Survival and persistence of foodborne pathogenic and indicator bacteria in spatially separated integrated crop-livestock farms
- 1Department of Agriculture, Food and Resource Sciences, University of Maryland Eastern Shore, Princess Anne, MD, United States
- 2USDA-ARS-Sustainable Agricultural Systems Laboratory, Beltsville, MD, United States
- 3U.S. Food and Drug Administration, College Park, MD, United States
- 4Department of Animal and Avian Sciences, and Center for Food Safety and Security Systems, University of Maryland, College Park, MD, United States
Certified organic production prohibits chemical fertilizer use but permits the application of biological soil amendments of animal origin (BSAAOs) to croplands for fertilizing soil. For fresh produce likely to be consumed uncooked, the United States Department of Agriculture National Organic Program (USDA-NOP) stipulates a 90-day withholding period between BSAAO application and crop harvest for produce not in direct contact with soil to reduce concerns about potential pathogen contamination from BSAAOs. For fresh produce crops in direct contact with soil this withholding period is 120-days. In this study, survival and pathogenesis of three foodborne pathogens and indicator bacteria in BSAAO-amended soils were analyzed with their potential to transfer to produce harvested from three organic, integrated crop-livestock farms (ICLFs) on the Maryland Eastern Shore. In total, 428 manure/compost, soil, untreated produce, and water samples were collected before/after BSAAO incorporation and monthly for 180-days. Samples were assessed for the presence of Listeria monocytogenes (L. monocytogenes), Salmonella, and Shiga toxin-producing E. coli (STEC) with associated virulence factor (VF) genes (STEC/VF-genes), and for aerobic plate count (APC), generic E. coli (gEC), and total coliform levels using standard methods and polymerase chain reaction (PCR). Overall, 18.4% (26/141) of produce samples were positive for at least one tested pathogen, with STEC/VF-genes being detected the most. Produce was contaminated with at least one pathogen and indicators 90- and 120-days post-BSAAO incorporation. Salmonella wasn’t detected in manure/compost samples, and 6.7% (1/15) and 66.7% (10/15) of manure/compost samples were positive for L. monocytogenes and STEC/VF-genes, respectively. In BSAAO-amended soils, 29.1% (74/254) were positive for at least one of the tested pathogens in this study. STEC/VF-genes were detected in 24.0% (61/254) of soils, while L. monocytogenes and Salmonella were detected in 4.3% (11/254) and 0.8% (2/254) of soils, respectively. These three pathogens were detected in soils directly after BSAAO incorporation but were followed by a reduced prevalence. However, pathogens were detected in soils 90- and/or 120-days post-application. The results of this study indicated pathogens associated with BSAAOs (contaminated manures/composts) can persist after soil incorporation and transfer to harvestable produce grown on ICLF’s on the Maryland Eastern Shore. Therefore, further research into withholding periods and other mitigation strategies is necessary to mitigate pathogen contamination risks on ICLFs.
Introduction
Integrated crop-livestock farm (ICLF) systems raise animals and grow crops in the same environment, utilizing the resulting products of one component for the successful growth of the other (Patterson et al., 2018; Pires et al., 2019). There are three categories of ICLFs based on the interaction between crop and livestock components: (1) Spatially separated (animals and crops are grown in separated areas), (2) Rotational (animals roam crop fields after harvest), and (3) Fully combined farming (animals and crops occupy the same space) (Salaheen et al., 2015; Peng et al., 2016). This complementary interaction between crops and animals on ICLFs provides environmental and financial benefits that do not accrue to the same extent in conventional or monocultural farming systems. On ICLFs, animal manures improve soil fertility for crop cultivation, while resulting crops are used as feed for animals, thus fostering a sustainable system by recycling on farm nutrients naturally (Patterson et al., 2018). Potential for increased total net farm revenue on ICLFs results from production of diverse products (crops, meats, dairy, eggs, etc.) and the recycling of on-farm resources (lower fertilizer, feed, pesticide, etc. costs). In contrast, conventional/monocultural farms generate fewer products and depend on synthetic pesticides, fertilizers, and antimicrobials that decrease biodiversity, pollute the environment, contaminate food, and contribute to resource depletion (Salaheen et al., 2015; Patterson et al., 2018; Pires et al., 2019; Sekaran et al., 2021).
The presence of animals, which are recognized natural carriers of foodborne pathogens, on ICLFs raise food safety concerns arising from potential cross-contamination between animal reservoirs and produce (Berry et al., 2013; Salaheen et al., 2016; Pires et al., 2019; Ekman et al., 2021; Nazareth et al., 2021). Animal manure on farms is of particular concern because it can directly or indirectly contaminate produce. Manures for soil amendments can directly contaminate soils and produce, and manure from animal enclosures, amended-soils, and stockpiles can indirectly contaminate fields, irrigation water, and produce due to runoff (Berry et al., 2013; Salaheen et al., 2015; Alegbeleye and Sant’Ana, 2020). Multiple studies have detected the foodborne pathogens Salmonella, Listeria monocytogenes (L. monocytogenes), or Shiga-toxin producing Escherichia coli (STEC) in pre-harvest soil, water, produce, fly, and animal feed, fecal, and hide samples collected from model and operating ICLF systems (Berry et al., 2013; Peng et al., 2016; Salaheen et al., 2016; Pires et al., 2019; Glaize et al., 2020; Ekman et al., 2021; Nazareth et al., 2021). Furthermore, crops and animals may share the same pre- and post-harvest facilities, therefore increasing the chances of cross-contamination between surfaces and tools when good agricultural practices (GAPs) and good handling practices (GHPs) are not followed (Salaheen et al., 2015; Peng et al., 2016).
Biological soil amendments of animal origin (BSAAOs) are essential to organic ICLFs and can be plant- or animal-based when derived from recycled plant matter or animal manures. Raw and composted manures from goats, cows, poultry, etc. are applied to agricultural soils to improve and sustain soil health (Ekman et al., 2021; Ramos et al., 2021). These manures are typically the amendment of choice because they are inexpensive, high in nutrient content, and thus improve several soil properties that include, but are not limited to, nutrients, biodiversity, organic matter, structure, and water infiltration and retention (Berry et al., 2013; Tran et al., 2020; Ekman et al., 2021; Ramos et al., 2021). Since organic farms are unable to utilize synthetic fertilizers and are required by the United States Department of Agriculture National Organic Program (USDA-NOP) to improve soil health over time, manure amendments are vital to a successful operation (Ramos et al., 2021). According to a recent survey of certified NOP producers, 46.8% used BSAAOs, and 58% utilized raw manures (Pires et al., 2018; Ramos et al., 2021). If contaminated, manures introduce pathogens into soils that contaminate crops through colonization of seedlings and transfer from soil to leaves through contact, splash, or dust (Ingham et al., 2005; Jacobsen and Bech, 2012; Ekman et al., 2021). Solomon et al. (2002) demonstrated that lettuce roots uptake E. coli and transfer to leaves, a mechanism that may apply to other plants and pathogens. High-risk produce and other crops with edible portions in proximity to manure-amended soils have an increased risk for contamination and causing foodborne illnesses. Fruits and vegetables typically consumed raw (e.g., leafy greens, tomatoes, bell peppers) are considered high-risk, and the lack of a pathogen killing step allows for survival of pathogens at consumption (Patterson et al., 2018; Nazareth et al., 2021). Foodborne illness cases associated with vegetables have been linked to on-farm aged and raw manures. Horse manure has been linked to vegetable contamination, while cattle manure (aged and raw) has been identified as contamination source in STEC O157:H7 outbreaks of lettuce (Tran et al., 2020; Ramos et al., 2021).
The United States Food and Drug Administration (FDA) Food Safety Modernization Act (FSMA) Produce Safety Rule (PSR) establishes preventive control methods for growing, harvesting, packaging, and holding of produce. To reduce transfer of pathogens from manure-amended soils to produce, the FDA-FSMA-PSR proposed a nine-month withholding period where farmers wait the allotted time between manure application and produce harvest (FDA, 2015). This lengthy withholding period would make using manure amendments difficult for farmers and, based on public comments from the agricultural industry, the FDA deferred action on the proposal until further research is conducted (Patterson et al., 2018; Ekman et al., 2021; Ramos et al., 2021). Currently, the FDA recommends farmers follow the USDA-NOP 90- and 120-day withholding period for low- and high-risk products, respectively (FDA, 2015; Ekman et al., 2021; Ramos et al., 2021). Although the USDA-NOP mandate is more convenient for farmers, it was developed based on the crop production cycle rather than scientifically-based assessments (Ramos et al., 2021). Multiple studies in laboratory or model ICLF systems detected pathogens in soils and produce after the minimum withholding period specified by the USDA-NOP. Studies described pathogen survival up to 332 days in inoculated manure-amended soils, and pathogen isolations from high-risk produce samples 168 days post-amendment (Islam et al., 2005; You et al., 2006; Ekman et al., 2021; Ramos et al., 2021). Though pathogens have shown increased persistence in laboratory studies, field studies have also found them present in soils and on produce after more than 120 days post-amendment (Tran et al., 2020; Ramos et al., 2021). Overall, studies suggest further research is needed to establish adequate protocols for on-farm manure handling and amendments.
Current trends surrounding consumer demand and foodborne illnesses highlight the need for research evaluating contamination risks associated with ICLFs. Over the past decade, there has been an increased demand for produce and organic, locally grown products. This is a result of the health benefits associated with fruits and vegetables, and the perception that organic, local products are safer due to fewer chemical inputs and less handling (Harvey et al., 2016; Carstens et al., 2019). Consequently, organic food market sales and the number of farmer’s markets in the United States have increased significantly from the 1990’s to 2019 (Herrero et al., 2010; USDA-AMS, 2015, 2019; Harvey et al., 2016; Peng et al., 2016; Bellemare and Nguyen, 2018; Kim et al., 2021). ICLFs are major suppliers for farmer’s markets, backyard stands, and community supported agriculture (CSAs), and even sell products at retail stores on occasion (Peng et al., 2016; Salaheen et al., 2016; Pires et al., 2019). This increasing popularity, in conjunction with the microbiological risks of BSAAOs can potentially trigger more foodborne illness cases and outbreaks.
Foodborne pathogens have been a recurring problem worldwide that negatively impact human health and parties involved in the production of foods. There are multiple pathways by which foods become contaminated, and research is necessary to further investigate and develop mitigation strategies. BSAAOs are essential to crop production but need further evaluation due to contamination risks associated with manures. Limited information is available on pathogen survival/transmission and the efficacy of the USDA-NOP 90- and 120-day rule on ICLFs. This study assessed the microbiological contamination of BSAAOs on ICLFs by enumerating total aerobic bacterial (APC), generic E. coli (gEC), and total coliform levels, while also testing for the presence of three major foodborne pathogens L. monocytogenes, Salmonella, and STEC serovars and associated virulence factor genes (STEC/VF-genes). Initial manures/composts, soils before (D0A) and after amendment (D0B) through day 180 (D180), and corresponding produce samples were evaluated to examine the pathogen load relative to the current withholding period.
Materials and methods
Farm selection criteria, description of farms, and study population
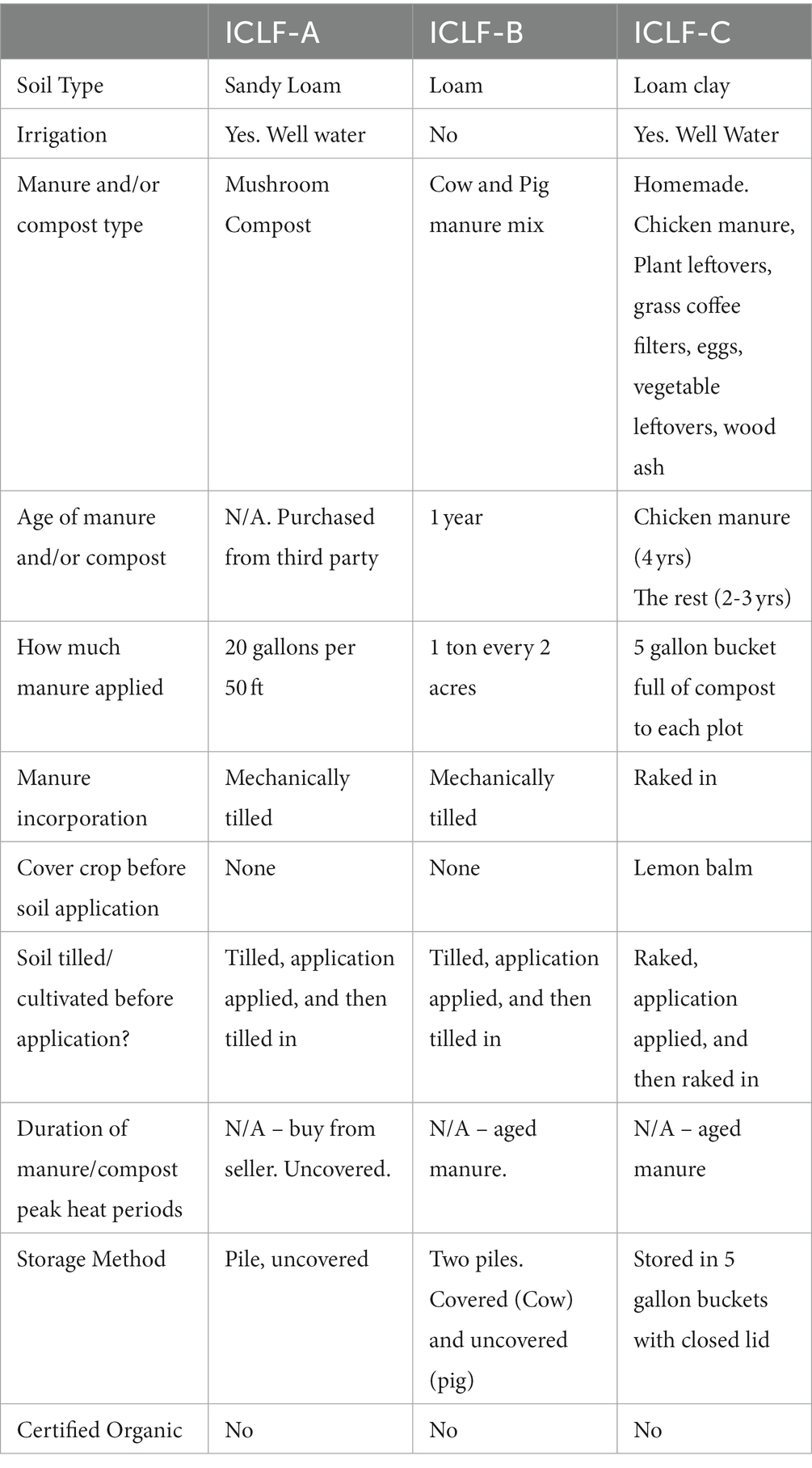
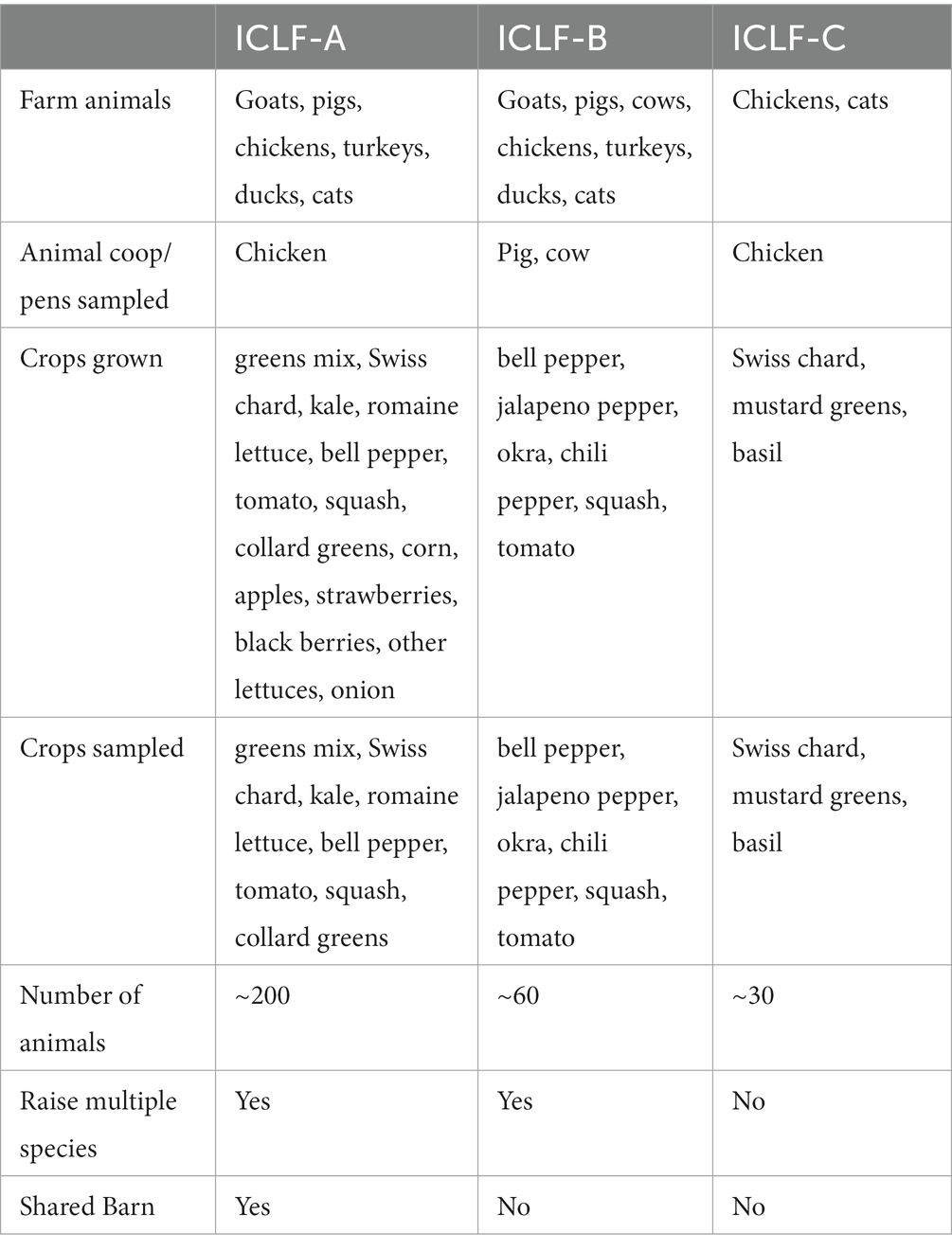
All recruited farms were located on the Eastern Shore of Maryland, representing the various types of environments, soils, produce, and management practices of fresh produce growers within the Mid-Atlantic Region of the United States. The Eastern Shore was selected as the study location because the number of ICLFs have recently increased in the Mid-Atlantic Region. In addition, multiple outbreaks have been linked to produce originating from the region, and the use of BSAAOs, such as poultry litter, which can contaminate crop fields, has become popular (Micallef et al., 2012; Gu et al., 2019; Peng et al., 2021). Locations of these farms were selected based on farmers/owners’ willingness to participate in the study and the farms meeting the criteria listed below. Recruited farms were contacted through a personal invitation by phone, email, and/or an in-person visit. Farms chosen for this experiment met the following criteria: (1) Organic (USDA-NOP certification not required), (2) ICLF that applied biological soil amendments (animal origin and/or plant based) to agricultural soils, (3) Grew high-risk fresh fruits and vegetables such as leafy greens, tomatoes, and peppers, (4) Generate and sell products locally (backyard stands, CSA, farmer’s markets, etc.), (5) Agreement to participate and ability to provide needed on-farm samples and farm management information. Three farms participated in this 10-month longitudinal field study by providing samples and farm information from March to December 2021. Farm information was obtained through a survey that assessed farm management practices including types of animals and produce grown, storage and application of manures, irrigation methods, etc. Farmers were provided soil health/fertility results of their agricultural soils before/after biological soil amendment application and had access to UMES’s Extension and Outreach Program network and resources for assistance if needed.
The number and type of samples collected at each sampling between pre-manure amendment (D0A) and day 180 (D180) varied between the three farms as a result of the number of plots and available produce on each farm. There was a total of 428 samples collected from the three ICLFs (15 manure/compost, 254 soil, 141 produce, and 18 water samples). The three farms were organic (not USDA-NOP certified), spatially separated ICLFs that applied biological soil amendments to agricultural fields and generated fresh produce along with animal products. The animals and manure/compost piles were in separate locations on the farm. Each ICLF grew produce such as leafy greens, tomatoes, and peppers, and had varying animals such as poultry, cows, goats, pigs, dogs, and cats. The three farms were given the labels ICLF-A, ICLF-B, and ICLF-C, so that farms retained anonymity. Unique biological soil amendments were applied at individual farms. ICLF-A applied commercially produced mushroom compost, ICLF-B applied farm-managed, aged cow/pig manure mix, and ICLF-C applied a farm-managed, aged chicken manure/vegetable material mix. For treatment, ICLF-B and ICLF-C aged the manures, while ICLF-A’s compost treatment was unavailable as it was obtained from a third party. All farms placed BSAAOs on soil surface and then mechanically tilled (ICLF-A and ICLF-B) or raked (ICLF-C) the amendment into the soil. ICLF-A and -C irrigated fields with well water, while ICLF-B did not irrigate agricultural fields. ICLF-A and ICLF-C used sprinkler and drip irrigation systems, respectively. ICLF-A, ICLF-B, and ICLF-C had sandy loam, loam, and clay loam soils, respectively. Detailed descriptions of each farm and its management practices are shown in Tables 1, 2.
During this study, samplings were conducted on farms from pre-manure amendment (D0A) to D180. Unexpectedly, collection of samples on certain sampling days was missed due to weather events or inability to contact farmers, and numbers may vary due to availability of samples. For example, on ICLF-B D180, soil samples were only collected from one plot as the others had been tilled because no crops were present near the end of the season. For each farm, samples were collected on D0A, D0B, and D180, but some collection days were missed between these periods. Sampling did not occur on ICLF-C for D30, and ICLF-B for D30, D60, and D150. All planned samplings were conducted on ICLF A. In addition, samples were also collected on D45 and D75 to ensure availability of produce data from each plot and farm. For example, on ICLF-C mustard greens were collected on D75 because produce would be heat injured and thus unavailable by D90 or D120. This study was conducted on working farms, contact with farmers was difficult at times, and sampling days were added to adjust for possible complications. Thus, the final sample set did not align with the original, balanced study design.
Sample collection
Sampling began in March 2021 and continued until December 2021. There was a total of 428 samples collected from three ICLFs located on the Eastern Shore of Maryland in this 10-month longitudinal study. Four types of samples were collected in this experiment: manure/compost, soil, produce, and water samples. Samples of on-farm stored manure/compost piles were collected before incorporation into soils. Soil samples were collected before (D0A) and post-soil amendment (D0B), and every 30 days (D30, D60, D90, D120, and D150) until D180. Crop samples were collected whenever they were available from the corresponding soil samples. All produce samples selected for analysis displayed no signs of damage, spoilage, or contamination (signs of bugs, pests, wild animals, feces could not be seen on selected samples), and appeared ripe and ready to eat. Before analysis, produce samples did not undergo any further processing after removal from soil. Samples were collected and processed as described in previous studies (Weller et al., 2015b; Gu et al., 2019; Glaize et al., 2020; Ramos et al., 2021). In brief, manure/compost samples were collected in replicates of 5 into 52 oz. Whirl-Pak bags (Nasco Whirl-Pak, Madison, WI) using a sterile, stainless-steel probe/scoop. Samples were collected in a composite manner at varying depths with at least 150 g per sample. Using a sterile, stainless-steel scoop, composite soil samples in replicates of 3 or 5 were collected from soil plots at a depth of ~7–15 cm from the soil surface into sterile Whirl-Pak bags. Soil samples were collected in a serpentine pattern with at least 150 g of soil collected per sample, and leafy greens and other vegetable samples (up to n = 11, depending on availability at time of collection) were aseptically harvested separately from the soils, and placed into sterile Whirl-Pak bags. Irrigation water samples were collected into sterilized Nalgene™ (Thermo Scientific, Waltham, MA) 1,000 mL bottles. Water was collected during summer and fall from farm well sources. Samples were collected from taps, which were sterilized with 70% ethanol and allowed to run for 2 min before sampling. All samples were placed on ice and transported to the Food Microbiology and Safety Laboratory at the University of Maryland Eastern Shore and processed within 48 h. To prevent contamination of samples, nitrile gloves, soil cores/scoops, and other sampling equipment were cleaned and sanitized (70% ethanol) before and after each sampling.
Sample processing, enumeration, enrichment, and pathogen isolation
Sample processing was done as described in previous studies (Pagadala et al., 2015; Ekman et al., 2021; Kim et al., 2021; Ramos et al., 2021). In brief, all samples were processed for the enumeration of total aerobic plate count (APC) and generic E. coli (gEC), total coliform levels, and for isolation of L. monocytogenes, Salmonella, and STEC/VF-genes. The sample processing methods were slightly different for stomach-able and non-stomach-able samples. Stomach-able samples could be blended to detach organisms such as soil, manure/compost, and leafy greens, whereas non-stomach-able samples could not be blended (such as bell peppers, tomatoes, squashes, chili peppers, jalapeno peppers, and okras).
APC, gEC and total coliform level enumeration
For stomach-able samples, each 25 g sample in a sterile Whirl-Pak bag containing 225 mL 0.1% peptone water (Fisher, Fair Lawn, NJ) was stomached (Seward Inc., Bohemia, NY) 2 min at 230 rpm. Then subsequent 10-fold serial dilutions were prepared. For non-stomach-able samples, produce samples and 0.1% sterile peptone water were added to Whirl-Pak bags in equal mass amounts and hand-massaged 2 min. From each primary sample suspension, a 1:5 dilution was prepared (2 mL product +8 mL peptone) to yield a 10−1 dilution, which was then used to prepare serial dilutions up to 10−7. 3M™ Petrifilm™ Aerobic Count Plate and E. coli/Coliform Count Petrifilms (3 M, St. Paul, MN) were used to enumerate their respective organisms. All samples were plated, incubated, and counted according to the manufacturer’s instructions.
Water samples
Water samples were evaluated for the presence of gEC and total coliforms using the Colilert* Quanti-Tray/2000 MPN system (IDEXX Laboratories, Westbrook, ME) according to the manufacturer’s instructions, with incubation at 35°C; results were interpreted based on the IDEXX MPN tables. None of the water samples yielded wells positive for gEC or coliforms, so water samples were not evaluated for the presence of pathogens.
Pathogen isolation
For soil, manure/compost, and stomach-able produce samples, 30 g of sample and 120 mL of primary enrichment broth were weighed into 24 oz. sterile Whirl-Pak bags. As for non-stomach-able produce samples, 30 mL from primary sample suspension used for Petrifilm analyzes were added to Whirl-Pak bags with 120 mL primary enrichment broth prepared according to manufacturer’s instructions.
Listeria monocytogenes (L. monocytogenes)
For L. monocytogenes enrichment and isolation, methods described in Ramos et al. (2021) were used with a few modifications. Samples were enriched with Listeria Enrichment broth (LEB) (Becton, Dickinson and Company, Sparks, MD), stomached for 2 min, and incubated at 37°C for 24 h. After incubation, 200 μL of samples were added to 5 mL of Frazer Broth (FM) (Oxoid Ltd., Hampshire, United Kingdom) and incubated for 48 h at 37°C. Resulting black tubes were then streak-plated onto Brilliance Listeria Agar (BLA) (Oxoid Ltd., Hampshire, United Kingdom) and incubated at 37°C for 48 h. Presumptive blue/green colonies with a halo were picked for confirmation.
Salmonella
For the Salmonella enrichment and isolation, samples were enriched in buffered peptone water (BPW) (Becton, Dickinson and Company, Sparks, MD), at 37°C for 24 h (Peng et al., 2016; Pires et al., 2019; Ramos et al., 2021; Protocol, TBD). After incubation, 100ul of sample was added to 10 mL Rappaport Vassiliadis (RV) broth (Becton, Dickinson and Company, Sparks, MD) and incubated for 24 h at 42°C for better Salmonella recovery as recommended by the Bacteriological Analytical Manual (BAM) (FDA, 1998; Hammack et al., 1999). Then samples were streak-plated onto Xylose Lysine Tergitol-4 (XLT4) agar (Becton, Dickinson and Company, Sparks, MD) and incubated at 37°C for 48 h. Presumptive black colonies were picked for confirmation.
Shiga-toxin producing E. coli and virulence factor genes
For STEC/VF-genes (Aditya et al., 2023), samples were enriched with Luria-Bertani broth (Becton, Dickinson and Company, Sparks, MD) supplemented with 5% sheep blood (Colorado Serum Co, Denver, CO), stomached for 2 min, and incubated for 24 h at 37°C. After incubation, samples were then streak-plated onto MacConkey with Sorbitol Agar (Becton, Dickinson and Company, Sparks, MD), and incubated for 24 h at 35°C with pink colonies presumed positive for STEC/VF-genes.
For all pathogens, at least three presumptive isolates were spread-plated onto Tryptic Soy Agar (TSA) (Becton, Dickinson and Company, Sparks, MD), and colonies were frozen at -80°C in cryovials containing TSB (Becton, Dickinson and Company, Sparks, MD) with 24% glycerol (Fisher, Fair Lawn, NJ).
Confirmation of presumptive pathogens
Listeria monocytogenes
All presumptive L. monocytogenes were confirmed using Polymerase chain reaction (PCR) targeting the hlyA as described by Aznar and Alarcon (2003), and DNA was extracted using InstaGene Matrix (Bio-Rad, Hercules, California) according to the manufacturer’s instructions. The primers (Invitrogen, Waltham, MA) for this reaction were: F, 5′-GAATGTAAACTTCGGCG-CAATCAG-3′ and R, 5′-GCCGTCGATGATTTGAACTTCATC-3. The final PCR reaction had a volume of 25 μL consisting of 12.5 μL GoTaq® Green Mastermix (Promega, Madison, WI), 8.5 μL PCR Water (Invitrogen, Waltham, MA), 0.5 μL of each primer (10 μM), and 3 μL of sample DNA. The PCR Thermocycler (Applied Biosystems, Waltham, MA) cycling conditions were set for 95°C for 5 min, then 30 cycles at 95°C for 45 s, 56°C for 45 s, 72°C for 60 s, and a final extension at 72°C for 7 min. The PCR products were separated by electrophoresis in a 1% agarose gel, and the gels were stained with ethidium bromide and viewed using a BioSpectrum® 310 Imaging System (UVP, LLC, Upland, CA). The hlyA target gene has a 388 bp fragment.
Salmonella spp.
To confirm frozen presumptive Salmonella isolates, TSB tubes were inoculated with 10 μL of freezer vial cultures and incubated overnight at 37°C for confirmation using the BAX® system PCR (Qualicon Diagnostic, Camarillo, CA) according to the manufacturer’s instructions. In brief, 5 μL of enriched samples and 200 μL of lysis reagent were added to cluster tubes that were heated at 37°C for 20 min, 95°C for 10 min, and then cooled in a cooling block (4°C) for 5 min. From this solution, 50 μL of lysate were added to PCR tubes supplied with BAX® PCR tablets that contain all the PCR reagents needed, including target-specific dye-labeled probes for fluorescent detection of Salmonella. These were then loaded and processed in the BAX® System Instrument, and results were interpreted based on methods detailed in the user guide.
Shiga-toxin producing E. coli and virulence factor genes
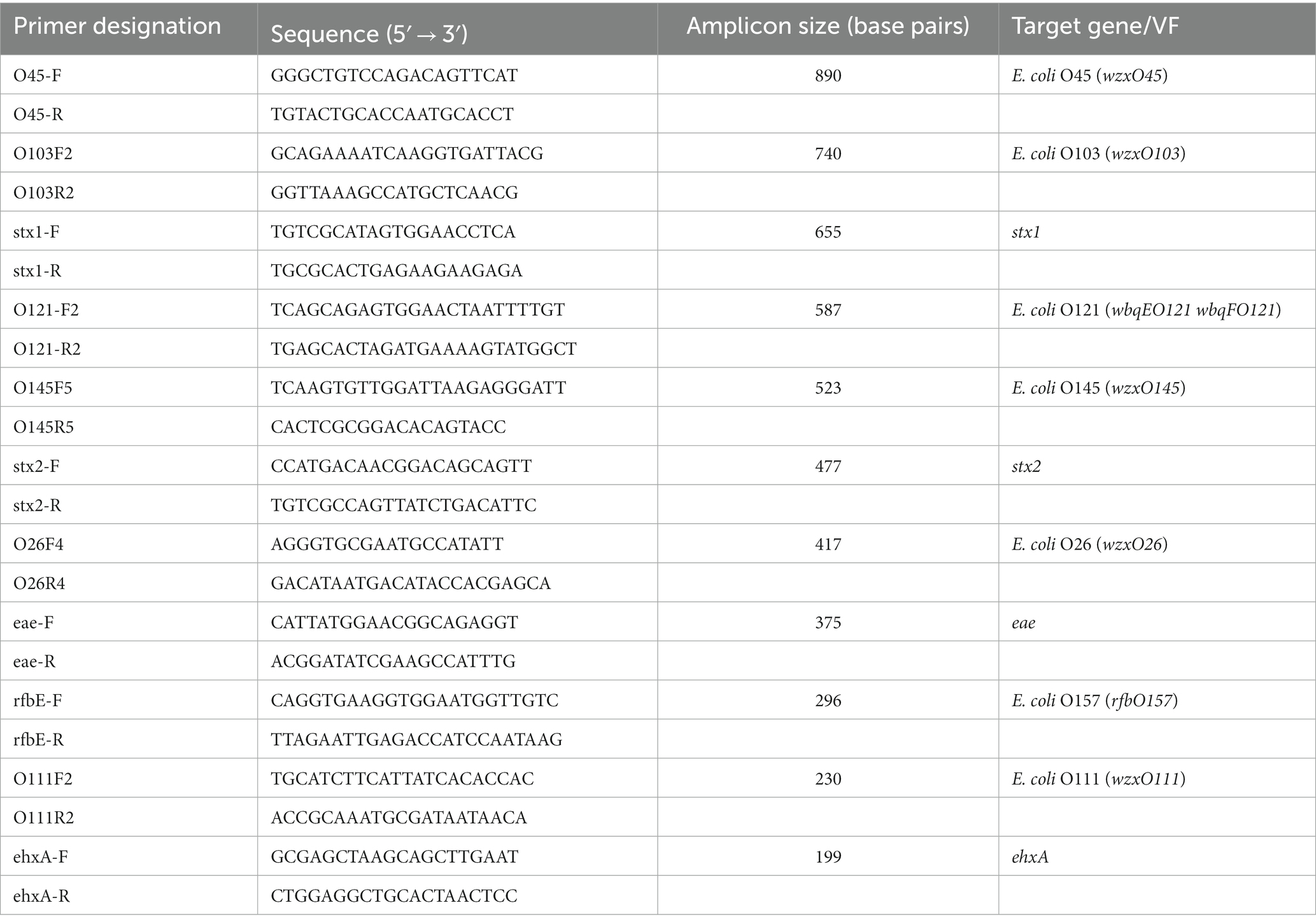
An 11-plex PCR protocol targeting seven major STEC serogroups and four VF genes was used to confirm presumptive isolates. The primers and protocols were derived from previously published studies (Bai et al., 2012; Haymaker et al., 2019). This assay targeted sequences unique to the “big six” (O45, O103, O121, O145, O26, O111) STEC serogroups, O157:H7, and STEC VF genes stx1, stx2, eae, and ehxA. These serogroups and VF-genes were selected because they have been frequently associated with and contribute to the development of STEC outbreaks and severe cases such as hemolytic uremic syndrome (HUS) and hemorrhagic colitis (Boerlin et al., 1999; Naseer et al., 2017; Galarce et al., 2019). Primer (Integrated DNA Technologies, Coralville, IA) sequences, amplicon size (base pair), and target genes are described in Table 3. Presumptive isolates from freezer vials were plated onto TSA for enrichment and incubated at 37°C overnight. DNA was extracted from the resulting cultures using InstaGene Matrix following the manufacturer’s instructions. The final volume of the PCR reaction was 20 μL consisting of 12.4 μL PCR water, 4 μL 5x MyTaq Reaction buffer (Meridian Bioscience, Cincinnati, OH), 1.2 μL concentration of MgCl2 (Meridian Bioscience, Cincinnati, OH), 0.4 μL MyTaq HS DNA polymerase (Meridian Bioscience, Cincinnati, OH), 1 μL of primer mix (concentration of 0.42 μM for O111and 0.21 μM for the other primers), and 1 μL of sample DNA. This PCR program was set for initial denaturation for 1 min at 95°C, then denaturation for 15 s at 95°C, annealing for 15 s at 62°C, extension for 90 s at 72°C. This was conducted for a total of 35 cycles. The PCR products were separated by electrophoresis in 1x LB buffer (Faster Better Media LLC, Baltimore, MD) on a 1% agarose gel with 1x GelRed® Nucleic Acid Gel Stain (Biotium Inc., Fremont, CA). Gels were viewed using UVP GelStudio™ Plus Touch (Analytik Jena US, Upland, CA) and Vision Works® Capture and Analysis Software (Analytik Jena US, Upland, CA).
Statistical analysis
Descriptive analysis was performed based on the methods described by Ramos et al. (2021). Prevalence of the three pathogens and gEC in soil, unadjusted for potential differences between farms, were determined by pooling sample data over farms. Where sufficient data was available, prevalence was also adjusted for spatial clustering on farms by fitting a mixed-effects logistic regression with categories such as season as the fixed effects and the farm as a random effect using the package lme4 in R. Unadjusted and, where appropriate, adjusted prevalence were determined for different season, farm, manure type, and production style categories. Point estimates and confidence intervals for the adjusted prevalence were determined by the coefficients of the model fit and their standard errors. Confidence intervals for unadjusted prevalence were determined nonparametrically by the Agressi-Coull method using the package PropCIs in R. Prevalence of the three pathogens and gEC in soil were plotted versus total coliform concentrations and the sampling timeline using the package ggplot in R. Mean log APC levels and their standard errors for both soil and produce were also plotted against the sampling timeline as well. The statistical significance of pairwise differences in prevalence among the different serovars and VF-genes were assessed using McNemer’s test. Differences were considered significant when the value of p was less than 0.05. All calculations were performed using R 4.1.2.
Results
Distribution of pathogens in produce samples
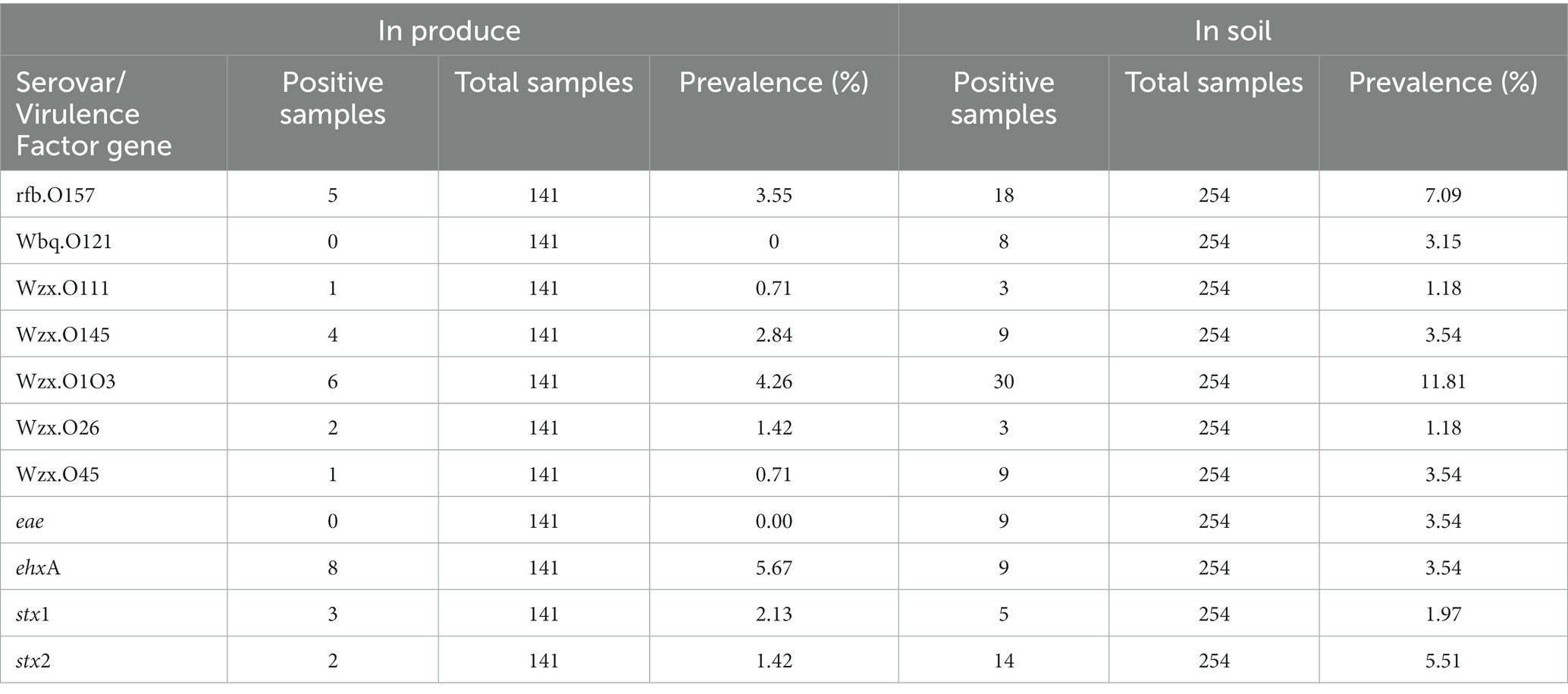
Of the 141 total produce samples, 18.4% (26/141) were positive for at least one of the tested pathogens with L. monocytogenes, Salmonella, and STEC/VF-genes being present in 3.5% (5), 0.7% (1), and 14.2% (20) samples, respectively (Table 4). Only one squash sample from ICLF-B on D90, amended with cow/pig manure mix was found to harbor L. monocytogenes. On ICLF-C, L. monocytogenes was detected on four mustard greens samples and Salmonella on one mustard green sample. All tested pathogens were present on D180 in soils that received an aged chicken manure/vegetable material mix. The most frequently detected pathogen in produce samples in this experiment was STEC as evidenced by the STEC/VF genes. On ICLF-A, STEC/VF-genes were isolated on D60, D75, D90, D150, and D180 from one squash, two tomatoes, three bell peppers, one greens mix, and three kale and five Swiss chard produce samples, respectively, from fields amended with commercially produced mushroom compost. Three bell peppers and two chili peppers obtained on D120 from ICLF-B were positive for STEC/VF-genes as well. No STEC/VF-genes were detected on E. coli isolates from produce collected from ICLF-C. The most frequently detected STEC serovars were O103, O157, and O145, while the most frequently detected VF-genes were ehxA, stx1, and stx2. Within each category, there were no significant differences (p < 0.05) among the serovars and VF-genes (Table 5). Multiple serovars and VF-genes were isolated from multiple samples. Only one (0.71%) produce sample tested positive for multiple pathogens. A mustard greens sample collected on D180 from ICLF-C was positive for L. monocytogenes and Salmonella. In terms of gEC, 3.5% (5) of produce samples from all farms were positive for gEC. Two were from ICLF-A on D30 (greens mix) and D180 (kale), two from ICLF-B on D90 (squash) and D180 (tomato), and one from ICLF-C on D75 (Swiss chard). The kale sample from ICLF-A was the only sample positive for both gEC and a pathogen (STEC and VF-genes). Mean APC Log CFU/g values peaked at D180. Furthermore, from the D90 sampling onward, APC levels remained 1 log CFU/g higher than D75 levels (Figure 1).
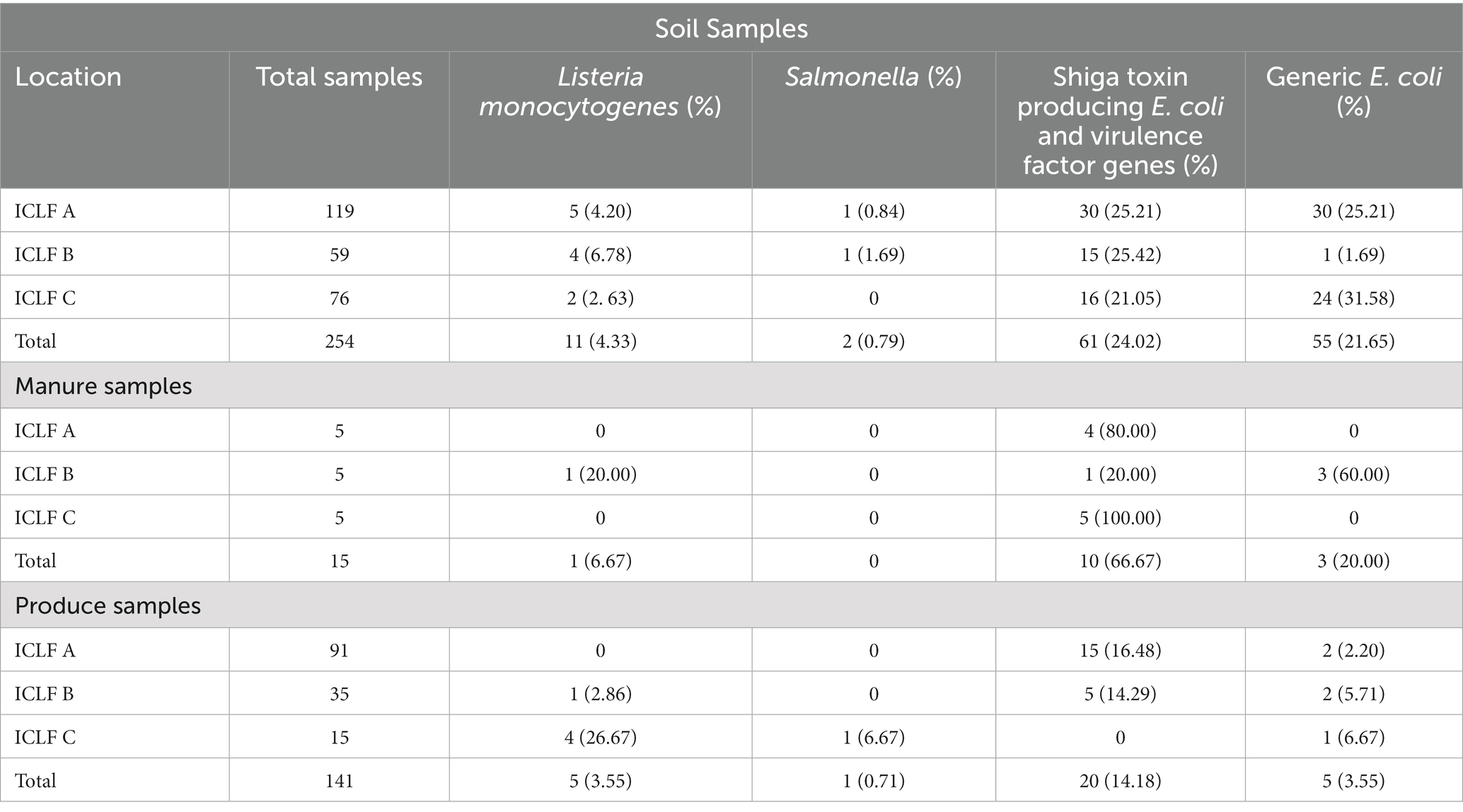
Table 4. Total samples collected and positive samples of three foodborne pathogens from soil, manure, and produce by farm and sample type.
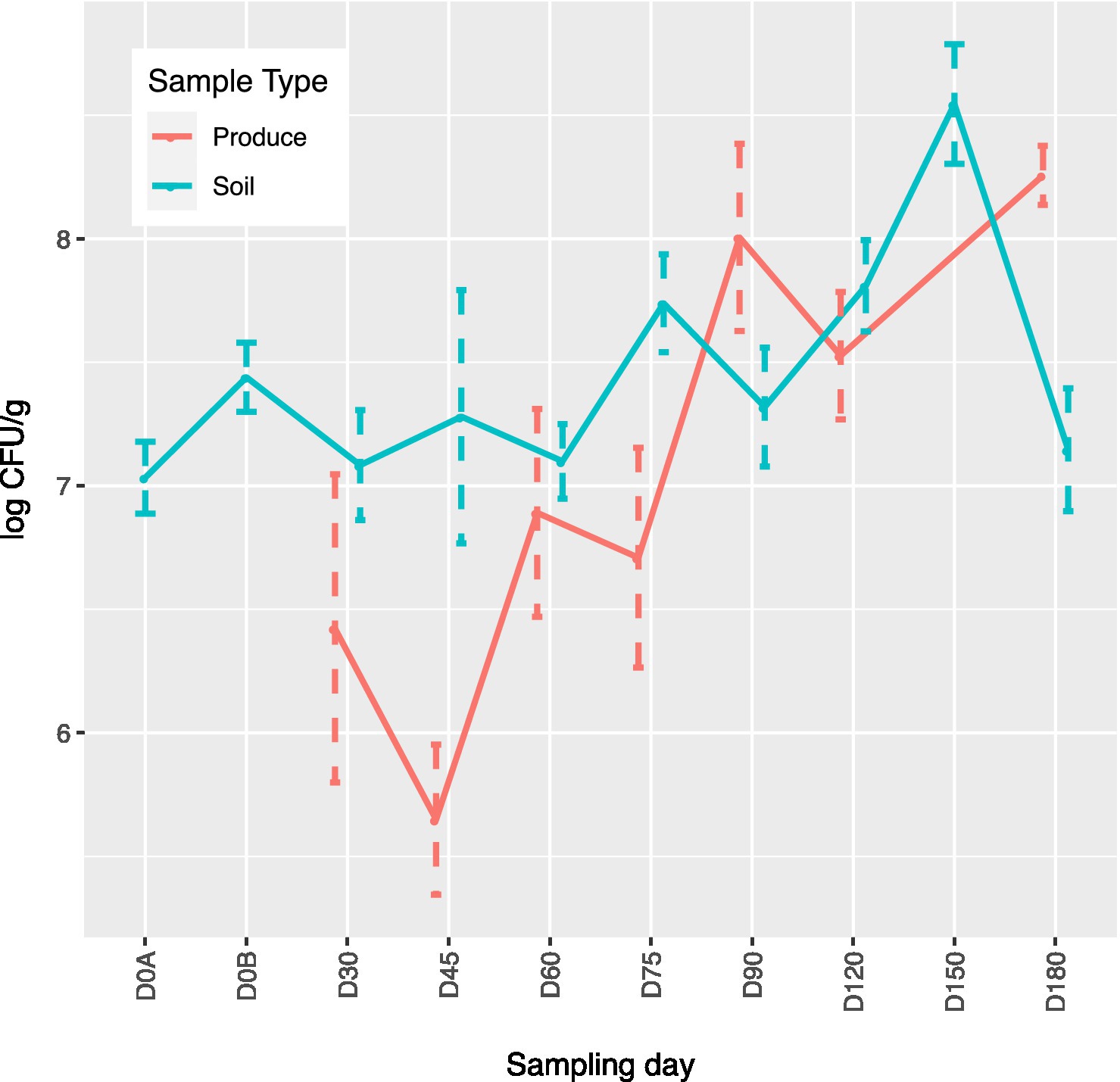
Figure 1. Aerobic plate count (APC) mean log CFU/g concentrations over time by sampling day for produce and soil samples. For produce, D150 values were not used due to technical error. D0A–before biological soil amendment of animal origin. D0B – after biological soil amendment of animal origin.
Prevalence of pathogens in biological soil amendment samples
There was a total of 15 manure/compost samples collected before application, and of these 73.3% (11/15) were positive for at least one pathogen (Table 6). The eleven positives consisted of four compost (36.4%) and seven (63.6%) aged manure samples. The four compost samples did not contain animal manures, while five (71.4%) and two (28.6%) of the aged-manure samples contained chicken manure (mixed with vegetable material) and cow/pig manure mixed, respectively. Furthermore, four (36.4%) of these 11 positives were produced commercially, and the remaining seven (63.6%) were managed on the farm. No samples were positive for Salmonella, and 6.7% (1/15) and 66.7% (10/15) of manure/compost samples were positive for L. monocytogenes and STEC/VF-genes, respectively. One manure sample from ICLF-B (aged cow/pig manure mix) was positive for L. monocytogenes, while four compost, one manure, and five manure samples were positive for STEC/VF-genes on ICLF-A (mushroom compost), ICLF-B, and ICLF-C (aged chicken manure/vegetable mix), respectively (Table 4). The most frequent serovar detected was O45 with four detections, while O103, O26, and O157 were each detected twice. The VF-gene stx1 was detected from four samples and stx2 from two. Three (20%) manure samples were positive for gEC all isolated from ICLF-B (Table 4). Only one of these samples was positive for gEC and pathogens. This sample was positive for gEC, L. monocytogenes, and STEC/VF-genes representing the only biological soil amendment sample positive for multiple pathogens.
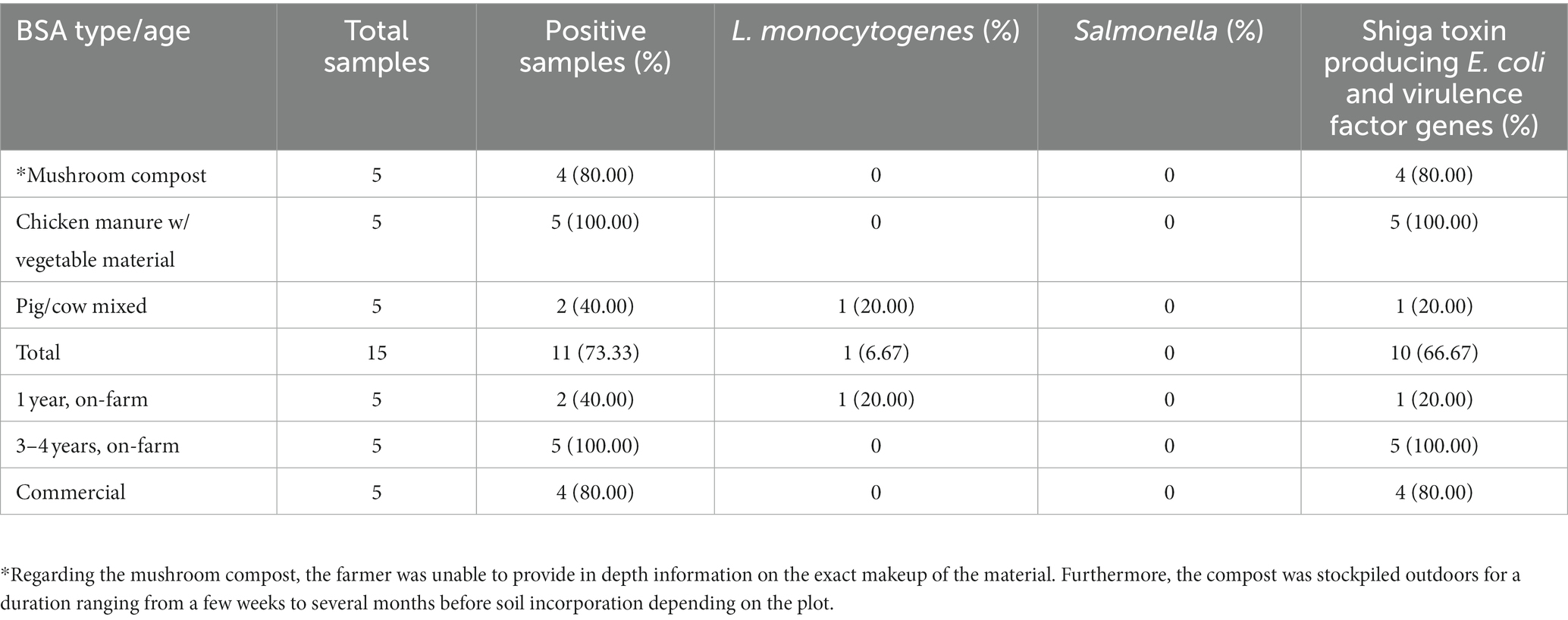
Table 6. Distribution of positive samples for foodborne pathogens in manure/compost samples by type, age/production style.
Prevalence of pathogens in soil samples
There was a total of 254 soil samples collected and 29.1% (74/254) of them were positive for at least one of the tested pathogens in this study. L. monocytogenes, Salmonella, and STEC/VF genes were detected in 4.3% (11/254), 0.8% (2/254), and 24.0% (61/254) of soil samples, respectively (Table 4). The most prevalent serovars detected were O103, O157, and O145/O45, while the most prevalent VF-gene detections were stx2, eae/ehxA, and stx1 (Table 5). Detection of O103 was significantly greater than O121, O111, O145, O26, and O45, and O157 was significantly greater than O111 and O26. There was a total of four (1.6%) samples from which multiple pathogens were isolated. Three were positive for L. monocytogenes and STEC/VF-genes, and the remaining was positive for Salmonella and STEC/VF-genes. Generic E. coli was isolated from 21.7% (55/254) of collected soils (Table 4). Of these, 38.2% (21/55) were positive for gEC and a tested pathogen. Two samples were positive for gEC, L. monocytogenes, and STEC/VF-genes, while the remaining 19 were positive for gEC and STEC/VF-genes.
Sufficient data was available for gEC to determine seasonal prevalence adjusted for clustering within farms with results displayed in Table 7. There was insufficient data available to estimate adjusted prevalence for the three pathogenic organisms. In the case of gEC, the adjusted prevalence values were consistently lower than the unadjusted prevalence values, which indicated some spatial clustering over farms. The total gEC adjusted prevalence decreased by 7.64% overall and by 7.1, 1.75, 10.02, and 8.69% in the fall, spring, summer, and winter, respectively. Due to adjustments to the original sampling plan, data was unevenly distributed between farms and seasons of this study, which may affect adjustments for spatial clustering within seasons. For L. monocytogenes, 81.2% (9/11) of detections occurred in the winter and spring months of sampling. Notably, 36.3% (4/11) of these L. monocytogenes positive soil samples occurred on ICLF-B, a farm that had 20% (1/5) manure samples positive for the same pathogen. For Salmonella, the two detections were split between the spring and summer months, one of each occurring on ICLF-A and ICLF-B. For STEC/VF-genes, 63.9% (39/61) of the detections occurred in the summer months, with 49.2% (30/61), 24.6% (15/61), and 26.2% (16/61) of positive samples from ICLF-A, ICLF-B, and ICLF-C, respectively. These three farms had at least one manure/compost sample positive for STEC/VF-genes. Similar to STEC/VF-gene positives, most of the gEC isolations occurred in the summer months at a detection rate of 76.4% (42/55). Surprisingly, the fewest detections occurred on ICLF-B, even though this farm had the only manure/compost samples positive for fecal contamination.
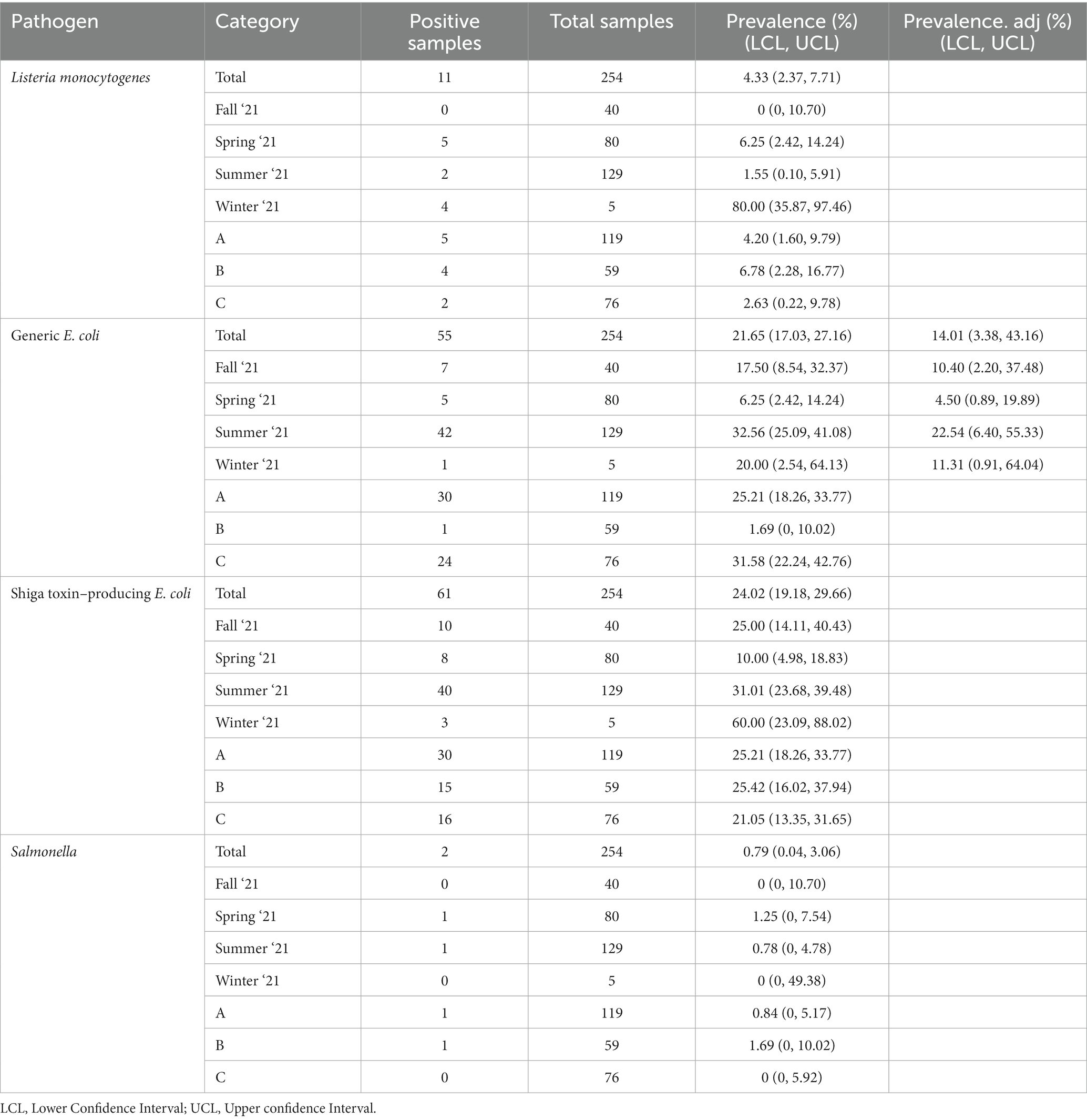
Table 7. Prevalence of foodborne pathogens and indicator organisms in soils based on observed values, adjusted for spatial clustering within farm overall, by season, and location.
The relationship between amendment type/production style and pathogen detections in soil were examined, along with the association of gEC and coliforms with pathogen prevalence as well. Compared to the 45.4% (5/11) of soils amended with mushroom compost, 54.5% (6/11) of soils amended with farm produced manures were positive for L. monocytogenes. The two Salmonella positive soil samples were each amended with a commercially produced mushroom compost and a farm produced cow/pig mixed manure. Soil samples amended with mushroom compost had the highest percentage of STEC/VF-gene positives at 49.2% (30/61), while the remaining 50.8% (31/61) of soil samples received amendments containing animal manure. Of these, 48.4% (15/31) and 51.6% (16/31) of soils received biological soil amendments (BSAs) containing cow/pig manure and chicken manure (mixed with vegetable material), respectively. The percentages of soils amended with commercially and farm produced manures/composts were nearly equal at 49.2% (30/61) and 50.8% (31/61), respectively. Fecal contamination was highest in soils amended with mushroom compost and commercially produced amendments, each with a prevalence of 54.5% (30/55). The unadjusted gEC prevalence of commercially-amended soils was 25.21% compared to the 18.52% of farm-amended soils, which suggests a stronger relationship between fecal contamination and soils amended with commercial biological soil amendments (Table 8).
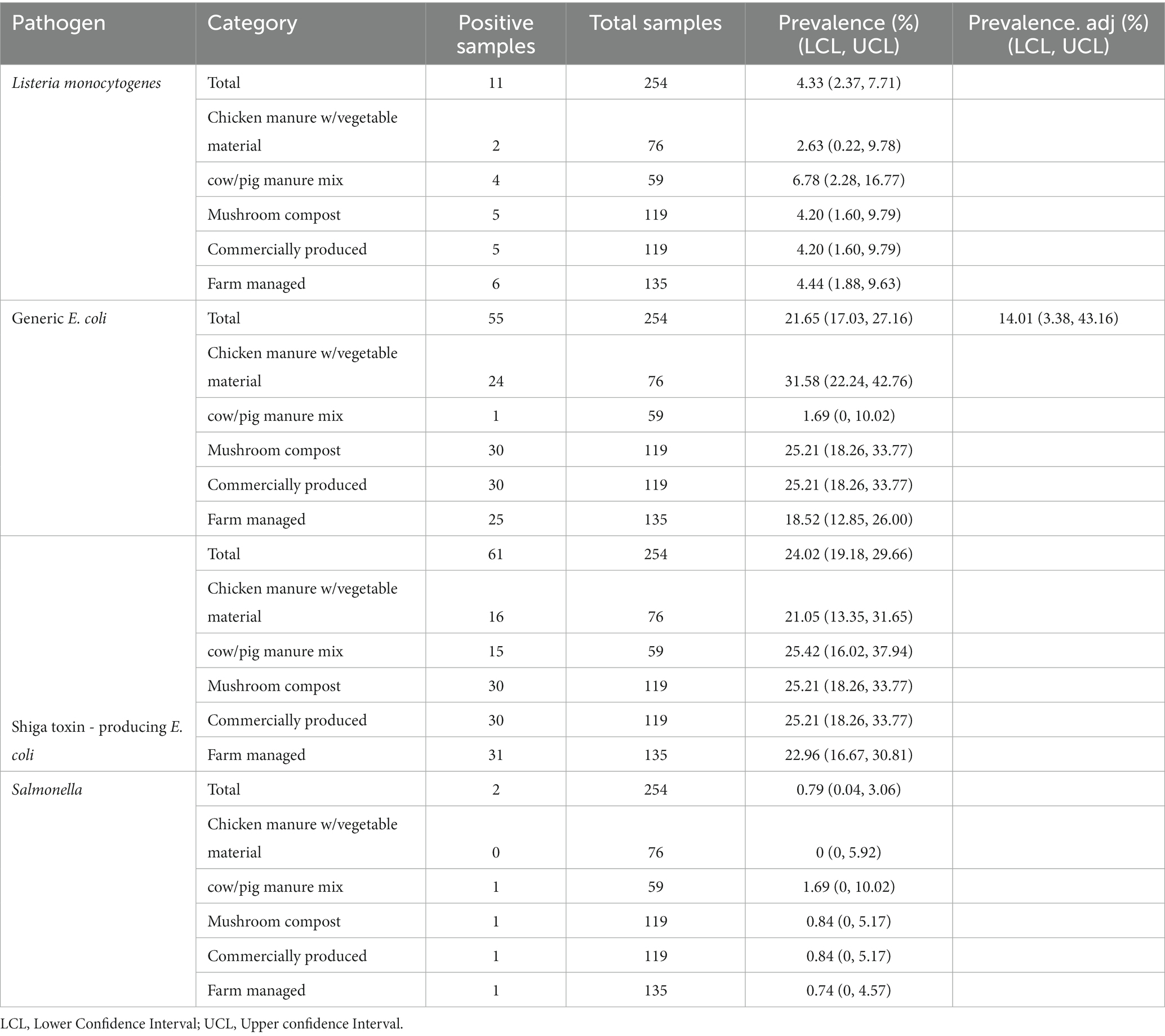
Table 8. Prevalence of foodborne pathogens and indicator organisms in soils based on observed values, adjusted for spatial clustering within overall samples, by manure source and production method.
In this study, the prevalence of the three pathogens increased as coliform concentrations in soil samples increased (Figure 2, Supplementary Tables 1–3). As coliform log count increased ≥3 log CFU/g, L. monocytogenes counts increased. As coliforms increased >4 log CFU/g, Salmonella counts increased, and as coliform counts increased to 4–5 log CFU/g, the prevalence of STEC/VF genes increased.
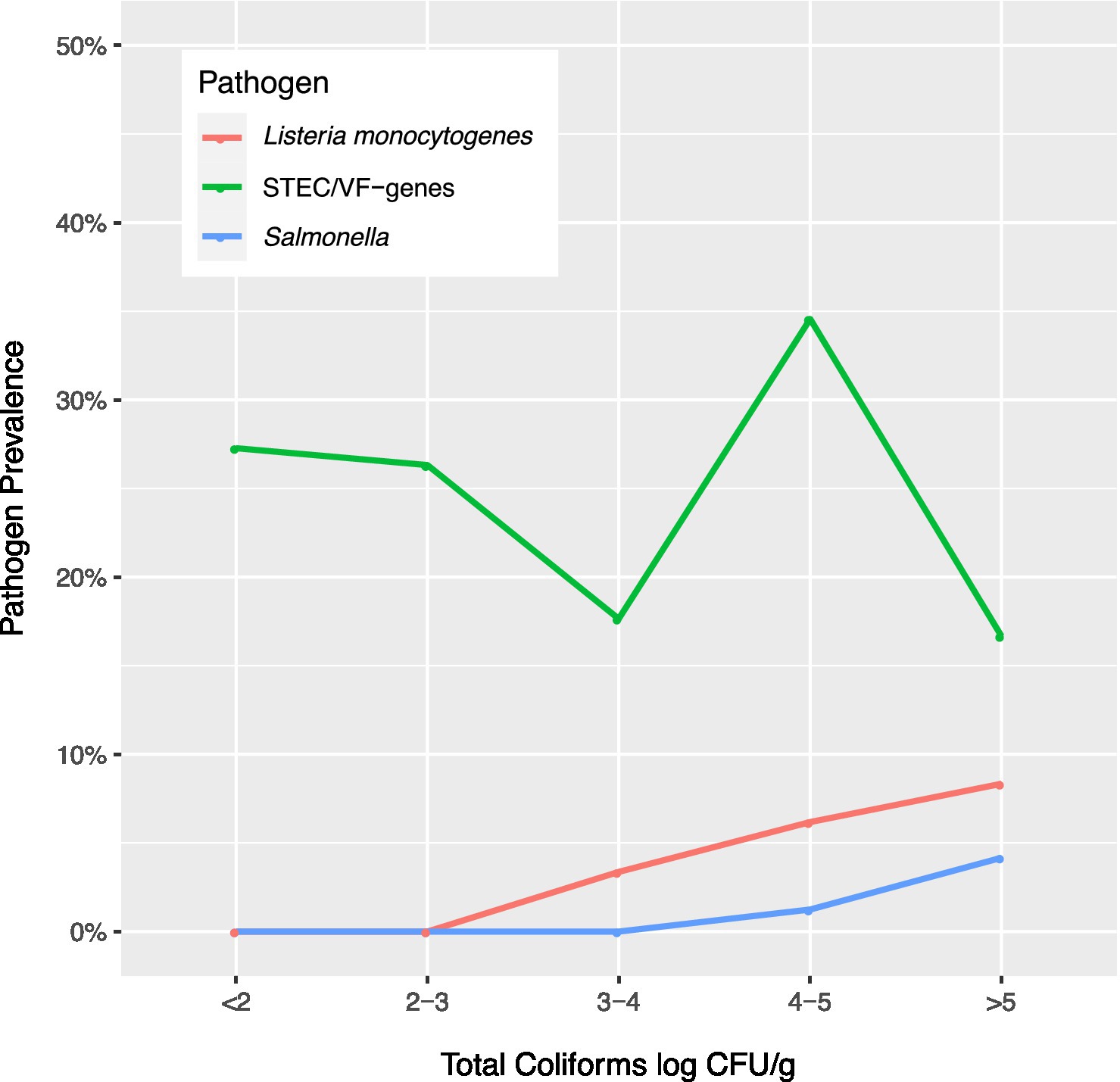
Figure 2. Prevalence of Listeria monocytogenes, Salmonella, and Shiga toxin-producing E. coli and virulence factor genes (STEC/VF-genes) relative to total coliform concentrations in soils.
Across all farms, for L. monocytogenes (Figure 3, Supplementary Table 4), biological soil amendments increased initial L. monocytogenes prevalence from 10 to 12.5%, but no additional detections were recovered until D75. Four of the five L. monocytogenes detections on D0A occurred in soils that were amended with manures/composts that had a L. monocytogenes positive baseline sample. On D75 and D90, 6.7 and 3.6% of samples were positive for L. monocytogenes; however, the pathogen was not isolated from D120 onwards. There were two Salmonella (Figure 4, Supplementary Table 5), detections in soil samples. One occurred in soil post-manure/compost application with pathogen levels going undetected until a spike on D120. This was followed by the pathogen going undetected for the remainder of the experiment. On D0A and D120, Salmonella was present in 2.5% of samples on both collection days. STEC/VF-genes (Figure 5, Supplementary Table 6) were present in soil samples on all days except for D45 and D60. The consistent persistence of the STEC/VF-genes may be due to all three farms having at least one soil and one manure/compost sample positive for STEC/VF-genes on D0B. Initially, the pathogen was detected in 17.5% of soil samples on D0B, and levels were reduced to 5.0% on D0A. The pathogen had a prevalence in 28.6% of samples on D90, and even spiked on D120 at 60.0%. Pathogen prevalence declined to 20.0% by D180, which was near the initial detection rate on D0B soil samples. For soils, mean APC log CFU/g levels increased after biological soil amendment incorporation and did not return to pre-application levels (Figure 1). Mean APC Log CFU/g concentrations peaked on D150; however, concentrations decreased 1 log CFU/g by D180. For gEC (Figure 6, Supplementary Table 7), the prevalence increased after amendment and peaked at D75. Fecal contamination was present every sampling day except for D30.
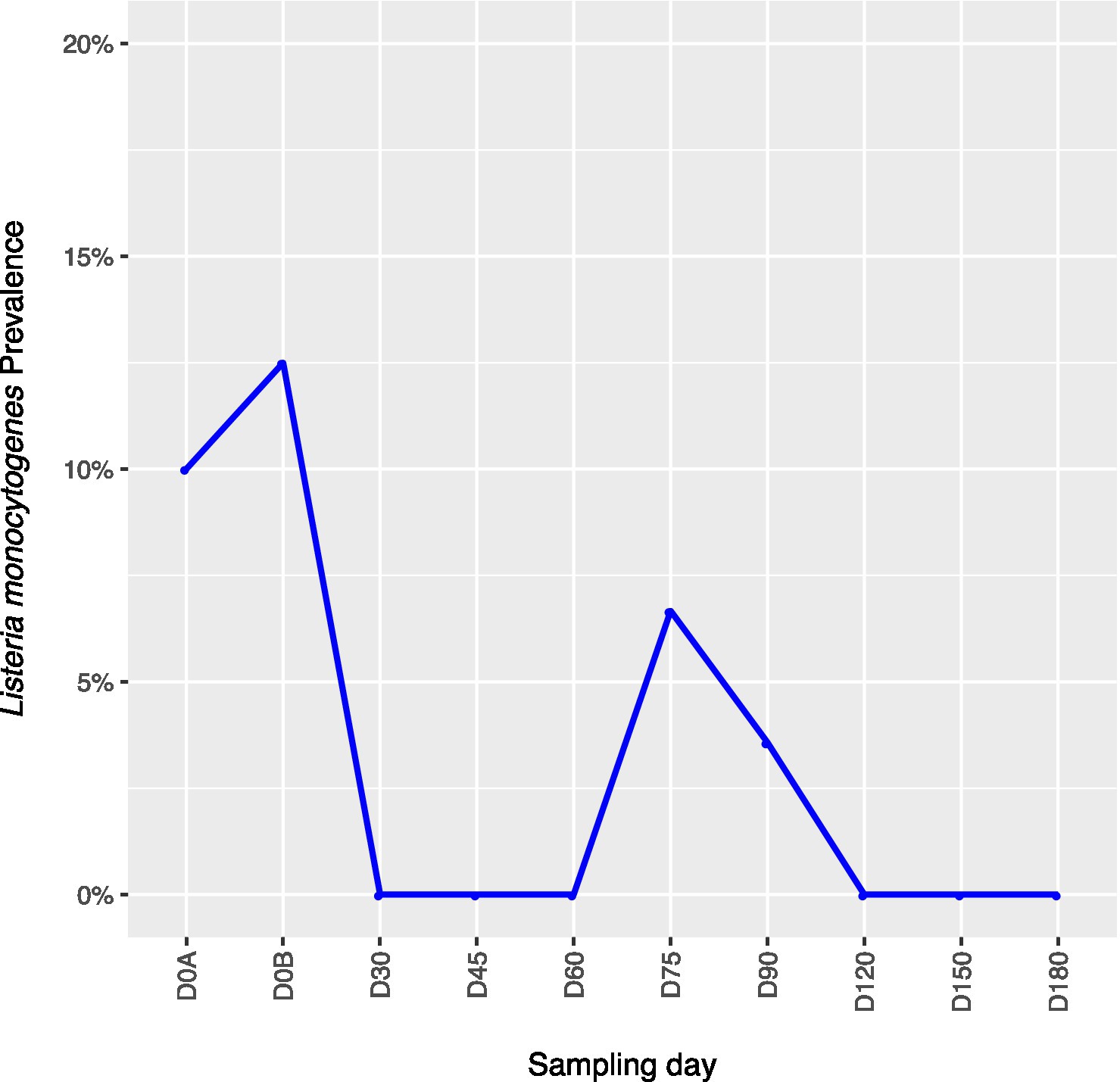
Figure 3. Listeria monocytogenes prevalence in soil samples relative to sampling day. D0A – before biological soil amendment of animal origin. D0B – after biological soil amendment of animal origin.
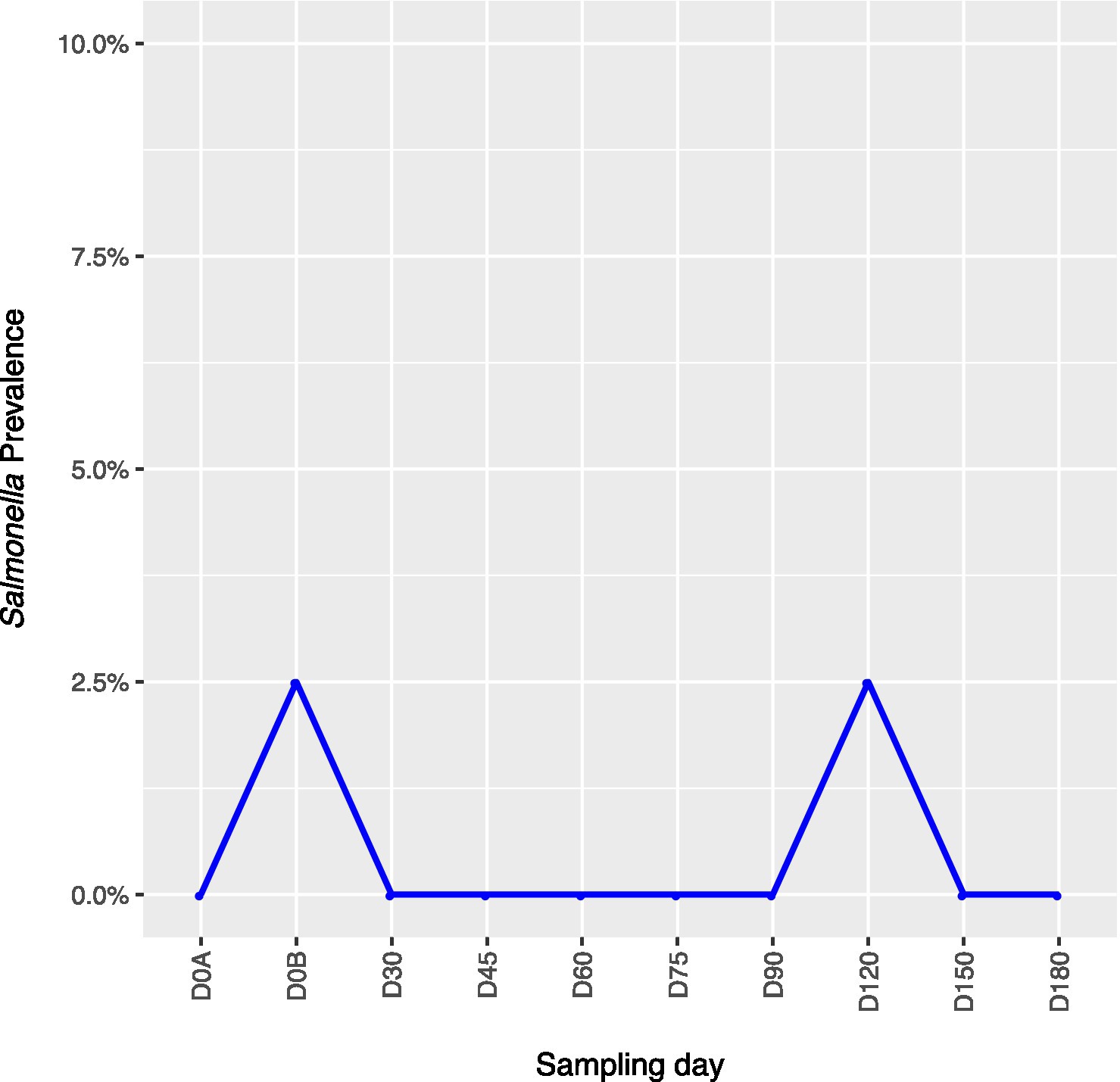
Figure 4. Salmonella prevalence in soil samples relative to sampling day. D0A – before biological soil amendment of animal origin. D0B – after biological soil amendment of animal origin.
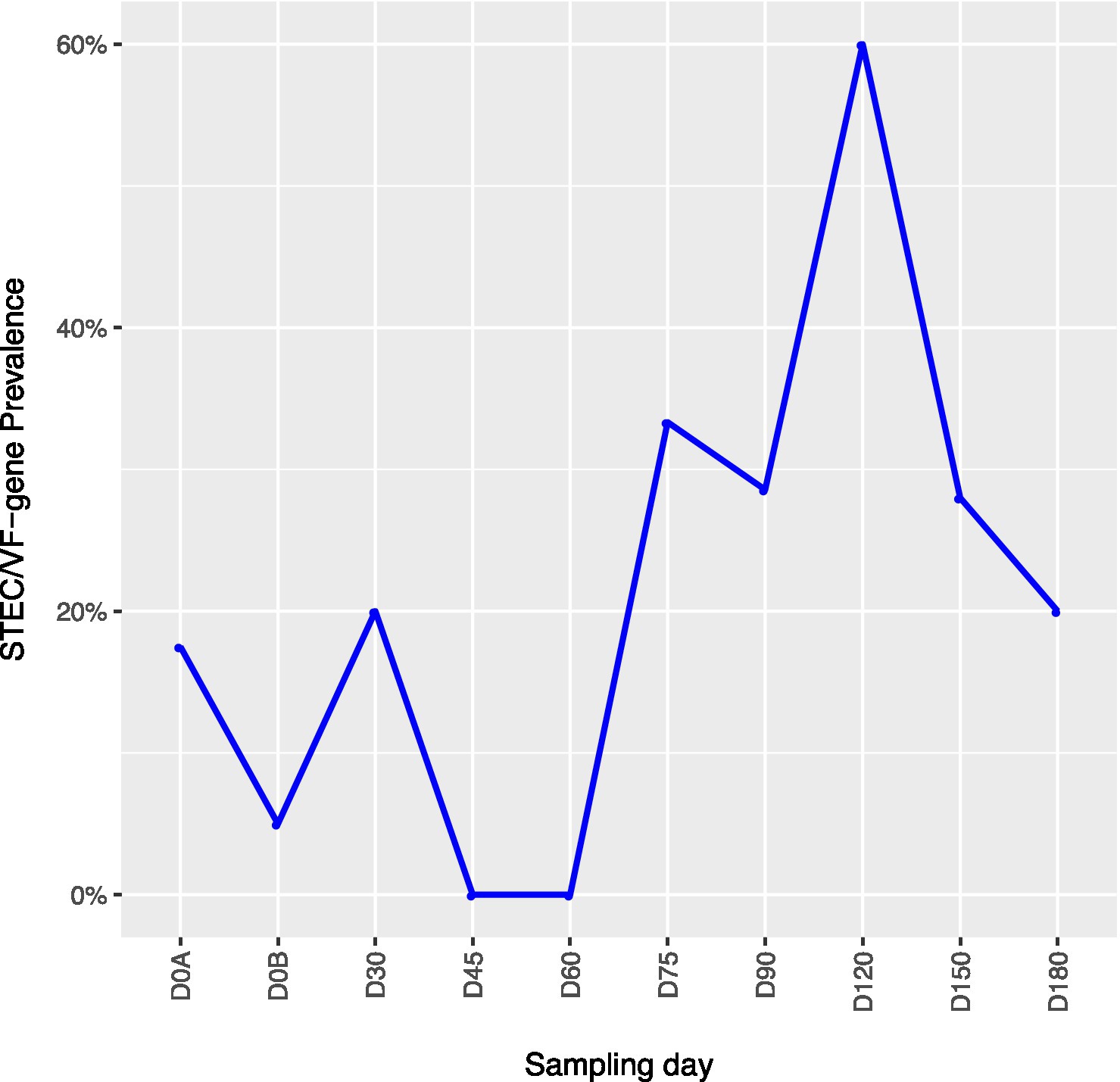
Figure 5. Shiga toxin-producing E. coli and virulence factor genes (STEC/VF-genes) prevalence in soil samples relative to sampling day. D0A – before biological soil amendment of animal origin. D0B – after biological soil amendment of animal origin.
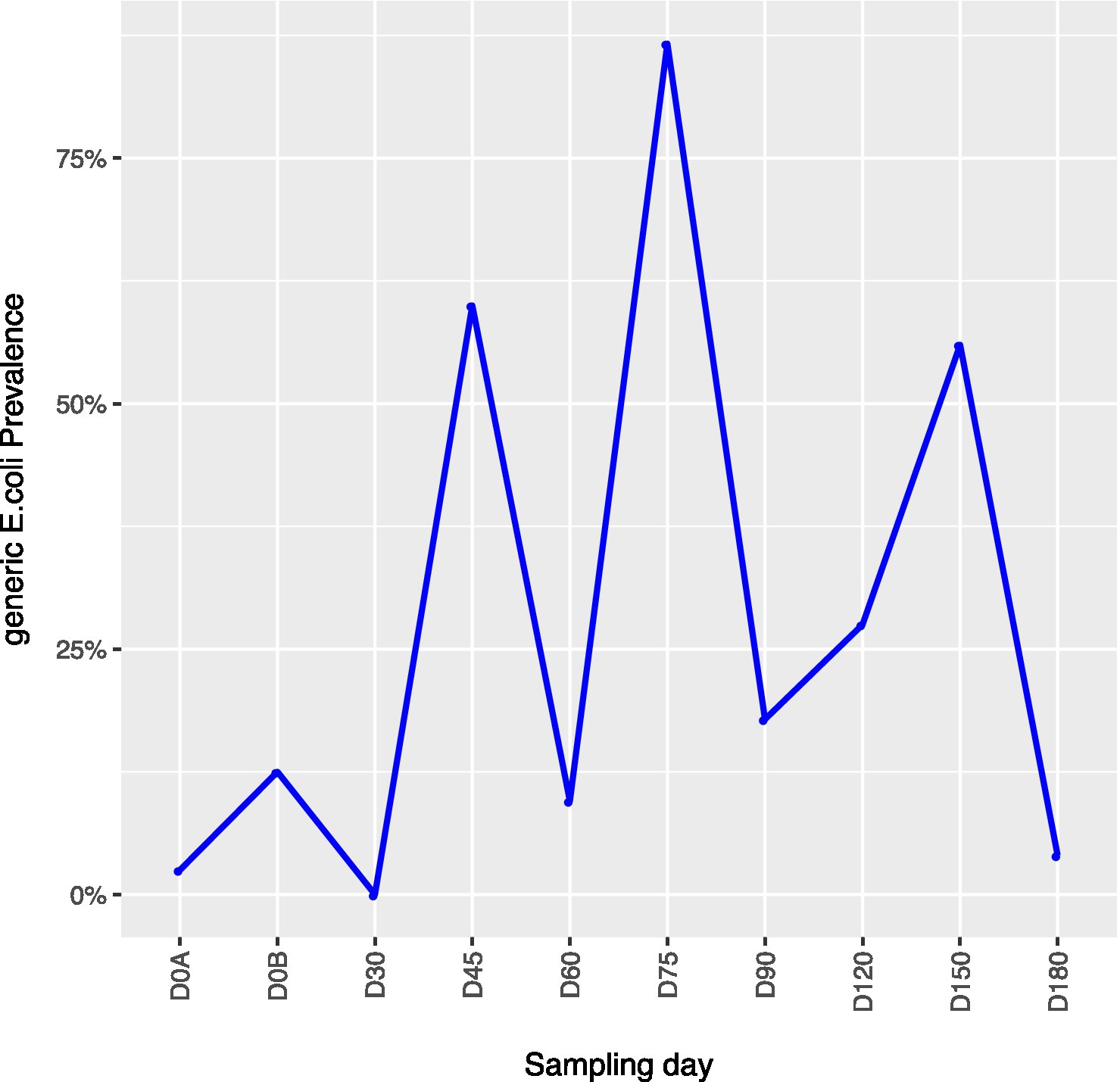
Figure 6. Generic E. coli concentrations in soil samples relative to sampling day. D0A – before biological soil amendment of animal origin. D0B – after biological soil amendment of animal origin.
Water samples
A total of 18 agricultural water samples were provided by two of the three farm locations in the summer and fall months of this study. ICLF-B did not irrigate their crops, so water samples were not collected from this farm. Well water was collected from ICLF-A and ICLF-B. All samples were negative for gEC and coliforms and therefore, there was no further analysis conducted on water samples.
Discussion
This study was unique in that it analyzed the presence of multiple pathogens and indicator organisms within working organic, small-scale ICLF (not NOP certified) environments. Products from these locations are typically sold at increasingly popular farmer’s markets, CSAs, and backyard stands. Therefore, the pathogen prevalence described in this study reflects natural contamination and risks associated with farming practices and the resulting produce cultivated in local ICLF environments on the Eastern Shore of Maryland. In other published studies, the experimental setups were limited for reasons which include: (1) Conducted in model (experimentally setup) systems, (2) Focused on fewer pathogens, one produce type, one BSAAO amendment type, and (3) Lacked an extended follow-up period of naturally contaminated, BSAAO-amended soils (Franz et al., 2011; Peng et al., 2016; Salaheen et al., 2016; Gu et al., 2018; Patterson et al., 2018; Pires et al., 2019; Sheng et al., 2019; Ekman et al., 2021). In our study, a variety of fresh produce commodities (leafy greens, peppers, tomatoes, and squash) were analyzed. Evaluated manures from multiple species including poultry, cattle, and pig, and mushroom compost were examined as well. Furthermore, manure/compost samples in this study were produced on-farm and commercially, while also representing different ages within participating ICLFs. Soils were amended with manures/composts from various sources and were evaluated up to 180 days post incorporation.
Contamination of produce
Overall, 18.4% of produce samples were positive for one of the tested bacterial pathogens in this experiment with at least one detection of each. Contamination was also found in produce from each of the three ICLF locations representing varying soil amendments, farm animals, and farm management practices. This percentage is notably higher compared to the 0.4% of pathogens detected in a longitudinal study conducted on USDA-NOP certified farms (Ramos et al., 2021). The increased pathogen prevalence in our study may be a combined result of sampling produce from uncertified farms and before the withholding period. Therefore, this notable discrepancy in pathogen detection indicates the importance of following USDA-NOP guidelines and the mandated withholding period between BSAAO application and produce harvest date. Specifically, our study found L. monocytogenes, Salmonella, STEC/VF-genes, and gEC in 3.5, 0.7, 14.2, and 3.5% of produce samples, respectively. The prevalence of these four microorganism categories on fresh produce have varied in multiple survey and longitudinal studies conducted in model and working farm studies replicating ICLF practices (Peng et al., 2016; Glaize et al., 2020; Ekman et al., 2021; Ramos et al., 2021).
In terms of the withholding period, multiple pathogens were isolated from fresh produce from D90 and D120 onward. L. monocytogenes was the second-highest detected pathogen among produce with isolations on D90 from one squash (ICLF-B) and D180 (ICLF-C) from four mustard greens samples. Ramos et al. (2021) found two leafy greens samples positive for L. monocytogenes on D90 and D120. Squash and leafy greens are both considered high-risk produce and under NOP regulations and should have been harvested after 120 days. Pathogen prevalence on D90 indicates the importance of waiting the extra time to allow for the further reduction of pathogens. Another study did not detect L. monocytogenes on mature lettuce plants harvested from amended-soils after the withholding period, but there were detections on immature lettuces harvested before, emphasizing the importance of the withholding period (Ekman et al., 2021). In New York, Weller et al. (2015b) also found low prevalence of L. monocytogenes on produce, where the pathogen was detected in 0.6% of spinach leaves. Soils, water, and other environmental reservoirs naturally harbor L. monocytogenes, so the presence of this pathogen at D90, D120, and D180 on produces may not be due to BSAs (Weller et al., 2015b; Ramos et al., 2021). Salmonella was the least detected pathogen and was isolated once on D180 from a mustard greens sample grown in soils amended with an aged chicken manure/vegetable material mix. This low detection rate was similar to other longitudinal studies where the pathogen was not detected at all after following the appropriate withholding period (Ekman et al., 2021; Ramos et al., 2021). Pagadala et al. (2015) did not isolate the pathogen from tomatoes either on small- to medium-sized conventional and organic farms. However, Salmonella had a higher number of detections in leafy greens, tomatoes, and peppers, from similar environments where soils received BSAs (Peng et al., 2016; Shah et al., 2019). Interestingly, Gu et al. (2018) found tomato leaves harvested from poultry litter-amended soils to be positive for Salmonella, while the tomato fruit itself was not. The contamination route was unclear but emphasizes the necessary precautions that should be taken when planting and harvesting crops in manure-amended soils. Factors like extreme weather events should be taken into consideration before comparing results from this study to others. On ICLF-C the D180 sampling took place after a flooding event occurred in the agricultural soils on the farm. The farmers knew it was protocol to not use or sell these products; however, we still sampled. The leafy greens positive for Listeria monocytogenes and Salmonella discussed earlier in this paragraph were taken after this flood, and one of these samples was even positive for both pathogens. Furthermore, agricultural soils on ICLF-C were less than 30 feet away from the on-farm chicken coop. These results indicate that weather events and soil proximity to animal-rearing facilities are important factors to be considered in agricultural production because they can lead to increased pathogen prevalence (Strawn et al., 2013a; Pagadala et al., 2015; Gu et al., 2018; Alegbeleye and Sant’Ana, 2020). Continued research and dissemination of findings in these areas are necessary to reduce contamination levels in agricultural environments.
The pathogen with the highest detection rate in produce was STEC/VF-genes, which were isolated from different produce samples multiple times on D90, D120, and D180. Other studies found that the percent prevalence of the pathogen remained notably lower or went undetected altogether, compared to the 14.2% of STEC/VF-gene positive produces in our study. Glaize et al. (2020) found 2.68% of produce samples positive for STEC, while Ramos et al. (2021) did not detect STEC on produce. Pagadala et al. (2015) did isolate VF-genes from tomatoes, but STEC could not be identified on any samples from small- and medium-sized farms. From the literature search, this appears to be the first study to analyze produce, soils, and manure/compost for the prevalence of STEC/VF-genes through an 11-plex PCR testing for genes unique to seven different serotypes and four different VF-genes. The increased range of STEC serovars/VF-genes tested for in our experiment may be the reason for this notably higher prevalence of STEC/VF-genes. This study found O103, O157 to be the top two serotypes, while O157 was not detected at all in other studies (Glaize et al., 2020; Ramos et al., 2021). This higher prevalence of O157 is of particular concern because this serovar has historically been a major health concern in the U.S. and is often associated with severe cases involving bloody diarrhea and HUS (Brooks et al., 2005; Conrad et al., 2014; Luna-Gierke et al., 2014). Although nonpathogenic, this study tested for gEC because it acts as an indicator organism for fecal contamination. Instead of testing for pathogenic E. coli, other studies tested for gEC as the organism mimics the transfer and survival of pathogenic E. coli and the detection of gEC is rapid and inexpensive (Berry et al., 2013; Hruby et al., 2018; Patterson et al., 2018; Ekman et al., 2021; Ramos et al., 2021). Generic E. coli had a lower percent prevalence on produce samples in our experiment compared to studies conducted in ICLF, model, organic, and conventional systems (Pagadala et al., 2015; Ekman et al., 2021; Ramos et al., 2021). In the present study, samples were not enriched before gEC analysis, whereas two of the previously cited sources did, which increases the detection probability of the organism through enrichment. This difference in methodology could explain the variances in gEC levels. Pagadala et al. (2015) also found gEC on 1.6% of organic tomatoes compared to the 6.9% of conventional tomatoes, indicating the risk of fecal contamination is not limited to organic or ICLF farming systems. On average, D90, D120, and D180 had the highest mean APC log CFU/g values for produce samples. Higher APC levels associated with these days are not surprising as fields most likely experience higher activity during these time periods. Since produce are harvestable there’s likely a higher influx/efflux of activity from farmers and wildlife (Weller et al., 2015a,b).
Pathogens in biological soil amendments
In this study, 73.3% of BSA samples were positive for at least one pathogen with an isolation occurring on each farm. This means that pathogens were detected in manures/composts from different species, production styles, and ages in small-scale, organic (not NOP certified) ICLFs. In our study, L. monocytogenes was detected in 6.7% of BSA samples compared to the 3.9% found in Ramos et al. (2021). The former isolated L. monocytogenes from a cow/pig manure mix, while the latter isolated the pathogen from ruminant and horse manures stored on ICLFs. In a model study, L. monocytogenes was also detected in bovine manures mimicking minimally managed manure piles typically found on small- to medium-scale organic ICLFs (Berry et al., 2013). The presence of pathogens in these minimally managed manure piles indicate the importance of properly storing and handling on-farm manure/composts. L. monocytogenes is often associated with cooler temperatures and this sole detection in our study took place a few weeks into spring. The ICLFs in this study applied BSA’s in March and April when mild temperatures may create a favorable environment for L. monocytogenes to survive or regrow in on-farm BSA stockpiles or agricultural soils (Berry et al., 2013; Strawn et al., 2013a; Ekman et al., 2021). Salmonella was not detected in manure/compost samples in this experiment, but it was detected in other studies across multiple species. In a multistate study, Salmonella was detected in 7.3% of cattle, poultry, and mixed manure samples (Ramos et al., 2021). Multiple studies in different regions detected the pathogen in poultry litter (dry, wet, stored), fecal and compost samples in percentages up to 26.6%. Sources of these sample were from poultry, cattle, sheep, and swine (Berry et al., 2013; Peng et al., 2016; Gu et al., 2018, 2019; Chen et al., 2019; Pires et al., 2019; Nazareth et al., 2021). Salmonella is often associated with cattle, and only one farm stored and applied cattle manure in this experiment, which might be the reason for its lack of detection (Nazareth et al., 2021; Ramos et al., 2021). Poultry manure is a popular biological soil amendment choice for cropland that has been identified as a common reservoir for Salmonella as well (Gu et al., 2018, 2019; Hruby et al., 2018; Phan-Thien et al., 2020). Poultry manure amendments were only tested from one farm location in this study as well. The lack of Salmonella detections in this study may reflect proper storage conditions on farms or factors unique to the region that successfully reduce the prevalence of this pathogen.
STEC/VF-genes were detected in 66.7% of manure/compost samples in this experiment with all biological soil amendment sample types (animal source and production style) being positive for Non-O157 STEC, while only commercially produced mushroom compost was positive for STEC O157. Non-O157 STEC were the serovars with the highest number of detections compared to STEC O157. This finding was similar to Ramos et al. (2021), which found 9.0 and 1.3% of manure and compost samples positive for non-O157 STEC and STEC O157, respectively. Non-O157 STEC was primarily found in ruminant manure and STEC O157 was found in poultry and mixed manure samples in the previously mentioned study. Although at a lower prevalence than our study, non-O157 STEC/VF-genes were also associated with litter, raw cattle/chicken manure, composted manures, and sheep fecal samples in comparable studies on diversified farms (Patterson et al., 2018; Glaize et al., 2020). STEC O157 was also detected in 15.4% of cattle manure samples on California Dairy farms by Chen et al. (2019). Nazareth et al. (2021) recovered STEC O157 from cattle fecal samples and minimally managed bovine manure piles. Although the pathogen was eventually reduced to undetectable levels in the piles, the presence of pathogens emphasizes the importance of proper manure/compost storage on farms. Generic E. coli was detected in 20% of samples, a percentage that was considerably lower than the STEC/VF-gene prevalence. In the literature search there did not appear to be studies that tested manure samples for gEC. Similar to these previous studies, pathogen prevalence in manure/compost in our study varied based on biological soil amendment source, type, and location. Our study had a notably higher percentage of samples positive for STEC/VF-genes, but our manure/compost sample collection was not as extensive occurring only once throughout this study. Samples were also collected in the spring, a time where cooler temperatures may not allow for the proper reduction of pathogens and create optimal conditions for the regrowth/proliferation of pathogens (Berry et al., 2013; Sharma and Reynnells, 2016; Alegbeleye and Sant’Ana, 2020).
Pathogen detections in soils
In this study, 29.1% of soil samples were positive for at least one of the tested pathogens, and STEC/VF-genes were detected at a considerably higher rate than Salmonella and L. monocytogenes. This overall pathogen prevalence was two times higher than the 12.9% in a similar study conducted by Ramos et al. (2021), which also detected STEC (non-O157 and O157 combined) at a higher rate than the other two pathogens. L. monocytogenes and Salmonella were detected in 5.0 and 0.8% of soil samples, respectively. The low rate of Salmonella detection in soil may be a result of the pathogen not being detected in biological soil amendments nor in D0A soil samples before amendment, whereas L. monocytogenes and STEC/VF-genes were detected in manure/compost and D0A soil samples. These pathogens were also detected at varying rates in other studies. In manure-amended soils, Ramos et al. (2021) found L. monocytogenes and Salmonella in similar percentages of 5.0 and 1.1% of soils across multiple regions, respectively. Multiple studies on produce farms in New York found soil samples to be positive for L. monocytogenes at higher percentages between 8 and 13%, while Salmonella was found at percentages between 0 and 5% (Strawn et al., 2013a,b; Weller et al., 2015a,b). Salmonella was also undetected on organic farms, which tend to rely on organic biological soil amendments, and conventional farms in the Mid-Atlantic region (Pagadala et al., 2015). The previously described New York experiments were conducted in experimental, conventional, or organic fields where practices such as manure-amendments, irrigation, and animal presence differed and were unique to each farm. Analysis of these experiments indicated that practices associated with farm soils that utilized manure-amendments or integrated crops and livestock increased prevalence of pathogens. Furthermore, Peng et al. (2016) described Salmonella to be found more frequently in mixed crop-livestock (MCL) farm environmental samples than conventional farm samples. In experimental plots in Virginia, naturally occurring Salmonella was also detected in rhizosphere soil samples amended with poultry litter, exemplifying the risk of pathogen contamination associated with the edible portions of produce in close contact with the soil (Gu et al., 2018).
The higher prevalence of STEC/VF-genes suggests a possible increased contamination risks associated with ICLF soils and the pathogen. However, this risk may be limited to the analyzed regions, farms, and farming practices because other studies that tested for the presence of multiple pathogens in soils within varying farm environments did not find STEC to have the highest prevalence (Strawn et al., 2013a; Pagadala et al., 2015; Weller et al., 2015a). The results from the present study are concerning as STEC/VF-genes were detected more frequently in 24.0% of amended soils with O103, O157 and stx2, eae/ehxA being two of the more frequently identified serovars and VF-genes, respectively. As mentioned earlier, STEC O157 is often associated with more severe cases of STEC infection, and the VF-genes are responsible for its pathogenicity (Brooks et al., 2005; Koitabashi et al., 2008; Conrad et al., 2014; Luna-Gierke et al., 2014). In New York produce farms, Weller et al. (2015a) and Strawn et al. (2013a) found STEC at a lower rate in soils under various management practices in 5 and 2% of samples, respectively. Similarly, Glaize et al. (2020) found 5.8% of soils in small- and large-scale North Carolina and Tennessee sustainable farming environments to be positive for STEC. In a multi-regional study, STEC O157 and STEC non-O157 were detected in 0.04 and 7.7% of manure-amended soils, respectively, on USDA-NOP certified farms (Ramos et al., 2021). In contrast to our results, STEC was not recovered in another study from soil samples in organic/conventional, small- to medium-sized farms in the mid-Atlantic regions of the United States (Pagadala et al., 2015). STEC/VF-genes were prevalent in manure/composts and soil samples on D0A, which might explain the increased and persistent detection of this pathogen throughout the experiment.
For L. monocytogenes and Salmonella, the pathogen prevalence increased after the incorporation of manures/composts into soils, which was similar to patterns observed in manure-amended soils within the same and other regions (Peng et al., 2021; Ramos et al., 2021). Despite these pathogens going undetected in the next sampling, L. monocytogenes and Salmonella were detected on one soil sample each on D90 and D120, respectively. Few studies appear to have addressed the persistence of L. monocytogenes in agricultural soils. One field study found L. monocytogenes in manure-amended soils to stabilize and persist in samples from D90 through D180 (Ramos et al., 2021). An experimental study using Listeria spp. to inoculate manures found the indicator organism to persist in manure amended soils for 50 days (Ekman et al., 2021). It should be noted that this study was conducted in Australia and was analyzing their withholding period of 45 and 90 days for their low- and high-risk products, respectively. As mentioned in the produce discussion, Listeria spp. are soilborne organisms, so the detections of the species and its pathogenic organisms are not unexpected as they naturally reside in soils. In field studies, Salmonella persistence in manure-amended soils varied with the pathogen surviving from 30 days to 7 months (Gu et al., 2019; Ramos et al., 2021). Using naturally contaminated poultry manure, Hruby et al. (2018) found Salmonella to persist 158 days after soil amendment in experimental plots. Furthermore, this study found the pathogen in 2011 and 2012 spring soil samples after manure-amendment, which emphasizes the survival ability of Salmonella in these soils. However, it should be noted that pathogen duration has been shown to be greater in experimental fields (Tran et al., 2020). Another experimental study found Salmonella to fall close to the limit of enumeration within the beginning weeks of the study; yet the pathogen could be detected in the later stages through enrichment where only a single bacterium is necessary (Ekman et al., 2021). The use of enrichment might be the reason for detections of L. monocytogenes and Salmonella on D90 and D120 in this current study after the post-amendment levels declined quickly. Two studies in experimental settings demonstrated that soils amended with poultry manure or litter improve the survival ability of Salmonella, which is another possible explanation for the detection of pathogens at these time intervals post amendment (Shah et al., 2019; Phan-Thien et al., 2020).
In contrast to the other pathogens in this study, STEC/VF-genes prevalence decreased after the incorporation of BSAs into soils. This reduction of pathogen load after manure incorporation has been previously observed in poultry litter amended soils with gEC and Salmonella, suggesting that certain amendments may have possible antimicrobial effects (Ekman et al., 2021). Following incorporation, the presence of STEC/VF-genes after BSA incorporation were detected more frequently than L. monocytogenes and Salmonella with multiple detections occurring from D90 to D180. In fact, the percentage peaked at D120 with half of the isolates being positive for STEC O157. Ramos et al. (2021) detected STEC O157 and Non-STEC O157 on D90 and D90 through 180, respectively, in amended-soils. Also, in experimental studies involving inoculations, STEC O157 survived up to 266 days in manure-amended soils (Franz et al., 2011). On the other hand, STEC was not detected at all in soils after sheep grazed produce fields before planting in a field study conducted on an organic ICLF in California (Patterson et al., 2018). In our study, gEC was detected in 38.2% of soils with detections occurring from D90 through D180. Ramos et al. (2021) and Patterson et al. (2018) found fecal contamination in 60.1 and 79.7% of samples, respectively, while Pagadala et al. (2015) found it in 8.9% of soils. Furthermore, the latter study found gEC counts to be higher in conventional than organic farms, which mainly rely on biological soil amendments. In terms of survival, Hruby et al. (2018) found gEC to survive up to 158 days after manure applications. Neher et al. (2019) found gEC to persist longer in poultry compost amended soils. In contrast to the previous discussed studies, the generalized gEC population model in Ekman et al. (2021) predicted that fecal contamination would be near the detection limit after 50 days post-manure incorporation. This quick decline may be due to the climate associated with Australia. Fecal contamination had consistent detections throughout our experiment; yet there were not enough samples with gEC in categories of higher levels. That is why the relationship between pathogen prevalence and total coliforms was evaluated instead. Although the confidence intervals were wide, the results were qualitatively similar to those found in Ramos et al. (2021) with total coliforms substituted for gEC.
For soils, mean APC log CFU/g values increased after the biological soil amendment incorporation and did not return to pre-application levels. Mean APC log CFU/g levels peaked on D150. However, levels had a 1 log CFU/g reduction by D180. In our study, there were pathogen detections and APC increases associated with soil samples collected from D90 through D180. These days when activity is likely higher and cross contamination could occur through vectors like workers, animals, bugs, irrigation, etc. may explain these results (Weller et al., 2015a,b). Two of the STEC/VF-gene detections that occurred on ICLF-C on D180 were identified after the flooding event mentioned in the produce discussion.
In this longitudinal study, analyzed pathogens and indicator organisms were prevalent in produce, manures/composts, and soils at varying rates, indicating the risks associated with storing and applying biological soil amendments on ICLFs in the Mid-Atlantic Region. There were similarities and differences between the prevalence of pathogens and indicator organisms that can be seen between this and other comparable studies. Before drawing comparisons and conclusions, factors such as farm management, geographical, meteorological, and experimental methods in the respective studies should be considered as well. For produce, contamination can be dependent on type, proximity to soil, consumption (raw or cooked), and irrigation methods. Plants have been recognized to have varying physical, chemical, microbial, and bioactive compound factors that can act as defense mechanisms against foodborne pathogens. These factors may affect the ability of foodborne pathogens to establish themselves on plant surfaces or to colonize plant tissues. The varying surfaces, barriers, microbiomes, inhibitory compounds, and injuries of different plants on different farms analyzed in this study affect the observed variation in pathogen prevalence (George and Brandl, 2021). Manure/composts contamination rates may vary due to manure source, animal diet, and rearing facilities. All farms in the present study applied manures/composts to soils; however, timing, rate, method, and source can affect persistence and transmission as well. Pathogen regrowth should not be eliminated since moisture, produce, and temperatures may create better conditions for bacteria to persist throughout the growing season (Tran et al., 2020; Ramos et al., 2021). In addition, the prevalence of pathogens on produce in these studies could result from other events such as wildlife intrusion, run-off, agricultural water, pets, and farmers (Weller et al., 2015a,b; Gu et al., 2019; Ramos et al., 2021). For example, all pathogens that were isolated from produce were not isolated from manures/composts that were applied to soils in the corresponding plots. This indicates that BSA’s may not have been the sole vector for contamination of produce. However, the isolation of pathogens from produce from biologically amended fields before and after the withholding period do implicate biological soil amendments as a potential source. Unfortunately, the original contamination source could not be confirmed as source tracking analyzes were not available for this study. There were other limitations in this study as well. Pathogen reduction levels over time could not be examined since this study was conducted on working farms and inoculation techniques were unavailable. Also, this study was conducted in one region for one season with an unbalanced study design; this limits the findings of this study to the unique agricultural practices at these respective locations. Although similar to comparable literature, enrichment and confirmation methods described here were unique to this study. Differences in these methodologies influence the detection probability of pathogens, which are not equivalent across all studies.
Water samples
In our study, gEC and coliforms were not detected in the tested well water samples, so we did not test for the prevalence of pathogens. Previous studies have found pathogens such as L. monocytogenes, Salmonella, and STEC to be present in irrigation water samples from different sources (Strawn et al., 2013a,b; Weller et al., 2015a,b; Gu et al., 2018, 2019; Haymaker et al., 2019). On the other hand, Ramos et al. (2021) did not find any water samples to be positive for pathogens either. In our study, unsterilized water samples were only collected from wells on two farm locations, and sample numbers were not comparable to previously cited references, which might explain the lack of detections. In addition, there was no enrichment step for this protocol. It’s possible that gEC and coliforms were present, but at a level lower than the detection limit. Study limitations include sampling scheme, location, sources, seasons, and time-period, so conclusions should be drawn carefully.
Conclusion
Samples were collected from working farms, so these results reflect natural contaminations and potential risks associated with organic fresh produce production. Salmonella and L. monocytogenes detections on produce were infrequent (prevalence of 0.7 and 3.5%, respectively), whereas STEC/VF-genes were present in 14.2% of samples. Salmonella was not detected in manure/compost, whereas L. monocytogenes and STEC/VF-genes were prevalent in 6.7 and 66.7% of BSA samples, respectively. All three pathogens were detected in soils, but similar to produce and manure/compost samples, STEC/VF-genes were the most prevalent at 24.0%. Although followed by reductions in prevalence, the three pathogens were also detected in soils immediately after manure/compost application. With regard to the NOP withholding period, multiple pathogens were isolated from produce, as well as soils, 90 and 120 days after application of BSAAOs. The ability of pathogens to persist longer than the withholding periods has been described in previous studies (Franz et al., 2011; Hruby et al., 2018; Gu et al., 2019; Ramos et al., 2021). Continued research is necessary to gain further knowledge of the complex interactions of pathogens in farming environments.
In addition to the USDA-NOP withholding periods there are other intervention strategies that need to be researched and disseminated to farmers and other related parties. Proper composting is a complex process that involves time, money, and attention, and as a result farmers often resort to stockpiling manure (known as ‘aged’ manure) over time, without exposure to lethal time–temperature regimes, and ultimately ineffective at pathogen inactivation (Berry et al., 2013; Alegbeleye and Sant’Ana, 2020). In our study, the three participating ICLFs all stored and aged their manure/composts with varying methods. While the management of amendments on ICLFs and the reduction of pathogens in manures/composts were not uniform at these study sites included in this study, results show the need for using best practices for on-site, on-farm, very small-scale production and storage of manure composts. These would provide farmers and small-scale, community growers with essential, practical approaches to manage BSAAOs to reduce pathogen loads and transfer to produce. Storage and application procedures also should reflect practices that are common and achievable for small-scale ICLFs. Another factor to consider is the distance of crop fields from manure/compost and animal rearing facilities on ICLFs. The California Leafy Greens Marketing Agreement (LGMA) recommended that edge of crop fields be 122 m from composting operations that utilize manure or animal products. LGMA also suggested edge of crop fields be 366 m and 1,610 m from concentrated animal feeding operations (CAFOs) with >1,000 and > 80,000 heads, respectively (Glaize et al., 2020; LGMA, 2021). In the present study, the distances between crop fields and composting/animal facilities varied across farms. If small-scale ICLFs lack the land to accommodate these recommendations, other mitigation strategies such as topography (uphill/downhill), vegetative buffers, and barriers can be used as well (LGMA, 2021).
Results from this study show that L. monocytogenes and Salmonella contamination of produce were relatively low when compared to STEC. The consistency of STEC/VF-gene detection throughout this study could be due to farming practices or regional and geographical conditions that allow for enhanced survival of this pathogen. Results here are limited in that they are unique to the study design, the management practices, and geographical locations of these farms. Tracing analysis was not performed in this study, thus cross-contamination of pathogens due to wildlife intrusion, irrigation, weather events, and workers cannot be eliminated as possible vectors. The BSAAOs provide important fertilizer inputs for organic farming systems, and data from this study can provide insights for producers and regulatory agencies to better understand and mitigate contamination risks associated with open-field sustainable, organic farming practices. Further research and proper dissemination of best practices concerning on-farm manure management and distances between crops and pathogen reservoirs are vital to the future safety of sustainable farming systems.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
BG: Writing – original draft, Writing – review & editing, Investigation, Methodology, Data curation, Validation, Visualization. PM: Writing – review & editing, Conceptualization, Methodology, Funding acquisition, Validation, Validation. AP-D: Writing – review & editing, Investigation, Methodology. MS: Writing – review & editing, Investigation, Methodology, Resources, Supervision, Visualization. FH: Writing – review & editing, Conceptualization, Funding acquisition, Visualization. JB: Writing – original draft, Writing – review & editing, Data curation, Software, Formal Analysis. DB: Writing – review & editing, Conceptualization, Funding acquisition, Project administration. SP: Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was funded by USDA-NIFA, Organic Transitions Program grant no. 2018511062880, USDA NIFA CBG 5–208070, USDA Evans Allen, and USDA ARS.
Acknowledgments
First, we would like to thank the farmers for their cooperation and participation throughout this study. We acknowledge Joan Meredith, Barbara Osei TuTu, Joey Haymaker, Annette Kenney, Edwina Barnett, and Enrique Escobar for the help and support in field and experimental work. We would also like to extend gratitude to Mohammad Ali for facilitating the enrollment of farmers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1287371/full#supplementary-material
References
Aditya, A., Tabashsum, Z., Alvarado Martinez, Z., Wei Tung, C., Suh, G., Nguyen, P., et al. (2023). Diarrheagenic Escherichia coli and their antibiotic resistance patterns in dairy farms and their microbial ecosystems. J. Food Prot. 86:100051. doi: 10.1016/j.jfp.2023.100051
Alegbeleye, O. O., and Sant’Ana, A. S. (2020). Manure-borne pathogens as an important source of water contamination: an update on the dynamics of pathogen survival/transport as well as practical risk mitigation strategies. Int. J. Hyg. Environ. Health 227:113524. doi: 10.1016/j.ijheh.2020.113524
Aznar, R., and Alarcon, B. (2003). PCR detection of Listeria monocytogenes: a study of multiple factors affecting sensitivity. J. Appl. Microbiol. 95, 958–966. doi: 10.1046/j.1365-2672.2003.02066.x
Bai, J., Paddock, Z. D., Shi, X., Li, S., An, B., and Nagaraja, T. G. (2012). Applicability of a multiplex PCR to detect the seven major Shiga toxin-producing Escherichia coli based on genes that code for serogroup-specific O-antigens and major virulence factors in cattle feces. Foodborne Pathog. Dis. 9, 541–548. doi: 10.1089/fpd.2011.1082
Bellemare, M. F., and Nguyen, N. (2018). Farmers markets and food-borne illness*. Am. J. Agric. Econ. 100, 676–690. doi: 10.1093/ajae/aay011
Berry, E. D., Millner, P. D., Wells, J. E., Kalchayanand, N., and Guerini, M. N. (2013). Fate of naturally occurring Escherichia coli O157:H7 and other zoonotic pathogens during minimally managed bovine feedlot manure composting processes. J. Food Prot. 76, 1308–1321. doi: 10.4315/0362-028X.JFP-12-364
Boerlin, P., McEwen, S. A., Boerlin-Petzold, F., Wilson, J. B., Johnson, R. P., and Gyles, C. L. (1999). Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37, 497–503. doi: 10.1128/JCM.37.3.497-503.1999
Brooks, J. T., Sowers, E. G., Wells, J. G., Greene, K. D., Griffin, P. M., Hoekstra, R. M., et al. (2005). Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192, 1422–1429. doi: 10.1086/466536
Carstens, C. K., Salazar, J. K., and Darkoh, C. (2019). Multistate outbreaks of foodborne illness in the United States associated with fresh produce from 2010 to 2017. Front. Microbiol. 10:2667. doi: 10.3389/fmicb.2019.02667
Chen, Z., Biswas, S., Aminabadi, P., Stackhouse, J. W., Jay-Russell, M. T., and Pandey, P. K. (2019). Prevalence of Escherichia coli O157 and Salmonella spp. in solid bovine manure in California using real-time quantitative PCR. Lett. Appl. Microbiol. 69, 23–29. doi: 10.1111/lam.13156
Conrad, C. C., Stanford, K., McAllister, T. A., Thomas, J., and Reuter, T. (2014). Further development of sample preparation and detection methods for O157 and the top 6 non-O157 STEC serogroups in cattle feces. J. Microbiol. Methods 105, 22–30. doi: 10.1016/j.mimet.2014.06.020
Ekman, J., Goldwater, A., Bradbury, M., Matthews, J., and Rogers, G. (2021). Persistence of human pathogens in manure-amended Australian soils used for production of leafy vegetables. Agriculture 11:14. doi: 10.3390/agriculture11010014
FDA. (1998). Bacteriological analytical manual ". 8th Edn. Washington, DC: Food and Drug Administration.
FDA. (2015). Standards for the growing, harvesting, packing, and holding of produce for human consumption. Federal Register The Daily Journal of the United States Government.
Franz, E., Van Hoek, A. H., Bouw, E., and Aarts, H. J. (2011). Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile. Appl. Environ. Microbiol. 77, 8088–8096. doi: 10.1128/AEM.00745-11
Galarce, N., Escobar, B., Sanchez, F., Paredes-Osses, E., Alegria-Moran, R., and Borie, C. (2019). Virulence genes, Shiga toxin subtypes, serogroups, and clonal relationship of Shiga toxin-producing Escherichia coli strains isolated from livestock and companion animals. Animals (Basel) 9:733. doi: 10.3390/ani9100733
George, A. S., and Brandl, M. T. (2021). Plant bioactive compounds as an intrinsic and sustainable tool to enhance the microbial safety of crops. Microorganisms 9:2485. doi: 10.3390/microorganisms9122485
Glaize, A., Gutierrez-Rodriguez, E., Hanning, I., Diaz-Sanchez, S., Gunter, C., Van Vliet, A. H. M., et al. (2020). Transmission of antimicrobial resistant non-O157 Escherichia coli at the interface of animal-fresh produce in sustainable farming environments. Int. J. Food Microbiol. 319:108472. doi: 10.1016/j.ijfoodmicro.2019.108472
Gu, G., Strawn, L. K., Oryang, D. O., Zheng, J., Reed, E. A., Ottesen, A. R., et al. (2018). Agricultural practices influence Salmonella contamination and survival in pre-harvest tomato production. Front. Microbiol. 9:2451. doi: 10.3389/fmicb.2018.02451
Gu, G., Strawn, L. K., Zheng, J., Reed, E. A., and Rideout, S. L. (2019). Diversity and dynamics of Salmonella enterica in water sources, poultry litters, and field soils amended with poultry litter in a major agricultural area of Virginia. Front. Microbiol. 10:2868. doi: 10.3389/fmicb.2019.02868
Hammack, T. S., Amaguaña, R. M., June, G. A., Sherrod, P. S., and Andrews, W. H. (1999). Relative effectiveness of selenite Cystine broth, Tetrathionate broth, and Rappaport-Vassiliadis medium for the recovery of Salmonella spp. from foods with a low microbial load. J. Food Prot. 62, 16–21. doi: 10.4315/0362-028X-62.1.16
Harvey, R. R., Zakhour, C. M., and Gould, L. H. (2016). Foodborne disease outbreaks associated with organic foods in the United States. J. Food Prot. 79, 1953–1958. doi: 10.4315/0362-028X.JFP-16-204
Haymaker, J., Sharma, M., Parveen, S., Hashem, F., May, E. B., Handy, E. T., et al. (2019). Prevalence of Shiga-toxigenic and atypical enteropathogenic Escherichia coli in untreated surface water and reclaimed water in the mid-Atlantic U.S. Environ. Res. 172, 630–636. doi: 10.1016/j.envres.2019.02.019
Herrero, M., Thornton, P. K., Notenbaert, A. M., Wood, S., Msangi, S., Freeman, H. A., et al. (2010). Smart investments in sustainable food production: revisiting mixed crop-livestock systems. Science 327, 822–825. doi: 10.1126/science.1183725
Hruby, C. E., Soupir, M. L., Moorman, T. B., Pederson, C., and Kanwar, R. (2018). Salmonella and fecal indicator bacteria survival in soils amended with poultry manure. Water Air Soil Pollut. 229:32. doi: 10.1007/s11270-017-3667-z
Ingham, S. C., Fanslau, M. A., Engel, R. A., Breuer, J. R., Breuer, J. E., Wright, T. H., et al. (2005). Evaluation of fertilization-to-planting and fertilization-to-harvest intervals for safe use of noncomposted bovine manure in Wisconsin vegetable production. J. Food Prot. 68, 1134–1142. doi: 10.4315/0362-028x-68.6.1134
Islam, M., Doyle, M. P., Phatak, S. C., Millner, P., and Jiang, X. (2005). Survival of Escherichia coli O157:H7 in soil and on carrots and onions grown in fields treated with contaminated manure composts or irrigation water. Food Microbiol. 22, 63–70. doi: 10.1016/j.fm.2004.04.007
Jacobsen, C. S., and Bech, T. B. (2012). Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res. Int. 45, 557–566. doi: 10.1016/j.foodres.2011.07.026
Kim, C., Fatani, A., Almuqati, R., Rahemi, A., Abujamous, A., Wynn, C., et al. (2021). Prevalence and antimicrobial resistance of foodborne pathogens in value-added commodities procured from farmers’ markets in Central Virginia. J. Food Saf. 41:e12931. doi: 10.1111/jfs.12931
Koitabashi, T., Cui, S., Kamruzzaman, M., and Nishibuchi, M. (2008). Isolation and characterization of the Shiga toxin gene (stx)-bearing Escherichia coli O157 and non-O157 from retail meats in Shandong Province, China, and characterization of the O157-derived stx2 phages. J. Food Prot. 71, 706–713. doi: 10.4315/0362-028x-71.4.706
LGMA. (2021). Commodity specific food safety guidelines for the production and harvest of lettuce and leafy greens. CalifoCalifornia LGMA: California Leafy Greens Handler Marketing Agreement. Available at: https://lgma-assets.sfo2.digitaloceanspaces.com/downloads/August-2021-CA-LGMA-Metrics_FINAL-v20211208_A11Y.pdf (Accessed August 4, 2023).
Luna-Gierke, R. E., Griffin, P. M., Gould, L. H., Herman, K., Bopp, C. A., Strockbine, N., et al. (2014). Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol. Infect. 142, 2270–2280. doi: 10.1017/S0950268813003233
Micallef, S. A., Rosenberg Goldstein, R. E., George, A., Kleinfelter, L., Boyer, M. S., McLaughlin, C. R., et al. (2012). Occurrence and antibiotic resistance of multiple Salmonella serotypes recovered from water, sediment and soil on mid-Atlantic tomato farms. Environ. Res. 114, 31–39. doi: 10.1016/j.envres.2012.02.005
Naseer, U., Lobersli, I., Hindrum, M., Bruvik, T., and Brandal, L. T. (2017). Virulence factors of Shiga toxin-producing Escherichia coli and the risk of developing haemolytic uraemic syndrome in Norway, 1992-2013. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1613–1620. doi: 10.1007/s10096-017-2974-z
Nazareth, J., Shaw, A., Delate, K., and Turnbull, R. (2021). Food safety considerations in integrated organic crop–livestock systems: prevalence of Salmonella spp. and E. coli O157:H7 in organically raised cattle and organic feed. Renew. Agric. Food Syst. 36, 8–16. doi: 10.1017/S1742170519000292
Neher, D. A., Cutler, A. J., Weicht, T. R., Sharma, M., and Millner, P. D. (2019). Composts of poultry litter or dairy manure differentially affect survival of enteric bacteria in fields with spinach. J. Appl. Microbiol. 126, 1910–1922. doi: 10.1111/jam.14268
Pagadala, S., Marine, S. C., Micallef, S. A., Wang, F., Pahl, D. M., Melendez, M. V., et al. (2015). Assessment of region, farming system, irrigation source and sampling time as food safety risk factors for tomatoes. Int. J. Food Microbiol. 196, 98–108. doi: 10.1016/j.ijfoodmicro.2014.12.005
Patterson, L., Navarro-Gonzalez, N., Jay-Russell, M. T., Aminabadi, P., Antaki-Zukoski, E., and Pires, A. F. A. (2018). Persistence of Escherichia coli in the soil of an organic mixed crop-livestock farm that integrates sheep grazing within vegetable fields. Zoonoses Public Health 65, 887–896. doi: 10.1111/zph.12503
Peng, M., Salaheen, S., Almario, J. A., Tesfaye, B., Buchanan, R., and Biswas, D. (2016). Prevalence and antibiotic resistance pattern of Salmonella serovars in integrated crop-livestock farms and their products sold in local markets. Environ. Microbiol. 18, 1654–1665. doi: 10.1111/1462-2920.13265
Peng, M., Tabashsum, Z., Millner, P., Parveen, S., and Biswas, D. (2021). Influence of manure application on the soil bacterial microbiome in integrated crop-livestock farms in Maryland. Microorganisms 9:2586. doi: 10.3390/microorganisms9122586
Phan-Thien, K., Metaferia, M. H., Bell, T. L., Bradbury, M. I., Sassi, H. P., Van Ogtrop, F. F., et al. (2020). Effect of soil type and temperature on survival of Salmonella enterica in poultry manure-amended soils. Lett. Appl. Microbiol. 71, 210–217. doi: 10.1111/lam.13302
Pires, A., Millner, P., Baron, J., and Jay-Russell, M. (2018). Assessment of current practices of organic farmers regarding biological soil amendments of animal origin in a multi-regional US study. Food Prot. Trends 88, 347–362.
Pires, A. F. A., Patterson, L., Kukielka, E. A., Aminabadi, P., Navarro-Gonzalez, N., and Jay-Russell, M. T. (2019). Prevalence and risk factors associated with Campylobacter spp. and Salmonella enterica in livestock raised on diversified small-scale farms in California. Epidemiol. Infect. 147:e321. doi: 10.1017/S095026881900205X
Ramos, T. D. M., Jay-Russell, M. T., Millner, P. D., Baron, J. N., Stover, J., Pagliari, P., et al. (2021). Survival and persistence of foodborne pathogens in manure-amended soils and prevalence on fresh produce in certified organic farms: a multi-regional baseline analysis. Front. Sustain. Food Syst. 5:4767. doi: 10.3389/fsufs.2021.674767
Salaheen, S., Chowdhury, N., Hanning, I., and Biswas, D. (2015). Zoonotic bacterial pathogens and mixed crop-livestock farming. Poult. Sci. 94, 1398–1410. doi: 10.3382/ps/peu055
Salaheen, S., Peng, M., and Biswas, D. (2016). Ecological dynamics of Campylobacter in integrated mixed crop-livestock farms and its prevalence and survival ability in post-harvest products. Zoonoses Public Health 63, 641–650. doi: 10.1111/zph.12279
Sekaran, U., Lai, L., Ussiri, D. A. N., Kumar, S., and Clay, S. (2021). Role of integrated crop-livestock systems in improving agriculture production and addressing food security–a review. J. Agric. Food Res. 5:100190. doi: 10.1016/j.jafr.2021.100190
Shah, M. K., Bradshaw, R., Nyarko, E., Handy, E. T., East, C., Millner, P. D., et al. (2019). Salmonella enterica in soils amended with heat-treated poultry pellets survived longer than bacteria in unamended soils and more readily transferred to and persisted on spinach. Appl. Environ. Microbiol. 85:7112. doi: 10.1128/AEM.00334-19
Sharma, M., and Reynnells, R. (2016). Importance of soil amendments: survival of bacterial pathogens in manure and compost used as organic fertilizers. Microbiol. Spectr. 4:15. doi: 10.1128/microbiolspec.PFS-0010-2015
Sheng, L., Shen, X., Benedict, C., Su, Y., Tsai, H. C., Schacht, E., et al. (2019). Microbial safety of dairy manure fertilizer application in raspberry production. Front. Microbiol. 10:2276. doi: 10.3389/fmicb.2019.02276
Solomon, E. B., Yaron, S., and Matthews, K. R. (2002). Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl. Environ. Microbiol. 68, 397–400. doi: 10.1128/AEM.68.1.397-400.2002
Strawn, L. K., Fortes, E. D., Bihn, E. A., Nightingale, K. K., Grohn, Y. T., Worobo, R. W., et al. (2013a). Landscape and meteorological factors affecting prevalence of three food-borne pathogens in fruit and vegetable farms. Appl. Environ. Microbiol. 79, 588–600. doi: 10.1128/AEM.02491-12
Strawn, L. K., Grohn, Y. T., Warchocki, S., Worobo, R. W., Bihn, E. A., and Wiedmann, M. (2013b). Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl. Environ. Microbiol. 79, 7618–7627. doi: 10.1128/AEM.02831-13
Tran, D. T. Q., Bradbury, M. I., Ogtrop, F. F. V., Bozkurt, H., Jones, B. J., and Mc, C. R. (2020). Environmental drivers for persistence of Escherichia coli and Salmonella in manure-amended soils: a meta-analysis. J. Food Prot. 83, 1268–1277. doi: 10.4315/0362-028X.JFP-19-460
USDA-AMS. (2015). Farmers markets and local food marketing. Available at: http://www.ams.usda.gov/AMSv1.0/ams.
USDA-AMS. (2019). National farmers market directory. USDA AMS. Available at: https://www.ams.usda.gov/local-food-directories/farmersmarkets.
Weller, D., Wiedmann, M., and Strawn, L. K. (2015a). Irrigation is significantly associated with an increased prevalence of Listeria monocytogenes in produce production environments in New York state. J. Food Prot. 78, 1132–1141. doi: 10.4315/0362-028X.JFP-14-584
Weller, D., Wiedmann, M., and Strawn, L. K. (2015b). Spatial and temporal factors associated with an increased prevalence of Listeria monocytogenes in spinach fields in New York state. Appl. Environ. Microbiol. 81, 6059–6069. doi: 10.1128/AEM.01286-15
Keywords: Listeria monocytogenes, Salmonella, STEC, biological soil amendments, fresh produce
Citation: Goodwyn B, Millner P, Punchihewage-Don AJ, Schwarz M, Hashem F, Bowers J, Biswas D and Parveen S (2023) Survival and persistence of foodborne pathogenic and indicator bacteria in spatially separated integrated crop-livestock farms. Front. Sustain. Food Syst. 7:1287371. doi: 10.3389/fsufs.2023.1287371
Edited by:
Hamada Imtara, Arab American University, PalestineReviewed by:
Khaled M. Sawalha, Al-Quds University, PalestineAmine El Bouzidi, Mohamed Premier University, Morocco
Nabil Radouane, Mohammed VI Polytechnic University, Morocco
Copyright © 2023 Goodwyn, Millner, Punchihewage-Don, Schwarz, Hashem, Bowers, Biswas and Parveen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salina Parveen, sparveen@umes.edu
 Brian Goodwyn
Brian Goodwyn Patricia Millner
Patricia Millner Anuradha Jeewantha Punchihewage-Don
Anuradha Jeewantha Punchihewage-Don Melinda Schwarz1
Melinda Schwarz1  John Bowers
John Bowers Debabrata Biswas
Debabrata Biswas Salina Parveen
Salina Parveen