MHC-II presentation by oral Langerhans cells impacts intraepithelial Tc17 abundance and Candida albicans oral infection via CD4 T cells
- 1Division of Basic Sciences, Department of Diagnostic and Biological Sciences, School of Dentistry, University of Minnesota, Minneapolis, MN, United States
- 2Division of Periodontology, Department of Developmental and Surgical Sciences, School of Dentistry, University of Minnesota, Minneapolis, MN, United States
In a murine model (LCΔMHC-II) designed to abolish MHC-II expression in Langerhans cells (LCs), ∼18% of oral LCs retain MHC-II, yet oral mucosal CD4 T cells numbers are unaffected. In LCΔMHC-II mice, we now show that oral intraepithelial conventional CD8αβ T cell numbers expand 30-fold. Antibody-mediated ablation of CD4 T cells in wild-type mice also resulted in CD8αβ T cell expansion in the oral mucosa. Therefore, we hypothesize that MHC class II molecules uniquely expressed on Langerhans cells mediate the suppression of intraepithelial resident-memory CD8 T cell numbers via a CD4 T cell-dependent mechanism. The expanded oral CD8 T cells co-expressed CD69 and CD103 and the majority produced IL-17A [CD8 T cytotoxic (Tc)17 cells] with a minority expressing IFN-γ (Tc1 cells). These oral CD8 T cells showed broad T cell receptor Vβ gene usage indicating responsiveness to diverse oral antigens. Generally supporting Tc17 cells, transforming growth factor-β1 (TGF-β1) increased 4-fold in the oral mucosa. Surprisingly, blocking TGF-β1 signaling with the TGF-R1 kinase inhibitor, LY364947, did not reduce Tc17 or Tc1 numbers. Nonetheless, LY364947 increased γδ T cell numbers and decreased CD49a expression on Tc1 cells. Although IL-17A-expressing γδ T cells were reduced by 30%, LCΔMHC-II mice displayed greater resistance to Candida albicans in early stages of oral infection. These findings suggest that modulating MHC-II expression in oral LC may be an effective strategy against fungal infections at mucosal surfaces counteracted by IL-17A-dependent mechanisms.
Introduction
The gut, skin, and oral cavity are major physical and immunological barriers to infection by pathogens and sites of immune encounters with beneficial microbiota. These barrier tissues have distinct assemblages of immune cells each adapted to respond in an appropriate manner to site-specific challenges. The stratified squamous epithelia of the skin and oral mucosa contain a unique population of innate immune cells called Langerhans cells (LCs), which capture antigens on major histocompatibility complex class II molecules (MHC-II) and then present to antigen-experienced CD4 T cells locally and to naïve CD4 T cells in draining lymph nodes (1, 2). LCs also direct CD4 T helper 17 (Th17) responses against extracellular pathogens (3, 4) and, potentially regulate specific immunity (5, 6). Although skin-resident and oral LCs occupy a similar niche in stratified epithelial barriers, have similar transcriptomic profiles and immunologic functions, they have distinctive ontogenetic differences (2, 7). Skin-resident LCs are involved in many skin disorders and contribute actively to defense against cutaneous infections with Candida albicans (1, 3).
Our understanding of the acute immune response to C. albicans comes mainly from murine studies (8, 9). Mice lacking T cell receptors appear highly susceptible to C. albicans infection implicating CD4 and/or CD8 T cells in facilitating resistance (8). In humans, C. albicans-specific memory T cells are largely Th17, and in mice IL-17A is critical for C. albicans clearance (9–11). IL-17A stimulates epithelial cells to produce antimicrobial peptides (12) and the neutrophil-recruiting chemokines C-X-C Motif Chemokine Ligand 8 (CXCL8) and granulocyte-monocyte colony stimulating factor (GM-CSF) (13). In mouse models of oral candidiasis, Th17 cells control initiation of local infection while Th1 cells prevent systemic spread (11, 14, 15). In addition to Th17 cells, innate cells of lymphoid origin can produce IL-17A, including γδ T, natural killer T, innate lymphoid cell type 3, TCR-β+ “natural” Th17 cells, and a population of major histocompatibility complex class I (MHC-I)-restricted CD8 T cells (Tc17) (8, 16). Tc17 cells produce reduced amounts of prototypical IFN-γ and cytotoxic molecules (17) and function as a potential novel source of IL-17A in the presence of low numbers of Th17 cells.
The ontogeny of Tc17 cells in vivo is not yet fully understood, but in vitro work suggests the critical importance of TGF-β and IL-6 in the differentiation of naïve CD8 T cells into Tc17 cells (17). For example, allochthonous microorganisms entering the cutaneous microbial flora increase Tc17 cell abundance, conferring IL-17A-dependent protection against C. albicans in a skin infection model (18). Moreover, in a CD4−/− mouse model that mimics the depressed CD4 T cell status of HIV-positive individuals, Tc17 cells were numerous and helped confer resistance to C. albicans in an oropharyngeal candidiasis infection model (19). In the oral epithelial lining, therefore, Tc17 cells appear to deliver potent anti-fungal immunity by providing compensatory IL-17A in immunocompromised CD4 T cell-deficient individuals. Interestingly, cultured Tc17 cells have a central memory phenotype and become plastic, shifting into the prototypical cytotoxic type 1 CD8 T cells (Tc1) when adoptively transferred and subsequently challenged in vivo (20).
In both humans and mice, oral dendritic cell subsets in direct innate and memory immune responses against C. albicans (16). These cell populations may contribute to defense against opportunistic oropharyngeal candidiasis affecting the oral mucosa, tongue, and the lining of the esophagus. In the development of a protective mucosal Tc17 population against C. albicans infection, the roles of mucosal antigen presenting cells including conventional dendritic cells, subepithelial langerin+ dendritic cells, and oral intraepithelial LCs have yet to be defined. LCs drive antigen-specific Th17 responses against C. albicans in the skin and against a periodontal pathogen in the oral cavity (3, 4). We hypothesized that MHC class II molecules uniquely expressed on LCs mediate the suppression of intraepithelial resident-memory CD8 T cell numbers that primarily express IL-17A to affect the susceptibility to C. albicans infection. Functional redundancy seems to occur in oral LC antigen presentation. An 80% reduction in cells expressing MHC-II has no impact on the development of an oral Th17 response in either specific-pathogen free conditions or in a ligature-induced periodontitis model (21). Interestingly, reduced numbers of MHC-II-expressing oral LCs increased oral CD8 T cells by a striking 30-fold (21). To better understand their function, we further characterize these CD8 T cells to be IL-17A-expressing Tc17 cells with a resident-memory phenotype localized within the suprabasal layer of the oral mucosa epithelium. This dramatic increase in oral Tc17 numbers regulated by MHC class II expression on LCs translated into increased protection against oropharyngeal candidiasis induced by C. albicans infection.
Materials and methods
Mice
Mice were housed in AAALAC approved specific-pathogen free conditions at the Animal Facilities of the University of Minnesota and their use approved by the Institutional Animal Care and Use Committee (Protocol 2110-39519A). All mice used are on the C57BL/6J genetic background. Generation of LCWT, LCΔMHC-II and IL-17ACre-RFP reporter mice have been described elsewhere (21, 22). A heterozygous breeding strategy was used to produce sibling experimental LCWT and LCΔMHC-II animals and genotypes were confirmed using PCR to detect segregation of the huLang-Cre transgene amongst littermates (21). Experiments were performed on age-matched (8–12 week) mice of both sexes unless specifically stated otherwise. Mice were co-housed in microisolator cages with a 12-h day/night cycle, standard chow, and water ad libitum.
Preparation of single cell suspensions from mouse tissues and flow cytometry
Mice were routinely injected retroorbitally with 1.25 µg of rat anti-mouse CD45 mAb conjugated to FITC (BioLegend; clone 30-F11) 3 min prior to euthanasia to discriminate blood-resident immune cells from interstitial immune cells as previously described (23).
Oral mucosa was harvested as previously described, judiciously avoiding potential nasal associated lymphoid tissue contamination (23). In some experiments, oral mucosa was subsequently treated with Dispase II to separate the epithelium from the underlying lamina propria and tissues processed separately (21). Briefly, oral mucosa was incubated in 2.2 U/ml Dispase II (Roche) at 37 °C with gentle agitation for 50 min. Fine tipped jewelry tweezers were then used to peel away epithelial sheets from the lamina propria. Minced oral mucosa/lamina propria and epithelial sheets were further treated with collagenase D (Roche) and DNase I (Sigma-Aldrich) before processing tissues to obtain single cell suspensions for flow cytometry as previously described (23).
A scalpel was used to manually remove and separate the dermis and epidermis from both the dorsal and ventral surfaces of the left ear for skin samples. Excised lungs were first rinsed in ice cold PBS. Skin and lung specimens were finely minced and then digested with collagenase D (2 mg/ml) and DNase I (1 mg/ml) at 37 °C for 60 min before processing to obtain single cell suspensions using standard procedures.
Cervical lymph nodes and spleen were harvested and processed to produce single cell suspensions using standard techniques as previously described (23). Single cell suspensions in all flow cytometry experiments were first stained with viability dye Zombie Aqua (BioLegend) followed by incubation with rat anti-mouse CD16/CD32 mAb (eBioscience; clone 93) to block Fc receptors. Cells were then incubated with combinations of the following rat/hamster anti-mouse fluorochrome-conjugated mAbs (BioLegend): CD45 (clone 30-F11), CD3 (clone 17A2), CD4 (clone RM4-5), CD8α (clone 53-6.7), CD8β (clone YTS156.7.7), CD90.2 (clone 30-H11), TCRβ (clone H57-597), TCRγ/δ (clone GL3), CD25 (eBioscience; clone PC61), CD44 (clone IM7), CD69 (clone H1.2F3), CD103 (clone 2E7), CD49a (clone HMa1). In experiments to identify potential mucosal-associated invariant T (MAIT) cells, single cell suspensions were additionally stained with major histocompatibility complex (MHC) class I-related protein 1 (MR1) tetramers, MR1 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) or MR1 6-formyl pterin (6-FP) purchased from the NIH tetramer facility based at Emory University, Atlanta, Georgia. Staining was performed in accordance with NIH tetramer facility instructions. In flow cytometry experiments using mucosal tissue, cells were pre-filtered through 70 µm cell strainers to remove cell clumps/debris. All cells were acquired on an LSR II flow cytometer (BD Biosciences) equipped with 4 lasers. Fluorescence emissions were analyzed using FlowJo 10.7.1 (Tree Star). To account for sample loss during mAb staining and/or variability in processed tissue size, immune cell populations of interest were normalized to 100,000 live non-immune cells when appropriate.
Depletion of CD4 T cells
Eight-10 week old IL-17ACre-RFP reporter mice were injected i.p. with 200 µg anti-CD4 mAb (BioXCell; clone GK1.5) or Rat IgG2b κ control (BioXCell; #BE0090) 4 times, 5 days apart. On day 20, mice were injected with FITC-labeled anti-CD45 mAb, sacrificed 3 min later, and oral mucosal tissue processed to generate single cell suspensions as described above. As a tissue lacking LCs (control tissue), the left lung was perfused with PBS, minced, treated with collagenase D and DNAse I and the resulting single cell suspensions further treated with ACK (Lonza) to lyse red blood cells. All single cell suspensions were subsequently stained with mAbs to identify CD4 T cells and CD8 T cells by flow cytometry as described above.
T cell phenotyping and TCR Vβ usage
To determine the phenotype of T cells, single cell suspensions were cultured in complete EHAA (Irvine Scientific) at 37 °C and polyclonally stimulated for 6 h with PMA-Ionomycin in the presence of brefeldin A as previously described (23). Cells were then incubated with Zombie Aqua, anti-mouse CD16/CD32 antibody and surface stained with mAbs as described above. Cells were fixed and made permeable using BD Cytoperm/Cytofix Plus kit reagents according to the manufacturer's instructions (BD Biosciences). Permeabilized cells were subsequently stained with anti-mouse IL-17A (eBioscience; clone eBio17B7) and IFN-γ (BioLegend; clone XMG1.2) fluorochrome-conjugated mAbs.
TCR Vβ gene usage was determined using a mouse Vβ TCR screening panel kit that targets the use of 17 different TCR Vβ alleles (BD Biosciences). Oral mucosa from 12 LCΔMHC-II mice was harvested, treated with Dispase II to separate the epithelium from the underlying lamina propria and single cell suspensions produced from pooled epithelial sheets. Cells were stained with mAbs to specifically identify CD4 T and CD8 T cells, then washed and stained with FITC-conjugated mAbs that recognize the 16 different TCRs in C57BL/6 mice following the manufacturer's instructions. In some experiments, cells were additionally stained with rat anti-mouse CD49a mAb prior to use of the Vβ TCR panel. Since the Vβ 17a gene is not expressed in the C57BL/6J mouse, the anti-Vβ 17a mAb can be viewed as a negative staining control.
Preparation of oral mucosa and epithelial sheets from tongue for confocal microscopy
Oral mucosal tissue was harvested as previously described (23), subsequently embedded in O.C.T. medium (Sakura Finetek) and 10 µm frozen cryostat sections cut and fixed with 4% paraformaldehyde. Sections were briefly re-hydrated in PBS and then blocked with PBS supplemented with 5% (v/v) rat serum and 5% (v/v) FBS for 60 min at room temperature and then incubated overnight at 4°C with 10 µg/ml Alexa Fluor 647 anti-mouse CD8α mAb (BioLegend; clone 53-6.7) in PBS 5% (v/v) rat serum. After two washes with PBS 0.1% (v/v) Tween 20, sections were counterstained with DAPI (Life Technologies) and imaged at 10–40× using a LSM 700 Zeiss confocal laser scanning microscope (Carl Zeiss).
Tongues were excised and much of the underlying musculature removed and discarded. Processed tongues were then treated with 2.2 U/ml Dispase II (Roche) for 50 min at 37 °C to allow the epithelium to be detached as a sheet from the remaining muscle. Epithelial sheets were fixed with 4% paraformaldehyde, re-hydrated in PBS, and then blocked with PBS supplemented with 5% (v/v) rat serum, 5% (v/v) FBS, and 0.1% (v/v) Tween 20 for 60 min at room temperature. Epithelial sheets were then incubated overnight at 4 °C with 10 µg/ml Alexa Fluor 647 anti-mouse/human CD207 mAb (Dendritics; clone 929F3.01) and 10 µg/ml Spark YG 570 anti-mouse CD8α mAb (BioLegend; clone 53-6.7) in PBS 5% (v/v) rat serum, and 0.1% (v/v) Tween 20. After two washes with PBS 0.1% (v/v) Tween 20, sections were counterstained with DAPI (Life Technologies), mounted in 100% glycerol and imaged at 10–40× using a LSM 700 Zeiss confocal laser scanning microscope (Carl Zeiss). Fiji software (Image J 2.1.0) was used to create single 2D images from captured Z stacks. Adjoining images were manually stitched together using Adobe Photoshop (Adobe Inc).
qPCR analysis of oral mucosa tissue
Total RNA was extracted from oral mucosa using a Master Pure Complete DNA and RNA Purification kit (Epicentre Technologies) and contaminating DNA removed following treatment of samples with AmbionTM Turbo DNase (ThermoFischer Scientific). RNA was reverse transcribed into cDNA using the ProtoScript II First Strand cDNA Synthesis Kit (New England BioLabs Inc.) according to the manufacturer's instructions.
cDNA samples were analyzed in triplicate for each gene of interest using a MX3000P Stratagene Real Time qPCR instrument (Agilent Technologies). Each qPCR reaction (final volume, 25 μl) contained 12.5 μl of SYBR Green Supermix (Bio-Rad Laboratories), 1 ml of each primer (0.3 μm), 5.5 μl of Nuclease-free water and 20 ng/5 μl of cDNA. A zero-template control was included in each assay. The thermo-cycling program consisted of one hold at 95 °C for 10 min followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, 30 s at 72 °C. A melting curve protocol was followed to determine the specificity of the product and to rule out primer-dimer formation or potential amplification of contaminating DNA. All qPCR primers used flanked introns. Primer sequence, binding position within cDNA, intron flanked and expected amplicon size are given in Supplementary Table S1.
From a panel of 5 potential housekeeping genes, hydroxymethylbilane synthase was determined to be the most stable in our samples as described (24). For the purposes of our analysis, a Cq value ≥35 was treated as gene expression not detected. The ΔΔCq method (25) was used to analyze the raw Cq values following normalization to expression of the housekeeping gene, hydroxymethylbilane synthase. Fold change (2-ΔΔCq) in gene expression was determined by comparison of the mean of the normalized expression values, between LCΔMHC-II and control LCWT groups, using the ΔΔCq method (25). Unpaired two-tailed Student's t-test was used to analyze the difference in the expression level of the tested genes between the groups. Genes with a statistical significance of ±2-fold change and a p value <0.05 were considered as differentially regulated.
Inhibition of TGFβ signaling using LY364947
The inhibitor of TGFβ receptor signaling (26) LY364947 (Selleckchem) was dissolved in DMSO to a concentration of 20 mg/ml, and aliquots stored at −80 °C. Working LY364947 solutions (0.4 mg/ml in PBS 2% DMSO) were prepared fresh from −80 °C stocks on the day on injection. LY364947 was injected intraperitoneally once a day for 7 days at a dose of 4 mg per kg body weight and mice were euthanized 24 h following the final LY364947 injection. Control mice received similar volumes of PBS 2% DMSO vehicle over the same injection regimen. Oral mucosa was harvested, and single cell suspensions produced from epithelial sheets for analysis by flow cytometry.
Oral infection of mice with Candida albicans and bioluminescence imaging
Mice were orally infected with C. albicans using an established co-infection model without immunosuppression (27). Streptococcus oralis 34, a gift from Anna Dongari-Bagtzoglou (University of Connecticut), was maintained on BHI broth agar plates (Research Products International). C. albicans strain DSY4976 stably expressing a red-shifted firefly luciferase gene and the non-transformed parental strain DSY49709 (28) were a gift from Dominique Sanglard (University of Lausanne) and were maintained on Yeast Peptone Dextrose (YPD) (1% yeast extract Fluka Analytical; 2% Bacto Peptone BD Biosciences; 2% Dextrose Sigma Aldrich) agar plates (Research Products International). Single colonies of C. albicans and S. oralis were utilized to establish liquid cultures for inoculum production. S. oralis was grown o/n in brain-heart infusion (BHI) broth (37 °C, 5% CO2, static) and cfu determined by optical density at 600 nm (OD600 nm reading of 1 = 1 × 108 cfu/ml). C. albicans was grown o/n in YPD broth in a shaking incubator at 28 °C to maintain growth as yeast cells. C. albicans and S. oralis were collected by centrifugation (537 g for 10 min at room temperature) and the cell pellets washed twice with room temperature Hank's Balanced Salt Solution (HBSS) (Gibco Life Technologies). After the final wash, S. oralis pellets were resuspended in HBSS at a concentration of 10 × 109 cfu/ml. Final C. albicans pellets were resuspended in a small volume of HBSS, spores counted, and the concentration adjusted to 2.4 × 109 cfu/ml in HBSS. The inoculum was prepared by mixing an equal volume of C. albicans and S. oralis in a 1.5 ml tube and placing the tube in a 30 °C heat block until inoculation.
Mice were anesthetized with a ketamine/xylazine mixture and placed in a supine position. Fifty µl of inoculum was applied to the applicator tip of a calcium alginate swab (Puritan) and the swab placed in the oral cavity on top of the tongue. Swabs were removed at 60 min. When required, bioluminescence was captured in live mice using an in vivo imaging system (IVIS) Spectrum (PerkinElmer). Briefly, 10 µl of D-Luciferin (PerkinElmer) substrate at 1.5 mg/ml was placed into the oral cavity of isoflurane-anesthetized mice and bioluminescence signal captured at 5 min using the following IVIS Spectrum settings: FOV = D; F1; Bin = 4; 5-min exposure. Images were analyzed using Living Image software (PerkinElmer). Region of interest (ROI) was established as 30 × 30 pixels (1.95 cm2) and a minimum threshold of 600 counts was set for all experiments. Radiance efficiency was captured for each ROI and expressed as total flux (photons/s). Background bioluminescence (∼3 × 104 total flux photons/s) was established in mice orally infected with the non-luciferase parental C. albicans strain DSY49709.
Statistical analysis
Data was analyzed and plotted using Prism 8 software (GraphPad Software) and displayed as means ± SEM or individual data points in plots showing correlations. Significance between two groups was determined by unpaired two-tailed Student's t-test or Mann–Whitney U-test when data did not show Gaussian distribution (Shapiro–Wilk normality test). Correlations were determined from computed Pearson correlation coefficients. p values less than 0.05 were considered significant. Biological sex was not considered a significant variable except in our oropharyngeal candidiasis infection model.
Results
CD8 T cells are located predominately in the suprabasal layer of the oral mucosa epithelium in LCΔMHC-II mice
Using flow cytometry, we showed CD8 T cells to be significantly increased in the oral mucosa of mice (LCΔMHC-II) engineered to ablate MHC-II on LCs (21). In LCΔMHC-II mice the enhanced number of CD8α+ cells were located within the stratified mucosal epithelium and almost exclusively within the suprabasal layer (Figure 1A). To quantify CD8 T cell distribution in the mucosal tissue, we separated the stratified epithelium from the underlying lamina propria. Virtually all CD8 T cells (96.9%) from LCΔMHC-II mice are located within the stratified mucosal epithelium (Figure 1B; Supplementary Figure S1), consistent with our immunohistology observations. In LCWT mice there were insufficient CD8 T cells to perform a similar analysis.
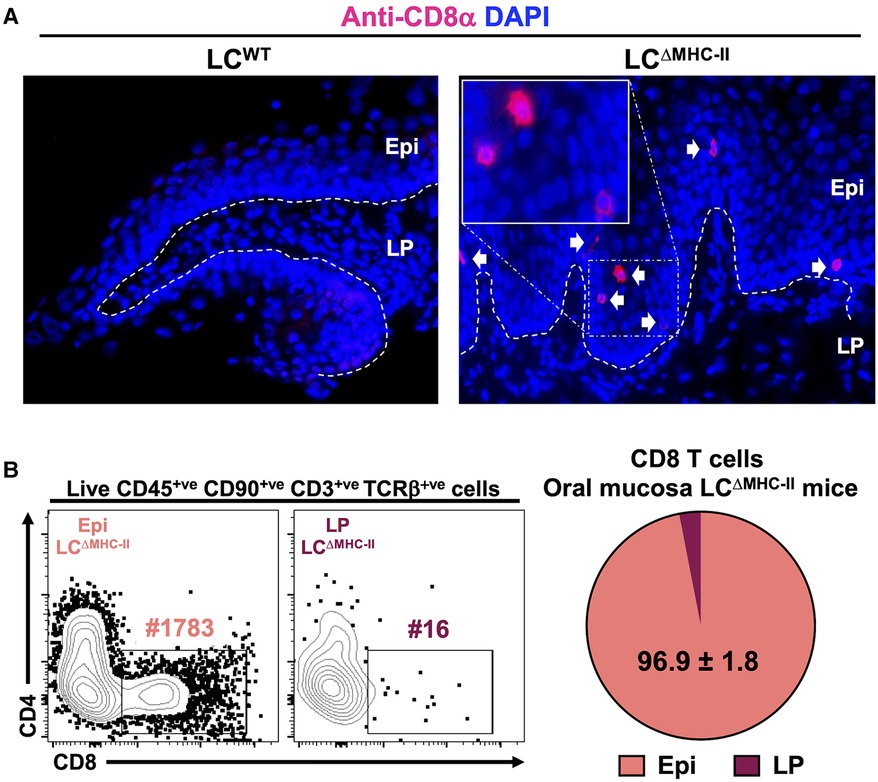
Figure 1. CD8 T cells in the oral mucosa of LCΔMHC-II mice reside predominantly within the suprabasal epithelial layer. (A) Ten µm oral mucosa frozen sections were stained with DAPI and rat anti-mouse CD8α mAb conjugated to Alexa Fluor 647. Representative images obtained from a LCΔMHC-II mouse and a huLang Cre negative “WT” littermate control (LCWT) are shown. White arrows indicate CD8 T cells located within the oral mucosal epithelium. Segmented white line separates Epi (epithelium) from LP (lamina propria). Scale bar is 50 µm. (B) Oral mucosa from LCΔMHC-II mice were treated with Dispase II to separate the tissue specimen into epithelium and underlining lamina propria components. Single cell suspensions were obtained from the two tissue fractions and stained with rat anti-mouse mAbs for flow cytometry. Live CD8 T cells were identified as Zombie Aqualo, CD45+, CD90.2+, CD3+, TCRβ+, CD8α+ and CD4−. Representative flow cytometry plots from Epi and LP samples are shown including the number of CD8 T cells (#) for each sample. Summary data from 4 LCΔMHC-II mice are displayed showing the mean percentage ± SEM of CD8 T cells found in the Epi or LP.
CD8 T cell numbers are increased in the skin of LCΔMHC-II mice but not in lung
Since CD8 T cells proximal to LCs are increased in the oral mucosa of LCΔMHC-II mice, we analyzed CD8 T cell numbers in other peripheral tissues where LCs reside. We hypothesize that LCs unable to display antigen on MHC-II molecules may be associated with the increase in CD8 T cells. To test this hypothesis, we compared lung, a mucosal tissue that does not ordinarily harbor LCs, with glabrous skin that possesses a LC-rich stratified epithelium (1). Congruent with our oral mucosa analysis (21), the number of CD4 T cells found in either the lung or the skin were similar when LCΔMHC-II mice were compared with LCWT littermate controls, although CD4 T cells were about 7-fold more numerous in the lung than in skin (Figure 2). In contrast, CD8 T cells were significantly more numerous in the skin of LCΔMHC-II than LCWT mice. In the same LCΔMHC-II mice, CD8 T cell numbers were not elevated in the lungs when compared to the LCWT mice (Figure 2). We also examined the stratified epithelial cervical-vaginal tissue, which contains LCs (29), but the vascularity of the tissue and epithelial changes with different stages of estrus compromised the normalization of CD8 T cell numbers across samples based on the live non-immune cell population (data not shown).
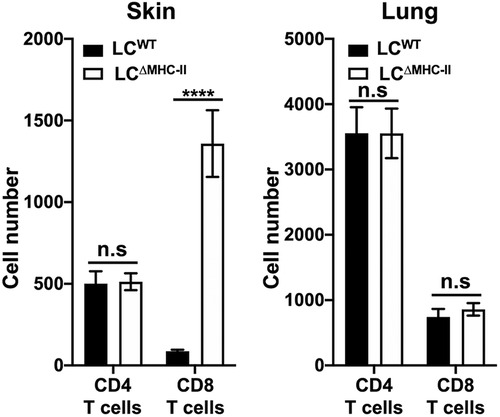
Figure 2. CD8 T cell numbers are increased in skin but not lungs of LCΔMHC-II mice. LCΔMHC-II mice or LCWT littermate controls received 1.25 µg of rat anti-mouse CD45 conjugated to FITC intravenously 3 min prior to sacrifice to exclude blood-resident immune cells. Single cell-suspensions from left lung and ear skin peels were stained with rat anti-mouse mAbs to identify CD4 T cells and CD8 T cells by flow cytometry. Live interstitial CD8 T cells and CD4 T cells were identified as Zombie Aqualo, CD45:FITC−, CD45:PE+, CD3+, TCRβ+, CD8α+, CD4− and Zombie Aqualo, CD45:FITC−, CD45:PE+, CD3+, TCRβ+, CD8α− and CD4+, respectively. Numbers of CD4 T cells and CD8 T cells, normalized to 100,000 live non-immune cells, are presented for lung and skin tissues. Data are from 3 experiments with at least 8 mice per tissue type and are plotted with means ± SEM. Means were compared using an unpaired two-tailed Student's t-test. ****p < 0.0001, n.s., not significant.
Oral mucosal CD8 T cells in LCΔMHC-II mice display an activated, tissue-resident memory phenotype and exhibit a broad TCR Vβ repertoire
T cells in the periphery usually express cell surface markers characterizing them as either effector memory or tissue memory. CD69 is often used as a marker of memory and in conjunction with the alpha E integrin, CD103, has been used to distinguish tissue-resident memory T cells from effector memory T cells (30) including those T cells residing within the oral mucosa (31, 32). Tissue-resident Tc1 CD8 T cells are also marked by the alpha 1 integrin, CD49a (33, 34). To determine whether the oral CD8 T cells have an effector or tissue-resident memory phenotype, we analyzed expression of CD69 and CD103 on CD8 T cells isolated from the spleen and the oral mucosa of LCΔMHC-II mice. Additionally, we analyzed CD49a on oral CD8 T cells. The spleen provides a surrogate CD8 T cell comparison since LCWT mice have too few CD8 T cells in the oral mucosa to analyze. We expected that the spleen would contain sufficient circulating effector memory cells (CD69+ve) to analyze but tissue-resident CD8 T cells (CD69+ve CD103+ve) would be absent (Figure 3A—left panel). In the oral mucosa of LCΔMHC-II mice, greater than 90% of CD8 T cells are CD69+ve CD103+ve double positive, consistent with a tissue-resident memory phenotype (Figure 3A—center panel). CD103 expression is consistent with the epithelial location of the CD8 T cells (30). We also identified a CD103+ve CD49a+ve CD8 T cell population, but interestingly also an atypical CD103+ve CD49a−ve population at a similar frequency (Figure 3A—right panel).
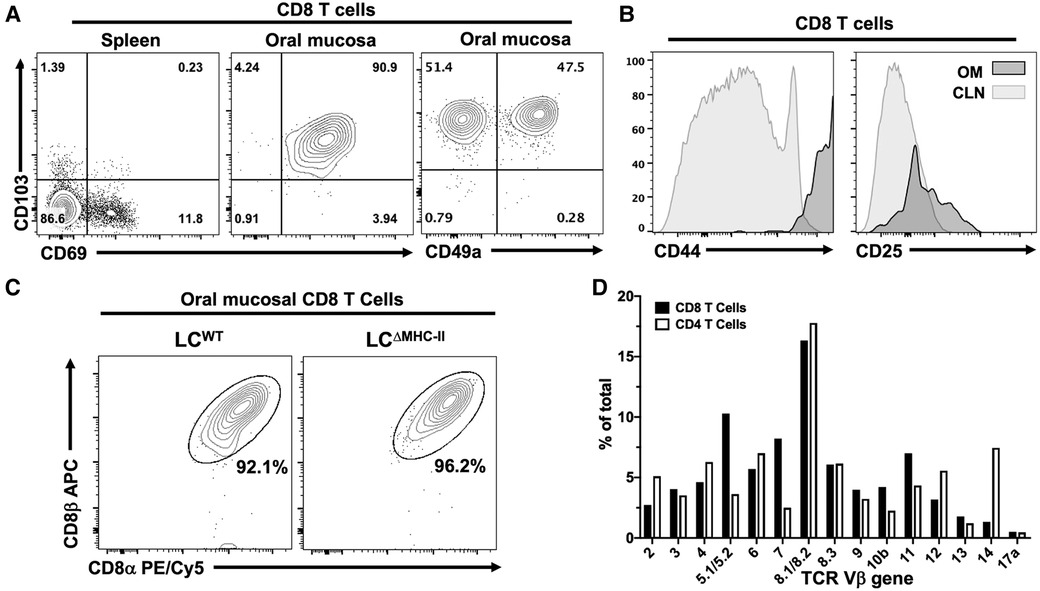
Figure 3. CD8 T cells in the oral mucosa of LCΔMHC-II mice have an activated tissue-resident memory phenotype, express the CD8 α/β heterodimer and have broad TCR Vβ usage. (A) Representative flow cytometry plots showing CD103, CD69 and CD49a expression on CD8 T cells isolated from tissues of a LCΔMHC-II mouse. Numbers indicate % of cells within each quadrant gate. (B) Representative flow cytometry histogram plots showing CD44 or CD25 expression on CD8 T cells isolated from cervical lymph nodes (CLN) or oral mucosa (OM) of a LCΔMHC-II mouse. (C) Representative flow cytometry plots showing CD8α and CD8β on CD8 T cells isolated from OM. Interstitial CD8 T cells were identified as Zombie Aqualo, CD45:FITC−, CD45:PE+, CD3+, TCRβ+, CD4−. Cells were obtained from a single LCΔMHC-II animal or pooled from 4 LCWT littermate mice. (D) Pooled cells from the epithelial sheets of 12 LCΔMHC-II mice were stained to identify CD4 T cells and CD8 T cells and subsequently stained with one of 15 different TCR Vβ mAbs. One aliquot remained unstained to set negative TCR-β gates for both CD4 T cells and CD8 T cells. CD4 T cells were identified as Zombie Aqualo, CD45+, CD90.2+, CD3+, CD4+, CD8α−; CD8 T cells were identified as Zombie Aqualo, CD45+, CD90.2+, CD3+, CD4−, CD8α+. The frequency of TCR Vβ usage amongst CD4 T cells and CD8 T cells is shown.
Oral mucosa CD8 T cells from LCΔMHC-II mice were also examined for expression of two activation markers, CD44 and CD25 (Figure 3B). These CD8 T cells appeared highly activated, expressing uniformly elevated levels of CD44 compared to CD8 T cells from cervical lymph nodes. Compared to cervical lymph node CD8 T cells, CD25 expression was only slightly elevated in the oral mucosal population.
The MHC-I co-receptor, CD8, can exist as αα homodimers or αβ heterodimers. In mice, CD8αα T cells are predominately located within the mucosal epithelial lining of the small intestine (35). Given the mucosal residency of these CD8αα T cells, we asked whether CD8 T cells found in the oral mucosa of LCΔMHC-II also express the CD8αα homodimer (Figure 3C). In the oral mucosa of both LCΔMHC-II and LCWT mice, however, CD8 T cells almost exclusively express the αβ heterodimer.
Mucosal-associated invariant T (MAIT) cells are another unconventional T cell subset that can express CD8 (36). Utilizing major histocompatibility complex I-related protein 1 (MR1)-specific tetramers, MAIT cells did not appear to reside in the CD8 T cell compartment of the oral mucosa of LCΔMHC-II mice (Supplementary Figure S2A). Nonetheless, CD4−ve CD8−ve MAIT cells were identified in small numbers within the oral mucosa of both LCΔMHC-II and LCWT mice (Supplementary Figure S2B).
To explore the clonality of the CD8αβ T cells in the oral mucosa of LCΔMHC-II mice, we next examined their TCR Vβ usage. A limited repertoire of Vβ gene usage would indicate that the CD8 T cells recognized and have responded to a narrow range of antigens. The CD8 αβ T cell population appeared in contrast to be polyclonal, displaying a broad repertoire of TCR molecules with Vβ usage ranging from 16.33 to 1.33% for the clones tested (Figure 3D). As anticipated, TCR Vβ usage was also broad in the CD4 T cell compartment. Vβ gene 17a is not expressed in the C57BL/6J mouse and at <0.5% can be considered background detection.
CD8 T cells in the oral mucosa of LCΔMHC-II mice express IL-17A
Classic Tc1 CD8 T cells typically express IFN-γ when IL-12 drives differentiation (37). However, the population of CD103+ve CD49a−ve CD8 T cells we identified (Figure 3A) is a phenotype associated with IL-17A expression (34). Consistent with our earlier published work (21), few CD8 T cells localize in the oral mucosa of LCWT mice and these cells predominately expressed IFN-γ and not IL-17A (Figure 4A). In contrast, a plurality of CD8 T cells from LCΔMHC-II mice expressed IL-17A (Tc17), with smaller numbers expressing IFN-γ (Tc1) or both proinflammatory cytokines (Tc1/17) (Figure 4A). All three CD8 T cell phenotypes were significantly elevated in LCΔMHC-II compared to LCWT mice. LCΔMHC-II mice show >30-fold more Tc17 cells than LCWT mice. Interestingly, Tc1/17 cells numbers were also increased.
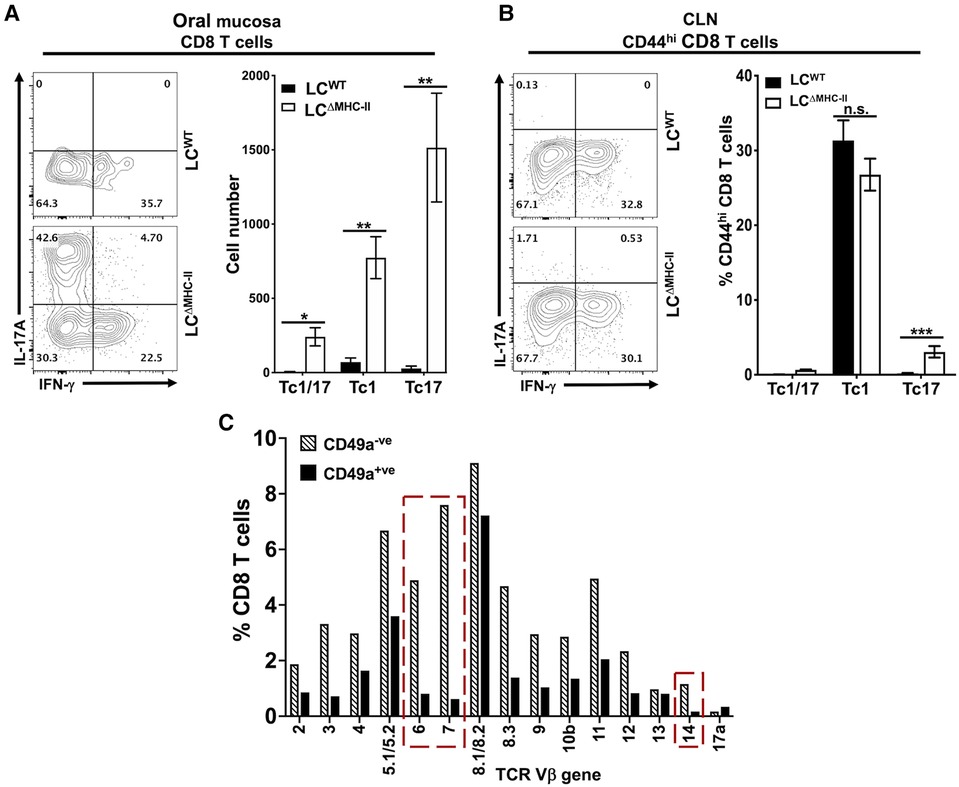
Figure 4. CD8 T cells in the oral mucosa of LCΔMHC-II mice predominately express IL-17A. Single cell suspensions were activated with PMA/ionomycin in the presence of brefeldin A, surface stained to identify CD8 T cells and then stained intracellularly with rat anti-mouse IL-17A and IFN-γ mAbs (A and B). Representative flow cytometry plots showing expression of IL-17A and IFN-γ are displayed. Numbers indicate % of cells within each quadrant gate. (A) Summary data from 3 independent experiments totaling at least 12 mice per group showing means ± SEM of Tc1 (IFN-γ+), Tc17 (IL-17A+) or Tc1/17 (IFN-γ+ and IL-17A+) CD8 T cell numbers normalized to 100,000 live non-immune cells. An unpaired two-tailed Student's t-test was used to test significance. *p < 0.05, **p < 0.01. (B) Summary data from 2 independent experiments of 8 mice per group showing means ± SEM of the relative frequency of Tc1, Tc17 and Tc1/17 CD8+ CD44hi T cells in cervical lymph nodes. Unpaired Student's t-test was used to test significance. ***p < 0.001. n.s., not significant. (C) TCR Vβ usage was determined as described in Figure 3D. The frequency of TCR Vβ usage amongst CD49a+ and CD49a− CD8 T cells is shown. Red dashed boxes highlight differences between CD49a+ and CD49a− CD8 T cells of 5X or greater.
LCΔMHC-II mice could fail to differentiate robust numbers of Tc1 cells. To consider this possibility, we examined the phenotype of CD8 T cells found in the cervical lymph nodes, which drain the oral mucosa and head region. To exclude the majority naïve CD8 T cells, only CD8 T cells that displayed an activated memory (CD44hi) phenotype were analyzed (Figure 4B). In cervical lymph nodes, there was no significant difference in the frequency of Tc1 cells found in LCΔMHC-II mice and LCWT mice. Both strains were found to have around 30% Tc1 cells, indicating that there is no defect in Tc1 cell generation in LCΔMHC-II mice. We did observe a small, but significant, increase in the frequency of Tc17 cells. The relative frequencies of Tc1 and Tc17 cells in cervical lymph nodes and in oral mucosa of LCΔMHC-II mice, however, are very different.
CD8 T cells in LCΔMHC-II mice generally displayed broad TCR Vβ usage (Figure 3D). CD8 T cells could also be separated into two distinct populations based on CD49a expression (Figure 3A). To determine differences in TCR Vβ usage between these two CD8 T cell populations, we used the anti-TCR Vβ mAb panel (Figure 4C). The Vβ genes 6, 7 and 14 showed frequencies that were at least 5-times greater amongst CD49a− CD8 T cells than CD49a+ CD8 T cells, suggesting a differential response to antigens. Consistent with this observation and given the tight association of CD49a expression with Tc1 cells (33, 34), CD49a+ CD8 T cells were almost exclusively negative for IL-17A expression (data not shown).
Intraepithelial CD4 T cells suppress CD8 T cell numbers in the oral mucosa
Given the effect of MHC-II presentation by LCs on intraepithelial CD8 T cell numbers, we tested the contribution of CD4 T cells on the control of CD8 T cell numbers in LC MHC-II competent mice. Global ablation of CD4 T cells in IL-17ACre-RFP reporter mice (Figure 5A) with an anti-CD4 mAb (38–40) resulted in increased intraepithelial CD8 T cell numbers by >14-fold (Figure 5B). Interestingly, 25%–66% of these CD8 T cells in the oral mucosa expressed IL-17A (Tc17) as measured by proxy tdTomato expression (Figure 5C). In contrast, CD8 T cells in the lungs of these mice did not increase or express IL-17A despite a >99% reduction in CD4 T cells (data not shown).
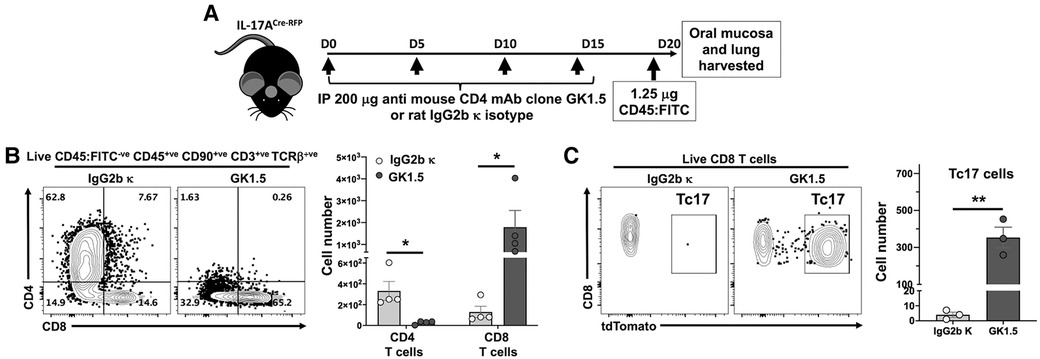
Figure 5. Depletion of CD4 T cells results in expansion of both CD8 T cells and Tc17 in the oral mucosa. (A) Experimental design to deplete CD4 T cells. 8–10 weeks old IL-17ACre-RFP mice were injected i.p. with 200 µg anti-CD4 mAb (Clone GK1.5; BioXCell) or Rat IgG2b κ control (#BE0090; BioXCell) 4 times, 5 days apart. On day 20 (D20) mice were intravenously injected with FITC-labeled anti-CD45 mAb and sacrificed 3 min later. Oral mucosa epithelial peels were treated with Dispase II, collagenase D and DNAse I and single cell suspensions stained to identify CD4 T cells and CD8 T cells by flow cytometry. (B) Typical flow cytometry plots showing live oral mucosa interstitial CD4 T cells and CD8 T cells from a GK1.5 or Rat IgG2b κ treated animal. Summary data shows mean ± SEM of CD4 T cell and CD8 T cell numbers. (C) Typical flow cytometry plots showing tdTomato expression (IL-17A proxy) in live oral mucosal interstitial CD8 T cells from a GK1.5 or Rat IgG2b κ treated animal. Mean ± SEM of Tc17 cells is plotted. Summary data compared by Student's two-tailed t-test. *p < 0.05; **p < 0.01.
γδ T cells correlate with CD8 T cell numbers and are suppressed in the oral mucosa of LCΔMHC-II mice
CD4 T cell abundance and Th17 frequency are not altered in the oral mucosa of LCΔMHC-II mice compared to LCWT mice (21). Given the increase in LCΔMHC-II Tc17 cells, we asked whether γδ T cells, which primarily express IL-17A, are also increased. Although the percentage of γδ T cells expressing IL-17A was ∼80% in both LCΔMHC-II and LCWT mice (data not shown), the number of oral mucosal γδ T cells expressing IL-17A was lower in LCΔMHC-II mice (Figure 6A).
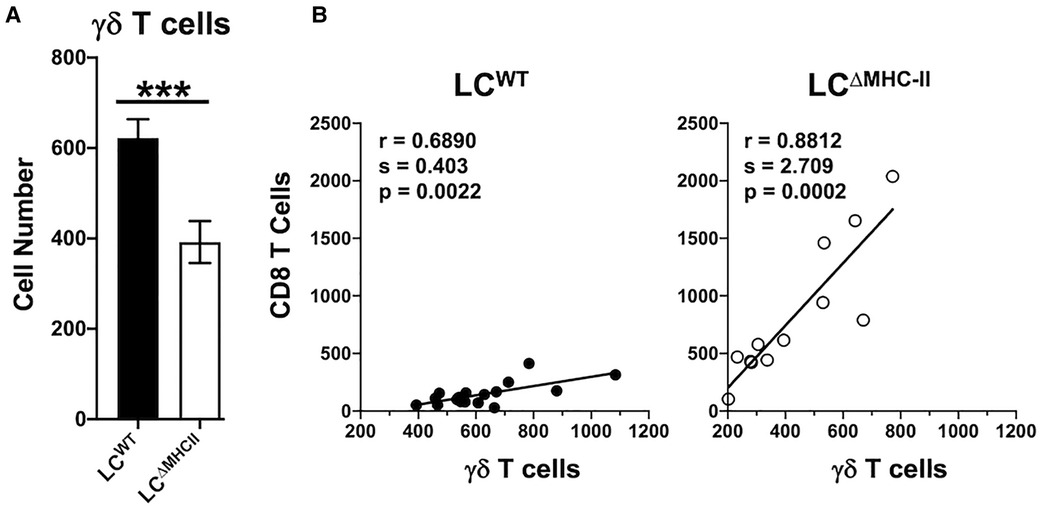
Figure 6. Decreased γδ T cells in the oral mucosa of LCΔMHC-II mice correlate with CD8 T cell numbers. (A) Oral mucosa from LCΔMHC-II mice or LCWT littermate controls was processed, and single cell suspensions stained with rat anti-mouse mAbs to identify γδ T cells by flow cytometry. Live γδ T cells were identified as Zombie Aqualo, CD45+, CD3hi, TCRγ/δ+, TCRβ−, CD8α−, CD4− cells. Summary data of γδ T cell numbers, normalized to 100,000 live non-immune cells, is presented. Data are from 3 independent experiments with at least 12 mice per group are plotted as means ± SEM. Means were analyzed using an unpaired two-tailed Student's t-test. ***p < 0.001. (B) Correlation between CD8 T cells and γδ T cells numbers identified in oral mucosal tissue of LCΔMHC-II mice or LCWT littermate controls by flow cytometry as described in A. Data are from 3 independent experiments with at least 12 mice per group. Simple Linear Regression was utilized to fit the slope(s) to the data. The significance (p value), of the calculated Pearson correlation coefficient (r) is reported.
CD8 T cell and γδ T cell numbers seemed differentially impacted in the oral mucosa of LCΔMHC-II mice. We next sought a relationship between CD8 T cell and γδ T cell numbers. Cell numbers were normalized across samples to live non-immune cells (primarily keratinocytes) to control for differences in sample processing (41). Interestingly, despite the low number of γδ T cells in the oral mucosa of LCΔMHC-II mice, CD8 T cell and γδ T cell numbers showed a positive and statistically significant correlation (Figure 6B). A positive correlation was also seen in the oral mucosa of LCWT mice, albeit with a smaller slope value than in LCΔMHC-II mice.
Expression of TGF-β1 is significantly elevated in the oral mucosa of LCΔMHC-II mice
Tc17 and Th17 cells have similar IL-6 and TGF-β1 requirements to drive polarization from naïve T cells in lymph nodes or maintenance and clonal expansion in the periphery (42–44). Furthermore, Tc17 cells maintain all the transcriptional circuitry needed to convert to a Tc1 phenotype where sustained IL-6-driven expression of Signal Transducer and Activator of Transcription 3 (STAT3) and Retinoic acid-related Orphan Receptor gamma t (RORγt) overrides the Tc1 program (42). We used qPCR analysis to examine whether LCΔMHC-II mice have elevated IL-6 or TGF-β1 expression in oral mucosa (Figure 7A). Here, we found that TGF-β1 gene expression was 4-times higher than LCWT mice, while IL-6 gene expression was similar in the two strains (Figure 7B).
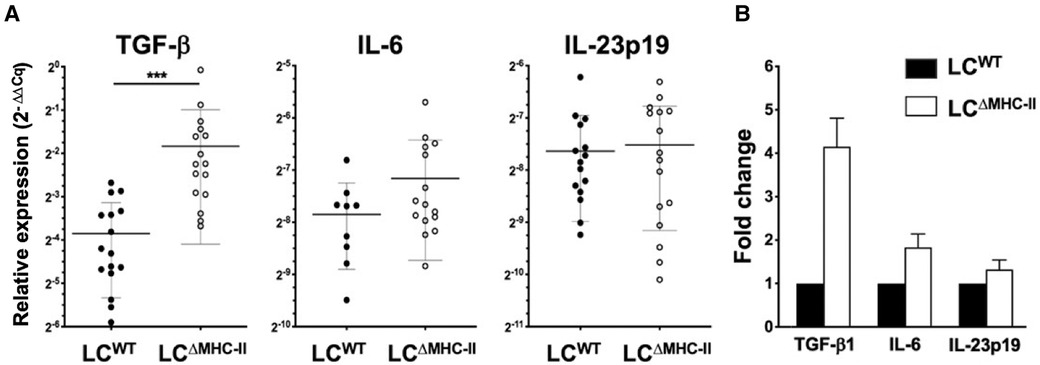
Figure 7. Expression TGF-β1 is significantly elevated in the oral mucosa of LCΔMHC-II mice. Total RNA extracted from the oral mucosa of age-matched (8–9 week-old) LCΔMHC-II mice or LCWT littermate controls was converted into cDNA and qPCR analysis performed. The ΔΔCq method was used to analyze the raw qPCR data (Cq values) following normalization to expression of the housekeeping gene, hydroxymethylbilane synthase. (A) Summary data of relative TGF-β1, IL-6 and IL-23p19 mRNA expression levels. Individual data points are plotted along with mean ± SEM. Data is from three independent experiments involving at least 9 mice. A two-tailed Student's t-test was used to test significance. ***p < 0.001; ****p < 0.0001. (B) Relative fold changes of TGF-β1, IL-6, and IL-23p19 mRNA in oral mucosal samples from LCΔMHC-II and WT mice.
Given increased levels of TGF-β1 expression in the oral mucosa, we extended the analysis to a broader set of genes that might be involved in sustaining Tc17 and Tc1 cells. Specifically, the global expression of IL-23p19 (Figure 7A), IL-12p35 and IFN-γ (data not shown) was similar in the two strains. Gene expression of IL-21 and IL-17A was not detected (Cq > 35) in the oral mucosa of either mouse strain. The global expression of the Treg-associated genes, IL-10 and EBI-3 were also similar in the oral mucosa of LCΔMHC-II and LCWT mice (data not shown).
TGF-β1 signaling is not required to maintain oral Tc17 cells in LCΔMHC-II mice
TGF-β1 is essential in vitro to differentiate naïve CD8 T cells towards a Tc17 phenotype (17, 45). To investigate the in vivo requirement for TGF-β1 in Tc17 differentiation and/or maintenance, we disrupted TGF-β-dependent signaling in LCΔMHC-II mice with the kinase inhibitor LY364947 (26). LY364947 treatment did not alter the frequency of Tc17 cells in the oral mucosa when compared to mice treated with vehicle control (Figure 8A). Additionally, LY364947 treatment did not impact the frequencies of Tc1 cells or CD8 T cells not expressing IL-17A or IFN-γ.
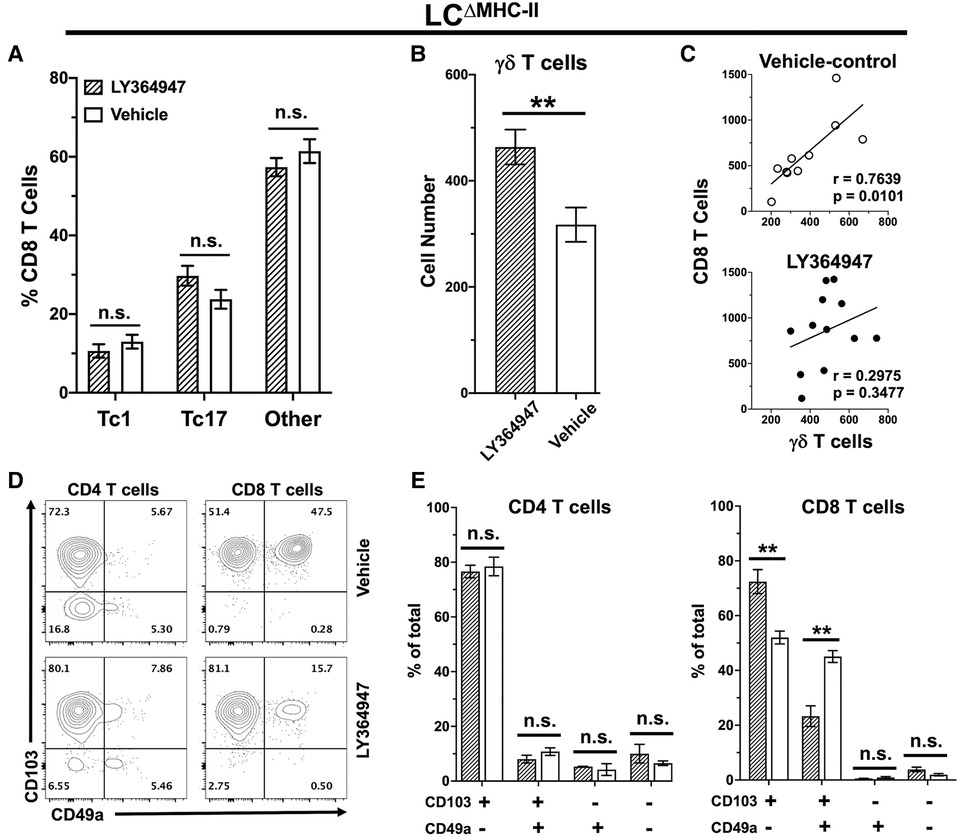
Figure 8. TGF-R1 kinase inhibition impacts γδ T cells and CD49a+ CD8 T cells but not Tc17 in LCΔMHC-II mice. LCΔMHC-II mice received vehicle control (2% DMSO in PBS) or 4 mg/kg LY364947 (2% DMSO in PBS) intraperitoneally daily for 7 days. Single cell suspensions from oral mucosa were analyzed by flow cytometry. (A) Summary data showing mean frequency ± SEM of Tc1, Tc17 and IL-17A/IFN-γ non-expressing CD8 T cells (other). Data are from 2 independent experiments with at least 8 mice per group. Means were analyzed using unpaired two-tailed Student's t-test. **p < 0.01, n.s., not significant. (B) Summary data from 3 independent experiments with at least 10 mice per treatment group showing mean number ± SEM of γδ T cells. Live γδ T cells were Zombie Aqualo, CD45+, CD3hi, TCR γ/δ+, TCRβ−, CD8α−, CD4−. Cell number was normalized to 100,000 live non-immune cells. Means were compared by unpaired two-tailed Student's t-test. (C) Correlation between CD8 T cell and γδ T cell numbers. Simple linear regression was used to fit the best slope to the data. Significance (p value) of the calculated Pearson correlation coefficient (r) is shown. (D) Representative flow plots showing CD103 and CD49a on oral mucosal CD4 and CD8 T cells following LY364947 or vehicle treatment. (E) Summary data showing frequency of CD103 and CD49a on CD4 or CD8 T cells. Data are from 3 to 4 mice. Means were analyzed by unpaired two-tailed Student's t-test. **p < 0.01, n.s., not significant.
We also questioned whether TGF-β1 signaling might be involved in the decreased numbers of γδ T cells in the oral mucosa of LCΔMHC-II mice (Figure 6A). Inhibition of TGF-β1 signaling with LY364947 significantly increased the number of γδ T cells in LCΔMHC-II mice (Figure 8B). TGF-β1 signaling appears to selectively impact γδ T cells and not CD8 T cells since the correlation between γδ T cell and CD8 T cell numbers in the oral mucosa is maintained in the vehicle control group but is lost in mice treated with LY364947 (Figure 8C).
The expression of CD49a and CD103 in CD8 T cells is reported to depend on TGF-β1 signaling (46, 47). Here, we determined the frequency of CD49a and CD103 expression on both CD4 T cells and CD8 T cells isolated from the oral mucosa of LCΔMHC-II mice treated with LY364947 or the vehicle control (Figure 8D). Inhibition of TGF-β1 signaling with LY364947 did not impact CD4 T cells (Figure 8E). However, in the CD103+ve subpopulation of CD8 T cells there was a significant decrease in the frequency of CD49a+ve cells and a concomitant increase in the frequency of the CD49a−ve cells in LY364947 treated LCΔMHC-II mice indicating that CD49a expression, but not CD103, was sensitive to TGF-β1 signaling disruption (Figure 8E). Moreover, the MFI of the CD103 signal remained unchanged (data not shown).
LCΔMHC-II mice have enhanced resistance to oral Candida albicans infection
Innate and adaptive immunity to C. albicans is significantly compromised at mucosal surfaces when IL-17A or its signaling pathway are abrogated (8, 14). IL-17A contributes to C. albicans resistance by inducing epithelial cells to express antimicrobial peptides and neutrophil chemotactic chemokine CXCL8 and GM-CSF (13). C. albicans in the murine oral cavity typically shows tropism for the dorsal surfaces of the tongue, therefore, we compared the number of LCs and CD8 T cells in the dorsal epithelial sheets of LCΔMHC-II and LCWT (Figure 9A). Despite similar number of LCs, the number of CD8α+ve cells were significantly higher in LCΔMHC-II mice.
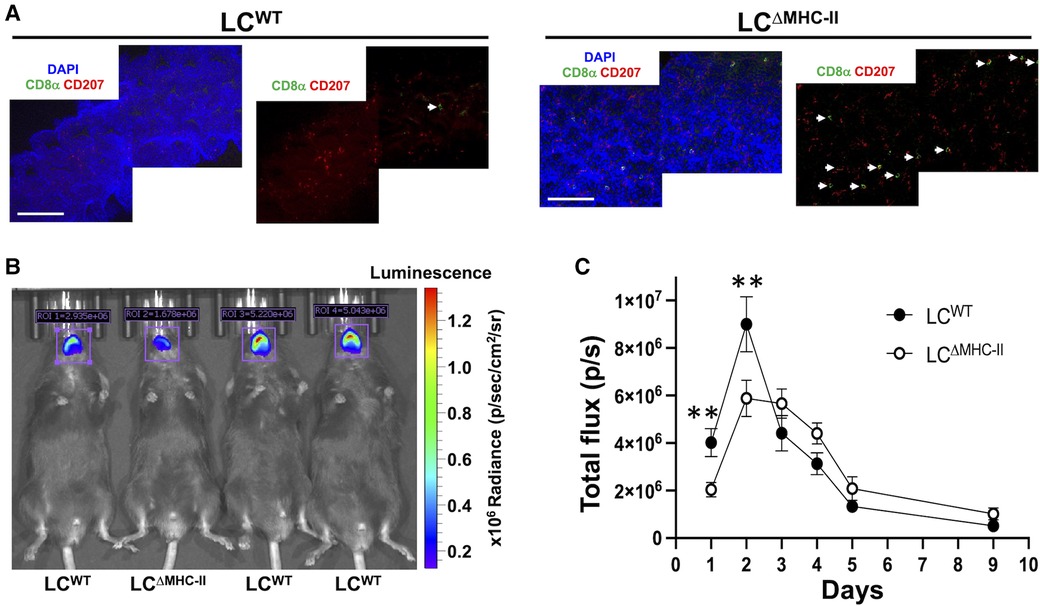
Figure 9. LCΔMHC-II mice have enhanced CD8 T cell numbers in tongue epithelium and increased resistant to oral candidiasis. (A) Epithelial sheets from tongues of LCWT and LCΔMHC-II mice were stained with DAPI (blue), rat anti-mouse CD8α mAb (green) and rat anti-mouse CD207 mAb (red). Representative confocal microscope images (40×) obtained from a LCΔMHC-II mouse and a LCWT are shown. White arrows indicate CD8α+ cells. Scale bar is 100 µm. (B,C) Age matched male sibling mice were co-infected with S. oralis 34 and C. albicans strain DSY4976 that expresses a red-shifted firefly luciferase gene. Mice were not immunosuppressed. Bioluminescence was measured on days 1, 2, 3, 4, 5, and 9 following oral administration of D-Luciferin. (B) Superimposed photograph with detected bioluminescence captured on day 1 using the IVIS Spectrum in vivo imaging system. Regions of interest (ROI) used to quantify bioluminescence are indicated along with the total flux in photons per s detected within the ROI. (C) Summary data from 4 independent experiments showing bioluminescence (total flux p/s) detected in the oral cavity of mice over 9 days. N = 16 (LCWT) and N = 13 (LCΔMHC-II). Mann–Whitney U-test was used to compare ranks at each day. **p < 0.01.
Given the increased numbers of Tc17 cells, we tested LCΔMHC-II mice for resistance to oral infection with C. albicans. We utilized an established C. albicans and Streptococcus oralis co-infection model that avoids the confounding effects of artificially induced immunosuppression to establish oral infection (27). For this work, we employed a C. albicans strain engineered to express a red-shifted firefly luciferase gene and measured in vivo the emitted bioluminescence as a proxy for C. albicans biomass over time (28) (Figure 9B). Although the numbers of Tc17 in the oral mucosa of male and female LCΔMHC-II mice are similar, we found that male mice of either strain were significantly more susceptible to C. albicans infection than their female counterparts (data not shown). Therefore, we elected to work with male mice in further experiments. While the burden for both mouse strains peaked at day 2, LCΔMHC-II mice had significantly less C. albicans burden than LCWT mice at day 1 and day 2 time points (Figure 9C). Interestingly, LCWT mice apparently cleared C. albicans faster than LCΔMHC-II mice from day 2 to day 3. However, from day 3 until the end of the experiment at day 9 C. albicans decreased at a similar rate in both mouse strains.
Discussion
In the oral mucosa of LCΔMHC-II mice engineered to develop LCs unable to express MHC-II we discovered strikingly high numbers of CD8 T cells (21). This work characterizes those CD8 T cells as intraepithelial tissue-resident CD8αβ T cells, the majority of which express IL-17A and not IFN-γ. These characteristics marks them as atypical Tc17 cells. In addition, the intraepithelial CD8αβ T cells are not mucosal-associated invariant T cells (MAIT cells) because they not do not recognize MR1 tetramers. Given the importance of IL-17A to mucosal immunity, particularly against C. albicans (8, 10, 13, 14), we also determined that LCΔMHC-II mice displayed enhanced resistance to oropharyngeal candidiasis.
A global MHC-II knockout mouse has been available for over 20 years (48), and the associated defect in the CD4 T cell compartment is well characterized. Perhaps not surprising, relatively few studies have characterized these mice for defects in the CD8 T cell compartment (49–51). In a model of human papillomavirus induced carcinoma, increased CD8 T cell numbers found in carcinomas of MHC-II knockout mice contributed to better control of tumor growth (51). Whether these cells were Tc1 or Tc17 was not determined, but the authors speculated that increased CD8 T cells were a compensatory effect for reduced numbers of tumor-infiltrating Th1 cells. In contrast, although oral Th1 and Helios+ Treg CD4 T cell compartments are reshaped, Th17 cell number is not impacted in LCΔMHC-II mice (21). This finding indicates that increased Tc17 cells are not a compensation for a loss in Th17 cell numbers. Interestingly, Th17-mediated protection against Citrobacter rodentium intestinal infections is abrogated in a bone marrow chimeric mouse model where MHC-II presentation is absent only on hematopoietic cells (50). Coincidentally, the number of Tc1 and Tc17 cells found in the gut of these mice was substantially increased. Unexpected expansion or control of Tc17 cells has also been described in situations of both CD4 T cell deficiency and the complete absence of CD4 T cell help (19, 52–56). CD4 Treg cells play an active role suppressing CD8 T cells either through mechanisms involving local MHC-II antigen presentation or through T cell receptor-independent processes (57). Certainly, our CD4 T cell ablation data supports the notion that a CD4 T cell population, possibility CD4 Treg cells, are regulating CD8 T cells via LCs in tissues where LCs, CD4 T cells and CD8 T cells co-exist. This contention is bolstered by data that showed CD8 T cell numbers in LCΔMHC-II mice were also dramatically increased in LC-rich tissue such as skin but not in the lung where LCs are not usually found. In addition, we found no increase in CD8 T cell numbers in lung despite near complete ablation of CD4 T cells.
CD8 T cells are a minority CD3+ T cell in the oral mucosa of humans and mice at steady state (21, 31, 58). Unlike gut intraepithelial CD8 T cells, most oral CD8 T cells that express CD69 are negative for CD103 indicating that they are likely recirculating effector memory T cells and not bona fide residents of the oral mucosa (31, 58). In stark contrast to these findings, greater than 90% of CD8 T cells in the oral mucosa of LCΔMHC-II mice co-expressed CD69 and CD103. Together with uniformly high CD44 expression and their intraepithelial location, these cells have the signature of conventional tissue-resident memory CD8 T cells (30). Furthermore, the relative paucity of Tc17 cells over Tc1 cells in the cervical lymph nodes that drain the oral mucosa suggests that most Tc17 cells generated in the LCΔMHC-II mouse do not enter the recirculating effector memory population.
TGF-β1 was significantly elevated in the oral mucosa of LCΔMHC-II mice and could potentially contribute to supporting the expansion of CD8 T cells. TGF-β signaling has been shown to be important for in vitro expression of CD103 on CD8 T cells and the in vivo maintenance of CD103+ intraepithelial gut resident CD8 T cells (59) and tissue-resident CD8 T cells in the skin (60). When inhibiting TGF-β signaling with LY364947 in vivo, the surface expression of CD103 on T cells and the frequency of CD103+ CD8 T cells did not change in the oral mucosa of LCΔMHC-II mice. Several factors may explain the apparent discrepancy between our data and that of Mackay et al. (59). First, there may be a different TGF-β threshold requirement for CD103 expression by CD8 T cells that ultimately migrate to and then take up residency in the gut or oral mucosa. Second, we used pharmacological intervention and not a genetic model to disrupt TGF-β signaling. In considering these discrepancies, it is also important to reiterate that TGF-β1 expression was elevated in the oral mucosa of LCΔMHC-II mice.
Consistent with the finding that CD49a expression on Tc1 cells in vitro appears to be dependent on TGF-β (46), our in vivo data reveals a significant reduction in the frequency of intraepithelial CD49a+ Tc1 cells in the oral mucosa of LCΔMHC-II mice. When TGF-β1 signaling was genetically abrogated or neutralized in a model of graft vs. host disease, the frequency of Tc17 cells did not change in agreement with our LY364947 treatment findings (61, 62). Taken together, we contend that oral intraepithelial CD8 T cells, and specifically Tc17 cells in LCΔMHC-II mice do not require TGF-β for maintenance of tissue residency.
Tc17 cells can contribute to inflammatory diseases, are associated with poor prognosis in some cancers, but also contribute to repair at barrier tissues like the skin, and via specific vaccines offer resistance to viral, bacterial and fungal pathogens (63). Unlike in psoriasis where Tc17 cell numbers are increased (33, 64), LCΔMHC-II mice do not exhibit pathological changes at oral mucosal surfaces or skin despite the high numbers of Tc17. Tc17 cells in LCΔMHC-II mice, therefore, do not seem to be overtly pathogenic. In this respect, LCΔMHC-II mice are similar to wild-type mice exposed to the commensal Staphylococcus epidermidis where antigen-specific skin resident Tc1 and Tc17 cell numbers are boosted without an associated inflammatory response (18, 65). Interestingly, exposure of skin to S. epidermidis offered prophylactic protection against C. albicans in a murine dermal infection model that was directly attributable to IL-17A expressed by Tc17 cells (18). Barrier resistance to C. albicans at both mucosal and skin surfaces critically depends on IL-17A promoting expression of antimicrobial peptides (12, 15) and eliciting recruitment of C. albicans-killing neutrophils (3, 10, 13, 14, 66, 67). Although TCR-independent bystander activation (68) cannot be excluded, IL-17A expression and protection against C. albicans in LCΔMHC-II mice could be driven by the presentation of antigens derived from members of the oral microbial community through TCR cognate interaction. Skin resistance to C. albicans infection induced by S. epidermidis seems to support a mechanism independent of C. albicans antigens (18). Broad TCR Vβ gene usage by the CD8 T cells is consistent with this viewpoint. Regardless of the mechanism, resistance to C. albicans in LCΔMHC-II mice warrants further investigation.
The oral mucosa is typically quiescent despite constant exposure to antigens derived from ingested food and a shedding oral microbiome. CD4+ FoxP3+ Treg cells comprise 30%–50% of the CD4 T cells found in the mouse oral mucosa at steady state, potentially implicating Treg cells in keeping this tissue quiescent (21, 69). In cancer, antigens presented by intratumoral dendritic cells drive Treg cells to actively suppress CD8 T cells (57, 70). Reduced MHC-II antigen presentation capacity by LCs in LCΔMHC-II mice might blunt the suppressive function of oral Treg cells via immunosuppressive cytokines IL-10 and/or IL-35 (71–73). Any reduction in local immunosuppression may, therefore, allow the expansion of tissue-resident CD8 T cells.
In summary, we report here that reduced frequency of oral LCs expressing MHC-II results in increased intraepithelial tissue-resident Tc17 cells in oral mucosa. Although to a lesser extent, classic IFN-γ expressing Tc1 cells also increased. Manipulating MHC-II dependent interactions with LCs in barrier tissues could be exploited to boost defense against diseases that benefit from increased Tc1 or Tc17 responses. Further work is required to delineate the exact mechanism by which LCs dampen CD8 T cells via CD4 T cells at barrier tissues. Here it would be of interest to determine if CD8 T cells in the oral mucosa respond to stable members of the oral microbial community and whether these CD8 T cells recognize formylated peptides presented on non-classical MHC-I molecules (65).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
PB: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Visualization. LF: Writing – review & editing, Data curation, Investigation, Visualization. PP: Data curation, Investigation, Validation, Writing – original draft, Methodology, Visualization. MC: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Resources, Writing – original draft, Writing – review & editing, Methodology, Supervision, Validation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
Funding was provided by NIH/NIDCR grants 1R21DE029289-01A1 [MC] and 1R03DE025882-01 [PDB-E].
Acknowledgments
We thank Drs. Anna Dongari-Bagtzoglou (University of Connecticut) and Dominique Sanglard (University of Lausanne) for the kind gifts of Streptococcus oralis 34 and Candida albicans, respectively and Dr. Weihua Guan at the Biostatistical Design and Analysis Center (BDAC) core, Clinical and Translational Science Institute for statistical support. The NIH tetramer facility at Emory University provided us with MR1 tetramer reagents. The University of Minnesota Institute for Molecular Virology is acknowledged for providing flow cytometry and confocal microscopy equipment. The University Imaging Center is acknowledged for providing bioluminescent imaging resources. We thank Dr. Mark Herzberg for editorial comments and suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/froh.2024.1408255/full#supplementary-material
Supplementary Figure S1
Identification of immune cell subsets in mouse oral mucosa by flow cytometry. Flow cytometry gating strategy utilized to identify immune cell subsets in total oral mucosa, epithelial sheets from oral mucosa, or lamina propria from oral mucosa. Single cell suspensions from an oral mucosal epithelium sheet from a LC∆MHC-II mouse were stained with Zombie Aqua to distinguish live from dead cells, Fc receptors blocked with rat anti-mouse CD16/32 mAb followed by staining with a panel of rat anti-mouse mAbs against cell surface markers. Typical flow cytometry plots obtained are shown. Percentage of cells within specific gates are indicated. The population of live non-immune cells used for normalization purposes is indicated.
Supplementary Figure S2
IL-17A expressing CD8 T cells in LC∆MH-II mice are not MAIT cells. Oral mucosa was harvested from LC∆MH-II mice or pooled from LCWT littermate controls. Single cell suspensions were stained with MR1 tetramers MR1 5-OP-RU or MR1 6-FP and then surface stained to identify CD8 T cells, CD4 T cells and CD3+ double negative (DN) T cells. Live CD8 T cells were identified as Zombie Aqualo, CD45+, CD3+, TCRb+, CD8a+, CD4-; Live CD4 T cells as Zombie Aqualo, CD45+, CD3+, TCRb+, CD8a-, CD4+; Live CD3+ DN T cells as Zombie Aqualo, CD45+, CD3+, TCRb+, CD8a-, CD4-. (A) Representative flow cytometry dot plots for CD4 T cells (left panels) and CD8 T cells (right panels) for both the MAIT MR1 tetramer (MR1 5-OP-RU) and the MR1 negative control tetramer (MR1 6-FP). Gates, drawn to detect potential MAIT cells, are based off the negative control samples stained with MR1 6-FP. Numbers indicate percentage of cells within the drawn gates. (B) Representative flow cytometry dot plots for MR1 5-OP-RU and MR1 6-FP stained CD3+ DN T cells.
Abbreviations
5-OP-RU, 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil; 6-FP, 6-formyl pterin; BHI, Brain-Heart Infusion; IVIS, in vivo imaging system; LCs, Langerhans cells; MHC-I, major histocompatibility complex class I; MHC-II, Major histocompatibility complex class II; RORγt, retinoic acid-related Orphan Receptor gamma t; STAT3, Signal Transducer and Activator of Transcription 3; TCR, T cell receptor; Th1, CD4 T helper 1 lymphocytes; Th17, CD4 T helper 17 lymphocytes; Treg, CD4 T regulatory lymphocytes; Tc1, CD8 T cytotoxic 1 cells; Tc17, CD8 T cytotoxic 17 cells; γδ T cells, gamma-delta T cells; TGF-β1, transforming growth factor-beta 1; TGF-R1, transforming growth factor-receptor 1; YPD, Yeast Peptone Dextrose.
References
1. Kaplan DH. Ontogeny and function of murine epidermal langerhans cells. Nat Immunol. (2017) 18(10):1068–75. doi: 10.1038/ni.3815
2. Hovav AH. Mucosal and skin langerhans cells—nurture calls. Trends Immunol. (2018) 39(10):788–800. doi: 10.1016/j.it.2018.08.007
3. Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, et al. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. (2011) 35(2):260–72. doi: 10.1016/j.immuni.2011.06.005
4. Bittner-Eddy PD, Fischer LA, Kaplan DH, Thieu K, Costalonga M. Mucosal langerhans cells promote differentiation of Th17 cells in a murine model of periodontitis but are not required for Porphyromonas Gingivalis-driven alveolar bone destruction. J Immunol. (2016) 197(4):1435–46. doi: 10.4049/jimmunol.1502693
5. Heyman O, Koren N, Mizraji G, Capucha T, Wald S, Nassar M, et al. Impaired differentiation of langerhans cells in the murine oral epithelium adjacent to Titanium dental implants. Front Immunol. (2018) 9:1712. doi: 10.3389/fimmu.2018.01712
6. Arizon M, Nudel I, Segev H, Mizraji G, Elnekave M, Furmanov K, et al. Langerhans cells down-regulate inflammation-driven alveolar bone loss. Proc Natl Acad Sci U S A. (2012) 109(18):7043–8. doi: 10.1073/pnas.1116770109
7. Capucha T, Mizraji G, Segev H, Blecher-Gonen R, Winter D, Khalaileh A, et al. Distinct murine mucosal langerhans cell subsets develop from pre-dendritic cells and monocytes. Immunity. (2015) 43(2):369–81. doi: 10.1016/j.immuni.2015.06.017
8. Conti HR, Gaffen SL. Il-17-mediated immunity to the opportunistic fungal pathogen Candida albicans. J Immunol. (2015) 195(3):780–8. doi: 10.4049/jimmunol.1500909
9. Liu F, Fan X, Auclair S, Ferguson M, Sun J, Soong L, et al. Sequential dysfunction and progressive depletion of Candida albicans-specific CD4 T cell response in hiv-1 infection. PLoS Pathog. (2016) 12(6):e1005663. doi: 10.1371/journal.ppat.1005663
10. Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17a for systemic anti-Candida albicans host defense in mice. J Infect Dis. (2004) 190(3):624–31. doi: 10.1086/422329
11. Gaffen SL, Moutsopoulos NM. Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci Immunol. (2020) 5(43):1–12. doi: 10.1126/sciimmunol.aau4594
12. Conti HR, Bruno VM, Childs EE, Daugherty S, Hunter JP, Mengesha BG, et al. Il-17 receptor signaling in oral epithelial cells is critical for protection against oropharyngeal candidiasis. Cell Host Microbe. (2016) 20(5):606–17. doi: 10.1016/j.chom.2016.10.001
13. McGeachy MJ, Cua DJ, Gaffen SL. The Il-17 family of cytokines in health and disease. Immunity. (2019) 50(4):892–906. doi: 10.1016/j.immuni.2019.03.021
14. Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, et al. Th17 cells and Il-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. (2009) 206(2):299–311. doi: 10.1084/jem.20081463
15. Conti HR, Gaffen SL. Host responses to Candida albicans: th17 cells and mucosal candidiasis. Microbes Infect. (2010) 12(7):518–27. doi: 10.1016/j.micinf.2010.03.013
16. Sparber F, Dolowschiak T, Mertens S, Lauener L, Clausen BE, Joller N, et al. Langerin+ dcs regulate innate Il-17 production in the oral Mucosa during Candida albicans-mediated infection. PLoS Pathog. (2018) 14(5):e1007069. doi: 10.1371/journal.ppat.1007069
17. Huber M, Heink S, Grothe H, Guralnik A, Reinhard K, Elflein K, et al. A Th17-like developmental process leads to Cd8(+) Tc17 cells with reduced cytotoxic activity. Eur J Immunol. (2009) 39(7):1716–25. doi: 10.1002/eji.200939412
18. Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. (2015) 520:104–8. doi: 10.1038/nature14052
19. Hernandez-Santos N, Huppler AR, Peterson AC, Khader SA, McKenna KC, Gaffen SL. Th17 cells confer long-term adaptive immunity to oral mucosal Candida albicans infections. Mucosal Immunol. (2013) 6(5):900–10. doi: 10.1038/mi.2012.128
20. Flores-Santibanez F, Cuadra B, Fernandez D, Rosemblatt MV, Nunez S, Cruz P, et al. In vitro-generated Tc17 cells present a memory phenotype and serve as a reservoir of Tc1 cells in vivo. Front Immunol. (2018) 9:209. doi: 10.3389/fimmu.2018.00209
21. Fischer LA, Bittner-Eddy PD, Costalonga M. MHC-II expression on oral langerhans cells differentially regulates mucosal CD4 and CD8 T cells. J Invest Dermatol. (2023) 144:573–84. doi: 10.1016/j.jid.2023.09.277
22. Bittner-Eddy PD, Fischer LA, Costalonga M. Cre-loxp reporter mouse reveals stochastic activity of the Foxp3 promoter. Front Immunol. (2019) 10:2228. doi: 10.3389/fimmu.2019.02228
23. Bittner-Eddy PD, Fischer LA, Tu AA, Allman DA, Costalonga M. Discriminating between interstitial and circulating leukocytes in tissues of the murine oral mucosa avoiding nasal-associated lymphoid tissue contamination. Front Immunol. (2017) 8:1398. doi: 10.3389/fimmu.2017.01398
24. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–excel-based tool using pair-wise correlations. Biotechnol Lett. (2004) 26(6):509–15. doi: 10.1023/b:bile.0000019559.84305.47
25. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta C(T)) method. Methods. (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
26. Li HY, Wang Y, Heap CR, King CH, Mundla SR, Voss M, et al. Dihydropyrrolopyrazole transforming growth factor-beta type I receptor kinase domain inhibitors: a novel benzimidazole series with selectivity versus transforming growth factor-beta type ii receptor kinase and mixed lineage kinase-7. J Med Chem. (2006) 49(6):2138–42. doi: 10.1021/jm058209g
27. Xu H, Sobue T, Bertolini M, Thompson A, Dongari-Bagtzoglou A. Streptococcus Oralis and Candida albicans synergistically activate mu-calpain to degrade E-cadherin from oral epithelial junctions. J Infect Dis. (2016) 214(6):925–34. doi: 10.1093/infdis/jiw201
28. Dorsaz S, Coste AT, Sanglard D. Red-shifted firefly luciferase optimized for Candida albicans in vivo bioluminescence imaging. Front Microbiol. (2017) 8:1478. doi: 10.3389/fmicb.2017.01478
29. Iijima N, Linehan MM, Zamora M, Butkus D, Dunn R, Kehry MR, et al. Dendritic cells and B cells maximize mucosal Th1 memory response to herpes simplex virus. J Exp Med. (2008) 205(13):3041–52. doi: 10.1084/jem.20082039
30. Masopust D, Soerens AG. Tissue-resident T cells and other resident leukocytes. Annu Rev Immunol. (2019) 37:521–46. doi: 10.1146/annurev-immunol-042617-053214
31. Mahanonda R, Champaiboon C, Subbalekha K, Sa-Ard-Iam N, Yongyuth A, Isaraphithakkul B, et al. Memory T cell subsets in healthy gingiva and periodontitis tissues. J Periodontol. (2018) 89(9):1121–30. doi: 10.1002/JPER.17-0674
32. Deli F, Romano F, Gualini G, Mariani GM, Sala I, Veneziano F, et al. Resident memory T cells: possible players in periodontal disease recurrence. J Periodontal Res. (2020) 55(2):324–30. doi: 10.1111/jre.12709
33. Cheuk S, Schlums H, Gallais Serezal I, Martini E, Chiang SC, Marquardt N, et al. Cd49a expression defines tissue-resident Cd8(+) T cells poised for cytotoxic function in human skin. Immunity. (2017) 46(2):287–300. doi: 10.1016/j.immuni.2017.01.009
34. Tokura Y, Phadungsaksawasdi P, Kurihara K, Fujiyama T, Honda T. Pathophysiology of skin resident memory T cells. Front Immunol. (2020) 11:618897. doi: 10.3389/fimmu.2020.618897
35. Cheroutre H, Lambolez F. Doubting the TCR coreceptor function of Cd8alphaalpha. Immunity. (2008) 28(2):149–59. doi: 10.1016/j.immuni.2008.01.005
36. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using Mr1 tetramers. J Exp Med. (2015) 212(7):1095–108. doi: 10.1084/jem.20142110
37. Shrikant PA, Rao R, Li Q, Kesterson J, Eppolito C, Mischo A, et al. Regulating functional cell fates in CD8 T cells. Immunol Res. (2010) 46(1–3):12–22. doi: 10.1007/s12026-009-8130-9
38. Vanpouille-Box C, Diamond JM, Pilones KA, Zavadil J, Babb JS, Formenti SC, et al. Tgfbeta is a master regulator of radiation therapy-induced antitumor immunity. Cancer Res. (2015) 75(11):2232–42. doi: 10.1158/0008-5472.CAN-14-3511
39. Moynihan KD, Opel CF, Szeto GL, Tzeng A, Zhu EF, Engreitz JM, et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat Med. (2016) 22(12):1402–10. doi: 10.1038/nm.4200
40. Kim J, Jeong Ryu S, Oh K, Ju JM, Yeong Jeon J, Nam G, et al. Memory programming in Cd8(+) T-cell differentiation is intrinsic and is not determined by Cd4 help. Nat Commun. (2015) 6:7994. doi: 10.1038/ncomms8994
41. Bittner-Eddy PD, Fischer LA, Costalonga M. Transient expression of Il-17a in Foxp3 fate-tracked cells in Porphyromonas Gingivalis-mediated oral dysbiosis. Front Immunol. (2020) 11:677. doi: 10.3389/fimmu.2020.00677
42. Ciucci T, Vacchio MS, Bosselut R. A Stat3-dependent transcriptional circuitry inhibits cytotoxic gene expression in T cells. Proc Natl Acad Sci U S A. (2017) 114(50):13236–41. doi: 10.1073/pnas.1711160114
43. Veldhoen M, Stockinger B. Tgfbeta1, a “jack of all trades”: the link with pro-inflammatory Il-17-producing T cells. Trends Immunol. (2006) 27(8):358–61. doi: 10.1016/j.it.2006.06.001
44. Yen HR, Harris TJ, Wada S, Grosso JF, Getnet D, Goldberg MV, et al. Tc17 CD8 T cells: functional plasticity and subset diversity. J Immunol. (2009) 183(11):7161–8. doi: 10.4049/jimmunol.0900368
45. Chen HW, Tsai JP, Yao TY, Hsieh CL, Chen IH, Liu SJ. Tgf-beta and il-21 cooperatively stimulate activated Cd8(+) T cells to differentiate into Tc17 cells. Immunol Lett. (2016) 174:23–7. doi: 10.1016/j.imlet.2016.04.006
46. Bromley SK, Akbaba H, Mani V, Mora-Buch R, Chasse AY, Sama A, et al. Cd49a regulates cutaneous resident memory Cd8(+) T cell persistence and response. Cell Rep. (2020) 32(9):108085. doi: 10.1016/j.celrep.2020.108085
47. Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. (2012) 188(10):4866–75. doi: 10.4049/jimmunol.1200402
48. Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, et al. Mice lacking all conventional mhc class ii genes. Proc Natl Acad Sci U S A. (1999) 96(18):10338–43. doi: 10.1073/pnas.96.18.10338
49. Do JS, Valujskikh A, Vignali DA, Fairchild RL, Min B. Unexpected role for mhc ii-peptide complexes in shaping CD8 T-cell expansion and differentiation in vivo. Proc Natl Acad Sci U S A. (2012) 109(31):12698–703. doi: 10.1073/pnas.1207219109
50. Rubino SJ, Geddes K, Magalhaes JG, Streutker C, Philpott DJ, Girardin SE. Constitutive induction of intestinal Tc17 cells in the absence of hematopoietic cell-specific mhc class ii expression. Eur J Immunol. (2013) 43(11):2896–906. doi: 10.1002/eji.201243028
51. Chaoul N, Tang A, Desrues B, Oberkampf M, Fayolle C, Ladant D, et al. Lack of mhc class ii molecules favors Cd8(+) T-cell infiltration into tumors associated with an increased control of tumor growth. Oncoimmunology. (2018) 7(3):e1404213. doi: 10.1080/2162402X.2017.1404213
52. Nanjappa SG, Heninger E, Wuthrich M, Sullivan T, Klein B. Protective antifungal memory Cd8(+) T cells are maintained in the absence of Cd4(+) T cell help and cognate antigen in mice. J Clin Invest. (2012) 122(3):987–99. doi: 10.1172/JCI58762
53. Tsai JP, Lee MH, Hsu SC, Chen MY, Liu SJ, Chang JT, et al. Cd4+ T cells disarm or delete cytotoxic T lymphocytes under Il-17-polarizing conditions. J Immunol. (2012) 189(4):1671–9. doi: 10.4049/jimmunol.1103447
54. Marquis M, Lewandowski D, Dugas V, Aumont F, Senechal S, Jolicoeur P, et al. Cd8+ T cells but not polymorphonuclear leukocytes are required to limit chronic oral carriage of Candida albicans in transgenic mice expressing human immunodeficiency virus type 1. Infect Immun. (2006) 74(4):2382–91. doi: 10.1128/IAI.74.4.2382-2391.2006
55. Kader M, Bixler S, Piatak M, Lifson J, Mattapallil JJ. Anti-retroviral therapy fails to restore the severe th-17: tc-17 imbalance observed in peripheral blood during simian immunodeficiency virus infection. J Med Primatol. (2009) 38(Suppl 1):32–8. doi: 10.1111/j.1600-0684.2009.00373.x
56. Nigam P, Kwa S, Velu V, Amara RR. Loss of Il-17-producing CD8 T cells during late chronic stage of pathogenic simian immunodeficiency virus infection. J Immunol. (2011) 186(2):745–53. doi: 10.4049/jimmunol.1002807
57. McRitchie BR, Akkaya B. Exhaust the exhausters: targeting regulatory T cells in the tumor microenvironment. Front Immunol. (2022) 13:940052. doi: 10.3389/fimmu.2022.940052
58. Park JY, Chung H, Choi Y, Park JH. Phenotype and tissue residency of lymphocytes in the murine oral mucosa. Front Immunol. (2017) 8:250. doi: 10.3389/fimmu.2017.00250
59. Mackay LK, Stock AT, Ma JZ, Jones CM, Kent SJ, Mueller SN, et al. Long-lived epithelial immunity by tissue-resident memory T (trm) cells in the absence of persisting local antigen presentation. Proc Natl Acad Sci U S A. (2012) 109(18):7037–42. doi: 10.1073/pnas.1202288109
60. Mohammed J, Beura LK, Bobr A, Astry B, Chicoine B, Kashem SW, et al. Stromal cells control the epithelial residence of dcs and memory T cells by regulated activation of tgf-beta. Nat Immunol. (2016) 17(4):414–21. doi: 10.1038/ni.3396
61. Gartlan KH, Markey KA, Varelias A, Bunting MD, Koyama M, Kuns RD, et al. Tc17 cells are a proinflammatory, plastic lineage of pathogenic Cd8+ T cells that induce gvhd without antileukemic effects. Blood. (2015) 126(13):1609–20. doi: 10.1182/blood-2015-01-622662
62. Dwivedi VP, Tousif S, Bhattacharya D, Prasad DV, Van Kaer L, Das J, et al. Transforming growth factor-Beta protein inversely regulates in vivo differentiation of interleukin-17 (Il-17)-producing Cd4+ and Cd8+ T cells. J Biol Chem. (2012) 287(5):2943–7. doi: 10.1074/jbc.C111.327627
63. Luckel C, Picard FSR, Huber M. Tc17 biology and function: novel concepts. Eur J Immunol. (2020) 50(9):1257–67. doi: 10.1002/eji.202048627
64. Di Meglio P, Villanova F, Navarini AA, Mylonas A, Tosi I, Nestle FO, et al. Targeting Cd8(+) T cells prevents psoriasis development. J Allergy Clin Immunol. (2016) 138(1):274–6. doi: 10.1016/j.jaci.2015.10.046
65. Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. (2018) 172(4):784–96.e18. doi: 10.1016/j.cell.2017.12.033
66. Park CO, Fu X, Jiang X, Pan Y, Teague JE, Collins N, et al. Staged development of long-lived T-cell receptor alphabeta T(H)17 resident memory T-cell population to Candida albicans after skin infection. J Allergy Clin Immunol. (2018) 142(2):647–62. doi: 10.1016/j.jaci.2017.09.042
67. Kashem SW, Igyarto BZ, Gerami-Nejad M, Kumamoto Y, Mohammed JA, Jarrett E, et al. Candida albicans morphology and dendritic cell subsets determine T helper cell differentiation. Immunity. (2015) 42(2):356–66. doi: 10.1016/j.immuni.2015.01.008
68. Lee H, Jeong S, Shin EC. Significance of bystander T cell activation in microbial infection. Nat Immunol. (2022) 23(1):13–22. doi: 10.1038/s41590-021-00985-3
69. Park JY, Chung H, DiPalma DT, Tai X, Park JH. Immune quiescence in the oral mucosa is maintained by a uniquely large population of highly activated Foxp3(+) regulatory T cells. Mucosal Immunol. (2018) 11(4):1092–102. doi: 10.1038/s41385-018-0027-2
70. Bauer CA, Kim EY, Marangoni F, Carrizosa E, Claudio NM, Mempel TR. Dynamic treg interactions with intratumoral apcs promote local ctl dysfunction. J Clin Invest. (2014) 124(6):2425–40. doi: 10.1172/JCI66375
71. Turnis ME, Sawant DV, Szymczak-Workman AL, Andrews LP, Delgoffe GM, Yano H, et al. Interleukin-35 limits anti-tumor immunity. Immunity. (2016) 44(2):316–29. doi: 10.1016/j.immuni.2016.01.013
72. Sawant DV, Yano H, Chikina M, Zhang Q, Liao M, Liu C, et al. Adaptive plasticity of il-10(+) and il-35(+) T(reg) cells cooperatively promotes tumor T cell exhaustion. Nat Immunol. (2019) 20(6):724–35. doi: 10.1038/s41590-019-0346-9
Keywords: Langerhans cells, Candida albicans, T cytotoxic 17 cells (Tc17), CD4 T cells, MHC-II antigen presentation, oral mucosa
Citation: Bittner-Eddy PD, Fischer LA, Parachuru PV and Costalonga M (2024) MHC-II presentation by oral Langerhans cells impacts intraepithelial Tc17 abundance and Candida albicans oral infection via CD4 T cells. Front. Oral. Health 5:1408255. doi: 10.3389/froh.2024.1408255
Received: 27 March 2024; Accepted: 13 May 2024;
Published: 30 May 2024.
Edited by:
Hatice Hasturk, The Forsyth Institute, United StatesReviewed by:
Agnes Schröder, University Hospital Regensburg, GermanyTomoki Maekawa, Niigata University, Japan
Carolina Isabel Rojas, University of Chile, Chile
© 2024 Bittner-Eddy, Fischer, Parachuru and Costalonga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimo Costalonga, costa002@umn.edu
†ORCID:
Peter D. Bittner-Eddy
orcid.org/0009-0003-0429-0965
Lori A. Fischer
orcid.org/0009-0004-5327-3683
Praveen Venkata Parachuru
orcid.org/0000-0002-1301-5047
Massimo Costalonga
orcid.org/0000-0001-5922-3709
 Peter D. Bittner-Eddy
Peter D. Bittner-Eddy Lori A. Fischer
Lori A. Fischer Praveen Venkata Parachuru
Praveen Venkata Parachuru Massimo Costalonga
Massimo Costalonga