Long non-coding RNA Loc105611671 promotes the proliferation of ovarian granulosa cells and steroid hormone production upregulation of CDC42
- 1College of Animal Science and Technology, Shihezi University, Shihezi, Xinjiang, China
- 2School of Pharmacy, Shihezi University, Shihezi, Xinjiang, China
Granulosa cells (GCs) are essential for follicular development, and long non-coding RNAs (LncRNAs) are known to support the maintenance of this process and hormone synthesis in mammals. Nevertheless, the regulatory roles of these lncRNAs within sheep follicular GCs remain largely unexplored. This study delved into the influence of a Loc105611671, on the proliferation and steroid hormone synthesis of sheep ovarian GCs and the associated target genes in vitro. Cell Counting Kit-8 (CCK-8) gain-of-function experiments indicated that overexpression of Loc105611671 significantly boosted GCs proliferation, along with estrogen (E2) and progesterone (P4) levels. Further mechanistic scrutiny revealed that Loc105611671 is primarily localized within the cytoplasm of ovarian granulosa cells and engages in molecular interplay with CDC42. This interaction results in the upregulation of CDC42 protein expression. Moreover, it was discerned that increased CDC42 levels contribute to augmented proliferation of follicular granulosa cells and the secretion of E2 and P4. Experiments involving co-transfection elucidated that the concurrent overexpression of CDC42 and Loc105611671 acted synergistically to potentiate these effects. These findings provide insights into the molecular underpinnings of fecundity in ovine species and may inform future strategies for enhancing reproductive outcomes.
1 Introduction
GCs proliferation and growth are pivotal to the process of follicular development (1). This dynamic sequence begins with oocyte growth, followed by the recruitment of GCs (2, 3). As they progress toward ovulation, GCs undergo morphological changes and proliferate (4). As the predominant cell type within the follicle, GCs not only regulate their own proliferation but also contribute to follicle development by synthesizing hormones (E2, P4) (5, 6) and growth factors (7, 8). The process begins with the transformation of cholesterol, during which the STAR protein plays a pivotal role in translocating cholesterol to the inner mitochondrial membrane. At this juncture, the enzyme P450scc (Cyp19a1 gene product) metabolizes cholesterol into pregnenolone (9). E2 production within GCs is facilitated by the enzymatic actions of P450arom aromatase and 17β-HSD (10). With the development and maturation of the follicle, E2 levels rise, consequently stimulating GCs proliferation and differentiation. This not only impacts the quality and maturation of the oocyte but also plays a role in follicular evolution (11, 12). Furthermore, E2 facilitates the production of P4 by activating the P4 receptor, thus influencing GC proliferation and follicular development (13). The concentration of P4 fluctuates throughout the stages of follicular development, peaking during the maturation phase (14).
LncRNAs are a diverse class of RNAs over 200 bp in length, including intronic, intergenic, and antisense variants (15). These lncRNAs are known to play essential regulatory roles in mammalian reproduction, participating in cell proliferation (16), apoptosis (17), follicle development (18), oocyte maturation (19), and steroid hormone synthesis (20), underscoring their significance in reproductive biology. Recent studies indicate that lncRNAs contribute to reproductive processes by interacting with proteins and other RNAs. For example, lncRNA PVT1 induces GC apoptosis by upregulating Foxo3a levels (21), while lncRNA RP11-552 M11.4 collaborates with BRCA2 to stimulate GC proliferation and prevent apoptosis (22). Despite these findings, research on mammalian reproduction has largely cantered on the discovery of novel lncRNAs (23–26), with limited investigation into the specific functions and mechanisms of lncRNAs, particularly in sheep ovarian GCs.
Our prior study revealed differential expression of Loc105611671 in Qira black sheep during the pre-estrus and estrus phases (27), suggesting it may influence sheep reproductive capacities by regulating GC functions. Yet, the exact role of Loc105611671 in sheep follicular GC regulation is still to be elucidated. To this end, we probed the impact of Loc105611671 on GC proliferation and steroid hormone secretion by developing an in vitro cultured follicular GC model. Our study is designed to elucidate the complex regulatory mechanisms lncRNAs exert on sheep follicular development. Concurrently, it may lay the groundwork for identifying novel therapeutic approaches to reproductive disorders such as polycystic ovarian syndrome (PCOS), a condition that can result in ovulatory failure.
2 Materials and methods
2.1 Isolation and culture of GC
During the peak breeding season (August to October), healthy sheep ovaries from animals aged 1 to 1.5 years were sourced from a local abattoir in Shihezi, Xinjiang Uygur Autonomous Region, China. Mature dominant follicles were carefully selected, their follicular fluid aspirated and collected into Petri dishes containing (Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 [DMEM/F12] (Gibco, France) medium. Oocytes were meticulously picked using a mouth pipette. The GCs were then transferred to erythrocyte lysis buffer to eliminate any red blood cells. The pelleted cells were washed twice with DMEM/F12 medium, cultured into Petri dishes enriched with 10% fetal bovine serum [FBS] (Gibco, France), 100 IU/mL penicillin, and 100 μg/mL streptomycin) aseptic culture at 37°C in a 5% CO2 atmosphere. After 48 h, non-adherent cells were removed by gentle medium replacement.
2.2 Quantitative reverse transcription-polymerase reaction (qRT-PCR)
Total RNA was extracted from cells using the TRIzol (Invitrogen, USA) assay. After the RNA samples were reverse transcribed into cDNA using the TransScript® First-Strand cDNA Synthesis SuperMix kit (Transgen, China) for quality assessment, they were assayed for gene expression by using the Perfectstart Green qPCR SuperMix PCR kit (Transgen, China) according to the user’s manual and a Roche Light Cycler 480 (Roche, Switzerland) to detect gene expression. The housekeeping gene, GAPDH, was used as an internal reference. Data were analyzed using the 2-ΔΔCt method. All primers used in this study are listed in Supplementary Table S1.
2.3 Plasmid construction and transfection of GCs
GCs were cultured to 60–70% confluence and then transfected, or co-transfected, with Lipofectamine 2000 (Invitrogen, USA) for 72 h. The overexpression construct for Loc105611671 was synthesized by cloning the full-length sequence into the EcoRI/BamHI sites of the pCDNA3.1-EGFP vector (denoted as LV-Loc105611671). Similarly, the CDC42 overexpression plasmid was produced by inserting the CDC42 coding sequence into the same sites of the pCDNA3.1-EGFP vector (denoted as LV-CDC42). An empty pCDNA3.1-EGFP vector served as the control (denoted as LV-EGFP). The primers utilized for cloning are listed in Table 1.
2.4 RNA-protein pull-down assay
To synthesize biotinylated transcripts, we used the T7 in vitro transcription kit mMESSAGE mMACHINE® Kit (cat. AM1344, Invitrogen, USA) for transcription; the amplification primers are listed in Table 1. After purification using the RNeasy Mini Kit (cat. 74,104, QIAGEN, Germany), biotinylated RNA was added to the cell lysates. After treatment with RNase-free DNase I, the biotin-labeled Loc105611671 was denatured for 3 min at 95°C, incubated on ice for 1 min, and rested at room temperature for 30 min to restore the secondary structure of the RNA. The RNA was then incubated with Streptavidin Magnetic Beads (cat. 21,344, Thermo, USA) for 1 h at room temperature and stirred in a clean test tube to form a magnetic bead-RNA mixture. Next, the extracted total GCs protein was added to the magnetic bead-RNA mixture and incubated for 1 h at room temperature with rotation to generate the magnetic bead-RNA-protein complexes. After washing and elution, RNA-bound proteins were collected and subsequently separated using SDS-PAGE elution and silver staining.
2.5 Cell proliferation analysis
For CCK-8 analysis (Transgen, China), post-transfection pellet cells were inoculated into 96-well plates (5 × 103 cells per well). At 24, 48, and 72 h, 10 μL of CCK-8 reagent was added to each well and then cultured for 3 h. All experiments were performed in triplicate. The absorbance of each well was then measured at 450 nm using an enzyme marker (Thermo Fisher, USA).
2.6 Determination of steroid hormone concentrations by ELISA
At the indicated time points, post-transfection cell culture supernatants were collected using sheep enzyme E2 (sensitivity: less than 1.0 pg./mL, specificity: no cross-reactivity with other soluble structural analogs, reproducibility: intra-plate coefficient of variation less than 9%, inter-plate coefficient of variation less than 11%, cat. JL17550), P4 (sensitivity: less than 0.1 ng/mL, specificity: no cross-reactivity with other soluble structural analogs, repeatability: intra-plate coefficient of variation less than 9%, inter-plate coefficient of variation less than 11%, cat. JL22308) ELISA was determined by absorbance at 450 nm by an enzyme labeller (Thermo Fisher, USA). Sheep E2 ELISA kit and P4 ELISA kit were purchased from Jianglai Biotechnology Co Ltd. (Jianglai, China).
2.7 Subcellular localization
For nuclear and cytoplasmic RNA isolation, the nuclear and cytoplasmic fractions were collected and extracted using the nuclear/cytoplasmic separation kit (cat. HR0241, Biolab, China) according to the manufacturer’s instructions. The expression of Loc105611671 and CDC42 in cytoplasmic and nuclear RNA of GCs was measured using qRT-PCR as mentioned above. XIST and ACTB were used as positive controls for the nucleus and cytoplasm, respectively.
2.8 Western blot analysis
Total proteins were extracted from the cells using RIPA lysis buffer (containing 1% PMSF) and then denatured by heating at 100°C. Equal amounts of protein were separated by SDS-PAGE and then transferred to PVDF membranes. After blocking with 5% skimmed milk and incubation with primary and secondary antibodies, immunoreactive bands on the membrane were reacted with ECL solution and detected by chemiluminescence imaging using a Tennant chemiluminescence imager (Tanon, China). The primary antibodies used in this study were anti-GAPDH (1:5000, cat. GTX100118, GeneTex, USA), anti-CDC42 (1:1000, cat. GTX134588, GeneTex, USA) and goat an-ti-rabbit (1:10000, cat. YK2231, Y&K Bio, China).
2.9 Dual luciferase reporter gene assay
293 T cells were cultured in 96-well plates (5 × 103 cells per well). When the cell density reached 50–70%, the transfection plasmids were cotransfected individually into the cells. After 48 h of transfection, luciferase activity was measured using a dual luciferase reporter gene assay system (BioTek, USA).
2.10 Statistical analysis
All experimental data were analyzed using GraphPad Prism 8.0 software (GraphPad Inc) or SPSS 26.0 system (SPSS Inc). An independent samples t-test was used to analyze the statistical differences between the two groups. Each experiment was repeated three times. p-values less than 0.05 were considered statistically significant as follows: “*” p < 0.05, “**” p < 0.01, and “***” p < 0.001.
3 Results
3.1 Overexpression of Loc105611671 enhances GCs proliferation
To elucidate the regulatory effects of Loc105611671 on GCs in vitro, we used overexpression plasmids to overexpress Loc105611671. The proliferation rate of GCs was analyzed using CCK-8 assay. The efficiency of Loc105611671 overexpression based on the pCDNA3.1-EGFP vector was confirmed using qRT-PCR (Figures 1A–C). The results demonstrated that Loc105611671 upregulation significantly enhanced GC proliferation at 24 h (0.747 ± 0.046 vs. 0.928 ± 0.026, p < 0.01) and at 48 h (1.088 ± 0.047 vs. 1.504 ± 0.032, p < 0.001) compared with the control. After 72 h (1.445 ± 0.204 vs. 1.617 ± 0.077, p > 0.05), the proliferation rates between the test and control groups began to align (Figure 1D). Additionally, the overexpression of Loc105611671 notably upregulated the mRNA expression levels of CDK1 and PCNA, both positive cell cycle regulators, while downregulating P21, a cell cycle inhibitor (Figure 1E). These results indicate that Loc105611671 plays a pivotal role in the proliferation of GCs.
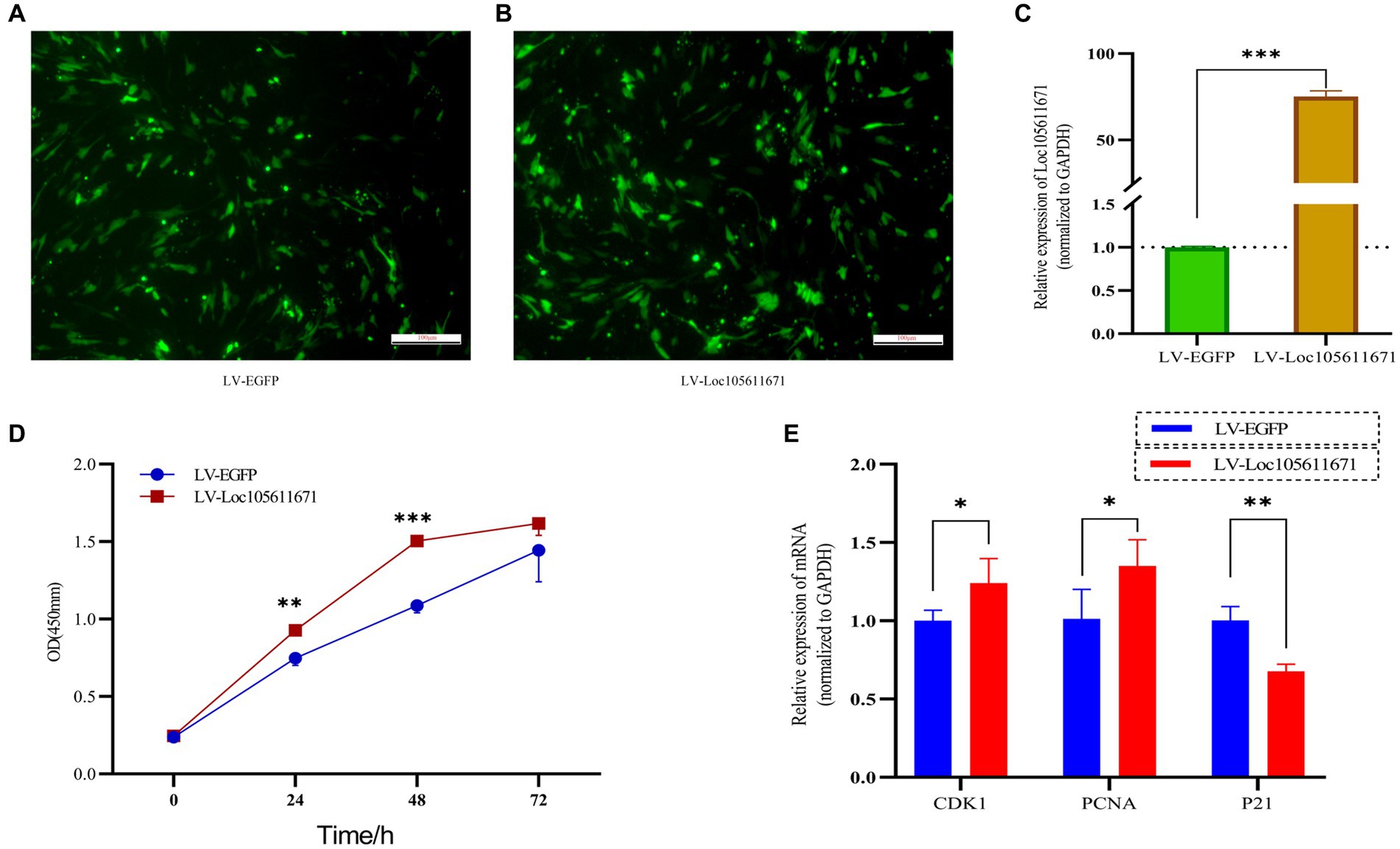
Figure 1. Overexpression of Loc10561167 enhances GCs proliferation. (A–C) The qRT-PCR method was utilized to assess Loc105611671 expression levels in GCs following transfection with LV-Loc105611671 vs. LV-EGFP (n = 4). (D) CCK-8 was employed to assess the growth level resulting from Loc105611671 overexpression in GCs (n = 3). (E) Transfection of GCs with LV-Loc105611671 was carried out for 24 h, after which mRNA levels of CDK1, PCNA, and P21 were measured (n = 4). Data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
3.2 Loc105611671 promotes E2 and P4 synthesis in GCs
To determine the effect of Loc105611671 expression on steroid hormone production, we performed ELISA to measure the E2 and P4 levels in ovarian GCs 24 h and 48 h after transfection with LV-Loc105611671. A significant elevation in E2 secretion was detected at 24 h (107.695 ± 2.022 vs. 121.170 ± 2.242 pg./mL, p < 0.01) and 48 h (93.653 ± 3.718 vs. 116.064 ± 0.638 pg./mL, p < 0.001) in the LV-Loc105611671 group compared to controls (Figures 2A,B). Similarly, P4 secretion levels were significantly increased following Loc105611671 overexpression at 24 h (7.032 ± 0.344 vs. 8.822 ± 0.678 ng/mL, p < 0.05) and at 48 h (7.211 ± 0.401 vs. 8.581 ± 0.228 ng/mL, p < 0.01) (Figures 2C,D). Moreover, a marked upregulation was observed in the mRNA expression of STAR, Cyp11a1, and Cyp19a1, genes involved in steroidogenesis (Figure 2E). These data suggest that Loc105611671 plays an important role in steroid hormone synthesis.
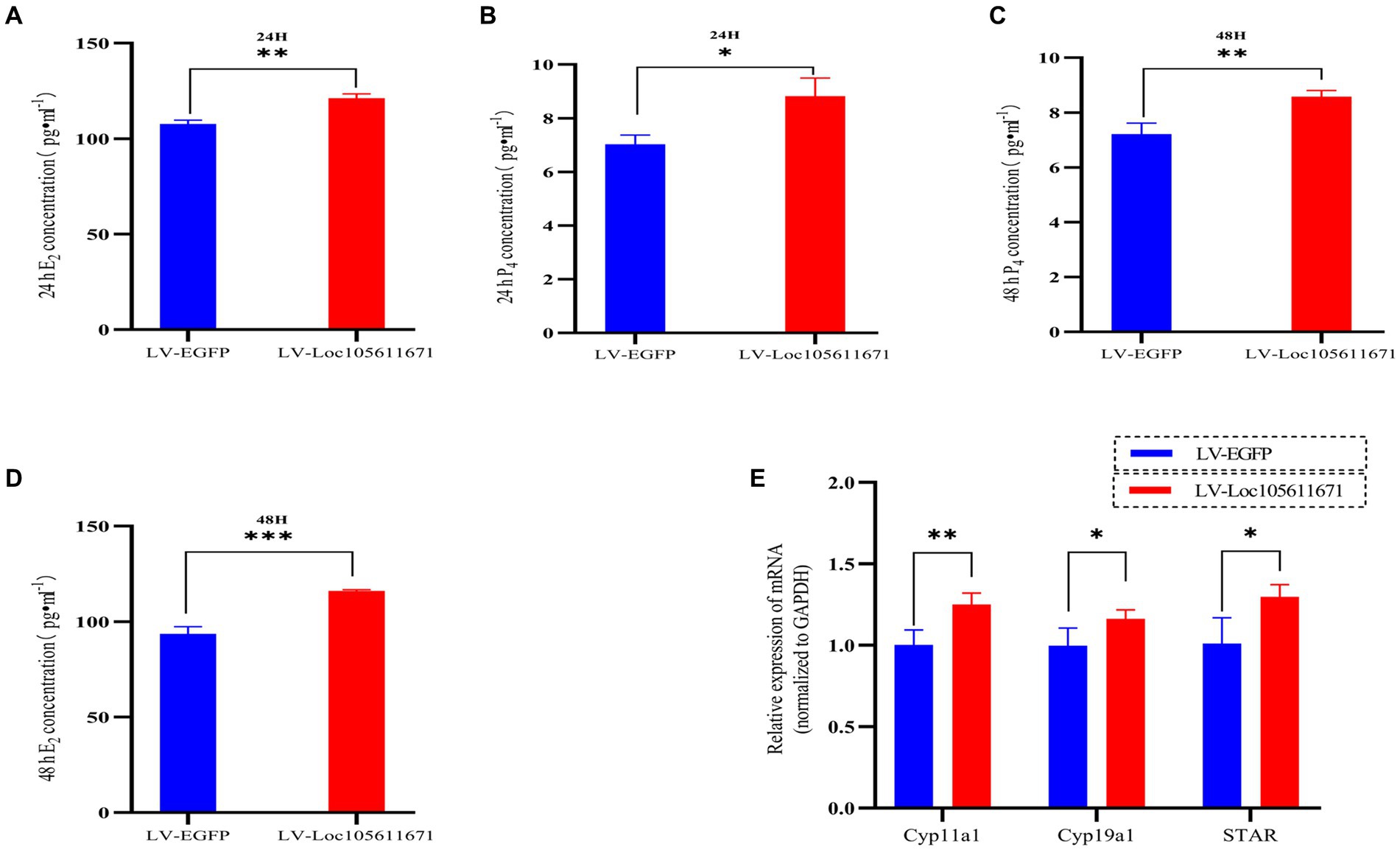
Figure 2. Loc105611671 regulates steroid secretion through GCs. The 24 h (A,C) and 48 h (B,D) E2 and P4 concentrations in cell supernatants were detected using ELISA (n = 3); (E) qRT-PCR was used to detect the expression levels of the genes important for steroid hormone synthesis, STAR, Cyp11a1, and Cyp19a1 mRNA expression (n = 4). Data represent the mean ± SD. *p < 0.05, **p < 0.01, or ***p < 0.001.
3.3 RNA pull-down assay to identify Loc105611671 binding proteins
Long non-coding RNAs (lncRNAs) enact their biological roles through interactions with diverse biomolecules, and these interactions are often influenced by the RNA’s subcellular localization. According to the iLoc-LncRNA database,1 Loc105611671 is predominantly found in the cytoplasm, a finding further substantiated by nucleoplasmic separation experiments (Figures 3B,C). To explore the secondary structure of Loc105611671, we utilized the RNAfold tool,2 which predicted Y-shaped structures in both minimally free energy (MFE) and centroid modes (Figure 3A). The cytoplasmic localization of Loc105611671, suggestive of its potential for protein interaction, prompted us to perform an RNA pull-down assay followed by mass spectrometry (Figure 3D), which identified 132 putative binding proteins (PBPs) (Figure 3E; Supplementary Table S2).
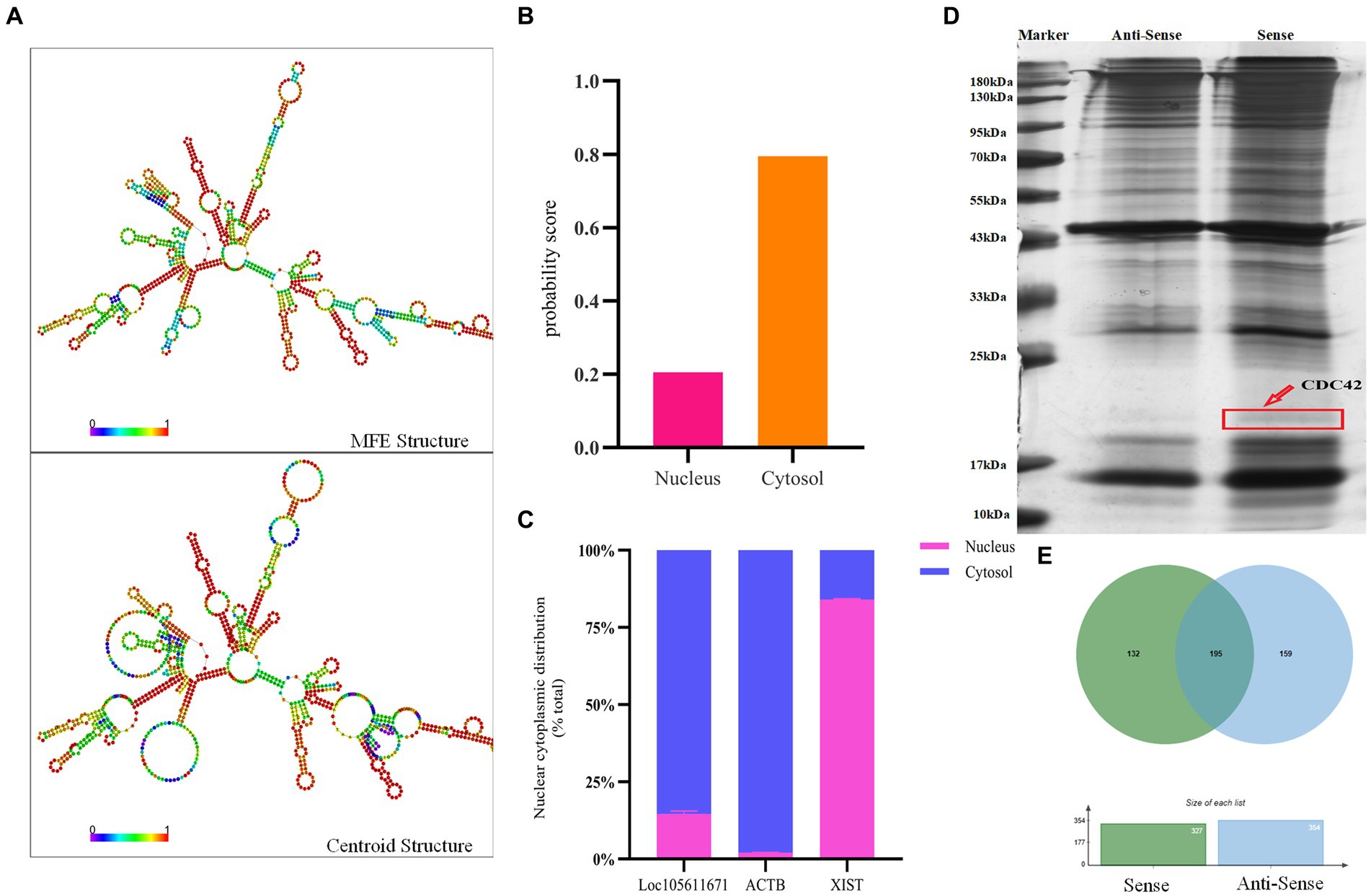
Figure 3. RNA pull-down captures Loc105611671 binding protein. (A) Secondary structure of Loc105611671 transcripts in MFE and Centroid modes predicted by the online tool RNAfold. (B) Loc105611671 subcellular localization in granule cells predicted by the online tool iLoc-LncRNA. (C) Subcellular localization of Loc105611671 (n = 3). (D) Silver-stained SDS-PAGE gels for proteins extracted from biotin-labeled Loc105611671 and antisense RNA, and two lanes for mass spectrometry. (E) Venn diagram of the results of mass spectrometry analysis.
Further analysis of Loc105611671 PBPs using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed enrichment in biological processes (BPs) such as metabolism, biological regulation, process regulation, and gene expression (Figure 4A). Molecular functions (MF) were predominantly associated with protein binding, nucleic acid binding, catalytic activity, RNA binding, and nucleotide binding (Figure 4B). Cellular components (CC) included intracellular organelles, the cytoplasm, and protein-containing complexes (Figure 4C). KEGG enrichment analysis identified 14 signaling pathways potentially linked to GC proliferation and hormone production, including PI3K-Akt signaling pathway, Wnt signaling pathway, GnRH signaling pathway, MAPK signaling pathway, oocyte meiosis and Cell cycle, etc. (Figure 4D).
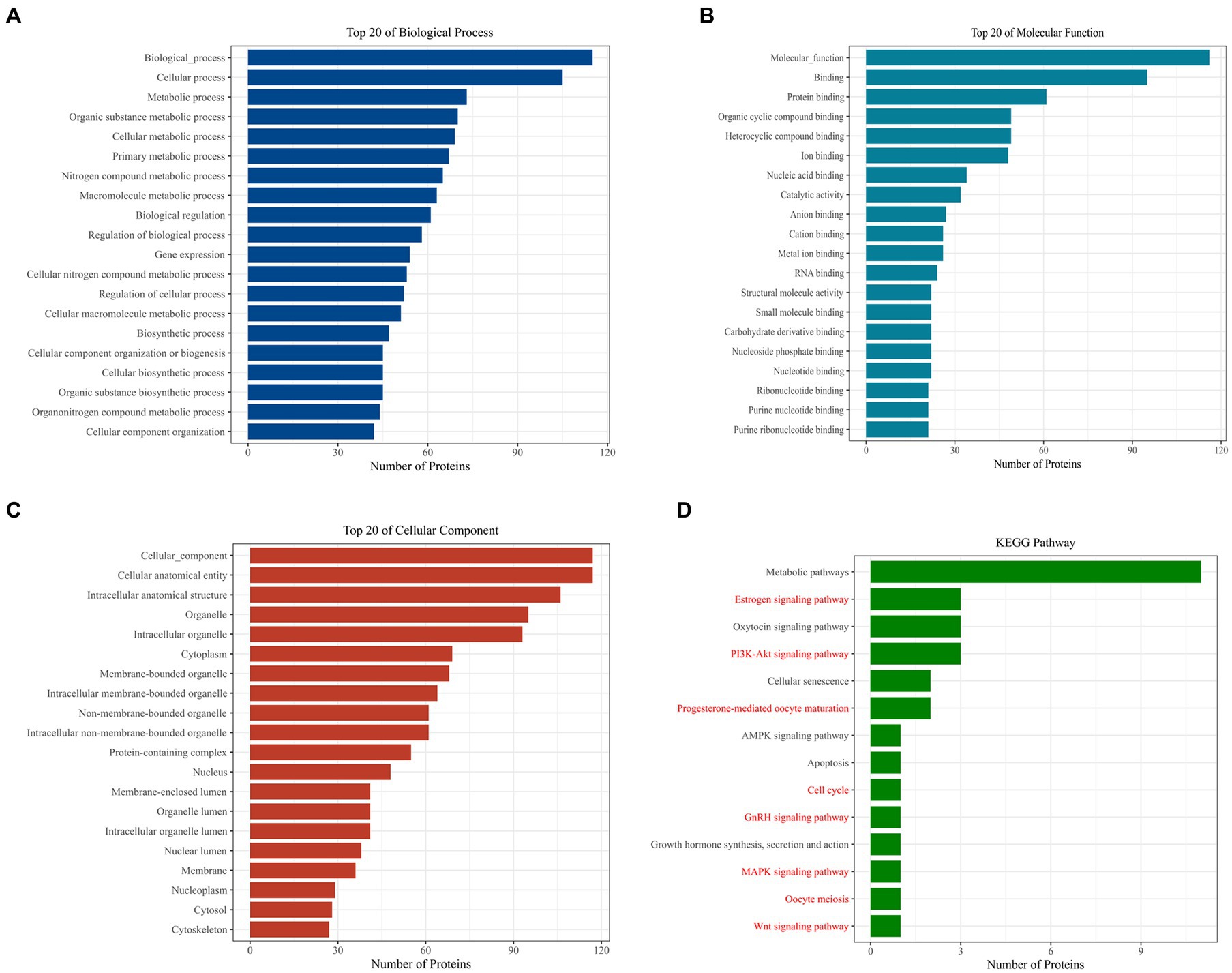
Figure 4. Bioinformatics analysis to identify proteins. GO analysis of Loc105611671 potential binding proteins (A–C). KEGG analysis of Loc105611671 potential binding proteins (D), The red pathway is more common in mammalian reproduction studies.
3.4 Direct interaction between Loc105611671 and CDC42 protein with positive regulatory effects
KEGG analysis identified six proteins as candidate binding proteins, of which CDC42 was involved in signaling pathways significant in cell proliferation and hormone production (Supplementary Table S3). Subsequent functional queries of these RNA-binding proteins revealed that CDC42 participates in the regulation of cell proliferation, differentiation, and apoptosis. Therefore, CDC42, which showed the highest reliability, was selected for subsequent studies. Subsequently, we used the online tool IntaRNA3 to predict a possible interaction between Loc105611671 and CDC42 3’UTR. This interaction was confirmed using a dual-luciferase reporter gene assay (Figure 5B). Nucleocytoplasmic separation of CDC42 demonstrated its presence in both the cytoplasm and nucleus, predominantly in the cytoplasm, providing additional support for the intracellular interaction with Loc105611671 (Figure 5A).
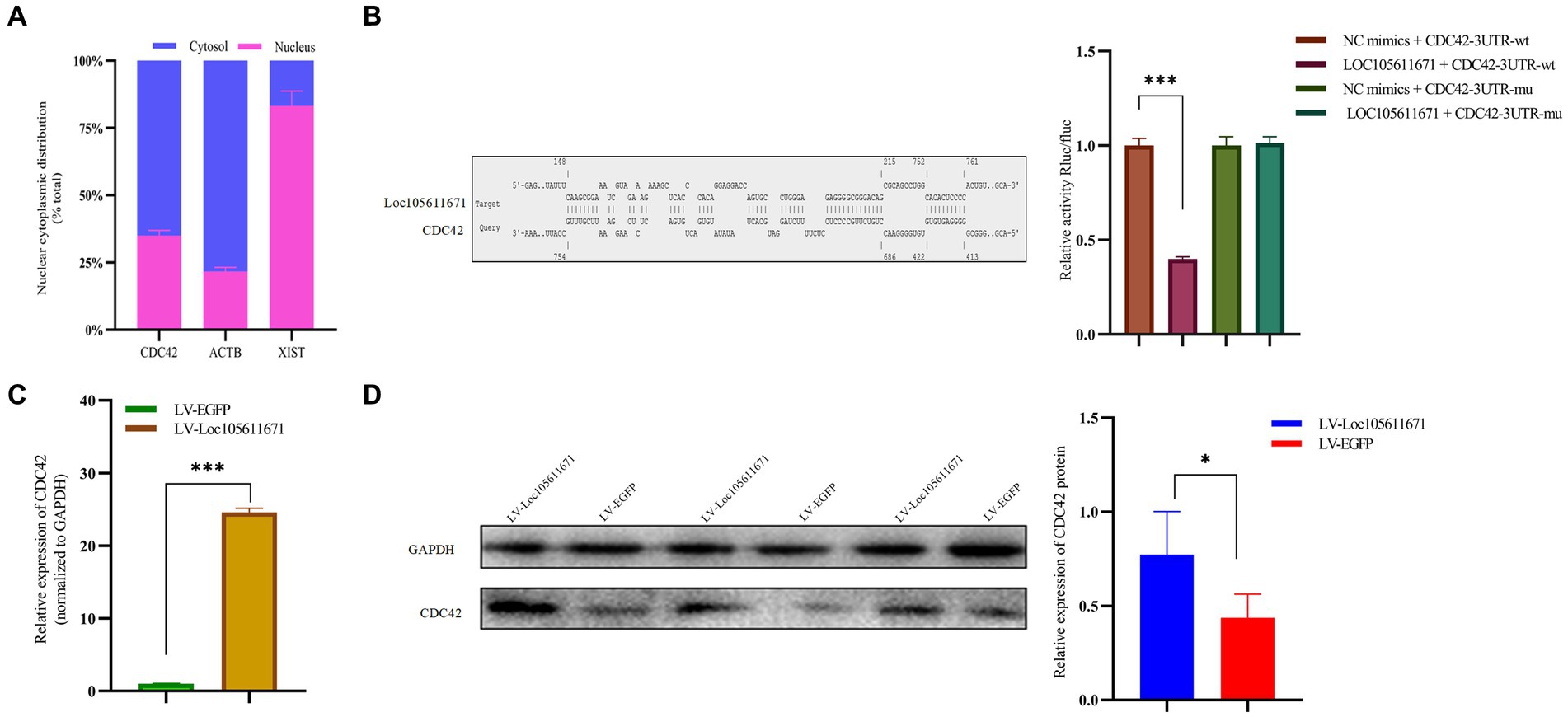
Figure 5. Loc105611671 interacts with CDC42 and positively regulates its protein and mRNA expression levels. (A) Subcellular localization of CDC42 was determined (n = 3). (B) The binding of Loc105611671 to the 3’UTR of CDC42 mRNA was demonstrated using the online IntaRNA tool and dual luciferase reproter gene assay (n = 3). (C,D) following Loc105611671 overexpression, significant increase in protein and mRNA expression levels of CDC42 was observed in GCs (n = 3). Data represent the mean ± SD. *p < 0.05, ***p < 0.001.
To investigate the promotion of GC proliferation and steroid hormone production through interaction with CDC42, we examined whether Loc105611671 would affect the expression of CDC42. Our findings revealed that overexpression of Loc105611671 significantly upregulated CDC42 protein and mRNA levels in GCs (Figures 5C,D). These outcomes indicate that Loc105611671 directly interacts with CDC42, augmenting its expression, and suggesting that CDC42 is a downstream effector of Loc105611671.
3.5 Manipulation of CDC42 expression regulates GCs proliferation
CDC42 is highly expressed in the oocytes of activated follicles, and its overexpression promotes the growth of primordial follicles in mouse ovaries (28). We hypothesized that CDC42 would act as a crucial downstream regulator of Loc105611671 and play a role in the promotion of GC proliferation. To verify this hypothesis, we elevated CDC42 expression levels in GCs (Figures 6A–C). CCK-8 results showed that GCs proliferated at a faster rate after transfection with CDC42 than the controls did at 24 h (0.306 ± 0.038 vs. 0.388 ± 0.008, p < 0.05), (0.559 ± 0.044 vs. 0.683 ± 0.059, p < 0.05) 48 h, and 72 h (0.971 ± 0.101 vs. 1.458 ± 0.126, p < 0.01) (Figure 6D). In addition, CDC42 overexpression significantly increased the mRNA level of PCNA but had no significant effect on CDK1 expression, and the expression of the negative regulator of proliferation, P21, was significantly downregulated (Figure 6E). These results suggest that Loc105611671 plays a crucial role in follicular development by promoting GC proliferation.
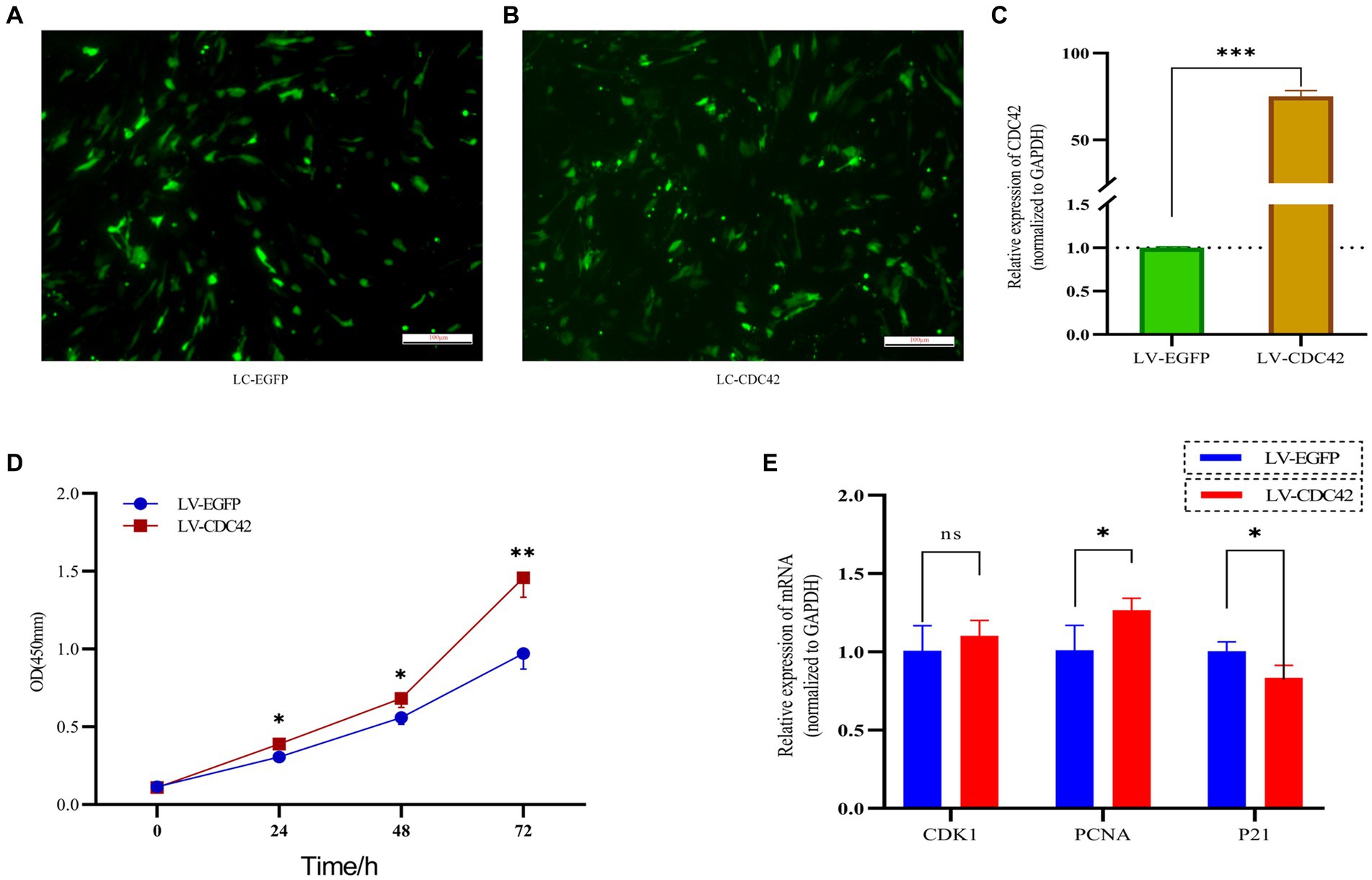
Figure 6. Overexpression of CDC42 promotes GCs proliferation. (A–C) Overexpression of CDC42 transfection efficiency, qRT-PCR to detect the relative expression level of CDC42 in GCs, LV-CDC42 vs. LV-EGFP (n = 4). (D) Growth levels of CDC42 overexpressing GCs were detected by CCK-8 (n = 3). (E) GCs were transfected with LV-CDC42 for 24 h and mRNA levels of CDK1, PCNA, and P21 were determined (n = 4). Data represent the mean ± SD. nsp > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001.
3.6 CDC42 is involved in the regulation of steroid hormone production in GCs
We further investigated the effect of CDC42 in GCs on E2 and P4 hormone production. The results showed that overexpression of CDC42 significantly increased the level of E2 secretion at both 24 h (93.908 ± 7.179 vs. 108.513 ± 1.617 pg./mL, p < 0.05) and 48 h (101.583 ± 2.082 vs. 110.048 ± 2.013997 pg./mL, p < 0.01) (Figures 7A,B). Similarly, P4 secretion was significantly increased at both 24 h (7.183 ± 0.345 vs. 8.312 ± 0.352 ng/mL, p < 0.05) and 48 h (7.489 ± 0.258 vs. 8.543 ± 0.128 ng/mL, p < 0.01) (Figures 7C,D). Furthermore, the elevated CDC42 expression markedly increased Cyp19a1 and STAR mRNA levels, without affecting Cyp11a1 expression (Figure 7E). In conclusion, the gain-of-function experiments demonstrated that high expression of CDC42 in transfected GCs could promote E2 and P4 hormone production.
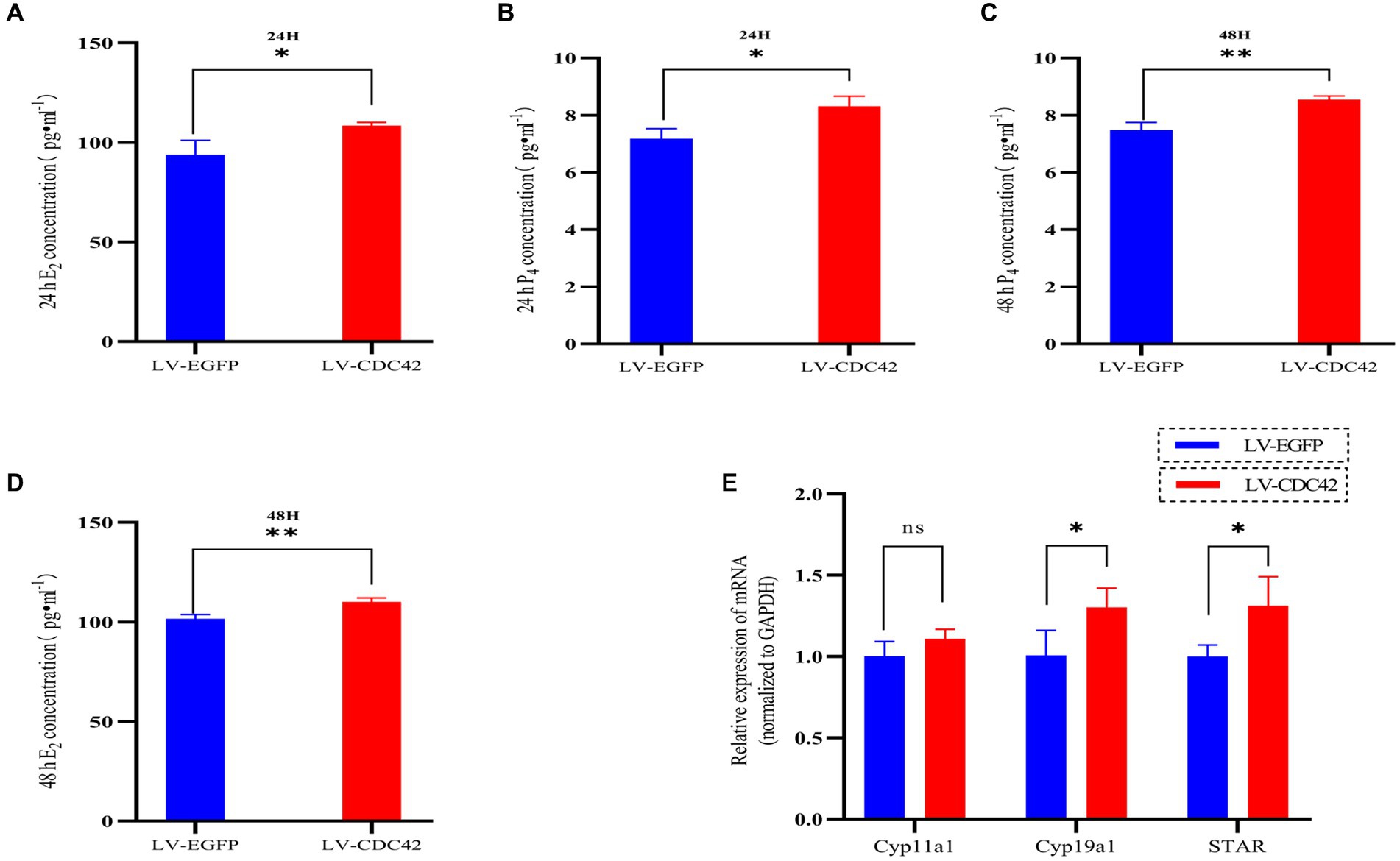
Figure 7. CDC42 is implicated in regulating steroid hormone production in GCs. The concentrations of E2 and P4 in cell supernatants were detected via ELISA for 24 h (A,C) and 48 h (B,D) (n = 3). (E) Gene expression analysis of key steroid hormone synthesis genes was performed using qRT-PCR (n = 4). Data represent the mean ± SD. nsp > 0.05, *p < 0.05, **p < 0.01.
3.7 Loc105611671 promotes GCs proliferation and E2 and P4 hormone production via CDC42
Considering that Loc105611671 directly to CDC42 protein and has similar biological functions in regulating GCs proliferation and E2 and P4 hormone production. Therefore, it was hypothesized that Loc105611671 plays a proliferative role in GCs by interacting with CDC42. To test this hypothesis, we performed co-transfection experiments (Figures 8A–C) and examined its biological functions. The results showed that simultaneous overexpression of Loc105611671 and CDC42 was observed at 24 h (0.281 ± 0.039 vs. 0.691 ± 0.042, p < 0.001), 48 h (0.596 ± 0.047 vs. 0.778 ± 0.034, p < 0.01) and 72 h (1.173 ± 0.039 vs. 1.440 ± 0.082, p < 0.01) significantly promoted granulocyte proliferation (Figure 8D). Meanwhile, E2 and P4 hormone production assays showed that overexpression of Loc105611671 and CDC42 significantly increased E2 secretion at 24 h (104.610 ± 0.996 vs. 122.285 ± 2.098 pg./mL, p < 0.001) and 48 h (104.916 ± 2.3697 vs. 121.101 ± 6.721 pg./mL, p < 0.05) (Figures 8F,G). Similarly, the level of P4 hormone production was increased at 24 h (7.908 ± 0.059 vs. 9.216 ± 0.144 ng/mL, p < 0.001) and 48 h (6.742 ± 0.546 vs. 9.388 ± 0.419 ng/mL, p < 0.01) (Figures 8H,I). In addition, mRNA levels of proliferation-related genes were significantly elevated after cotransfection, and the expression of genes related to the process of steroid hormone synthesis was also promoted (Figures 8E,J). Collectively, these data strongly suggest that Loc105611671 regulates GC cell proliferation and E2 and P4 hormone production by targeting CDC42.
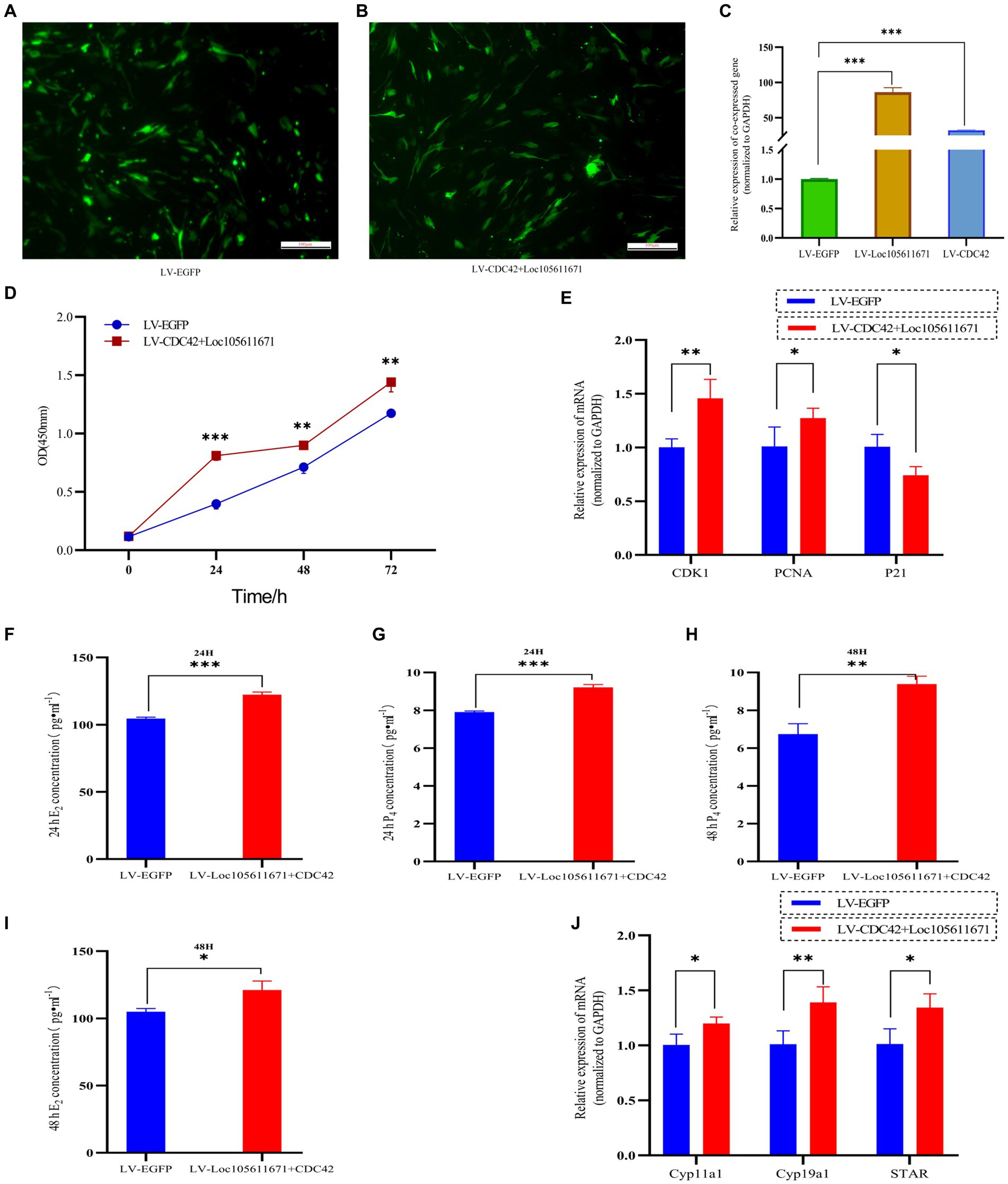
Figure 8. Loc105611671 regulates GCs proliferation and E2 and P4 hormone production by promoting increased CDC42. (A–C) qRT-PCR was performed to detect the relative expression levels of co-transfected target plasmids in GCs (n = 4). (D) CCK-8 assay to detect the growth level of co-transfected LV-CDC42 + Loc105611671 granulocytes (n = 3). (E) Determination of mRNA levels of CDK1, PCNA and P21 in co-transfected GCs for 24 h (n = 3). E2 and P4 concentrations (n = 3) in GCs after 24 h (F,H) 48 h (G,I) of co-transfection. (J) mRNA expression levels of Cyp11a1, Cyp19a1, and STAR, genes important for steroid hormone synthesis, were detected by qRT-PCR (n = 4). Data represent the mean ± SD. *p < 0.05, **p < 0.01, or ***p < 0.001.
4 Discussion
In this investigation, our findings indicate that Loc105611671 plays a crucial role in promoting the proliferation of sheep ovarian GCs and the biosynthesis of E2 and P4. Loc105611671 enhances GC proliferation and hormone secretion by modulating CDC42 expression and directly interacting with it.
GCs as the ovarian’s primary functional units, are instrumental in follicular growth and maturation, offering nutritional support and secreting hormones imperative for follicular development (5, 7, 29). Notably, Loc105611671 was identified in the follicles of Qira Black sheep during estrus, implicating a regulatory effect on follicular development through GC modulation (27). Consequently, there is a compelling need for further research into Loc105611671 roles in GC proliferation and steroid hormone production. The functionality and mechanistic action of Loc105611671 in sheep GCs, however, have remained uncharted. Bridging this knowledge gap, we isolated GCs from ovine follicles to explore the impact of Loc105611671 overexpression on their proliferation and steroidogenesis. Functional assays revealed that elevated Loc105611671 levels not only stimulated GCs proliferation but also upregulated the cell cycle regulators CDK1 and PCNA and downregulated P21. Intriguingly, these effects attenuated after 72 h, potentially due to the dilution of exogenous genes amidst ongoing GCs division. Correspondingly, Loc105611671 overexpression significantly boosted E2 and P4 concentrations and the expression of STAR, Cyp11a1, and Cyp19a1, highlighting its role in promoting steroidogenesis. In conclusion, these findings suggest that Loc105611671 is an important regulator of follicular development.
LncRNA function correlates with its subcellular localization (30). Nucleoplasmic separation experiments have demonstrated that Loc105611671 is enriched in the cytoplasm of GCs, suggesting its protein-binding capacity. This regulatory network of LncRNAs has been identified in the literature on GCs’ functional mechanisms. For example, lncRNA ZNF674-AS1 affects GC proliferation by interacting with ALDOA (31). lnc-GULP1-2:1 was also found to bind directly to COL3A1 and affect GC proliferation by regulating COL3A1 expression and localization (32). In this study, we used RNA pull-down, mass spectrometry, and dual-luciferase gene assay experiments to further reveal that CDC42 directly binds to and interacts with Loc105611671 in GCs. To our knowledge, we are the first to report CDC42 acting as an lncRNA-binding protein in sheep GCs. However, understanding the detailed protein-binding motifs in Loc105611671 requires further characterization.
CDC42 is a member of the Rho GTPase family and is involved in various cellular functions and signaling pathways, including cell proliferation (33), apoptosis (34), and the cell cycle (35). In particular, CDC42 plays a crucial role in establishing mammalian oocyte polarity. For example, CDC42 deficiency disrupts oocyte maturation during development in vitro (36). The subcellular localization of CDC42 during primordial follicle activation is of interest. In dormant primordial follicles, CDC42 is specifically expressed in oocyte cytoplasm. When primordial follicles were activated, CDC42 expression on the oocyte membrane increased significantly (28). Furthermore, CDC42 interacts with epidermal growth factor EGF to improve primordial follicle activation in mice by regulating the PI3K signaling pathway (37). In this study, elevated levels of CDC42 were associated with enhanced cell proliferation, potentially contributing to improved GCs function. These findings are consistent with those in the literature on various animal species (33, 38, 39). As CDC42 can induce steroid hormone activity, an interactive relationship is implied between CDC42 expression and steroid hormone production (40, 41). Therefore, in this study, we investigated steroid hormone synthesis. The upregulation of CDC42 resulted in a significant increase in E2 and P4 levels and STAR and Cyp19a1 expression, indicating that CDC42 promotes steroid hormone production in GCs. Subsequently, In vivo co-transfection experiments affirmed that Loc105611671 and CDC42 co-overexpression synergistically intensified GC proliferation and hormone secretion beyond their individual effects.
Unfortunately, although our findings have demonstrated that Loc105611671 plays a positive regulatory role in follicular granulosa cell proliferation through CDC42, the specific molecular mechanisms behind this phenomenon remain unknown. CDC42 is a crucial kinase within the MAPK signaling pathway and it influences a range of cellular behaviors. Therefore, our future research will concentrate on investigating how the Loc105611671-CDC42-regulated MAPK signaling pathway impacts follicular granulosa cells.
5 Conclusion
In conclusion, our study establishes that Loc105611671 enhances follicular granulosa cell proliferation and steroidogenesis through its interaction with CDC42. This novel mechanism of lncRNA-protein interaction deepens our understanding of the physiological roles played by lncRNAs in coordinating GCs function and the complex follicular maturation process. Moreover, these findings lay the groundwork for the development of innovative therapeutic strategies targeting reproductive diseases.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
JW: Conceptualization, Data curation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. HC: Investigation, Software, Supervision, Writing – original draft. YZ: Conceptualization, Investigation, Writing – review & editing. HS: Investigation, Methodology, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China 31660643.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1366759/full#supplementary-material
Footnotes
1. ^http://lin-group.cn/server/iLoc-LncRNA/home.php
2. ^http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi
References
1. Feng, GH, Liu, J, Lu, ZT, Li, YK, Deng, M, Liu, GB, et al. miR-450-5p and miR-202-5p synergistically regulate follicle development in black goat. Int J Mol Sci. (2023) 24:20. doi: 10.3390/ijms24010401
2. Jolly, PD, Smith, PR, Heath, DA, Hudson, NL, Lun, S, Still, LA, et al. Morphological evidence of apoptosis and the prevalence of apoptotic versus mitotic cells in the Membrana granulosa of ovarian follicles during spontaneous and induced atresia in ewes. Biol Reprod. (1997) 56:837–46. doi: 10.1095/biolreprod56.4.837
3. Matsuda, F, Inoue, N, Manabe, N, and Ohkura, S. Follicular growth and atresia in mammalian ovaries: regulation by survival and death of granulosa cells. J Reprod Dev. (2012) 58:44–50. doi: 10.1262/jrd.2011-012
4. Lintern-Moore, S, and Moore, GP. The initiation of follicle and oocyte growth in the mouse ovary. Biol Reprod. (1979) 20:773–8. doi: 10.1095/biolreprod20.4.773
5. Chauvin, S, Cohen-Tannoudji, J, and Guigon, CJ. Estradiol signaling at the heart of Folliculogenesis: its potential deregulation in human ovarian pathologies. Int J Mol Sci. (2022) 23:20. doi: 10.3390/ijms23010512
6. Chou, CH, and Chen, MJ. The effect of steroid hormones on ovarian follicle development In: G Litwack, editor. Ovarian cycle. Vitamins and hormones, vol. 107. San Diego: Elsevier Academic Press Inc. (2018). 155–75.
7. Monte, APO, Santos, JM, Menezes, VG, Gouveia, BB, Lins, T, Barberino, RS, et al. Growth differentiation Factor-9 improves development, mitochondrial activity and meiotic resumption of sheep oocytes after in vitro culture of secondary follicles. Reprod Domest Anim. (2019) 54:1169–76. doi: 10.1111/rda.13485
8. Richani, D, and Gilchrist, RB. The epidermal growth factor network: role in oocyte growth, maturation and developmental competence. Hum Reprod Update. (2018) 24:1–14. doi: 10.1093/humupd/dmx029
9. Fuentes, N, and Silveyra, P. Estrogen receptor signaling mechanisms. Adv Protein Chem Struct Biol. (2019) 116:135–70. doi: 10.1016/bs.apcsb.2019.01.001
10. Simpson, ER, Clyne, C, Rubin, G, Boon, WC, Robertson, K, Britt, K, et al. Aromatase – a brief overview. Annu Rev Physiol. (2002) 64:93–127. doi: 10.1146/annurev.physiol.64.081601.142703
11. Engelhardt, H, Smith, KB, McNeilly, AS, and Baird, DT. Expression of messenger ribonucleic acid for inhibin subunits and ovarian secretion of inhibin and estradiol at various stages of the sheep estrous cycle. Biol Reprod. (1993) 49:281–94. doi: 10.1095/biolreprod49.2.281
12. Pierre, A, Mayeur, A, Marie, C, Cluzet, V, Chauvin, J, Frydman, N, et al. Estradiol regulates mRNA levels of estrogen receptor Beta 4 and Beta 5 isoforms and modulates human granulosa cell apoptosis. Int J Mol Sci. (2021) 22:16. doi: 10.3390/ijms22095046
13. Ting, AY, Xu, J, and Stouffer, RL. Differential effects of estrogen and progesterone on development of primate secondary follicles in a steroid-depleted milieu in vitro. Hum Reprod. (2015) 30:1907–17. doi: 10.1093/humrep/dev119
14. Thierry, H, van Dessel, T, Schipper, I, Pache, TD, van Geldorp, H, de Jong, FH, et al. Normal human follicle development: an evaluation of correlations with Oestradiol, androstenedione and progesterone levels in individual follicles. Clin Endocrinol. (1996) 44:191–8. doi: 10.1046/j.1365-2265.1996.662483.x
15. Statello, L, Guo, CJ, Chen, LL, and Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
16. Yao, XL, El-Samahy, MA, Li, XD, Bao, YJ, Guo, JH, Yang, F, et al. LncRNA-412.25 activates the LIF/STAT 3 signaling pathway in ovarian granulosa cells of Hu sheep by sponging miR-346. FASEB J. (2022) 36:e22467. doi: 10.1096/fj.202200632R
17. Wang, Y, Guo, YX, Duan, CH, Yang, RC, Zhang, LC, Liu, YQ, et al. Long non-coding RNA GDAR regulates ovine granulosa cells apoptosis by affecting the expression of apoptosis-related genes. Int J Mol Sci. (2022) 23:20. doi: 10.3390/ijms23095183
18. Li, N, Zhou, YQ, Cai, JL, Wang, YF, Zhou, XF, Hu, MT, et al. A novel trans-acting lncRNA of ACTG1 that induces the remodeling of ovarian follicles. Int J Biol Macromol. (2023) 242:125170. doi: 10.1016/j.ijbiomac.2023.125170
19. Zhou, M, Liu, XQ, Qiukai, E, Shang, YX, Zhang, XQ, Liu, ST, et al. Long non-coding RNA Xist regulates oocyte loss via suppressing miR-23b-3p/miR-29a-3p maturation and upregulating STX17 in perinatal mouse ovaries. Cell Death Dis. (2021) 12:14. doi: 10.1038/s41419-021-03831-4
20. Wang, Y, Guo, YX, Duan, CH, Li, JJ, Ji, SK, Yan, HH, et al. LncGSAR controls ovarian granulosa cell steroidogenesis via sponging miR-125b to activate SCAP/SREBP pathway. Int J Mol Sci. (2022) 23:20. doi: 10.3390/ijms232012132
21. Wang, F, Chen, XM, Sun, B, Ma, YJ, Niu, WB, Zhai, J, et al. Hypermethylation-mediated downregulation of lncRNA PVT1 promotes granulosa cell apoptosis in premature ovarian insufficiency via interacting with Foxo3a. J Cell Physiol. (2021) 236:5162–75. doi: 10.1002/jcp.30222
22. Huang, KJ, Geng, JS, and Wang, J. Long non-coding RNA RP11-552m11.4 promotes cells proliferation, migration and invasion by targeting Brca2 in ovarian Cancer. Cancer Sci. (2018) 109:1428–46. doi: 10.1111/cas.13552
23. Huang, WL, Zhang, XX, Li, A, Xie, LL, and Miao, XY. Differential regulation of MRNAs and lncRNAs related to lipid metabolism in Duolang and small tail Han sheep. Sci Rep. (2022) 12:12. doi: 10.1038/s41598-022-15318-z
24. La, YF, Tang, JS, He, XY, Di, R, Wang, XY, Liu, QY, et al. Identification and characterization of mRNAs and lncRNAs in the uterus of Polytocous and Monotocous small tail Han sheep (Ovis Aries). PeerJ. (2019) 7:e6938. doi: 10.7717/peerj.6938
25. Wang, JL, Chen, HY, and Zeng, XC. Identification of hub genes associated with follicle development in multiple births sheep by WGCNA. Front Vet Sci. (2022) 9:18. doi: 10.3389/fvets.2022.1057282
26. Yao, XL, Yang, F, El-Samahy, MA, Liu, B, Zhao, BR, Gao, XX, et al. Identification and characterization of unique and common lncRNAs and mRNAs in the pituitary, ovary, and uterus of Hu sheep with different prolificacy. Genomics. (2022) 114:110511. doi: 10.1016/j.ygeno.2022.110511
27. Chen, X, Chen, HY, Jiang, S, Shen, H, and Zeng, XC. Identification of lncRNA expression in the estrous cycle of Qira black sheep and its combination with Mirna analysis. Kafkas Univ Vet Fak Derg. (2021) 27:733–40. doi: 10.9775/kvfd.2021.26203
28. Yan, H, Zhang, JW, Wen, J, Wang, YB, Niu, WB, Teng, Z, et al. CDC42 controls the activation of primordial follicles by regulating PI3k signaling in mouse oocytes. BMC Biol. (2018) 16:16. doi: 10.1186/s12915-018-0541-4
29. Wang, P, Li, WT, Liu, ZY, He, XY, Hong, QH, Lan, R, et al. Identification of Wnt4 alternative splicing patterns and effects on proliferation of granulosa cells in goat. Int J Biol Macromol. (2022) 223:1230–42. doi: 10.1016/j.ijbiomac.2022.11.083
30. Bridges, MC, Daulagala, AC, and Kourtidis, A. LNCcation: lncRNA localization and function. J Cell Biol. (2021) 220:17. doi: 10.1083/jcb.202009045
31. Li, D, Wang, XY, Li, GY, Dang, YJ, Zhao, SD, and Qin, YY. lncRNA ZNF674-AS1 regulates granulosa cell glycolysis and proliferation by interacting with ALDOA. Cell Death Discov. (2021) 7:107. doi: 10.1038/s41420-021-00493-1
32. Yao, GD, Kong, Y, Yang, G, Kong, DQ, Xu, YJ, He, JH, et al. Lnc-Gulp1-2:1 affects granulosa cell proliferation by regulating Col3a1 expression and localization. J Ovarian Res. (2021) 14:10. doi: 10.1186/s13048-021-00769-1
33. Gao, M, Liu, LY, Li, SL, Zhang, XD, Chang, ZW, and Zhang, MZ. Inhibition of cell proliferation and metastasis of human hepatocellular carcinoma by miR-137 is regulated by CDC42. Oncol Rep. (2015) 34:2523–32. doi: 10.3892/or.2015.4261
34. Chen, HL, Yang, YF, Wang, YL, Li, Y, He, YM, Duan, JX, et al. Phospholipase C inhibits apoptosis of porcine primary granulosa cells cultured in vitro. J Ovarian Res. (2019) 12:10. doi: 10.1186/s13048-019-0567-4
35. Luo, N, Guo, J, Chen, L, Yang, W, Qu, X, and Cheng, Z. Arhgap10, downregulated in ovarian Cancer, suppresses Tumorigenicity of ovarian Cancer cells. Cell Death Dis. (2016) 7:10. doi: 10.1038/cddis.2015.401
36. Zhang, JQ, Ma, RJ, Li, L, Wang, LN, Hou, XJ, Han, LS, et al. Intersectin 2 controls actin cap formation and meiotic division in mouse oocytes through the Cdc42 pathway. FASEB J. (2017) 31:4277–85. doi: 10.1096/fj.201700179R
37. Zhang, JW, Yan, L, Wang, YB, Zhang, S, Xu, XQ, Dai, YL, et al. In vivo and in vitro activation of dormant primordial follicles by EGF treatment in mouse and human. Clin Transl Med. (2020) 10:e182. doi: 10.1002/ctm2.182
38. Tian, YQ, Li, XL, Wang, WJ, Hao, HS, Zou, HY, Pang, YW, et al. Knockdown of bone morphogenetic protein 4 gene induces apoptosis and inhibits proliferation of bovine cumulus cells. Theriogenology. (2022) 188:28–36. doi: 10.1016/j.theriogenology.2022.05.015
39. Xu, RF, Qin, N, Xu, XX, Sun, X, Chen, XX, and Zhao, JH. Inhibitory effect of SLIT2 on granulosa cell proliferation mediated by the CDC42-PAKs-ERK1/2 MAPK pathway in the Prehierarchical follicles of the chicken ovary. Sci Rep. (2018) 8:16. doi: 10.1038/s41598-018-27601-z
40. Rivera, HM, and Bethea, CL. Ovarian steroids increase Spinogenetic proteins in the macaque dorsal raphe. Neuroscience. (2012) 208:27–40. doi: 10.1016/j.neuroscience.2012.02.002
Keywords: sheep, ovaries, Loc105611671, RNA-RNA interaction, CDC42
Citation: Wang J, Chen H, Zhang Y, Shen H and Zeng X (2024) Long non-coding RNA Loc105611671 promotes the proliferation of ovarian granulosa cells and steroid hormone production upregulation of CDC42. Front. Vet. Sci. 11:1366759. doi: 10.3389/fvets.2024.1366759
Edited by:
Ning Song, Anhui Agricultural University, ChinaCopyright © 2024 Wang, Chen, Zhang, Shen and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiancun Zeng, zengxiancun@163.com
 Jinglei Wang
Jinglei Wang Hanying Chen2
Hanying Chen2 