Study on the microbial diversity of ear canal secretions from patients with otomycosis
- Department of Otorhinolaryngology, Hubei NO.3 People's Hospital of Jianghan University, Wuhan, China
Otomycosis is caused by fungi, which usually cause discharge and additional discomfort. The highest incidence of otomycosis occurs in summer. To better treat this disease, it is necessary to study the microbial diversity of otomycosis secretions. In this regard, this study used high-throughput sequencing technology to determine the microbial diversity of the ear canal secretions of six typical patients with otomycosis in Wuhan via internal transcribed spacer (ITS) and 16S rRNA analyses and proposed a reasonable clinical treatment plan. Six patients with otomycosis in the Department of Otorhinolaryngology, Hubei Third People's Hospital Affiliated with Jianghan University, were selected from June 2022 to August 2022. The results showed that Staphylococcus spp. (average abundance 29.05%) was the dominant bacteria and Aspergillus spp. (average abundance 90.68%) was the dominant fungus involved in otomycosis secretion. Aspergillus spp. can cause inflammation of the external auditory canal combined with bacterial infections such as Staphylococcus spp., which can cause discharge in the ear canal. High-throughput sequencing provides comprehensive information on the microbial community involved in otomycosis discharge and will aid in evaluating the efficacy of clinical treatment and medication.
1 Introduction
Otomycosis, also known as otitis externa mycotica, is caused by the invasion or massive reproduction of opportunistic fungi and the production of secretions, increasing the complexity of treatment (1). Otomycosis is a common inflammatory lesion in otolaryngology that accounts for 10%–20% of ear canal infections (2). Otomycosis is very common in tropical and subtropical regions where the temperature and humidity are high (3). Frequent ear picking (4), chronic otitis media (5), and the use of antibiotics (6) all increase the incidence of otomycosis. Otomycosis can also cause symptoms such as ear itching, hearing impairment, and ear pain (7).
A. niger or A. flavus complex in the genus Aspergillus spp., and Candida albicans, Candida parapsilosis in the genus Candida spp. have been reported to be common causative agents of otomycosis (8, 9). However, the fungal community involved in otomycosis varies with region, climate and patient immune status (10). For example, Aspergillus tubeingensis is the dominant and most common isolated species in Western China and Southern Ireland (11, 12), and A. tubingensis is most common in southern Iran (13). C.albicans is more common in immunocompromised patients with otomycosis than in immunocompromised patients without otomycosis (14). Fungal infection lesions can sometimes completely cover the eardrum, causing hearing loss that is difficult to heal on its own (15). Fungal infection is more persistent and easier to relapse, so effective treatment measures should be taken in a timely manner. When treating otomycosis, the first step is to diagnose the condition, which usually involves examining the ear canal to look for signs of fungal infection, such as redness, swelling, discharge, or specific types of fungus. After diagnosis, the first step is to clear the ear canal containing the fungus and secretions. Second, the selection of appropriate antifungal drugs is crucial for the treatment of otomycosis, usually based on the type of fungus (such as Candida spp. or Aspergillus spp.) and drug resistance. Clotrimazole (16), miconazole (17) and tolnaftate (18) are currently commonly used drugs for the treatment of otomycosis. In some cases, topical antifungal ear drops may be needed.
Clinically, determining what kind of opportunistic fungal infection is involved and whether bacteria are involved are highly important for the early diagnosis and treatment of otomycosis. At present, opportunistic fungi are mainly identified clinically by isolation, culture, and microscopic examination based on fungal morphology (19). However, the above methods have several limitations, such as equipment conditions, personnel level, and staining methods (20). Moreover, the culture process easily results in contamination by pathogenic bacteria, and traditional methods can identify only the main pathogenic bacteria. Therefore, it is difficult to accurately identify pathogenic bacteria based on the abovementioned traditional methods. In recent years, with the rise of high-throughput sequencing technology, culture-free methods have been gradually introduced for the identification of pathogenic microorganisms (21). To date, high-throughput sequencing has not been widely used in the study of otomycosis. In this study, high-throughput sequencing technology was used to study the fungal and bacterial communities involved in otomycosis to provide a reference for clinical treatment.
2 Materials and methods
2.1 Ethics and consent
Patients with otomycosis were recruited from June 2022 to August 2022 from the Department of Otorhinolaryngology, Hubei Third People's Hospital Affiliated with Jianghan University. This study was approved by the Ethics Committee of the Third People's Hospital of Hubei Province Affiliated with Jianghan University (No. 2023010), and written informed consent was obtained from each patient participating in this study.
2.2 Sample collection
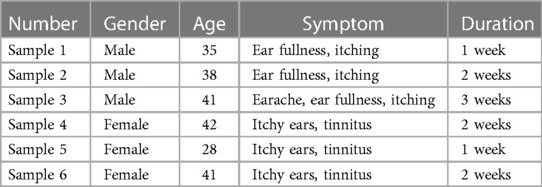
In this study, 6 patients with otomycosis who met the inclusion criteria (all had a history of ear picking, no otitis media, and no history of diabetes) were recruited. Otomycosis was diagnosed through ear canal observation, direct smears, and culture samples, and all 6 patients had ear canal discharge. The ear secretions from one ear were collected, and the patients were not treated with other drugs before collection (See Table 1 for details). Ear canal secretions were collected using sterile cotton swabs soaked in sterile saline, packed into sterile sampling tubes, and stored in a −80°C freezer, after which DNA extraction and polymerase chain reaction (PCR) amplification were performed.
2.3 DNA extraction, PCR amplification, library construction and sequencing
Fungal DNA and bacterial DNA were extracted from the samples using a FastDNA® Spin Kit for Soil (American Mpbio Corporation) (22) following the manufacturer's protocol to ensure that the DNA extracted from each sample met the requirements. DNA quality was checked using 2% agarose gel electrophoresis, and the concentration and purity of the DNA were detected using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Next, the ITS1F-ITS2R region of the fungal internal transcriptional spacer gene was amplified using primers (ITS 1F 5′-CTTGGTCATTTAGAGGAAGTAA-3′; ITS 2R 5′-GCTGCGTTCTTCATCGATGC-3′) (23); the V3-V4 region of the bacterial 16S rRNA gene was amplified using primers (338F 5′-ACTCCTACGGGAGGCAGCAG-3′; and 806R 5′-GGACTACHVGGGTWTCTAAT-3′) (24). Amplified products were purified using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) for library construction. The library was subsequently sequenced on the Illumina NextSeq 2000 platform of Shanghai Meiji Biomedical Technology Co., Ltd. The original data were uploaded to the NGDC database with the upload number CRA011427.
2.4 Data analysis
The bacterial diversity was clustered using the USEARCH11-uparse algorithm, the OTU sequence similarity was 0.97, and the species classification database was silva138/16s_bacteria, with a classification confidence of 0.7. The fungal diversity was clustered using the USEARCH11-uparse algorithm, the operational taxonomic unit (OTU) sequence similarity was 0.97, and the species classification database was unite8.0/its_fungi, with a classification confidence of 0.7. R software (version 4.1.1) was used to calculate the diversity of bacteria and fungi, and BugBase was used to predict the phenotype of bacteria in the samples (https://bugbase.cs.umn.edu/index.html).
3 Results and discussion
3.1 Sequencing results
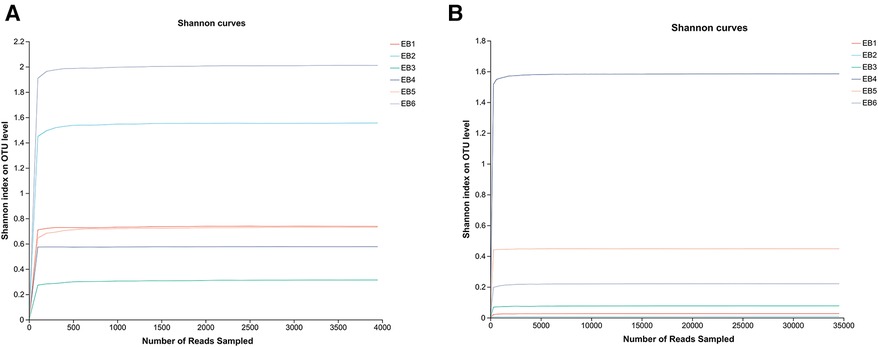
OTUs are used to classify and compare microorganisms, while the Shannon index is an ecological index that measures community diversity. In microbial community analysis, Shannon dilution curves are often used to evaluate the richness and evenness of species diversity in a sample. The OTU Shannon dilution curves of both fungi and bacteria in the otomycosis secretion samples tended to flatten (as shown in Figures 1A,B), indicating that the detection rate of microbial communities in the otomycosis secretion samples was close to saturation, and that the amount of the current sequencing could cover most of the species in the sample. This means that the sequencing performed was sufficient to cover most species in the sample, providing a reliable basis for subsequent analysis (25).
3.2 Bacterial diversity
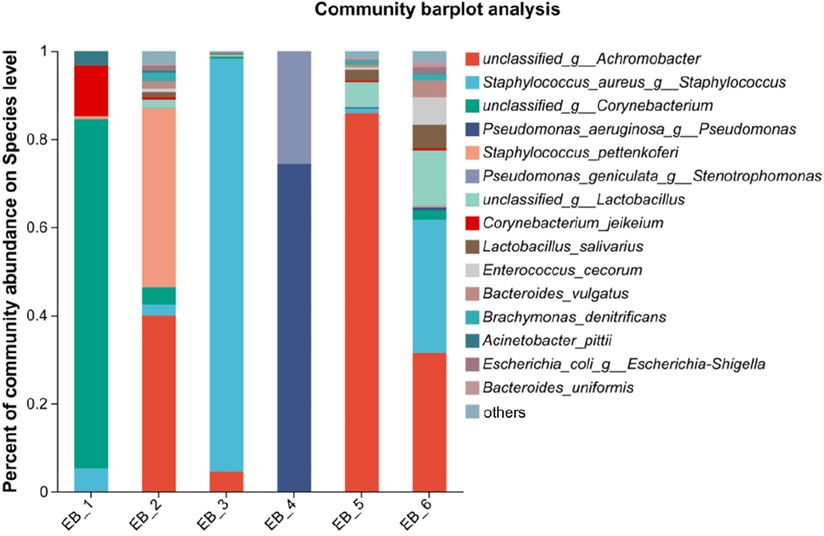
After quality control filtering and removal of chimeric sequences, a total of 257,429 sequences were generated, and 228,551 sequences were obtained after optimization, with an average length of 425 bp. These findings demonstrated that the sequencing depth was sufficient to provide reliable bacterial community analysis and was deep enough to reveal the microbial diversity in the samples (26). Approximately ≥97% of the sequences were clustered into one OTU, and the species OTUs with sequence numbers ≥5 in at least 3 samples were retained. The species OTUs with a total sequence ≥20 were retained, and the OTUs aligned to the mitochondrial sequence were removed and flattened according to the minimum sample sequence. A total of 29 OTUs were obtained. The dominant bacterial phyla (abundance >1%) were Proteobacteria, Firmicutes, and Actinobacteria, and the dominant genera were Staphylococcus spp., Achromobacter spp., Corynebacterium spp., Pseudomonas spp., and Lactobacillus spp. (as shown in Figure 2A). Among them, Staphylococcus spp. is a common pathogen and a pus-forming bacterium that was detected in 5 samples, and the average abundance was 29.05% (as shown in Figure 2B). The high abundances of Proteobacteria, Firmicutes, Actinobacteria, etc., indicate that these bacteria play a dominant role in the ear canal microbial community. Identification of dominant genera such as Staphylococcus spp., Achromobacter spp., Corynebacterium spp., Pseudomonas spp., and Lactobacillus spp. can aid in understanding the pathological mechanisms of ear canal infection (27). Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa, coagulase-negative Staphylococcus spp. and Klebsiella pneumoniae are common bacteria in the ear canal secretions of patients with suppurative otitis media (28); S. epidermidis is a normal bacterium in the ear canal (29); and the rest are pathogenic bacteria, which is similar to the results of this study. The high abundance of Staphylococcus spp. and other bacteria in ear canal secretions indicates their important role in ear canal infections. Identifying these bacteria can help develop targeted treatment plans (30). Additionally, possible bacterial infection should be considered in patients with otomycosis who clinically do not respond well to conventional antifungal drugs. The coexistence of bacteria and fungi may complicate treatment, so identification and treatment of these pathogenic bacteria should be considered during treatment (31).
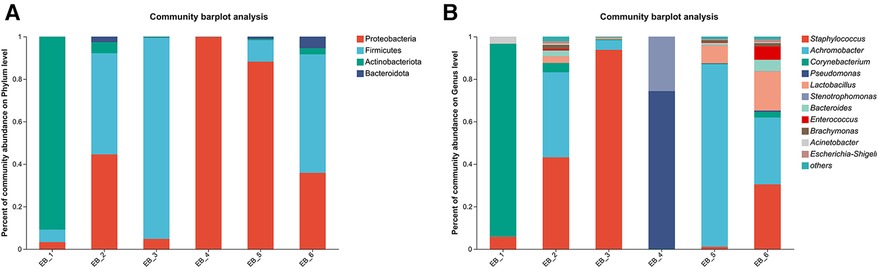
Figure 2. Phylum- and genus-level community structure map of otomycosis secretions. Different colors represent different phyla and genera, and the colored blocks on the right are sorted in descending order of abundance. (A) The community structure of the main bacteria at the phylum level. Proteobacteria, Firmicutes, and Actinobacteria were the dominant phyla. (B) The community structure of the main bacteria at the genus level. Staphylococcus spp., Achromobacter spp., Corynebacterium spp., Pseudomonas spp., and Lactobacillus spp. were the dominant genera.
At the species level (as shown in Figure 3), S. aureus was present in 3 out of the 6 samples, ranking second in average abundance (the average abundance was 22.12%). Chen et al. (5) showed that methicillin-resistant S. aureus is the main pathogen of otomycosis. Methicillin-resistant S. aureus is a major cause of bacterial infections (bacteremia, endocarditis, soft tissue infections, hospital-acquired infections, etc.) in hospital and community settings (32). P. aeruginosa was found in only sample 4. According to the analysis shown in Figure 4, P. aeruginosa significantly increased the diversity of fungi, increasing the complexity of clinical treatment. The next step should be to isolate and characterize the most abundant unidentified bacterial species. In addition, the interaction between bacteria and fungi is worthy of further exploration.
3.3 Fungal diversity
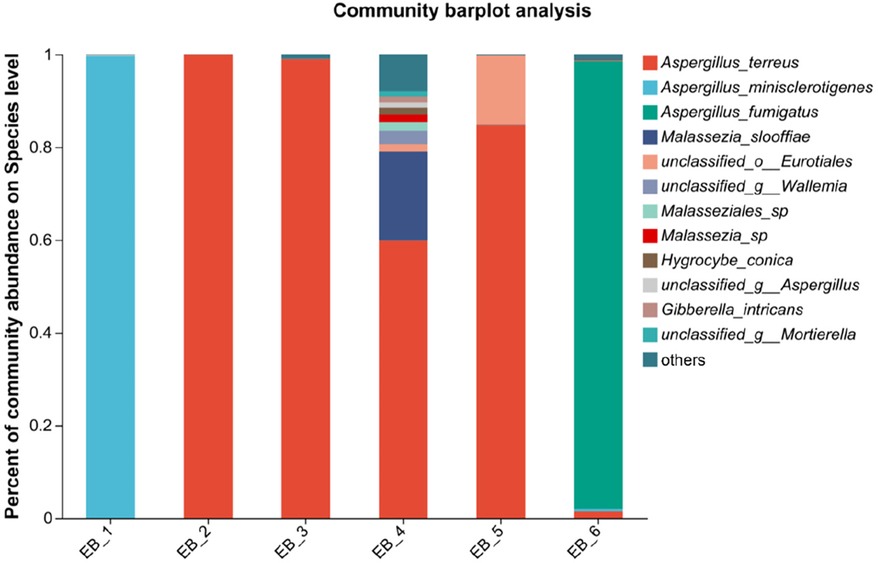
After quality control filtering and removal of chimeric sequences, a total of 252,964 sequences were generated, and 239,866 sequences were optimized, with an average length of 261 bp. ≥97% of the sequences were clustered into one OTU and smoothed according to the minimum sample sequence; a total of 80 OTUs were obtained. The dominant phylum was Ascomycota, and the dominant genus was Aspergillus spp.(average abundance 90.68%) (as shown in Figures 5A,B); these findings are consistent with the results of Gu et al. (7) on patients with otomycosis in Nanjing, Jiangsu Province, China. Aspergillus spp. is the dominant genus involved in otomycosis in southern China. This study highlights the importance of geographic location in determining human microbial community structure; the dominance of Aspergillus spp. in otomycosis in southern China may be related to environmental factors and ecosystem characteristics. According to recent studies, otomycoses are commonly caused by the A. niger complex and yeasts (Candida). Notably, the A. niger or A. flavus complex are the major fungal species causing otomycosis. The above studies highlight the complexity of otomycosis and the challenges in their treatment (33).
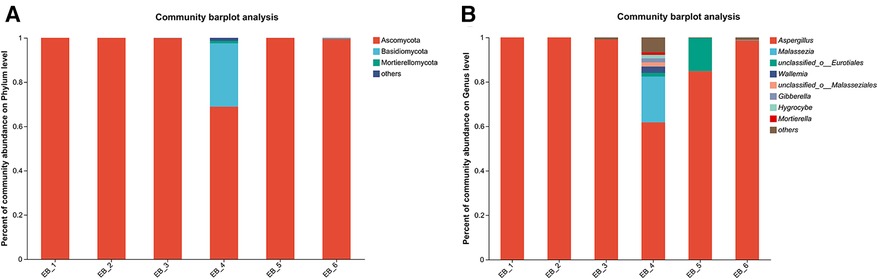
Figure 5. Phylum- and genus-level community structure diagram of otomycosis secretions. Different colors represent different phyla and genera, and the colored blocks on the right are sorted in descending order of abundance. (A) The community structure of the main fungi at the phylum level; the dominant phylum was Ascomycota. (B) The community structure of the main fungi at the genus level; the dominant genus was Aspergillus spp.
At the species level, four of the six samples were Aspergillus terreus (average abundance 57.48%), one was Aspergillus minisclerotigenes (average abundance 16.73%), and one was Aspergillus fumigatus (average abundance 16.10%) (as shown in Figure 4). The results of Zhang et al. (12) showed that the most common fungal inflammation in the external auditory canal was caused by A. terreus (50.9%), whereas A. fumigatus was less common (9.1%), similar to the results of this study. A. terreus occurs in a variety of environments and is the fourth leading cause of invasive and noninvasive aspergillosis (34). A. fumigatus is a saprophytic fungus found primarily in soil that can also cause invasive airborne infections. In immunocompromised patients, these infections are often fatal (35, 36). A. fumigatus occurs less frequently in otomycoses but remains an important pathogen. There are no reports about otomycosis caused by A. minisclerotigenes. Studies have shown that A. minisclerotigenes can cause fungal keratitis (37). This suggest that, although currently understudied, the impact of A. minisclerotigenes should also be paid attention to during the treatment of otomycosis. Therefore, if otomycosis is clinically diagnosed, the scabs and secretions in the external auditory canal of the patient should be thoroughly removed, and antifungal medication should be applied to allow the infection to act as an antifungal agent, thereby eliminating the symptoms of otomycosis. When treating otomycosis, it is important to consider that different Aspergillus spp. may have different susceptibilities to antifungal drugs. For different pathogenic bacteria, different therapeutic strategies should be adopted, and not all antifungal drugs can be used for the treatment of otomycosis; for example, among Aspergillus spp., only A. terreus is intrinsically resistant to amphotericin B (32). Next, the pathogenesis of otomycosis caused by A. minisclerotigenes and the effect of existing drugs on its treatment can be investigated.
3.4 Bacterial phenotype prediction
The results of the bacterial phenotype prediction showed that most of the bacteria were aerobic (aerobic bacteria), and the proportion of anaerobic bacteria was relatively low, which was in line with the actual environment of the ear canal. The microbial community of the ear canal is typically dominated by aerobic bacteria, consistent with the situation in which the ear canal is an aerated and drier environment. Aerobic bacteria grow and reproduce more easily in such an environment. The proportion of anaerobic bacteria in the ear canal was lower. This may be due to the natural physiological conditions of the ear canal that limit the growth of anaerobic bacteria (27). In addition, otomycosis secretions have a strong biofilm-forming ability (38), which is consistent with the dominance of Staphylococcus spp. in terms of bacterial diversity. A biofilm is a protective layer formed by microorganisms, such as bacteria and fungi, that helps them survive in harsh environments. This biofilm formation is particularly important in ear canal infections because it can lead to persistent and difficult-to-treat ear canal infections (39). Achromobacter spp. contributed 40.15% to the phenotype of bacterial biofilm formation. Corynebacterium spp. and Pseudomonas spp. contributed 15.97% and 9.32%, respectively, to the observed bacterial biofilm formation. The contribution of Acinetobacter spp. to biofilm formation was significant, consistent with its role in hospital-acquired infections. Achromobacter spp. are multidrug-resistant bacteria that can survive in hospital settings and cause serious infections, such as hospital-acquired pneumonia. Corynebacteria spp. and Pseudomonas spp. also play important roles in the formation of biofilms. The biofilm-forming ability of these bacteria is a key factor in their adaptation and survival in ear canal infections (40). In addition, most bacteria have a certain potential pathogenicity (mainly Achromobacter spp., 40.15%), which should receive increased attention in clinical treatment; that is, the harm caused by pathogenic bacteria cannot be ignored when treating fungal infections. Studies have shown that Achromobacter spp. can form biofilms (41, 42) and cause diseases, such as hospital-acquired pneumonia (HAP) (43), which is consistent with the predictive results of this phenotype. The potential pathogenicity of Achromobacter spp. needs to be taken seriously in clinical treatment. When treating otomycosis, doctors need to be aware of the harm caused by these bacteria. Because these bacteria may be resistant to multiple antibiotics, choosing the appropriate antibiotic and treatment regimen is critical for controlling and preventing the spread of infection (44).
4 Conclusion
In this study, high-throughput sequencing was used to identify the microbial community of the ear canal secretions of 6 patients with otomycosis of the external auditory canal in Wuhan, Hubei Province. The sequencing results showed that the dominant genus of fungi was Aspergillus spp., and the dominant genus of bacteria was Staphylococcus spp. The results provide comprehensive information on the microbial community of patients with otomycosis of the external auditory canal, which can provide a reference for clinical treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
This study was approved by the Ethics Committee of the Third People's Hospital of Hubei Province Affiliated to Jianghan University (No. 2023010), and written informed consent was obtained from each patient participating in this study.
Author contributions
ZC: Conceptualization, Data curation, Formal Analysis, Writing – original draft, Writing – review & editing. ZZ: Formal Analysis, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mao C, Li A, Hu J, Wang P, Peng D, Wang J, et al. Efficient and accurate diagnosis of otomycosis using an ensemble deep-learning model. Front Mol Biosci. (2022) 9:951432. doi: 10.3389/fmolb.2022.951432
2. Kiakojuri K, Mahdavi Omran S, Roodgari S, Taghizadeh Armaki M, Hedayati MT, Shokohi T, et al. Molecular identification and antifungal susceptibility of yeasts and molds isolated from patients with otomycosis. Mycopathologia. (2021) 186:245–57. doi: 10.1007/s11046-021-00537-1
3. Alarid-Coronel J, Celis-Aguilar E, Escobar-Aispuro L, Muñoz-Estrada V. Otomycosis in immunocompetent patients: clinical and mycological features. Our experience with 40 cases. Clin Otolaryngol. (2017) 43(1):373–7. doi: 10.1111/coa.12966
4. Huang W, Li Y, Huang J, Luo Y, Huang N. Endoscope ear pick: an emerging but neglected medical device. Front Med (Lausanne). (2022) 9:977554. doi: 10.3389/fmed.2022.977554
5. Chen C-H, Wang C-Y, Cheng M-Y, Hsih W-H, Tien N, Chou C-H, et al. Definite therapy of mixed infection alleviates refractory dilemma of adult chronic suppurative otitis media. J Microbiol Immunol Infect. (2022) 55:1283–92. doi: 10.1016/j.jmii.2022.07.014
6. Alshahni MM, Alshahni RZ, Fujisaki R, Tamura T, Shimizu Y, Yamanishi C, et al. A case of topical ofloxacin-induced otomycosis and literature review. Mycopathologia. (2021) 186:871–6. doi: 10.1007/s11046-021-00581-x
7. Gu X, Cheng X, Zhang J, She W. Identification of the fungal community in otomycosis by internal transcribed spacer sequencing. Front Microbiol. (2022) 13:820423. doi: 10.3389/fmicb.2022.820423
8. Tasić-Otašević S, Golubović M, Đenić S, Ignjatović A, Stalević M, Momčilović S, et al. Species distribution patterns and epidemiological characteristics of otomycosis in Southeastern Serbia. J Mycol Med. (2020) 30:101011. doi: 10.1016/j.mycmed.2020.101011
9. Sukumar B, Premamalini T, Shree SN, Kindo AJ. P022 luliconazole—a novel potent imidazole activity against Aspergillus niger and Aspergillus flavus causing otomycosis. Med Mycol. (2022) 60:22. doi: 10.1093/mmy/myac072.p022
10. Westby D, O'Connell N, Powell J, Fenton JE. The changing nature of paediatric otomycosis in the Mid-West of Ireland. J Laryngol Otol. (2020) 134:592–6. doi: 10.1017/s0022215120001164
11. Zhang LL, Wang X, Houbraken J, Mei H, Deng S. Molecular identification and in vitro antifungal susceptibility of Aspergillus isolates recovered from otomycosis patients in Western China. Mycopathologia. (2020) 185:527–35. doi: 10.1007/s11046-020-00448-7
12. Zhang S, Jin M, Hu S, Zhang Y, Zhou G. Administration of 1% topical voriconazole drops was effective and safe in the treatment of refractory otomycosis without tympanic membrane perforation. Ann Otol Rhinol Laryngol. (2020) 130:273–9. doi: 10.1177/0003489420946783
13. Javidnia J, Ghotbi Z, Ghojoghi A, Solhjoo K, Alshahni MM, Jeddi SA, et al. Otomycosis in the south of Iran with a high prevalence of tympanic membrane perforation: a hospital-based study. Mycopathologia. (2022) 187:225–33. doi: 10.1007/s11046-022-00626-9
14. Pooja G, Kirtivardhan V, O'Horo JC, Aditya S. Fungal culture diagnostic stewardship: an avenue for antimicrobial stewardship in the immunocompromised host. Open Forum Infect Dis. (2019) 6:148. doi: 10.1093/ofid/ofz360.342
15. Gheorghe DC, Niculescu A-G, Bîrcă AC, Grumezescu AM. Nanoparticles for the treatment of inner ear infections. Nanomaterials. (2021) 11:1311. doi: 10.3390/nano11051311
16. Kiakojuri K, Rajabnia R, Mahdavi Omran S, Pournajaf A, Karami M, Taghizadeh Armaki M. Role of clotrimazole in prevention of recurrent otomycosis. BioMed Res Int. (2019) 2019:5269535. doi: 10.1155/2019/5269535
17. Nemati S, Gerami H, Faghih Habibi A, Kazemnejad E, Shabani Asl N, Aghsaghloo V, et al. Sertaconazole versus clotrimazole and miconazole creams in the treatment of otomycosis: a placebo-controlled clinical trial. Iran J Otorhinolaryngol. (2022) 34:27–34. doi: 10.22038/ijorl.2021.54805.2872
18. Jimenez-Garcia L, Celis-Aguilar E, Díaz-Pavón G, Muñoz Estrada V, Castro-Urquizo Á, Hernández-Castillo N, et al. Efficacy of topical clotrimazole vs. topical tolnaftate in the treatment of otomycosis. A randomized controlled clinical trial. Braz J Otorhinolaryngol. (2020) 86:300–7. doi: 10.1016/j.bjorl.2018.12.007
19. de Souza Costa P, Prado A, Bagon NP, Negri M, Svidzinski TIE. Mixed fungal biofilms: from mycobiota to devices, a new challenge on clinical practice. Microorganisms. (2022) 10:1721. doi: 10.3390/microorganisms10091721
20. Lu G, Wang Z, Zhang B, Zhou Z, Hu D, Zhang D. Detecting forest musk deer abscess disease pathogens using 16S rRNA high-throughput sequencing technology 13:3142. Animals (Basel). (2023) 13(19):3142. doi: 10.3390/ani13193142
21. Miller S, Chiu C. The role of metagenomics and next-generation sequencing in infectious disease diagnosis. Clin Chem. (2021) 68:115–24. doi: 10.1093/clinchem/hvab173
22. Bou Orm E, Sauvagère S, Rocher J, Benezet J-C, Bayle S, Siatka C, et al. Estimating the bias related to DNA recovery from hemp stems for retting microbial community investigation. Appl Microbiol Biotechnol. (2023) 107:4665–81. doi: 10.1007/s00253-023-12582-5
23. Adams RI, Miletto M, Taylor WJ, Bruns TD. Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. (2013) 7:1262–73. doi: 10.1038/ismej.2013.28
24. Xu N, Tan G, Wang H, Gai X. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur J Soil Biol. (2016) 74:1–8. doi: 10.1016/j.ejsobi.2016.02.004
25. Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. (2001) 67:4399–406. doi: 10.1128/aem.67.10.4399-4406.2001
26. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the illumina HiSeq and MiSeq platforms. ISME J. (2012) 6:1621–4. doi: 10.1038/ismej.2012.8
27. Brook I. The role of bacterial interference in otitis, sinusitis and tonsillitis. Otolaryngol Head Neck Surg. (2005) 133:139–46. doi: 10.1016/j.otohns.2005.03.012
28. Kaźmierczak W, Janiak-Kiszka J, Budzyńska A, Nowaczewska M, Kaźmierczak H, Gospodarek-Komkowska E. Analysis of pathogens and antimicrobial treatment in different groups of patients with chronic otitis media. J Laryngol Otol. (2022) 136:219–22. doi: 10.1017/s0022215121003224
29. Méric G, Mageiros L, Pensar J, Laabei M, Yahara K, Pascoe B, et al. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat Commun. (2018) 9:5034. doi: 10.1038/s41467-018-07368-7
30. Post JC. Direct evidence of bacterial biofilms in otitis media. Laryngoscope. (2015) 125(9):2003–14. doi: 10.1002/lary.25291
31. Marom T, Nokso-Koivisto J, Chonmaitree T. Viral-bacterial interactions in acute otitis media. Curr Allergy Asthma Rep. (2012) 12:551–8. doi: 10.1007/s11882-012-0303-2
32. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. (2019) 17:203–18. doi: 10.1038/s41579-018-0147-4
33. Bojanović M, Ignjatović A, Stalević M, Arsić-Arsenijević V, Ranđelović M, Gerginić V, et al. Clinical presentations, cluster analysis and laboratory-based investigation of Aspergillus otomycosis—a single center experience. Journal of Fungi. (2022) 8:315. doi: 10.3390/jof8030315
34. Vahedi Shahandashti R, Lass-Flörl C. Antifungal resistance in Aspergillus terreus: a current scenario. Fungal Genet Biol. (2019) 131:103247. doi: 10.1016/j.fgb.2019.103247
35. Latgé J-P, Chamilos G. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev. (2019) 33. doi: 10.1128/cmr.00140-18
36. Arastehfar A, Carvalho A, Houbraken J, Lombardi L, Garcia-Rubio R, Jenks JD, et al. Aspergillus fumigatus and aspergillosis: from basics to clinics. Stud Mycol. (2021) 100:100115. doi: 10.1016/j.simyco.2021.100115
37. Karimizadeh EM, Eslampoor A, Dolatabadi S, Najafzadeh MJ, Houbraken J. First case of fungal keratitis due to Aspergillus minisclerotigenes in Iran. Current Medical Mycology. (2019) 5:45. doi: 10.18502/cmm.5.2.1162
38. Ballah FM, Islam MS, Rana ML, Ferdous FB, Ahmed R, Pramanik PK, et al. Phenotypic and genotypic detection of biofilm-forming Staphylococcus aureus from different food sources in Bangladesh. Biology (Basel). (2022) 11:949. doi: 10.3390/biology11070949
39. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. (1999) 284:1318–22. doi: 10.1126/science.284.5418.1318
40. Percival SL, Suleman L, Vuotto C, Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J Med Microbiol. (2015) 64:4. doi: 10.1099/jmm.0.000032
41. Cameron LC, Bonis B, Phan CQ, Kent LA, Lee AK, Hunter RC. A putative enoyl-CoA hydratase contributes to biofilm formation and the antibiotic tolerance of Achromobacter xylosoxidans. NPJ Biofilms Microbiomes. (2019) 5:20. doi: 10.1016/10.1038/s41522-019-0093-6
42. Sandri A, Saitta GM, Veschetti L, Boschi F, Passarelli Mantovani R, Carelli M, et al. In vivo inflammation caused by Achromobacter spp. cystic fibrosis clinical isolates exhibiting different pathogenic characteristics. Int J Mol Sci. (2023) 24:7432. doi: 10.3390/ijms24087432
43. Chao L, Fei P, Jun G, Weifeng Y, Yi J, Changting L, et al. Hospital acquired pneumonia due to Achromobacter spp. in a geriatric ward in China: clinical characteristic, genome variability, biofilm production, antibiotic resistance and integron in isolated strains. Front Microbiol. (2016) 7:621. doi: 10.3389/fmicb.2016.00621
Keywords: otomycosis, fungi, bacteria, microbial community, diversity
Citation: Chen Z and Zhao Z (2024) Study on the microbial diversity of ear canal secretions from patients with otomycosis. Front. Surg. 11:1277799. doi: 10.3389/fsurg.2024.1277799
Received: 15 August 2023; Accepted: 30 January 2024;
Published: 9 February 2024.
Edited by:
Suzana Otašević, University of Niš, SerbiaReviewed by:
Javad Javidnia, Mazandaran University of Medical Sciences, IranIman Haghani, Mazandaran University of Medical Sciences, Iran
© 2024 Chen and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhang Zhao zhaozhang19827@163.com
 Zhuxiang Chen
Zhuxiang Chen  Zhang Zhao
Zhang Zhao