Rhizospheric microorganisms: The gateway to a sustainable plant health
- Food Security and Safety Focus Area, Faculty of Natural and Agricultural Sciences, North-West University, Mmabatho, South Africa
Plant health is essential for food security, and constitutes a major predictor to safe and sustainable food systems. Over 40% of the global crops' productions are lost to pests, insects, diseases, and weeds, while the routinely used chemical-based pesticides to manage the menace also have detrimental effects on the microbial communities and ecosystem functioning. The rhizosphere serves as the microbial seed bank where microorganisms transform organic and inorganic substances in the rhizosphere into accessible plant nutrients as plants harbor diverse microorganisms such as fungi, bacteria, nematodes, viruses, and protists among others. Although, the pathogenic microbes initiate diseases by infiltrating the protective microbial barrier and plants' natural defense systems in the rhizosphere. Whereas, the process is often circumvented by the beneficial microorganisms which antagonize the pathogens to instill disease resistance. The management of plant health through approaches focused on disease prevention is instrumental to attaining sustainable food security, and safety. Therefore, an in-depth understanding of the evolving and succession of root microbiomes in response to crop development as discussed in this review opens up new-fangled possibilities for reaping the profit of beneficial root–microbiomes' interactions toward attaining sustainable plant health.
Introduction
Agricultural management practices, such as crop rotation, fertilizer application, and land tilling modify soil physical and chemical properties, amidst diverse interacting forces which shape soil microbial populations (Schmidt et al., 2019). The important plant microbiomes can be categorized into aboveground and belowground. The phyllosphere/phyllobiome characterized by the aboveground is made up of leaves, stems, and reproductive organs, while the rhizosphere/rhizobiome which consists of the belowground is the roots' part and the associated soil (Lucaciu et al., 2019). The interactions between plant roots and microorganisms have a direct impact on the soil, and the rhizosphere region is the home to diverse microorganisms (Adedeji and Babalola, 2020), and a “hot spot” of intensive microbial activity (Dai et al., 2019). The plant microbiome consists of a variety of microbes such as bacteria, fungi, protists, archaea, and viruses that reside on or inside their host plants (Zhang et al., 2021). Rhizosphere-associated microbes have an extensive variety of metabolic abilities and play an important role in the rhizosphere environment, such as organic matter breakdown and nutrient cycling, both of which benefit plant growth and health. More importantly, the microbial population aids plant development and responds to a range of environmental conditions (Dai et al., 2019).
Plants are key sources of energy input in a plant-soil subsystem, providing nutrients and carbon for heterotrophic microbial communities in the form of root exudates and litter deposits (Shanmugam and Kingery, 2018). Through rhizodeposition which is a major phenomenon that occurs in the rhizosphere, high- and low-molecular-weight compounds including secondary metabolites, organic acids, amino acids, sugars, vitamins, and polysaccharides are released into the surrounding soil in the form of exudates. This increases the richness of microbiological and functional diversity, leading to the formation of extremely lively and unique microbial communities linked with plant roots (Chukwuneme et al., 2021). Although the microbiome is made up of a variety of organisms, most research on microbial function in disease prevention and plant health has concentrated on fungi and bacteria (Xiong et al., 2020).
As reported in a recent study, soil and climatic characteristics are significant determinants of microbial community composition, variety, and metabolic potential in a range of the surroundings (Lladó et al., 2018). This is a result of the swift response of the microbes to abiotic and biotic changes. Hence, the structure and function of soil microbial communities are frequently utilized as pointers for determining soil fertility and quality (Enagbonma et al., 2020). Further insights into the essential functions of rhizosphere microbiomes and plant genetic characteristics under various situations have been accessed through the advent of OMICS technology which is based on Next-Generation Sequencing (NGS) (Pramanik et al., 2020), and provides the gateway to attaining sustainable plant health.
The plant rhizospheric zone is a niche of microbial activity based on the resistance of the root system against phytopathogens. The region provides a variety of benefits to plant health both directly and indirectly (Gupta et al., 2019). It is generally recognized that soil microbes serve as a primary nutrient reservoir for plants through their active participation in nutrient cycle processes such as nitrogen cycling, organic matter decomposition and mineral weathering, all of which influence plant growth (Jilling et al., 2018). Thus, plant roots and microbe interactions are critical for plant participation in the ecosystem's functioning. Understanding ecosystem responses to the changing environments require knowledge of microbial diversity distribution and functions. More so, root exudates contain an extensive variety of chemicals, including amino acids, carbohydrates, siderophores, and enzymes, that can reinforce the activities of the microbial community and significantly change the structure of the rhizospheric microbial community, contingent on the plant species and genotype (Wei et al., 2017). Plant-associated microbial communities promote plant development and resilience to diverse abiotic and biotic stressors. Soil microbial species through their roles in carbon sequestration and biogeochemical cycling play important roles in the mitigation of climate change that presently constitutes a global concern (Dubey et al., 2019). Improved ability to predict functionality from dynamic aberrations in root microbial composition is critical for building “designer” microbiome interventions and enhancing agricultural production and sustainability. This will necessitate a more comprehensive genomic and functional classification of plant-linked microbes, their genetic factor, and plant-microbe interactions for their exact useful roles (Richardson et al., 2021).
Beneficial, commensal, or pathogenic relationships exist between specific microorganisms and their plant hosts. Plant microbiomes, in many instances, show a beneficial role at the community level, protecting plants from pathogens, while also boosting growth, health, and providing plants with an adaptive edge (Zhang et al., 2021). Therefore, the interactions between plants and their microbiota are intricate and constantly changing (Fausto et al., 2018). This interaction and interdependence on the abiotic and biotic components of the soil environment can be explained by the plant-soil feedback mechanism, as shown by Shanmugam and Kingery (2018).
Plant defense system: Abating the invasion of pathogens by the rhizospheric microbiome
Plant diseases have caused serious economic and nutritional crises in the past and are still responsible for significant losses in global crop production today (Chun and Chandrasekaran, 2019). Among the factors that contribute to production loss, pests and diseases constitute major global damages (Ab-Rahman et al., 2018). As a result of the unrestrained and widespread use of artificial agrochemicals for plant protection and growth, environmental pollution has become a serious problem. Agrochemical use indefinitely has various negative consequences, including increasing resistance in plant pathogenic microorganisms, detrimental impacts on non-target organisms, and deterioration of soil health (Choudhary et al., 2017). When it comes to plant disease, there is a complex interplay between the host plant and the pathogen, and the resistance/susceptibility response might include a variety of factors (Kumar and Dubey, 2020). Disease suppressive soils are the most visible example of microbe-mediated plant defense against pathogen infection in agricultural soils Meanwhile, structures such as the physical and chemical barriers, quick active defenses, passive and delayed active defenses including the non-host resistance, all cumulate to plant defense systems. However, the rapid active defenses entail changes in cell wall reinforcement, membrane function, the hypersensitive reaction, and the initial oxidative burst which results in phytoalexins, and programmed cell death.
Systemic acquired resistance, pathogenesis-related gene expression, pathogen containment, and wound repair are all examples of delayed active defenses (SAR). Salicylic acid, which is required for defense against SAR and biotrophic pathogens, along with ethylene and jasmonic acid are both involved in defense against necrotrophic pathogens in addition to beneficial plant-microbe interactions, such as induced systemic resistance (ISR) and priming as examples of plant defense signaling molecules (Ab-Rahman et al., 2018). Our ability to control and modify the rhizosphere microbiota is currently restricted. Inoculation is the most direct technique to change the microbiome (Kumar and Dubey, 2020). For decades, commercially accessible products comprising of one or more kinds of fungi or bacteria have been available. The majority of these species, however, were obtained using typical culturing methods that do not mimic the chemical milieu of soil. Other options include adding organic materials to soils that contain diverse but undefined microbial mixes in the hopes of improving function and pathogen resistance. In this case, the capacity of beneficial inoculants could be limited to spread (Alori et al., 2017). Plant breeding or engineering features are other techniques to engineer the rhizosphere. We will be able to engineer root architecture, specific root exudates, or other plant qualities that support beneficial microbiomes into crops using CRISPR and other gene-editing technologies as we identify root architecture, specific root exudates, or other plant traits that support beneficial microbiomes. This technology has been effectively explored in safeguarding plants against both biotic and abiotic stress. An instance is the production of rice lines which has broad-spectrum resistance to Xanthomonas oryzae pv. oryzae that resulted in the modification of the promoter region of O. sativa SWEET11, O. sativa SWEET13, and O. sativa SWEET14 (Oliva et al., 2019), while other studies have also investigated the plant—microbe interactions with the use of CRISPR-Cas (Shelake et al., 2019). This method appears promising because it mimics the interactions that sustain beneficial microorganisms in natural systems and that have been designated through holobiont development (Wallenstein, 2017; Thomashow et al., 2019). Chemical pesticides are now widely employed to ensure long-term productivity and harvest, stressing the widespread usage of phytochemical pesticides. Nonetheless, these chemical pesticide tactics have negative consequences for crops, the environment, and the health of both farming communities and consumer populations. As a result, alternative and long-term disease methods are required. An alternative pest control strategy using bio-pesticides instead of synthetic or chemical pesticides poses a positive phenomenon in reducing pest threat with prevention of chemical residue deposition, reduced toxicity, combinatorial optimal attributes on efficacy, and reduced environmental impacts in the realm of humans and the environment affecting only the pests will positively have an impact (Chun and Chandrasekaran, 2019). In some recent studies, the success of biopesticde formulations from varying Trichoderma species were reported effective in the management of tomoto root rot diseases (Olowe et al., 2022), while Mishra et al. (2022) found that the use of 2,4- DAPG-producing P. fluorescens JM-1 suppressd F. monoliforme-causing ear rot diseases in maize and the PGP features of P. fluorescens JM-1 (IAA generation and solubilization phosphate and Zn) also aided in the enhancement of maize growth indices.
Given the importance of microorganisms in nutrient mineralization and solubilization, soil organic matter creation, and pathogen pressure, rhizosphere engineering would benefit from methodologies and tools that permit us to modify the microbiome (Wallenstein, 2017). The interaction between plants and rhizospheric bacteria has been thoroughly investigated for biogeochemical cycling activities, plant growth promotion, and biocontrol, all of which are critical for plant health. Hence the need for in-depth knowledge of microbial diversities in the plant rhizosphere to enable adequate deployment of plant beneficial microbes toward plant growth promotion (Olanrewaju et al., 2019). Variations in the physical environmental condition and structure of the host-associated rhizosphere microorganisms are frequently linked to plant pathogen invasion. Thus, understanding plant-pathogen infections in highly varied field conditions constitutes a critical challenge for food security and crop protection in the future (Wei et al., 2018). The pathogen's introduction has the potential to disrupt the plant's finely regulated relationships with its microbiome. Changes in the native microbial community structure and interactions that provide favorable conditions for the pathogen virulence led to fast declines in host health and productivity (Zhang et al., 2021). For that reason, the pathogen must compete with other bacterial species for colonization of the plant rhizosphere, which is the initial stage in infection. When a pathogen reaches a certain density, it activates its virulence genes and invades plant roots (Wei et al., 2018), which may infect host tissue and successfully exit the rhizosphere war zone, most soil-borne pathogens must grow saprophytically in the rhizosphere to influence their host or to attain adequate numbers on their host. The microbial population of the soil in which infection occurs has an impact on the pathogen's success. To some extent, typical agricultural soils have the ability to restrict a pathogen. This can be determined by comparing the severity of disease after pathogen inoculation in sterilized and non-sterilized soils (Berendsen et al., 2012). Recent research has found that microbiome functioning, including disease resistance, may result through the interaction of species instead of the existence of a specific feature or species. This have been shown as an emerging approach to overcoming the laboratory to field hurdles, since the use of microbial consortia entails the deployment of microbes with similar mode of action that are able to tolerate different environmental conditions and effective om the plant genotypes, thereby complementing each other. This has been effectively explored in tomato, maize and potato among other crops (Compant et al., 2019). Each stage of ecological succession is connected with certain features in the main species, putting the existing community arrangement in the context of a dynamic temporal procedure. When applied to the rhizosphere, ecological succession can offer a novel prognostic outline for species and trait occurrence, offer new entry points to engineer its ability to prevent diseases, and elucidate how a suppressive microbiome assembles (Hu et al., 2020).
The plant-soil feedback: Essential tools in microbiome management
The past investigations of plant-soil feedback have revealed the varying effect of different plant species on the biotic and abiotic soil conditions, which in turn affects the growths of other heterospecific and conspecific individuals in that soil (Kulmatiski and Kardol, 2008). Either positively or negatively, the plant-soil feedbacks can drive patterns of plant diversity, succession, and invasion, which can shape the community composition and ecosystem functioning. While the environmental forces that occur over the landscape include stress, disturbance, and competition, plants allocate resources to growth, reproduction, and maintenance in diverse ways to cope with these constraints (Beals et al., 2020).
One of the basic structures that allow plant stability against both biotic and abiotic pressures is the root system. Although scientists have given less attention to the study of plants root, roots have been found essential for plant productivity and adaptations. The root architecture is made up of structural features such as root length, length of lateral roots, and spread number that enable plants to exhibit plasticity in response to environmental changes, and the optimum performance of the root (Khan et al., 2016). This three-dimensional spatial organization of root structures is especially important for the function of roots in difficult situations. A deep root structure, for example, is required for plants to use water and nitrate in deeper soil layers, and hence useful for drought resistance, mostly in nitrogen-scarce circumstances. A shallow root structure with increased adventitious roots to promote foraging on the topsoil, on the other hand, is critical for crops to acquire comparatively steady nutrients like phosphorus. Developing cultivars with root architectures that are appropriate for the field conditions promises to be a long-term and cost-effective way to improve crop nutrient efficiency and stress tolerance (Yang et al., 2017). The advent of more detailed molecular and analytical tools which allow full explanation of the dynamics of rhizosphere and endosphere communities has sparked transformed interest in plant-microbe interactions.
Recent data has shown that root type and location influence the composition and diversity of the communities of bacterial, fungal, and actinobacterial, including specialized functional groups like arbuscular mycorrhizal fungi, across a wide range of plant species. Differences in root assembly and the quantity and composition of root exudates, as well as plant-microbe, microbe-microbe, and microbe–fauna interactions, all contribute to these variances (Richardson et al., 2021). The importance of interactions between plants and their phyllosphere microbes is gaining attention, as they stimulate plant development and contribute to pathogen defense, these may later alter the suitability of natural plant inhabitants along with the quality and production of agricultural products. Meanwhile, endophytic bacteria have been recognized for their ability to defend their hosts from pests, insects, and microbiological diseases, thereby acting as biocontrol agents (Fausto et al., 2018). Crop rotation may also improve microbial diversity in soil because various plant species favor distinct bacteria in the soil due to changes in root exudation. Intercropping, where diverse crops are planted together and the roots of different plant species interact directly, has a good impact on soil fertility in addition to microbial variety and activity (Mitter et al., 2019).
A greater understanding of the root microbiome succession, such as how the microbes vary in response to crop development, opens up new-fangled possibilities for reaping the advantages of beneficial root–microbial interactions (Richardson et al., 2021). Metagenomics has a large potential to equip with vital information on plant-microbe interactions, which is required for new technologies to boost agricultural output in the long run (Bramhachari et al., 2017).
Soil microbial communities play a contributory role in determining plant growth and soil fertility. The changes that occur in their structure and dynamics in response to various soil management approaches can provide information regarding soil quality and biological complexity (Fausto et al., 2018). Also, microbes such as Azotobacter spp., Azoarcus spp., Burkholderia sp., Actinorhizobium spp., Rhizobium spp., Gluconacetobacter sp., Herbaspirillum sp., Bacillus polymyxa, and especially Azospirillum sp., Enterobacter, and Pseudomonas spp. that stimulate nutrient acquisition and plant growth have traditionally been utilized in agriculture as single strains to counter nitrogen and phosphorus fertilizer inputs Thus, a variety of parameters, including tissue, genotype, physiological status, growth stage, soil ecological conditions, and agricultural practices influence the plant microbial colonization. Natural population studies, on the other hand, reveal that clusters of bacteria with specific functions play crucial roles in adhering and releasing inorganic nutrients to physical surfaces, along with breaking down organic wastes and integrating them into the soil (Backer et al., 2018; Arif et al., 2020; Ray et al., 2020). Soil microbes are regarded as a significant constituent of the complex interplay of elements that contribute to the environmental quality required for long-term, healthy food production (Barea, 2015).
Soil organic matter stabilization, decomposition, bioremediation, nutrient cycling, and soil aggregate production are all significant tasks carried out by soil microorganisms. This reaction with organic matter and inorganic fertilizers consequently determine the fitness of soil health for crop production (Dangi et al., 2020). Furthermore, plant beneficial microbes are essential in the development of plant growth stimulators, bio-pesticides, and bio-fertilizers to decrease agrochemical use. These microbes improve soil fertility, soil health, and plant growth in a variety of ways, including improving nutrition (recycling, nutrient mobilization, nitrogen fixation), regulating phytohormone levels (cytokinins, gibberellic acid, ethylene, and indole acetic acid, among others), suppressing soil-borne pathogens (via antibiotic production, cyanides, and siderophores), and environmental symbiosis (via siderophores). Microbes may improve environmental conditions and soil biochemicals, while generating plant tolerance to environmental challenges, as a result of these qualities (Sahib et al., 2020). Organic fertilizers have been proven to recover soil microbial biomass by providing carbon-rich chemicals to the normally carbon-limited microbial populations in agricultural soils. Change of practices in soil management, for example, organic fertilizer treatment, has been found to increase biochemical properties and soil microbial quickly after application. Although there are benefits to employing organic nitrogen in traditional farming where mineral manures are widely employed, this area has been scarcely investigated (Dangi et al., 2020). Breeding for increased plant response for enhanced interactions and colonization of the plant environment is a crucial feature for the more efficient and effective use of microorganisms in agriculture. Plant breeding has mostly focused on increasing harvest yield and developing pathogen resistance; however, these procedures have brought about a decrease in genetic diversity in modern crops (Mitter et al., 2019).
Interplay of the environment and species-specific microbiome on plant health
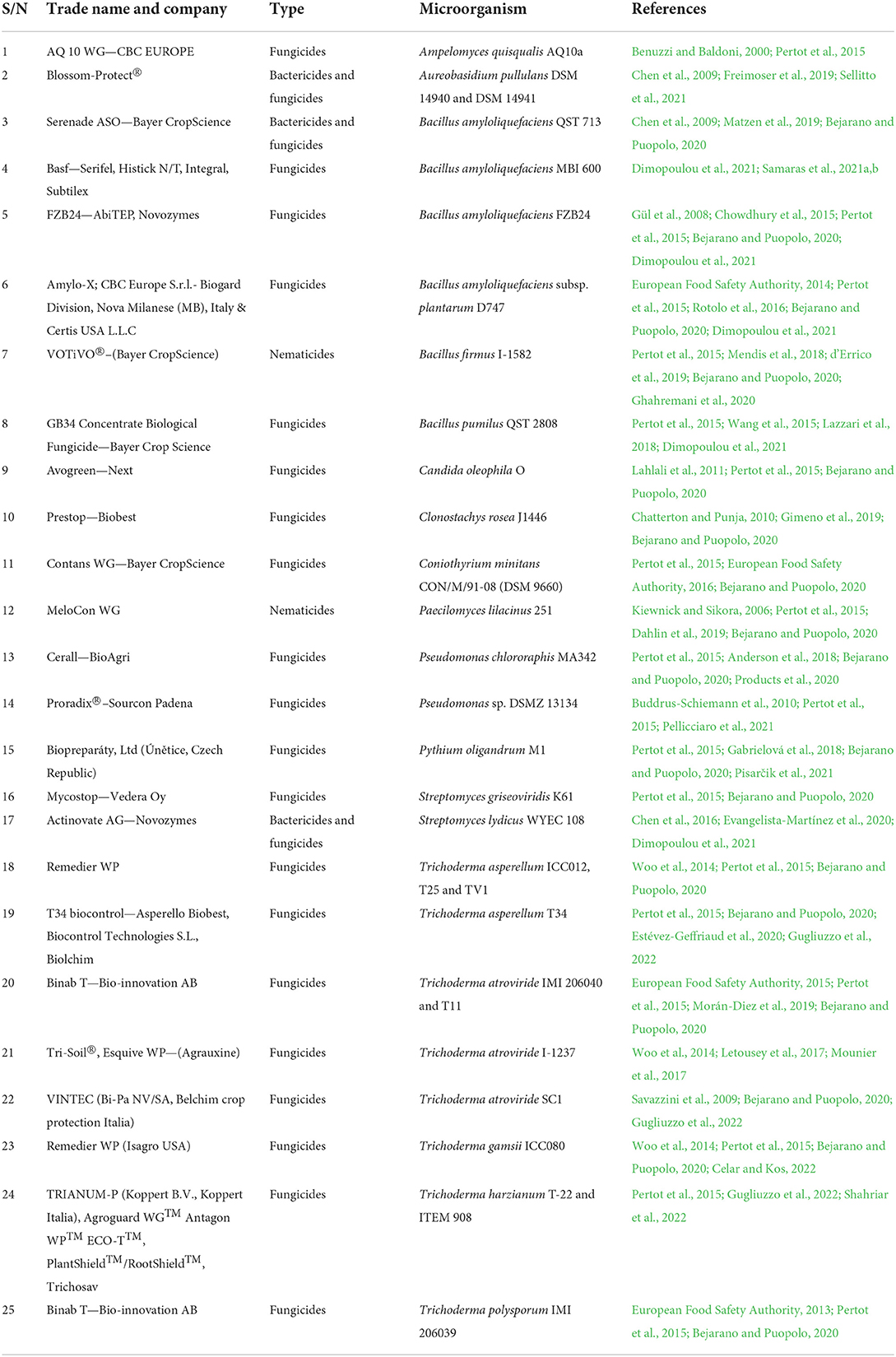
The potential of some soil microbes to promote plants' health and performance has been harnessed to boost pest management and agricultural yield by altering the soil microbiomes community (Howard et al., 2020). Since the era of next-generation sequencing, a renewed interest has sparked in the composition and role of microbial species diversity toward controlling plant productivity and soil health in broader ecological contexts (Sahib et al., 2020). As a result, a wide range of bacteria, fungi, and archaea components of the soil microbiome have received significantly greater attention in soil microbiology research (Barea, 2015). In line with some earlier studies, plant species in different successional phases possess different ecophysiological properties that could significantly impact the soil biological features, as in the case during early succession when fast-growing plant species produce a labile carbon substrate that favors bacterial-dominated communities. On the other hand, slow-growing plant species that control late-successional phases produce complex substrates (high in lignin) which favor the efficient lignin decomposing fungi (Shanmugam and Kingery, 2018). There is growing knowledge of the interactions between any consortium or inoculant strain and the receiving environment, such as abiotic soil conditions, the target plant and soil, their associated micro-and macrobiota, and more broadly ecological conditions that provide knowledge-based approaches for microbiome applications toward developing more targeted and-effective microbial applications (Mitter et al., 2019) (Figure 1).
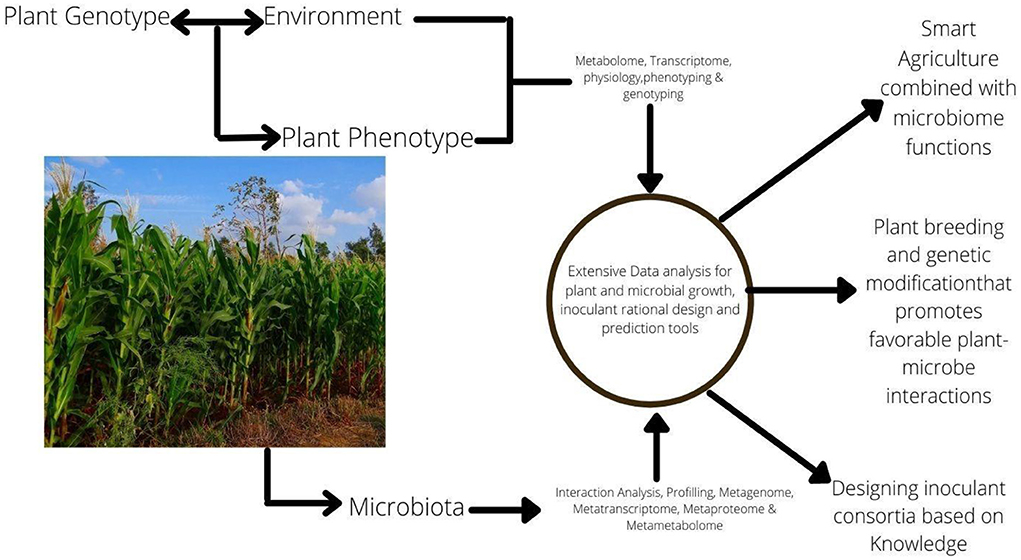
Figure 1. In-depth knowledge of the interactions of holobiont and their function leads to knowledge-based approaches for microbiome applications. Such as harnessing the potentials of the Plant—Soil—Microbes interaction in (i) the selections of plant type or varieties that favors beneficial plant-microbe interactions, (ii) microbial inoculant complementation, and (iii) agricultural management practices that enrich microbiota with desirable bacteria.
Plant-associated microbiomes can benefit their hosts, however, the rhizosphere's microbial makeup, along with colonization efficiency, are influenced by factors such as ecological, host's genetics, and physiological status (Rodriguez et al., 2019). To a greater extent, plant properties including biomass production, disease suppression, growth response, and flowering phenotype are all linked to specific plant microbiota. As a result, plant microbiota modification or the effects of farming management will influence plant characteristics and performance (Compant et al., 2019). The functional and mechanistic elements of the interactions between microorganisms, plants, the environment, and agricultural management methods are essential to realizing the predicted benefits of plant microbes in agriculture (Busby et al., 2017) (Figure 1).
Mechanism of action of beneficial microbes
Microbial biological control agents work in a variety of ways to protect crops against diseases. The many processes used by bio control agents to control plant diseases can be broadly characterized as direct antagonisms and Indirect antagonism.
Direct antagonism
Physical contact and/or a high degree of selectivity for infections by the bio control agent cause direct antagonism. It contains the following items:
a) Hyperparasitism—this interaction entails the tropic development of a biocontrol agent toward the target organism, coiling, and eventual attack and disintegration of the target pathogen's cell wall or membrane by enzyme activity. Fungi frequently engage in this type of interaction, e.g., Trichoderma (Junaid et al., 2013; Köhl et al., 2019a,b).
b) Direct Antibiosis—Antimicrobial metabolites are secondary metabolites that belong to a diverse category of organic, low-molecular-weight chemicals produced by bacteria and are toxic to other microorganisms' growth or metabolic processes (Köhl et al., 2019b). However, because of the modest quantities produced in comparison to other, less hazardous organic chemicals in the phytosphere, the effective quantities are difficult to quantify. A bio control agent that produces adequate amounts of antibiotics in the area of the plant pathogen is effective (Junaid et al., 2013). Biocontrol bacteria from the genera Agrobacterium, Bacillus, Pantoea, Pseudomonas, Serratia, Stenotrophomonas, Streptomyces, and others have been found to produce antimicrobial compounds, most of which have broad-spectrum activity (Köhl et al., 2019b).
Indirect antagonism
The capacity of biocontrol bacteria to boost plant development indirectly has piqued attention, in terms of getting a better understanding of some of the underlying mechanisms used by biocontrol bacteria and commercializing these bacteria instead of chemical pesticides (Admassie et al., 2020). It includes the following:
a) Competition—Soils and living plant surfaces are frequently nutrient-limited environments from a microbiological standpoint. As a result, a microorganism colonizing the phytosphere must efficiently compete for the available nutrients. Both biocontrol agents and infections battle for nutrients and space in the environment in order to develop themselves. This rivalry is thought to be an indirect interaction between the pathogen and the biocontrol agent, in which the pathogens are kept out of the site by depletion of the food supply and physical occupation (Junaid et al., 2013). For example, Bacillus spp. can control fungal pathogens by competition for space at the root surface and nutrients, particularly those produced as seed or root exudates, occurring in the rhizosphere (Pertot et al., 2015).
b) Induced Resistance and Priming—Plants defend themselves against infections through a number of physical and chemical processes. One of the most promising agronomic strategies for preventing biotic losses in crops is to improve resistance. Inducible resistance mechanisms supplement constitutive mechanisms like cuticles. Stimuli identified by particular recognition receptors activate induced plant defense mechanisms. MAMPs are microorganism-produced resistance-inducing stimuli. MAMPs with recognized receptors include glucan, chitin, and xylan, which are generated by Phytophthora megasperma and Trichoderma viride, respectively (Köhl et al., 2019b).
Bioprospecting rhizospheric microorganisms as biocontrol agents
Microbial-based solutions are currently one of the rapidly-growing agricultural sectors which are potential alternatives or complement the use of synthetic fertilizers and pesticides (Malusà et al., 2021). Microbial pesticides are biopesticides created from beneficial microorganisms that promote plant development, and nutrient utilization efficiency which results in higher agricultural yields (Asad, 2022). Furthermore, microbial pesticides have the potential to suppress pests and are one of the choice alternatives for plant disease management in recent times (Patil and Solanki, 2016). Since only about 1% of the candidate isolates retrieved from the studied samples could eventually become commercial products, many scientists currently explore approaches to improve the search and identification of suitable microbial strain that qualifies as potential biopesticides. However, recent approaches have embraced the use of DNA sequencing of representative samples to directly target activity-related genes (Glare et al., 2016). This method has been promising in unveiling the diversities of microbes associated with the rhizosphere for possible exploration of those with biocontrol functions. Hence, discovering, screening, identifying, and characterization of microorganisms with biocontrol potentials and the ability to colonize, persist, and adapt to a complex environment like the rhizosphere is an intriguing disease management method (Gomez-Lama Cabanas et al., 2018) (Figure 2). Identification and selection of a suitable sampling environment where prospective microbial candidates could be isolated is the first step in the development of microbial-based biopesticides. It is important to assess the biology and nature of the prevailing pest problem in the area where a microbial biopesticide is planned to be used.
Figure 2. Illustration of the process involved from discovery to application of potential microbial.
This assessment considers the biological principles of diseases and pests such as the host range, nature of the symptoms, the most vulnerable phases of the host, and the most suitable period of intervention (Mandakini and Manamgoda, 2021). Isolation using culture-dependent approaches have traditionally been used to exploit ambient microbiomes. High-throughput sequencing techniques that have been developed in recent decades have shown considerable promise in probing the intricate microbial networks found in plant microbiomes. The efficacy of the sequencing method in the identification of new and highly efficient microbial strains has been proved (Malusà et al., 2021).
Microorganisms exert biocontrol influence through a variety of processes, including antibiosis, parasitism, competition, and generation of plant systemic resistance. These properties promote the employment of biocontrol bioassays in screening for members with biopesticide potentials among a large number of microbial (Mandakini and Manamgoda, 2021). Microbiome modification has also recently been revealed as an effective method for screening microbial strains (Malusà et al., 2021), while the incorporation of molecular approaches into the screening process has also been equally beneficial in determining the full potential of the promising strains.
Formulation and application of microbial biopesticides
The first and most important step in the formulation of biocontrol agents is biomass generation. Culturing conditions affect the fitness and viability of microorganisms throughout formulation, storage period, application, as well as population densities upon harvest. A procedure that is peculiar to each microbial strain must be properly screened to enhance the performance of microbes as bioagents in the field (Bejarano and Puopolo, 2020). Once the biopesticide inoculum is generated, it must be kept stable during storage, distribution, and use at the target site. The formulation enables technical product stability, as well as persistence, safety, high efficacy, and user-friendliness. As a result, it is regarded as an important stage in the development of a high-quality and practical biopesticide. The active component (generated inoculum) is blended with inert ingredients like transporters, surface active ingredients (stickers, spreaders), and other additives during the formulation process (stabilizers, coloring agents, etc.) (Bejarano and Puopolo, 2020).
Biopesticides formulation becomes more efficient as production technology improves in the last few decades, this has therefore resulted in some breakthroughs recorded in the field of biopesticides. In the recent times, the application, persistence, and efficacy of biopesticides are enhanced with the use of prills and emulsions in its formulation. More so, the application through seed coating is appreciated for its appealing means of delivering biopesticides particularly in the soil. This method increasingly gains relevance as more agents are discovered to be rhizosphere colonizers or even capable of endophytic colonization (Glare et al., 2016). However, one of the difficult aspects of biopesticides use is that most application procedures were designed for synthetic pesticides rather than living organisms. Whereas, to ascertain the efficacy of the formulation produced, it must be successfully delivered to the target plant diseases under a favorable field conditions suitable for the disease expression (Mishra et al., 2019). Since the application should ideally bring the active chemicals in contact with the pest/disease and keeping them active for several weeks. The aboveground application has traditionally been carried out with spray applicators, while the below ground application of microbial agents with the use of specialized equipment with appropriate droplet size, and focused treatment, are receiving more attention (Glare et al., 2016). Although, the roles of biological formulations as biofertilizer which entails the improvement of resource acquisition and efficient use, and biopesticide which reducing losses due to pests and pathogens, has been embraced by the legislation and regulatory agencies (Malusà et al., 2021). However, the current legislative framework in place in many countries as regards the development and marketing of microbial-based goods makes it difficult to vigorously explore and take advantage of the multifunctionality of these beneficial microbes.
The beneficial microbes and soil health: Impacts on food safety
Soil health refers to the ability of soil to continue to operate as a vital living ecosystem that supports humans, animals, and plants, and it links agricultural and soil research to stakeholder demands, policy, and long-term supply chain management. Historically, soil assessments were focused on crop production (Lehmann et al., 2020), but existing safe agricultural practices emphasize the decrease of agrochemical usage in favor of different forms of eco-friendly crop management, like the use of abiotic and biotic stress-tolerant rootstocks or the improvement of productivity and plant health through soil management (Vida et al., 2020). Furthermore, the biotic stressors which consist of the fungal pathogens of maize such as Exerohillum turcicum causing Northern corn leaf blight (NCLB), Bipolaris maydis (Southern corn leaf blight) Rhizoctonia solani (Banded leaf and sheath blight), Colletotrichum graminicola (Colletotrichum graminicola) and Macrophomina phaseolina (Charcoal rot). Also, the bacteria diseases caused by the pathogens Dickeya zeae (Bacterial stalk rot), Clavibacter nebraskensis (Goss's bacterial wilt and blight), Xanthomonas vasicola (Bacterial leaf streak/spot) among many others reduce the grain quality and yield produced (Damicone et al., 2018; Begum et al., 2019; Zhu et al., 2021) (Table 1).
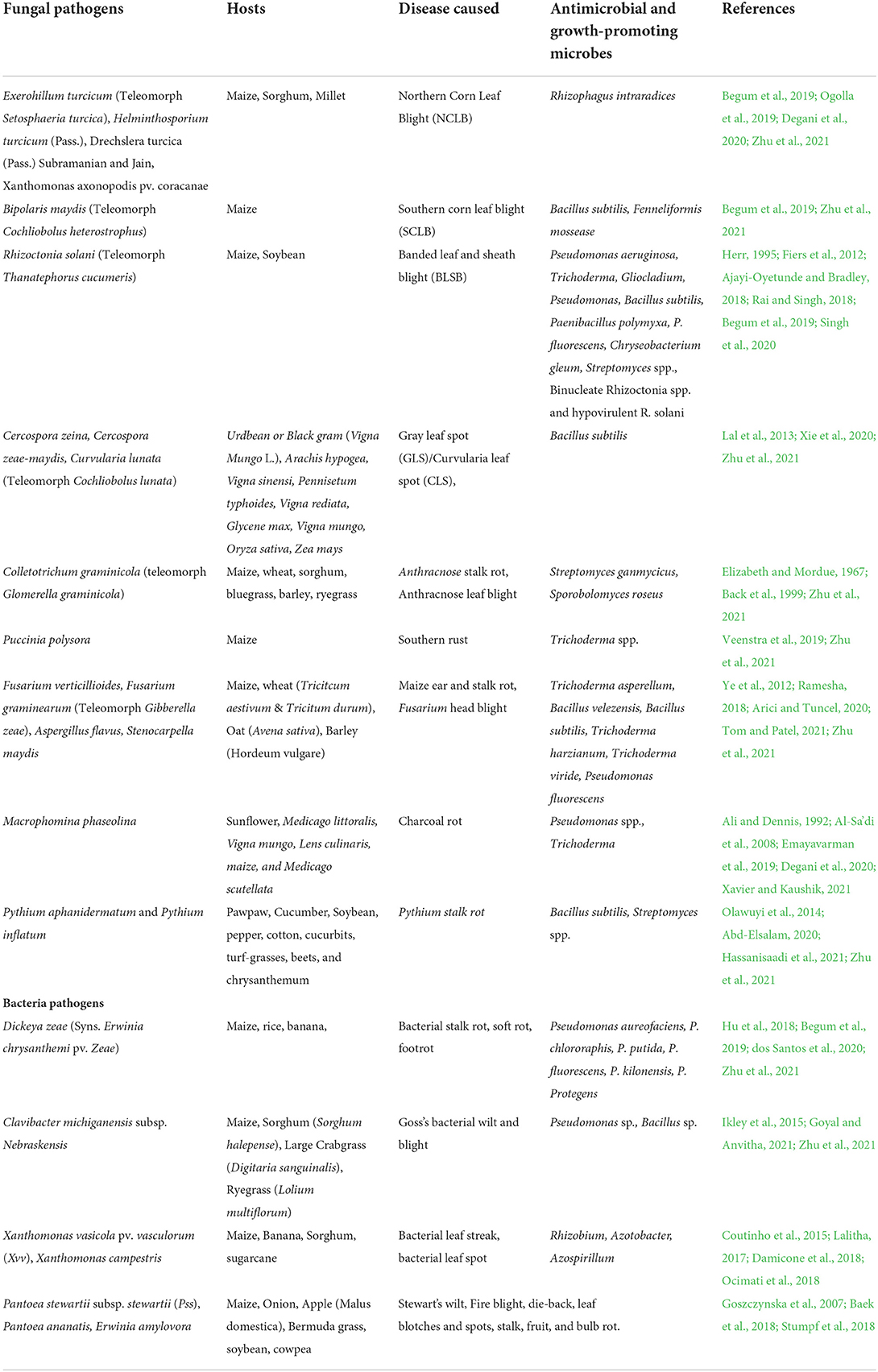
Table 1. Fungal and bacteria pathogens, diseases caused, and biological agents for managing agricultural crops.
Recent research has shown that using a beneficial microbe can improve agricultural output and plant health when produced under constrained environments. Varying antimicrobial and growth-promoting microorganisms have been investigated and developed from beneficial microbes such as Pseudomonas aureofaciens, Pseudomonas chlororaphis Bacillus sp., Rhizobium, Azotobacter, and Azospirillum among several others, and were effective in mitigating the debilitating effect of the pathogens (Lalitha, 2017; Begum et al., 2019; dos Santos et al., 2020) (Table 1).
The diversity of microorganisms in the rhizosphere zone aids plant growth by providing defense against pests and pathogens, facilitating nutrition uptake, and assisting plant tolerance to a variety of abiotic and biotic stimuli. Various types of abiotic stress include salinity, high temperature, and drought which results in negative impacts, for example, losses in crop productivity by disease susceptibility, nutrient acquisition, reducing water absorption and disturbing hormonal balance, and by affecting the photosynthetic capacity of the plant. However, because only around 1–5% of the bacteria on the planet are cultivable, the remaining 95–99% of germs are uncultivable; these helpful microbes are not fully utilized. For numerous years, understanding plant-microbe interactions have been a major emphasis of study (Kumar and Dubey, 2020). The employment of beneficial microorganisms (which improve the plant's health and quality, as well as assist in the reprocessing of crop leftovers with less ecological impact) is one of the most important techniques for long-term energy and food production (Dubey et al., 2019). Numerous soil microorganisms interact with each other and with plants in a variety of ways that aid in the maintenance and molding of various ecosystem components.
Healthy soils are the base of productive and sustainable agroecosystems, and they can be maintained by following basic soil health principles such as increasing plant diversity through crop rotation, protecting surface soils by growing more plants, minimal soil disturbance, polycultures, and cover crops, and enhancing soil microbiome, among others. Microbes aid to maintain soil health, improving root growth, water retention capacity, vital nutrient availability, carbon storage, cycling, filter contaminants, and conserving biodiversity (Dubey et al., 2019).
Beneficial microbes are opening up new avenues for long-term disease (pathogen) control, emphasizing the relevance of the plant microbes for crop productivity (Bonanomi et al., 2018). Since the 1980s, organic amendments have been used to manipulate the soil microbiota to minimize pathogen inoculum or virulence in favorable soil, albeit the efficiency of these methods varies depending on the pathogen/host system (De Corato, 2020). Compost, crop residues, peat, organic wastes, and biochar are examples of such organic soil additives (Akanmu et al., 2020; Chukwuka et al., 2020), although biofumigation is also used, for example, to reduce disease through soil fungistatic (Bonanomi et al., 2018). Organic manure (both animal dung and biomass) applied as a soil amendment, in contrast to issues linked with the use of agrochemicals, plays a vital role in the control of plant health and the recycling of soil nutrients (Akanmu et al., 2021). Organically altered soil has a potentially superior operational microbiome, with more positive microbial activities and functionally connected species than soils that have been fertilized with chemical fertilizers.
To better understand soil health, it will be required to characterize the functional groupings of microorganisms that are connected with specific organic amendments. Organic amendments including materials produced from composts, farmyard manure, crop residue and biochar among others, have been reported to modify soil structure and enables the efficient absorption and both nutrient and water retention in the soil (Akanmu et al., 2021). Organic amendments fine-tuned and optimized for specific soil/crop combinations, would create long-term microbial ecosystem services and soil health, resulting in yields that are equivalent to or greater than chemical fertilizers alone (Arif et al., 2020). Since the early 2000s, supplementing beneficial external microbiota with Composts, organic amendments, and biological control agents has appeared to be a potential strategy for boosting the suppression of favorable soils (De Corato, 2020). As a result, soil organic matter can be conserved with the help of organic amendments in agricultural systems, which is a recognized technique for reducing soil deterioration. Organic additions, on the other hand, have the potential to change the soil microbiome. The quantity and quality of organic matter input not only affect the soil's physicochemical properties, but also affects biotic factors related to the soil microbiota, such as microbial biomass and diversity, community structure, and soil activities, all of which have a direct impact on disease biological control (Vida et al., 2020).
Plants contribute to the stability of the rhizobiome by producing fixed carbon resources. Plant beneficial microbes, in turn, aid plants' growth by root modification, acquiring nutrients and protecting plants from infections, among other things. As a result, the rhizobiome plays a crucial role in sustaining the plant's health. Disassembling plant microbiomes and building synthetic microbial communities for reconstitution studies to examine interspecies and intraspecies interactions is a potential way to understand the reciprocal impacts of plants and their microbiota (Lucaciu et al., 2019). Fast-growing species were gradually substituted by stress-tolerant and hostile species as the plants grow. Using synthetic consortia derived from different plant growth phases shows that the high functional diversity of climax microbiomes contributes to increased resistance to pathogenic microbial invasion. Hence, root-associated microbes have the potential of improving plant efficiency and output in cropping systems, according to a growing body of studies (Ab-Rahman et al., 2018; Hu et al., 2020).
Current trends in the use of rhizospheric microorganisms
The use of rhizospheric microbes has been explored in the formulation of biopesticides, and the efficacy of the products against pest and diseases in a diverse type of crops have been proved. Newly discovered microbial species and strains are analyzed on a regular basis for their potential to serve as a biocontrol agent. Despite several typical drawbacks, the use of biocontrol agents still holds a lot of potential (Köhl et al., 2019a). In recent times, many biopesticide companies are entering into licensing agreements to market and sell microbial biocontrol products, while many international companies have ventured into the formulation and marketing of microbial biocontrol products (Marian and Shimizu, 2019). However, despite the large corpus of research on the bioagents' ability to protect crops, only a few microbes are recognized as active substances at the time of writing, and the majority of which are Trichoderma and Bacillus spp. strains (Table 2).
Conclusion and future prospects
Effective management of plant health is strategic to attaining sustainable food production and reducing hunger, thereby fulfilling an important Sustainable Development Goal (SDG) of 2030. To achieve this, an efficient disease and pest management approach focused on disease prevention need to be instituted. This could be achieved through the beneficial microbes' roles in the abatement of pathogenic invasion, support of plant-soil feedback, and modulating the species-specific rhizosphere microbiome interplay toward plants' wellness and productivity. Hence, the gateway to attaining safe and sustainable plant health is dependent on the efficient coordination of the rhizosphere microbiome in plant development and disease management, as well as their sustainable deployment as biofertilizers and biopesticides in managing plant health. Of recent. the breakthroughs in system biology, molecular biology, and associated computational technologies has enabled the eludication of the structural and functional characteristics of microorganisms with prospects in growth promotion and biocontrol potientials. This has therefore enabled the indepth understanding of the rhizospheric microbes, however, the functional dynamics among the antagonists opens up new possibilities for the design of microbial antagonist consortia which could be an effective broad spectrum microbial formulation against varying plant diseases. Although strain mixtures for the biocontrol of plant pathogens are already commercially available as described in the literature, employing taxonomically divergent but functionally complementary strains could be a promising approach to follow in the near future in the quest to design a standardized, multi-targeted, efficacious biocontrol strategy.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
OB recognizes National Research Fund (NRF South Africa) for grants (UID123634 and UID132595) that support work in her research group.
Acknowledgments
SD and AA appreciate the North-West University, South Africa for the respective Postgraduate and Postdoctoral privileges enjoyed in the University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd-Elsalam, K. A. (2020). Host Plants and Specificity of the Genus Pythium Pythium. Boca Raton, FL: CRC Press, 162–175.
Ab-Rahman, S. F. S., Singh, E., Pieterse, C. M., and Schenk, P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 267, 102–111. doi: 10.1016/j.plantsci.2017.11.012
Adedeji, A. A., and Babalola, O. O. (2020). Secondary metabolites as plant defensive strategy: a large role for small molecules in the near root region. Planta 252, 1–12. doi: 10.1007/s00425-020-03468-1
Admassie, M., Assefa, F., and Alemu, T. (2020). Different mechanism, application, methods and properties of plant growth promoting rhizosphere bacteria. Appl. Microbiol. 6, 177. doi: 10.35248/2471-9315.20.6.177
Ajayi-Oyetunde, O., and Bradley, C. (2018). Rhizoctonia solani: taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant Pathol. 67, 3–17. doi: 10.1111/ppa.12733
Akanmu, A., Sobowale, A., Abiala, M., Olawuyi, O., and Odebode, A. (2020). Efficacy of biochar in the management of Fusarium verticillioides Sacc. causing ear rot in Zea mays L. Biotechnol. Rep. 26, e00474. doi: 10.1016/j.btre.2020.e00474
Akanmu, A. O., Babalola, O. O., Venturi, V., Ayilara, M. S., Saanu, A. B., Amoo, A. E., et al. (2021). Plant disease management: leveraging on the plant-microbe-soil interface in the biorational use of organic amendments. Front. Plant Sci. 12, 1590. doi: 10.3389/fpls.2021.700507
Ali, S., and Dennis, J. (1992). Host range and physiologic specialisation of Macrophomina phaseolina isolated from field peas in South Australia. Aust. J. Exp. Agric. 32, 1121–1125. doi: 10.1071/EA9921121
Alori, E. T., Dare, M. O., and Babalola, O. O. (2017). “Microbial inoculants for soil quality and plant health,” in Sustainable Agriculture Reviews (Springer), 281–307.
Al-Sa'di, A., Drenth, A., Deadman, M., and Aitken, E. (2008). Genetic diversity, aggressiveness and metalaxyl sensitivity of Pythium aphanidermatum populations infecting cucumber in Oman. Plant Pathol. 57, 45–56. doi: 10.1111/j.1439-0434.2007.01318.x
Anderson, J. A., Staley, J., Challender, M., and Heuton, J. (2018). Safety of Pseudomonas chlororaphis as a gene source for genetically modified crops. Transgenic Res. 27, 103–113. doi: 10.1007/s11248-018-0061-6
Arici, S. E., and Tuncel, Z. N. (2020). Antifungal activity of useful microorganisms against a phytopathogenic fungus on maize. Emerg. Mater. Res. 9, 743–749. doi: 10.1680/jemmr.20.00121
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 38, 1385–1396. doi: 10.1016/j.tibtech.2020.04.015
Asad, S. A. (2022). Mechanisms of action and biocontrol potential of Trichoderma against fungal plant diseases-a review. Ecol. Complexity 49, 100978. doi: 10.1016/j.ecocom.2021.100978
Back, P., Landschoot, P., and Huff, D. (1999). Variation in pathogenicity, morphology, and RAPD marker profiles in Colletotrichum graminicola from turfgrasses. Crop Sci. 39, 1129–1135. doi: 10.2135/cropsci1999.0011183X003900040030x
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1473. doi: 10.3389/fpls.2018.01473
Baek, K. Y., Lee, H.-H., Son, G. J., Lee, P. A., Roy, N., Seo, Y.-S., et al. (2018). Specific and sensitive primers developed by comparative genomics to detect bacterial pathogens in grains. Plant Pathol. J. 34, 104. doi: 10.5423/PPJ.OA.11.2017.0250
Barea, J. (2015). Future challenges and perspectives for applying microbial biotechnology in sustainable agriculture based on a better understanding of plant-microbiome interactions. J. Soil Sci. Plant Nutr. 15, 261–282. doi: 10.4067/S0718-95162015005000021
Beals, K. K., Moore, J. A., Kivlin, S. N., Bayliss, S. L., Lumibao, C. Y., Moorhead, L. C., et al. (2020). Predicting plant-soil feedback in the field: meta-analysis reveals that competition and environmental stress differentially influence PSF. Front. Ecol. Evolut. 8, 191. doi: 10.3389/fevo.2020.00191
Begum, N., Qin, C., Ahanger, M. A., Raza, S., Khan, M. I., Ashraf, M., et al. (2019). Role of arbuscular mycorrhizal fungi in plant growth regulation: implications in abiotic stress tolerance. Front. Plant Sci. 10, 1068. doi: 10.3389/fpls.2019.01068
Bejarano, A., and Puopolo, G. (2020). “Bioformulation of microbial biocontrol agents for a sustainable agriculture,” in How Research Can Stimulate the Development of Commercial Biological Control Against Plant Diseases, eds A. De Cal, P. Melgarejo, and N. Magan (Cham: Springer). doi: 10.1007/978-3-030-53238-3_16
Benuzzi, M., and Baldoni, G. (2000). AQ 10, a new biofungicide based on Ampelomyces quisqualis for powdery mildew control on grapes. Inf. Fitopatol. 50, 33–36.
Berendsen, R. L., Pieterse, C. M., and Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trend Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bonanomi, G., Lorito, M., Vinale, F., and Woo, S. L. (2018). Organic amendments, beneficial microbes, and soil microbiota: toward a unified framework for disease suppression. Annu. Rev. Phytopathol. 56, 1–20. doi: 10.1146/annurev-phyto-080615-100046
Bramhachari, P. V., Nagaraju, G. P., and Kariali, E. (2017). “Metagenomic approaches in understanding the mechanism and function of PGPRs: perspectives for sustainable agriculture,” in Agriculturally Important Microbes for Sustainable Agriculture, eds M. P. Meena, J. Bisht, and A. Pattanayak (Singapore: Spinger), 163–182.
Buddrus-Schiemann, K., Schmid, M., Schreiner, K., Welzl, G., and Hartmann, A. (2010). Root colonization by Pseudomonas sp. DSMZ 13134 and impact on the indigenous rhizosphere bacterial community of barley. Microbial Ecol. 60, 381–393. doi: 10.1007/s00248-010-9720-8
Busby, P. E., Soman, C., Wagner, M. R., Friesen, M. L., Kremer, J., Bennett, A., et al. (2017). Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15, e2001793. doi: 10.1371/journal.pbio.2001793
Celar, F. A., and Kos, K. (2022). Compatibility of the commercial biological control agents Trichoderma asperellum (ICC 012) and Trichoderma gamsii (ICC 080) with selected herbicides. J. Plant Dis. Protection 129, 85–92. doi: 10.1007/s41348-021-00547-7
Chatterton, S., and Punja, Z. (2010). Factors influencing colonization of cucumber roots by Clonostachys rosea f. catenulata, a biological disease control agent. Biocontrol Sci. Technol. 20, 37–55. doi: 10.1080/09583150903350253
Chen, X., Pizzatti, C., Bonaldi, M., Saracchi, M., Erlacher, A., Kunova, A., et al. (2016). Biological control of lettuce drop and host plant colonization by rhizospheric and endophytic streptomycetes. Front. Microbiol. 7, 714. doi: 10.3389/fmicb.2016.00714
Chen, X.-H., Scholz, R., Borriss, M., Junge, H., Mögel, G., Kunz, S., et al. (2009). Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 140, 38–44. doi: 10.1016/j.jbiotec.2008.10.015
Choudhary, R. C., Kumaraswamy, R., Kumari, S., Sharma, S., Pal, A., Raliya, R., et al. (2017). Cu-chitosan nanoparticle boost defense responses and plant growth in maize (Zea mays L.). Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-08571-0
Chowdhury, S. P., Hartmann, A., Gao, X., and Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42-a review. Front. Microbiol. 6, 780. doi: 10.3389/fmicb.2015.00780
Chukwuka, K. S., Akanmu, A. O., Umukoro, B. O., Asemoloye, M. D., and Odebode, A. C. (2020). “Biochar: A vital source for sustainable agriculture,” in Biostimulants in Plant Science, eds S. M. Mirmajlessi and R. Radhakrishnan (London: IntechOpen). p. 51–74. doi: 10.5772/intechopen.86568
Chukwuneme, C. F., Ayangbenro, A. S., Babalola, O. O., and Kutu, F. R. (2021). Functional diversity of microbial communities in two contrasting maize rhizosphere soils. Rhizosphere 17, 100282. doi: 10.1016/j.rhisph.2020.100282
Chun, S. -C., and Chandrasekaran, M. (2019). Chitosan and chitosan nanoparticles induced expression of pathogenesis-related proteins genes enhances biotic stress tolerance in tomato. Int. J. Biol. Macromol. 125, 948–954. doi: 10.1016/j.ijbiomac.2018.12.167
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Coutinho, T., Van der Westhuizen, L., Roux, J., McFarlane, S., and Venter, S. (2015). Significant host jump of Xanthomonas vasicola from sugarcane to a Eucalyptus grandis clone in South Africa. Plant Pathol. 64, 576–581. doi: 10.1111/ppa.12298
Dahlin, P., Eder, R., Consoli, E., Krauss, J., and Kiewnick, S. (2019). Integrated control of Meloidogyne incognita in tomatoes using fluopyram and Purpureocillium lilacinum strain 251. Crop Prot. 124, 104874. doi: 10.1016/j.cropro.2019.104874
Dai, L., Zhang, G., Yu, Z., Ding, H., Xu, Y., and Zhang, Z. (2019). Effect of drought stress and developmental stages on microbial community structure and diversity in peanut rhizosphere soil. Int. J. Mol. Sci. 20, 2265. doi: 10.3390/ijms20092265
Damicone, J., Cevallos, F., and Olson, J. (2018). First report of bacterial leaf streak of corn caused by Xanthomonas vasicola pv. vasculorum in Oklahoma. Plant Dis. 102, 437–437. doi: 10.1094/PDIS-04-17-0523-PDN
Dangi, S., Gao, S., Duan, Y., and Wang, D. (2020). Soil microbial community structure affected by biochar and fertilizer sources. Appl. Soil Ecol. 150, 103452. doi: 10.1016/j.apsoil.2019.103452
De Corato, U. (2020). Soil microbiota manipulation and its role in suppressing soil-borne plant pathogens in organic farming systems under the light of microbiome-assisted strategies. Chem. Biol. Technol. Agric. 7, 1–26. doi: 10.1186/s40538-020-00183-7
Degani, O., Dor, S., Abraham, D., and Cohen, R. (2020). Interactions between Magnaporthiopsis maydis and Macrophomina phaseolina, the causes of wilt diseases in maize and cotton. Microorganisms 8, 249. doi: 10.3390/microorganisms8020249
d'Errico, G., Marra, R., Crescenzi, A., Davino, S. W., Fanigliulo, A., Woo, S. L., et al. (2019). Integrated management strategies of Meloidogyne incognita and Pseudopyrenochaeta lycopersici on tomato using a Bacillus firmus-based product and two synthetic nematicides in two consecutive crop cycles in greenhouse. Crop Prot. 122, 159–164. doi: 10.1016/j.cropro.2019.05.004
Dimopoulou, A., Theologidis, I., Varympopi, A., Papafotis, D., Mermigka, G., Tzima, A., et al. (2021). Shifting perspectives of translational research in bio-bactericides: reviewing the Bacillus amyloliquefaciens paradigm. Biology 10, 1202. doi: 10.3390/biology10111202
dos Santos, R. M., Diaz, P. A. E., Lobo, L. L. B., and Rigobelo, E. C. (2020). Use of plant growth-promoting rhizobacteria in maize and sugarcane: characteristics and applications. Front. Sustain. Food Syst. 4, 136. doi: 10.3389/fsufs.2020.00136
Dubey, A., Malla, M. A., Khan, F., Chowdhary, K., Yadav, S., Kumar, A., et al. (2019). Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers. Conserv. 28, 2405–2429. doi: 10.1007/s10531-019-01760-5
Elizabeth, J., and Mordue, M. (1967). Colletotrichum coccodes. CMI Descriptions of Pathogenic Fungi and Bacteria. No. 131. Commonwealth Mycological Institute, Kew, Surrey, England. doi: 10.1079/DFB/20056400131
Emayavarman, P., Renukadevi, P., Ravikesavan, R., and Nakkeeran, S. (2019). Antifungal efficacy and growth promotion by Trichoderma virens TRI 37 and Bacillus amyloloiquefaciens (VB7) against Macrophomina phaseolina-the maize charcoal rot pathogen. Int. J. Curr. Microbiol. App. Sci 8, 2682–2693. doi: 10.20546/ijcmas.2019.81
Enagbonma, B. J., Ajilogba, C. F., and Babalola, O. O. (2020). Metagenomic profiling of bacterial diversity and community structure in termite mounds and surrounding soils. Arch. Microbiol. 202, 2697–2709. doi: 10.1007/s00203-020-01994-w
Estévez-Geffriaud, V., Vicente, R., Vergara-Díaz, O., Narváez Reinaldo, J. J., and Trillas, M. I. (2020). Application of Trichoderma asperellum T34 on maize (Zea mays) seeds protects against drought stress. Planta 252, 1–12. doi: 10.1007/s00425-020-03404-3
European Food Safety Authority. (2013). Conclusion on the peer review of the pesticide risk assessment of the active substance Trichoderma polysporum strain IMI 206039. EFSA J. 11, 3035. doi: 10.2903/j.efsa.2013.3035
European Food Safety Authority. (2014). Conclusion on the peer review of the pesticide risk assessment of the active substance Bacillus amyloliquefaciens subsp. plantarum strain D747. EFSA J. 12, 3624. doi: 10.2903/j.efsa.2014.3624
European Food Safety Authority. (2015). Conclusion on the peer review of the pesticide risk assessment of the active substance Trichoderma atroviride, strains IMI-206040, T11. EFSA J. 13, 3056. doi: 10.2903/j.efsa.2015.3056
European Food Safety Authority. (2016). Peer review of the pesticide risk assessment of the active substance Coniothyrium minitans strain CON/M/91-08. EFSA J. 14, e04517. doi: 10.2903/j.efsa.2016.4517
Evangelista-Martínez, Z., Contreras-Leal, E. A., Corona-Pedraza, L. F., and Gastélum-Martínez, É. (2020). Biocontrol potential of Streptomyces sp. CACIS-1.5 CA against phytopathogenic fungi causing postharvest fruit diseases. Egypt. J. Biol. Pest Control 30, 1–10. doi: 10.1186/s41938-020-00319-9
Fausto, C., Mininni, A. N., Sofo, A., Crecchio, C., Scagliola, M., Dichio, B., et al. (2018). Olive orchard microbiome: characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol. Divers. 11, 597–610. doi: 10.1080/17550874.2019.1596172
Fiers, M., Edel-Hermann, V., Chatot, C., Le Hingrat, Y., Alabouvette, C., and Steinberg, C. (2012). Potato soil-borne diseases. A review. Agron. Sustain. Dev. 32, 93–132. doi: 10.1007/s13593-011-0035-z
Freimoser, F. M., Rueda-Mejia, M. P., Tilocca, B., and Migheli, Q. (2019). Biocontrol yeasts: mechanisms and applications. World J. Microbiol. Biotechnol. 35, 1–19. doi: 10.1007/s11274-019-2728-4
Gabrielová, A., Mencl, K., Suchánek, M., Klimeš, R., Hubka, V., and Kolarík, M. (2018). The oomycete Pythium oligandrum can suppress and kill the causative agents of dermatophytoses. Mycopathologia 183, 751–764. doi: 10.1007/s11046-018-0277-2
Ghahremani, Z., Escudero, N., Beltrán-Anadón, D., Saus, E., Cunquero, M., Andilla, J., et al. (2020). Bacillus firmus strain I-1582, a nematode antagonist by itself and through the plant. Front. Plant Sci. 11, 796. doi: 10.3389/fpls.2020.00796
Gimeno, A., Sohlberg, E., Pakula, T., Limnell, J., Keller, B., Laitila, A., et al. (2019). TaqMan qPCR for quantification of Clonostachys rosea used as a biological control agent against Fusarium graminearum. Front. Microbiol. 10, 1627. doi: 10.3389/fmicb.2019.01627
Glare, T. R., Gwynn, R. L., and Moran-Diez, M. E. (2016). Development of biopesticides and future opportunities. Microbial Based Biopesticides 211–221. doi: 10.1007/978-1-4939-6367-6_16
Gomez-Lama Cabanas, C., Legarda, G., Ruano-Rosa, D., Pizarro-Tobias, P., Valverde-Corredor, A., Niqui, J. L., et al. (2018). Indigenous Pseudomonas spp. strains from the olive (Olea europaea L.) rhizosphere as effective biocontrol agents against Verticillium dahliae: from the host roots to the bacterial genomes. Front. Microbiol. 9, 277. doi: 10.3389/fmicb.2018.00277
Goszczynska, T., Botha, W., Venter, S., and Coutinho, T. (2007). Isolation and identification of the causal agent of brown stalk rot, a new disease of maize in South Africa. Plant Dis. 91, 711–718. doi: 10.1094/PDIS-91-6-0711
Goyal, K., and Anvitha, D. (2021). Wilting: the root cause of amateur death. International J. Res. Educ. Sci. Methods 9, 634−638.
Gugliuzzo, A., Aiello, D., Biondi, A., Giurdanella, G., Siscaro, G., Zappalà, L., et al. (2022). Microbial mutualism suppression by Trichoderma and Bacillus species for controlling the invasive ambrosia beetle Xylosandrus compactus. Biol. Control 170, 104929. doi: 10.1016/j.biocontrol.2022.104929
Gül, A., Kidoglu, F., and Tüzel, Y. (2008). Effects of nutrition and Bacillus amyloliquefaciens on tomato (Solanum lycopersicum, L.) growing in perlite. Spanish J. Agric. Res. 6, 422–429. doi: 10.5424/sjar/2008063-335
Gupta, R., Singh, A., Srivastava, M., Shanker, K., and Pandey, R. (2019). Plant-microbe interactions endorse growth by uplifting microbial community structure of Bacopa monnieri rhizosphere under nematode stress. Microbiol. Res. 218, 87–96. doi: 10.1016/j.micres.2018.10.006
Hassanisaadi, M., Shahidi Bonjar, G. H., Hosseinipour, A., Abdolshahi, R., Ait Barka, E., and Saadoun, I. (2021). Biological control of Pythium aphanidermatum, the causal agent of tomato root rot by two streptomyces root symbionts. Agronomy 11, 846. doi: 10.3390/agronomy11050846
Herr, L. J. (1995). Biological control of Rhizoctonia solani by binucleate Rhizoctonia spp. and hypovirulent R. solani agents. Crop Protect. 14, 179–186. doi: 10.1016/0261-2194(95)00017-G
Howard, M. M., Muñoz, C. A., Kao-Kniffin, J., and Kessler, A. (2020). Soil microbiomes from fallow fields have species-specific effects on crop growth and pest resistance. Front. Plant Sci. 11, 1171. doi: 10.3389/fpls.2020.01171
Hu, J., Wei, Z., Kowalchuk, G. A., Xu, Y., Shen, Q., and Jousset, A. (2020). Rhizosphere microbiome functional diversity and pathogen invasion resistance build up during plant development. Environ. Microbiol. 22, 5005–5018. doi: 10.1111/1462-2920.15097
Hu, M., Li, J., Chen, R., Li, W., Feng, L., Shi, L., et al. (2018). Dickeya zeae strains isolated from rice, banana and clivia rot plants show great virulence differentials. BMC Microbiol. 18, 1–15. doi: 10.1186/s12866-018-1300-y
Ikley, J. T., Wise, K. A., and Johnson, W. G. (2015). Annual Ryegrass (Lolium multiflorum), Johnsongrass (Sorghum halepense), and Large Crabgrass (Digitaria sanguinalis) are alternative hosts for Clavibacter michiganensis subsp. nebraskensis, causal agent of goss's wilt of corn. Weed Sci. 63, 901–909. doi: 10.1614/WS-D-15-00028.1
Jilling, A., Keiluweit, M., Contosta, A. R., Frey, S., Schimel, J., Schnecker, J., et al. (2018). Minerals in the rhizosphere: overlooked mediators of soil nitrogen availability to plants and microbes. Biogeochemistry 139, 103–122. doi: 10.1007/s10533-018-0459-5
Junaid, J. M., Dar, N. A., Bhat, T. A., Bhat, A. H., and Bhat, M. A. (2013). Commercial biocontrol agents and their mechanism of action in the management of plant pathogens. Int. J. Modern Plant Anim. Sci. 1, 39–57.
Khan, M., Gemenet, D. C., and Villordon, A. (2016). Root system architecture and abiotic stress tolerance: current knowledge in root and tuber crops. Front. Plant Sci. 7, 1584. doi: 10.3389/fpls.2016.01584
Kiewnick, S., and Sikora, R. (2006). Biological control of the root-knot nematode Meloidogyne incognita by Paecilomyces lilacinus strain 251. Biol. Control 38, 179–187. doi: 10.1016/j.biocontrol.2005.12.006
Köhl, J., Booij, K., Kolnaar, R., and Ravensberg, W. J. (2019a). Ecological arguments to reconsider data requirements regarding the environmental fate of microbial biocontrol agents in the registration procedure in the European Union. Bio Control. 64, 469–487. doi: 10.1007/s10526-019-09964-y
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019b). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10, 845. doi: 10.3389/fpls.2019.00845
Kulmatiski, A., and Kardol, P. (2008). “Getting plant-soil feedbacks out of the greenhouse: experimental and conceptual approaches,” in Progress in Botany (Springer), 449–472.
Kumar, A., and Dubey, A. (2020). Rhizosphere microbiome: engineering bacterial competitiveness for enhancing crop production. J. Adv. Res. 24, 337–352. doi: 10.1016/j.jare.2020.04.014
Lahlali, R., Raffaele, B., and Jijakli, M. (2011). UV protectants for Candida oleophila (strain O), a biocontrol agent of postharvest fruit diseases. Plant Pathol. 60, 288–295. doi: 10.1111/j.1365-3059.2010.02368.x
Lal, M., Kumar, S., Ali, M., Khan, A., Singh, V., and Murti, S. (2013). Host range, susceptibility period of Curvularia lunata causing leaf spot of black gram and germplasm screening. Agriways 1, 142–146.
Lalitha, S. (2017). “Plant growth-promoting microbes: A boon for sustainable agriculture,” in Sustainable Agriculture towards Food Security, ed A. Dhanarajan (Singapore: Springer), 125–158.
Lazzari, V., Boebel, A., Guidi, C., Lazzati, S., Ricci, M., and Cantoni, A. (2018). “Bacillus pumilus QST2808 SC, new bio fungicide to control powdery mildew on grape and vegetables,” in Atti, Giornate Fitopatologiche, Chianciano Terme (SI), Italia, 6-9 marzo 2018, Vol. secondo, 59–66.
Lehmann, J., Bossio, D. A., Kögel-Knabner, I., and Rillig, M. C. (2020). The concept and future prospects of soil health. Nat. Rev. Earth Environ. 1, 544–553. doi: 10.1038/s43017-020-0080-8
Letousey, P., Heysch, P., Martin, A., Cadiou, M., Boulisset, F., Mounier, E., et al. (2017). Tri-Soil®, a new biofungicide based on Trichoderma atroviride I-1237 to control soilborne diseases in vegetable crop production. Paper presented at the 6e COMAPPI, Conférence sur les Moyens Alternatifs de Protection pour une Production Intégrée, Lille, France, 21-23 mars 2017.
Lladó, S., López-Mondéjar, R., and Baldrian, P. (2018). Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 102, 4331–4338. doi: 10.1007/s00253-018-8950-4
Lucaciu, R., Pelikan, C., Gerner, S. M., Zioutis, C., Köstlbacher, S., Marx, H., et al. (2019). A bioinformatics guide to plant microbiome analysis. Front. Plant Sci. 10, 1313. doi: 10.3389/fpls.2019.01313
Malusà, E., Berg, G., Biere, A., Bohr, A., Canfora, L., Jungblut, A. D., et al. (2021). A holistic approach for enhancing the efficacy of soil microbial inoculants in agriculture:: from lab to field scale. Global J. Agric. Innov. 176, 190. doi: 10.15377/2409-9813.2021.08.14
Mandakini, H. T., and Manamgoda, D. S. (2021). “Microbial biopesticides: Development and application,” in Microbial Technology for Sustainable Environment, eds P. Bhatt, S. Gangola, D. Udayanga, and G. Kumar (Singapore: Springer), 167–189.
Marian, M., and Shimizu, M. (2019). Improving performance of microbial biocontrol agents against plant diseases. J. General Plant Pathol. 85, 329–336. doi: 10.1007/s10327-019-00866-6
Matzen, N., Heick, T. M., and Jørgensen, L. N. (2019). Control of powdery mildew (Blumeria graminis spp.) in cereals by Serenade® ASO (Bacillus amyloliquefaciens (former subtilis) strain QST 713). Biol. Control 139, 104067. doi: 10.1016/j.biocontrol.2019.104067
Mendis, H. C., Thomas, V. P., Schwientek, P., Salamzade, R., Chien, J.-T., Waidyarathne, P., et al. (2018). Strain-specific quantification of root colonization by plant growth promoting rhizobacteria Bacillus firmus I-1582 and Bacillus amyloliquefaciens QST713 in non-sterile soil and field conditions. PLoS ONE 13, e0193119. doi: 10.1371/journal.pone.0193119
Mishra, A., Arshi, A., Mishra, S. P., and Bala, M. (2019). “Microbe-based biopesticide formulation: a tool for crop protection and sustainable agriculture development. Microbial Technology for the Welfare of Society (Springer), 125–145.
Mishra, J., Mishra, I., and Arora, N. K. (2022). 2, 4-Diacetylphloroglucinol producing Pseudomonas fluorescens JM-1 for management of ear rot disease caused by Fusarium moniliforme in Zea mays L. 3 Biotech 12, 1–15. doi: 10.1007/s13205-022-03201-7
Mitter, B., Brader, G., Pfaffenbichler, N., and Sessitsch, A. (2019). Next generation microbiome applications for crop production-limitations and the need of knowledge-based solutions. Curr. Opin. Microbiol. 49, 59–65. doi: 10.1016/j.mib.2019.10.006
Morán-Diez, M. E., Carrero-Carrón, I., Rubio, M. B., Jiménez-Díaz, R. M., Monte, E., and Hermosa, R. (2019). Transcriptomic analysis of Trichoderma atroviride overgrowing plant-wilting Verticillium dahliae reveals the role of a new M14 metallocarboxypeptidase CPA1 in biocontrol. Front. Microbiol. 10, 1120. doi: 10.3389/fmicb.2019.01120
Mounier, E., Heysch, P., Cortes, F., Cadiou, M., and Pajot, E. (2017). Trichoderma atroviride, strain I-1237, reduces the impact of Pythium spp. in carrot crop production. Acta Hortic. 1153, 169–174. doi: 10.17660/ActaHortic.2017.1153.24
Ocimati, W., Were, E., Groot, J. C., Tittonell, P., Nakato, G. V., and Blomme, G. (2018). Risks posed by intercrops and weeds as alternative hosts to Xanthomonas campestris pv. musacearum in banana fields. Front. Plant Sci. 9, 1471. doi: 10.3389/fpls.2018.01471
Ogolla, M., Muraya, M., and Onyango, B. (2019). Incidence and severity of turcicum leaf blight caused by Exserohilum turcicum (pass.) Leonard and Suggs) on sorghum populations in different regions of Tharaka Nithi County, Kenya. J. Sci. Eng. Res. 6, 104–111.
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., and Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103, 1155–1166. doi: 10.1007/s00253-018-9556-6
Olawuyi, O., Odebode, A., Oyewole, I., Akanmu, A., and Afolabi, O. (2014). Effect of arbuscular mycorrhizal fungi on Pythium aphanidermatum causing foot rot disease on pawpaw (Carica papaya L.) seedlings. Arch. Phytopathol. Plant Protect. 47, 185–193. doi: 10.1080/03235408.2013.806079
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Olowe, O. M., Nicola, L., Asemoloye, M. D., Akanmu, A. O., and Babalola, O. O. (2022). Trichoderma: potential bio-resource for the management of tomato root rot diseases in Africa. Microbiol. Res. 257, 126978. doi: 10.1016/j.micres.2022.126978
Patil, H. J., and Solanki, M. K. (2016). “Microbial inoculant: modern era of fertilizers and pesticides,” in Microbial Inoculants in Sustainable Agricultural Productivity, eds D. P. Singh, H. B. Singh, and R. Prabha (New Delhi: Springer), 319–343.
Pellicciaro, M., Lione, G., Giordano, L., and Gonthier, P. (2021). Biocontrol potential of Pseudomonas protegens against Heterobasidion species attacking conifers in Europe. Biol. Control 157, 104583. doi: 10.1016/j.biocontrol.2021.104583
Pertot, I., Alaboubette, C., Hinarejos, E., and Franca, S. (2015). Mini Papaer: The Use of Microbial Biocontrol Agents against Soil-Borne Diseases. Eip-Agri, Agriculture and Innovation, Bruselas, Bélgica. Available online at: http://ec.europa.eu/eip/agriculture/sites/agrieip/files/8_eip_sbd_mp_biocontrol_final.pdf
Pisarčik, M., Hakl, J., Szabó, O., and Hrevušová, Z. (2021). Efficacy of variable timing of Pythium oligandrum applications on red clover grown under field conditions. Crop Prot. 149, 105780. doi: 10.1016/j.cropro.2021.105780
Pramanik, K., Das, A., Banerjee, J., Das, A., Chatterjee, S., Sharma, R., et al. (2020). Metagenomic insights into rhizospheric microbiome profiling in lentil cultivars unveils differential microbial nitrogen and phosphorus metabolism under rice-fallow ecology. Int. J. Mol. Sci. 21, 8895. doi: 10.3390/ijms21238895
Products, E. P. o. P. P. R. t., Hernandez-Jerez, A. F., Adriaanse, P., Aldrich, A., Berny, P., and Pelkonen, O. (2020). Statement on the translocation potential by Pseudomonas chlororaphis MA342 in plants after seed treatment of cereals and peas and assessment of the risk to humans. EFSA J. 18, e06276. doi: 10.2903/j.efsa.2020.6276
Rai, D., and Singh, S. (2018). Is banded leaf and sheath blight a potential threat to maize cultivation in Bihar? Int. J. Curr. Microbiol. App. Sci. 7, 671–683. doi: 10.20546/ijcmas.2018.711.080
Ramesha, V. (2018). Effect of different bio-control agents against stalk rot of maize caused by Fusarium solani and their identification. Pharma. Innov. J. 7, 559–564. Available online at: https://www.thepharmajournal.com/archives/2018/vol7issue11/PartJ/7-11-51-249.pdf
Ray, P., Lakshmanan, V., Labbé, J. L., and Craven, K. D. (2020). Microbe to microbiome: a paradigm shift in the application of microorganisms for sustainable agriculture. Front. Microbiol. 11, 3323. doi: 10.3389/fmicb.2020.622926
Richardson, A. E., Kawasaki, A., Condron, L. M., Ryan, P. R., and Gupta, V. V. (2021). “Root microbiome structure and microbial succession in the rhizosphere,” in Rhizosphere Biology: Interactions Between Microbes and Plants (Singapore: Springer), 109–128.
Rodriguez, P. A., Rothballer, M., Chowdhury, S. P., Nussbaumer, T., Gutjahr, C., and Falter-Braun, P. (2019). Systems biology of plant-microbiome interactions. Mol. Plant 12, 804–821. doi: 10.1016/j.molp.2019.05.006
Rotolo, C., De Miccolis Angelini, R. M., Pollastro, S., and Faretra, F. (2016). A TaqMan-based qPCR assay for quantitative detection of the biocontrol agents Bacillus subtilis strain QST713 and Bacillus amyloliquefaciens subsp. plantarum strain D747. BioControl 61, 91–101. doi: 10.1007/s10526-015-9701-4
Sahib, M. R., Pervaiz, Z. H., Williams, M. A., Saleem, M., and DeBolt, S. (2020). Rhizobacterial species richness improves sorghum growth and soil nutrient synergism in a nutrient-poor greenhouse soil. Sci. Rep. 10, 1–13. doi: 10.1038/s41598-020-72516-3
Samaras, A., Karaoglanidis, G. S., and Tzelepis, G. (2021a). Insights into the multitrophic interactions between the biocontrol agent Bacillus subtilis MBI 600, the pathogen Botrytis cinerea and their plant host. Microbiol. Res. 248, 126752. doi: 10.1016/j.micres.2021.126752
Samaras, A., Nikolaidis, M., Antequera-Gómez, M. L., Cámara-Almirón, J., Romero, D., Moschakis, T., et al. (2021b). Whole genome sequencing and root colonization studies reveal novel insights in the biocontrol potential and growth promotion by Bacillus subtilis MBI 600 on cucumber. Front. Microbiol. 11, 600393. doi: 10.3389/fmicb.2020.600393
Savazzini, F., Longa, C. M. O., and Pertot, I. (2009). Impact of the biocontrol agent Trichoderma atroviride SC1 on soil microbial communities of a vineyard in northern Italy. Soil Biol. Biochem. 41, 1457–1465. doi: 10.1016/j.soilbio.2009.03.027
Schmidt, J. E., Kent, A. D., Brisson, V. L., and Gaudin, A. C. (2019). Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 7, 1–18. doi: 10.1186/s40168-019-0756-9
Sellitto, V. M., Zara, S., Fracchetti, F., Capozzi, V., and Nardi, T. (2021). Microbial biocontrol as an alternative to synthetic fungicides: boundaries between pre-and postharvest applications on vegetables and fruits. Fermentation 7, 60. doi: 10.3390/fermentation7020060
Shahriar, S. A., Islam, M. N., Chun, C. N. W., Kaur, P., Rahim, M. A., Islam, M. M., et al. (2022). Microbial metabolomics interaction and ecological challenges of Trichoderma species as biocontrol inoculant in crop rhizosphere. Agronomy 12, 900. doi: 10.3390/agronomy12040900
Shanmugam, S. G., and Kingery, W. L. (2018). Changes in soil microbial community structure in relation to plant succession and soil properties during 4000 years of pedogenesis. Eur. J. Soil Biol. 88, 80–88. doi: 10.1016/j.ejsobi.2018.07.003
Shelake, R. M., Pramanik, D., and Kim, J. Y. (2019). Exploration of plant-microbe interactions for sustainable agriculture in CRISPR era. Microorganisms 7, 269. doi: 10.3390/microorganisms7080269
Singh, S., Singh, U. B., Malviya, D., Paul, S., Sahu, P. K., Trivedi, M., et al. (2020). Seed biopriming with microbial inoculant triggers local and systemic defense responses against Rhizoctonia solani causing banded leaf and sheath blight in maize (Zea mays L.). Int. J. Environ. Res. Public Health 17, 1396. doi: 10.3390/ijerph17041396
Stumpf, S., Kvitko, B., Gitaitis, R., and Dutta, B. (2018). Isolation and characterization of novel Pantoea stewartii subsp. indologenes strains exhibiting center rot in onion. Plant Dis. 102, 727–733. doi: 10.1094/PDIS-08-17-1321-RE
Thomashow, L. S., Kwak, Y. S., and Weller, D. M. (2019). Root- associated microbes in sustainable agriculture: models, metabolites and mechanisms. Pest Manag. Sci. 75, 2360–2367. doi: 10.1002/ps.5406
Tom, A., and Patel, S. (2021). Dynamic association of Fusarium verticillioides with maize and its biological control. Pharma. Innov. J. 10, 1010–1019. Available online at: https://www.thepharmajournal.com/archives/2021/vol10issue7/PartM/10-7-112-132.pdf
Veenstra, A., Rafudeen, M. S., and Murray, S. L. (2019). Trichoderma asperellum isolated from African maize seed directly inhibits Fusarium verticillioides growth in vitro. Euro. J. Plant Pathol. 153, 279–283. doi: 10.1007/s10658-018-1530-8
Vida, C., de Vicente, A., and Cazorla, F. M. (2020). The role of organic amendments to soil for crop protection: induction of suppression of soilborne pathogens. Ann. Appl. Biol. 176, 1–15. doi: 10.1111/aab.12555
Wallenstein, M. D. (2017). Managing and manipulating the rhizosphere microbiome for plant health: a systems approach. Rhizosphere. 3, 230–232. doi: 10.1016/j.rhisph.2017.04.004
Wang, T., Liang, Y., Wu, M., Chen, Z., Lin, J., and Yang, L. (2015). Natural products from Bacillus subtilis with antimicrobial properties. Chin. J. Chem. Eng. 23, 744–754. doi: 10.1016/j.cjche.2014.05.020
Wei, Z., Hu, J., Yin, S., Xu, Y., Jousset, A., Shen, Q., et al. (2018). Ralstonia solanacearum pathogen disrupts bacterial rhizosphere microbiome during an invasion. Soil Biol. Biochem. 118, 8–17. doi: 10.1016/j.soilbio.2017.11.012
Wei, Z., Hu, X., Li, X., Zhang, Y., Jiang, L., Li, J., et al. (2017). The rhizospheric microbial community structure and diversity of deciduous and evergreen forests in Taihu Lake area, China. PLoS ONE 12, e0174411. doi: 10.1371/journal.pone.0174411
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126. doi: 10.2174/1874437001408010071
Xavier, F., and Kaushik, M. (2021). Biological management of charcoal rot of maize caused by Macrophomina phaseolina by using Trichoderma: A. Pharma. Innov. J. 10, 417–41. Available online at: https://www.thepharmajournal.com/archives/2021/vol10issue6/PartF/10-5-184-453.pdf
Xie, S., Liu, J., Gu, S., Chen, X., Jiang, H., and Ding, T. (2020). Antifungal activity of volatile compounds produced by endophytic Bacillus subtilis DZSY21 against Curvularia lunata. Ann. Microbiol. 70, 1–10. doi: 10.1186/s13213-020-01553-0
Xiong, W., Song, Y., Yang, K., Gu, Y., Wei, Z., Kowalchuk, G. A., et al. (2020). Rhizosphere protists are key determinants of plant health. Microbiome 8, 1–9. doi: 10.1186/s40168-020-00799-9
Yang, Y., Zhao, Q., Li, X., Ai, W., Liu, D., Qi, W., et al. (2017). Characterization of genetic basis on synergistic interactions between root architecture and biological nitrogen fixation in soybean. Front. Plant Sci. 8, 1466. doi: 10.3389/fpls.2017.01466
Ye, Y.-f., Li, Q.-q., Gang, F., Yuan, G.-q., Miao, J.-h., and Wei, L. (2012). Identification of antifungal substance (Iturin A2) produced by Bacillus subtilis B47 and its effect on southern corn leaf blight. J. Integr. Agric. 11, 90–99. doi: 10.1016/S1671-2927(12)60786-X
Zhang, Y., Trivedi, P., Xu, J., Roper, M. C., and Wang, N. (2021). The citrus microbiome: from structure and function to microbiome engineering and beyond. Phytobiomes J. 5, 249–262. doi: 10.1094/PBIOMES-11-20-0084-RVW
Keywords: microbial community, food security, food safety, plant-soil-microbe association, rhizosphere engineering, plant disease management
Citation: Dlamini SP, Akanmu AO and Babalola OO (2022) Rhizospheric microorganisms: The gateway to a sustainable plant health. Front. Sustain. Food Syst. 6:925802. doi: 10.3389/fsufs.2022.925802
Received: 21 April 2022; Accepted: 07 July 2022;
Published: 04 August 2022.
Edited by:
Vineet Kumar, Lovely Professional University, IndiaReviewed by:
Jamilah Syafawati Yaacob, University of Malaya, MalaysiaEzekiel Mugendi Njeru, Kenyatta University, Kenya
Copyright © 2022 Dlamini, Akanmu and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola Oluranti Babalola, olubukola.babalola@nwu.ac.za
 Siphiwe Prudence Dlamini
Siphiwe Prudence Dlamini Akinlolu Olalekan Akanmu
Akinlolu Olalekan Akanmu Olubukola Oluranti Babalola
Olubukola Oluranti Babalola