Decoding chikungunya virus non-structural protein 3 interacting partners in THP-1 derived infected macrophages through proteomic profiling
- Vector-Borne Disease Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, India
Introduction: Chikungunya virus (CHIKV), an alphavirus transmitted by mosquitoes, has instigated several epidemics in recent years, sparking intensive efforts to understand its biology. Despite progress, the understanding of CHIKV’s molecular interactions with host cell constituents, especially in susceptible cells such as macrophages remains limited.
Methods: We used a mass spectrometry platform to characterize the interactions between CHIKV-nsP3, a viral nonstructural protein, and proteins in human THP-1 macrophage cells.
Results and Discussion: Our findings revealed 196 high-confidence interactions primarily involving nsP3. Further, the sub-cellular localization, and pathways these interacting proteins might be involved in were deduced using computational methods. The interacting partners were further incorporated into a comprehensive host-virus interaction network derived from extensive literature on alphavirus-host interactions. Collectively, this study offers the first interaction map between CHIKV nsP3 protein and THP-1 cells, illuminating new probable roles of host cell proteins in CHIKV’s replication cycle.
1 Introduction
Chikungunya disease (CHIKD) is an arthropod borne viral disease known to cause epidemics of fever associated with arthralgia. The causal agent, chikungunya virus (CHIKV) is an alphavirus, belonging to the family Togaviridae and contains a positive sense single stranded RNA genome consisting of two open reading frames flanked by untranslated regions (3`UTR& 5`UTR). The two open reading frames encode three structural (capsid, envelope E1 and E2) and four non-structural proteins (nsP1, nsP2, nsP3 and nsP4). Structural proteins are mainly involved in the formation of viral particles while non-structural proteins play an important role in viral genome replication by the formation of replication complex. Biochemical functions of nsPs are described: nsP1 shows methyl and guanyl transferase activity, nsP2 protein exhibits helicase and protease functions, nsP4 is known as RNA-dependent RNA polymerase while nsP3 protein has diverse biochemical functions and plays an essential role in viral RNA replication (1–3).
The CHIKV-nsP3 protein comprises of three domains: macrodomain, AUD domain, and hypervariable domain. The macrodomain is located at the N-terminal region of the protein, exhibits ADP ribosyl hydrolase activity and is crucial for the initiation of the virus replication. The alphavirus unique domain (AUD) lies adjacent to the macrodomain and is involved in viral assembly. The hypervariable region is present at the C-terminal region of the protein and plays an important role in determining the pathogenesis of the virus through interacting with host specific factors (4–7).
Different cell types are targeted by CHIKV throughout the course of infection, such as fibroblast, endothelial and epithelial cells, monocytes, and macrophages. Monocytes and macrophages play a major role during the pathogenesis of CHIKV and are amongst the crucial targets of CHIKV infection. The macrophages get activated upon infection, which leads to the production of a high amount of proinflammatory cytokines such as IL-1β IL-6, and IFN-1 expression. Simultaneously, these cells are recruited at the infection site to promote the inflammatory process. Several molecular alterations such as changes in global transcript level, protein expression level, and post-translational modification occur in the host during the virus infection (8, 9).
Proteomics along with bioinformatics analysis has advanced in recent times to provide important insights into cellular pathways and their signaling mechanisms. We conducted the present study to identify those host cellular factors present in infected human macrophages that interact with CHIKV nsP3. We performed co-immunoprecipitation of CHIKV infected THP-1 derived macrophages with recombinant nsP3 antibody followed by mass spectrometry (MS). We obtained the host partners of THP-1 derived macrophages that are interacting with CHIKV-nsP3 and subjected them to several bioinformatics analyses such as pathway analysis, sub-cellular localization analysis, protein-protein interaction networks, to understand the role of these molecules in CHIKV replication. Our study has thrown light on possible roles of those interacting partners of nsP3 that are present in the immune cells such as macrophages during CHIKV infection.
2 Materials and methods
2.1 Generation of plasmid constructs
To express CHIKV-nsP3 protein, nsP3 gene was amplified from CHIKV RNA using the primers (Forward: 5′-GTAATGGGATGTGCACCGTCGTACCGG-3′), (Reverse: 5′-TTACTCGTCGTCCGTGTCTG-3′) and cloned it in pET 28a vector with N-terminal His-tag for protein expression and purification in Escherichia coli (E.coli) codon plus cells. To confirm the positive constructs, RT PCR was done with total RNA using gene-specific primers with the Prime Script One-Step RT PCR kit (Takara, Japan).
2.2 Cell lines and viruses
THP-1, a human acute monocytic leukemia cell line obtained from ATCC (TIB-202), were cultured in Roswell Park Memorial Institute (RPMI) medium (Cat no: AL011S, HiMedia, India) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and 1% L-glutamine at 37°C with 5% CO2. The THP1 monocytes were further activated with phorbol 12-myristate 13-acetate (PMA) at a working concentration of 20 ng/ml in RPMI media. Post 48 hours of treatment with PMA, the adherent THP1 derived macrophages were washed with RPMI complete media and further used for experiments. The Vero cell line (ATCC-CCL-81) was maintained in Dulbecco’s Modified Eagle Media (DMEM) (Cat no: AL007A, HiMedia, India) supplemented with 10% FBS, penicillin/streptomycin at 37°C and 5% CO2. The CHIKV was isolated and purified from patient sera collected during an outbreak and propagated in both C6/36 and Vero cell lines (Accession no. JF950631.1) (10).
2.3 Virus infection
THP-1 cells were grown in 6-well plates in RPMI medium. Cells were infected with CHIKV strain (at a multiplicity of infection (MoI) 1. The virus was diluted in sera and antibiotic-free RPMI medium, and cells were incubated at 37°C. After 2hpi, cells were washed with phosphate-buffered saline (PBS) and grown in complete RPMI media. The cells and media were collected at different time points, and cell pellets were washed once with PBS and resuspended in RIPA lysis buffer for western blot.
2.4 Protein purification and antibody generation
CHIKV nsP3 protein was synthesized and purified based on methods outlined in previous publications (11, 12). Full length CHIKV-nsP3 gene was amplified from CHIKV-RNA and cloned in pet28a vector with N -terminal His tag for expression and purification in BL21 E. coli cells. Clones were transformed into E. coli BL21 (DE3 strain). CHIKV-nsP3 expressing E. coli cells were grown at 37°C and induced with 1 mM IPTG. Following cell lysis, the lysates were processed through Ni-NTA affinity column chromatography. CHIKV-nsP3 protein was eluted using elution tris buffer with 5% glycerol and varying concentrations of imidazole. Elutions were dialyzed against tris buffer and 5% glycerol and purified by Ion exchange chromatography where the bound protein was washed with Buffer A (10 mM NaCl, 25 mM Tris and 3 mM β-mercaptoethanol) followed by elution in Buffer B (1 M NaCl, 25 mM Tris and 3 mM β-mercaptoethanol). Eluted fractions containing purified CHIKV-nsP3 was collected and concentration was measured by Bradford protein assay (Bio-Rad, USA) using BSA as standard. The purified nsP3 protein was confirmed via Western blot and prepped for different experiments. In-house polyclonal antibody generation was done using lab rabbits. Briefly, an emulsion of purified CHIKV-nsP3 protein (100μg per rabbit) and Freund’s complete adjuvant was prepared and injected into rabbits. Booster doses were prepared by mixing the CHIKV-nsP3 protein with Freund’s incomplete adjuvant and were administered at regular intervals. Post each booster Bleeds were systematically collected from the rabbits and centrifuged to isolate the sera. Sera containing anti-CHIKV nsP3Ab was further purified by protein-A and protein-G beads. Specifically, we pooled the rabbit sera (2, 3 and 4) and subjected to our Protein A and G beads. The volume-based approach was used to ensure complete capture of all IgG antibodies. After incubation, antibodies were washed with wash buffer and were further eluted from the column using 0.1M glycine (pH 2.0) and neutralized in phosphate buffer. The purified anti nsP3 polyclonal antibody was further used for our in vitro study. Cross reactivity and Efficacy of purified antibody was checked by Western blot assay as well as IFA assay. The raised antibody was checked against the purified CHIKV-nsP3 protein as well as against a cell lysate to confirm antibody reactivity and specificity. Time kinetic study also confirmed the sensitivity of the purified antibody. Further before carrying out CoIP experiment, we optimized the specificity of the antibody in infected lysate and used IgG agarose bead as a negative control to filter the binding of non-specific proteins both in uninfected and infected samples. CoIP experiments were performed using purified antibody. The animals used in the study were approved by the institutional animal ethics committee (ICGEB/IAEC/02042019/VBD-4).
2.5 Co-immunoprecipitation of THP-1 lysate with purified nsP3
To understand the interacting host partners of CHIKV nsP3, we performed CO-IP of infected THP-1 lysate with purified CHIKV-nsp3 antibody (generated in-house) using Pierce co-immunoprecipitation kit (Thermo Scientific, USA) as per the manufacturer’s instructions. Briefly, 100 µl Protein A beads were incubated with CHIKV-nsP3 specific antibody overnight at 4°C. Post washing, antibody labeled beads were further incubated with infected THP-1 lysate (1mg of total protein concentration) overnight at 4°C. Further, beads were washed with IP wash buffer followed by three sets of elution using elution buffer. These Co-IP elutes were analyzed using mass spectrometry.
2.6 Sample preparation for mass spectrometry
Co-IP elutes were processed with in-solution trypsin digestion. Firstly, Co-IP elutes were adjusted with 50 mM ammonium bicarbonate buffer pH 8.0. Further, it was reduced with 10mM Dithiothreitol (DTT) for an hour at Room Temperature and alkylated with 40mM iodoacetamide (IAA) for an hour. Protein elutes were incubated in a water bath at 37°C for 16 hours for digestion using trypsin-Gold (Promega) at 1:50 proportion. The trypsin digestion reaction was stopped using formic acid (0.1%). The digested peptides were concentrated in speed-vac and analyzed by LC-MS/MS.
2.7 Mass spectrometry
LC-MS/MS analysis was performed Orbitrap Velos Pro MS coupled to Easy n-LC 1000 (Thermo Fisher Scientific). Tryptic digested peptide mixtures were first loaded onto a reverse-phase C-18 pre-column (Acclaim PepMap, 75 μm x 2 cm, 3μm, 100A°, Thermo Fisher Scientific) and then separated using an analytical column (Acclaim PepMap, 50μm x 15 cm, 2μm, 100A°) using a gradient of 5% solvent B (0.1% formic acid in 95/5 acetonitrile/water) to 35% solvent B in 35 min. The eluted peptides were injected into the mass spectrometer and MS1 data were acquired using a mass range from 350-2000 Da in Full scan mode at 60000 resolution. The top 15 precursors were allowed to fragment using CID (collision-induced dissociation) in the ion trap with a collision energy of 35. The raw data were analyzed using Proteome Discoverer 1.4 with the SEQUEST algorithm against Uniprot database. The identification of peptides was validated using Percolator at 5% FDR.
2.8 Western blot
Protein sample concentration was determined by BCA reagent and plotted against bovine serum albumin (BSA) standards of known concentrations. An equal amount of protein samples was loaded in 10% SDS-PAGE gels and run at 100V till the dye front reached the other end. Proteins were then transferred onto the nitrocellulose membrane. The membrane was then blocked with PBS (pH 7.4) + 5% BSA for 1hr at RT. The membrane was then incubated with primary antibodies (1:2,000 dilution) in PBS+2.5% BSA overnight at 4°C. The membrane was then washed with PBS+ 1% Tween-20 for 10 minutes and repeated thrice. It was then incubated with anti-mouse HRP secondary antibodies (1:5,000 dilution) in PBST+2.5% BSA for 1hr at RT. The membrane was then washed with PBS+ 0.1% Tween-20 for 10 min repeated thrice and visualized in Chemidoc MP (Bio-Rad).
2.9 Confocal imaging
The protocol of confocal imaging in THP-1-derived macrophage cells was previously published (9). Briefly, cells were seeded on glass coverslips in 12-well plates with 20ng/ml PMA concentration for its activation. Post activation, fixing was done with 4% paraformaldehyde in PBS for 30min and further permeabilized in 100% chilled methanol at 4˚C for 1hr. After washing with PBS, cells were incubated with 1%BSA and 0.3% Triton X-100 in PBS, Blocking was done for 45min using blocking buffer, followed by overnight incubation of CHIKV nsP3 primary antibody (in house generated, 1:100 dilution) in blocking buffer. Post-washing, Alexa Fluor 594 conjugated secondary antibodies (Invitrogen) were added for 1hr. Then the coverslips were washed three times with PBS and stained with DAPI (Invitrogen). Images of the sample were taken using a Nikon confocal microscope.
2.10 Bioinformatics analyses
The schematic workflow of bioinformatics analysis of MS data is shown in Supplementary Figure 2A. Proteins/peptides satisfying high FDR confidence were taken forth to statistical analysis in XLSTAT. Common proteins between CHIKV uninfected and infected libraries were excluded and unique proteins with high FDR were further accessed for functional annotation and subcellular localization.
2.11 Subcellular localization and functional annotation
Subcellular localization of the unique host proteins of nsP3-IP was identified using the web tool DeepLoc-1.0 (13) (http://www.cbs.dtu.dk/services/DeepLoc/) and extracting the information from the Human Protein Atlas database (https://www.proteinatlas.org) (14, 15). DeepLoc-1.0 was trained on experimentally verified Uniprot proteins and used Neural Network methods to predict the subcellular localization of the proteins. The proteins were classified into their respective pathways using different pathway analysis servers i.e., ShinyGO (http://bioinformatics.sdstate.edu/go/), KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/), FLAME (16) and Metascape tool (https://metascape.org/gp/index.html). The multiple gene sets testing and p-value adjustment were done using the Benjamini–Hochberg test. Significant pathways were selected with corrected p-values< 0.05.
2.12 Protein interaction network
The STRING database was used to fetch the host virus protein–protein interaction (PPI) network (https://string-db.org) (17). The 196 significant human protein along with nsP3 viral protein from the pull down study were further taken for this analysis. According to their organellar localization, the proteins in the network were grouped. Cytoscape 3.0 was used to visualize the network (https://cytoscape.org) (18).
3 Results
3.1 Expression profiling of CHIKV-nsP3 protein in CHIKV infected THP-1 derived macrophages
First, we profiled the expression of CHIKV-nsP3 protein in infected macrophages over a period of 48 hours to understand the expression of viral protein in relation to viral replication. We infected the THP1- derived macrophages with CHIKV (MoI 1) and observed the infection over time duration of 48 hours post infection. We observed 65-70% of cells to be infected at 24hpi that increased over the infection time points. For the purpose of detecting CHIKV-nsP3 in the infected cells, we purified CHIKV-nsP3 using previously published protocol (Ramesh et al.) (12), and raised antibodies in rabbit. The antibodies were further validated for their specificity both using recombinant CHIKV-nsP3 protein and cell lysate in uninfected and infected conditions (Figure 1A). When CHIKV-nsP3 expression was profiled in THP-1 derived macrophages during different time points, we observed that nsP3 protein expression began to increase from 12hpi and peaked at 24hpi, after which there was a significant reduction of CHIKV-nsP3 post 36hpi (Figure 1B). The increased expression of CHIKV-nsP3 at 24hpi was further validated using confocal imaging (Figure 1C). Based on the above observation of nsP3 protein expression at 24hpi in THP-1 derived macrophages, we performed a Co-IP study followed by mass spectrometry at this time point.
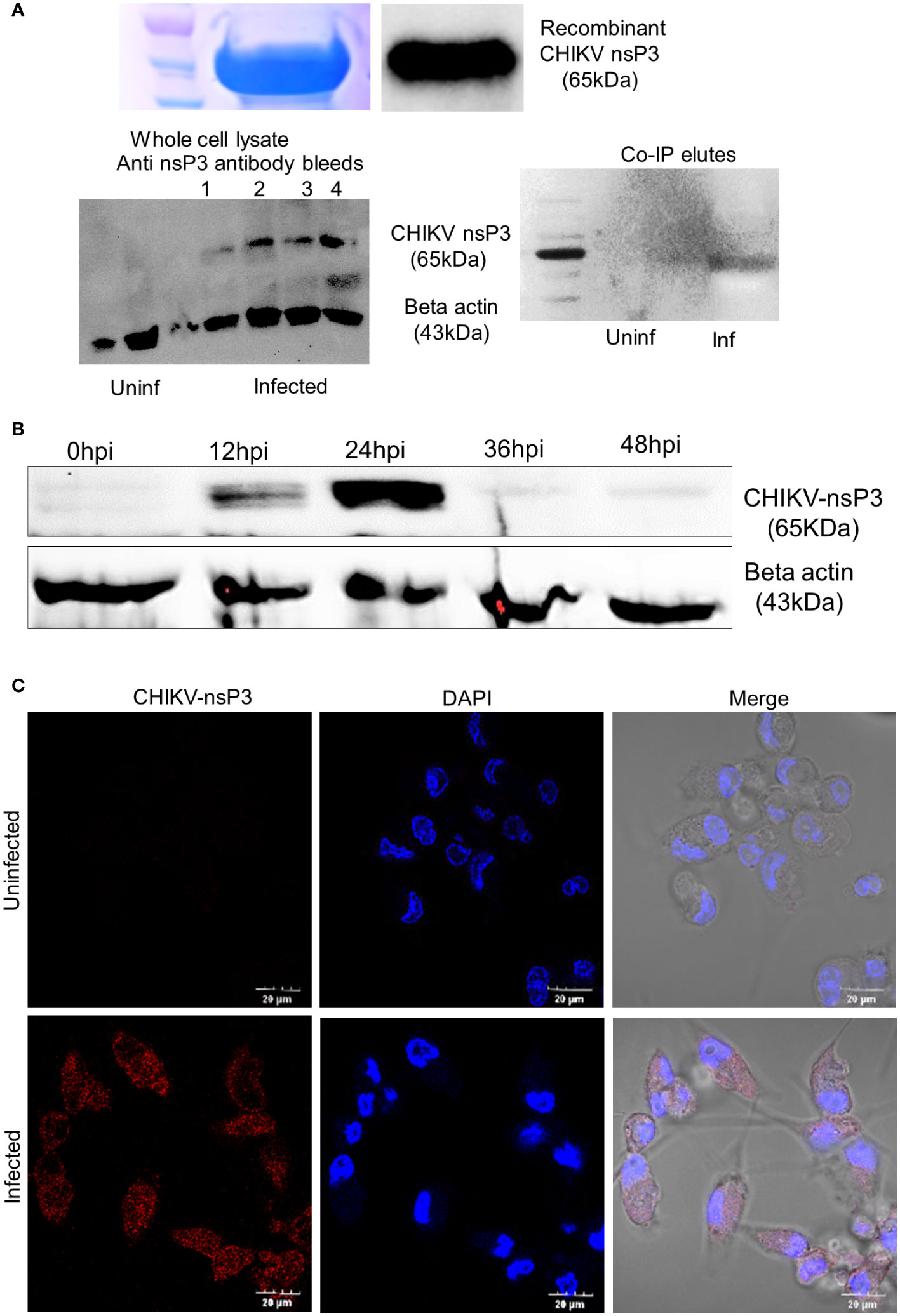
Figure 1 Antibody characterization and Protein expression profiling of CHIKV-nsP3 in CHIKV (MOI1) infected THP-1 derived macrophages. (A) Anti nsP3 antibody validation in recombinant protein, infected lysate and 24 hour post infection (24hpi) of the CoIP lysate using western blotting. (B) Inverted image for expression level of protein CHIKV-nsP3 in CHIKV infected human macrophages at different time points i.e., 0hpi, 12hpi, 24hpi, 36hpi and 48hpi using western blot. Actin was used as housekeeping control. Full western blot image is available as Supplementary Figure 6. (C) THP-1 derived macrophages were infected with CHIKV at MoI 1. Post 24hpi, cells were stained with DAPI (blue), and anti-CHIKV-nsP3 (red) and subjected to confocal imaging. Quantifications are measured in at least 6 different images per sample using NIS image processing software. All images are from live cells. Scale bar 20 μm.
3.2 Identification of CHIKV-nsP3 interacting host factors and their subcellular localization
To identify the interacting partners of CHIKV-nsP3, we performed co-immunoprecipitation of THP-1 infected lysate with CHIKV-nsP3 antibody. IgG antibody was used as a negative control to filter the binding of non-specific proteins both in uninfected and infected samples. The analysis revealed a total of 562 proteins to be present in the infected triplicate samples, while 692 proteins were found in uninfected triplicate samples, as shown in Figures 2A, B and Supplementary Table 1. Analysis of these proteins further revealed a total 366 host proteins to be common between the uninfected and infected samples. Further filtration of the proteins on the basis of high FDR and minimum peptide number two yielded a total of 196 proteins that were unique in CHIKV-nsP3 CO-IP samples (Figure 2B).
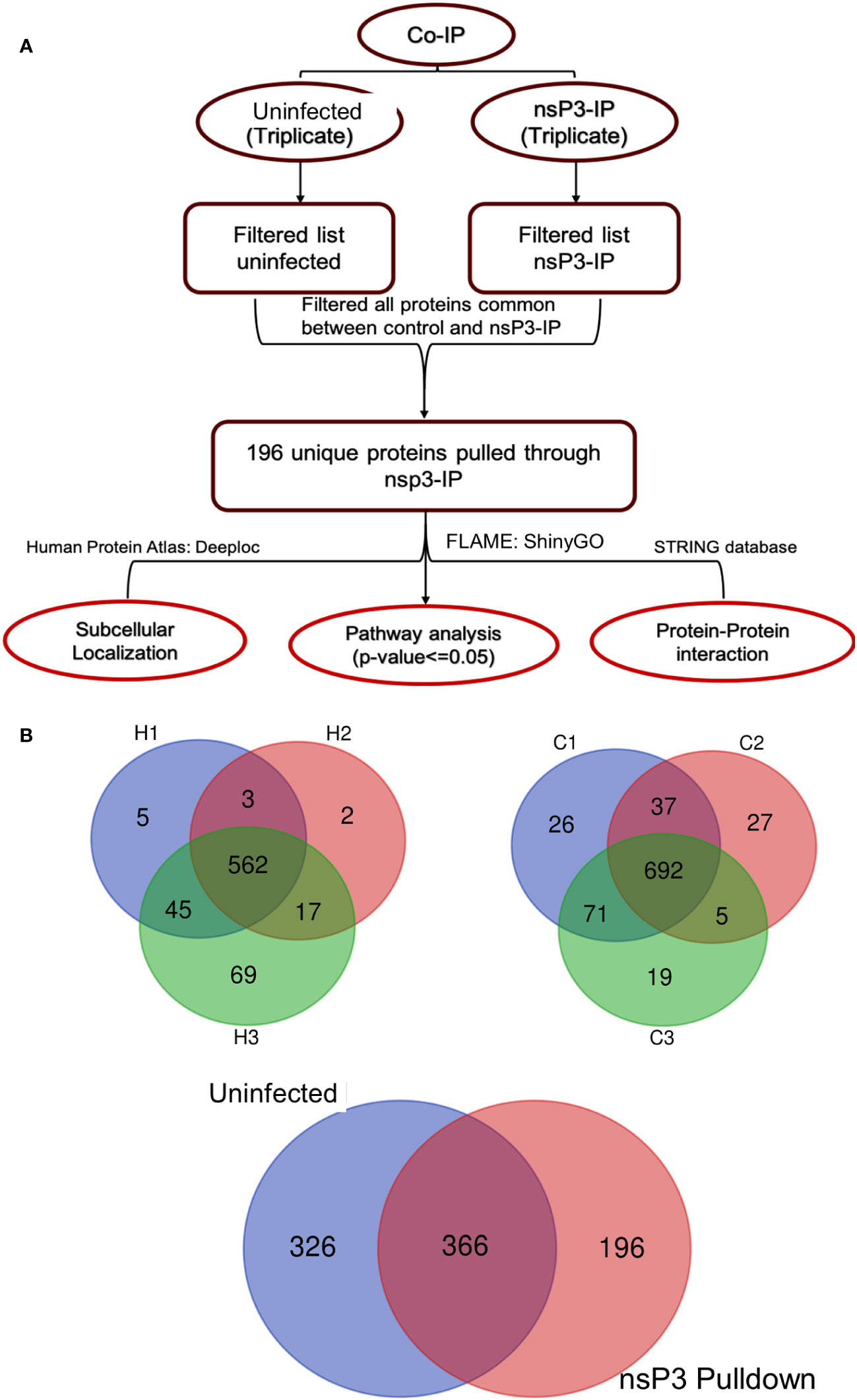
Figure 2 Identification of CHIKV-nsP3 interacting host factors. (A) The workflow of bioinformatics analysis of MS data. (B) Three-layer Venn diagrams are representing common and unique identified proteins across the uninfected and CHIKV infected cells in triplicate datasets. C1, C2, and C3 are the uninfected triplicate samples and H1, H2 and H3 are CHIKV infected triplicate samples. The overlapping regions correspond to the number of proteins present at more than one time. 692 protein were common across all the uninfected samples while 562 were common in infected samples. Proteins for which peaks were present at least in duplicates were taken for downstream analysis. 326 proteins were cumulatively unique to uninfected samples and 196 to infected samples.
The sub-cellular localization of the 196 interacting proteins was ascertained using the DeepLoc1.0 tool (http://www.cbs.dtu.dk/services/DeepLoc/) (19) and the Human Protein Atlas Database (Supplementary Table 2). Based on localization, these proteins were broadly classified into the cytoplasm, nucleus, endo membranal system, and extracellular matrix or secretory. It was observed that 96 (49%) resided in the cytoplasm while 48 proteins (24.4%) were localized in the nucleus. Further, 58 proteins (30%) were found to reside in the endo membranal system (Figure 3) (15). A total of 8 proteins were secreted and resided in the extracellular matrix. Some of the proteins were found to present in multiple cellular locations. For instance, ENSG00000033867 (SLC4A7) was present in plasma membrane, cytosol and focal adhesion sites. ENSG00000064763 (FAR2) was present in Golgi apparatus and nucleoli. Likewise, ENSG00000067066 (SP100) and ENSG00000068366 (ACSL4) were present in nuclear bodies, nucleoplasm, Golgi apparatus, and mitochondria respectively (Supplementary Table 2). It was interesting to note that in our confocal analysis, the majority of CHIKV nsP3 was uniformly localized in the cytoplasm while, some small clusters were found to be localized near the plasma membrane.
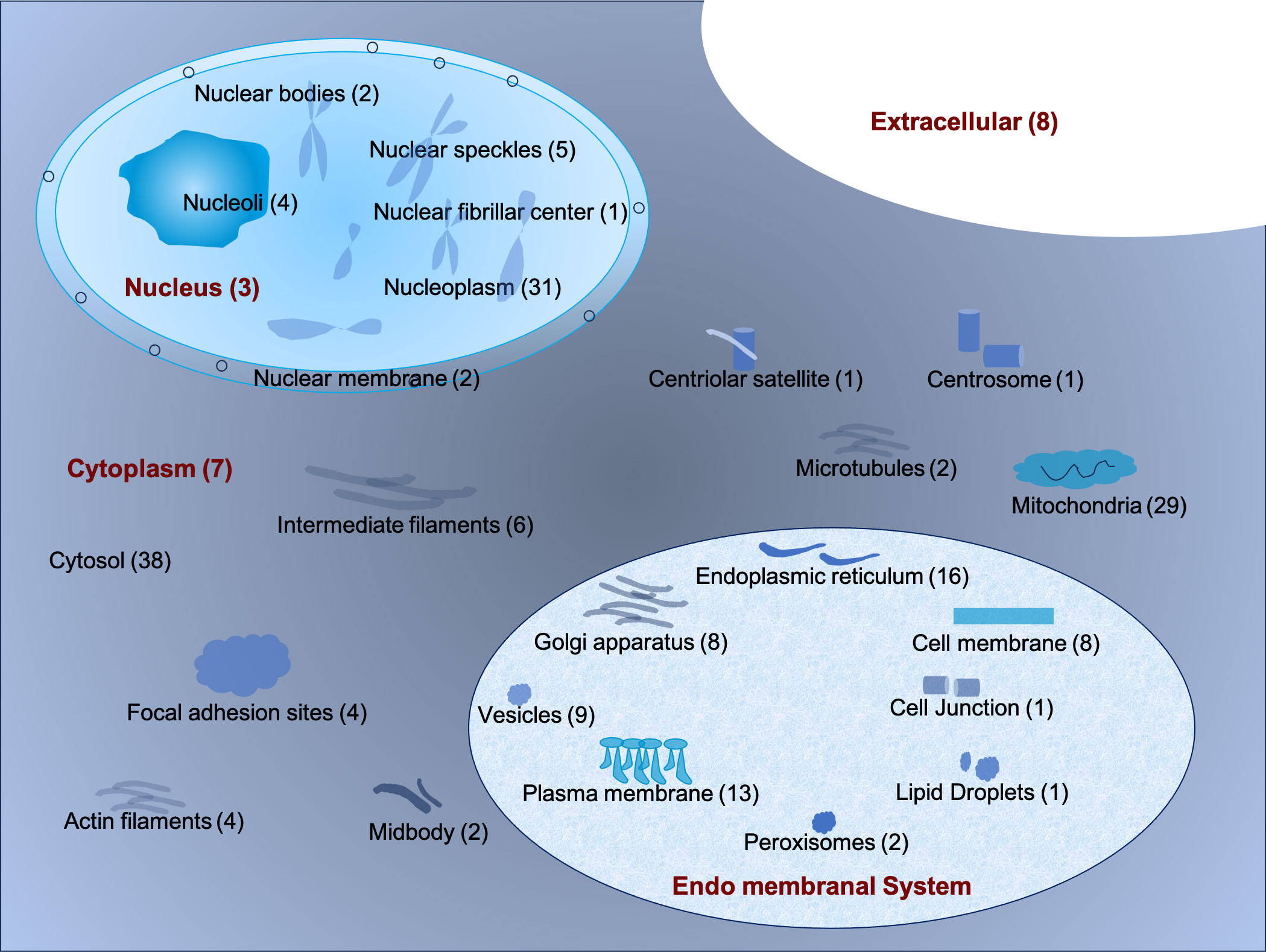
Figure 3 The schematic representation of the sub-cellular localization of interacting host proteins with CHIKV-nsP3 using DeepLoc 1.0 tool and Human Protein Atlas Database.
3.3 Functional annotation and pathway analysis of the interacting partners
The CHIKV-nsP3 interacting proteins were further classified into different pathways using FLAME and ShinyGO servers. A total of 88 pathways (Supplementary Table 3) were found significant based on corrected P-value≤ 0.05. Metabolic pathways were the most significant pathways with a maximum number of proteins involved in the process followed by pathways of neurodegeneration. A total of six pathways were involved in metabolic processes which included the TCA cycle, glycolysis/gluconeogenesis, oxidative phosphorylation, ribosome, pyruvate metabolism, and cellular senescence while five pathways were involved in the cell signaling process which includes cGMP-PKG signaling pathway, calcium signaling, glucagon signaling pathway and mRNA surveillance pathway (Figure 4A). An additional enrichment analysis was also performed using metascape software to get specific protein-wise functional involvement in biological process (Figure 4C, Supplementary Table 4). The analysis revealed that viral infection pathways, endosomal pathways, diabetic cardiomyopathy, systemic lupus erythematosus, cellular respiration were the most significant pathways at single protein analysis level. Top 20 pathways is shown in Figure 4C.
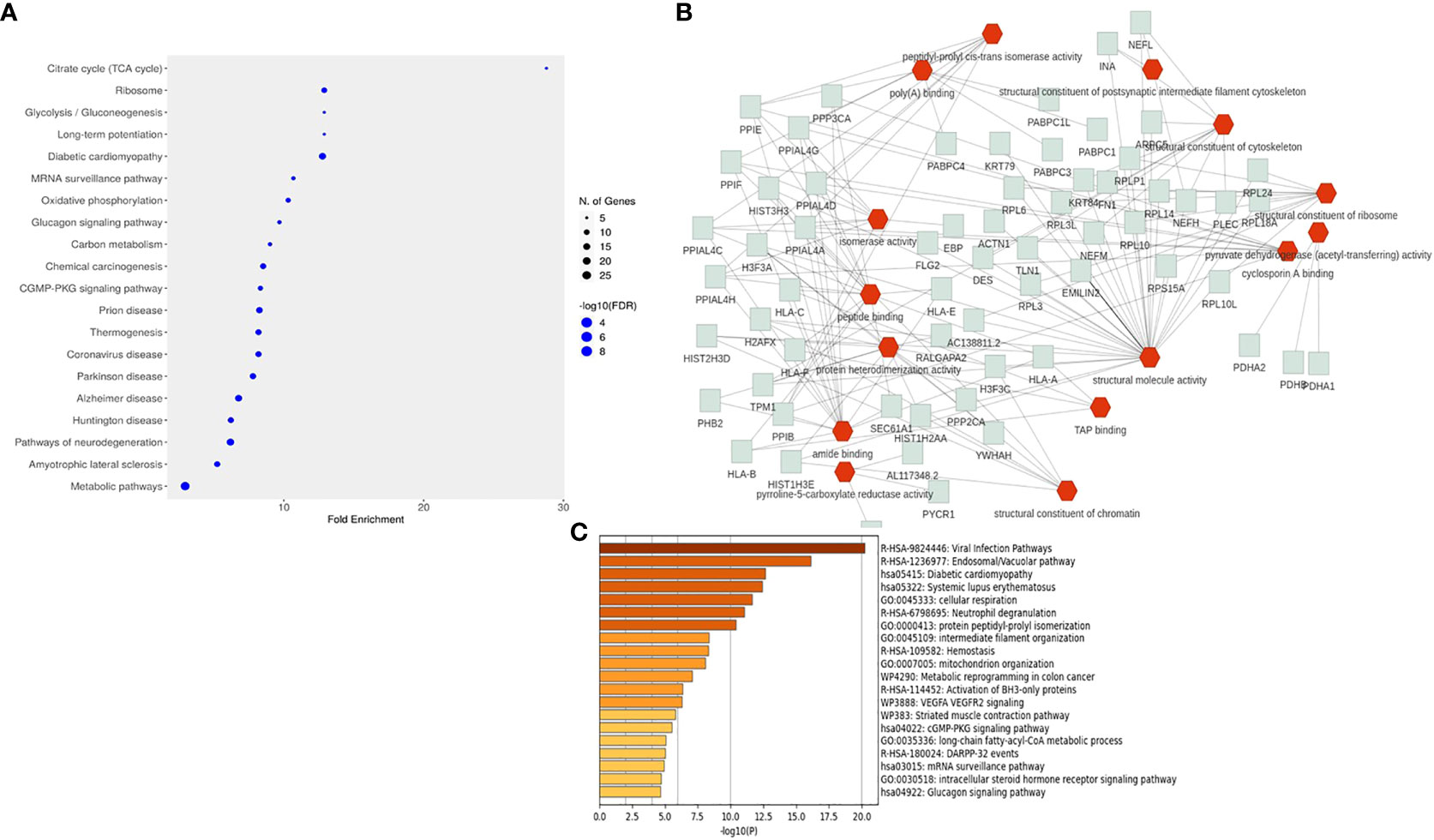
Figure 4 Pathways enrichment analysis of interacting host proteins with CHIKV-nsP3. (A) Dot plot representation of the top 20 significant pathways from the enrichment analysis. (B) The interactive network chart explains the biological processes of interacting partners with CHIKV-nsP3. Red edges show the biological processes while green edges show proteins related to these processes. (C) Enrichment analysis performed using metascape software. Bar graph is depicting top 20 pathways derived from the current study.
Protein set enrichment analysis of 196 interacting proteins revealed that the majority of proteins were involved in metabolic processes and cellular organization. Functional attributes of these proteins further showed that they were involved in binding activities like protein binding, DNA binding, chromatin binding, and ion binding activity while some have enzymatic activity such as hydrolase and transferase (Figure 4B). The analysis further revealed that SLC25A5, ATP2A2, TPM1, NCLN, SURF4, PAM16, COA3, DSTN, NETH, and TLN1 proteins were involved in neurofilament bundle assembly, intermediate filament organization, mitochondrial organization and organelle organization. Human leukocyte antigen (HLA) protein family members were majorly participating in positive regulation of T cell-mediated cytotoxicity, protection from natural killer cell-mediated cytotoxicity, antigen processing, and presentation of endogenous peptide antigen via MHC class I and Ib via ER pathway. APCS and DNAJA1 proteins formed separate clusters for protein binding process. HPX, CTSS, C3, FN1, MMP14, SLC18A3, and PYCR1 were found to regulate the L-proline biosynthetic process, organonitrogen compound biosynthetic process, and organonitrogen compound metabolic process.
3.4 Protein-protein interaction network associated with viral protein nsp3
All 196 proteins were assessed by PPI of which 110 proteins were identified in the resulting network. The disconnected nodes have been omitted from the analysis. The analysis revealed several clusters connecting into nsp3 by nine proteins namely fibronectin-1 (FN1), H3 Clustered Histone 13 (H3C13), Poly(A) Binding Protein Cytoplasmic 1 (PABPC1), Poly(A) Binding Protein Cytoplasmic 4 (PABPC4), and DnaJ Heat Shock Protein Family (Hsp40) Member A1 (DNAJA1), large ribosomal subunit protein 24 (RPL24), Ribosomal Protein L6 (RPL6), and OCIA domain-containing protein 1(OCIAD1) were then mapped into the PPI network using literature evidence from STRING database. Earlier studies on nsP3 interaction unveiled that G3BP interacting with CHIKV nsP3 protein and play a crucial role in regulating viral replication as well as cellular stress (20, 21) and was one of the most well studied interacting partner of alphavirus nsP3. However, in the present interaction study, we could not identify both G3BP1 and G3BP2 in the list of proteins derived from the MS analysis probably due to the choice of cells that were taken in the study. Nevertheless, when we created the PPI with all the interacting partners using the current data, we observed that six of the interacting partners mentioned above also directly interacted with G3BP1 and G3BP2 (Figure 5). Essentially, these six proteins formed the primary interactors with several of the other proteins interacting with these proteins along with CHIKV nsP3 and formed clusters based on their functions (Figure 5). The biggest cluster proteins (far left of the network) were primarily those that interacted with FN1 and PABPC1 proteins containing alpha-actin family which plays crucial role in cell division, migration, junction formation, chromatin remodeling, transcriptional regulation, vesicle trafficking, and cell shape regulation, human leukocyte antigen family that help at different sites with beta-2-microglobulin, an alpha-beta T-cell receptor, and inhibitory molecules, Pyrroline-5-carboxylate reductase which catalyzes the NADPH specific reduction of pyrroline-5-carboxylate to generate proline, and deformed epidermal autoregulatory factor that is essential for early embryonic and neuronal development etc. The middle cluster of Figure 5 was found to comprise L ribosomal proteins which plays vital role in cell proliferation regulation and in some instances as inducers of cell death. Protein phosphatase play key roles in setting the levels of tyrosine, serine and threonine phosphorylation in cells, thus participating in the regulation of many physiological processes, including cell growth, tissue differentiation and inter-cellular communication, solute carrier family transport a wide array of molecules, including sugars, amino acids, vitamins, nucleotides, metals, inorganic ions, organic anions, oligopeptides, and drugs, Prohibitin 2 protein that regulates various biological processes, including cell cycle, proliferation, apoptosis, transcription, signal transduction, and mitochondrial ridge morphogenesis, Cytochrome C1 plays a role in the electron transfer during oxidative phosphorylation, ADP ribosylation factor 3 controls invasion modality and metastasis by regulating N-cadherin levels, Resistance to inhibitors of cholinesterase-8A which is critical for growth factor receptor-induced actin cytoskeletal reorganization. This cluster is predominantly carrying interacting partners of PABPC1, PABPC4, and DNAJA1. The right cluster comprised of proteins of histone cluster family which are basic nuclear proteins, responsible for the nucleosome structure of the chromosomal fiber, Damage-specific DNA binding protein acts as a sensor of damage to maintain the balance between genome integrity and cell cycle progression, and Mini chromosome Maintenance Complex Component 7, a DNA helicase essential for genomic DNA replication were found to be interacting majorly with H3F3A and H3F3B.
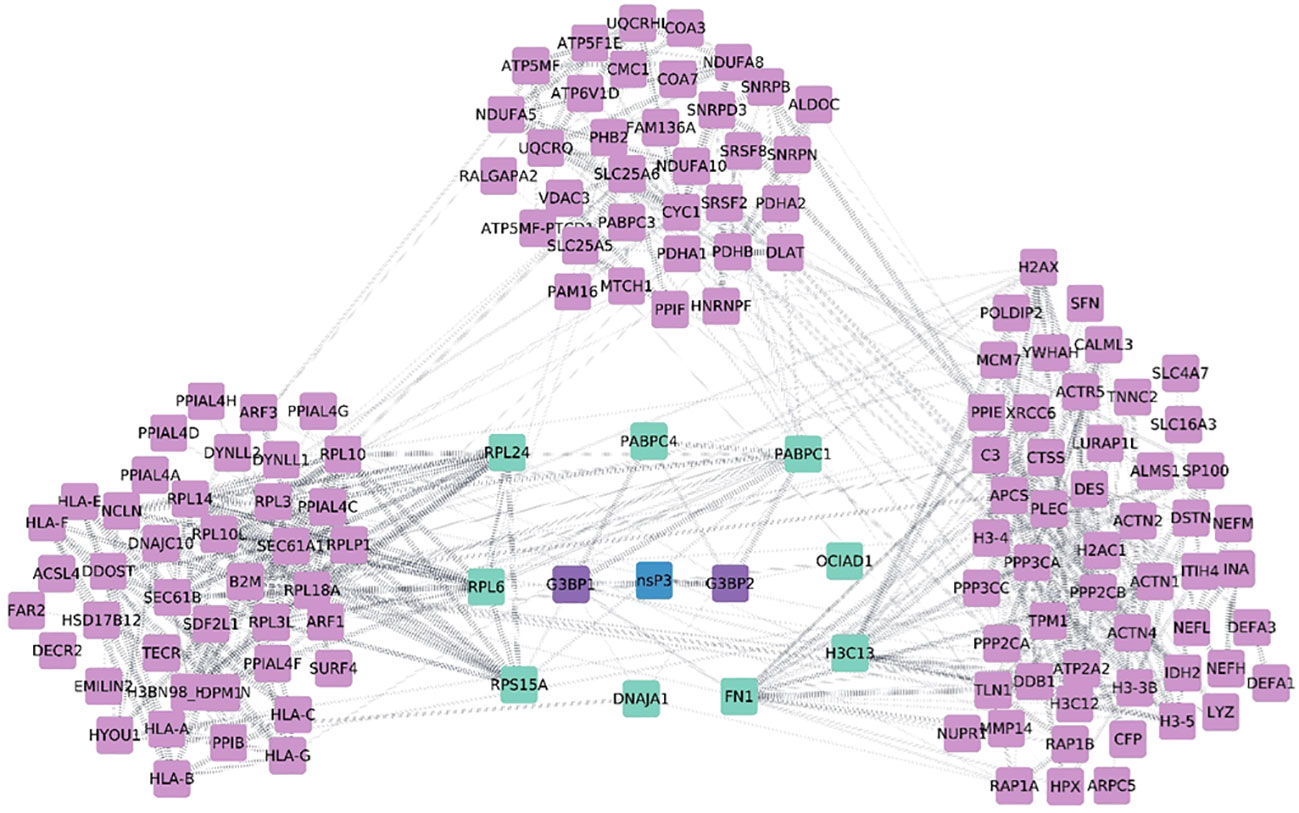
Figure 5 Diagrammatic representation of protein-protein interaction network from nsp3 Co-IP studies. The network was visualized using Cytoscape 3.0. CHIKV-nsP3 was highlighted in teal blue color while human proteins from the study were in sky blue and purple color. Proteins named G3BP1 and G3BP2 were intermediate proteins between viral proteins as per previous studies (experimentally validated). These two proteins are marked in blue color and their edges are highlighted in blue color with dotted lines. Thicker nodes are showing strong experimental evidence of interaction.
4 Discussion
Macrophages are susceptible to getting infected with CHIKV and hold a critical function in the progression of infection owing to their role in the host’s defense against the virus. Depending upon the stimulatory environment, M1/M2 dichotomy produces the two activated states of macrophages exhibiting unique phenotypic and functional characteristics (22). Pro inflammatory conditions drive THP1 derived macrophages towards M1 phenotype while anti-inflammatory cytokine presence leads to the differentiation of cell population towards M2 phenotype. M1 macrophages are stimulated by TNFα, IFNγ, and LPS during pathogenesis and secrete cytokines such as TNFα, IL-6, inducible nitric oxide synthase (iNOS) and chemokines. The alternatively activated M2 macrophages is stimulated by IL-4 and produces anti-inflammatory factor such as IL-10, TGFβ, VEGF with the ultimate aim in wound repairing (23) and is also known to be involved in the pathogenesis of osteoarthritis. Identifying host proteins such as alpha defensin 1 to be interacting with CHIKV nsP3 in the present study throw light to the possible role of these molecules in promoting polarizing the macrophages by activating NF-κB pathway and thus attenuates chronic arthritis symptoms caused during CHIKV infection (24). Similarly, we identified another protein, namely, Prohibitin, that is known to act as a crucial regulator in the functional plasticity of macrophages to be an interacting partner with the viral protein (25). Our study can be described as the first to identify interacting partners in macrophages with CHIKV-nsP3 during CHIKV infection. Previous studies have reported interactions with model cell lines such as Vero, HEK, and BHK-1, and provide an overall interaction pattern and do not highlight specific cell type interactions (5, 26).
Recent studies on the global transcriptome analysis of macrophages infected with CHIKV have highlighted the perturbance of immune signaling and metabolic pathways upon CHIKV infection. The study reported that CHIKV induced major alterations in the transcriptome of infected macrophages at 24hpi with a robust production of chemokines and cytokines. This possibly is a strategy employed by the virus towards its survival by using host metabolism and defense mechanism (9); a feature corroborated in the current study with respect to host-viral protein.
Cell membranes are critical for all positive sense stranded RNA virus and exploitation of the membrane for the initial interaction of the virus with the host cell serves as a foremost tactic for establishing infection. Studies on DENV virus report a significant alteration in the lipid metabolism within host cells to promote viral replication (27, 28). In the present study we identified Fatty acyl-CoA as an interacting partner, known to regulate assembly of replication complex. A previous study on CHIKV reported that expression of fatty acid synthase (FASN) and stearoyl-CoA desaturase (SCD1) are required for optimal CHIKV genome replication (29). FASN also plays a pivotal role for various RNA viruses and its enzymatic activity regulates crucial metabolic processes including Respiratory Syncytial Virus (30), hepatitis C virus (HCV) and dengue virus (DENV) (31, 32). However, in the current study we couldn’t observe the two lipogenic enzymes as nsP3 interacting partners in THP1 derived macrophages. In our study, cGMP-PKG signaling pathways were found to be modulated which directly regulate the NF Kappa beta pathway, facilitating the activation of innate immune responses (33). Surfeit 4, another interacting partner, is a well-known protein in HCV infection. It has been observed to interact with NS4B and plays a pivotal role in the regulation of HCV replication through the formation of DM vesicles (34). Exploring its mechanism in relation to CHIKV infection needs further investigation in this direction.
Sub-cellular localization of the nsP3 interacting proteins unveiled that the majority of cellular proteins interact with CHIKV-nsP3 in the cytoplasm and our study is also consistent with these reports (12, 20, 26, 35). One of the main reasons for a large number of interacting host proteins with nsP3 is the presence of the hypervariable domain which is intrinsically disordered and binds to several crucial members of the cellular proteins such as the SH3 domain, G3BP and its homolog Rin in mosquitoes, NAP1 family. These three groups of proteins are essential in viral replication (26). G3BP proteins interact with specific repeating amino acid sequences present in the disordered region of the C terminal hypervariable domain. Knockout of G3BP results in the abolishment of CHIKV replication. The HVD domain also plays a major role in virus adaptability to replication in different hosts such as mosquitoes and vertebrates (6). In our study, we couldn’t observe any interaction between G3BP and CHIKV nsP3 protein. This lack of interaction could be due to low expression of G3BP proteins in THP1-derived macrophages. Notably, THP1-derived macrophages showed permissiveness to CHIKV infection, while earlier reports suggested that the G3BP1 protein is predominantly found in non-permissive macrophages (36, 37). Further in-depth studies are needed to shed more light on these observations. CHIKV requires macrophage as an obligate host for its replication. Interacting partner H+ ATPase is important for the maintenance of endosomal and lysosomal activity that the virus requires to establish a successful replication in the macrophage (38). Other interacting partners such as the PABPs have previously been reported to be essential in viral replication. It is known that viruses are dependent on host cellular systems using PABPs to aid replication and protein synthesis. Even without specific nuclear signals, PABPs can shuttle between the nucleus and cytosol. Viruses can regulate the host translation initiation process by cleaving PABPs or relocating them into the nucleus. Also, they can interfere with stress granules containing PABPs (39). Previous reports demonstrated that CHIKV interacts with HSP70 for its entry into the mosquito cell (40) and HSP40 is a crucial partner of HSP70. Hence, we hypothesize that CHIKV nsP3 may interact with both PABPs and HSP40, potentially impacting the viral replication.
Our PPI landscape revealed certain proteins that indirectly interact with CHIKV-nsP3. Earlier studies on rotavirus demonstrated the interaction of viral nsP3 with viral mRNAs, eIF4G, and a cellular protein called RoXaN. These interactions help in suppressing the host translation machinery by displacing the cytoplasmic poly(A) binding protein (PABP-C1) (41). Similarly, in coronavirus, viral nsP3 dislodged PABP-C4 from the translation system by interacting with several cellular factors such as eIF4G and RoXaN (42). Altogether, understanding the protein-protein interactions network in CHIKV infections has provided insights into the functional role of the interacting partners, and future investigations are required to decipher the mechanisms used by the virus to infect and evade the macrophage immune system.
5 Limitations
While this study provides valuable insights into the molecular interactions between CHIKV and host cells, several limitations should be acknowledged. First, the high-confidence interactions identified between CHIKV-nsP3 and human proteins require further validation to confirm their biological significance. Also, we acknowledge that the use of mitogen-induced THP-1 derived macrophages may not be the ideal alternative to hPBMC-derived macrophages as there is very less secretion of IL-6, 8 and 10 during inflammatory condition. Furthermore, the identified host pathways involved in CHIKV replication are based on proteomics data, which may not capture post-translational modifications or dynamic changes in protein-protein interactions that could be crucial for understanding the host-virus interplay.
6 Conclusion
This study demonstrates the power of proteomic techniques in elucidating the intricate relationship between chikungunya virus (CHIKV) and host cells, particularly human THP-1 cells. The proteomic profile of CHIKV-infected THP-1 cells was analyzed using LC-MS-based mass spectrometry, revealing the involvement of the CHIKV-nsP3 protein in various host pathways and its interaction with macrophage lysates. Critical cellular pathways implicated in viral replication were identified, including metabolic pathway, cGMP-PKG signaling pathway, calcium signaling, and mRNA surveillance pathway. Notably, host cell metabolism and particularly the TCA cycle were found to be pivotal for virus replication. In addition, the cGMP-PKG signaling pathway modulates the innate immune response, while calcium, an intracellular messenger, is implicated in multiple stages of viral infection. Further investigation confirmed the localization of the nsP3 protein in the cytoplasm and its interaction with host cell proteins, including PABPs and HSP40, disrupting cellular processes and potentially affecting viral replication. While CHIKV was found to modulate the RNAi pathway through its nsP3 and nsP2 proteins, no direct interactions between the nsP3 protein and core RNAi proteins were found in human macrophages. Overall, this study provides valuable insights into the molecular interactions between CHIKV and its host cells, opening new avenues for targeted antiviral strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRIDE, PXD045252.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
SS: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. PS: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. NM: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. SC: Data curation, Formal analysis, Software, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a financial grant from the Department of Biotechnology, Government of India (BT/PR14725/AGR/36/672/2010) to SS.
Acknowledgments
We thank Mrs. Purnima Kumar for her assistance in confocal microscopy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2024.1310161/full#supplementary-material
References
1. Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol. (2015) 96. doi: 10.1099/jgv.0.000249
2. Jagadesh A, Jayaram A, Babu N, Mudgal PP, Sudandiradas R, Sheik S, et al. Current status of chikungunya in India. Front Microbiol. (2021) 12. doi: 10.3389/fmicb.2021.695173
3. Mathur K, Anand A, Dubey SK, Sanan-Mishra N, Bhatnagar RK, Sunil S. Analysis of chikungunya virus proteins reveals that non-structural proteins nsP2 and nsP3 exhibit RNA interference (RNAi) suppressor activity. Sci Rep. (2016) 6:38065. doi: 10.1038/srep38065
4. Foy NJ, Akhrymuk M, Akhrymuk I, Atasheva S, Bopda-Waffo A, Frolov I, et al. Hypervariable domains of nsP3 proteins of new world and old world alphaviruses mediate formation of distinct, virus-specific protein complexes. J Virol. (2013) 87. doi: 10.1128/JVI.02853-12
5. Lukash T, Agback T, Dominguez F, Shiliaev N, Meshram C, Frolova EI, et al. Structural and functional characterization of host FHL1 protein interaction with hypervariable domain of chikungunya virus nsP3 protein. J Virol. (2020) 95. doi: 10.1128/JVI.01672-20
6. Kim DY, Reynaud JM, Rasalouskaya A, Akhrymuk I, Mobley JA, Frolov I, et al. New world and old world alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PloS Pathog. (2016) 12. doi: 10.1371/journal.ppat.1005810
7. Rathore APS, Haystead T, Das PK, Merits A, Ng ML, Vasudevan SG. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antiviral Res. (2014) 103:7–16. doi: 10.1016/j.antiviral.2013.12.010
8. Nyman TA, Matikainen S. Proteomics to study macrophage response to viral infection. J Proteomics. (2018) 180:99–107. doi: 10.1016/j.jprot.2017.06.018
9. Srivastava P, Chaudhary S, Malhotra S, Varma B, Sunil S. Transcriptome analysis of human macrophages upon chikungunya virus (CHIKV) infection reveals regulation of distinct signaling and metabolic pathways during the early and late stages of infection. Heliyon. (2023) 9:E17158. doi: 10.1016/j.heliyon.2023.e17158
10. Shrinet J, Jain S, Sharma A, Singh SS, Mathur K, Rana V, et al. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J. (2012) 9:100. doi: 10.1186/1743-422X-9-100
11. George A, Amrutha MS, Srivastava P, Sai VVR, Sunil S, Srinivasan R. Label-free detection of chikungunya non-structural protein 3 using electrochemical impedance spectroscopy. J Electrochem Soc. (2019) 166:B1356. doi: 10.1149/2.1081914jes
12. Kumar R, Srivastava P, Mathur K, Shrinet J, Dubey SK, Chinnappan M, et al. Chikungunya virus non-structural protein nsP3 interacts with Aedes aEgypti DEAD-box helicase RM62F. Virusdisease. (2021) 32:657–65. doi: 10.1007/s13337-021-00734-y
13. Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. (2017) 33:4049. doi: 10.1093/bioinformatics/btx548
14. Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Sci (1979). (2017) 357:6352. doi: 10.1126/science.aan2507
15. Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. A subcellular map of the human proteome. Sci (1979). (2017) 356:6340. doi: 10.1126/science.aal3321
16. Thanati F, Karatzas E, Baltoumas FA, Stravopodis DJ, Eliopoulos AG, Pavlopoulos GA. Flame: A web tool for functional and literature enrichment analysis of multiple gene lists. Biol (Basel). (2021) 10. doi: 10.1101/2021.06.02.446692
17. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. Erratum: The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49:D605–12. doi: 10.1093/nar/gkaa1074
18. Excoffier L, Gouy A, Daub JT, Shannon P, Markiel A, Ozier O, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Nucleic Acids Res. (2017) 13. doi: 10.1101/gr.1239303
19. Kumar-Singh A, Shrinet J, Parniewska MM, Fuxe J, Dobra K, Hjerpe A. Mapping the interactome of the nuclear heparan sulfate proteoglycan syndecan-1 in mesothelioma cells. Biomolecules. (2020) 10:1034. doi: 10.3390/biom10071034
20. Fros JJ, Domeradzka NE, Baggen J, Geertsema C, Flipse J, Vlak JM, et al. Chikungunya Virus nsP3 Blocks Stress Granule Assembly by Recruitment of G3BP into Cytoplasmic Foci. J Virol. (2012) 86. doi: 10.1128/JVI.01506-12
21. Götte B, Liu L, McInerney GM. The enigmatic alphavirus non-structural protein 3 (nsP3) revealing its secrets at last. Viruses. (2018) 10:105. doi: 10.3390/v10030105
22. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. (2008) 8:958–69. doi: 10.1038/nri2448
23. Murray PJ. Macrophage polarization. Annu Rev Physiol. (2017) 79:541–66. doi: 10.1146/annurev-physiol-022516-034339
24. Xie JW, Wang Y, Xiao K, Xu H, Luo ZY, Li L, et al. Alpha defensin-1 attenuates surgically induced osteoarthritis in association with promoting M1 to M2 macrophage polarization. Osteoarthritis Cartilage. (2021) 29:1048–59. doi: 10.1016/j.joca.2021.04.006
25. Xu YXZ, Ande SR, Ikeogu NM, Zhou K, Uzonna JE, Mishra S. Prohibitin plays a role in the functional plasticity of macrophages. Mol Immunol. (2022) 144:152–65. doi: 10.1016/j.molimm.2022.02.014
26. Meshram CD, Agback P, Shiliaev N, Urakova N, Mobley JA, Agback T, et al. Multiple host factors interact with the hypervariable domain of chikungunya virus nsP3 and determine viral replication in cell-specific mode. J Virol. (2018) 92. doi: 10.1128/JVI.00838-18
27. Zhang J, Diaz A, Mao L, Ahlquist P, Wang X. Host acyl coenzyme A binding protein regulates replication complex assembly and activity of a positive-strand RNA virus. J Virol. (2012) 86. doi: 10.1128/JVI.06701-11
28. St Clair LA, Mills SA, Lian E, Soma PS, Nag A, Montgomery C, et al. Acyl-coa thioesterases: A rheostat that controls activated fatty acids modulates dengue virus serotype 2 replication. Viruses. (2022) 14:240. doi: 10.3390/v14020240
29. Bakhache W, Neyret A, McKellar J, Clop C, Bernard E, Weger-Lucarelli J, et al. Fatty acid synthase and stearoyl-CoA desaturase-1 are conserved druggable cofactors of Old World Alphavirus genome replication. Antiviral Res. (2019) 172:104642. doi: 10.1016/j.antiviral.2019.104642
30. Ohol YM, Wang Z, Kemble G, Duke G. Direct inhibition of cellular fatty acid synthase impairs replication of respiratory syncytial virus and other respiratory viruses. PloS One. (2015) 10. doi: 10.1371/journal.pone.0144648
31. Huang JT, Tseng CP, Liao MH, Lu SC, Yeh WZ, Sakamoto N, et al. Hepatitis C virus replication is modulated by the interaction of nonstructural protein NS5B and fatty acid synthase. J Virol. (2013) 87. doi: 10.1128/JVI.02526-12
32. Tongluan N, Ramphan S, Wintachai P, Jaresitthikunchai J, Khongwichit S, Wikan N, et al. Involvement of fatty acid synthase in dengue virus infection. Virol J. (2017) 14:28. doi: 10.1186/s12985-017-0685-9
33. Kanoh H, Iwashita S, Kuraishi T, Goto A, Fuse N, Ueno H, et al. cGMP signaling pathway that modulates NF-κB activation in innate immune responses. iScience. (2021) 24:103473. doi: 10.1016/j.isci.2021.103473
34. Kong L, Aoyagi H, Yang Z, Ouyang T, Matsuda M, Fujimoto A, et al. Surfeit 4 contributes to the replication of hepatitis C virus using double-membrane vesicles. J Virol. (2020) 94. doi: 10.1128/JVI.00858-19
35. Gao Y, Goonawardane N, Ward J, Tuplin A, Harris M. Multiple roles of the non-structural protein 3 (nsP3) alphavirus unique domain (AUD) during Chikungunya virus genome replication and transcription. PloS Pathog. (2019) 15. doi: 10.1371/journal.ppat.1007239
36. Felipe VLJ, Paula AV, Silvio UI. Chikungunya virus infection induces differential inflammatory and antiviral responses in human monocytes and monocyte-derived macrophages. Acta Trop. (2020) 211:105619. doi: 10.1016/j.actatropica.2020.105619
37. Page C, François C, Goëb V, Duverlie G. Human parvovirus B19 and autoimmune diseases. Review of the literature and pathophysiological hypotheses. J Clin Virol. (2015) 72:69–74. doi: 10.1016/j.jcv.2015.09.007
38. Geisow MJ, D’Arcy Hart P, Young MR. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: Studies by fluorescence spectroscopy. London, England: J Cell Biol (1981) 89:645–52. doi: 10.1083/jcb.89.3.645
39. Gao J, Tang YD, Hu W, Zheng C. When poly(A) binding proteins meet viral infections, including SARS-coV-2. J Virol. (2022) 96:e00136–22. doi: 10.1128/jvi.00136-22
40. Ghosh A, Desai A, Ravi V, Narayanappa G, Tyagi BK. Chikungunya virus interacts with heat shock cognate 70 protein to facilitate its entry into mosquito cell line. Intervirology. (2018) 60:247–62. doi: 10.1159/000489308
41. Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, et al. Nuclear Localization of Cytoplasmic Poly(A)-Binding Protein upon Rotavirus Infection Involves the Interaction of NSP3 with eIF4G and RoXaN. J Virol. (2008) 82. doi: 10.1128/JVI.00872-08
Keywords: chikungunya virus (CHIKV), macrophages, non-structural protein 3, proteome, THP1
Citation: Srivastava P, Mishra N, Chaudhary S and Sunil S (2024) Decoding chikungunya virus non-structural protein 3 interacting partners in THP-1 derived infected macrophages through proteomic profiling. Front. Virol. 4:1310161. doi: 10.3389/fviro.2024.1310161
Received: 09 October 2023; Accepted: 28 February 2024;
Published: 14 March 2024.
Edited by:
Perumal Arumugam Desingu, Indian Institute of Science (IISc), IndiaReviewed by:
Abhinav Kaushik, Harvard University, United StatesMaia Kavanagh Williamson, LifeArc, United Kingdom
Copyright © 2024 Srivastava, Mishra, Chaudhary and Sunil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sujatha Sunil, sujatha@icgeb.res.in
†These authors have contributed equally to this work
 Priyanshu Srivastava
Priyanshu Srivastava Nimisha Mishra
Nimisha Mishra Sakshi Chaudhary
Sakshi Chaudhary Sujatha Sunil
Sujatha Sunil