Transcriptome Differences in Pig Tracheal Epithelial Cells in Response to Pasteurella Multocida Infection
- 1State Key Laboratory of Agricultural Microbiology, College of Animal Science and Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2Key Laboratory of Preventive Veterinary Medicine in Hubei Province, The Cooperative Innovation Centre for Sustainable Pig Production, Wuhan, China
- 3Key Laboratory of Prevention and Control Agents for Animal Bacteriosis (Ministry of Agriculture), Institute of Animal Husbandry and Veterinary Science, Hubei Academy of Agricultural Sciences, Wuhan, China
Pasteurella multocida generally colonizes mammalian/bird respiratory tracts and mainly causes respiratory disorders in both humans and animals. To date, the effects of P. multocida infection on the respiratory epithelial barriers and molecules in host respiratory epithelial cells in their response to P. multocida infection are still not well-known. In this study, we used newborn pig tracheal epithelial (NPTr) cells as an in vitro model to investigate the effect of P. multocida infection on host respiratory epithelial barriers. By detecting the transepithelial electrical resistance (TEER) values of NPTr cells and the expression of several known molecules associated with cell adherens and junctions, we found that P. multocida infection disrupted the barrier functions of NPTr cells. By performing RNA sequencing (RNA-Seq), we determined 30 differentially expressed genes (DEGs), including the vascular endothelial growth factor A (VEGFA) encoding gene VEGFA, which participated in biological processes (GO:0034330, GO:0045216, and GO:0098609) closely related to epithelial adhesion and barrier functions. These 30 DEGs participated in 22 significant signaling pathways with a p-value < 0.05, including the transforming growth factor (TGF)-beta signaling pathway (KEGG ID: ssc04350), hypoxia-inducible factor 1 (HIF-1) signaling pathway (KEGG ID: ssc04066), epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance (KEGG ID: ssc01521), tumor necrosis factor (TNF) signaling pathway (KEGG ID: ssc04668), and mitogen-activated protein kinase (MAPK) signaling pathway (KEGG ID: ssc04010), which are reported to have roles in contributing to the production of inflammatory factors as well as the regulation of epithelial adhesion and barrier function in other tissues and organisms. The results presented in this study may help improve our understanding of the pathogenesis of P. multocida.
Introduction
The zoonotic pathogen Pasteurella multocida is the causative agent of multiple diseases in a variety of domestic and wild animals and in humans (1). According to the capsular antigens, P. multocida strains are classified into five serogroups (A, B, D, E, and F) (1–3). In pigs, P. multocida strains, in particular those belonging to serogroups A and D, are commonly associated with the development of respiratory disorders, such as progressive atrophic rhinitis (PAR) and pneumonia (4). P. multocida also plays a crucial role in porcine respiratory disease complex (PRDC) and hemorrhagic septicemia (5). This pathogen is proposed to be a threat of high impact to the pig industry, as evidenced by its prevalence: 8.0% in diseased pigs with pneumonia or PAR in China, 10.3–15.6% in pigs with pneumonia in Korea, and 15.6% of isolated respiratory pathogens in the United States (6). However, current knowledge about the pathogenesis of swine is limited.
In mammals, respiratory tracts such as the trachea and bronchi are areas where P. multocida strains commonly colonize (7). The mammalian respiratory epithelium forms the first line of defense against the invasion and stimuli of respiratory pathogens (8, 9). As an important respiratory pathogen, P. multocida infection might induce the dysfunction of the host epithelial barriers, thereby contributing to the bacterial invasion and the development of many inflammatory disorders of the airways and lungs (9). However, the effects of P. multocida infection on the epithelial barriers and the responses of mammalian respiratory epithelial cells to P. multocida infection are still not well-known. Recently, several technologies such as RNA sequencing (RNA-Seq) have emerged as powerful tools to analyze gene expression and have been widely used in “pathogen–host interaction” studies (10). To elucidate the genes in mammalian tracheal epithelial cells in response to P. multocida infection, we used pig tracheal epithelial cells as a model and performed RNA-Seq to identify the differentially expressed genes (DEGs) in the cells during bacterial infection.
Materials and Methods
Bacterial Strains, Cells, and Growth Conditions
P. multocida HN05 is a serogroup D strain which was isolated from the trachea of a pig with respiratory disorders in Hunan Province, China, in 2010. We have determined its whole genome sequence and deposited it to GenBank with the accession number PPVF00000000 (1). Strain HN05 was cultured in tryptic soy broth (TSB) medium (Becton, Dickinson and Company, Sparks, MD, USA) supplemented with 5% bovine serum at 37°C for 8–12 h. Newborn pig tracheal epithelial (NPTr) cells (established following serial culture of primary cells derived from tracheal tissues and were kindly gifted by Prof. Hongbo Zhou at Huazhong Agricultural University, Wuhan, China) (11) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, ThermoFisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco, ThermoFisher, Waltham, MA, USA) under 5% CO2 atmosphere at 37°C.
Cell Culture and Bacterial Infection
Monolayer cells (2.5 × 106/2 ml per well) in each well of a six-well plate (Corning, Corning, NY, USA) were washed with phosphate-buffered saline (PBS) three times and were incubated with fresh DMEM (2 ml/well). P. multocida HN05 (2.5 × 108/100 μl per well) was inoculated into three wells of the plate, while PBS (100 μl/well) was added into the other three wells. The plate was incubated under 5% CO2 atmosphere at 37°C for 4 h. The medium in each well of the plate was discarded and the cells were washed with PBS, followed by the addition of 1 ml Trizol into each well of the plate. This bacterial infection assay was performed three times at separate time points. In each independent experiment, three wells of cells were treated with either P. multocida or PBS. RNAs extracted from bacterial infected cells and/or PBS-treated cells in these three wells were pooled and were regarded as one sample for further use.
RNA Isolation, Construction and Sequencing of Transcriptome Library
Total RNAs from the bacterial infected cells and the PBS-treated cells were isolated using the Trizol reagent protocol (Invitrogen, ThermoFisher, Waltham, MA, USA). The quantity and quality of the extracted RNAs were checked by using electrophoresis on 1% agarose gel and a NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 System (Agilent Technologies, Santa Clara, CA, USA). After quality evaluation, a total of 1 μg RNA per sample was used as the input material for the RNA sample preparations. Sequencing libraries were generated using a NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, Ipswich, MA, USA) following the manufacturer's recommendations. Library preparations were sequenced on an Illumina Novaseq platform at Novogene Co., Ltd. (Beijing, China) using the paired-end 150-bp sequencing protocol. After sequencing, raw data in fastq format were firstly processed through in-house perl scripts. In this step, clean data were obtained by removing reads containing adapters, reads containing ploy-N, and low-quality reads from raw data. At the same time, the Q20, Q30, and GC contents of the clean data were calculated. All the downstream analyses were based on clean data with high quality. Paired-end clean reads were aligned to the reference genome (GenBank accession no. NC_010443.5) using Hisat2 v2.0.5. The mapped reads of each sample were assembled by StringTie (v1.3.3b) (12) in a reference-based approach. The data generated in this study have been deposited to NCBI's Sequence Read Archive (SRA) database, with accession number SRR14026556.
Differential Expression Analysis
The gene expression of the cells incubated with P. multocida and/or PBS was analyzed using the DESeq2 R package (1.16.1). The resulting p-values were adjusted using the Benjamini and Hochberg approach for controlling the false discovery rate. Genes with an adjusted p-value < 0.05 found by DESeq2 were considered as differentially expressed.
Quantitative Real-Time PCR
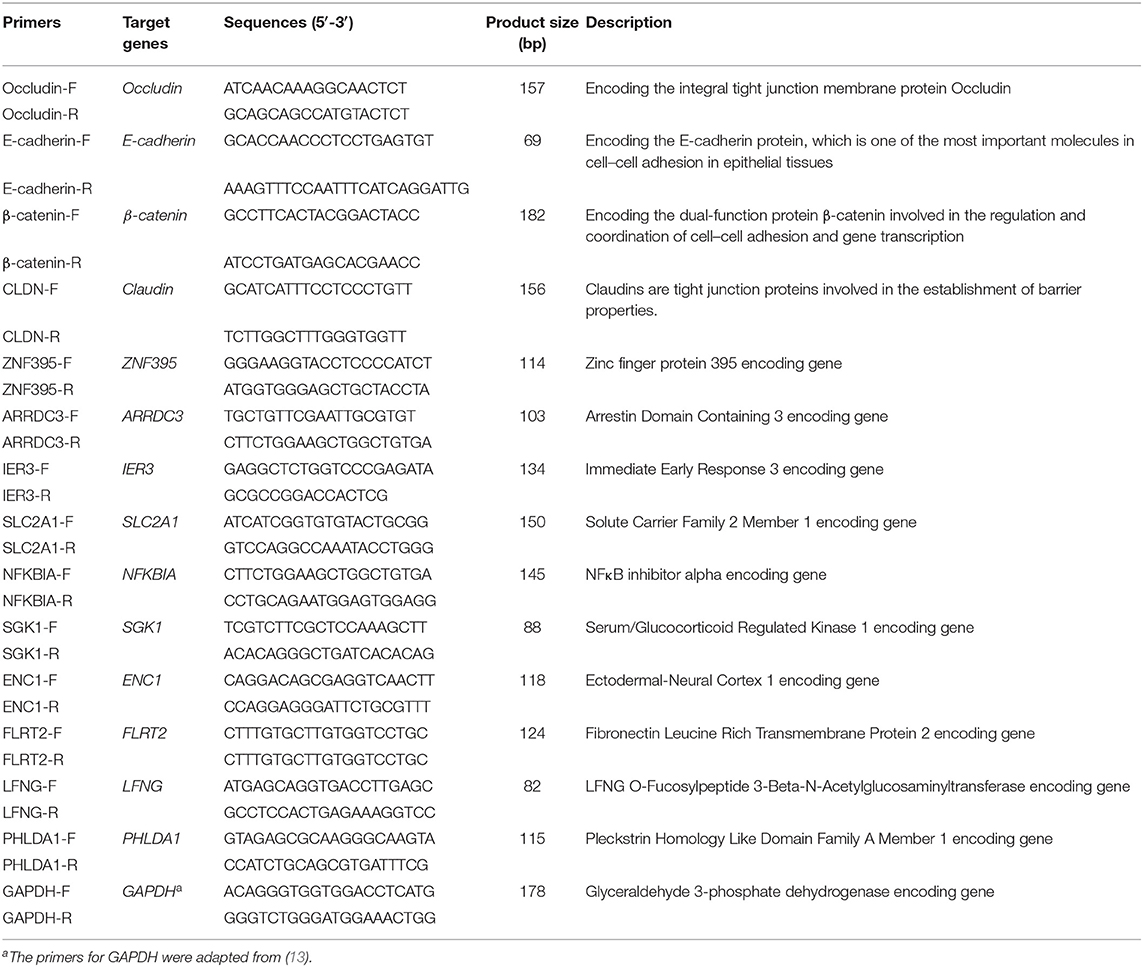
To validate the results of RNA-Seq, NPTr cells were treated as described in section Cell culture and bacterial infection, and total RNAs were extracted from both the bacterial infected cells and the PBS-treated cells. Complementary DNAs (cDNAs) were synthesized using a PrimeScript™ RT Master Mix Kit (TaKaRa, Kusatsu, Japan). The transcription of selected genes in different groups of cells was detected by quantitative real-time PCR (qPCR) using the primers listed in Table 1. The relative transcription levels of the genes are shown as a ratio of the target gene to the reference gene using the formula 2−(ΔΔCt) (14).
Electric Cell–Substrate Impedance Sensing
The putative effect of P. multocida on the barrier function of NPTr cells was determined with the electric cell–substrate impedance sensing (ECIS) technology, as described previously (15). Briefly, approximately 7 × 104 NPTr cells were seeded on collagen-coated, gold-plated electrodes in 96-well chamber slides (96W1E+), linked to the ECIS Zθ equipment (Applied BioPhysics, Troy, NY, USA), and continuously cultured until confluence was reached. The transepithelial electrical resistance (TEER) value was recorded to reflect the barrier function of the cells. After stable maximal TEER was reached, P. multocida HN05 (~108 CFU) was added to the cells and the changes in the TEER values were recorded automatically by the ECIS system.
Immunofluorescence Microscopy
Immunofluorescence microscopy analysis was performed following a previously described method (16). Briefly, the NPTr monolayers in each well (~2 × 105 cells per well) of a 12-well plate (Corning, Corning, NY, USA) were treated with either P. multocida HN05 at 100 MOI (multiplicity of infection) or 100 μl PBS (mock). The plate was incubated at 37°C for 4 h. Next, P. multocida-infected cells and mock cells were washed with PBS three times and fixed with 4% paraformaldehyde (PFA) for 20 min. Afterwards, PFA was removed and the samples washed three times with PBS. The samples were then permeabilized with 0.1% Triton X-100 for 10 min, washed three times with PBS, and were further blocked with 1% bovine serum albumin (BSA) for 30 min. Next, 22 mg/ml glucine was added to the samples, which were then incubated with a primary antibody and a secondary antibody consecutively for 1 h each. After washing with PBS, the nuclei were stained and embedded in Antifade Mounting Medium with DAPI (4′,6-diamidino-2-phenylindole; Beyotime, Shanghai, China). The samples were finally stored at 4°C for further analysis.
The P. multocida-infected samples and control samples were incubated with β-tubulin antibody (1:200; Proteintech, Wuhan, China) as the primary antibody at 4°C overnight, followed by staining with Cy3-labeled antibody against β-tubulin (1:100; Proteintech, Wuhan, China) as the secondary antibody. The samples were analyzed using a Nikon A1 HD25 confocal laser scanning microscope. Images were analyzed using NIS-Elements Viewer 4.20 software (Nikon, Tokyo, Japan), while imaging analysis of the cell thickness was calculated using ImageJ software.
Western Blot
NPTr monolayer cells were treated with either P. multocida or PBS, as described above in section Cell culture and bacterial infection. Challenged cells were lysed in RIPA buffer (Beyotime, Shanghai, China) with a protease inhibitor cocktail (Sigma-Aldrich, Burlington, MA, USA), sonicated, and then centrifuged at 10,000 × g for 10 min at 4°C. The insoluble debris was removed and the protein concentration in the supernatant measured using a BCA protein assay kit (Beyotime, Shanghai, China). The cell lysates were then separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred into polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). The blots were blocked in 5% BSA in Tris-buffered saline with Tween 20 (TBST) for 2 h at room temperature and then incubated overnight with either β-catenin polyclonal antibody, E-cadherin polyclonal antibody, or β-actin antibodies (all from Proteintech, Wuhan, China). After washing, the blots were incubated with species-specific horseradish peroxidase-conjugated antibodies and finally visualized with enhanced chemiluminescence (ECL) reagents (Beyotime, Shanghai, China). All Western blots were densitometrically quantified using ImageJ software, and the results were analyzed as the relative immunoreactivity of each protein normalized to the respective loading control.
Statistical Analysis
All data are displayed as the mean ± standard error of mean (SEM) and were evaluated using unpaired, two-tailed Student's t-test. GraphPad Prism 8.0 software was used for all statistical analyses. Significant differences were considered at p < 0.05.
Results
P. multocida Infection Induces the Disruption of the Barrier Functions of NPTr Cells
To explore the effects of P. multocida infection on the barrier functions of the cells, we used an ECIS system to monitor changes in the TEER values. The results revealed that infection of P. multocida induced a significant decrease in the TEER values of the cells (Figure 1A). Analysis of the microtubule structure showed a lower density of microtubules at the cell distal area in P. multocida-infected cells compared to that of the control cells (Figure 1B). Since adherens and tight junctions are crucial for epithelial adhesion and barrier function in many tissues and organisms (17), we detected the transcription of several genes (Occludin, E-cadherin, β-catenin, and Claudin) associated with cell adherens and junctions by qPCR. The results revealed that P. multocida infection induced decreased expressions of these molecules (Figure 1C). The Western blot assay also showed that the expressions of E-cadherin and β-catenin were downregulated post-P. multocida infection (Figures 1D,E). All of the above findings indicated that P. multocida infection resulted in the dysfunction of the NPTr barrier functions.
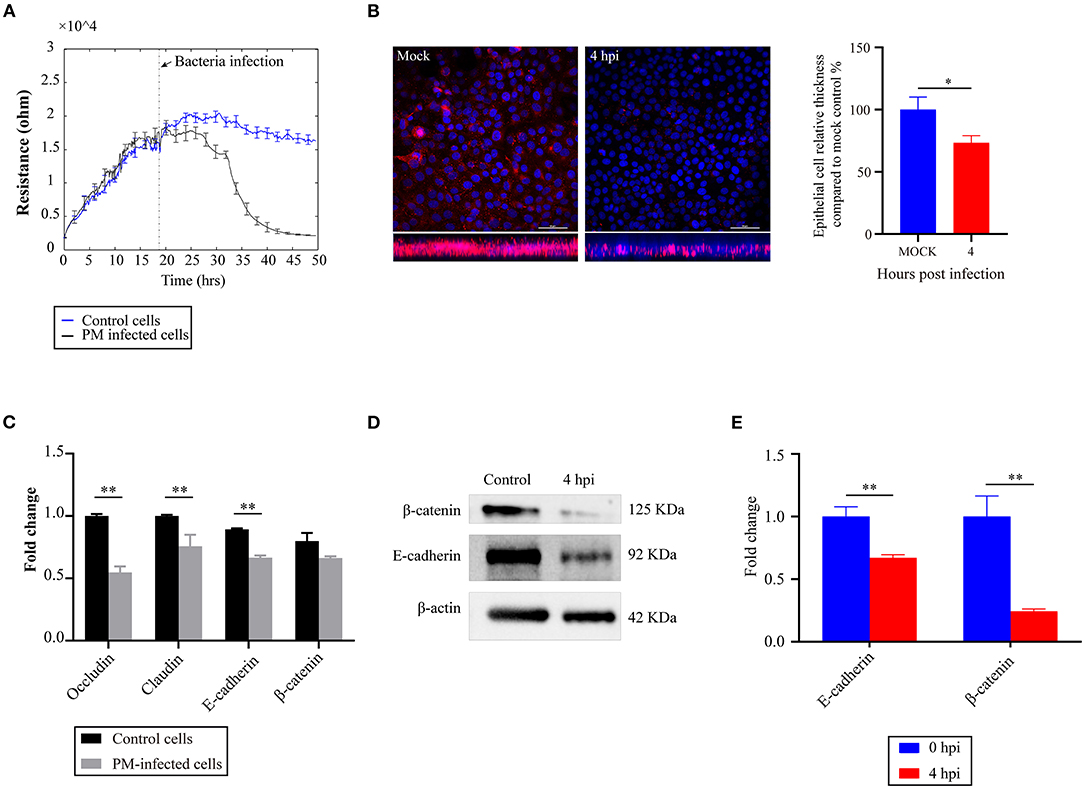
Figure 1. Effects of Pasteurella multocida infection on the barrier functions of newborn pig tracheal epithelial (NPTr) cells. (A) Changes in the transepithelial electrical resistance (TEER) values of the NPTr cells during P. multocida infection as monitored by an electric cell–substrate impedance sensing (ECIS) system. (B) Analysis of the density of microtubules at the cell distal area in P. multocida-infected cells compared to the control cells. Bacteria- or PBS-treated cells were stained with antibodies to β-tubulin (in pink under immunofluorescence microscopy). Left shows the vertical sections of the immunostained samples, while the column chart in the right reveals the relative thickness of the epithelial cell layer compared to mock cells. Scale bars, 50 μm. (C) Transcription of selected molecules (Occludin, E-cadherin, β-catenin, and Claudin) associated with cell adherens and junctions in different groups of cells as determined by qPCR. (D) Expressions of E-cadherin and β-catenin in different groups of cells as determined by Western blot. (E) Densitometrically quantified results of Western blot using ImageJ software. Data are presented as the as mean ± SEM. *p < 0.05; **p < 0.01. PM, Pasteurella multocida; hpi, hours post-infection.
Transcriptome Sequence Data From RNA-Seq
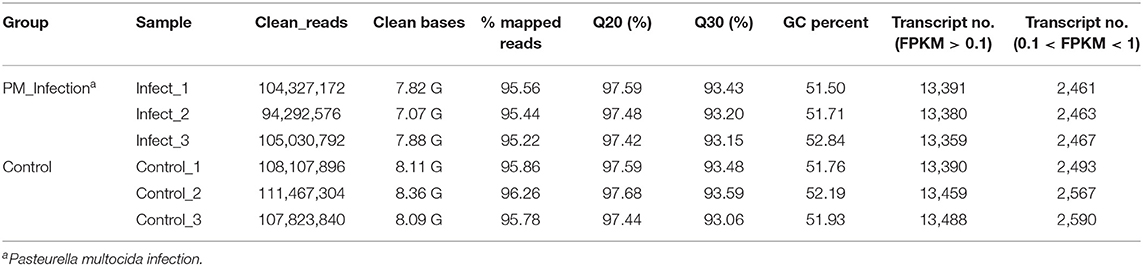
To find the key molecules or pathways in NPTr cells mediating the dysfunction of the cell barrier functions during P. multocida infection, transcriptome sequencing was performed. In total, six cDNA libraries from two groups (PM_infection and Control) were sequenced, which yielded a total of 63.10 million 150-bp paired-end clean reads, varying from approximately 9.43 million to 11.15 million reads for each of the samples (Table 2). More than 97.42% had quality scores at the Q20 level, and more than 93.06% had quality scores at the Q30 level. On average, approximately 95.69% of the clean reads were mapped to the reference genome (GenBank accession no. NC_010443.5) (Table 2).
Calculation of the values of fragments per kilobase of transcript per million mapped reads (FPKM) identified an aggregate of 40,130 expressed genes in P. multocida-infected cells and 40,337 expressed genes in the control cells, with the FPKM threshold of 0.1 (Table 2). The FPKM density in the bacterial infection group and the control group exhibited a similar skewed distribution, and approximately 18.38–19.20% of the expressed genes were lowly expressed (0.1 < FPKM < 1) (Figure 2A). Compared to the control cells, a total of 7,036 DEGs were determined in P. multocida-infected NPTr cells [|log2(FoldChange)| > 1, padj < 0.05], including 3,613 downregulated DEGs and 3,423 upregulated ones (Figures 2B,C).
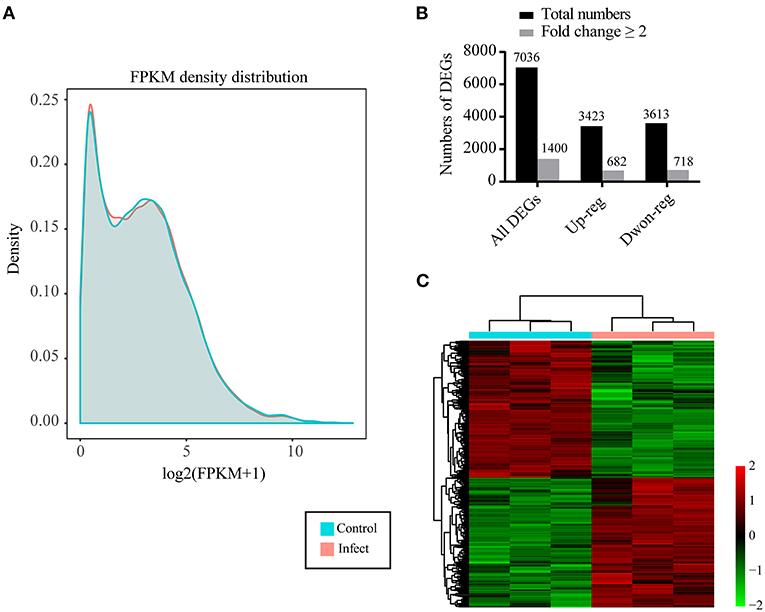
Figure 2. Characteristics of the differentially expressed genes (DEGs) in newborn pig tracheal epithelial (NPTr) cells infected by Pasteurella multocida compared to the control cells. (A) Density plot of a gene's log2(FPKM + 1) distribution visualized by CummeRbund. The X-axis represents the log2(FPKM + 1) of the genes, while the Y-axis represents the distribution density. Different groups are shown in different colors. (B) Number of total DEGs, upregulated DEGs, and downregulated DEGs in P. multocida-infected cells compared to the control cells. (C) Heatmap showing the distribution of the upregulated and downregulated DEGs in P. multocida-infected cells compared to the control cells. PM, Pasteurella multocida.
Validation of DEGs
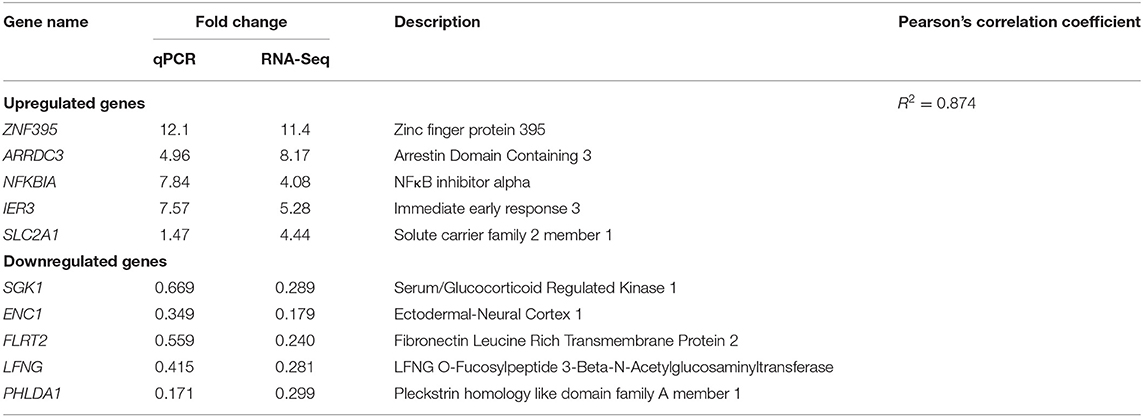
To validate the results of RNA-Seq, we randomly selected 10 genes, namely, five upregulated genes (ZNF395, ARRDC3, NFKBIA, IER3, and SLC2A1) and five downregulated genes (SGK1, ENC1, FLRT2, LFNG, and PHLDA1) in RNA-Seq, for qPCR detection (Table 3). The gene transcription patterns detected by qPCR were similar to those obtained from the RNA-Seq results (Table 3).
DEGs Participate in the Dysfunction of Epithelial Barriers Formed by NPTr
To understand the putative functions of the DEGs, the 6,565 DEGs determined in P. multocida-infected cells compared to the control cells were mapped to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) databases for analyses. GO analysis determined 71 enriched significant biological processes in bacterial infected cells compared to the control cells (padj < 0.05; Supplementary Table 1). Among these enriched biological processes, three (GO:0034330, GO:0045216, and GO:0098609) were closely related to epithelial adhesion and barrier functions, and they were involved in the participation of 30 DEGs (Figures 3A,B). According to the KEGG analysis, these 30 DEGs participated in 22 significant signaling pathways with a p-value < 0.05 (Figure 4A and Supplementary Table 2). Among these 22 significant KEGG signaling pathways, the transforming growth factor (TGF)-beta signaling pathway (KEGG ID: ssc04350), hypoxia-inducible factor 1 (HIF-1) signaling pathway (KEGG ID: ssc04066), epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance (KEGG ID: ssc01521), tumor necrosis factor (TNF) signaling pathway (KEGG ID: ssc04668), and mitogen-activated protein kinase (MAPK) have been shown in previous studies to have roles in the regulation of epithelial adhesion and barrier function in other tissues and organisms (18–21). Therefore, we checked the transcription of the key genes involved in these pathways. The results revealed that the key genes positively regulating the pathways showed significantly increased levels of transcription in bacterial infected cells compared to those in control cells (Figure 4B). In contrast, the key genes (HIF-1αN) negatively regulating the pathways displayed significantly decreased levels of transcription in bacterial infected cells compared to those in control cells (Figure 4B).
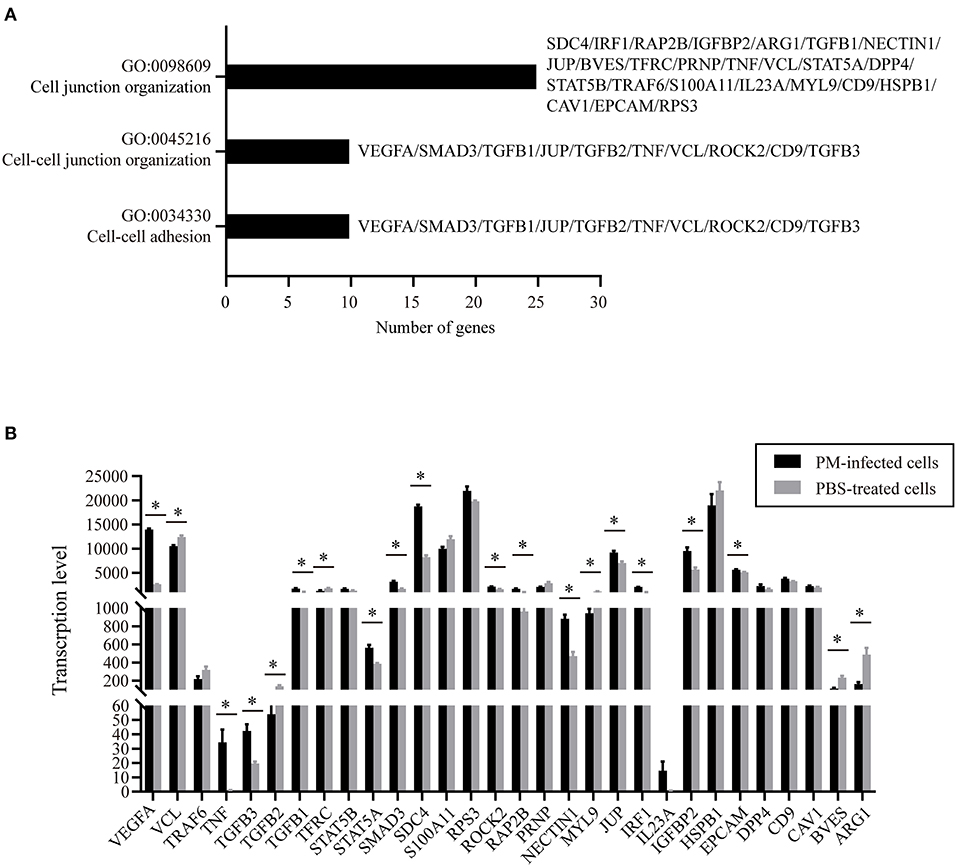
Figure 3. Gene ontology (GO) biological processes and differentially expressed genes (DEGs) closely related to epithelial adhesion and barrier functions. (A) Three biological processes (GO:0034330, GO:0045216, and GO:0098609) that are closely related to epithelial adhesion and barrier functions determined by GO analysis. (B) Transcription of the DEGs participating in the three biological processes in P. multocida-infected and PBS-treated cells as determined by RNA-Seq. PM, Pasteurella multocida. *p < 0.05.
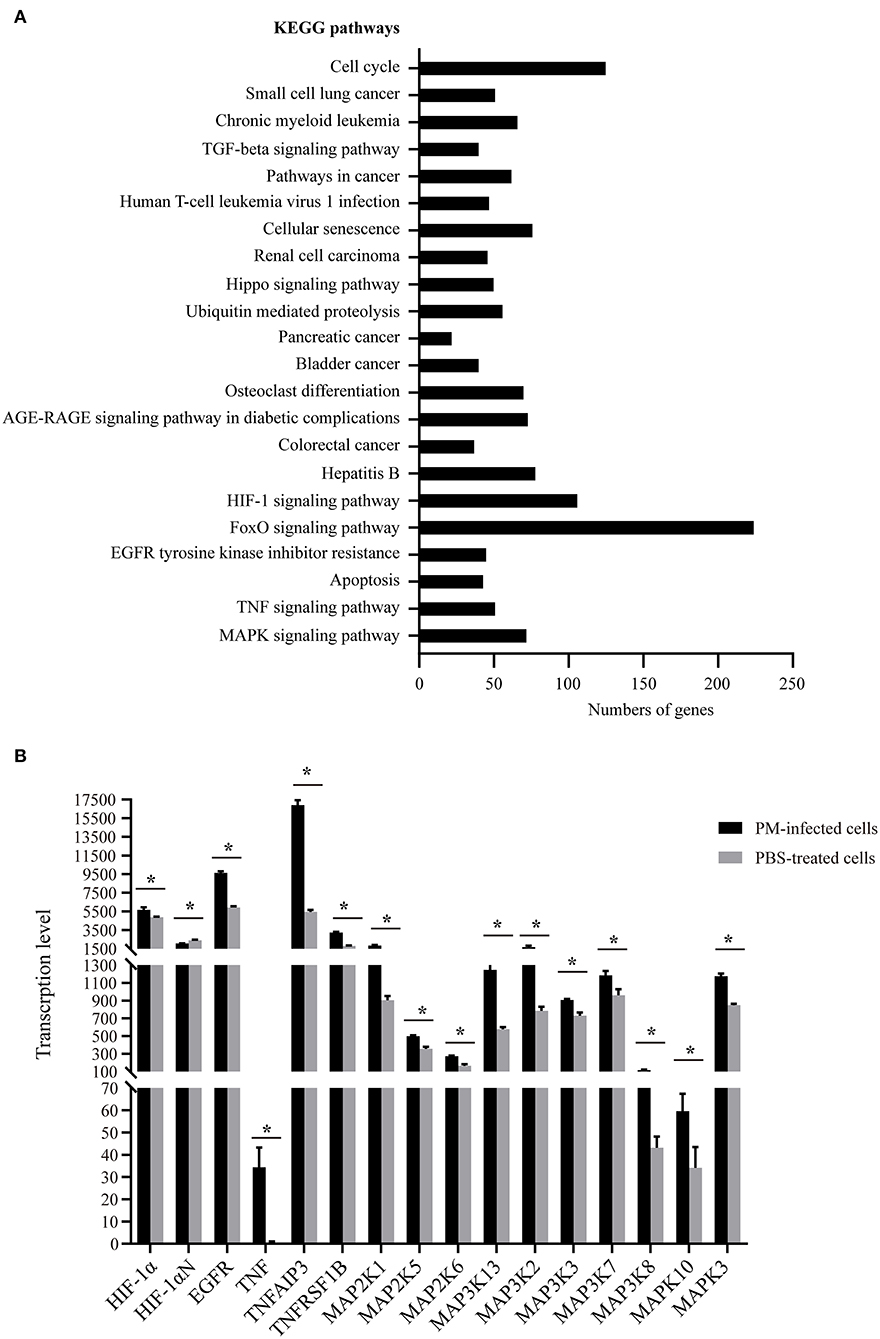
Figure 4. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the 30 differentially expressed genes (DEGs) closely related to epithelial adhesion and barrier functions. (A) The 22 significant signaling pathways with a p-value < 0.05 that the 30 DEGs participate in. (B) Transcription of the key genes involved in selected KEGG pathways (KEGG IDs: ssc04350, ssc04066, ssc01521, ssc04668, and ssc04010) that have roles in the regulation of epithelial adhesion and barrier functions in other tissues and organisms as determined by RNA-Seq. PM, Pasteurella multocida. *p < 0.05.
Discussion
As a leading cause of human and animal respiratory disorders, P. multocida and its interaction with respiratory epithelial cells are important for pathogenesis. However, knowledge about the influence of P. multocida on the respiratory epithelial barrier and the interaction between the bacteria and epithelial cells is still limited. In the present study, we used NPTr cells as a model and found that infection of P. multocida led to decreases in the TEER values of the cells. For the culture models of the endothelial and/or epithelial monolayers, TEER is a widely accepted quantitative technique to measure the integrity of tight junction dynamics (22, 23). A decreased TEER value reflects the dysfunction of the endothelial or epithelial barriers in cell culture models (23, 24). The above finding is suggestive of P. multocida infection disrupting the host's respiratory barrier formed by epithelial cells. To verify this result, we investigated the thickness of the epithelial cell layer post-P. multocida infection through immunofluorescence microscopy analysis. In this experiment, treated cells were stained with antibodies to β-tubulin (in pink under immunofluorescence microscopy), and the results revealed that P. multocida infection led to a decrease of β-tubulin. It is known that β-tubulin is a major component of microtubules that perform many functions, including intracellular transport and the generation and maintenance of cellular morphology, in eukaryotic cells (25). In many studies, a change in microtubules (indicated by β-tubulin) has been used as an agent to indicate endothelial barrier functions (26, 27), and a decreased density of microtubules (indicated by β-tubulin) suggests an increased endothelial cell permeability (27, 28). Consistently, a decreased density of microtubules (indicated by β-tubulin) in P. multocida-infected cells may also indicate decreased endothelial barrier functions. We also detected the transcription of several known genes that encode proteins playing roles in the maintenance of the barrier functions of epithelial cells, such as Occludin, E-cadherin, β-catenin, and Claudin (17). The results revealed that the transcription of these genes decreased significantly during P. multocida infection. The Western blot assays also confirmed the decreased expressions of E-cadherin and β-catenin from the protein level (we did not determine the expressions of Occludin and Claudin because we did not have their antibodies at hand during the revision). All of the above findings suggest that P. multocida infection disrupts the host respiratory epithelial barrier. This process might be beneficial for the invasion of P. multocida and the development of pneumonic pasteurellosis, as well as the other inflammatory disorders of the airways and lungs (9).
To find out possible molecules in host cells that contribute to the dysfunction of the respiratory barrier during bacterial infection, we determined DEGs in P. multocida-infected cells compared to the PBS-treated cells by using RNA-Seq. Our further analyses finally determined 30 DEGs participating in three biological processes (GO:0034330, GO:0045216, and GO:0098609) that were closely related to epithelial adhesion and barrier functions. Among these genes, VEGFA is the most noteworthy one. VEGFA encodes vascular endothelial growth factor A (VEGF-A), which is known for its functions in angiogenesis, stimulating endothelial cell proliferation and migration, and increasing vascular permeability (29). An increased expression of VEGF-A has been used as an important indicator of the dysfunction of the blood–brain barrier and intestinal barrier (30, 31). Here, we found that the transcription of VEGFA significantly increased in P. multocida-infected NPTr cells, suggesting that VEGF-A might also play a role in mediating the dysfunction of the tracheal epithelial barrier after P. multocida infection. In other barrier models, e.g., in blood–brain barrier models, it has been shown that the production of inflammatory factors contributes to the dysfunction of the endothelial and/or epithelial barriers after pathogen infection and the activation of many signaling pathways such as TNF signaling, TGF-beta signaling, SMAD3 signaling, STAT5 signaling, HIF-1 signaling pathway, EGFR tyrosine kinase inhibitor resistance, and MAPK signaling pathway, all of which are involved in regulating the production of inflammatory factors and/or VEGF-A (32–37). The RNA-Seq results shown herein revealed that the transcription of many key factors involved in the above-mentioned signaling pathways significantly increased in P. multocida-infected cells, indicating that the activation of these signals might also contribute to the production of inflammatory factors, which may finally be beneficial for the dysfunction of the tracheal epithelial barrier after P. multocida infection.
In summary, we reported the possible effects of P. multocida infection on the tracheal epithelial barrier in the present study. Our findings indicate that P. multocida infection could induce the dysfunction of the tracheal epithelial barrier and activate several signals playing important roles in inducing the production of inflammatory factors and VEGF-A, an important marker commonly used to indicate the dysfunction of endothelial or epithelial barriers. The dysfunction of the tracheal epithelial barrier and the activation of these pro-inflammatory signals might be beneficial for P. multocida invasion and the development of many inflammatory disorders of the airways and lungs. Our results shown herein will provide more knowledge about the pathogenesis of P. multocida.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra, SRR14026556.
Author Contributions
ZP, HC, and BW contributed to conception and design of the study. XuW, FW, LL, WL, SL, and LH performed the experiments. FW and ZP performed the statistical analysis. ZP and XuW wrote the first draft of the manuscript. ZP, XiW, HC, and BW revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported in part by China Postdoctoral Science Foundation (grant no. 2020T130232) and the Agricultural Science and Technology Innovation Program of Hubei Province (grant no. 2018skjcx05). The funders have no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely acknowledge Professor Hongbo Zhou and Dr. Lu Li at Huazhong Agricultural University (Wuhan, China) for the gift of NPTr cells. We also thank the staff at Novogene (Beijing, China) for the technical support for the RNA-Seq.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.682514/full#supplementary-material
Supplementary Table 1. Differentially expressed genes involved in 71 enriched significant biological processes in P. multocida-infected NPTr cells compared to PBS-treated control cells (padj <0.05).
Supplementary Table 2. Significant KEGG signaling pathways with a p-value < 0.05 that differentially expressed genes closely related to the epithelial adhesion and barrier function participate in.
References
1. Peng Z, Wang X, Zhou R, Chen H, Wilson BA, Wu B. Pasteurella multocida: genotypes and genomics. Microbiol Mol Biol Rev. (2019) 83:e00014–9. doi: 10.1128/MMBR.00014-19
2. Carter GR. Studies on Pasteurella multocida. I. A hemagglutination test for the identification of serological types. Am J Vet Res. (1955) 16:481–4.
3. Rimler RB, Rhoades KR. Serogroup F. A new capsule serogroup of Pasteurella multocida. J Clin Microbiol. (1987) 25:615–8. doi: 10.1128/jcm.25.4.615-618.1987
4. Peng Z, Wang H, Liang W, Chen Y, Tang X, Chen H, et al. A capsule/lipopolysaccharide/MLST genotype D/L6/ST11 of Pasteurella multocida is likely to be strongly associated with swine respiratory disease in China. Arch Microbiol. (2018) 200:107–18. doi: 10.1007/s00203-017-1421-y
5. Wilkie IW, Harper M, Boyce JD, Adler B. Pasteurella multocida: diseases and pathogenesis. Curr Top Microbiol Immunol. (2012) 361:1–22. doi: 10.1007/82_2012_216
6. Kim J, Kim JW, Oh SI, So B, Kim WI, Kim HY. Characterisation of Pasteurella multocida isolates from pigs with pneumonia in Korea. BMC Vet Res. (2019) 15:119. doi: 10.1186/s12917-019-1861-5
7. Wilson BA, Ho M. Pasteurella multocida: from zoonosis to cellular microbiology. Clin Microbiol Rev. (2013) 26:631–55. doi: 10.1128/CMR.00024-13
8. Invernizzi R, Lloyd CM, Molyneaux PL. Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology. (2020) 160:171–82. doi: 10.1111/imm.13195
9. Hiemstra PS, McCray PB Jr., Bals R. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur Respir J. (2015) 45:1150–62. doi: 10.1183/09031936.00141514
10. Chitarra CS, Oliveira Filho JX, Morés N, Silva M, Cândido SL, Cezarino PG, et al. Identification of Pasteurella multocida transcribed genes in porcine lungs through RNAseq. Microb Pathog. (2018) 122:180–3. doi: 10.1016/j.micpath.2018.06.014
11. Zhang S, Wang R, Su H, Wang B, Sizhu S, Lei Z, et al. Sus scrofa miR-204 and miR-4331 negatively regulate swine H1N1/2009 influenza A virus replication by targeting viral HA and NS, respectively. Int J Mol Sci. (2017) 18:749. doi: 10.3390/ijms18040749
12. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. (2015) 33:290–5. doi: 10.1038/nbt.3122
13. Chen Z, Chen H, Li X, Yuan Q, Su J, Yang L, et al. Fumonisin B(1) damages the barrier functions of porcine intestinal epithelial cells in vitro. J Biochem Mol Toxicol. (2019) 33:e22397. doi: 10.1002/jbt.22397
14. Li P, He F, Wu C, Zhao G, Hardwidge PR, Li N, et al. Transcriptomic analysis of chicken lungs infected with avian and bovine Pasteurella multocida serotype A. Front Vet Sci. (2020) 7:452. doi: 10.3389/fvets.2020.00452
15. Szulcek R, Bogaard HJ, van Nieuw Amerongen GP. Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J Vis Exp. (2014) 85:51300. doi: 10.3791/51300
16. Su A, Tong J, Fu Y, Müller S, Weldearegay YB, Becher P, et al. Infection of bovine well-differentiated airway epithelial cells by Pasteurella multocida: actions and counteractions in the bacteria-host interactions. Vet Res. (2020) 51:140. doi: 10.1186/s13567-020-00861-2
17. Niessen CM. Tight junctions/adherens junctions: basic structure and function. J Invest Dermatol. (2007) 127:2525–32. doi: 10.1038/sj.jid.5700865
18. Wu S, Wang N, Li J, Wang G, Seto SW, Chang D, et al. Ligustilide ameliorates the permeability of the blood-brain barrier model in vitro during oxygen-glucose deprivation injury through HIF/VEGF Pathway. J Cardiovasc Pharmacol. (2019) 73:316–25. doi: 10.1097/FJC.0000000000000664
19. Chen L, Liu W, Wang P, Xue Y, Su Q, Zeng C, et al. Endophilin-1 regulates blood-brain barrier permeability via EGFR-JNK signaling pathway. Brain Res. (2015) 1606:44–53. doi: 10.1016/j.brainres.2015.02.032
20. Coelho-Santos V, Leitão RA, Cardoso FL, Palmela I, Rito M, Barbosa M, et al. The TNF-α/NF-κB signaling pathway has a key role in methamphetamine-induced blood-brain barrier dysfunction. J Cereb Blood Flow Metab. (2015) 35:1260–71. doi: 10.1038/jcbfm.2015.59
21. Lan YL, Wang X, Zou YJ, Xing JS, Lou JC, Zou S, et al. Bazedoxifene protects cerebral autoregulation after traumatic brain injury and attenuates impairments in blood-brain barrier damage: involvement of anti-inflammatory pathways by blocking MAPK signaling. Inflamm Res. (2019) 68:311–23. doi: 10.1007/s00011-019-01217-z
22. Chen S, Einspanier R, Schoen J. Transepithelial electrical resistance (TEER): a functional parameter to monitor the quality of oviduct epithelial cells cultured on filter supports. Histochem Cell Biol. (2015) 144:509–15. doi: 10.1007/s00418-015-1351-1
23. Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ. TEER measurement techniques for in vitro barrier model systems. J Lab Autom. (2015) 20:107–26. doi: 10.1177/2211068214561025
24. Li F, Du P, Yang W, Huang D, Nie S, Xie M. Polysaccharide from the seeds of Plantago asiatica L. alleviates nonylphenol induced intestinal barrier injury by regulating tight junctions in human Caco-2 cell line. Int J Biol Macromol. (2020) 164:2134–40. doi: 10.1016/j.ijbiomac.2020.07.259
25. Lee WH, Kim JY, Kim YS, Song HJ, Song KJ, Song JW, et al. Upregulation of class II beta-tubulin expression in differentiating keratinocytes. J Invest Dermatol. (2005) 124:291–7. doi: 10.1111/j.0022-202X.2004.23506.x
26. Tian X, Tian Y, Moldobaeva N, Sarich N, Birukova AA. Microtubule dynamics control HGF-induced lung endothelial barrier enhancement. PLoS ONE. (2014) 9:e105912. doi: 10.1371/journal.pone.0105912
27. Petrache I, Birukova A, Ramirez SI, Garcia JG, Verin AD. The role of the microtubules in tumor necrosis factor-alpha-induced endothelial cell permeability. Am J Respir Cell Mol Biol. (2003) 28:574–81. doi: 10.1165/rcmb.2002-0075OC
28. Finamore A, Massimi M, Conti Devirgiliis L, Mengheri E. Zinc deficiency induces membrane barrier damage and increases neutrophil transmigration in Caco-2 cells. J Nutr. (2008) 138:1664–70. doi: 10.1093/jn/138.9.1664
29. Lange C, Storkebaum E, de Almodóvar CR, Dewerchin M, Carmeliet P. Vascular endothelial growth factor: a neurovascular target in neurological diseases. Nat Rev Neurol. (2016) 12:439–54. doi: 10.1038/nrneurol.2016.88
30. Huang J, Kelly CP, Bakirtzi K, Villafuerte Gálvez JA, Lyras D, Mileto SJ, et al. Clostridium difficile toxins induce VEGF-A and vascular permeability to promote disease pathogenesis. Nat Microbiol. (2019) 4:269–79. doi: 10.1038/s41564-018-0300-x
31. Argaw AT, Asp L, Zhang J, Navrazhina K, Pham T, Mariani JN, et al. Astrocyte-derived VEGF-A drives blood-brain barrier disruption in CNS inflammatory disease. J Clin Invest. (2012) 122:2454–68. doi: 10.1172/JCI60842
32. McMillin MA, Frampton GA, Seiwell AP, Patel NS, Jacobs AN, DeMorrow S. TGFβ1 exacerbates blood-brain barrier permeability in a mouse model of hepatic encephalopathy via upregulation of MMP9 and downregulation of claudin-5. Lab Invest. (2015) 95:903–13. doi: 10.1038/labinvest.2015.70
33. Ronaldson PT, Demarco KM, Sanchez-Covarrubias L, Solinsky CM, Davis TP. Transforming growth factor-beta signaling alters substrate permeability and tight junction protein expression at the blood-brain barrier during inflammatory pain. J Cereb Blood Flow Metab. (2009) 29:1084–98. doi: 10.1038/jcbfm.2009.32
34. van de Beek de Beek D, Brouwer M, Hasbun R, Koedel U, Whitney CG, Wijdicks E. Community-acquired bacterial meningitis. Nat Rev Dis Primers. (2016) 2:16074. doi: 10.1038/nrdp.2016.74
35. Kim KS. Investigating Bacterial penetration of the blood-brain barrier for the pathogenesis, prevention, and therapy of bacterial meningitis. ACS Infect Dis. (2020) 6:34–42. doi: 10.1021/acsinfecdis.9b00319
36. Le Guennec L, Coureuil M, Nassif X, Bourdoulous S. Strategies used by bacterial pathogens to cross the blood-brain barrier. Cell Microbiol. (2020) 22:e13132. doi: 10.1111/cmi.13132
Keywords: Pasteurella multocida, new-born pig tracheal epithelial cells, respiratory epithelial barriers, dysfunction, RNA-Seq
Citation: Wang X, Wang F, Lin L, Liang W, Liu S, Hua L, Wang X, Chen H, Peng Z and Wu B (2021) Transcriptome Differences in Pig Tracheal Epithelial Cells in Response to Pasteurella Multocida Infection. Front. Vet. Sci. 8:682514. doi: 10.3389/fvets.2021.682514
Received: 18 March 2021; Accepted: 26 July 2021;
Published: 19 August 2021.
Edited by:
Dirk Werling, Royal Veterinary College (RVC), United KingdomReviewed by:
Abdelaziz Ed-Dra, Zhejiang University, ChinaJochen Meens, University of Veterinary Medicine Hannover, Germany
Copyright © 2021 Wang, Wang, Lin, Liang, Liu, Hua, Wang, Chen, Peng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong Peng, pengzhong@mail.hzau.edu.cn; orcid.org/0000-0001-5249-328X; Bin Wu, wub@mail.hzau.edu.cn
 Xueying Wang1,2
Xueying Wang1,2  Fei Wang
Fei Wang Wan Liang
Wan Liang Zhong Peng
Zhong Peng Bin Wu
Bin Wu