- 1Graduate School of Agricultural Science, Hokkaido University Kita-9, Sapporo, Japan
- 2Graduate School of Life Science, Hokkaido University Kita-10 Nishi-8, Sapporo, Japan
Although bacterial population behavior has been investigated in a variety of foods in the past 40 years, it is difficult to obtain desired information from the mere juxtaposition of experimental data. We predicted the changes in the number of bacteria and visualize the effects of pH, aw, and temperature using a data mining approach. Population growth and inactivation data on eight pathogenic and food spoilage bacteria under 5,025 environmental conditions were obtained from the ComBase database (www.combase.cc), including 15 food categories, and temperatures ranging from 0°C to 25°C. The eXtreme gradient boosting tree was used to predict population behavior. The root mean square error of the observed and predicted values was 1.23 log CFU/g. The data mining model extracted the growth inhibition for the investigated bacteria against aw, temperature, and pH using the SHapley Additive eXplanations value. A data mining approach provides information concerning bacterial population behavior and how food ecosystems affect bacterial growth and inactivation.
1 Introduction
Different types of microorganisms are present in food. Some of these cause foodborne illness and food spoilage. To control food pathogens and spoilage bacteria, various preservation techniques have been developed to prevent harmful bacteria from growing during processing, distribution, and storage. Many factors influence the microbial response in food ecosystems. For instance, temperature, pH, water activity (aw), antimicrobial additives, and gas components can affect bacterial population behavior (Leistner, 2000; Doyle et al., 2019), even though the effects on bacterial growth and inactivation vary by bacterial species or genus. Adjusting the various environmental conditions in food enables the suppression of bacterial growth and food spoilage (Gould, 1996; Leistner, 2000). Thus, appropriate microbiological control can help prevent food loss and improve food safety.
To quantify and evaluate bacterial growth for control, many studies on microbiological response in food have been conducted since Roberts & Jarvis (1983) introduced predictive microbiology, which originated from the research by Bigelow (1921), Bigelow & Esty (1920), and Esty & Meyer (1922). Each experimental data on bacterial growth and inactivation was obtained by counting the number of colonies on the culture plate as viable cell counts or by measuring optical density for the cell density over time under controlled conditions, such as temperature, pH, and aw. Microbial responses in food have been explained by mathematical models, the main exploratory variables of which are temperature, pH, and aw (Ross and McMeekin, 1994; Jagannath and Tsuchido, 2003). Over the last 40 years, experimental data on microbial responses to the food environment have been collected by research institutions, universities, and companies according to their objectives. The accumulated data are stored in databases such as the ComBase database (www.combase.cc), which was developed to provide easy access to microbiological data in research establishments and publications produced by different laboratories (Baranyi et al., 2004). Currently, every effort is vital for collecting data with similar species or conditions through literature or database to assess product safety. A comprehensive statistical analysis is needed for understanding bacterial population behavior regardless of food and bacteria.
Studies have been conducted to understand the global trends of microbial responses from the accumulated data. In predictive microbiology, studies have used meta-analysis, which is a method to amalgamate, summarize, or review previous quantitative research for identifying trends with a statistical model of specific foods and bacteria [e.g., evaluating inactivation of Escherichia coli in fermented meat (McQuestin et al., 2009), meta-analysis for quantitative microbiological risk assessments and benchmarking data (den Besten and Zwietering, 2012), growth and inactivation of Listeria monocytogenes in milk or non-thermal inactivation of Listeria monocytogenes in fermented sausages (Mataragas et al., 2015)]. Statistical models generally require analysts to specify the functional form between explanatory variables and response variables (Hochachka et al., 2007). Studies using meta-analysis have provided results that suit some objectives, such as trends in maximum growth rate or bacterial growth/inactivation by foods. However, a meta-analysis with statistical models alone is not necessarily systematic and tends to be fragmentary in terms of cross-food/bacteria analysis. Bacterial growth is affected by not only factors such as temperature, pH, and aw, but also cell density (Koutsoumanis and Sofos, 2005; Skandamis et al., 2007; Bidlas et al., 2008), characteristics of each food, and gaseous atmosphere (Doyle et al., 2019). Identifying complex relationships between food and bacteria requires the development of complex mathematical formulas with high-dimensional variables. To predict the bacterial population change and to explore the influence of each factor on microbial response using big data, a non-parametric approach, which needs to develop no hypothesis based on domain knowledge, is considered useful (Deringer et al., 2021).
Data mining is an effective method for analyzing large amounts of accumulated data. Data mining is the secondary analysis of a large database to identify and interpret hidden patterns (Hand, 1998). The recent accumulation of big data has promoted the development of databases in various fields. Consequently, data mining has been employed in many fields such as agriculture (Cortez et al., 2009; Gulyaeva et al., 2020), ecology (Hochachka et al., 2007; Ross et al., 2018), healthcare and medicine (Cios and William Moore, 2002; Delen et al., 2005; Koh and Tan, 2005; Mohanty et al., 2022), and food quality (Cortez et al., 2009; Jiménez-Carvelo et al., 2019; Nychas et al., 2021). Using machine learning, the relationship between the response and function can be determined empirically from the data. This approach can discover new knowledge of these patterns. To the best of our knowledge, only one data mining study has been conducted in the field of predicting bacterial population behavior. Hiura et al. (2021) predicted the bacterial behavior of Listeria monocytogenes using the ComBase database of microbial responses to food environments. The ComBase database contains information about bacteria and foods, such as the name of the bacterial genus or species and the category or name of the medium or food. Such information enables us to extract the comprehensive characteristics of bacterial responses, such as the association between food ecosystems and bacterial population changes, by analyzing the data reported in previous studies. In this manner, exploring the whole trend of interactions between microbial growth and inactivation and conditions from accumulated data would have an advantage in the comparison and evaluation of bacterial population changes in various bacteria and different foods.
In the present study, the objective was to not only develop a single machine learning model for predicting population behavior of food-related bacteria in various kinds of food, but to also visualize the effect of pH, aw, and temperature using a data mining approach. Data regarding the change in viable cell number over time were used for eight foodborne and food spoilage bacteria: “Aeromonas hydrophila,” “Bacillus cereus,” “Escherichia coli,” “Listeria monocytogenes,” “Pseudomonas spp.,” “Staphylococcus aureus,” “Salmonella spp.,” and “Yersinia enterocolitica.” The microbial responses to the food environment were collected from the ComBase database. The collected data included population behavior based on 15 food categories —“beef,” “culture medium,” “pork,” “poultry,” “seafood/fish,” “vegetable or fruit and their products,” “water,” “dessert food,” “milk,” “sausage,” “cheese,” “eggs and egg product,” “juice and beverage,” “sauce/dressing,” and “bread”— with temperature ranging 0°C–25°C. Data mining and machine learning approaches provide information concerning population behavior and its effects on food ecosystems.
2 Materials and methods
2.1 Data selection from ComBase database
The ComBase database contains quantified microbial responses to food with approximately 60,000 records collected from various research establishments and publications. Changes in the bacterial density over time were recorded for each experimental condition. The dataset of a change in bacterial density over time in ComBase contains “Record ID,” “Organism,” “Food category,” “Food name,” “Temperature,” “pH,” “aw,” “Conditions,” “Time,” and “Viable cell counts”. Each dataset of changes in a bacterial population is assigned a “Record ID,” which allows us to recognize one series of experiments on population behavior.
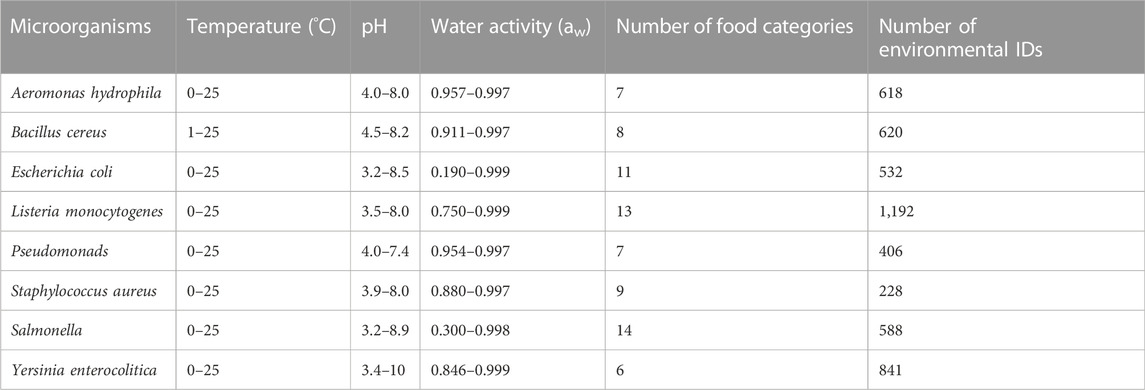
In this study, we investigated changes in the populations of eight pathogenic and food spoilage bacteria: A. hydrophila, B. cereus, E. coli, L. monocytogenes, Pseudomonas spp. (Pseudomonads), S. aureus, Salmonella spp. (Salmonella), and Y. enterocolitica. These bacteria are known to cause food spoilage and foodborne illnesses. Fifteen kinds of food categories were included “beef,” “culture medium,” “pork,” “poultry,” “seafood/fish,” “vegetable or fruit and their products,” “water,” “dessert food,” “milk,” “sausage,” “cheese,” “eggs and egg product,” “juice and beverage,” “sauce/dressing,” and “bread.” The data used for model development and evaluation were those with temperatures ranging from 0°C to 25°C and containing greater than or equal to four observed values in each series of experiments on bacterial population behavior. In addition, records for which viable counts at 0 h were unavailable were excluded, because the objective values,
2.2 Data preprocessing
In the present study, we set the change ratio of viable counts as the objective variable to predict bacterial behavior to evaluate both the increase and decrease in the bacterial population. For each Record ID, the cell concentration was transformed to a common logarithm of the change ratio of viable counts to the initial cell number
where
2.3 Model development
2.3.1 eXtreme gradient-boosting tree (XGBoost) model
The XGBoost was first proposed by Chen and Guestrin in 2016. XGboost extends the concept of the Gradient Boosting Decision Tree (GBDT). The GBDT is an iterative decision tree that includes multiple decision trees (Friedman, 2001). The GBDT is a tree-based ensemble technique that uses a decision tree as the base model, and gradient boosting trains it sequentially by adding each base model and fixing the errors generated by the previous tree model. The GBDT method has been widely employed in machine learning and data mining studies (Chang et al., 2018; Nguyen et al., 2019; Rodrigo et al., 2021; Shehadeh et al., 2021). XGBoost was used in the present study because it has several advantages in terms of fewer requirements for feature engineering, allowing steps such as handling missing values without specific processing, and variables without normalization and scaling (Wang et al., 2020; Mohanty et al., 2022). The XGBoost models were built using the XGBoost (Version 1.5.0) Python Package (https://xgboost.readthedocs.io/en/latest/python/index.html).
2.3.2 Modeling procedure
We aimed to develop a machine learning model for predicting bacterial responses to various food environments, characterized by controlling factors such as temperature, pH, and aw. Eight input variables that included five numerical data types—temperature (°C), pH, aw, time (h), and initial cell number (log CFU/g)—and three categorical data types—food category, food name, and organism—were used to develop a model to predict change ratio of a bacterial population.
There were several steps to divide the imbalanced whole dataset into training and test dataset. First, the whole dataset was separated by “Microorganisms.” Second, the dataset separated by “Microorganisms” was separated by “Food category.” Third, the dataset separated by “Microorganisms” and “Food category” was randomly divided into 9:1 without overlapping with the experimental conditions in the training and test datasets. Thus, the imbalanced dataset was separated into the training and test dataset. The training dataset was used to build a model for predicting bacterial responses to various food environments, while its hyperparameters were optimized. The test dataset was used to evaluate the performance of the tuned model.
Prior to training the predicting model, the hyperparameters of the XGBoost model used in this study were determined by a 5-fold cross-validation and grid search. Cross-validation validates the model performance using only the training dataset under an arbitrary hyperparameter set. It attempts to avoid overfitting which deteriorates the performance on unknown data (i.e., test dataset). In this method, the training dataset was divided into 5-fold (4-fold of training data and 1-fold of validation data) and then the training data was used to train a model, and validation data was used to verify the performance. Repeating this validation cycle by swapping the validation data with the training data, the performance of the model was validated. A grid search was conducted by selecting each hyperparameter value from a pre-defined range, and thus the highest performing (i.e., optimal) hyperparameters are determined. The XGBoost model hyperparameters were set in some ranges (Supplementary Table S1) and optimized as follows: a maximum tree depth of 9, min_child_weight of 1, gamma of 0.3, subsample of 0.6, colsample_bytree of 0.6, and reg_alpha of 100.
2.4 Evaluation of model accuracy
The prediction accuracy of the developed model was evaluated using 542 test datasets of environmental ID that were not used in model development. The coefficient of determination (R2) and root mean square error (RMSE) were calculated for all test data, each organism, and each food category, as an index to evaluate the accuracy of the model. The R2 and RMSE values are given by Eqs 2, 3, respectively:
where
2.5 Two-dimensional (2D) plot visualization of bacterial behaviors
Using the developed model, the
We then compared our 2D color map with the data in the literature on growth/no-growth experiments, which was not recorded in ComBase. The data used for external validation were selected, considering that the experimental conditions are simple enough to describe bacterial behavior by eight explanatory variables. As a representative inactivation process, we cite a study of growth/no growth experiments of L. monocytogenes in broth (Koutsoumanis et al., 2004). The growth/no-growth L. monocytogenes were experimentally observed in a culture medium after 30 days at 25 °C (a), with pH 5.47–5.58 (b) and aw of 0.965–0.967 (c) after 30 days.
2.6 Interpretation of machine learning model
2.6.1 Feature importance
The feature importance was calculated to interpret the developed model from the process of model development. This allowed us to understand how each explanatory variable contributed to the predicted performance during the training of the XGBoost algorithm. The importance of the features was evaluated using gain, which is an index that shows the usefulness of a feature in constructing a tree-based model. A higher value indicates that the feature significantly affects the predicted
2.6.2 SHapley Additive eXplanations (SHAP) value
As another approach for model interpretation, we used the SHAP framework proposed by Lundberg and Lee (2017). SHAP is a new and flexible method that addresses the machine learning system as a so-called “black-box model” by providing an interpretation of how strongly the features affect the predicted outcome. Although the feature importance employed by XGBoost is positioned as the global explanation of the model, SHAP can directly measure a local feature explanation for a single sample, which could otherwise go unnoticed (Moncada-Torres et al., 2021). Because SHAP approaches are model-agnostic, they are used with various model types and in many fields of study (Lundberg and Lee, 2017; Agius et al., 2020; Mangalathu et al., 2020; Ndraha et al., 2021; Rodrigo et al., 2021; Yang and Liu, 2021; Zoabi et al., 2021).
The SHAP value for a single feature within a single sample describes the extent to which that feature contributes to the predicted output. A higher SHAP value indicates that a feature has a larger impact on the predicted
SHAP values were calculated using TreeSHAP (Lundberg et al., 2018), and a variant of SHAP, which was developed for tree-based machine learning models, such as XGBoost, as incorporated in the SHAP (Version 0.40.0) Python Package (https://shap.readthedocs.io/en/latest/index.html). All pre-processing steps, model development, and statistical analyses were performed using Python (version 3. 8. 12).
3 Results
3.1 Evaluation of model accuracy
By developing a single machine learning model, our data-mining approach meets in rough agreement with respect to prediction accuracy under various types of microorganisms and food categories. Figure 1 represents overall predicted results including all types of microorganisms and food categories, in which the R2 and RMSE values obtained were 0.76 and 1.23, respectively. The accuracy evaluated is consistently convincing compared with that of Hiura et al. (2021) (0.75 for R2 and 1.02 for RMSE, respectively). Followed by this, results divided by each microorganism, and by each food category are shown in Figures 2, 3, respectively. For each organism in Figure 2, the RMSE values were 1.35, 1.41, 1.42, 1.20, 1.42, 1.03, 1.07, and 1.08, for A. hydrophila, B. cereus, E. coli, L. monocytogenes, Pseudomonads, S. aureus, Salmonella, and Y. enterocolitica, respectively. In Figure 3, the RMSE values for the culture medium, beef, pork, poultry, sausage, eggs, seafood, milk, cheese, vegetables or fruits, bread, dessert food, beverage, water, and sauce/dressing were 1.21, 1.24, 1.29, 1.52, 1.43, 1.11, 1.06, 1.17, 1.51, 1.31, 0.60, 0.98, 1.29, 1.33, and 1.89, respectively. These results show that the developed model responds flexibly to various environmental conditions in different amounts of data. Note that the R2 and RMSE in sauce/dressing [Figure 3 (o)] are comparably worse than other organisms and food categories.
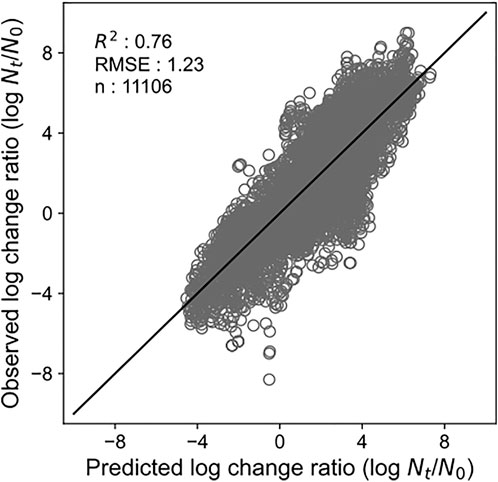
FIGURE 1. Comparison of the predicted and observed log change ratio for all the test data. The solid line represents residuals = 0.
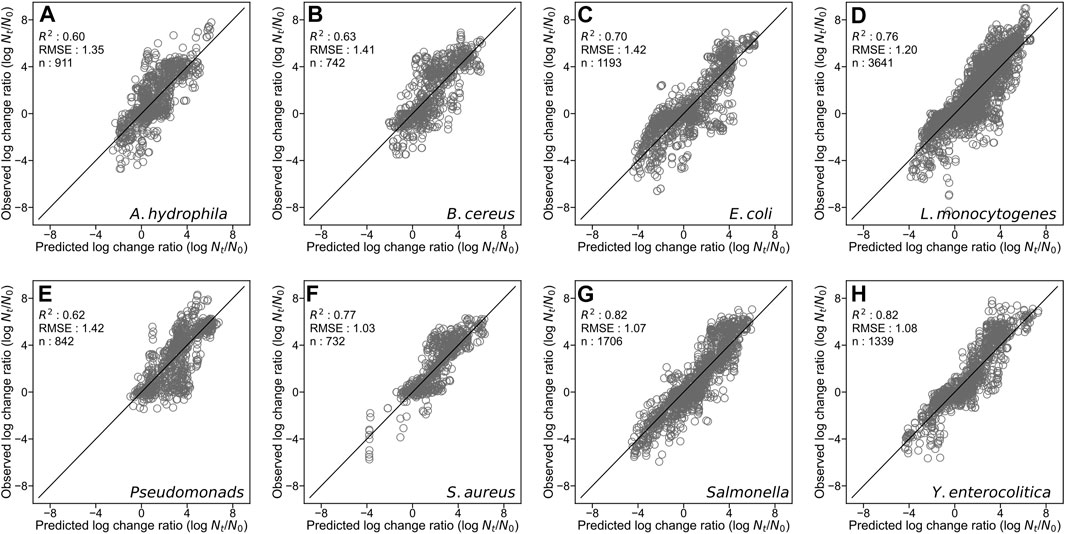
FIGURE 2. Comparison of the predicted and observed log change ratio for test data of Aeromonas hydrophila (A), Bacillus cereus (B), Escherichia coli (C), Listeria monocytogenes (D), Pseudomonads (E), Staphylococcus aureus (F), Salmonella (G), and Yersinia enterocolitica (H). The solid line represents residuals = 0.
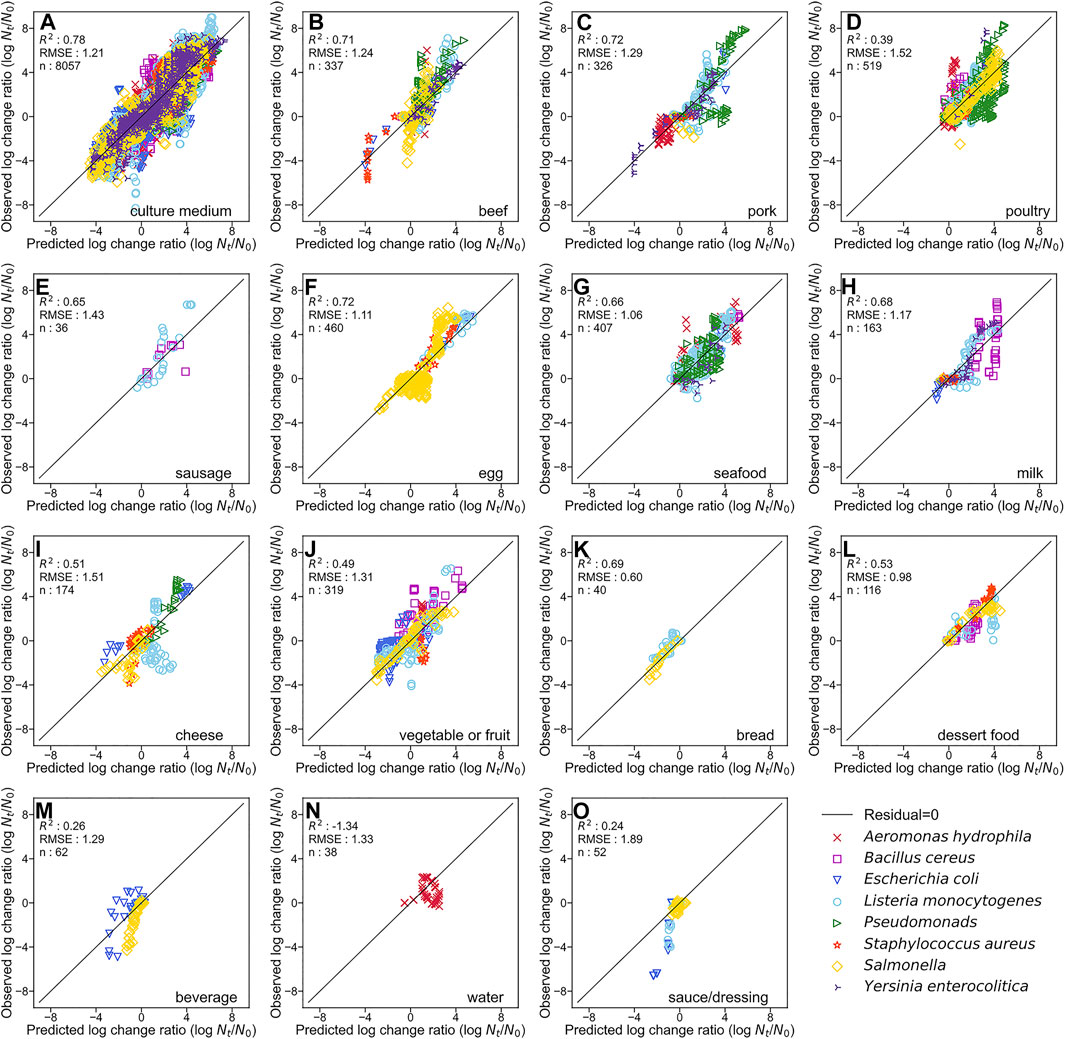
FIGURE 3. Comparison of the predicted and observed log change ratio for test data of culture medium (A), beef (B), pork (C), poultry (D), sausage (E), egg (F), seafood (G), milk (H), cheese (I), vegetable or fruit (J), bread (K), dessert food (L), beverage (M), water (N), and sauce/dressing (O). The solid line represents residuals = 0.
3.2 2D color plot visualization of bacterial behaviors
We introduced new 2D visualizations to illustrate the bacterial growth/survival ratio using combinations of temperature, pH, and aw. Figure 4 shows a color map of eight bacterial behaviors in the broth after 10 days when the initial number was 4 log CFU/g. The limiting aw value of growth for S. aureus was 0.90, whereas that for most microorganisms was an aw value of 0.91 or above. The minimum pH value of B. cereus for growth was estimated to be 5.0, whereas that of many other organisms was 4.0–4.5. All eight bacteria grew when the pH was greater than 5.5. Some examples of 2D color maps, such as temperature–aw and pH–temperature, can be found in the Supplementary Information. We compared the observed growth/no-growth experiment of L. monocytogenes in a culture medium at 25°C (Koutsoumanis et al., 2004) and our 2D map prediction. As shown in Figure 5, the validity of the 2D plots was visually confirmed.
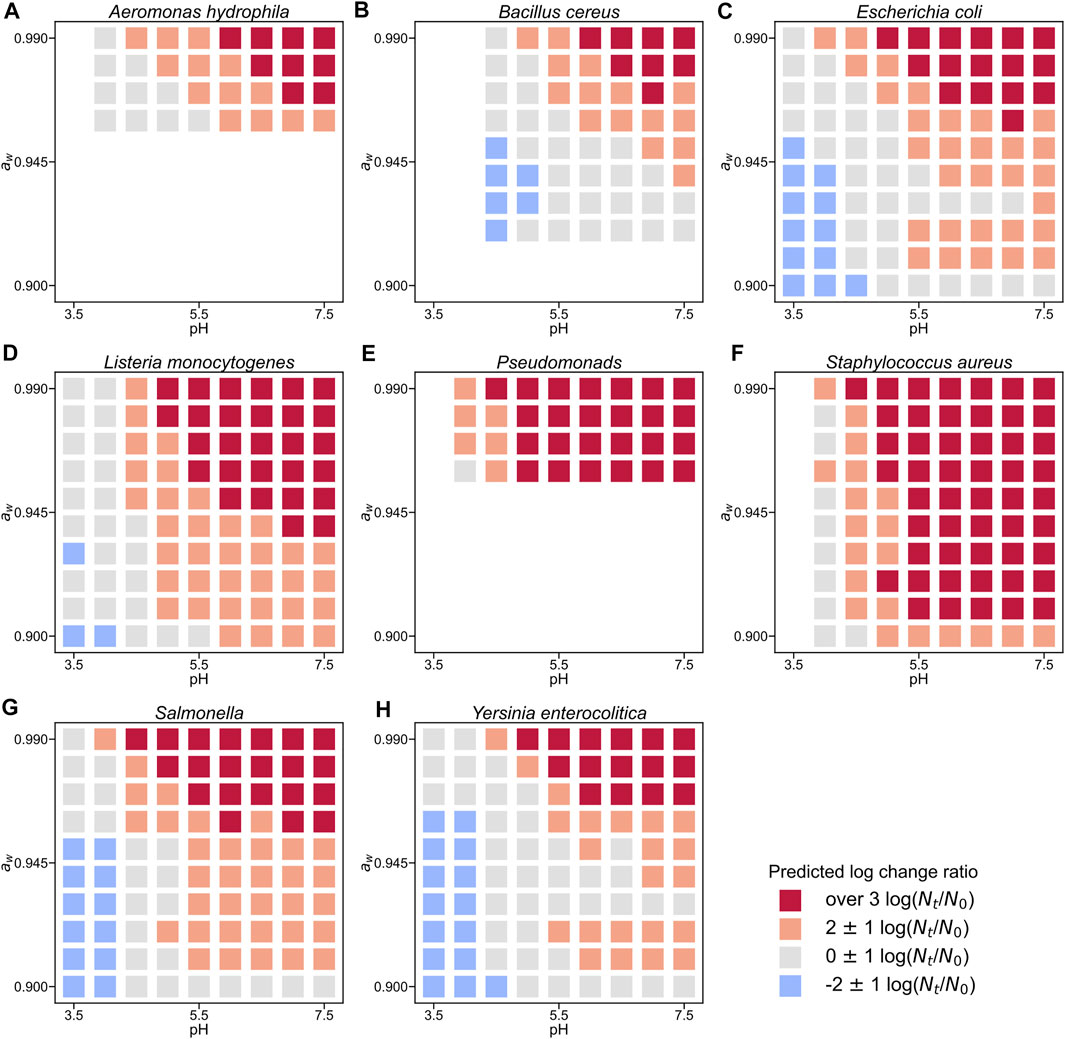
FIGURE 4. Change ratio from initial cell counts in broth at 20°C with an initial concentration of 4 log CFU/g after 10 days for Aeromonas hydrophila (A), Bacillus cereus (B), Escherichia coli (C), Listeria monocytogenes (D), Pseudomonads (E), Staphylococcus aureus (F), Salmonella (G), and Yersinia enterocolitica (H). Each square plot represents the value of the log change ratio (
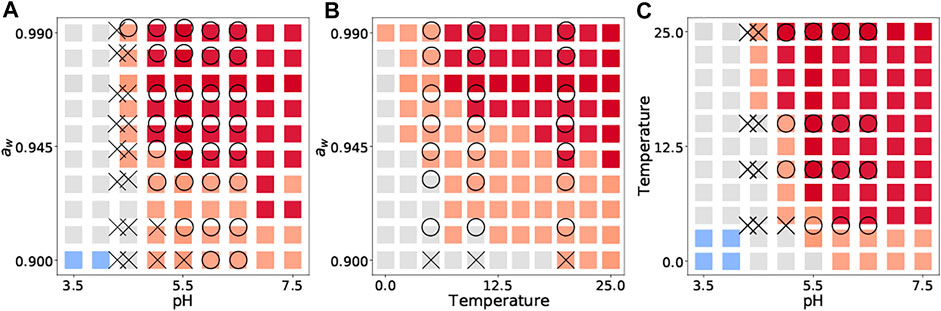
FIGURE 5. Comparison between observed growth (
3.3 Interpretation of the model
3.3.1 Feature importance
We calculated the feature importance to obtain the explanatory variable that was important in terms of contribution to the prediction performance in model development. Figure 6 shows the feature importance of the developed XGBoost model. The importance of each feature represents the ratio of the importance of each feature when the sum of all feature importance values is 1. “Initial cell number,” “Time,” and “aw” contributed the most to model development and to almost the same extent. A categorical variable “Organism” representing the name of bacteria contributed to model development mostly to the same extent as the numerical variables “pH” and “Temperature.” Information regarding food, such as food category and name, also contributed to model development. All features contributed to the model development to some extent.
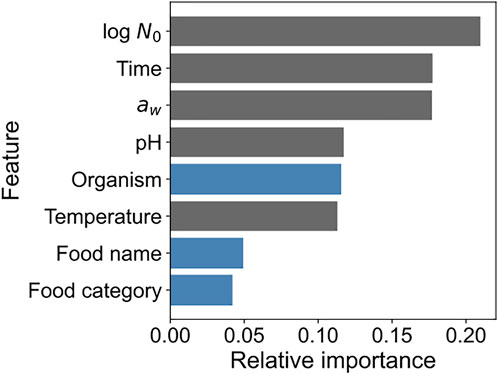
FIGURE 6. Feature importance of the developed XGBoost model. The x-axis indicates the relative importance, and the y-axis indicates the feature names. Blue and gray bars indicate categorical and numerical variables, respectively.
3.3.2 SHAP value
To see the model interpretability in a deeper perspective (i.e., the relationship between each environmental condition and the bacterial growth), we introduced the SHAP framework. The SHAP values for the three environmental features were calculated to determine the contribution of the environmental factors to bacterial growth. The SHAP value explains the contribution of each variable to the predicted
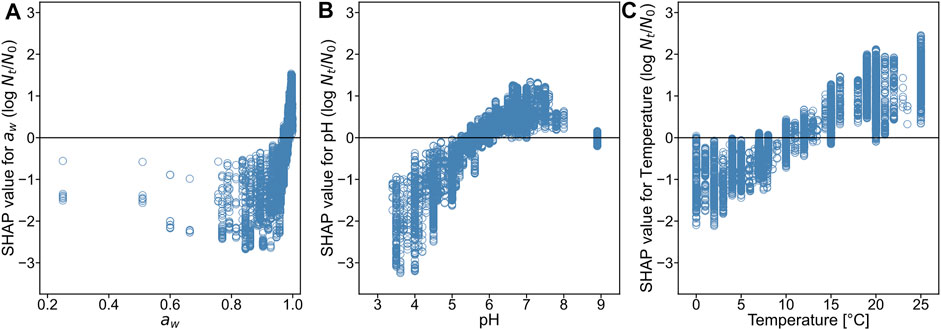
FIGURE 7. SHapley Additive eXplanations (SHAP) dependency plots for water activity (A), pH (B), and temperature (C).
4 Discussion
In the present study, we demonstrated the application of a data mining approach to predict bacterial population behavior using the ComBase database (Figures 1–3) and visualized these as 2D maps (Figure 4). Categorical data such as organism, food category, and food name also contributed to the construction of the model to some extent in the developed model (Figure 6). In addition, we demonstrated the environmental effects on the growth of the bacterial population (Figure 7). The data mining approach allowed us to model and reveal the multidimensional relationship between bacterial population behavior and the food environment. We showed that a data-driven approach to analyzing accumulated data could be useful for addressing food safety issues.
Although the dataset used in this study consisted of numerical and categorical variables, using a machine learning algorithm enabled us to predict bacterial population behavior using a single predictive model (Figure 1). Unlike numerical variables, categorical variables (e.g., food category, food name, and organism) must be replaced with numerical data, such as dummy variables for numerical operations (Palaniappan and Awang, 2008; Kim and Hong, 2017). Statistical modeling makes it difficult to consider categorical data when multiple conditions exist, such as food names and environmental conditions (Kim and Hong, 2017; Hiura et al., 2021). In contrast, the machine learning model enabled the description of the relationship between the food environment and bacteria, which is difficult for statistical models or human hand definition owing to the high dimensionality. Additionally, we succeeded in extending the model proposed by Hiura et al. (2021) to include eight bacterial species and 15 food categories.
We visualized the bacterial population behavior based on the idea of panoramic evaluation of the whole trend of the microbial response to various conditions. Our 2D map visualization showed a combination of factors that prevented bacterial growth (Figure 4). Compared to the literature data, our 2D color map can describe trends of population behavior of Listeria monocytogenes to the same extent (Figure 5), which supports the validity of our color map. Similar to our study, Ratkowsky & Ross (1995) proposed a growth/no-growth interface model. The growth/no growth interface models estimate the probability of bacterial growth and find combinations of factors preventing growth. The growth/no growth interface has been widely used in previous studies in predictive microbiology (Tienungoon et al., 2000; McKellar and Lu, 2001; Le Marc et al., 2005; Polese et al., 2011; Coroller et al., 2012; Kuroda et al., 2019). This approach was used to determine whether bacteria can grow easily under a wide range of experimental conditions. However, this interface cannot express the details of the bacterial population density. In the present study, we succeeded in evaluating not only whether there was growth or not, but also the change in bacterial population density (Figure 4). Our visualizing method helps us understand the bacterial concentration in various conditions at glance. Our visualization methods can be useful for developing processes that provide information for realistic estimations of food safety risks. Thus, our 2D map models are important for the dissemination of food safety regulations.
The SHAP value describes the contribution of each explanatory variable to each predicted
Although our study uses data-driven methods to analyze the experimental data in the ComBase database with some advantages and expectations, it also has some limitations. Our model is not assumed to predict the ecology of microorganisms, because the ComBase database mainly focuses on bacterial growth and inactivation of only one type of bacterial species for each experiment for simplification. In the future, the competitive microbial condition would be analyzed with the dataset containing the ecology of microorganisms by data driven methods.
5 Conclusion
Data mining predicted the population behavior of eight foodborne pathogens and spoilage bacteria in the 15 food environments. In addition, growth inhibition owing to the food environment was quantitatively evaluated using data-driven methods. Our approach enabled us to extract useful information regarding food safety from a large amount of experimental data. The bacterial population behavior predicted by this procedure can provide guidelines for determining food processing and storage conditions. The main findings of this study support the data mining approach as valuable in the field of food microbiology.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
JH, JS, SN, SK, and KK conceptualized the study. JH, JS, SN, and KK designed the computation. JH and JS analyzed the data. JH wrote the python script and the first draft of the manuscript. All authors reviewed the manuscript.
Funding
This work was also supported by Kieikai Research Foundation and Tobe Maki scholarship Foundation (to KK), the Japan Science and Technology Agency Center of Innovation Program (grant number: JPMJCE1301), CREST (JPMJCR20H4), and JSPS KAKENHI (JP20H00425, JP21K19813) (to SN).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frfst.2022.979028/full#supplementary-material
References
Adams, M. R., and Nicolaides, L. (1997). Review of the sensitivity of different foodborne pathogens to fermentation. Food control. 8 (5–6), 227–239. doi:10.1016/s0956-7135(97)00016-9
Agius, R., Brieghel, C., Andersen, M. A., Pearson, A. T., Ledergerber, B., Cozzi-Lepri, A., et al. (2020). Machine learning can identify newly diagnosed patients with CLL at high risk of infection. Nat. Commun. 11 (1), 363–417. doi:10.1038/s41467-019-14225-8
Baranyi, J., Tamplin, M., and Ross, T. (2004). The ComBase initiative. Microbiol. Aust. 25 (3), 32. doi:10.1071/ma04332
Bidlas, E., Du, T., and Lambert, R. J. W. (2008). An explanation for the effect of inoculum size on MIC and the growth/no growth interface. Int. J. Food Microbiol. 126 (1–2), 140–152. doi:10.1016/j.ijfoodmicro.2008.05.023
Chang, Y. C., Chang, K. H., and Wu, G. J. (2018). Application of eXtreme gradient boosting trees in the construction of credit risk assessment models for financial institutions. Appl. Soft Comput. 73, 914–920. doi:10.1016/j.asoc.2018.09.029
Chen, T., and Guestrin, C. (2016). XGBoost: A scalable tree boosting system. Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM, New York, NY, USA. 2016, 785–794. doi:10.1145/2939672.2939785
Cios, K. J., and William Moore, G. (2002). Uniqueness of medical data mining. Artif. Intell. Med. 26 (1–2), 1–24. doi:10.1016/S0933-3657(02)00049-0
Coroller, L., Kan-King-Yu, D., Leguerinel, I., Mafart, P., and Membre, J. M. (2012). Modelling of growth, growth/no-growth interface and nonthermal inactivation areas of Listeria in foods. Int. J. Food Microbiol. 152 (3), 139–152. doi:10.1016/j.ijfoodmicro.2011.09.023
Cortez, P., Cerdeira, A., Almeida, F., Matos, T., and Reis, J. (2009). Modeling wine preferences by data mining from physicochemical properties. Decis. Support Syst. 47 (4), 547–553. doi:10.1016/j.dss.2009.05.016
Delen, D., Walker, G., and Kadam, A. (2005). Predicting breast cancer survivability: A comparison of three data mining methods. Artif. Intell. Med. 34 (2), 113–127. doi:10.1016/j.artmed.2004.07.002
den Besten, H. M. W., and Zwietering, M. H. (2012). Meta-analysis for quantitative microbiological risk assessments and benchmarking data. Trends Food Sci. Technol., 34–39. doi:10.1016/j.tifs.2011.12.004
Deringer, V. L., Bartok, A. P., Bernstein, N., Wilkins, D. M., Ceriotti, M., and Csanyi, G. (2021). Gaussian process regression for materials and molecules. Chem. Rev. 121, 10073–10141. doi:10.1021/acs.chemrev.1c00022
Doyle, M. P., Diez-Gonzalez, F., and Hill, C. (2019). Food microbiology: Fundamentals and frontiers. Food Microbiol. Fundam. Front. doi:10.1002/9781683670476
Friedman, J. H. (2001). Greedy function approximation: A gradient boosting machine. Ann. Stat. 29 (5), 1189–1232. doi:10.1214/aos/1013203451
Gould, G. W. (1996). Methods for preservation and extension of shelf life. Int. J. Food Microbiol. 33 (1), 51–64. doi:10.1016/0168-1605(96)01133-6
Gulyaeva, M., Huettmann, F., Shestopalov, A., Okamatsu, M., Matsuno, K., Chu, D. H., et al. (2020). Data mining and model-predicting a global disease reservoir for low-pathogenic Avian Influenza (A) in the wider Pacific rim using big data sets. Sci. Rep. 10 (1), 16817–16911. doi:10.1038/s41598-020-73664-2
Hand, D. J. (1998). Data mining: Statistics and more? Am. Stat. 52 (2), 112–118. doi:10.1080/00031305.1998.10480549
Hiura, S., Koseki, S, and Koyama, K. (2021). Prediction of population behavior of Listeria monocytogenes in food using machine learning and a microbial growth and survival database. Sci. Rep. 11 (1), 10613. doi:10.1038/s41598-021-90164-z
Hochachka, W. M., Caruana, R., Fink, D., Munson, A., Riedewald, M., Sorokina, D., et al. (2007). Data-mining discovery of pattern and process in ecological systems. J. Wildl. Manage. 71 (7), 2427. doi:10.2193/2006-503
Jagannath, A., and Tsuchido, T. (2003). ‘Predictive microbiology: A review’, biocontrol science. Japan: The Society for Antibacterial and Antifungal Agents, 1–7. doi:10.4265/bio.8.1
Jay, J. M., Loessner, M. J., and Golden, D. A. (2008a). “Intrinsic and extrinsic parameters of foods that affect microbial growth,” in Modern food microbiology (Springer US), 39–59. doi:10.1007/0-387-23413-6_3
Jay, J. M., Loessner, M. J., and Golden, D. A. (2008b). “Protection of foods with low-temperatures, and characteristics of psychrotrophic microorganisms,” in Modern food microbiology (Springer US), 395–413. doi:10.1007/0-387-23413-6_16
Jiménez-Carvelo, A. M., et al. (2019). Alternative data mining/machine learning methods for the analytical evaluation of food quality and authenticity – a review. Food Res. Int. Elsevier Ltd, 25–39. doi:10.1016/j.foodres.2019.03.063
Kim, K., Hong, J. S, Yun, K. Y., Han, S. E., Kim, E. S., Kwon, B. S., et al. (2017). Laparoscopically assisted suprapubic surgery for adnexal tumors under epidural anesthesia. Minim. Invasive Ther. Allied Technol. 98, 39–43. doi:10.1080/13645706.2016.1223695
Koh, H. C., and Tan, G. (2005). Data mining applications in healthcare. J. Healthc. Inf. Manag. 19 (2), 64–72. doi:10.4314/ijonas.v5i1.49926
Koutsoumanis, K. P., Kendall, P. A., and Sofos, J. N. (2004). A comparative study on growth limits of Listeria monocytogenes as affected by temperature, pH and aw when grown in suspension or on a solid surface. Food Microbiol. 21 (4), 415–422. doi:10.1016/j.fm.2003.11.003
Koutsoumanis, K. P., and Sofos, J. N. (2005). Effect of inoculum size on the combined temperature, pH and aw limits for growth of Listeria monocytogenes. Int. J. Food Microbiol. 104 (1), 83–91. doi:10.1016/j.ijfoodmicro.2005.01.010
Kuroda, S., Okuda, H., Ishida, W., and Koseki, S. (2019). Modeling growth limits of Bacillus spp. spores by using deep-learning algorithm. Food Microbiol. 78, 38–45. doi:10.1016/j.fm.2018.09.013
Le Marc, Y., Pin, C., and Baranyi, J. (2005). “Methods to determine the growth domain in a multidimensional environmental space,” in International journal of food microbiology (Elsevier), 3–12. doi:10.1016/j.ijfoodmicro.2004.10.003
Leistner, L. (2000). “Basic aspects of food preservation by hurdle technology,” in International journal of food microbiology (Elsevier), 181–186. doi:10.1016/S0168-1605(00)00161-6
Lundberg, S. M., Erion, G. G., and Lee, S.-I. (2018). Consistent individualized feature attribution for tree ensembles. Available at:https://arxiv.org/abs/1802.03888v3 (Accessed: January 21, 2022).
Lundberg, S. M., and Lee, S. I. (2017). “A unified approach to interpreting model predictions,” in Advances in neural information processing systems, 4766–4775. Available at:https://github.com/slundberg/shap (Accessed: September 28, 2021).
Mangalathu, S., Hwang, S. H., and Jeon, J. S. (2020). Failure mode and effects analysis of RC members based on machine-learning-based SHapley Additive exPlanations (SHAP) approach. Eng. Struct. 219, 110927. doi:10.1016/j.engstruct.2020.110927
Mataragas, M., Rantsioua, K., Alessandria, V., and CocoLin, L. (2015). Estimating the non-thermal inactivation of Listeria monocytogenes in fermented sausages relative to temperature, pH and water activity. Meat Sci. 100, 171–178. doi:10.1016/j.meatsci.2014.10.016
McKellar, R. C., and Lu, X. (2001). A probability of growth model for Escherichia coli O157:H7 as a function of temperature, pH, acetic acid, and salt. J. Food Prot. 64 (12), 1922–1928. doi:10.4315/0362-028X-64.12.1922
McQuestin, O. J., Shadbolt, C. T., and Ross, T. (2009). Quantification of the relative effects of temperature, pH, and water activity on inactivation of Escherichia coli in fermented meat by meta-analysis. Appl. Environ. Microbiol. 75 (22), 6963–6972. doi:10.1128/AEM.00291-09
Mohanty, S. D., Lekan, D., McCoy, T. P., Jenkins, M., and Manda, P. (2022). Machine learning for predicting readmission risk among the frail: Explainable AI for healthcare. Patterns 3 (1), 100395. doi:10.1016/j.patter.2021.100395
Moncada-Torres, A., van Maaren, M. C., Hendriks, M. P., Siesling, S., and Geleijnse, G. (2021). Explainable machine learning can outperform Cox regression predictions and provide insights in breast cancer survival. Sci. Rep. 11 (1), 6968–7013. doi:10.1038/s41598-021-86327-7
Ndraha, N., Hsiao, H. I., Hsieh, Y. Z., and Pradhan, A. K. (2021). Predictive models for the effect of environmental factors on the abundance of Vibrio parahaemolyticus in oyster farms in Taiwan using extreme gradient boosting. Food control. 130, 108353. doi:10.1016/j.foodcont.2021.108353
Nguyen, M., Long, S. W., McDermott, P. F., Olsen, R. J., Olson, R., Stevens, R. L., et al. (2019). Using machine learning to predict antimicrobial MICs and associated genomic features for nontyphoidal Salmonella. J. Clin. Microbiol. 57 (2), 012600–e1318. doi:10.1128/JCM.01260-18
Nychas, G.-J., Sims, E., Tsakanikas, P., and Mohareb, F. (2021). Data science in the food industry. Annu. Rev. Biomed. Data Sci. 4 (1), 341–367. doi:10.1146/annurev-biodatasci-020221-123602
Palaniappan, S., and Awang, R. (2008). Intelligent heart disease prediction system using data mining techniques. AICCSA 08 - 6th IEEE/ACS International Conference on Computer Systems and Applications. IEEE, Doha, Qatar 108–115. doi:10.1109/AICCSA.2008.449352431 March 2008 - 04 April 2008
Polese, P., Del Torre, M., Spaziani, M., and Stecchini, M. L. (2011). A simplified approach for modelling the bacterial growth/no growth boundary. Food Microbiol. 28 (3), 384–391. doi:10.1016/j.fm.2010.09.011
Rodrigo, H., Beukes, E. W., Andersson, G., and Manchaiah, V. (2021). Exploratory data mining techniques (decision tree models) for examining the impact of internet-based cognitive behavioral therapy for tinnitus: Machine learning approach. J. Med. Internet Res. 23 (11), e28999. doi:10.2196/28999
Ross, S. R. P. J., Friedman, N. R., Dudley, K. L., Yoshimura, M., Yoshida, T., and Economo, E. P. (2018). Listening to ecosystems: Data-rich acoustic monitoring through landscape-scale sensor networks. Ecol. Res. 33 (1), 135–147. doi:10.1007/s11284-017-1509-5
Ross, T., and McMeekin, T. A. (1994). Predictive microbiology. Int. J. Food Microbiol. 23 (3–4), 241–264. doi:10.1016/0168-1605(94)90155-4
Shehadeh, A., Alshboul, O., Al Mamlook, R. E., and Hamedat, O. (2021). Machine learning models for predicting the residual value of heavy construction equipment: An evaluation of modified decision tree, LightGBM, and XGBoost regression. Automation Constr. 129, 103827. doi:10.1016/j.autcon.2021.103827
Skandamis, P. N., Stopforth, J. D., Kendall, P. A., Belk, K. E., Scanga, J. A., Smith, G. C., et al. (2007). Modeling the effect of inoculum size and acid adaptation on growth/no growth interface of Escherichia coli O157:H7. Int. J. Food Microbiol. 120 (3), 237–249. doi:10.1016/j.ijfoodmicro.2007.08.028
Tienungoon, S., Ratkowsky, D. A., McMeekin, T. A., and Ross, T. (2000). Growth limits of Listeria monocytogenes as a function of temperature, pH, NaCl, and lactic acid. Appl. Environ. Microbiol. 66 (11), 4979–4987. doi:10.1128/AEM.66.11.4979-4987.2000
Wang, L., Wang, X., Chen, A., Jin, X., and Che, H. (2020). Prediction of type 2 diabetes risk and its effect evaluation based on the xgboost model. Healthc. Switz. 8 (3), 247. doi:10.3390/healthcare8030247
Yang, S., and Liu, M. (2021). Mining meta-indicators of university ranking: A machine learning approach based on SHAP. Available at: https://arxiv.org/abs/2111.12526v1 (Accessed: January 7, 2022).
Keywords: databases, predictive microbiology, data-driven methods, SHapley Additive, population behavior
Citation: Hosoe J, Sunagawa J, Nakaoka S, Koseki S and Koyama K (2022) Data mining for prediction and interpretation of bacterial population behavior in food. Front. Food. Sci. Technol. 2:979028. doi: 10.3389/frfst.2022.979028
Received: 27 June 2022; Accepted: 05 December 2022;
Published: 15 December 2022.
Edited by:
Gopalan Sivaraman, Central Institute of Fisheries Technology (ICAR), IndiaReviewed by:
Qingli Dong, University of Shanghai for Science and Technology, ChinaDonald W. Schaffner, Rutgers, The State University of New Jersey, United States
Copyright © 2022 Hosoe, Sunagawa, Nakaoka, Koseki and Koyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kento Koyama, a2tveWFtYUBhZ3IuaG9rdWRhaS5hYy5qcA==
†These authors have contributed equally to this work
 Junpei Hosoe
Junpei Hosoe Junya Sunagawa
Junya Sunagawa Shinji Nakaoka
Shinji Nakaoka Shige Koseki
Shige Koseki Kento Koyama
Kento Koyama