Genomics-assisted speed breeding for crop improvement: present and future
- 1Laboratory for Biotechnology, Institute of Field and Vegetable Crops, National Institute of the Republic of Serbia, Novi Sad, Serbia
- 2Legumes Department, Institute of Field and Vegetable Crops, National Institute of the Republic of Serbia, Novi Sad, Serbia
Global agricultural productivity and food security are threatened by climate change, the growing world population, and the difficulties posed by the pandemic era. To overcome these challenges and meet food requirements, breeders have applied and implemented different advanced techniques that accelerate plant development and increase crop selection effectiveness. However, only two or three generations could be advanced annually using these approaches. Speed breeding (SB) is an innovative and promising technology to develop new varieties in a shorter time, utilizing the manipulation of controlled environmental conditions. This strategy can reduce the generation length from 2.5 to 5 times compared to traditional methods and accelerate generation advancement and crop improvement, accommodating multiple generations of crops per year. Beside long breeding cycles, SB can address other challenges related to traditional breeding, such as response to environmental conditions, disease and pest management, genetic uniformity, and improving resource efficiency. Combining genomic approaches such as marker-assisted selection, genomic selection, and genome editing with SB offers the capacity to further enhance breeding efficiency by reducing breeding cycle time, enabling early phenotypic assessment, efficient resource utilization, and increasing selection accuracy and genetic gain per year. Genomics-assisted SB holds the potential to revolutionize plant breeding by significantly accelerating the identification and selection of desirable genetic traits, expediting the development of improved crop varieties crucial for addressing global agricultural challenges.
1 Introduction
Global climate change is a leading aspect threatening agricultural productivity worldwide, along with the challenges of meeting the food requirements of the world’s expanding population. Increased variations in rainfall and raised global temperature, along with increasing unpredictability in growing conditions, have caused the emergence, changed distribution and prevalence of pests and diseases. Abiotic and biotic stresses and loss of agricultural land caused by climate changes have direct or indirect negative impacts on agricultural production, usually causing yield losses (Hoegh-Guldberg et al., 2019; Paul et al., 2019; Yu et al., 2021). Climate change has likely already affected global food production by reducing the yields of the top ten global crops, including barley, cassava, maize, oil palm, rapeseed, rice, sorghum, soybean, sugarcane and wheat, by 3–12% by mid-century and 11–25% by century’s end, under a vigorous warming scenario (Ray et al., 2019). The study also found that impacts are mostly negative in Europe, Southern Africa and Australia (Ray et al., 2019). Thus, efficient systems for delivering improved varieties are crucial for successful adaptation and keeping up with shifting environmental conditions. On the other side, global population growth increased demand for food production and expanded the need for crops with improved nutritional profiles to address malnutrition and dietary deficiencies and to adapt to changing dietary preferences (Fróna et al., 2019). Also, expanding population increased urbanization and put pressure on available agricultural land. Furthermore, in the pandemic era, food security has continued to decline, affecting millions worldwide. The pandemic has affected disruptions in supply chains, labor shortages, and economic challenges, which contributed to rising food prices, affecting affordability for vulnerable populations (FAO, IFAD, UNICEF, WFP and WHO, 2021). Thus, the pandemic underscored the importance of building resilient and sustainable food systems to withstand increased global food insecurity. Therefore, to overcome these challenges and contribute to long-term global food security, the development of superior crop varieties is highly required (Lenaerts et al., 2019; Kondić-Špika et al., 2022b; Cvejić et al., 2023; Potts et al., 2023).
The major bottleneck in the process of breeding is long generation time. For most crop plants, conventional breeding often takes more than a decade to develop and release new cultivars (Jähne et al., 2020). Following the crossing of selected parent plants, 4–6 successive generations are typically required to reach a fixed homozygous state for the identification of a superior genotype. This process also involves multi-year testing in replicated field trials at multiple locations for the detection of candidate genotypes across a wide range of conditions (Voss-Fels et al., 2019). The development of hybrids in cross-pollinated crops follows similar timetables, wherein up to 10 years can pass between parental selection and inbreeding (Shimelis and Laing, 2012). Shortening breeding cycles is crucial to address the challenges of traditional breeding, particularly the prolonged generation times, allowing for faster and more efficient development of crop varieties in response to evolving agricultural needs.
It is considered that shortening the selection cycle’s duration has the biggest effect on genetic gain when considering the cost–benefit ratio (Bonnecarrere et al., 2019). Variable agroecological conditions usually allow only one crop cycle per year, while in some tropical conditions, it is possible to obtain two generations per year (Laux et al., 2010). Thus, it has become imperative to develop and exploit new breeding technologies to ensure the rapid production of improved cultivars and accelerate genetic gain for important traits. Over time, various plant breeding approaches were used to advance generation and to fasten the breeding cycle, such as optimization of the traditional selection method - single seed descent (SSD) method utilized during breeding for the development of homozygous lines, use of off-season nursery for growing of two or more generations per year in contrasting environments (shuttle breeding) (Ortiz et al., 2007), in vitro/embryo culture (Kondić-Špika et al., 2022a), double haploid (DH) technique (Kondić-Špika et al., 2008; Kondic-Špika et al., 2011; De La Fuente et al., 2020), marker-assisted selection (MAS; Bernardo, 2016), and the use of genetic engineering or genome editing (Gaba et al., 2021; Varshney et al., 2021b). However, even with these methods, only two to three generations per year could be advanced (Fang et al., 2021). Moreover, off-season nurseries are often expensive, and logistically difficult to manage, with possible genetic material loss during transportation and the brief intervals between crop cycles; hence, they do not ensure successful seed production and are not appropriate for large-scale. In addition, the DH technique is not accessible for a variety of crops and often demands highly qualified personnel, specialized equipment and financial resources for in vitro culture. Furthermore, transgenic or genome-edited crops could be doubtful options because of political legislation or public safety concerns.
Speed breeding (SB) represents a promising next-generation breeding technology for growing plants under controlled and manipulable conditions to accelerate breeding programs by decreasing generation time and resources and increasing the number of generations per year (Watson et al., 2018; Bhatta et al., 2021). This approach holds significant importance for overcoming traditional breeding limitations, particularly for crops with long breeding cycles. Compared to the usual field and greenhouse conditions, SB can reduce generation length from 2.5 to 5 times. Also, it is assumed that obtaining up to five generations per year in an SB system could approximately double annual genetic gain compared to breeding programs that use off-season nurseries (Jähne et al., 2020). Beside enhancing breeding efficiency, SB rapidly shortens breeding cycles, providing a timely response to emerging challenges such as climate variability and specific agricultural needs. This agility in breeding, achieved through SB, ensures the timely development of resilient and high-yielding crop varieties, contributing to global food security and sustainability (Gudi et al., 2022).
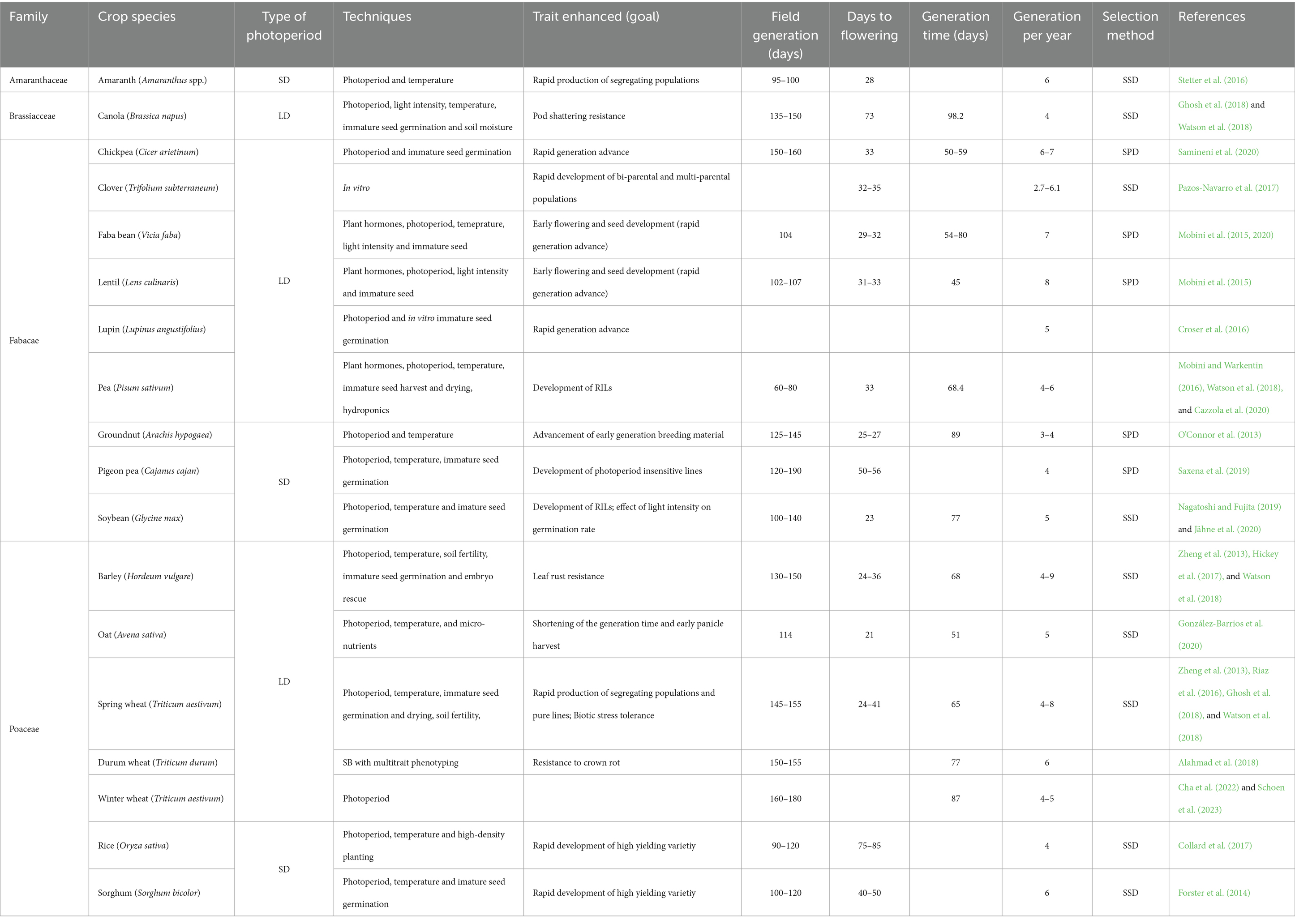
SB promotes rapid growth and development of plants from the vegetative to the reproductive stage, typically in controlled environments such as growth chambers or greenhouses, manipulating the major parameters required by the plants (photoperiod, quality and intensity of light, temperature, and humidity; Figure 1). In general, plants can be categorized into three groups based on how the length of the day influences flowering and reproductive processes of the plant (photoperiodism): short-day plants (SDP) typically flower when the duration of daylight is shorter than a critical length, long-day plants (LDP) flower when the duration of daylight exceeds a critical length, and day-neutral plants (DNP) that are less influenced by the photoperiod and tend to flower independently of day length (Ghosh et al., 2018). SB was first proposed as a strategy for LDP by extending the photoperiod to about 22 h per day (Watson et al., 2018). Extended photoperiods cause early flowering due to the conversion of phytochromes from their active to inactive forms, using specific light intensities. Exposition to prolong daily light reduces the generation time of LDP, while in SDP and photoperiod-sensitive crops, this approach would inhibit flowering. Consequently, the protocols for short-day crops (soybean, rice, and amaranth) were proposed, varying light intensities to create the ideal optimal light conditions for each crop (Jähne et al., 2020). In numerous crops, early flowering and seed formation have been found to be successfully promoted by applying light intensity ranging from 360–650 μmol/m2/s within the PAR range (Ghosh et al., 2018; Watson et al., 2018), enabling SB procedure. Beside light intensity, light quality is another major element with a direct effect on different plant growth activities such as photosynthetic and transpiration rates, stomatal conductance, and the level of intercellular CO₂ (Yang et al., 2017). Considering that the key factor in controlling a plant’s ability to flower is the red to far-red (R:FR) ratio, altering this ratio may cause different responses in flowering. Therefore, light spectrum and light quality are essential for the optimization of SB protocols. Different lighting sources can be used in the growth chambers to create PAR (photosynthetically active radiation) range: light-emitting diodes (LEDs), metal halide bulbs combined with LEDs, halogen or sodium vapor lamps (SVLs) (Ghosh et al., 2018). Furthermore, it is important to consider the different reactions of plant species to the wavelength spectra emitted by different lighting sources. It has been shown that early flowering in legumes, including lentil, pea, and chickpea, can be induced by applying blue and far-red light spectrums (Mobini et al., 2015). Photoperiod, light intensity and quality are important factors in flowering regulation, while maintaining consistent temperature and humidity levels can contribute to faster growth. Ideal soil and air temperatures are essential to achieving satisfactory germination, seedling establishment and proper vegetative and reproductive growth (Shendekar et al., 2023). Several main crops, such as soybean, canola, wheat, barley, and maize, have distinct temperature requirements for different phases of growth. For example, they usually need 12–30°C for seed germination and 25–30°C for overall growth and flowering (Wanga et al., 2021). Moreover, winter wheat requires vernalization or cold temperature stress for the transition from vegetative to the reproductive stage (Zheng et al., 2023), while in certain crops, temperatures higher than 33°C can result in reduced pollen viability and increased male-sterility. Also, there has been a considerable variance in the responses of different photoperiod-sensitive crops to temperature regimes that affect their transition from the vegetative to the reproductive stage (Yang et al., 2014). Therefore, the optimal temperature regime, considering both maximum and minimum temperatures, should be used for each crop in SB protocols. Different systems can be used to control temperature, such as foggers, solar air power batteries, fan and pad cooling systems (Wanga et al., 2021). Furthermore, it was also demonstrated that variations in soil moisture can have a significant impact on various aspects of plant growth and development, including plant height, days to flowering, maturity, and seed set (Hussain et al., 2018), which is beneficial for SB. In different crops, such as wheat, barley, canola, and chickpea, grain filling and maturation were accelerated after flowering by gradually lowering the soil’s moisture level (Watson et al., 2018). In general, for most of the crops, humidity of 60–70% is suitable, while crops adapted to dry zones require less humidity (Ghosh et al., 2018).

Figure 1. Manipulation of different environmental factors in speed breeding (this figure has been designed using icons from Flaticon.com).
Optimization of all these conditions enhances the rate of photosynthesis and other physiological and metabolic processes, which stimulates early flowering, biomass accumulation and early seed development to reduce generation time, allowing multiple generations to be grown in a shorter period (Ghosh et al., 2018; Shendekar et al., 2023). Understanding the different requirements for photoperiod and light parameters of different crop species is crucial for optimizing growth conditions and achieving rapid generation turnover in SB approaches and could be a limiting factor for successful implementation of SB. Thus, there is a need for protocol optimization on a crop-specific and variety-specific level (Jähne et al., 2020; Pandey et al., 2022) to achieve optimal growth and development, with greater uniformity. Due to genotypic variation in terms of growth characteristics, photoperiod sensitivities, and responses to environmental conditions, optimizing protocols based on genotype ensures that the specific needs and preferences of each plant variety are met. In this way, the full potential of each genotype is harnessed, maximizing the efficiency and success of the approach and enhancing reproducibility. Moreover, optimizing protocols and providing the ideal conditions can maximize resource efficiency, such as energy, space, and time.
Besides optimizing the plant development environment, SB can also be accomplished through other means, such as stress treatments, immature seed harvesting, treatments with plant growth regulators (PGRs), high-density planting, embryo rescue, increasing CO2 concentrations, genetic engineering targeting the flowering pathway or grafting juvenile plants to mature rootstocks (Richard et al., 2015; Ghosh et al., 2018; Hickey et al., 2019; Pandey et al., 2022). Under conditions of moisture stress, many crop species exhibit early flowering and seed production (Shavrukov et al., 2017). This approach is used in SB through the application of both drought and flooding stress to cause flowering for early seed sets. In addition, it is considered that immature seed harvesting, avoiding the ripening stage, could significantly shorten the generation time when combined with single-seed descent (SSD) (Chahal and Gosal, 2002). Immature seed harvesting involves picking immature pods and drying seeds under artificial conditions for a few days before shelling and sowing, allowing plants to cycle even faster from seed to seed (Jan et al., 2022). In several crop species, plant nutrition or treatments with plant growth regulators (PGRs) have been effectively employed to expedite development and initiate flowering and seed formation, particularly in techniques like organ tissue culture (Bonea, 2022). Observing different reactions to PGRs can be important in controlled conditions where certain crops, such as lentils and faba beans, have demonstrated the capacity to produce up to eight generations per year (Trnka et al., 2019). High-density planting is a cost-effective strategy for SB that considers planting crops with more plants closer together, which outperforms traditional methods in terms of yield potential. It is a useful technique since it not only stimulates early flowering and maturity, and shortens crop cycles, but also makes it possible to maintain the large population size needed for advanced selections (Sinha et al., 2021). The specific plant species can affect the effectivness of high-density planting. The effect of high-density planting on flowering is not consistently detected across studies and genotype variations considerably alter plant responses (Fukushima et al., 2011). In addition, it has been found that crop plants respond to elevated CO2 by transitioning from the vegetative to the reproductive phase. Although the reaction may range throughout crop species and within genotypes of the same species, elevated CO2 levels might expedite the transition from vegetative to reproductive stages and boost plant development (Jagadish et al., 2016).
2 Development of SB protocols for main crop species
The SB approach involves growing plants in controlled environments with specific conditions that influence the acceleration of various physiological processes in plants and quick-generation cycling. This offers flexibility for integration into a larger-scale screening of plant populations in the shortest amount of time and space. The main advantage of SB technology over other breeding methodologies aimed at reducing generation turnover time and accelerating the process of creating new varieties is its suitability for application to diverse germplasm without special requirements regarding in vitro culturing.
Different SB approaches have been successfully used to develop and standardize protocols for major crops, including commercially and scientifically significant cereal, legume and oilseed species, which covered SDP, DNP and LDP (Table 1; Ghosh et al., 2018; Watson et al., 2018; Samantara et al., 2022; Shendekar et al., 2023). This enabled the achievement of even 9 generations within a year for some crops. In this way, homozygosity can be reached in a year or two, providing a great opportunity to develop varieties in a short period of time and to deal with challenging food security (Hickey et al., 2019).
2.1 Wheat
As one of the most widely cultivated grain crops that feed approximately 35% of the world’s population, wheat has consistently played a key role in global food security strategy programs. Its grain serves as a crucial source of protein, dietary fibres, B vitamins, and minerals in the human diet (Shewry and Hey, 2015). Despite the existence of five domesticated Triticum taxa (diploid T. monococcum, tetraploid T. dicoccon and T. durum, and hexaploid T. aestivum and T. spelta), global wheat production is predominantly centered on bread and durum wheat, comprising 90–95% and 5–10% of the total, respectively (Shewry, 2009; Shewry and Hey, 2015). Bread wheat is primarily used as flour for various flatbreads and pastries, whereas durum wheat stands out as the key ingredient in the production of pasta, couscous, and bulgur. Traditional cereal breeding methodologies have brought many significant improvements regarding wheat yield and quality (Hristov et al., 2010; Mladenov et al., 2011). Furthermore, the introduction of the new breeding tools relying on the molecular and phenotyping high-throughput approaches enabled accurate pyramidization of genes with the potential to result in varieties with stable production in the environment with different combinations of abiotic and biotic stresses (Kondić-Špika et al., 2023). However, the long breeding cycle presents one of the main constraints in achieving revolutionary progress in wheat production, impeding the timely development and deployment of improved cultivars. Traditional wheat breeding involves a series of steps, with each phase demanding considerable time. The delayed release of resilient and high-yielding wheat varieties hampers the ability to adapt quickly to evolving challenges posed by climate variability, emerging pests, and shifting consumer preferences. In contrast, approaches like SB enable the acceleration of the breeding cycle, allowing rapid progress, facilitating the development of wheat varieties that can address contemporary agricultural demands and contribute significantly to global food security. History of the development of wheat SB protocols, can be traced to the mid-1980s, when joint efforts by NASA and Utah State University to assess the possibility of growing rapid cycling wheat under constant light on space stations, resulted in the creation of the dwarf wheat line USU-Apogee (Bugbee and Koerner, 1997). Later on, at the beginning of the 2000s, researchers at the University of Queensland introduced the term ‘speed breeding’ for a set of improved methods to hasten wheat breeding (Hickey et al., 2019). So far, several protocols have been developed for the SB of spring wheat. The group of authors developed a very effective protocol for rapid generation cycles based on culturing young embryos and optimizing plant growth conditions that allowed the production of up to eight generations of spring wheat per year (Zheng et al., 2013). Afterwards, to make SB technically simpler and capable of processing a larger number of genotypes, researchers from the United Kingdom and Australia developed two SB protocols for spring bread wheat and durum wheat that involve the use of extended photoperiod and high light levels to accelerate plant development, followed by immature seed harvesting and planting developing kernels (Ghosh et al., 2018; Watson et al., 2018). According to the first protocol, plants grown in a controlled environment room with an extended photoperiod (22 h light/2 h dark) were compared with the plants grown in glasshouses with no supplementary light or heating during the spring and early summer. The authors concluded that plants grown under the SB protocol progressed to flowering in 35–39 days, approximately half the time of those grown in glasshouse conditions. Under SB protocol, a slight decrease was observed in seed number per spike, while wheat plants produced a healthy number of spikes per plant, and crosses produced viable seeds. The second protocol included the use of a temperature-controlled glasshouse fitted with high-pressure sodium lamps aiming to extend the photoperiod to a day length of 22 h. Here, control treatment plants were grown in a glasshouse using a natural 12-h control photoperiod. Both control and treatment used the same temperature regime: 22°C day and 17°C night. As a result, flowering was significantly reduced relative to control plants (22 ± 2 days), and wheat plants produced significantly more spikes than those in day-neutral conditions. Further, the authors recorded an additional reduction in the breeding cycle when wheat seeds were harvested before maturity, 14 days post-anthesis, followed by 4-day cold treatment, without the need for labor-intensive embryo rescue (Watson et al., 2018). The development of SB protocols for winter wheat varieties addressed challenges to optimizing controlled environments for accelerated growth due to vernalization requirements for the transition from the vegetative to reproductive phase, making it complicated to synchronize growth cycles in SB. Therefore, SB protocols for winter wheat were modified by adding one step that involved seed exposure to low temperatures and optimal results were obtained at 10°C (Zheng et al., 2023). However, this methodology was not successful when applied to mature seeds because of the postharvest dormancy phenomenon that exists among winter genotypes, while it gave very promising results with the use of the embryo rescue technique when it was possible to produce up to 6 generations of wheat accessions per year.
In conclusion, originating from efforts from more than 40 years ago, SB protocols for spring wheat have evolved, enabling the production of up to eight generations per year. The protocols have been refined for simplicity and efficiency, involving extended photoperiods, resulting in a remarkable reduction in time to flowering and an increase in spike production. Despite challenges in adapting SB to winter wheat, modifications involving vernalization have shown promise.
2.2 Maize
Maize (Zea mays L.) represents the major source of food in the world owing to its large growing areas and productivity, and it is also the basic fodder and important raw material for industrial processing (Muntean et al., 2022). On a global level, maize ranks first in terms of production quantities, followed by rice, wheat, barley and sorghum (FAO, 2022). Although it originated in tropical regions from the short-day species Teosinte (Z. mays ssp. parviglumis), during centuries of cultivation and artificial selection, maize was spread to higher latitudes and adapted to grow under long-day conditions (Brambilla et al., 2017; Mikić et al., 2017; Osnato et al., 2022). While studying maize sensitivity to photoperiod changes under controlled conditions, it was concluded that long photoperiods in photoperiod-sensitive maize lines repressed the development of the tassels, delayed flowering time and increased the period between pollen shed and silking (Chen et al., 2015). Similarly, tropical maize lines showed a delay in flowering transitions under long-day conditions, compared to those adapted to temperate zones for more than 3 weeks (Alter et al., 2016). The same authors observed strong differences among regulated genes between temperate and tropical lines indicating the complexity of flowering time regulation in maize. Thus, traditional breeding cycles for maize can be protracted due to the dependence on specific photoperiod conditions for flowering. Considering the maize requirement of a short day for induction of the reproductive phase, SB may have potential applications for accelerating its vegetative growth (Watson et al., 2018). Although many crop-specific SB protocols in growth chambers or glasshouses have been established for different species, they have not yet been developed for maize (Singh et al., 2021). Besides its photoperiod reaction, there are also other challenges to SB implementation in maize. Being a tall crop with a canopy that can reach a height of 2.5 m, maize needs more space and bigger chambers for growth and development (Singh et al., 2021). Another approach that can shorten the maize breeding cycle is double-haploid (DH) technology (Farooqi et al., 2022). Great success in the application of DHs has been achieved in maize, which was evidenced by the development of parental lines in the majority of the seed companies (Shendekar et al., 2023). When applying DH technology, the time required to develop completely homozygous maize inbred lines is greatly reduced to 2–3 generations (Mitiku, 2022). Coupled with other approaches, like MAS and off-season nurseries, DH technology can enhance breeding efficiency and genetic gain.
To sum up, although SB protocols have been established for other species, maize faces challenges in implementing photoperiod-based growth protocols. So far, DH technology has been successful in shortening the maize breeding cycle to 2–3 generations, reducing the time required to develop homozygous maize inbred lines.
2.3 Rapeseed
Rapeseed (Brassica napus L.) is an important oil crop grown in over 50 countries worldwide (Yadava et al., 2012). It is also called oilseed rape or, in the case of varieties with low erucic acid content, canola. It is an allotetraploid crop derived from hybridization between Brassica rapa L. (AA) and Brassica oleracea L. (CC). Besides its oil and protein-rich meal, it considerably participates in global biofuel production (Zhang et al., 2023). Compared to other major crops, rapeseed yield per hectare is the lowest, reaching ~3,210 kg/ha in Europe in the period between 2015 and 2020 (Zheng et al., 2022). Hence, one of the major challenges in the current unpredictable climates as well as the reduction in the available crop production area is meeting the world’s demand for edible oil. In conventional breeding, with the application of greenhouse cultivation, two to three generations per year can be obtained in major oil crops including rapeseed. By achieving multiple generations annually, breeders can expedite the selection and advancement of desirable traits in rapeseed varieties. This enhanced breeding efficiency not only accelerates the overall breeding process but also contributes to the development of improved rapeseed cultivars with enhanced yield, oil quality, and resilience to environmental challenges. The introduction of SB in rapeseed, a LDP, enables speeding up the breeding process by obtaining even four to five generations per year, depending on the genotype and the growth conditions (Song et al., 2022). A detailed protocol for the initiation of setting up SB systems in several crops, including rapeseed, was provided, showing that phenotyping of some traits, such as pod shattering, can be performed in SB systems in adult plants (Ghosh et al., 2018). According to the results, a 22 h photoperiod led to shortening the time to flowering, duration of flowering, days till drying off, and time to harvest, compared to a 16 h photoperiod. This SB regimen negatively affected the number of seeds per pot and thousand-grain weight, while germination of the harvested seeds was not affected or even was slightly higher in SB conditions. Furthermore, it was reported that seed production (g per plant) was similar between SB conditions in which a 22 h photoperiod was used and day-neutral conditions with a 12 h photoperiod, in 7 rapeseed cultivars (Watson et al., 2018). Recently, a comprehensive SB (CSB) approach was proposed, with the following steps: (1) vernalization of the germinated seeds, (2) high-density seedling culture, and (3) accelerated flowering and maturation with the optimized light regime (Song et al., 2022). For the majority of examined semi-winter and winter canola varieties, the authors observed acceleration in reaching key growth stages, however, a winter canola variety Darmor-bzh did not flower. Thus, the authors had to improve the CSB protocol for this variety by adding 500 μmol/m2/s far-red light. This showed the importance of optimizing the growth conditions on a genotype-specific level in the SB protocol. Furthermore, the implementation of CSB in combination with marker-assisted and phenotypic selection, as reported (Wang et al., 2023), represents an innovative and comprehensive approach to crop improvement. This hybrid approach harnessed the power of accelerated plant growth through CSB while leveraging molecular markers for precise trait selection and enabled the accelerated creation of six introgression lines with different combinations of genes associated with abiotic and biotic stress resistance, high oleic acid content traits and early maturity. This integrated strategy not only expedites the breeding process but also allows for the targeted incorporation of multiple beneficial characteristics by efficiently addressing diverse stressors and quality traits.
In short, the introduction of SB, particularly in LDP, can accelerate the breeding process by obtaining four to five generations per year in rapeseed, depending on genotypes and growth conditions. Comprehensive SB (CSB) approaches, incorporating vernalization, high-density seedling culture, and optimized light regimes, offer further advancements. The integrated approach of CSB and MAS expedites the breeding process but also enables targeted incorporation of multiple beneficial traits.
2.4 Sunflower
Sunflower (Helianthus annuus) is the fourth most important oil crop used mainly for human consumption, and just 10% for biodiesel production and other industrial purposes (Radanović et al., 2023). Sunflowers, characterized by their out-crossing nature, require homozygous parents to produce hybrids, a process that requires demanding backcrossing and selection efforts. Achieving homozygosity in parental lines is crucial to ensuring genetic uniformity and stability in the resulting hybrids. Due to the out-crossing behavior of sunflowers, attaining homozygosity involves multiple generations of backcrossing to eliminate genetic heterogeneity. This process may require up to eight generations of carefully controlled crosses, followed by stringent selection for desired traits. Despite being an important crop, there are no available SB protocols for acceleration of the breeding process of this SDP. To speed up the creation of new, superior sunflower parental lines, a reliable and effective doubled haploid (DH) induction technique would be a useful resource. However, there are a lot of obstacles to creating a successful protocol, such as sunflower’s resistance to in vitro culture regeneration as well as long fresh seed dormancy, which creates a production bottleneck for DH (Miladinović et al., 2019; Mabuza et al., 2023). Several techniques, including anther culture, microspore culture, interspecific hybridization, and embryo rescue, have been used to create a sunflower DH induction regimen, but none of them have proven effective or dependable. Low crossability, low seed set, low germination and regeneration, and a high albinism rate are the primary obstacles (Kaya, 2014; Mabuza et al., 2023). As a result, different strategies are required to get over these restrictions and raise the production efficiency of sunflower DH. Promising results have been obtained by authors who managed to generate haploid embryos by pollen irradiation and pollination of female flowers with irradiated pollen (Aktaş et al., 2023). Another approach could be using immature embryo culture to shorten the generation time in breeding programs. This technique allows the production of fertile plants from immature embryos and enables the production of three to four generations per year, in contrast to one generation per year with conventional breeding (Vasić and Vasiljević, 1994; Dağüstü et al., 2012). In addition, the development of new breeding techniques, such as genome editing, in combination with different -omics tools could provide new perspectives for speeding up sunflower breeding (Miladinović et al., 2019). Using this technique, multiple genes can be individually engineered at the same time, which in combination with different prediction models, could speed up the modification of linked genes or QTLs that are usually difficult to segregate (Flavell, 2010; Miladinović et al., 2021). The first steps in that direction were made with the recent development and optimization of protocols for genome editing in sunflowers (Yildirim et al., 2022, 2023). Further development of high-throughput and sequence-based genotyping, along with high-throughput and precision phenotyping, should provide a complete set of tools for developing tools for wider application of SB in sunflower improvement programs (Cvejić et al., 2023).
In general, the absence of SB protocols for sunflowers adds to the complexity of their breeding programs. The conventional DH induction techniques face obstacles such as in vitro culture resistance and long seed dormancy. Despite attempts with various techniques, significant challenges persist, including low crossability, seed set, germination rates, and high albinism. Recent advancements, including pollen irradiation for haploid embryo generation and immature embryo culture, show promise for overcoming some limitations. Moreover, the integration of genome editing and -omics tools opens new avenues for accelerating sunflower breeding.
2.5 Soybean
Soybean (Glycine max L.) is the most important protein cropand is used for animal feed and human nutrition. In addition to the high content of protein and oil, soybeans also contain numerous compounds with a positive effect on health, such as phytoestrogens and antioxidants. The global production of soybean was the fourth highest of all crops and over the past two decades, worldwide soybean production and the area harvested have increased more rapidly compared to other staple crops (FAO, 2022). Soybean breeding is a long and complex process that involves the crossing, selection, and evaluation of different traits, such as yield, maturity, quality, and stress tolerance. Achieving multiple generations per year for soybean is significant for accelerating genetic improvement to meet the increasing global demand for protein and oil, as well as adapting to the changing climate and environmental conditions. Different strategies were explored to hasten soybean breeding, keeping in mind that uninterrupted periods of darkness and high temperatures are needed for the induction of flowering, accelerating the growth and development of short-day plants (Mao et al., 2017; Miladinović et al., 2018). Approaches, such as immature embryo culture or DH, could shorten one generation time to 65–70 days in soybean (Rosenberg and Rinne, 1988), while the use of off-season nurseries doubled the rate of generation advancement (Gai et al., 2015). On the other hand, SB techniques that use artificial, controlled environments with varying photoperiod, temperature, or CO2 concentration could produce five generations per year in soybeans, compared to one or two generations in the field (Nagatoshi and Fujita, 2019; Jähne et al., 2020; Jan et al., 2022). Specificly, using CO2-supplemented growth chambers and fluorescent lamps accelerated soybean breeding of elite Japanese cultivar Enrei by applying a protocol (14 h light, 30°C day/25°C night, CO2 supplementation 400–600 p.p.m.) that reduced the vegetation period from 102 to 132 days reported in the field to 70 days enabling five generations per year (Nagatoshi and Fujita, 2019). This method also increased the crossing efficiency of soybeans. CO2 supplementation enhanced the growth and productivity of plants, while specific light and temperature conditions shortened the days to flowering, and the reproductive period was significantly shortened by the reaping and planting of immature seeds. Furthermore, a SB protocol for soybeans based on LEDs providing a high-throughput, rapid SSD system was developed (Jähne et al., 2020). A 10 h photoperiod using a blue-light-enriched, far-red-deprived light spectrum enabled soybean to mature within 77 days after sowing, allowing the development of five generations per year. The authors proposed a possible improvement of the protocol by increasing the CO2 level, under the condition that a higher photosynthesis rate is achieved through more intense lighting. The additional speed should be weighed against the extra costs of the system setup and operational costs. In the experiments, far-red and blue light treatment at night in most cases caused highly heterogeneous flowering time among soybean genotypes, showing the different responses of cultivars from different maturity groups. Although far-red light (>700 nm) did not affect flowering time, the authors concluded that it caused elongated petioles, which in turn caused lodging. Thus, far-red light should be avoided to grow robust soybean plants suitable for high-throughput systems, while low red/blue light ratio, with green or cool white LEDs (4,000 K) should be included for optimal visual observations. They proposed that light intensity should be ~500 μmol/m2s at 50 cm distance from the light source to achieve fast generation times using a moderate budget. Other authors also defined the growth conditions for SB system using LED light source, advancing one generation of soybean within 73 days, enabling the growth of five generations per year (Lee et al., 2023). One of the challenges related to SB approaches that use artificial greenhouses is their limited scale and high cost. Thus, a cost-saving SB system established by integrating off-site nurseries and fresh-seeding methods under natural conditions was proposed, accomplishing at least four generations per year, which facilitates and accelerates soybean improvement at a low cost (Fang et al., 2021). This methodology was combined with MAS to predict the optimal adaptation region of the advanced generation lines based on the maturity genes E1-E4 instead of phenotype identification that could facilitate the breeding process for the target region (Fang et al., 2021). Combining SB with MAS using molecular markers for precise adaptation region prediction, streamlined the breeding process by eliminating the need for time-consuming phenotype identification. This not only accelerates breeding cycles but also enhances efficiency and resource utilization.
In conclusion, SB techniques utilizing controlled environments with manipulated photoperiod, temperature, and CO2 concentration offer a promising avenue for accelerating soybean breeding. Achieving up to five generations per year, in contrast to traditional methods, significantly reduces the vegetation period. While challenges related to artificial greenhouses, including limited scale and high costs, persist, innovative solutions, such as integrating off-site nurseries and fresh-seeding methods under natural conditions, have been proposed. Additionally, combining SB with MAS streamlines the breeding process, eliminating the need for time-consuming phenotype identification and enhancing overall efficiency and resource utilization.
3 Integration of SB with genomic approaches
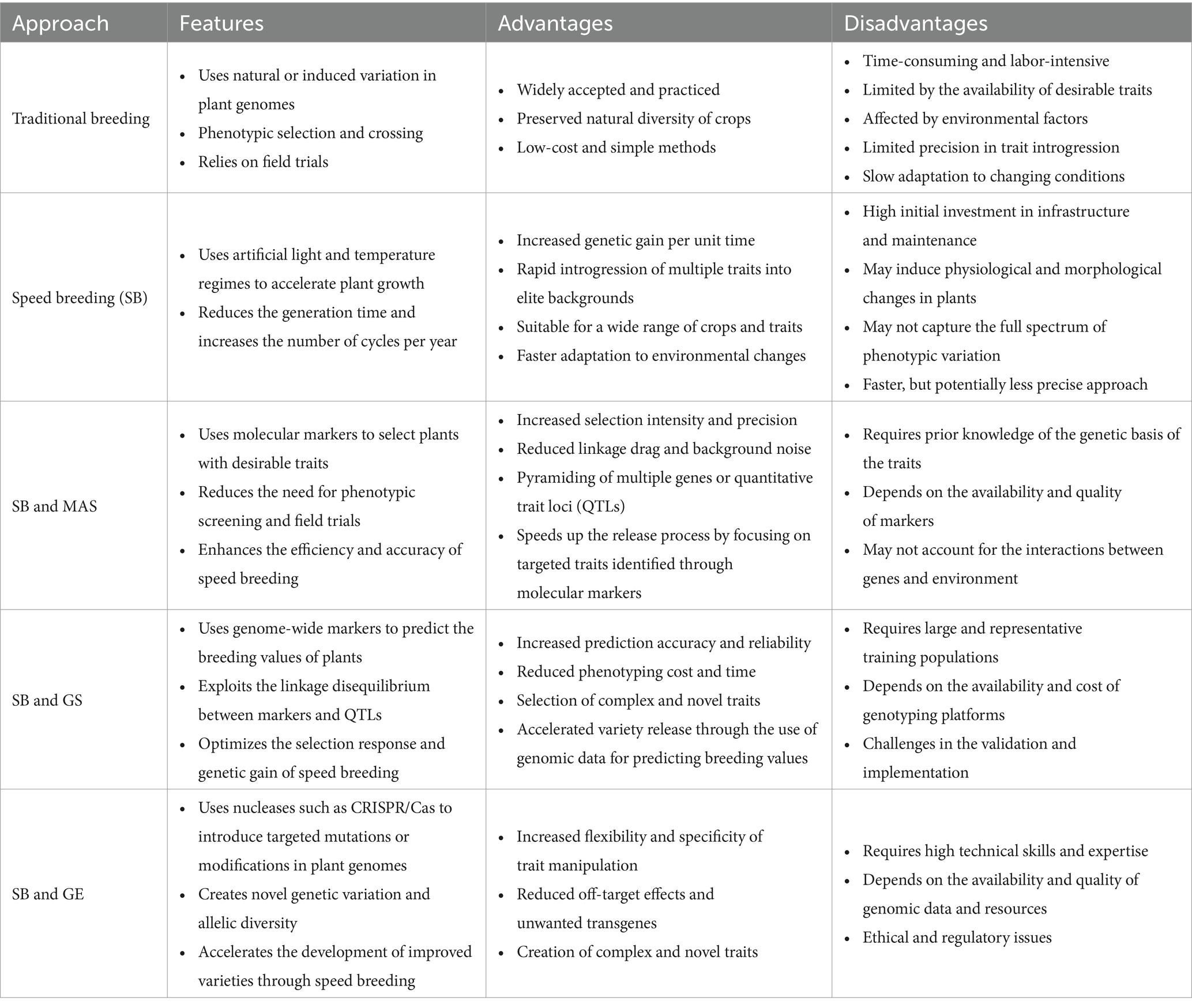
Standardized SB systems have the potential to significantly reduce the duration of main breeding processes for developing new varieties or to accelerate plant development for other plant research purposes, such as crossing, backcrossing, generation advancement, phenotyping adult plant traits, rapid gene identification, development of mapping populations, pyramiding target traits, mutant studies and genetic transformation experiments (Figure 2; Ghosh et al., 2018; Watson et al., 2018; Hickey et al., 2019; Ahmar et al., 2020; Bhatta et al., 2021; Varshney et al., 2021a). Different breeding selection methods, such as single seed descent (SSD), single pod descent (SPD), single plant selection (SPS), and clonal selection can be integrated into SB to produce a fixed population at a much faster and more affordable rate resulting in accelerated development and release of new cultivars (Hickey et al., 2017; Watson et al., 2018). With these methods, researchers can effectively advance offspring under high-density planting within controlled conditions, while also requiring less labour and cultivation space during initial generations (Table 2). It could also expand the possibilities for maintaining genetic diversity within a crop breeding program and be extended to field environments. In addition to directly accelerating some breeding activities, the rapid development of mapping populations through SB has also stimulated research studies that elucidate associations between genes and traits for breeding purposes (Samantara et al., 2022).
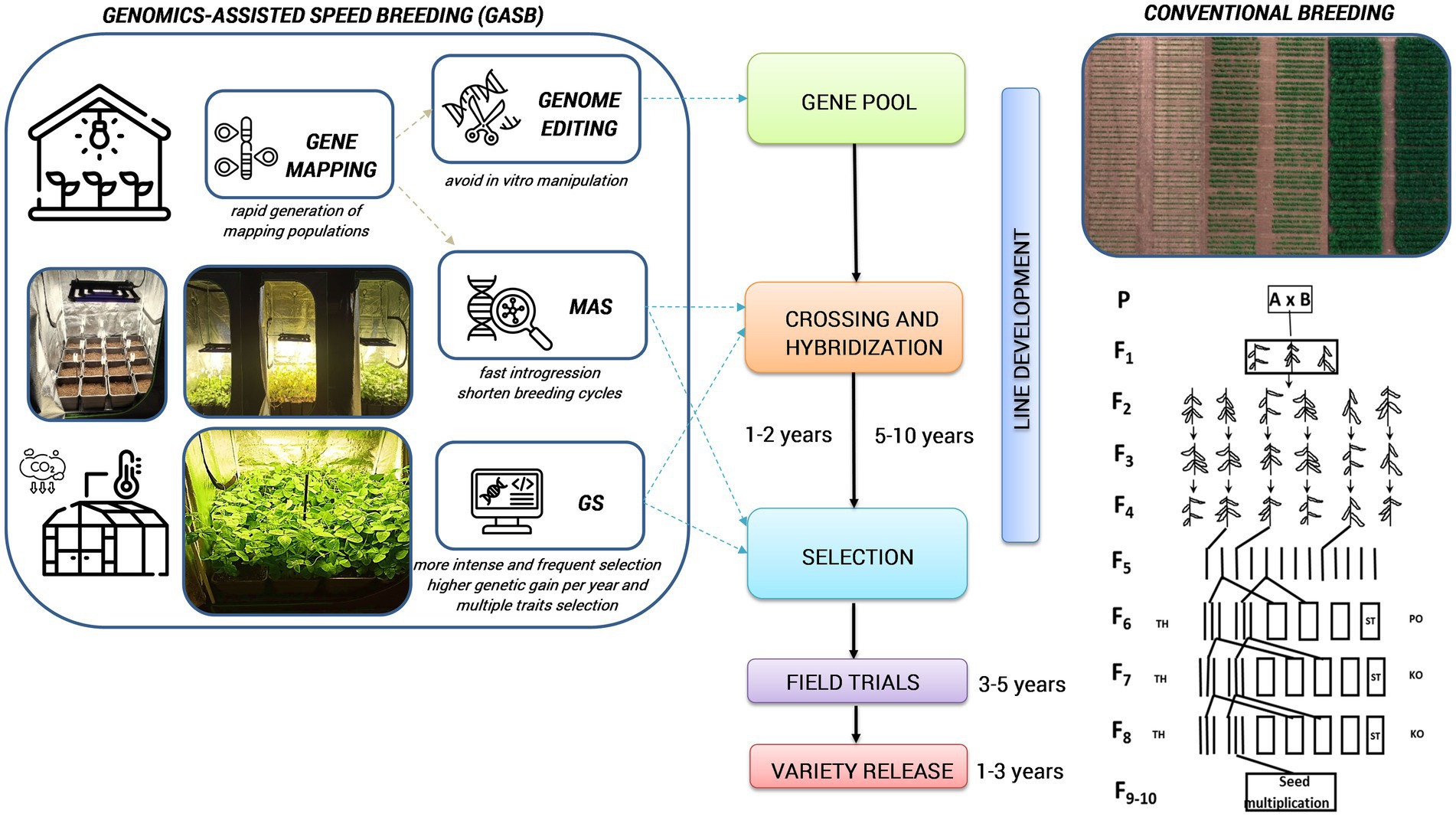
Figure 2. Integrated GASB to accelerate variety release compared to the conventional breeding. MAS, marker-assisted selection; GS, genomic selection; (this figure has been designed using icons from Flaticon.com).
Integration of fast-forward genomic breeding techniques with SB (genomics-assisted speed breeding [GASB]) has a great potential to enable further advancement of crop improvement, by leveraging the benefits of all approaches in terms of improving selection efficiency and reducing generation time. Combining the SB approach with genomics and phenomics may encompass candidate gene discovery, MAS, marker-assisted backcrossing (MABC), genomic selection (GS), genome editing, or metabolic pathway editing for desired traits followed by high-throughput phenotyping (Figure 2). Moreover, GASB can speed up the creation of new variability for crop improvement by quickly manipulating the target region in the genome by minimizing the time, field space and overall resources (Varshney et al., 2021b). These genomic tools circumvent laborious phenotyping and facilitate multi-trait selection under growth chambers (Hickey et al., 2019). Furthermore, by combining metabolomic and SB, a robust and quick risk assessment of gene-edited crops can be achieved over several generations (Razzaq et al., 2021).
3.1 Gene mapping and MAS
Gene mapping provides the identification of major genes and quantitative trait loci (QTLs) associated with phenotypic variation, enabling better insight into the genetic control of agronomically important traits and suppling fundamental information for MAS, gene cloning, and genome structure studies (Brbaklić et al., 2015; Dimitrijevic and Horn, 2018; Gupta et al., 2019; Trkulja et al., 2019). The process relies on the comprehensive genotyping and phenotyping of various genetic materials, followed by appropriate statistical analysis for the detection of genes and QTLs. The most common materials used for trait dissection in gene mapping are biparental populations (F2, doubled haploids, backcross and recombinant inbred lines), multiple parental populations (multi-parent advanced generation inter-cross - MAGIC populations) and nested association mapping (NAM) populations (Li et al., 2010; Huang et al., 2015; Chidzanga et al., 2022). The availability of new sequencing and genotyping technologies together with the development of high-throughput phenotyping approaches, facilitate the evaluation of genetic material for agronomic traits in multiple environments and seasons. However, the production of diverse mapping populations requires substantial amount of time for the selection of parents, their crossing and the evaluation of the variability among obtained lines. All these activities can be performed by SB, producing large and genetically diverse mapping populations with reduced time and overall cost (Watson et al., 2018). This discovery has led to significant advancements in the domains of MAS and QTL analysis (Potts et al., 2023).
Moreover, superior genes or alleles for the traits of interest can be identified through genome-wide association studies or pan-genomics, enabling a reduction in time needed for gene discovery and increasing mapping resolution power (Varshney et al., 2021a). Identified molecular markers that are linked to the gene or QTL, controlling a particular trait of interest, could be used for MAS to increase the efficiency of selection, especially of traits that are under the influence of environmental factors (Collard et al., 2005). MAS is the most effective when targeting a small number of genes with a large effect, such as resistance genes (Chhetri et al., 2017). Identification of target traits and transfer to desirable genotypes by conventional breeding, besides being less efficient than MAS, is also more labour-intensive and time-consuming (Table 2). The advancements and integration of gene mapping techniques and MAS with SB methods could overcome the bottleneck of prolonged crop breeding cycles, providing better exploitation of genetic resources in the development of climate-resilient varieties and general improvement of food security (Watson et al., 2018; Raza et al., 2023). Different MAS approaches, such as MABC, gene pyramiding, marker-assisted recurrent selection (MARS) and genomic selection could be accelerated by GASB which reduces generation turnover time and enables the growth of multiple generations within a single year. Integration of SB techniques with backcrossing and MABC could speed up the transfer of desirable traits from donor to recipient parent. In conventional backcrossing, for the recovery of the recurrent parent, more than six generations are required, while applying DNA markers in MABC can be reduced to three generations (Tanksley et al., 1989). By integrating MAS with SB, the hst1 gene, conferring salt tolerance, was transferred into the high-yielding rice genotype (Rana et al., 2019). In 17 months and six generations, the authors succeeded in developing the BC3F3 population with the desired homozygous allele, through three backcrosses, followed by two cycles of self-fertilization. Whole-genome sequencing of advanced progeny showed that 93.5% of the BC3F2 genome was similar to the recipient parent, while 2.7% was fixed, being donor parent homozygous alleles.
In addition, integration of SB with gene mapping could be particularly convenient to facilitate the discovery and later on quick insertion of genes for resistance or tolerance to biotic and abiotic stress, which may enable an urgent response to environmental stress by providing the development of resistant genotypes for various crops (Devi et al., 2023). Furthermore, SB could hasten the exploration and utilization of allelic diversity in landraces and wild relatives of crops that present valuable sources of resistance to various abiotic and biotic factors, ultimately contributing to the diversification and strengthening of crop resilience (Anđelković et al., 2020; Pour-Aboughadareh et al., 2021). An integrated approach was also used to introgress some valuable alleles from wild relatives in lentils (Lulsdorf and Banniza, 2018). Also, the discovery of new sources of disease resistance using GASB was well demonstrated in the study of Riaz et al. (2017), who were screening Vavilov wheat collection using SB and molecular markers linked to the known leaf rust resistance genes. The authors did rapid phenotyping under accelerated controlled conditions at the seedling and adult stages. Based on the results, lines carrying known genes for leaf rust resistance and lines with potentially novel sources of resistance were identified. Another successful example of the integration of SB with MAS was introgression of imidazolinone Group 2 herbicide tolerance into chickpeas (Croser et al., 2021). The authors developed KASP markers to discriminate between homozygous tolerant and heterozygous intolerant genotypes in the F2 generation. Only homozygous tolerant plants were processed for F4 generation, which led to saving cost and time. Moreover, Australian wheat researchers have implemented a multi-trait approach, including QTL analysis, to improve yield and stability under water and heat stress (Christopher et al., 2015). In this study, SB approach coupled with molecular markers, novel phenotyping techniques, and NAM population, identified valuable traits such as stay-green and root characteristics, validated known QTLs and discovered new ones for these traits.
3.2 Genomic selection
Unlike traditional MAS, which is mostly based on a few major-effect loci to perform selection, genomic selection simultaneously uses genome-wide DNA markers to assess genomic estimated breeding values (GEBVs) for complex traits (Meuwissen et al., 2001). This approach was developed predominantly to understand quantitative traits, controlled by many loci with a small effect. The effects of all loci are estimated in large training populations by establishing prediction models that combine genome-wide DNA markers with extensive phenotypic data. After the development of the prediction model, the breeding values of candidate breeding lines are assessed based on estimated marker effects and genotypic profiles of candidates. The lines with higher GEBV will be selected for the next generation. The advantage of genomic selection over other breeding methods is in rapid screening of elite germplasm, acceleration of crop breeding cycles, and time and resource utilization, especially for traits measured late in the variety development process or that are costly to phenotype, such as yield (Crossa et al., 2017; Đorđević et al., 2019). Lines can be selected and used as parents early in the process of variety development, and multiple breeding cycles based on GEBV can be achieved in the same amount of time as a single cycle of traditional breeding only through assessing the genetic potential of genotypes that have not been tested before.
While genomic prediction contributes to a faster annual rate of genetic gain, its greatest benefit is thought to come from combining it with other technologies that shorten generation cycle times (Hickey et al., 2017; Gorjanc et al., 2018). An integrated approach that combines SB and genomic selection assumes parent selection based on GEBVs followed by a selection of progenies generated by SB. To encourage quick breeding cycling, this process is performed numerous times. Given that SB can significantly shorten generation time (Watson et al., 2018), using genomic selection for choosing the parents in each generation could significantly increase the genetic gain (Pandey et al., 2022). Although the next-generation sequencing technology enabled the practical application of genomic selection, the biggest challenge for implementing this approach is still genome sequencing costs (Bhat et al., 2016). One way to reduce the costs would be to use genomic selection every other generation or to choose candidates only if they meet certain requirements for traits like disease resistance that can be accurately phenotyped throughout SB (Riaz et al., 2016). Moreover, this may also lead to a reduction in inbreeding when compared to phenotypic or genomic selection (Jighly et al., 2019). It is considered that this scheme combined with multiple traits, such as normalized difference vegetation index and canopy temperature and with available high-throughput phenotyping platforms could yield higher gains, enabling early-stage selection (Crain et al., 2018). Also, SB coupled with artificial intelligence (AI) can significantly facilitate understanding of genomic architecture by using a machine learning approach for genomic selection model development (Niazian and Niedbała, 2020). The prospect of combining GS and SB to accelerate genetic gain in crops was recommended in more recent studies (Hickey et al., 2019; Bohra et al., 2020; Krishnappa et al., 2021).
A simulation study suggested that combining genomic selection with SB, known as ‘Speed GS’, for traits with different genetic architecture and heritability, can maximize genetic gain per unit time and reduce generation time in comparison with conventional breeding (Jighly et al., 2019). This approach has also been demonstrated in spring wheat to increase the genetic gain of complex traits (Voss-Fels et al., 2019). SB was used for the phenotyping of specific traits in the training population of wheat and selection candidate development and phenotyping steps. Speed GS was used to predict grain yield across multiple environments and to shorten the breeding cycle through the rapid generation of inbred lines (Watson et al., 2019). The authors incorporated four speed-breeding traits from a training population into multivariate genomic selection models, which significantly increased the predictive ability of yield compared to univariate GS models.
3.3 Genome editing
Genetic engineering primarily entails the insertion or deletion of a gene or gene segment in the recepient crop using biotechnology, providing several benefits over traditional breeding methods (Rai, 2022). With the further improvement of these technologies, and the emergence of genome editing (GE) technology, crop scientists have the option of making changes at a targeted location. Genome editing techniques exploiting programmable nucleases including transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeat (CRISPR)-Cas-associated nucleases have been used in crop improvement to a certain extent, but there is a significant room for improvement. The most commonly used GE technique is CRISPR-Cas which utilizes non-homologous end joining and homology-directed repair for DNA repair, as well as single-base editing enzymes (Pickar-Oliver and Gersbach, 2019). There are six types of Cas proteins, of which Cas9 is the most exploited one in plant breeding thanks to its accuracy, affordability, and ease of use (Aksoy et al., 2022). CRISPR-Cas9 system relies on guide RNA (gRNA) for guidance and targeting specificity, however, it still has some limitations such as Cas9-related toxicity, possible off-targets, and restrictions in target sites (Zhao et al., 2020; Varshney et al., 2021a). Some other systems such as CRISPR from Prevotella and Francisella 1 (Cpf1) show promise for use in GE as they exhibit higher efficiency rates compared to Cas9.
The advantage of using CRISPR/Cas systems in GE is the option of targeting multiple sites thus enabling simultaneous pyramiding of the desired genes. This fastens up breeding programs and in combination with SB, it can lead to a significant reduction in time to create plant material with improved resistance to pathogens or with improved quality parameters (Table 2). This way edited plants can be grown in SB conditions to quickly produce edited seeds, which can speed up homozygosity and the rate at which genetic gain can occur. Genome editing can produce desirable lines that can be preselected at the T1 generation, and rigorous evaluation can be done at the T2 generation to ensure that off-target genotypes are eliminated (Samantara et al., 2022). However, there is still a labour-intensive step in tissue culture laboratories that needs to be performed to obtain edited plants. In a recently proposed ExpressEdit approach that integrates gene editing and SB, plant shoot apical meristems could be directly treated using techniques like particle bombardment with the Cas9 gene and sgRNA (single guide RNA) sequences to get around the bottleneck of traditional tissue culture and plant regeneration in the lab. Plants that lack Cas9 but carry the new trait could be identified by screening their progeny for the new trait (e.g., disease resistance). On the other hand, Cas9 might stay in “CRISPR-ready” plants, which could undergo additional rounds of editing by using sgRNA to target distinct gene targets (Hickey et al., 2019). ExpressEdit can also be used in conjunction with MAS and high-throughput phenotyping to increase breeding process efficiency. This will facilitate the rapid development of the mutant variety or a variety carrying a mutant allele (i.e., 5–6 years instead of 8–10 years) (Voss-Fels et al., 2019). The integration of SB with genome editing has been proven in Brassica napus, B. oleracea, tomato and soybean (Yang et al., 2017; Murovec et al., 2018; Bao et al., 2020; Liu et al., 2021).
Another drawback to fully exploiting genome editing and SB integration in breeding programs is the existence of strict GMO (Genetically Modified Organism) regulations in many countries (Table 2). Thus, the exploitation of DNA-free GE approaches may show promising potential. In some countries like the EU, these crops would still fall within the strict GMO regulations. However, there are now countries that do not concentrate on the process but rather on the product, and thus CRISPR/Cas system should have a very bright future in wider application in crop breeding and production.
3.4 Recent high-throughput genotyping based approaches
Current advances in next-generation sequencing technologies and omics methods have made it possible to access the genome sequences and transcriptomes of multiple plant species, significantly transforming plant breeding. With the public availability of plant genomes, there is a chance to identify candidate genes, QTLs, and associated genome-wide molecular markers, enabling high-throughput marker-assisted breeding (Naqvi et al., 2022). Pan-genomic research has been initiated as a result of the development of next-generation sequencing technologies, providing a great platform for studying genetic diversity, examining multiple genomes at once, determining crop evolution and adaptation as well as getting a better understanding of genome functions that may be used in crop breeding (Zhao et al., 2018). The term pan-genome refers to a species’ whole gene pool that is present in different individuals. Recently, pan-genomes of some key crops have been constructed and this concept is applied to agricultural plant research (Hurgobin et al., 2018). The integration of pan-genomes with SB and genome editing could accelerate trait-specific breeding. Crop pan-genomes will make better breeding schemes easier to implement, especially for complex genome crops where they will help identify shared or distinct gene combinations for various traits (Naqvi et al., 2022).
The accessibility of whole genome sequencing data for a large number of lines for a given crop enabled haplotype identification. A haplotype represents a group of polymorphic markers that are inherited together in progeny with minimum recombination (Garg, 2021). In rice and numerous other species, a number of haplotypes linked to increased yield have been discovered (Sharma et al., 2023). Haplotype breeding can be achieved by combining desirable haplotypes into the genetic background of crops. This introgression takes a long time due to the requirement of more breeding cycles and long generation time for the crop. However, an integrated approach that combines haplotype breeding and SB and GS has the potential to accelerate genetic gain (Sharma et al., 2023; Shendekar et al., 2023).
4 SB technology: current status, challenges and future perspectives
In the present fast-changing climate and challenging global crises, the traditional slow breeding procedures and programs are not adequate and suitable for quick responses and solutions that the present and future agriculture needs. To be efficient and flexible enough to address all these challenges, plant breeding needs prompt and precise techniques to be incorporated into crop improvement and the development of climate-resilient, adaptive and stable crop varieties. So far, many of these technologies such as shuttle breeding, in vitro/embryo culture, DH technology, MAS, genetic engineering, and genome editing are known and have already been introduced into breeding programs worldwide. SB is just one of them and should be integrated with all other approaches and innovations, to offer breeders a contemporary and powerful technology applicable to solving a variety of problems as fast as possible. This results in more effective and expedited plant breeding programs that maintain crop yields and guarantee food security and promote sustainable agriculture. Breeders can quickly create new plant varieties better adapted to environmental challenges like drought, heat or salinity. Moreover, by creating crop varieties with improved resilience to pests and diseases and increased nutrient efficiency, SB lessens dependency on chemical inputs like fertilizers and pesticides.
Currently, the efficient SB protocols are available for a limited number of cereal, legume, vegetable and oilseed crops (Ghosh et al., 2018; Watson et al., 2018). Also, using this technology several varieties have been released so far in wheat, barley, pea, canola, rice and tomato (Shendekar et al., 2023). For wider practical breeding outcomes and realization of the SB potential to substantially accelerate genetic gain, the optimization of SB protocols for all important crop species is necessary, as well as further modifications of the existing protocols based on local needs/innovations. SB is based on the induction of early flowering in photoperiod-responsive crops (Pandey et al., 2022). However, the differences in photoperiodic requirements between different crops could make standardization of SB protocols very difficult (Jackson and Jackson, 2009). Although SB is a valuable tool to accelerate the conventional breeding process, controlled and intensive growth conditions during cycles could lead to limited plant growth and consequently lower seed yield due to the crossover interaction between genotypes and growth systems (Tanaka et al., 2016; Sharma et al., 2019; González-Barrios et al., 2020). These variations may have an impact on the stability and uniformity of crops, raising concerns about the consistency of crop performance across various environments. Hence, an additional effort should be made to determine optimal growth conditions for both, crop species and sometimes also different genotypes within the species, to mitigate the negative effect that intensive growth conditions could have on plants (Pandey et al., 2022). For example, excessive photoperiod can slow down plant growth and elevate stress hormone levels. Thus, it is required to precisely balance the need to accelerate growth with the need to prevent stress-induced responses. The limited seed yields could also lead to the loss of some breeding populations and a decrease in genetic variability, which could pose a problem, especially in crops where the SSD method is used in breeding (Saxena et al., 2019). This issue could be partially overcome by preserving backup seeds from each individual through each generation (Pandey et al., 2022). Additionally, some of the major challenges in SB include disease and pest control, as well as tracking individuals for gene discovery (Potts et al., 2023).
Moreover, rigorous testing and evaluation of new varieties in field conditions are still necessary to ensure suitability for commercial cultivation. Thus, SB technology needs to be combined with effective field trials or high-throughput phenotyping platforms (HTPP) to enhance the crop improvement process. For accurate measurement, the SB facility should be supplied with automated platforms, hyperspectral sensors, high-end cameras, lasers, thermometers, lux meters, humidity meters, etc., (Sharma et al., 2023). Thanks to the availability of enhanced phenotyping tools such as thermal imaging, 3D imaging, magnetic resonance imaging, computed tomography, and imaging spectroscopy, almost any plant trait can be measured (Shendekar et al., 2023). Accurate phenotyping can facilitate faster improvement of target traits involved in biotic and abiotic stress, addressing the problem of global food security in the future. A combination of multi trait phenotyping and SB was applied for the promotion of root adaptation in water limited environments in wheat (Christopher et al., 2015). Moreover, high-throughput phenotyping and SB technology were used together in barley for introgression of disease resistance to leaf rust, net and spot forms of blotch (Hickey et al., 2017). Possible future directions for SB include exploiting the genetic diversity and novel alleles present in the wild relatives and landraces of crop plants by using SB and molecular methods to introduce them into elite cultivars. This will enhance the genetic variation and adaptability of crop plants to changing environments. Furthermore, high-throughput phenotyping platforms that can capture the dynamic responses of plants to different stress factors under SB conditions should be developed. This will enable the identification of novel traits and genes associated with stress tolerance and yield potential. In this way, dynamic SB programs that can adapt to the changing climate and consumer preferences by incorporating diverse germplasm and novel traits should be developed. Also, Artificial Intelligence (AI) and machine learning have opened up new possibilities for SB, including the ability to make precise decisions and handle large and complex datasets, which could facilitate the discovery of new patterns, relationships, and insights that can guide the breeding decisions and strategies and could provide new insights into how plants function in extreme climates (Rai, 2022).
Finally, SB technology as such requires specific expertise, effective plant phenotyping facilities, appropriate infrastructure and continuous financial support for research and development (Shimelis et al., 2019). Training personnel for specific knowledge needed for SB is one of the aspects that needs to be focused on in the future. The introduction of SB into the breeding programs requires investment in growth chambers with adequate light and temperature conditions, which could be relatively high (Pandey et al., 2022). Maintaining controlled environmental conditions also requires higher energy consumption, which further increases the total cost of SB applications. A potential solution to this problem could be the use of sustainable energy sources, such as solar panels, and the use of energy-efficient LEDs in growth chambers (Yao et al., 2017). With the help of LED lighting systems, precise control of the length and intensity of the light, which is beneficial for photosynthesis, growth and crop development is possible (Jähne et al., 2020). Therefore, further technological innovations are needed to reduce prices for the facilities and to make it affordable for a wider community, small breeding companies and research institutions for better exploitation of the technology. Although these requirements are not limiting factors for the technology application in developed countries, they seriously limit the use of SB, remaining a challenge in developing countries due to their limited infrastructure, poor expertise and insufficient governmental support, as described by several authors (Chiurugwi et al., 2019; Wanga et al., 2021; Samantara et al., 2022). Thus, international collaborations and partnerships of multi-disciplinary teams are needed for faster knowledge and SB technology transfer not only into basic and applied research but also into breeding companies to accelerate the application of SB technology worldwide.
5 Conclusion
Recent advances in SB techniques provide a potential alternative for reducing the amount of time, space, and resources needed to develop and release high-performing cultivars. SB can be achieved by distinct approaches, impacting different phases of plant breeding by accelerating the breeding cycle. SB protocols for numerous economically significant species have been developed. However, further optimization and application of SB should be broadened to other crops with lengthy generation times or that are challenging to breed. The further acceleration of the rate of genetic gain in breeding programs and the development of stable and high-performing crop varieties for addressing global challenges such as changing climate and food security could be achieved through GASB. This integrated approach maximizes the benefits of each technique and produces a comprehensive framework for modern plant breeding. However, future evaluation and research should be focused on the potential detrimental effects of SB conditions on phenotypic and genetic changes and agronomic performance and stability of speed-bred lines under field conditions. Other challenges, such as the lack of qualified personnel, necessary infrastructure, consistent water and electricity supplies, and high costs have limited the application of SB, especially in developing countries, and particularly in public plant breeding programs. Therefore, efforts should be redirected to capacity building, technology transfer, and financing of SB coupled with modern breeding approaches to facilitate crop improvement programs.
Author contributions
MĆ: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. DM: Writing – original draft, Writing – review & editing, Funding acquisition. VĐ: Writing – original draft, Writing – review & editing, Funding acquisition. DT: Writing – original draft, Writing – review & editing. AR: Writing – original draft, Writing – review & editing. SG: Writing – original draft, Writing – review & editing. AK-Š: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia, grant number 451-03-66/2024-03/200032, by the Science Fund of the Republic of Serbia through IDEAS project “Creating Climate Smart Sunflower for Future Challenges” (SMARTSUN), grant number 7732457, by the Science Fund of the Republic of Serbia through PRISMA project “Soybean Yield Prediction Using Multi-omics Data Integration” (SoyPredict), grant number 6788, and by the European Commission through Twinning Western Balkans project CROPINNO, grant number 101059784, by Center of Excellence for Innovations in Breeding of Climate-Resilient Crops - Climate Crops and Center of Excellence for Legumes, Institute of Field and Vegetable Crops, Novi Sad, Serbia.
Acknowledgments
We acknowledge MSc Predrag Ranđelović from the Institute of Field and Vegetable Crops, National Institute of the Republic of Serbia for providing the photos in Figure 2.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmar, S., Gill, R. A., Jung, K. H., Faheem, A., Qasim, M. U., Mubeen, M., et al. (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int. J. Mol. Sci. 21:2590. doi: 10.3390/ijms21072590
Aksoy, E., Yildirim, K., Kavas, M., Kayihan, C., Yerlikaya, B. A., Calik, I., et al. (2022). General guidelines for CRISPR/Cas-based genome editing in plants. Mol. Biol. Rep. 49, 12151–12164. doi: 10.1007/s11033-022-07773-8
Aktaş, Y. E., Aydin, Y., and Altinkut Uncuoglu, A. (2023). Induction of haploid plants for speed-up breedıng in sunflower (Helianthus annuus L.) by pollen irradiation. Gen. Appl. 7, 35–44. doi: 10.31383/ga.vol7iss1ga05
Alahmad, S., Dinglasan, E., Leung, K. M., Riaz, A., Derbal, N., Voss-Fels, K. P., et al. (2018). Speed breeding for multiple quantitative traits in durum wheat. Plant Methods 14:36. doi: 10.1186/s13007-018-0302-y
Alter, P., Bircheneder, S., Zhou, L. Z., Schlüter, U., Gahrtz, M., Sonnewald, U., et al. (2016). Flowering time-regulated genes in maize include the transcription factor ZmMADS1. Plant Physiol. 172, 389–404. doi: 10.1104/pp.16.00285
Anđelković, V., Cvejić, S., Jocić, S., Kondić-Špika, A., Marjanović Jeromela, A., Mikić, S., et al. (2020). Use of plant genetic resources in crop improvement–example of Serbia. Genet. Resour. Crop. Evol. 67, 1935–1948. doi: 10.1007/s10722-020-01029-9
Bao, A., Zhang, C., Huang, Y., Chen, H., Zhou, X., and Cao, D. (2020). Genome editing technology and application in soybean improvement. Oil Crop Sci. 5, 31–40. doi: 10.1016/j.ocsci.2020.03.001
Bernardo, R. (2016). Bandwagons I, too, have known. Theo. Appl. Genet. 129, 2323–2332. doi: 10.1007/S00122-016-2772-5
Bhat, J. A., Ali, S., Salgotra, R. K., Mir, Z. A., Dutta, S., Jadon, V., et al. (2016). Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 7:221. doi: 10.3389/fgene.2016.00221
Bhatta, M., Sandro, P., Smith, M. R., Delaney, O., Voss-Fels, K. P., Gutierrez, L., et al. (2021). Need for speed: manipulating plant growth to accelerate breeding cycles. Curr. Opin. Plant Biol. 60:101986. doi: 10.1016/j.pbi.2020.101986
Bohra, A., Saxena, K. B., Varshney, R. K., and Saxena, R. K. (2020). Genomics-assisted breeding for Pigeonpea improvement. Theor. Appl. Genet. 133, 1721–1737. doi: 10.1007/s00122-020-03563-7
Bonea, D. (2022). Speed breeding and its importance for the improvement of agricultural crops. AAMC. 52, 59–66. doi: 10.52846/aamc.v52i1.1314
Bonnecarrere, V., Rosas, J., and Ferraro, B. (2019). Economic impact of marker-assisted selection and rapid generation advance on breeding programs. Euphytica 215:197. doi: 10.1007/s10681-019-2529-8
Brambilla, V., Gomez-Ariza, J., Cerise, M., and Fornara, F. (2017). The importance of being on time: regulatory networks controlling photoperiodic flowering in cereals. Front. Plant Sci. 8:665. doi: 10.3389/fpls.2017.00665
Brbaklić, L., Trkulja, D., Kondić-Špika, A., Hristov, N., Denčić, S., Mikić, S., et al. (2015). Genetic associations in the detection of QTLs for wheat spike-related traits. Pesqui. Agropecu. Brasileira. 50, 149–159. doi: 10.1590/S0100-204X2015000200007
Bugbee, B., and Koerner, G. (1997). Yield comparisons and unique characteristics of the dwarf wheat cultivar ‘USU-apogee’. Adv. Space Res. 20, 1891–1894. doi: 10.1016/s0273-1177(97)00856-9
Cazzola, F., Bermejo, C. J., Guindon, M. F., and Cointry, E. (2020). Speed breeding in pea (Pisum sativum L.), an efficient and simple system to accelerate breeding programs. Euphytica 216:178. doi: 10.1007/s10681-020-02715-6
Cha, J.-K., O'Connor, K., Alahmad, S., Lee, J.-H., Dinglasan, E., Park, H., et al. (2022). Speed vernalization to accelerate generation advance in winter cereal crops. Mol. Plant 15, 1300–1309. doi: 10.1016/j.molp.2022.06.012
Chahal, G. S., and Gosal, S. S. (2002). Principles and Procedures of Plant Breeding: Biotechnological and Conventional Approaches. Alpha Science International. Boca Raton, FL: CRC Press, 604
Chen, Q., Zhong, H., Fan, X. W., and Li, Y. Z. (2015). An insight into the sensitivity of maize to photoperiod changes under controlled conditions. Plant, Cell Environ. 38, 1479–1489. doi: 10.1111/pce.12361
Chhetri, M., Bariana, H., Wong, D., Sohail, Y., Hayden, M., and Bansal, U. (2017). Development of robust molecular markers for marker-assisted selection of leaf rust resistance gene Lr23 in common and durum wheat breeding programs. Mol. Breeding. 37, 1–8. doi: 10.1007/s11032-017-0628-6
Chidzanga, C., Mullan, D., Roy, S., Baumann, U., and Garcia, M. (2022). Nested association mapping-based GWAS for grain yield and related traits in wheat grown under diverse Australian environments. Theor. Appl. Genet. 135, 4437–4456. doi: 10.1007/s00122-022-04230-9
Chiurugwi, T., Kemp, S., Powell, W., and Hickey, L. T. (2019). Speed breeding orphan crops. Theor. Appl. Genet. 132, 607–616. doi: 10.1007/s00122-018-3202-7
Christopher, J., Richard, C., Chenu, K., Christopher, M., Borrell, A., and Hickey, L. (2015). Integrating rapid phenotyping and speed breeding to improve stay-green and root adaptation of wheat in changing, water-limited, Australian environments. Procedia Environ. Sci. 29, 175–176. doi: 10.1016/j.proenv.2015.07.246
Collard, B. C. Y., Beredo, J. C., Lenaerts, B., Mendoza, R., Santelices, R., Lopena, V., et al. (2017). Revisiting rice breeding methods–evaluating the use of rapid generation advance (RGA) for routine rice breeding. Plant. Prod. Sci. 20, 337–352. doi: 10.1080/1343943X.2017.1391705
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., and Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Crain, J., Mondal, S., Rutkoski, J., Singh, R. P., and Poland, J. (2018). Combining high-throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. Plant Genome U.S. 11, 1–14. doi: 10.3835/plantgenome2017.05.0043
Croser, J., Mao, D., Dron, N., Michelmore, S., McMurray, L., Preston, C., et al. (2021). Evidence for the application of emerging technologies to accelerate crop improvement – a collaborative pipeline to Introgress herbicide tolerance into chickpea. Front. Plant Sci. 12:779122. doi: 10.3389/fpls.2021.779122
Croser, J. S., Pazos-Navarro, M., Bennett, R. G., Tschirren, S., Edwards, K., Erskine, W., et al. (2016). Time to flowering of temperate pulses in vivo and generation turnover in vivo–in vitro of narrow-leaf lupin accelerated by low red to far-red ratio and high intensity in the far-red region. Plant Cell Tissue Organ Cult. 127, 591–599. doi: 10.1007/s11240-016-1092-4
Crossa, J., Pérez-Rodríguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., De Los Campos, G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011
Cvejić, S., Jocić, S., Mitrović, B., Bekavac, G., Mirosavljević, M., Marjanović Jeromela, A., et al. (2023). “Innovative approaches in breeding of climate-resilient crops” in Climate Change and Agriculture: Perspectives, Sustainability and Resilience Ed. N. Benkeblia, (New Jersey, USA: John Wiley & Sons Ltd.) 111–156.
Dağüstü, N., Bayram, G., Sincik, M., and Bayraktaroğlu, M. (2012). The short breeding cycle protocol effective on diverse genotypes of sunflower (Helianthus annuus L.). Turk. J. Field Crops. 17, 124–128.
De La Fuente, G. N., Frei, U. K., Trampe, B., Ren, J., Bohn, M., Yana, N., et al. (2020). A diallel analysis of a maize donor population response to in vivo maternal haploid induction: II. Haploid male fertility. Crop. Sci. 60, 873–882. doi: 10.1002/CSC2.20021
Devi, O. B., Haokip, S. W., and Singh, Y. S. (2023). Speed breeding: an effective strategy for breeding of crop new kinds and its future prospects. Vigyan Varta. 4, 83–86.
Dimitrijevic, A., and Horn, R. (2018). Sunflower hybrid breeding: from markers to genomic selection. Front. Plant Sci. 8:2238. doi: 10.3389/fpls.2017.02238
Đorđević, V., Ćeran, M., Miladinović, J., Balešević-Tubić, S., Petrović, K., Miladinov, Z., et al. (2019). Exploring the performance of genomic prediction models for soybean yield using different validation approaches. Mol. Breeding. 39:74. doi: 10.1007/s11032-019-0983-6
Fang, Y., Wang, L., Sapey, E., Fu, S., Wu, T., Zeng, H., et al. (2021). Speed-breeding system in soybean: integrating off-site generation advancement, fresh seeding, and marker-assisted selection. Front. Plant Sci. 12:717077. doi: 10.3389/fpls.2021.717077
FAO, IFAD, UNICEF, WFP and WHO (2021). “The state of food security and nutrition in the world 2021” in Transforming Food Systems for Food Security, Improved Nutrition and Affordable Healthy Diets for All. Ed. C. Holleman (Rome: FAO)
Farooqi, M. Q. U., Nawaz, G., Wani, S. H., Choudhary, J. R., Rana, M., Sah, R. P., et al. (2022). Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.). Front. Plant Sci. 13:965878. doi: 10.3389/fpls.2022.965878
Flavell, R. (2010). From genomics to crop breeding. Nat. Biotechnol. 28, 144–145. doi: 10.1038/nbt0210-144
Forster, B. P., Till, B. J., Ghanim, A. M. A., Huynh, H. O. A., Burstmayr, H., and Caligari, P. D. S. (2014). Accelerated plant breeding. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 9, 1–16. doi: 10.1079/PAVSNNR20149043
Fróna, D., Szenderák, J., and Harangi-Rákos, M. (2019). The challenge of feeding the world. Sustain. For. 11:5816. doi: 10.3390/su11205816
Fukushima, A., Shiratsuchi, H., Yamaguchi, H., and Fukuda, A. (2011). Effects of nitrogen application and planting density on morphological traits, dry matter production and yield of large grain type rice variety bekoaoba and strategies for super high-yielding rice in the tohoku region of Japan. Plant Prod. Sci. 14, 56–63. doi: 10.1626/pps.14.56
Gaba, Y., Pareek, A., and Singla-Pareek, S. L. (2021). Raising climate-resilient crops: journey from the conventional breeding to new breeding approaches. Curr. Genomics 22, 450–467. doi: 10.2174/1389202922666210928151247
Gai, J., Liu, K., and Zhao, J. (2015). A review on advances in science and technology in Chinese seed industry. Sci. Agric. Sin. 48, 3303–3315.
Garg, S. (2021). Computational methods for chromosome-scale haplotype reconstruction. Genome Biol. 22:101. doi: 10.1186/s13059-021-02328-9
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suárez, M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protocol. 13, 2944–2963. doi: 10.1038/s41596-018-0072-z
González-Barrios, P., Bhatta, M., Halley, M., Sandro, P., and Gutiérrez, L. (2020). Speed breeding and early panicle harvest accelerates oat (Avena sativa L.) breeding cycles. Crop Sci. 61, 320–330. doi: 10.1002/csc2.20269
Gorjanc, G., Gaynor, R. C., and Hickey, J. M. (2018). Optimal cross selection for long-term genetic gain in two-part programs with rapid recurrent genomic selection. Theor. Appl. Genet. 131, 1953–1966. doi: 10.1007/s00122-018-3125-3
Gudi, S., Kumar, P., Singh, S., Tanin, M. J., and Sharma, A. (2022). Strategies for accelerating genetic gains in crop plants: special focus on speed breeding. Physiol. Mol. Biol. Plants 28, 1921–1938. doi: 10.1007/s12298-022-01247-8
Gupta, P. K., Kulwal, P. L., and Jaiswal, V. (2019). Association mapping in plants in the post-GWAS genomics era. Adv. Genet. 104, 75–154. doi: 10.1016/bs.adgen.2018.12.001
Hickey, L. T., Germa, S. E., Diaz, J. E., Ziems, L. A., Fowler, R. A., Platz, G. J., et al. (2017). Speed breeding for multiple disease resistance in barley. Euphytica 213, 1–14. doi: 10.1007/s10681-016-1803-2
Hickey, L. T., Hafeez, A. N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hoegh-Guldberg, O., Jacob, D., Taylor, M., Guillén Bolaños, T., Bindi, M., Brown, S., et al. (2019). The human imperative of stabilizing global climate change at 1.5 C. Science 365:6974. doi: 10.1126/science.aaw6974
Hristov, N., Mladenov, N., Đurić, V., Kondić-Špika, A., and Marjanović-Jeromela, A. (2010). Improvement of wheat quality in cultivars released in Serbia during the 20th century. Cereal Res. Commun. 38, 111–121. doi: 10.1556/CRC.37.2009.4.9
Huang, B. E., Verbyla, K. L., Verbyla, A. P., Raghavan, C., Singh, V. K., Gaur, P., et al. (2015). MAGIC populations in crops: current status and future prospects. Theor. Appl. Genet. 128, 999–1017. doi: 10.1007/s00122-015-2506-0
Hurgobin, B., Golicz, A. A., Bayer, P. E., Chan, C. K. K., Tirnaz, S., Dolatabadian, A., et al. (2018). Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 16, 1265–1274. doi: 10.1111/pbi.12867
Hussain, H. A., Hussain, S., Khaliq, A., Ashraf, U., Anjum, S. A., Men, S., et al. (2018). Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Front. Plant Sci. 9:393. doi: 10.3389/fpls.2018.00393
Jackson, S. D., and Jackson, S. D. (2009). Plant responses to photoperiod. New Phytol. 181, 517–531. doi: 10.1111/j.1469-8137.2008.02681.x
Jagadish, S. V. K., Bahuguna, R. N., Djanaguiraman, M., Gamuyao, R., Prasad, P. V. V., and Craufurd, P. Q. (2016). Implications of high temperature and elevated CO2 on flowering time in plants. Front. Plant Sci. 7:913. doi: 10.3389/fpls.2016.00913
Jähne, F., Hahn, V., Würschum, T., and Leiser, W. L. (2020). Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 133, 2335–2342. doi: 10.1007/s00122-020-03601-4
Jan, S. A., Tabassum, R., and Bashir, H. (2022). Speed breeding methods for soybean improvement: recent advances. J. Nutr. Health Food Eng. 12, 41–42. doi: 10.15406/jnhfe.2022.12.00354
Jighly, A., Lin, Z., Pembleton, L. W., Cogan, N. O. I., Spangenberg, G. C., Hayes, B. J., et al. (2019). Boosting genetic gain in Allogamous crops via speed breeding and genomic selection. Front. Plant Sci. 10:1364. doi: 10.3389/fpls.2019.01364
Kaya, Y. (2014). “Sunflower” in Alien Gene Transfer in Crop Plants. eds. A. Pratap and J. Kumar, vol. 2 (New York, NY: Springer)
Kondić-Špika, A., Glogovac, S., Trkulja, D., Marjanović Jeromela, A., and Marjanović, M. (2022a). Response of wheat genotypes to excess boron estimated by in vitro culture. Genetika 54, 907–919. doi: 10.2298/GENSR2202907K
Kondić-Špika, A., Kobiljski, B., and Hristov, N. (2008). Efficiency of anther culture technique in the production of wheat double haploids. Zbornik Matice Srpske za Prirodne Nauke. 115, 35–40. doi: 10.2298/ZMSPN0815035K
Kondić-Špika, A., Mikić, S., Mirosavljević, M., Trkulja, D., Marjanović Jeromela, A., Rajković, D., et al. (2022b). Crop breeding for a changing climate in the Pannonian region: towards integration of modern phenotyping tools. J. Exp. Bot. 73, 5089–5110. doi: 10.1093/jxb/erac181
Kondić-Špika, A., Trkulja, D., Brbaklić, L., Mikić, S., Glogovac, S., Johansson, E., et al. (2023). “Marker-assisted selection for the improvement of cereals and pseudocereals” in Developing Sustainable and Health Promoting Cereals and Pseudocereals Eds. M. Rakszegi, M. Papageorgiou, and J. M. Rocha (Cambridge, Massachusetts, United States: Academic Press), 253–283.
Kondic-Špika, A., Vukosavljev, M., Kobiljski, B., and Hristov, N. (2011). Relationships among androgenetic components in wheat and their responses to the environment. J. Biol. Res.-Thessalon. 16, 217–223.
Krishnappa, G., Savadi, S., Tyagi, B. S., Singh, S. K., Mamrutha, H. M., Kumar, S., et al. (2021). Integrated genomic selection for rapid improvement of crops. Genomics 113, 1070–1086. doi: 10.1016/j.ygeno.2021.02.007
Laux, P., Jäckel, G., Tingem, R. M., and Kunstmann, H. (2010). Impact of climate change on agricultural productivity under rainfed conditions in Cameroon – a method to improve attainable crop yields by planting date adaptations. Agric. For. Meteorol. 150, 1258–1271. doi: 10.1016/j.agrformet.2010.05.008
Lee, D., Han, K., Kim, J. H., Jun, T.-H., and Lee, J. S. (2023). Development of speed-breeding system for Korean soybean varieties [Glycine max (L.) Merr] using LED light source. Plant Breed. Biotech. 11, 49–55. doi: 10.9787/PBB.2023.11.1.49
Lenaerts, B., Collard, B. C. Y., and Demont, M. (2019). Review: improving global food security through accelerated plant breeding. Plant Sci. 287:110207. doi: 10.1016/j.plantsci.2019.110207
Li, H., Hearne, S., Bänziger, M., Li, Z., and Wang, J. (2010). Statistical properties of QTL linkage mapping in biparental genetic populations. Heredity 105, 257–267. doi: 10.1038/hdy.2010.56
Liu, Y., Shi, Y., Su, D., Wang, L., and Zhengguo, L. (2021). SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic. Res. 8:3. doi: 10.1038/S41438-020-00431-9/42041746/41438_2020_ARTICLE_431.PDF
Lulsdorf, M. M., and Banniza, S. (2018). Rapid generation cycling of an F2 population derived from a cross between Lens culinaris Medik. And Lens ervoides (Brign.) Grande after aphanomyces root rot selection. Plant Breed. 137, 486–491. doi: 10.1111/pbr.12612
Mabuza, L. M., Mchunu, N. P., Crampton, B. G., and Swanevelder, D. Z. H. (2023). Accelerated breeding for Helianthus annuus (sunflower) through doubled Haploidy: an insight on past and future prospects in the era of genome editing. Plan. Theory 12:485. doi: 10.3390/plants12030485
Mao, T., Li, J., Wen, Z., Wu, T., Wu, C., Sun, S., et al. (2017). Association mapping of loci controlling genetic and environmental interaction of soybean flowering time under various photo-thermal conditions. BMC Genomics 18:415. doi: 10.1186/s12864-017-3778-3
Meuwissen, T. H. E., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. doi: 10.1093/genetics/157.4.1819
Mikić, S., Kondić-Špika, A., Brbaklić, L., Stanisavljević, D., Ćeran, M., Trkulja, D., et al. (2017). Molecular and phenotypic characterisation of diverse temperate maize inbred lines in Southeast Europe. Zemdirbyste-Agriculture 104, 31–40. doi: 10.13080/z-a.2017.104.005
Miladinović, D., Antunes, D., Yildirim, K., Bakhsh, A., Cvejić, S., Kondić-Špika, A., et al. (2021). Targeted plant improvement through genome editing: from laboratory to field. Plant Cell Rep. 40, 935–951. doi: 10.1007/s00299-020-02655-4
Miladinović, Ј., Ćeran, М., Đorđević, V., Balešević-Tubić, S., Petrović, K., Đukić, V., et al. (2018). Allelic variation and distribution of the major maturity genes in different soybean collections. Front. Plant Sci. 9:1286. doi: 10.3389/fpls.2018.01286
Miladinović, D., Hladni, N., Radanović, A., Jocić, S., and Cvejić, S. (2019). “Sunflower and climate change: possibilities of adaptation through breeding and genomic selection” in Genomic Designing of Climate-Smart Oilseed Crops (Cham, Switzerland: Springer International Publishing), 173–238.
Mitiku, T. (2022). Review on haploid and double haploid maize (Zea mays) breeding technology. J. Agric. Sc. Food Technol. 8, 052–058. doi: 10.17352/2455-815X.000145
Mladenov, N., Hristov, N., Kondic-Spika, A., Đurić, V., Jevtić, R., and Mladenov, V. (2011). Breeding progress in grain yield of winter wheat cultivars grown at different nitrogen levels in semiarid conditions. Breeding Sci. 61, 260–268. doi: 10.1270/jsbbs.61.260
Mobini, S., Khazaei, H., Warkentin, T. D., and Vandenberg, A. (2020). Shortening the generation cycle in Faba bean (Vicia faba) by application of cytokinin and cold stress to assist speed breeding. Plant Breed. 139, 1181–1189. doi: 10.1111/pbr.12868
Mobini, S. H., Lulsdorf, M., Warkentin, T. D., and Vandenberg, A. (2015). Plant growth regulators improve in vitro flowering and rapid generation advancement in lentil and faba bean. In Vitro Cell Dev. Biol. Plant 51, 71–79. doi: 10.1007/s11627-014-9647-8
Mobini, S. H., and Warkentin, T. D. (2016). A simple and efficient method of in vivo rapid generation technology in pea (Pisum sativum L.). In Vitro Cell Dev. Biol. Plant 52, 530–536. doi: 10.1007/s11627-016-9772-7
Muntean, L., Ona, A., Berindean, I., Racz, I., and Muntean, S. (2022). Maize breeding: from domestication to genomic tools. Agronomy 12:2365. doi: 10.3390/agronomy12102365
Murovec, J., Guček, K., Bohanec, B., Avbelj, M., and Jerala, R. (2018). DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front. Plant Sci. 9:1594. doi: 10.3389/fpls.2018.01594
Nagatoshi, Y., and Fujita, Y. (2019). Accelerating soybean breeding in a CO2-supplemented growth chamber. Plant Cell Physiol. 60, 77–84. doi: 10.1093/pcp/pcy189
Naqvi, R. Z., Siddiqui, H. A., Mahmood, M. A., Najeebullah, S., Ehsan, A., Azhar, M., et al. (2022). Smart breeding approaches in post-genomics era for developing climate-resilient food crops. Front. Plant Sci. 13:972164. doi: 10.3389/fpls.2022.972164
Niazian, M., and Niedbała, G. (2020). Machine learning for plant breeding and biotechnology. Agriculture 10:436. doi: 10.3390/agriculture10100436
O’Connor, D. J., Wright, G. C., Dieters, M. J., George, D. L., Hunter, M. N., Tatnell, J. R., et al. (2013). Development and application of speed breeding technologies in a commercial peanut breeding program. Peanut Sci. 40, 107–114. doi: 10.3146/PS12-12.1
Ortiz, R., Trethowan, R., Ferrara, G. O., Iwanaga, M., Dodds, J. H., Crouch, J. H., et al. (2007). High yield potential, shuttle breeding, genetic diversity, and a new international wheat improvement strategy. Euphytica 157, 365–384. doi: 10.1007/s10681-007-9375-9
Osnato, M., Cota, I., Nebhnani, P., Cereijo, U., and Pelaz, S. (2022). Photoperiod control of plant growth: flowering time genes beyond flowering. Front. Plant Sci. 12:805635. doi: 10.3389/fpls.2021.805635
Pandey, S., Singh, A., Parida, S. K., and Prasad, M. (2022). Combining speed breeding with traditional and genomics-assisted breeding for crop improvement. Plant Breed. 141, 301–313. doi: 10.1111/pbr.13012
Paul, K., Pauk, J., Kondic-Spika, A., Grausgruber, H., Allahverdiyev, T., Sass, L., et al. (2019). Co-occurrence of mild salinity and drought synergistically enhances biomass and grain retardation in wheat. Front. Plant Sci. 10:501. doi: 10.3389/fpls.2019.00501
Pazos-Navarro, M., Castello, M., Bennett, R. G., Nichols, P., and Croser, J. (2017). In vitro-assisted single-seed descent for breeding-cycle compression in subterranean clover (Trifolium subterraneum L.). Crop Pasture Sci. 68, 958–966. doi: 10.1071/CP17067
Pickar-Oliver, A., and Gersbach, C. A. (2019). The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Bio. 20, 490–507. doi: 10.1038/s41580-019-0131-5
Potts, J., Jangra, S., Michael, V. N., and Wu, X. (2023). Speed breeding for crop improvement and food security. Crops 3, 276–291. doi: 10.3390/crops3040025
Pour-Aboughadareh, A., Kianersi, F., Poczai, P., and Moradkhani, H. (2021). Potential of wild relatives of wheat: ideal genetic resources for future breeding programs. Agronomy 11:1656. doi: 10.3390/agronomy11081656
Radanović, A., Cvejić, S., Jocković, M., Dedić, B., Jocić, S., and Miladinović, D. (2023). “Conventional and molecular breeding for sunflower nutrition quality improvement” in Advanced Crop Improvement, Volume 2: Case Studies of Economically Important Crops Eds. A. Raina, M. R. Wani, R. A. Laskar, N. Tomlekova, and S. Khan. (Cham: Springer International Publishing), 351–391.
Rai, K. K. (2022). Integrating speed breeding with artificial intelligence for developing climate-smart crops. Mol. Biol. Rep. 49, 11385–11402. doi: 10.1007/s11033-022-07769-4
Rana, M. M., Takamatsu, T., Baslam, M., Kaneko, K., Itoh, K., Harada, N., et al. (2019). Salt tolerance improvement in rice through efficient SNP marker-assisted selection coupled with speed-breeding. Int. J. Mol. Sci. 20:2585. doi: 10.3390/ijms20102585
Ray, D. K., West, P. C., Clark, M., Gerber, J. S., Prishchepov, A. V., and Chatterjee, S. (2019). Climate change has likely already affected global food production. PLoS One 14:e0217148. doi: 10.1371/journal.pone.0217148
Raza, A., Mubarik, M. S., Sharif, R., Habib, M., Jabeen, W., Zhang, C., et al. (2023). Developing drought smart, ready to grow future crops. Plant Genome U.S. 16:e20279. doi: 10.1002/tpg2.20279
Razzaq, A., Kaur, P., Akhter, N., Wani, S. H., and Saleem, F. (2021). Next-generation breeding strategies for climate-ready crops. Front. Plant Sci. 12:620420. doi: 10.3389/fpls.2021.620420
Riaz, A., Athiyannan, N., Periyannan, S., Afanasenko, O., Mitrofanova, O., Aitken, E. A. B., et al. (2017). Mining Vavilov’s treasure chest of wheat diversity for adult plant resistance to Puccinia triticina. Plant Dis. 101, 317–323. doi: 10.1094/PDIS-05-16-0614-RE
Riaz, A., Periyannan, S., Aitken, E., and Hickey, L. (2016). A rapid phenotyping method for adult plant resistance to leaf rust in wheat. Plant Methods 12:17. doi: 10.1186/s13007-016-0117-7
Richard, C. A., Hickey, L. T., Fletcher, S., Jennings, R., Chenu, K., and Christopher, J. T. (2015). Highthroughput phenotyping of seminal root traits in wheat. Plant Methods 11:13. doi: 10.1186/s13007-015-0055-9
Rosenberg, L. A., and Rinne, R. W. (1988). Changes in seed constituents during germination and seedling growth of precociously matured soybean seeds (Glycine max (L.) Merr.). Ann. Bot. 60, 705–712.
Samantara, K., Bohra, A., Mohapatra, S. R., Prihatini, R., Asibe, F., Singh, L., et al. (2022). Breeding more crops in less time: a perspective on speed breeding. Biology 11:275. doi: 10.3390/biology11020275
Samineni, S., Sen, M., Sajja, S. B., and Gaur, P. M. (2020). Rapid generation advance (RGA) in chickpea to produce up to seven generations per year and enable speed breeding. Crop J. 8, 164–169. doi: 10.1016/j.cj.2019.08.003
Saxena, K. B., Saxena, R. K., Hickey, L. T., and Varshney, R. K. (2019). Can a speed breeding approach accelerate genetic gain in pigeonpea? Euphytica 215:202. doi: 10.1007/s10681-019-2520-4
Schoen, A., Wallace, S., Holbert, M. F., Brown-Guidera, G., Harrison, S., Murphy, P., et al. (2023). Reducing the generation time in winter wheat cultivars using speed breeding. Crop Sci. 63, 2079–2090. doi: 10.1002/csc2.20989
Sharma, A., Jones, J. B., and White, F. F. (2019). Recent advances in developing disease resistance in plants. F1000Research. 8:1934. doi: 10.12688/f1000research.20179.1
Sharma, S., Kumar, A., Dhakte, P., Raturi, G., Vishwakarma, G., Barbadikar, K. M., et al. (2023). Speed breeding opportunities and challenges for crop improvement. J. Plant Growth Regul. 42, 46–59. doi: 10.1007/s00344-021-10551-8
Shavrukov, Y., Kurishbayev, A., Jatayev, S., Shvidchenko, V., Zotova, L., Koekemoer, F., et al. (2017). Early flowering as a drought escape mechanism in plants: how can it aid wheat production? Front. Plant Sci. 8:1950. doi: 10.3389/fpls.2017.01950
Shendekar, S., Kute, N., Madhu, B., Gadpayale, D., Meshram, M., Basavaraj, P. S., et al. (2023). Unlocking crop potential: speed breeding and its synergies with modern breeding techniques. Biol Forum Int J. 15, 89–100.
Shewry, P. R., and Hey, S. J. (2015). The contribution of wheat to human diet and health. Food Energy Secur. 4, 178–202. doi: 10.1002/fes3.64
Shimelis, H., Gwata, E. T., and Laing, M. D. (2019). “Crop improvement for agricultural transformation in southern Africa” in Transforming Agriculture in Southern Africa (London, UK: Routledge), 97–103.
Shimelis, H., and Laing, M. (2012). Timelines in conventional crop improvement: pre-breeding and breeding procedures. Aust. J. Crop. Sci. 6, 1542–1549.
Singh, I., Sheorani, S., Kumar, B., Kumar, K., and Rakshit, S. (2021). Speed breeding in maize (Zea mays) Vis-à-Vis in other crops: status and prospects. Indian J. Agr. Sci. 91, 1267–1273. doi: 10.56093/ijas.v91i9.116059
Sinha, P., Singh, V. K., Bohra, A., Kumar, A., Reif, J. C., and Varshney, R. K. (2021). Genomics and breeding innovations for enhancing genetic gain for climate resilience and nutrition traits. Theor. Appl. Genet. 134, 1829–1843. doi: 10.1007/s00122-021-03847-6
Song, Y., Duan, X., Wang, P., Li, X., Yuan, X., Wang, Z., et al. (2022). Comprehensive speed breeding: a high-throughput and rapid generation system for long-day crops. Plant Biotechnol. J. 20, 13–15. doi: 10.1111/pbi.13726
Stetter, M. G., Zeitler, L., Steinhaus, A., Kroener, K., Biljecki, M., and Schmid, K. J. (2016). Crossing methods and cultivation conditions for rapid production of segregating populations in three grain amaranth species. Front. Plant Sci. 7:816. doi: 10.3389/fpls.2016.00816
Tanaka, J., Hayashi, T., and Iwata, H. (2016). A practical, rapid generation-advancement system for rice breeding using simplified biotron breeding system. Breed. Sci. 66, 542–551. doi: 10.1270/jsbbs.15038
Tanksley, S., Young, N., Paterson, A., and Bonierable, M. W. (1989). RFLP mapping in plant breeding: new tools for an old science. Nat. Biotechnol. 7, 257–264. doi: 10.1038/nbt0389-257
Trkulja, D., Kondić-Špika, A., Brbaklić, L., Kobiljski, B., Mikić, S., Mirosavljević, M., et al. (2019). Genetic structure and allelic richness of the wheat core collection for association mapping of yield. Zemdirbyste-Agriculture 106, 257–264. doi: 10.13080/z-a.2019.106.033
Trnka, M., Feng, S., Semenov, M. A., Olesen, J. E., Kersebaum, K. C., Rötter, R. P., et al. (2019). Mitigation efforts will not fully alleviate the increase in water scarcity occurrence probability in wheat-producing areas. Sci. Adv. 5:eaau2406. doi: 10.1126/sciadv.aau2406
Varshney, R. K., Bohra, A., Roorkiwal, M., Barmukh, R., Cowling, W. A., Chitikineni, A., et al. (2021a). Fast-forward breeding for a food-secure world. Trends Genet. 37, 1124–1136. doi: 10.1016/j.tig.2021.08.002
Varshney, R. K., Bohra, A., Yu, J., Graner, A., Zhang, Q., and Sorrells, M. E. (2021b). Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 26, 631–649. doi: 10.1016/j.tplants.2021.03.010
Vasić, D., and Vasiljević, L. (1994). Mogućnost skraćenja ciklusa selekcije suncokreta primenom kulture embriona. Selekcija i semenarstvo (Plant Breed. Seed Product.) 1, 103–106.
Voss-Fels, K. P., Stahl, A., Wittkop, B., Lichthardt, C., Nagler, S., Rose, T., et al. (2019). Breeding improves wheat productivity under contrasting agrochemical input levels. Nat. Plants. 5, 706–714. doi: 10.1038/s41477-019-0445-5
Wang, Z., Wang, F., Yu, Z., Shi, X., Zhou, X., Wang, P., et al. (2023). Pyramiding of multiple genes generates rapeseed introgression lines with clubroot and herbicide resistance, high oleic acid content, and early maturity. The Crop J. 11, 895–903. doi: 10.1016/j.cj.2022.10.009
Wanga, M. A., Shimelis, H., Mark, J. M., and Laing, D. (2021). Opportunities and challenges of speed breeding: a review. Plant Breed. 140, 185–194. doi: 10.1111/pbr.12909
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat Plants. 4, 23–29. doi: 10.1038/s41477-017-0083-8
Watson, A., Hickey, L. T., Christopher, J., Rutkoski, J., Poland, J., and Hayes, B. J. (2019). Multivariate genomic selection and potential of rapid indirect selection with speed breeding in spring wheat. Crop Sci. Breed. Gen. 59, 1945–1959. doi: 10.2135/cropsci2018.12.0757
Yadava, D. K., Vasudev, S., Singh, N., Mohapatra, T., and Prabhu, K. V. (2012). “Breeding major oil crops: present status and future research needs” in Technological Innovations in Major World oil Crops, Ed. S.K. Gupta. vol. 1 (New York, NY: Springer), 17–51.
Yang, S., Weers, B. D., Morishige, D. T., and Mullet, J. E. (2014). CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14:148. doi: 10.1186/1471-2229-14-148
Yang, H., Wu, J.-J., Tang, T., Liu, K.-D., and Dai, C. (2017). CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7:7489. doi: 10.1038/s41598-017-07871-9
Yao, Y., Zhang, P., Liu, H., Lu, Z., and Yan, G. (2017). A fully in vitro protocol towards large scale production of recombinant inbred lines in wheat (Triticum aestivum L.). Plant Cell Tiss. Org. Culture (PCTOC) 128, 655–661. doi: 10.1007/s11240-016-1145-8
Yildirim, K., Kucuk, I. S., Miladinović, D., and Saraç, Ç. G. (2023). Development CRISPR/CAS9-Mediated Resistance in Sunflower Against O. Cumana. Proceedings, 5th International Symposium on Broomrape in Sunflower, 1–3 November 2023, Antalya, pp. 24–25.
Yildirim, K., Sevgen, İ., Kondić-Špika, A., Cvejić, S., Jocić, S., and Miladinović, D. (2022). The First Report on Efficient CRISPR-Based Protocol for Sunflower. Proceedings, 20th International Sunflower Conference, 20–23 June 2022, Novi Sad, Serbia, pp. 186.
Yu, C., Miao, R., and Khanna, M. (2021). Maladaptation of U.S. corn and soybeans to a changing climate. Sci. Rep. 11:12351. doi: 10.1038/s41598-021-91192-5
Zhang, T., Yu, S., Pan, Y., Li, H., Liu, X., and Cao, J. (2023). Properties of texturized protein and performance of different protein sources in the extrusion process: a review. Food Res. Int. 174:113588. doi: 10.1016/j.foodres.2023.113588
Zhao, J., Fang, H., and Zhang, D. (2020). Expanding application of CRISPR-Cas9 system in microorganisms. Synthetic Syst. Biotechnol. 5, 269–276. doi: 10.1016/j.synbio.2020.08.001
Zhao, Q., Feng, Q., Lu, H., Li, Y., Wang, A., Tian, Q., et al. (2018). Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284. doi: 10.1038/s41588-018-0041-z
Zheng, Z., Gao, S., Wang, H., and Liu, C. (2023). Shortening generation times for winter cereals by vernalizing seedlings from young embryos at 10 degree Celsius. Plant Breed. 142, 202–210. doi: 10.1111/pbr.13074
Zheng, M., Terzaghi, W., Wang, H., and Hua, W. (2022). Integrated strategies for increasing rapeseed yield. Trends Plant Sci. 27, 742–745. doi: 10.1016/j.tplants.2022.03.008
Keywords: food security, speed breeding, marker-assisted selection, genomic selection, genome editing, genomics-assisted speed breeding (GASB)
Citation: Ćeran M, Miladinović D, Đorđević V, Trkulja D, Radanović A, Glogovac S and Kondić-Špika A (2024) Genomics-assisted speed breeding for crop improvement: present and future. Front. Sustain. Food Syst. 8:1383302. doi: 10.3389/fsufs.2024.1383302
Edited by:
Muhammad Saqlain Zaheer, Khwaja Fareed University of Engineering and Information Technology (KFUEIT), PakistanReviewed by:
Awais Riaz, University of Arkansas, United StatesAkhtar Hameed, Muhammad Nawaz Shareef University of Agriculture, Pakistan
Copyright © 2024 Ćeran, Miladinović, Đorđević, Trkulja, Radanović, Glogovac and Kondić-Špika. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina Ćeran, marina.ceran@ifvcns.ns.ac.rs
 Marina Ćeran
Marina Ćeran Dragana Miladinović
Dragana Miladinović Vuk Đorđević
Vuk Đorđević Dragana Trkulja
Dragana Trkulja Aleksandra Radanović
Aleksandra Radanović Svetlana Glogovac1
Svetlana Glogovac1  Ankica Kondić-Špika
Ankica Kondić-Špika