Alterations in the Gut Microbial Composition and Diversity of Tibetan Sheep Infected With Echinococcus granulosus
- 1College of Life Science, Anqing Normal University, Anqing, China
- 2Research Center of Aquatic Organism Conservation and Water Ecosystem Restoration in Anhui Province, Anqing Normal University, Anqing, China
- 3Jilin Agricultural Science and Technology University, Key Lab of Preventive Veterinary Medicine in Jilin Province, Jilin, China
Hydatidosis/cystic echinococcosis (CE) caused by Echinococcus granulosus is a parasitic zoonotic disease worldwide, threatening animal health and production and public health safety. However, it is still unclear that whether E. granulosus infection can result in the alteration of gut microbiota in Tibetan sheep. Therefore, a study was designed to investigate the influences of E. granulosus infection on gut microbiota of Tibetan sheep. A total of 10 ovine small intestinal contents (five from healthy and five from infected) were obtained and subjected to high-throughput sequencing by MiSeq platform. A total of 2,395,641 sequences and 585 operational taxonomic units (OTUs) were identified. Firmicutes and Proteobacteria were the most dominant phyla in all samples. Moreover, the proportions of Armatimonadetes and Firmicutes in the infected Tibetan sheep were significantly decreased, whereas Actinobacteria, Chloroflexi, and Acidobacteria had significantly increased. At the genus level, the Christensenellaceae_R-7_group and Ruminococcaceae_NK4A214_group were the predominant bacterial genera in all the samples. Furthermore, the healthy Tibetan sheep exhibited higher abundances of Intestinimonas, Butyrivibrio, Pseudobutyrivibrio, Ruminococcaceae, Eubacterium_coprostanoligenes_group, Oxobacter, Prevotella_1, Ruminiclostridium_6, Coprococcus_1, Ruminococcus, Lachnospiraceae_UCG-002, Olsenella, and Acetitomaculum, whereas Kocuria, Clostridium_sensu_stricto_1, Slackia, Achromobacter, and Stenotrophomonas levels were lower. In conclusion, our results conveyed an information that E. granulosus infection may cause an increase in pathogenic bacteria and a decrease in beneficial bacteria. Additionally, a significant dynamical change in gut microbiota could be associated with E. granulosus infection.
Introduction
Tibetan sheep is an ancient species of the Qinghai-Tibet plateau that prevails from central Kazakhstan to Shanxi province in China and from Altai mountains to Himalaya. It is the largest prevailing wild-type sheep in the world and has adapted to hypoxic conditions (3,500–5,000 m above sea level) and low temperature of the area (1). This sheep is a primary source of income, leather, milk, and meat for the local herdsmen (2). The abundant herbage resources in the Tibetan plateau have provided subsistence conditions for this sheep. However, this region has a higher incidence of echinococcosis in sheep and their herdsmen probably due to prevailing substandard hygienic practices (3).
Hydatidosis/cystic echinococcosis (CE) is a worldwide zoonosis caused by Echinococcus granulosus sensu lato that causes health and economic losses, especially in the areas of Central Asia, western China, southern Europe, North and Central Africa, and south-western Latin America (4). In the People's Republic of China, it is mainly prevalent in the western parts of the country including Xinjiang, Qinghai, Gansu, Tibet, and Sichuan provinces (5). In pastoral areas, the human infection rate can reach 50% (6). Echinococcus granulosus mainly infects the liver but may also infect the lungs, heart, brain, and intestines in the hosts, resulting in rashes, fever, abdominal pain, diarrhea, and even death (5).
The intestine colonizes a great variety of microbes including bacteria, protozoa, and fungi (7). Gut microbiota plays important roles in metabolism, nutrient absorption, and mucosal immunity (8). The variation in the normal gut microbiota can influence metabolic activities and health of the host (9). The composition of gut microbiota is influenced by several extrinsic and intrinsic factors, including food type, environment, species, age, and disease (10). Therefore, the richness and diversity of gut microbiota can indicate host health status and indirectly of various host disease situations. E. granulosus can inhabit the small intestine and liver of the host. However, little is known about the characteristics of gut microbiota in Tibetan sheep infected with E. granulosus. Therefore, the objective of the present study was to compare and analyze the differences in gut microbiota in healthy and E. granulosus-infected sheep.
Materials and Methods
Sample Acquisition
A total of 10 (five healthy and five E. granulosus-infected) 1-year-old Tibetan sheep were selected from a commercial feedlot farm at Tibet, China. The infected sheep was diagnosed by a professional veterinarian and determined by molecular biology. The ratio of females to males in both groups was 2:3. The selected Tibetan sheep were fed on free-range grassland and self-propagated via the commercial farm. All the selected sheep possessed a similar genetic background, and no other disease was observed prior to the sample collection. All the sheep were euthanized, and the contents were obtained from intermediate areas of the duodenum, ileum, and jejunum of the control and the infected groups. The collected intestinal contents were transported immediately in sterile plastic bags and stored at −80°C until further analysis. Moreover, the diseased liver and lungs were collected for microscopy and DNA extraction.
DNA Extraction and PCR Amplification of E. granulosus
For molecular confirmation, the total genomic DNA of E. granulosus was isolated using the TIANamp Genomic DNA Kit according to the manufacturer's instructions. Moreover, the specific primers (forward: 5′-ATTATAGAAAATTTTCGTTTTACACGC-3′ and reverse: 5′-AAGCATGATGCAAAAGGCAAATAAACC-3′) were synthesized to amplify the fragment of the cox1 of mitochondrial gene. The design of the primer was based on previous research and synthesized by Jinsirui Biotechnology Co., Ltd. (Nanjing, China) (11). The PCR-amplified products were analyzed through 1.5% agarose gel by following electrophoresis and the Hi-TIANgel Midi Purification Kit. Subsequently, the PCR products were delivered to the Qingke Biotech Company (Wuhan, China) for sequencing analysis and subjected to BLAST in NCBI. Based on sequencing results, a phylogenetic tree was developed by using MEGA 7 software to determine the conformation of parasitic species.
Microbial Genomic DNA Extraction
DNA of each sample was extracted using QIAamp DNA Mini Kit following the manufacturer's instructions. To assess the extraction quality of DNA, 0.8% (w/v) agarose gel electrophoresis was used. Moreover, the concentration of the DNA was quantified by using a NanoDropTM spectrophotometer.
Amplification and Sequencing of 16S rRNA Gene
Specific gene primers (338F: ACTCCTACGGGAGGCAGCA and 806R: GGACTACHVGGGTWTCTAAT) were produced. PCR amplification products were assessed through gel electrophoresis. AxyPrep DNA Gel Extraction Kit was used to recycle target fragment. The Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Massachusetts, USA) was used for fluorescent quantitation of PCR amplification recovery products on a microplate reader according to preliminary quantitative results of electrophoresis. Sequencing libraries were constructed using TruSeq Nano DNA LT Library Prep Kit (Illumina, USA) following the manufacturer's specification. The End Repair Mix2 was used to repair the sequence ends of the amplified products. A magnetic bead screening system was used to remove the self-connected fragments in the linker followed by the purification of the library system. PCR was done to amplify the obtained DNA fragments for enriching sequence library templates. The AMPure XP Beads were used to repurify the enriched library product.
Before sequencing, the quality of libraries was detected on Agilent Bioanalyzer, and the qualified libraries should only have one peak and no linker. Moreover, the libraries were quantified via using Quant-iT™ PicoGreen™ dsDNA Assay Kit, and library concentrations above 2 nM were finally selected. The selected sequencing library was diluted by gradient and mixed in proportion. The mixed libraries were subjected to 2 × 250-bp paired-end sequencing using MiSeq Reagent Kit V3 (600 cycles) on the MiSeq sequencing machine.
Statistical and Bioinformatics Analysis
The original 16S rRNA data files were subjected to initial quality screen and formal analysis by QIIME™ software (version 1.9.1). Also, short and low-quality sequences (<200 bp) were removed. The clustering program VSEARCH (1.9.6.) was used to merge the sequences and partition operational taxonomic unit (OTU) at ≥97% sequence similarity. A confidence threshold of 0.8 was used to generate a sequence of each OTU as per the Ribosomal Database Project (RDP). Furthermore, the MUSCLE software was used for multiple sequence alignments and phylogenetic analysis of different OTUs. The sparse curves and four diversity indexes (Chao1, ACE, Simpson, and Shannon) were used for assessing sequencing depth and alpha diversity, respectively. R (v3.0.3) and GraphPad Prism (version 6.0c) were used to statistically analyze the data. A value of p ≤ 0.05 was considered statistically significant. The standard deviation values were presented as means ± SD.
Results
Clinical and Molecular Examination
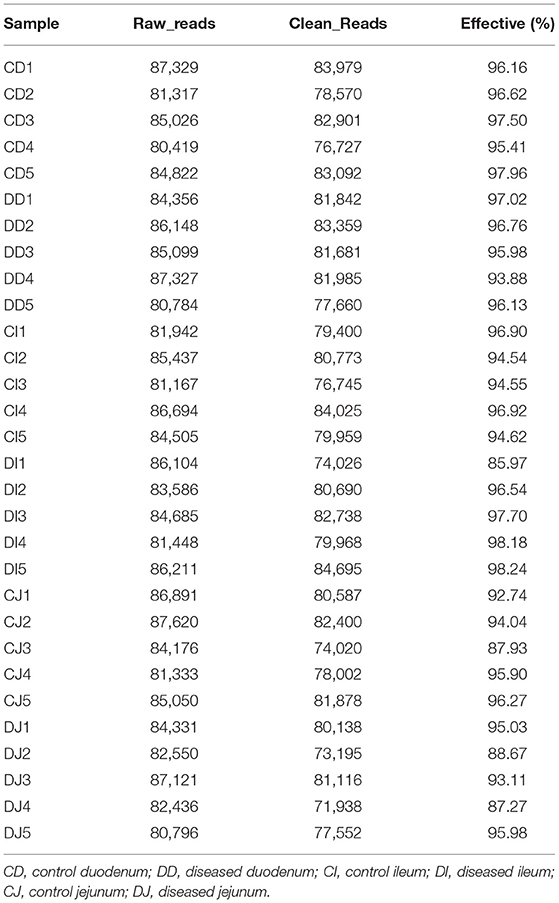
The visual assessment revealed that the healthy Tibetan sheep possessed an active mental state and appetite. Conversely, the infected sheep showed dispiritedness, decreased appetite, and dyspnea. The infected sheep also showed signs of pain and dodge when touched on the abdomen. Moreover, hydatid sacs in the liver and lungs were observed in the infected sheep on postmortem examination (Figures 1A,B). Additionally, we also observed obvious parasite morphology through the microscope (Figures 1C,D). Phylogenetic analysis of collected samples revealed high homology with the E. granulosus under the statistical evaluation assessed using 1,000 bootstraps values (Figure 1E).
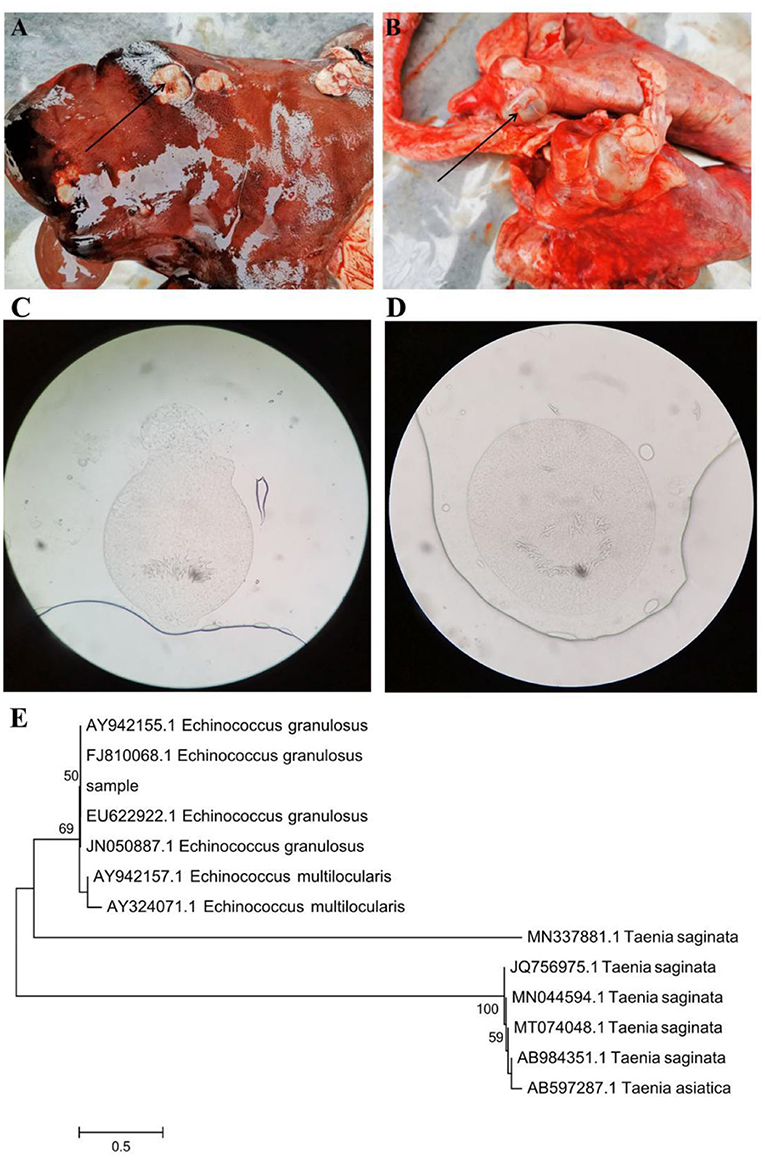
Figure 1. Gross examination of the liver (A) and lung (B) in the hydatid-infected Tibetan sheep. The black arrows indicate hydatid sacs. Microscopic observation and phylogenetic analysis. (C,D) Morphological observation of Echinococcus granulosus. (E) A phylogenetic tree constructed by using the neighbor-joining method.
Sequence Analyses
In this study, a total of 418,913, 423,714, 419,745, 422,034, 425,070, and 417,234 raw sequences were acquired from CD (control duodenum), DD (E. granulosus-infected duodenum), CI (control ileum), DI (E. granulosus-infected ileum), CJ (control jejunum), and DJ (E. granulosus-infected jejunum), respectively (Table 1). After optimizing the original data, 2,395,641 valid sequences were acquired from all the samples (Table 1). Moreover, the rarefaction curve (Shannon and Chao1 curves) for all samples extended all the way to the right end of the x-axis, indicating that the present sequencing depth was sufficient to reflect the diversity of microorganisms contained in all groups (Figures 2A,B). Following taxonomic assignment, a total of 21,568 OTUs (CD = 5,188, DD = 4,226, CI = 4,518, DI = 2,692, CJ = 2,668, and DJ = 2,276) were recognized, and 585 OTUs were common in all the samples (Figures 2C–F). Furthermore, the quantity of unique OTUs in the CD, DD, CI, DI, CJ, and DJ was 3,443, 2,481, 3,305, 1,479, 1,222, and 1,614, respectively (Figures 2C–E).
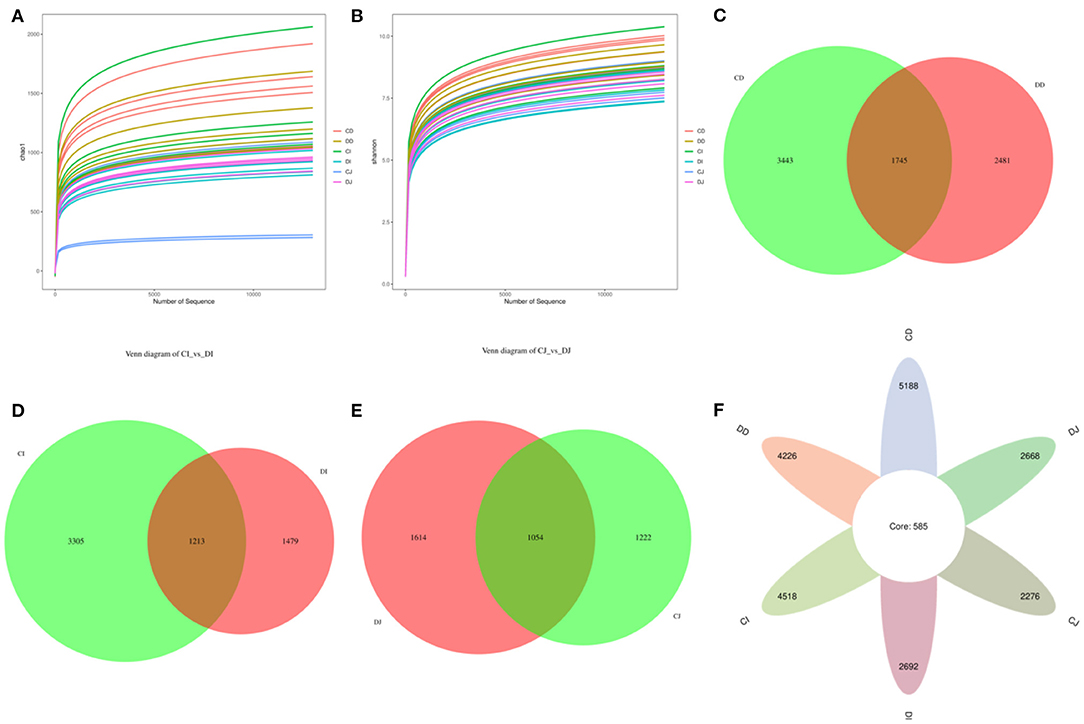
Figure 2. Feasibility analysis and venn diagrams. The rarefaction curves (A,B) were used to evaluate the adequacy of sequencing for each sample. Each curve indicates a sample. (C) Venn diagrams of the OUTs distribution in the CD and DD. (D) Venn diagrams of the OUTs distribution in the CI and DI. (E) Venn diagrams of the OUTs distribution in the CJ and DJ. (F) Venn diagrams for core OTUs compositions.
Alterations in the Gut Microbial Diversities
In the present study, Good's coverage estimates were approximately 100% for all the samples, exhibiting excellent coverage (Figure 3A). The average of Chao1 index in the control (CD, CJ, and CI) and E. granulosus-infected (DD, DJ, and DI) groups varied from 972.40 to 1,563.00 and 719.60 to 1,308.60, respectively (Figure 3B). Moreover, the duodenum possessed the highest Chao1 and Shannon indices as compared to the jejunum and ileum. The average of Chao1 indices in CD, CI, and CJ groups (1,563.00, 1,374.00, and 972.40, respectively) was higher than that in DD, DI, and DJ groups (1,308.60, 918.40, and 719.60, respectively), and a statistically non-significant difference (P > 0.05) was found between these groups (Figure 3B). The gut microbial abundance values did not differ significantly between the control and E. granulosus-infected groups by Chao1 index. Similarly, the average of Simpson and Shannon indices of the control group was higher than that of the E. granulosus-infected group, whereas no obvious difference was found between the two groups (Figures 3C,D). A non-significant difference (P > 0.05) in the gut microbial evenness was found in the E. granulosus-infected and control groups. PCoA plots, which reflect the difference and similarity between groups and individuals, were generated to assess the gut bacterial beta diversity. The beta diversity analysis indicated that the individuals in all groups were clustered together, suggesting that the differences in the principal compositions of gut microbial community of the different groups were insignificant (Figures 3E,F).
Figure 3. The differences of gut microbial diversities between healthy and hydatid-infected Tibetan sheep were non-significant. The alpha diversity of intestinal microbial community can be evaluated by the (A) Good's coverage, (B) Chao1, (C) Simpson, and (D) Shannon. (E) PCoA map based on weighted uniFrac distance (F) PCoA map based on unweighted uniFrac distance.
Changes in the Composition of Gut Bacterial Community
The composition and structure of gut microbiota in various intestinal segments (duodenum, jejunum, and ileum) were analyzed at different taxonomical levels, respectively (Figure 4). At the phylum level, Firmicutes (69.54, 69.63%) and Proteobacteria (10.60, 12.35%) were dominant in the duodenum of CD and DD groups, and the sum of abundances was more than 80% (Figure 4A). In the CI, DI, CJ, and DJ groups, the most significant bacteria at phylum level were Firmicutes (85.07, 72.79, 75.67, and 78.17%), Patescibacteria (5.41, 5.09, 10.32, and 6.92%), Proteobacteria (1.41, 12.32, 4.89, and 2.22%), and Actinobacteria (4.96, 5.28, 6.42, and 9.77%) (Figure 4A). Christensenellaceae_R-7_group (12.78, 15.51, 20.34, and 28.84%), Ruminococcaceae_NK4A214_group (5.31, 7.91, 12.08, and 14.45%), Firmicutes_unclassified (6.25, 5.17, 6.91, and 7.58%), and Candidatus_Saccharimonas (5.12, 5.17, 10.30, and 6.90%) were the four most dominant genera in the CD, DD, CJ, and DJ groups (Figure 4B). Moreover, the most abundant genera were Christensenellaceae_R-7_group (20.74%), Ruminococcaceae_NK4A214_group (12.95%), Firmicutes_unclassified (6.33%), and Romboutsia (8.16%) in the CI group, while Ruminococcaceae_NK4A214_group (23.78%), Firmicutes_unclassified (11.75%), Romboutsia (6.74%), and Pseudomonas (8.59%) were observed as predominant in the DI groups. Interestingly, Christensenellaceae_R-7_group was constantly the most preponderant bacterium in all the samples. Moreover, the primary composition of gut bacterial community in different intestinal samples could also be found in the heatmap (Figure 5).
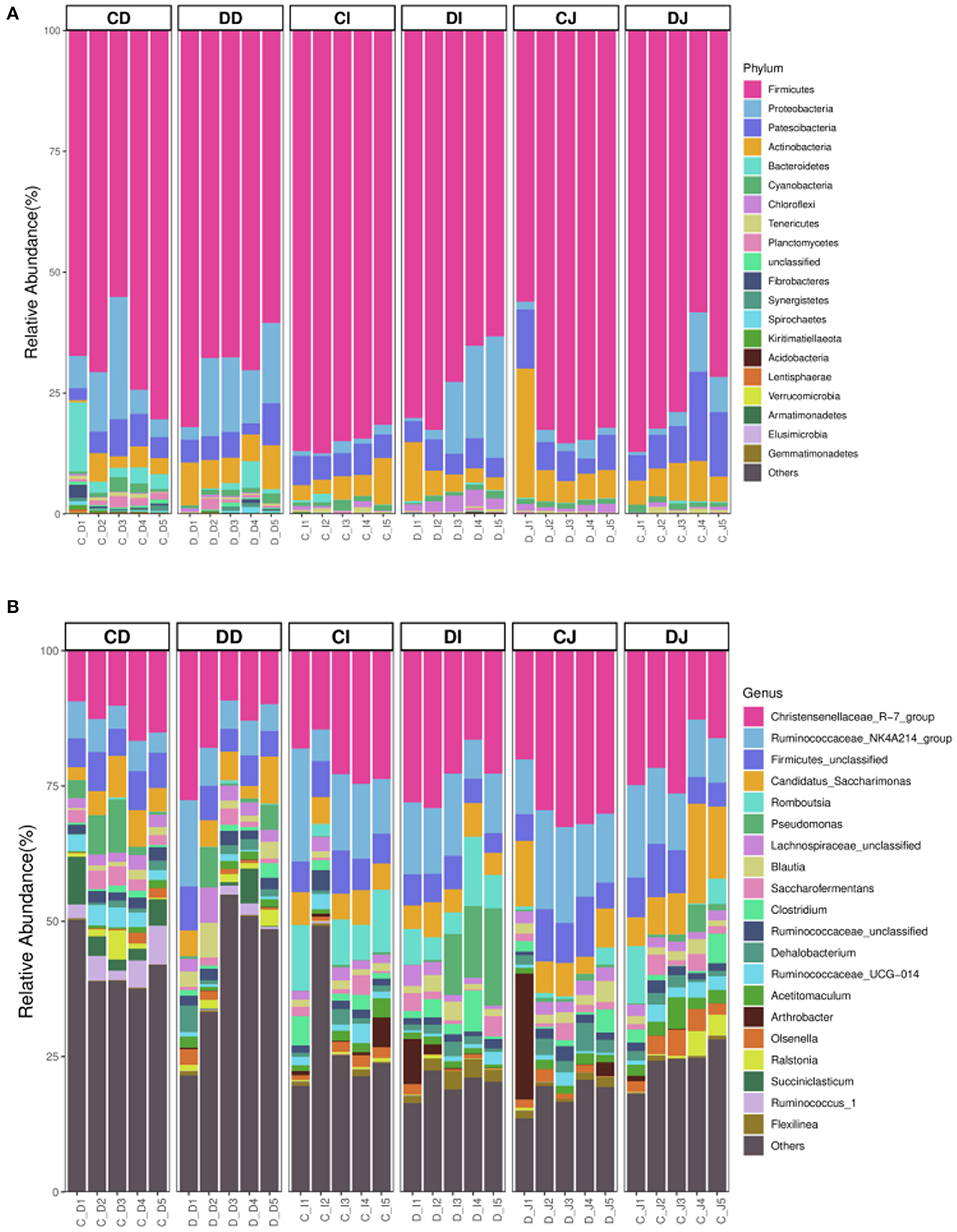
Figure 4. The relative richness of the gut microbiota in healthy and Echinococcus granulosus infected Tibetan sheep. (A) The top 20 dominant phylum of the Tibetan-sheep gut microbiota. (B) The top 20 major genera of the Tibetan-sheep gut microbiota.
The comparison of gut microbiota between the control (CD, CI, and CJ) and E. granulosus-infected (DD, DI, and DJ) groups indicated that the abundance of Armatimonadetes at the phylum level in the CD group was distinctly higher than in the DD group, while the Actinobacteria content was lower (P < 0.05) (Figure 6A). Chloroflexi and Acidobacteria in the CI group were distinctly lower than in the DI group, whereas the Firmicutes level was higher (P < 0.05 or P < 0.01) (Figure 6A). Furthermore, the relative abundance of Chloroflexi was significantly more dominant in the DJ group than the CJ group (P < 0.05) (Figure 6A). At the genus level, Intestinimonas, Butyrivibrio, Pseudobutyrivibrio, Ruminococcaceae_UCG-014, Ruminococcus_1, Oxobacter, Prevotella_1, Ruminococcaceae_UCG-013, and Ruminiclostridium_6 were distinctly higher in the CD group (P < 0.05) than in the DD group, while the Kocuria, Clostridium_sensu_stricto_1, Slackia, and Achromobacter levels were lower (P < 0.05 or P < 0.01) (Figure 6B). Meanwhile, Eubacterium_coprostanoligenes_group, Coprococcus_1, Ruminococcus, Lachnospiraceae_UCG-002, Ruminococcus_gauvreauii_group, Olsenella, and Ruminococcus_1 were significantly higher in the CI group than in the DI group, while the Clostridium_sensu_stricto_8 and Stenotrophomonas contents were lower (P < 0.05) (Figure 6B). Furthermore, a comparison of the CI and DI groups displayed a significant increase (P < 0.05) in the abundance of Acetitomaculum, Olsenella, Ruminococcus_2, Lachnospiraceae_UCG-002, Eubacterium_coprostanoligenes_group, Lachnospiraceae_FE2018_group, Coprococcus_1, and Ruminococcaceae_UCG-013 (P < 0.05) (Figure 6B).
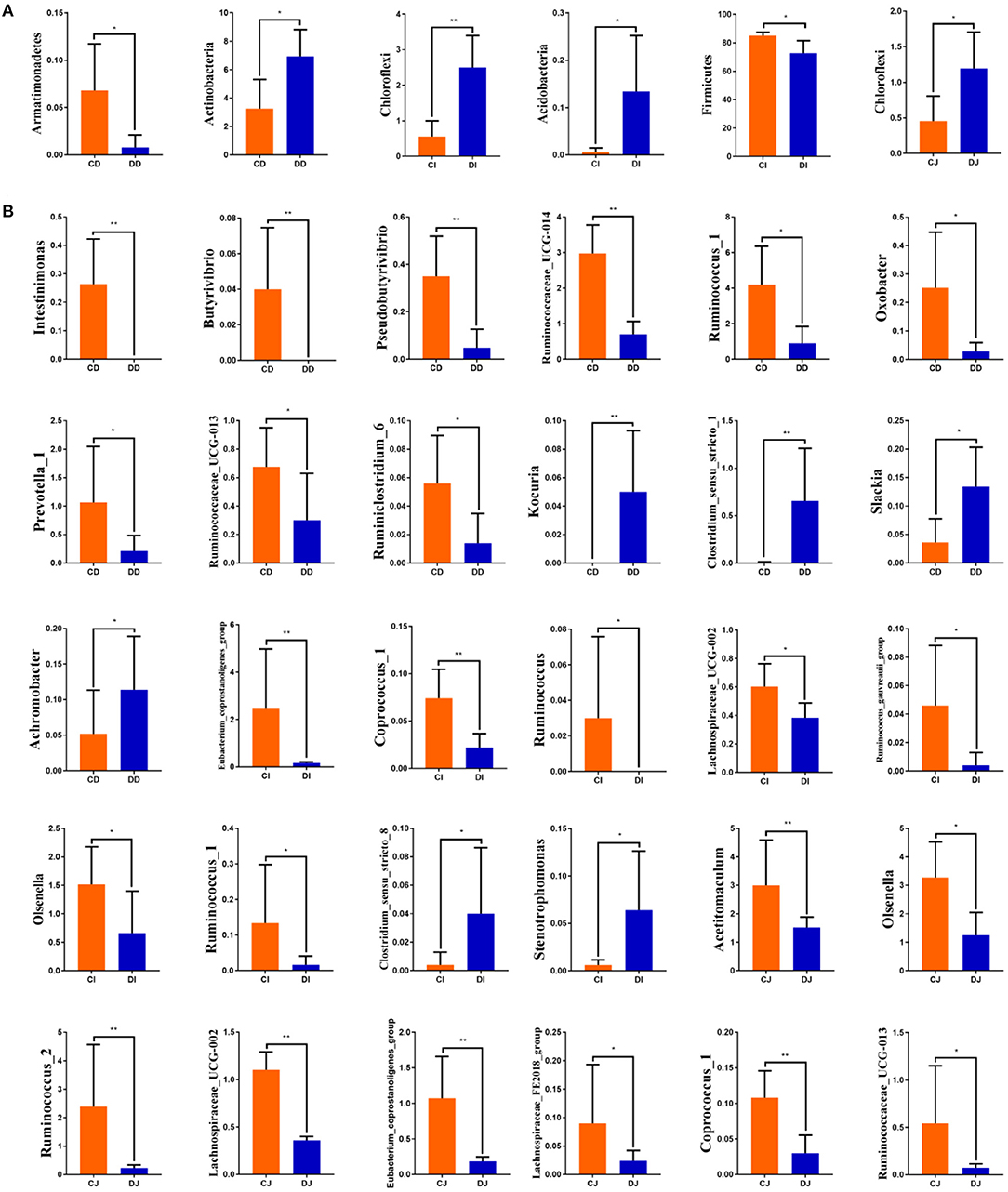
Figure 6. Significant changes in the compositions of gut microbial community at the phylum (A) and genus (B) levels.
Discussion
Gut microbial community is a dynamic and complicated system that significantly influences the host physiology (12). Moreover, increasing evidence indicated that gut microbial community poses a barrier for the host against colonization and invasion of the pathogenic bacterium (13, 14). Therefore, the analysis and investigation of gut microbiota possess significance in preventing and treating certain diseases. To date, numerous research have investigated the relationship of microbial community structure and multiple diseases including diarrhea, diabetes, asthma, and obesity (9, 10). However, to our best understanding, only one study focused on the impact of E. granulosus infection on gut microbiota (15). In this study, we analyzed the intestinal microbiota composition in healthy and infected Tibetan sheep by high-throughput sequencing techniques.
Although, most of the research on gut microbiota employs fecal samples, the diversity of gut microbial community cannot be completely reflected by these samples (12). Therefore, intestinal samples were collected to evaluate the changes of gut microbiota. We observed that the number of OTUs and the alpha diversity indexes were lower in the E. granulosus-infected group than the control group, indicating that E. granulosus infection caused a downward trend in the abundance and diversity of gut microbiota. Similarly, He et al. also indicated that parasitic infection decreased the diversity of gut microbiota in piglets (16). The gut microbiota is an important barrier for the host against the invasion of pathogenic bacteria, which in turn depends on the normal gut microbial composition and diversity (17). Therefore, the lower diversity and richness of gut microbiota could increase the risk of infections caused by secondary pathogenic bacteria (14). Numerous studies revealed that the intestinal function was positively related to gut microbial abundance, and the higher gut microbial richness and diversity favor nutrient absorption and conducting complex physiological functions (18, 19). It has been demonstrated that parasites can cause weight loss and malnutrition of the host by affecting the intestinal absorption function (20). Therefore, the influence of parasitic infection on intestinal functions may be mediated by affecting gut microbial composition and structure.
The gut microbial community is a special ecosystem in the intestine consisting of various microbes that interact as commensals, pathogens, and/or opportunistic pathogens (21, 22). The interaction among various types of bacteria not only promotes metabolism and nutrient absorption but also contributes to the immune system maturation against infection, hence decreasing the risk of disease (23). Generally, the composition of gut microbiota in ruminants is affected by multiple factors, such as host age, nutrition, sex, stress, disease, and growing environment (24). This study revealed that Firmicutes and Proteobacteria were the dominant phyla in all the samples, regardless of the health status. Moreover, those phyla were also observed to be widely distributed in goats, yak, and cattle, indicating their importance in intestinal ecology and function (25, 26). Interestingly, although E. granulosus infection cannot alter the diversity of dominant bacterial phyla in Tibetan sheep, the percentage of some bacteria was altered dramatically. Compared with the CI group, the ratio of Proteobacteria and Actinobacteria in the gut microbiota of the DI group was increased, while the proportion of Firmicutes was decreased. It is known that Firmicutes mainly consists of many gram-positive bacteria including Lactococcus, Listeria, Bacillus, and Lactobacillus (27). Previous research has shown that Firmicutes plays a key role in the digestion of proteins and carbohydrates (28). Therefore, the abundance of Firmicutes in the gut environment is conducive to meet the energy and nutritional demands in animals (29). Furthermore, Lactobacillus, Listeria, and Lactococcus in the Firmicutes are considered as beneficial bacteria, which play their roles in maintaining gut flora balance and preventing pathogenic invasion (30, 31). Proteobacteria comprises great amounts of (gram-negative) pathogenic bacteria, e.g., Salmonella, Helicobacter pylori, Vibrio cholera, and Escherichia coli, and is the largest phylum (32, 33). The abovementioned pathogenic bacteria cause vomiting, diarrhea, gastritis, gastrointestinal ulcers, and even death, which seriously threaten the health of animals (34). Consequently, the higher percentage of Proteobacteria in the microbial community may increase the incidence of host. A previous study has indicated that the Actinobacteria content was noticeably increased in sheep diarrhea (26). Moreover, the synergy between Actinobacteria and host can influence pathogenic interactions in the intestine (35). Those results revealed distinct alterations in the relative richness of preponderant bacterial phyla of Tibetan sheep, which further implied its gut microbial alterations.
At the genus level, the percentage of Ruminococcaceae, Ruminococcus, Lachnospiraceae, Coprococcus, Acetitomaculum, Olsenella, Oxobacter, Ruminiclostridium, Intestinimonas, Butyrivibrio, Pseudobutyrivibrio, Eubacterium_coprostanoligenes, and Prevotella in E. granulosus-infected Tibetan sheep was obviously reduced as compared to control Tibetan sheep. Ruminococcaceae, a potential intestinal probiotic, is beneficial to degrade cellulose and starch and negatively correlated with liver cirrhosis and non-alcoholic fatty liver (36). It is reported that Ruminococcus plays a crucial role in degrading cellulose and produces small-chain fatty acid, e.g., formic, lactic, and acetic acids (37). Moreover, Ruminiclostridium, Lachnospiraceae, Coprococcus, Acetitomaculum, Olsenella, and Oxobacter can also produce short-chain fatty acids (38–42). Previous studies have revealed that short-chain fatty acid plays a key role in regulating gut microbial balance and maintaining the morphology and functionality of intestinal epithelial cells (43, 44). Lactic acid ameliorates digestive enzymes' activity and possesses bacteriostatic effects via regulating the gastrointestinal pH (30). Therefore, the higher abundances of Ruminiclostridium, Ruminococcus, Lachnospiraceae, Coprococcus, Acetitomaculum, Olsenella, and Oxobacter contribute to improve the growth and reduce gastrointestinal bacterial diseases in animals. It is known that Intestinimonas and Pseudobutyrivibrio produce butyrate and are essential for host health (45). Butyrate can decrease appetite and activate brown adipose tissue through the brain–gut axis, resulting in reducing cardiovascular disease and diabetes caused by obesity (46, 47). Currently, butyrate-producing bacteria are considered potential probiotics for treating and alleviating inflammatory bowel disease due to their anti-inflammatory and immunomodulatory functions (48). Eubacterium_coprostanoligenes is an anaerobe and possesses the ability to lower cholesterol (49). Remarkably, the abundance of Eubacterium_coprostanoligenes is negatively associated with the severity of anxiety (24). Butyrivibrio and Prevotella principally participate in the digestion and decomposition of cellulose and carbohydrate (50). Moreover, they can produce short-chain fatty acid (51). By contrast, the percentages of Clostridium, Kocuria, Slackia, Achromobacter, and Stenotrophomonas were significantly higher in infected Tibetan sheep. Clostridiae cause toxemia and diarrhea in ruminants (52). Moreover, it has been reported that Clostridium contributes to the occurrence of necrotic enteritis in infants (53). Previous studies have indicated that Kocuria can cause catheter-related bacteremia peritonitis in humans (54, 55) Slackia can lead to empyema and acute respiratory distress syndrome (56). Moreover, Shao and Zhu showed that the level of Slackia was dramatically increased in humans exposed to various metals for a long time (57). Achromobacter and Stenotrophomonas are the emerging pathogens, closely related to cystic fibrosis and bacteremia (58, 59). Our results revealed that E. granulosus infection could cause distinct dynamic changes in gut microbial community via increasing the proportion of pathogenic and beneficial bacteria. Previous research indicated that E. granulosus infections could impair intestinal mucosa and intestinal barrier function and alter intestinal mucosal immunity (60). Moreover, gut microbial community has also been demonstrated to play key roles in intestinal permeability and immune system maturation (61, 62). Thus, gut microbial dysbiosis may influence the immunity and intestinal barrier function, which in turn increases the risk for other diseases. Some opportunistic pathogens, such as bacteria, fungi, and viruses, are residing as part of normal gut microbiota but may take opportunity to result in diseases in gut microbial dysbiosis and immunocompromised situations (63, 64). Remarkably, this study also conveyed an important message that hydatidosis may be prevented through improving the quantity of beneficial bacteria in the intestine.
In conclusion, the present study investigated the influence of E. granulosus infection on the gut microbiota of Tibetan sheep. Results demonstrated that E. granulosus infection significantly altered the gut microbial composition, characterized by a decreased percentage of beneficial to pathogenic bacteria. Remarkably, there were several limitations in the present study, including a small sample size, individual variation, external environment, and failure to investigate the influence of intestinal fungal communities and viruses on E. granulosus infection.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA657975.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of the Anqing Normal University.
Author Contributions
ZL conceived and designed the experiments. ZL and BY contributed to the sample collection, reagents, materials, and analysis tools. All authors contributed to the article and approved the submitted version.
Funding
The Provincial Natural Science Research Project (no. KJ2017A357) at Anhui University, the Key Research and Development Plan of Anhui Province (no. 201904e01020013), the Science and Technology Development at Jilin Province, China (20180520040JH), and the Scientific Research Project of Education Department of Jilin Province (JJKH20210410KJ) supported this work through a planning grant program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Liu Q, Ma R, Zhao Q, Shang L, Cai J, Wang X, et al. Seroprevalence of Toxoplasma gondii infection in Tibetan sheep in Northwestern China. J Parasitol. (2010) 96:1222–3. doi: 10.1645/GE-2601.1
2. Li K, Mehmood K, Zhang H, Jiang X, Shahzad M, Dong XQ, et al. Characterization of fungus microbial diversity in healthy and diarrheal yaks in Gannan region of Tibet Autonomous Prefecture. Acta Trop. (2018) 182:14–26. doi: 10.1016/j.actatropica.2018.02.017
3. Wang ZH, Wang XM, Liu XQ. Echinococcosis in China, a review of the epidemiology of Echinococcus spp. Ecohealth. (2008) 5:115–26. doi: 10.1007/s10393-008-0174-0
4. Tamarozzi F, Legnardi M, Fittipaldo A, Drigo M, Cassini R. Epidemiological distribution of Echinococcus granulosus s.l. infection in human and domestic animal hosts in European Mediterranean and Balkan countries: a systematic review. PLoS Negl. Trop. Dis. (2020) 14:e0008519. doi: 10.1371/journal.pntd.0008519
5. Woolsey ID, Miller AL. Echinococcus granulosus sensu lato and Echinococcus multilocularis: a review. Res. Vet. Sci. (2020) 135:517–22. doi: 10.1016/j.rvsc.2020.11.010
6. Yang YR, Clements ACA, Gray DJ, Atkinson JM, Williams GM, Barnes Tamsin S, et al. Impact of anthropogenic and natural environmental changes on Echinococcus transmission in Ningxia Hui Autonomous Region, the People's Republic of China. Parasit Vectors. (2012) 5:146. doi: 10.1186/1756-3305-5-146
8. Shapira M. Gut microbiotas and host evolution: scaling up symbiosis. Trends Ecol Evol. (2016) 31:539–49. doi: 10.1016/j.tree.2016.03.006
9. Phimister EG, Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
10. Paola MD, Filippo CD, Cavalieri D, Ramazzotti M, Poullet JB, Massart S, et al. PP90 impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. (2010) 107:14691–6. doi: 10.1073/pnas.1005963107
11. Šnábel V, Kuzmina T, Antipov A, Yemets O, Cavallero S, Miterpáková M, et al. Molecular study of Echinococcus granulosus cestodes in Ukraine and the first genetic identification of Echinococcus granulosus sensu stricto (G1 genotype) in the country. Acta Parasitol. (2021). doi: 10.1007/s11686-021-00450-z. [Epub ahead of print].
12. Zeng Y, Zeng D, Zhang Y, Ni X, Tang Y, Zhu H, et al. Characterization of the cellulolytic bacteria communities along the gastrointestinal tract of Chinese Mongolian sheep by using PCR-DGGE and real-time PCR analysis. World J Microbiol Biotechnol. (2015) 31:1103–13. doi: 10.1007/s11274-015-1860-z
13. Elson CO, Alexander KL. Host-microbiota interactions in the intestine. Dig Dis. (2015) 33:131–6. doi: 10.1159/000369534
14. Wang Y, Li A, Liu J, Mehmood K, Wangdui B, Shi H, et al. L. pseudomesenteroides and L. johnsonii isolated from yaks in Tibet modulate gut microbiota in mice to ameliorate enteroinvasive Escherichia coli-induced diarrhea. Microb Pathog. (2019) 132:1–9. doi: 10.1016/j.micpath.2019.04.020
15. Bao J, Zheng H, Wang Y, Zheng X, He L, Qi W, et al. Echinococcus granulosus infection results in an increase in Eisenbergiella and Parabacteroides genera in the gut of mice. Front Microbiol. (2018) 9:2890. doi: 10.3389/fmicb.2018.02890
16. He K, Yan WC, Sun CY, Liu J, Bai RZ, Wang TQ, et al. Alterations in the diversity and composition of gut microbiota in weaned piglets infected with Balantioides coli. Vet Parasitol. (2020) 288:109298. doi: 10.1016/j.vetpar.2020.109298
17. Tanoue T, Umesaki Y, Honda K. Immune responses to gut microbiota-commensals and pathogens. Gut Microbes. (2010) 1:224–33. doi: 10.4161/gmic.1.4.12613
18. Dias J, Marcondes MI, Motta de Souza S, Cardoso da Mata E Silva B, Fontes Noronha M, Tassinari Resende R, et al. Bacterial community dynamics across the gastrointestinal tracts of dairy calves during preweaning development. Appl. Environ. Microbiol. (2018) 84:e02675-17. doi: 10.1128/AEM.02675-17
19. Li BB, Zhang K, Li C, Wang XL, Chen YL, Yang YX. Characterization and comparison of microbiota in the gastrointestinal tracts of the goat (Capra hircus) during preweaning development. Front Microbiol. (2019) 10:2125. doi: 10.3389/fmicb.2019.02125
20. Hughes S, Kelly P. Interactions of malnutrition and immune impairment, with specific reference to immunity against parasites. Parasite Immunol. (2006) 28:577–88. doi: 10.1111/j.1365-3024.2006.00897.x
21. Abudabos A, Alyemni A, Dafalla Y, Khan R. Effect of organic acid blend and bacillus subtilis alone or in combination on growth traits, blood biochemical and antioxidant status in broilers exposed to salmonella typhimurium challenge during the starter phase. J Appl Anim Res. (2017) 45:538–42. doi: 10.1080/09712119.2016.1219665
22. Zhang H, Rehman MU, Li K, Luo HQ, Lan YF, Nabi F, et al. Antimicrobial resistance of Escherichia coli isolated from tibetan piglets suffering from white score diarrhea. Pak Vet J. (2016) 37:43–6.
23. Li A, Wang Y, Pei L, Mehmood K, Li K, Qamar H, et al. Influence of dietary supplementation with Bacillus velezensis on intestinal microbial diversity of mice. Microb Pathog. (2019) 136:103671. doi: 10.1016/j.micpath.2019.103671
24. Chen Y-H, Bai J, Wu D, Yu S-F, Qiang X-L, Bai H, et al. Association between fecal microbiota and generalized anxiety disorder: severity and early treatment response. J Affect Disord. (2019) 59:56–66. doi: 10.1016/j.jad.2019.08.014
25. Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. (2013) 7:1069–79. doi: 10.1038/ismej.2013.2
26. Wang Y, Zhang H, Zhu L, Xu Y, Liu N, Sun X, et al. Dynamic distribution of gut microbiota in goats at different ages and health states. Front Microbiol. (2018) 9:2509. doi: 10.3389/fmicb.2018.02509
27. Garneau J, Tremblay D, Moineau S. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology. (2008) 373:298–309. doi: 10.1016/j.virol.2007.12.002
28. Cheryl S, Greg W, Jeffrey S. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: regulation by carbon source and oxygen. J Bacteriol. (2006) 188:4663–72. doi: 10.1128/JB.00125-06
29. Sun B, Wang X, Bernstein S, Huffman M, Xia D, Gu Z, et al. Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Sci Rep. (2016) 6:26035. doi: 10.1038/srep26035
30. Wang Y, Li A, Jiang X, Zhang H, Mehmood K, Zhang L, et al. Probiotic potential of Leuconostoc pseudomesenteroides and Lactobacillus strains isolated from Yaks. Front Microbiol. (2018) 9:2987. doi: 10.3389/fmicb.2018.02987
31. Li A, Wang Y, Li Z, Qamar H, Mehmood K, Zhang LH, et al. Probiotics isolated from yaks improves the growth performance, antioxidant activity, and cytokines related to immunity and inflammation in mice. Microb Cell Fact. (2019) 18:112. doi: 10.1186/s12934-019-1161-6
32. Yang H, Xiao Y, Gui G, Li J, Wang J, Li D. Microbial community and short-chain fatty acid profile in gastrointestinal tract of goose. Poult Sci. (2018) 97:1420–8. doi: 10.3382/ps/pex438
33. Huang Z, Kraus V. Does lipopolysaccharide-mediated inflammation have a role in OA. Nat Rev Rheumatol. (2016) 12:123–9. doi: 10.1038/nrrheum.2015.158
34. Wang ZJ, Chen XF, Zhang ZX, Li YC, Deng J, Tu J, et al. Effects of anti-Helicobacter pylori concomitant therapy and probiotic supplementation on the throat and gut microbiota in humans. Microb Pathog. (2017) 109:156–61. doi: 10.1016/j.micpath.2017.05.035
35. Miao V, Davies J. Actinobacteria: the good, the bad, and the ugly. Antonie Van Leeuwenhoek. (2010) 98:143–50. doi: 10.1007/s10482-010-9440-6
36. Shang Q, Shan X, Cai C, Hao J, Li G, Yu G. Dietary fucoidan modulates the gut microbiota in mice by increasing the abundance of Lactobacillus and Ruminococcaceae. Food Funct. (2018) 9:655. doi: 10.1039/c7fo90052j
37. Berg Miller M, Antonopoulos D, Rincon M, Band M, Bari A, Akraiko T, et al. Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS ONE. (2009) 4:e6650. doi: 10.1371/journal.pone.0006650
38. Zhuang P, Zhang Y, Shou QY, Li HY, Zhu Y, He LL, et al. Eicosapentaenoic and docosahexaenoic acids differentially alter gut microbiome and reverse high-fat diet-induced insulin resistance. Mol. Nutr. Food. Res. (2020). 64:e1900946. doi: 10.1002/mnfr.201900946
39. Bengelsdorf FR, Poehlein A, Schiel-Bengelsdorf B, Daniel R, Dürre P. Genome sequence of the acetogenic bacterium Oxobacter pfennigii DSM 3222T. Genome Announc. (2015) 3:e01408-15. doi: 10.1128/genomeA.01408-15
40. Laskar M, Awata T, Kasai T, Katayama A. Anaerobic dechlorination by a humin-dependent pentachlorophenol-dechlorinating consortium under autotrophic conditions induced by homoacetogenesis. Int J Environ Res Public Health. (2019) 16:2873. doi: 10.3390/ijerph16162873
41. Greening RC, Leedle JA. Enrichment and isolation of Acetitomaculum ruminis, gen. nov, sp nov: acetogenic bacteria from the bovine rumen. Arch Microbiol. (1989) 151:399–406. doi: 10.1007/BF00416597
42. Spalinger MR, Kasper S, Chassard C, Raselli T, Frey-Wagner I, Gottier C, et al. PTPN2 controls differentiation of CD4+ T cells and limits intestinal inflammation and intestinal dysbiosis. Mucosal Immunol. (2015) 8:918–29. doi: 10.1038/mi.2014.122
43. Tan J, Mckenzie C, Potamitis M, Thorburn A, Mackay C, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. (2014) 121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
44. Li Q, Chen H, Zhang M, Wu T, Liu R, et al. Altered short chain fatty acid profiles induced by dietary fiber intervention regulate AMPK levels and intestinal homeostasis. Food Funct. (2019) 10:7174–87. doi: 10.1039/c9fo01465a
45. Cai W, Xu JX, Li G, Liu T, Guo XL, Wang HJ, et al. Ethanol extract of propolis prevents high-fat diet-induced insulin resistance and obesity in association with modulation of gut microbiota in mice. Food. Res. Int. (2020) 130:108939. doi: 10.1016/j.foodres.2019.108939
46. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. (2013) 498:99–103. doi: 10.1038/nature12198
47. Bui TP, Ritari J, Boeren S, de Waard P, Plugge CM, de Vos WM. Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat Commun. (2015) 6:10062. doi: 10.1038/ncomms10062
48. Zhang QP, Wu YQ, Wang J, Wu GJ, Long WM, Xue ZS. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci Rep. (2016) 6:27572. doi: 10.1038/srep27572
49. Freier TA, Beitz DC, Li L, Hartman PA. Characterization of Eubacterium coprostanoligenes sp. nov, a cholesterol-reducing anaerobe. Int J Syst Bacteriol. (1994) 44:137–42. doi: 10.1099/00207713-44-1-137
50. Lourenco JM, Kieran TJ, Seidel DS, Glenn TC, Silveira MF, Callaway TR, et al. Comparison of the ruminal and fecal microbiotas in beef calves supplemented or not with concentrate. PLoS ONE. (2020) 15:e0231533. doi: 10.1371/journal.pone.0231533
51. Liu C, Li X, Chen Y, Cheng Z, Duan Q, et al. Age-related response of rumen microbiota to mineral salt and effects of their interactions on enteric methane emissions in cattle. Microb Ecol. (2017) 73:590–601. doi: 10.1007/s00248-016-0888-4
52. Lewis CJ, Naylor RD. Sudden death in sheep associated with Clostridium sordellii. Vet Rec. (1998) 142:417–21. doi: 10.1136/vr.142.16.417
53. Zhou Y, Shan G, Sodergren E, Weinstock G, Walker WA, Gregory KE. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: a case-control study. PLoS ONE. (2015) 10:e0118632. doi: 10.1371/journal.pone.0118632
54. Lee JY, Kim SH, Jeong HS, Oh Seung H, Kim HR, Kim YH. Two cases of peritonitis caused by Kocuria marina in patients undergoing continuous ambulatory peritoneal dialysis. J Clin Microbiol. (2009) 47:3376–8. doi: 10.1128/JCM.00847-09
55. Dunn R, Bares S, David MZ. Central venous catheter-related bacteremia caused by Kocuria kristinae: case report and review of the literature. Ann Clin Microbiol Antimicrob. (2011) 10:31. doi: 10.1186/1476-0711-10-31
56. Man MY, Shum HP, Wu A. A case of severe empyema with acute respiratory distress syndrome caused by, Slackia exigua, requiring veno-venous extracorporeal membrane oxygenation. Anaerobe. (2017) 48:7–11. doi: 10.1016/j.anaerobe.2017.06.017
57. Shao MM, Zhu Y. Long-term metal exposure changes gut microbiota of residents surrounding a mining and smelting area. Sci Rep. (2020) 10:4453. doi: 10.1038/s41598-020-61143-7
58. Garrigos T, Neuwirth C, Chapuis A, Bador J, Amoureux L, et al. Development of a database for the rapid and accurate routine identification of Schromobacter species by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Clin Microbiol Infect. (2020) 27:126.e1–5. doi: 10.1016/j.cmi.2020.03.031
59. Muder RR, Harris AP, Muller S, Edmond M, Chow JW, Papadakis K, et al. Bacteremia due to Stenotrophomonas (Xanthomonas) maltophilia: a prospective, multicenter study of 91 episodes. Clin Infect Dis. (1996) 22:508–12. doi: 10.1093/clinids/22.3.508
60. Ma H, Zhang H, Apaer S, Ma H, Tuxun T, Li H, et al. Impact of intestine mucosal immune barrier in sheep naturally infected with Echinococcus granulosus. Int J Clin Exp Pathol. (2016) 9:7069–77.
61. Hu L, Geng S, Li Y, Cheng S, Fu X, Yue X, et al. Exogenous fecal microbiota transplantation from local adult pigs to crossbred newborn piglets. Front Microbiol. (2017) 8:2663. doi: 10.3389/fmicb.2017.02663
62. Liu J, Wang H, Li L, Miao C, Zhang Y, Zhou B. Intestinal barrier damage involved in intestinal microflora changes in fluoride-induced mice. Chemosphere. (2019) 234:409–18. doi: 10.1016/j.chemosphere.2019.06.080
63. Li A, Liu B, Li F, He Y, Wang L, Fakhar-E-Alam K, et al. Integrated bacterial and fungal diversity analysis reveals the gut microbial alterations in diarrheic giraffes. Front Microbiol. (2021) 12:712092. doi: 10.3389/fmicb.2021.712092
Keywords: gut microbiota, cystic echinococcosis, Tibetan sheep, Echinococcus granulosus, hydatidosis
Citation: Liu Z and Yin B (2022) Alterations in the Gut Microbial Composition and Diversity of Tibetan Sheep Infected With Echinococcus granulosus. Front. Vet. Sci. 8:778789. doi: 10.3389/fvets.2021.778789
Received: 17 September 2021; Accepted: 02 December 2021;
Published: 13 January 2022.
Edited by:
Indranil Samanta, West Bengal University of Animal and Fishery Sciences, IndiaReviewed by:
Aruna Pal, West Bengal University of Animal and Fishery Sciences, IndiaKun Li, Nanjing Agricultural University, China
Copyright © 2022 Liu and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Liu, lzg12021@163.com; Baishuang Yin, ybs3421@126.com
 Zhigang Liu
Zhigang Liu Baishuang Yin
Baishuang Yin