A novel multiplex qPCR assay for clinical diagnosis of non-human malaria parasites-Plasmodium knowlesi and Plasmodium cynomolgi
- Parasite-Host Biology Group, ICMR–National Institute of Malaria Research, New Delhi, India
Introduction: The imminent risk of zoonoses of non-human malaria parasites is not far from reality in India, as has been observed in the case of Plasmodium knowlesi (Pk), and so is possible with P. cynomolgi (Pc), already reported from South East Asian countries. Therefore, a novel multiplex qPCR assay was developed and evaluated for detection of non-human malaria parasites- Pk and Pc in populations at risk.
Methods: The qPCR primers were designed in-house with fluorescence labeled probes (HEX for Pk and FAM for Pc). DNA samples of Pk and Pc were used as templates and further the qPCR assay was evaluated in 250 symptomatic and asymptomatic suspected human blood samples from malaria endemic areas of North Eastern states of India.
Results: The qPCR assay successfully amplified the target 18S rRNA gene segment from Pk and Pc and was highly specific for Pk and Pc parasites only, as no cross reactivity was observed with P. falciparum (Pf), P. vivax (Pv), P. malariae (Pm), and P. ovale (Po). Standard curves were generated to estimate the limit of detection (LOD) of Pk and Pc parasites DNA (0.00275 & 0.075 ng/μl, respectively). Due to COVID-19 pandemic situation during 2020–21, the sample accessibility was difficult, however, we managed to collect 250 samples. The samples were tested for Pf and Pv using conventional PCR- 14 Pf and 11 Pv infections were observed, but no Pk and Pc infections were detected. For Pk infections, previously reported conventional PCR was also performed, but no Pk infection was detected.
Discussion: The multiplex qPCR assay was observed to be robust, quick, cost-effective and highly sensitive as compared to the currently available conventional PCR methods. Further validation of the multiplex qPCR assay in field setting is desirable, especially from the high-risk populations. We anticipate that the multiplex qPCR assay would prove to be a useful tool in mass screening and surveillance programs for detection of non-human malaria parasites toward the control and elimination of malaria from India by 2030.
Introduction
In Indian context, major human malaria parasites are P. falciparum (Pf), P. vivax (Pv), P. malariae (Pm), P. ovale (Po) and P. knowlesi (Pk) (1–7). Moreover, non-human malaria causing Plasmodium species- P. cynomolgi (Pc) with established zoonosis in humans has become a cause for concern, at least in the South East Asian countries (8). The risk of expansion of non-human malaria in humans is increasing gradually, majorly due to deforestation, substantial changes in the ecology, host & vector availability and adaptive changes in the parasites (9). It would not be surprising that non-human primate Plasmodium parasites- Pk and Pc might be in circulation in the Indian populations, but they are rendered elusive owing to the probable misdiagnosis by routine microscopy and lack of robust diagnostic tools to detect sparsely distributed Pk and Pc infections (10, 11). There are reports of more than 30 species of non-human primate Plasmodium spp. and seven of them including Pk, Pc and others- P. brasilianum, P. eylesi (Pe), P. inui (Pi), P. schwetzi (Ps), and P. simium had been observed as transmissible to humans (10, 12–18). The major hosts of Pk, Pc, Pi, P. fieldi and P. coatneyi are the non-human primates Macaca fascicularis (long-tailed macaques), Macaca nemestrina (the pig-tailed macaque), Trachypithecus obscuras (dusky leaf monkey or spectacled langur) and Presbytis melalophus (banded leaf monkey) (19, 20). These non-human primate species are prevalent in South East Asia and India as well (21).
The mosquito vectors of human and non-human primate malaria are Anopheles minimus, An. dirus, An. sundaicus, An. sinensis and An. maculates are commonly found in geographically specific regions of India (1, 5, 22–25). Natural infections of Pk and Pc have been previously reported in macaque monkeys and humans from Malaysia (26, 27). North-Eastern states of India are in proximity to such regions where non-human malaria parasites might be in circulation and humans frequently travel on both sides. India has all the suitable vectors and hosts for non-human Plasmodium species; the geographical and climatic conditions are also conducive for the proliferation of Pk and Pc (28).
Pc is phenotypically and phylogenetically similar to Pv; thereby making the identification of Pv and Pc quite difficult in blood slides during routine microscopy. Often, routine microscopy of Pk, Pc and Pm can lead to misdiagnosis by the microscopists in primary health centers of the remote areas. In these circumstances, it becomes imperative to accurately estimate and understand the burden and transmission dynamics of non-human Plasmodium spp.- especially Pk and Pc in India human populations.
The current study presents in-house development of a rapid, sensitive and species-specific multiplex qPCR assay targeting the 18S rRNA, for detection of Pk and Pc. Multiplexing for multiple Plasmodium parasites (Pk and Pc) in single tube would be efficient to save resources during any kind of mass screening programmes and would also save on precious biological samples. The qPCR assay for Pk was also compared to well-known established PCR assays to detect Pk infections in humans (29).
Materials and methods
Design of in-house multiplex qPCR assay
The gene sequences of Pk and Pc 18S rRNA gene were extracted from Reference GenBank accession numbers (Pk-LC483580.1 and Pc-KU708868.1) for design of in-house multiplex qPCR forward and reverse primers and the fluorescence labeled probes (HEX for Pk and FAM for Pc) using online tool (https://biosearchtech.com) (Table 1). The probes were BLAST searched (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and observed to be species-specific for Pk and Pc. The oligonucleotides and probes were commercially synthesized from GCC Biotech (I) Pvt. Ltd., India. The final reaction volume (20 μl) constituted of 4 μl of genomic DNA, 500 nM of each species-specific primers and 400 nM of each probe, 10 μl of 2X Platinum multiplex PCR master mix (Applied Biosystems; Thermo Fisher Scientific, USA). The qPCR conditions were- initial denaturation 95°C for 5 min. followed by 40 cycles of denaturation at-95°C for 15 s. and primer annealing-extension at 53°C for 1 min. The amplifications were performed in BioRad CFX96 Connect Real-Time PCR System, USA.

Table 1. List of qPCR primers and respective probes for amplification of Pk and Pc 18S rRNA target regions.
Sensitivity and specificity of qPCR assay
The DNA samples of Pk and Pc parasites were obtained from CSIR-Central Drug Research Institute, Lucknow, India and the yields were observed to be 27 and 7.5 ng/μl, respectively (30). Further, the Pk and Pc genomic DNAs were 10-fold serially diluted four times to estimate the limit of detection (LOD), and each dilution was tested in triplicate. To rule out the cross-reactivity with other species of Plasmodium (Pf, Pv, Pm, Po) the assay was validated in 250 blood samples collected from suspected endemic areas of North Eastern States of India. The specificity of Pk was also cross checked using a conventional reported PCR method (29).
Human blood sample collection and DNA isolation
During 2020-21, the COVID-19 pandemic severely hampered the sample collection. However, a total of 250 samples were collected from north eastern state of India from mass surveys to identify symptomatic and asymptomatic-suspected malaria patients with ethical approval from the Institutional ethics committee (IEC No. ECR/NIMR/EC/ 2019/332). Blood smears were made and blood samples were also collected on Whatman 3MM filter paper for detection of non-human Pk and Pc parasites by microscopy and molecular methods. DNA was isolated from the punched spots using QIAGEN kit as per manufacturer's instructions. All the 250 blood samples were tested for detection of Pf and Pv using nested PCR methods (31).
Results
The qPCR assay was designed for detection of 18S rRNA gene segment of the Pk and Pc parasites. The in-house designed primers and probes were used for the amplification of the 18S rRNA gene segments of the Pk and Pc, respectively. Standard curves from the serial dilutions were generated for each parasite (Pk and Pc) with estimated R2 values of 0.98 and 0.99, respectively (Figures 1A, C). To assess the limit of detection (LOD) of parasites DNA, the Pk and Pc DNA were serially diluted and subjected to qPCR assay. The LOD of parasites DNA in multiplex qPCR assays were observed to be 0.00275 and 0.075 ng/μl of Pk and Pc genomic DNA, respectively (Figures 1B, D). The associated Cq values corresponding to the minimal detection limits were 28.50 and 31.5 for Pk and Pc, respectively (Figures 1B, D). The in-house designed qPCR primers and probes were observed to species-specific for Pk and Pc in the multiplex format, as the 18S rRNA primers and probes specific for Pk and Pc did not amplify any of the human malaria parasites DNA (Pf, Pv, Pm and Po) (Figure 2); thus, validating the multiplex qPCR assay to be species-specific for Pk and Pc.
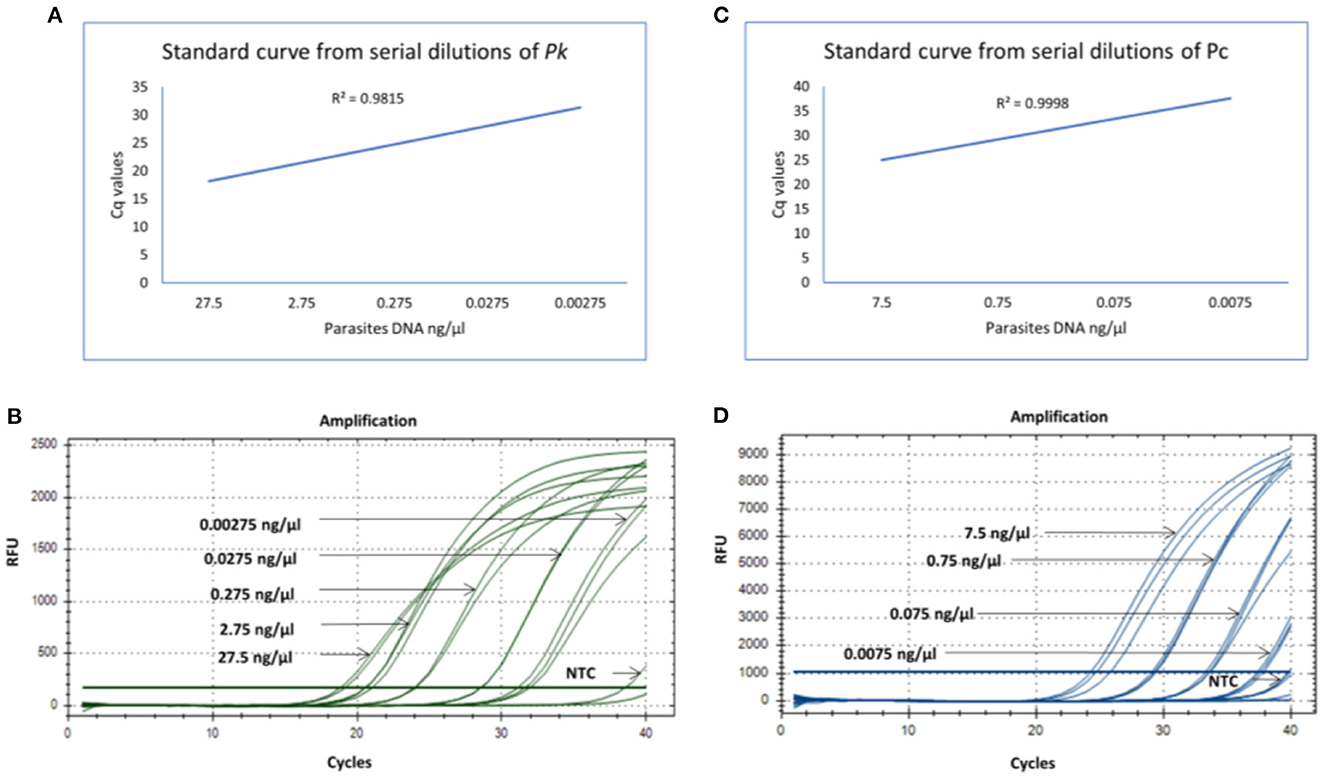
Figure 1. (A) Standard curve generated from 10-fold serial dilutions from known concentrations of parasites DNA (Pk) in the qPCR assay. (B) qPCR amplification cycles for Pk serial dilutions with fluoroscent probe HEX (green). (C) Standard curve generated from 10-fold serial dilutions from known concentrations of parasites DNA (Pc) in the qPCR assay. (D) qPCR amplification cycles for Pc serial dilutions with fluoroscent probe FAM (blue).
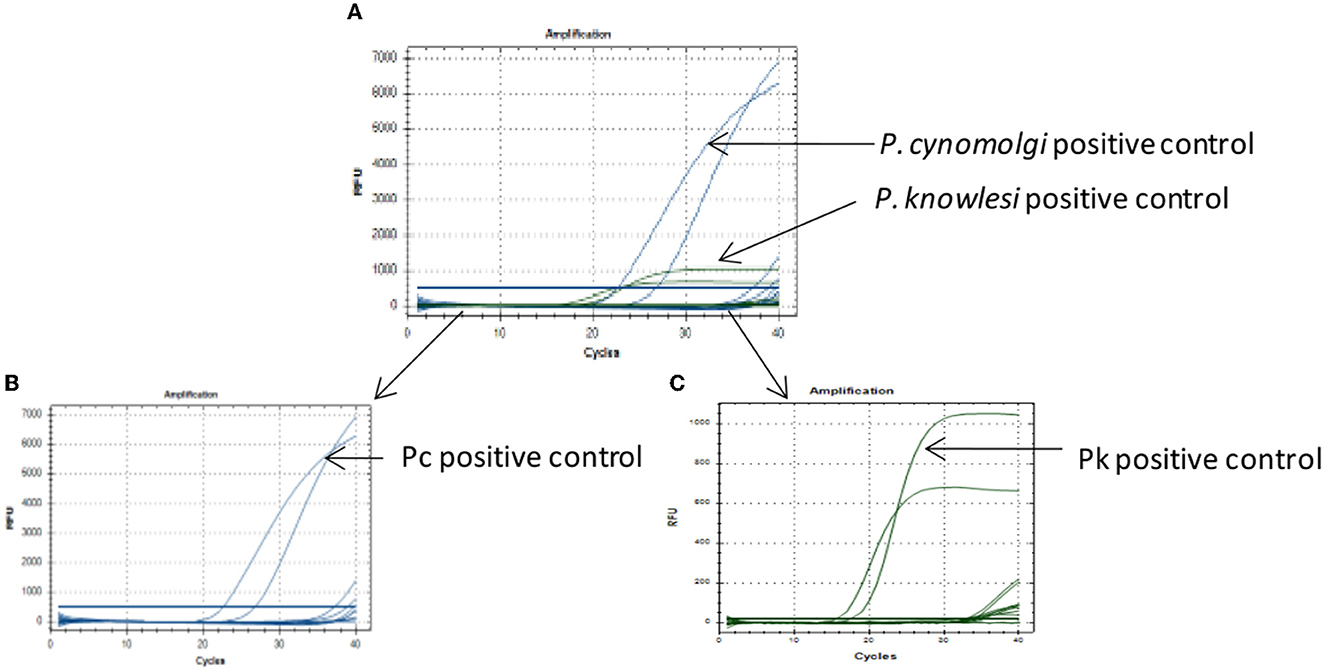
Figure 2. Specificity of the qPCR assay. (A) Multiplex qPCR assay for detection of Pk and Pc parasites DNA as positive controls, while templates from Pf, Pv, Pm and Po parasites DNA did not amplify. (B) Selected fluorophore probe FAM (blue) for P.cynomolgi showed good amplification but no amplification for Pf, Pv, Pm, and Po. (C) Selected fluorophore probe HEX (green) for P. knowlesi showed good amplification but no amplification for Pf, Pv, Pm, and Po.
Microscopy was used for detection of malaria parasites and further, the multiplex qPCR primers and probes were evaluated with 250 human blood samples collected from suspected areas from North Eastern states of India. We did not observe any Pk and Pc positive infection from these samples. However, we did observe Pf and Pv infections using microscopy and previously reported primers for detection of Pf and Pv parasites (31) in these samples as listed in Table 2. 27/250 samples were febrile (symptomatic) at the time of sample collection; males and females were in approximately equal proportion of the total number of samples. 14/250 samples were found to be positive for Pf infections; while 11/250 were found positive for Pv infections. However, none of the samples were observed positive for Pk and Pc parasites, either by the in-house developed qPCR assay as well as with established conventional PCR primers described previously for detection of Pk (29). In the absence of any positive sample for Pk at the least even by the conventional PCR primers reported previously, the qPCR primers for Pk would be considered highly sensitive, however further validation of the primers shall be assessed with Pk positive samples from other sites in future.
Discussion
The non-human Plasmodium parasites Pk, Pc, P. fieldi and Pi have been reported from Malaysia; while Pk and Pc were also reported from India as well (4–8). The non-human malaria parasite Pk in the human host and vector have been reported from different states of India such as Bihar, Delhi, Andaman & Nicobar Islands and Uttar Pradesh (1, 2, 5, 7). In this study, we developed in-house multiplex qPCR assay for detection of non-human malaria parasite species Pk and Pc. This diagnostic tool shall prove to be critical in detection, identification, surveillance and monitoring of the non-human malaria parasite Pk and Pc, ultimately contributing toward the control and elimination of malaria by 2030 from India.
The lowest limit of detection from genomic DNA of Pk and Pc parasites DNA was found to be 0.00275 and 0.075 ng/μl, respectively (Figures 1B, D). The probes were also BLAST searched and found to species-specific for Pk and Pc only. Further, no cross-reactivity was observed with any of the human malaria parasites- Pf, Pv, Po and Pm, proving them to be species-specific (Figure 2). Therefore, the in-house developed species-specific multiplex qPCR assay is highly specific and sensitive for detection of Pk and Pc. The assay was further evaluated for detection of Pk and Pc in 250 human blood samples collected from highly malaria endemic areas of North Eastern states of India and yielded no positive infection for Pk and Pc.
We anticipate that the qPCR assay would prove to be a useful tool for detection of Pk and Pc infections in the vector mosquitoes also (An. dirus, An. minimus, An. sundaicus, An. sinensis and An. maculates) as well as in their natural hosts- non-human primates (1, 5, 21, 23–25, 28). The multiplex qPCR assay for detection of Pk and Pc parasites is robust, quick, cost-effective, sensitive and species-specific to undertake investigations particularly focused on the transmission dynamics of these non-human malaria parasites in India. However, further studies with larger number of samples are needed to validate the usefulness of the qPCR assay for Pk and Pc.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by Institutional Ethics Committee ICMR-National Institute of Malaria Research, Delhi, India (IEC No. ECR/NIMR/EC/ 2019/332). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
RD conceived the study, designed and performed the experiments, analyzed the data, and wrote the manuscript. KV analyzed the data and wrote the manuscript. KP analyzed the data and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was funded by Department of Science and Technology-Science and Engineering Research Board (DST-SERB), New Delhi, India (EEQ/2019/000416).
Acknowledgments
The Pk and Pc parasite DNA were kindly received from CSIR-CDRI, Lucknow, India. The study is approved by the Research Integrity Committee (RIC) committee, ICMR-NIMR (RIC-71/2022). We are thankful to the ICMR-National Institute of Malaria Research, New Delhi, India for providing laboratory facilities. The designed primers and probes of the qPCR assay have been submitted for a patent in ICMR, New Delhi, India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Singh S, Prakash A, Yadav RN, Mohapatra PK, Sarma NP, Sarma DK, et al. Anopheles (Cellia) maculatus group: its spatial distribution and molecular characterization of member species in north-east India. Acta Trop. (2012) 124:62–70. doi: 10.1016/j.actatropica.2012.06.011
2. Tyagi RK, Das MK, Singh SS, Sharma YD. Discordance in drug resistance-associated mutation patterns in marker genes of Plasmodium falciparum and Plasmodium knowlesi during coinfections. Chemother J Antimicrob. (2013) 183:1081–8. doi: 10.1093/jac/dks508
3. Alias H, Surin J, Mahmud R, Shafie A, Mohd Zin J, Mohamad Nor M, et al. Spatial distribution of malaria in Peninsular Malaysia from 2000 to 2009. Parasites Vectors. (2014) 7:186. doi: 10.1186/1756-3305-7-186
4. Dixit J, Zachariah A, PK S, Chandramohan B, Shanmuganatham V, Karanth KP, et al. Reinvestigating the status of malaria parasite (Plasmodium sp.) in Indian non-human primates. PLOS Neglect Trop Dis. (2018) 12:e0006801. doi: 10.1371/journal.pntd.0006801
5. Vidhya PT, Sunish IP, Maile A, Zahid AK. Anopheles sundaicus Mosquitoes as Vector for Plasmodium knowlesi, Andaman and Nicobar Islands, India. Emerg Infect Dis. (2019) 25:817. doi: 10.3201/eid2504.181668
6. Kaur C, Pramanik A, Kumari K, Mandage R, Dinda AK, Sankar J, et al. Renal detection of Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi in malaria associated acute kidney injury: a retrospective case–control study. BMC Res Notes. (2020) 13:37. doi: 10.1186/s13104-020-4900-1
7. Chaturvedi R, Deora N, Bhandari D, Parvez S, Sinha A, Sharma A, et al. Trends of neglected Plasmodium species infection in humans over the past century in India. One Health. (2021) 186:100190. doi: 10.1016/j.onehlt.2020.100190
8. Yap NJ, Hossain H, Nada-Raja T, Ngui R, Muslim A, Hoh BP, et al. Natural Human Infections with Plasmodium cynomolgi, P. inui, and 4 other Simian Malaria Parasites, Malaysia. Emerg Infect Dis. (2021) 216:2187. doi: 10.3201/eid2708.204502
9. Lee KS, Divis PC, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. (2011). Plasmodium knowlesi: Reservoir Hosts and Tracking the Emergence in Humans and Macaques. PLOS Pathogens. 7:e1002015. doi: 10.1371/journal.ppat.1002015
10. Ramasamy R. Zoonotic malaria-global overview and research and policy needs. Front Public Health. (2014) 190:123. doi: 10.3389/fpubh.2014.00123
11. Mewara A, Sehgal R. Guest commentary: Plasmodium knowlesi-need to diagnose in India. Trop Parasitol. (2017) 189:2.
12. Coatney GR, Elder HA, Contacos PG, Getz ME, Greenland R, Rossan RN, et al. (1961). Transmission of the M strain of Plasmodium cynomolgi to man. Am J. Trop Med Hyg 199:673–8. doi: 10.4269/ajtmh.1961.10.673
13. Schmidt LH, Greenland R, Genther CS. The transmission of Plasmodium cynomolgi to man. Am J. Trop Med Hyg. (1961) 195:679–88. doi: 10.4269/ajtmh.1961.10.679
14. Contacos PG, Lunn JS, Coatney GR, Kilpatrick JW, Jones FE. Quartan-type malaria parasite of new world monkeys transmissible to man. Science. (1963) 142:676. doi: 10.1126/science.142.3593.676-a
15. Coatney GR, Chin W, Contacos PG, King HK. Plasmodium inui, a quartan-type malaria parasite of Old World monkeys transmissible to man. Parasitol J. (1966) 198:660–3. doi: 10.2307/3276423
16. Deane Lm Fau-Deane MP, Deane Mp Fau-Ferreira Neto J, Ferreira Neto J. Studies on transmission of simian malaria and on a natural infection of man with Plasmodium simium in Brazil. Bull World Health Organ. (1966) 194:805.
17. Contacos Pg Fau-Coatney GR, Coatney Gr Fau-Orihel TC, Orihel Tc Fau-Collins WE, Collins We Fau-Chin W, Chin W, Fau-Jeter MH, et al. Transmission of Plasmodium schwetzi from the chimpanzee to man by mosquito bite. Am J. Trop Med Hyg. (1970) 196:190–95. doi: 10.4269/ajtmh.1970.19.190
18. Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. (2004) 192:2211. doi: 10.3201/eid1012.040293
19. Moyes CL, Henry AJ, Golding N, Huang Z, Singh B, Baird JK, et al. Defining the geographical range of the Plasmodium knowlesi reservoir. PLOS Neglect Trop Dis. (2014) 8:e2780. doi: 10.1371/journal.pntd.0002780
20. Nakaviroj S, Kobasa T, Teeranaipong P, Putaporntip C, Jongwutiwes S. An autochthonous case of severe Plasmodium knowlesi malaria in Thailand. Am J Trop Med Hyg. (2015) 201:569. doi: 10.4269/ajtmh.14-0610
21. Subbarao SK. Centenary celebrations article: Plasmodium knowlesi: from macaque monkeys to humans in South-east Asia and the risk of its spread in India. Dis J Parasit. (2011) 35:87–93. doi: 10.1007/s12639-011-0085-9
22. SA K, Dutta P, Borah J, Mahanta J. Survey of new mosquito species of Meghalaya, India. Biol J. Environ. (2013) 34:191–95.
23. Zomuanpuii R, Fau-Guruswami G, Guruswami G, Fau-Nachimuthu SK, Nachimuthu SK. A three year study on distribution and ecology of Anophelines in Thenzawl, Mizoram, India. Biol J Environ. (2014) 204:369.
24. Dev V, Manguin S. Biology, distribution and control of Anopheles (Cellia) minimus in the context of malaria transmission in northeastern India. Parasites Vectors. (2016) 9:585. doi: 10.1186/s13071-016-1878-6
25. Sindhania A, Das MK, Sharma G, Surendran SN, Kaushal BR, Lohani HP, et al. Molecular forms of Anopheles subpictus and Anopheles sundaicus in the Indian subcontinent. Malar J. (2020) 19:1–17. doi: 10.1186/s12936-020-03492-2
26. Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar. J. (2014) 13:1–7. doi: 10.1186/1475-2875-13-68
27. Sutton PL, Luo Z, Divis PC, Friedrich VK, Conway DJ, Singh B, et al. Characterizing the genetic diversity of the monkey malaria parasite Plasmodium cynomolgi. Infect Genet Evol. (2016) 40:243–52. doi: 10.1016/j.meegid.2016.03.009
28. Prakash A, Walton C, Bhattacharyya DR, O'Loughlin S, Mohapatra PK, Mahanta J, et al. Molecular characterization and species identification of the Anopheles dirus and An. minimus complexes in north-east India using r-DNA ITS-2. Acta Trop. (2006) 100:156–61. doi: 10.1016/j.actatropica.2006.09.009
29. Imwong M, Tanomsing N, Pukrittayakamee S, Day NP, White NJ, Snounou G. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. Microbiol J. Clin. (2009) 47:4173–5. doi: 10.1128/JCM.00811-09
30. Kamau E, Alemayehu S, Feghali KC, Saunders D, Ockenhouse CF, et al. Multiplex qPCR for detection and absolute quantification of malaria. PLoS ONE. (2013) 8:e71539. doi: 10.1371/journal.pone.0071539
Keywords: Plasmodium knowlesi, Plasmodium cynomolgi, diagnostics, multiplex qPCR, non-human malaria
Citation: Das R, Vashisht K and Pandey KC (2023) A novel multiplex qPCR assay for clinical diagnosis of non-human malaria parasites-Plasmodium knowlesi and Plasmodium cynomolgi. Front. Vet. Sci. 10:1127273. doi: 10.3389/fvets.2023.1127273
Received: 19 December 2022; Accepted: 06 January 2023;
Published: 26 January 2023.
Edited by:
Ariel L. Rivas, University of New Mexico, United StatesReviewed by:
Byoung-Kuk NA, Gyeongsang National University, Republic of KoreaDhaneswar Prusty, Central University of Rajasthan, India
Yash Gupta, Mayo Clinic Florida, United States
Copyright © 2023 Das, Vashisht and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ram Das,  ramdas9@gmail.com
ramdas9@gmail.com
 Ram Das
Ram Das Kapil Vashisht
Kapil Vashisht Kailash C. Pandey
Kailash C. Pandey