A comprehensive review of small regulatory RNAs in Brucella spp.
- Center for One Health Research, Department of Biomedical Sciences and Pathobiology, VA-MD College of Veterinary Medicine, Virginia Tech, Blacksburg, VA, United States
Brucella spp. are Gram-negative bacteria that naturally infect a variety of domesticated and wild animals, often resulting in abortions and sterility. Humans exposed to these animals or animal products can also develop debilitating, flu-like disease. The brucellae are intracellular pathogens that reside predominantly within immune cells, typically macrophages, where they replicate in a specialized compartment. This capacity of Brucella to survive and replicate within macrophages is essential to their ability to cause disease. In recent years, several groups have identified and characterized small regulatory RNAs (sRNAs) as critical factors in the control of Brucella physiology within macrophages and overall disease virulence. sRNAs are generally <300 nucleotides in length, and these independent sRNA transcripts are encoded either next to (i.e., cis-encoded) or at a distant location to (i.e., trans-encoded) the genes that they regulate. Trans-encoded sRNAs interact with the mRNA transcripts through short stretches of imperfect base pairing that often require the RNA chaperone Hfq to facilitate sRNA-mRNA interaction. In many instances, these sRNA-mRNA interactions inhibit gene expression, usually by occluding the ribosome-binding site (RBS) and/or by decreasing the stability of the mRNA, leading to degradation of the transcript. A number of sRNAs have been predicted and authenticated in Brucella strains, and a variety of approaches, techniques, and means of validation have been employed in these efforts. Nonetheless, some important issues and considerations regarding the study of sRNA regulation in Brucella need to be addressed. For example, the lack of uniform sRNA nomenclature in Brucella has led to difficulty in comparisons of sRNAs across the different Brucella species, and there exist multiple names in the literature for what are functionally the same sRNA. Moreover, even though bona fide sRNAs have been discovered in Brucella, scant functional information is known about the regulatory activities of these sRNAs, or the extent to which these sRNAs are required for the intracellular life and/or host colonization by the brucellae. Therefore, this review summarizes the historical context of Hfq and sRNAs in Brucella; our current understanding of Brucella sRNAs; and some future perspectives and considerations for the field of sRNA biology in the brucellae.
Review
Brucella species naturally infect a wide range of wild and domesticated animals, including cattle, goats, and swine. In these animals, infection results in the disease brucellosis, which is often characterized by reproductive pathologies, such as abortions and sterility (1). Brucella species are additionally zoonotic pathogens, as they are efficiently transmitted from animals to humans, and the resulting infections are characterized by a debilitating, flu-like illness that often presents with a characteristic relapsing fever (2). While rare, complications of chronic human brucellosis can be fatal due to infective endocarditis. More common, non-lethal symptoms, such as arthritis and neurological symptoms, also contribute to the morbidity of humans suffering from chronic brucellosis (3). Additionally, given their small size, low infectious dose, and easy aerosolization, some Brucella species are considered potential agents of biological weaponry (4).
As the brucellae are intracellular pathogens that replicate in specialized niches within mesenchymal cells, they must carefully coordinate expression of genes encoding numerous metabolic, virulence-associated, and cell division proteins to successfully persist within their hosts (5). Despite the importance of Brucella temporo-spatially regulating transcription and translation in order to replicate and result in virulent brucellosis, small regulatory RNAs (sRNAs) remain poorly characterized within the brucellae. Here, we review the identified and confirmed sRNAs within Brucella, discuss their known or speculated connection with development of virulent brucellosis, and outline a few of the critical questions that remain to be addressed regarding sRNAs and Brucella.
What are small RNAs?
Bacterial small regulatory RNAs (sRNAs) are single-stranded RNA molecules that range between 50 and 300 nucleotides long. sRNAs are conceptually divided into two distinct classes, cis-encoded and trans-encoded, based on their genetic location in relationship to their functional site (6). Cis-encoded sRNAs are encoded on the DNA strand immediately opposite of their regulatory target and engage in extensive RNA-RNA complementary base-pairing with the mRNA transcript of the regulated protein. These sRNA-mRNA interactions commonly serve to inhibit protein translation through occlusion of ribosome binding (7). Trans-encoded sRNAs are encoded in an area of the genome independent of their regulatory target(s), and are often located between protein-encoding open reading frames. Trans-encoded sRNAs typically act through short segments known as seed-regions that form in the secondary structure of the sRNA (8, 9). The exposed single strand of nucleotides in the seed region can then bind with mRNA target, resulting in the regulation. Such interactions between trans-encoded sRNAs and their mRNA targets are often mediated by the RNA chaperone protein Hfq, which is discussed more extensively below (10).
Trans-encoded sRNA regulation can either positively or negatively impact protein translation. Some trans-encoded sRNAs increase protein levels through stabilization of the mRNA transcript through inhibition of RNase activity resulting in less transcript turnover and more protein translation or by restructuring the mRNA transcript to reveal the ribosome binding site (11). Alternatively, sRNA binding can negatively impact gene expression. For instance, the sRNA-mRNA binding can create a double-stranded RNA molecule that is targeted for degradation by RNases, or restructures the mRNA transcript to occlude the ribosome binding site and thus inhibit translation (6). Regardless of the exact mechanism, in the archetypical scenario sRNAs act as a layer of post-transcriptional regulation to fine-tune protein translation.
Hfq: The riboregulator
First described as a host-factor of Escherichia coli required for the replication of the RNA Qβ phage, Hfq has proven to be an RNA binding protein deeply conserved across bacteria (12). Hfq is also considered a major virulence factor in many bacteria, as Δhfq mutants exhibit pleiotropic phenotypes, including delayed growth rate, small colony phenotypes, increased sensitivities to a variety of stresses, and decreased virulence (13–16). Structurally, Hfq is an Sm-like family protein that is amazingly found in all three domains of life, showcasing the evolutionary role this RNA chaperone encompasses (17–19). Hfq forms a homohexamer structure that reveals 3 binding faces: the distal face, the proximal face, and the outer ring or rim surface (20). Each binding face has specific affinity for properties of either sRNAs or mRNAs. The distal face of Hfq preferentially binds single stranded ARN or ARNN motifs, (R signifies a purine and N represents any base), commonly found on mRNA sequences. The proximal face has high affinity for RNA molecules enriched with Uracil residues, which are commonly found in Rho-independent terminator sequences of sRNAs, followed by stem loop structures (21). The rim or lateral side of Hfq was recently found to be another important interaction face that binds UA-rich RNA molecules through positively charged residues (22). Hfq facilitates RNA regulation by increasing local concentrations of sRNA and mRNA targets, and can induce structural changes to assist in sRNA binding (23). By spatially concentrating sRNA and mRNAs, Hfq facilitates the imperfect base-pairing and regulatory actions of the sRNA.
It is important to note that many of biophysical properties of Hfq have been determined from studies involving Gammaproteobacteria, such as Escherichia coli. Comparative studies of the Hfq of E. coli to that of Caulobacter crescentus, an alphaproteobacterium more closely related to Brucella, reveals significant biophysical differences between E. coli and C. crescentus in Hfq-RNA interactions at the C-terminal domain (24, 25). Hfq proteins in Brucella strains discussed in this review (i.e., B. abortus, B. melitensis, and B. suis) are 100% identical at the amino acid level, and interestingly, Hfq proteins in both C. crescentus and Brucella are approximately 20% shorter than the E. coli Hfq protein due to truncations in the C-terminal portion of the proteins (Figure 1). These differences in the C-terminal regions of Hfq in C. crescentus and E. coli are responsible for the distinctive binding properties of each protein, but to date, careful biophysical studies of the Brucella Hfq protein have not been performed (24, 25). Additionally, the Hfq protein of Brucella spp. has not yet been characterized by either crystallography or cryoelectron microscopy to allow molecular docking analysis. Given the amino acid conservation, we would tentatively suggest that the Brucella Hfq acts in a manner more similar to that of C. crescentus than E. coli, but this remains speculative until further data are available.

Figure 1. The protein sequence alignment of Hfq for Escherichia coli, Caulobacter crescentus and classical Brucella species. Amino acid differences between C. crescentus and Brucella are highlighted in yellow. Alignment was performed in the UniProt platform using the Clustal Omega algorithm (26, 27).
Nonetheless, the variability of Hfq biophysical binding characteristics and amino acid sequence differences underscore the evolutionary specialization of Hfq proteins to fine-tune bacterial regulatory networks to meet changing environments. This observation is borne out in the links between Hfq and virulence in numerous bacteria from across multiple orders, including Pseudomonas aeruginosa (13), Vibrio cholerae (15), Salmonella typhimurium (28), Francisella tularensis (29), Neisseria meningitidis (30), Yersinia pestis (14), and Yersinia pseudotuberculosis (31). Hfq is additionally important for the successful colonization of hosts in other Alphaproteobacteria, such as the plant symbiont Sinorhizobium (32) or plant pathogen Agrobacterium (33). Importantly, the link between Hfq and bacterial virulence was first described B. abortus, as a B. abortus Δhfq strain was shown to be highly attenuated in both cellular and animal models of infection (16).
sRNAs in Brucella
Brucella is an intracellular pathogen that resides within macrophages and dendric cells in a specialized compartment called the Brucella-containing vacuole (BCV) (34, 35). The BCV is trafficked through the macrophage where the bacteria are exposed to a variety of stresses and signals, and the brucellae must sense and adapt to these harsh conditions in order to survive and replicate within the host cells (5). Central to the adaptation of Brucella species to their intracellular environment is the use of sRNAs to quickly and efficiently alter gene expression and protein levels in order to cope with the onslaught of cellular defense mechanisms, as outlined in the following sections. Before proceeding to the discussion of sRNAs, it is important to note that the nomenclature and species status within the genus Brucella is fairly unique. The debates regarding Brucella nomenclature have been expertly reviewed elsewhere (36, 37). The three Brucella species examined in this review, B. abortus, B. melitensis, and B. suis, are approximately 97% similar at the nucleotide level and show a closed pangenome (38–40). As such, while we present and discuss the sRNAs in the particular species in which they were identified, it is highly likely that a sRNA found in one species is also produced in the other two species, and we will note specific cases where this cross-species conservation has been experimentally validated.
The first sRNAs confirmed in B. abortus were called AbcR1 and AbcR2 due to their homology to the Agrobacterium tumefaciens sRNAs, AbcR1 and AbcR2, which exhibit regulatory activities toward genes encoding of ABC-type transport systems (41). These sRNAs are conserved among many members of the Rhizobiales, which includes bacteria such as Agrobacterium spp., Sinorhizobium spp., and Brucella spp. (42). The AbcR sRNAs were identified in a study analyzing a B. abortus hfq deletion strain, and in that study, two proteins, BAB1_1794 and BAB2_0612, were shown to be highly over-produced when hfq was deleted (43). These two proteins are orthologs of the A. tumefaciens proteins Atu2422 and Atu1879, respectively, and it had been reported that the production of Atu2422 and Atu1879 was regulated by AbcR1 and AbcR2 in A. tumefaciens (41). Bioinformatic analyses revealed that potential AbcR1 and AbcR2 orthologs were encoded in the B. abortus genome, and subsequent northern blot analyses determined that bona fide AbcR sRNAs were indeed produced by B. abortus (43). Importantly, the AbcR sRNAs are fully conserved in B. abortus 2308, B. melitensis 16M, and B. suis 1330, as northern blot analyses demonstrated that all three strains produce AbcR1 and AbcR2. Mutational analyses revealed that AbcR1 and AbcR2 are together essential for Brucella's virulence in macrophages and in a mouse model of infection. Deletion of either abcR gene individually did not alter the virulence of B. abortus, but deletion of both abcR1 and abcR2 resulted in significant attenuation in both macrophages and mice. These data indicated that AbcR1 and AbcR2 perform redundant functions. In order to identify the regulatory targets of the AbcR sRNAs in B. abortus, transcriptomic and proteomic analyses were performed, and these experiments demonstrated that 25 transcripts were over-expressed and 16 proteins over-produced in the abcR1 abcR2 deletion strain compared to the parental strain. It was shown that deletion of abcR1 and abcR2 leads to increased stability of target mRNAs, indicating that the AbcR sRNAs facilitate the degradation of these mRNAs. Of note, the genes exhibiting dysregulation in the abcR1 abcR2 mutant encode components of ABC-type transport systems, including BAB1_1794, BAB2_0612, and BAB2_0879. The periplasmic-binding proteins BAB1_1794 and BAB2_0879 are part of transport systems involved in the importation of the non-proteogenic amino acid GABA in B. abortus, but neither protein is required for the full virulence of B. abortus (44, 45). While the function of BAB2_0612 remains unknown, it is clear that BAB2_0612 is required for B. abortus virulence, as a bab2_0612 deletion strain is highly attenuated in a mouse model of infection (45).
Following the initial characterization of the Brucella AbcR sRNAs, a mechanistic study defined the specific elements of AbcR1 and AbcR2 that control the expression of specific subsets of genes, as well as virulence (45). It was shown that two distinct regulatory motifs, called M1 and M2, are present in both AbcR1 and AbcR2, but strikingly, the M2 motif is alone responsible for controlling the virulence-linked gene expression pathways of the AbcR sRNAs. The M1 and M2 seed regions, CUCCCA and GUUCCC, respectively, interact with complementary sequences in target mRNAs, leading to degradation of the mRNAs, and direct binding of AbcR1 and AbcR2 to the BAB1_0879 mRNA was experimentally validated. Interestingly, the M2-targeted BAB2_0612 mRNA, but not BAB1_0879, is required for the full virulence of B. abortus. As noted above, the physiological activity of BAB2_0612 is currently unknown, but it is clear that AbcR-regulated BAB2_0612 by means of the M2 seed region is required for B. abortus virulence. Regarding the M1 and M2 motifs, these regulatory regions are entirely conversed in the AbcR1 and AbcR2 sRNAs of B. abortus, B. melitensis and B. suis. While the AbcR sRNA regulatory circuit has been illuminated in recent years, there are still several important questions that remain about this important pathway. First, the identity of a transcriptional regulator of abcR1 expression has not been determined. Sheehan et al. identified the LysR-type transcriptional regulator, VtlR, as an activator of abcR2 expression, and VtlR is crucial for B. abortus virulence (56). However, the transcriptional regulator of abcR1 is not known. Is abcR1 constitutively expressed, or is abcR1 expression under the control of a specific regulator or regulators? Second, the RNase (or RNases) involved in AbcR-mediated degradation of target mRNAs has not been described. Specific RNases have been studied in Brucella strains, but to date, none of the studied RNases have been linked to AbcR regulatory events (53, 55). Finally, what are the dynamics of abcR expression during infection of macrophages and animals? This information will be extremely valuable for the complete elucidation of the mechanism(s) of AbcR-linked virulence in Brucella.
Since the initial discovery of sRNAs in Brucella, many bioinformatics prediction programs have been implemented to search for additional sRNAs. Many pipelines have predicted sRNAs in several different Brucella species. Although many of these findings remain as predictions, some sRNA candidates have been validated, but very few have been functionally characterized. Regarding prediction vs. authentication, this review focuses on Brucella sRNAs whose existence have been confirmed using northern blot analysis (i.e., the gold standard) or RT-PCR analysis, and a summary of bona fide Brucella sRNAs is reported in Table 1. While many of the prediction approaches and large-scale transcriptomic analyses provide vast lists of putative sRNAs, rigorous validation experiments are needed to substantiate the authenticity of these sRNAs.
Dong et al. (46) used the bioinformatics program SIPHT to search for putative rho-independent terminators within intergenic regions (i.e., known sRNA signature elements of many bacterial sRNAs), paired with the NAPP database (http://rssf.i2bc.paris-saclay.fr/NAPP/index.php), which predicts bacterial sRNA elements, to predict 129 sRNA candidates that were limited to intergenic regions (46, 57). These sRNA candidates were organized using a BASRC nomenclature for B. abortus sRNA candidate followed by chromosome number and candidate number. Of these 129 sRNA candidates, only 7 out of 20 were verified using RT-PCR. The regulatory targets of those 7 sRNAs were predicted using another bioinformatics software, sTarPicker (58). The predicted sRNA-target regulatory interactions were then tested using a two-plasmid lacZ reporter system in E. coli. This is a commonly used approach in the field that relies on a low-copy plasmid carrying a gene reporter (e.g., GFP, lacZ) that is fused to the regulatory region (e.g., 5'-untranslated region) of a putative sRNA-targeted gene, and a high-copy plasmid from which the gene encoding the sRNA of interest is expressed (reference to one prominent system used in the field: (59)). Using this type of system, the activity of the gene reporter can be monitored in the presence and absence of the sRNA to gain insight into the regulatory activity of a sRNA on a specific target mRNA. In the study by 44, the two-plasmid reporter system demonstrated that expression of BASRCI408, BASRCI385, BASRCI414, BASRCI153, or BASRCII26 led to reduced β-galactosidase activity when lacZ was fused to BAB1_2002, BAB1_0472, BAB1_0854, BAB1_1361, and BAB2_0187, respectively, indicating that these sRNAs negatively regulate their specific target genes. In the end, further investigation on the regulatory mechanism of these sRNAs is needed to gain insight into the regulatory activities these sRNAs, and importantly, more work is needed to understand the role of these sRNAs in Brucella physiology and virulence.
The Dong et al. (46) study led to the further characterization of one sRNA candidate that, after further characterization, was named BsrH for Brucella sRNA regulating HemH (47). BsrH is a cis-encoded sRNA antisense of hemH (bab2_0075 / bab_rs26720), which encodes ferrochelatase, an enzyme involved in heme biosynthesis. Importantly, HemH is critical for the full virulence of B. abortus in cellular and mouse models of infection (60). The production of BsrH was validated using RT-PCR, and BsrH levels are increased in response to acidic and oxidative stress. The regulation of HemH by BsrH was tested using a two-plasmid reporter system in E. coli, and these experiments demonstrated that over-expression BsrH reduced β-galactosidase activity of a hemH-lacZ fusion, indicating that BsrH negatively regulates hemH. However, no direct binding studies between BsrH and HemH were reported. Even though BsrH controls the expression of hemH, over-expression of BsrH had no impact on the virulence of B. abortus in vitro or in vivo. Interestingly, in many Gammaproteobacteria, iron homeostasis is coordinated by sRNAs (e.g., RhyB, PrrF), but to date, analogous iron-linked sRNAs have not been identified in the Alphaproteobacteria (61, 62). As such, BsrH is a very exciting example of a potential connection between iron homeostasis and sRNAs in the Alphaproteobacteria, and more work is warranted to define BsrH in Brucella, as well as BsrH orthologs in other closely related bacteria.
In the previously described study by Dong et al. (46), the authors described the identification of 129 putative sRNAs, but in that study, only 7 (of 20 assessed) of the sRNAs were authenticated by RT-PCR. In a subsequent study, the authors verified another 43 sRNAs (of 109 tested) via RT-PCR from (48). Over-expression of one of these sRNAs, named BASI74 led to decreased survival in a macrophage model of infection. The target prediction program, sTarPicker, was used to predict regulatory targets, of which four mRNAs exhibited elevated levels when BASI74 was over-expressed in the wild-type background, indicating that BASI74 positively regulates these four genes: (BAB1_0847: hypothetical protein; BAB1_1154: SAM-dependent methyltransferase; BAB1_1335: hypothetical protein; and BAB1_1361: hypothetical protein) (58). However, the levels of the target mRNAs were unaffected in the basI74 deletion strain. It is possible that BASI74 functions to control gene expression without interfering with the stead-sate levels of the mRNAs, as bacterial sRNAs can regulate the translation of mRNAs into proteins without affecting the stability of the mRNA (6). Overall, more work is needed to determine the regulatory and functional activities of BASI74 in B. abortus.
Wang et al. also employed another computational approach that limited the search to intergenic regions of the B. melitensis genome (49). Using RNAMotif and sRNAPredict programs to predict possible promoters and terminators within intergenic regions, 21 candidate Brucella sRNAs named BSR for Brucella small-noncoding sRNA followed by the gene number of the downstream protein-encoding gene were predicted (63, 64). Northern blot analyses were used to confirm 8 of the predicted sRNAs, and some of these confirmed sRNAs exhibited diminished levels in a B. melitensis hfq mutant, indicating that these sRNAs require Hfq for their stability. These sRNAs were: BSR0709, BSR0653, BSR1350, BSR0739, BSR1073, BSR0626, BSR0602, and BSR1141. One Hfq-dependent sRNA, BSR0602, showed the highest abundance during stationary phase of growth, and BSR0602 levels are elevated when the bacteria are exposed to acidic, oxidative, and heat stress. TargetRNA was used to predict targets of BSR0602, leading to the hypothesis that BSR0602 regulates the expression of BMEI0106, a global transcriptional regulator of the GntR family (65). qRT-PCR analyses revealed increased expression of BMEI0106 in a bsr0602 deletion strain, and expression was almost abolished in a bsr0602 overexpression strain. Additionally, a two-plasmid reporter system in E. coli confirmed the negative regulation of BMEI0106 by BSR0602, as well as demonstrated the motif within BSR0602 that binds to the bmeI0106 mRNA. This study determined that a bmeI0106 deletion strain is attenuated in a mouse model of infection, and while a bsr0602 deletion strain exhibited similar infectivity as the wild-type strain, a bsr0602 over-expression strain demonstrated decreased fitness compared to the wild-type strain in an in vivo competition experiment. These data indicate that the BSR0602 regulatory pathway is required for the full virulence of B. melitensis.
Wang et al. provided further characterization of BSR1141 in a subsequent report (50), and here, it was reported that BSR1141 expression is induced in response to stress conditions, including acidic and oxidative stress. In the same vein, a bsr1141 deletion strain exhibits reduced survival in both oxidative and low pH environments, and this phenotype is genetically complemented when bsr1141 is supplied in trans on a plasmid. Importantly, the bsr1141 deletion strain is attenuated in experimentally infected mice, indicating that BSR1141 is required for B. melitensis virulence. Using bioinformatic approaches coupled with qRT-PCR analysis, it was determined that BSR1141 is linked to the proper expression of more than 40 individual genes; however, it is not clear how many of these putative targets are directly controlled by BSR1141. Of interest, it appears that BSR1141 positively regulates the expression of virB2, which encodes part of the critical VirB type IV secretion system (66), and this regulatory event may, in part, explain the attenuation of the bsr1141 deletion strain. Indeed, mutation of virB2 leads to attenuation of Brucella strains in cellular and animal models of infection, and thus, the decrease in virB2 expression in the absence of BSR1141 may contribute to the decreased virulence of the bsr1141 deletion strain (67). Nonetheless, more work is needed to fully define the regulatory activity of BSR1141.
Saadeh et. al took an alternative approach to find sRNAs in B. suis (51). This group utilized a co-immunoprecipitation approach with a chromosomally epitope-tagged (i.e., 3xFLAG tag) version of the RNA chaperone Hfq as bait to enrich for sRNAs, and following precipitation of the RNAs, strand-specific cDNA library generation and deep sequencing was performed. Instead of using prediction programs, this group mapped the transcriptomic data to the B. suis genome, concentrating on intergenic regions of the chromosomes. This technique led to the discovery of 33 candidate Hfq-associated sRNAs, of which 10 were confirmed using northern blot analysis. Target prediction analyses using TargetRNA determined putative regulatory associations for many of these sRNAs, but no validation was performed to authenticate the sRNA-target regulatory links (65). Overall, this work identified several novel Hfq-associated sRNAs, and additionally work is needed to define: 1 the regulatory activity of the sRNAs; and 2 the role of the sRNA in the physiology of Brucella.
Another strand-specific deep-sequencing approach focused on intergenic regions was used to identify 1321 sRNA candidates in B. melitensis (52). One candidate, BSR0441 was chosen for further characterization due to its predicted targets involvement in virulence, and the expression of BSR0441 was confirmed by RT-PCR. Deletion of bsr0441 leads to decreased survival in murine macrophages at certain time points (i.e., 8-and 48-hours post-infection, but not 24-hours post-infection), indicating that BSR0441 may be important for the ability of B. melitensis to survive and replicate in macrophages. Regulatory targets of BSR0441 were predicted using TargetRNA2 (68); however, no direct assessment of BSR0441-target regulatory activity was performed. Rather, the expression of putative target mRNAs was analyzed during B. melitensis infection of macrophages and mice, and therefore, it is difficult to discern the impact of the sRNA on gene expression vs. the impact on the infection environment. In the end, further investigation is needed to define the regulatory mechanism of BSR0441.
In a study exploring RNaseIII, the investigators employed a high-throughput sequencing approach to identify 126 sRNAs in B. melitensis strain 027 (53). The goal of the study was to identify sRNAs that may be targeted by RNaseIII for regulatory events, and to this end, a representative sRNA, BM-sr00117, was confirmed via northern blot analysis. However, no further investigation involving this sRNA was performed. To further understand how BM-sr00117contributes to the biology of Brucella, more work is needed.
Xu et al. (54) predicted 14 sRNAs in B. melitensis M28 using two bioinformatic prediction programs, Softberry (http://www.softberry.com/) and Arnold (http://rssf.i2bc.paris-saclay.fr/toolbox/arnold/), that focused on conserved promoter and terminator elements within intergenic regions (54). Among these predicted sRNAs, seven were confirmed via RT-PCR and northern blot analyses, and these new sRNAs were named Bmsr for Brucella melitensis small RNA. One sRNA, Bmsr1 was studied further, and it was determined that deletion of bmsr1 leads to reduced intracellular survival in RAW264.7 cells compared to the parental B. melitensis strain. Moreover, the bmsr1 deletion strain was attenuated in a mouse model of infection, and taken together, these data indicate that Bmsr1 is important for the full virulence of B. melitensis. TargetRNA2 combined with iTRAQ proteomic analysis was used to develop of list of putative Bmsr1 regulatory targets, and interestingly, the bmsr1 deletion strain showed lower levels of several VirB type IV secretion system proteins, as well as a reduction in levels of VjbR, a LuxR-type regulator. Both the VirB system and VjbR are well-document virulence factors in Brucella strains, and thus, dysregulation of these important virulence determinants likely plays a role in the attenuation of the bmsr1 deletion strain (66, 69). While these findings are interesting and exciting, it is not clear how Bmsr1 is connected to the production of the VirB and VjbR proteins. Is this a direct regulatory event, or is this regulation indirect? Given the prominent roles played by the VirB system and VjbR, additional research is needed to characterize Bmsr1-mediated control of these systems.
Recent work investigating the endoribonuclease RNaseE uncovered determined that a sRNA named Bsr4, for Brucella small RNA, is dysregulated when RNaseE activity is compromised (55). Bsr4 had previously been identified in B. melitensis and in B. suis where it was called BSR1141 and BSnc118, respectively, and the authors designated the sRNA as Bsr4 as a means of clarifying the nomenclature of Brucella sRNAs (described in more detail in the following section) (49–51). Bsr4 was found to be highly abundant in a rnaseE-trunc mutant strain, indicating RNaseE aids in the degradation of Bsr4. Sheehan et al. (56) also confirmed that Bsr4 is dependent on the RNA chaperone Hfq for full stability, but bsr4 is dispensable for Brucella infection, as the bsr4 deletion strain exhibited no difference in the ability of the bacteria to survive and replicate in murine macrophages or to chronically infect mice. To date, the regulatory activity of Bsr4 remain uncharacterized.
Contribution of sRNAs to Brucella biology and future directions
Even though many sRNAs that have been predicted in Brucella spp., only a few have been validated and further investigated. An overview of specific Brucella sRNAs and their regulatory targets is depicted in Figure 2. Many of these sRNAs are required for virulence and regulate transcripts that are key virulence factors of Brucella, and thus, sRNAs heavily contribute to the adaption process once inside the macrophage. However, many questions are still left unanswered regarding the authenticity of predicted regulatory targets of these sRNAs, and to answer this, experimental approaches need to be employed to validate Brucella sRNA target mRNA predictions.
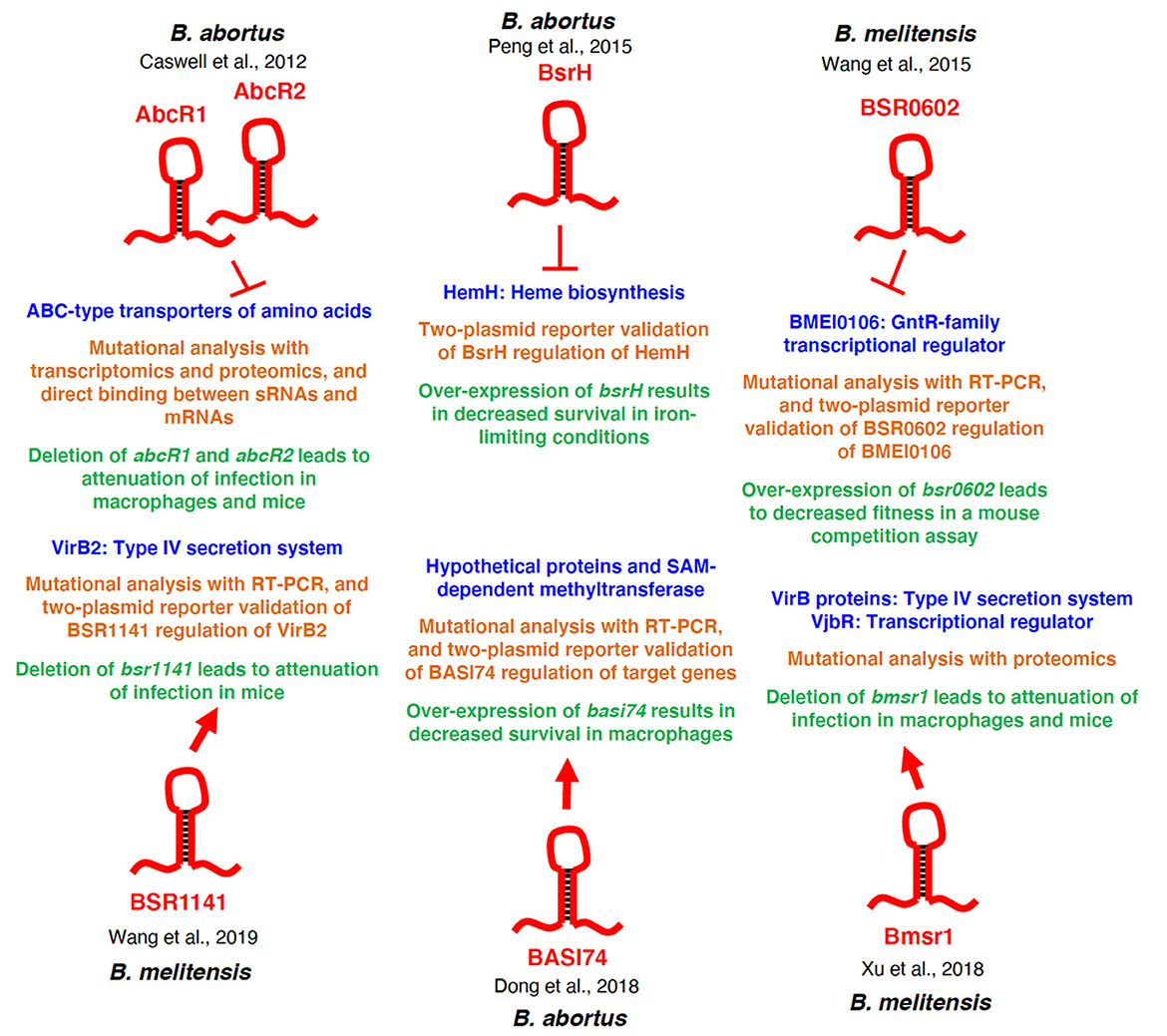
Figure 2. Regulatory overview of Brucella sRNAs. The specific sRNAs are depicted in red, and the regulatory targets/target systems are depicted in blue. The methods used to characterize the sRNA-target relationship are shown in orange, and the physiological consequences of deletion or over-expression of the sRNA is shown in green. Additionally, the Brucella species in which each sRNA was studied is indicated for each sRNA. Arrows indicate activation of expression, and perpendicular lines indicate repression of gene expression.
In addition to the characterization of sRNA-mRNA regulatory activities, very little is known about how expression of Brucella sRNAs is controlled. What transcriptional regulators are orchestrating sRNAs levels in Brucella? What of these transcriptional regulators and/or the sRNAs themselves sensing and responding to in order to appropriately coordinate gene expression during infection? What are the mechanisms and processes of sRNA turnover in the brucellae? These are fundamental and extremely exciting questions that remain to be answered.
As highlighted in the previous paragraphs, there is a wide variation in nomenclature used in the Brucella sRNA field. This is completely understandable given that we all try our best to organize and collate data in a useful manner, but the lab-to-lab nomenclature of Brucella sRNAs is almost untranslatable. This can be extremely confusing for other researchers, even those very intimately connected to the field. As such, our group has developed a “Rosetta Stone of Brucella sRNAs” that we will continue to maintain as new sRNAs are identified and characterize, and the database is freely available on our laboratory website: http://caswelllab.com/. Our proposal is that designations be assigned as Brucella sRNA or “Bsr” regardless of the species in which the sRNA is identified. Given the high degree of similarity among Brucella species, it is highly likely that one sRNA in one species is also produced by other species (38–40). Admittedly, there may be sRNAs identified that are species-specific, and this should be noted accordingly and thoroughly investigated, as these findings could have implications in the speciation of Brucella and/or the natural host preference of Brucella strains. Nonetheless, the field will greatly benefit from an organized and systematic approach t o Brucella sRNA nomenclature, and we hope that others will join us in organizing a more clear and interpretable approach to designating sRNAs.
In the end, Brucella sRNAs represent an open frontier of research, full of exciting and presumably highly specialized regulatory mechanisms, and thus, much remains to be elucidated about sRNAs-mediated pathways in the brucellae.
The orientation of the sRNA gene is denoted with a red arrow.
Gene locus tags have been provided, and these locus tags are for three Brucella reference strains, even if a specific sRNA was identified in another Brucella strain. The reference strains and the web links to their genome information is as follows: B. melitensis 16M (https://www.ncbi.nlm.nih.gov/genome/943?genome_assembly_id=300462), B. suis 1330 (https://www.ncbi.nlm.nih.gov/genome/806?genome_assembly_id=300358), and B. abortus 2308 (https://www.ncbi.nlm.nih.gov/genome/520?genome_assembly_id=167411).
Author contributions
KK and CC conceived the plan for the review. KK wrote the initial draft of the manuscript. MC and CC reviewed and edited the text. CC designed and constructed the figures. All authors contributed to the article and approved the submitted version.
Funding
The National Institute of Allergy and Infectious Diseases has supported Brucella small RNA research in the Caswell laboratory (AI117648 and AI149124). MC is supported by the Animal Model Research for Veterinarians Program (T32OD028239) at Virginia-Maryland College of Veterinary Medicine.
Acknowledgments
We would like to thank Tristan Stoyanof for critically evaluating the manuscript.
Conflict of interest
The authors declare that this work was carried out in the absence of commercial and financial relationships that could be seen as potential conflicts of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Olsen SC, Palmer MV. Advancement of knowledge of Brucella over the past 50 years. Vet Pathol. (2014) 51:1076–89. doi: 10.1177/0300985814540545
2. Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. The new global map of human brucellosis. Lancet Infect Dis. (2006) 6:91–9. doi: 10.1016/S1473-3099(06)70382-6
3. Dean AS, Crump L, Greter H, Hattendorf J, Schelling E, Zinsstag J, et al. Clinical manifestations of human brucellosis: a systematic review and meta-analysis. PLoS Negl Trop Dis. (2012) 6:e1929. doi: 10.1371/journal.pntd.0001929
4. Pappas G, Panagopoulou P, Christou L, Akritidis N. Brucella as a biological weapon. Cell Mol Life Sci. (2006) 63:2229–36. doi: 10.1007/s00018-006-6311-4
5. Roop RM 2nd, Barton IS, Hopersberger D, Martin DW. Uncovering the hidden credentials of Brucella virulence. Microbiol. Mol. Biol. Rev. (2021) 85:e00021–19. doi: 10.1128/MMBR.00021-19
6. Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. (2009) 57:1099–109. doi: 10.1016/j.cell.2009.01.043
7. Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. (2007) 10:134–9. doi: 10.1016/j.mib.2007.03.010
8. Balbontin R, Fiorini F, Figueroa-Bossi N, Casadesús J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol Microbiol. (2010) 78:380–94. doi: 10.1111/j.1365-2958.2010.07342.x
9. Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol. (2006) 61:1013–22. doi: 10.1111/j.1365-2958.2006.05288.x
10. Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. (2007) 10:125–33. doi: 10.1016/j.mib.2007.03.015
11. Storz G, Vogel J, Wassarman KM. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol Cell. (2011) 43:880–91. doi: 10.1016/j.molcel.2011.08.022
12. August JT, Eoyang L, Fernandez DE, Hasegawa MT, Kuo S, Rensing CH, et al. Phage-specific and host proteins in the replication of bacteriophage RNA. Fed Pro. (1970) 29:1170–5.
13. Sonnleither E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jäger KE, et al. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Pathog Microb. (2003) 35:217–28. doi: 10.1016/S0882-4010(03)00149-9
14. Geng J, Song Y, Yang L, Feng Y, Qiu Y, Li G, et al. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS ONE. (2009) 4:e6213. doi: 10.1371/journal.pone.0006213
15. Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol Microbiol. (2004) 53:345–54. doi: 10.1111/j.1365-2958.2004.04142.x
16. Robertson GT, Roop RM. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. (1999) 34:690–700. doi: 10.1046/j.1365-2958.1999.01629.x
17. Nielsen JS, Bøggild A, Andersen CB, Nielsen G, Boysen A, Brodersen DE, et al. An Hfq-like protein in archaea: crystal structure and functional characterization of the Sm protein from Methanococcus Jannaschii. RNA. (2007) 13:2213–23. doi: 10.1261/rna.689007
18. Reeves WH, Narain S, Satoh M. Henry Kunkel, Stephanie Smith, clinical immunology, and split genes. Lupus. (2003) 12:213–7. doi: 10.1191/0961203303lu360xx
19. Wilusz CJ, Wilusz J. Eukaryotic Lsm proteins: lessons from bacteria. Nat Struct Mol Biol. (2005) 12:1031–6. doi: 10.1038/nsmb1037
20. Park S, Prévost K, Heideman EM, Carrier MC, Azam MS, Reyer MA, et al. Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. Elife. (2021) 10:e64207. doi: 10.7554/eLife.64207
21. Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. (2011) 9:578–89. doi: 10.1038/nrmicro2615
22. Dimastrogiovanni D, Fröhlich KS, Bandyra KJ, Bruce HA, Hohensee S, Vogel J, Luisi BF. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife. (2014) 3:e05375. doi: 10.7554/eLife.05375
23. Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. (2011) 17:1544–50. doi: 10.1261/rna.2767211
24. Santiago-Frangos A, Woodson SA. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev RNA. (2018) 9:e1475. doi: 10.1002/wrna.1475
25. Cai H, Roca J, Zhao YF, Woodson SA. Dynamic refolding of OxyS sRNA by the Hfq RNA chaperone. J Mol Biol. (2022) 434:167776. doi: 10.1016/j.jmb.2022.167776
26. Sievers F, Higgins DG. Clustal omega for making accurate alignments of many protein sequences. Protein Sci. (2018) 27:135–45. doi: 10.1002/pro.3290
27. UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. (2021) 49:D480–D489.
28. Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. (2007) 63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x
29. Meibom KL, Forslund AL, Kuoppa K, Alkhuder K, Dubail I, Dupuis M, et al. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infect Immun. (2009) 77:1866–80. doi: 10.1128/IAI.01496-08
30. Fantappiè L, Metruccio MM, Seib KL, Oriente F, Cartocci E, Ferlicca F, et al. The RNA chaperone Hfq is involved in stress response and virulence in Neisseria meningitidis and is a pleiotropic regulator of protein expression. Infect Immun. (2009) 77:1842–53. doi: 10.1128/IAI.01216-08
31. Schiano CA, Bellows LE, Lathem WW. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect Immun. (2010) 78:2034–44. doi: 10.1128/IAI.01046-09
32. Gao M, Barnett MJ, Long SR, Teplitski M. Role of the Sinorhizobium meliloti global regulator Hfq in gene regulation and symbiosis. Mol Plant Microbe Interact. (2010) 23:355–65. doi: 10.1094/MPMI-23-4-0355
33. Moller P, Overlöper A, Förstner KU, Wen TN, Sharma CM, Lai EM, et al. Profound impact of Hfq on nutrient acquisition, metabolism and motility in the plant pathogen Agrobacterium tumefaciens. PLoS One. (2014) 9:e110427. doi: 10.1371/journal.pone.0110427
34. Celli, J., Chastellier, D. E., Franchini, C., Pizarro-Cerda, D. M., and Moreno, J. E., and Gorvel, J. P. (2003) Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198, 545–556. doi: 10.1084/jem.20030088
35. Pizarro-Cerda J, Méresse S, Parton RG, van der Goot G, Sola-Landa A, Lopez-Goñi I, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. (1998) 66:5711–24. doi: 10.1128/IAI.66.12.5711-5724.1998
36. Moreno E. The one hundred year journey of the genus Brucella (Meyer and Shaw 1920). FEMS Microbiol. Rev. (2021) 45:fuaa045. doi: 10.1093/femsre/fuaa045
37. Moreno E, Blasco JM, Letesson JJ, Gorvel JP, Moriyón I. Pathogenicity and its implications in taxonomy: the Brucella and Ochrobactrum case. Pathogens. (2022) 11:377. doi: 10.3390/pathogens11030377
38. Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H, et al. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivison of the class Proteobacteria. J Bacteriol. (1990) 172:3569–7356. doi: 10.1128/jb.172.7.3569-3576.1990
39. Verger JM, Grimont F, Grimont PA, Grayon M. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int J Syst Evo. (1985) 35:292–5. doi: 10.1099/00207713-35-3-292
40. Suárez-Esquivel M, Chaves-Olarte E, Moreno E, Guzmán-Verri C. Brucella genomics: macro and micro evolution. Int J Mol Sci. (2020) 21:7749. doi: 10.3390/ijms21207749
41. Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol. (2011) 80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x
42. Becker A, Overlöper A, Schlüter JP, Reinkensmeier J, Robledo M, Giegerich R, et al. Riboregulation in plant-associated α-proteobacteria. RNA Biol. (2014) 11:550–62. doi: 10.4161/rna.29625
43. Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, et al. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol. (2012) 85:345–60. doi: 10.1111/j.1365-2958.2012.08117.x
44. Budnick JA, Sheehan LM, Benton AH, Pitzer JE, Kang L, Michalak P, et al. Characterizing the transport and utilization of the neurotransmitter GABA in the bacterial pathogen Brucella abortus. PLoS ONE. (2020) 15:e0237371. doi: 10.1371/journal.pone.0237371
45. Sheehan LM, Caswell CC. A 6-Nucleotide Regulatory Motif within the AbcR Small RNAs of Brucella abortus Mediates Host-Pathogen Interactions. MBio. (2017) 8:e00473–17. doi: 10.1128/mBio.00473-17
46. Dong H, Peng X, Wang N, Wu Q. Identification of novel sRNAs in Brucella abortus 2308. FEMS Microbiol Lett. (2014) 354:119–25. doi: 10.1111/1574-6968.12433
47. Peng X, Dong H, Wu Q. A new cis-encoded sRNA, BsrH, regulating the expression of hemH gene in Brucella abortus 2308. FEMS Microbiol Lett. (2015) 362:1–7. doi: 10.1093/femsle/fnu017
48. Dong H, Peng X, Liu Y, Wu T, Wang X, De Y, et al. BASI74, a Virulence-Related sRNA in Brucella abortus. Front Microbiol. (2018) 9:2173. doi: 10.3389/fmicb.2018.02173
49. Wang Y, Ke Y, Xu J, Wang L, Wang T, Liang H, et al. Identification of a Novel Small Non-Coding RNA Modulating the Intracellular Survival of Brucella melitensis. Front Microbiol. (2015) 6:164. doi: 10.3389/fmicb.2015.00164
50. Wang Y, Ke Y, Duan C, Ma X, Hao Q, Song L, et al. A small non-coding RNA facilitates Brucella melitensis intracellular survival by regulating the expression of virulence factor. Int J Med Microbiol. (2019) 309:225–31. doi: 10.1016/j.ijmm.2019.04.002
51. Saadeh B, Caswell CC, Chao Y, Berta P, Wattam AR, Roop RM, et al. Transcriptome-Wide Identification of Hfq-Associated RNAs in Brucella suis by Deep Sequencing. J Bacteriol. (2016) 198:427–35. doi: 10.1128/JB.00711-15
52. Zhong Z, Xu X, Li X, Liu S, Lei S, Yang M, et al. Large-scale identification of small noncoding RNA with strand-specific deep sequencing and characterization of a novel virulence-related sRNA in Brucella melitensis. Sci Rep. (2016) 6:25123. doi: 10.1038/srep25123
53. Wu CX, Xu XJ, Zheng K, Liu F, Yang XD, Chen CF, et al. Characterization of ribonuclease III from Brucella. Gene. (2016) 579:183–92. doi: 10.1016/j.gene.2015.12.068
54. Xu D, Song J, Li G, Cai W, Zong S, Li Z, et al. A novel small RNA Bmsr1 enhances virulence in Brucella melitensis M28. Vet Microbiol. (2018) 223:1–8. doi: 10.1016/j.vetmic.2018.07.007
55. Sheehan LM, Budnick JA, Fyffe-Blair J, King KA, Settlage RE, Caswell CC, et al. The Endoribonuclease RNase E Coordinates Expression of mRNAs and Small Regulatory RNAs and Is Critical for the Virulence of Brucella abortus. J Bacteriol. (2020) 202:e00240–20. doi: 10.1128/JB.00240-20
56. Sheehan LM, Budnick JA, Blanchard C, Dunman PM, Caswell CC. A LysR-family transcriptional regulator required for virulence in Brucella abortus is highly conserved among the α-proteobacteria. Mol Microbiol. (2015) 98:318–28. doi: 10.1111/mmi.13123
57. Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE. (2008) 3:e3197. doi: 10.1371/journal.pone.0003197
58. Ying X, Cao Y, Wu J, Liu Q, Cha L, Li W, et al. sTarPicker: a method for efficient prediction of bacterial sRNA targets based on a two-step model for hybridization. PLoS ONE. (2011) 6:e22705. doi: 10.1371/journal.pone.0022705
59. Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. (2007) 35:1018–37. doi: 10.1093/nar/gkl1040
60. Almiron M, Martínez M, Sanjuan N, Ugalde RA. Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect Immun. (2001) 69:6225–30. doi: 10.1128/IAI.69.10.6225-6230.2001
61. Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. (2002) 99:4620–5. doi: 10.1073/pnas.032066599
62. Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA. (2004) 101:9792–7. doi: 10.1073/pnas.0403423101
63. Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ, et al. Prediction of rho-independent transcriptional terminators in Escherichia coil. Nucleic Acid Res. (2001) 29:3583–94. doi: 10.1093/nar/29.17.3583
64. Livny J, Fogel MA, Davis BM, Waldor MK. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res. (2005) 33:4096–105. doi: 10.1093/nar/gki715
65. Tjaden B. TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res. (2008) 36(wed server issue):W109-113. doi: 10.1093/nar/gkn264
66. Jong DE, Tsolis MF. Brucellosis and type IV secretion. Future Microbiol. (2012) 7:47–58. doi: 10.2217/fmb.11.136
67. Hartigh DEN, Sun AB, Sondervan YH, Heuvelmans D, Reinders N, Ficht MO, et al. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect Immun. (2004) 72:5143–9. doi: 10.1128/IAI.72.9.5143-5149.2004
68. Kery MB, Feldman M, Livny J, Tjaden B. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. (2014) 42(web server issue):W124–129. doi: 10.1093/nar/gku317
Keywords: Brucella species, small RNAs (sRNAs), virulence, intracellular bacteria, Alphaproteobacteria
Citation: King KA, Caudill MT and Caswell CC (2022) A comprehensive review of small regulatory RNAs in Brucella spp. Front. Vet. Sci. 9:1026220. doi: 10.3389/fvets.2022.1026220
Received: 23 August 2022; Accepted: 15 November 2022;
Published: 01 December 2022.
Edited by:
Subhash Verma, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaReviewed by:
Roy Martin Roop II, East Carolina University, United StatesDhriti Sinha, University of Texas Health Science Center at Houston, United States
Copyright © 2022 King, Caudill and Caswell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clayton C. Caswell, caswellc@vt.edu
 Kellie A. King
Kellie A. King Mitchell T. Caudill
Mitchell T. Caudill  Clayton C. Caswell
Clayton C. Caswell