Loci Associated With Antibody Response in Feral Swine (Sus scrofa) Infected With Brucella suis
- 1United States Department of Agriculture, Animal and Plant Health Inspection Service, Wildlife Services, National Wildlife Research Center, Fort Collins, CO, United States
- 2Department of Animal Sciences, Colorado State University, Fort Collins, CO, United States
- 3United States Department of Agriculture, Animal and Plant Health Inspection Service, Wildlife Services, National Feral Swine Damage Management Program, Fort Collins, CO, United States
- 4United States Department of Agriculture, Agricultural Research Service, Infectious Bacterial Diseases, National Animal Disease Center, Ames, IA, United States
- 5United States Department of Agriculture, Animal and Plant Health Inspection Service, Wildlife Services, Raleigh, NC, United States
- 6United States Department of Agriculture, Animal and Plant Health Inspection Service, Veterinary Services, Center for Epidemiology and Animal Health, Fort Collins, CO, United States
Feral swine (Sus scrofa) are a destructive invasive species widespread throughout the United States that disrupt ecosystems, damage crops, and carry pathogens of concern for the health of domestic stock and humans including Brucella suis—the causative organism for swine brucellosis. In domestic swine, brucellosis results in reproductive failure due to abortions and infertility. Contact with infected feral swine poses spillover risks to domestic pigs as well as humans, companion animals, wildlife, and other livestock. Genetic factors influence the outcome of infectious diseases; therefore, genome wide association studies (GWAS) of differential immune responses among feral swine can provide an understanding of disease dynamics and inform management to prevent the spillover of brucellosis from feral swine to domestic pigs. We sought to identify loci associated with differential antibody responses among feral swine naturally infected with B. suis using a case-control GWAS. Tissue, serum, and genotype data (68,516 bi-allelic single nucleotide polymorphisms) collected from 47 feral swine were analyzed in this study. The 47 feral swine were culture positive for Brucella spp. Of these 47, 16 were antibody positive (cases) whereas 31 were antibody negative (controls). Single-locus GWAS were performed using efficient mixed-model association eXpedited (EMMAX) methodology with three genetic models: additive, dominant, and recessive. Eight loci associated with seroconversion were identified on chromosome 4, 8, 9, 10, 12, and 18. Subsequent bioinformatic analyses revealed nine putative candidate genes related to immune function, most notably phagocytosis and induction of an inflammatory response. Identified loci and putative candidate genes may play an important role in host immune responses to B. suis infection, characterized by a detectable bacterial presence yet a differential antibody response. Given that antibody tests are used to evaluate brucellosis infection in domestic pigs and for disease surveillance in invasive feral swine, additional studies are needed to fully understand the genetic component of the response to B. suis infection and to more effectively translate estimates of Brucella spp. antibody prevalence among feral swine to disease control management action.
Introduction
In the United States (U.S.) there are ~6 million invasive feral swine (Sus scrofa), which are defined as any released or escaped domestic pigs, Eurasian wild boars, or hybrids of the two (1, 2). Both the abundance of feral swine and extent of the geographic range have increased rapidly over the past 30 years due to the high reproductive potential of the species, limited predation pressure, abundance of food (both native flora and fauna and agricultural products), and human-mediated introduction into uninvaded habitats (3, 4). As range and abundance have increased, so too have the economic and ecological costs of feral swine. Feral swine negatively impact the aesthetic and cultural value of landscapes, with costs to tourism and silvicultural sectors; however, direct costs to agriculture are most notable. In a survey of 10 of the 38 states with established populations, feral swine cause an estimated $190 million in crop damages annually (5). Feral swine also damage pastures through rooting and trampling behaviors that kill desired plant species and allow unpalatable species to quickly spread (6). Among the broader costs associated with the expansion of this invasive species, feral swine serve as an important reservoir for a number of pathogens (e.g., Brucella spp., pseudorabies virus, and trichinella) with the potential for spillover to livestock, humans, companion animals, and wildlife (7–9).
Of the pathogens commonly detected in feral swine, swine brucellosis is among the most important, in large part because of its broad host specificity (10). Brucella spp. are facultative intracellular bacteria that primarily infect phagocytic cells following infection of the host. Typical pathogen recognition involves actin cytoskeletal remodeling in which membrane protrusions extend and uptake the stimulatory particle, thus generating a phagosome (11). The phagosome typically matures and fuses with the lysosome to create a phagolysosome, which destroys microorganisms. However, Brucella spp. have been shown to disrupt this process within the host cell by modifying the original phagosome into a membrane bound vesicle, referred to as the Brucella-containing vacuole [BCV; (12)], which prevents lysosome fusion. The BCV moves along the endocytic pathway developing membrane markers associated with both the late endosome and the endoplasmic reticulum. The Type IV secretion system (T4SS) Vir B has been demonstrated to use effector proteins to modulate secretory trafficking and promote bacterial pathogenesis (13, 14). Knowledge of the infection pathway and kinetics of Brucella spp. is crucial for interpreting diagnostic results and understanding host pathophysiology.
Several species of Brucella have been isolated from feral swine, including suis, abortus, and microti (15–17), respectively. However, Brucella suis is the only species to cause systemic or generalized infection in swine and can lead to reproductive failure (18). Through federal, state, tribal, and industry partnerships, brucellosis was eradicated from domestic pigs in the U.S. in 2011 (19–21). However, brucellosis remains prevalent among feral swine, as disease surveillance efforts throughout the invaded range within the U.S. documented an apparent Brucella spp. antibody prevalence of 4.3% (8). Prevalence rates vary geographically, with highest antibody prevalence observed among Hawaii, South Carolina, and Alabama [14.4, 11.6, and 10.8%, respectively; (21)]. The high prevalence of B. suis in feral swine poses a risk of reemergence of this bacterial pathogen in domestic pigs. Approximately 36.5% of commercial swine facilities, maintaining 11.3% of the nation's pork inventory, are located in regions where feral swine are present (22). Further, monitoring of feral swine fitted with Global Positioning System (GPS) telemetry collars has demonstrated direct interactions with livestock, with pasture-raised pigs at greatest risk for B. suis exposure (23). United States pork production and processing is estimated to contribute $39 billion to the gross domestic product (24); however, reemergence of B. suis among commercial swine could result in tremendous economic losses and trade restrictions for pork producers (25).
Although swine are the primary host for B. suis, this bacterium can infect a number of other species, including cattle (26). Brucella suis generally causes few clinical signs and is not believed to be transmitted among cattle (27). However, infection of lactating cows results in shedding of bacteria in milk and the development of antibodies, which cannot be differentiated from B. abortus antibodies with current diagnostic serological tests (28). Thus, bacterial spillover from feral swine to cattle creates significant diagnostic challenges in addition to posing a public health risk via consumption of raw milk (27). Furthermore, as a zoonotic pathogen, B. suis has significant public health implications with human cases of brucellosis in the U.S. most often associated with exposure through feral swine hunting and field dressing (29).
Immunity against intracellular pathogens, including Brucella spp. relies on the induction of cell-mediated immunity, primarily interferon gamma (IFN-γ)-producing CD4+ T cells, or T helper 1 (TH1) responses. These responses can be measured from blood samples; however, they require cellular isolation, in vitro antigen stimulation, and can take days to obtain results. Therefore, cell-mediated responses are not frequently used for diagnostic purposes. Rather for Brucella suis, as with other Brucella spp., infection is diagnosed using antigen and antibody assays. However, culture and serological diagnostics for B. suis often produce conflicting results (9, 28, 30, 31). Brucella lipopolysaccharides (LPS) are generally less toxic than those of other gram-negative bacteria, which are potent stimulators of innate immune responses through pattern-recognition receptors (32). However, the O-polysaccharide of the Brucella LPS is immunodominant for humoral responses during infection in most natural hosts. Therefore, the O-polysaccharide is the primary antigen used in most serologic tests for detecting infection with B. abortus, B. melitensis, or B. suis (33). In infected swine, brucellosis serologic tests have lower sensitivity than in other host species such that serologic testing is usually evaluated on a herd, rather than an individual basis (26). Low sensitivity of the assay may be due to the fact that most serologic tests were developed and validated for cattle infected with B. abortus and there may be structural differences between B. abortus and B. suis LPS (34).
Genetic variation has been documented to play a role in immune kinetics for numerous species and diseases (35–39). For example, genetic variation in host resistance or tolerance has been reported (scientifically and anecdotally) in swine for African swine fever, foot-and-mouth disease, atrophic rhinitis, pseudorabies, and brucellosis (36, 39). Similarly, we sought to evaluate whether genetic factors may define variation in seroconversion to B. suis infection with implications for the reliability of antibody tests used for monitoring disease prevalence among feral swine (9). We conducted case-control genome-wide association studies (GWAS) in which allele frequencies of single nucleotide polymorphisms (SNP) spanning the genome were evaluated to identify loci associated with B. suis seroconversion in wild-caught, naturally infected feral swine.
Materials and Methods
Sample Collection
Tissue (submandibular, parotid, medial retropharyngeal, tracheobronchial, gastrohepatic, axillary or inguinal lymph nodes, spleen, and reproductive tract) and serum samples were collected from 376 feral swine at two Texas abattoirs in 2015 as described by Pedersen et al. (9). Tissue cultures were conducted at the U.S. Department of Agriculture's Agricultural Research Service in Ames, Iowa and at the College of Veterinary Medicine at Texas A&M University in College Station, Texas. If any of the tissue samples contained Brucella spp., the animal was considered culture positive. In accordance with established protocols, the following eight independent serological assays were completed for each animal at the National Veterinary Services Laboratories (NVSL): (1) buffered antigen plate agglutination test (BAPA), (2) competitive enzyme-linked immunosorbent assay (cELISA), (3) complement fixation, (4) fluorescence polarization assay (FPA), (5) the rivanol test, (6) plate agglutination, (7) tube agglutination, and (8) card test. Given the limitations of these serological assays, feral swine were considered seropositive for B. suis if two or more of these assays were positive (9). Forty-nine feral swine were culture positive for Brucella spp. and, of these, 16 were considered antibody positive (Table 1). Similarly, individuals with one or zero positive results from the 8 serological assays were classified as negative. In this study, animals that were both culture positive and seropositive were defined as cases whereas animals that were culture positive but seronegative were defined as controls.
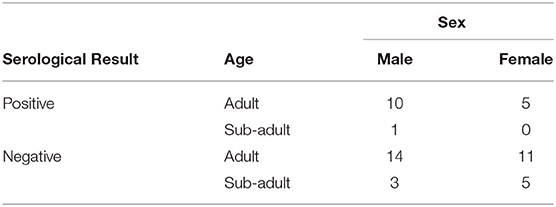
Table 1. Feral swine samples collected at two Texas abattoirs that were culture positive for B. suis.
Misclassification of the serological status of an animal could result from two potential sources of error—error due to diagnostic test performance or the delay in immune response once infected resulting in a false negative result. The probability of an individual being misclassified as negative when positive due to diagnostic test performance was calculated as the combined probability of seven or eight false negative serological results using the lowest previously published sensitivity value for each serological diagnostic test (40, 41). False negative error resulting from the delay in immune response was assessed using data describing the serum antibody response of an infected animal to the eight diagnostic tests [complete methods available in Supplementary Material; (42, 43)]. Bayesian generalized additive models were fit to these serological response data for each of the eight diagnostic tests resulting in the predicted increase and variation in serological response by day after infection. Using these posterior predictions for serological response, two simulations were conducted to determine the probability that an individual animal will test positive. First, the probability of at least two positive test results for each day post-infection were simulated. Second, because the time of infection is unknown for sampled animals in this study the probability that our sample included serological true positive and false negative animals was conservatively calculated assuming infection occurred within 120 days of being sampled.
Genotype Data
Bi-allelic SNP genotypes for feral swine were generated using the GeneSeek Genomic Profiler for Porcine bead array [68,516 loci; Illumina BeadChip microarrays (San Diego, California) licensed exclusive to GeneSeek, Neogen Corporation (Lincoln, Nebraska); (44)]. Single nucleotide polymorphisms were mapped to the Sscrofa11.1 reference genome assembly (45) and non-autosomal loci were removed, leaving 62,128 loci available for analysis. We then used SNP & Variation Suite (SVS; Golden Helix, Bozeman, Montana) and PLINK 1.9 (46) to implement standard quality control measures for GWAS analysis of SNP genotypes, specifically pruning loci with call rates ≤0.90, minor allele frequency <0.01, Hardy-Weinberg Equilibrium <1 × 10−6, and removing samples with call rates ≤0.90 and heterozygosity rate ±3 standard deviations from the mean. After quality control, 47 swine (16 cases [seropositive]/31 [seronegative] controls) and 53,162 SNP were retained for analysis. As an additional quality control step, we screened these 47 individuals for close familial relationships (identical by descent [pi-hat] estimate >0.375). Finding no dyads that exceeded this threshold, all 47 individuals were retained.
Statistical Analysis
A single-locus mixed model GWAS was evaluated using the efficient mixed-model association eXpedited (EMMAX) methodology in SVS (Golden Helix, Bozeman, Montana). Association analyses were conducted using three genetic models: additive, dominant, and recessive. Given a bi-allelic locus with two alleles (A and a), the additive model assumes that there is a linear increase in disease risk with each copy of the A allele; thus, the increase in disease risk from aa to Aa would again be doubled among AA homozygotes. The recessive model assumes that two copies of the A allele increases the risk of disease whereas the dominant model assumes that one or more A alleles increases the risk of disease (47, 48). The mixed model was as follows:
where, y was a vector of observations, X was a matrix of fixed effects, β was a vector of fixed effects to be estimated, Z was a matrix relating random effects to the observations in y, μ was a vector of random effects to be estimated, and ε was a vector of residual errors. It was assumed that = and = ; therefore, = (49). K was a pre-computed genomic relationship matrix to account for population structure. Age (sub-adult vs. adult), sex, facility, and month in which the samples were collected were examined as potential fixed effects using an Akaike Information Criterion (AIC) approach (50). After screening fixed effects with AIC, only age was retained for additional model development. Association test statistics can be inflated by underlying population stratification (i.e., genetic structure associated with systematic differences in allele frequencies among genetic (sub)populations included in the sample) (51); therefore, genomic inflation factors (λ) were estimated to ensure that each model sufficiently corrected for population stratification. The genomic inflation factor is expressed as the median of the observed distribution of the test statistic divided by median of the expected distribution of the test statistic, where λ < 1.01 suggests small test statistic inflation, λ < 1.05 suggests moderate test statistic inflation, and λ > 1.1 suggests highly inflated test statistics (52). Our genomic inflation factors were 1.03, 1.00, and 1.04 for the additive, dominant, and recessive models, respectively; therefore, the models were deemed appropriate for subsequent analyses.
Single nucleotide polymorphisms were considered associated with B. suis seroconversion when their unadjusted P < 5 × 10−5, moderately associated when their unadjusted P-value fell between 5 × 10−7 and 1 × 10−5, and strongly associated when unadjusted P < 5 × 10−7 (53). To further examine the association between these loci and B. suis seroconversion, an odds ratio (OR) and 95% confidence interval were calculated for the major allele at each locus (54) as follows:
where OR represents the odds ratio for the major allele at a given locus, A represents the number of major alleles within the cases, D represents the number of minor alleles within the control group, B represents the number of minor alleles within the cases, and C represents the number of major alleles in controls (55, 56).
Functional Annotation
Reference sequence (RefSeq) gene transcripts, annotated by the National Center for Biotechnology Information (NCBI; Sus scrofa annotation release 106), were used to identify genes within 2 Mb (1 Mb upstream and 1 Mb downstream) of the candidate SNP (57–59). This interval was approximately twice the average haplotype block size (394.88 kb) for swine (60). The Pig Quantitative Trait Locus Database (Pig QTLdb; https://www.animalgenome.org/cgi-bin/QTLdb/SS/index) was queried to identify traits that were previously associated with the genes of interest. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database (61) was used to identify disease-related pathways within S. scrofa that contained the candidate genes.
Results
Evaluation of the Misclassification of Serological Status
The combined probability of eight false negative serological results [P(x1,.,x8) = 2.03 × 10−6] or seven false negative results and one positive result [P(x1,.,x7) = 2.98 × 10−4] was low. The probability of accurately classifying an animal as serologically positive increased logistically with time since infection, approaching 1 at ~32 days post-infection. Assuming B. suis animals in this study were infected within 120 days of sampling, the probability that the sample included an animal misclassified as negative (false negative) was 0.13.
Additive Model
Three loci on chromosomes 9, 10, and 18 were associated with B. suis seroconversion (Figure 1). The odds ratio for the major allele at each locus was 0.10, 0.19, and 0.22 for rs339122633 (G), rs81477530 (C), and rs81469187 (A), respectively (Table 2). This suggests that the major alleles, in these three cases, were associated with a decreased likelihood of B. suis seroconversion. The 2 megabase (Mb) regions encompassing these three loci contained 26 annotated genes and 14 uncharacterized genes. A thorough review of candidate gene function, infection kinetics, and the immunologic response to B. suis infection reduced the number of putative candidate genes from 40 to three:(1) acyloxyacyl hydrolase (AOAH), (2) engulfment and cell motility 1 (ELMO1), and (3) prostaglandin synthase 2 (PTGS2). Locus rs339122633 was located within an intron of Hemicentin 1 (HMCN1); however, based on our current understanding of the functions of HMCN1, there is no obvious link to B. suis infection.
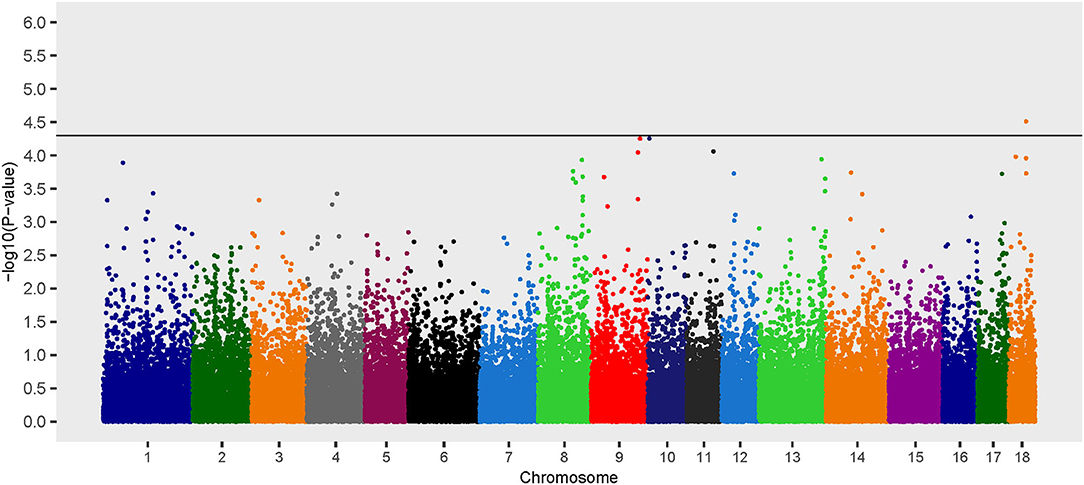
Figure 1. Manhattan plot of genome-wide association study for seroconversion following Brucella suis infection in feral swine using the EMMAX-GRM additive model. The black line [–log10(P-value) = 4.3] denotes an association with B. suis.
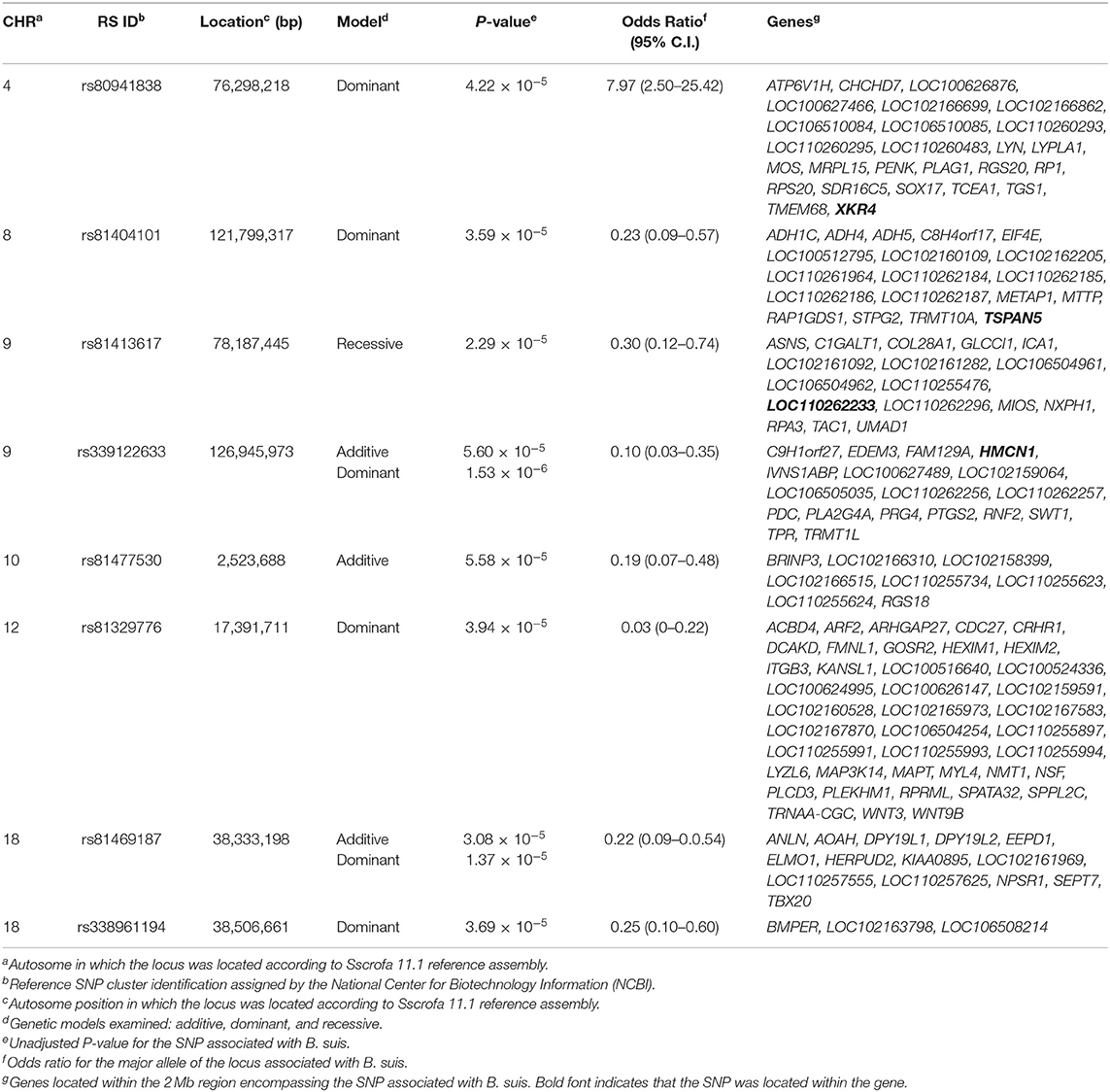
Table 2. Single nucleotide polymorphisms associated with seroconversion following Brucella suis infection in feral swine sampled at two Texas slaughterhouses.
AOAH
Expressed by monocytes, macrophages, neutrophils, and dendritic cells (62), the AOAH enzyme (encoded by the AOAH gene) removes secondary fatty acyl chains from lipopolysaccharides (LPS) on the outer membrane of Gram-negative bacteria, rendering the target bacterium immunologically inert (63, 64). In general, exposure to LPS induces a robust inflammatory response. This is followed by a period of tolerance that is believed to have evolved to minimize inflammation-induced damage during recovery from microbe exposure (64, 65). Once AOAH deacylates lipid A, the bioreactive center of the LPS, the ability to elicit an inflammatory response is greatly reduced (66, 67). Removal of fatty acid chains renders the LPS biologically inactive and reestablishes sensitivity for subsequent infections (64, 68).
As this SNP was associated with a decreased likelihood of seroconversion, modification of AOAH function may result in a phenotype in which the LPS of B. suis is not effectively processed in porcine phagocytes. Thus, LPS antigens, such as the O side-chain, may not be expressed as well with major histocompatibility complex (MHC) II molecules on the surface of antigen presenting cells, making them less available for immune recognition. As cell wall-derived polysaccharides and glycol lipids are not readily digested by lysososomal enzymes, they can be retained for long periods of time inside macrophages (69). Others have reported that Brucella LPS can form ternary complexes with MHC class II molecules that are sequestered in the macrodomains at the cell surface, which prevents immunologic presentation (70). The phenotype associated with the SNP may lead to more ternary complexes that decrease immunologic presentation by phagocytic cells, thereby facilitating reduced humoral responses.
ELMO1
The Engulfment and Cell Motility 1 gene encodes ELMO1, a host-signaling molecule in phagocytic and T cells that modulates cellular activities through activation of small Rac GTPases (71, 72). Rac cycles between active and inactive states based on binding of guanosine diphosphate (GDP) from guanine nucleotide exchange factors (GEF). The cytosolic region of brain angiogenesis inhibitor 1 (BAII, a receptor that recognizes the core carbohydrate of lipopolysaccharide) interacts with ELMO1 leading to Rac activation (72). This suggests a role for ELMO1 in innate immunity detection and response to bacterial pathogens (73). ELMO1 stimulated responses occur largely from within the cell after phagocytosis engulfment of the target, resulting in an amplified signal from the concentration of bacterial pathogen-associated molecular patterns within phagosomes (73).
In lymphocytes, ELMO1 appears to primarily function by regulating Dock2, a GEF for activating Rac GTPase (71). The Dock2.ELMO1 complex is essential for chemokine dependent migration of primary T and B cells and for key steps in the interactions between T cells and target cells. ELMO1 regulates polarization and migration in response to chemokine signals, but a lack of ELMO1 does not appear to impair normal homeostatic migration of peripheral T cells (71). After activation of the T cell receptor, a filamentous actin ring is part of the immunological synapse that forms between a T cell and the surface of a target cell. This provides a structural framework for effector function including directional secretion of cytokines and cytolytic factors (74). The Dock2.ELMO1 complex and Rac activation are important in formation of the filamentous actin ring as part of the T cell activation process. Dock2 has also been shown to be critical for T cell cytotoxicity (74), which may be important because of the intracellular localization of Brucella spp. Regulators of Rac GTPase signaling, such as ELMO1 and Dock2, continue to be of interest for understanding their roles in regulating T cell activation and function. As this SNP was also associated with a decreased likelihood of seroconversion, ELMO1 may function to decrease the humoral response by reducing internalization of Brucella spp. into phagocytic cells, reducing degradation of bacterial antigens in phagosomes, and/or decreasing pro-inflammatory immune responses following infection.
PTGS2
The Prostaglandin-Endoperoxide Synthase 2 (PTGS2) gene encodes the PTGS2 enzyme that catalyzes the breakdown of arachidonic acid from cell wall phospholipids to bioactive eicosanoid compounds (75, 76). PTGS2 is expressed by a wide range of cells including macrophages, fibroblasts, vascular endothelial cells, and smooth muscle cells (77) and exposure of immune cells (dendritic or monocytes) to B. abortus or its LPS increases expression of prostaglandins (78, 79). Previous studies suggest that increased prostaglandin concentrations favor prolonged survival of Brucella within host cells. Cyclooxygenase inhibition reduces Brucella colonization in a murine model (78) and has been shown to have suppressive effects on several other immune-modulating effectors.
Prostaglandins are also potent immune modulators for T cells and can block T cell proliferation and promote cytokines associated with TH-2 responses (80, 81). As previous studies suggest that Brucella uses the prostaglandin pathway to subvert immune responses and enhance persistence in infected cells, it would be logical to hypothesize PTGS2 affects immune responses through inhibiting the synthesis of prostaglandins, thereby reducing stimulation of TH-2 responses associated with antibody production. As animals with this SNP were culture positive, it would suggest that factors other than prostaglandin synthesis have critical roles in in vivo clearance of B. suis in pigs.
Dominant Model
Six loci on chromosomes 4, 8, 9, 12, and 18 were associated with B. suis seroconversion (Figure 2). Of the six loci, two were previously identified using the additive model: rs339122633 (PTGS2) and rs81469187 (AOAH). Three of the four SNP identified exclusively using the dominant model had OR < 1 which suggests that the major alleles at these loci (rs81404101 [major allele G], rs81329776 [major allele C], and rs338961194 [major allele G]) were associated with a decreased likelihood of B. suis seroconversion (Table 2). In contrast, the major allele (T nucleotide) for rs80941838 on chromosome 4 had an OR > 1 which suggests an increased likelihood of B. suis seroconversion. The two Mb genomic regions surrounding the significant SNP contained 55 annotated genes and 33 uncharacterized genes. Five putative candidate genes were identified after an extensive review of gene function and brucellosis infection: (1) Integrin Subunit Beta 3 (ITGB3), (2) Lysozyme Like 6 (LYZL6), (3) Mitogen-Activated Protein Kinase 14 (MAP3K14), (4) Tetraspanin 5 (TSPAN5), and (5) LYN Proto-oncogene, Src Family Tyrosine Kinase (LYN).
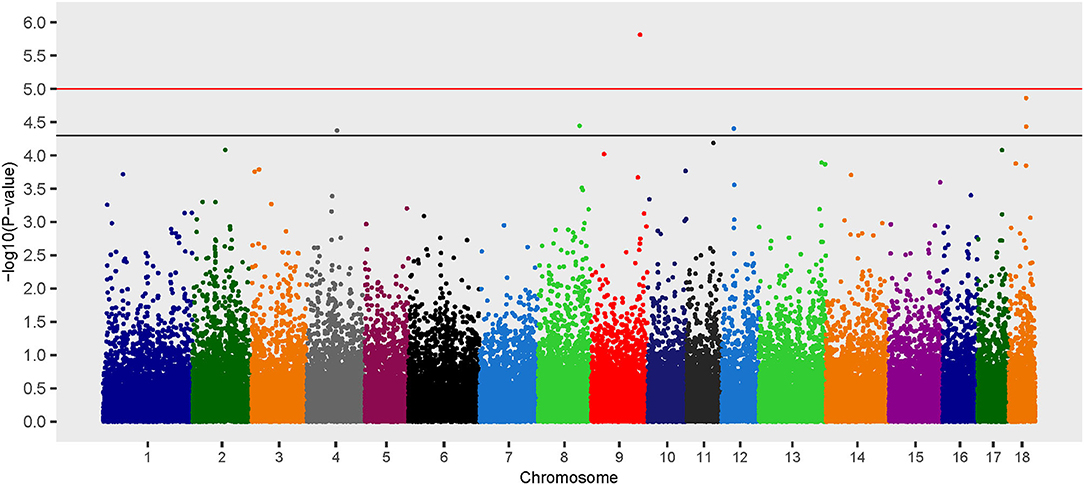
Figure 2. Manhattan plot of genome-wide association study for seroconversion following Brucella suis infection in feral swine using the EMMAX-GRM dominant model. The black line [–log10(P-value) = 4.3] denotes an association and the red line [–log10(P-value) = 5.0] denotes a moderate association with B. suis according to guidelines set forth by the Wellcome Trust Case Control Consortium (53).
ITGB3
Integrins are transmembrane receptors important for cell adhesion and can be exploited by a number of pathogens for binding to host cells and internalization (82). ITGB3, encoded by the ITGB3 gene, is a fibrinogen and vitronectin receptor that is expressed on platelets and monocytes with diverse roles in cell migration, adhesion, and signaling (83–85). ITGB3 (also known as αVβ3) can bind to adhesive proteins resulting in endothelial cell migration, angiogenesis, and TGF-B1 signaling (86).
Prostaglandin E2 suppresses the expression and activity of ITGB3 in human endometrial epithelial and stromal cells (87), suggesting a possible link with Prostaglandin-Endoperoxide Synthase 2, another gene identified in the current study. ITGB3 is also a marker and regulator of cellular senescence, a process that prevents propagation of damaged cells in tissue. In human primary fibroblasts under in vitro conditions, ITGB3 accelerates the onset of senescence by activating transforming growth factor beta [TGF-B; (88)]. Although senescent cells are metabolically active, the immune system senses these cells and eliminates them. The observed SNP could also have influenced N-glycosylation sites as these sites play a key role in regulating adhesive functions of integrins (89). Loss of certain individual N-glycan sites either reduce or enhance integrin activation, indicating that N-linked glycosylation can exert both positive and negative effects on integrin function.
Although the connection between ITGB3 and reduced humoral responses was not evident, a possible link is with prostaglandin synthase, cellular senescence with immunologic elimination of targeted cells, or the role of this integrin in cell-to-cell adhesion and communication are all possible mechanisms.
LYZL6
Lysozymes are proteins that catalyze the hydrolysis of peptidoglycan on the bacterial cell wall. This results in bacteriolysis and the release of bacterial products, including cell wall peptidoglycan, which activates pattern recognition receptors in host cells (90, 91). Phagocytosis is an important innate immune function, which results in the fusion of bacteria within phagosomes with lysosomes and leads to acidification and degradation through proteases and oxidants. Through a poorly understood mechanism, Brucella spp. that are localized within phagosomes prevent fusion with lysosomes (92, 93). However, almost 90% of internalized bacteria in Brucella-containing phagosomes are killed, most likely through cellular processes such as acidification and oxidation that results from fusion with lysosomes (94). In antigen-presenting cells (especially dendritic cells), the bactericidal process could lead to expression of cleaved bacterial products on the phagocytic cell surface through exogenous antigen processing.
A probable hypothesis for the reduced likelihood of seroconversion associated with LYZL6 would be that the compromised lysozyme results in reduced hydrolysis of the bacterial cell wall. This would result in peptidoglycan not being cleaved for antigen presentation and stimulation of cellular activation processes that result from interaction with pathogen pattern receptors.
MAP3K14
Mitogen-activated protein kinase 14 (MAP3K14) encodes NF-κB inducing kinase, a family of transcription factors that play important roles in the regulation of various cellular processes including cell growth, cell survival, cell development, and many aspects of immune function [e.g., immune responses and inflammation; (95–97)]. There are two major signaling pathways by which NF-κB activation can occur: canonical and non-canonical. Both are involved in immune stimulation and regulation; however, the non-canonical pathway functions include formation and architecture of secondary lymphoid organs, humoral immunity, dendritic cell maturation, and osteoclast and T cell differentiation. Additionally, the role of the non-canonical pathway in inflammatory diseases has been studied including rheumatoid arthritis, systemic lupus erythematosus, nephropathy, metabolic inflammation, and multiple sclerosis [reviewed in (98)].
While both pathways function in immunity and inflammation, the non-canonical pathway appears to be more selective than the canonical pathway, which could be partly attributed to the restricted type of receptors that can trigger its activation. However, certain pathogens have been shown to induce activation of the non-canonical NF-κB pathway including influenza virus (99), vesicular stomatitis virus (100), respiratory syncytial virus (101), and various herpesviruses (102–104). Some bacteria, such as Helicobacter pylori (105) and Legionella pneumophila (106), are also capable of activating the non-canonical pathway. Manipulation or modulation of host cell signaling serves as a virulence mechanism, which could enhance pathogen survival. Although some studies have shown the role of the canonical NF-κB pathway during Brucella infection (107, 108), there is no available information on the role of the non-canonical pathway during Brucella infection.
TSPAN5
Tetraspanins are conserved proteins that span the membrane of eukaryotic cells as membrane scaffolds, bringing together surface molecules such as integrins and cell-specific receptors into plasma membrane microdomains (109–112). Through their function as molecular scaffolds, tetraspanins contribute to development, reproduction, intracellular trafficking, and immunity (113, 114).
Our analysis identified a SNP in Tetraspanin 5, a gene that encodes TSPAN5 - a broadly distributed protein with reported physiologic functions in neurons, cartilage, osteoclasts, and the cardiovascular system (115–118). TSPAN5 is a member of a subgroup of tetraspanins (TspanC8) that interact closely with ADAM10 and regulate several functional aspects. Exit of ADAM10 from the endoplasmic reticulum, trafficking to either late endosomes or the plasma membrane, and specificity of ADAM10 relative to positive or negative regulation of Notch signaling is all modulated by TSPAN5 (111). ADAM10 is a transmembrane metalloproteinase that is responsible for cleaving off the ectodomain of various transmembrane proteins, which allows the intracellular domain to enter the cell nucleus and modulate gene expression, including those of cytokines (111, 119). Dendritic cells from mice in which ADAM10 has been knocked out have dramatic reductions in IgE production and do not develop significant TH2 immune responses (120). A role for ADAM metalloproteinases, sometimes through activation of Notch 1, has been demonstrated in both B cells (thymocyte and B cell development, function, antigen presentation), T cells (proliferation, activation, expression of CD44 adhesion molecules) and activation of NK cells suggesting an important role for this class of proteases in immune function [reviewed in (121)]. The role of TSPAN5 in reduced humoral responses in feral swine after Brucella infection is not yet known, but it could be hypothesized that the effect may have been mediated through reduced activation of ADAM metalloproteinases.
LYN
Tyrosine-protein kinase Lyn, encoded by the LYN gene, is a member of the Src family of intracellular membrane-associated tyrosine kinases (SFK). Regulation of Lyn signaling is mediated by protein interactions through its Src homology (SH) domains, SH2/SH3, and its phosphorylation status. Although originally identified within the hematopoietic compartment, Lyn is expressed in many tissues and transmits signals from a variety of receptors, including the B cell receptor (122, 123). Downstream, Lyn can phosphorylate various targets including immunoreceptor tyrosine-based inhibitory/activation motifs (ITIM/ITAM) (124, 125), PI3 kinase (126), STAT5 (127), and MAP kinases, among others, resulting in both positive and negative regulatory signals. Lyn deficiency and Lyn overexpression both result in defects in the myeloid and lymphoid systems (122, 123, 128, 129).
In B cells, Lyn is the main SFK and has both positive and negative roles in B cell receptor signaling (124, 130). Within the BCR complex, Lyn is associated with IgM/IgD as well as CD19, and it is rapidly phosphorylated upon BCR cross-linking. Lyn can also positively and negatively influence toll-like receptor 4 (TLR4) signaling, a pattern recognition receptor, which recognizes bacterial LPS and plays a critical role in the initiation of innate immune responses (131). Lyn acts on TLR4 signaling, in part, by interfering with the activity of interferon regulatory factor-5 (IRF5), a molecule central to the downstream signaling of TLRs (132). In murine models, macrophages deletion of Lyn is associated with greater production of TNF-α, IL-6, and type I interferons after LPS stimulation (133).
Our data would suggest that the LYN gene may be involved in regulation of the humoral response to Brucella infection. Due to the role of Tyrosine-protein kinase Lyn in both BCR and TLR4 signaling, its effects could be mediated directly on B cells or through activation of innate immune danger signals. The crucial function of Lyn as both an activator and regulator of B cell responses, makes it an interesting target given the observed humoral phenotype in this study.
Recessive Model
The recessive model revealed one locus associated with B. suis seroconversion (Figure 3). The odds ratio for the major allele (nucleotide T) at this locus (rs81413617) was 0.30, which indicates that this allele is associated with a decreased likelihood of B. suis seroconversion (Table 2). Seven uncharacterized genes as well as 10 annotated genes were located within the two Mb genomic region encompassing this SNP. The SNP, rs81413617, was located on chromosome 9 within an uncharacterized gene LOC110262233; however, one of the genes located 0.35 Mb upstream of rs81413617 was linked to immune response: Collagen Type XXVIII Alpha 1 Chain (COL28A1).
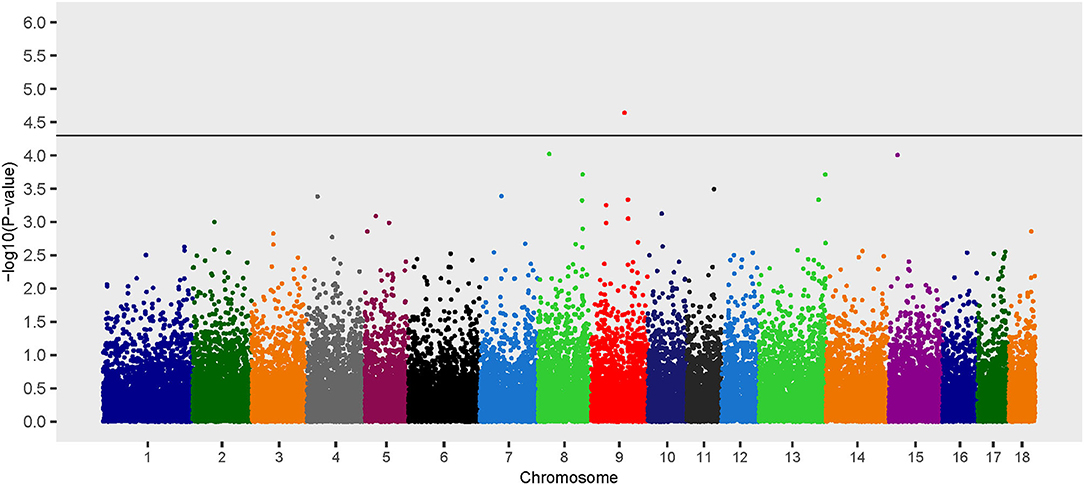
Figure 3. Manhattan plot of genome-wide association study for seroconversion following Brucella suis infection in feral swine using the EMMAX-GRM recessive model. The black line [–log10(P-value) = 4.3] denotes an association with B. suis.
COL28A1
The collagen superfamily covers a variety of subclasses and collagens that can be found in all tissues throughout the vertebral body and are important for tissue integrity (134). COL28A1 collagen is believed to have greatest expression in the nervous system and encodes a von Willebrand factor A domain that facilitates protein-protein interactions. Genome-wide association analyses have previously identified an association between this type of collagen with Interleukin 1 beta (IL-1β) secretion in African Americans following smallpox vaccination (135) and resistance to clinical mastitis in cattle (136). The gene was also found to be down regulated in a rat model of lipopolysaccharide-induced epididymitis (137) and upregulated in a bleomycin lung injury murine model (138).
Current knowledge does not readily explain how COL28A1 would influence humoral responses to B. suis in pigs; however, the possible tie to IL-1β secretion is of interest. IL-1β plays a crucial role in modulating host immune responses to inhaled pathogens through expression of chemokines and adhesion molecules, enhancing phagocytic activities of neutrophils and monocytic cells, and increasing production of reactive oxygen species (139, 140). In addition, mice lacking IL-1β receptors have increased pulmonary colonization after intratracheal infection with Brucella abortus (141).
Discussion
In evaluating the physiological response of feral swine to B. suis infection with case-control GWAS, we identified eight variants and 145 positional candidate genes suggesting seroconversion following B. suis infection of feral swine may be under polygenic control. A review of the current understanding of gene function and infection kinetics reduced the number of plausible candidate genes involved in the immunologic response of feral swine to B. suis from 145 to nine. This aligns with previous studies that evaluated the genetic architecture of immune response in domestic swine (39, 142, 143). The nine genes that were thoroughly explored were all related to immune function—most notably phagocytosis and induction of an inflammatory response. Brucella spp. are intracellular bacteria that utilize various methods to evade and modulate the host immune response. The identified loci and putative candidate genes may play an important role in host immune responses to B. suis infection, characterized by a detectable bacterial presence yet a differential antibody response.
The candidate loci mapped to non-coding regions of the swine genome, suggesting involvement in regulation of gene expression. Non-coding variants can modulate transcription factor binding within promoter and enhancer regions, methylation, targeted recruitment of transcriptional activators and repressors, gene splicing, microRNA (miRNA) binding to 3′UTR, and expression of long non-coding RNAs [lncRNA; (144–147)]. Studies suggest that changes in gene expression can dysregulate immune pathways that are fundamental in disease outcomes (146, 148). For instance, SNP found within cytokine promoters have been linked to susceptibility to Mycobacterium tuberculosis—a pathogenic intracellular bacteria that causes tuberculosis (149).
Several genes associated with host resistance or susceptibility to Brucella spp. have been identified in cattle, goats, and humans. For example, previous studies have reported associations between SNP in natural resistance-associated macrophage protein 1 (NRAMP1) and B. abortus resistance or susceptibility in cattle (150–152). Another gene previously linked to B. abortus in cattle is Toll-like receptor 4 [TLR4; (152)]. In goats, Rossi et al. (153, 154) associated an intronic polymorphism (rs657542977) in protein tyrosine phosphatase receptor type T (PTPRT) and an insertion/deletion polymorphism (InDels; rs660531540) in interferon regulatory factor 3 (IRF3) with host resistance. A study of human patients infected with Brucella revealed a SNP within interferon gamma (IFNG) associated with increased susceptibility to B. melitensis (155). However, brucellosis is caused by multiple species of Brucella that exhibit preferential host specificity [e.g., B. abortus [cattle, bison, and buffalo], B. melitensis [sheep and goats], B. suis [pigs], B. ovis [sheep], B. canis [dogs], B. neotomae [mice], B. pinnipedia [seals, sea lions, walruses], and B. cetacea [whales, porpoises and dolphins]; (156, 157)]. Therefore, brucellosis disease dynamics may differ in swine relative to other species due differences among hosts and Brucella spp.
Although genes previously associated with Brucella spp. resistance/susceptibility are conserved across species, we did not find associations of these genes in our survey of naturally infected feral swine. This discordance may be due to the fact that our analysis focused on the seroconversion of culture-positive animals as opposed to resistant (non-infected) or susceptible (infected) phenotypes such as IFN-γ responses. Also, the genotyping array used in this study did not contain SNP within three of the five genes previously associated with brucellosis infections. Future research could use targeted resequencing to evaluate the influence of genes previously associated with immune response to Brucella spp. that were not effectively evaluated with this SNP array.
With this analysis we identified statistical associations between loci and seroconversion in infected feral swine; however, GWAS are ineffective for determining the causal variant and the mechanism in which the variant affects biological pathways contributing to seroconversion (158, 159). Further, our limited sample represented naturally infected animals opportunistically identified from a broader disease surveillance study (9). Therefore, follow-up experimental infection studies are needed to fully elucidate the genetic drivers of differential host responses to swine brucellosis and provide additional understanding of epidemiological processes.
Although brucellosis was eradicated from U.S. commercial swine, in some regions of the U.S. there has been a shift from biosecure commercial operations to pasture systems, thus, presenting an increased opportunity for disease spillover through contact with feral swine. Re-emergence of B. suis in domestic pigs would have enduring effects on production systems and the pork export market. Currently, the prevalence of brucellosis among feral swine, and the concomitant risk of disease transmission to domestic pigs, is interpreted through serological assays as a component of a nationwide disease surveillance program. The underestimation of infection using routine serological diagnostics is problematic and creates substantial challenges in estimating true disease prevalence, which is critical for effective management (160). Understanding genetic determinants of seroconversion following infection with B. suis across the invaded range would enable better interpretation of serologic diagnostic results, optimal allocation of surveillance efforts, and focused elimination efforts on feral swine populations that pose the greatest risk of spillover to domestic pigs (161).
Data Availability Statement
Data presented in this study are available through the European Variation Archive under accession: PRJEB40856.
Ethics Statement
Ethical review and approval was not required for the animal study because previously published data was used (9) and genotyping was conducted ancillary to sample collection.
Author Contributions
CP and TS contributed to the development of the idea, collection of the data, data analysis and interpretation, and preparation of the document. VB and SS contributed to the development of the idea, data analysis and interpretation, and preparation of the document. SO and PB contributed to data analysis and interpretation and preparation of the document. KP contributed to collection of the data, data analysis and interpretation, and preparation of the document. RM contributed to data analysis and preparation of the document. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the U.S. Department of Agriculture, National Feral Swine Damage Management Program, and the National Wildlife Research Center.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Antoinette Piaggio, Holly Neibergs, and Milton Thomas for feedback on the manuscript. The findings and conclusions in this publication are those of the authors and should not be construed to represent any official USDA or U.S. Government determination or policy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.554674/full#supplementary-material
References
1. Lewis JS, Corn JL, Mayer JJ, Jordan TR, Farnsworth ML, Burdett CL, et al. Historical, current, and potential population size estimates of invasive wild pigs (Sus scrofa) in the United States. Biol Invasions. (2019) 21:2373–84. doi: 10.1007/s10530-019-01983-1
2. Smyser TJ, Tabak MA, Slootmaker C, Robeson MS II, Miller RS, Bosse M, et al. Rapid expansion of an invasive ungulate driven by bridgehead populations of admixed wild and domestic lineages. Mol Ecol. (2020) 29:1103–19. doi: 10.1111/mec.15392
3. Tabak MA, Piaggio AJ, Miller RS, Sweitzer RA, Ernest HB. Anthropogenic factors predict movement of an invasive species. Ecosphere. (2017) 8:e01844. doi: 10.1002/ecs2.1844
4. Hernández FA, Sayler KA, Bounds C, Milleson MP, Carr AN, Wisely SM. Evidence of pseudorabies virus shedding in feral swine (Sus scrofa) populations of Florida, USA. J Wildl Dis. (2018) 4:45–53. doi: 10.7589/2017-04-071
5. Anderson A, Slootmaker C, Harper E, Holderieath J, Shwiff SA. Economic estimates of feral swine damage and control in 11 US states. Crop Prot. (2016) 89:89–94. doi: 10.1016/j.cropro.2016.06.023
6. Bankovich B, Boughton E, Boughton R, Avery ML, Wisely SM. Plant community shifts caused by feral swine rooting devalue Florida rangeland. Agric Ecosyst Environ. (2016) 15:45–54. doi: 10.1016/j.agee.2015.12.027
7. Hutton T, Deliberto T, Owen S, Morrison B. Disease Risks Associated with Increasing Feral Swine Numbers and Distribution in the United States. Midwest Association of Fish and Wildlife Agencies, Wildlife and Fish Health Commission, 15. (2006). Available online at: https://www.aphis.usda.gov/wildlife_damage/nwdp/pdf/Hutton_Pig_Paper_177657_7.pdf
8. Bevins SN, Pedersen K, Lutman MW, Gidlewski T, Deliberto TJ. Consequences associated with the recent range expansion of nonnative feral swine. BioScience. (2014) 64:291–9. doi: 10.1093/biosci/biu015
9. Pedersen K, Bauer NE, Olsen S, Arenas-Gamboa AM, Henry AC, Sibley TD, et al. Identification of Brucella spp. in feral swine (Sus scrofa) at abattoirs in Texas, USA. Zoonoses Public Health. (2017) 64:647–54. doi: 10.1111/zph.12359
10. Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D, et al. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med. (2011) 102:118–31. doi: 10.1016/j.prevetmed.2011.04.007
11. Pauwels AM, Trost M, Beyaert R, Hoffmann E. Patterns, receptors, and signals: Regulation of phagosome maturation. Trends in Immunol. (2017) 38:407–22. doi: 10.1016/j.it.2017.03.006
12. Celli J. The intracellular lifecycle of Brucella spp. Microbiol Spectr. (2019) 7:1–17. doi: 10.1128/microbiolspec.BAI-0006-2019
13. Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. (2010) 12:1203–12. doi: 10.1111/j.1462-5822.2010.01499.x
14. Myeni S, Child R, Ng TW, Kupko JJ III, Wehrly TD, Porcella SF, et al. Brucella modulates secretory trafficking via multiple type IV secretion effector proteins. PLoS Pathog. (2013) 9:e1003556. doi: 10.1371/journal.ppat.1003556
15. Stoffregen WC, Olsen SC, Wheeler CJ, Bricker BJ, Palmer MV, Jensen AE, et al. Diagnostic characterization of a feral swine herd enzootically infected with Brucella. J Vet Diagn Invest. (2007) 19:227–37. doi: 10.1177/104063870701900301
16. Higgins J, Stuber T, Quance C, Edwards WH, Tiller RV, Linfield T, et al. Molecular epidemiology of Brucella abortus isolates from cattle, elk, and bison in the United States, 1998 to 2011. Appl Environ Microbiol. (2012) 78:3674–84. doi: 10.1128/AEM.00045-12
17. Rónai Z, Kreizinger Z, Dán Á, Drees K, Foster JT, Bányai K, et al. First isolation and characterization of Brucella microti from wild boar. BMC Vet Res. (2015) 11:147. doi: 10.1186/s12917-015-0456-z
18. Olsen SC, Palmer MV. Advancement of knowledge of Brucella over the past 50 years. Vet Pathol. (2014) 51:1076–89. doi: 10.1177/0300985814540545
20. USDA APHIS. What Is Swine Brucellosis? (2018). Available online at: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/swine-disease-information/swine-brucellosis (acessed April 1, 2020).
21. Pedersen K, Bevins SN, Schmit BS, Lutman MW, Milleson MP, Turnage CT, et al. Apparent prevalence of swine brucellosis in feral swine in the United States. Hum-Wildl Interact. (2012) 6:38–47.
22. Miller RS, Sweeney SJ, Slootmaker C, Grear DA, DiSalvo PA, Kiser D, et al. Cross-species transmission potential between wild pigs, livestock, poultry, wildlife, and humans: Implications for disease risk management in North America. Sci Rep. (2017) 7:7821. doi: 10.1038/s41598-017-07336-z
23. Wyckoff AC, Henke SC, Campbell TA, Hewitt DG, Vercauteren KC. Feral swine contact with domestic swine: a serological survey and assessment of potential for disease transmission. J Wildl Dis. (2009) 45:422–9. doi: 10.7589/0090-3558-45.2.422
24. National Pork Producer's Council (n.d) Pork facts. Available online at: http://nppc.org/pork-facts/ (accessed April 1, 2020).
25. Witmer GW, Sanders RB, Taft AC. “Feral swine – are they a disease threat to livestock in the United States?” In: The 10th Wildlife Damage Management Conference Proceedings: Hot Springs, AR, April 6-9. Hot Springs, AR: The 10th Wildlife Damage Management (2003). p. 316–25.
26. Olsen SC, Tatum FM. Swine brucellosis: current perspectives. Vet Med Res Rep. (2017) 8:1–12. doi: 10.2147/VMRR.S91360
27. Ewalt DR, Payeur JB, Rhyan JC, Geer PL. Brucella suis biovar 1 in naturally infected cattle: a bacteriological, serological, and histological study. J Vet Diagn Invest. (1997) 9:417–20. doi: 10.1177/104063879700900414
28. Musser JM, Schwartz AL, Srinath I, Waldrup KA. Use of serology and bacterial culture to determine prevalence of Brucella spp. in feral swine (Sus scrofa) in proximity to a beef cattle herd positive for Brucella suis and Brucella abortus. J Wildl Dis. (2013) 49:215–20. doi: 10.7589/2012-06-169
29. Giurgiutiu D, Banis C, Hunt E, Mincer J, Nicolardi C, Weltman A, et al. Brucella suis infection associated with feral swine hunting-Three States, 2007-2008. MMWR Surveill Summ. (2009) 58:618–21. doi: 10.1016/j.tmaid.2017.03.006
30. Ferris RA, Schoenbaum MA, Crawford RP. Comparison of serologic tests and bacteriologic culture for detection of brucellosis in swine from naturally infected herds. J Am Vet Med Assoc. (1995) 207:1332–3.
31. Pedersen K, Quance CR, Robbe-Austerman S, Piaggio AJ, Bevins SN, Goldstein SM, et al. Identification of Brucella suis from feral swine in selected states in the USA. J Wildl Dis. (2014) 50:171–9. doi: 10.7589/2013-09-235
32. Kianmehr Z, Ardestani SK, Soleimanjahi H, Fotouhi F, Alamian S, Ahmadian S. Comparison of biological and immunological characterization of Lipopolysaccharides from Brucella abortus RB51 and S19. Jundishapur J Microbiol. (2015) 8:e24853. doi: 10.5812/jjm.24853
33. Lee JJ, Simborio HL, Bernardo-Reyes AW, Kim DG, Hop HT, Min W, et al. Immunoproteomic identification of immunodominant antigens independent of the time of infection in Brucella abortus 2308-challenged cattle. Vet Res. (2015) 46:17. doi: 10.1186/s13567-015-0147-6
34. Cloeckaert A, Weynants V, Godfroid J, Verger JM, Grayon M, Zygmunt MS. O-Polysaccharide epitopic heterogeneity at the surface of Brucella spp. studied by enzyme-linked immunosorbent assay and flow cytometry. Clin Diagn Lab Immunol. (1998) 5:862–70. doi: 10.1128/CDLI.5.6.862-870.1998
35. Frank SA. Genetic variability of hosts. In: Frank SA, editor. Immunology and Evolution of Infectious Disease. New Jersey, NJ: Princeton University Press (2002). p. 111–23. doi: 10.1515/9780691220161-009
36. Bishop SC, De Jong M, Gray D. Opportunities for incorporating genetic elements into the management of farm animal diseases: policy issues. In: Food and Agriculture Organization of the United Nations (FAO) Study Paper. Food and Agriculture Organization of the United Nations (FAO) (2002). 18:1–36. Available online at: http://www.fao.org/3/a1250e/annexes/Thematic%20Studies/bsp18e.pdf
37. Bishop SC. Disease resistance: genetics. In: Pond WG, Bell AW, editors. Encyclopedia of Animal Science. New York, NY: Marcel Dekker, Inc. (2005) 288–90. doi: 10.1201/9781482276664-87
38. Nicholas FW. Animal breeding and disease. Philos Trans R Soc Lond B Biol Sci. (2005) 360:1529–36. doi: 10.1098/rstb.2005.1674
39. Garry Adams L, Schutta CJ. Natural resistance against brucellosis: a review. Open Vet Sci J. (2010) 4:61–7. doi: 10.2174/1874318801004010061
40. Fosgate GT, Adesiyun AA, Hird DW, Johnson WO, Hietala SK, Schurig GG, et al. Comparison of serologic tests for detection of Brucella infections in cattle and water buffalo (Bubalus bubalis). Am J Vet Res. (2002) 63:1598–605. doi: 10.2460/ajvr.2002.63.1598
41. Nielsen K. Diagnosis of brucellosis by serology. Vet Microbiol. (2002) 90:447–59. doi: 10.1016/S0378-1135(02)00229-8
42. Jungersen G, Sørensen V, Giese S, Stack J, Riber U. Differentiation between serological responses to Brucella suis and Yersinia enterocolitica serotype O[ratio]9 after natural or experimental infection in pigs. Epidemiol Infect. (2006) 134:347–57. doi: 10.1017/S095026880500511X
43. Brown V.R., Bowen R.A., Ledesma N., Hartwig A., Gordy P., Pierce C.F., et al. (2020). Pathogenesis and Immune Responses of Heritage Breed Pigs to Experimental Inoculation with Brucella suis (Manuscript submitted for publication). Department of Biomedical Sciences, Colorado State University, Fort Collins, CO, United States.
44. Ramos AM, Crooijmans RP, Affara NA, Amaral AJ, Archibald AL, Beever JE, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE. (2009) 4:e6524. doi: 10.1371/journal.pone.0006524
45. Warr A, Affara N, Aken B, Beiki H, Bickhart DM, Billis K, et al. An improved pig reference genome sequence to enable pig genetics and genomics research. Gigascience. (2020). 9:giaa051. doi: 10.1093/gigascience/giaa051
46. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience. (2015) 4:7. doi: 10.1186/s13742-015-0047-8
47. Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. (2011) 6:122–33. doi: 10.1038/nprot.2010.182
48. Bush WS, Moore JH. Genome-wide association studies. PLOS Comput Biol. (2012) 8:e1002822. doi: 10.1371/journal.pcbi.1002822
49. Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. Variance component model to account for sample structure in genome-wide assocation studies. Nature Genet. (2010) 42:348–54. doi: 10.1038/ng.548
50. Bozdogan H. Model selection and Akaike's information criterion (AIC): the general theory and its analytical extensions. Psychometrika. (1987) 52:345–70. doi: 10.1007/BF02294361
51. Devlin B, Roeder K. Genomic control for association studies. Biometrics. (1999) 55:997–1004. doi: 10.1111/j.0006-341X.1999.00997.x
52. Aulchenko YS. GWA analysis in presence of stratification: theory. In: GenABEL Tutorial. GenABEL project contributors (2015). p. 137–66.
53. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common disease and 3,000 shared controls. Nature. (2007) 447:661. doi: 10.1038/nature05911
54. Morris JA, Gardner MJ. Calculating confidence intervals for relative risks (odds ratios) and standardised ratios and rates. Brit Med J. (1988) 296:1313–6. doi: 10.1136/bmj.296.6632.1313
55. Altman DG. Comparing groups – categorical data. In: Altman DG, editor. Practical Statistics for Medical Research. Florida, FL: CRC Press (1991). p. 229–76. doi: 10.1201/9780429258589
56. Kiser JN, White SN, Johnson KA, Hoff JL, Taylor JF, Neibergs HL. Identification of loci associated with susceptibility to Mycobacterium avium subspecies paratuberculosis (MAP) tissue infection in cattle. J Anim Sci. (2017) 95:1080–91. doi: 10.2527/jas.2016.1152
57. Onteru SK, Gorbach DM, Young JM, Garrick DJ, Dekkers JCM, Rothschild MF. Whole genome association studies of residual feed intake and related traits in the pig. PLoS ONE. (2013) 8:e61756. doi: 10.1371/journal.pone.0061756
58. Horodyska J, Hamill RM, Varley PF, Reyer H, Wimmers K. Genome-wide association analysis and functional annotation of positional candidate genes for feed conversion efficiency and growth rate in pigs. PLoS ONE. (2017) 12:e0173482. doi: 10.1371/journal.pone.0173482
59. Tang Z, Xu J, Yin L, Yin D, Zhu M, Yu M, et al. Genome-wide association study reveals candidate genes for growth relevant traits in pigs. Front Genet. (2019) 10:302. doi: 10.3389/fgene.2019.00302
60. Veroneze R, Lopes PS, Guimarães SEF, Silva FF, Lopes MS, Harlizius B, et al. Linkage disequilibrium and haplotype block structure in six commercial pig lines. J Anim Sci. (2013) 91:3493–501. doi: 10.2527/jas.2012-6052
61. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (1999) 27:29–34. doi: 10.1093/nar/27.1.29
62. Staab JF, Ginkel DL, Rosenberg GB, Munford RS. Saposin-like domain influences the intracellular localization, stability, and catalytic activity of human acylosyacyl hydrolase. J Biol Chem. (1994) 269:23736–42.
63. Feulner JA, Lu M, Shelton JM, Zhang M, Richardson JA, Munford RS. Identification of acycloxyacyl hydrolase, a lipopolysaccharide-detoxifying enzyme, in the murine urinary tract. Infect Immun. (2004) 72:3171–8. doi: 10.1128/IAI.72.6.3171-3178.2004
64. Gorelik A, Illes K, Nagar B. Crystal structure of the mammalian lipopolysacharide detoxifier. Proc Nat Acad Sci USA. (2018) 115:E896–905. doi: 10.1073/pnas.1719834115
65. Lu M, Varley AW, Ohta S, Hardwick J, Munford RS. Host inactivation of bacterial lipopolysaccharide prevents prolonged microbial tolerance following Gram-negative bacterial infection. Cell Host Microbe. (2008) 4:293–302. doi: 10.1016/j.chom.2008.06.009
66. Hagen FS, Grant FJ, Kuijper JL, Slaughter CA, Moomaw CR, Orth K, et al. Expression and characterization of recombinant human acyloxyacyl hydrolase, a leukocyte enzyme that deacylates bacterial lipopolysaccharides. Biochem. (1991) 30:8415–23. doi: 10.1021/bi00098a020
67. Lu M, Zhang M, Takashima A, Weiss J, Apicella MA, Li XH, et al. Lipopolysaccharide deacylation by an endogenous lipase controls innate antibody responses to Gram-negative bacteria. Nat Immunol. (2005) 6:989–94. doi: 10.1038/ni1246
68. Zou B, Jiang W, Han H, Li J, Mao W, Tang Z, et al. Acyloxyacyl hydrolase promotes the resolution of lipopolysaccharide-induced acute lung injury. PLoS Pathogen. (2017) 13:e1006436. doi: 10.1371/journal.ppat.1006436
69. Leyva-Cobian FR, Unanue ER. Intracellular interference with antigen presentation. J Immunol. (1988) 141:1445–50.
70. Forestier C, Moreno E, Méresse S, Phalipon A, Olive D, Sansonetti P, et al. Interaction of Brucella abortus Lipopolysaccharide with major histocompatibility complex class II molecules in B lymphocytes. Infect Immun. (1999) 67:4048–54. doi: 10.1128/IAI.67.8.4048-4054.1999
71. Stevenson C, de la Rosa G, Anderson CS, Murphy PS, Capece T, Kim M, et al. Essential role of Elmo1 in Dock2-dependent lymphocyte migration. J Immun. (2014) 192:6062–70. doi: 10.4049/jimmunol.1303348
72. Sarkar A, Tindle C, Pranadinata RF, Reed S, Eckmann L, Stappenbeck TS, et al. ELMO1 regulates autophagy induction and bacterial clearance during enteric infection. J Infect Dis. (2017) 216:1655–66. doi: 10.1093/infdis/jix528
73. McCormick BA. ELMO1: more than just a director of phagocytosis. Cell Mol Gastroenterol Hepatol. (2015) 1:262–3. doi: 10.1016/j.jcmgh.2015.04.002
74. Franciszkiewicz K, Le Floc'h A, Boutet M, Vergnon I, Schmitt A, Mami-Chouaib F. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res. (2013) 73:617–28. doi: 10.1158/0008-5472.CAN-12-2569
75. Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: Structural, cellular, and molecular biology. Annu Rev Biochem. (2000) 69:145–82. doi: 10.1146/annurev.biochem.69.1.145
76. Cox DG, Pontes C, Guino E, Navarro M, Osorio A, Canzian F, et al. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Brit J Cancer. (2004) 91:339–43. doi: 10.1038/sj.bjc.6601906
77. Rocca B, Spain LM, Pure E, Langenbach R, Patrono C, Fitzgerald GA. Distinct roles of prostaglandin H synthases 1 and 2 in T-cell development. J Clin Invest. (1999) 103:1469–77. doi: 10.1172/JCI6400
78. Gagnaire A, Gorvel L, Papadopoulos A, Von Bargen K, Mège JL, Gorvel JP. COX-2 inhibition reduces Brucella bacterial burden in draining lymph nodes. Front Microbiol. (2016) 7:1987. doi: 10.3389/fmicb.2016.01987
79. López-Urrutia L, Alonso A, Bayón Y, Nieto ML, Orduña A, Crespo MS. Brucella lipopolysaccharides induce cyclooxygenase-2 expression in monocytic cells. Biochem Biophys Res Commun. (2001) 289:372–5. doi: 10.1006/bbrc.2001.5995
80. Harris SG, Padilla J, Koumas L, Ray D, Phipps RP. Prostaglandins as modulators of immunity. Trends Immunol. (2002) 23:144–50. doi: 10.1016/S1471-4906(01)02154-8
81. Woolard MD, Wilson JE, Hensley LL, Jania LA, Kawula TH, Drake JR, et al. Francisella tularensis-infected macrophages release prostaglandin E2 that blocks T cell proliferation and promotes a Th2-like response. J Immunol. (2007) 178:2065–74. doi: 10.4049/jimmunol.178.4.2065
82. Scibelli A, Roperto S, Manna L, Pavone LM, Tafuri S, Della Morte R, et al. Engagement of integrins as a celular route of invasion by bacterial pathogens. Vet J. (2007) 173:482–91. doi: 10.1016/j.tvjl.2006.01.010
83. Switala-Jelen K, Dabrowska K, Opolski A, Lipinska L, Nowaczyk M, Gorski A. The biological functions of β3 integrins. Folia Biol. (2004) 50:143–52.
84. Langsenlehner U, Renner W, Yazdani-Biuki B, Eder T, Wascher TC, Paulweber B, et al. Integrin alpha-2 and beta-3 gene polymorphisms and breast cancer risk. Breast Cancer Res Treat. (2006) 97:67–72. doi: 10.1007/s10549-005-9089-4
85. Thompson EE, Pan L, Ostrovnaya I, Weiss LA, Gern JE, Lemanske RF Jr, et al. Integrin β3 genotype influences asthma and allergy phenotypes in the first 6 years of life. J Allergy Clin Immuno. (2007) 119:1423–9. doi: 10.1016/j.jaci.2007.03.029
86. Rivera-Soto R, Dissinger NJ, Damania B. Kaposi's sarcoma-associated herpesvirus viral interleukin-6 signaling upregulates integrin β3 levels and is dependent on STAT3. J Virol. (2020) 94:e01384–19. doi: 10.1128/JVI.01384-19
87. Lee J, Banu SK, Burghardt RC, Starzinski-Powitz A, Arosh JA. Selective inhibition of prostaglandin E2 receptors EP2 and EP4 inhibits adhesion of human endometriotic epithelial and stromal cells through suppression of integrin-mediated mechanisms. Biol Reprod. (2013) 88:1–11. doi: 10.1095/biolreprod.112.100883
88. Rapisarda V, Borghesan M, Miguela V, Encheva V, Snijders AP, Lujambio A, et al. Integrin beta 3 regulates cellular senescence by activating the TGF-β pathway. Cell Rep. (2017) 18:2480–93. doi: 10.1016/j.celrep.2017.02.012
89. Cai X, Thinn AM, Wang Z, Shan H, Zhu J. The importance of N-glycosylation on β 3 integrin ligand binding and conformational regulation. Sci Rep. (2017) 7:4656. doi: 10.1038/s41598-017-04844-w
90. Zhang K, Gao R, Zhang H, Cai X, Shen C, Wu C, et al. Molecular cloning and characterization of three novel lysozyme-like genes, predominantly expressed in the male reproductive system of humans, belonging to the C-type lysozyme/alpha-lactalbumin family. Biol Repro. (2005) 73:1064–71. doi: 10.1095/biolreprod.105.041889
91. Ragland SA, Criss AK. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. (2017) 13:e1006512. doi: 10.1371/journal.ppat.1006512
92. Pizarro-Cerdá J, Moreno E, Gorvel JP. Invasion and intracellular trafficking of Brucella abortus in nonphagocytic cells. Microbes Infect. (2000) 2:829–35. doi: 10.1016/S1286-4579(00)90368-X
93. Omotade TO, Roy CR. Manipulation of host cell organelles by intracellular pathogens. In: Cossart P, Roy CR, Sansonetti P, editors. Bacteria and Intracellularity. New Jersey, NJ: John Wiley and Sons (2019). p. 179–96. doi: 10.1128/9781683670261.ch13
94. Case ED, Samuel JE. Contrasting lifestyles within the host cell. In: Kudva IT, Cornick NA, Plummer PJ, Zhang Q, Nicholson TL, Bannantine JP, Bellaire BH, editors. Virulence Mechanisms of Bacterial Pathogens. New Jersey, NJ: John Wiley and Sons (2016). p. 667–92. doi: 10.1128/9781555819286.ch23
95. Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. (2008) 132:344–62. doi: 10.1016/j.cell.2008.01.020
97. Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol. (2009) 27:693–733. doi: 10.1146/annurev.immunol.021908.132641
98. Sun SC. The non-canonical NF-κB pathway in immunity and inflammation. Nat Rev Immunol. (2017) 17:545–58. doi: 10.1038/nri.2017.52
99. Rckle A, Haasbach E, Julkunen I, Planz O, Ehrhardt C, Ludwig S. The NS1 protein of influenza a virus blocks RIG-I-mediated activation of the noncanonical NF-B pathway and p52/RelB-dependent gene expression in lung epithelial cells. J Virol. (2012) 86:10211–7. doi: 10.1128/JVI.00323-12
100. Jin J, Hu H, Li HS, Yu J, Xiao Y, Brittain GC, et al. Noncanonical NF-κB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. (2014) 40:342–54. doi: 10.1016/j.immuni.2014.02.006
101. Liu P, Li K, Garofalo RP, Brasier AR. Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-κB2 complexes via a novel retinoic acid-inducible gene-I· NF-κB-inducing kinase signaling pathway. J Biol Chem. (2008) 283:23169–78. doi: 10.1074/jbc.M802729200
102. Matta H, Chaudhary PM. Activation of alternative NF-κB pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein (vFLIP). Proc Natl Acad Sci USA. (2004) 101:9399–404. doi: 10.1073/pnas.0308016101
103. Cho IR, Jeong S, Jhun BH, Lee B, Kwak YT, Lee SH, et al. Activation of non-canonical NF-kB pthway mediated by STP-A11, an oncoprotein of Herpesvirus saimiri. Virology. (2007) 359:37–45. doi: 10.1016/j.virol.2006.09.001
104. de Jong SJ, Albrecht JC, Giehler F, Kieser A, Sticht H, Biesinger B. Noncanonical NF-κB activation by the oncoprotein Tio occurs through a nonconserved TRAF3-binding motif. Sci Signal. (2013) 6:ra27. doi: 10.1126/scisignal.2003309
105. Yanai A, Maeda S, Hikiba Y, Shibata W, Ohmae T, Hirata Y, et al. Clinical relevance of Helicobacter pylori sabA genotype in Japanese clinical isolates. J Gastroenterol Hepatol. (2007) 22:2228–32. doi: 10.1111/j.1440-1746.2007.04831.x
106. Ge J, Xu H, Li T, Zhou Y, Zhang Z, Li S, et al. A Legionella type IV effector activates the NF-κB pathway by phosphorylating the IκB family of inhibitors. Proc Natl Acad Sci USA. (2009) 106:13725–30. doi: 10.1073/pnas.0907200106
107. Hop HT, Reyes AW, Huy TX, Arayan LT, Min W, Lee HJ, et al. Activation of NF-kB-mediated TNF-induced antimicrobial immunity is required for the efficient Brucella abortus clearance in RAW 264.7 cells. Front Cell Infect Microbiol. (2017) 7:437. doi: 10.3389/fcimb.2017.00437
108. Hop HT, Arayan LT, Reyes AW, Huy TX, Min WG, Lee HJ, et al. Heat-stress-modulated induction of NF-κB leads to brucellacidal pro-inflammatory defense against Brucella abortus infection in murine macrophages and in a mouse model. BMC Microbiol. (2018) 18:44. doi: 10.1186/s12866-018-1185-9
109. Bassani S, Cingolani LA. Tetraspanins: interactions and interplay with integrins. Int J Biochem Cell Biol. (2012) 44:703–8. doi: 10.1016/j.biocel.2012.01.020
110. Zhou J, Fujiwara T, Ye S, Li X, Zhao H. Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits osteoclastogenesis in vitro. Calcif Tissue Int. (2014) 95:209–17. doi: 10.1007/s00223-014-9883-2
111. Saint-Pol J, Billard M, Dornier E, Eschenbrenner E, Danglot L, Boucheix C, et al. New insights into the tetraspanin Tspan5 using novel monoclonal antibodies. J Bio Chem. (2017) 292:9551–66. doi: 10.1074/jbc.M116.765669
112. Vences-Catalán F, Levy S. Immune targeting of tetraspanins involved in cell invasion and metastasis. Front Immunol. (2018) 9:1277. doi: 10.3389/fimmu.2018.01277
113. Termini CM, Gillette JM. Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol. (2017) 5:34. doi: 10.3389/fcell.2017.00034
114. Harada Y, Suzuki T, Fukushige T, Kizuka Y, Yagi H, Yamamoto M, et al. Generation of the heterogeneity of extracellular vesicles by membrane organization and sorting machineries. Biochim Biophys Acta Gen Subj. (2019) 1863:681–91. doi: 10.1016/j.bbagen.2019.01.015
115. Di Cesare P, Hauser N, Lehman D, Pasumarti S, Paulsson M. Cartilage oligomeric matrix protein (COMP) is an abundant component of tendon. FEBS Lett. (1994) 354:237–40. doi: 10.1016/0014-5793(94)01134-6
116. Juenger CH, Holst MI, Duffe K, Jankowski J, Baader SL. Tetraspanin-5 (Tm4sf9) mRNA expression parallels neuronal maturation in the cerebellum of normal and L7En-2 transgenic mice. J Comp Neurol. (2005) 483:318–28. doi: 10.1002/cne.20439
117. Acharya C, Yik JH, Kishore A, Van Dinh V, Di Cesare PE, Haudenschild DR. Cartilage oligomeric matrix protein and its binding partners in the cartilage extracellular matrix: interaction, regulation and role in condrogenesis. Matrix Biol. (2014) 37:102–11. doi: 10.1016/j.matbio.2014.06.001
118. Fu Y, Kong W. Cartilage oligomeric matrix proteins: matricellular and matricrine signaling in cardiovascular homeostasis and disease. Curr Vasc Pharmacol. (2017) 15:186–96. doi: 10.2174/1570161115666170201121232
119. Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. (2009) 15:2288–99. doi: 10.2174/138161209788682442
120. Damle SR, Martin RK, Cockburn CL, Lownik JC, Carlyon JA, Smith AD, et al. ADAM 10 and Notch1 on murine dendritic cells control the development of type 2 immunity and IgE production. Allergy. (2018) 73:125–36. doi: 10.1111/all.13261
121. Lambrecht BN, Vanderkerken M, Hammad H. The emerging role of ADAM metalloproteinases in immunity. Nat Rev Immunol. (2018) 18:745–58. doi: 10.1038/s41577-018-0068-5
122. Hibbs ML, Tarlinton DM, Armes J, Grail D, Hodgson G, Maglitto R, et al. Multiple defects in the immune system of Lyn-deficient mice, culminating in autoimmune disease. Cell. (1995) 83:301–11. doi: 10.1016/0092-8674(95)90171-X
123. Hibbs ML, Harder KW, Armes J, Kountouri N, Quilici C, Casagranda F, et al. Sustained activation of Lyn tyrosine kinase in vivo leads to autoimmunity. J Exp Med. (2002) 196:1593–604. doi: 10.1084/jem.20020515
124. Xu Y, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM. Lyn tyrosine kinase: accentuating the positive and the negative. Immunity. (2005) 22:9–18. doi: 10.1016/S1074-7613(04)00381-4
125. Scapini P, Pereira S, Zhang H, Lowell CA. Multiple roles of Lyn kinase in myeloid cell signaling and function. Immunol Rev. (2009) 228:23–40. doi: 10.1111/j.1600-065X.2008.00758.x
126. Ingley E, Sarna MK, Beaumont JG, Tilbrook PA, Tsai S, Takemoto Y, et al. HS1 interacts with Lyn and is critical for erythropoietin-induced differentiation of erythroid cells. J Biol Chem. (2000) 275:7887–93. doi: 10.1074/jbc.275.11.7887
127. Chin H, Arai A, Wakao H, Kamiyama R, Miyasaka N, Miura O. Lyn physically associates with the erythropoietin receptor and may play a role in activation of the Stat5 pathway. Blood. (1998) 91:3734–45. doi: 10.1182/blood.V91.10.3734
128. Tilbrook PA, Ingley E, Williams JH, Hibbs ML, Klinken SP. Lyn tyrosine kinase is essential for erythropoietin-induced differentiation of J2E erythroid cells. EMBO J. (1997) 16:1610–9. doi: 10.1093/emboj/16.7.1610
129. Harder KW, Parsons LM, Armes J, Evans N, Kountouri N, Clark R, et al. Gain-and loss-of-function Lyn mutant mice define a critical inhibitory role for Lyn in the myeloid lineage. Immunity. (2001) 15:603–15. doi: 10.1016/S1074-7613(01)00208-4
130. Gauld SB, Cambier JC. Src-family kinases in B-cell development and signaling. Oncogene. (2004) 23:8001–6. doi: 10.1038/sj.onc.1208075
131. Borzȩcka-Solarz K, Dembińska J, Hromada-Judycka A, Traczyk G, Ciesielska A, Ziemlińska E, et al. Association of Lyn kinase with membrane rafts determines its negative influence on LPS-induced signaling. Mol Biol Cell. (2017) 28:1147–59. doi: 10.1091/mbc.E16-09-0632
132. Ban T, Sato GR, Nishiyama A, Akiyama A, Takasuna M, Umehara M, et al. Lyn kinase suppresses the transcriptional activity of IRF5 in the TLR-MyD88 pathway to restrain the development of autoimmunity. Immunity. (2016). 45:319–32. doi: 10.1016/j.immuni.2016.07.015
133. Keck S, Freudenberg M, Huber M. Activation of murine macrophages via TLR2 and TLR4 is negatively regulated by a Lyn/PI3K module and promoted by SHIP1. J Immun. (2010) 184:5809–18. doi: 10.4049/jimmunol.0901423
134. Veit G, Kobbe B, Keene DR, Paulsson M, Koch M, Wagener R. Collagen XXVIII, a novel von Willebrand factor A domain-containing protein with many imperfections in the collagenous domain. J Bio Chem. (2006) 281:3494–504. doi: 10.1074/jbc.M509333200
135. Kennedy RB, Ovsyannikova IG, Pankratz VS, Haralambieva I, Vierkant R, Poland G. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Hum Genet. (2012) 131:1403–21. doi: 10.1007/s00439-012-1174-2
136. Cai Z, Guldbrandtsen B, Lund MS, Sahana G. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genomics. (2018) 19:656. doi: 10.1186/s12864-018-5050-x
137. Song X, Lin NH, Wang YL, Chen B, Wang HX, Hu K. Comprehensive transcriptome analysis based on RNA sequencing identifies critical genes for lipopolysaccharide-induced epididymitis in a rat model. Asian J Androl. (2019) 21:605–11. doi: 10.4103/aja.aja_21_19
138. Schiller HB, Fernandez IE, Burgstaller G, Schaab C, Scheltema RA, Schwarzmayr T, et al. Time-and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol Syst Biol. (2015) 11:819. doi: 10.15252/msb.20156123
139. Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. (2011) 22:189–95. doi: 10.1016/j.cytogfr.2011.10.001
140. Pinkerton JW, Kim RY, Robertson AAB, Hirota JA, Wood LG, Knight DA, et al. Inflammasomes in the lung. Mol Immunol. (2017) 86:44–55. doi: 10.1016/j.molimm.2017.01.014
141. Hielpos MS, Ferrero MC, Fernández AG, Falivene J, Vanzulli S, Comerci DJ, et al. Btp proteins from Brucella abortus modulate the lung innate immune response to infection by the respiratory route. Front Immunol. (2017) 8:1011. doi: 10.3389/fimmu.2017.01011
142. Edfors-Lilja I, Wattrang E, Marklund L, Moller M, Andersson-Eklund L, Andersson L, et al. Mapping quantitative trait loci for immune capacity in the pig. J Immunol. (1998) 161:829–35.
143. Lee YM, Alam M, Choi B, Kim KS, Kim JJ. A whole genome association study to detect single nucleotide polymorphisms for blood components (immunity) in a cross between Korean native pig and Yorkshire. Asian-Australas J Anim Sci. (2012) 25:1674–80. doi: 10.5713/ajas.2012.12503
144. Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta Mol Basis Dis. (2014) 1842:1910–22. doi: 10.1016/j.bbadis.2014.03.011
145. Zhang F, Lupski JR. Non-coding genetic variants in human disease. Hum Mol Genet. (2014) 24:R102–R110. doi: 10.1093/hmg/ddv259
146. Rojano E, Seoane P, Ranea JA, Perkins JR. Regulatory variants: from detection to predicting impact. Brief Bioinform. (2018) 20:1639–54. doi: 10.1093/bib/bby039
147. Ramsuran V, Ewy R, Nguyen H, Kulkarni S. Variation in the untranslated genome and susceptibility to infections. Front Immunol. (2018) 9:2046. doi: 10.3389/fimmu.2018.02046
148. Trossbach SV, Hecher L, Schafflick D, Deenen R, Popa O, Lautwein T, et al. Dysregulation of a specific immune-related network of genes biologically defines a subset of schizophrenia. Transl Psychiatry. (2019) 9:1–6. doi: 10.1038/s41398-019-0486-6
149. Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. (2003) 16:463–96. doi: 10.1128/CMR.16.3.463-496.2003
150. Adams LG, Templeton JW. Genetic resistance to bacterial diseases of animals. Rev Sci Tech. (1998) 17:200–19. doi: 10.20506/rst.17.1.1085
151. Barthel R, Feng J, Piedrahita JA, McMurray DN, Templeton JW, Adams GL. Stable transfection of bovine Nramp1 gene into murine RAW264.7 cells: effect on Brucella abortus survival. Infect Immun. (2001) 69:3110–9. doi: 10.1128/IAI.69.5.3110-3119.2001
152. Prakash O, Kumar A, Sonwane A, Rathore R, Singh RV, Chauhan A, et al. Polymorphism of cytokine and innate immunity genes associated with bovine brucellosis in cattle. Mol Biol Rep. (2014) 41:2815–25. doi: 10.1007/s11033-014-3136-3
153. Rossi UA, Hasenauer FC, Caffaro ME, Neumann R, Salatin A, Poli MA, et al. A haplotype at intron 8 of PTPRT gene is associated with resistance to Brucella infection in Argentinian creole goats. Vet Microbiol. (2017) 207:133–7. doi: 10.1016/j.vetmic.2017.06.001
154. Rossi UA, Hasenauer FC, Caffaro ME, Raschia MA, Maurizio E, Cortez HS, et al. Association of an IRF3 putative functional uORF variant with resistance to Brucella infection: A candidate gene based analysis of InDel polymorphisms in goats. Cytokine. (2019) 115:109–15. doi: 10.1016/j.cyto.2018.11.024
155. Bravo MJ, de Dios Colmenero J, Alonso A, Caballero A. Polymorphisms of the interferon gamma and interleukin 10 genes in human brucellosis. Eur J Immunogenet. (2003) 3:433–5. doi: 10.1111/j.1365-2370.2003.00419.x
156. Spickler AR, Roth JA, Galyon J, Lofstedt J. Emerging and Exotic Diseases of Animals. Iowa, IA: Center for Food Security and Public Health, Iowa State University (2010).
157. O'Callaghan D, Whatmore AM. Brucella genomics as we enter the multi-genome era. Brief Funct Genomics. (2011) 10:334–41. doi: 10.1093/bfgp/elr026
158. van der Sijde MR, Ng A, Fu J. Systems genetics: From GWAS to disease pathways. Biochim Biophys Acta Mol Basis Dis. (2014) 1842:1903–9. doi: 10.1016/j.bbadis.2014.04.025
159. Gallagher MD, Chen-Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genet. (2018) 102:717–30. doi: 10.1016/j.ajhg.2018.04.002
160. Pepin KM, Kay SL, Golas BD, Shriner SS, Gilbert AT, Miller RS, et al. Inferring infection hazard in wildlife populations by linking data across individual and population scales. Ecol Lett. (2017) 20:275–92. doi: 10.1111/ele.12732
Keywords: Brucella suis, brucellosis, disease spillover, feral swine, GWAS, SNP, Sus scrofa
Citation: Pierce CF, Brown VR, Olsen SC, Boggiatto P, Pedersen K, Miller RS, Speidel SE and Smyser TJ (2020) Loci Associated With Antibody Response in Feral Swine (Sus scrofa) Infected With Brucella suis. Front. Vet. Sci. 7:554674. doi: 10.3389/fvets.2020.554674
Received: 22 April 2020; Accepted: 23 October 2020;
Published: 25 November 2020.
Edited by:
Armanda Bastos, University of Pretoria, South AfricaReviewed by:
Gerald Reiner, University of Giessen, GermanyKarren M. Plain, The University of Sydney, Australia
Copyright © 2020 Pierce, Brown, Olsen, Boggiatto, Pedersen, Miller, Speidel and Smyser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Courtney F. Pierce, courtney.pierce@usda.gov
 Courtney F. Pierce
Courtney F. Pierce Vienna R. Brown3
Vienna R. Brown3  Paola Boggiatto
Paola Boggiatto