Constructing Micro-Landscapes: Management and Selection Practices on Microbial Communities in a Traditional Fermented Beverage
- 1Instituto de Investigaciones en Ecosistemas y Sustentabilidad, Universidad Nacional Autónoma de México, Campus Morelia, Morelia, Mexico
- 2Instituto de Ecología, A.C. – INECOL, Veracruz, Mexico
Colonche is a traditional beverage produced in Mexico by the fermentation of fruits of several cacti species. In the Meridional Central Plateau region of Mexico, where this study was conducted, it is mainly produced with fruits of Opuntia streptacantha; there, the producers perform spontaneous fermentation and/or fermentations through inoculums. Several factors can change the microbial community structure and dynamics through the fermentation process, but little attention has been directed to evaluate what type and extent of change the human practices have over the microbial communities. This study aims to assess the microbiota under spontaneous and inoculated fermentation techniques, the microorganisms present in the inoculums and containers, and the changes of microbiota during the process of producing colonche with different techniques. We used next-generation sequencing of the V3-V4 regions of the 16S rRNA gene and the ITS2, to characterize bacterial and fungal diversity associated with the different fermentation techniques. We identified 701 bacterial and 203 fungal amplicon sequence variants (ASVs) belonging to 173 bacterial and 187 fungal genera. The alpha and beta diversity analysis confirmed that both types of fermentation practices displayed differences in richness, diversity, and community structure. Richness of bacteria in spontaneous fermentation (0D = 136 ± 0.433) was higher than in the inoculated samples (0D = 128 ± 0.929), while fungal richness in the inoculated samples (0D = 32 ± 0.539) was higher than in spontaneous samples (0D = 19 ± 0.917). We identified bacterial groups like Lactobacillus, Leuconostoc, Pediococcus and the Saccharomyces yeast shared in ferments managed with different practices; these organisms are commonly related to the quality of the fermentation process. We identified that clay pots, where spontaneous fermentation is carried out, have an outstanding diversity of fungal and bacterial richness involved in fermentation, being valuable reservoirs of microorganisms for future fermentations. The inoculums displayed the lowest richness and diversity of bacterial and fungal communities suggesting unconscious selection on specific microbial consortia. The beta diversity analysis identified an overlap in microbial communities for both types of fermentation practices, which might reflect a shared composition of microorganisms occurring in the Opuntia streptacantha substrate. The variation in the spontaneous bacterial community is consistent with alpha diversity data, while fungal communities showed less differences among treatments, probably due to the high abundance and dominance of Saccharomyces. This information illustrates how traditional management guides selection and may drive changes in the microbial consortia to produce unique fermented beverages through specific fermentation practices. Although further studies are needed to analyze more specifically the advantages of each fermentation type over the quality of the product, our current analysis supports the role of traditional knowledge driving it and the relevance of plans for its conservation.
Introduction
Throughout history, human societies have developed knowledge and techniques to use a plethora of species of plants, animals, fungi and other organisms from different ecosystems to satisfy nutritional needs (Campbell-Platt, 1994; Tamang et al., 2016, 2020, 2021). Interactions between people and nature commonly include a high diversity of management practices through which people adequate organisms and other components of ecosystems to secure their livelihoods; these practices are context-dependent on local conditions, outstandingly human culture and environment (Jones et al., 1997; Odling-Smee et al., 2003; Anderson, 2005). Traditional or local knowledge commonly includes complex bodies of information on the ecological context, relationships and behavior of the elements used as food, extensive repertoires of preparation procedures, as well as about their relationships with customs, taboos, rituals, and other cultural aspects (Nabhan, 2010; Tamang, 2010; Ratcliffe et al., 2019). All these biocultural facets can be visualized through the diversity of food products and practices related to it, and provide basic notions about what is edible, where and when edible elements are available, the way these should be harvested, and how they can be improved by cooking, roasting, fermenting, or making it harmless (Lévi-Strauss, 2012; Ratcliffe et al., 2019; Tamang et al., 2020, 2021; Tsafrakidou et al., 2020; Fernández-Llamazares et al., 2021; Gadaga et al., 2021; Kennedy et al., 2021).
Fermentation practices are part of the local knowledge and food systems, directed to procure and improve human health and wellbeing, but fermented products have changed the human food supply worldwide (Harris et al., 1989; Kuhnlein and Receveur, 1996; Steinkraus, 1996; Harris, 1998; Quave and Pieroni, 2014; Svanberg, 2015; Sõukand et al., 2015; Flachs and Orkin, 2019; He et al., 2019). Fermentation can contribute to construct sustainable food systems, diversify food production, and procure safety, security and sovereignty in human communities around the world (Johns and Sthapit, 2004; Marshall and Mejia, 2011; Ojeda-Linares et al., 2021). It allows preserving and improving nutritional value of food and conferring desirable properties to the final products such as textures and sensorial properties, which are completely unlike to those of the starting materials (Smid and Hugenholtz, 2010; Smid and Kleerebezem, 2014). The microbial communities of bacteria, yeasts, and molds play a key role determining the quality of the fermented products, influencing their acidity, flavor, texture, nutritional value and other health benefits (Forssten et al., 2011; Todorov and Holzapfel, 2015; Gutiérrez-Uribe et al., 2017; El Sheikha and Hu, 2020). Changes in the microbial composition may be caused by variation in environmental factors, thus organoleptic and physicochemical characteristics of a fermented product could be modified and driven by managing fermentation environments through human practices (Escalante et al., 2016; Rebollar et al., 2017). The persons that perform and drive changes and follow the fermentation process in traditional contexts are recognized as traditional fermentation managers (Nabhan, 2010; Flachs and Orkin, 2019; Ojeda-Linares et al., 2020).
A common practice used to produce traditional ferments is the “spontaneous” fermentation, which involves microorganisms occurring in a local environment and is influenced by temperature, substrate type, techniques and tools employed, and other cultural factors that shape the composition and dynamics of the microbiota (De Vuyst and Vancanneyt, 2007; Vogelmann et al., 2009; Chaves-López et al., 2020). Due to the influence of all these factors, this type of fermentation is commonly assumed to be unpredictable, producing outcomes of inconsistent attributes; local people commonly have knowledge about how environmental and technological factors may influence the product and carry out practices to manage them, looking for decreasing the unpredictability of the resulting product. The other general practice is the use of starter cultures, which are characteristic for numerous fermented products around the world. Inoculation refreshment favors a periodically stable microbial community which provides an easy way to achieve optimal fermentations and desirable products (De Vuyst and Vancanneyt, 2007; De Vuyst et al., 2009; Brandt, 2014; Harth et al., 2016; Mukisa et al., 2017). Therefore, it is thought that this type of practice is associated with a set of selective forces favoring a specific community of microorganisms, guiding the dynamics and structure of the microbiota through the fermentation process (Vogelmann et al., 2009; Vogelmann and Hertel, 2011; Gibbons and Rinker, 2015; Liu et al., 2021).
The influence of human management over composition, structure and dynamics of microbial communities has been insufficiently analyzed, even when humans historically have developed numerous management techniques to direct, diversify and innovate the quality or to prevent the spoilage of fermented products around the world (Tamang and Fleet, 2009; Tamang et al., 2016). Ethnozymological studies have documented a wide range of activities that producers perform to adequate fermented products to their purposes; for instance, boiling the substrates, adding salt, plant, or animal products with antiseptic or flavoring roles are common practices (Quave and Pieroni, 2014; Hong et al., 2015; Sõukand et al., 2015; Pieroni et al., 2017; Álvarez-Ríos et al., 2020; Ojeda-Linares et al., 2021). Documenting the known-how of the management over the microbial communities in fermented products has high importance to understand the implications of human practices on the structure and dynamics of microbial communities during fermentation and to recover old human experience for innovation. In addition, this information provides elements to analyze possible processes of domestication at species and/or community levels.
Management includes a broad spectrum of practices that favor phenotypic attributes of organisms like size, flavors, colors, nutritional values, attributes to satisfy customs, rituals, ornamental, and other cultural purposes (Anderson, 2005; Casas et al., 2007). Through time, management has determined differences between wild and managed populations, the latter having higher frequencies of human-favored phenotypes. Such processes have been especially assessed in plant and animal populations where specific visible phenotypes are favored (Casas et al., 2007; Blancas et al., 2010). At ecosystem level, management practices may be directed to change species richness, diversity, and structure of communities and this is also an expression of domestication, the landscape domestication (Casas et al., 1997; Smith, 2007; Albuquerque et al., 2019; Clement et al., 2020, 2021; Franco-Moraes et al., 2021). But, although the processes involved in ecosystem management are common and widespread throughout the world, these have been less studied than domestication at species populations level and even fewer are studies documenting management and domestication at micro-landscape level.
In this study we explored the general hypothesis that the composition and structure of the microbiota involved in fermentation would differ according to management practices. We expected to find a higher microbial diversity in techniques involving spontaneous fermentation than in those using inoculums, which would be favoring reduced consortiums of microorganisms due to continuous human selection. Such differences would also influence the dynamics of change through the fermentation of communities with higher and lower diversity toward less and more predictable products, respectively. Characterizing and analyzing the type and extent of change of microbial communities according to management practices are conceptually relevant to construct and test hypotheses about how these processes and techniques lead to domestication of microorganism consortiums and, probably, of specific lineages. Also, to design innovative strategies to improve food quality. Understanding how certain management practices do help to predict how diversity, composition, and richness of microbiota can be and change, would support principles of selection and management programs of microbial communities to obtain desirable products.
Such hypothesis has been narrowly explored, particularly because testing it requires methods and study systems able to compare and measure across confidently. A traditional fermented product prepared with the same substrate and different management practices represents an optimal study system for such purpose, while using High Throughput Sequencing (HTS) techniques allow detecting or rejecting associations between microbial composition and management practices. Currently, HTS allow glimpsing the composition and structure of microbial communities in different ecosystems (Metzker, 2010; Caporaso et al., 2011) and fermenting landscapes are not the exception (Foligné and Pot, 2013; De Filippis et al., 2017; Sha et al., 2018; Astudillo-Melgar et al., 2019).
To address our hypothesis, we selected the traditional fermented beverage called colonche. Colonche is a group of traditional Mexican beverages prepared since pre-Columbian times by the fermentation of fruits of several cactus species, including several species of Opuntia prickly pears and columnar cacti (Ojeda-Linares et al., 2020). We conducted our study in the Meridional Central Plateau region of Mexico, where the main substrate for colonche production is Opuntia streptacantha fruit. For its production, prickly pear fruits are gathered in wild and cultivated populations and stored in plastic trays almost every day from August to October, when fruits are available. Local people carry out different fermentation practices, based on spontaneous or inoculated procedures. To produce colonche through spontaneous fermentation, they harvest and peel the Opuntia fruits in situ, and then place them inside a clay pot where fermentation occurs for about 12 h. In this area the colonche production is mainly performed by old men (60 ± 5 years old). The inoculated fermentation is performed by firstly boiling the fruits to concentrate their juice; then, the producers add the starter cultures from previous batches and store the fermenting juice in plastic containers for 4 h (Ojeda-Linares et al., 2020). The production of inoculated colonche in the area is mainly performed by young women (32 ± 4 years old), and the production represents a recent recovery of this beverage by the younglings. Therefore, an analysis of the microbiota associated with this fermented beverage prepared with the same substrate under different fermentation practices is possible and would allow documenting how management over the fermentation process influences the microbial communities.
Nowadays, there is a worrying erosion of local food systems, causing the disappearance of traditional food products worldwide (Pingali, 2007; Turner and Turner, 2007; Johns et al., 2013; Albert et al., 2015; Hernández-Santana and Narchi, 2018; Akinola et al., 2020). Fermented products have been poorly considered in the analysis of erosion of local food systems, even when some of them have almost disappeared. Nonetheless, a renewed interest in their production has motivated studies and actions to guarantee its permanence (Madej et al., 2014; Svanberg, 2015; Cano and Suárez, 2020; Ojeda-Linares et al., 2020, 2021). In México, it has been recorded the loss of some traditional fermented beverages, even though the main substrates are available in the nearby, but the transmission of knowledge has been lost (Ojeda-Linares et al., 2021). The vanishing of fermented products not only favors disappearing fragments of foodscapes, but also the traditional ecological knowledge over fermentation practices, and therefore the efforts that humans have historically engineered to shape microbial communities and the evolutionary processes involved in making unique products.
This study aims to describe the microbial community of colonche under different management practices and highlights the relevance of traditional knowledge to construct the micro-landscapes and, in general, to preserve unique products that are part of a local food systems. We aspire to contribute to a deeper understanding of the processes that can be involved in the selection and domestication of microbial consortia and specific lineages of the microbiota. We also aspire to contribute to the conservation of traditional knowledge over fermentation practices and the establishment of a microbial panel helpful to determine the geographical origin of the product and supports the relevance and the way of its conservation.
Materials and Methods
Study Area and Sampling
We studied colonche samples prepared by the fermentation of O. streptacantha fruits. We collected nine samples from spontaneous fermentations in the village of Laguna de Guadalupe (LG) in the state of Guanajuato, México from three different producers that performs similar practices. Also, nine samples of inoculated colonche produced by three producers in Mexquitic de Carmona (MC) in the neighboring state of San Luis Potosi (Figures 1A,E). We in addition collected samples of residuals from two clay pots one day before producers began spontaneous colonche fermentation. These clay pots have been used for almost 80 years for colonche production and are stored indoors when fruits of O. streptacantha are not available. Before the fermentation starts, the producers clean them only with water (no soap is added since according to people it changes the flavor of the product; Figure 1B). In addition, we collected and analyzed two samples of the inoculum, which were saved from the last production season, stored in a fridge and ready to use in the following batches (Figure 1C) and two samples of the cooked cactus prickly pear fruit juice before fermentation in the inoculated treatment (Figure 1D). The cooked samples represent a treatment that producers of the locality of Mexquitic performed because it is presumed to give a longer shelf life to colonche, since it can be stored in this way before the inoculum is added.
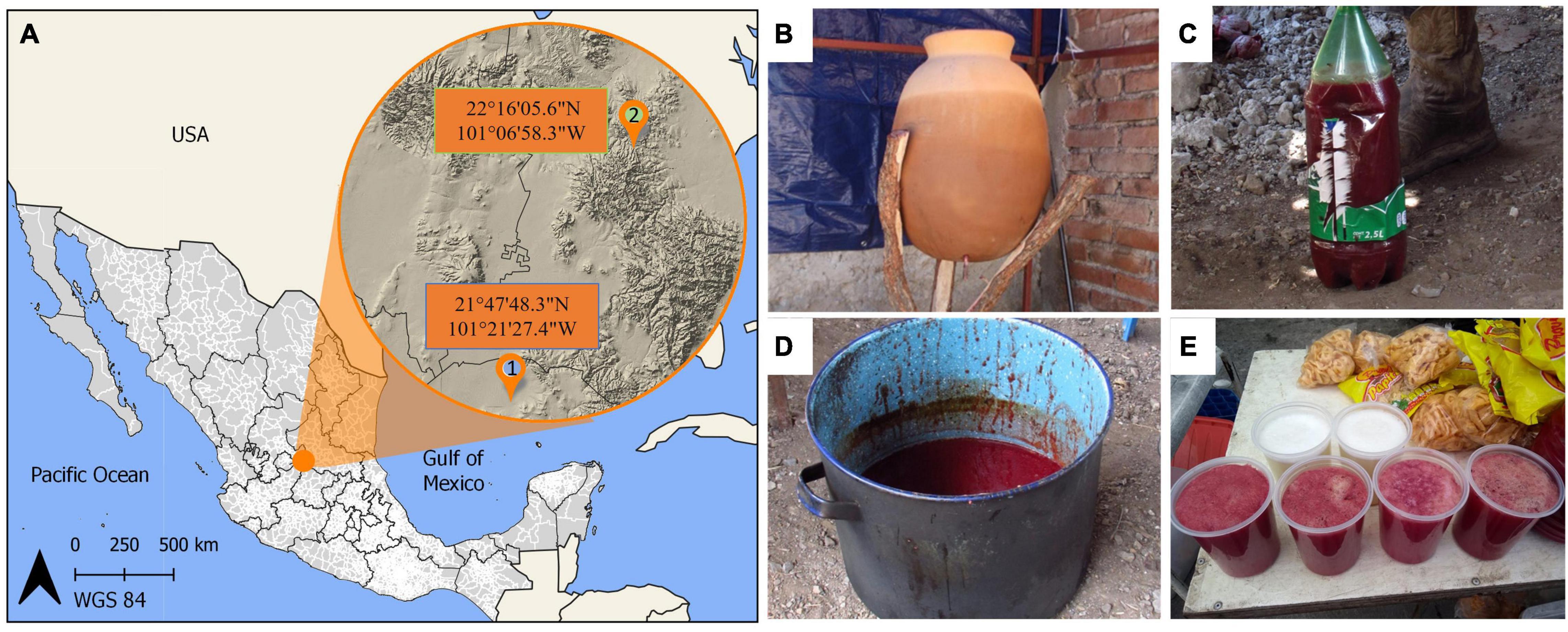
Figure 1. Sampling sites and colonche samples (A1) the locality of Laguna de Guadalupe in Guanajuato state; (A2), Mexquitic de Carmona in San Luis Potosí. (B) Claypots where spontaneous fermentation occurs; (C) Inoculum stored from the last season; (D) Cooked and boiled Cactus prickly pear fruits where sugars are concentrated; (E) Inoculated samples ready to sell in the local markets.
DNA Extraction and Sequencing
Genomic DNA was extracted for all the samples with the ZR Soil Microbe DNA MiniPrep kit (Zymo Research) following the manufacturer’s protocol. DNA was quantified using Qubit Fluorometric Quantitation (Thermo Fisher Scientific) and its quality was assessed on a 1% agarose gel. The obtained high-quality DNA samples were stored at −20°C util the library preparation. The libraries and targeted metagenomic sequencing were performed at ZymmoBIOMICS Service (Irvine, CA). Libraries of 16S rDNA and ITS amplicons were sequenced on the Illumina® MiSeq™ platform 2 × 300 bp paired end. For bacteria, the V3-V4 regions of the 16S rRNA were amplified as has been reported in previous works (Caporaso et al., 2011; Li and Yue, 2016). For fungi, the internal transcribed spacer (ITS) was amplified using the ITS2 primers (Yao et al., 2010; Badotti et al., 2017; Baldrian et al., 2021).
Microbiome Analysis in the Colonche Samples
The 16S rRNA and ITS amplicon sequences were processed using QIIME 2™ (Caporaso et al., 2010; Bolyen et al., 2019), version qiime2-2018.8. For all data, the DADA2 (Callahan et al., 2016) pipeline was used for quality filtering, removal of chimeric sequences, merge paired end reads and generate a table of amplicon sequence variants of each dataset (ASVs, Callahan et al., 2017). To eliminate the bases corresponding to the primers, the first 16 bases of the forward reads and 24 bases of the reverse reads of the 16S rRNA sequences were trimmed. From the ITS sequences, 22 and 24 bases of forward and reverse reads were clipped, respectively. Furthermore, to remove low-quality regions of the sequences, each read was clipped to some position where the quality decreases and was determined with the interactive quality plots of QIIME 2™ (Bolyen et al., 2019). The 16S rRNA sequences were truncated to 315 bases for forward reads and 250 bases for reverse reads, and the ITS sequences were truncated to 310 bases and 215 bases for forward and reverse reads, respectively.
Taxonomy was assigned to the representative unique sequences from each ASVs based on the Naïve Bayes classifier (Bokulich et al., 2018) and annotated according to the Greengenes 13_8 database (DeSantis et al., 2006); for the 16S rRNA data and from the ITS data the UNITE database v8.0 (Nilsson et al., 2019) was used. For each dataset, the ASVs represented by less than 10 sequences were removed in all samples, and the ASVs that were classified as plant chloroplast, mitochondria, archaea, unassigned sequences, or only assigned to phylum level were also eliminated. For diversity and statistical analysis, the ASVs tables of bacteria and fungi were normalized by their relative abundance (Fitzpatrick et al., 2018; Solís-García et al., 2021).
Diversity and Statistical Data Analysis
Alpha diversity was evaluated with the Hill numbers (qD), where the order q determines the sensitivity of the measure to the relative abundance (Chao and Chiu, 2016). 0D is equivalent to the richness; the measure 1D counts the ASVs proportionally to their abundances and corresponds to the effective numbers of common ASVs; the measure for 2D considers the most abundant ASVs and can be interpreted as the effective number of dominant ASVs (Chao et al., 2014; Montes-Carreto et al., 2021). The comparisons of the diversity measures among colonche management practices were made under the same sample coverage and the no overlap in the 95% CI values indicates significant differences (Cumming et al., 2007). Sample coverage values, qD, and their confidence intervals were calculated with the iNEXT R package 3.5.3 (Hsieh et al., 2016), using as an endpoint the maximum number of sequences in each treatment.
For beta diversity analyses, each phylogenetic tree was constructed with the ASVs of bacteria or fungi of the inoculated and spontaneous colonche management practices. The sequences alignment was made with MAFFT, the trees were inferred with FastTree and then rooted at its midpoint. The ASVs tables of bacteria and fungi log2-transformed and the dissimilarity matrices of the Unifrac weighted and unweighted distances were calculated with the phyloseq (McMurdie and Holmes, 2013) and vegan packages (Oksanen et al., 2013). The phylogenetic dissimilarities of the bacterial and fungal communities were represented with the ordination method of non-metric multidimensional scaling (NMDS) based on the Unifrac weighted and unweighted distances (Lozupone et al., 2011). To compare the structure of each community of the inoculated and spontaneous colonche management practices a permutational multivariate analysis of variance (PERMANOVA) was computed using the vegan package with 999 permutations.
Significant differences in the relative abundance of each taxon between the inoculated and spontaneous colonche management practices were evaluated through the Mann-Whitney-Wilcoxon test with an FDR correction in R v.3.5.2 software, considering as a significant values P < 0.05. The shared bacteria and fungi ASVs among both treatments were plotted as a Venn diagram with the VennDiagram package (Chen and Boutros, 2011).
Results
Preprocessing and Taxonomic Classification
To visualize the changes in the bacterial communities between fermentation management practices we obtained pair-end sequencing of the V3-V4 region of the 16SrRNA leads to a total of 2,398,545 reads, with an average of 28,769.79 (± 33,516.46) reads per sampling, then, after filtering the ASVs table, a total of 701 in all 16S samples, belonging to 173 bacterial genera (Supplementary Table 1). For fungal communities by ITS sequence, a total of 1,505,383 pair-end were obtained with an average of 62,724.291 (± 23,303.49) reads for all the sampling which composed a total of 203 ASVs; then, after filtering the table with 0.01% of relative abundance, 201 ASVs were obtained, belonging to 187 yeast genera (Supplementary Table 1). The bacterial and fungal communities of each colonche management practices had a sample coverage of 100% suggesting that the most of the ASVs were captured (Supplementary Figure 1A). Also, rarefaction curves for all the samples reached the plateau indicating that the sequencing depth was sufficient to cover most of the bacterial and fungal diversity in each colonche management practices (Supplementary Figure 1B).
Colonche Microbiome Is Partially Consistent With the Management Hypothesis
To analyze our hypothesis, the alpha diversity was estimated using the Hill numbers (qD). For the ASVs of the bacterial community (Supplementary Table 2), the clay pots samples showed a higher richness (0D = 543 ± 0.121), followed by the spontaneous samples (0D = 136 ± 0.433), the inoculated samples (0D = 128 ± 0.929), the cooked samples (0D = 113.00 ± 3.485), and being the inoculum samples (0D = 47 ± 0.163) the ones with the lowest richness (Figure 2A). Also, the clay pot samples were the most diverse (1D = 131.575 ± 1.412, 2D = 38.087 ± 1.212) compared with the other colonche management practices (Figures 2B,C and Supplementary Table 2). The diversity values of the effective number of common and dominant ASVs were significantly higher in the inoculated samples (1D = 18.277 ± 0.929, 2D = 11.507 ± 0.04) contrarily to the spontaneous samples (1D = 11.081 ± 0.049, 2D = 4.33 ± 0.019; Figures 2B,C). Thus, there is a reduction in richness from spontaneous to inoculated fermentation practices in the colonche microbiome, which is consistent with our hypothesis. The richness of the ASVs of the fungal community (Figure 2D and Supplementary Table 2) showed a different pattern, the clay pots also display the highest values (0D = 159 ± 0.151), followed by the inoculated samples (0D = 32 ± 0.539), then the cooked samples (0D = 31 ± 0.458), the spontaneous samples (0D = 19 ± 0.917), and finally the inoculum with the less richness (0D = 3 ± 0.151). Diversity of clay pots samples (1D = 28.873 ± 0.214, 2D = 14.787 ± 0.145) was higher than that of the other colonche management practices (Figures 2E,F). In contrast, diversity of common and dominant fungal ASVs was significantly higher in the spontaneous samples (1D = 2.667 ± 0.004, 2D = 2.442 ± 0.004) than in the inoculated samples (1D = 1.826 ± 0.007, 2D = 1.346 ± 0.004; Figures 2E,F).
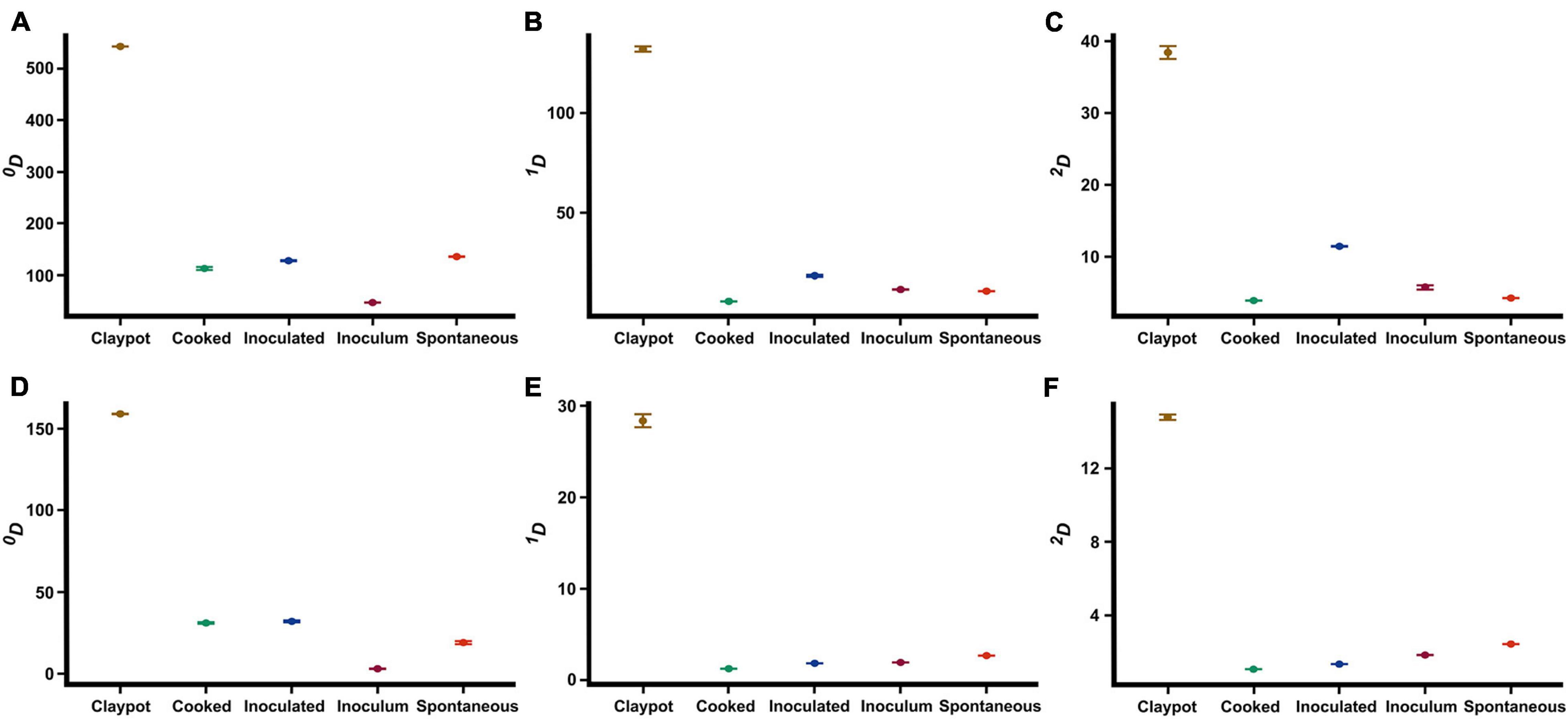
Figure 2. Comparison of the richness and alpha diversity of the bacterial (A–C) and fungal communities (D–F) between the different colonche management practices estimated with the Hill numbers (qD). Interval plots of richness (A,D), effective number of common amplicon sequence variants (ASVs) (B,E), and effective number of dominant ASVs.
Colonche Microbial Differences Between Spontaneous and Inoculated Fermentation Practices
To characterize the differences between spontaneous and inoculated colonche microbiome, the beta diversity of bacterial and fungal communities was represented with a NMDS using weighted and unweighted Unifrac distances (Figure 3 and Supplementary Figure 2). Although phylogenetic dissimilarities between the bacterial groups in spontaneous and inoculated fermentations by the unweighted Unifrac distance allow visualizing the splitting of both bacterial communities (p < 0.036), there is an inoculated sample that is closer to the spontaneous samples, and the spontaneous fermented samples are more disperse and less clustered (Figure 3A and Supplementary Table 3). The weighted Unifrac distances partially allow discriminating fungal community (p < 0.047), it can be seen that the spontaneous bacterial communities are clustered, while the inoculated samples are more variable and there are samples that overlap with the spontaneous treatment (Figure 3B and Supplementary Table 4). Both results are supported by PERMANOVA analysis (999 permutations). Summarizing, the overlap in microbial communities for both types of fermentation practices might reflect a common shared composition of microorganisms occurring in the Opuntia streptacantha substrate. The variation in the spontaneous bacterial community is consistent with the data of alpha diversity, while for fungal communities there are less differences among treatments, which might reflect the high abundance and dominance of Saccharomyces genus, as referred to for alpha diversity (Figure 2B).
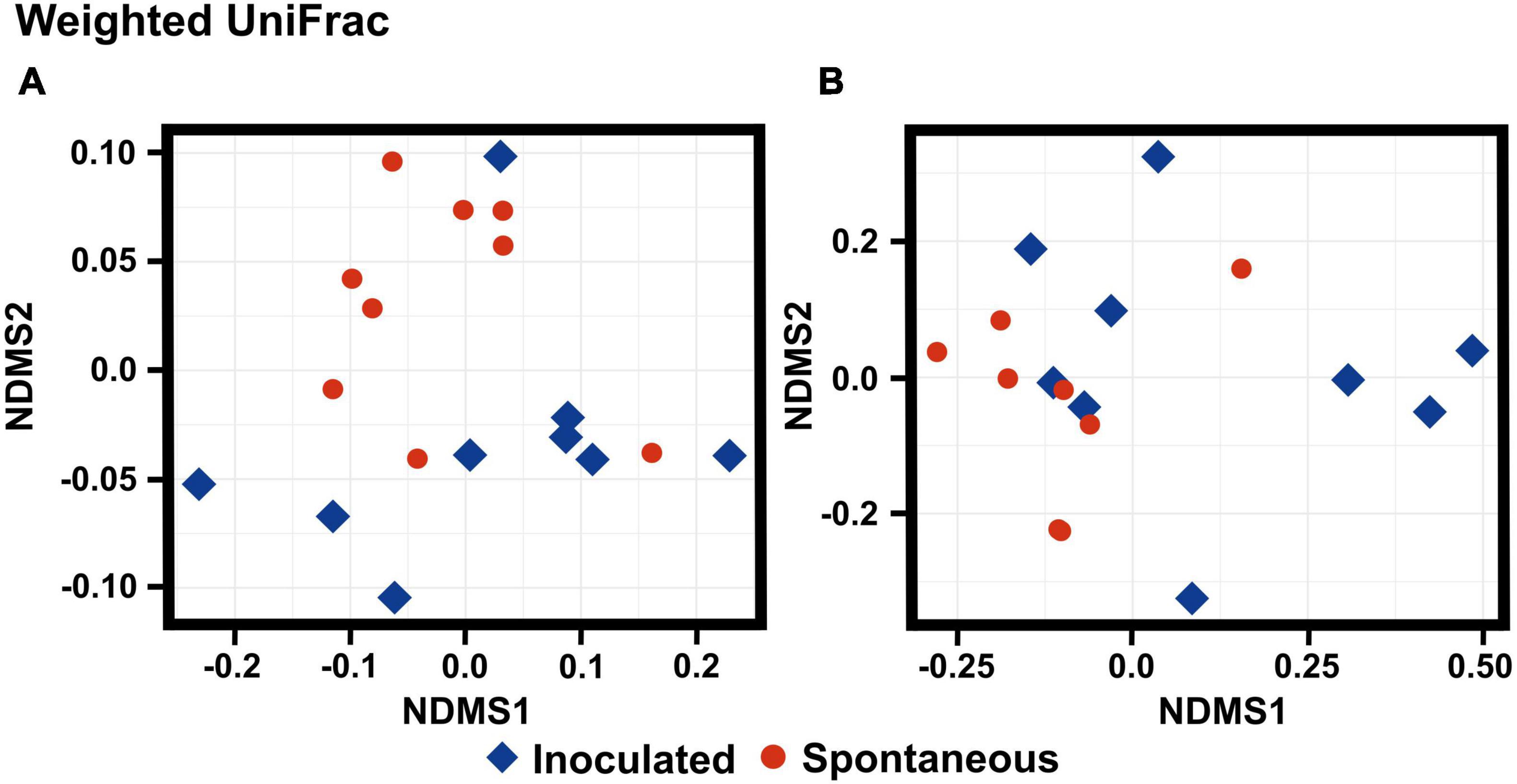
Figure 3. Non-metric multidimensional scaling (NMDS) plots-based (A) on UniFrac weighted distance of the bacterial community structure; (B) an Unifrac weighted distances of the fungal community structure associated with inoculated and spontaneous fermentation practices in the colonche production.
Colonche Microbial Genus Composition in the Different Fermentation Practices
The Microbial Community in Spontaneous Fermentation: Clay Pots and Spontaneous Colonche
Besides different diversity levels, we found that clay pots have a high richness in the composition of bacteria and fungi at the genus level (Figure 2). Among the most abundant bacteria genera occurring in the clay pots are: Arthrobacter (14.92%), Elstera (10.12%), Alkanindiges (7.72%), Massilia (6.92%), Pseudomonas (5.18%), Nocardioides (1.79%), Roseomonas (1.62%), Rhizobium (1.40%), Blastococcus (1.39%), Achromobacter (1.20%), Brevundimonas (1.15%). While for fungal community, Penicillium (38.15%), Cladosporium (9.70%), Didymella (9.60%), Wickerhamomyces (8.90%), Naganishia, (7.86%), Alternaria (6.42%), Rhodotorula (4.06%), Fusarium (3.02%), Saitozyma (1.62%), Aureobasidium (1.38%), Vishniacozyma (1.14%), Cystofilobasidium (1.01%) and although Saccharomyces (0.42%) ends up as the most dominant yeast in the spontaneous fermentation, its abundance is lower in the clay pots (Figure 4).
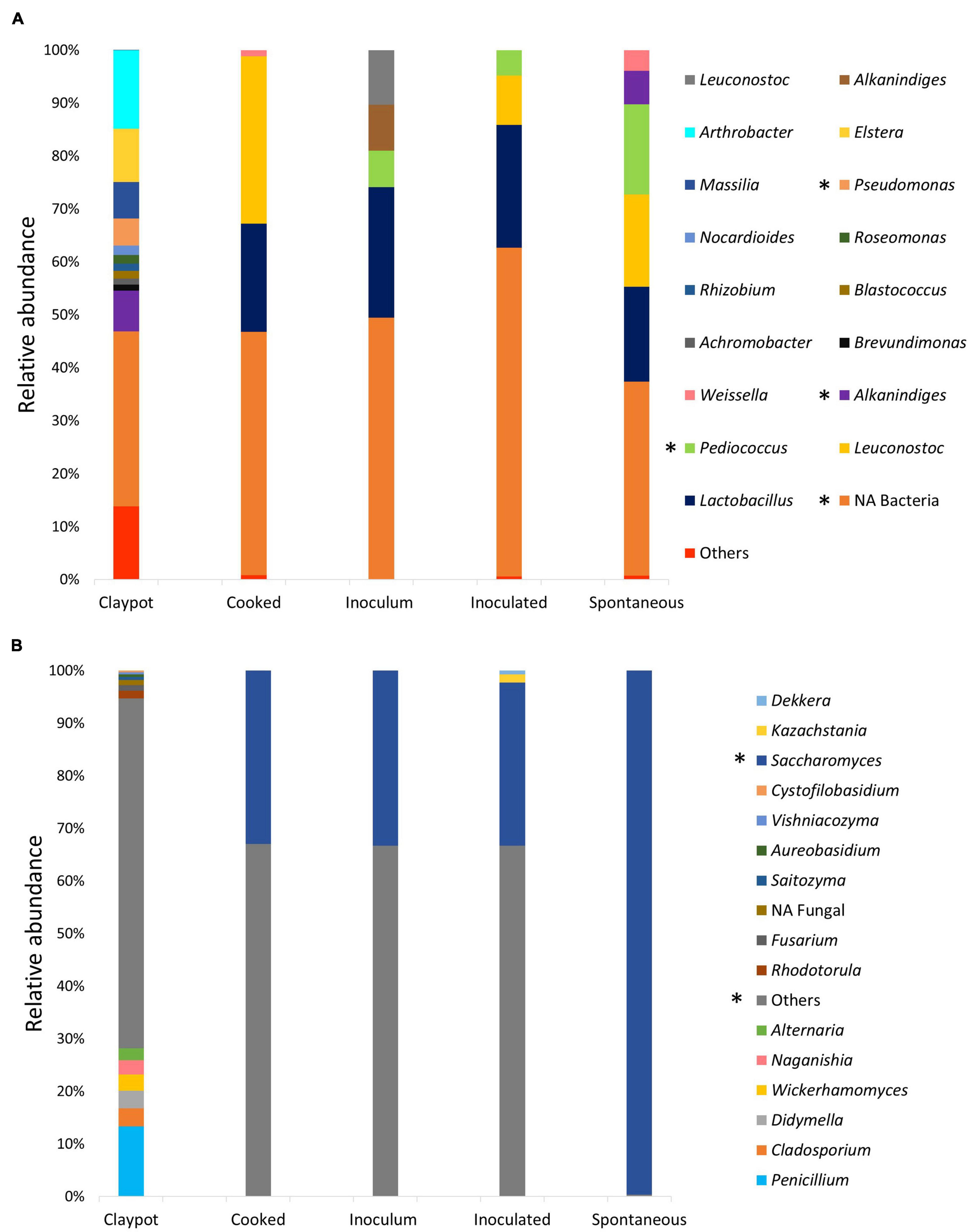
Figure 4. Composition of the (A) Bacterial Genera composition of the community associated with the management practices and colonche production stages and, (B) the fungal Genera composition. Low abundance taxonomic groups (relative abundance < 1%) were reported as others. NA mean not assigned. The asterisk indicates significant differences in taxa relative abundance between management practices.
At the end of the spontaneous fermentation process, 10 fungal ASVs (Aureobasidium, Candida, Cladosporium, Pichia, Saccharomyces, Torulaspora, Rhodotorula, Wickerhamomyces, one unidentified taxon and one not assigned fungal) are shared within the clay pots. This group is mainly involved in the final stages of the fermentation process of the spontaneous fermentation. Four groups are exclusive of the spontaneous fermented colonche (Dekkera, Kazachstania, Kluyveromyces and Knufia) and 45 are unique for clay pots. This behavior is similar in the bacterial community, in which 6 genera (Massilia, Elizabethkingia, Clostridium, Bacillus, Alkanindiges and one not assigned taxon) are shared with the spontaneous final product and the clay pots. For instance, 12 genera (Weissella, Tanticharoenia, Tatumella, Streptococcus, Rheinheimera, Pediococcus, Lactococcus, Lactobacillus, Gluconacetobacter, Faecalibacterium, Dorea and Aeromonas) are present in the final product of spontaneous fermentation, being particularly remarkable Lactobacillus and Leuconostoc, which are present in higher abundance (Figure 4). Finally, 71 ASVs are exclusive of the microbiota communities of the clay pots.
The Microbial Community in the Inoculated Fermentation: Inoculums, Cooked and Inoculated Colonche
It can be visualized that the inoculums are the samples with the lowest richness of bacteria and yeasts; for instance, we identified only 7 bacteria and 2 yeast genera, mainly dominated by multiple taxa of Lactic Acid Bacteria (LAB) as: Lactobacillus (24.60%), Leuconostoc (10.34%), Alkanindiges (8.65%), Pediococus (6.92%), while yeasts are dominated by Saccharomyces (99.9%) and Kazachstania (0.01%). Surprisingly, there were a higher number of ASVs for the cooked samples with 32 bacteria genera (Supplementary Table 5) and 21 yeasts, contrasting with the final product of the inoculated fermentation, which had fewer ASVs (Supplementary Table 6).
As mentioned above, in the inoculated fermentation process all microbial ASVs of the inoculums are shared with the inoculated final product. Also, Alkanindiges, Clostridium Corynebacterium, Gluconacetobacter, Lactobacillus, Leuconostoc, Massilia, Pediococcus, Sphingomonas, Staphylococcus, Weissella and one not assigned taxon are shared between the cooked and the inoculated samples. Dorea and Elstera are the only two exclusive genera of the inoculated samples and 18 genera (Actinomycetospora, Actinotelluria, Albidovulum, Arthrobacter, Blastococcus, Dietzia, Gaiella, Helcobacillus, Micrococcus, Microlunatus, Nocardioides, Oceaniovalibus, Ochrobactrum, Roseomonas, Solirubrobacter, Sporosarcina, Streptomyces and Tanticharoenia) are exclusive of the cooked samples.
For the fungal community we registered 9 genera being commonly shared between the cooked treatment and inoculated samples (Candida, Cladosporium, Dipodascus, Hanseniaspora, Kazachstania, Kluyveromyces, Saccharomyces, Torulaspora and one unidentified taxon). The genera Alternaria, Arthrinium, Cryptococcus, Didymella, Filobasidium, Fusarium, Mucor, Naganishia, Rhodotorula and Starmerella are exclusive of cooked samples and only Aureobasidium, Dekkera, Issatchenkia and Pichia are unique for the final product of inoculated fermentation.
The Microbial Composition Between Spontaneous and Inoculated Fermentations
For bacterial communities we found that 12 ASVs are shared among the fermentation practices, like Lactobacillus (p < 1e-1), Alkanindiges, Clostridium, Dorea, Faecalibacterium, Gluconacetobacter, Lactococcus, Leuconostoc (p < 1e-1), Pediococcus (p < 1e-1), Tatumella, Tanticharoenia, and Weisella (p < 1e-1). For instance, 6 genera are exclusive to spontaneous treatment like Aeromonas, Bacillus, Elizabethkingia, Massilia, Rheinheimera and Streptococcus and 4 are exclusive of the inoculated samples (Staphylococcus, Sphingomona, Corynebacterium, Elstera) (Figure 5A). In the fungal communities, we identified 11 genera shared among samples under different fermentation practices: Aureobasidium, Candida, Cladosporium, Dekkera (p < 1e-1), Hanseniaspora, Kazachstania (p < 1e-6), Kluyveromyces, Pichia, Saccharomyces, Torulaspora and other unidentified taxa. The inoculated samples displayed 4 exclusive ASVs (Wickerhamiella, Issatchenkia, Dipodascus and Clavispora), while the spontaneous fermentation samples registered 3 unique ASVs (Wickerhamomyces, Knufia and one not assigned genus) (Figure 5B).
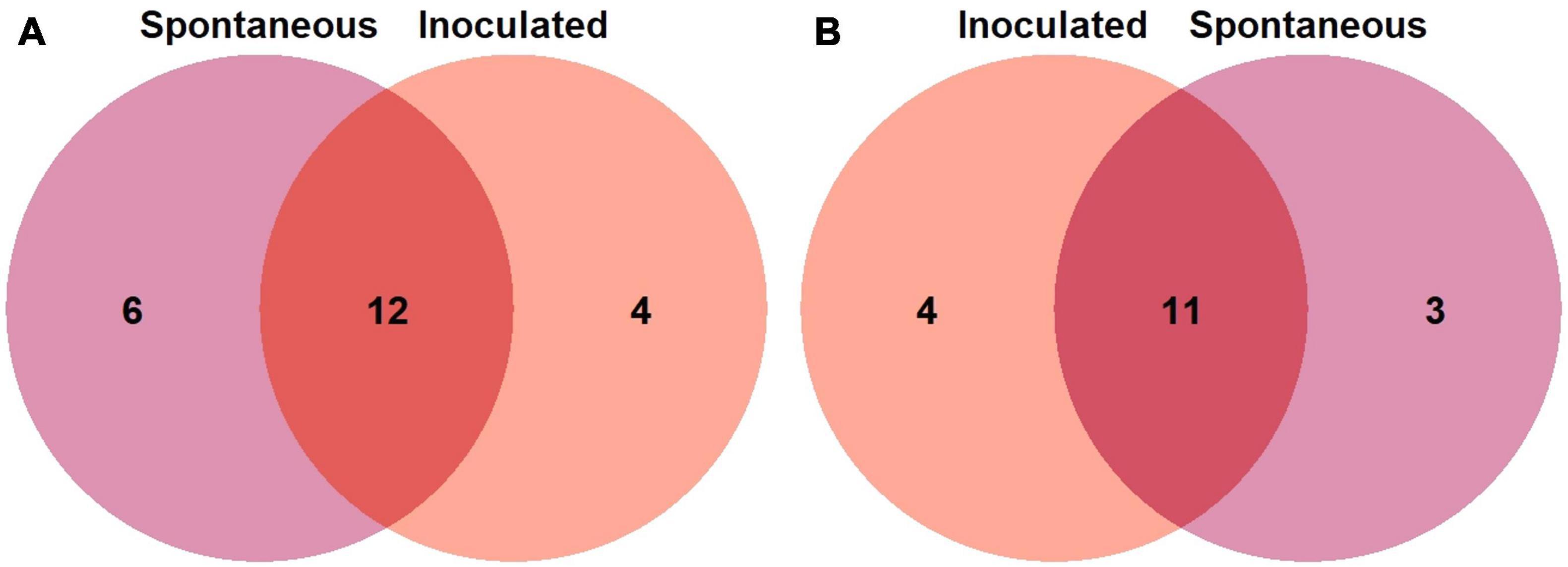
Figure 5. Venn diagrams of the number of unique and shared amplicon sequence variants (ASVs) of the (A) bacterial and (B) fungal community associated with the fermentation practices of colonche samples inoculated (red) and spontaneous (purple).
Discussion
Differences Between Spontaneous vs. Inoculated Fermentations
Our results confirm that spontaneous and inoculated fermentation practices significantly differ in the composition of microbial assemblages and the relative abundance of bacterial and fungal taxa in fermenting communities. These results are in concordance with most of the literature reporting that spontaneous fermentations exhibit a higher number of microbial species than inoculated products (Tamang et al., 1988; Raspor et al., 2002; Haruta et al., 2006; Domizio et al., 2007; Alves et al., 2010; Hong et al., 2015; Anagnostopoulos et al., 2020; Lu et al., 2021). These results confirm the reduction of diversity hypothesis, as can be visualized in Figure 2, where the lowest values of diversity for both bacteria and fungi were recorded in the inoculums and the reduction at the final composition for bacterial communities occurs in the inoculated samples. Nevertheless, this relation is not unidirectional for fungi, for which we observed small differences, contrarily to what has been previously reported (Raspor et al., 2002).
Fermentation traditionally occurs spontaneously and is initiated by a diverse community of indigenous yeasts and bacteria associated to the plants, the environment, or the fermentation facilities where it is produced (Pretorius et al., 1999; Motarjemi, 2002; Bokulich et al., 2013; Bokulich and Mills, 2013; da Silva Vale et al., 2021). In different traditional fermented beverages, the yeast genera Hanseniaspora, Pichia, Metschnikowia, Candida, Torulaspora, Rhodotorula, Cryptococcus, Lachancea, Zygosaccharomyces have been recorded as the most common at the initial stages of the successional process of fermentation, while strains of Saccharomyces cerevisiae become dominant at the last stages (Krieger-Weber et al., 2020). Although successional dynamics were not assessed in our current analysis, it could be considered that the fermentative environment as the clay pots where spontaneous fermentation occurs and the cooked samples before the inoculation, displayed a higher richness at both initial stages. This information also confirms that there is a marked dominance of the genus Saccharomyces at the final stages of the fermentation, and it occurs independently if it is spontaneous or inoculated fermented colonche. This can be explained because of the Saccharomyces capacity to quickly adapt to variable environmental conditions (Legras et al., 2018).
Our results confirm that the non-Saccharomyces yeast group comprises a variety of oxidative, weakly fermentative and strongly fermentative yeasts of genera like Rhodotorula, Cryptococcus, Hanseniaspora, Candida, Pichia, Issatchenkia, Metschnikowia, Lachancea, Zygosaccharomyces, Starmerella, Torulaspora and others (Barata et al., 2012), which are found at different frequencies at the clay pots and the cooked samples. This group has low fermentation power and are sensitive to prevailing anoxic conditions and increasing ethanol levels (Martinez et al., 2013; Quirós et al., 2014; Morales et al., 2015). Depending on the fermentative capacity and metabolic activity of the individual species, non-Saccharomyces yeasts are able to maintain their viability until the middle of fermentation before starting to decline (Xufre et al., 2006; Renouf et al., 2007; Andorrà et al., 2008; Zott et al., 2008, 2010; Bagheri et al., 2015; Sha et al., 2017). Nevertheless, the current results showed the dominance of Saccharomyces over the non-Saccharomyces groups in the final products of both types of fermentations. A similar observation was reported for other fermented beverages around the world (Tamang et al., 1988; Lyu et al., 2013; Yang et al., 2016; Sha et al., 2017).
In both types of fermentation, we identified constantly the Pediococcus genus. Some species of this genus are associated with spoilage of fermented beverages, especially beers and wines (Mokoena, 2017). It has been associated with the synthesis of excessive diacetyl, exopolysaccharides, and biogenic amines, all of which have a detrimental impact on the quality of the product (Wade et al., 2019). However, recent research has supported the contention that Pediococcus spp. can grow in wines considered to be microbiologically stable. In fact, the presence of Pediococcus spp. in wines not always lead to spoilage, and new findings have suggested potential uses for Pediococcus spp. to contribute to desirable characteristics of wines under certain circumstances. As Pediococcus was one of the most common genera identified in colonche samples, further studies through High-performance liquid chromatography (HPLC) should be addressed to characterize if the high abundance is associated with the spoilage or if it brings desirable characteristics to colonche and the distinction in the aromatic profile between fermentation practices.
The presence of yeast genera like Wickerhamiella, Clavispora, Dipodascus and Issatchenkia, has been described previously in pulque fermentation (Rocha-Arriaga et al., 2020). For instance, the differences of the fungal community between the inoculated fermentation are due to the common use of pulque to improve their fermentation or the use of the same containers for pulque production. Torulaspora is one of the few microorganisms previously identified in colonche samples (Ulloa and Herrera, 1978). By the current sampling we identified its presence in clay pots and in the cooked samples, which might represent the initial stages of fermentation. However, at the end of the fermentation under both types of practices low numbers of this genus were identified, and such low population density may be due to strict anaerobic conditions and low invertase activity (Visser et al., 1990; Hanl et al., 2005), mainly promoted by Saccharomyces species.
Kluyveromyces was also found in the colonche samples, it is a genus that has been isolated from a great variety of habitats and has been described to have high metabolic diversity and a substantial degree of intraspecific polymorphism. Therefore, several different biotechnological applications have been investigated with this yeast: production of enzymes, single-cell protein, aromatic compounds, and other applications (Varela et al., 2017). However, it cannot grow under strictly anaerobic conditions and the ethanol production is almost exclusively linked to oxygen limitation (Visser et al., 1990; van Dijken et al., 1993; Bellaver et al., 2004). In this sense, the higher abundance of Kluyveromyces genus in cooked and inoculated colonche samples might be due to a low alcohol content, but further studies are still needed to address the relationship between physicochemical attributes and the presence of microbial communities.
Kazachstania genus is typically encountered in the plant substrate, for instance, it is associated with grapevines and, in a low frequency, in grape must. Although it has not yet been characterized in cactus prickly pear fruits, this yeast genus provides positive aroma attributes that should be explored further. Its low presence in the final inoculated colonche product might be due to the dominance of the Saccharomyces which display a higher abundance in this treatment. Rhodotorula is also a common basidiomycete yeast that has been reported in several dairy products, it has a weak fermentative capacity, and has also been found associated with insects as vectors (Fonseca and Inácio, 2006; Kemler et al., 2017). The presence of this basidiomycete yeast might reflect the presence of insects closer to the clay pots and the low cautions in the storage of the cooked substrate. Nevertheless, this yeast was not found at the end of fermentation under both treatments, suggesting that hygienic practices are performed or inhibited during fermentation. In general, fungi held more stable ranges of diversity indexes in most of the samples analyzed, a similar pattern previously reported in beverages like pulque (Rocha-Arriaga et al., 2020).
Multiple strains of LAB from the Lactobacillus genus were registered in this study, Lactobacillus was the most abundant genus of bacteria, found abundantly (∼ 50%) in all phases of colonche fermentation except in clay pots. LAB are found in decomposing plant material and fruits, in dairy products, fermented meat and fish, cereals, beets, pickled vegetables, sourdough, silages, fermented beverages, juices, sewage and in cavities of humans and animals (König et al., 2009; Devi et al., 2013; Liu et al., 2014; Mokoena, 2017). LAB genera include Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, Aerococcus, Alloiococcus, Carnobacterium, Dolosigranulum, Enterococcus, Oenococcus, Tetragenococcus, Vagococcus and Weissella (Khalid, 2011), with Lactobacillus being the largest genus, including more than 100 species that are abundant in carbohydrate-rich substances and most of them are the most common and shared among samples of colonche in both fermentations process. It has been characterized that some LAB are used as probiotics, due to their health benefits (Ljungh and Wadstrom, 2006; De Leblanc et al., 2007; Azaïs-Braesco et al., 2010; Evivie et al., 2017). Although the main aim of this work was not to identify possible probiotics of LAB genera in colonche, we suggest that further studies in this direction may contribute to improve intestinal microbiota and human health through this beverage.
Our current study identified that 12 bacterial and 11 fungal ASVs are shared between spontaneous and inoculated colonche samples. Nevertheless, only the bacterial genera Lactobacillus, Leuconostoc, and Pediococcus and the Saccharomyces yeasts were the most abundant for both fermentation practices and appear in the inoculum and in the cooked samples. We propose as a first exploratory analysis that these 4 genera might reflect the microbial core of colonche because they appear common and abundantly in all the colonche samples. These genera are common throughout the fermentation of several fermented beverages and are closely related to food quality (Zhu et al., 2020; An et al., 2021; Ban et al., 2022). Nevertheless, further analysis as functional assignation and metaproteomics under a network approach can give insights of the functional microorganism’s core and the relevance to achieve a predictable product.
The Clay Pots: Not a Starter Culture but a Practical Reservoir for Future Fermentations
The clay pots displayed some microorganism’s genera associated with pathogenic activity, as are the cases of some strains of Clostridium, Pseudomonas, and some species of the genus Pediococcus. Nevertheless, through the fermentation stages these pathogenic strains disappear, perhaps because of the multiple strains of Lactobacillus in the spontaneous fermentation process. LAB have been applied in food preservation, partly due to their antimicrobial properties and because their capability to acidify the environment, making it harmless for other bacterial groups (Cizeikiene et al., 2013; Arena et al., 2018; Singh, 2018). Bacteriocins are a group of potent antimicrobial peptides produced by some microorganisms including LAB, primarily active against closely related organisms, mostly Gram-positive bacteria to gain competitive advantage for nutrients (Parada et al., 2007; Zacharof and Lovitt, 2012; Mokoena, 2017; Timothy et al., 2021). In fermented foods, LAB display various antimicrobial activities, through production of various metabolites, including lactic acid, hydrogen peroxide, and bacteriocins. Therefore, the high presence of Lactobacillus might reflect the drastic changes in the composition from the clay pot samples through the final product, thus inhibiting the pathogens in the environment.
As mentioned above, clay pots displayed the highest richness of bacterial and fungal genera, including several non-Saccharomyces genera, some beneficial bacterial groups as LAB and some detrimental bacterial groups associated with the spoilage of the beverage. Nevertheless, some of these microorganisms contribute to subsequent microbial growth and brings a stable community, for instance, 10 fungal ASVs and 6 bacterial genera are shared between the clay pots and the spontaneous colonche. In this sense, a stable consortium as an inoculum might not be required in spontaneous fermentation due to the presence of microbial diversity in the clay pots that can promote a starter community for fermentation process. Further studies should consider including other microbial inputs, for example the cactus pear fruits that are exposed to a variety of microorganisms derived from the environment (e.g., surfaces and soil), producer’s hands, and tools that can contribute to microbial communities and the quality of colonche as has been performed with cocoa fermentation (Schwan and Wheals, 2004; Lefeber et al., 2010; Figueroa-Hernández et al., 2019; Viesser et al., 2021), in which these factors contribute to the quality of the product.
The production of spontaneous colonche is mostly performed in clay pots, nevertheless, sometimes producers store the peeled cactus prickly pear fruits in plastic containers and then the fruits are placed inside the clay pots (Ojeda-Linares et al., 2020). Although we did not characterize the microbial and the physicochemical differences between the containers, it has been recorded that the material of the fermentation containers plays a significant role changing the abundance of specific genus as Lactococcus and Pedioccous and can affect pH values favoring a quicker fermentation (Liu et al., 2019). It is possible that the presence of these genera is favored by a previous storage in plastic containers and changes in the pH values might also change the dynamic in the clay pot by acidifying the medium. Though, further analysis should be performed to characterize the effect of the containers for colonche production.
The Inoculums: Insights for Unconscious Microorganisms’ Selection
The producers of traditional fermented beverages select inoculums as a practical way to acquire an optimal fermentation capacity and their capability to give consistent quality and aroma compositions through the fermentation process. As documented before, the selection of inoculums is related to a community of bacteria and yeasts that improves the fermentation dynamics and is mainly characterized by a reduction of diversity (Alves et al., 2010; Anagnostopoulos et al., 2020; Lu et al., 2021). In the colonche production, cinaiste is the common name of the inoculum, it is stored from previous batches year after year, and our data indicate a marked dominance of S. cerevisiae strains and a higher number of reads compared with the rest of samples analyzed. The large inoculum of active S. cerevisiae cells in most cases ensures a rapid dominance of a single strain and will therefore likely reduce any impact that the natural microbiota may have if spontaneous fermentation is allowed. In addition, the inoculum shows the lowest bacterial diversity, which is consistent with a reduced richness in starter cultures, a pattern that has been previously documented for the selection of starter cultures in other traditional fermented products (Orji et al., 2003; Zorba et al., 2003; Freire et al., 2015; Fagbemigun et al., 2021; Lima et al., 2021). Our results therefore expand the documentation of the deliberate selection of microbial consortia to acquire an optimal fermented product.
Selection of microorganisms is an attempt to enhance a fermentative environment, such type of efforts to transform the environment are proposed to play an important and underappreciated role in shaping biotic communities and evolutionary processes (Jones et al., 1997; Odling-Smee et al., 2003; Anderson, 2005). In this context, the studies of management of microenvironments can make relevant contributions to analyze general processes of domestication of organisms and their environments. Morphological and physiological changes in plants and animals show evidence of domestication and provide insights of the specific human practices that produce them. Analogous changes in microorganisms are not easy to identify. In microorganisms as yeasts, features like the overexpression of metabolic routes like maltotriose and lactate, the loss of sexual reproduction and the decreasing survival in nature, have been considered significant traits to analyze domestication (Gibbons and Rinker, 2015; Gallone et al., 2016; Gibbons, 2019), but there is still a long way to explore in this direction. However, the characterization of the practices performed to produce colonche, particularly the selection of inoculums and their specific consortia, provide a signature of deliberate human management and shaping of the fermenting microenvironments.
Summarizing, our results confirm that the choice of fermentation practice and management practices as cooking the fruits or storing batches year after year influence the composition, structure, and dynamics of microbiota communities in the different stages of colonche fermentation. Coexistence of the different fermentation practices and management techniques favors the context to maintain and diversify microbial communities involved in the colonche production. As well, through the carefully reproduction of inoculums, producers promote and direct selection over microbial communities to guide the process of fermentation. The clay pots are important reservoirs of diversity, but they are not commonly considered relevant in fermentation studies. Although the most dominant bacterial and fungal ASVs were not dominant in the clay pot, its presence might highlight the importance at the beginning of the fermentation and the practices performed to clean it are also relevant to maintain the microbial community. Finally, characterizing the management practices that the producers perform to obtain specific traditional products gives insights about the knowledge that humans have to reshape biotic communities and micro-landscapes to obtain a quality product at the end of both fermentation types. Thus, it is relevant to conserve these techniques of micro-biocultural heritage, ensure its maintenance and, in some contexts, recover it in the context of the foodscape diversity (Mintz and Du Bois, 2002; Alexiades, 2003, 2009; Marshall and Mejia, 2011; Rocha et al., 2015).
Conclusion
The characterization of the microbial communities related to the production of a traditional fermented beverage prepared with fruits of O. streptacantha under spontaneous and inoculated fermentation practices, confirmed that the fermentation practices affect the species richness, diversity, and the community structure through spontaneous and inoculated fermentation practices. However, differences are slightly significant for the final products for fungal and bacterial communities, which might be the result of the increasing relative abundance of LAB and the outstanding abundance of Saccharomyces genus in final products of both fermentation types. The current information corroborates that there is a continuous selection on microbial communities through the elaboration and management of inoculums. This practice reduces the microbial community and, in the case studied, traditional fermenters select mostly LAB and Saccharomyces genera to begin their new batches of colonche year after year. Diverse microorganisms live in the container where fermentation occurs, nevertheless, trough the fermentation dynamics the diversity changes, and a stable community is reached. Although the current research was not directed to assess the differences in the colonche quality, the microbial composition at the end of the fermentation and the demand of the consumers for this product supports that both types of fermentation practices yield a good product. However, further analysis as the characterization of microbial functionalities and the physicochemical attributes might help to improve the fermentation practices. Altogether, these results provide the potential of the traditional knowledge and management over microorganisms to produce fermented beverages, thus, the importance to maintain these products, the practices and the diversity associated over the fermentation process. These results are relevant to ensure food security and safety in localities where food availability is constrained.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA781103.
Author Contributions
CO-L conceived, designed, conducted, collected data, and wrote the manuscript. IS-G contributed to analyze the data, constructed figures, and drafted the manuscript. AC conceived, designed, advised, funded, drafted, and reviewed all versions of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research received financial support from the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, DGAPA, UNAM (IN206217 and IN206520), CONACYT (A1-S-14306).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Posgrado en Ciencias Biológicas, UNAM and the Consejo Nacional de Ciencia y Tecnología CONACYT for supporting studies of Linares César. The Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (IN206217 and IN206520), CONACYT (A1-S-14306) for financial support of the research. We also acknowledge the support of LANASE-UNAM and the following projects 2018-LN293701, 2019-LN299033, 2020-LN314852, 2021-LN31581 for the access to sequencing facilities. We especially thank people from the Altiplano region for their hospitality, generosity, and participation in the study. Most importantly for colonche preservation and sharing. Likewise, we recognize the heaviest labor to the colonche fest organizers to diffuse their cultural heritage.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.821268/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves of the sample coverage for (A) bacterial and, (B) fungal community. Rarefaction curves of the observed ASVs of the (C) bacterial and (D) fungal community associated with the colonche stages and fermentation practices.
Supplementary Figure 2 | Non-metric multidimensional scaling (NMDS) plots-based (A) on UniFrac unweighted distance of the bacterial community structure; (B) and the fungal community structure associated with inoculated and spontaneous fermentation practices in the colonche production.
References
Akinola, R., Pereira, L. M., Mabhaudhi, T., de Bruin, F. M., and Rusch, L. (2020). A review of indigenous food crops in Africa and the implications for more sustainable and healthy food systems. Sustainability 12:3493. doi: 10.3390/su12083493
Albert, S., Aswani, S., Fisher, P. L., and Albert, J. (2015). Keeping food on the table: human responses and changing coastal fisheries in Solomon Islands. PLoS One 10:e0130800. doi: 10.1371/journal.pone.0130800
Albuquerque, U. P., do Nascimento, A. L. B., da Silva, Chaves, L., Feitosa, I. S., de Moura, J. M. B., et al. (2019). A brief introduction to niche construction theory for ecologists and conservationists. Biol. Conserv. 237, 50–56. doi: 10.1016/j.biocon.2019.06.018
Alexiades, M. (2009). “The cultural and economic globalisation of traditional environmental knowledge systems,” in Landscape, process and power: re-evaluating traditional environmental knowledge, ed. H. Serena (Oxford: Berghanh), 68–90.
Alexiades, M. N. (2003). Ethnobotany in the third millennium: expectations and unresolved issues. Delpinoa 45, 15–28.
Álvarez-Ríos, G. D., Figueredo-Urbina, C. J., and Casas, A. (2020). Physical, Chemical, and Microbiological Characteristics of Pulque: management of a Fermented Beverage in Michoacán, Mexico. Foods 9:361. doi: 10.3390/foods9030361
Alves, J. A., Lima, L. C. D. O., Dias, D. R., Nunes, C. A., and Schwan, R. F. (2010). Effects of spontaneous and inoculated fermentation on the volatile profile of lychee (Litchi chinensis Sonn) fermented beverages. Int. J. Food Sci. Technol. 45, 2358–2365. doi: 10.1111/j.1365-2621.2010.02409.x
An, F., Li, M., Zhao, Y., Zhang, Y., Mu, D., Hu, X., et al. (2021). Metatranscriptome-based investigation of flavor-producing core microbiota in different fermentation stages of Dajiang, a traditional fermented soybean paste of Northeast China. Food Chem. 343:128509. doi: 10.1016/j.foodchem.2020.128509
Anagnostopoulos, D. A., Kamilari, E., and Tsaltas, D. (2020). Evolution of bacterial communities, physicochemical changes and sensorial attributes of natural whole and cracked picual table olives during spontaneous and inoculated fermentation. Front. Microbiol. 11:1128. doi: 10.3389/fmicb.2020.01128
Andorrà, I., Landi, S., Mas, A., Guillamón, J. M., and Esteve-Zarzoso, B. (2008). Effect of oenological practices on microbial populations using culture-independent techniques. Food Microbiol. 25, 849–856. doi: 10.1016/j.fm.2008.05.005
Arena, M. P., Capozzi, V., Russo, P., Drider, D., and Fiocco, D. (2018). Immunobiosis and probiosis: antimicrobial activity of lactic acid bacteria with a focus on their antiviral and antifungal properties. Appl. Microbiol. Biotechnol. 102, 9949–9958. doi: 10.1007/s00253-018-9403-9
Astudillo-Melgar, F., Ochoa-Leyva, A., Utrilla, J., and Huerta-Beristain, G. (2019). Bacterial diversity and population dynamics during the fermentation of palm wine from Guerrero Mexico. Front. Microbiol. 10:531. doi: 10.3389/fmicb.2019.00531
Azaïs-Braesco, V., Bresson, J. L., Guarner, F., and Corthier, G. (2010). Not all lactic acid bacteria are probiotics,…but some are. Br. J. Nutr. 103, 1079–1081. doi: 10.1017/S0007114510000723
Badotti, F., de Oliveira, F. S., Garcia, C. F., Vaz, A. B. M., Fonseca, P. L. C., Nahum, L. A., et al. (2017). Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 17:42. doi: 10.1186/s12866-017-0958-x
Bagheri, B., Bauer, F. F., and Setati, M. E. (2015). The diversity and dynamics of indigenous yeast communities in grape must from vineyards employing different agronomic practices and their influence on wine fermentation. South Afr. J. Enol. Vitic. 36, 243–251.
Baldrian, P., Větrovský, T., Lepinay, C., and Kohout, P. (2021). High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Divers. 1, 1–9. doi: 10.1007/s13225-021-00472-y
Ban, S., Chen, L., Fu, S., and Xu, Y. (2022). Modelling and predicting population of core fungi through processing parameters in spontaneous starter (Daqu) fermentation. Int. J. Food Microbiol. 363:109493. doi: 10.1016/j.ijfoodmicro.2021.109493
Barata, A., Malfeito-Ferreira, M., and Loureiro, V. (2012). The microbial ecology of wine grape berries. Int. J. Food Microbiol. 153, 243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025
Bellaver, L. H., de Carvalho, N. M. B., Abrahão-Neto, J., and Gombert, A. K. (2004). Ethanol formation and enzyme activities around glucose-6-phosphate in Kluyveromyces marxianus CBS 6556 exposed to glucose or lactose excess. FEMS Yeast Res. 4, 691–698. doi: 10.1016/j.femsyr.2004.01.004
Blancas, J., Casas, A., Rangel-Landa, S., Moreno-Calles, A., Torres, I., Pérez-Negrón, E., et al. (2010). Plant Management in the Tehuacán-Cuicatlán Valley, Mexico 1. Econ. Bot. 64, 287–302. doi: 10.1007/s12231-010-9133-0
Bokulich, N. A., and Mills, D. A. (2013). Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 79, 5214–5223. doi: 10.1128/AEM.00934-13
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bokulich, N. A., Ohta, M., Richardson, P. M., and Mills, D. A. (2013). Monitoring seasonal changes in winery-resident microbiota. PLoS One 8:e66437. doi: 10.1371/journal.pone.0066437
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brandt, M. J. (2014). Starter cultures for cereal based foods. Food Microbiol. 37, 41–43. doi: 10.1016/j.fm.2013.06.007
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Campbell-Platt, G. (1994). Fermented foods: a world perspective. Food Res. Int. 27, 253–257. doi: 10.1016/0963-9969(94)90093-0
Cano, A. N. H., and Suárez, M. E. (2020). Ethnobiology of algarroba beer, the ancestral fermented beverage of the Wichí people of the Gran Chaco I: a detailed recipe and a thorough analysis of the process. J. Ethn. Food. 7:4.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Casas, A., Caballero, J., Mapes, C., and Zárate, S. (1997). Manejo de la vegetación, domesticación de plantas y origen de la agricultura en Mesoamérica. Bol. Soc. Bot. México 61, 31–47. doi: 10.17129/botsci.1537
Casas, A., Otero-Arnaiz, A., Pérez-Negrón, E., and Valiente-Banuet, A. (2007). In situ management and domestication of plants in Mesoamerica. Ann. Bot. 100, 1101–1115. doi: 10.1093/aob/mcm126
Chao, A., and Chiu, C. H. (2016). “Species richness: estimation and comparison,” in Wiley StatsRef: Statistics Reference Online, eds N. Balakrishnan, T. Colton, B. Everitt, W. Piegorsch, F. Ruggeri, and J. L. Teugels (Chichester: John Wiley & Sons, Ltd), 1–26. doi: 10.1002/9781118445112.stat03432.pub2
Chao, A., Chiu, C. H., and Jost, L. (2014). Unifying species diversity, phylogenetic diversity, functional diversity, and related similarity and differentiation measures through Hill numbers. Annu. Rev. Ecol. Evol. Syst. 45, 297–324. doi: 10.1146/annurev-ecolsys-120213-091540
Chaves-López, C., Rossi, C., Maggio, F., Paparella, A., and Serio, A. (2020). Changes occurring in spontaneous maize fermentation: an overview. Fermentation 6:36. doi: 10.3390/fermentation6010036
Chen, H., and Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12:35. doi: 10.1186/1471-2105-12-35
Cizeikiene, D., Juodeikiene, G., Paskevicius, A., and Bartkiene, E. (2013). Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 31, 539–545. doi: 10.1016/j.foodcont.2012.12.004
Clement, C. R., Casas, A., Parra-Rondinel, F. A., Levis, C., Peroni, N., Hanazaki, N., et al. (2021). Disentangling domestication from food production systems in the neotropics. Quaternary 4:4. doi: 10.3390/quat4010004
Clement, C. R., Levis, C., Franco-Moraes, J., and Junqueira, A. B. (2020). “Domesticated nature: the culturally constructed niche of humanity,” in Participatory biodiversity conservation Concepts, Experiences, and Perspectives, ed. C. Baldauf (Cham: Springer), 35–51. doi: 10.1007/978-3-030-41686-7_3
Cumming, G., Fidler, F., and Vaux, D. L. (2007). Error bars in experimental biology. J. Cell Biol. 177, 7–11. doi: 10.1083/jcb.200611141
da Silva Vale, A., de Melo Pereira, G. V., de Carvalho Neto, D. P., Sorto, R. D., Goés-Neto, A., Kato, R., et al. (2021). Facility-specific ‘house’microbiome ensures the maintenance of functional microbial communities into coffee beans fermentation: implications for source tracking. Environ. Microbiol. Rep. 13, 470–481. doi: 10.1111/1758-2229.12921
De Filippis, F., Parente, E., and Ercolini, D. (2017). Metagenomics insights into food fermentations. Microb. Biotechnol. 10, 91–102. doi: 10.1111/1751-7915.12421
De Leblanc, A. D. M., Matar, C., and Perdigón, G. (2007). The application of probiotics in cancer. Br. J. Nutr. 98, S105–S110. doi: 10.1017/S0007114507839602
De Vuyst, L., and Vancanneyt, M. (2007). Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24, 120–127. doi: 10.1016/j.fm.2006.07.005
De Vuyst, L., Vrancken, G., Ravyts, F., Rimaux, T., and Weckx, S. (2009). Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 26, 666–675. doi: 10.1016/j.fm.2009.07.012
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Devi, M., Rebecca, L. J., and Sumathy, S. (2013). Bactericidal activity of the lactic acid bacteria Lactobacillus delbreukii. J. Chem. Pharm. Res. 5, 176–180.
Domizio, P., Lencioni, L., Ciani, M., Di Blasi, S., Pontremolesi, C. D., and Sabatelli, M. P. (2007). Spontaneous and inoculated yeast populations dynamics and their effect on organoleptic characters of Vinsanto wine under different process conditions. Int. J. Food Microbiol. 115, 281–289. doi: 10.1016/j.ijfoodmicro.2006.10.052
El Sheikha, A. F., and Hu, D. M. (2020). Molecular techniques reveal more secrets of fermented foods. Crit. Rev. Food Sci. Nutr. 60, 11–32. doi: 10.1080/10408398.2018.1506906
Escalante, A., López Soto, D. R., Velázquez Gutiérrez, J. E., Giles-Gómez, M., Bolívar, F., and López-Munguía, A. (2016). Pulque, a traditional Mexican alcoholic fermented beverage: historical, microbiological, and technical aspects. Front. Microbiol. 7:1026. doi: 10.3389/fmicb.2016.01026
Evivie, S. E., Huo, G. C., Igene, J. O., and Bian, X. (2017). Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr. Res. 61:1318034. doi: 10.1080/16546628.2017.1318034
Fagbemigun, O., Rösch, N., and Sung, C. G. (2021). Nutritional and sensory qualities of indigenous and starter culture fermented milk from Nigeria. Int. J. Sci. Technol. 9, 7–9.
Fernández-Llamazares, Á, Lepofsky, D., Lertzman, K., Armstrong, C. G., Brondizio, E. S., Gavin, M. C., et al. (2021). Scientists’ Warning to Humanity on Threats to Indigenous and Local Knowledge Systems. J. Ethnobiol. 41, 144–169. doi: 10.2993/0278-0771-41.2.144
Figueroa-Hernández, C., Mota-Gutierrez, J., Ferrocino, I., Hernández-Estrada, Z. J., González-Ríos, O., Cocolin, L., et al. (2019). The challenges and perspectives of the selection of starter cultures for fermented cocoa beans. Int. J. Food Microbiol. 301, 41–50. doi: 10.1016/j.ijfoodmicro.2019.05.002
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. U. S. A. 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Flachs, A., and Orkin, J. D. (2019). Fermentation and the ethnobiology of microbial entanglement. Ethnobiol. Lett. 10, 35–39. https://www.jstor.org/stable/26910053
Foligné, B., and Pot, B. (2013). Probiotics from research to market: the possibilities, risks and challenges. Curr. Opin. Microbiol. 16, 284–292. doi: 10.1016/j.mib.2013.06.008
Fonseca, A., and Inácio, J. (2006). “Phylloplane yeasts,” in Biodiversity and ecophysiology of yeasts, eds G. Péter and C. Rosa (Berlin: Springer), 263–301. doi: 10.1007/3-540-30985-3_13
Forssten, S. D., Sindelar, C. W., and Ouwehand, A. C. (2011). Probiotics from an industrial perspective. Anaerobe 17, 410–413. doi: 10.1016/j.anaerobe.2011.04.014
Franco-Moraes, J., Clement, C. R., de Oliveira, J. C., and de Oliveira, A. A. (2021). A framework for identifying and integrating sociocultural and environmental elements of indigenous peoples’ and local communities’ landscape transformations. Perspect. Ecol. Conserv. 19, 143–152. doi: 10.1016/J.PECON.2021.02.008
Freire, A. L., Ramos, C. L., and Schwan, R. F. (2015). Microbiological and chemical parameters during cassava based-substrate fermentation using potential starter cultures of lactic acid bacteria and yeast. Food Res. Int. 76, 787–795. doi: 10.1016/j.foodres.2015.07.041
Gadaga, T. H., Lehohla, M., and Ntuli, V. (2021). Traditional fermented foods of Lesotho. J. Microbiol. Biotechnol. Food Sci. 2021, 2387–2391.
Gallone, B., Steensels, J., Prahl, T., Soriaga, L., Saels, V., Herrera-Malaver, B., et al. (2016). Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397–1410. doi: 10.1016/j.cell.2016.08.020
Gibbons, J. G., and Rinker, D. C. (2015). The genomics of microbial domestication in the fermented food environment. Curr. Opin. Genet. Dev. 35, 1–8. doi: 10.1016/j.gde.2015.07.003
Gutiérrez-Uribe, J. A., Figueroa, L. M., Martín-del-Campo, S. T., and Escalante, A. (2017). “Pulque,” in Fermented Foods in Health and Disease Prevention, eds J. Frias, C. Martinez-Villaluenga, and E. Peñas (Cambridge: Academic Press), 543–556. doi: 10.1016/B978-0-12-802309-9.00023-6
Hanl, L., Sommer, P., and Arneborg, N. (2005). The effect of decreasing oxygen feed rates on growth and metabolism of Torulaspora delbrueckii. Appl. Microbiol. Biotechnol. 67, 113–118. doi: 10.1007/s00253-004-1695-2
Harris, L. J. (1998). “The microbiology of vegetable fermentations,” in Microbiology of fermented foods (Boston: Springer), 45–72. doi: 10.1007/978-1-4613-0309-1_2
Harris, L. J., Daeschel, M. A., Stiles, M. E., and Klaenhammer, T. R. (1989). Antimicrobial Activity of Lactic Acid Bacteria Against Listeria monocytogene s. J. Food Prot. 52, 384–387. doi: 10.4315/0362-028X-52.6.384
Harth, H., Van Kerrebroeck, S., and De Vuyst, L. (2016). Community dynamics and metabolite target analysis of spontaneous, backslopped barley sourdough fermentations under laboratory and bakery conditions. Int. J. Food Microbiol. 228, 22–32. doi: 10.1016/j.ijfoodmicro.2016.04.011
Haruta, S., Ueno, S., Egawa, I., Hashiguchi, K., Fujii, A., Nagano, M., et al. (2006). Succession of bacterial and fungal communities during a traditional pot fermentation of rice vinegar assessed by PCR-mediated denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 109, 79–87. doi: 10.1016/j.ijfoodmicro.2006.01.015
He, J., Zhang, R., Lei, Q., Chen, G., Li, K., Ahmed, S., et al. (2019). Diversity, knowledge, and valuation of plants used as fermentation starters for traditional glutinous rice wine by Dong communities in Southeast Guizhou, China. J. Ethnobiol. Ethnomed. 15:20. doi: 10.1186/s13002-019-0299-y
Hernández-Santana, G., and Narchi, N. E. (2018). “The seri traditional food system: cultural heritage, dietary change, and the (Re) awakening of dietary resilience among coastal hunter-gatherers in the Mexican Northwest,” in In Coastal Heritage and Cultural Resilience, ed. L. L. Price (Cham: Springer), 135–182. doi: 10.1007/978-3-319-99025-5_7
Hong, L., Zhuo, J., Lei, Q., Zhou, J., Ahmed, S., Wang, C., et al. (2015). Ethnobotany of wild plants used for starting fermented beverages in Shui communities of southwest China. J. Ethnobiol. Ethnomed. 11:42. doi: 10.1186/s13002-015-0028-0
Hsieh, T. C., Ma, K. H., and Chao, A. (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (H ill numbers). Methods Ecol. Evol. 7, 1451–1456. doi: 10.1111/2041-210X.12613
Johns, T., and Sthapit, B. R. (2004). Biocultural diversity in the sustainability of developing-country food systems. Food Nutr. Bull. 25, 143–155. doi: 10.1177/156482650402500207
Johns, T., Powell, B., Maundu, P., and Eyzaguirre, P. B. (2013). Agricultural biodiversity as a link between traditional food systems and contemporary development, social integrity and ecological health. J. Sci. Food Agric. 93, 3433–3442. doi: 10.1002/jsfa.6351
Jones, C. G., Lawton, J. H., and Shachak, M. (1997). Positive and negative effects of organisms as physical ecosystem engineers. Ecology 78, 1946–1957. doi: 10.1890/0012-96581997078[1946:PANEOO]2.0.CO;2
Kemler, M., Witfeld, F., and Yurkov, A. (2017). “Phylloplane yeasts in temperate climates,” in Yeasts in Natural Ecosystems: Diversity, eds P. Buzzini, M. A. Lachance, and A. M. Yurkov (Cham: Springer), 171–197. doi: 10.1007/978-3-319-62683-3_6
Kennedy, G., Kanter, R., Chotiboriboon, S., Covic, N., Delormier, T., Longvah, T., et al. (2021). Traditional and Indigenous Fruits and Vegetables for Food System Transformation. Curr. Dev. Nutr. 5:nzab092. doi: 10.1093/cdn/nzab092
König, H., Unden, G., and Fröhlich, J. (2009). Biology of Microorganisms on Grapes, in Must and in Wine. Heidelberg: Springer, 3–30. doi: 10.1007/978-3-319-60021-5
Krieger-Weber, S., Heras, J. M., and Suarez, C. (2020). Lactobacillus plantarum, a new biological tool to control malolactic fermentation: a review and an outlook. Beverages 6:23. doi: 10.3390/beverages6020023
Kuhnlein, H. V., and Receveur, O. (1996). Dietary change and traditional food systems of indigenous peoples. Annu. Rev. Nutr. 16, 417–442. doi: 10.1146/annurev.nu.16.070196.002221
Lefeber, T., Janssens, M., Camu, N., and De Vuyst, L. (2010). Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 76, 7708–7716. doi: 10.1128/AEM.01206-10
Legras, J. L., Galeote, V., Bigey, F., Camarasa, C., Marsit, S., Nidelet, T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727. doi: 10.1093/molbev/msy066
Lévi-Strauss, C. (2012). “The culinary triangle,” in Food and Culture, eds C. Counihan and P. Van Esterik (Oxfordshire: Routledge), 54–61.
Li, X., and Yue, X. (2016). Analysis of the microbiota in naturally fermented cabbage of Northeastern part of China by high-throughput sequencing of the V3-V4 regions of the 16S rRNA gene. Int. J. Agric. Biol. 18, 1153–1158. doi: 10.17957/IJAB/15.0219
Lima, C. O. D. C., Vaz, A. B., De Castro, G. M., Lobo, F., Solar, R., Rodrigues, C., et al. (2021). Integrating microbial metagenomics and physicochemical parameters and a new perspective on starter culture for fine cocoa fermentation. Food Microbiol. 93:103608. doi: 10.1016/j.fm.2020.103608
Liu, D., Legras, J. L., Zhang, P., Chen, D., and Howell, K. (2021). Diversity and dynamics of fungi during spontaneous fermentations and association with unique aroma profiles in wine. Int. J. Food Microbiol. 338:108983. doi: 10.1016/j.ijfoodmicro.2020.108983
Liu, L., She, X., Qian, Y., Li, Y., Tao, Y., Che, Z., et al. (2019). Effect of different fermenting containers on the deterioration of Sichuan pickle. LWT 111, 829–836. doi: 10.1016/j.lwt.2019.05.024
Liu, W., Pang, H., Zhang, H., and Cai, Y. (2014). “Biodiversity of lactic acid bacteria,” in Lactic Acid Bacteria, eds H. Zhang and Y. Cai (Dordrecht: Springer), 103–203. doi: 10.1007/978-94-017-8841-0_2
Ljungh, A., and Wadstrom, T. (2006). Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 7, 73–90.
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Lu, Y., Guan, X., Li, R., Wang, J., Liu, Y., Ma, Y., et al. (2021). Comparative study of microbial communities and volatile profiles during the inoculated and spontaneous fermentation of persimmon wine. Process Biochem. 100, 49–58. doi: 10.1016/j.procbio.2020.09.023
Lyu, C., Chen, C., Ge, F., Liu, D., Zhao, S., and Chen, D. (2013). A preliminary metagenomic study of puer tea during pile fermentation. J. Sci. Food Agric. 93, 3165–3174. doi: 10.1002/jsfa.6149
Madej, T., Pirożnikow, E., Dumanowski, J., and Łuczaj, Ł (2014). Juniper beer in Poland: the story of the revival of a traditional beverage. J. Ethnobiol. 34, 84–103. doi: 10.2993/0278-0771-34.1.84
Marshall, E., and Mejia, D. (2011). Traditional Fermented Food and Beverages for Improved Livelihoods. Rome: FAO, 21.
Martinez, F. A. C., Balciunas, E. M., Salgado, J. M., Gonzalez, J. M. D., Converti, A., and de Souza Oliveira, R. P. (2013). Lactic acid properties, applications and production: a review. Trends Food Sci. Technol. 30, 70–83. doi: 10.1016/j.tifs.2012.11.007
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Metzker, M. L. (2010). Sequencing technologies: the next generation. Nat. Rev. Genet. 11, 31–46. doi: 10.1038/nrg2626
Mintz, S. W., and Du Bois, C. M. (2002). The anthropology of food and eating. Annu. Rev. Anthropol. 31, 99–119. doi: 10.1146/annurev.anthro.32.032702.131011
Mokoena, M. P. (2017). Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules 22:1255. doi: 10.3390/molecules22081255
Montes-Carreto, L. M., Aguirre-Noyola, J. L., Solís-García, I. A., Ortega, J., Martinez-Romero, E., and Guerrero, J. A. (2021). Diverse methanogens, bacteria and tannase genes in the feces of the endangered volcano rabbit (Romerolagus diazi). PeerJ 9:e11942. doi: 10.7717/peerj.11942
Morales, P., Rojas, V., Quirós, M., and Gonzalez, R. (2015). The impact of oxygen on the final alcohol content of wine fermented by a mixed starter culture. Appl. Microbiol. Biotechnol. 99, 3993–4003. doi: 10.1007/s00253-014-6321-3
Motarjemi, Y. (2002). Impact of small scale fermentation technology on food safety in developing countries. Int. J. Food Microbiol. 75, 213–229. doi: 10.1016/S0168-1605(01)00709-7
Mukisa, I. M., Ntaate, D., and Byakika, S. (2017). Application of starter cultures in the production of Enturire–a traditional sorghum-based alcoholic beverage. Food Sci. Nutr. 5, 609–616. doi: 10.1002/fsn3.438
Nabhan, G. P. (2010). Ethnobiology for a diverse world: microbial ethnobiology and the loss of distinctive food cultures. J. Ethnobiol. 30, 181–183. doi: 10.2993/0278-0771-30.2.181
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Odling-Smee, F. J., Laland, K. N., and Feldman, W. (2003). Niche construction. Monographs in Population Biology. Princeton: Princeton University Press, 37. doi: 10.1353/pbm.2003.0003
Ojeda-Linares, C. I., Vallejo, M., Lappe-Oliveras, P., and Casas, A. (2020). Traditional management of microorganisms in fermented beverages from cactus fruits in Mexico: an ethnobiological approach. J. Ethnobiol. Ethnomed. 16:1. doi: 10.1186/s13002-019-0351-y
Ojeda-Linares, C., Álvarez-Ríos, G. D., Figueredo-Urbina, C. J., Islas, L. A., Lappe-Oliveras, P., Nabhan, G. P., et al. (2021). Traditional fermented beverages of Mexico: a biocultural unseen foodscape. Foods 10:2390. doi: 10.3390/foods10102390
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R. B., et al. (2013). Package ‘vegan’. Commun. Ecol. Package Ver. 2, 1–295. http://cran.r-project.org/
Orji, M. U., Mbata, T. I., Aniche, G. N., and Ahonkhai, I. (2003). The use of starter cultures to produce ‘Pito’, a Nigerian fermented alcoholic beverage. World J. Microbiol. Biotechnol. 19, 733–736. doi: 10.1023/A:1025172506965
Parada, J. L., Caron, C. R., Medeiros, A. B. P., and Soccol, C. R. (2007). Bacteriocins from lactic acid bacteria: purification, properties and use as biopreservatives. Braz. Arch. Biol. Technol. 50, 512–542. doi: 10.1590/S1516-89132007000300018
Pieroni, A., Sõukand, R., Quave, C. L., Hajdari, A., and Mustafa, B. (2017). Traditional food uses of wild plants among the Gorani of South Kosovo. Appetite 108, 83–92. doi: 10.1016/j.appet.2016.09.024
Pingali, P. (2007). Westernization of Asian diets and the transformation of food systems: implications for research and policy. Food Policy 32, 281–298. doi: 10.1016/j.foodpol.2006.08.001
Pretorius, I. S., Van der Westhuizen, T. J., and Augustyn, O. P. H. (1999). Yeast biodiversity in vineyards and wineries and its importance to the South African wine industry. A review. South Afr. J. Enol. Vitic. 20, 61–70. doi: 10.21548/20-2-2234
Quave, C. L., and Pieroni, A. (2014). Fermented foods for food security and food sovereignty in the Balkans: a case study of the Gorani people of Northeastern Albania. J. Ethnobiol. 34, 28–43. doi: 10.2993/0278-0771-34.1.28
Quirós, M., Rojas, V., Gonzalez, R., and Morales, P. (2014). Selection of non-Saccharomyces yeast strains for reducing alcohol levels in wine by sugar respiration. Int. J. Food Microbiol. 181, 85–91. doi: 10.1016/j.ijfoodmicro.2014.04.024
Raspor, P., Cus, F., Jemec, K. P., Zagorc, T., Cadez, N., and Nemanic, J. (2002). Yeast population dynamics in spontaneous and inoculated alcoholic fermentations of Zametovka must. Food Technol. Biotechnol. 40, 95–102. http://www.ftb.com.hr
Ratcliffe, E., Baxter, W. L., and Martin, N. (2019). Consumption rituals relating to food and drink: a review and research agenda. Appetite 134, 86–93. doi: 10.1016/j.appet.2018.12.021
Rebollar, E. A., Sandoval-Castellanos, E., Roessler, K., Gaut, B. S., Alcaraz, L. D., Benítez, M., et al. (2017). Seasonal changes in a maize-based polyculture of central Mexico reshape the co-occurrence networks of soil bacterial communities. Front. Microbiol. 8:2478. doi: 10.3389/fmicb.2017.02478
Renouf, V., Claisse, O., and Lonvaud-Funel, A. (2007). Inventory and monitoring of wine microbial consortia. Appl. Microbiol. Biotechnol. 75, 149–164. doi: 10.1007/s00253-006-0798-3
Rocha, J. A., Boscolo, O. H., and Fernandes, L. R. R. D. M. V. (2015). Ethnobotany: a instrument for valorisation and identification of potential for the protection of traditional knowledge. Interações 16, 67–74. doi: 10.1590/151870122015105
Rocha-Arriaga, C., Espinal-Centeno, A., Martinez-Sánchez, S., Caballero-Pérez, J., Alcaraz, L. D., and Cruz-Ramírez, A. (2020). Deep microbial community profiling along the fermentation process of pulque, a biocultural resource of Mexico. Microbiol. Res. 241:126593. doi: 10.1016/j.micres.2020.126593
Schwan, R. F., and Wheals, A. E. (2004). The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 44, 205–221. doi: 10.1080/10408690490464104
Sha, S. P., Jani, K., Sharma, A., Anupma, A., Pradhan, P., Shouche, Y., et al. (2017). Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci. Rep. 7:10967. doi: 10.1038/s41598-017-11609-y
Sha, S. P., Suryavanshi, M. V., Jani, K., Sharma, A., Shouche, Y. S., and Tamang, J. P. (2018). Diversity of yeasts and molds by culture-dependent and culture-independent methods for mycobiome surveillance of traditionally prepared dried starters for the production of Indian alcoholic beverages. Front. Microbiol. 9:2237. doi: 10.3389/fmicb.2018.02237
Singh, V. P. (2018). Recent approaches in food bio-preservation-a review. Open Vet. J. 8, 104–111. doi: 10.4314/ovj.v8i1.16
Smid, E. J., and Hugenholtz, J. (2010). Functional genomics for food fermentation processes. Annu. Rev. Food Sci. Technol. 1, 497–519. doi: 10.1146/annurev.food.102308.124143
Smid, E. J., and Kleerebezem, M. (2014). Production of aroma compounds in lactic fermentations. Annu. Rev. Food Sci. Technol. 5, 313–326. doi: 10.1146/annurev-food-030713-092339
Smith, B. D. (2007). Niche construction and the behavioral context of plant and animal domestication. Evol. Anthropol. 16, 188–199. doi: 10.1002/evan.20135
Solís-García, I. A., Ceballos-Luna, O., Cortazar-Murillo, E. M., Desgarennes, D., Garay-Serrano, E., Patiño-Conde, V., et al. (2021). Phytophthora root rot modifies the composition of the avocado rhizosphere microbiome and increases the abundance of opportunistic fungal pathogens. Front. Microbiol. 11:3484. doi: 10.3389/fmicb.2020.574110
Sõukand, R., Pieroni, A., Biró, M., Dénes, A., Dogan, Y., Hajdari, A., et al. (2015). An ethnobotanical perspective on traditional fermented plant foods and beverages in Eastern Europe. J. Ethnopharmacol. 170, 284–296. doi: 10.1016/j.jep.2015.05.018
Steinkraus, K. H. (1996). Handbook of Indigenous Fermented Food, 2nd Edn. New York: Marcel Dekker, Inc.
Svanberg, I. (2015). Ræstur fiskur: air-dried fermented fish the Faroese way. J. Ethnobiol. Ethnomed. 11:76. doi: 10.1186/s13002-015-0064-9
Tamang, J. P. (2010). “Diversity of fermented beverages,” in Fermented Foods and Beverages of the World, eds J. P. Tamang and K. Kailasapathy (New York: CRC Press), 85–125. doi: 10.1007/s00253-016-7941-6
Tamang, J. P., Cotter, P. D., Endo, A., Han, N. S., Kort, R., Liu, S. Q., et al. (2020). Fermented foods in a global age: east meets West. Compr. Rev. Food Sci. Food Saf. 19, 184–217. doi: 10.1111/1541-4337.12520
Tamang, J. P., Jeyaram, K., Rai, A. K., and Mukherjee, P. K. (2021). Diversity of beneficial microorganisms and their functionalities in community-specific ethnic fermented foods of the Eastern Himalayas. Food Res. Int. 148:110633. doi: 10.1016/j.foodres.2021.110633
Tamang, J. P., Sarkar, P. K., and Hesseltine, C. W. (1988). Traditional fermented foods and beverages of Darjeeling and Sikkim–a review. J. Sci. Food Agric. 44, 375–385. doi: 10.1002/jsfa.2740440410
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016). Review: diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.1007/BF00437031
Tamang, J. P., and Fleet, G. H. (2009). “Yeasts diversity in fermented foods and beverages,” in Yeasts Biotechnology: Diversity and Applications, eds T. Satyanarayana and G. Kunze (New York: Springer), 169–198. doi: 10.1007/978-1-4020-8292-4_9
Timothy, B., Iliyasu, A. H., and Anvikar, A. R. (2021). Bacteriocins of lactic acid bacteria and their industrial application. Curr. Top. Lact. Acid Bact. Probiotics 7, 1–13. doi: 10.35732/ctlabp.2021.7.1.1
Todorov, S. D., and Holzapfel, W. H. (2015). “Traditional cereal fermented foods as sources of functional microorganisms,” in Advances in fermented foods and beverages, ed. W. Holzapfel (Sawston: Woodhead Publishing), 123–153. doi: 10.1016/B978-1-78242-015-6.00006-2
Tsafrakidou, P., Michaelidou, A. M., and Biliaderis, G. C. (2020). Fermented cereal-based products: nutritional aspects, possible impact on gut microbiota and health implications. Foods 9:734. doi: 10.3390/foods9060734
Turner, N. J., and Turner, K. L. (2007). Traditional food systems, erosion and renewal in Northwestern North America. Indian J. Tradit. Knowl. 6, 57–68. http://hdl.handle.net/123456789/828
Ulloa, M., and Herrera, T. (1978). Torulopsis taboadae, una nueva especie de levadura aislada del colonche de Zacatecas. México. Sci. Fungorum 12, 5–12.
van Dijken, J. P., Weusthuis, R. A., and Pronk, J. T. (1993). Kinetics of growth and sugar consumption in yeasts. Antonie van leeuwenhoek 63, 343–352. doi: 10.1007/BF00871229
Varela, J. A., Gethins, L., Stanton, C., Ross, P., and Morrissey, J. P. (2017). “Applications of Kluyveromyces marxianus in Biotechnology,” in Yeast diversity in human welfare, eds T. Satyanarayana and G. Kunze (Singapore: Springer), 439–453. doi: 10.1007/978-981-10-2621-8_17
Viesser, J. A., de Melo Pereira, G. V., de Carvalho Neto, D. P., Favero, G. R., de Carvalho, J. C., Goés-Neto, A., et al. (2021). Global cocoa fermentation microbiome: revealing new taxa and microbial functions by next generation sequencing technologies. World J. Microbiol. Biotechnol. 37:118. doi: 10.1007/s11274-021-03079-2
Visser, W., Scheffers, W. A., Batenburg-van, der Vegte, W. H., and van Dijken, J. P. (1990). Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56, 3785–3792. doi: 10.1128/aem.56.12.3785-3792.1990
Vogelmann, S. A., and Hertel, C. (2011). Impact of ecological factors on the stability of microbial associations in sourdough fermentation. Food Microbiol. 28, 583–589. doi: 10.1016/j.fm.2010.11.010
Vogelmann, S. A., Seitter, M., Singer, U., Brandt, M. J., and Hertel, C. (2009). Adaptability of lactic acid bacteria and yeasts to sourdoughs prepared from cereals, pseudocereals and cassava and use of competitive strains as starters. Int. J. Food Microbiol. 130, 205–212. doi: 10.1016/j.ijfoodmicro.2009.01.020
Wade, M. E., Strickland, M. T., Osborne, J. P., and Edwards, C. G. (2019). Role of Pediococcus in winemaking. Aust. J. Grape Wine Res. 25, 7–24. doi: 10.1111/ajgw.12366
Xufre, A., Albergaria, H., Inácio, J., Spencer-Martins, I., and Gírio, F. (2006). Application of fluorescence in situ hybridisation (FISH) to the analysis of yeast population dynamics in winery and laboratory grape must fermentations. Int. J. Food Microbiol. 108, 376–384. doi: 10.1016/j.ijfoodmicro.2006.01.025
Yang, L., Yang, H. L., Tu, Z. C., and Wang, X. L. (2016). High-throughput sequencing of microbial community diversity and dynamics during douchi fermentation. PLoS One 11:e0168166. doi: 10.1371/journal.pone.0168166
Yao, H., Song, J., Liu, C., Luo, K., Han, J., Li, Y., et al. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One 5:e13102. doi: 10.1371/journal.pone.0013102
Zacharof, M. P., and Lovitt, R. W. (2012). Bacteriocins produced by lactic acid bacteria a review article. Apcbee Procedia 2, 50–56. doi: 10.1016/j.apcbee.2012.06.010
Zhu, M. Z., Li, N., Zhou, F., Ouyang, J., Lu, D. M., Xu, W., et al. (2020). Microbial bioconversion of the chemical components in dark tea. Food Chem. 312:126043. doi: 10.1016/j.foodchem.2019.126043
Zorba, M., Hancioglu, O., Genc, M., Karapinar, M., and Ova, G. (2003). The use of starter cultures in the fermentation of boza, a traditional Turkish beverage. Process Biochem. 38, 1405–1411. doi: 10.1016/S0032-9592(03)00033-5
Zott, K., Claisse, O., Lucas, P., Coulon, J., Lonvaud-Funel, A., and Masneuf-Pomarede, I. (2010). Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol. 27, 559–567. doi: 10.1016/j.fm.2010.01.006
Keywords: microbiota, management, ferments, ethnozymology, colonche, landscape domestication
Citation: Ojeda-Linares CI, Solís-García IA and Casas A (2022) Constructing Micro-Landscapes: Management and Selection Practices on Microbial Communities in a Traditional Fermented Beverage. Front. Ecol. Evol. 10:821268. doi: 10.3389/fevo.2022.821268
Received: 24 November 2021; Accepted: 19 January 2022;
Published: 10 February 2022.
Edited by:
Ulysses Paulino Albuquerque, Federal University of Pernambuco, BrazilReviewed by:
Melissa Fontes Landell, Federal University of Alagoas, BrazilBaiba Pruse, Ca’ Foscari University of Venice, Italy
Marcos Antonio De Morais Jr., Federal University of Pernambuco, Brazil
Copyright © 2022 Ojeda-Linares, Solís-García and Casas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alejandro Casas, acasas@cieco.unam.mx
 César Iván Ojeda-Linares1
César Iván Ojeda-Linares1  Itzel Anayansi Solís-García
Itzel Anayansi Solís-García Alejandro Casas
Alejandro Casas