Molecular epidemiology and characterization of antimicrobial-resistant Staphylococcus haemolyticus strains isolated from dairy cattle milk in Northwest, China
- 1Key Laboratory of New Animal Drug Project, Gansu Province/Key Laboratory of Veterinary Pharmaceutical Development, Ministry of Agriculture and Rural Affairs, Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Sciences (CAAS), Lanzhou, China
- 2Lanzhou Center for Animal Disease Control and Prevention, Lanzhou, China
Introduction: Non-aureus Staphylococcus (NAS) species are currently the most commonly identified microbial agents causing sub-clinical infections of the udder and are also deemed as opportunistic pathogens of clinical mastitis in dairy cattle. More than 10 NAS species have been identified and studied but little is known about S. haemolyticus in accordance with dairy mastitis. The present study focused on the molecular epidemiology and genotypic characterization of S. haemolyticus isolated from dairy cattle milk in Northwest, China.
Methods: In this study, a total of 356 milk samples were collected from large dairy farms in three provinces in Northwest, China. The bacterial isolation and presumptive identification were done by microbiological and biochemical methods following the molecular confirmation by 16S rRNA gene sequencing. The antimicrobial susceptibility testing (AST) was done by Kirby-Bauer disk diffusion assay and antibiotic-resistance genes (ARGs) were identified by PCR. The phylogenetic grouping and sequence typing was done by Pulsed Field Gel Electrophoresis (PFGE) and Multi-Locus Sequence Typing (MLST) respectively.
Results: In total, 39/356 (11.0%) were identified as positive for S. haemolyticus. The overall prevalence of other Staphylococcus species was noted to be 39.6% (141/356), while the species distribution was as follows: S. aureus 14.9%, S. sciuri 10.4%, S. saprophyticus 7.6%, S. chromogenes 4.2%, S. simulans 1.4%, and S. epidermidis 1.1%. The antimicrobial susceptibility of 39 S. haemolyticus strains exhibited higher resistance to erythromycin (92.3%) followed by trimethoprim-sulfamethoxazole (51.3%), ciprofloxacin (43.6%), florfenicol (30.8%), cefoxitin (28.2%), and gentamicin (23.1%). All of the S. haemolyticus strains were susceptible to tetracycline, vancomycin, and linezolid. The overall percentage of multi-drug resistant (MDR) S. haemolyticus strains was noted to be 46.15% (18/39). Among ARGs, mphC was identified as predominant (82.05%), followed by ermB (33.33%), floR (30.77%), gyrA (30.77%), sul1 (28.21%), ermA (23.08%), aadD (12.82%), grlA (12.82%), aacA-aphD (10.26%), sul2 (10.26%), dfrA (7.69%), and dfrG (5.13%). The PFGE categorized 39 S. haemolyticus strains into A-H phylogenetic groups while the MLST categorized strains into eight STs with ST8 being the most predominant while other STs identified were ST3, ST11, ST22, ST32, ST19, ST16, and ST7.
Conclusion: These findings provided new insights into our understanding of the epidemiology and genetic characteristics of S. haemolyticus in dairy farms to inform interventions limiting the spread of AMR in dairy production.
1 Introduction
Bovine mastitis is the most common and economically deadly disease impacting the dairy sector all over the world (Gussmann et al., 2019). The incidence of clinical mastitis (CM) on large dairy farms in China could reach up to 3.3% per month (Dallago et al., 2021). Clinical mastitis is caused by numerous pathogens, but Staphylococcus species classified as coagulase-positive Staphylococcus including S. aureus and coagulase-negative Staphylococcus (CNS), also known as non-aureus Staphylococcus (NAS), are the most common pathogens in lactating dairy cows (De Buck et al., 2021). NAS is one of the most important and widespread groups of pathogens including numerous species such as S. chromogenes, S. haemolyticus, S. sciuri, S. saprophyticus, S. simulans, S. succinus, and S. epidermidis (Wald et al., 2019). NAS pathogens are not only opportunistic in animals and humans but are also widely distributed in the environment such as dust, soil, water, and air (Wu et al., 2021). Although NAS species are considered to be a pathogen of low clinical incidence, it has recently been identified as the relatively common causative agent of bovine mastitis in many countries including China (Al-Harbi et al., 2021). Intra-mammary infection (IMI) with NAS species is often associated with asymptomatic mastitis leading to an increase in somatic cell count and sometimes severe mastitis resulting in reduced milk yield (Adkins et al., 2018). Among NAS, S. haemolyticus ranked second in terms of bacterial recovery and is one of the important opportunistic pathogenic bacteria (Jenkins et al., 2019).
Mastitis remains one of the most common reasons for antimicrobial, especially antibiotics, use in dairy herds worldwide (Hogeveen et al., 2011). The higher prevalence of multidrug-resistant strains in livestock has led to the extensive use of antimicrobials to treat multiple diseases on dairy farms in China (Tang et al., 2017). Staphylococcus species including NAS can develop resistance to multiple antimicrobials, thus reducing the effectiveness of antimicrobial therapy (Jenkins et al., 2019). The most common antimicrobial resistance genes (ARGs) detected among different classes of antimicrobials include β-lactam-resistance genes (blaZ and mecA), tetracycline-resistance genes (tetO, tetK, tetM, and tetL), aminoglycoside-resistance genes (aadD, aphA3, and aacA-aphD), and macrolide-lincosamide resistance genes (ermA, ermB, ermC, ermT, msrA, mphC, and lnuA) (Pizauro et al., 2019; Qu et al., 2019; Pérez et al., 2020; Walid et al., 2021). Multidrug-resistant NAS strains are increasingly reported, and the growing resistance of NAS agents to antimicrobials also limits the choice of drugs for treatment purposes (Schoenfelder et al., 2017). NAS species are also recognized as reservoirs of drug-resistance genes, highlighting their threat to public health (Osman et al., 2017). Molecular epidemiological studies by multiplex PCR, pulse field gel electrophoresis (PFGE), 16S rRNA gene sequencing, and multi-locus sequence typing (MLST) are important in understanding the source and route of transmission of these pathogens (Kim et al., 2019; Lin et al., 2022).
Because of the ubiquitous nature and acquisition of ARGs by these microbial agents, it is important to understand their epidemiology through the molecular approaches mentioned above (Srednik et al., 2017; Benites et al., 2021). The Northwest region of China is an important agricultural and pastoral region with much potential for the dairy industry in Gansu, Ningxia, Qinghai, and Xinjiang provinces, and the status of AMR among NAS, particularly S. haemolyticus isolated from the dairy herd is still unknown in the northwest region of China. Therefore, the present study was designed to determine the molecular epidemiology and antimicrobial resistance traits of S. haemolyticus isolated from dairy origin in Northwest, China. This study also focused on the genotypic characterization of S. haemolyticus strains concerning phylogenetic lineage and sequence types (STs).
2 Materials and methods
2.1 Materials
Antibiotic disks used for AST were purchased from Shanghai Kejia Drug Testing Equipment Co., Ltd; SmaI, XbaI, proteinase K, and La Taq DNA polymerase from Takara Co., Ltd; Lysozyme and lysostaphin from Sigma Aldrich, Beijing, China; Mueller-Hinton (MH) and Mannitol Salt agar from Guangdong Huankai Microbial Technology Co., Ltd; Agarose from Shanghai Shenggong Biological Co., Ltd; SeaKem Gold Agarose from Lonza Co., Ltd; and bacterial genomic DNA extraction kit from Tiangen Biochemical Technology Co., Ltd. The quality control strain used in this experiment was Staphylococcus haemolyticus ATCC25923, obtained from a culture bank.
2.2 Sample collection and transportation
A total of n = 356 milk samples were collected from five large dairy farms in five cities of three provinces [Gansu (Lanzhou, LZ n= 96; Zhangye, ZY n = 57), Qinghai (Xining, XN n = 48; Yushu, YS n = 71), Ningxia, NX n = 84] in the Northwest region of China. The milk samples were drawn directly from the animal teats into sterile 50 mL tubes. Before taking the sample, the animal udder was washed with lukewarm water and dried with a clean towel. The teats of animals were swabbed with 0.5% iodine solution (Merck, Germany), and the hands of the milking personnel were also washed with an antiseptic solution. The first few streaks of milk were discarded, and the middle streaks were collected as adopted by Aqib et al., 2021. The collected milk samples were stored at 4°C and transported to the microbiology laboratory for further processing.
2.3 Isolation and identification of Staphylococcus species
In this study, 100 μL of collected milk samples were cultured on blood agar supplemented with 5% sheep blood. The agar plates were incubated at 37°C for 24 hours and hemolysis patterns were observed. Further, isolates were purified, and differentiated by culturing on mannitol salt agar following the prior incubation conditions, and presumptive identification was done by microbiological examination based on colony morphology, gram staining, catalase, and coagulase tests as done previously (Gizaw et al., 2020). At the same time, all isolates were identified by the automatic biochemical identification VITEK-2 system (BioMérieux, France) according to the prior recommendations (Chajęcka-Wierzchowska et al., 2023). Furthermore, the molecular confirmation of the target specie, S. haemolyticus strains was done by 16S rRNA gene sequencing according to the previously described protocol (Qu et al., 2019). Briefly, the genomic DNA was extracted using genomic DNA Extraction Kit (Tiangen Biochemical Technology Co., Ltd) and PCR was performed using the primers mentioned in Table S1. The PCR reaction mixture (25 μL) comprised 12.5 μL Master Mix, 1 μL of forward and reverse primer each, 1 μL of extracted DNA, and 9.5 μL of deionized water. The PCR amplification conditions were adjusted as initial denaturation at 95°C for 5 minutes, followed by 35 cycles of complete denaturation at 95°C for 30 seconds, elongation at 55°C for 30 seconds, and extension at 74°C for 1 minute with a final extension at 74°C for 5 minutes. The positive (S. haemolyticus ATCC25923) and negative (without genomic DNA) control PCR were done to validate the results. The PCR product was visualized on 1% agarose gel under the GelDoc XR system and purified with Wizard® genomic DNA purification kit (Promega, USA) following the guidelines of the manufacturer and then sent elsewhere (Beijing Huada Gene Technology Co., Ltd) for sequencing analysis. The sequencing data were analyzed by checking the sequence similarity index ≥99.0% and submitted to the National Center for Biotechnology Information (NCBI) GenBank (https://blast.ncbi.nlm.nih.gov/) under the accession numbers from OQ652547 to OQ652584 and OQ787674.
2.4 Antimicrobial susceptibility testing of S. haemolyticus strains
The antimicrobial susceptibility of the 16S rRNA gene confirming S. haemolyticus strains was assessed on the Mueller Hinton agar (MHA) by Kirby-Bauer disk diffusion (KBDD) assay following the EUCAST guidelines against nine antimicrobial agents belonging to nine different classes. The tested antibiotics included were cefoxitin (CFX, 30μg), ciprofloxacin (CIP, 5μg), gentamicin (GEN, 10μg), tetracycline (TET, 30μg), trimethoprim-sulfamethoxazole (SXT, 1.25-23.75μg), florfenicol (FFC, 30μg), erythromycin (ERY, 15μg), and linezolid (LZD, 10μg). The AST of vancomycin was done by MIC broth micro-dilution assay according to the protocol described in the EUCAST guidelines (EUCAST, 2019). Briefly, inoculum for KBDD was prepared by adjusting the turbidity at 0.5 McFarland Standard and swabbing was done on MHA under sterile conditions. Antibiotic disks were placed on agar surface with sterile forcep at the proper distance, and plates were incubated at 35 ± 1°C for 18 ± 2h. After incubation, the zone of inhibition was measured and compared with EUCAST clinical breakpoints (EUCAST, 2019).
2.5 Identification of ARGs
All of the 39 strains were screened for the detection of ARGs by PCR. A total of 30 ARGs against nine antibiotics belonged to nine different antimicrobial classes; ermA, ermB, ermC, ermF, erm(33), mphC, and msrB for erythromycin (macrolides, lincosamides, and streptogramin B); vanA and vanB for vancomycin (glycopeptide); cfr for linezolid (oxazolidinones); cfxA for cefoxitin (cephalosporin); tetM, tetO, tetL, and tetK for tetracycline (tetracycline’s); fexA and floR for florfenicol (amphenicol); aacA-aphD and aadD for gentamicin (aminoglycosides); gyrA, gyrB, grlA, and grlB for ciprofloxacin (fluoroquinolones); and sul1, sul2, sul3, dfrA, dfrD, dfrG, and dfrK for trimethoprim-sulfamethoxazole (sulfonamides) were screened by PCR amplification using the primer mentioned previously (Table S2). The genomic DNA was extracted using the DNA extraction kit and PCR reaction mixture (25 µL) comprising 12.5 µL Master Mix, 1 µL of each primer, 1 µL genomic DNA, and 9.5 µL of deionized water. The PCR amplification conditions were adjusted as 95°C for 5 min for initial denaturation, followed by 30 cycles at 95°C for 30 s, 30 cycles of annealing at various temperatures for 30 s (Table S2), initial extension at 72°C for 1 min, and followed by final elongation at 72°C for 10 min. The PCR product was run on 1% agarose gel at 80V and 130mA for 1 hour for visualization under UV light by ethidium bromide staining.
2.6 Pulsed-field gel electrophoresis
All 39 S. haemolyticus strains were also subjected to PFGE analysis to determine their phylogenetic grouping as described previously (Kim et al., 2019). Briefly, the bacterial culture grown overnight was embedded with low-melting SKG agarose gel, and then blocks were placed in 1×TE buffer containing 10 mg/L lysostaphin and incubated at 37°C for 3.5h. Subsequently, the blocks were digested with SmaI at 30°C for 3h. Then, the blocks were loaded into 1% SKG gel, and gel electrophoresis was performed in 0.5×TBE buffer for 18h with a 120° change angle, 6 V/cm voltage, and switching time of 5-40 sec. Salmonella H9812 (XbaI digestion) was used as size markers. After electrophoresis, the gel was stained with ethidium bromide (1: 10000) for 30 min, decolored in deionized water for 30 min, and photographed under a gel imaging system. The phylogenetic analysis of the PFGE band patterns was done by PyElph software and compared with the standard (Pavel and Vasile, 2012).
2.7 Multi-locus sequence typing
MLST analysis was done based on primer sequences published on the MLST website (https://pubmlst.org/shaemolytic/) for seven housekeeping genes of S. haemolyticus including arc haemo, SH1200, hemH, leuB, SH1431, cfxE, and riboseABC. The seven housekeeping genes were PCR amplified and sequenced using the genomic DNA of S. haemolyticus as a template. The sequencing data were submitted to the MLST database (http://www.pubmlst.net/databases/) to determine the STs of S. haemolyticus. The alignment of obtained sequences was done using ChromasPro computer software and the phylogenetic tree was made by the Neighbor-Joining Method in the MEGA-X software.
3 Results
3.1 Prevalence of S. haemolyticus and other Staphylococcus species
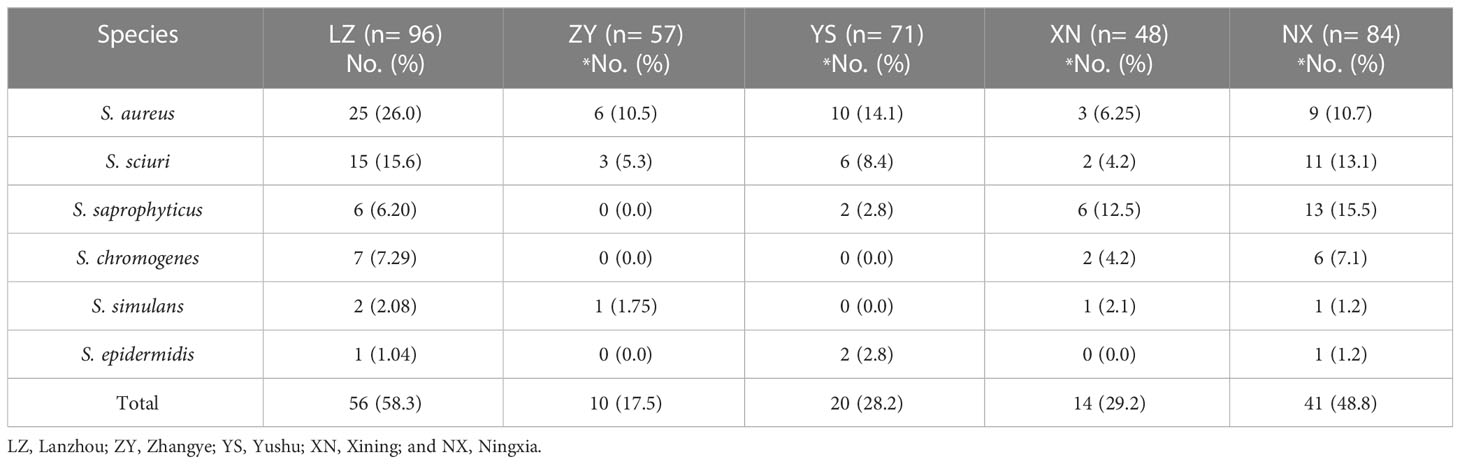
For the current study, a total of 356 milk samples were collected from different dairy farms in different cities (Lanzhou, Zhangye, Xining, Yushu, and Ningxia) in Northwest, China. In the present study, 180/356 (50.6%) isolates were recovered and identified as Staphylococcus species. Moreover, the prevalence of S. aureus was noted to be 14.9% (53/356) and NAS was 35.7% (127/356) (Figure 1A). Among NAS, S. haemolyticus was identified as the predominant specie 11.0% (39/356), followed by S. sciuri 10.4% (37/356), S. saprophyticus 7.6% (27/356), S. chromogenes 4.2% (15/356), S. simulans 1.4% (5/356), and S. epidermidis 1.1% (4/356) (Figure 1B). The prevalence of S. haemolyticus was higher (14.6%; 7/48) in Xining, followed by Zhangye (14.0%; 8/57), Lanzhou (12.5%; 12/96), Ningxia (8.3%; 7/84), and was lower in Yushu 7.04% (5/71) (Figure 1C). The overall prevalence of all other Staphylococcus species was noted to be higher in Lanzhou (58.3%; 56/96), followed by Ningxia 48.8% (41/84), Xining 29.2% (14/48), Yushu 28.2% (20/71), and Zhangye 17.5% (10/57). Moreover, the prevalence of S. aureus was noted to be 26.0%, 10.5%, and 14.1% in Lanzhou, Zhangye, and Yushu, respectively, while S. saprophyticus was higher in Xining (12.5%) and Ningxia (15.5%). The prevalence of other Staphylococcus species among different sampling cities is presented in Table 1.
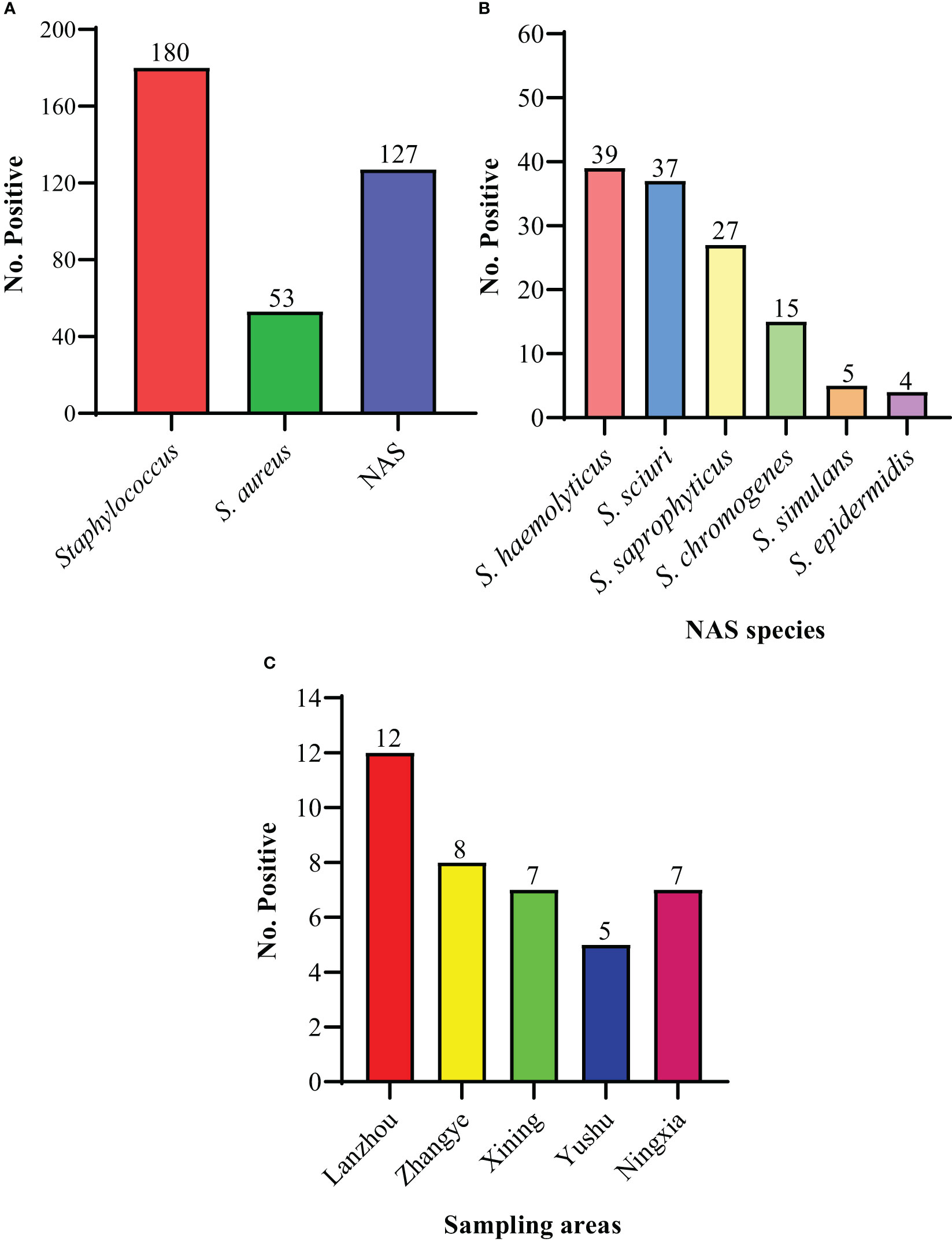
Figure 1 The prevalence of isolated species. (A) The overall prevalence of Staphylococcus, S. aureus, and non-aureus Staphylococcus (NAS) species. (B) The individual distribution of NAS species. (C) The distribution of 39 S. haemolyticus strains within sampling cities.
3.2 Phenotypic AMR characteristics of 39 S. haemolyticus strains
Most of the S. haemolyticus strains showed resistance to ERY (92.3%, 36/39), followed by SXT (51.3%, 20/39), CIP (43.6%, 17/39), FFC (30.8%, 12/39), FOX (28.2%, 11/39), and GEN (23.1%, 9/39). However, all strains were susceptible to TET, VAN, and LZD. Moreover, none of the strains showed intermediate susceptibility to all antibiotics (Figure 2). S. haemolyticus strains from LZ, YS, and NX showed 100% resistance to ERY, which was noted to be lower in other cities (85.7%, ZY, and 75.0%, XN). The AMR against SXT was noted at 50%, 42.8%, 87.5%, 20.0%, and 42.8% by S. haemolyticus strains from LZ, ZY, XN, YS, and NX, respectively (Table 2). Moreover, none of the strains from ZY and NX showed resistance to FOX with a similar trend from XN and NX against GEN. It was observed that 80% of the strains from Yushu City were resistant to multiple antimicrobials (FOX+GEN+CIP+FFC) and 20% to SXT. All of the strains from all sampling cities were susceptible to TET, VAN, and LZD. The overall AMR was noted to be higher in strains from YS, followed by NX, LZ, XN, and ZY (Table 2).
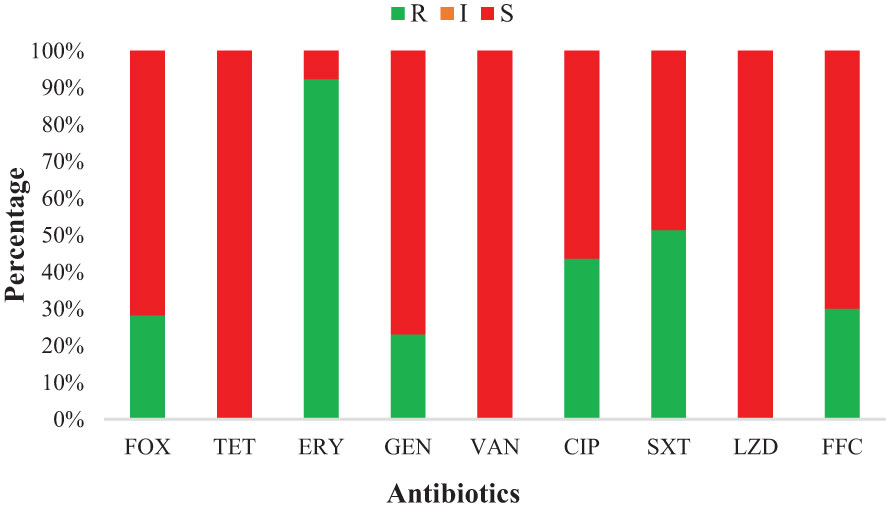
Figure 2 Antimicrobial susceptibility of various antibiotics against S. haemolyticus strains of dairy origin: FOX, Cefoxitin; ERY, Erythromycin; TET, Tetracycline; GEN, Gentamicin; CIP, Ciprofloxacin; VAN, Vancomycin; SXT, Trimethoprim-Sulfamethoxazole; FFC, Florfenicol; and LZD, Linezolid.
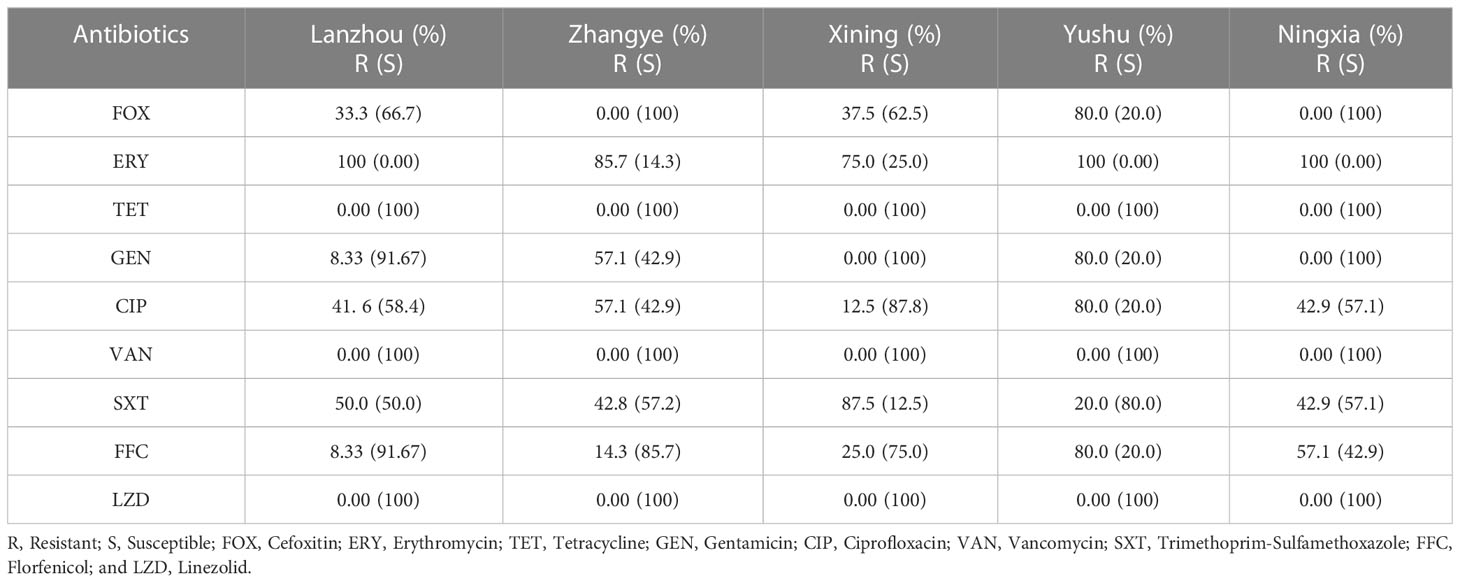
Table 2 The percentage distribution of antimicrobial-resistant and susceptible strains among different sampling cities.
3.3 Multi-drug resistant S. haemolyticus strains
The current study depicted 46.15% (18/39) MDR S. haemolyticus strains that showed resistance to at least one antimicrobial agent from ≥3 classes of antimicrobials. Among MDR strains, 30.77% (12/39) were resistant to three antibiotic classes, followed by 10.26% (4/39) resistant to five classes, and 5.13% (2/39) resistant to four classes. None of the strains (0.0%) showed resistance to all six classes of antibiotics (Figure 3A). The percentage of MDR strains was noted to be highest at 12.82% (5/39) from Lanzhou, followed by an equal percentage of 10.26% (4/39) from Zhangye and Yushu. The MDR strains were noted to be 7.69% (3/39) from Ningxia and 5.13% (2/39) from Xining (Figure 3B).
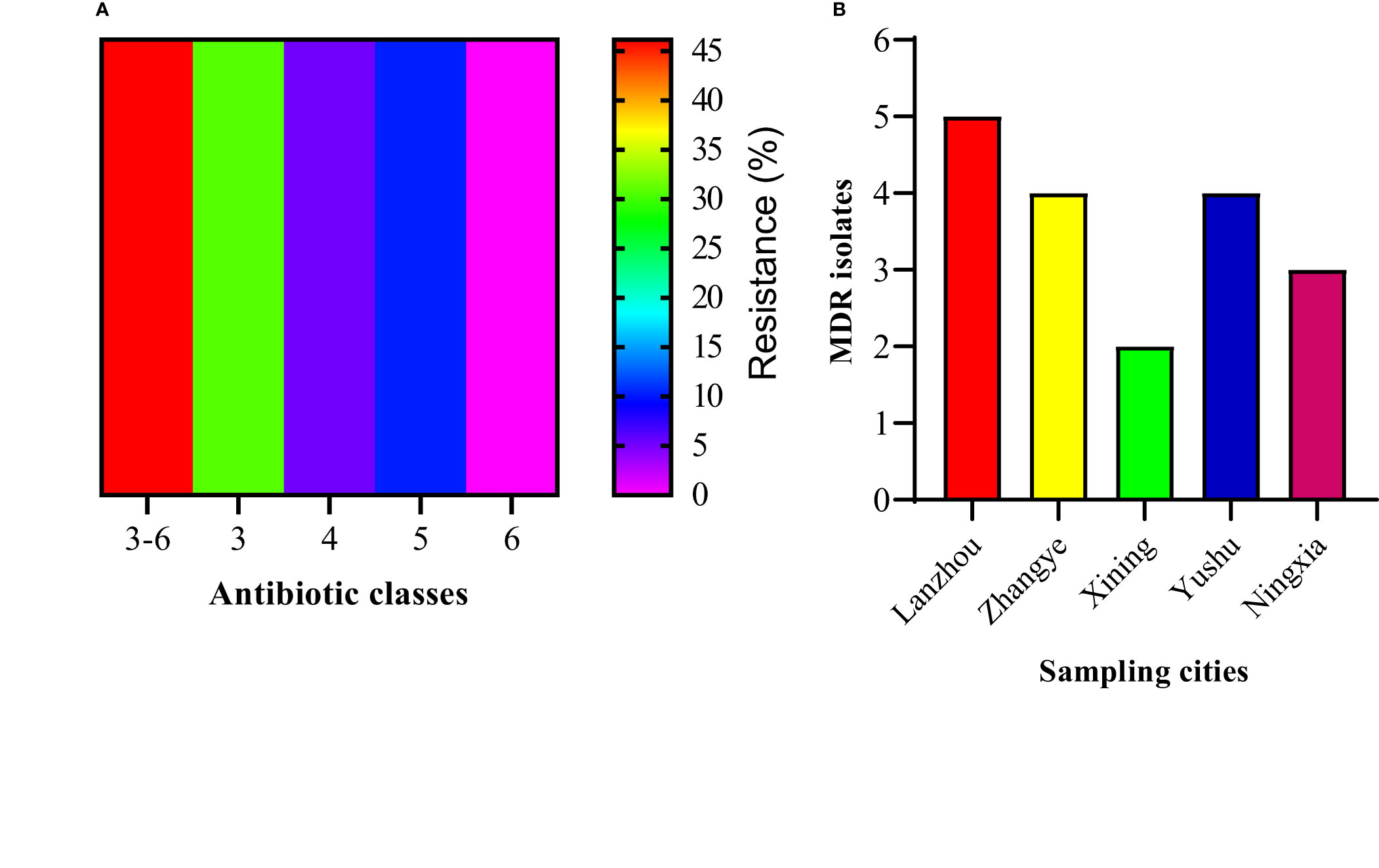
Figure 3 The percentage of MDR S. haemolyticus. (A) The percentage of S. haemolyticus strains showing resistance to three, four, five, six, and three to six classes of antibiotics. (B) The percentage distribution of MDR S. haemolyticus strains among different sampling sites.
3.4 Genotypic AMR characteristics of S. haemolyticus strains
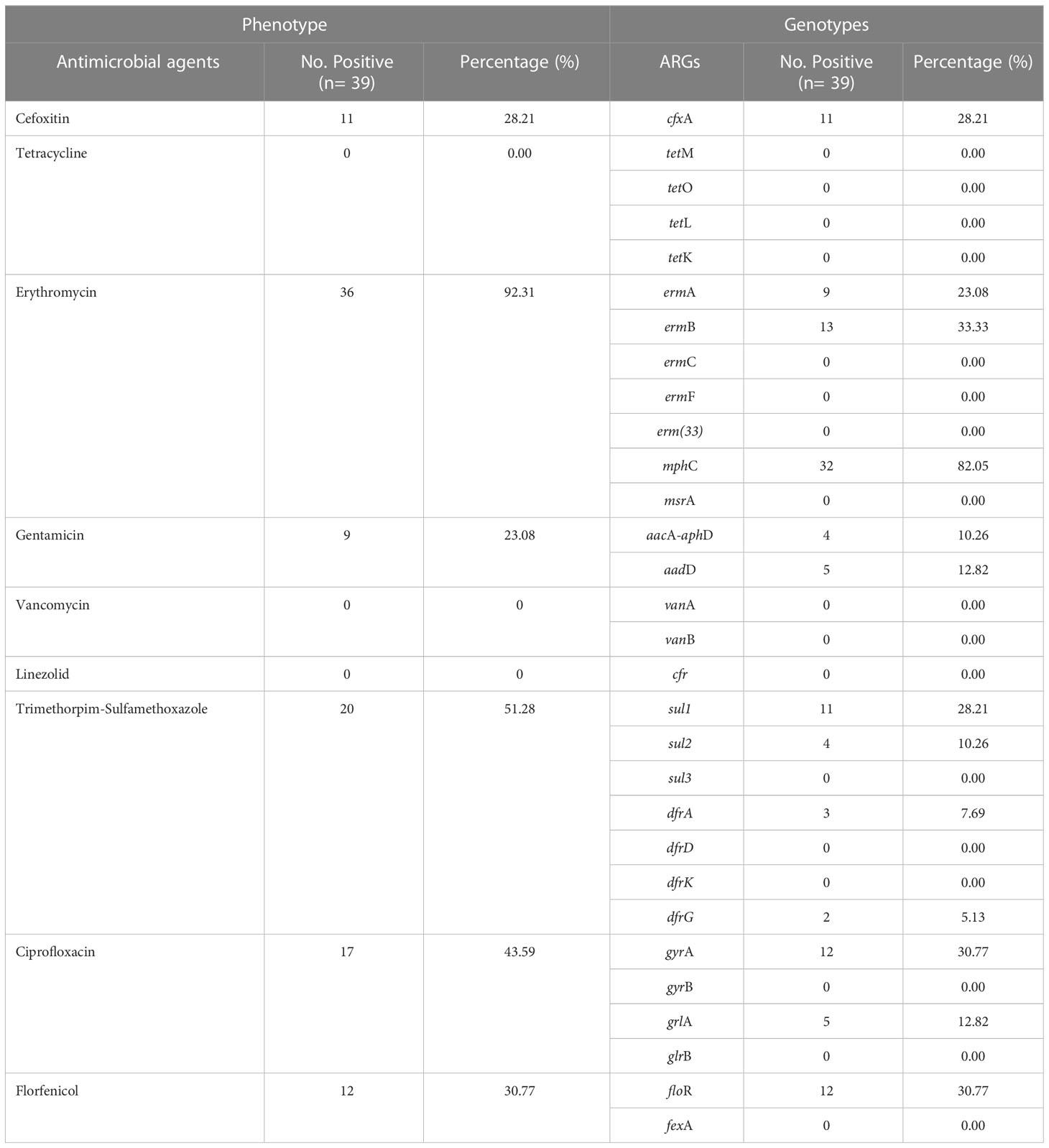
A total of 30 ARGs belonged to nine different antimicrobials and classes; ermA, ermB, ermC, ermF, erm(33), mphC, and msrB for erythromycin (macrolides, lincosamides, and streptogramin B); vanA and vanB for vancomycin (glycopeptide); cfr for linezolid (oxazolidinones); cfxA for cefoxitin (cephalosporin); tetM, tetO, tetL, and tetK for tetracycline (tetracycline’s); fexA and floR for florfenicol (amphenicol); aacA-aphD and aadD for gentamicin (aminoglycosides); gyrA, gyrB, grlA, and grlB for ciprofloxacin (fluoroquinolones); and sul1, sul2, sul3, dfrA, dfrD, dfrG, and dfrK for trimethoprim-sulfamethoxazole (sulfonamides) were screened. Among 30 ARGs, 12 genes from different classes and multiple genes acquiring spectrum were identified. Among macrolides, lincosamides, and streptogramin B resistance genes, mphC was most predominant (82.05%), followed by ermB (33.33%) and ermA (23.08%), while ermC, ermF, erm(33), and msrB genes were not identified. Among amphenicol-resistance genes, only the floR gene was harbored by 30.77% of strains. Among the aminoglycosides, the aadD gene was a little more prevalent (12.82%) than aacA-aphD (10.26%). The prevalence of the gyrA gene was noted to be higher (30.77%) compared to grlA (12.82%), with no identification of gyrB and grlB among the fluoroquinolones. However, none of the S. haemolyticus strains was harboring the glycopeptides, oxazolidinones, and tetracycline-resistance genes. Moreover, among sulfonamides, sul1 (28.21%) was predominant, followed by sul2 (10.26%), dfrA (7.69%), and dfrG (5.13%), while none of the strains was carrying the sul3, dfrD, and dfrK genes (Figure 4A). All of the S. haemolyticus strains were carrying the multiple genes spectrum, and most of the strains (43.6%, 17/39) were carrying the three different patterns of two ARGs: mphC+ermB (23.1%, 9/39), mphC+sul1 (10.3%, 4/39), and ermB+sul2 (10.3%, 4/39), followed by three different patterns of three ARGs; floR+dfrA+ermA (7.7%, 3/39), mphC+cfxA+grlA (12.8%, 5/39), and mphC+aadD+gyrA (5/39). Moreover, 7.69% (3/39), 5.13% (2/39), and 10.3% (4/39) were carrying the single pattern of four, five, and seven ARGs (Figure 4B).
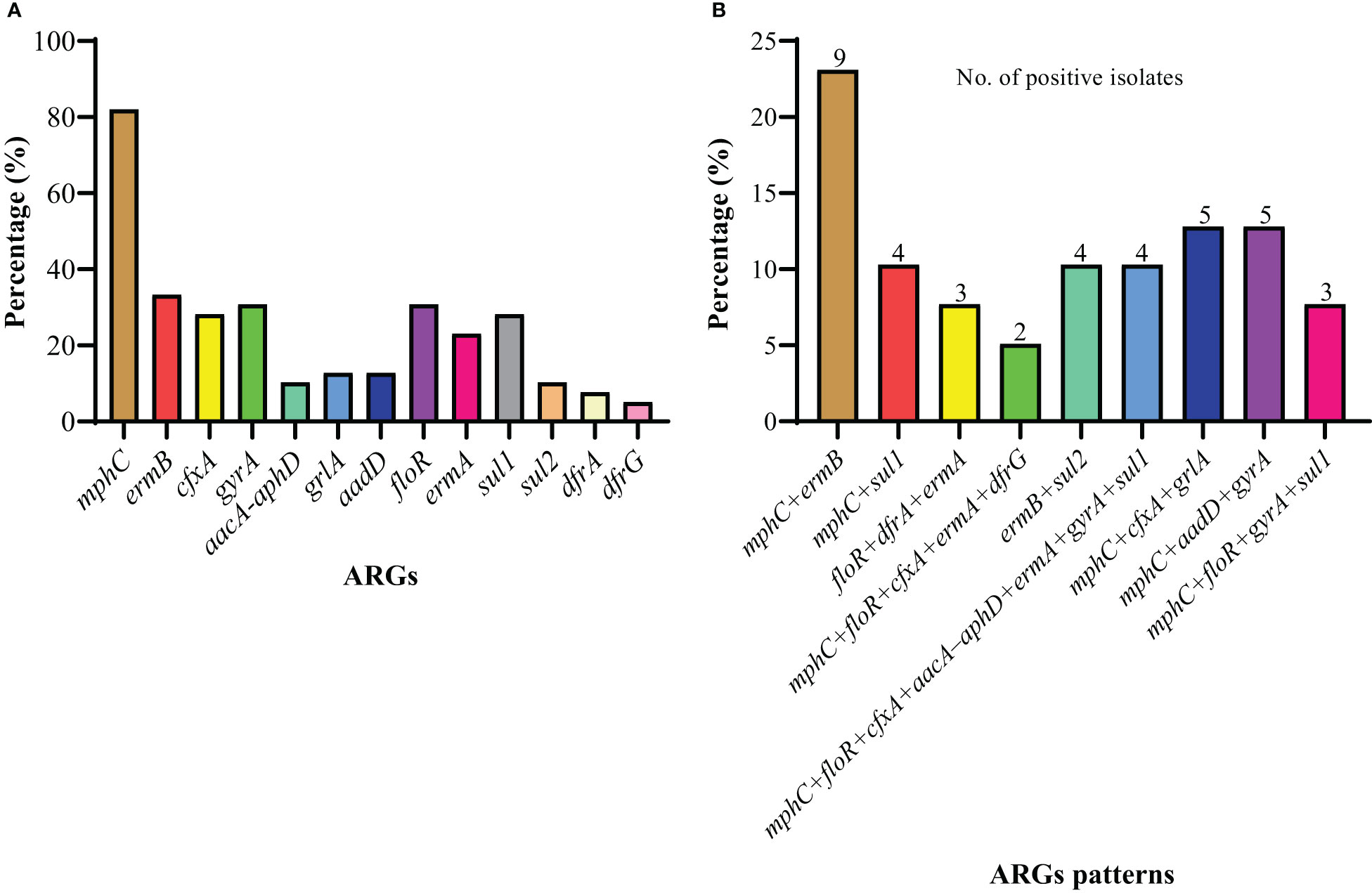
Figure 4 The distribution of ARGs among 39 S. haemolyticus strains. (A) The individual gene distribution (B) The different patterns of ARGs possessed by strains.
3.5 The comparison between phenotypic and genotypic resistance profile
The comparison in the phenotypic and genotypic expression of 39 S. haemolyticus strains is presented in Table 3. This comparative analysis was done with the perspective to analyze the expressions of S. haemolyticus strains and whether the strains showing resistance phenotypically were also carrying the ARGs or not and vice versa. The study noted unique phenotypic and genotypic expressions for all antibiotics except erythromycin. The number of phenotypically resistant strains was noted at 36/39 (92.31%) while the genotypic analysis showed that few strains were carrying the multiple-resistance genes. All of the strains showed no phenotypic and genotypic resistance to tetracycline, vancomycin, and linezolid.
3.6 The phylogenetic grouping of S. haemolyticus strains
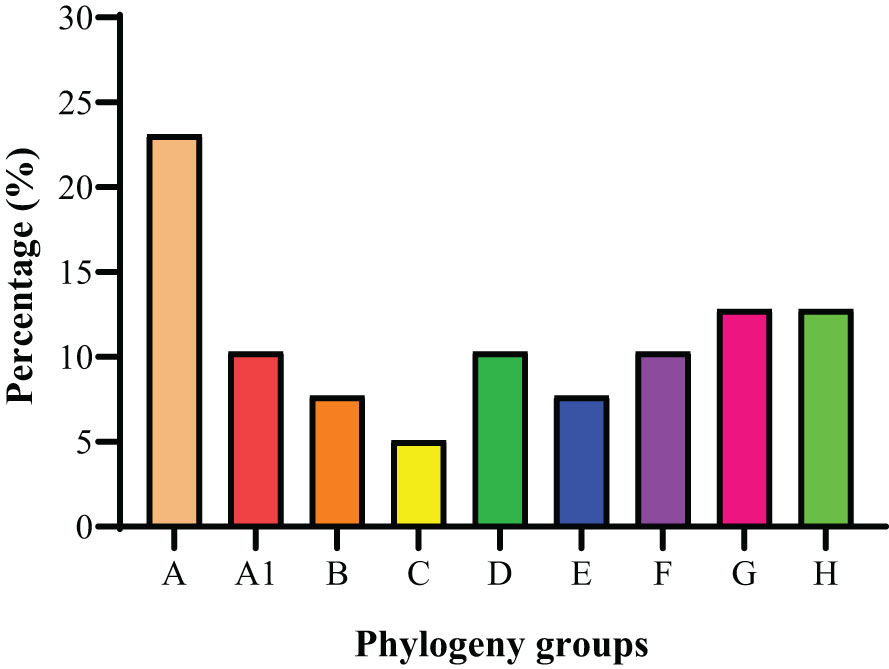
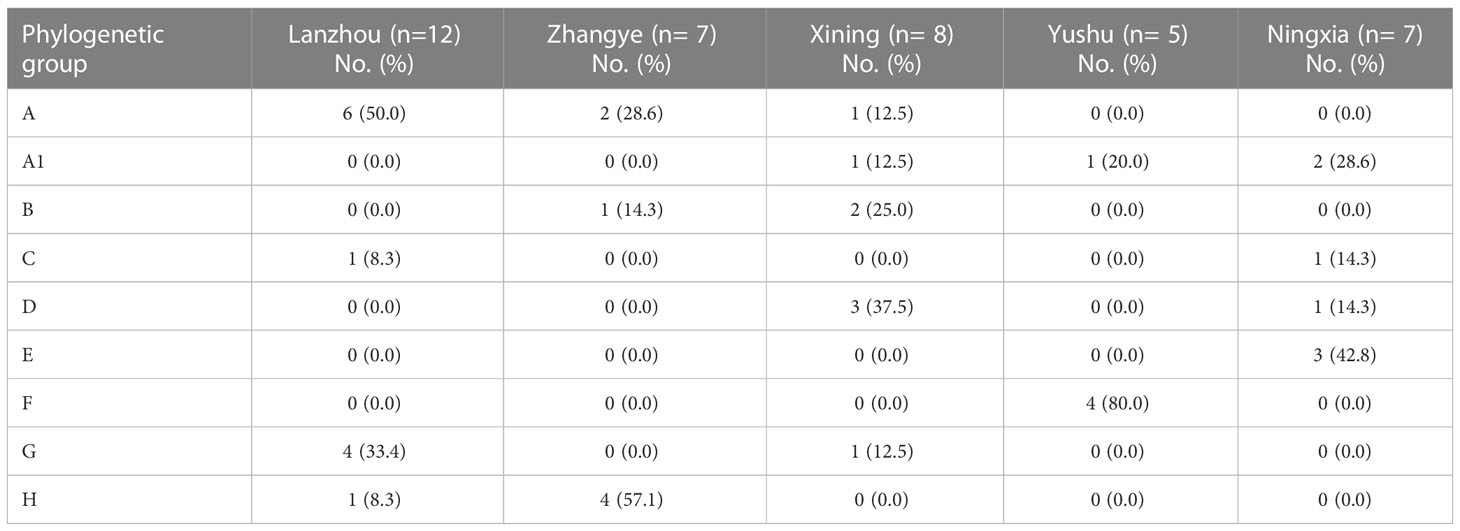
The phylogenetic grouping of 39 S. haemolyticus strains determined by PFGE generated by SmaI digestion and multiple PFGE patterns was observed among the strains. The BioNumerics cluster analysis software was used to construct the phylogenetic tree (Figure S1). The 39 S. haemolyticus were classified into eight different phylogenetic groups A, A1, B, C, D, E, F, G, and H. It was observed that most of the S. haemolyticus strains (23.1%, 9/39) belonged to group A and the least belonged (5.1%, 2/39) to group C. Moreover, 10.3% (4/39) were classified A1, D, and F, each, and 7.7% (3/39) into groups B and E, each. The percentage of strains categorized under groups G and H was noted to be 12.8% (5/39) in each group (Figure 5). In the phylogenetic grouping of strains among the different sampling cities, most of the strains from Lanzhou, Zhangye, Xining, Yushu, and Ningxia belonged to groups A, H, D, F, and E, respectively. Moreover, the phylogenetic grouping of S. haemolyticus strains from different cities was as follows: Lanzhou (A, C, G, and H), Zhnagye (A, B, and H), Xining (A, A1, B, D, and G), Yushu (A1 and F), and Ningxia (A1, C, D, and E) (Table 4).
3.7 Multi-locus sequence typing
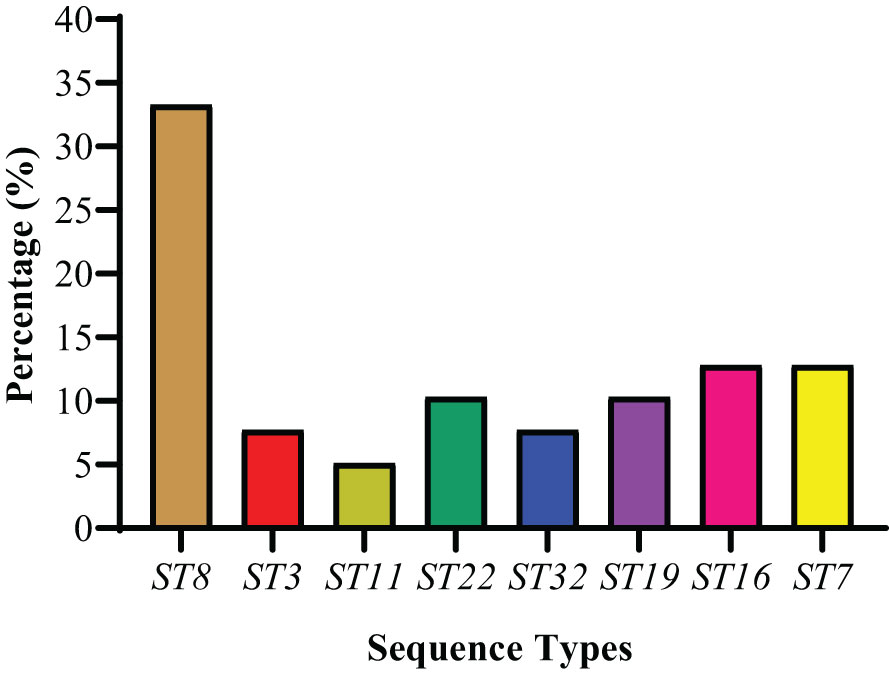
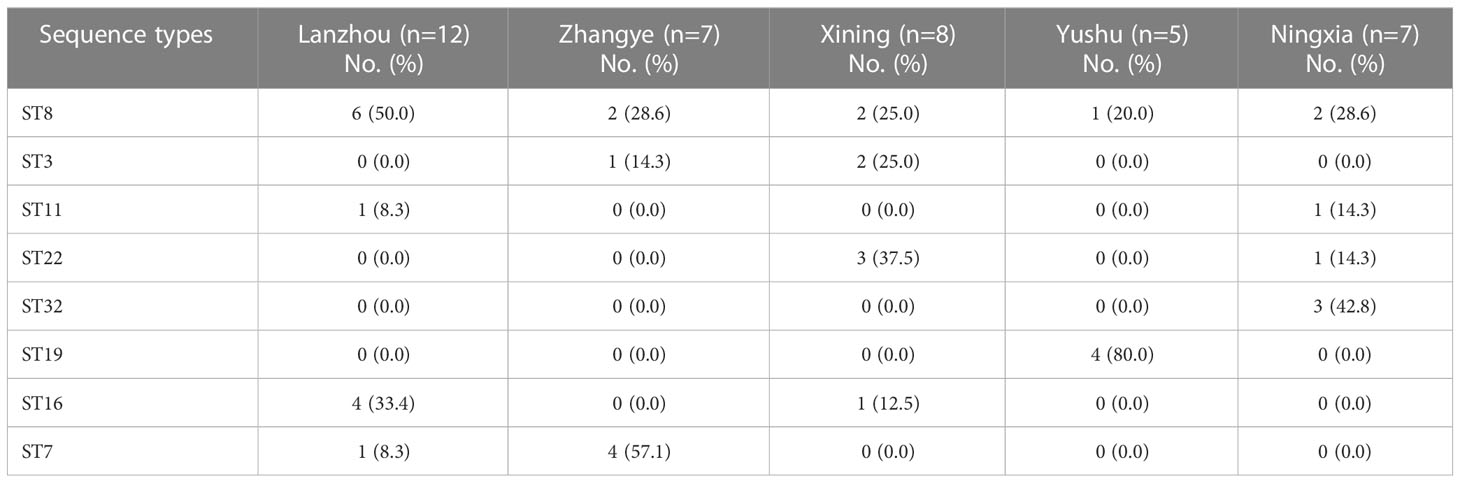
The MLST was done by the amplification and sequencing of seven housekeeping genes. Thirty-nine S. haemolyticus strains were categorized into eight different STs (ST8, ST3, ST11, ST22, ST32, ST19, ST16, and ST7). Among STs, ST8 was found to be dominant, accounting for 33.3% (13/39) (Figure 6). The percentage distribution of ST22 and ST19 were noted as similar (10.3%, 4/39 each). However, 5.1% (2/39) of strains were categorized as ST11, while the percentage of other STs, ST3, and ST32 were noted as 7.7% (3/39), followed by ST16 and ST7 as 12.8% (5/39) each (Figure 6). The detailed ST distribution in different sampling cities is given in Table 5. The dominant STs from Lanzhou, Zhangye, Xining, Yushu, and Ningxia were classified as ST8, ST7, ST22, ST19, and ST32, respectively. Moreover, the strains from Yushu only belonged to ST8 and ST19. The distribution of other STs identified in other cities was as follows: Lanzhou (ST11, ST16, ST7), Zhnagye (ST8, ST3), Xining (ST8, ST3, ST16), and Ningxia (ST8, ST11, ST22) (Table 5).
3.8 The association between STs, phylogenetic groups, and ARGs patterns
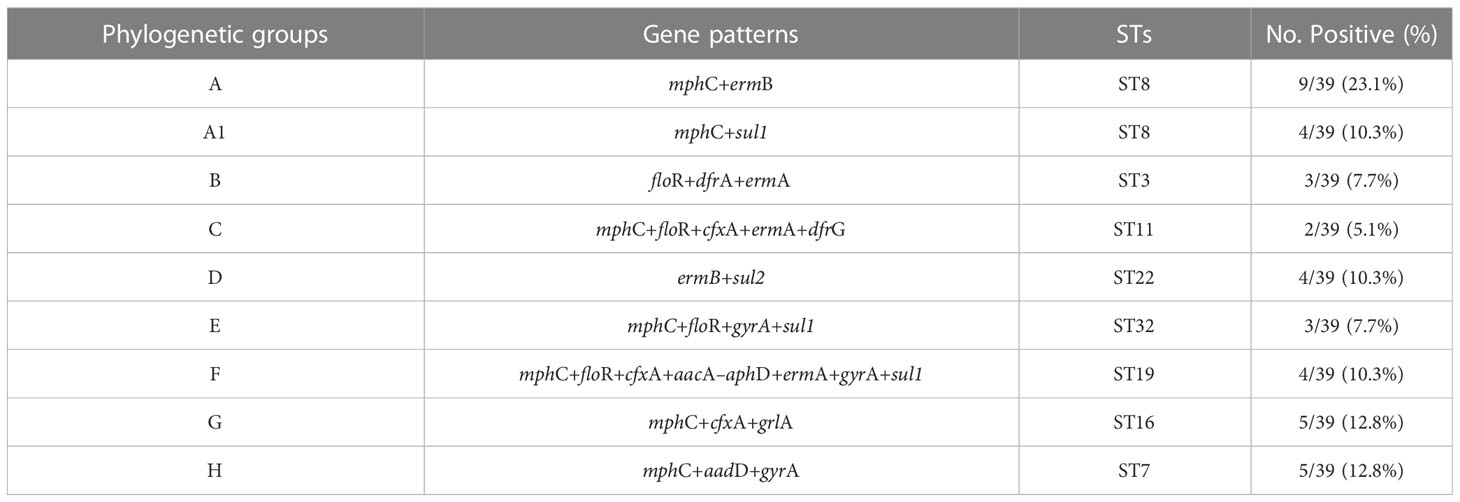
Most of the S. haemolyticus strains belonged to ST8 (13/39), and the phylogenetic group A+A1 was carrying the mphC+ermB and mphC+sul1 gene patterns. Moreover, 10.3% of strains belonged to phylogenetic groups D and F with ST22 and ST19 carrying the ermB+sul2 and mphC+floR+cfxA+aacA–aphD+ermA+gyrA+sul1 gene patterns, respectively. The least number of strains, 2/39 (5.1%) classified under ST11 and group C, were carrying the mphC+floR+cfxA+ermA+dfrG resistance gene pattern. However, an equal number of strains 5/39 (12.8%) that were classified into ST16 and ST7 belonging to phylogenetic groups G and H harbored the mphC+cfxA+grlA and mphC+aadD+gyrA gene patterns, respectively (Table 6).
4 Discussion
As per our knowledge and literature search, the current study is the first study reporting the prevalence and characterization of S. haemolyticus strains isolated from dairy cattle milk in Northwest, China, with a large sample size (n= 356). In the present study, 180/356 (50.6%) isolates were recovered and identified as Staphylococcus species confirming them as the major pathogen of dairy mastitis in China (Gao et al., 2017; He et al., 2020; Song et al., 2020). The prevalence of S. aureus was noted to be 14.9% (53/356) and NAS was 35.7% (127/356). Among the NAS, the prevalence of S. haemolyticus was noted to be 11.0% (39/356), while other NAS species were as follows: S. sciuri 10.4% (37/356), S. saprophyticus 7.6% (27/356), S. chromogenes 4.2% (15/356), S. simulans 1.4% (5/356), and S. epidermidis 1.1% (4/356) based on the bacteriological examination by the VITEK-2 system, as done previously (Gizaw et al., 2020; Chajęcka-Wierzchowska et al., 2023). Further confirmation of the targeted specie of S. haemolyticus strains was done by 16S rRNA gene sequencing, which is known to be a reliable molecular technique for microbial identification (Johnson et al., 2019; Han et al., 2020). The present study identified 39/356 (11.0%) strains as S. haemolyticus, which is consistent with previous findings (Klimiene et al., 2016). The current study identified S. haemolyticus, S. sciuri, S. saprophyticus, S. chromogenes, S. simulans, and S. epidermidis as the most frequently isolated NAS species from mastitic milk of dairy cows, which is consistent with previous studies from other regions of China (Qu et al., 2019) and other countries (Vanderhaeghen et al., 2014; Goetz et al., 2017; Jenkins et al., 2019). Moreover, in this study, the prevalence of S. aureus was noted to be 14.9% (53/356), which is higher than other Staphylococcus species. These results are consistent with a previously reported study that S. aureus is the major pathogen among Staphylococcus species associated with cow mastitis (Cortimiglia et al., 2016). However, many previous studies have reported the prevalence and characterization of S. aureus isolated from cow milk from different provinces in Northwest, China (Li et al., 2015; Li and Zhao, 2018; Dan et al., 2019; Kou et al., 2021; Shi et al., 2021). Our study only focused on the epidemiology of NAS, while other factors such as lactation stage, milking practices, and severity of disease-related factors such as somatic cell count were not recorded. The previous studies hypothesized that prior mentioned factors along with immunity level, nutrition, and farm management practices may be the predisposing factors for opportunistic pathogens such as NAS to invade and proliferate within the udder, leading to inflammation with clinical signs (Pal et al., 2019; Qu et al., 2019; Libera et al., 2021). Except for these factors, NAS species are also found to be ubiquitous in environmental settings such as soil, dust, and farm environment which may be the possible cause of udder infection by environmental opportunistic pathogens such as NAS (Schoenfelder et al., 2017). Adkins et al. (2018) previously documented that the intra-mammary (IM) infection of NAS species leads to subclinical mastitis with an increase in somatic cell count and later to clinical mastitis resulting in reduced milk yield. The control of NAS is essential and complicated because too many species are responsible for this problem. However, the implementation of proper mastitis prevention and control measures is guided by the National Mastitis Council (2017) to control the IM infections caused by NAS in dairy cows.
In this study, we analyzed the susceptibility of 39 S. haemolyticus strains of dairy origin against the nine antimicrobial agents, which are cefoxitin, tetracycline, vancomycin, linezolid, erythromycin, gentamicin, ciprofloxacin, trimethoprim-sulfamethoxazole, and florfenicol. Overall, most of the strains were found to be resistant to erythromycin (92.3%), followed by trimethoprim-sulfamethoxazole (51.3%), ciprofloxacin (43.6%), florfenicol (30.8%), cefoxitin (28.2%), and gentamicin (23.1%). The increased resistance to these antibiotics by Staphylococcus species has also been reported previously because of the increased use of these antibiotics in the treatment of NAS infections (Botrel et al., 2010; Persson et al., 2011; Qian et al., 2015; Kim et al., 2019). A similar study conducted by Gizaw et al. (2020) using archived isolates from cattle, farm environment, and personals noted a 3.6% resistance to ciprofloxacin, 12.5% to kanamycin, and 55.4% to erythromycin, which is much lower than the current findings, indicating the extensive use of these antibiotics at the sampled dairy farms in Northwest, China. Moreover, in the current study, the cefoxitin resistance was noted to be lower compared to the previous findings (Gizaw et al., 2020). The variations in antimicrobial resistance in different studies may be due to the highly genetically heterogeneous nature of bacterial isolates, which may exhibit varying degrees of phenotypic and genotypic expressions (Pacha et al., 2021). Moreover, another reason for the difference in susceptibilities might be the difference in susceptibility assay, Staphylococcus specie, and isolation source because according to our knowledge, none of the studies specifically check the antimicrobial response of S. haemolyticus strains isolated from dairy milk in China. Furthermore, all of the 39 S. haemolyticus strains were 100% susceptible to tetracycline, vancomycin, and linezolid, which is consistent with previous studies conducted by Kim et al. (2019) and Mello et al. (2020). Moreover, in this study, 46.15% of strains were MDR S. haemolyticus with 30.77% resistant to three antimicrobial classes, followed by 10.26% and 5.13% resistance to four and five classes, respectively. The irregular and non-justified use of antibiotics in livestock, especially in dairy production systems leads to the emergence of bacterial strains resistant to multiple antibiotics, which poses a potential threat to animal and human health (Phophi et al., 2019; Attallah et al., 2021). This misuse of antimicrobials put unnatural selective pressure on bacteria, which accelerates the evolution of resistant strains (Fair and Tor, 2014). Therefore, the justified use of antimicrobials in dairy production must be ensured and monitored through surveillance studies to get updated information regarding antimicrobial resistance. Moreover, the antimicrobial resistance can be lowered by restricting the use of antimicrobials as a feed supplement in production systems and finding alternative sources such as herbal medicine, essential oils, and traditional Chinese veterinary medicine (Lu et al., 2008; Yang et al., 2019; Cheng and Han, 2020; Ajose et al., 2022).
Previously, it was documented that food animals, especially cattle, were recognized as the reservoir of drug-resistant bacteria carrying ARGs, which can be transmitted to humans via the food web (Bennani et al., 2020; Sivagami et al., 2020). To understand the mechanism of gene transfer and the emergence of multi-drug resistant strains within and across the bacterial species, the knowledge of ARGs carried by pathogens is essential (Tóth et al., 2020). Therefore, the present study investigated the ARGs carried by S. haemolyticus strains isolated from dairy cattle milk; 30 ARGs belonging to nine antimicrobial classes were screened. Among the 30 ARGs, 12 resistance genes were identified among 39 S. haemolyticus strains. Among the macrolides, lincosamides, and streptogramin B resistance genes, mphC was most predominant, followed by ermB and ermA, while ermC, ermF, erm(33), and msrB genes were not identified. The erm family of gene codes for methylases confers resistance to streptogramin B (Petinaki and Papagiannitsis, 2019), macrolides (Feßler et al., 2018b), and lincosamides (Feßler et al., 2018a), while the mphC gene code for phosphotransferase confers resistance only to macrolides (Chajęcka-Wierzchowska et al., 2023). Other macrolide genes such as msrA and B gene codes for transporter proteins that confer resistance to both streptogramin B and macrolides (Quezada-Aguiluz et al., 2022) were not identified in this study. A similar study done by Qu et al. (2019) also detected that the abovementioned genes carried by Staphylococcus species conferred resistance to macrolides, lincosamides, and streptogramin B with an additional lnuA gene. Among the cephalosporins, the cfxA gene was identified, which is also in line with the findings of Qu et al. (2019) and other previous studies (El-Ashker et al., 2020; Jose et al., 2020). The higher frequency of these genes from dairy isolates indicates the potential use of cephalosporins for the treatment of clinical mastitis on dairy farms in China. Previously, Qian et al. (2015) reported that β-lactam antimicrobial agents such as penicillin, cephalosporin, and monobactams were extensively used for the treatment of bovine mastitis caused by Staphylococcus species. Among the amphenicol ARGs, only the floR gene was harbored by S. haemolyticus strains, which is in accordance with the previously published study by Wu et al. (2021). Florfenicol is one of the most commonly used antimicrobials in the dairy sector because of its wide range of antimicrobial activity (Kawalek et al., 2016). Florfenicol acts by binding with the 50S ribosomal subunit, resulting in the inhibition of protein synthesis. floR gene encodes for membrane-associated proteins that confer resistance to florfenicol (Shore et al., 2016). Another study conducted by Li et al. (2020) confirmed that floR was found to be the predominant gene conferring resistance to florfenicol in different animal-derived bacteria. Among the aminoglycosides, both aadD and aacA-aphD genes were identified, which is in line with previously published studies (Qu et al., 2019; Kowalewicz et al., 2023). The aacA-aphD gene code for a multifunctional enzyme confers resistance to multiple aminoglycosides such as amikacin, gentamicin, tobramycin, and kanamycin (Schwarz et al., 2018), while aadD gene codes for adenyltransferase enzyme confer resistance to neomycin, kanamycin, tobramycin, and paromomycin (Batool et al., 2021). The prevalence of the gyrA gene was noted to be higher compared to grlA among the fluoroquinolones that confer resistance to ciprofloxacin, as studied previously (Bonsaglia et al., 2018; Pham et al., 2019). The acquisition of gyrA/B and grlA/B genes by Staphylococcus species confers resistance to fluoroquinolones because they code for two enzymes, DNA gyrase and topoisomerase IV, respectively, which are required by the bacteria during DNA replication (Schwarz et al., 2018; Lapointe et al., 2021). However, none of the S. haemolyticus strains was harboring the glycopeptides, oxazolidinones, and tetracycline-resistance genes, which is contrary to the findings of Qu et al. (2019) but inconsistent with the findings of Liu et al. (2022) for tetracycline. Among sulfonamides, sul1 was predominant, followed by sul2, dfrA, and dfrG, while none of the strains was carrying the sul3, dfrD, and dfrK genes. A study conducted by Nurjadi et al. (2021) also identified that dfrA, dfrD, dfrG, and dfrK genes confer resistance to trimethoprim, and Khan et al. (2022) detected sul1, sul2, and sul3 genes that confer resistance to sulfonamides among the Staphylococcus species. The dfr genes confer resistance to trimethoprim by modifications in the target dihydrofolate reductase enzyme (Nurjadi et al., 2021). All of the S. haemolyticus strains were carrying the multiple gene patterns that are mentioned in Figure 5B, which follows many gene patterns identified in a previous study (Qu et al., 2019). This could be explained by the fact that a single resistance phenotype can be mediated by multiple resistance genes (Varela et al., 2021). The presence of diverse ARGs in mastitic milk of dairy cows poses a potential threat to the transmission of these genes from animal to human microbiota through horizontal gene transfer (HGT) by mobile genetic elements (Argudín et al., 2017; Hu et al., 2017). Therefore, it is necessary to monitor antimicrobial resistance, and their transmission mechanisms are of much significance to guide the rational use of antimicrobials in dairy production.
The PFGE and MLST methods are still used for the genotypic characterization of bacterial isolates and to track the dissemination of infections with limitations (Neoh et al., 2019; He and Reed, 2020; Pizauro et al., 2021). Therefore, the present study used these methods for the genotypic characterization of 39 S. haemolyticus strains and grouped strains into eight phylogenetic groups and sequence types. Based on PFGE, most of the strains belonged to group A (9/39), followed by G (5/39), H (5/39), A1 (4/39), D (4/39), F (4/39), B (3/39), E (3/39), and C (2/39). Most of the strains belonging to group A are consistent with the previous findings of Qu et al. (2019). Previously, a study conducted by Naushad et al. (2016) also grouped NAS species into five: A, B, C, D, and E phylogenetic groups based on whole genome sequencing (WGS). It was observed that phylogenetic groups were sharing a common cluster among 39 S. haemolyticus strains isolated from the close locality. The study noted that the farms in a close geographical area may share the same genotypes, suggesting clonal transmission between dairy farms (Rainard et al., 2018). Notably, the MLST results were found to be consistent with the results of PFGE as each ST type corresponded to a single PFGE type. Among the STs, eight unrelated STs were identified with ST8 being predominant, followed by ST7, ST16, ST22, ST19, ST3, ST32, and ST11. Previously, multiple studies from human and animal settings reported ST8 as an emerging sporadic clone among Staphylococcus species from different regions of mainland China (Li et al., 2018; Liu et al., 2018; Dong et al., 2020). The other identified STs in this study have also been reported previously among the Staphylococcus species from China: ST7 (Ou et al., 2020; Zhu et al., 2022; Gu et al., 2023), ST 3 (Chang et al., 2022; Lin et al., 2022), ST 16 (Du et al., 2013), ST22 (Zhou et al., 2022; Gu et al., 2023), and ST19 (Jiang et al., 2016). As per our literature search, ST11 and ST32 were not reported previously among the Staphylococcus genus from China, while ST32 was reported in the S. epidermidis strain from the USA (Mendes et al., 2012). The presence of common STs in both animal and human strains is indicative of in-between species transfer of genetic heritability from both perspectives. The capability of a few pathogens from one host specie to another specie poses a potential hazard to human health and the food chain (Richardson et al., 2018). ST11 is reported in China in gram-negative bacteria, e.g., K. pneumonia and S. enteritidis strains (Li et al., 2021; Zhang et al., 2021). The identification of ST11 among S. haemolyticus strains in our study may indicate the possible transfer of genes from one bacterial specie to another specie in different ecological niches through horizontal gene transfer mechanisms such as transduction, conjugation, and transformation (Bolotin and Hershberg, 2017; Richardson et al., 2018). There is a need to conduct in-depth studies to understand more about virulence determinants, plasmid typing, serotyping, and gene transfer mechanisms within one health framework to understand the genetic basis of antimicrobial resistance spread and pathogenicity mechanisms using advanced characterization techniques such as whole genome sequencing (WGS).
5 Conclusions
The study noted a relatively high prevalence of S. haemolyticus and other Staphylococcus species at sampled dairy farms in Northwest, China, highlighting the importance of emerging antimicrobial-resistant pathogens affecting dairy herds. The antimicrobial susceptibility showed higher resistance to erythromycin, trimethoprim-sulfamethoxazole, gentamicin, cefoxitin, ciprofloxacin, and florfenicol, suggesting that the use of these antimicrobials at dairy farms needs to be closely monitored. However, susceptibility to tetracycline, vancomycin, and linezolid could be the antimicrobials used to treat mastitis caused by this pathogen. Most of the strains carrying the multiple resistance genes pose a potential threat to public health via the consumption of contaminated milk. The phylogenetic analysis classified strains from groups A-H, while MLST detected eight STs, with ST8 being the most predominant. These findings provided new insights into our understanding of the epidemiology and genetic characteristics of S. haemolyticus in dairy farms to inform interventions limiting the spread of AMR in dairy production.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the animal study because the study did not involve any invasive procedure on animals. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MS: Writing-original draft; JX and XM: Investigation and Methodology; ZW and XH: Data curation and Formal analysis; ZH, RS, and HZ: Writing-review and editing; WP: Conceptualization, Investigation, Funding acquisition, and Supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (25-LZIHPS-03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1183390/full#supplementary-material
References
Adkins, P., Dufour, S., Spain, J., Calcutt, M., Reilly, T., Stewart, G., et al. (2018). Molecular characterization of non-aureus staphylococcus spp. from heifer intramammary infections and body sites. J. Dairy Sci. 101 (6), 5388–5403. doi: 10.3168/jds.2017-13910
Ajose, D. J., Oluwarinde, B. O., Abolarinwa, T. O., Fri, J., Montso, K. P., Fayemi, O. E., et al. (2022). Combating bovine mastitis in the dairy sector in an era of antimicrobial resistance: ethno-veterinary medicinal option as a viable alternative approach. Front. Veterinary Sci. 9, 287. doi: 10.3389/fvets.2022.800322
Al-Harbi, H., Ranjbar, S., Moore, R. J., Alawneh, J. I. (2021). Bacteria isolated from milk of dairy cows with and without clinical mastitis in different regions of Australia and their AMR profiles. Front. Veterinary Sci. 8, 743725. doi: 10.3389/fvets.2021.743725
Aqib, A. I., Saqib, M., Khan, S. R., Ahmad, T., Shah, S. A. R., Naseer, M. A., et al. (2021). Non-steroidal anti-inflammatory drugs, plant extracts, and characterized microparticles to modulate antimicrobial resistance of epidemic mecA positive S. aureus of dairy origin. Applied Nanoscience 11, 553–563.
Argudín, M. A., Deplano, A., Meghraoui, A., Dodémont, M., Heinrichs, A., Denis, O., et al. (2017). Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics 6 (2), 12. doi: 10.3390/antibiotics6020012
Attallah, N. G., Negm, W. A., Elekhnawy, E., Elmongy, E. I., Altwaijry, N., El-Haroun, H., et al. (2021). Elucidation of phytochemical content of cupressus macrocarpa leaves: in vitro and in vivo antibacterial effect against methicillin-resistant staphylococcus aureus clinical isolates. Antibiotics 10 (8), 890. doi: 10.3390/antibiotics10080890
Batool, N., Shamim, A., Chaurasia, A. K., Kim, K. K. (2021). Genome-wide analysis of staphylococcus aureus sequence type 72 isolates provides insights into resistance against antimicrobial agents and virulence potential. Front. Microbiol. 11, 613800. doi: 10.3389/fmicb.2020.613800
Benites, N. R., Hora, A. S., Mello, P., Laes, M., Brandão, P. E., Melville, P. A., et al. (2021). Genotypic evaluation of antimicrobial resistance in staphylococcus spp. isolated from bovine clinical mastitis. Arquivo Brasileiro Medicina Veterinária e Zootecnia 73, 302–310. doi: 10.1590/1678-4162-11984
Bennani, H., Mateus, A., Mays, N., Eastmure, E., Stärk, K. D., Häsler, B. (2020). Overview of evidence of antimicrobial use and antimicrobial resistance in the food chain. Antibiotics 9 (2), 49. doi: 10.3390/antibiotics9020049
Bolotin, E., Hershberg, R. (2017). Horizontally acquired genes are often shared between closely related bacterial species. Front. Microbiol. 8, 1536. doi: 10.3389/fmicb.2017.01536
Bonsaglia, E., Silva, N., Rossi, B., Camargo, C., Dantas, S., Langoni, H., et al. (2018). Molecular epidemiology of methicillin-susceptible staphylococcus aureus (MSSA) isolated from milk of cows with subclinical mastitis. Microbial Pathogenesis 124, 130–135. doi: 10.1016/j.micpath.2018.08.031
Botrel, M.-A., Haenni, M., Morignat, E., Sulpice, P., Madec, J.-Y., Calavas, D. (2010). Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in rhône-alpes, France. Foodborne Pathog. Dis. 7 (5), 479–487. doi: 10.1089/fpd.2009.0425
Chajęcka-Wierzchowska, W., Gajewska, J., Zadernowska, A., Randazzo, C. L., Caggia, C. (2023). A comprehensive study on antibiotic resistance among coagulase-negative staphylococci (CoNS) strains isolated from ready-to-Eat food served in bars and restaurants. Foods 12 (3), 514. doi: 10.3390/foods12030514
Chang, S.-C., Lin, L.-C., Lu, J.-J. (2022). An lnu (A)-carrying multi-resistance plasmid derived from sequence type 3 methicillin-resistant staphylococcus lugdunensis may contribute to antimicrobial resistance in staphylococci. Antimicrobial Agents Chemotherapy 66 (8), e00197–e00122. doi: 10.1128/aac.00197-22
Cheng, W. N., Han, S. G. (2020). Bovine mastitis: risk factors, therapeutic strategies, and alternative treatments–a review. Asian-Australasian J. Anim. Sci. 33 (11), 1699. doi: 10.5713/ajas.20.0156
Cortimiglia, C., Luini, M., Bianchini, V., Marzagalli, L., Vezzoli, F., Avisani, D., et al. (2016). Prevalence of staphylococcus aureus and of methicillin-resistant s. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy region (Northern Italy). Epidemiol. Infect. 144 (14), 3046–3051. doi: 10.1017/S0950268816001576
Dallago, G. M., Wade, K. M., Cue, R. I., McClure, J. T., Lacroix, R., Pellerin, D., et al. (2021). Keeping dairy cows for longer: a critical literature review on dairy cow longevity in high milk-producing countries. Animals 11 (3), 808. doi: 10.3390/IECA2020-08827
Dan, M., Yehui, W., Qingling, M., Jun, Q., Xingxing, Z., Shuai, M., et al. (2019). Antimicrobial resistance, virulence gene profile and molecular typing of staphylococcus aureus isolates from dairy cows in xinjiang province, northwest China. J. Global Antimicrobial Resistance 16, 98–104. doi: 10.1016/j.jgar.2018.08.024
De Buck, J., Ha, V., Naushad, S., Nobrega, D. B., Luby, C., Middleton, J. R., et al. (2021). Non-aureus staphylococci and bovine udder health: current understanding and knowledge gaps. Front. Veterinary Sci. 8, 658031. doi: 10.3389/fvets.2021.658031
Dong, Q., Liu, Y., Li, W., Chen, M., Li, W., Wang, X., et al. (2020). Phenotypic and molecular characteristics of community-associated staphylococcus aureus infection in neonates. Infect. Drug Resistance 13, 4589–4600. doi: 10.2147/IDR.S284781
Du, X., Zhu, Y., Song, Y., Li, T., Luo, T., Sun, G., et al. (2013). Molecular analysis of staphylococcus epidermidis strains isolated from community and hospital environments in China. PloS One 8 (5), e62742. doi: 10.1371/journal.pone.0062742
El-Ashker, M., Gwida, M., Monecke, S., Ehricht, R., Elsayed, M., El-Gohary, F., et al. (2020). Microarray-based detection of resistance genes in coagulase-negative staphylococci isolated from cattle and buffalo with mastitis in Egypt. Trop. Anim. Health Production 52, 3855–3862. doi: 10.1007/s11250-020-02424-1
EUCAST (2019). Breakpoint tables for interpretation of MICs and zone diameters. Available at: https://www.eucast.org/ast_of_bacteria (Accessed 10 Jan, 2023).
Fair, R. J., Tor, Y. (2014). Antibiotics and bacterial resistance in the 21st century. Perspect. Medicinal Chem. 6, S14459. doi: 10.4137/PMC.S14459
Feßler, A. T., Wang, Y., Wu, C., Schwarz, S. (2018a). Mobile lincosamide resistance genes in staphylococci. Plasmid 99, 22–31. doi: 10.1016/j.plasmid.2018.06.002
Feßler, A. T., Wang, Y., Wu, C., Schwarz, S. (2018b). Mobile macrolide resistance genes in staphylococci. Plasmid 99, 2–10. doi: 10.1016/j.plasmid.2018.05.001
Gao, J., Barkema, H. W., Zhang, L., Liu, G., Deng, Z., Cai, L., et al. (2017). Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J. Dairy Sci. 100 (6), 4797–4806. doi: 10.3168/jds.2016-12334
Gizaw, F., Kekeba, T., Teshome, F., Kebede, M., Abreham, T., Hayishe, H., et al. (2020). Distribution and antimicrobial resistance profile of coagulase-negative staphylococci from cattle, equipment, and personnel on dairy farm and abattoir settings. Heliyon 6 (3), e03606. doi: 10.1016/j.heliyon.2020.e03606
Goetz, C., Tremblay, Y. D., Lamarche, D., Blondeau, A., Gaudreau, A. M., Labrie, J., et al. (2017). Coagulase-negative staphylococci species affect biofilm formation of other coagulase-negative and coagulase-positive staphylococci. J. Dairy Sci. 100 (8), 6454–6464. doi: 10.3168/jds.2017-12629
Gu, J., Shen, S., Xiong, M., Zhao, J., Tian, H., Xiao, X., et al. (2023). ST7 becomes one of the most common staphylococcus aureus clones after the COVID-19 epidemic in the city of wuhan, China. Infect. Drug Resistance 16, 843–852. doi: 10.2147/IDR.S401069
Gussmann, M., Steeneveld, W., Kirkeby, C., Hogeveen, H., Farre, M., Halasa, T. (2019). Economic and epidemiological impact of different intervention strategies for subclinical and clinical mastitis. Preventive Veterinary Med. 166, 78–85. doi: 10.1016/j.prevetmed.2019.03.001
Han, D., Gao, P., Li, R., Tan, P., Xie, J., Zhang, R., et al. (2020). Multicenter assessment of microbial community profiling using 16S rRNA gene sequencing and shotgun metagenomic sequencing. J. Adv. Res. 26, 111–121. doi: 10.1016/j.jare.2020.07.010
He, W., Ma, S., Lei, L., He, J., Li, X., Tao, J., et al. (2020). Prevalence, etiology, and economic impact of clinical mastitis on large dairy farms in China. Veterinary Microbiol. 242, 108570. doi: 10.1016/j.vetmic.2019.108570
He, Y., Reed, S. (2020). Pulsed-field gel electrophoresis typing of staphylococcus aureus strains. Methicillin-Resistant Staphylococcus Aureus (MRSA) Protocols: Cutting-Edge Technol. Advancements 2069, 79–88. doi: 10.1007/978-1-4939-9849-4_5
Hogan, J., Smith, K. “Using bulk tank milk cultures in a dairy practice,” in: workshop mastitis microbiology diagnostics, national mastitis council(Arlington).
Hogeveen, H., Pyorala, S., Waller, K. P., Hogan, J. S., Lam, T. J., Oliver, S. P., et al. (2011). “Current status and future challenges in mastitis research,” in Proceedings of the 50th annual meeting of the national mastitis council (Arlington, USA: Wageningen University & Research, Library), 36–48.
Hu, Y., Gao, G. F., Zhu, B. (2017). The antibiotic resistome: gene flow in environments, animals and human beings. Front. Med. 11, 161–168. doi: 10.1007/s11684-017-0531-x
Jenkins, S. N., Okello, E., Rossitto, P. V., Lehenbauer, T. W., Champagne, J., Penedo, M. C., et al. (2019). Molecular epidemiology of coagulase-negative staphylococcus species isolated at different lactation stages from dairy cattle in the united states. PeerJ 7, e6749. doi: 10.7717/peerj.6749
Jiang, H., Chen, M., Li, T., Liu, H., Gong, Y., Li, M. (2016). Molecular characterization of streptococcus agalactiae causing community-and hospital-acquired infections in shanghai, China. Front. Microbiol. 7, 1308. doi: 10.3389/fmicb.2016.01308
Johnson, J. S., Spakowicz, D. J., Hong, B.-Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10 (1), 5029. doi: 10.1038/s41467-019-13036-1
Jose, J., Sankar, S., Mani, B. K., Unnikrishnan, M., Bosewell, A., Mini, M. (2020). Phenotypic and genotypic characterization of mecA and blaZ gene mediated antibiotic resistance in bovine mastitis due to staphylococcus spp. Indian J. Anim. Res. 54 (1), 125–127. doi: 10.18805/ijar.B-3726
Kawalek, J., Howard, K., Jones, Y., Scott, M., Myers, M. (2016). Depletion of florfenicol in lactating dairy cows after intramammary and subcutaneous administration. J. Veterinary Pharmacol. Ther. 39 (6), 602–611. doi: 10.1111/jvp.12315
Khan, S. B., Khan, M. A., Ahmad, I., Khan, F. A., Khan, H., Khan, S. A. (2022). Prevalence of antibiotic resistant genes in staphylococcus aureus isolated from bovine mastitis. Pakistan J. Zoology 54 (5), 2239. doi: 10.17582/journal.pjz/20200403170447
Kim, S.-J., Moon, D. C., Park, S.-C., Kang, H. Y., Na, S. H., Lim, S.-K. (2019). Antimicrobial resistance and genetic characterization of coagulase-negative staphylococci from bovine mastitis milk samples in Korea. J. Dairy Sci. 102 (12), 11439–11448. doi: 10.3168/jds.2019-17028
Klimiene, I., Virgailis, M., Pavilonis, A., Siugzdiniene, R., Mockeliunas, R., Ruzauskas, M. (2016). Phenotypical and genotypical antimicrobial resistance of coagulase-negative staphylococci isolated from cow mastitis. Polish J. Veterinary Sci. 19 (3), 639–646. doi: 10.1515/pjvs-2016-0080
Kou, X., Cai, H., Huang, S., Ni, Y., Luo, B., Qian, H., et al. (2021). Prevalence and characteristics of staphylococcus aureus isolated from retail raw milk in northern xinjiang, China. Front. Microbiol. 12, 705947. doi: 10.3389/fmicb.2021.705947
Kowalewicz, C., Timmermans, M., Fretin, D., Wattiau, P., Boland, C. (2023). An in-house 45-plex array for the detection of antimicrobial resistance genes in gram-positive bacteria. MicrobiologyOpen 12 (1), e1341. doi: 10.1002/mbo3.1341
Lapointe, G., Skepper, C. K., Holder, L. M., Armstrong, D., Bellamacina, C., Blais, J., et al. (2021). Discovery and optimization of DNA gyrase and topoisomerase IV inhibitors with potent activity against fluoroquinolone-resistant gram-positive bacteria. J. Medicinal Chem. 64 (9), 6329–6357. doi: 10.1021/acs.jmedchem.1c00375
Li, C., Zhang, Z., Xu, X., He, S., Zhao, X., Cui, Y., et al. (2021). Molecular characterization of cephalosporin-resistant salmonella enteritidis ST11 isolates carrying bla CTX-m from children with diarrhea. Foodborne Pathog. Dis. 18 (10), 702–711. doi: 10.1089/fpd.2020.2878
Li, L., Zhao, X. (2018). Characterization of the resistance class 1 integrons in staphylococcus aureus isolates from milk of lactating dairy cattle in northwestern China. BMC Veterinary Res. 14 (1), 1–7. doi: 10.1186/s12917-018-1376-5
Li, L., Zhou, L., Wang, L., Xue, H., Zhao, X. (2015). Characterization of methicillin-resistant and-susceptible staphylococcal isolates from bovine milk in northwestern China. PloS One 10 (3), e0116699. doi: 10.1371/journal.pone.0116699
Li, X., Zhou, Y., Zhan, X., Huang, W., Wang, X. (2018). Breast milk is a potential reservoir for livestock-associated staphylococcus aureus and community-associated staphylococcus aureus in shanghai, China. Front. Microbiol. 8, 2639. doi: 10.3389/fmicb.2017.02639
Li, P., Zhu, T., Zhou, D., Lu, W., Liu, H., Sun, Z., et al. (2020). Analysis of resistance to florfenicol and the related mechanism of dissemination in different animal-derived bacteria. Front. Cell. Infect. Microbiol. 10, 369. doi: 10.3389/fcimb.2020.00369
Libera, K., Konieczny, K., Witkowska, K., Żurek, K., Szumacher-Strabel, M., Cieslak, A., et al. (2021). The association between selected dietary minerals and mastitis in dairy cows–a review. Animals 11 (8), 2330. doi: 10.3390/ani11082330
Lin, L.-C., Liu, T.-P., Chang, S.-C., Lu, J.-J. (2022). Characterization of new staphylococcus haemolyticus ST42 populations in northern Taiwan. Microbial Drug Resistance 28 (1), 56–62. doi: 10.1089/mdr.2019.0459
Liu, H., Dong, L., Zhao, Y., Meng, L., Wang, J., Wang, C., et al. (2022). Antimicrobial susceptibility, and molecular characterization of staphylococcus aureus isolated from different raw milk samples in China. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.840670
Liu, Y., Du, F.-l., Liu, P.-p., Mei, Y.-f., Wan, L.-g., Wei, D.-d., et al. (2018). Molecular epidemiology and virulence features of staphylococcus aureus bloodstream isolates in a regional burn center in china 2012–2016. Microbial Drug Resistance 24 (9), 1354–1360. doi: 10.1089/mdr.2017.0209
Lu, Y., Hu, Y.-L., Kong, X.-F., Wang, D.-Y. (2008). Selection of component drug in activating blood flow and removing blood stasis of Chinese herbal medicinal formula for dairy cow mastitis by hemorheological method. J. Ethnopharmacology 116 (2), 313–317. doi: 10.1016/j.jep.2007.11.031
Mello, P. L., Riboli, D. F. M., Martins, L., Brito, M. A. V. P., Victória, C., Calixto Romero, L., et al. (2020). Staphylococcus spp. isolated from bovine subclinical mastitis in different regions of Brazil: molecular typing and biofilm gene expression analysis by RT-qPCR. Antibiotics 9 (12), 888. doi: 10.3390/antibiotics9120888
Mendes, R. E., Deshpande, L. M., Costello, A. J., Farrell, D. J. (2012). Molecular epidemiology of staphylococcus epidermidis clinical isolates from US hospitals. Antimicrobial Agents chemotherapy 56 (9), 4656–4661. doi: 10.1128/AAC.00279-12
National Mastitis Council (2017). Laboratory handbook on bovine mastitis. Available at: https://www.nmconline.org/publications/ (Accessed 10 Jan, 2023).
Naushad, S., Barkema, H. W., Luby, C., Condas, L. A., Nobrega, D. B., Carson, D. A., et al. (2016). Comprehensive phylogenetic analysis of bovine non-aureus staphylococci species based on whole-genome sequencing. Front. Microbiol. 7, 1990. doi: 10.3389/fmicb.2016.01990
Neoh, H.-m., Tan, X.-E., Sapri, H. F., Tan, T. L. (2019). Pulsed-field gel electrophoresis (PFGE): a review of the “gold standard” for bacteria typing and current alternatives. Infection Genet. Evol. 74, 103935. doi: 10.1016/j.meegid.2019.103935
Nurjadi, D., Zizmann, E., Chanthalangsy, Q., Heeg, K., Boutin, S. (2021). Integrative analysis of whole genome sequencing and phenotypic resistance toward prediction of trimethoprim-sulfamethoxazole resistance in staphylococcus aureus. Front. Microbiol. 11, 607842. doi: 10.3389/fmicb.2020.607842
Osman, K., Alvarez-Ordóñez, A., Ruiz, L., Badr, J., ElHofy, F., Al-Maary, K. S., et al. (2017). Antimicrobial resistance and virulence characterization of staphylococcus aureus and coagulase-negative staphylococci from imported beef meat. Ann. Clin. Microbiol. Antimicrobials 16 (1), 1–10. doi: 10.1186/s12941-017-0210-4
Ou, C., Shang, D., Yang, J., Chen, B., Chang, J., Jin, F., et al. (2020). Prevalence of multidrug-resistant staphylococcus aureus isolates with strong biofilm formation ability among animal-based food in shanghai. Food Control 112, 107106. doi: 10.1016/j.foodcont.2020.107106
Pacha, P., Munoz, M., González-Rocha, G., San Martín, I., Quezada-Aguiluz, M., Aguayo-Reyes, A., et al. (2021). Molecular diversity of staphylococcus aureus and the role of milking equipment adherences or biofilm as a source for bulk tank milk contamination. J. Dairy Sci. 104 (3), 3522–3531. doi: 10.3168/jds.2020-19121
Pal, M., Regasa, A., Gizaw, F. (2019). Etiology, pathogenesis, risk factors, diagnosis and management of bovine mastitis: a comprehensive review. Int. J. 6, 40–55.
Pavel, A. B., Vasile, C. I. (2012). PyElph-a software tool for gel images analysis and phylogenetics. BMC Bioinf. 13 (1), 1–6. doi: 10.1186/1471-2105-13-9
Pérez, V. K. C., da Costa, G. M., Guimaraes, A. S., Heinemann, M. B., Lage, A. P., Dorneles, E. M. S. (2020). Relationship between virulence factors and antimicrobial resistance in staphylococcus aureus from bovine mastitis. J. Global Antimicrobial Resistance 22, 792–802. doi: 10.1016/j.jgar.2020.06.010
Persson, Y., Nyman, A.-K. J., Grönlund-Andersson, U. (2011). Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Veterinaria Scandinavica 53 (1), 1–8. doi: 10.1186/1751-0147-53-36
Petinaki, E., Papagiannitsis, C. (2019). Resistance of staphylococci to macrolides-Lincosamides-Streptogramins b (MLS. Staphylococcus aureus 117, 117–133. doi: 10.5772/intechopen.75192
Pham, T. D., Ziora, Z. M., Blaskovich, M. A. (2019). Quinolone antibiotics. Medchemcomm 10 (10), 1719–1739. doi: 10.1039/C9MD00120D
Phophi, L., Petzer, I.-M., Qekwana, D. N. (2019). Antimicrobial resistance patterns and biofilm formation of coagulase-negative staphylococcus species isolated from subclinical mastitis cow milk samples submitted to the onderstepoort milk laboratory. BMC Veterinary Res. 15 (1), 1–9. doi: 10.1186/s12917-019-2175-3
Pizauro, L. J. L., de Almeida, C. C., Silva, S. R., MacInnes, J. I., Kropinski, A. M., Zafalon, L. F., et al. (2021). Genomic comparisons and phylogenetic analysis of mastitis-related staphylococci with a focus on adhesion, biofilm, and related regulatory genes. Sci. Rep. 11 (1), 17392. doi: 10.1038/s41598-021-96842-2
Pizauro, L. J., de Almeida, C. C., Soltes, G. A., Slavic, D., de Ávila, F. A., Zafalon, L. F., et al. (2019). Detection of antibiotic resistance, mecA, and virulence genes in coagulase-negative staphylococcus spp. from buffalo milk and the milking environment. J. Dairy Sci. 102 (12), 11459–11464. doi: 10.3168/jds.2018-15920
Qian, M., Tang, S., Wu, C., Wang, Y., He, T., Chen, T., et al. (2015). Synergy between baicalein and penicillins against penicillinase-producing staphylococcus aureus. Int. J. Med. Microbiol. 305 (6), 501–504. doi: 10.1016/j.ijmm.2015.05.001
Qu, Y., Zhao, H., Nobrega, D. B., Cobo, E. R., Han, B., Zhao, Z., et al. (2019). Molecular epidemiology and distribution of antimicrobial resistance genes of staphylococcus species isolated from Chinese dairy cows with clinical mastitis. J. Dairy Sci. 102 (2), 1571–1583. doi: 10.3168/jds.2018-15136
Quezada-Aguiluz, M., Aguayo-Reyes, A., Carrasco, C., Mejías, D., Saavedra, P., Mella-Montecinos, S., et al. (2022). Phenotypic and genotypic characterization of macrolide, lincosamide and streptogramin b resistance among clinical methicillin-resistant s taphylococcus aureus isolates in Chile. Antibiotics 11 (8), 1000. doi: 10.3390/antibiotics11081000
Rainard, P., Foucras, G., Fitzgerald, J. R., Watts, J., Koop, G., Middleton, J. (2018). Knowledge gaps and research priorities in staphylococcus aureus mastitis control. Transboundary Emerging Dis. 65, 149–165. doi: 10.1111/tbed.12698
Richardson, E. J., Bacigalupe, R., Harrison, E. M., Weinert, L. A., Lycett, S., Vrieling, M., et al. (2018). Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2 (9), 1468–1478. doi: 10.1038/s41559-018-0617-0
Schoenfelder, S. M., Dong, Y., Feßler, A. T., Schwarz, S., Schoen, C., Köck, R., et al. (2017). Antibiotic resistance profiles of coagulase-negative staphylococci in livestock environments. Veterinary Microbiol. 200, 79–87. doi: 10.1016/j.vetmic.2016.04.019
Schwarz, S., Fessler, A. T., Loncaric, I., Wu, C., Kadlec, K., Wang, Y., et al. (2018). Antimicrobial resistance among staphylococci of animal origin. Microbiol. Spectr. 6 (4), 6–4. doi: 10.1128/9781555819804.ch7
Shi, C., Yu, Z., Ho, H., Wang, J., Wu, W., Xing, M., et al. (2021). Occurrence, antimicrobial resistance patterns, and genetic characterization of staphylococcus aureus isolated from raw milk in the dairy farms over two seasons in China. Microbial Drug Resistance 27 (1), 99–110. doi: 10.1089/mdr.2019.0358
Shore, A. C., Lazaris, A., Kinnevey, P. M., Brennan, O. M., Brennan, G. I., O'Connell, B., et al. (2016). First report of cfr-carrying plasmids in the pandemic sequence type 22 methicillin-resistant staphylococcus aureus staphylococcal cassette chromosome mec type IV clone. Antimicrobial Agents Chemotherapy 60 (5), 3007–3015. doi: 10.1128/AAC.02949-15
Sivagami, K., Vignesh, V. J., Srinivasan, R., Divyapriya, G., Nambi, I. M. (2020). Antibiotic usage, residues and resistance genes from food animals to human and environment: an Indian scenario. J. Environ. Chem. Eng. 8 (1), 102221. doi: 10.1016/j.jece.2018.02.029
Song, X., Huang, X., Xu, H., Zhang, C., Chen, S., Liu, F., et al. (2020). The prevalence of pathogens causing bovine mastitis and their associated risk factors in 15 large dairy farms in China: an observational study. Veterinary Microbiol. 247, 108757. doi: 10.1016/j.vetmic.2020.108757
Srednik, M. E., Tremblay, Y. D., Labrie, J., Archambault, M., Jacques, M., Fernández Cirelli, A., et al. (2017). Biofilm formation and antimicrobial resistance genes of coagulase-negative staphylococci isolated from cows with mastitis in Argentina. FEMS Microbiol. Lett. 364 (8), fnx001. doi: 10.1093/femsle/fnx001
Tang, K. L., Caffrey, N. P., Nóbrega, D. B., Cork, S. C., Ronksley, P. E., Barkema, H. W., et al. (2017). Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planetary Health 1 (8), e316–e327. doi: 10.1016/S2542-5196(17)30141-9
Tóth, A. G., Csabai, I., Krikó, E., Tőzsér, D., Maróti, G., Patai, Á.V., et al. (2020). Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 10 (1), 7464. doi: 10.1038/s41598-020-63675-4
Vanderhaeghen, W., Piepers, S., Leroy, F., Van Coillie, E., Haesebrouck, F., De Vliegher, S. (2014). Invited review: effect, persistence, and virulence of coagulase-negative staphylococcus species associated with ruminant udder health. J. Dairy Sci. 97 (9), 5275–5293. doi: 10.3168/jds.2013-7775
Varela, M. F., Stephen, J., Lekshmi, M., Ojha, M., Wenzel, N., Sanford, L. M., et al. (2021). Bacterial resistance to antimicrobial agents. Antibiotics 10 (5), 593. doi: 10.3390/antibiotics10050593
Wald, R., Hess, C., Urbantke, V., Wittek, T., Baumgartner, M. (2019). Characterization of staphylococcus species isolated from bovine quarter milk samples. Animals 9 (5), 200. doi: 10.3390/ani9050200
Walid, M., Salama, A., Elnahiriry, S., Abdeen, E., Hadad, G., Abo-Shama, U. (2021). Antibiogram and antibiotic resistance genes among coagulase-negative staphylococci recovered from bovine mastitis. Adv. Anim. Vet. Sci. 9 (8), 1267–1274. doi: 10.17582/journal.aavs/2021/9.8.1267.1274
Wu, C., Zhang, X., Liang, J., Li, Q., Lin, H., Lin, C., et al. (2021). Characterization of florfenicol resistance genes in the coagulase-negative staphylococcus (CoNS) isolates and genomic features of a multidrug-resistant staphylococcus lentus strain H29. Antimicrobial Resistance Infect. Control 10 (1), 1–10. doi: 10.1186/s13756-020-00869-5
Yang, W.-T., Ke, C.-Y., Wu, W.-T., Lee, R.-P., Tseng, Y.-H. (2019). Effective treatment of bovine mastitis with intramammary infusion of angelica dahurica and rheum officinale extracts. Evidence-Based Complementary Altern. Med. 2019, 1–11. doi: 10.1155/2019/7242705
Zhang, M., Li, J., Lu, Y., Wu, W., Wu, J., Xu, Y., et al. (2021). Expanding of ST11 carbapenemase-producing klebsiella pneumoniae subclones in a Chinese hospital, shenzhen, China. Infect. Drug Resistance 14, 1415–1422. doi: 10.2147/IDR.S299478
Zhou, W., Jin, Y., Liu, X., Chen, Y., Shen, P., Xiao, Y. (2022). Comparison of genetic features and evolution of global and chinese strains of community-associated methicillin-resistant staphylococcus aureus ST22. Microbiol. Spectr. 10 (1), e02037–e02021. doi: 10.1128/spectrum.02037-21
Keywords: dairy mastitis, Staphylococcus haemolytius, antimicrobial resistance, phylogeny, sequence typing, multi-drug resistance (MDR)
Citation: Shoaib M, Xu J, Meng X, Wu Z, Hou X, He Z, Shang R, Zhang H and Pu W (2023) Molecular epidemiology and characterization of antimicrobial-resistant Staphylococcus haemolyticus strains isolated from dairy cattle milk in Northwest, China. Front. Cell. Infect. Microbiol. 13:1183390. doi: 10.3389/fcimb.2023.1183390
Received: 10 March 2023; Accepted: 27 April 2023;
Published: 17 May 2023.
Edited by:
Rima El-Herte, Creighton University, United StatesReviewed by:
Agnes Agunos, Public Health Agency of Canada (PHAC), CanadaMaja Kosecka-Strojek, Jagiellonian University, Poland
Copyright © 2023 Shoaib, Xu, Meng, Wu, Hou, He, Shang, Zhang and Pu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanxia Pu, puwanxia@caas.cn
 Muhammad Shoaib
Muhammad Shoaib Jie Xu1
Jie Xu1  Wanxia Pu
Wanxia Pu