ABCC9 Is Downregulated and Prone to Microsatellite Instability on ABCC9tetra in Canine Breast Cancer
- College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
Tumorigenesis is associated with metabolic abnormalities and genomic instability. Microsatellite mutations, including microsatellite instability (MSI) and loss of heterozygosity (LOH), are associated with the functional impairment of some tumor-related genes. To investigate the role of MSI and LOH in sporadic breast tumors in canines, 22 tumors DNA samples and their adjacent normal tissues were evaluated using polyacrylamide gel electrophoresis and silver staining for 58 microsatellites. Quantitative real-time polymerase chain reaction, promoter methylation analysis and immunohistochemical staining were used to quantify gene expression. The results revealed that a total of 14 tumors (6 benign tumors and 8 breast cancers) exhibited instability as MSI-Low tumors. Most of the microsatellite loci possessed a single occurrence of mutations. The maximum number of MSI mutations on loci was observed in tumors with a lower degree of differentiation. Among the unstable markers, FH2060 (4/22), ABCC9tetra (4/22) and SCN11A (6/22) were high-frequency mutation sites, whereas FH2060 was a high-frequency LOH site (4/22). The ABCC9tetra locus was mutated only in cancerous tissue, although it was excluded by transcription. The corresponding genes and proteins were significantly downregulated in malignant tissues, particularly in tumors with MSI. Furthermore, the promoter methylation results of the adenosine triphosphate binding cassette subfamily C member 9 (ABCC9) showed that there was a high level of methylation in breast tissues, but only one case showed a significant elevation compared with the control. In conclusion, MSI-Low or MSI-Stable is characteristic of most sporadic mammary tumors. Genes associated with tumorigenesis are more likely to develop MSI. ABCC9 protein and transcription abnormalities may be associated with ABCC9tetra instability.
Introduction
Tumorigenesis is a complex multistep process associated with metabolic abnormalities and genomic instability (1). Studies have shown that tumor cells differ significantly from normal cells in terms of ion channel expression activity and membrane potential (2, 3). Through electrochemical synapse ionic coupling networks, tumor cells can induce or inhibit the occurrence and metastasis of tumors (4). The adenosine triphosphate (ATP)-binding cassette subfamily C, member 9 (ABCC9) can be matched with potassium channel proteins Kir6.1 (KCNJ8) or Kir6.2 (KCNJ11) to assemble ATP sensitive K+ channels (KATP) in the heart, pancreaticislets, skeletal muscle and smooth muscle (5). The KATP channel is controlled by G proteins and allows potassium to flow into the cell. Previous studies have found that blocking the activity of KATP channels can significantly inhibit the proliferation of glioma and xenografted cells, inhibit the cell cycle at the G0/G1 phase, and induce apoptosis (6, 7). In contrast, the opening of KATP located on the mitochondrial membrane can attenuate cell apoptosis by maintaining the mitochondrial membrane potential (8).
As short tandem repeat DNA motifs (1–6 bp), microsatellites (MS) are ubiquitous in the eukaryotic genome, and the mutational rate of insertions/deletions in MS sequences is 10–100 times higher than that of traditional gene coding sequences. In 1993, cancer geneticists first discovered loss of heterozygosity (LOH) and microsatellite instability (MSI) in colorectal tumor tissues as a result of DNA mismatch-repair pathway obstruction, revealing a new pathway for oncogenesis (9). A previous study revealed that MSI is associated with clinical and pathological features in tumor tissues (10). Patients with the MSI-positive phenotype have a more robust T lymphocyte response than microsatellite-stable (MSS) cancer patients (11, 12). In addition, recent studies have shown that the diagnosis of MSI is tissue-specific, with varying frequency and prognostic values across multiple cancer types (13–15).
Mammary tumors as the common disease in female dogs. The MSI in canine mammary tumors (CMTs) has not been well-studied. Therefore, the aim of this trial was to investigated the relationship between MSI and tumor formation by screening MS loci in CMTs.
Methods
Material Collection and Histopathology Examinations
Twenty-two CMTs from different breeds of female dogs were provided by the Teaching Hospital of Nanjing Agricultural University. Procedures were approved by the Animal Ethics Committee of Nanjing Agricultural University (NJAU - 20171019, 10 October 2017). Experiment operates were performed under the Guidelines for Care and Use of Laboratory Animals of Jiangsu province (SYXK2017 - 0027). The mean age of the 22 canine patients was 9.77 ± 0.50 years, and the main breed was poodles (7/22, 31.8%). The adjacent normal and mammary gland tumors were excised; half of the samples were fixed in 10% formalin solution, and the remaining samples were stored at −80°C for further DNA and RNA extraction. The fixed samples were processed in a series of graded ethanol solutions and cleared with xylene. The samples were then embedded in paraffin, sectioned at 4 μm thickness, and stained with hematoxylin and eosin. Each stained tumor and its matched non-neoplastic tissue were examined using light microscopy.
DNA Extraction and Microsatellite Locus Identification
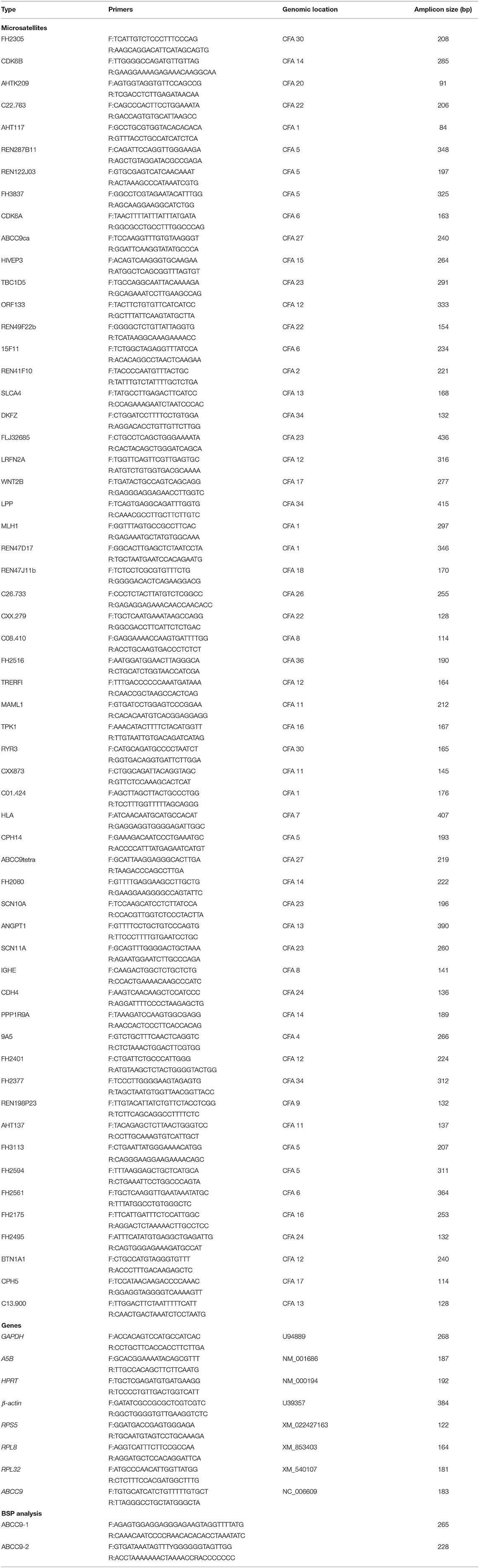
DNA was isolated using the Animal Tissues/Cells Genomic DNA Extraction Kit (Solarbio Science & Technology Co., Beijing, China). Based on the instructions, 25 mg of tissue sample was used. The concentration and purity were estimated using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The polymerase chain reaction (PCR) was performed using 500 ng of total DNA and TaKaRa Premix Taq™ according to the manufacturer's recommendations (Takara Co., Otsu, Japan). Genomic microsatellite loci were identified as described in our previous study (16), Table 1 shows the 58 pairs of primers used in this research. The cycle conditions were as follows: an initial incubation of 94°C for 5 min followed by 30 cycles of 30 s at 94°C, 30 s at their Tm (56–60°C), 30 s at 72°C, and finally extension at 72°C for 10 min. PCR amplified fragments were separated by 10% denatured polyacrylamide gel electrophoresis for 8 h at 100 V; mutations were observed by silver staining.
MSI was defined as addition or deletion of fragments to one or both tumor DNA alleles compared with normal tissues; LOH was defined as a reduction in the DNA signal intensity of tumor allele at least 50% (17). Positive cases were repeated three times to confirm the results. For MSI identification, mutation products were purified and cloned into the pMD19-T vector (Takara Co., Otsu, Japan) and sequencing. Sequence alignments were conducted using DNAMAN software v9.0.1.
RNA Extraction and mRNA Analysis
Total RNA was isolated using the Total RNA Extraction Kit (Solarbio Science & Technology Co., Beijing, China). Based on the instructions, 100 mg of tissue sample was used. The size of the RNA samples was estimated using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States). Reverse transcription was performed using 800 ng of total RNA treated with DNase I and PrimeScript RT Master Mix Perfect Real Time according to the manufacturer's instructions (Takara Co., Otsu, Japan).
The mRNA levels of MSI-mutated adjacent genes were detected by quantitative real-time PCR (QRT-PCR). Target sequences were amplified using Green Fast qPCR Mix (Takara Co., Otsu, Japan) and analyzed with an ABI 7300 instrument (Applied Biosystems, Foster City, CA, USA). The primer information is presented in Table 1. The cycle conditions were as follows: 95°C for 15 s followed by 95°C for 5 s and 60°C for 31 s for 40 cycles. The specific of each gene primer was confirmed by melting curve performance and gel electrophoresis. Results were presented as CT mean values of three technique replicates.
Reference genes were evaluated using geNorm v3.5 and NormFinder v0.953. Finally, the geometric means of A5B, RPL8, GAPDH, RPL32, RPS5, β-actin, and HRPT were used for normalization.
DNA Methylation Analysis
DNA methylation was measured using next-generation sequencing based bisulfite sequencing PCR (18). First, DNA modification with sodium bisulfite of 6 canine breast cancers and matched samples was performed using an EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer's protocol. The sequence included 2,000 bp upstream of the ABCC9 transcription start site and 1,000 bp downstream (a total of 3 kb). Elution products were then used as templates for PCR amplification with 35 cycles using the KAPA 2G Robust HotStart PCR Kit (Kapa Biosystems, Wilmington, MA, USA). The primers for BSP were designed using online MethPrimer software (Table 1). The bisulfite sequencing PCR products of each sample were pooled equally, 5'-phosphorylated, 3'-dA-tailed and ligated to a barcoded adapter using T4 DNA ligase (Thermo Fisher Scientific, Foster city, CA, USA). The barcoded libraries were then prepared and sequenced on an Illumina platform. Using the clean sequencing reads directly aligned to the target sequences, Bsmap v2.73 software was used with the default parameters. Methylation level was defined as the fraction of “C” read counts in the total read counts of both “C” and “T” for each covered C site. According to the method of Lister (19), each methylation context calculates the probability mass function, and only those CGs covered by at least 200 reads in one sample were considered for testing.
Immunohistochemical Analysis
Tissue sections were taken from 22 CMTs along with adjacent controls after fixation in 4% paraformaldehyde, dehydration, and embedding in paraffin. The expression of ABCC9 (1:200, Affinity Biosciences Cat# DF9255) in the breast was examined using an SP immunohistochemistry kit (Sangon Biotech Co., Shanghai, China) according to the manufacturer's instructions. A semiquantitative determination was conducted with Image J software to detect protein expression. The immunohistochemical staining intensity was expressed in average optical density (AOD) units, AOD = integrated optical density (IOD)/Area; five fields were randomly selected in a blinded manner, counted for the signal density of tissue areas, and then statistically analyzed.
Data Analysis and Statistics
The statistical analyses were conducted with GraphPad Prism software version 8.0 and SPSS version 21.0. A genome map of microsatellite loci was constructed using the MapChart program. The comparison of results between MSI/LOH and tumor type and methylation data were performed using Fisher's exact test. The Mann–Whitney U-test was used to analyze vs. benign and breast cancer groups. The relative mRNA expression levels of ABCC9 in tumors and matched normal tissues were calculated using the 2−ΔΔCt method. The t-test was performed to compare the relative mRNA level and protein expression between the two groups. The results are presented as the mean ± SD. The statistical significance was set at p < 0.05 for all analyses.
Results
The Pathological Identification of CMTs
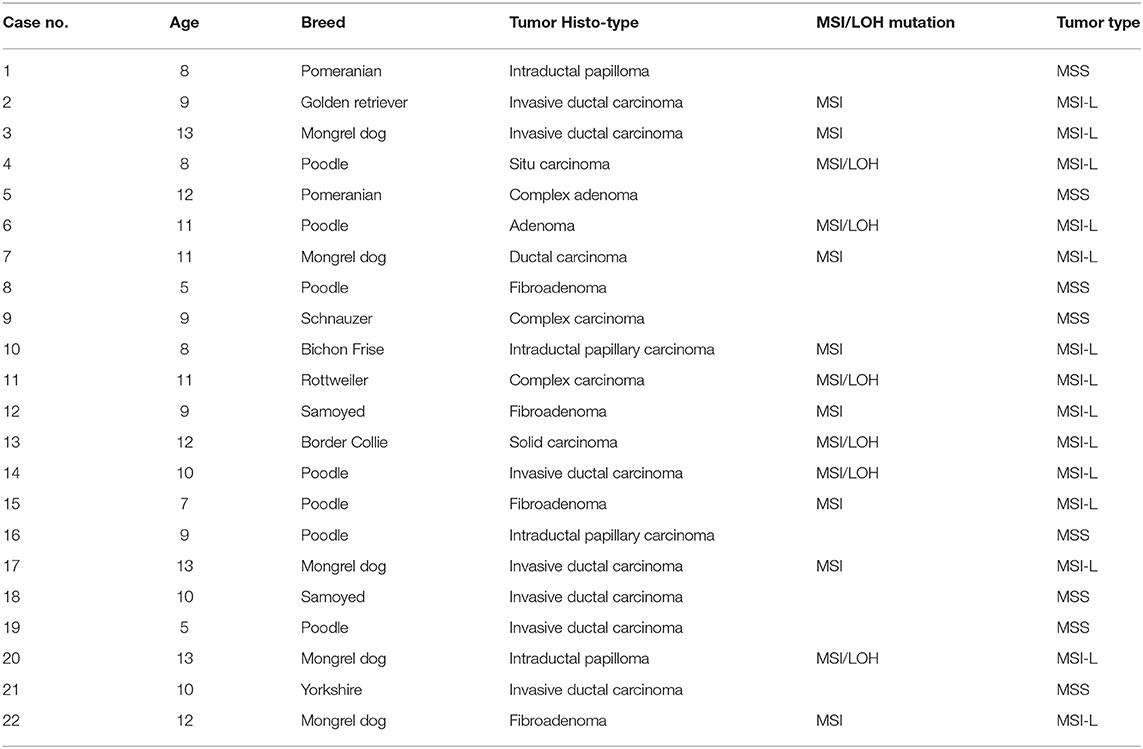
The 22 CMTs were classified as either benign (8/22, 36.4%) or malignant (14/22, 63.6%) (Figure 1). Based on the predominant cell type, the benign tumors were subclassified as fibroadenoma (4/8, 50%), complex adenoma (1/8, 12.5%), adenoma (1/8, 12.5%), or intraductal papilloma (2/8, 25%). The malignant tumors were subclassified into invasive ductal carcinoma (7/14, 50%), situ carcinoma (1/14, 7.1%), ductal carcinoma (1/14, 7.1%), complex carcinoma (2/14, 14.3%), intraductal papillary carcinoma (2/14, 14.3%), or solid carcinoma (1/14, 7.1%).
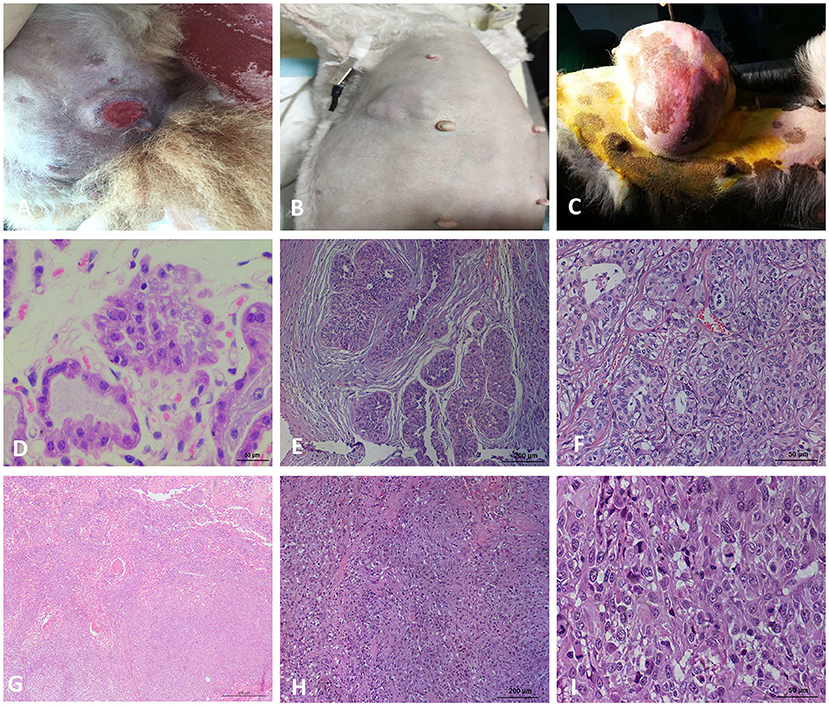
Figure 1. Macroscopic observation and HE staining of CMTs. (A) Macroscopic observation of CMT, the skin surface of the tumor ruptured. (B) Macroscopic observation of CMT, a cauliflow-like mass in mammary gland with obviously boundary and hard texture. (C) Macroscopic observation of CMT, the tumor located on the mammary tissue with a great size. (D) HE staining of the breast lobules in a normal dog (400×). (E) HE staining of mammary gland adenoma (200×), the capsule is intact and tumor cells grow in the enlarged lumen. (F) HE staining of mammary gland adenoma (400×), Adenoma arising in the glandular tissue, myoepithelial cells are inconspicuous, the islands of neoplastic cells are separated by a fine fibrovascular connective. (G) HE staining of solid carcinoma (200×). (H) HE staining for ductal carcinoma (200×), tumor cells invaded the connective tissue, glandular ducts were disappeared. (I) HE staining for ductal carcinoma (400×), tumor cells are pleomorphic and mitotic.
Malignant Tumors Have More MS Mutation Loci Than Benign Tumors
Using the panel of 58 MS markers, a LOH/MSI analysis between tumor tissues and their matched non-neoplastic tissues was carry out, the variation in the electropherogram of MS makers was described in Figure 2A. Differential bands were extracted and sequenced (Figure 2B). The sequencing result verified that the mutation form of MS in CMTs mainly included the insertion or deletion of nucleic acid fragments in repeated sequences. In addition, point mutations were also discovered in flank conserved sequences of MSI loci. Based on the National Cancer Institute guidelines (20), 14 tumors (14/22, 63.6%) were defined as MSI-L (MSI-Low), and 8 tumors were defined as MSS (MSI-Stable) (Table 2). Of the MSI-L tumors, 5 were diagnosed as benign tumors, and 9 were diagnosed as breast cancers. In addition, we found that the phenomenon of LOH was present in 6 MSI-L tumors (6/14, 42.9%), of which 2 tumors were benign and 4 tumors were malignant. There was no evidence of a difference in mutation rates between MSI and LOH in benign or malignant tumors (Fisher's test, P > 0.05). However, the histological type was significantly correlated with the number of MSI loci. Malignant tumors had more MS mutation loci than benign tumors (P < 0.05) (Figure 2C). Case 13 had the highest frequency of MSI (10/58, 17.2%) in this study, which was defined by pathological grading as grade III.
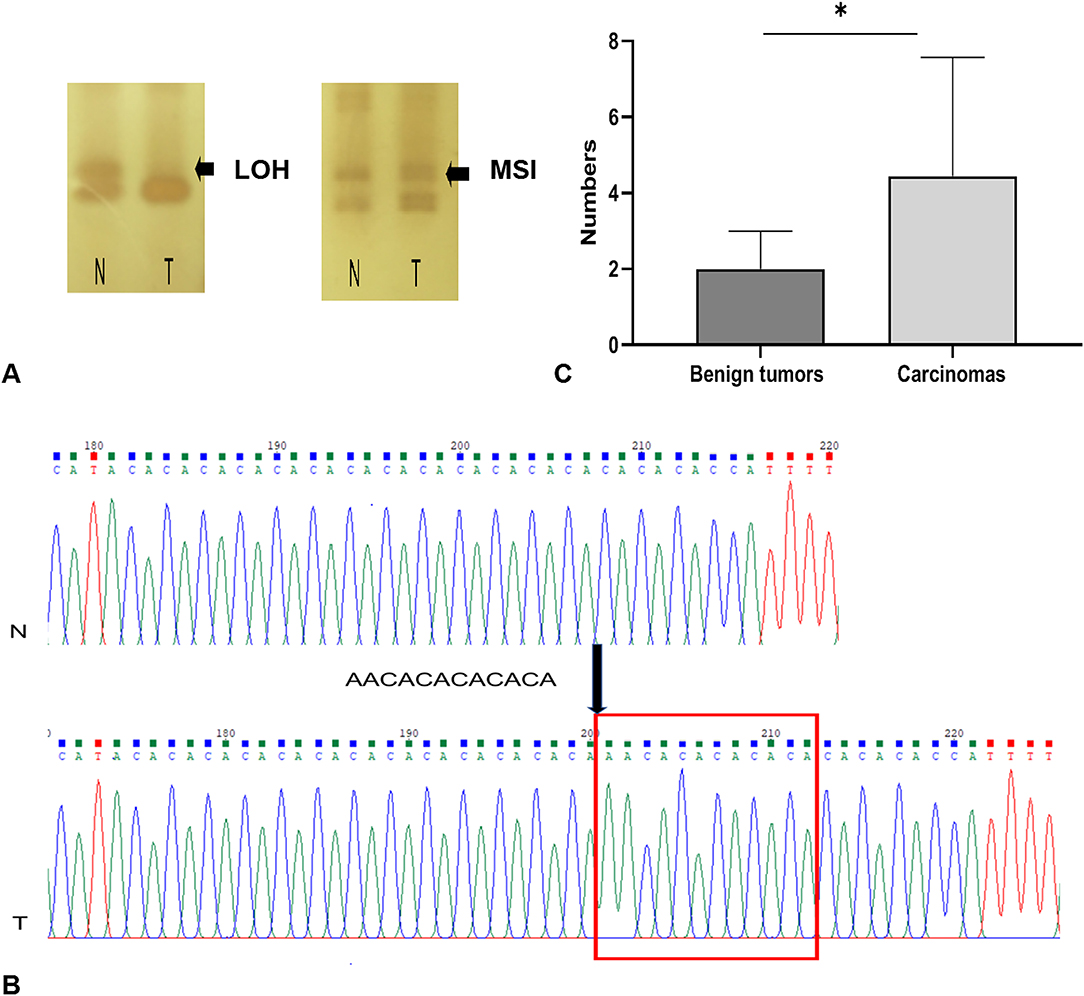
Figure 2. MSI and LOH occurring in CMTs. (A) MSI and LOH detection in denatured polyacrylamide gels. (B) The sequencing result of MSI locus, there was a repetitive fragment insertion in tumors. (C) Carcinomas have more mutation loci than benign tumors. *indicates a significant difference between the two groups, P < 0.05.
Tetranucleotide Microsatellites Are Prone to Instability in CMTs
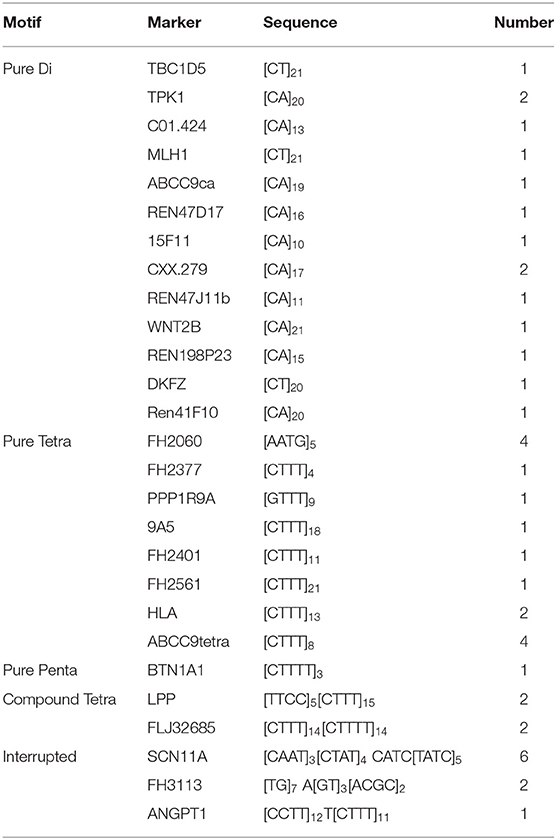
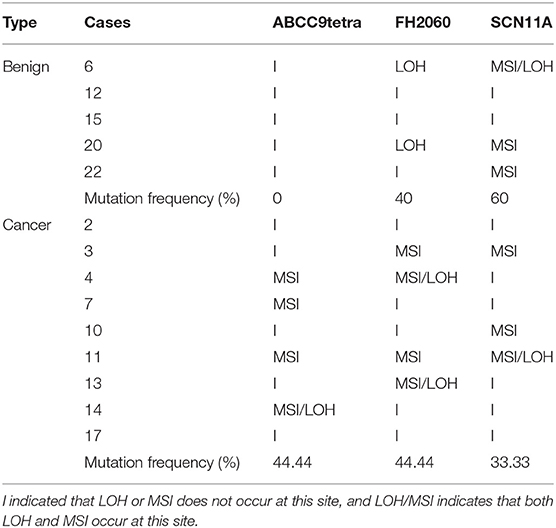
A total of 44 aberrations of MSI were found at 27 MS loci (27/58, 46.5%), which were distributed across 17 chromosomes (Figure 3). The classification of mutated MS markers in this study was shown in Table 3. In addition to dinucleotide [CA]n, tetranucleotide [CTTT]n and more complex types of microsatellite loci also has a high mutation frequency in this research. Among them, most of MS loci were only mutated once (1/14, 7.1%). The interrupted marker SCN11A (6/14, 42.9%) and tetranucleotide markers FH2060 (4/14, 28.6%) and ABCC9tetra (4/14, 28.6%) were loci with high mutation rate from the result. Moreover, the phenomenon of LOH was also observed on FH2060 (4/6, 66.75%), SCN11A (2/6, 33.3%), ABCC9tetra (1/6, 16.7%) and PPP1RA (1/6, 16.7%). Table 4 shows the mutation results for ABCC9tetra, FH2060 and SCN11A markers in CMTs. There were five tumor cases had at least two loci mutated as MSI or LOH for ABCC9tetra, FH2060 and SCN11A. Because of the locus of ABCC9tetra was only mutated in malignant group, the relationship between ABCC9tetra and breast cancer were studied.
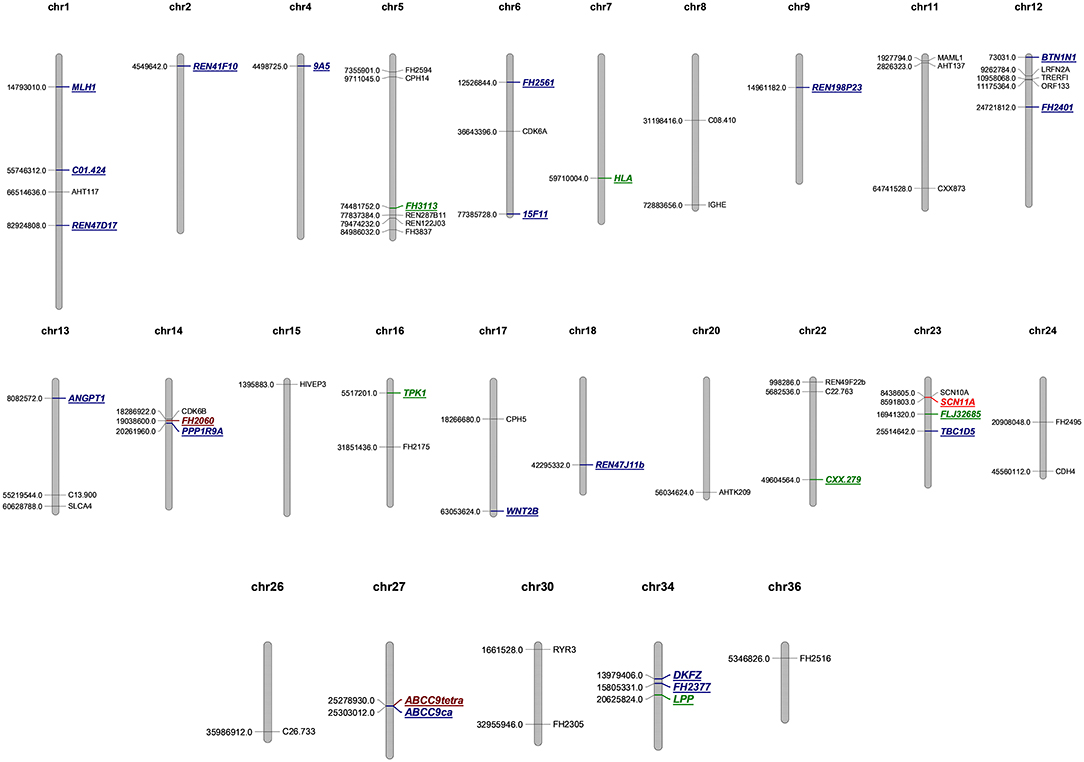
Figure 3. Genome map of microsatellite loci in this study. MSS are depicted in black, the single mutation of microsatellite loci is labeled blue, twice mutation is green, four times mutation is brown, and six is marked in red.
ABCC9 Is Downregulated in Canine Breast Cancer
NCBI revealed that ABCC9tetra was located in the intron region of ABCC9, and the mutation in this locus did not cause a frameshift mutation in open reading frame. But the result of QRT–PCR showed that the mRNA level of ABCC9 was significantly downregulated in the malignant group (P < 0.05) (Figure 4A). And the result of immunohistochemistry was similar to it. The AOD value showed that the expression of ABCC9 protein in malignant tumors was significantly lower than that in para-cancer tissues and benign tumors (P < 0.05) (Figure 4B). Strongly positive cells can be observed in normal and para-cancer tissues (Figure 4C) and even in benign tumors (Figure 4D). However, the number of ABCC9 positive cells was significant decreased in malignant tumors (Figure 4E). Moreover, the expression of ABCC9 protein may be related to the cellular composition and pathological grading. In the tumor sample of grade III, ABCC9 strongly positive cells almost disappeared, and were only weakly or micro-expressed in cells (Figure 4F). In addition, ABCC9 protein expression and mRNA levels were significantly reduced in tumor samples with ABCC9tetra locus instability (P < 0.05).
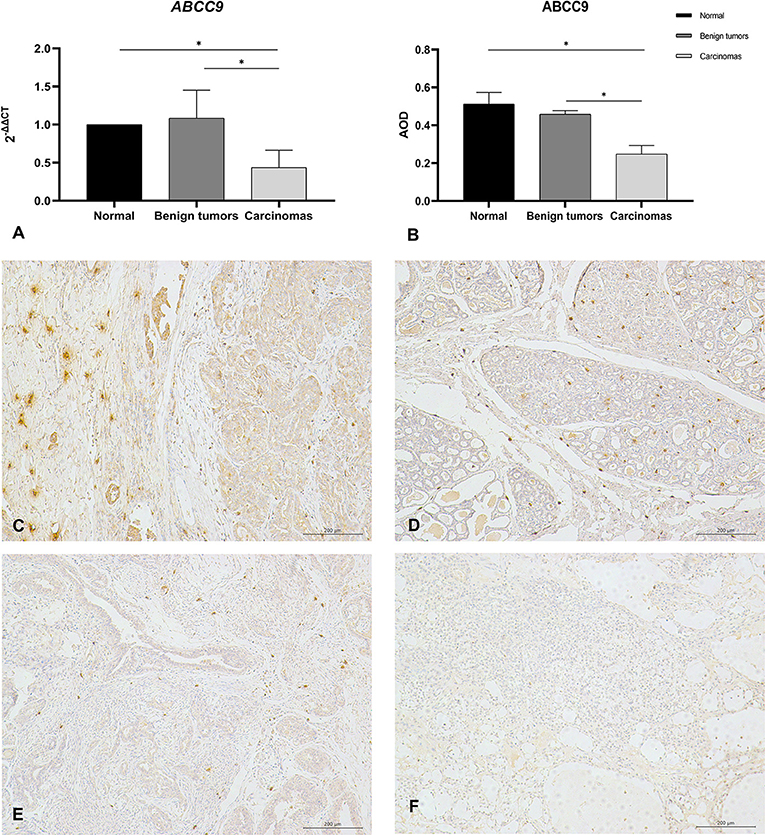
Figure 4. The mRNA level and protein expression of ABCC9. (A) The mRNA level of ABCC9, the mRNA level was expression by 2−ΔΔCt. (B) Average optical density values of ABCC9 protein in CMTs. AOD = IOD/Area, all of data are shown as means ± SD, *indicates that there is significant difference between two groups (P < 0.05). (C) IHC staining of ABCC9 in paracancer tissue (200×). (D) IHC staining of ABCC9 in benign tumor (200×). (E) IHC staining of ABCC9 in complex breast cancer (200×). (F) No strong positive staining of ABCC9 in higher malignancy cells (200×). Strongly positive cells can be observed in para-cancer tissues and benign tumors, but was significant decreased in malignant tumors.
Total of 6 tumor sample with ABCC9 mRNA levels significantly reduced were tested by methylation analysis. MathPrimer software detected a 703 bp CpG island in ABCC9 5′UTR (GC = 65.4%, and Obs/Exp ratio = 0.92). The methylation results of ABCC9 promoter CpG island revealed that high levels of methylation occurred at multiple sites in cancer tissues, but no new methylation sites were formed (Figure 5A). There was no significant difference in methylation level of each site (Figure 5C). And only one cancer sample showed significantly higher promoter methylation level than the control tissue (P < 0.05), with both MSI and LOH (Figure 5B).
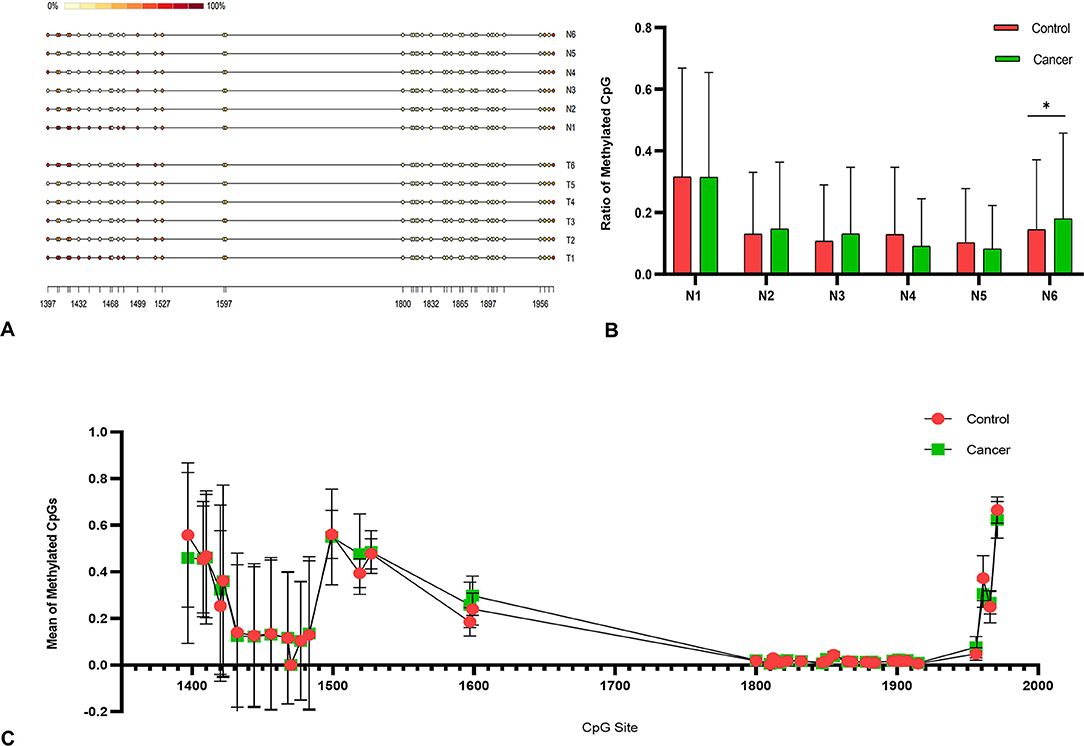
Figure 5. Methylation analysis of ABCC9 promoter. (A) Methylation analysis of ABCC9 CpG island. (B) Methylation CpG levels of six samples. (C) Methylation levels in average CpG sites. *indicates a significant difference between the two groups, P < 0.05.
Discussion
Genomic instability is a hallmark of tumors, and tumor tissue has a higher mutation rate than non-tumor tissue. Study showed that the sensitivity of MSI detection is not limited by tumor heterogeneity or normal tissue contamination when large resection tissues are used (21). The most endorsed explanation of MS mutagenesis is the slip strand mispairing model, and repeated numbers of motifs are highly polymorphic among individuals. A previous study of an MS mutation model showed that deletion is produced by the misalignment loop in the template chain, and insertion is subsequently produced in the nascent chain (22, 23). According to the sequence alignment analysis, we found that MS mutations mainly included the insertion or deletion of repeat sequences and point mutations of flanking conserved sequences. In addition, in the same MS locus, the forms of the mutations were differed among the samples. This phenomenon may be due to the mutation of MS occurring at different stages of tumor cell replication, whereas the point mutation may be caused by the suppression of mismatch repair genes. The length and unit type of MS and DNA shape are the main factors influencing DNA fragility and have the greatest influence on the mutation rate (24). In addition to dinucleotides, tetranucleotides and interrupted MS also showed frequent mutations in our research, which confirmed the susceptibility of the DNA structure to mutation.
The guidelines of the National Cancer Institute suggest that MS that display instability at ≥ 2 loci or instability at ≥ 30–40% of loci (more than five loci) be defined as MSI-High (MSI-H). If all tumor MS loci are comparable to their normal specimen, the tumor is classified as MSS. The range between MSS and MSI-H is defined as MSI-L (20). To date, tumors with an MSI-H frequency of 0% and tumors with MSI mutations all exhibited the MSI-L type, which is consistent with studies by Eldama'ria and Ando (17, 25). Work by Dustin showed that 800 loci are required to achieve diagnostic sensitivity and specificity for HBC, and diagnosis using predefined microsatellite locus panels is challenging (26). Overall, 31 MS loci were stable, and 27 MS loci exhibited MSI. Different cancer types exhibited distinct patterns of MS mutations. It appears that for breast tumors, the instability event may have a more neutral fitness effect, resulting in fewer recurrent mutation loci.
Although there was no significant difference in the frequency of MSI or LOH between benign and malignant tumors, malignant tumors had more MSI mutation loci than benign tumors. Of the 23 that we previously reported (4 benign and 19 malignant tumors), ABCC9tetra, FLJ32685, SCN11A and 9A5 loci showed a higher incidence of instability events in most canine breast cancers (16). In the present work, ABCC9tetra (4/22, 18.2%) and SCN11A (6/22, 27.3%) loci also exhibited higher mutation rates in CMTs. Our newly discovered high-frequency MSI locus, FH2060 (4/22, 18.2%), also had the highest LOH frequency (4/22, 18.2%). This phenomenon is potentially caused by selective pressures in tumor evolution (14). Biological pressures are involved in the selection of MS mutations, and some specific MS may be subject to positive or negative selection through changes in gene expression or function that result in more malignant transformation such as proliferation and metastasis (27, 28). Furthermore, a previous study showed that LOH can confer a growth advantage in tumor cells, and the tumor suppressor genes BRCA1 and BRCA2 loci are frequently altered due to allelic imbalance during carcinogenesis in the breast (29). Therefore, we suspected that the MSI locus was involved in the formation of breast tumors and began to explore the genes adjacent to the MSI locus.
Cancer genome sequencing has revealed that regional autosomal differential mutation rates at megabase resolution are related to changes in the timing of DNA replication or in gene expression and are less correlated with cancer type (30). The effect of DNA damage on highly expressed genes is limited to the MS within a specific gene in a specific tissue. In our results, ABCC9tetra, an MS locus, was mutated only in malignant tumors. The expression of adjacent gene ABCC9 was significantly decreased in the malignant tumor group. It is worth noting that the ABCC9 protein is involved in bioelectric control. ABCC9 can couples with potassium channel proteins KCNJ8 or KCNJ11 to form the KATP channel. The KATP channels were located on cell membranes and mitochondrial membranes. Past studies have shown that the channels formed by different combinations of KCNJ8, KCNJ11, ABCC8, and ABCC9 vary based on tissue localization (31). Immunohistochemistry reflected that ABCC9 was overexpressed in both normal and paracancerous tissues and in benign tumors, indicating that it is involved in the assembly of the KATP channel in the breast.
The ionic concentrations of Na+, K+, Ca2+, and Cl− are regulated by ion channels. In this study, ABCC9 on cell membranes and the mRNA level of ABCC9 were significantly decreased in malignant tissues. Furthermore, a negative correlation was observed between ABCC9 expression and cancer grading, with positive cells basically disappearing in cancer samples of grade III. This relationship may be due to the inhibition of the KATP channel in cancer tissue. The cytoplasm of depolarized cells is more positively charged relative to the extracellular space and has a less negative Vmem (32). Inhibition of potassium influx can lead to continuous depolarization of cells, which can induce mitosis and promote the proliferation of cancer cells (33, 34). Furthermore, a study of cardiac ischemia-reperfusion injuries revealed that the opening of mitoKATP channels could inhibit the depolarization of the mitochondrial membrane and protect against apoptosis in its early stages (35).
In addition, many studies have shown that ABCC9 can be used as a biomarker for cancers. The enrichment analysis of gastric cancer found that ABCC9 was involved in ATPase activity, transmembrane transport, and ABC transporters (36). Another study on the methylation pattern of breast cancer revealed that ABCC9 is a potential grade III biomarker of breast cancer in white individuals. However, in our study, only one case of cancer showed a significant increase in promoter CpG islands, which could not explain the reduced gene expression.
In conclusion, CMT is a highly heterogeneous disease with multiple genetic and epigenetic alterations. Malignant tumors have more unstable loci than benign tumors, which may be related to altered gene expression. ABCC9 is significantly downregulated in breast cancer and ABCC9tetra is particularly prone to mutation. In the future, additional studies on the regulation of ABCC9 protein in cancer cells are needed.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Nanjing Agricultural University (NJAU-20171019, 10 October 2017). Experiment operates were performed under the Guidelines for Care and Use of Laboratory Animals of Jiangsu province (SYXK2017-0027). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
PH and DWY: conceptualization. PH: methodology, software, formal analysis, resources, data curation, and writing-original draft preparation. SQW and XJH: validation. KYS: investigation. DWY: writing-review, editing and visualization. DJY: supervision, project administration, and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
Natural Science Foundation of Jiangsu Province (BK20130686), National Natural Science Foundation of China (30871847), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.819293/full#supplementary-material
References
1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
2. Ko JH, Ko EA, Gu W, Lim I, Bang H, Zhou T. Expression profiling of ion channel genes predicts clinical outcome in breast cancer. Mol Cancer. (2013) 12:106. doi: 10.1186/1476-4598-12-106
3. Prevarskaya N, Skryma R, Shuba Y. Ion channels and the hallmarks of cancer. Trends Mol Med. (2010) 16:107–21. doi: 10.1016/j.molmed.2010.01.005
4. Tuszynski J, Tilli TM, Levin M. Ion channel and neurotransmitter modulators as electroceutical approaches to the control of cancer. Curr Pharm Des. (2017) 23:4827–41. doi: 10.2174/1381612823666170530105837
5. Olson TM, Terzic A. Human K(ATP) channelopathies: diseases of metabolic homeostasis. Pflugers Arch. (2010) 460:295–306. doi: 10.1007/s00424-009-0771-y
6. Ru Q, Tian X, Wu YX, Wu RH, Pi MS, Li CY. Voltage-gated and ATP-sensitive K+ channels are associated with cell proliferation and tumorigenesis of human glioma. Oncol Rep. (2014) 31:842–8. doi: 10.3892/or.2013.2875
7. Huang L, Li B, Li W, Guo H, Zou F. ATP-sensitive potassium channels control glioma cells proliferation by regulating ERK activity. Carcinogenesis. (2009) 30:737–44. doi: 10.1093/carcin/bgp034
8. Liu X, Sun K, Song A, Zhang X, Zhang X, He X. Curcumin inhibits proliferation of gastric cancer cells by impairing ATP-sensitive potassium channel opening. World J Surg Oncol. (2014) 12:389. doi: 10.1186/1477-7819-12-389
9. Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. (1993) 260:816–9. doi: 10.1126/science.8484122
10. Patino GA, Isom LL. Electrophysiology and beyond: multiple roles of Na+ channel β subunits in development and disease. Neurosci Lett. (2010) 486:53–9. doi: 10.1016/j.neulet.2010.06.050
11. Mandal R, Samstein RM, Lee KW, Havel JJ, Wang H, Krishna C, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. (2019) 364:485–91. doi: 10.1126/science.aau0447
12. Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. (2017) 551:517–20. doi: 10.1038/nature24473
13. Baudrin LG, Deleuze JF, How-Kit A. Molecular and computational methods for the detection of microsatellite instability in cancer. Front Oncol. (2018) 8:621. doi: 10.3389/fonc.2018.00621
14. Hause RJ, Pritchard CC, Shendure J, Salipante SJ. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med. (2016) 22:1342–50. doi: 10.1038/nm.4191
15. Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, et al. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. (2006) 8:305–11. doi: 10.2353/jmoldx.2006.050092
16. Khand FM, Yao DW, Hao P, Wu XQ, Kamboh AA, Yang DJ. Microsatellite instability and MMR genes abnormalities in canine mammary gland tumors. Diagnostics (Basel). (2020) 10:104. doi: 10.3390/diagnostics10020104
17. de Vargas Wolfgramm E, Alves LN, Stur E, Tovar TT, De Nadai Sartori MP, de Castro Neto AK, et al. Analysis of genome instability in breast cancer. Mol Biol Rep. (2013) 40:2139–44. doi: 10.1007/s11033-012-2272-x
18. Pan X, Gong D, Nguyen DN, Zhang X, Hu Q, Lu H, et al. Early microbial colonization affects DNA methylation of genes related to intestinal immunity and metabolism in preterm pigs. DNA Res. (2018) 25:287–96. doi: 10.1093/dnares/dsy001
19. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. (2009) 462:315–22. doi: 10.1038/nature08514
20. Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A national cancer institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. (1998) 58:5248–57.
21. Danjoux M, Guimbaud R, Al Saati T, Meggetto F, Carrère N, Portier G, et al. Contribution of microdissection for the detection of microsatellite instability in colorectal cancer. Hum Pathol. (2006) 37:361–8. doi: 10.1016/j.humpath.2005.06.022
22. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. (2004) 5:435–45. doi: 10.1038/nrg1348
23. Maruvka YE, Mouw KW, Karlic R, Parasuraman P, Kamburov A, Polak P, et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat Biotechnol. (2017) 35:951–9. doi: 10.1038/nbt.3966
24. Fujimoto A, Fujita M, Hasegawa T, Wong JH, Maejima K, Oku-Sasaki A, et al. Comprehensive analysis of indels in whole-genome microsatellite regions and microsatellite instability across 21 cancer types. Genome Res. (2020) 30:334–46. doi: 10.1101/gr.255026.119
25. Ando Y, Iwase H, Ichihara S, Toyoshima S, Nakamura T, Yamashita H, et al. Loss of heterozygosity and microsatellite instability in ductal carcinoma in situ of the breast. Cancer Letters. (2000) 156:207–14. doi: 10.1016/s0304-3835(00)00467-5
26. Long DR, Waalkes A, Panicker VP, Hause RJ, Salipante SJ. Identifying optimal loci for the molecular diagnosis of microsatellite instability. Clin Chem. (2020) 66:1310–8. doi: 10.1093/clinchem/hvaa177
27. Chen H, Maxwell C, Connell M. The generation, detection, and prevention of genomic instability during cancer progression and metastasis. Cancer Metastasis. (2015) 20:15–38.
28. Charames GS, Bapat B. Genomic instability and cancer. Curr Mol Med. (2003) 3:589–96. doi: 10.2174/1566524033479456
29. Maxwell KN, Wubbenhorst B, Wenz BM, De Sloover D, Pluta J, Emery L, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun. (2017) 8:319. doi: 10.1038/s41467-017-00388-9
30. Supek F, Lehner B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature. (2015) 521:81–4. doi: 10.1038/nature14173
31. Akrouh A, Halcomb SE, Nichols CG, Sala-Rabanal M. Molecular biology of K(ATP) channels and implications for health and disease. IUBMB Life. (2009) 61:971–8. doi: 10.1002/iub.246
32. Blackiston DJ, McLaughlin KA, Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. (2009) 8:3527–36. doi: 10.4161/cc.8.21.9888
33. Fukushiro-Lopes DF, Hegel AD, Rao V, Wyatt D, Baker A, Breuer EK, et al. Preclinical study of a Kv11.1 potassium channel activator as antineoplastic approach for breast cancer. Oncotarget. (2018) 9:3321–37. doi: 10.18632/oncotarget.22925
34. Zhang P, Yang X, Yin Q, Yi J, Shen W, Zhao L, et al. Inhibition of SK4 potassium channels suppresses cell proliferation, migration and the epithelial-mesenchymal transition in triple-negative breast cancer cells. PLoS ONE. (2016) 11:e0154471. doi: 10.1371/journal.pone.0154471
35. Jia D. The protective effect of mitochondrial ATP-sensitive K+ channel opener, nicorandil, combined with Na+/Ca2+ exchange blocker KB-R7943 on myocardial ischemia-reperfusion injury in rat. Cell Biochem Biophys. (2011) 60:219–24. doi: 10.1007/s12013-010-9142-8
Keywords: microsatellite instability, canine breast cancer, oncogenesis, the adenosine triphosphate binding cassette subfamily C member 9, loss of heterozygosity
Citation: Hao P, Song KY, Wang SQ, Huang XJ, Yao DW and Yang DJ (2022) ABCC9 Is Downregulated and Prone to Microsatellite Instability on ABCC9tetra in Canine Breast Cancer. Front. Vet. Sci. 8:819293. doi: 10.3389/fvets.2021.819293
Received: 21 November 2021; Accepted: 06 December 2021;
Published: 07 January 2022.
Edited by:
Hui Zhang, South China Agricultural University, ChinaReviewed by:
Meng-yao Guo, Northeast Agricultural University, ChinaXiaobing Li, Yunnan Agricultural University, China
Copyright © 2022 Hao, Song, Wang, Huang, Yao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Da-wei Yao, yaodawei@njau.edu.cn; De-ji Yang, djyang@njau.edu.cn
 Pan Hao
Pan Hao  Da-wei Yao
Da-wei Yao De-ji Yang
De-ji Yang