Expression of Fox genes in the cephalochordate Branchiostoma lanceolatum
- Centre National De La Recherche Scientifique, UPMC université Paris 06, UMR 7232, BIOM, Observatoire Océanologique de Banyuls sur Mer, Banyuls sur Mer, France
Forkhead box (Fox) genes code for transcription factors that play important roles in different biological processes. They are found in a wide variety of organisms and appeared in unicellular eukaryotes. In metazoans, the gene family includes many members that can be subdivided into 24 classes. Cephalochordates are key organisms to understand the functional evolution of gene families in the chordate lineage due to their phylogenetic position as an early divergent chordate, their simple anatomy and genome structure. In the genome of the cephalochordate amphioxus Branchiostoma floridae, 32 Fox genes were identified, with at least one member for each of the classes that were present in the ancestor of bilaterians. In this work we describe the expression pattern of 13 of these genes during the embryonic development of the Mediterranean amphioxus, Branchiostoma lanceolatum. We found that FoxK and FoxM genes present an ubiquitous expression while all the others show specific expression patterns restricted to diverse embryonic territories. Many of these expression patterns are conserved with vertebrates, suggesting that the main functions of Fox genes in chordates were present in their common ancestor.
Introduction
Forkhead box (Fox) transcription factors originated early during evolution and are specific to opisthokonts. They are present in fungi as well as in metazoans (Mazet et al., 2006; Larroux et al., 2008; Shimeld et al., 2010a) in which they play essential roles during embryonic development (Carlsson and Mahlapuu, 2002; Tuteja and Kaestner, 2007a,b; Benayoun et al., 2011). Fox proteins possess a helix-turn-helix DNA-binding domain called the forkhead domain which corresponds to a conserved region of approximately 110 amino acids (Weigel and Jackle, 1990; Clark et al., 1993). A molecular phylogeny-based classification of the Fox gene family allowed to propose its subdivision into 24 classes (ranged from FoxA to FoxS and including subfamilies that were recently subdivided: FoxJ (FoxJ1 and FoxJ2), FoxL (FoxL1 and FoxL2), and FoxN (FoxN1/4 and FoxN2/3) (Mazet et al., 2003). Many Fox gene losses or duplications occurred in different bilaterian clades, affecting different Fox classes. For example, FoxAB is found in cephalochordates and in the sea urchin but not in tunicates or vertebrates (Tu et al., 2006; Yu et al., 2008a), and families R and S are vertebrate-specific (Wotton and Shimeld, 2006; Shimeld et al., 2010b). Using phylogenetic analyses, it has been proposed that 22 Fox gene families were already present in the bilaterian ancestor (Shimeld et al., 2010b).
Cephalochordates (i.e., amphioxus) belong to the chordate phylum together with tunicates and their sister group, the vertebrates. They present morphological, developmental, and genomic characteristics that are proposed to be very similar to the ancestral state in the chordate clade, making amphioxus a key model system to understand chordate evolution (Bertrand and Escriva, 2011, 2014). Interestingly, it has been shown that amphioxus is the only living bilaterian possessing at least one member of each of the 22 Fox gene families proposed to have been present in Urbilateria (Yu et al., 2008a). Thus, the study of Fox genes in this cephalochordate may shed light on the functional evolutionary history of this transcription factor gene family. Past studies using genomic data from the Caribbean cephalochordate Branchiostoma floridae described the presence of 32 Fox genes in this species (Yu et al., 2008a) and the expression pattern of 11 of these genes was previously described: FoxAa and FoxAb (formerly named AmHNF3-1 and AmHNF3-2, respectively) (Shimeld, 1997), FoxB (Mazet and Shimeld, 2002), FoxC (Mazet et al., 2006), FoxD (Yu et al., 2002b), FoxE4 (Yu et al., 2002a), FoxF (Mazet et al., 2006; Onimaru et al., 2011), FoxG (Toresson et al., 1998), FoxL1 (Mazet et al., 2006), FoxN1/4a (Bajoghli et al., 2009), FoxQ1 and FoxQ2 (Yu et al., 2003; Mazet et al., 2006). In this work we searched for Fox sequences in the transcriptome of the Mediterranean amphioxus Branchiostoma lanceolatum. We found 28 Fox sequences and we describe here the spatiotemporal expression pattern of 13 Fox genes during embryonic development, including seven previously described in B. floridae and six for which expression was not known. We show that in B. lanceolatum some Fox genes exhibit ubiquitous expression as FoxK and FoxM, while the others show specific and dynamic expression patterns restricted to diverse embryonic territories. These expression patterns suggest that Fox genes are performing both general and specific functions during amphioxus embryonic development, most of them being probably ancestral in the chordate clade.
Materials and Methods
Phylogenetic Analysis
All reference sequences, except for B. lanceolatum, were obtained from Genbank or from Fritzenwanker et al. (2014) The multiple alignment was performed only for the conserved Forkhead amino acid domain sequences using the MUSCLE module implemented in MEGA 6 and manually refined in its interface (Tamura et al., 2013). The best fit substitution model for phylogenetic reconstruction was estimated using MEGA 6 (Tamura et al., 2011). Bayesian inference (BI) tree was inferred using MrBayes 3.2 (Ronquist et al., 2012), with the model recommended by MEGA 6 under the Akaike information criterion (RtRev+Γ), at the CIPRES Science Gateway V. 3.1 (Miller et al., 2015). Two independent runs were performed, each with four chains and 1 million generations. A burn-in of 25% was used and a 50 majority-rule consensus tree was calculated for the remaining trees.
Cloning and Expression Study
B. lanceolatum Fox sequences were recovered from its reference transcriptome (Oulion et al., 2012) by TBLASTN using sequences from B. floridae as queries. Specific primers were then designed for RT-PCR amplification from total RNA. Primer sequences are as follow:
FoxA_a_5′ AAGTCGCCGGTGTACGAGATG
FoxA_a_3′ GTATTATAGAGACGAAGGTTG
FoxA_b_5′ CATTTCCTCAGAACAGACATG
FoxA_b_3′ TCCTAAAGACTCCCAACAACA
FoxAB_5′ CAGTGTGAGGTGAACATCATG
FoxAB_3′ CGATTGACAGGTTGATAGAAC
FoxB_5′ ACAACAGGACCCTGACTCGT
FoxB_3′ GCATTCCCTGACGTCTTGA
FoxC_5′ AACCGTCCCGTTTTCCTCATG
FoxC_3′ CAGTTTTGATTCGTAAGGACT
FoxD_5′ ACAGCTGTGGAGTGGACACTT
FoxD_3′ CACGAGACATGTAAGTCTCCG
FoxEa_5′ AACCAACCCCGTACCAGCATG
FoxEa_3′ ATATGACACGGACACTGAACT
FoxG_5′ ACGCACATTAGCACAGTTCG
FoxG_3′ ACTTGACCCTGGCTTGACAC
FoxJ1_5′ TACAGACAACTGTAAACCATG
FoxJ1_3′ TTGTAATGCAGGGTGGGGCCT
FoxK_5′ GGAAGGCGGAGTTGGACAATG
FoxK_3′ CCGGACACGTCCTGCACCTGT
FoxM_5′ AGGAGAGTGTGACAAACCATG
FoxM_3′ TTCTCAGCTATTCAGTAATAC
FoxN1/4a_5′ GCGCACCGAGTATCGTTCTGA
FoxN1/4a_3′ ACATAGGTAGGACTATGTACT
FoxN2/3_5′ CAGTAAACACGAGCAGACATG
FoxN2/3_3′ AGCTGAAGACAATGATGATCC
A mix of total mRNA of B. lanceolatum extracted from embryos at different developmental stages was used as a template for retro-transcription. Amplification was performed using Advantage 2 Polymerase kit (Clontech) and a touch-down PCR program with annealing temperature ranging from 65 to 40°C. Amplified fragments were cloned using the pGEM-T Easy system (Promega) and sub-cloned in pBluescript II KS+ for probe synthesis.
Whole Mount In situ Hybridization
Probes were synthesized using the DIG labeling system (Roche) after plasmid linearization with the appropriate enzymes. Ripe animals of B. lanceolatum were collected in Argelès-sur-Mer (France), and gametes were obtained by heat stimulation (Fuentes et al., 2004, 2007). In vitro fertilization was undertaken in Petri dishes filled with filtered sea water. Fixation and whole mount in situ hybridization were performed as described in Somorjai et al. (2008).
Results
Molecular Phylogenetic Analysis of B. lanceolatum Fox Gene Sequences
We looked for Fox gene sequences in the reference transcriptome of B. lanceolatum (Oulion et al., 2012). The sequences that were recovered were used to conduct a phylogenetic tree reconstruction presented in Figure 1. We showed that B. lanceolatum possesses at least 28 Fox genes, each of them being orthologous to one of the 32 genes described in B. floridae and corresponding to at least one member of each of the 22 families present in the bilaterian ancestor (Yu et al., 2008a). Specific duplications, that occurred in the cephalochordate clade at least in the ancestor of B. floridae and B. lanceolatum, gave rise to three members in the FoxQ2 group (FoxQ2a, FoxQ2b, FoxQ2c), two members in the FoxN1/4 group (FoxN1/4a and FoxN1/4b), and two genes in the FoxE group (FoxEa and FoxEc). We then analyzed the expression pattern during B. lanceolatum embryonic development of 13 of these 28 Fox genes corresponding to those showing a higher expression level in the transcriptome (Oulion et al., 2012).
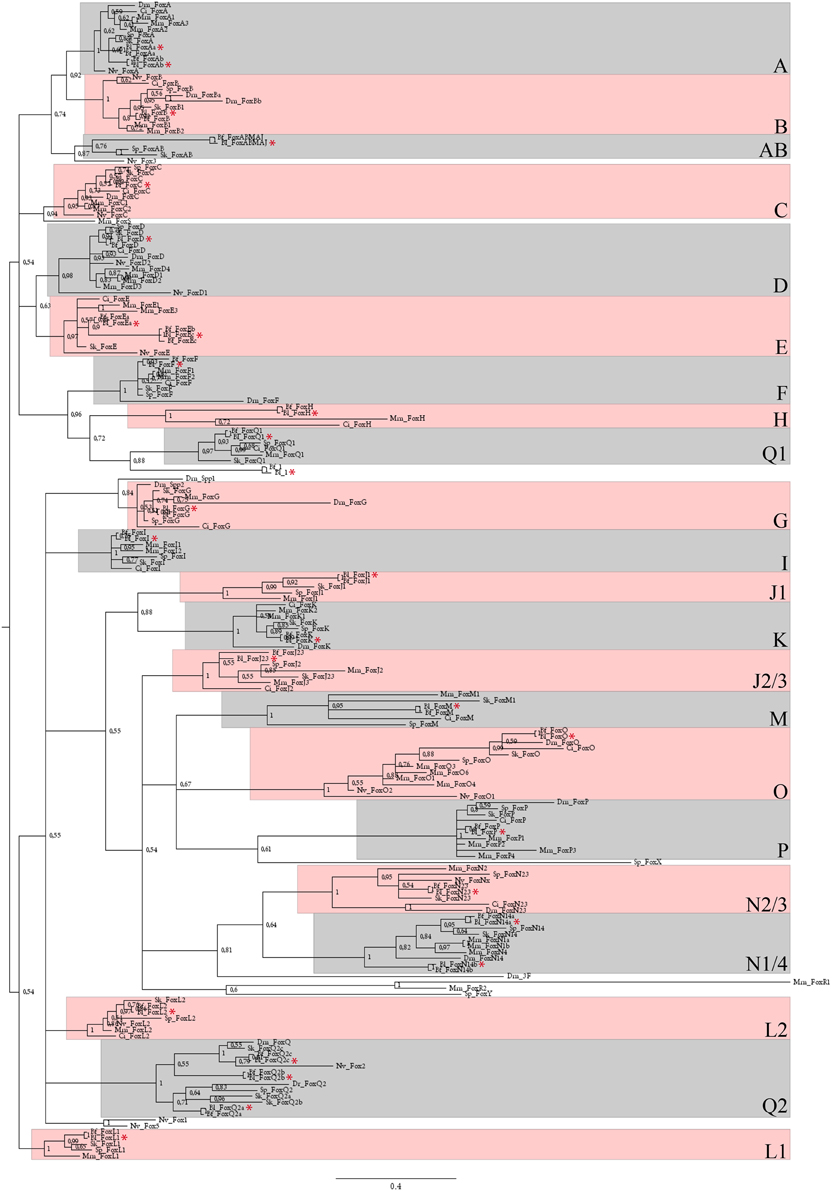
Figure 1. Phylogenetic analysis of B. lanceolatum Fox genes. Unrooted 50 majority-rule consensus Bayesian inference tree based on the amino acid sequences of the forkhead domain. Posterior probablilities are shown at each node. The different paralogy groups are colored in pink or light blue boxes. Divergent sequences appeared outside these boxes. Only one amphioxus Fox gene, named Fox1 (Yu et al., 2008a), that probably originated by a specific duplication and fast evolutionary rate in cephalochordates, localizes outside these paralogy groups. Abbreviations: Dm, Drosophila melanogaster; Mm, Mus musculus; Dr, Danio rerio; Ci, Ciona intestinalis; Sp, Strongylocentrotus purpuratus; Sk, Saccoglossus kowalevskii; Nv, Nematostella vectensis; Bf, Branchiostoma floridae; Bl, Branchiostoma lanceolatum. Red stars indicate Bl sequences. Scale bar represents 0.4 amino acid substitution per site.
FoxAa and FoxAb
FoxAa (formerly named AmHNF3-1) (Shimeld, 1997) was first expressed at the gastrula stage in the anterior ventral endoderm and in the mesendodermal layer of the dorsal blastoporal lip (Figures 2A,B). At the late gastrula stage, we detected transcripts in the axial dorsal mesendoderm corresponding to the presumptive notochord territory, as well as in mesendoderm cells of the archenteron floor (Figures 2C,D). Expression in the axial mesoderm and endoderm persisted through mid-late neurula stage (Figures 2E,F). Later on, at late neurula stage before the mouth opens, the expression in the notochord was restricted to the most anterior and posterior tips of the embryo, while the endodermal expression was restricted to the middle region of the gut (Figure 2G). At the larva stage, the expression at the anterior tip of the notochord and in the tailbud was still observed and we detected a diffuse expression in the gut (Figure 2H).
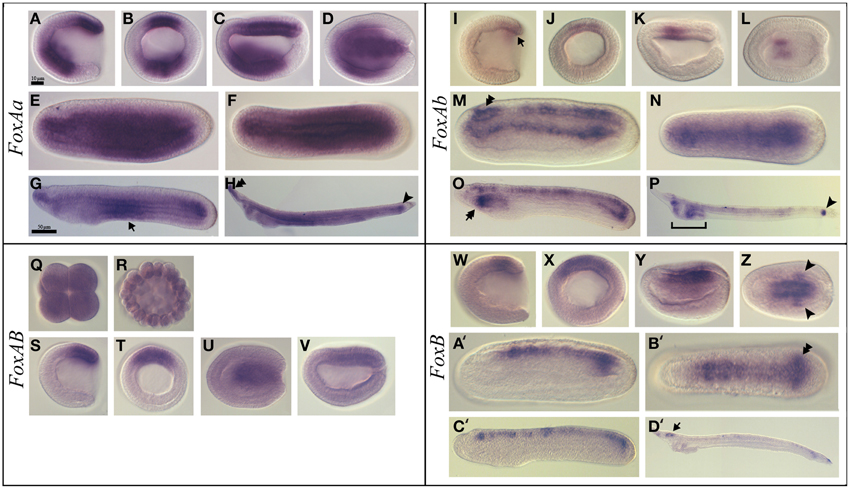
Figure 2. Expression of B. lanceolatum FoxAa, FoxAb, FoxAB, and FoxB. In all the panels except (B, J, Q, R, T, X) anterior is to the left. In lateral and blastoporal views dorsal is to the top. FoxAa expression pattern (A–H). Gastrula lateral (A) and blasporal (B) views. Late gastrula lateral (C) and dorsal (D) views. Mid-late neurula lateral (E) and dorsal (F) views. In the late neurula lateral view (G) arrow marks the endodermal expression in the middle region. In the larva stage lateral view (H), the double arrowhead indicates the expression in the anterior tip of the notochord and the arrowhead marks the expression in the tailbud. FoxAb expression pattern (I–P). In the gastrula lateral (I) and blastoporal (J) views the arrow indicates the expression in the mesendodermal part of the dorsal blastoporal lip. Late gastrula lateral (K) and dorsal (L) views. In the mid-late neurula lateral (M) and dorsal (N) views the double arrowhead marks the expression in the cerebral vesicle. In the late neurula lateral view (O), the double arrow marks the expression in the most anterior part of the pharynx. In larva lateral view (P) the arrowhead indicates the expression in the tailbud. FoxAB expression pattern (Q–V). Eight-cell stage (Q). Blastula stage (R). Gastrula lateral (S) and blasporal (T) views. Late gastrula lateral (U) and dorsal (V) views. FoxB expression pattern (W–D'). Gastrula lateral (W) and blastoporal (X) views. Early neurula lateral view (Y). In the early neurula dorsal (Z) view the arrowhead indicates the two expression patches in the posterior paraxial mesendoderm. Mid-late neurula lateral (A') and dorsal (B') views. The double arrowhead marks the expression in the newly formed somites. Late neurula lateral view (C'). In larva lateral view (D') the arrow indicates the expression in the cerebral vesicle. Scale bar: 10 μm (A–F), (I–N), (Q-V), (W-B'), and 50 μm (G,H), (O,P), (C',D').
FoxAb (formerly named AmHNF3-2) (Shimeld, 1997) expression was first detected at the gastrula stage as a weak signal in the mesendodermal part of the dorsal blastoporal lip (Figures 2I,J). At the late gastrula stage, we detected expression in the central paraxial mesoderm on both sides of the notochord anlagen (Figures 2K,L). At the mid-late neurula stage transcripts were detected in the neural tube, including the cerebral vesicle, and in the dorsal part of the endoderm (Figures 2M,N). At the late neurula stage, before the mouth opens, FoxAb was expressed in the neural tube and in the most anterior part of the pharynx. In the posterior region, expression was detected in the tailbud and in the dorsal midline of the gut (Figure 2O). At the larva stage, we observed expression in the pharynx, in the preoral pit, in the club-shaped gland and in the tailbud. At this stage, the expression in the neural tube gets restricted to some neurons and to the posterior part of the cerebral vesicle (Figure 2P and Figure S1A).
FoxAB
FoxAB transcripts were detected as a weak and ubiquitous signal from the eight-cell stage to the blastula stage (Figures 2Q,R). This ubiquitous expression was confirmed by the presence of reads in transcriptome analyses (data not shown). At the gastrula stage we observed a strong specific expression in the dorsal blastoporal lip, the amphioxus putative organizer (Figures 2S,T). At the late gastrula stage, expression gets restricted to the presumptive notochord territory (Figures 2U,V). No expression could be detected by in situ hybridization in later stages.
FoxB
FoxB expression was first detected dorsally, both in the ectoderm and in the mesendoderm, as a weak signal in mid gastrula stage embryos (Figures 2W,X). Later on, in early neurula stage embryos, a signal could be observed in the neural plate on either side of the midline, as well as in two patches in the posterior paraxial mesendoderm (Figures 2Y,Z). During the late neurula stage, expression was detected in the most posterior paraxial mesoderm that give rise to the newly formed somites and in the neural tube posterior to the cerebral vesicle (Figures 2A',B'). Then, FoxB expression in the mesoderm faded away in late neurulae (Figure 2C') and get later restricted to the cerebral vesicle and to some neurons along the neural tube in larvae (Figure 2D' and Figure S1B).
FoxC
FoxC was expressed at the gastrula stage in the dorsal paraxial mesendoderm (Figures 3A,B). Later on, at the late gastrula stage, expression was detected in the region that gives rise to the three most anterior somites (Figures 3C,D). In mid-late neurulae, the transcripts remained all along the body in the somites and a new expression domain appeared in the anterior endoderm at the level where the first gill slit opens (Figure 3E). At the late neurula stage, the expression persisted in the pharynx and somites and was also detected in the club-shaped gland anlagen (Figures 3F,G). At the larva stage a diffuse expression was observed in the somites as well as in the preoral pit, in the club-shaped gland and in the first gill slit (Figure 3H and Figure S1C).
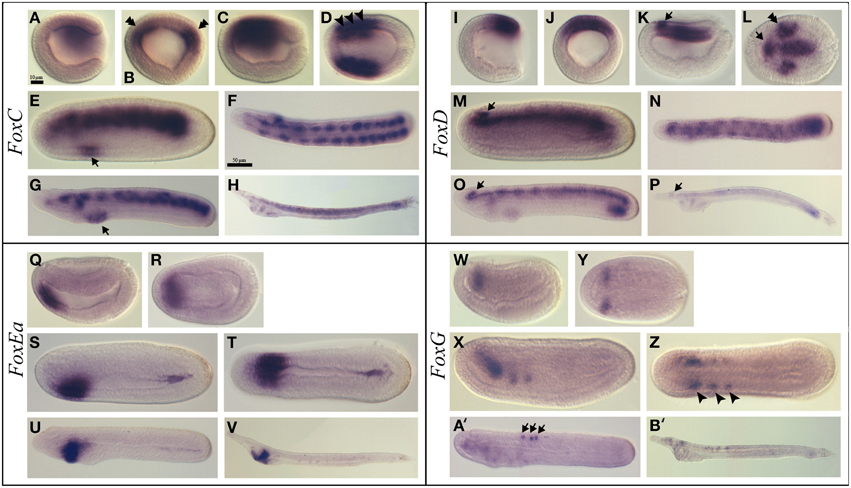
Figure 3. Expression of B. lanceolatum FoxC, FoxD, FoxEa, and FoxG. In all the panels except (B,J), anterior is to the left. In lateral and blastoporal views dorsal is to the top. FoxC expression pattern (A–H). Gastrula lateral (A) and blastoporal (B) views. The double arrowhead indicates the expression in the paraxial mesoderm. Late gastrula lateral (C) and dorsal (D) views. The arrowheads marks the region that will give rise to the three most anterior somites. In mid-late neurula lateral view (E) the arrow indicates a new expression domain in the anterior endoderm. Late neurula dorsal (F) and lateral (G) views. The arrow marks the expression domain in the pharynx. Larva lateral view (H). FoxD expression pattern (I–P). Gastrula lateral (I) and blasporal (J) views. Late gastrula lateral (K) and dorsal (L) views. The arrow indicates the expression in the anterior region of the neural plate and the double arrowhead marks the expression in the paraxial dorsal mesendoderm. Mid-late neurula lateral view (M). Late neurula dorsal (N) and lateral (O) views. Larva lateral view (P). In (M, O, P) the arrows indicate the expression domain in the cerebral vesicle. FoxE expression pattern (Q–V). Early neurula lateral (Q) and dorsal (R) views. Mid-late neurula lateral (S) and dorsal (T) views. Late neurula lateral view (U). Larva lateral view (V). FoxG expression pattern (W–B'). Early neurula lateral (W) and dorsal (Y) views. Mid-late neurula lateral (X) and dorsal (Z) views. The arrowhead indicates the expression in the three most anterior somites. In the late neurula stage lateral view (A') the arrows mark the neurons within the neural tube. Larva stage lateral view (B'). Scale bar: 10 μm (A–E), (I–L), (Q–T), (W–Z), and 50 μm (F–H), (N–P), (U,V), (A',B').
FoxD
FoxD transcripts were first detected at the gastrula stage in the dorsal blastoporal lip (Figures 3I,J). Then, at the late gastrula stage, FoxD was expressed in the dorsal axial mesendoderm, in part of the dorsal paraxial mesendoderm as two patches on both sides of the midline and in the anterior region of the neural plate (Figures 3K,L). At the mid-late neurula stage, the notochord and the somites, as well as the cerebral vesicle, were labeled (Figure 3M). At the late neurula stage, before the mouth opens, transcripts were detected in the paraxial somitic mesoderm, in the notochord, in the cerebral vesicle and in the posterior endoderm (Figures 3N,O). A faint labeling was also detected at this stage in the first gill slit and in the club-shaped gland anlagens. At the larva stage, we observed a low expression level in the cerebral vesicle, in the preoral pit, in the club-shaped gland, in the first gill slit, in the notochord and in the posterior part of the gut. We also observed an anterior to posterior gradient of expression in the somites (Figure 3P and Figure S1D).
FoxEa
FoxEa (formerly named FoxE4 in B. floridae) expression was first detected at early neurula stage in the antero-ventral mesendoderm (Figures 3Q,R). Later on, at the mid-late neurula stage, FoxEa transcripts were detected ventrally in the endoderm with a higher expression level on the right side of the pharynx (Figures 3S,T), and a slight expression domain in the posterior gut was also visible. At the late neurula stage, FoxEa transcripts remained ventrally in the pharyngeal endoderm on the right side (Figure 3U). Finally, at the larva stage, transcripts were detected in the club-shaped gland (Figure 3V and Figure S1E).
FoxG
FoxG expression was first observed at the neurula stage in the anterior region of the first somites (Figures 3W,Y). At the late neurula stage, FoxG was expressed in the anterior ventral region of the three most anterior somites (Figures 3X,Z). Later on, in late neurula before the mouth opens, a neural expression appeared in some individual neurons within the neural tube, while the expression observed in the first somites disappeared (Figure 3A'). This expression persisted in the larva stage embryos in which FoxG was also detected in some neurons of the cerebral vesicle (Figure 3B' and Figure S1F).
FoxJ1
FoxJ1 showed a dynamic expression pattern. Expression began during gastrulation and was detected in the ectoderm except the ectoderm around the blastopore (Figures 4A,B). Later on, at the late gastrula stage, this expression pattern persisted in the ectoderm that give rise to the epidermis (Figures 4C,D). At the mid-late neurula stage, we detected transcripts in the neural tube while the expression in the epidermis was completely lost (Figures 4E,F). This neural tube expression was no more observed in late neurula stage embryos before the mouth opens (data not show), however at the larva stage we observed expression at the anterior tip of the embryo and in the pharynx at the level of the preoral pit and of the first gill slit (Figure 4G and Figure S1G).
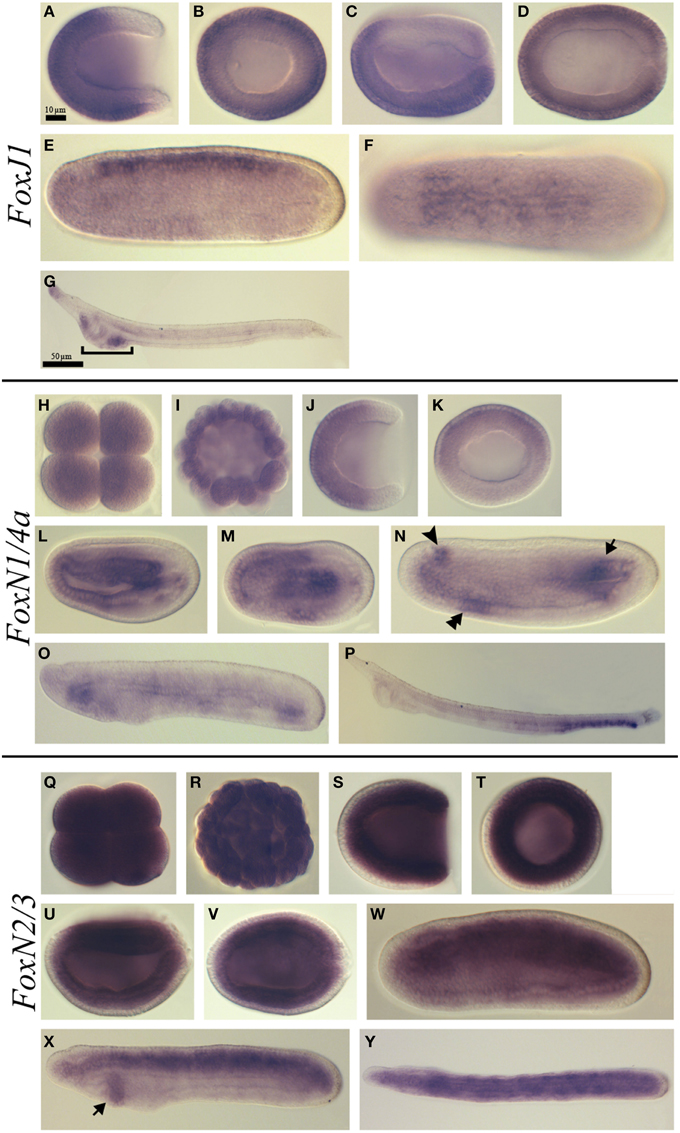
Figure 4. Expression of B. lanceolatum FoxJ1, FoxN1/4a, and FoxN2/3. In all the panels except (B, H, I, K, Q, R, T) anterior is to the left. In lateral and blastoporal views dorsal is to the top. FoxJ1 expression pattern (A–G). Gastrula lateral (A) and blasporal (B) views. Late gastrula lateral (C) and dorsal (D) views. Mid-late neurula lateral (E) and dorsal (F) views. In the larva lateral view (G) the bracket indicates the pharyngeal region. FoxN1/4a expression pattern (H–P). Eight-cell stage (H). Blastula stage (I). Gastrula lateral (J) and blastoporal views (K). Early neurula lateral (L) and dorsal (M) views. In the mid-late neurula lateral view (N), the arrowhead, double arrowhead and arrow mark the three main expression domains: at the level of the cerebral vesicle, in the anterior ventral endoderm and in the posterior mesoderm, respectively. Late neurula stage lateral view (O). Larva stage lateral view (P). FoxN2/3 expression pattern (Q–Y). Eight-cell stage (Q). Blastula stage (R). Gastrula lateral (S) and blasporal (T) views. Late gastrula lateral (U) and dorsal (V) views. Mid-late neurula lateral view (W). Late neurula lateral (X) and dorsal (Y) views. The arrow in (X) indicates the expression domain in the pharyngeal endoderm. Scale bar: 10 μm (A–F), (H–N), (Q–W), and 50 μm (G), (O–P), (X,Y).
FoxK
FoxK was ubiquitously expressed from the eight-cell stage to the blastula stage (Figures S2A,B). At the gastrula stage, the expression became restricted to the mesendoderm (Figures S2C,D), and by the late gastrula stage transcripts were detected mostly in the dorsal mesoderm (Figures S2E,F). At the mid-late neurula stage, we detected a stronger expression in the most anterior region of the embryo (Figures S2G,H). Transcripts were then detected in the whole embryo at the late neurula stage with a stronger expression in the anterior tip (Figures S2I,J). Finally, at the larva stage, we observed a ubiquitous expression with a higher level at the anterior tip and in the pharynx (Figure S2K).
FoxM
FoxM transcripts were detected ubiquitously during the whole embryonic development, from the eight-cell stage until the mid-late neurula stage except in the epidermis (Figures S2L–S). Later on, at late neurula stage, FoxM expression could not be detected anymore by in situ hybridization (Figure S2T).
FoxN1/4a
Ubiquitous FoxN1/4a expression was detected from the eight-cell stage until the blastula stage (Figures 4H,I). At the gastrula stage, a signal was detected in the anterior ectoderm (Figures 4J,K). Later on, at the early neurula stage, we observed transcripts in the anterior endoderm as well as in the axial central mesoderm (Figures 4L,M). At the mid-late neurula stage, we detected three major expression domains: one anterior, at the level of the cerebral vesicle, a second one in the anterior ventral endoderm and a third one in the posterior mesoderm (Figure 4N). At the late neurula stage before the mouth opens, we observed expression in the anterior and posterior endoderm (Figure 4O). Finally, at the larva stage, we detected expression in the posterior region of the gut and in the anus (Figure 4P).
FoxN2/3
Ubiquitous expression of FoxN2/3 was observed from the eight-cell stage (Figure 4Q) to the blastula stage (Figure 4R). Then, at the gastrula stage, the expression was restricted to the mesendoderm (Figures 4S,T). At the late gastrula stage, the expression remained strong in the mesendoderm but started to become lower in the ventral part (Figures 4U,V). By the mid-late neurula stage, FoxN2/3 transcripts were detected in the mesoderm and in the neural tube (Figure 4W). At the late neurula stage, before the mouth opens, the expression was mainly detected in the paraxial mesoderm (somites) and in the notochord. A new expression domain also appeared at this stage in the pharyngeal endoderm (Figures 4X,Y). At the larva stage, we did not detect any specific signal using in situ hybridization.
Discussion
Fox Genes Expression in Cephalochordate Species
The complete or partial embryonic expression patterns of FoxAa, FoxAb, FoxB, FoxC, FoxD, FoxEa, FoxG, and FoxN1/4a were previously described in B. floridae and/or B. belcheri (Shimeld, 1997; Terazawa and Satoh, 1997; Toresson et al., 1998; Mazet and Shimeld, 2002; Yu et al., 2002a,b; Mazet et al., 2006; Bajoghli et al., 2009). These genes overwhelmingly show a similar embryonic expression to what we observed in B. lanceolatum, as we have previously noticed for other important developmental genes (Somorjai et al., 2008). However, our work brings some new information.
First, in contrast to what has been described in B. floridae, we showed that FoxAa and FoxAb have different expression patterns. Indeed, in B. floridae, FoxAb in situ hybridization data showed that it has a similar expression to FoxAa at early stages whereas expression was no more detected after the eight somites stage (Shimeld, 1997). Here we showed that although both genes were expressed in the mesendodermal part of the dorsal blastoporal lip at the gastrula stage, the overall expression patterns are consistently different between the two genes and we observed a restricted expression of FoxAb from the gastrula to the larva stage. These discrepancies might be explained by the fact that the level of expression of FoxAb is very low. Indeed, staining of embryos hybridized to FoxAb took very long suggesting a low expression level. Thus, the staining time used in B. floridae might have been too short to detect expression in late stage embryos. Moreover, the expression we observed for FoxAa in B. lanceolatum is different from what was observed in B. floridae but similar to what has been described in B. belcheri (Terazawa and Satoh, 1997). Indeed, as in B. belcheri, FoxAa was not expressed in the central nervous system of B. lanceolatum. On the other hand, FoxAb showed a very specific expression in the ventral part of the neural tube in neurula stage embryos, which has been proposed to be homologous to the vertebrate floor plate. Vertebrates have three FoxA group paralogous genes that are expressed in the organizer, the notochord, the floor plate and the endoderm (Friedman and Kaestner, 2006). In Ciona (Di Gregorio et al., 2001), Ci-fkh is also expressed in the notochord, the floor plate and the endoderm. The data we obtained in B. lanceolatum suggest that the expression of FoxA in the chordate ancestor was similar to what is observed in tunicates and that independent sub-functionalizations occurred in cephalochordates after specific gene duplication and in vertebrates after the two rounds of whole genome duplications.
Concerning FoxB, expression in B. floridae was first detected in neurulae with five somites (Mazet and Shimeld, 2002). Here we showed that in B. lanceolatum FoxB expression could be observed in gastrula embryos in the dorsal posterior mesendoderm and ectoderm. Then, in neurulae, we detected expression in the neural plate similar to B. floridae, as well as an expression in the most posterior somites that was not previously described. This expression in the neural plate/neural tube and in the lastly formed somites persisted until the late neurula stage. Interestingly, in amphioxus three different somitic populations have been described (Bertrand et al., 2011). The first, most anterior, population forms under the control of the FGF signal and the two posterior populations forms independently of the FGF signal. Several genes are expressed specifically in these three somitic populations but only one gene, Mox,(Minguillon and Garcia-Fernandez, 2002) is expressed in the second and third populations. The present data suggest that FoxB also plays a role in the formation of these somitic population since it is also expressed in the two most-posterior somitic populations.
In B. floridae, FoxC has been described as being firstly expressed in the mesoderm of neurulae but its expression was described only in one developmental stage (Mazet et al., 2006). Here we showed that expression starts much earlier, at the gastrula stage, in the dorsal paraxial mesendoderm, the presumptive somitic mesoderm territory. Expression persisted in the paraxial mesoderm/somites until the larva stage, and at the late neurula stage we started to observe expression in the club-shaped gland anlagen and at the place where the first gill slit opens. These data suggest a major ancestral role of FoxC during somitogenesis which would have been conserved in vertebrates (Kume et al., 2001; Wilm et al., 2004; Wotton et al., 2008) and lost in tunicates in which FoxC is expressed in neural and palp cells (Imai et al., 2006).
FoxD and FoxEa expression in B. lanceolatum was very similar to previous descriptions in B. floridae (Yu et al., 2002a,b). However we noticed expression in some specific regions of the pharynx in late neurulae and larvae for FoxD, and a transient expression in mid-late and late neurula stage embryos in the posterior endoderm for FoxEa that were not described in the Caribbean species.
FoxG, previously known as Brain Factor 1 (BF-1), was described in B. floridae as a gene that is ventrally expressed in the cerebral vesicle and in the anterior-most portion of the first somite pair (Toresson et al., 1998). Our results showed a conserved expression pattern in the cerebral vesicle area in B. lanceolatum. However, mesoderm expression is not only limited to the first somite pair but the first three somite pairs exhibit the same pattern at the neurula stage suggesting that this gene might play a role during anterior somitogenesis. This result highlights the functional differences between the formation of the anterior somites which is under the control of the FGF signaling pathway and the formation of the most posterior somites which is not FGF-dependent (Bertrand et al., 2011). Moreover, expression is localized in the ventral part of these three most anterior somites which will give rise to the perivisceral coelom, suggesting a function of FoxG in the establishment of the somitic compartments.
FoxJ1 and the Formation of Motile Cilia
FoxJ1 orthologs were identified in many eumetazoans as well as in sponges (Larroux et al., 2006) and choanoflagellates (King et al., 2008). In vertebrates, FoxJ1 plays an essential role in the generation of motile cilia and in mediating Left/Right asymmetry (Chen et al., 1998; Brody et al., 2000; Yu et al., 2008b). It has also recently been shown that misexpression of FoxJ1 from placozoans, echinoderms and platyhelminthes in zebrafish embryos induces the expression of ciliary genes, whereas the inactivation of FoxJ1 in the flatworm Schmidtea mediterranea impairs the normal differentiation of motile cilia, suggesting a conserved function in metazoans (Vij et al., 2012). This conserved function is also supported by the embryonic expression of FoxJ1 in different phyla (Choi et al., 2006; Tu et al., 2006; Fritzenwanker et al., 2014). In B. lanceolatum, we showed that FoxJ1 is first expressed in the ectoderm of the gastrulae, excluding the blastoporal region and the presumptive neural plate, at the time at which motile cilia start to grow. Then, in neurulae, expression was lost in the epidermis and appeared in the closed neural tube. At the larva stage, expression was restricted to the anterior tip of the animal and to the ciliated preoral pit and first gill slit. This expression pattern suggests that in amphioxus FoxJ1 might also play a role in the formation of motile cilia. However, other cells, like the epithelial gut cells, also harbor motile cilia and do not express FoxJ1, suggesting that other genes might also be implicated in ciliogenesis in these embryonic structures.
FoxAB
In B. lanceolatum, FoxAB was transiently expressed in the organizer at the gastrula stage and in the presumptive notochord later on. No expression could be detected in mid-neurulae or larvae. FoxAB family genes were described in hemichordates (Fritzenwanker et al., 2014), sea urchin (Tu et al., 2006) and cnidarians and are absent in vertebrates and tunicates, the two other chordate clades (Yu et al., 2008a). In the hemichordate Saccoglossus kowalevskii, FoxAB is expressed in the ectoderm and the mouth perforates through the ring expressing this gene in the ventral side (Fritzenwanker et al., 2014). In bryozoans, FoxAB also shows an ectodermal expression (Fuchs et al., 2011). Therefore, it is still difficult to propose any scenario for the evolution of the function of FoxAB family genes in bilaterians. FoxAB could have been recruited for the patterning of the notochord field in the ancestor of chordates, but the absence of genes of this family in tunicates and vertebrates make this hypothesis unlikely.
FoxK and FoxM Ubiquitous Expression
We detected a ubiquitous expression of FoxK starting at the eight-cell stage until the larva stage. In other bilaterians data are scarce. In vertebrates, there are two paralogs in the FoxK family, FoxK1 and FoxK2. In mouse, the study of the function of FoxK1 during embryonic development was undertaken showing that the gene is involved in myogenic differentiation (Bassel-Duby et al., 1994). In Ciona intestinalis (Imai et al., 2004) as in the hemichordate S. kowalevskii (Fritzenwanker et al., 2014), the expression of FoxK is quite ubiquitous as observed for B. lanceolatum. Finally, studies in Drosophila have shown that FoxK is involved in the differentiation of midgut in the fly embryo (Casas-Tinto et al., 2008). Altogether these data do not allow us to infer any putative ancestral function for FoxK family genes and further studies are required in different animal phyla.
FoxM expression is also ubiquitous in B. lanceolatum and was first detected as early as the eight-cell stage. Then the expression level continuously decreased while development proceeds and became undetectable by in situ hybridization at the late neurula stage. In Xenopus, FoxM1 is maternally expressed and transcripts are thereafter detected in the neuroectoderm (Pohl et al., 2005). Moreover this gene has been shown to be important for early neuronal differentiation (Ueno et al., 2008). In mouse, FoxM1 is expressed in dividing cells and knock-out animals exhibit embryonic lethal phenotype due to many malformations affecting different organs such as the liver, the heart, the lung, or the vasculature (Kalin et al., 2011). As for FoxK, the data available up to now do not give us any indication on the putative ancestral function of genes belonging to the FoxM family.
FoxN1/4a and FoxN2/3 Expression
In all vertebrates studied so far, FoxN1 plays an essential role in thymus development (Ma et al., 2012; Neves et al., 2012; Lee et al., 2013; Romano et al., 2013). Moreover, in mammals, FoxN1 is essential for hair formation whereas it is also expressed in chick during feather development (Darnell et al., 2014). Although mammal and fish FoxN1s are able to activate the expression of hair keratin genes, FoxN1/4 from amphioxus is not because its N-terminal region of the forkhead domain is different compared with vertebrates (Schlake et al., 2000). On the other hand, FoxN4 is expressed in the nervous system, including retina, during vertebrate development (Danilova et al., 2004; Kelly et al., 2007; Boije et al., 2013). Outside vertebrates, embryonic expression has been described in S. kowalevskii (Fritzenwanker et al., 2014) and in a single developmetal stage of B. floridae (Bajoghli et al., 2009). In the hemichordate, expression of FoxN1/4 is ubiquitous during early development and is thereafter observed in the ectoderm. In B. lanceolatum, the expression of FoxN1/4a was very dynamic with a maternal ubiquitous expression followed by restricted expression in the ectoderm at the gastrula stage, in the endoderm and axial mesoderm in neurulae, in the cerebral vesicle, the pharynx and the posterior somites later on, and, finally, in the posterior gut of the larvae. These data suggest that FoxN1 and FoxN4 probably acquired new functions in vertebrates, and analysis of the expression of FoxN1/4 family genes in tunicates will be needed to better understand this point. Interestingly, the gut of amphioxus larva and adult is considered as a major organ for immunity and FoxN1/4a might, as vertebrates FoxN1, play a role in the control of immune system function in amphioxus. However, further functional studies are required to test this hypothesis.
In vertebrates, FoxN3 is important for craniofacial and eye development (Schuff et al., 2007; Samaan et al., 2010; Schmidt et al., 2011). In Xenopus, FoxN3 is expressed in neural crest and eye field whereas FoxN2 is expressed early in the eye field and then in branchial arches, retina and vagal ganglion (Schuff et al., 2006). In mouse, FoxN2 is expressed in craniofacial, limb, nervous system and somitic tissues (Tribioli et al., 2002). In Ciona intestinalis, expression of FoxN2/3 is quite ubiquitous during early development and becomes more intense in the sensory vesicle, the mesenchyme, the notochord and the palps after gastrulation (Imai et al., 2004). In sea urchin FoxN2/3 is expressed in the non-skeletogenic mesoderm and, later on, in the endoderm and it has been shown that FoxN2/3 function is important for ingression and for the expression of genes coding for proteins of the skeletal matrix (Rho and Mcclay, 2011). Here, we show that FoxN2/3 in amphioxus was ubiquitously expressed at early stages. Then, at the gastrula stage, its expression was restricted to the endomesoderm and later on we observed a specific expression in the somites. Altogether, this suggests a conserved role of FoxN2/3 in the development of mesoderm in deuterostomes, although genes of this family seem to have acquired specific functions in each chordate lineage.
Conclusions
Analyzing the expression of Fox genes in the Mediterranean amphioxus, B. lanceolatum showed us several points. First, as previously described for other gene families (Somorjai et al., 2008), the expression of orthologous genes in different amphioxus species shows a high degree of stasis. However, differences may be found that can easily be explained by variation in experimental sensitivity. And, second, the comparative analyzes of the expression of amphioxus Fox genes with other metazoans and particularly chordates have shown a high degree of conservation for some genes (e.g., FoxC, FoxD), but also divergent patterns in others (e.g., FoxM, FoxN1/4a). This indicates that Fox genes were necessary for essential functions in metazoans but they were also instrumental for the evolution of new functions. Further studies in amphioxus and other metazoans, and particularly functional studies, will be extremely important in the future to establish the complete picture of Fox genes expression and function and their role in the evolution of animals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DA holds a fellowship from CONICYT “Becas Chile.” Part of this study was supported by the EXOMOD grant from the CNRS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fevo.2015.00080
References
Bajoghli, B., Aghaallaei, N., Hess, I., Rode, I., Netuschil, N., Tay, B. H., et al. (2009). Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 138, 186–197. doi: 10.1016/j.cell.2009.04.017
Bassel-Duby, R., Hernandez, M. D., Yang, Q., Rochelle, J. M., Seldin, M. F., and Williams, R. S. (1994). Myocyte nuclear factor, a novel winged-helix transcription factor under both developmental and neural regulation in striated myocytes. Mol. Cell Biol. 14, 4596–4605.
Benayoun, B. A., Caburet, S., and Veitia, R. A. (2011). Forkhead transcription factors: key players in health and disease. Trends Genet. 27, 224–232. doi: 10.1016/j.tig.2011.03.003
Bertrand, S., Camasses, A., Somorjai, I., Belgacem, M. R., Chabrol, O., Escande, M. L., et al. (2011). Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc. Natl. Acad. Sci. U.S.A. 108, 9160–9165. doi: 10.1073/pnas.1014235108
Bertrand, S., and Escriva, H. (2011). Evolutionary crossroads in developmental biology: amphioxus. Development 138, 4819–4830. doi: 10.1242/dev.066720
Bertrand, S., and Escriva, H. (2014). “Chordates: The acquisition of an axial backbone,” The Tree of Life, eds P. Vargas and R. Zardoya (Sunderland, MA: Sinauer Associates, Inc), 460–468.
Boije, H., Shirazi Fard, S., Ring, H., and Hallbook, F. (2013). Forkheadbox N4 (FoxN4) triggers context-dependent differentiation in the developing chick retina and neural tube. Differentiation 85, 11–19. doi: 10.1016/j.diff.2012.12.002
Brody, S. L., Yan, X. H., Wuerffel, M. K., Song, S. K., and Shapiro, S. D. (2000). Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 23, 45–51. doi: 10.1165/ajrcmb.23.1.4070
Carlsson, P., and Mahlapuu, M. (2002). Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250, 1–23. doi: 10.1006/dbio.2002.0780
Casas-Tinto, S., Gomez-Velazquez, M., Granadino, B., and Fernandez-Funez, P. (2008). FoxK mediates TGF-beta signalling during midgut differentiation in flies. J. Cell Biol. 183, 1049–1060. doi: 10.1083/jcb.200808149
Chen, J., Knowles, H. J., Hebert, J. L., and Hackett, B. P. (1998). Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J. Clin. Invest. 102, 1077–1082. doi: 10.1172/JCI4786
Choi, V. M., Harland, R. M., and Khokha, M. K. (2006). Developmental expression of FoxJ1.2, FoxJ2, and FoxQ1 in Xenopus tropicalis. Gene Expr. Patterns 6, 443–447. doi: 10.1016/j.modgep.2005.11.007
Clark, K. L., Halay, E. D., Lai, E., and Burley, S. K. (1993). Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364, 412–420. doi: 10.1038/364412a0
Danilova, N., Visel, A., Willett, C. E., and Steiner, L. A. (2004). Expression of the winged helix/forkhead gene, foxn4, during zebrafish development. Brain Res. Dev. Brain Res. 153, 115–119. doi: 10.1016/j.devbrainres.2004.05.014
Darnell, D. K., Zhang, L. S., Hannenhalli, S., and Yaklichkin, S. Y. (2014). Developmental expression of chicken FOXN1 and putative target genes during feather development. Int. J. Dev. Biol. 58, 57–64. doi: 10.1387/ijdb.130023sy
Di Gregorio, A., Corbo, J. C., and Levine, M. (2001). The regulation of forkhead/HNF-3beta expression in the Ciona embryo. Dev. Biol. 229, 31–43. doi: 10.1006/dbio.2000.9964
Friedman, J. R., and Kaestner, K. H. (2006). The Foxa family of transcription factors in development and metabolism. Cell Mol. Life Sci. 63, 2317–2328. doi: 10.1007/s00018-006-6095-6
Fritzenwanker, J. H., Gerhart, J., Freeman, R. M. Jr., and Lowe, C. J. (2014). The Fox/Forkhead transcription factor family of the hemichordate Saccoglossus kowalevskii. Evodevo 5:17. doi: 10.1186/2041-9139-5-17
Fuchs, J., Martindale, M. Q., and Hejnol, A. (2011). Gene expression in bryozoan larvae suggest a fundamental importance of pre-patterned blastemic cells in the bryozoan life-cycle. Evodevo 2:13. doi: 10.1186/2041-9139-2-13
Fuentes, M., Benito, E., Bertrand, S., Paris, M., Mignardot, A., Godoy, L., et al. (2007). Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum). J. Exp. Zool. B Mol. Dev. Evol. 308, 484–493. doi: 10.1002/jez.b.21179
Fuentes, M., Schubert, M., Dalfo, D., Candiani, S., Benito, E., Gardenyes, J., et al. (2004). Preliminary observations on the spawning conditions of the European amphioxus (Branchiostoma lanceolatum) in captivity. J. Exp. Zool. B Mol. Dev. Evol. 302, 384–391. doi: 10.1002/jez.b.20025
Imai, K. S., Hino, K., Yagi, K., Satoh, N., and Satou, Y. (2004). Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047–4058. doi: 10.1242/dev.01270
Imai, K. S., Levine, M., Satoh, N., and Satou, Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183–1187. doi: 10.1126/science.1123404
Kalin, T. V., Ustiyan, V., and Kalinichenko, V. V. (2011). Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle 10, 396–405. doi: 10.4161/cc.10.3.14709
Kelly, L. E., Nekkalapudi, S., and El-Hodiri, H. M. (2007). Expression of the forkhead transcription factor FoxN4 in progenitor cells in the developing Xenopus laevis retina and brain. Gene Expr. Patterns 7, 233–238. doi: 10.1016/j.modgep.2006.10.003
King, N., Westbrook, M. J., Young, S. L., Kuo, A., Abedin, M., Chapman, J., et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788. doi: 10.1038/nature06617
Kume, T., Jiang, H., Topczewska, J. M., and Hogan, B. L. (2001). The murine winged helix transcription factors, Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes Dev. 15, 2470–2482. doi: 10.1101/gad.907301
Larroux, C., Fahey, B., Liubicich, D., Hinman, V. F., Gauthier, M., Gongora, M., et al. (2006). Developmental expression of transcription factor genes in a demosponge: insights into the origin of metazoan multicellularity. Evol. Dev. 8, 150–173. doi: 10.1111/j.1525-142X.2006.00086.x
Larroux, C., Luke, G. N., Koopman, P., Rokhsar, D. S., Shimeld, S. M., and Degnan, B. M. (2008). Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25, 980–996. doi: 10.1093/molbev/msn047
Lee, Y. H., Williams, A., Hong, C. S., You, Y., Senoo, M., and Saint-Jeannet, J. P. (2013). Early development of the thymus in Xenopus laevis. Dev. Dyn. 242, 164–178. doi: 10.1002/dvdy.23905
Ma, D., Wang, L., Wang, S., Gao, Y., Wei, Y., and Liu, F. (2012). Foxn1 maintains thymic epithelial cells to support T-cell development via mcm2 in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 109, 21040–21045. doi: 10.1073/pnas.1217021110
Mazet, F., Amemiya, C. T., and Shimeld, S. M. (2006). An ancient Fox gene cluster in bilaterian animals. Curr. Biol. 16, R314–R316. doi: 10.1016/j.cub.2006.03.088
Mazet, F., and Shimeld, S. M. (2002). The evolution of chordate neural segmentation. Dev. Biol. 251, 258–270. doi: 10.1006/dbio.2002.0831
Mazet, F., Yu, J. K., Liberles, D. A., Holland, L. Z., and Shimeld, S. M. (2003). Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene 316, 79–89. doi: 10.1016/S0378-1119(03)00741-8
Miller, M. A., Schwartz, T., Pickett, B. E., He, S., Klem, E. B., Scheuermann, R. H., et al. (2015). A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. Online 11, 43–48. doi: 10.4137/EBO.S21501
Minguillon, C., and Garcia-Fernandez, J. (2002). The single amphioxus Mox gene: insights into the functional evolution of Mox genes, somites, and the asymmetry of amphioxus somitogenesis. Dev. Biol. 246, 455–465. doi: 10.1006/dbio.2002.0660
Neves, H., Dupin, E., Parreira, L., and Le Douarin, N. M. (2012). Modulation of Bmp4 signalling in the epithelial-mesenchymal interactions that take place in early thymus and parathyroid development in avian embryos. Dev. Biol. 361, 208–219. doi: 10.1016/j.ydbio.2011.10.022
Onimaru, K., Shoguchi, E., Kuratani, S., and Tanaka, M. (2011). Development and evolution of the lateral plate mesoderm: comparative analysis of amphioxus and lamprey with implications for the acquisition of paired fins. Dev. Biol. 359, 124–136. doi: 10.1016/j.ydbio.2011.08.003
Oulion, S., Bertrand, S., Belgacem, M. R., Le Petillon, Y., and Escriva, H. (2012). Sequencing and analysis of the Mediterranean amphioxus (Branchiostoma lanceolatum) transcriptome. PLoS ONE 7:e36554. doi: 10.1371/journal.pone.0036554
Pohl, B. S., Rossner, A., and Knochel, W. (2005). The Fox gene family in Xenopus laevis:FoxI2, FoxM1 and FoxP1 in early development. Int. J. Dev. Biol. 49, 53–58. doi: 10.1387/ijdb.051977bp
Rho, H. K., and Mcclay, D. R. (2011). The control of foxN2/3 expression in sea urchin embryos and its function in the skeletogenic gene regulatory network. Development 138, 937–945. doi: 10.1242/dev.058396
Romano, R., Palamaro, L., Fusco, A., Giardino, G., Gallo, V., Del Vecchio, L., et al. (2013). FOXN1: a master regulator gene of thymic epithelial development program. Front. Immunol. 4:187. doi: 10.3389/fimmu.2013.00187
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Samaan, G., Yugo, D., Rajagopalan, S., Wall, J., Donnell, R., Goldowitz, D., et al. (2010). Foxn3 is essential for craniofacial development in mice and a putative candidate involved in human congenital craniofacial defects. Biochem. Biophys. Res. Commun. 400, 60–65. doi: 10.1016/j.bbrc.2010.07.142
Schlake, T., Schorpp, M., Maul-Pavicic, A., Malashenko, A. M., and Boehm, T. (2000). Forkhead/winged-helix transcription factor Whn regulates hair keratin gene expression: molecular analysis of the nude skin phenotype. Dev. Dyn. 217, 368–376. doi: 10.1002/(SICI)1097-0177(200004)217:4<368::AID-DVDY4>3.0.CO;2-Z
Schmidt, J., Schuff, M., and Olsson, L. (2011). A role for FoxN3 in the development of cranial cartilages and muscles in Xenopus laevis (Amphibia: Anura: Pipidae) with special emphasis on the novel rostral cartilages. J. Anat. 218, 226–242. doi: 10.1111/j.1469-7580.2010.01315.x
Schuff, M., Rossner, A., Donow, C., and Knochel, W. (2006). Temporal and spatial expression patterns of FoxN genes in Xenopus laevis embryos. Int. J. Dev. Biol. 50, 429–434. doi: 10.1387/ijdb.052126ms
Schuff, M., Rossner, A., Wacker, S. A., Donow, C., Gessert, S., and Knochel, W. (2007). FoxN3 is required for craniofacial and eye development of Xenopus laevis. Dev. Dyn. 236, 226–239. doi: 10.1002/dvdy.21007
Shimeld, S. M., Boyle, M. J., Brunet, T., Luke, G. N., and Seaver, E. C. (2010a). Clustered Fox genes in lophotrochozoans and the evolution of the bilaterian Fox gene cluster. Dev. Biol. 340, 234–248. doi: 10.1016/j.ydbio.2010.01.015
Shimeld, S. M., Degnan, B., and Luke, G. N. (2010b). Evolutionary genomics of the Fox genes: origin of gene families and the ancestry of gene clusters. Genomics 95, 256–260. doi: 10.1016/j.ygeno.2009.08.002
Shimeld, S. M. (1997). Characterisation of amphioxus HNF-3 genes: conserved expression in the notochord and floor plate. Dev. Biol. 183, 74–85. doi: 10.1006/dbio.1996.8481
Somorjai, I., Bertrand, S., Camasses, A., Haguenauer, A., and Escriva, H. (2008). Evidence for stasis and not genetic piracy in developmental expression patterns of Branchiostoma lanceolatum and Branchiostoma floridae, two amphioxus species that have evolved independently over the course of 200 Myr. Dev. Genes Evol. 218, 703–713. doi: 10.1007/s00427-008-0256-6
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Terazawa, K., and Satoh, N. (1997). Formation of the chordamesoderm in the amphioxus embryo: analysis with Brachyury and fork head/HNF-3 genes. Dev. Genes Evol. 207, 1–11. doi: 10.1007/s004270050086
Toresson, H., Martinez-Barbera, J. P., Bardsley, A., Caubit, X., and Krauss, S. (1998). Conservation of BF-1 expression in amphioxus and zebrafish suggests evolutionary ancestry of anterior cell types that contribute to the vertebrate telencephalon. Dev. Genes Evol. 208, 431–439. doi: 10.1007/s004270050200
Tribioli, C., Robledo, R. F., and Lufkin, T. (2002). The murine fork head gene Foxn2 is expressed in craniofacial, limb, CNS and somitic tissues during embryogenesis. Mech. Dev. 118, 161–163. doi: 10.1016/S0925-4773(02)00220-4
Tu, Q., Brown, C. T., Davidson, E. H., and Oliveri, P. (2006). Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev. Biol. 300, 49–62. doi: 10.1016/j.ydbio.2006.09.031
Tuteja, G., and Kaestner, K. H. (2007a). Forkhead transcription factors II. Cell 131, 192. doi: 10.1016/j.cell.2007.09.016
Tuteja, G., and Kaestner, K. H. (2007b). SnapShot: forkhead transcription factors I. Cell 130, 1160. doi: 10.1016/j.cell.2007.09.005
Ueno, H., Nakajo, N., Watanabe, M., Isoda, M., and Sagata, N. (2008). FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development 135, 2023–2030. doi: 10.1242/dev.019893
Vij, S., Rink, J. C., Ho, H. K., Babu, D., Eitel, M., Narasimhan, V., et al. (2012). Evolutionarily ancient association of the FoxJ1 transcription factor with the motile ciliogenic program. PLoS Genet 8:e1003019. doi: 10.1371/journal.pgen.1003019
Weigel, D., and Jackle, H. (1990). The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell 63, 455–456. doi: 10.1016/0092-8674(90)90439-L
Wilm, B., James, R. G., Schultheiss, T. M., and Hogan, B. L. (2004). The forkhead genes, Foxc1 and Foxc2, regulate paraxial versus intermediate mesoderm cell fate. Dev. Biol. 271, 176–189. doi: 10.1016/j.ydbio.2004.03.034
Wotton, K. R., Mazet, F., and Shimeld, S. M. (2008). Expression of FoxC, FoxF, FoxL1, and FoxQ1 genes in the dogfish Scyliorhinus canicula defines ancient and derived roles for Fox genes in vertebrate development. Dev. Dyn. 237, 1590–1603. doi: 10.1002/dvdy.21553
Wotton, K. R., and Shimeld, S. M. (2006). Comparative genomics of vertebrate Fox cluster loci. BMC Genomics 7:271. doi: 10.1186/1471-2164-7-271
Yu, J. K., Holland, L. Z., Jamrich, M., Blitz, I. L., and Hollan, N. D. (2002a). AmphiFoxE4, an amphioxus winged helix/forkhead gene encoding a protein closely related to vertebrate thyroid transcription factor-2: expression during pharyngeal development. Evol. Dev. 4, 9–15. doi: 10.1046/j.1525-142x.2002.01057.x
Yu, J. K., Holland, N. D., and Holland, L. Z. (2002b). An amphioxus winged helix/forkhead gene, AmphiFoxD: insights into vertebrate neural crest evolution. Dev. Dyn. 225, 289–297. doi: 10.1002/dvdy.10173
Yu, J. K., Holland, N. D., and Holland, L. Z. (2003). AmphiFoxQ2, a novel winged helix/forkhead gene, exclusively marks the anterior end of the amphioxus embryo. Dev. Genes Evol. 213, 102–105. doi: 10.1007/s00427-003-0302-3
Yu, J. K., Mazet, F., Chen, Y. T., Huang, S. W., Jung, K. C., and Shimeld, S. M. (2008a). The Fox genes of Branchiostoma floridae. Dev. Genes Evol. 218, 629–638. doi: 10.1007/s00427-008-0229-9
Keywords: Fox genes, amphioxus, Evo-Devo, chordates, embryonic development
Citation: Aldea D, Leon A, Bertrand S and Escriva H (2015) Expression of Fox genes in the cephalochordate Branchiostoma lanceolatum. Front. Ecol. Evol. 3:80. doi: 10.3389/fevo.2015.00080
Received: 29 May 2015; Accepted: 07 July 2015;
Published: 28 July 2015.
Edited by:
Naoki Osada, Hokkaido University, JapanCopyright © 2015 Aldea, Leon, Bertrand and Escriva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie Bertrand and Hector Escriva, Laboratoire Arago, Avenue du Fontaulé, 66650 Banyuls-sur-Mer, France, stephanie.bertrand@obs-banyuls.fr; hescriva@obs-banyuls.fr
 Daniel Aldea
Daniel Aldea Anthony Leon
Anthony Leon Stephanie Bertrand
Stephanie Bertrand Hector Escriva
Hector Escriva