Specific Detection and Differentiation Between Brucella melitensis and Brucella abortus by a Duplex Recombinase Polymerase Amplification Assay
- 1State Key Laboratory of Veterinary Etiological Biology, Key Laboratory of Grazing Animal Diseases of Ministry of Agriculture, Key Laboratory of Veterinary Public Health of Ministry of Agriculture, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2Kassala Veterinary Research Laboratory, Central Veterinary Research Laboratory, Animal Resources Research Corporation, Khartoum, Sudan
Brucellosis is a highly contagious zoonosis caused by a species under the genus Brucella. A duplex recombinase polymerase amplification (Duplex RPA) assay for the specific detection of Brucella melitensis and Brucella abortus was developed in this study. Primers were designed targeting hypothetical protein genes and membrane transporter genes of B. melitensis and B. abortus, respectively. The newly developed assay was validated for its analytical sensitivity and specificity. Different samples were collected from the Qinghai, Inner Mongolia, and Xinjiang provinces. After DNA extraction, the samples were analyzed by Duplex RPA, real-time PCR, and multiplex AMOS PCR to estimate the prevalence of brucellosis in sheep and yak in West China. The analytical sensitivities of Duplex RPA were 9 × 102 plasmid copies of B. melitensis and 9 × 101 plasmid copies of B. abortus, but by mixing the reaction tubes after 4 min of incubation, the sensitivities were 4 × 100 and 5 × 100 copies of B. melitensis and B. abortus, respectively. There was no cross-reactivity with Brucella suis, Chlamydia abortus, Salmonella typhimurium, Escherichia coli, and Toxoplasma gondii. The screening of field samples by Duplex RPA revealed that the prevalence of B. melitensis in sheep and yak was 75.8% and the prevalence of B. abortus was 4.8%. Multiplex AMOS PCR showed that the prevalence of B. melitensis was 19.3%, and that of B. abortus was 4.8%. It was concluded that the developed Duplex RPA is sensitive and specific to the detection of and differentiation between B. melitensis and B. abortus which will be useful in epidemiological surveillance and in the clinical settings.
Introduction
Brucellosis is a contagious zoonotic disease caused by different species under the genus Brucella (1). Brucella melitensis, Brucella abortus, and Brucella suis are responsible for severe human infection in addition to economic losses in livestock due to abortions and loss of fertility (2). Two species B. melitensis and B. abortus are considered the most important species that cause illness in humans and domestic animals in many countries throughout the world. Three biovars of B. melitensis mainly infect sheep and goats, while the preferred hosts of seven biovars of B. abortus are cattle and buffaloes (3). The main clinical manifestations of B. melitensis infection in sheep and goats and B. abortus in cattle are abortion and stillbirth in females and orchitis and loss of fertility in males (4). The distribution of B. melitensis has long been associated with the Mediterranean littoral, however, it is now known to be more widely distributed, with only North America, North Europe, South-East Asia and Oceania being spared (5). B. abortus is distributed in many African, European, Asian, and American countries (6). Identification of Brucella organisms at the species and biotype level, mainly relies on culture, isolation, and subsequent identification by morphology, biochemical tests, reaction with monospecific antisera, sensitivity to dyes, and phage lysis (7). Several molecular methods have been developed to detect Brucella at the species level despite the high sequence homology between different species (8). The first species-specific PCR to be developed was multiplex AMOS PCR which can detect B. melitensis, B. abortus biovar 1, 2, and 4, B. ovis, and B. suis biovar 1 (9). Later Bruce-Ladder multiplex PCR was developed to identify 10 species in addition to vaccine strains of B. abortus and B. melitensis (10). Real-time PCR and multiplex PCR microarrays have been established to identify species (11, 12). Real-time and lateral flow dipstick RPAs have been developed for the detection of Brucella spp. (13). Because of the high prevalence of B. melitensis and B. abortus in many countries throughout the world, their economic impact is due to losses of productive livestock. In addition to their zoonotic potential, we developed a novel Duplex RPA for the species specific detection of B. melitensis and B. abortus and to differentiate between them, and then validated the assay in field samples compared to real-time PCR and multiplex AMOS PCR. Another target of this work was to estimate the prevalence of brucellosis outbreaks in the Qinghai, Inner Mongolia, and Xinjiang provinces in West China by cross-sectional molecular detection by Duplex RPA, real-time PCR, and multiplex AMOS PCR.
Materials and Methods
Bacterial Strains
B. melitensis biovar 3, B. melitensis biovar 1 M5 vaccine strain, B. abortus S19, and B. suis biovar 1 S2 vaccine were used as reference strains. The Reference DNA was obtained from Lanzhou Veterinary Research Institute and Harbin Veterinary Research Institute and was confirmed by AMOS PCR (9). The concentration of B. melitensis biovar 3 DNA was 10 ng/μL, that of B. melitensis biovar 1 M5 strain was 13 ng/μL, that of B. abortus S19 was 14 ng/μL, and that of B. suis biovar 1 S2 vaccine strain was 11 ng/μL. The purity of DNA samples measured by the ratio A260/A280 was 1.8–2.0.
Collection of Samples and DNA Extraction
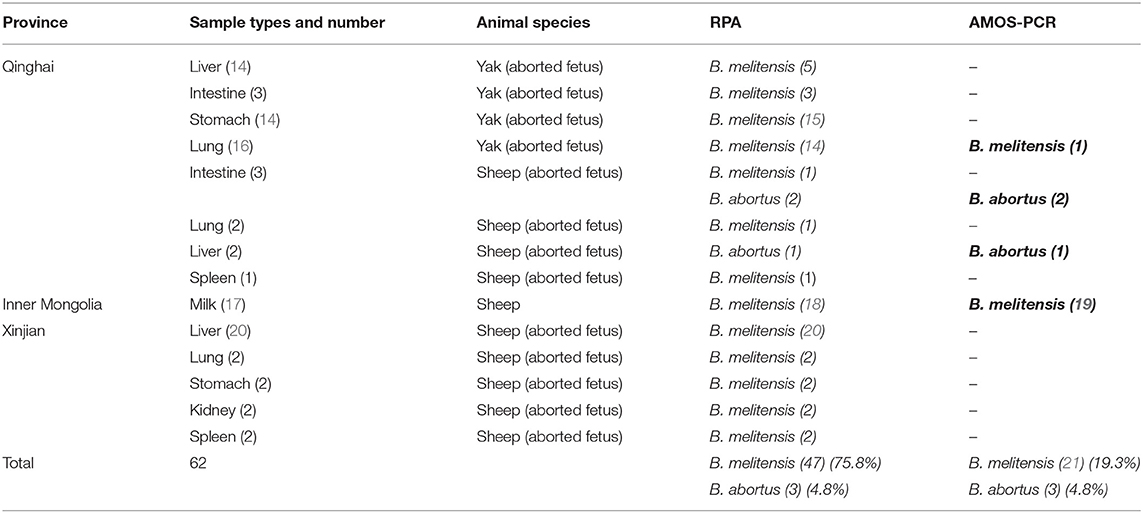
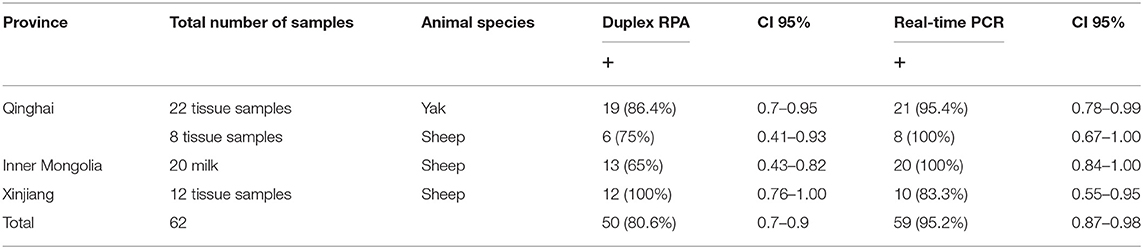
Sixty-two different samples from sheep and yak were collected from the Qinghai, Inner Mongolia, and Xinjiang provinces. Tissue samples were collected in sterile plastic bags and milk samples were collected in sterile vials. The number and types of samples from each province are described in Table 1. Genomic DNA was extracted using the TIANamp Genomic DNA kit (TIANGEN Biotech, Beijing, China). Tissue samples were cut into small pieces and ground in a mortar and pestle. Then small portions of grinded tissues (>10 mg) were lysed by the addition of 200 μL of GA buffer, 20 μL of proteinase K (provided in the kit), and RNase (100 mg/mL), and were incubated in a water bath at 56°C for 20–30 min until completely lysed. The next steps were carried out following the kit manufacturer's instructions. Milk samples (10 mL) were centrifuged at 3,000 RPM for 5 min, and fluid between the supernatant and deposit was pipetted and discarded. The supernatant and deposit were mixed together, transferred to 1.5 mL tubes, and then lysed with the addition of 200 μL GA buffer, 20 μL proteinase K, and RNase (100 mg/mL), and were incubated at 56°C for 10 min. The remaining steps were performed as outlined in the kit manual. The extracted DNA samples were quantified using NanoDrop (Infinite 200 PRO, TECAN, Groedig, Austria). DNA samples were stored at −20°C until analysis.
Selection of Specific Sequences and Bioinformatics Analysis
According to the previous studies of (9, 15, 20), the specific sequence regions of B. melitensis (Accession number: CP007763.1 of B. melitensis 16 M strain) of the hypothetical protein gene and B. abortus (Accession number: AE017224.1 of B. abortus strain 9-941) of the membrane transporter gene was selected and retrieved from the NCBI GenBank database (http://www.ncbi.nih.gov). The similarities between each sequence and other sequences were determined by the Basic Local Aligned Sequence Tool (BLAST). Multiple sequence alignment of each B. melitensis and B. abortus sequence was carried out by the MegAlign software (DNASTAR Lasergene, Madison, Wisconsin USA).
Primers Design
Primers were designed according to the appendix in the TwistAmp™ reaction kit manuals (http://www.twistdx.co.uk/images/uploads/docs/Appendix.pdf). Unlike PCR, the RPA primers should be 30–35 nucleotides in length, with GC content between 40 and 60%, no shorter than 30 nucleotides and longer than 45 nucleotides, no long tracks of guanines at the 5' end, Gibbs free energy (-_ΔG) should be between −4 and −5 kcal/mol for both the 5' and 3' ends of the primers. The species-specific primer pairs were designed by the Oligo Primers analysis software (Version 6.31 Molecular Biology Insights, Inc., USA), the parameters in the software such as primer length, and amplified sequence length range were adjusted, the primers were synthesized by (Tsingke Biological Technology, Xian, China).
Duplex Recombinase Polymerase Amplification (Duplex RPA) Development and Primary Experimentation
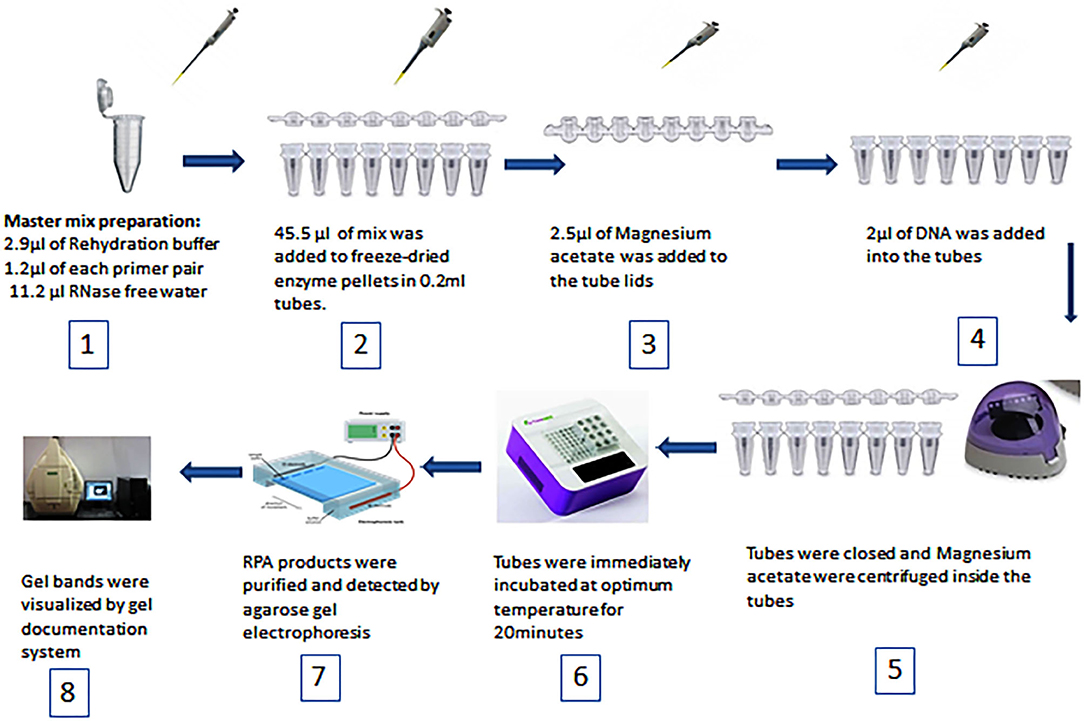
Duplex RPA was performed with a volume of 50 μL using a TwistAmp Basic kit (TwistDx, Cambridge, United Kingdom). The master mix was composed of 29.5 μL of RPA rehydration buffer, 1.2 μL (5 μM) of each primer, 11.2 μL of RNase free water (TAKARA Clontech, Shiga, Japan), 2.5 μL (280 mM) of magnesium acetate, and 2 μL of the DNA template. All reagents were prepared in 1.5 mL tubes except magnesium acetate and the DNA template. Then 45.5 μL of master mix was added to freeze-dried enzyme pellets in 0.2 mL reaction tubes each containing dried enzyme pellets. Magnesium acetate was pipetted into the tube lids. Subsequently 2 μL of the standard or genomic DNA template was added to each tube. The tubes were closed, the magnesium acetate was centrifuged into the tubes using a Minispin centrifuge, and the tubes were immediately placed in a dry bath heat block (TIANGEN Biotech, Beijing, China). To select the optimum temperature, the reaction tubes were incubated at 37, 38, and 39°C for 20 min. After incubation, the RPA reaction products were purified by the TIANquick Midi Purification Kit (TIANGEN Biotech, Beijing, China). The purification procedure was carried out according to the steps outlined in the kit manual. Purified RPA products were loaded in a 1.5–2% agarose gel and agarose gel electrophoresis was carried out for 15–30 min. The specific bands were visualized by a gel documentation system (BIORAD, Hercules, California, USA). The steps of RPA are demonstrated in Figure 1. The total time of the RPA reaction was between 45 and 60 min.
To examine candidate primers and optimal temperature, genomic DNA of B. melitensis biovar 3 and B. abortus strain 19 were used as positive control and B. suis biovar 1 S2 vaccine was used as negative control in addition to RNase free water as a non-template control.
Construction of Positive Control Plasmid DNA
The positive control plasmid DNA was constructed by ligation of a specific sequence of B. melitensis and B. abortus to the plasmid vector pMD-19 (TAKARA Clontech, Shiga, Japan). After purification of the RPA product, agarose gel electrophoresis, and the visualization of the bands, specific bands of B. melitensis and B. abortus were cut and subjected to gel DNA extraction by an AxyPrep DNA gel extraction kit (Axygen Bioscience, California, USA). After extraction from the gel, DNA was ligated to the plasmid vector by the addition of 4 μL of DNA to 1 μL of vector and 5 μL of 2 × solution (provided with plasmid vector) and then incubated at 16°C overnight. The transformation was performed after the incubation of DNA, plasmid vector, and 2 × solution mixtures by the addition of 5 μL of mixture to 50 μL of Escherichia coli DH5α competent cells in a 1.5 mL tube. The tube was then incubated on ice for 30 min, placed in a water bath at 42°C for 60–90 s, and placed again on ice for 3 min. One milliliter of Luria-Bertani (LB) broth was added to the tube which was then incubated in a shaker incubator at 37°C for 45 min. Luria Bertani (LB) agar plates containing 100 μg/mL ampicillin were inoculated with 100 μL of the cultured LB broth. The plates were incubated at 37°C for 18–24 h. Individual colonies (10, 13) were cultured into 1 mL of LB broth and incubated at 37°C in a shaker incubator. The plasmid DNA was extracted from the cultured broth by the TIAN prep Rapid Mini Plasmid Kit (TIANGEN Biotech, Beijing, China). The extracted plasmid was quantified by NanoDrop (Infinite 200 PRO, TECAN, Groedig, Austria). The copy numbers were calculated by the equation: number of copies = (amount * 6.022 × 1023)/(length * 1 × 109 * 650). The quantified plasmid was serially diluted 10-fold with elution buffer supplied in the extraction kit and analyzed by RPA to determine analytical sensitivity (detection limit).
Analytical Sensitivity of Duplex RPA
To determine the analytical sensitivity (detection limit) of Duplex RPA, serial dilutions of the constructed plasmid of B. melitensis ranging from 9 × 107 and 9 × 101 copy numbers per reaction and B. abortus ranging from 9 × 106 and 9 × 101 copy numbers per reaction were analyzed at 38°C for 20 min. Two microliters of each dilution were added to a single 0.2 mL tube, and in the last one 2 μL of RNase free water was added instead of the DNA template, which was considered a non-template control. Every run was repeated three times. To increase the sensitivity of Duplex RPA, further evaluation was performed by testing other 10-fold dilutions of the constructed plasmids of B. melitensis ranging from 4 × 105 and 4 × 100 copy numbers per reaction and B. abortus ranging from 5 × 106 and 5 × 100 per reaction. Each 0.2 mL tube received 2 μL of DNA of each dilution and the last tube received 2 μL of RNase free water added to be considered as a non-template control. The reaction tubes (0.2 mL) were removed from the dry bath heat block after 4 min from the beginning and were mixed by inversion 10 times and then placed back into the dry bath heat block. The reaction tubes were incubated at 38°C for 20 min. Every run was repeated three times.
Analytical Specificity of Duplex RPA
To determine the analytical specificity, 2 μL of DNA of B. melitensis and B. melitensis M5, B. abortus S19, B. abortus, and B. suis S2 was first confirmed by AMOS PCR (9) and analyzed by Duplex RPA at an optimal temperature for 20 min. Other DNA samples (2 μL) of organisms such as Chlamydia abortus, Salmonella typhimurium, E. coli, and Toxoplasma gondii were analyzed by Duplex RPA to determine analytical specificity.
Real-Time PCR
The primer pair and probe of the real-time PCR assay were published by (14). Real-time PCR was prepared in a total volume of 20 μL. The total mix contained 10 μL of TaqMan Master mix (AceQs qPCR Probe Master Mix, Vanzyme, Nanjing, China), 0.4 μL of each primer, 0.2 μL of probe, 7 μL of RNase free water, and 2 μL of the DNA template. The reaction conditions were 95°C for 5 min followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The reaction was incubated in a real-time PCR thermocycler (BIORAD, Hercules, California, USA). The primer pair and probe used in the real-time PCR are presented in Table 2.
Multiplex AMOS PCR
The AMOS (abortus-melitensis-ovis-suis) (9, 16) PCR was performed using five sets of primers (Table 2). The total reaction mix consisted of: 25 μL of 2XTaq master mix (TIANGEN Biotech, Beijing, China), 0.2 μM of each primer and RNase free water (TAKARA Clontech, Shiga, Japan), and 2 μL of the DNA template. The PCR conditions were as follows: initial denaturation at 96°C for 5 min followed by 40 cycles of 95°C denaturation for 1 min, 55.5°C annealing for 2 min and 72°C extension for 2 min, and a final extension at 72°C for 5 min. Species-specific bands were visualized after agarose gel electrophoresis.
Screening of Field Samples With Duplex RPA and AMOS PCR
The collected field samples were simultaneously screened by Duplex RPA, real-time PCR, and AMOS PCR for the species-specific detection of B. melitensis and B. abortus. RPA and PCR procedures were performed as described above.
Statistical Analysis
The results of the field sample screening by Duplex RPA and real-Time PCR were analyzed by Microsoft Excel version 2010 to estimate the prevalence of brucellosis in aborted and seropositive animals. The upper and lower limits of prevalence at 95% confidence intervals were calculated by Wilson score intervals using the online calculator available on the website: http://epitools.ausvet.com.au/content.php?page=CIProportion.
Results
Detection of Bacterial Strains by AMOS PCR
Bacterial strains showed positive AMOS PCR results characterized by a specific band for each species (Figure 2). B. melitensis yielded a band of 730 bp, B. abortus had a 498 bp band, and B. suis produced a 285 bp band.
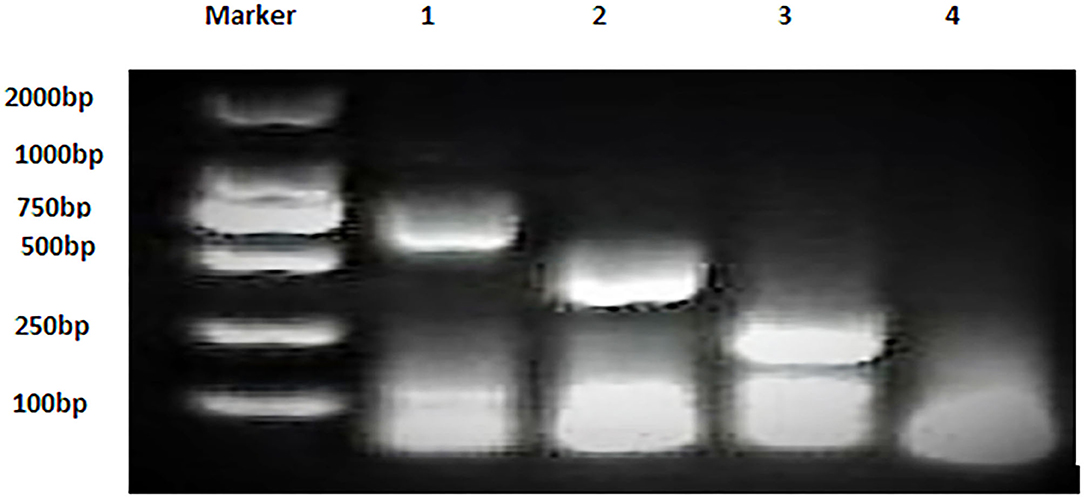
Figure 2. Multiplex AMOS PCR of Brucella spp. strains. Lane 1: B. melitensis 730 bp, Lane 2: B. abortus 498 bp, Lane 3: B. suis 285 bp, and Lane 4: non-template control RNase free water.
Sequence Alignments and Primer Pairs
The results obtained from BLAST revealed the high similarity of each sequence of B. melitensis and B. abortus to different strains of B. melitensis and B. abortus. The selected B. melitensis sequence showed some similarity with Brucella ceti and B. suis. Thus, the selected primers were located in the B. melitensis specific region. The selected sequence region of B. abortus did not show any similarities with other species. The alignment of partial sequences from B. melitensis is demonstrated in Figure 3. The partial sequence of B. abortus and its similar sequence alignments are shown in Figure 4. The primer pairs of B. melitensis and B. abortus are presented in Table 1. Forward and reverse primer locations are indicated in each sequence of B. melitensis and B. abortus by black highlights in Figures 3, 4.
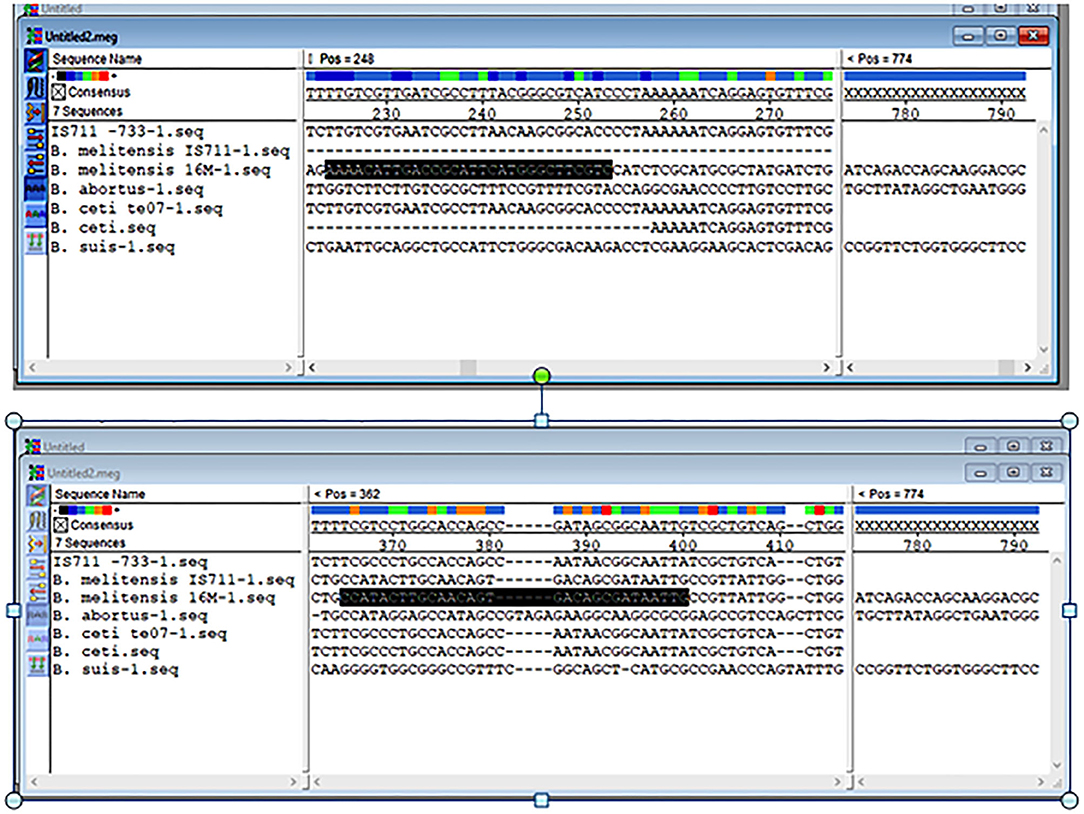
Figure 3. Alignments of the partial sequence of B. melitensis IS711 and hypothetical protein coding region. The multiple sequences have been aligned by the Clustal W method.
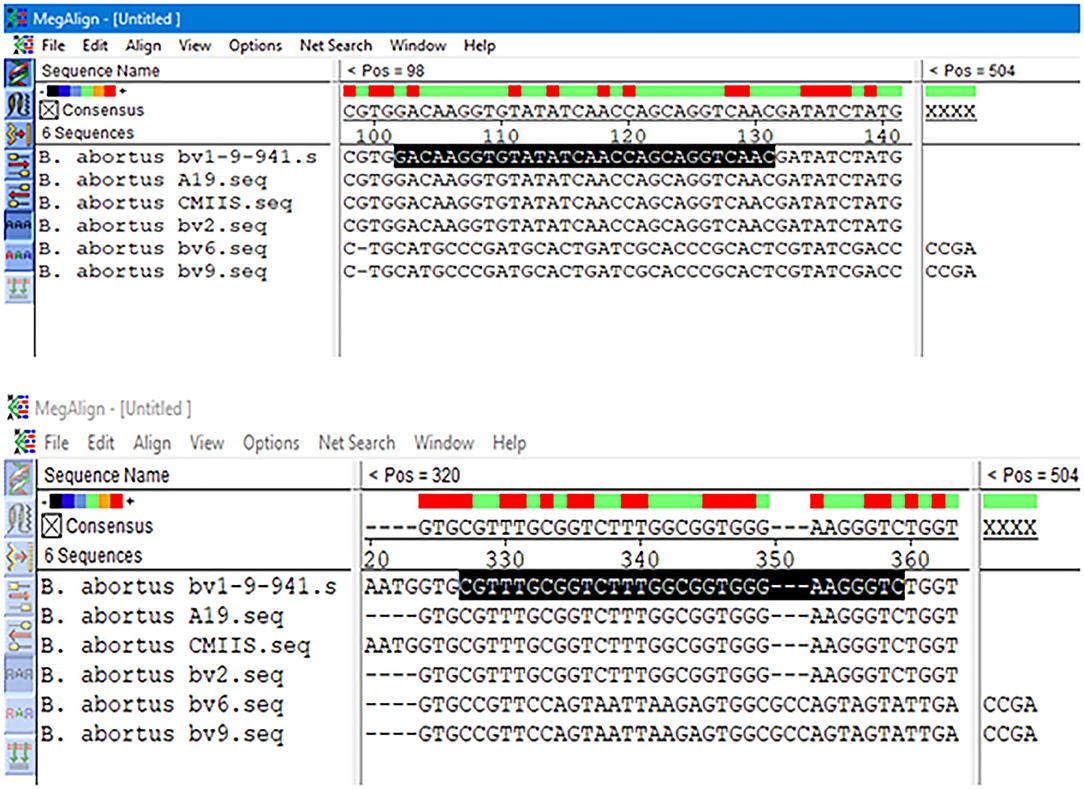
Figure 4. Alignments of B. abortus outer membrane transporter gene coding region different partial sequences. The alignment tool was Clustal W.
Duplex RPA Optimal Conditions
Duplex RPA can be performed at reaction temperatures ranging from 37 to 39°C. There was no difference in the results between 37, 38, and 39°C. The selected reaction conditions were 38°C for 20 min. The selected temperature was arbitrarily selected. The selected primers of B. melitensis yielded a 167 bp band, while B. abortus yielded a 235 bp band.
Analytical Sensitivity and Specificity
The analytical sensitivity of Duplex RPA by amplifying different serial dilutions of plasmids bearing target sequences of B. melitensis and B. abortus was 9 × 102 copies of the constructed plasmid of B. melitensis and 9 × 101 copies of B. abortus plasmid (Figures 5A, 6A). The test could detect B. melitensis biovar 3, B. melitensis M5, and B. abortus A19 (Figures 6, 7A). The mixing of the RPA reaction 4 min after starting increased the sensitivity to 4 × 100 and 5 × 100 copies of B. melitensis and B. abortus, respectively (Figures 5B, 6B). There was no cross-reactivity of the developed RPA with B. suis S2 and other bacteria, including C. abortus, S. typhimurium, E. coli, and the parasite T. gondii (Figure 7B).
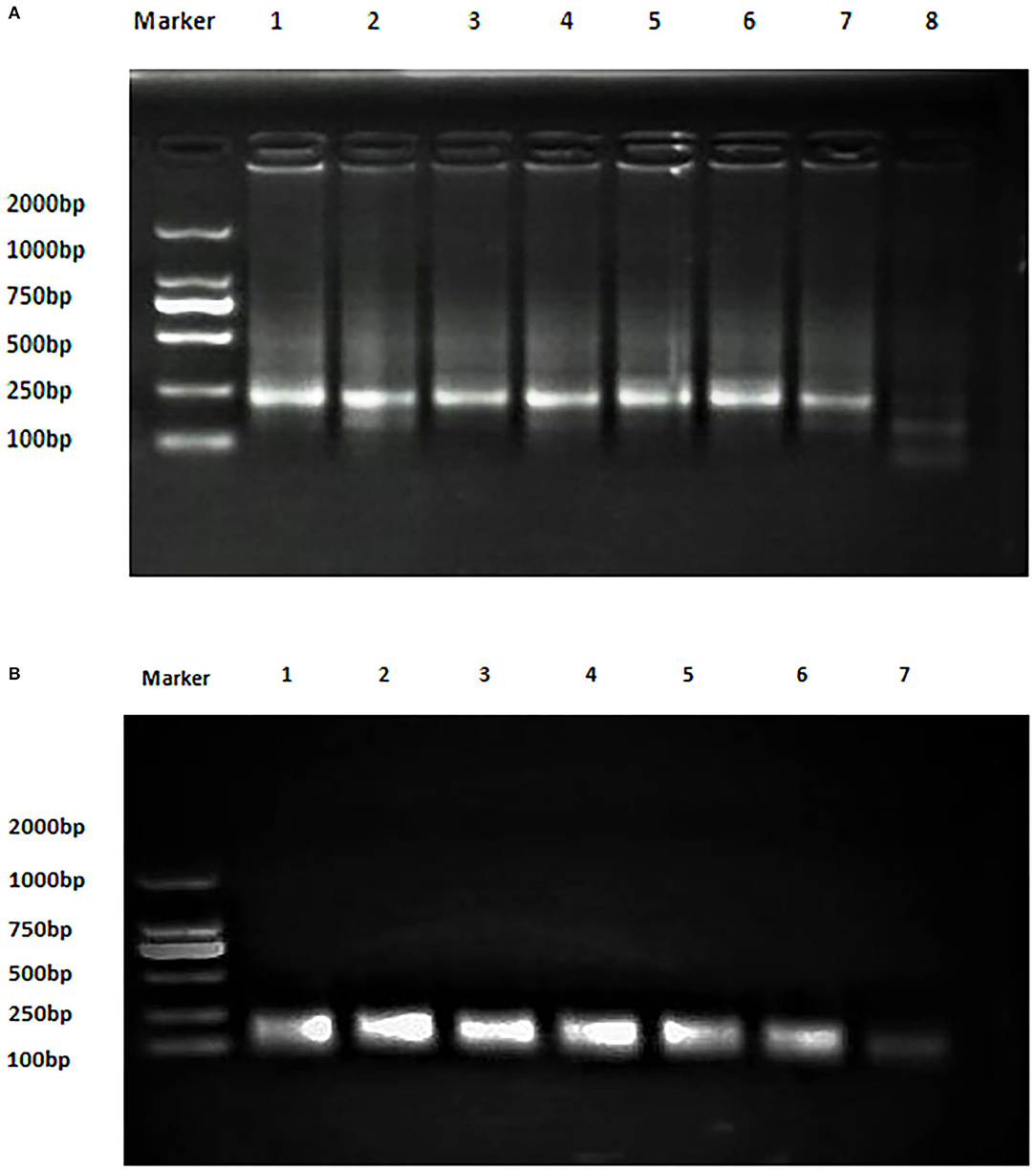
Figure 5. Analytical sensitivity of B. melitensis and B. abortus RPA: (A) B. abortus melitensis plasmid serial dilutions, DNA marker 2,000 bp. Lane 1: 9 × 107, Lane 7: 9 × 101 considered negative. (B) B. abortus plasmid serial dilutions, DNA marker 2,000 bp, Lane 1: 9 × 106 and Lane 7 is the non-template control (RNase free water).
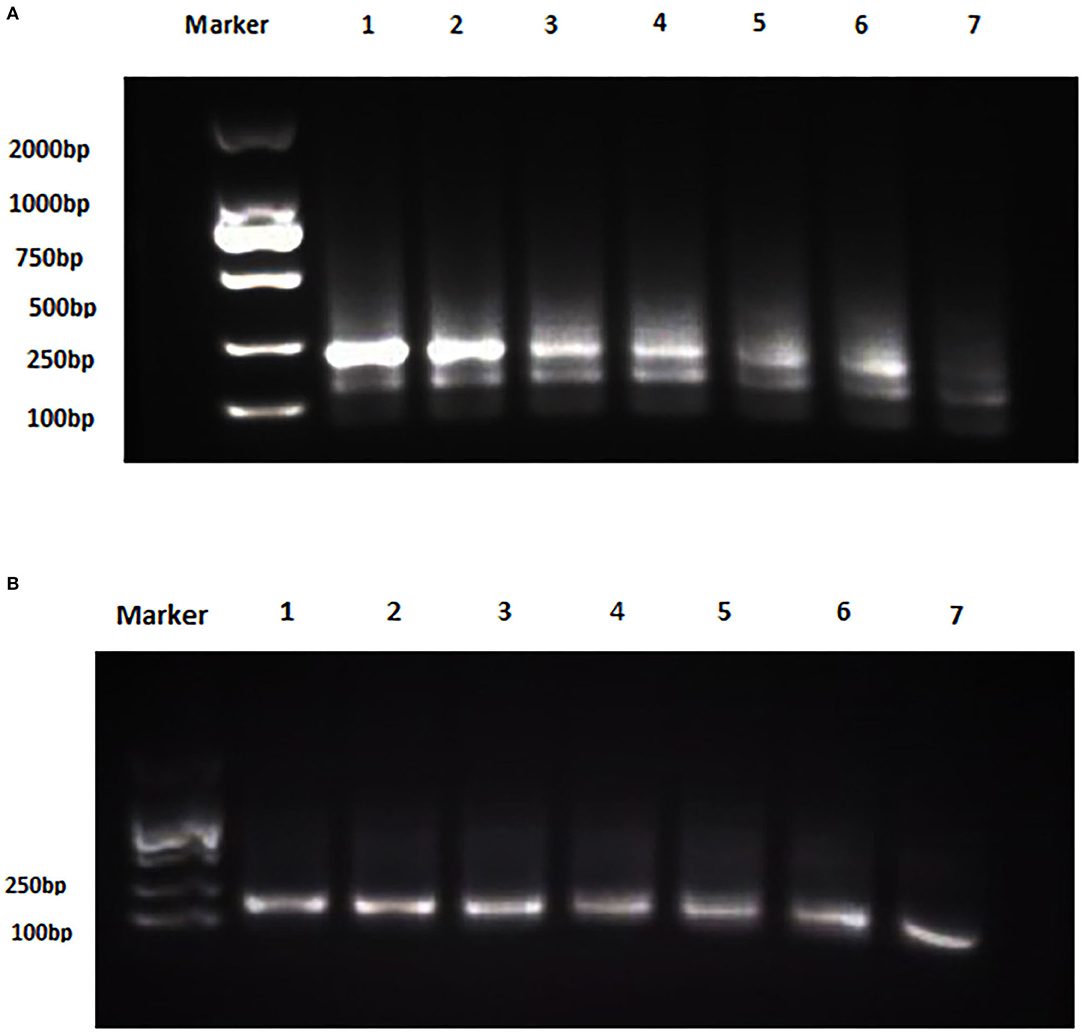
Figure 6. Analytical sensitivity of B. melitensis and B. abortus RPA: (A) B. melitensis plasmid serial dilutions, DNA marker 2,000 bp. Lane 1: 4 × 105, Lane 6: 4 × 100, and Lane 7 is the non-template control (RNase free water). (B) B. abortus plasmid serial dilutions, DNA marker 2,000 bp, Lane 1: 5 × 105 and Lane 7: 5 × 100 , and Lane 8 is the non-template control (RNase free water).
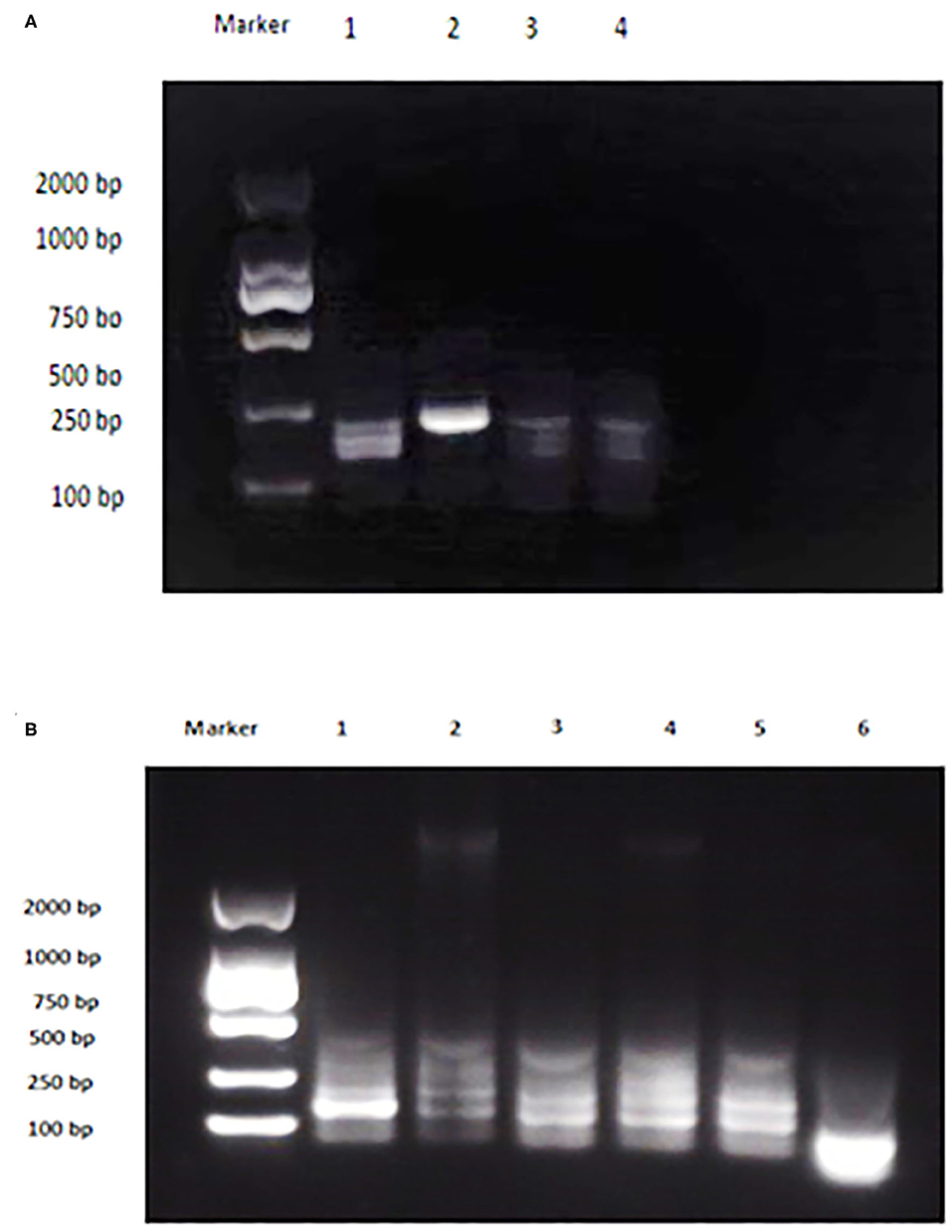
Figure 7. Analytical specificity of Duplex RPA. (A) DNA marker 2,000 bp, Lane 1: B. melitensis bv. 3, Lane 2: B. abortus A 19, Lane 3: B. suis S2, Lane 4: non-template control. (B) DNA marker 2,000 bp, Lane 1: B. melitensis M5, Lane 2: C. abortus, Lane 3: S. typhimurium, Lane 4: E. coli, Lane 5: T. gondii, Lane 6: non-template control (RNase free water).
Results of Screening Field Samples
Different types of sheep and yak samples from the Qinghai, Inner Mongolia, and Xingjian provinces showed a high prevalence of brucellosis, with prevalence rates of 80.6% by duplex RPA and 95.2% by real-time PCR (Table 3). The incidence of B. melitensis was 47 (75.8%), and that of B. abortus was 3 (4.8%). The incidence results obtained by AMOS PCR were 12 (19.3%) for B. melitensis and 3 (4.8%) for B. abortus (Table 1). A sheep was found to be infected with B. melitensis and B. abortus, and yak were found to be infected with B. abortus and B. melitensis (Table 1). These results reveal the transmission of B. abortus to sheep and B. melitensis to yak.
Discussion
Brucellosis is a re-emerging zoonotic disease caused by the closely related species of the genus Brucella. B. melitensis and B. abortus were the first described species in 1887 and 1895 A.D., respectively (22). Livestock species (cattle, sheep, goats, swine, and camel) can be infected with B. melitensis, B. abortus, and B. suis, which are responsible for severe human infection (23). Cattle can be infected by B. abortus and transiently infected with B. suis and more commonly by B. melitensis when they come into direct contact with infected pigs, goats, and sheep in common pastures and at shared water sources. B. abortus, B. melitensis, and B. suis can be transmitted by cow's milk and cause a serious public health threat (24). The newly developed Duplex RPA could detect either B. melitensis or B. abortus with a reaction time of 20 min; additionally it did not require sophisticated equipment instead employing a water bath or a heat block dry bath. The previously developed types of PCR, i.e., AMOS PCR, Bruce-Ladder PCR, and multiplex real-time PCR, were successfully capable of detecting and differentiating between different species under the genus Brucella. Furthermore, these techniques can differentiate between field strains and vaccine strains (10, 19), but these techniques require 2–4 h of reaction time and require a thermocycler. Similar to PCR and multiplex PCR, Duplex RPA was used for end-point detection by agarose gel electrophoresis, which increased the time of the test. Therefore, the developed Duplex RPA cannot be considered a rapid test. Other types of PCR have been developed for species-specific detection and differentiation between field and vaccine strains (B. melitensis, B. suis, and B. abortus-specific PCR) (18, 21, 25). One of the limitations of developed RPA is the inability to differentiate vaccinated animals from naturally infected animals. Generally, RPA is considered a newly invented technique that can rapidly amplify DNA or RNA within 10–20 min (26). Similar to PCR and real-time PCR, multiplexing in RPA is applicable. Therefore, many sensitive and specific multiplexed RPA assays have been developed for the diagnosis of many pathogens (27). The sensitivity of RPA can be increased by mixing during incubation, which is supported by a selection of short fragments of the target sequence that leads to swift amplification (28, 29). In multiplex lateral flow RPA for the detection of many intestinal protozoa, the molecular sensitivities are 403 synthetic gene copies per reaction of the Giardia, 425 of the Cryptosporidium, and 368 of the Entamoeba target (30). The developed multiplex RPA for the detection of methicillin-resistant Staphylococcus aureus, Salmonella enterica, and Neisseria gonorrhoeae showed limits of detection of 10 colony-forming units for methicillin-resistant S. aureus and S. enterica and 100 colony-forming units for N. gonorrhoeae (30). The detection sensitivity of multiplex RPA for the detection of Campylobacter coli and Campylobacter jejuni was 1 CFU/reaction in pure culture. In the case of food applications, the detection limit of C. coli and C. jejuni using the RPA assay were 1 CFU/mL from chicken broth, 103 CFU/g from egg and chicken meat samples without a pre-enrichment procedure, and 1 CFU/g from 24 h enriched egg and chicken meat samples (17). Similar to the previously developed multiplex RPA assays, the developed Duplex RPA is highly sensitive especially after mixing components after 4 min of incubation. However, the end-point detection of most of the multiplex RPA assays depend on real-time fluorescence detection or lateral flow immunochromatographic strips, which make it a rapid technique.
In western China, bacterial isolation and identification, AMOS PCR and multi-locus variable number tandem repeat analysis found that B. melitensis biovar 3 was the dominant causative agent of sheep brucellosis, while B. abortus biovars 1 and 3 were found in infected yaks (31, 32). This study agrees with a previous study reporting the isolation of B. melitensis from yak in Qinghai province, west of China (33). The importance of Brucella species identification, in addition to the determination of the epidemiological situation, is also essential for vaccine-type selection, for example; if B. melitensis is endemic to a region, B. melitensis vaccines such as B. melitensis Rev.1 or B. melitensis M5 can be used; in contrast, B. abortus strain 19 can only be used areas in which B. abortus is endemic. There was high variation between the results obtained by RPA and AMOS PCR, which may be attributed to the higher sensitivity of RPA and its ability to amplify DNA in the presence of PCR inhibitors (26); conversely, AMOS PCR amplifies long fragments DNA of B. melitensis and B. abortus which are 730 bp and 498 bp, respectively, influencing the sensitivity (34). In addition, AMOS multiplex PCR can only detect all biovars of B. melitensis, biovars 1, 2, and 4 of B. abortus, B. suis biovar 1, and B. ovis (9). The cost estimation of one reaction of RPA is 8 Euros while, that of AMOS PCR is 1 Euro and that of real-time PCR is 3 Euros. The high cost of the RPA reaction is attributed to the production of RPA reagents by one company (35).
From previous studies, the species and biovars isolated from western China were B. melitensis biovars 1 and 3, B. abortus biovars 1 and 3, and B. suis biovar 3 (33), which explains the variation of the results between AMOS-PCR, real-time PCR, and Duplex RPA. Through our work, RPA products can be purified, sequenced, or applied for ligation to plasmid vectors and cloning. This study has limitations, mainly in analyzing an inadequate number of Brucella species and strains and other genetically related bacteria. Therefore, this work must be continued by analyzing other species and biovars of B. melitensis, B. abortus, and B. suis. Further, this study can be considered as preliminary and key for the development of real-time or lateral flow RPA for the detection of two or three species of Brucella or the differentiation between field and vaccine strains. Additionally, RPA can be developed for the simultaneous detection of many bacteria species that can cause abortion, such as C. abortus, Coxiella burnetti, and Brucella spp.
It is concluded that Duplex RPA is an isothermal assay for specific detection and differentiation between B. melitensis and B. abortus that is highly sensitive and specific. The assay takes less time (20 min incubation) than multiplex AMOS PCR and real-time PCR. The test sensitivity can be increased by mixing the reaction components. The developed RPA is more sensitive than multiplex AMOS-PCR. Because the test still has limitations characterized by using agarose gel electrophoresis, this could further be improved by the application of a real-time exoprobe and a lateral flow dipstick for end-point detection. These characteristics demonstrate that Duplex RPA is a sensitive, specific, and direct detection technique for the species-specific detection of brucellosis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
All animals were handled, samples collected and processed, and all techniques carried out in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People's Republic of China. The Animal Administration and Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences approved the study (Permit No. LVRIAEC-2014-009). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
GM, XC, JZ, and BF designed the study and wrote the paper. GM performed all the experiments, ZLi performed PCR, real-time PCR, and results analysis. ZLo participated in the collection and preparation of samples. NZ participated in article editing and proofreading. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Gansu Province Scientific and Technical Supporting Program (No. 1304NKCA162), the Gansu Province Agricultural Biotechnology Research and Application Development Project (No. GNSW-2013-27), and the innovation fund of Chinese Academy of Agricultural Sciences (CAAS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the team members of the Livestock and Poultry Zoonotic Diseases—Lanzhou Veterinary Research Institute—Chinese Academy of Agricultural Sciences for their cooperation. Special thanks to Dr. Niyokwishimira Alfred for helping in the sequence alignment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.539679/full#supplementary-material
References
1. Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D, et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med. (2011) 102:118–31. doi: 10.1016/j.prevetmed.2011.04.007
3. McDermott JJ, Arimi SM. Brucellosis in sub-Saharan Africa: Epidemiology, control and impact. Vet Microbiol. (2002) 90:111–34. doi: 10.1016/S0378-1135(02)00249-3
4. Megid J, Mathias L, Robles C. Clinical manifestations of brucelosis in domestic animals and humans. Open Vet Sci. (2010) 4:119–26. doi: 10.2174/1874318801004010119
5. Benkirane A. Ovine and caprine brucellosis: world distribution and control/eradication strategies in West Asia/North Africa region. Small Ruminant Res. (2006) 62:19–25. doi: 10.1016/j.smallrumres.2005.07.032
6. Corbel MJ. Brucellosis: an overview. Conference, International Jerusalem. Emerg. Zoonoses. (1997) 3:213–21.
7. Padilla Poester F, Nielsen K, Ernesto Samartino L, Ling Yu W. Diagnosis of brucellosis. Open Vet Sci J. (2010) 4:46–60. doi: 10.2174/1874318801004010046
8. Asfaw Geresu M, Mamo Kassa G. A review on diagnostic methods of brucellosis. J Vet Sci Technol. (2015) 7:323. doi: 10.4172/2157-7579.1000323
9. Bricker BJ, Halling SM. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol. (1994) 32:2660–6. doi: 10.1128/JCM.32.11.2660-2666.1994
10. López-Goñi I, García-Yoldi D, Marín CM, de Miguel MJ, Barquero-Calvo E, Guzmán-Verri C, et al. New Bruce-ladder multiplex PCR assay for the biovar typing of Brucella suis and the discrimination of Brucella suis and Brucella canis. Vet Microbiol. (2011) 154:152–5. doi: 10.1016/j.vetmic.2011.06.035
11. Probert WS, Schrader Kimmi N., Khuong NY, Bystrom SL, Graves MH. Real-time multiplex PCR assay for detection of Brucella spp. J. Clin Microbiol. (2004) 42:1290–3. doi: 10.1128/JCM.42.3.1290
12. Schmoock G, Ehricht R, Melzer F, Elschner M, Tomaso H, Neubauer H, et al. Development of a diagnostic multiplex polymerase chain reaction microarray assay to detect and differentiate Brucella spp. Diag Microbiol Infect Dis. (2011) 71:341–53. doi: 10.1016/j.diagmicrobio.2011.08.013
13. Gumaa MM, Cao X, Li Z, Lou Z, Zhang N, Zhang Z, et al. Establishment of a recombinase polymerase amplification (RPA) assay for the detection of Brucella spp. infection. Mol Cell Probes. (2019) 47:101434. doi: 10.1016/j.mcp.2019.101434
14. Bounaadja L, Albert D, Chénais B, Hénault S, Zygmunt MS, Poliak S, et al. Real-time PCR for identification of Brucella spp.: a comparative study of IS711, bcsp31 and per target genes. Vet Microbiol. (2009) 137:156–64. doi: 10.1016/j.vetmic.2008.12.023
15. Kang S-I Her M, Kim JY, Lee JJ, Lee K, Sung SR, et al. Rapid and specific identification of Brucella abortus using the loop-mediated isothermal amplification (LAMP) assay. Comp Immunol Microbiol Infect Dis. (2015) 40:1–6. doi: 10.1016/j.cimid.2015.03.001
16. Weiner M, Iwaniak W, Szulowski K. Comparison of PCR-based AMOS, bruce-ladder, and MLVA assays for typing of Brucella species. Bull Vet Inst Pulawy. (2011) 55:630.
17. Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Microchim Acta. (2014) 181:1715–23. doi: 10.1007/s00604-014-1198-5
18. Alvarez LP, Marcellino RB, Martinez A, Robles CA. Duplex PCR for the diagnosis of Brucella melitensis and its differentiation from the REV-1 vaccine strain. Small Rumin Res. (2017) 146:1–4. doi: 10.1016/j.smallrumres.2016.11.004
19. Bricker BJ, Ewalt DR, Olsen SC, Jensen AE. Evaluation of the Brucella abortus species–specific polymerase chain reaction assay, an improved version of the Brucella AMOS polymerase chain reaction assay for cattle. J Vet Diagn Invest. (2003) 15:374–8. doi: 10.1177/104063870301500413
20. Ratushna VG, Sturgill DM, Ramamoorthy S, Reichow SA, He Y, Lathigra R, et al. Molecular targets for rapid identification of Brucella spp. BMC Microbiol. (2006) 6:13. doi: 10.1186/1471-2180-6-13
21. Redkar R, Rose S, Bricker B, DelVecchio V. Real-time detection of Brucella abortus, Brucella melitensis and Brucella suis. Mol Cell Probes. (2001) 15:43–52. doi: 10.1006/mcpr.2000.0338
22. Seleem MN, Boyle SM, Sriranganathan N. Brucellosis: a re-emerging zoonosis. Vet Microbiol. (2010) 140:392–8. doi: 10.1016/j.vetmic.2009.06.021
23. Nicoletti P. A short history of brucellosis. Vet Microbiol. (2002) 90:5–9. doi: 10.1016/S0378-1135(02)00209-2
24. Acha PN, Szyfres B. Zoonoses, and Communicable Diseases Common to Man and Animals: Volume Bacterioses I, and Mycoses. 3rd ed. PAHO Scientific and Technical Publication No. 580. Washington, DC: Pan American Health Organisation (2005). p. 378.
25. Nan W, Tan P, Wang Y, Xu Z, Mao K, Peng D, et al. Duplex PCR for differentiation of the vaccine strain Brucella suis S2 and B. suis biovar 1 from other strains of Brucella spp. Vet J. (2014) 201:427–8. doi: 10.1016/j.tvjl.2014.05.033
26. Kaden R, Ferrari S, Alm E, Wahab T. A novel real-time PCR assay for specific detection of Brucella melitensis. BMC Infect Dis. (2017) 17:1–6. doi: 10.1186/s12879-017-2327-7
27. Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem. (2016) 62:947–58. doi: 10.1373/clinchem.2015.245829
28. Mohamed AG, Ramadan KM, Monem HA, Toukhy Essam EL, Khairy EA. Amos PCR as a rapid screening method for differentiation of infected and vaccinated cattle and sheep with brucellosis. Glob Vet. (2013) 11:748–56. doi: 10.5829/idosi.gv.2013.10.6.73194
29. Lillis L, Siverson J, Lee A, Cantera J, Parker M, Piepenburg O, et al. Factors influencing Recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol Cell Probes. (2016) 30:74–8. doi: 10.1016/j.mcp.2016.01.009
30. Crannell Z, Castellanos-Gonzalez A, Nair G, Mejia R, White AC, Richards-Kortum R. Multiplexed recombinase polymerase amplification assay to detect intestinal protozoa. Anal Chem. (2016) 88:1610–6. doi: 10.1021/acs.analchem.5b03267
31. Kim JY, Lee JL. Development of a multiplex real-time recombinase polymerase amplification (RPA) assay for rapid quantitative detection of Campylobacter coli and jejuni from eggs and chicken products. Food Control. (2017) 73:1247–55. doi: 10.1016/j.foodcont.2016.10.041
32. Cao X, Li Z, Liu Z, Fu B, Liu Y, Shang Y, et al. Molecular epidemiological characterization of Brucella isolates from sheep and yaks in northwest China. Transbound Emerg Dis. (2018) 65:e425–33. doi: 10.1111/tbed.12777
33. Cao X, Shang Y, Liu Y, Li Z, Jing Z. Genetic characterization of animal Brucella isolates from Northwest Region in China. Biomed Res Int. (2018) 2018:2186027. doi: 10.1155/2018/2186027
34. Mayer-Scholl A, Draeger A, Göllner C, Scholz HC, Nöckler K. Advancement of a multiplex PCR for the differentiation of all currently described Brucella species. J Microbiol Methods. (2010) 80:112–4. doi: 10.1016/j.mimet.2009.10.015
Keywords: Duplex RPA, B. melitensis, B. abortus, real-time PCR, AMOS multiplex PCR
Citation: Gumaa MM, Li Z, Cao X, Zhang N, Lou Z, Zhou J and Fu B (2020) Specific Detection and Differentiation Between Brucella melitensis and Brucella abortus by a Duplex Recombinase Polymerase Amplification Assay. Front. Vet. Sci. 7:539679. doi: 10.3389/fvets.2020.539679
Received: 15 July 2020; Accepted: 12 October 2020;
Published: 25 November 2020.
Edited by:
Kai Zhou, First Affiliated Hospital of Southern University of Science and Technology, ChinaReviewed by:
Faham Khamesipour, Sabzevar University of Medical Sciences, IranZeliang Chen, Shenyang Agricultural University, China
Copyright © 2020 Gumaa, Li, Cao, Zhang, Lou, Zhou and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jizhang Zhou, zhoujizhang@caas.cn; Baoquan Fu, fubaoquan@163.com
 M. M. Gumaa
M. M. Gumaa Zhaocai Li
Zhaocai Li Xiaoan Cao
Xiaoan Cao Nianzhang Zhang
Nianzhang Zhang Zhongzi Lou
Zhongzi Lou Jizhang Zhou
Jizhang Zhou Baoquan Fu
Baoquan Fu