Presence of foodborne pathogens and survival of generic Escherichia coli in an organic integrated crop-livestock system
- 1Department of Population Health and Reproduction, School of Veterinary Medicine, University of California-Davis, Davis, CA, United States
- 2Western Center for Food Safety, University of California-Davis, Davis, CA, United States
- 3Department of Plant Sciences, University of California-Davis, Davis, CA, United States
- 4Michael Fields Agricultural Institute, East Troy, WI, United States
Introduction: Integrated crop-livestock systems (ICLS) use animals to graze crop residues or cover crops before planting fresh produce and provide ecosystem services to support organic vegetable production. However, there is a risk of foodborne pathogen transfer to fresh produce because grazing may introduce enteric foodborne pathogens into the soil via animal feces, which may subsequently be transferred to the produce.
Methods: To examine the effect of cover crop use and the risk of cover crop grazing on the contamination of soil and produce by foodborne pathogens in ICLS, a three-year (2019–2021) experimental study was conducted in organically managed plots, which were assigned three different treatments (fallow without cover crop or grazing, cover crop without grazing, or cover crop with grazing by sheep) in a maize/tomato rotation. During the three years of the experiment, a total of 184 pre- and post-graze fecal samples and 96 samples of tomatoes were collected to test for foodborne pathogens (Escherichia coli O157, non-O157 Shiga toxin-producing Escherichia coli (STEC), and Listeria (L.) monocytogenes). Soil samples were collected monthly until 126–171 days after grazing (824 in total) to examine the presence of foodborne pathogens, and generic E. coli (MPN/g) was quantified to compare its persistence among the three treatments.
Results and Discussion: We did not detect any foodborne pathogens from harvested tomatoes in 2020 and 2021. One non-O157 STEC positive soil sample (0.1%, 1/824) was detected in the fallow treatment, and one L. monocytogenes-positive (1.1%, 1/92) was detected from the post-graze fecal samples. When assessing proportions of generic E. coli positive and counts of generic E. coli in the soil samples using mixed effect zero-inflated negative binomial models, soil samples collected in the graze cover crop treatment plot showed significant increases in the counts of generic E. coli until 61–82 days post grazing, but no difference was observed after 96–123 days, compared to the baseline of the fallow treatment. Findings from generic E. coli counts support the use of the United States Department of Agriculture (USDA) National Organic Program (NOP) 90- or 120-day interval rule between applying raw manure and harvesting in organic farming into ICLS. Additionally, we confirmed that commercial organic compost application before cover crop seeding in the winter had no significant effect on the proportions and counts of generic E. coli in the soil of the following growing seasons. This longitudinal field trial confirmed that the effect of sheep grazing on foodborne pathogen contamination in ICLS is minimal but further studies comparing the genetic associations between fecal and soil samples would be necessary to distinguish the source of foodborne pathogen contamination.
1 Introduction
Integrated crop-livestock systems (ICLS) offer promising alternatives to intensive specialized farming by diversifying cropping systems to reconnect nutrient cycles and build sustainability and resilience (Bell and Moore, 2012; Garrett et al., 2020). One form of ICLS involves grazing of cover crops in between cash crops on a single field (Hilimire, 2011; Moraine et al., 2014). Winter cover crops are planted during the inter-crop period to keep soil covered in presence of living roots to build soil health, conserve natural resources, provide nutrients to the next crop and lower the incidence of pest and disease (Franzluebbers et al., 2011; Schipanski et al., 2014). Raw (i.e., untreated) manure deposited in the field during grazing of cover crop biomass benefits soil fertility by providing labile and organic nutrients for the following crop (Medina-Roldán et al., 2008; Da Silva et al., 2014). Thus, the soil is fertilized by connecting nutrient cycling, and biodiversity is enhanced through the use of cover crops and grazing, which could also help with weed and pest management (Lemaire et al., 2014; Garrett et al., 2017).
Recently, both organic and conventional agriculture have adopted ICLS as a part of regenerative agriculture practices. In particular, organic agriculture frequently follows ICLS as it prohibits the use of synthetic fertilizers and pesticides, and therefore uses natural soil amendments (i.e., compost and animal manure) instead (USDA, 2000). However, livestock can naturally carry enteric foodborne pathogens (e.g., Shiga toxin-producing Escherichia coli (STEC), Salmonella spp.), which can shed in feces (Hutchison et al., 2005). In ICLS, application of manure during livestock grazing increases the risk of foodborne pathogen transfer to fresh produce from contaminated soils (Ingham et al., 2004; Franz et al., 2005; Jacobsen and Bech, 2012). Due to the risk of transferring foodborne pathogens to fresh produce from untreated manure and animal feces, the Food and Drug Administration (FDA) Food Safety Modernization Act (FSMA) addresses concerns about raw manure application, grazing animals, and wildlife intrusion in produce farms (FDA, 2015). The FSMA does not object to the United States Department of Agriculture (USDA) National Organic Program (NOP) standards regarding application of raw manure, which entails that untreated animal manure must be incorporated into the soil at least 90 days prior to harvest for crops whose edible portions are not in contact with soil, or 120 days for those whose edible portions are in direct contact with soil (USDA-AMS NOP, 2011b). However, due to the lack of sufficient science-based data, the FDA has deferred its final decision regarding the wait time between raw manure application and harvest and has not yet specified any criteria for animal grazing. Instead, farmers are encouraged to voluntarily follow the USDA-NOP standards.
Among foodborne outbreaks reported by the Centers for Disease Control and Prevention (CDC) over the last decade, fruits and vegetable row crops (e.g., leafy greens, tomatoes, and melons), often consumed raw or minimally processed, were the most common sources (Carstens et al., 2019; Centers for Disease Control and Prevention, 2022). In recent years, more foodborne outbreaks were associated with organic foods in the US, and the majority of them were attributed to produce items (Harvey et al., 2016). Among foodborne pathogens, STEC including Escherichia coli O157 (E. coli O157) and Listeria monocytogenes (L. monocytogenes) were predominantly present in vegetable row crops and were responsible for high hospitalization rates, although Salmonella caused the most multistate outbreaks when all sources of food were included (Centers for Disease Control and Prevention, 2022). In agricultural environments, feces from ruminants (cattle, sheep, and goats) are generally considered a potential source of contamination of STEC since they are natural reservoirs of the bacteria (Hutchison et al., 2005; Franz and Van Bruggen, 2008; Jay-Russell, 2013). Due to the sparse distribution of STEC in natural environments, the quantification of generic E. coli has commonly been used as an indicator of fecal contamination and as a surrogate of STEC in soil when studying foodborne pathogens (Park et al., 2015; Allende et al., 2017; Patterson et al., 2018). Soils, once exposed, are also known to typically harbor L. monocytogenes for extended periods when compared to STEC, even at lower temperatures (Nicholson et al., 2005; Ivanek et al., 2006).
Research is currently limited regarding the persistence and prevalence of foodborne pathogens in the soil and produce after grazing by livestock in ICLS systems. Most of the recent studies have been conducted on livestock manure application without having animals directly on the soil (Sharma et al., 2019; Ramos et al., 2021; Pires et al., 2023). Regardless, most growers are encouraged to follow the NOP 90- to 120- day interval rules for grazing. Previous observational study done in California repeatedly measured generic E. coli in a commercial organic ICLS farm grazed by sheep, and showed that the mean concentration of generic E. coli decreased under the limit of detection in the soil by 84 to 111 days post-graze (Patterson et al., 2018). However, this study was performed on a commercial organic farm with variable grazing times among sub-fields and the absence of control or baseline samples, and without testing for pathogenic E. coli.
The aim of this study is to examine the effect of cover crop use and the risk of grazing on the contamination of soil and produce by foodborne pathogens in an experimental ICLS field. The presence of foodborne pathogens (E. coli O157, non-O157 STEC, and L. monocytogenes) in fecal, soil, and produce samples was examined in three different treatments (fallow, cover crop without grazing, cover crop with grazing by sheep) through a three-year experimental study in an ICLS. Additionally, generic E. coli in the soil, as an indicator of fecal contamination and a surrogate of pathogenic E. coli, was quantified monthly to compare its persistence among the three treatments. Sheep were used for grazing in this experimental study, since California has a large population of sheep and lambs (i.e., approximately 570,000), which are widely used as grazing animals on crop, vineyard, and orchard fields (Hoar et al., 2013; USDA National Agricultural Statistics Service, 2019; Brewer et al., 2023).
2 Materials and methods
2.1 Study design
A replicated field trial was conducted over three-years (2019–2021) in a certified organic field at the Russell Ranch Sustainable Agriculture Facility, University of California, Davis (UC Davis) (38° 32′ 36.87″, −121° 52′ 11.89″). Soils are classified as Yolo silt loam and Rincon silty clay loam, though textural analysis showed this site has a clay loam/clay texture in the top 30 cm with 15–40% sand, 20–40% silt and 33–48% clay, and about 1% total carbon at the start of the experiment. The experiment was setup as a randomized complete block design with four replicates per treatment – fallow, cover crop tilled without grazing (non-graze CC), and cover crop grazed by sheep (graze CC). Neither cover crop seeding nor sheep grazing were conducted in the fallow treatment, which served as a control. Each block replicate consisted of 9 beds measuring 13.5 m wide (i.e., 1.5 m per one bed). The three treatments were randomly allocated within 3 bed sections of each block (Supplement 1A). In 2019 and 2021, commercially composted poultry litter was applied to all plots before cover crop mixtures were seeded. In 2020, the effect of compost versus non-compost was compared by dividing the block into two parts (i.e., split-plot design) (Supplement 1B). Either the top or bottom part of each block measuring 83.2 m in length was assigned as the area in which compost was not applied.
Sheep grazing was initiated when the cover crops grew up to 25 cm and soil moisture conditions allowed. Sheep grazed the field until the sward height was 10 cm (USDA-Natural Resources Conservation Service, 2013). Depending on the cover crop growth and cash crop establishment (i.e., seeding corn or transplanting of tomatoes), grazing events were planned once or twice per growing season. Electrical fences were installed around the areas corresponding to the graze CC treatment the day before the sheep were transported from the barn to the field. The sheep flock used for grazing comprised a mix of breeds (e.g., Suffolk, Hampshire, Dorset, and crossbreed sheep), with ages ranging from 1 to 6 years old. The total number of sheep and grazing length varied from year to year, depending on cover crop biomass; 120 sheep grazed once (April 8th–15th, 2019), 80 sheep grazed twice (February 10th–12th and March 19th–20th, 2020), and 49 sheep grazed twice (March 2nd–4th and April 5th –7th, 2021) in the first, second, and third growing season, respectively. Sheep grazed rotationally, starting from block 1 and moving toward block 4 with the intention of implementing a uniform grazing intensity across the blocks. Sheep were provided ad libitum water, and overnight lights to protect them from wildlife, with always one person watching over the field at night. This study was approved by the Institutional Animal Care and Use Committee of the University of California, Davis (IACUC #21028).
After the final grazing event each year, both graze CC and non-graze CC were terminated before seeding or transplanting the cash crops. Table 1 summarizes the types of cash crop planted, the cover crop mix seeded during winter in the field, and the specific soil sampling dates for each year of the experiment. All crops in rotations were irrigated and managed according to organic certification.
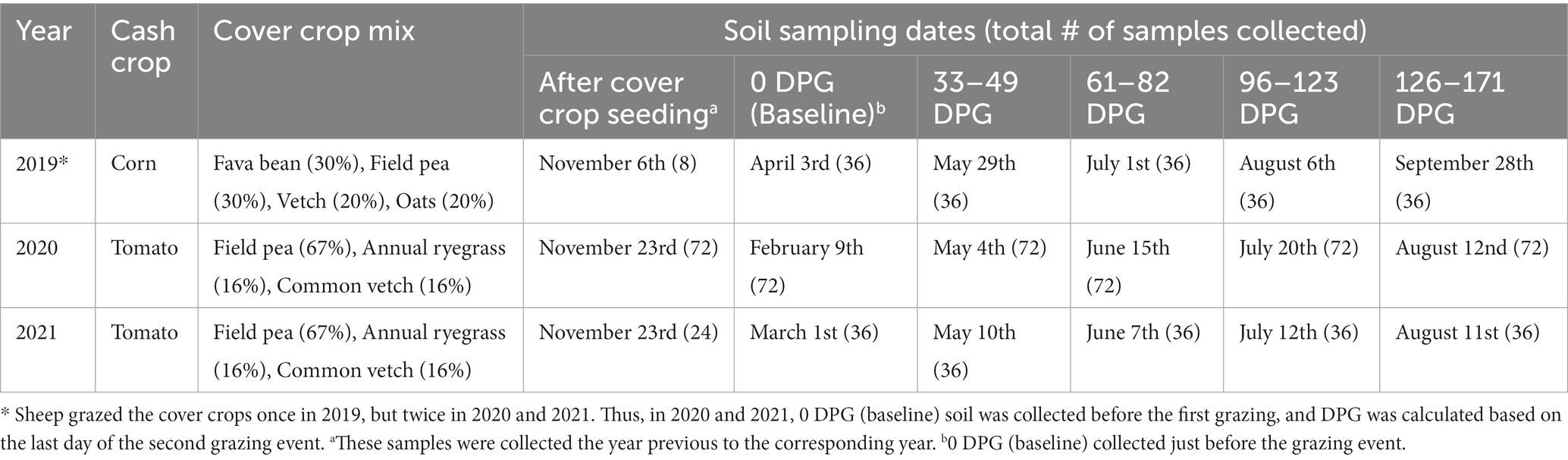
Table 1. Types of cash crop planted, cover crop mix, and dates of soil sampling (days post grazing (DPG)) in each year in the organic integrated crop-livestock experiment (2019–2021).
2.2 Sample collection
Soil samples were collected six times during each growing season: after compost application and cover crop seeding in November, just before grazing (0-Days Post-Graze), and monthly after the grazing event until approximately 120–150 Days Post-Graze (DPG). When grazing was conducted twice in the growing season, DPG was calculated based on the last day of the second grazing event. A total of 824 soil samples were collected over the 3 years, and the number of soil samples collected varied depending on the year and sampling dates (Table 1) with 36 soil samples (i.e., 12 samples per treatment) collected on each sampling date in 2019 and 2021, and 72 soil samples (i.e., 12 samples per treatment and compost application) collected on each sampling date in 2021.
Soil sampling was only conducted in the middle bed of each treatment within a block, with the two side beds serving as a buffer (Supplement 1). A stainless-steel soil core sampling probe (2.54 cm diameter) was used to collect the soil samples, and samples were placed in sterile Whirl-Pak bags (Nasco, Modesto, CA). Gloves were changed and the soil core was sanitized with 70% ethanol between each treatment and block. Three soil samples were taken per beds (12 beds × 3 composites), representing one-third of each bed as subplots (i.e., 36 subplots in 2019 and 2021, and 72 subplots in 2020). Each composite was comprised of 5 soil cores (15 cm in depth) collected from 5 separate loci spaced evenly from the top to the bottom of each bed.
To determine whether foodborne pathogens (i.e., non-O157 STEC, E. coli O157:H7, and L. monocytogenes) were present in the feces of grazing sheep, pre- and post-graze fecal samples were collected from the sheep before they were moved from the barn to the experimental field, and after sheep were removed from the graze CC plots. Individual fresh fecal pats (approximately 30 g of fecal material) were scooped from the barn or field ground using gloves aseptically and placed into a sterile Whirl-pack. A total number of 40–96 fecal samples per year were collected depending on the number of sheep grazing. The sheep used for the experiment were separated from the other sheep in the barn 1 week before they were moved to graze on the field, and their diet mostly consisted of alfalfa hay with a grain ration (rolled barley – 34%, rolled corn – 32%, soybean – 24%, molasses 8%, and fat – 2%).
For produce, 24 samples of tomatoes (<100 g per sample) were collected during the 96–123 DPG and 126–171 DPG periods each year (July 20th and August 12nd in 2020, and July 12nd and August 11st in 2021) for a total of 96 samples. As was done for soil sampling, tomatoes were collected from the middle bed of each treatment within a block for foodborne pathogen testing. In the first year (2019), corn for animal feed was planted, so no produce samples were collected.
Two bottles of irrigation water samples (1 L each) were collected from the water tap connected to the irrigation lines at the field for each growing season to test whether the foodborne pathogens (i.e., non-O157 STEC, E. coli O157:H7, and L. monocytogenes) or generic E. coli were from the water source.
All samples (i.e., soil, feces, produce, and water) were transported to the laboratory in a cooler with ice packs and processed within 24 h of collection.
2.3 Sample preparation and enrichment
The microbial analyses for the isolation of non-O157 STEC, E. coli O157:H7, L. monocytogenes, and generic E. coli have been previously described in recent publications (Patterson et al., 2018; Ramos et al., 2021; Pires et al., 2023). Briefly, for each soil sample, a Tryptic Soy Broth (TSB) enrichment (BD BactoTM, Heidelberg, Germany) was carried out to detect generic E. coli, non-O157 STEC, and E. coli O157:H7, and a Listeria Enrichment Broth (LEB) enrichment (Neogen Culture Media, Lansing, MI, United States) was done to detect L. monocytogenes. All the media were prepared following the manufacturers’ instructions. Thirty grams of soil were placed in a pre-refrigerated 24 oz. Whirl-Pak bag filled with 270 mL of TSB or LEB (corresponding to a 1:10 dilution) and manually homogenized. For the TSB reservoir, eight wells of a 48-well reservoir (E&K Scientific, Santa Clara, CA, United States) were filled with 5 mL of the sample in TSB medium, and it was serially diluted up to 10−6 with 4 replications per sample (Atwill et al., 2015). For the TSB enrichment, samples were incubated at 25°C for 2 h followed by 42°C for 8 h with 50 rpm shaking, then held at 6°C with no shaking in a Multitron programmable shaking incubator (Eppendorf, Hauppauge, NY, United States) until the isolation step. For the LEB enrichment, samples were incubated for 18 h at 30°C with 100 rpm shaking, then held at 6°C with no shaking until the isolation step.
For the fecal and produce samples, all the incubation steps were the same as for the soil samples. However, 10 g of feces from each sample were placed into a Whirl-Pak bag containing 90 mL of TSB or LEB. Each produce sample (1–2 tomatoes/bag, ~100 g) was weighed and placed into 100 mL of Buffered Peptone Water (BPW) (Hardy Diagnostics, Santa Maria, CA, United States), and manually massaged for 1 min until homogenized. Then, 100 mL of BPW from each bag was transferred to 100 mL of 2 times the concentration of TSB (60 g of media in 1 L of purified autoclaved water) used for the soil samples (Ramos et al., 2021; Pires et al., 2023).
For water samples, 1 L of water was poured through a sterile 0.45 mm nitrocellulose filter assembly using standard membrane filtration techniques and placed in 100 mL of TSB or LEB for the further isolation steps (Partyka et al., 2018).
2.4 Enumeration and isolation of generic Escherichia coli in soil
To measure the concentration (Most Probable Number, MPN/g) and determine the presence of generic E.coli in the soil and produce samples, 4 μL from each dilution well of the TSB reservoir with four replicates per sample as well as 10 μL from the TSB bag were streaked onto CHROMagar E. coli (ECC) (CHROMagar Microbiology, Paris, France) followed by incubation for 24 h at 37°C (Atwill et al., 2015; Ramos et al., 2021). Up to four colonies per sample that presented E. coli-like features (i.e., blue colonies) were re-streaked onto secondary and tertiary CHROMagar ECC. At least two presumptive positive pure isolates per sample were subsequently subjected to a standard polymerase chain reaction (PCR) assay targeting the uspA (universal stress protein A) gene (Nyström and Neidhardt, 1992). MPN series cell densities were calculated based on dilution to extinction using an MPN Calculator (Avineon, Inc. and US Environmental Protection Agency, 2013). The limit of quantification (LOQ) was 0.089 MPN/g for the positive samples.
2.5 Non-O157 STEC, Escherichia coli O157:H7, and Listeria monocytogenes isolation
Detection of non-O157 STEC, E. coli O157:H7, and L. monocytogenes was done for all soil, pre- and post-graze fecal, produce, and water samples. To detect non-O157 STEC, 1 mL of pre-enriched sample in TSB enrichment was added to a tube containing 9 mL of modified enterohemorrhagic E.coli (mEHEC) selective media (Biocontrol, Bellevue, WA, United States), and incubated for 12 h at 42°C (Cooley et al., 2013, 2014). Ten μL of the mEHEC solution was then streaked onto ChromSTEC agar (CHROMagarTM, Paris, France) followed by incubation for 24 h at 37°C. Up to six presumptive positive isolates corresponding to purple colonies that fluoresced under ultraviolet light were re-streaked on to secondary and tertiary ChromSTEC agar. A final assumed pure colony was streaked onto Tryptic Soy Agar (TSA), followed by incubation at 37°C overnight. Isolates were confirmed as non-0157 STEC using a PCR assay targeting the stx1 and stx2 genes (Paton and Paton, 1998).
To detect E. coli O157:H7, immunomagnetic separation (IMS) using Dyna anti-E. coli O157 beads was performed on TSB enrichment with the automated Dyna Bead Retriever (Invitrogen, Carlsbad, CA United States) to concentrate E. coli O157:H7 cells. An IMS product of 50 μL washed beads was planted onto both Rainbow agar (Biolog, Hayward, CA, United States) with novobiocin (20 mg/L) and tellurite (0.8 mg/L) (MP Biomedicals, Solon, OH, United States), and MacConkey II Agar with sorbitol supplemented with potassium tellurite (2.5 mg/L) and cefixime (0.05 mg/L) (CT-SMAC). Both sets of plates were incubated for 24 h at 37°C. Up to two presumptive positive isolates corresponding to dark blue and gray colonies to Rainbow agar, and to brown colonies on CT-SMAC were cross-streaked on to the other agar plates (i.e., CT-SMAC to Rainbow, or Rainbow to CT-SMAC). To confirm isolates as E. coli O157:H7, a PCR assay targeting the eaeA gene was conducted after transferring onto TSA (Paton and Paton, 1998).
As with E. coli O157:H7, IMS was also performed with pre-enriched samples in LEB enrichment to concentrate L. monocytogenes cells using Dyna anti-Listeria beads. An IMS product of 30 μL washed beads added to Brilliance Listeria Agar (BLA) with BLA selective and differential supplements (Oxoid, Hants, United Kingdom). A 100 μL product of washed beads was added to 5 mL of Fraser Broth (BD, Sparks, MD). Both BLA agar plates and Fraser Broth were incubated at 37°C for 48 h (Cooley et al., 2014). Presumptive positive isolates corresponding to blue colonies with a halo around them in BLA were re-streaked onto secondary BLA and the process was repeated for tertiary isolation. A 10 μL loop of Fraser Broth, which changed to black color after the incubation, was added to BLA and streaked by tertiary isolation as well. To confirm isolates as L. monocytogenes, a PCR assay targeting the hylA gene was done after transferring onto TSA (Kawasaki et al., 2005).
For the gene confirmation (uspA for generic E. coli, stx1 and stx2 for non-O157 STEC, eaeA for E. coli O157:H7, and hylA for L. monocytogenes), the results of the PCR assays were analyzed by agarose gel electrophoresis, running for 60 min with 100 voltages.
2.6 Statistical analyses
Descriptive analysis was used to summarize the proportions of foodborne pathogens (E. coli O157:H7, non-O157 STEC, and L. monocytogenes) in soil, feces, and produce samples. To compare the proportions of foodborne pathogens in feces, a two-proportion z-test was used because pre- and post-graze samples were not collected from the same individuals. For soil samples, proportions and concentrations of generic E. coli positive samples were also summarized; the proportion of samples positive for generic E. coli (%) was calculated as the number of positives out of the total number of collected samples in each treatment group (i.e., fallow, non-graze CC, and graze CC), on each sampling day, in each year (i.e., 12 samples/sampling day). The concentrations of generic E. coli in the soil (log10 MPN of dry weight of soil/100 g) were reported as median and range (min–max) for each treatment group, sampling day, and year, after transforming the values under LOQ as LOQ/2 (i.e., 0.0445 MPN/g).
For modeling, the outcome variable – generic E. coli concentration (MPN/100 g) in the soil – was log10-transformed and rounded off to the nearest integer, if the data were non-zero, so that it could be treated as count data. Due to the high proportion (58.3%) of zero data (i.e., negative soil samples for the presence of generic E. coli) and over-dispersion (mean = 0.8, sd = 1.3) of generic E. coli count, mixed effect zero-inflated negative binomial models (ZINB) were used to assess the effects of the treatment, sampling day, year, and two-way interaction term between treatment and sampling day. ZINB models have two components, a zero-inflated component and conditional component; the zero-inflated part models the probability of observing zero in generic E. coli as in logistic regression, and the conditional part has a continuous count outcome corresponding to the log count of generic E. coli with a negative binomial distribution. All explanatory variables included in the models were categorical. The models were run using the glmmTMB package in R software (Brooks et al., 2017). To account for the repeated collection of soil samples from the same subplot over time each year, subplot was added as a random effect to all models. Correlations between the repeated measurements from the same subplots in each year were also considered, and a covariance structure of first-order autoregressive (AR1) was examined. Only variables that had a p-value <0.2 in univariable analyses were kept for the multivariable analyses. Forward stepwise model selection was conducted manually based on Akaike information criteria (AIC) values. The data collected before each growing season (November of the correspondent year) were reported separately and not included in the analyses as the model aimed to examine the counts of generic E. coli following grazing events. The effect of compost was examined in a separate model, in addition to the other predictor variables (i.e., treatment, sampling day, year) using the subset of 2020 data (i.e., 360 soil samples). All statistical analyses were performed using R (version 4.2.0), with the significance level set at 0.05.
3 Results
3.1 Presence of foodborne pathogens and generic Escherichia coli in the soil
In all 3 years of this experiment, one soil sample (0.1%, 1/824) tested positive for non-O157 STEC. It was collected from the fallow treatment (block 3) on 96–123 DPG in 2019. None of the other soil samples collected during the 3 years of the experiment tested positive for foodborne pathogens (i.e., E. coli O157:H7, non-O157 STEC, and L. monocytogenes).
As for generic E. coli, 41.7% (225/540) of the soil samples tested positive. Among the generic E. coli positive samples, 56.9% (128/225) were collected from the graze CC treatment group. The median concentration of generic E. coli (log10 MPN/100 g of dry weight of soil) in the graze CC treatment was 1.02 (0.65–7.53) for the 3 years. In the graze CC treatment, 33–49 DPG or/and 61–82 DPG showed the highest proportions of generic E. coli positives each year (Figure 1). In particular, 100% (12/12) of the soil samples collected at 61–82 DPG in 2019 and at 33–49 DPG in 2020 were generic E. coli positive. The median concentrations of generic E. coli in the graze CC treatment were the highest on 33–49 DPG in all 3 years (Table 2). Proportions of soil samples positive for generic E. coli in the non-graze CC treatment (23.3%, 42/180) was lower than that in the fallow treatment (30.6%, 55/180) over the 3 years (Figure 1), and the median concentrations of generic E. coli from both treatments were below one in all treatment years (Table 2). The range of generic E. coli was larger in the fallow (0.65–5.77) than the non-graze CC treatment (0.65–4.73).
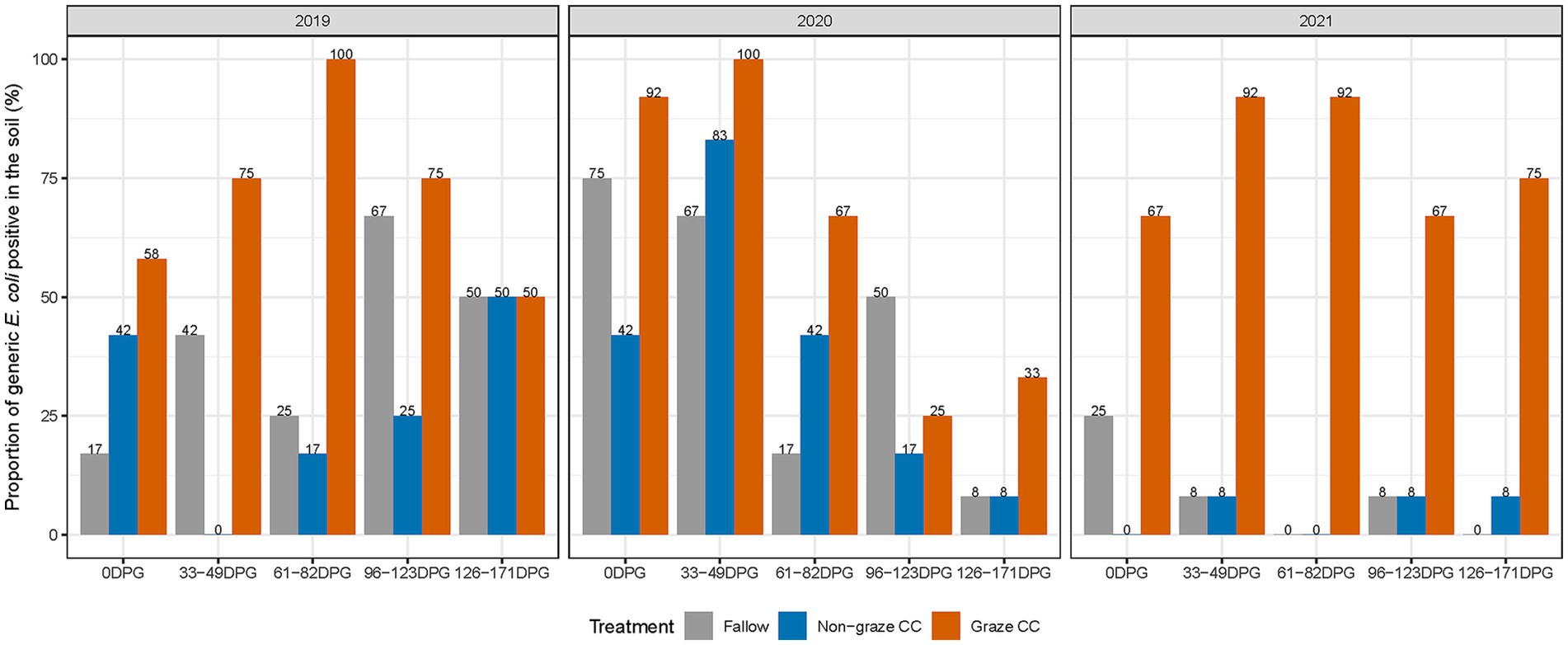
Figure 1. Proportion of generic E. coli positive (%) soil samples in each treatment by sampling day and year in the organic integrated crop-livestock experiment (2019–2021). Twelve soil samples were collected per treatment group (fallow, non-graze CC, graze CC) on 0, 33–49, 61–82, 96–123, and 126–171-days post grazing (DPG) in each year (Samples collected from the non-composted area in 2020 were not included).
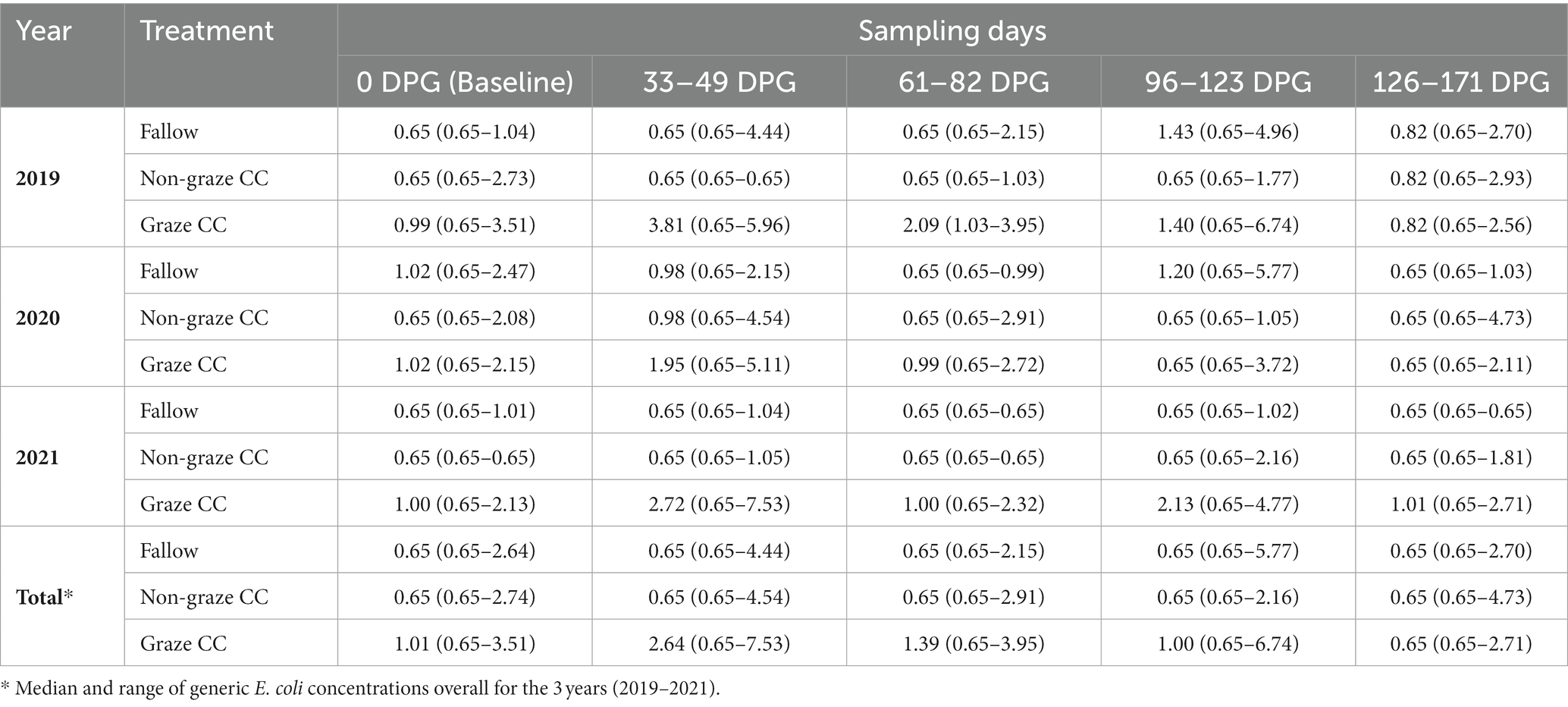
Table 2. Median and range (min – max) of generic E. coli concentration in the soil samples (log10 MPN/100 g of dry weight of soil) for each treatment and sampling day (0, 33–49, 61–82, 96–123, and 126–171-days post grazing (DPG)) in the organic integrated crop-livestock experiment (2019–2021).
Before growing seasons, when examining the soil samples collected in November after cover crop seeding (i.e., 104 samples), none of them were generic E. coli positive in 2019, but 30.6% (22/72) in 2020 and 70.8% (17/24) in 2021 were positive (Table 3). There was a continuous increase in the proportion of generic E. coli-positive soil samples in each treatment group, and the graze CC treatment had the highest increase (i.e., 50% in 2020, and 100% in 2021). The median concentration of generic E. coli (log10 MPN/100 g) in each treatment also increased from 2020 to 2021; the median concentrations changed from 0.65 (0.65–3.37) to 0.98 (0.65–4.54) in the fallow treatment, from 0.65 (0.65–4.67) to 1.19 (0.65–2.89) in the non-graze CC treatment, and from 0.31 (0.65–2.09) to 0.98 (0.98–2.69) in the graze CC treatment.
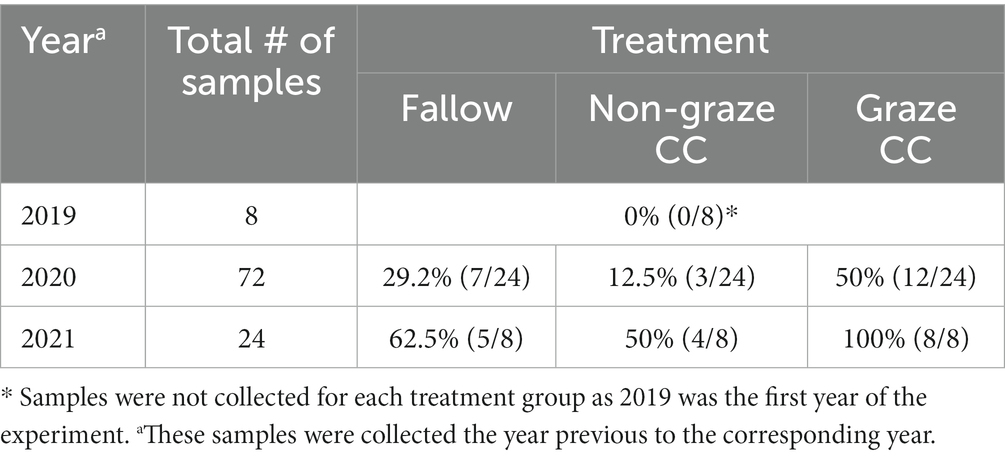
Table 3. Proportion of generic E. coli positive soil samples (%) collected in November after cover crop seeding in the organic integrated crop-livestock experiment (2019–2021).
3.2 Effects of treatment, sampling day, and year on generic Escherichia coli in the soil
The univariable analyses results for the count of generic E. coli (log10 MPN/100 g rounded off to the nearest integer) from the mixed-effect zero-inflated negative binomial models are summarized in Table 4. None of the variables showed significant associations with the probability of zero count in generic E. coli (i.e., zero-inflated components). Thus, only conditional components (log10 count of generic E. coli) of the models were included. Year, sampling day, and treatment were all significantly associated with the count of generic E. coli in the soil at the significance level of 0.2 in the univariable analyses. Compared to 2021, 2019 and 2020 had a higher risk of having positive generic E. coli counts in soil. The count of generic E. coli in soil significantly increased at 33–49 DPG (RR = 2.08, p < 0.001) compared to the baseline (i.e., before grazing). The graze CC treatment had significantly higher counts of generic E. coli (RR = 3.07, p < 0.001), and the non-graze CC treatment had significantly lower counts (RR = 0.72, p = 0.09), when compared to the fallow treatment. The interaction between sampling day and treatment was also significantly associated with the count of generic E. coli (p < 0.001).
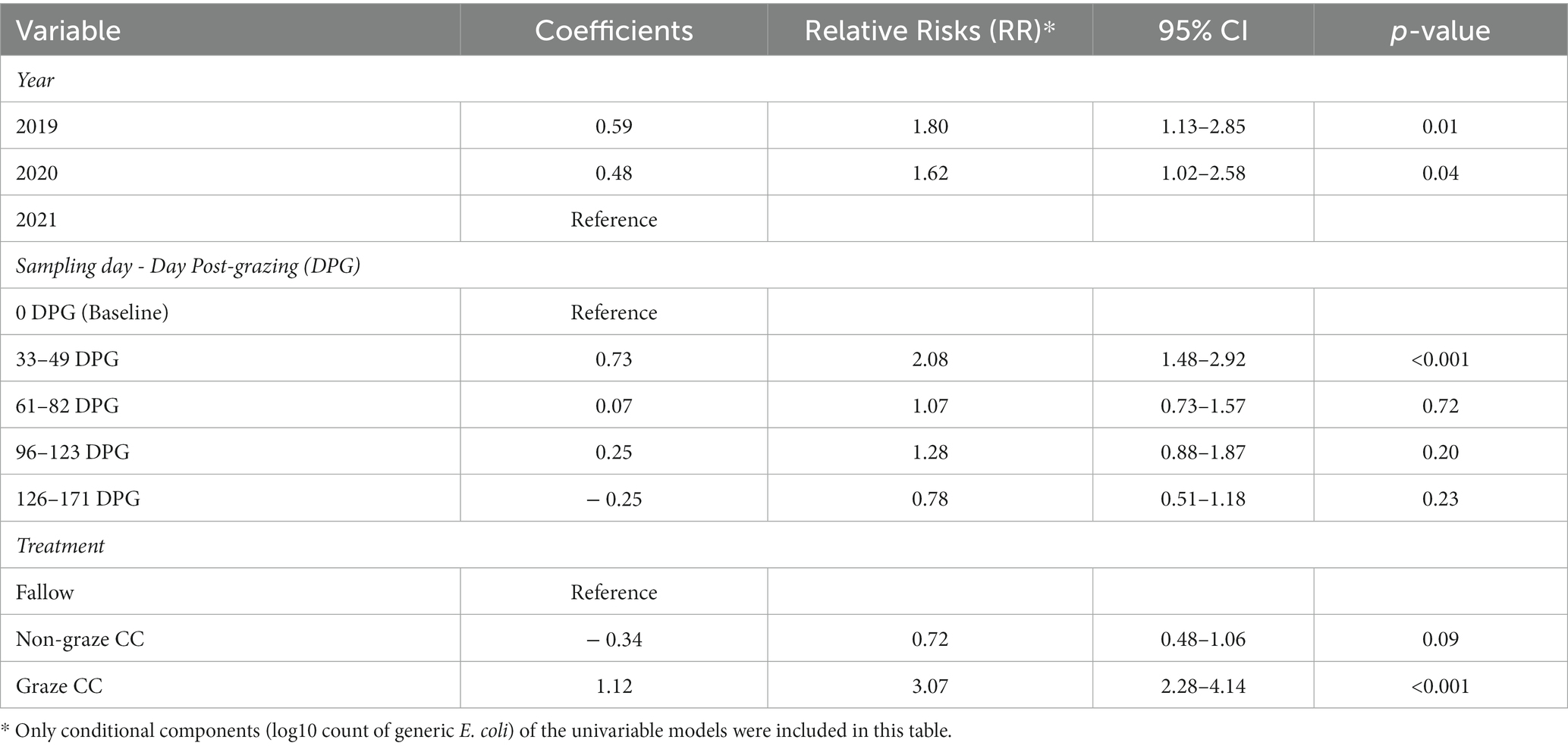
Table 4. Univariable analyses results for the counts of generic E. coli in soil samples from the organic integrated crop-livestock experiment (2019–2021) using mixed-effect zero-inflated negative binomial models with subplot as a random effect.
The final multivariable mixed-effect zero-inflated negative binomial model included all three explanatory variables (year, sampling day, and treatment), as well as the interaction between sampling day and treatment (Table 5). The correlation (AR1) between residuals across sampling days in the subplots was 0.88. The zero-inflated component of the final model contained the treatment effect, which was the only significant factor among the three explanatory variables; graze CC had a significantly lower probability of having a zero count of generic E. coli (i.e., absence of generic E. coli) compared to the fallow (OR = 0.09, 95% CI = 0.01–0.54) and non-graze CC treatments (OR = 0.04, 95% CI = 0.007–0.27). The conditional component (i.e., count part) of the final model indicated that soil samples collected at 96–123 DPG had significantly higher expected counts of generic E. coli compared to the baseline (RR = 2.95, 95% CI = 1.48–5.89) in the fallow treatment. Whereas no significant differences were observed between 2019 and 2020 or between 2020 and 2021, 2019 had higher expected counts of generic E. coli in the soil (RR = 1.70, 95% CI = 1.15–2.49) than 2021. The effect of treatment depended on the sampling day. No significant difference was observed among three treatments at baseline, but soil samples collected from the graze CC at 33–49 DPG (RR = 2.28, 95% CI = 0.99–5.25) and 61–82 DPG (RR = 3.44, 95% CI = 1.17–10.07) had higher expected counts of generic E. coli than those collected at baseline. However, this difference was not significant after 96 DPG. Soil samples collected from the non-graze CC at 96–123 DPG showed significantly lower expected counts of generic E. coli than those collected at baseline in fallow (RR = 0.21, 95% CI = 0.06–0.69), whereas those from the fallow treatment at 96–123 DPG showed significantly higher expected counts of generic E. coli compared to the baseline (RR = 2.95, 95% CI = 1.48–5.89).
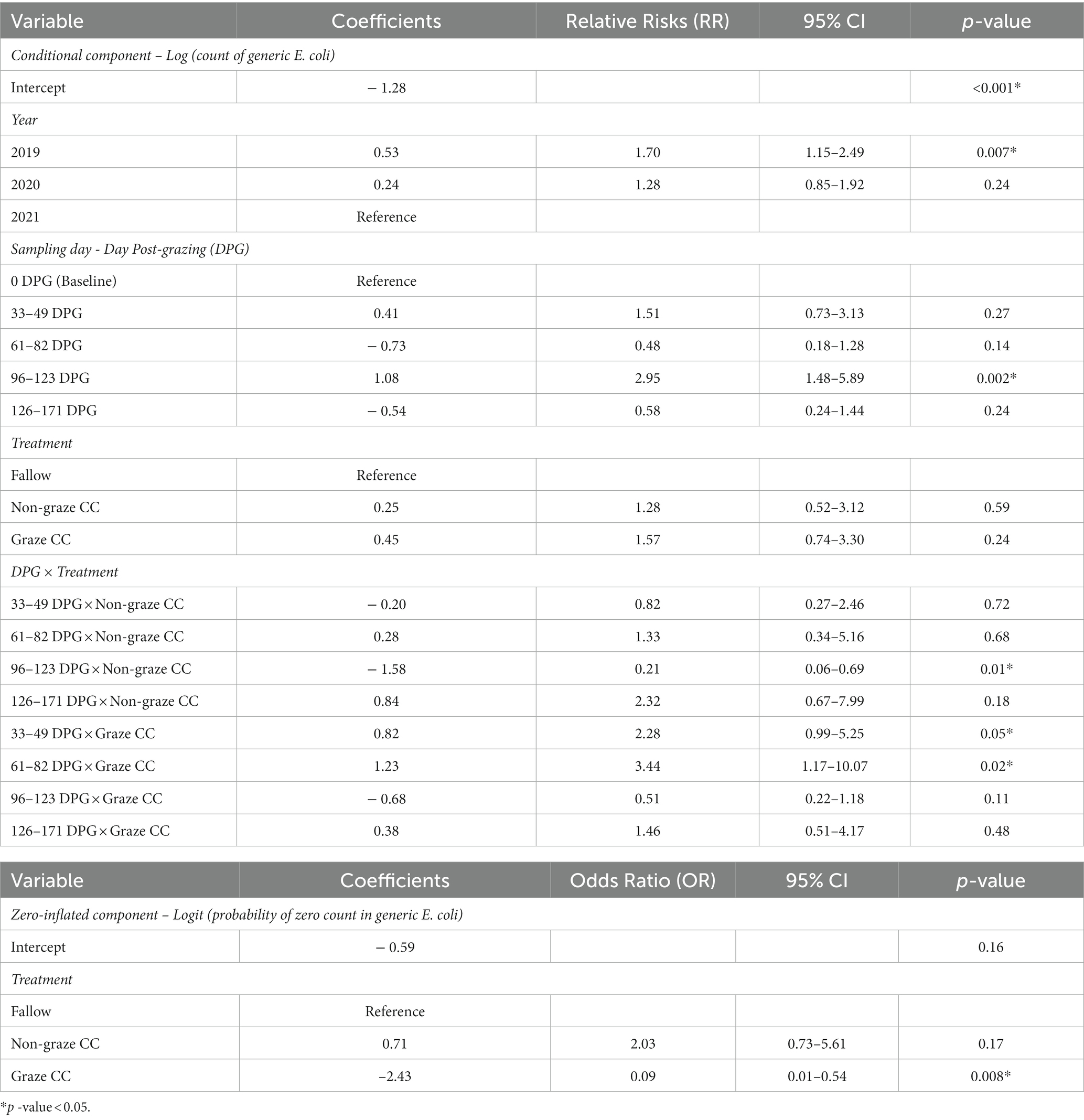
Table 5. Multivariable analyses results for the counts of generic E. coli in soil samples from the organic integrated crop-livestock experiment (2019–2021) using a mixed-effect zero-inflated negative binomial model with subplot as a random effect.
3.3 Compost effect on generic Escherichia coli in the soil
When examining the compost effect using the subset of 2020 data (i.e., 360 soil samples), similar patterns in the proportion of generic E. coli positive soil samples by sampling day were observed between soil amended with compost and without compost (Figure 2). The proportion of generic E. coli positive soil samples from the fallow treatment was 43.3% (26/60) in soil with compost and 28.3% (17/60) in soil without application of compost. In contrast, the differences in the proportions of generic E. coli positive between soil with and without compost in the non-graze CC (3.3%) and graze CC treatments (3.4%) were smaller than that observed for the fallow treatment (15%); the proportions of generic E. coli positive in non-graze CC were 38.3% (23/60) in soil with compost and 41.6% (25/60) in soil without compost, and those in the graze CC were 63.3% (38/60) in soil with compost and 66.7% (40/60) in soil without compost. The proportions and median concentrations of generic E. coli positive for the fallow treatment were higher in composted soil on 0 DPG and 33–49 DPG than in non-composted soil (Table 6). In the univariable analysis of the count of generic E. coli using mixed-effect zero-inflated negative binomial models with subplots as a random effect, compost application was not significantly associated with the count of generic E. coli in soil (RR = 0.9, 95% CI = 0.68–1.19). No further multivariable analyses were therefore conducted.
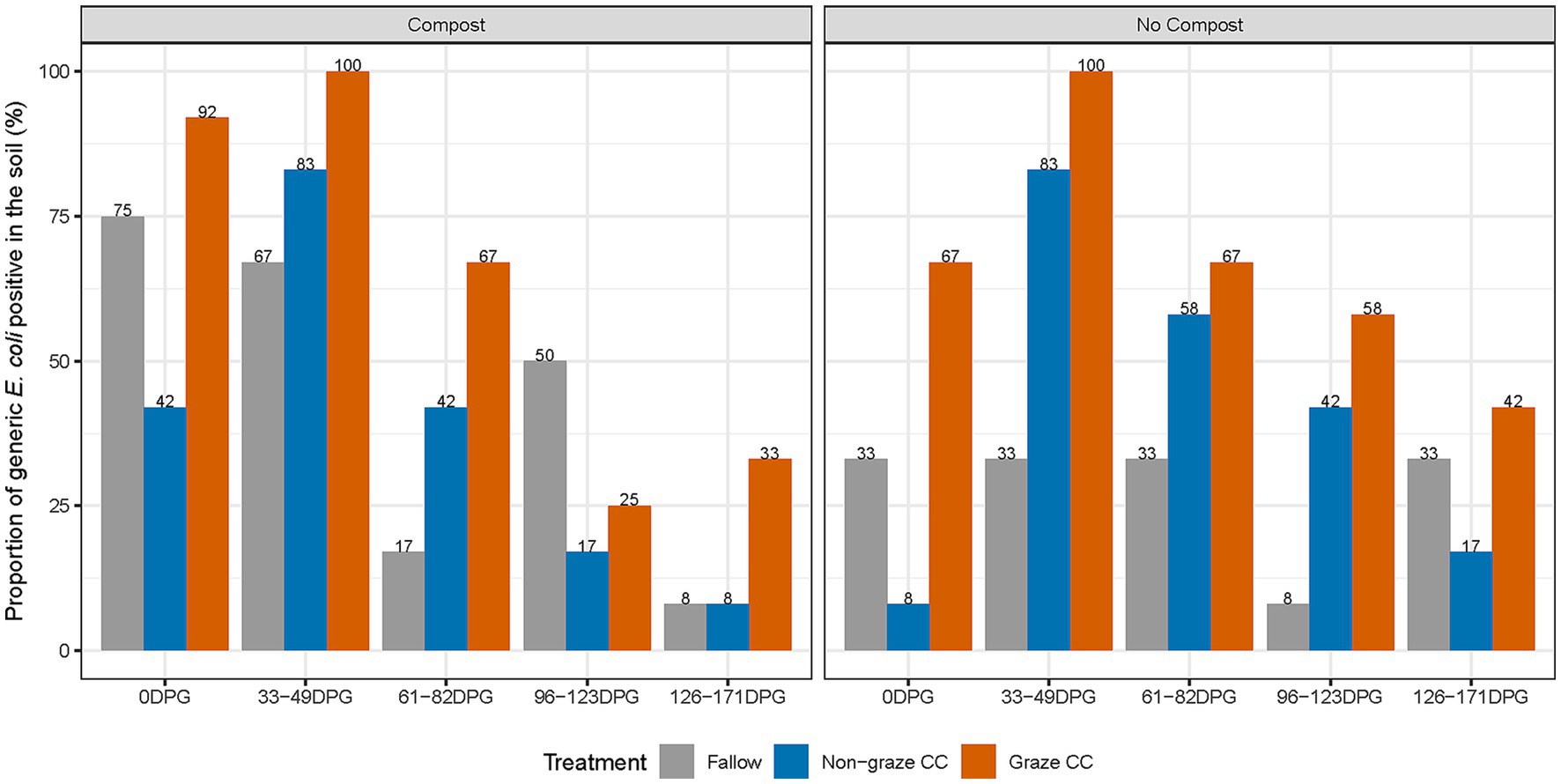
Figure 2. Proportion of generic E. coli positive (%) soil samples from the organic integrated crop-livestock experiment by treatment, sampling day, and compost application in 2020.
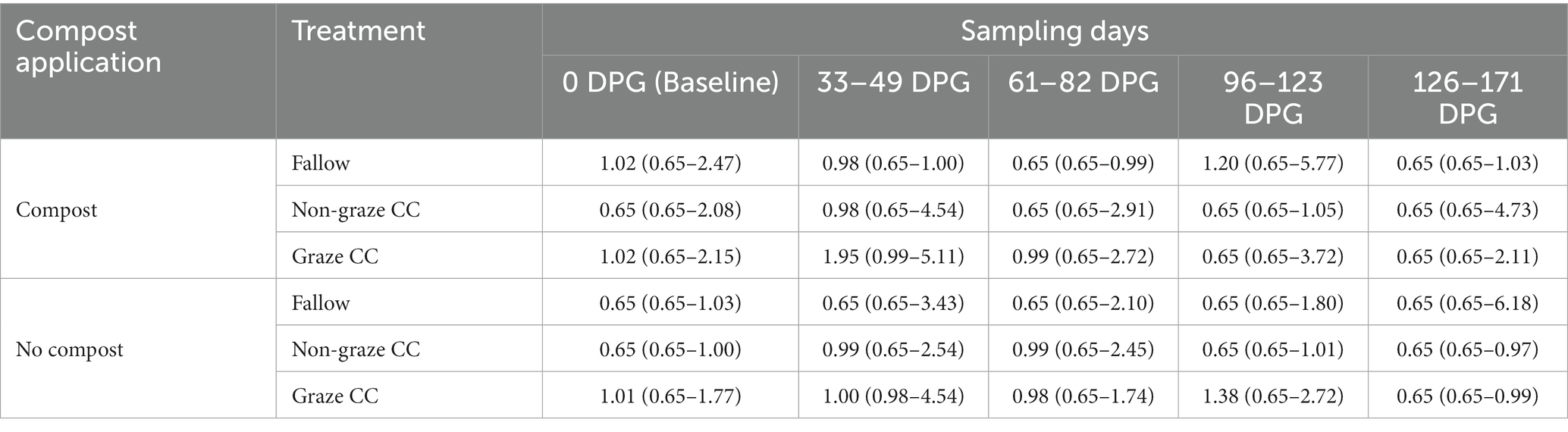
Table 6. Median and range (min – max) of generic E. coli concentrations in the soil samples (log10 MPN/100 g) from the organic integrated crop-livestock experiment by treatment, sampling day, and compost application in 2020.
3.4 Foodborne pathogens in fecal, produce, and water samples
Of the 92 pre-graze fecal samples collected from the sheep before being transported to the experimental field, 23.9% (22/92) were positive for non-O157 STEC. Similarly, of the 92 post-graze fecal samples collected from the field after the sheep were removed, 26.1% (24/92) were positive for non-O157 STEC, with no significant difference in the prevalence of non-O157 STEC compared to pre-graze fecal samples (p = 0.86). None of the pre- or post-graze fecal samples tested positive for E. coli O157:H7, whereas one post-graze fecal sample (1.1%, 1/92) tested positive for L. monocytogenes in 2019 (Table 7). None of the tomato samples or water samples tested positive for generic E. coli, non-O157 STEC, E. coli O157:H7, or L. monocytogenes.
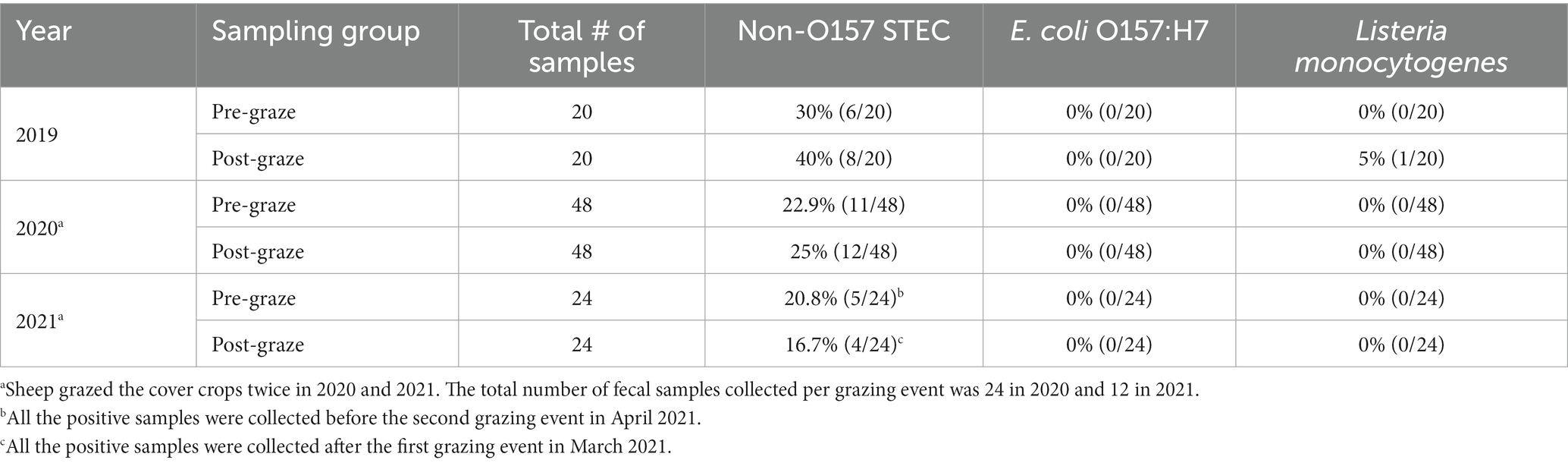
Table 7. Prevalence of foodborne pathogens (non-O157 STEC, E. coli O157:H7, and L. monocytogenes) in pre- and post-graze fecal samples in the organic integrated crop-livestock experiment (2019–2021).
4 Discussion
In this ICLS experimental study, we did not detect any foodborne pathogens (E. coli O157, non-O157 STEC, and L. monocytogenes) from harvested tomatoes or from the soil collected after grazing. Additionally, there was no difference among treatments after 96–123 DPG in the proportions of generic E. coli positive soil samples or in the counts of generic E. coli in the soil. These results support the use of the NOP 90- to 120-day interval rule between manure application and harvesting of produce in ICLS. Finally, we confirmed that compost application before cover crop seeding in the winter had no significant effect on the proportions of generic E. coli positive soil samples and counts of generic E. coli in the soil during the following growing season.
A single soil sample tested positive for non-O157 STEC during the three-year study; it was collected from the fallow treatment plot at 96–123 DPG in 2019. Wildlife (i.e., coyotes, rodents, or birds) around the field may be a potential source of non-O157 STEC contamination in the fallow treatment (Kilonzo et al., 2013; Jay-Russell et al., 2014; Atwill et al., 2015), as we observed some coyotes at night while watching over the grazing sheep, and egrets and rodents in the field multiple times when sampling. In fact, this soil sample was collected in August, which is typically well past the corn crop harvesting dates. Therefore, this could be a contributing factor to wildlife being attracted to the area. Moreover, generic E. coli concentrations spiked in the fallow treatment at 96–123 DPG in 2019, although the median concentration it was below LOQ up to 61–82 DPG, which supports the hypothesis of wildlife intrusion into the field. However, to confirm the contamination source of these positive samples, further molecular analysis of the isolates is necessary to compare relatedness of generic E. coli strains between soil and fecal samples from grazed sheep.
As for the effect of sheep grazing on the prevalence or persistence of foodborne pathogens in soil, we did not observe any foodborne pathogens in soil samples from the grazed fields. On the other hand, we isolated non-O157 STEC from pre- and post-graze sheep fecal samples. The intermittent shedding of non-O157 STEC could be explained by diet changes or transportation stress of the sheep to the trial fields. A few studies reported that diet components, abrupt diet changes, or the stress due to transport influenced E. coli O157 shedding in cattle feces (Callaway et al., 2003; Bach et al., 2004; Jacob et al., 2008). However, no significant difference in the proportion of non-O157 STEC positive between pre- and post-graze fecal samples was observed in the present study. This may be due to the short grazing period (i.e., three to eight days each growing season) to assess the effect of diet change. Furthermore, the single L. monocytogenes positive fecal sample detected after the four-day grazing. L. monocytogenes is an important foodborne pathogen that can be isolated from ruminant feces; a lower prevalence has been reported in healthy sheep and farm environment (e.g., feed, soil, and water) compared to cattle herds and its environment (e.g., feed, soil, and water) (Nightingale et al., 2004; Hurtado et al., 2017). The transmission dynamics between ruminants and its environment remains unclear, but natural environmental sources such as water or soil, or management practices (contaminated feed) are potential routes of transmission (Bagatella et al., 2022). Previous studies reported that water irrigation sources, higher rainfall, recent worker activity in fields, or observation of wildlife around fields were all significant risk factors for the higher prevalence of L. monocytogenes in produce fields (Strawn et al., 2013; Weller et al., 2015; Harrand et al., 2020). Additionally, L. monocytogenes is known to be naturally harbored in soils for long periods, which can make it difficult to distinguish the source of pathogen contamination (Linke et al., 2014). Possibly, since none of the pre-graze fecal and soil samples tested positive for L. monocytogenes in this study, diet components, such as moisture contents or pH of the cover crops in the field, could be relevant to presence of L. monocytogenes in post-graze fecal samples (Ivanek et al., 2006).
More than 50% of generic E. coli positive samples were associated with grazing. The graze CC treatment showed the highest proportion of generic E. coli positive in the soil at 33–49 DPG or/and 61–82 DPG, with the highest concentration (log10 MPN/100 g) at 33–49 DPG. This corresponded to the results from our previous observational ICLS study done in a working farm which showed the highest mean generic E. coli concentration (i.e., 3.7 log10 MPN/g) in the grazed soil on 48 DPG (Patterson et al., 2018). Interestingly, we found that the fallow treatment had a significantly higher probability of having generic E. coli in the soil than the non-graze CC treatment, although the overall concentrations of generic E. coli in both treatments were very low. The only difference between the two treatments was the use of cover crops during winter before the growing seasons in the non-graze CC treatment; the two treatment groups had otherwise the same environmental conditions. These results may support the antibacterial effect of cover crops, especially in reducing generic E. coli contamination of the soil. Cover crops are known to suppress soil-borne plant pathogens (Liu et al., 2021), but studies are limited regarding their effects on the survival of bacterial foodborne pathogens. Results from a study examining the influence of cover crops (i.e., hairy vetch, rye, and crimson clover) on the dynamics of E. coli and Listeria innocua in the soil used for vegetable production showed some attenuation effects on E. coli in the soil, although the results differed depending on the species of cover crop, year, and sampling day (Reed-Jones et al., 2016). Similarly, an experimental study using three different chopped cover crops (i.e., buckwheat, mustard greens, and sun hemp) in contaminated soil showed significant reduction in the concentration of generic E. coli within 30 to 40 days after the tillage when compared to the soil without cover crop (Zhao et al., 2023). This antimicrobial effect was linked to the secretion of secondary metabolites by cover crops, which functions as a defense mechanism against pathogen contamination (Guerrieri et al., 2019).
We observed interaction between treatment and sampling day in defining the counts of generic E. coli in the soil. In particular, the counts of generic E. coli of the graze CC treatment differed significantly over time with the higher risk of generic E. coli contamination in the soil by 61–82 DPG but not after 96 DPG, compared to the fallow treatment. The non-graze CC treatment showed significantly lower counts of generic E. coli at 96–123 DPG, but this is most likely due to the high generic E. coli value in the fallow treatment (i.e., reference treatment group of the model) at 96–123 DPG; when comparing the sampling day effect within the non-graze CC treatment, there was no significant difference observed in the counts of generic E. coli in the soil across all other sampling days compared to the baseline of the non-graze CC treatment. Therefore, the graze CC was the only treatment that had significant changes over time in the counts of generic E. coli in the soil. The findings from our previous observational study were similar to the present study, as we found that the mean concentration of generic E. coli decreased under the limit of detection (i.e., 2.3 MPN/g) in the soil 84–111 days after grazing (Patterson et al., 2018). These results from both studies show that the waiting time between grazing and harvesting of 90–120 days in the NOP standards would be appropriate for grazing events on cover crops in ICLS.
Applying commercially produced composted organic poultry manure in the field before growing cover crops did not change the concentrations of generic E. coli over time in comparison to non-composted areas. Although we did not test the commercial compost to ensure that it met the required quality criteria from the U.S. Environmental Protection Agency (i.e., less than 1,000 MPN/g fecal coliforms) (U.S. Environmental Protection Agency, 2006), the organic compost product would have followed the NOP guidance for treatment, which requires composted animal material to be maintained at a temperature of minimum 55°C for 3 days in static aerated piles or for 15 days using windrow composting system (USDA-AMS NOP, 2011a). This process ensures the inactivation of pathogens through elevated temperatures (Berry et al., 2013).
Generic E. coli was used as a surrogate for pathogenic E. coli, as well as an indicator of fecal contamination in this study, which has been generally accepted in the scientific community due to the low and heterogeneous distribution of pathogenic E. coli in nature (Mubiru et al., 2000; Park et al., 2015; Allende et al., 2017). However, studies have shown conflicting results on similarities between the survival of generic E. coli and pathogenic E. coli in soil. A meta-analysis found that the average decline rate of commensal E. coli concentrations (log CFU/day) in soil was significantly lower than that of pathogenic E. coli (Franz et al., 2014). In contrast, a predictive model assessing the survival of E. coli O157 and generic E. coli in amended soils under various environmental conditions in the mid-Atlantic US showed similar declining rates between them (Pang et al., 2020). In addition, a random forest model predicting survival duration of generic E. coli and attenuated E. coli O157 under various weather and spatiotemporal predictors investigated through 12 different field trials in the Mid-Atlantic US also found that the factors (i.e., site, year, season) exhibiting the greatest prediction were similar for both pathogens (Sharma et al., 2019). Therefore, although soil characteristics should be further included in the models in future studies, using generic E. coli as a proxy of pathogenic E. coli still seems acceptable.
This study had some important strengths. First, it was a three-year longitudinal field trial investigating natural contamination of soil and produce by foodborne pathogens through sheep grazing following real ICLS schedules. Its results therefore better reflect real-world field conditions than other laboratory or greenhouse experimental studies. Several studies pointed out that the survival of E. coli under laboratory conditions did not mimic real-world conditions, and that directly applying the results to natural field conditions may not be suitable (Franz et al., 2014; Çekiç et al., 2017). Second, the design of this study was experimental with randomly assigned treatments in the same field, guaranteeing internal validity among treatment groups by providing comparable environmental conditions (e.g., weather, location of the field). Lastly, this study adds to the scarce published scientific literature assessing the risks of ICLS using sheep on microbial contamination of soil and produce (Hoar et al., 2013).
This study also had some limitations, as we could not assess the effects of soil characteristics such as pH, temperature, or moisture that may have differed among treatment groups, and several previous experimental studies showed that those characteristics had significant impacts on the survival of foodborne or indicator pathogens (Franz et al., 2014; Park et al., 2016). Validating this study’s results in other regions of the US is necessary before they can be generalized, as a single field in California may not be representative of other organic ICLS in the US. Several studies have indeed shown that the survival of pathogens in manure-amended soil was associated with various spatiotemporal and weather factors (Park et al., 2015; Sharma et al., 2019). In addition, there might be a cumulative effect of continuous ICLS farming in the same field over time, which should be investigated further, as we observed that the proportion of generic E. coli positive soil samples collected in November continuously increased every year from 2019 to 2021.
5 Conclusion
Our three-year experimental study confirmed that the effect of sheep grazing on foodborne pathogen contamination in ICLS is minimal, and the concentration of generic E. coli in the grazed CC soil decreased and comparable to the fallow or non-graze CC treatments by 96–123 days post grazing. This supports the application of the NOP 90–120 days rule regarding the time-interval between raw manure application and harvesting to ICLS. In addition, we observed that compost application before cover crop seeding in the winter did not affect the generic E. coli concentration in the soil during the following growing season. Further studies comparing the genetic associations between the foodborne pathogens isolated in fecal and soil samples would be necessary to identify the source of foodborne pathogen contamination. Finally, the effect of meteorological and environmental factors should be further investigated in other regions of the US.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of the University of California, Davis. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SC: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. MTJ-R: Conceptualization, Funding acquisition, Methodology, Resources, Writing – review & editing. CC-K: Methodology, Writing – review & editing. JDF: Formal analysis, Writing – review & editing. VH: Data curation, Methodology, Writing – review & editing. PA: Data curation, Methodology, Writing – review & editing. SRW: Conceptualization, Methodology, Writing – review & editing. ACMG: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. NT: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. AP: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Project or Publication was made possible by the U.S. Department of Agriculture’s (USDA) Agricultural Marketing Service through grants AM190100XXXXG08. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the USDA.
Acknowledgments
We acknowledge Amanda Taylor, James Stover, Thais de Melo Ramos, Nicholas La, Olivia Thorn, and Thomas Luder for assistance in the laboratory and field work. We would like to thank Gaudin’s lab personnel for the assistance with field work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1343101/full#supplementary-material
References
Allende, A., Castro-Ibáñez, I., Lindqvist, R., Gil, M. I., Uyttendaele, M., and Jacxsens, L. (2017). Quantitative contamination assessment of Escherichia coli in baby spinach primary production in Spain: effects of weather conditions and agricultural practices. Int. J. Food Microbiol. 257, 238–246. doi: 10.1016/j.ijfoodmicro.2017.06.027
Atwill, E. R., Chase, J. A., Oryang, D., Bond, R. F., Koike, S. T., Cahn, M. D., et al. (2015). Transfer of Escherichia coli O157:H7 from simulated wildlife scat onto Romaine lettuce during foliar irrigation. J. Food Prot. 78, 240–247. doi: 10.4315/0362-028X.JFP-14-277
Avineon, Inc. and US Environmental Protection Agency (2013). Most probable number (MPN) calculator version 2.0.
Bach, S. J., McALLISTER, T. A., Mears, G. J., and Schwartzkopf-Genswein, K. S. (2004). Long-haul transport and lack of preconditioning increases fecal shedding of Escherichia coli and Escherichia coli O157:H7 by calves†. J. Food Prot. 67, 672–678. doi: 10.4315/0362-028X-67.4.672
Bagatella, S., Tavares-Gomes, L., and Oevermann, A. (2022). Listeria monocytogenes at the interface between ruminants and humans: a comparative pathology and pathogenesis review. Vet. Pathol. 59, 186–210. doi: 10.1177/03009858211052659
Bell, L. W., and Moore, A. D. (2012). Integrated crop–livestock systems in Australian agriculture: trends, drivers and implications. Agric. Syst. 111, 1–12. doi: 10.1016/j.agsy.2012.04.003
Berry, E. D., Millner, P. D., Wells, J. E., Kalchayanand, N., and Guerini, M. N. (2013). Fate of naturally occurring Escherichia coli O157:H7 and other zoonotic pathogens during minimally managed bovine feedlot manure composting processes. J. Food Prot. 76, 1308–1321. doi: 10.4315/0362-028X.JFP-12-364
Brewer, K. M., Muñoz-Araya, M., Martinez, I., Marshall, K. N., and Gaudin, A. C. (2023). Long-term integrated crop-livestock grazing stimulates soil ecosystem carbon flux, increasing subsoil carbon storage in California perennial agroecosystems. Geoderma 438:116598. doi: 10.1016/j.geoderma.2023.116598
Brooks, M., Kristensen, K., Benthem, K. J., Magnusson, A., Berg, C. W., Nielsen, A., et al. (2017). glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9:378. doi: 10.32614/RJ-2017-066
Callaway, T. R., Elder, R. O., Keen, J. E., Anderson, R. C., and Nisbet, D. J. (2003). Forage feeding to reduce Preharvest Escherichia coli populations in cattle, a review. J. Dairy Sci. 86, 852–860. doi: 10.3168/jds.S0022-0302(03)73668-6
Carstens, C. K., Salazar, J. K., and Darkoh, C. (2019). Multistate outbreaks of foodborne illness in the United States associated with fresh produce from 2010 to 2017. Front. Microbiol. 10:2667. doi: 10.3389/fmicb.2019.02667
Çekiç, S. K., De, J., Jubair, M., and Schneider, K. R. (2017). Persistence of indigenous Escherichia coli in raw bovine manure-amended soil. J. Food Prot. 80, 1562–1573. doi: 10.4315/0362-028X.JFP-17-033
Centers for Disease Control and Prevention (2022). Summary of possible multistate enteric (intestinal) disease outbreaks in 2017–2020. Available at: https://www.cdc.gov/foodsafety/outbreaks/multistate-outbreaks/annual-summaries/annual-summaries-2017-2020.html (Accessed September 27, 2022).
Cooley, M. B., Jay-Russell, M., Atwill, E. R., Carychao, D., Nguyen, K., Quiñones, B., et al. (2013). Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS One 8:e65716. doi: 10.1371/journal.pone.0065716
Cooley, M. B., Quiñones, B., Oryang, D., Mandrell, R. E., and Gorski, L. (2014). Prevalence of Shiga toxin producing Escherichia coli, Salmonella enterica, and Listeria monocytogenes at public access watershed sites in a California central coast agricultural region. Front. Cell. Infect. Microbiol. 4:30. doi: 10.3389/fcimb.2014.00030
Da Silva, F. D., Amado, T. J. C., Bredemeier, C., Bremm, C., Anghinoni, I., and Carvalho, P. C. D. F. (2014). Pasture grazing intensity and presence or absence of cattle dung input and its relationships to soybean nutrition and yield in integrated crop–livestock systems under no-till. Eur. J. Agron. 57, 84–91. doi: 10.1016/j.eja.2013.10.009
FDA (2015). FSMA final rule on produce safety - standards for the growing, harvesting, packing, and holding of produce for human consumption. Available at: https://www.federalregister.gov/documents/2015/11/27/2015-28159/standards-for-the-growing-harvesting-packing-and-holding-of-produce-for-human-consumption
Franz, E., Schijven, J., de Roda Husman, A. M., and Blaak, H. (2014). Meta-regression analysis of commensal and pathogenic Escherichia coli survival in soil and water. Environ. Sci. Technol. 48, 6763–6771. doi: 10.1021/es501677c
Franz, E., and Van Bruggen, A. H. C. (2008). Ecology of E. coli O157:H7 and Salmonella enterica in the primary vegetable production chain. Crit. Rev. Microbiol. 34, 143–161. doi: 10.1080/10408410802357432
Franz, E., van Diepeningen, A. D., de Vos, O. J., and van Bruggen, A. H. C. (2005). Effects of cattle feeding regimen and soil management type on the fate of Escherichia coli O157:H7 and Salmonella enterica Serovar typhimurium in manure, manure-amended soil, and lettuce. Appl. Environ. Microbiol. 71, 6165–6174. doi: 10.1128/AEM.71.10.6165-6174.2005
Franzluebbers, A. J., Sulc, R. M., and Russelle, M. P. (2011). “Opportunities and challenges for integrating north-American crop and livestock systems” in Grassland productivity and ecosystem services. eds. G. Lemaire, J. Hodgson, and A. Chabbi (UK: CABI), 208–218.
Garrett, R. D., Niles, M. T., Gil, J. D. B., Gaudin, A., Chaplin-Kramer, R., Assmann, A., et al. (2017). Social and ecological analysis of commercial integrated crop livestock systems: current knowledge and remaining uncertainty. Agric. Syst. 155, 136–146. doi: 10.1016/j.agsy.2017.05.003
Garrett, R. D., Ryschawy, J., Bell, L. W., Cortner, O., Ferreira, J., Garik, A. V. N., et al. (2020). Drivers of decoupling and recoupling of crop and livestock systems at farm and territorial scales. Ecol. Soc. 25:art24. doi: 10.5751/ES-11412-250124
Guerrieri, A., Dong, L., and Bouwmeester, H. J. (2019). Role and exploitation of underground chemical signaling in plants. Pest Manag. Sci. 75, 2455–2463. doi: 10.1002/ps.5507
Harrand, A. S., Strawn, L. K., Illas-Ortiz, P. M., Wiedmann, M., and Weller, D. L. (2020). Listeria monocytogenes prevalence varies more within fields than between fields or over time on conventionally farmed New York produce fields. J. Food Prot. 83, 1958–1966. doi: 10.4315/JFP-20-120
Harvey, R. R., Zakhour, C. M., and Gould, L. H. (2016). Foodborne disease outbreaks associated with organic foods in the United States. J. Food Prot. 79, 1953–1958. doi: 10.4315/0362-028X.JFP-16-204
Hilimire, K. (2011). Integrated crop/livestock agriculture in the United States: a review. J. Sustain. Agric. 35, 376–393. doi: 10.1080/10440046.2011.562042
Hoar, B. R., Atwill, E. R., Carlton, L., Celis, J., Carabez, J., and Nguyen, T. (2013). Buffers between grazing sheep and leafy crops augment food safety. Calif. Agric. 67, 104–109. doi: 10.3733/ca.v067n02p104
Hurtado, A., Ocejo, M., and Oporto, B. (2017). Salmonella spp. and Listeria monocytogenes shedding in domestic ruminants and characterization of potentially pathogenic strains. Vet. Microbiol. 210, 71–76. doi: 10.1016/j.vetmic.2017.09.003
Hutchison, M. L., Walters, L. D., Avery, S. M., Munro, F., and Moore, A. (2005). Analyses of livestock production, waste storage, and pathogen levels and Prevalences in farm manures. Appl. Environ. Microbiol. 71, 1231–1236. doi: 10.1128/AEM.71.3.1231-1236.2005
Ingham, S. C., Losinski, J. A., Andrews, M. P., Breuer, J. E., Breuer, J. R., Wood, T. M., et al. (2004). Escherichia coli contamination of vegetables grown in soils fertilized with noncomposted bovine manure: garden-scale studies. Appl. Environ. Microbiol. 70, 6420–6427. doi: 10.1128/AEM.70.11.6420-6427.2004
Ivanek, R., Gröhn, Y. T., and Wiedmann, M. (2006). Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog. Dis. 3, 319–336. doi: 10.1089/fpd.2006.3.319
Jacob, M. E., Fox, J. T., Drouillard, J. S., Renter, D. G., and Nagaraja, T. G. (2008). Effects of dried distillers’ grain on fecal prevalence and growth of Escherichia coli O157 in batch culture fermentations from cattle. Appl. Environ. Microbiol. 74, 38–43. doi: 10.1128/AEM.01842-07
Jacobsen, C. S., and Bech, T. B. (2012). Soil survival of Salmonella and transfer to freshwater and fresh produce. Food Res. Int. 45, 557–566. doi: 10.1016/j.foodres.2011.07.026
Jay-Russell, M. T. (2013). What is the risk from wild animals in food-borne pathogen contamination of plants? CABI Rev. 2013, 1–16. doi: 10.1079/PAVSNNR20138040
Jay-Russell, M. T., Hake, A. F., Bengson, Y., Thiptara, A., and Nguyen, T. (2014). Prevalence and characterization of Escherichia coli and Salmonella strains isolated from stray dog and coyote feces in a major leafy greens production region at the United States-Mexico border. PLoS One 9:e113433. doi: 10.1371/journal.pone.0113433
Kawasaki, S., Horikoshi, N., Okada, Y., Takeshita, K., Sameshima, T., and Kawamoto, S. (2005). Multiplex PCR for simultaneous detection of Salmonella spp., Listeria monocytogenes, and Escherichia coli O157:H7 in meat samples. J. Food Prot. 68, 551–556. doi: 10.4315/0362-028X-68.3.551
Kilonzo, C., Li, X., Vivas, E. J., Jay-Russell, M. T., Fernandez, K. L., and Atwill, E. R. (2013). Fecal shedding of zoonotic food-borne pathogens by wild rodents in a major agricultural region of the Central California coast. Appl. Environ. Microbiol. 79, 6337–6344. doi: 10.1128/AEM.01503-13
Lemaire, G., Franzluebbers, A., Carvalho, P. C. D. F., and Dedieu, B. (2014). Integrated crop–livestock systems: strategies to achieve synergy between agricultural production and environmental quality. Agric. Ecosyst. Environ. 190, 4–8. doi: 10.1016/j.agee.2013.08.009
Linke, K., Rückerl, I., Brugger, K., Karpiskova, R., Walland, J., Muri-Klinger, S., et al. (2014). Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 80, 5583–5592. doi: 10.1128/AEM.01018-14
Liu, X., Hannula, S. E., Li, X., Hundscheid, M. P. J., Klein Gunnewiek, P. J. A., Clocchiatti, A., et al. (2021). Decomposing cover crops modify root-associated microbiome composition and disease tolerance of cash crop seedlings. Soil Biol. Biochem. 160:108343. doi: 10.1016/j.soilbio.2021.108343
Medina-Roldán, E., Arredondo, J. T., Huber-Sannwald, E., Chapa-Vargas, L., and Olalde-Portugal, V. (2008). Grazing effects on fungal root symbionts and carbon and nitrogen storage in a shortgrass steppe in Central Mexico. J. Arid Environ. 72, 546–556. doi: 10.1016/j.jaridenv.2007.07.005
Moraine, M., Duru, M., Nicholas, P., Leterme, P., and Therond, O. (2014). Farming system design for innovative crop-livestock integration in Europe. Animal 8, 1204–1217. doi: 10.1017/S1751731114001189
Mubiru, D. N., Coyne, M. S., and Grove, J. H. (2000). Mortality of Escherichia coli O157:H7 in two soils with different physical and chemical properties. J. Environ. Qual. 29, 1821–1825. doi: 10.2134/jeq2000.00472425002900060012x
Nicholson, F. A., Groves, S. J., and Chambers, B. J. (2005). Pathogen survival during livestock manure storage and following land application. Bioresour. Technol. 96, 135–143. doi: 10.1016/j.biortech.2004.02.030
Nightingale, K. K., Schukken, Y. H., Nightingale, C. R., Fortes, E. D., Ho, A. J., Her, Z., et al. (2004). Ecology and transmission of Listeria monocytogenes infecting ruminants and in the farm environment. Appl. Environ. Microbiol. 70, 4458–4467. doi: 10.1128/AEM.70.8.4458-4467.2004
Nyström, T., and Neidhardt, F. C. (1992). Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 6, 3187–3198. doi: 10.1111/j.1365-2958.1992.tb01774.x
Pang, H., Mokhtari, A., Chen, Y., Oryang, D., Ingram, D. T., Sharma, M., et al. (2020). A predictive model for survival of Escherichia coli O157:H7 and Generic E. coli in soil amended with untreated animal manure. Risk Anal. 40, 1367–1382. doi: 10.1111/risa.13491
Park, S., Navratil, S., Gregory, A., Bauer, A., Srinath, I., Szonyi, B., et al. (2015). Multifactorial effects of ambient temperature, precipitation, farm management, and environmental factors determine the level of generic Escherichia coli contamination on Preharvested spinach. Appl. Environ. Microbiol. 81, 2635–2650. doi: 10.1128/AEM.03793-14
Park, Y., Pachepsky, Y., Shelton, D., Jeong, J., and Whelan, G. (2016). Survival of manure-borne Escherichia coli and fecal coliforms in soil: temperature dependence as affected by site-specific factors. J. Environ. Qual. 45, 949–957. doi: 10.2134/jeq2015.08.0427
Partyka, M. L., Bond, R. F., Chase, J. A., and Atwill, E. R. (2018). Spatial and temporal variability of bacterial indicators and pathogens in six California reservoirs during extreme drought. Water Res. 129, 436–446. doi: 10.1016/j.watres.2017.11.038
Paton, A. W., and Paton, J. C. (1998). Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, Enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36, 598–602. doi: 10.1128/JCM.36.2.598-602.1998
Patterson, L., Navarro-Gonzalez, N., Jay-Russell, M. T., Aminabadi, P., Antaki-Zukoski, E., and Pires, A. F. A. (2018). Persistence of Escherichia coli in the soil of an organic mixed crop-livestock farm that integrates sheep grazing within vegetable fields. Zoonoses Public Health 65, 887–896. doi: 10.1111/zph.12503
Pires, A. F. A., Ramos, T. D. M., Baron, J. N., Millner, P. D., Pagliari, P. H., Hutchinson, M., et al. (2023). Risk factors associated with the prevalence of Shiga-toxin-producing Escherichia coli in manured soils on certified organic farms in four regions of the USA. Front. Sustain. Food Syst. 7:1125996. doi: 10.3389/fsufs.2023.1125996
Ramos, T. D. M., Jay-Russell, M. T., Millner, P. D., Baron, J. N., Stover, J., Pagliari, P., et al. (2021). Survival and persistence of foodborne pathogens in manure-amended soils and prevalence on fresh produce in certified organic farms: a multi-regional baseline analysis. Front. Sustain. Food Syst. 5:674767. doi: 10.3389/fsufs.2021.674767
Reed-Jones, N. L., Marine, S. C., Everts, K. L., and Micallef, S. A. (2016). Effects of cover crop species and season on population dynamics of Escherichia coli and Listeria innocua in soil. Appl. Environ. Microbiol. 82, 1767–1777. doi: 10.1128/AEM.03712-15
Schipanski, M. E., Barbercheck, M., Douglas, M. R., Finney, D. M., Haider, K., Kaye, J. P., et al. (2014). A framework for evaluating ecosystem services provided by cover crops in agroecosystems. Agric. Syst. 125, 12–22. doi: 10.1016/j.agsy.2013.11.004
Sharma, M., Millner, P. D., Hashem, F., Vinyard, B. T., East, C. L., Handy, E. T., et al. (2019). Survival of Escherichia coli in manure-amended soils is affected by spatiotemporal, agricultural, and weather factors in the mid-Atlantic United States. Appl. Environ. Microbiol. 85, e02392–e02318. doi: 10.1128/AEM.02392-18
Strawn, L. K., Gröhn, Y. T., Warchocki, S., Worobo, R. W., Bihn, E. A., and Wiedmann, M. (2013). Risk factors associated with Salmonella and Listeria monocytogenes contamination of produce fields. Appl. Environ. Microbiol. 79, 7618–7627. doi: 10.1128/AEM.02831-13
U.S. Environmental Protection Agency (2006). Method 1681: Fecal coliforms in sewage sludge (biosolids) by multiple-tube fermentation using A-1 medium. Available at: https://www.epa.gov/sites/default/files/2015-08/documents/method_1681_2006.pdf.
USDA (2000). National Organic Program Part 205 subpart G the National List of allowed and prohibited substances. Available at: https://www.ams.usda.gov/rules-regulations/organic/national-list (Accessed August 28, 2023).
USDA National Agricultural Statistics Service (2019). Pacific region sheep and goats. Available at: https://ucanr.edu/sites/nichemarketing/files/341648.pdf.
USDA-AMS NOP (2011a). Guidance compost and Vermicompost in organic crop production. Available at: https://www.ams.usda.gov/sites/default/files/media/5021.pdf.
USDA-AMS NOP (2011b). Guidance processed animal manures in organic crop production. Available at: https://www.ams.usda.gov/sites/default/files/media/5006.pdf.
Weller, D., Wiedmann, M., and Strawn, L. K. (2015). Irrigation is significantly associated with an increased prevalence of Listeria monocytogenes in produce production environments in New York state. J. Food Prot. 78, 1132–1141. doi: 10.4315/0362-028X.JFP-14-584
Keywords: grazing, food safety, fresh produce, sustainable agriculture, soil, sheep
Citation: Cheong S, Jay-Russell MT, Chandler-Khayd C, Di Francesco J, Haghani V, Aminanadi P, Williams SR, Gaudin ACM, Tautges N and Pires AFA (2024) Presence of foodborne pathogens and survival of generic Escherichia coli in an organic integrated crop-livestock system. Front. Sustain. Food Syst. 8:1343101. doi: 10.3389/fsufs.2024.1343101
Edited by:
Vasco Augusto Pilão Cadavez, Agricultural College of Bragança, PortugalReviewed by:
Laurent Guillier, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceGuodong Zhang, United States Food and Drug Administration, United States
Arícia Possas, University of Cordoba, Spain
Copyright © 2024 Cheong, Jay-Russell, Chandler-Khayd, Di Francesco, Haghani, Aminanadi, Williams, Gaudin, Tautges and Pires. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alda F. A. Pires, apires@ucdavis.edu
 Sejin Cheong
Sejin Cheong Michele T. Jay-Russell
Michele T. Jay-Russell Carolyn Chandler-Khayd
Carolyn Chandler-Khayd Juliette Di Francesco
Juliette Di Francesco Viktoria Haghani
Viktoria Haghani Peiman Aminanadi
Peiman Aminanadi Sequoia R. Williams
Sequoia R. Williams Amélie C. M. Gaudin
Amélie C. M. Gaudin Nicole Tautges4
Nicole Tautges4  Alda F. A. Pires
Alda F. A. Pires