The True Host/s of Picobirnaviruses
- 1Department of Biomedical Sciences, Ross University School of Veterinary Medicine, Basseterre, Saint Kitts and Nevis
- 2College of Animal Biotechnology, Guru Angad Dev Veterinary and Animal Science University, Ludhiana, India
Picobirnaviruses (PBVs) are bisegmented double-stranded RNA viruses that have been detected in a wide variety of animal species including invertebrates and in environmental samples. Since PBVs are ubiquitous in feces/gut contents of humans and other animals with or without diarrhea, they were considered as opportunistic enteric pathogens of mammals and avian species. However, the virus remains to be propagated in animal cell cultures, or in gnotobiotic animals. Recently, the classically defined prokaryotic motif, the ribosomal binding site sequence, has been identified upstream of putative open reading frame/s in PBV and PBV-like sequences from humans, various animals, and environmental samples, suggesting that PBVs might be prokaryotic viruses. On the other hand, based on the detection of some novel PBV-like RNA-dependent RNA polymerase sequences that use the alternative mitochondrial genetic code (that of mold or invertebrates) for translation, and principal component analysis of codon usage bias for these sequences, it has been proposed that PBVs might be fungal viruses with a lifestyle reminiscent of mitoviruses. These contradicting observations warrant further studies to ascertain the true host/s of PBVs, which still remains controversial. In this minireview, we have focused on the various findings that have raised a debate on the true host/s of PBVs.
Introduction
Picobirnaviruses (PBVs) are bisegmented double-stranded RNA viruses that belong to the sole genus Picobirnavirus within the family Picobiraviridae (1). Picobirnaviruses have been widely reported in fecal samples/gut contents of humans and various animal species with, or without diarrhea (1–3). Traditionally, PBVs are considered as opportunistic enteric pathogens of mammals and avian species (1–5). On the other hand, PBVs have also been detected in invertebrates and environmental samples (6–9). During the past few years, the whole genomes, or at least the complete/nearly complete gene segment-2 sequences of several PBV strains from humans, different animal species and environmental samples have been obtained using next generation sequencing technologies, or a modified non-specific primer-based amplification method (1, 4, 6–8, 10–20). Analyses of the expanded repertoire of diverse full-length/nearly full-length PBV sequences revealed remarkable features in the PBV genome including those that suggest that PBVs might be actually prokaryotic or fungal viruses (1, 4, 6–8, 10–21). In this minireview, we have discussed the various findings that have raised a debate on the true hosts of PBVs.
Picobirnavirus Morphology and Genome
Picobirnaviruses are spherical, non-enveloped viruses with a diameter of ~33–37 nm (1). Since PBVs are non-cultivable, information on the virus capsid is based on those of recombinant virion-like particles (22). PBV possess a simple icosahedral capsid that is composed of 60 symmetric dimers (22). The PBV capsid contains two segments of dsRNA, designated as gene segment-1 (~2.2–2.7 kb in size) and −2 (~1.2–1.9 Kb in size) (Figure 1) (1–3, 5). Because of their small size (“pico” in Spanish) and bi-segmented (“bi” in Latin) nature of the viral genome, the viruses were named “Picobirnaviruses” (23). However, PBVs with monopartite genomes have also been reported in a few studies (6, 24, 25).
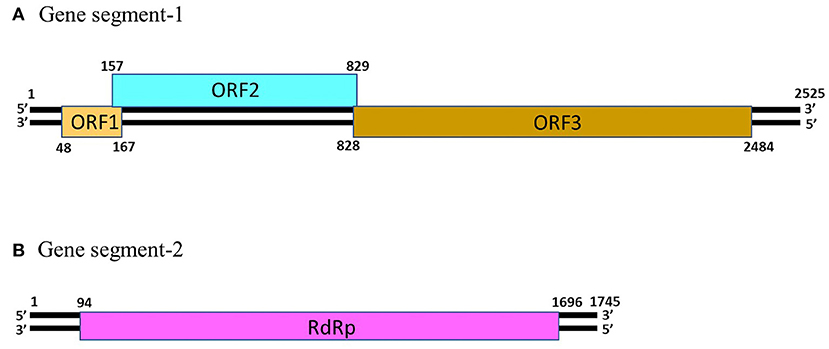
Figure 1. The genomic organization of human picobirnavirus genogroup-I strain Hy005102. (A) Gene segment-1 (GenBank accession number AB186897) of PBV strain Hy005102 consists of 3 putative open reading frames (ORF), designated as ORF1, ORF2 and ORF3. The ORF3 codes for a precursor of the viral capsid protein. (B) Gene segment-2 (GenBank accession number AB186898) of PBV strain Hy005102 possess a single ORF that encodes the viral RNA-dependent RNA polymerase (RdRp).
The gene segment-1 of PBVs consists of 2 or 3 open reading frames (ORFs), designated as ORF1, ORF2, and ORF3 from the 5′- end (Figure 1) (1, 13). The ORF3 codes for a precursor of the capsid protein which undergoes autocatalytic cleavage to generate the mature capsid protein, and a charged peptide that is believed to interact electrostatically with the viral RNA (22). The ORF2 encodes a protein characterized by repeats of the ExxRxNxxxE motif (1, 26). However, the function/s of this protein is not yet known. On the other hand, the functionality, or even the presence of ORF1 in gene segment-1 of PBVs remain to be elucidated (1, 13, 26). The PBV gene segment-2 possess a single large ORF that encodes the RNA-dependent RNA polymerase (RdRp) (Figure 1) (1–3, 5). The 5′- (GUAAA) and 3′- (ACUGC) termini sequences appear to be conserved in the gene segment 2 of PBVs (1, 10, 14–20). The PBV RdRp catalyzes RNA synthesis with both single-stranded RNA and dsRNA templates, and transcription occurs in a semi-conservation manner (27). During encapsidation, the RdRp is believed to form a complex with the viral genome (27).
Picobirnaviruses exhibit high genetic diversity within and between host species (1–5, 10–18, 24, 28–30). A viral metagenomics study in diarrheic free-ranging wolves has provided evidence for genetic reassortment events among PBVs (31). Most studies on genetic diversity of PBVs are based on gene segment-2/RdRp sequences (2–5). The phylogenetic analysis of PBV RdRp sequences has been shown in Figure 2. Majority of the PBV RdRp sequences reported so far use the standard genetic code for translation, whilst, recently, some novel PBV-like RdRp gene sequences that use an alternative mitochondrial genetic code have been detected in bats, humans, invertebrates, and a mongoose (6, 11, 12, 14). Picobirnaviruses using the standard genetic code cluster separately from PBVs using the alternative mitochondrial genetic code (12, 14). However, three PBV-like RdRp sequences (from a bat, a mongoose and a myriapod) that use an alternative mitochondrial genetic code were found to cluster with PBVs using the standard genetic code (12, 14). These unique PBV-like RdRp sequences have been discussed in the section “evidence that picobirnaviruses might infect fungi” of the review article. Within the cluster of PBV RdRp sequences using the standard genetic code, PBVs are further classified into two genogroups (genogroup-I (GI) and GII), although several PBVs that could not be classified into either genogroup have also been reported (1–5, 10–20, 24, 29, 30). Picobirnavirus GI strains have been more frequently detected compared to GII strains (2, 3, 5).
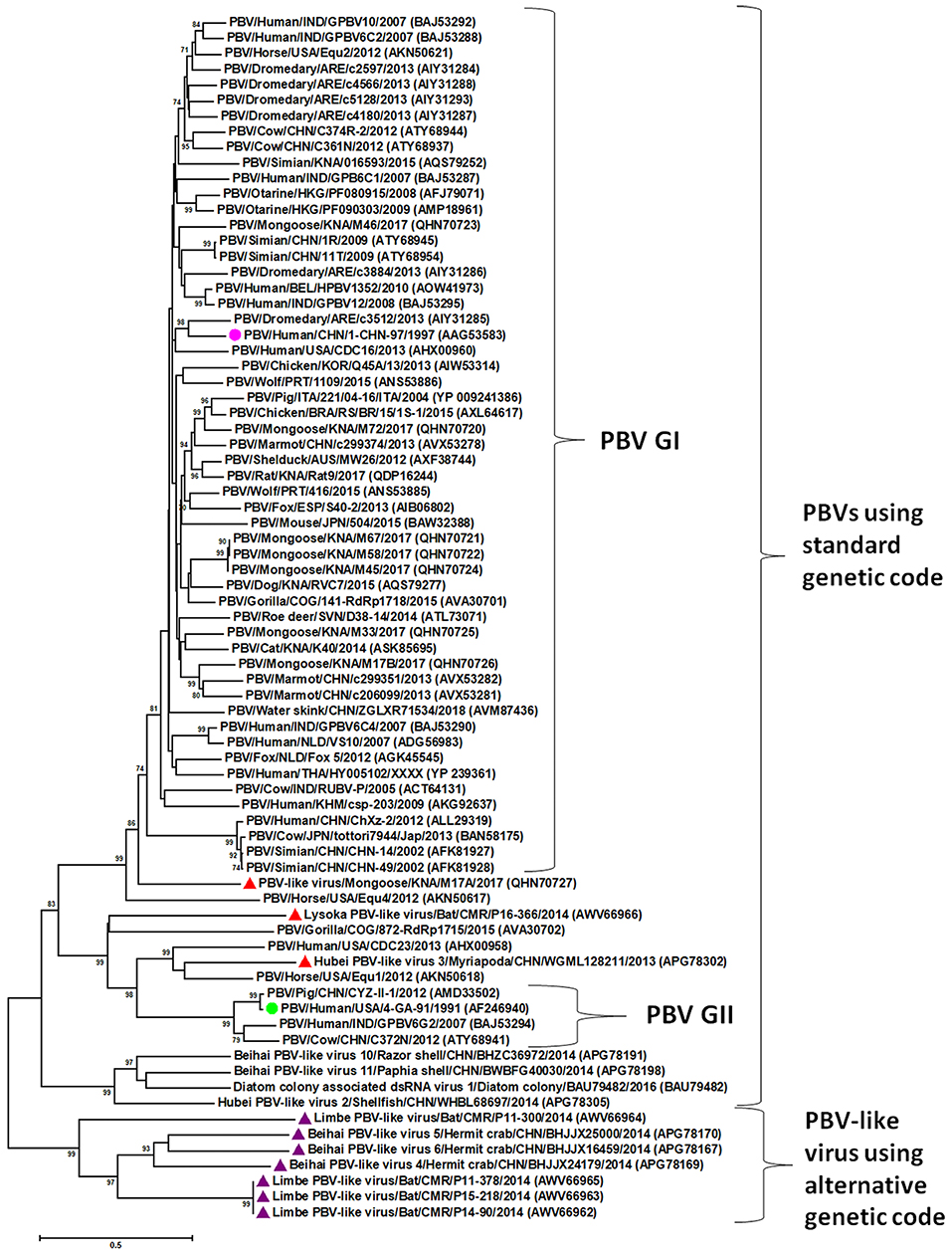
Figure 2. Phylogenetic analysis of the picobirnavirus (PBV) and PBV-like RNA-dependent RNA polymerase (RdRp) sequences. The phylogenetic tree was constructed by the Maximum Likelihood method using the MEGA6 software. Phylogenetic distances were measured using the LG + G model of substitution. The tree was statistically supported by bootstrapping with 500 replicates. Bootstrap values <70% are not shown. Scale bar, 0.5 substitutions per amino acid. The name of the PBV strain includes virus/host of detection/country/common name/date of collection. GenBank accession numbers are shown in parentheses. Pink circle: prototype PBV genogroup-I (GI) strain; green circle: prototype PBV GII strain; purple triangles: PBV-like viruses that use an alternative mitochondrial genetic code to translate the RdRp and cluster separately from PBVs using the standard genetic code; red triangles: PBV-like strains that use an alternative mitochondrial genetic code to translate the RdRp, yet cluster within PBVs using standard genetic code.
Picobirnavirus Infection in Humans and Animals
Picobirnavirus infection in humans and animals have been excellently reviewed by Ganesh et al. (2) and Malik et al. (3). Picobirnaviruses have been detected in sporadic cases of diarrhea as well as associated with outbreaks of gastroenteritis in humans and in a wide variety of animals (1–3, 5). They are often reported in coinfection with other enteric pathogens (1–5, 32–34). On the other hand, PBVs have also been frequently detected in apparently healthy humans and animals without diarrhea (1–3, 5, 14, 15, 18).
Based on studies in immunocompromised and immunosuppressed humans, PBVs were considered as opportunistic enteric pathogens (2, 3). PBVs have been reported more frequently in HIV-infected patients with diarrhea than those without diarrhea (35–38), and in fecal samples from kidney transplant patients (39, 40). In organ transplant recipients, PBVs were predictive of the occurrence of severe enteric graft-vs.-host disease (GVHD), and correlated with the high levels of GVHD severity markers in feces (41). Recently, it was demonstrated that pregnant women with type 1 diabetes (T1D) are more likely to harbor PBVs than those without T1D (42).
Prolonged fecal shedding of PBVs, characterized by alternating periods of high-, low-, and no- virus detection, have been observed in asymptomatic animals, and in healthy and HIV-infected humans (35, 38, 43–50). Various factors, such as age, stress, physiological status and environmental conditions have been proposed to influence the PBV shedding patterns in infected humans and animals. In studies monitoring PBV shedding patterns in animals, highest excretion rates were observed during the lactogenic period in pigs and sheep, whilst increased viral shedding occurred at a young age in rabbits (weaned), broilers (aged 2–7 weeks) and rhea (~3 weeks of age) (44, 46, 49, 51, 52). Based on these observations, it has been suggested that animals could acquire PBV infection early in life, followed by establishment of persistent infection, with infected adults serving as asymptomatic carriers (2, 3, 5).
Although mostly reported in feces/gut contents, PBVs have also been detected, albeit rarely, in the respiratory tract of humans (from individuals with unexplained respiratory disease in Netherlands and cases of severe acute respiratory infection in Uganda) and animals (asymptomatic cattle, monkeys and pigs) and in the plasma of a febrile horse, suggesting an expanded tissue tropism of the virus (10, 24, 53–55).
PBVs exhibit high genetic diversity between and within host species, and phylogenetically, species specific clustering patterns have not been observed so far (1, 4, 10–18, 24, 28–31). Nevertheless, based on sequence identities and phylogenetic analysis, interspecies transmission events including zoonoses have been proposed for PBVs, although several of these studies were based on partial gene segment-2 sequences and are not conclusive (2–5, 11, 40, 43, 53, 56–60).
Evidence That Picobirnaviruses Might Infect Prokaryotes
One of the intriguing recent findings on PBVs has been the identification of a classically defined prokaryotic motif, the ribosomal binding site (RBS) sequence (also known as Shine-Dalgarno sequence), upstream of putative ORF/s in PBV gene segment-1 and−2 sequences (Table 1) (13, 21). In prokaryotes, the RBS sequence (AGGAGG), or its subset (4-, 5-, or 6- mer of AGGAGG) enables the mRNA to bind to ribosome, resulting in initiation of translation, and is mostly located anywhere between −18 and −4 nucleotides upstream from the start codon (21, 61–63). Certain viruses that infect prokaryotes have been shown to be highly enriched for RBS sequences, such as the bacteriophages with segmented dsRNA genomes of family Cystoviridae (13, 21, 64, 65). Similar to prokaryotic mRNAs and the cystoviruses, the RBS sequence, or its subset has been found to be conserved upstream of putative ORF/s (putative ORF1, 2 and 3 in gene segment-1 and putative ORF for RdRp in gene segment-2) in published PBV sequences from humans, animals and environmental samples (4, 7, 8, 11–18, 21). The presence of the conserved prokaryotic RBS sequence upstream of the putative start codon/s in representative PBV gene segment-1 sequences, gene segment-2 sequences of PBV GI and GII strains, and PBV-like RdRp sequences are shown in Table 1. In fact, PBVs exhibited an enrichment level for RBS sequences that was higher than those observed in any known prokaryotic viral family (21). Based on these observations, it has been proposed that PBVs might be actually prokaryotic viruses (13, 21). Since not all prokaryotic viruses appear to retain the prokaryotic RBS sequence, and not every bacterial phylum exhibits a high level of enrichment for RBS sequences, it has been hypothesized that viruses enriched for RBS sequences, such as PBVs, were more likely to infect bacteria that highly conserves the RBS sequences for its own genes (21).
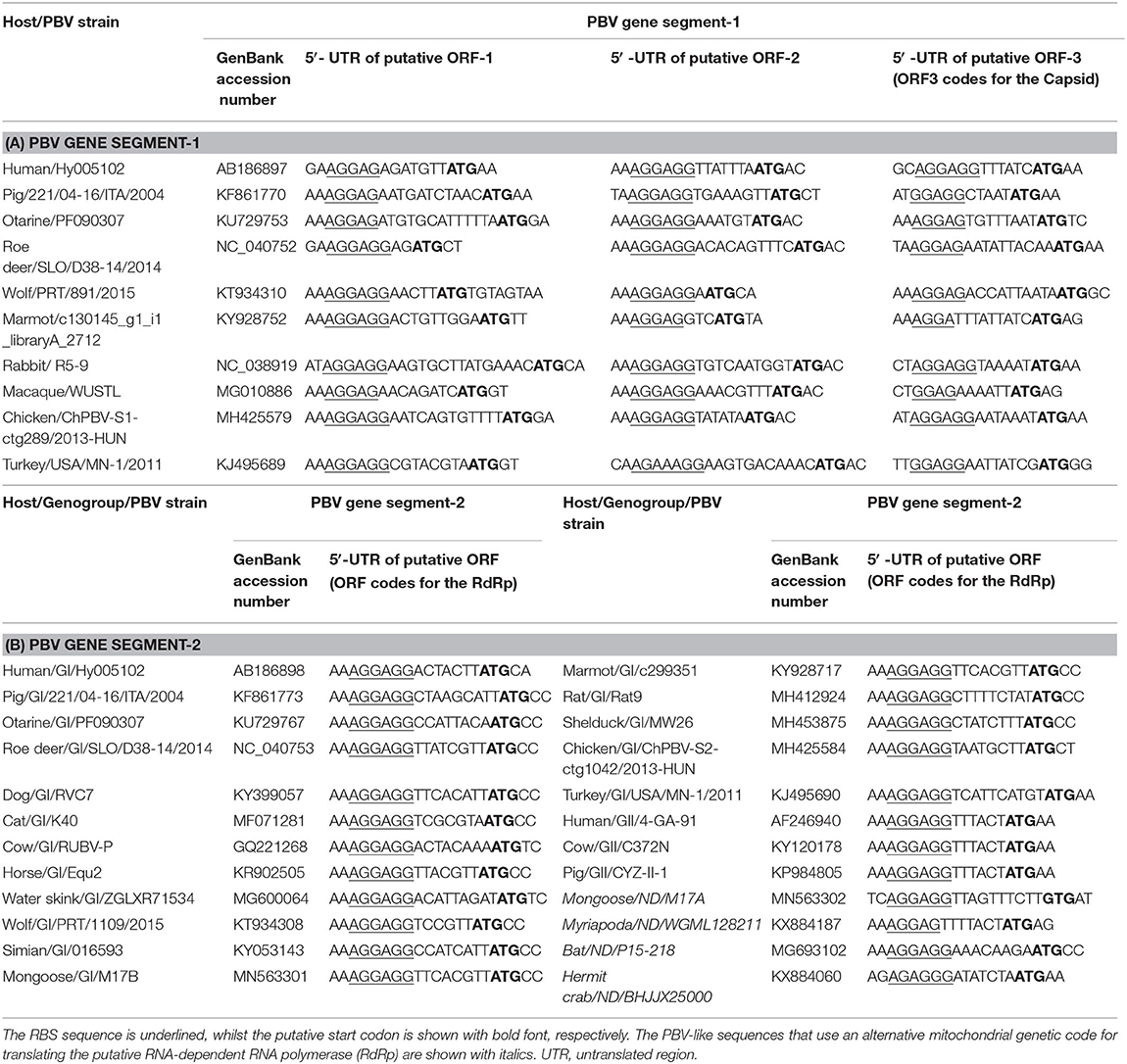
Table 1. The location of the prokaryotic ribosomal binding site (RBS) sequence upstream of the putative open reading frame/s (ORF) in picobirnavirus (PBV) and PBV-like sequences.
Supporting the hypothesis on prokaryotic hosts, PBVs remain to be successfully propagated in eukaryotic cell cultures (1). However, previous experiences with adapting enteric viruses, especially noroviruses to cell culture have been extremely challenging (66). Furthermore, the lack of a cell culture platform in itself does not rule out the possibility that PBVs are animal viruses. Recently, attempts were made to cultivate PBVs in prokaryotic cells by inoculating 3% cloacal suspension from a PBV positive chicken into brain heart infusion broth (13). The in vitro cultures were maintained under aerobic and anaerobic conditions for 2 weeks, and regularly monitored for PBV RNA by RT-qPCR assay. There was no evidence for amplification of PBVs during the study period. Since the sample was collected and frozen long-term to analyze viral nucleic acid rather than maintain bacterial diversity or retain viral infectivity, it might have been possible that the number of cultivable, intact bacteria as well as the low initial infective particles were significantly decreased. However, in the same study, expression analysis in Escherichia coli using 6xHis-tagged recombinant PBV segment-1 and western blot assay revealed the in vivo functionality of PBV segment-1 sequences containing the RBS motif in a bacterial system (13). The ubiquitous nature of PBVs, especially high prevalence of the virus in environmental samples including sewage water and detection of viral RNA in feces/gut contents from a wide variety of animal species including atypical hosts such as reptiles and invertebrates, and persistent fecal shedding by asymptomatic animals, suggest that PBVs might be prokaryotic viruses of the gut microbiome (1–3, 6–9, 43–52).
On the other hand, certain observations with PBVs are reminiscent of eukaryotic viruses, such as (i) viremia in a sick horse and respiratory tract infections in cattle, humans, monkeys and pigs, (ii) immune response in a rabbit that was temporally associated with PBV excretion, and (iii) autoproteolytic processing of the PBV capsid protein and liposome-perforating capacity of viral particles (10, 22, 24, 53–55, 67). However, similar findings have also been reported for bacteriophages:(i) bacteriophages have been detected in blood (phagemia) and respiratory samples, (ii) immune responses have been raised against prokaryotic viruses, and (iii) autoproteolytic capacities have been demonstrated for bacteriophages (68–72).
Evidence That Picobirnaviruses Might Infect Fungi
The recent detection of unique PBV-like sequences that lack a putative ORF for RdRp using the standard genetic code, but use an alternative mitochondrial genetic code for translation has further complicated the ongoing debate on true hosts of PBVs (6, 11, 12, 14). These PBV RdRp-like sequences have been detected in bats, humans, invertebrates (crustaceans and myriapods), and a mongoose (6, 11, 12, 14). The human PBV-like RdRp sequences were closely related (99% sequence identities) to that of a bat PBV-like RdRp sequence detected in the same region (11). The PBV-like RdRp sequences were found to translate the putative RdRp using the invertebrate mitochondrial genetic code (transl_table=5, NCBI genetic codes, www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi#SG5) as well as the mold mitochondrial genetic code (transl_table=4, NCBI genetic codes, www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi#SG4) (6, 11, 12, 14). Based on phylogenetic analysis of the PBV- and PBV-like RdRp sequences, except for three PBV-like strains (Lysoka PBV-like virus/Bat/CMR/P16-366/2014, Hubei PBV-like virus 3/Myriapoda/CHN/WGML128211/2013, and PBV-like virus/Mongoose/KNA/M17A/2017), the PBV-like RdRp sequences formed a separate cluster that was distinct from the PBVs using standard genetic code including PBV GI and GII strains (Figure 2) (6, 11, 12, 14). The metagenomics data pool reporting these PBV-like RdRp sequences did not reveal any PBV-like capsid sequences (6, 12). Based on these observations, it has been proposed that the PBV-like strains using the alternative mitochondrial genetic code might have a lifestyle that is reminiscent of mitoviruses (12).
Mitoviruses (genus Mitovirus, family Narnaviridae) are plus-stranded RNA virus-like elements that replicate in the fungal mitochondria, although they have also been reported in plants and invertebrates (thought to be derived from fungal symbionts) (73–75). Mitoviruses lack a capsid, and the viral genome consists of a single long ORF that encodes a deduced protein with the conserved motifs of a viral RdRp. Principal component analysis of the codon usage bias revealed close clustering of the PBV-like RdRp sequences from bats and invertebrates with those of mitoviruses using the mitochondrial genetic code, corroborating the hypothesis that PBVs might behave like mitoviruses (12). By phylogenetic analysis of RdRp sequences from different virus families, PBVs and partitiviruses (known to infect fungi and plants, and share similarities in capsid architecture and genome organization with PBVs) were found to constitute the partitivirus-picobirnavirus clade, which, interestingly, also consisted of some naked RNA replicons that reproduce in algae mitochondria or chloroplasts, translate using a mitochondrial genetic code, and exhibit mitovirus-like behavior (76). Taken together, these findings suggested that PBVs might be fungal viruses.
On the other hand, at least three PBV-like RdRp sequences (P16-366, WGML128211, and M17A from a bat, myriapod and mongoose, respectively) that use the alternative mitochondrial genetic code (that of mold or invertebrate) for translation were found to cluster within PBVs using the standard genetic code (Figure 2) (6, 12, 14). Furthermore, a capsid sequence was identified for the bat PBV-like strain P16-366 (12). Based on phylogenetic analysis of RdRp sequences, it has been hypothesized that the dsRNA viruses of the partitivirus-picobirnavirus clade might have evolved through reassortment events involving gene segments encoding, respectively, a dsRNA virus capsid protein related to those of the major clade of dsRNA viruses (cystoviruses, totiviruses, and reoviruses) and a positive sense RNA virus RdRp (possibly from a naked RNA replicon within the partitivirus-picobirnavirus clade) (76), which might offer a possible explanation for the origin of P16-366. Although the presence or absence of a capsid sequence could not be determined for the mongoose PBV-like strain M17A, the RdRp sequence of M17A retained the various features (5′- terminal nucleotide sequence and the three motifs (DFXKFD, SGSGGT and GDD) in putative RdRp) that are conserved in gene segment-2 of PBVs, and phylogenetically, clustered near PBV GI strains (14). Further analyses are required to decipher the complex evolution of these PBV-like strains.
Limitations and Future Scope of Research
Studies so far could not establish a consistent association between PBV detection and diarrhea in humans and animals (1, 5). Moreover, there are a few reports on detection of PBVs in respiratory samples and in a serum sample from mammals (10, 24, 53–55). To date, PBVs remain to be successfully propagated in mammalian cell culture systems and/or gnotobiotic animal models (1). As a result, the tissue tropism and pathogenesis of PBVs in mammals remain to be elucidated so far, necessitating further research toward establishing mammalian cell culture systems, intestinal organoids, and/or gnotobiotic animal models that would support the propagation of the virus. On the other hand, the recent speculations that PBVs might actually infect bacteria or fungi were based on analyses of PBV and PBV-like sequences (7, 8, 12–14, 21, 76). To date, PBVs have not yet been isolated from cultured bacterial or fungal cells. In order to conclusively establish that PBVs are prokaryotic or mycoviruses, future research should focus on successful propagation of PBVs in various prokaryotic and fungal cell culture systems, especially those derived from the gut microbiome of mammals.
Conclusions
Since PBVs have been mostly detected in feces/gut contents of animals and humans with or without diarrhea, they were considered as opportunistic enteric pathogens of mammals (2, 3). However, the identification of the prokaryotic RBS sequence upstream of putative ORF/s in PBV and PBV-like sequences indicate that PBVs might actually infect bacteria (7, 8, 13, 21). Furthermore, by phylogenetic analysis of viral RdRp sequences, the partitivirus-picobirnavirus clade has been hypothesized to have originated in an as-yet-undiscovered lineage of prokaryotic RNA viruses (76). On the other hand, detection of some novel PBV-like RdRp sequences that use the alternative mitochondrial genetic code (that of mold or invertebrates) for translation has raised the speculation that PBVs might be fungal viruses with a mitovirus-like lifestyle (6, 12, 14). Based on these observations, it might be possible that PBVs actually infect the gut microbiome of mammals and not mammalian cells. Taken together, these contradicting findings warrant further studies to ascertain the true host/s of PBVs, which still remains controversial. Until the true host/s of PBVs are proven, caution should be exercised during interpretation of PBV-related data in animals and humans, especially those on interspecies transmission events.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by the One Health Center for Zoonoses and Tropical Veterinary Medicine, Ross University School of Veterinary Medicine, St. Kitts and Nevis.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Delmas B, Attoui H, Ghosh S, Malik YS, Mundt E, Vakharia VN. Ictv virus taxonomy profile: picobirnaviridae. J Gen Virol. (2019) 100:133–4. doi: 10.1099/jgv.0.001186
2. Ganesh B, Masachessi G, Mladenova Z. Animal picobirnavirus. VirusDisease. (2014) 25:223–38. doi: 10.1007/s13337-014-0207-y
3. Malik YS, Kumar N, Sharma K, Dhama K, Shabbir MZ, Ganesh B, et al. Epidemiology, phylogeny, and evolution of emerging enteric picobirnaviruses of animal origin and their relationship to human strains. Biomed Res Int. (2014) 2014:780752. doi: 10.1155/2014/780752
4. Joycelyn SJ, Ng A, Kleymann A, Malik YS, Kobayashi N, Ghosh S. High detection rates and genetic diversity of picobirnaviruses (PBVs) in pigs on St. Kitts Island: identification of a porcine PBV strain closely related to simian and human PBVs. Infect Genet Evol. (2020) 84:102383. doi: 10.1016/j.meegid.2020.104383
5. Kumar N, Mascarenhas JDAP, Ghosh S, Masachessi G, da Silva Bandeira R, Nates S V, et al. Picobirnavirus. In: Malik YS, Singh RK, Dhama K, editors. Animal-Origin Viral Zoonoses. Singapore: Springer (2020). p. 291–312.
6. Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, et al. Redefining the invertebrate RNA virosphere. Nature. (2016) 540:539–43. doi: 10.1038/nature20167
7. Guajardo-Leiva S, Chnaiderman J, Gaggero A, Díez B. Metagenomic insights into the sewage RNA virosphere of a large city. Viruses. (2020) 12:1050. doi: 10.3390/v12091050
8. Adriaenssens EM, Farkas K, Harrison C, Jones DL, Allison HE, McCarthy AJ. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. mSystems. (2018) 3:e00025-18. doi: 10.1128/msystems.00025-18
9. Bell N, Khamrin P, Kumthip K, Rojjanadumrongkul K, Nantachit N, Maneekarn N, et al. Molecular detection and characterization of picobirnavirus in environmental water in Thailand. Clin Lab. (2020) 66:855–8. doi: 10.7754/CLIN.LAB.2019.191013
10. Woo PCY, Teng JLL, Bai R, Tang Y, Wong AYP, Li KSM, et al. Novel picobirnaviruses in respiratory and alimentary tracts of cattle and monkeys with large intra-and inter-host diversity. Viruses. (2019) 11:574. doi: 10.3390/v11060574
11. Yinda CK, Vanhulle E, Conceição-Neto N, Beller L, Deboutte W, Shi C, et al. Gut virome analysis of cameroonians reveals high diversity of enteric viruses, including potential interspecies transmitted viruses. mSphere. (2019) 4:e00585-18. doi: 10.1128/msphere.00585-18
12. Yinda CK, Ghogomu SM, Conceição-Neto N, Beller L, Deboutte W, Vanhulle E, et al. Cameroonian fruit bats harbor divergent viruses, including rotavirus H, bastroviruses, and picobirnaviruses using an alternative genetic code. Virus Evol. (2018) 4: vey008. doi: 10.1093/ve/vey008
13. Boros Á, Polgár B, Pankovics P, Fenyvesi H, Engelmann P, Phan TG, et al. Multiple divergent picobirnaviruses with functional prokaryotic Shine-Dalgarno ribosome binding sites present in cloacal sample of a diarrheic chicken. Virology. (2018) 525:62–72. doi: 10.1016/j.virol.2018.09.008
14. Kleymann A, Becker AAMJ, Malik YS, Kobayashi N, Ghosh S. Detection and molecular characterization of picobirnaviruses (PBVs) in the mongoose: identification of a novel PBV using an alternative genetic code. Viruses. (2020) 12:99. doi: 10.3390/v12010099
15. Ghosh S, Shiokawa K, Aung MS, Malik YS, Kobayashi N. High detection rates of picobirnaviruses in free roaming rats (Rattus spp.): molecular characterization of complete gene segment-2. Infect Genet Evol. (2018) 65:131–5. doi: 10.1016/j.meegid.2018.07.024
16. Navarro R, Yibin C, Nair R, Peda A, Aung MS, Ketzis J, et al. Molecular characterization of complete genomic segment-2 of picobirnavirus strains detected in a cat and a dog. Infect Genet Evol. (2017) 54:200–4. doi: 10.1016/j.meegid.2017.07.006
17. Ghosh S, Kobayashi N, Nagashima S, Naik TN. Molecular characterization of full-length genomic segment 2 of a bovine picobirnavirus (PBV) strain: evidence for high genetic diversity with genogroup I PBVs. J Gen Virol. (2009) 90:2519–24. doi: 10.1099/vir.0.013987-0
18. Gallagher CA, Navarro R, Cruz K, Aung MS, Ng A, Bajak E, et al. Detection of picobirnaviruses in vervet monkeys (Chlorocebus sabaeus): molecular characterization of complete genomic segment-2. Virus Res. (2017) 230:13–8. doi: 10.1016/j.virusres.2016.12.021
19. Wakuda M, Pongsuwanna Y, Taniguchi K. Complete nucleotide sequences of two RNA segments of human picobirnavirus. J Virol Methods. (2005) 126:165–9. doi: 10.1016/j.jviromet.2005.02.010
20. Rosen BI, Fang ZY, Glass RI, Monroe SS. Cloning of human picobirnavirus genomic segments and development of an RT-PCR detection assay. Virology. (2000) 277:316–29. doi: 10.1006/viro.2000.0594
21. Krishnamurthy SR, Wang D. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology. (2018) 516:108–14. doi: 10.1016/j.virol.2018.01.006
22. Duquerroy S, Da Costa B, Henry C, Vigouroux A, Libersou S, Lepault J, et al. The picobirnavirus crystal structure provides functional insights into virion assembly and cell entry. EMBO J. (2009) 28:1655–65. doi: 10.1038/emboj.2009.109
23. Malik YS, Ghosh S. Etymologia: picobirnavirus. Emerg Infect Dis. (2020) 26:89. doi: 10.3201/eid2601.et2601
24. Li L, Giannitti F, Low J, Keyes C, Ullmann LS, Deng X, et al. Exploring the virome of diseased horses. J Gen Virol. (2015) 96:2721–33. doi: 10.1099/vir.0.000199
25. Shi M, Lin XD, Chen X, Tian JH, Chen LJ, Li K, et al. The evolutionary history of vertebrate RNA viruses. Nature. (2018) 556:197–202. doi: 10.1038/s41586-018-0012-7
26. Da Costa B, Duquerroy S, Tarus B, Delmas B. Picobirnaviruses encode a protein with repeats of the ExxRxNxxxE motif. Virus Res. (2011) 158:251–6. doi: 10.1016/j.virusres.2011.02.018
27. Collier AM, Lyytinen OL, Guo YR, Toh Y, Poranen MM, Tao YJ. Initiation of RNA polymerization and polymerase encapsidation by a small dsRNA virus. PLoS Pathog. (2016) 12:e1005523. doi: 10.1371/journal.ppat.1005523
28. Knox MA, Gedye KR, Hayman DTS. The challenges of analysing highly diverse picobirnavirus sequence data. Viruses. (2018) 10:685. doi: 10.3390/v10120685
29. Woo PCY, Lau SKP, Teng JLL, Tsang AKL, Joseph M, Wong EYM, et al. Metagenomic analysis of viromes of dromedary camel fecal samples reveals large number and high diversity of circoviruses and picobirnaviruses. Virology. (2014) 471–473:117–25. doi: 10.1016/j.virol.2014.09.020
30. Duraisamy R, Akiana J, Davoust B, Mediannikov O, Michelle C, Robert C, et al. Detection of novel RNA viruses from free-living gorillas, Republic of the Congo: genetic diversity of picobirnaviruses. Virus Genes. (2018) 54:256–71. doi: 10.1007/s11262-018-1543-6
31. Conceição-Neto N, Mesquita JR, Zeller M, Yinda CK, Álvares F, Roque S, et al. Reassortment among picobirnaviruses found in wolves. Arch Virol. (2016) 161:2859–62. doi: 10.1007/s00705-016-2987-4
32. Karte C, Platje N, Bullermann J, Beer M, Höper D, Blome S. Re-emergence of porcine epidemic diarrhea virus in a piglet-producing farm in northwestern Germany in 2019. BMC Vet Res. (2020) 16:329. doi: 10.1186/s12917-020-02548-4
33. Kylla H, Dutta TK, Roychoudhury P, Subudhi PK. Coinfection of diarrheagenic bacterial and viral pathogens in piglets of Northeast region of India. Vet World. (2019) 12:224–30. doi: 10.14202/vetworld.2019.224-230
34. Mahar JE, Shi M, Hall RN, Strive T, Holmes EC. Comparative analysis of RNA virome composition in rabbits and associated ectoparasites. J Virol. (2020) 94:e02119-19. doi: 10.1128/jvi.02119-19
35. Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, et al. Enteric viruses and diarrhea in HIV-infected patients. Enteric Opportunistic Infections Working Group. N Engl J Med. (1993) 329:14–20.
36. Giordano MO, Martinez LC, Rinaldi D, Gúinard S, Naretto E, Casero R, et al. Detection of picobirnavirus in HIV-infected patients with diarrhea in Argentina. J Acquir Immune Defic Syndr Hum Retrovirol. (1998) 18:380–3. doi: 10.1097/00042560-199808010-00010
37. González GG, Pujol FH, Liprandi F, Deibis L, Ludert JE. Prevalence of enteric viruses in human immunodeficiency virus seropositive patients in Venezuela. J Med Virol. (1998) 55:288–92. doi: 10.1002/(SICI)1096-9071(199808)55:4<288::AID-JMV6>3.0.CO;2-X
38. Giordano MO, Martinez LC, Rinaldi D, Espul C, Martinez N, Isa MB, et al. Diarrhea and enteric emerging viruses in HIV-infected patients. AIDS Res Hum Retroviruses. (1999) 15:1427–32. doi: 10.1089/088922299309937
39. Valle MC, Martínez LC, Ferreyra LJ, Giordano MO, Isa MB, Paván JV, et al. Agentes virales asociados al sindrome diarreico en pacientes trasplantados renales [Viral agents related to diarrheic syndrome in kidney transplanted patients]. Medicina (B Aires). (2001) 61:179–82.
40. Giordano MO, Martinez LC, Masachessi G, Barril PA, Ferreyra LJ, Isa MB, et al. Evidence of closely related picobirnavirus strains circulating in humans and pigs in Argentina. J Infect. (2011) 62:45–51. doi: 10.1016/j.jinf.2010.09.031
41. Legoff J, Resche-Rigon M, Bouquet J, Robin M, Naccache SN, Mercier-Delarue S, et al. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat Med. (2017) 23:1080–5. doi: 10.1038/nm.4380
42. Kim KW, Allen DW, Briese T, Couper JJ, Barry SC, Colman PG, et al. Distinct gut virome profile of pregnant women with type 1 diabetes in the ENDIA study. Open Forum Infect Dis. (2019) 6: ofz025. doi: 10.1093/ofid/ofz025
43. Ganesh B, Bányai K, Martella V, Jakab F, Masachessi G, Kobayashi N. Picobirnavirus infections: viral persistence and zoonotic potential. Rev Med Virol. (2012) 22:245–56. doi: 10.1002/rmv.1707
44. Martínez LC, Masachessi G, Carruyo G, Ferreyra LJ, Barril PA, Isa MB, et al. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol. (2010) 10:984–8. doi: 10.1016/j.meegid.2010.06.004
45. Masachessi G, Martínez LC, Giordano MO, Barril PA, Isa BM, Ferreyra L, et al. Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol. (2007) 152:989–98. doi: 10.1007/s00705-006-0900-2
46. Masachessi G, Martinez LC, Ganesh B, Giordano MO, Barril PA, Isa MB, et al. Establishment and maintenance of persistent infection by picobirnavirus in greater rhea (Rhea Americana). Arch Virol. (2012) 157:2075–82. doi: 10.1007/s00705-012-1400-1
47. Masachessi G, Ganesh B, Martinez LC, Giordano MO, Barril PA, Isa MB, et al. Maintenance of picobirnavirus (PBV) infection in an adult orangutan (Pongo pygmaeus) and genetic diversity of excreted viral strains during a three-year period. Infect Genet Evol. (2015) 29:196–202. doi: 10.1016/j.meegid.2014.11.019
48. Haga IR, Martins SS, Hosomi ST, Vicentini F, Tanaka H, Gatti MSV. Identification of a bisegmented double-stranded RNA virus (Picobirnavirus) in faeces of giant anteaters (Myrmecophaga tridactyla). Vet J. (1999) 158:234–6. doi: 10.1053/tvjl.1999.0369
49. Kunz AF, Possatti F, de Freitas JA, Alfieri AA, Takiuchi E. High detection rate and genetic diversity of picobirnavirus in a sheep flock in Brazil. Virus Res. (2018) 255:10–3. doi: 10.1016/j.virusres.2018.06.016
50. Kapusinszky B, Minor P, Delwart E. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol. (2012) 50:3427–34. doi: 10.1128/JCM.01589-12
51. Ludert JE, Abdul-Latiff L, Liprandi A, Liprandi F. Identification of picobirnavirus, viruses with bisegmented double stranded rna, in rabbit faeces. Res Vet Sci. (1995) 59:222–5. doi: 10.1016/0034-5288(95)90006-3
52. Tamehiro CY, Alfieri AF, Médici KC, Alfieri AA. Segmented double-stranded genomic RNA viruses in fecal samples from broiler chicken. Brazilian J Microbiol. (2003) 34:349–53. doi: 10.1590/s1517-83822003000400013
53. Smits SL, Poon LLM, van Leeuwen M, Lau PN, Perera HKK, Peiris JSM, et al. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis. (2011) 17:2328–30. doi: 10.3201/eid1712.110934
54. Smits SL, van Leeuwen M, Schapendonk CME, Schürch AC, Bodewes R, Haagmans BL, et al. Picobirnaviruses in the human respiratory tract. Emerg Infect Dis. (2012) 18:1539–40. doi: 10.3201/eid1809.120507
55. Cummings MJ, Tokarz R, Bakamutumaho B, Kayiwa J, Byaruhanga T, Owor N, et al. Precision surveillance for viral respiratory pathogens: virome capture sequencing for the detection and genomic characterization of severe acute respiratory infection in Uganda. Clin Infect Dis. (2019) 68:1118–25. doi: 10.1093/cid/ciy656
56. Li W, Qiang X, Qin S, Huang Y, Hu Y, Bai B, et al. Virome diversity analysis reveals novel enteroviruses and a human picobirnavirus in stool samples from African green monkeys with diarrhea. Infect Genet Evol. (2020) 82: 104279. doi: 10.1016/j.meegid.2020.104279
57. Bányai K, Martella V, Bogdán Á, Forgách P, Jakab F, Meleg E, et al. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J Gen Virol. (2008) 89:534–9. doi: 10.1099/vir.0.83134-0
58. Chen M, Sun H, Lan D, Hua X, Cui L, Yuan C, et al. Molecular detection of genogroup I and II picobirnaviruses in pigs in China. Virus Genes. (2014) 48:553–6. doi: 10.1007/s11262-014-1058-8
59. Wang Y, Bányai K, Tu X, Jiang B. Simian genogroup I picobirnaviruses: prevalence, genetic diversity, and zoonotic potential. J Clin Microbiol. (2012) 50:2779–82. doi: 10.1128/JCM.00634-12
60. Ganesh B, Banyai K, Masachessi G, Mladenova Z, Nagashima S, Ghosh S, et al. Genogroup i picobirnavirus in diarrhoeic foals: can the horse serve as a natural reservoir for human infection? Vet Res. (2011) 42:52. doi: 10.1186/1297-9716-42-52
61. Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. (1975) 254:34–8. doi: 10.1038/254034a0
62. Ma J, Campbell A, Karlin S. Correlations between Shine-Dalgarno sequences and gene features and operon structures. J Bacteriol. (2002) 184:5733–45. doi: 10.1128/jb.184.20.5733-5745.2002
63. Steitz JA. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. (1969) 224:957–64. doi: 10.1038/224957a0
64. Osterman IA, Evfratov SA, Sergiev PV, Dontsova OA. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. (2013) 41:474–86. doi: 10.1093/nar/gks989
65. Mindich L. Bacteriophage φ6: a unique virus having a lipid-containing membrane and a genome composed of three dsRNA segments. Adv Virus Res. (1988) 35:137–76. doi: 10.1016/S0065-3527(08)60710-1
66. Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin SC, Ayyar BV, et al. Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges. Viruses. (2019) 11:638. doi: 10.3390/v11070638
67. Gallimore C, Lewis D, Brown D. Detection and characterization of a novel bisegmented double-stranded RNA virus (picobirnavirus) from rabbit faeces. Arch Virol. (1993) 133:63–73. doi: 10.1007/BF01309744
68. Blanco-Picazo P, Fernández-Orth D, Brown-Jaque M, Miró E, Espinal P, Rodríguez-Rubio L, et al. Unravelling the consequences of the bacteriophages in human samples. Sci Rep. (2020) 10:6737. doi: 10.1038/s41598-020-63432-7
69. Dabrowska K, Switała-Jelen K, Opolski A, Weber-Dabrowska B, Gorski A. A review: bacteriophage penetration in vertebrates. J Appl Microbiol. (2005) 98:7–13. doi: 10.1111/j.1365-2672.2004.02422.x
70. Lee CK, Bent SJ. Uncovering the hidden villain within the human respiratory microbiome. Diagnosis. (2014) 1:203–12. doi: 10.1515/dx-2014-0039
71. Górski A, Kniotek M, Perkowska-Ptasińska A, Mróz A, Przerwa A, Gorczyca W, et al. Bacteriophages and transplantation tolerance. Transplant Proc. (2006) 38:331–3. doi: 10.1016/j.transproceed.2005.12.073
72. Wang S, Chang JR, Dokland T. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. (2006) 348:133–40. doi: 10.1016/j.virol.2005.12.021
73. Nibert ML, Debat HJ, Manny AR, Grigoriev IV, De Fine Licht HH. Mitovirus and mitochondrial coding sequences from basal fungus entomophthora muscae. Viruses. (2019) 11:351. doi: 10.3390/v11040351
74. Shackelton LA, Holmes EC. The role of alternative genetic codes in viral evolution and emergence. J Theor Biol. (2008) 254:128–34. doi: 10.1016/j.jtbi.2008.05.024
75. Shahi S, Eusebio-Cope A, Kondo H, Hillman BI, Suzuki N. Investigation of host range of and host defense against a mitochondrially replicating mitovirus. J Virol. (2019) 93:e01503-18. doi: 10.1128/jvi.01503-18
Keywords: picobirnavirus, true host/s, opportunistic animal pathogen, prokaryotic virus, fungal virus
Citation: Ghosh S and Malik YS (2021) The True Host/s of Picobirnaviruses. Front. Vet. Sci. 7:615293. doi: 10.3389/fvets.2020.615293
Received: 08 October 2020; Accepted: 24 December 2020;
Published: 20 January 2021.
Edited by:
Subhash Verma, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaReviewed by:
Soma Chattopadhyay, Institute of Life Sciences (ILS), IndiaSanthamani Ramasamy, Public Health Research Institute (PHRI), United States
Copyright © 2021 Ghosh and Malik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Souvik Ghosh, souvikrota@gmail.com
 Souvik Ghosh
Souvik Ghosh Yashpal S. Malik
Yashpal S. Malik