenetic regulation. Front. Epigenet. Epigenom. 1:1245832. doi: 10.3389/freae.2023.1245832
Received: 23 June 2023; Accepted: 21 July 2023;
Published: 04 August 2023.
Edited by:
Sandipan Brahma, University of Nebraska Medical Center, United StatesReviewed by:
Sitaram Gayatri, Evozyne, Inc., United StatesJian Xu, University of Southern California, United States
Copyright © 2023 Pham, Tao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanzhong Yang, yyang@coh.org
 Hoang Quoc Hai Pham
Hoang Quoc Hai Pham Xiaoqun Tao1,2
Xiaoqun Tao1,2  Yanzhong Yang
Yanzhong Yang