Brief research report: OM-85 reduces SARS-COV-2 viral RNA expression in nasopharyngeal cells from COVID-19 patients
- 1Laboratory of Clinical and Experimental Immunology, Health and Life Science School – Pontifícia Universidade Católica do Rio Grande do Sul (PUCRS), Porto Alegre, Brazil
- 2Molecular and Functional Biology Research Center (CPBMF), PUCRS, Porto Alegre, Brazil
- 3Preclincal Research Department, OM-Pharma, Geneva, Switzerland
- 4School of Medicine, PUCRS, Porto Alegre, Brazil
OM-85 is a bacterial lysate from common respiratory tract pathogens, with an excellent safety profile, widely used to prevent recurrent respiratory tract infections. Several studies have been reporting the immunomodulating properties and antiviral roles of OM-85. The COVID-19 pandemic, originating in 2019, has presented a significant global public health crisis. While effective vaccines have been developed, vaccination rates vary considerably, and numerous concerning viral variants continue to emerge. The challenge persists in creating early interventions to halt the progression of the disease to its severe stages. To examine the therapeutic effect of OM-85 after SARS-CoV-2 infection and compared to recombinant human (rhINF-β) we collected nasopharyngeal cells from COVID-19 patients. The cells were treated ex-vivo with OM-85 or hrINF-β and the response was analyzed after 24h for gene expression by real-time PCR. We found that OM-85 decreased the SARS-CoV-2 N1 gene expression and increased RIG-I (DDX58) in these cells. The expression of ACE2 was undetected in these samples. These data support the antiviral effect of OM-85 against SARS-CoV-2.
1 Introduction
The COVID-19 pandemic has affected a huge number of people around the world. Although the development of vaccines and the adoption of efficient and sanitary measures, several variants are still very active and affect individuals. There is a need to develop new effective therapeutic approaches to minimize the burden of these severe respiratory viral infections and all associated sequelae. Treatments based on microbial-derived products, such as probiotics and postbiotics, can prevent and treat viral-induced respiratory diseases. Many of these studies have focused on the use of microbial therapies administered by the oral route, inducing the gut-lung axis into producing a potent host antiviral response. Recently, the delivery of these products through the airways is emerging as a safe and effective alternative strategy (1). OM-85 is a postbiotic product containing a standardized lysate obtained by chemical lysis of 21 microorganisms strains associated with respiratory infections. OM-85 contains different microbial products, such as bacterial metabolites, proteins, peptides, traces of fatty amino acids, and saccharides. This bacterial lysate is widely used to prevent recurrent respiratory infection when administered orally (2, 3). Likewise, the concept that OM-85 directly administrated into the airways modulates immune response has been previously explored to prevent asthma and respiratory syncytial virus (RSV) infection in mice (4, 5). The protective effects of OM-85 are mainly due to its modulation of both cellular and humoral responses (2, 6, 7), presenting an anti-inflammatory effect and improving antiviral response mediated by interferons. OM-85 can be effective in preventing severe coronavirus infection in mice when administered through the oral route (8). Additionally, the SARS-CoV-2 receptor ACE-2 is downregulated in the lungs of mice treated intranasally with OM-85 (5). Moreover, OM-85 decreased the expression of SARS-CoV-2 receptors angiotensin-converting enzyme 2 (ACE2), and transmembrane protease serine subtype 2 (TMPRSS2) in human bronchial epithelial cells. The infection by a SARS-CoV-2 spike protein pseudo-typed lentivirus was reduced in cells pretreated with OM-85. Most of these studies focus on the prophylactic effect of OM-85. In this study, we aimed to evaluate the effect of ex-vivo treatment with OM-85 in nasopharyngeal cells already infected with SARS-CoV-2 isolated from COVID-19 patients.
2 Methods
2.1 Ethics approval
The study design was approved by the local ethical committee under the protocol CAAE n° 30754220.3.0000.5336. Written informed consent was obtained from the patients before nasopharyngeal sample collection.
2.2 Nasopharyngeal sample collection and characterization
We collected nasopharyngeal samples from adult hospitalized COVID-19 patients positive for SARS-CoV-2. Nasopharyngeal lavage was performed instilled 10 ml of saline solution (0.9%) into the nostril using a sterile soft catheter (number 8) connected to a syringe. Viral presence was confirmed by RT-PCR within 48h before inclusion. Subject enrollment occurred from March to July 2021. The following variables were considered exclusion criteria: patients on mechanical ventilation; pregnant or breastfeeding women; patients with severe liver disease (alanine aminotransferase and/or aspartate aminotransferase > 5x normal); severe nephropathy (kidney transplantation or dialysis), HIV infection, cancer, hereditary angioedema, other immunodeficiencies, previous myocardial ischemic disease, previous thromboembolic disease. To differentiate epithelial/stromal cells (CD45- cells) from the hematopoietic immune cells (CD45+ cells) the sample was stained with anti-human CD45 FITC (1:200, #555482, clone HI30, BD Biosciences) and analyzed by Flow cytometry (FACS Canto II, BD Bioscience). Gating strategy for each panel is available on Supplementary Figure 1. To confirm the predominant epithelial cells phenotype, the processed sample was centrifuged and stained with hematoxylin and eosin for cytology analysis, as previously described in (9) (Panotico rápido, Laborclin) (Supplementary Figure 1).
2.3 Ex-vivo treatment with OM-85
To perform the ex-vivo treatment nasopharyngeal samples were centrifuged, washed with sterile PBS-EDTA (5mM) solution, and the cell pellet was suspended with Dulbecco’s Modified Eagle Medium (Nutrient Mixture F-12) (DMEM/F-12), 20mM HEPES (pH 7.4), 5% fetal bovine serum (FBS), and 0.06mM NaHCO3. Cells were seeded in 96-well-plates and treated with 10 µg/ml OM-85 concentrate (OM-Pharma) or 100 ηg/mL of recombinant human interferon beta (rhIFN-β) protein (#PHC4244, ThermoFisher Scientific), and with PBS as control, at BSL-3 level room. The concentration of 10 mg/ml of OM-85 was based on previous data with RSV we recently generated that maintain cell viability (4). After 24h, cells were collected for flow cytometry and gene expression analysis by qRT-PCR. We chose 24h of culture based on your previous study using the nasopharyngeal cells (10).
2.4 Flow cytometry
After ex-vivo treatment with OM-85 cells were stained with anti-human CD326 (EpCAM) FITC (1:200, # 324201, clone 9C4, BioLegend) followed by staining with anti-SARS-CoV-2 spike protein (1:100, # MA5-36247, clone HL6, Thermo Fisher Scientific) and secondary staining with anti-rabbit IgG H&L Cy3-conjugated (1:1000, # ab97075, Abcam) after FcBlock incubation. The cells were acquired at the FACS Canto II (BD Bioscience) the data were analyzed using FlowJo software (TreeStar).
2.5 Quantitative real-time PCR analysis
After ex-vivo treatment with OM-85, for viral gene expression, we used 2019-nCoV N1 and RNaseP primers and probes (QuatroG, #100199). We used following TaqMan primers and probes: (ACTB Hs03023943_g1(the housekeeping gene); IFNB1 Hs01077958_s1; IL1B Hs01555410_m1; IFIH1 Hs00223420_m1; DDX58 Hs01061436_m1; ISG15 Hs01921425_s1; OASL Hs00984387_m1; IL6 Hs00174131_m1; IL8 Hs00174103_m1; TNFA Hs00174128_m1. ACE2 Hs01085333_m1; TLR2 Hs02621280_s1; MAVS Hs00920075_m1; IRF3 Hs01547283_m1; TSLP Hs00263639_m1; MTOR Hs00234508_m1; IFNL1 Hs00601677_g1. ThermoFisher Scientific). PCR conditions followed the TaqMan™ Universal PCR Master Mix protocols (Thermo Fisher Scientific). Quantification of gene expression was conducted using StepOne™ (Applied Biosystems). The threshold cycle value (ΔCt) was obtained by subtracting the Ct value from the endogenous gene from the Ct value of the target gene. The gene expression was calculated according to the 2-ΔCt formula.
2.6 Enzyme-linked immunosorbent assays
The supernatant was collected for cytokine analysis. The following cytokines were measured by ELISA: IFN-β(#41415, pbl Assay Science), IL-6 (#KHC0061, Thermo Fisher Scientific), IL-8 (#KHC0081, Thermo Fisher Scientific), IL-10 (#BMS215-2, Thermo Fisher Scientific), and TNF-α (#KHC3011, Thermo Fisher Scientific). The assays were performed according to the manufacturer’s instructions and read at 490 nm wavelength, using an EZ Leia 400 microplate reader (Biochrom, Massachusetts, USA).
2.7 Statistical analysis
The data were summarized as mean and standard error of the mean (SEM). Univariate comparisons between groups were performed using the Mann-Whitney test, Wilcoxon test, and Kruskal-Wallis for multiple comparisons (with Dunn’s multiple comparison test as post hoc). The data were analyzed using IBM SPSS Statistics Version 25.0 (IBM Corp, NY, USA) and GraphPad Prism Version 8.0 (GraphPad Software, CA, USA).
3 Results
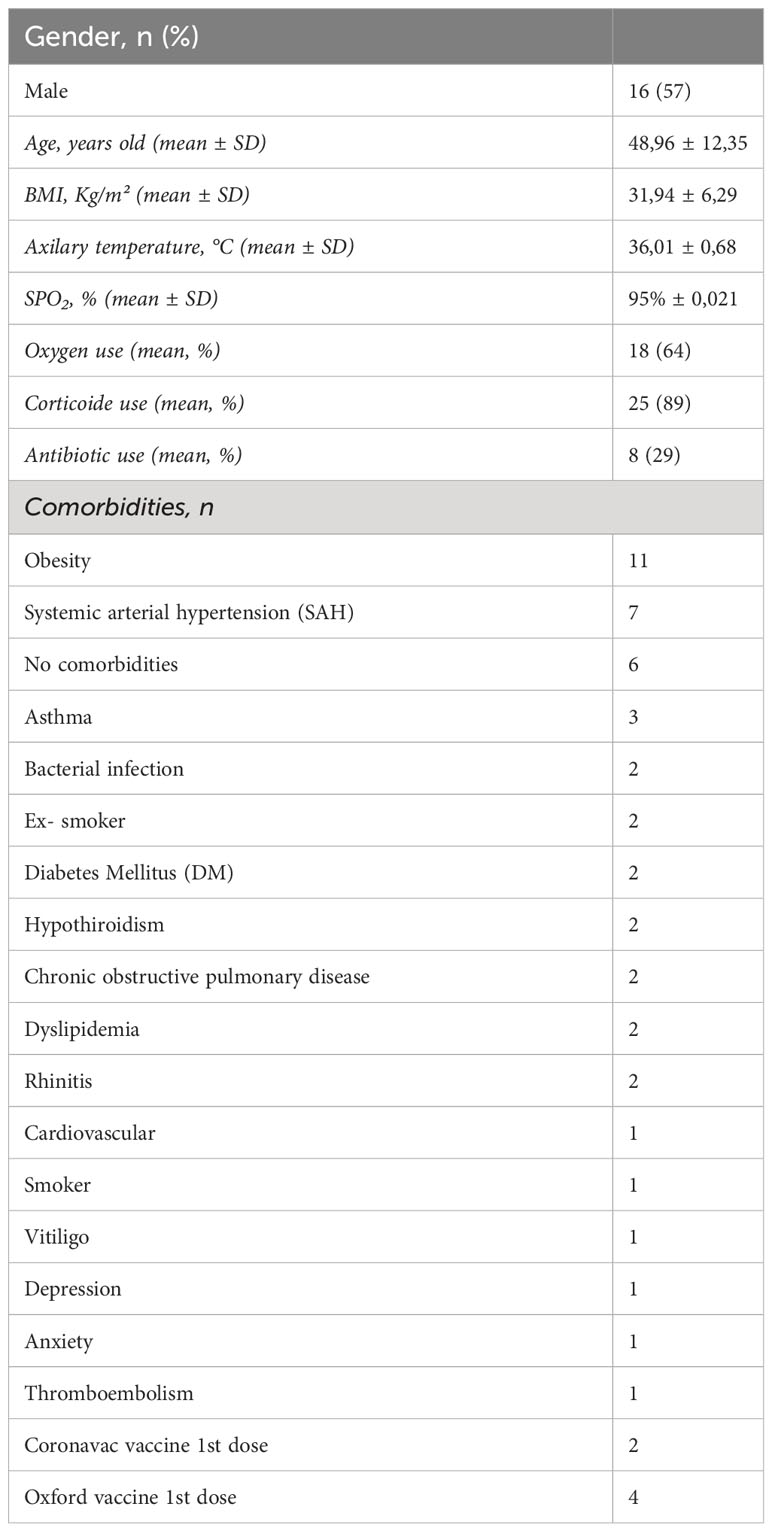
28 patients were enrolled to nasopharyngeal sample collection, 17 were males (57%), at a median age of 48.96 years (CI 95% 44.49-53.43); mean body mass index (BMI) 31.94Kg/m²; and 64% were on oxygen supplementation. Table 1 describes the clinical and demographic characteristics of the patients that the nasopharyngeal samples were collected. To first characterize our samples before treatment with OM-85, we used a combined analysis of flow cytometry and cytologic staining. We learned that the predominant cell population within our sample is CD45- cells (epithelial/stromal cells) and not hematopoietic immune cells (CD45+), similarly to we have previous published (9) (Supplementary Figure 1).
After OM-85 ex-vivo treatment the expression of SARS-CoV-2 spike protein on nasopharyngeal cells from COVID-19 patients is decreased (Figure 1A; Supplementary Figure 2). Interestingly, expression of viral N1 gene was significantly reduced in cells treated with OM-85 compared to untreated control (Figure 1B). The effect of OM-85 on SARS-CoV-2-infected human nasopharyngeal cells ex-vivo was similar to rhINF-β-treated cells (Figure 1C). To further examine these findings, we next analyzed the expression of antiviral response-associated genes, and their corresponding differential expression are shown in Figure 1D. OM-85 did neither modulated the expression of IFN-β (IFNB1 gene) (n=12; p=0.3804), nor the responsive genes ISG15 (n=26; p=0.7454) and MDA-5 (melanoma differentiation-associated protein 5) (IFIH1 gene) (n=12; p=0.5186). Considering its importance in the context of COVID-19, the Retinoic-acid inducible gene I (RIG-I) viral recognition pathway was also investigated and here, OM-85 treatment presented significantly two-fold increase of RIG-I expression (DDX58 gene) (n=8; p=0.0156). In contrast to RIG-I, OASL (2’-5’-oligoadenylate synthetase-like) (n=19; p=0.0289) had its expression significantly reduced after OM-85 treatment. There was no difference between genes associated with the inflammatory response, i.e., IL-6 (n=5; p=0.0625) and IL1-β (n=18; p=0.737). Gene expression of ACE-2, IL-8, MAVS, IRF-3, mTOR, IFN-λ, TRL2, and TLSP, were not detected in the samples, possibly due to the low expression of these genes by the time of harvesting. We did not find differences in cytokine levels (IFN-β, IL-6, IL-8, IL-10, and TNF-α) in the supernatant between untreated and treated cells (Table 2).
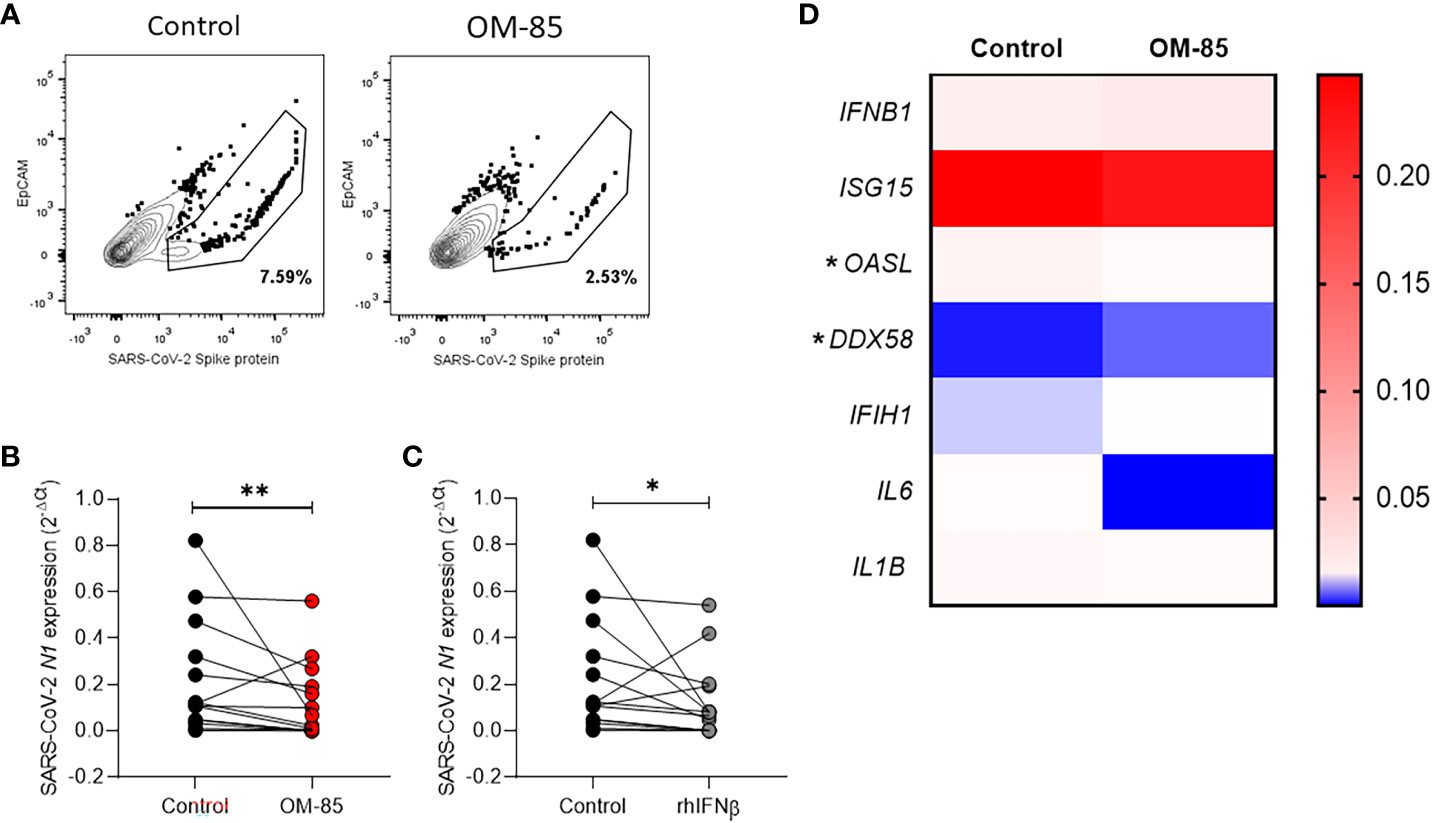
Figure 1 Ex-vivo OM-85 treatment on SARS-CoV-2 infected nasopharyngeal cells reduces the viral gene expression. Nasopharyngeal samples from adult hospitalized COVID-19 patients positive for SARS-CoV-2 were collected and processed to obtain the cells. Cells obtained from nasopharyngeal samples were culture and treated with OM-85 (10 µg/ml) or rhIFNβ (100 ηg/ml) and with PBS and control for 24h. (A) Cells were stained with anti-EpCAM (epithelial cells specific marker) and anti-SARS-CoV-2 Spike protein. The drawn gates show the abundance of Spike protein within the epithelial cell population. (B, C) Real-time PCR showing the SARS-CoV-2 N1 protein gene expression in nasopharyngeal samples treated with OM-85 or rhIFNβ for 24h. (D) Gene expression of cytokines and antiviral response-associated genes in nasopharyngeal samples treated with OM-85 for 24h. Data are presented as mean ± SEM. Statistical significance in (B, C) was determined by Wilcoxon test, and in (D) was determined by Mann-Whitney test *p <.05. **p <.01.
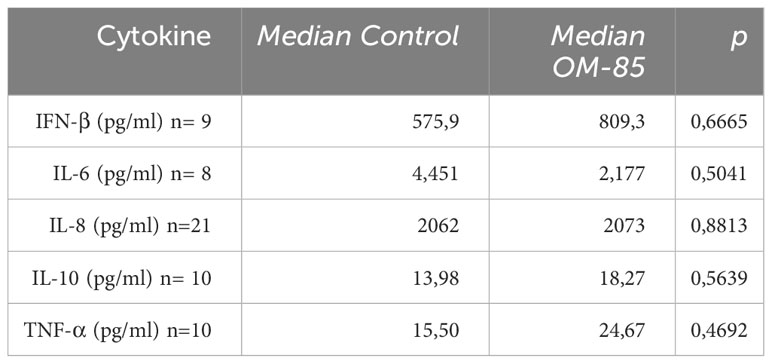
Table 2 Cytokine analysis in the supernatant of the treated nasopharyngeal cells measured by ELISA (Mann-Whitney test).
4 Discussion
Here we demonstrated that ex-vivo OM-85 treatment on SARS-CoV-2 infected nasopharyngeal cells reduced the viral gene expression. These data highlight a different therapeutic effect for OM-85 on the respiratory tract, a bacterial lysate that normally is used as a prophylactic postbiotic to prevent recurrent respiratory infection. While OM-85 did not significantly increase the production of IFN-β, in this time point analyzed, interestingly there was an increase in the expression of RIG-I and reduced the expression of OASL, confirming the modulation of the interferon pathway.
One strength of our study is that we tested the antiviral effect of OM-85 directly on airway cells freshly collected from COVID-19 patients. The epithelium in the upper respiratory tract is a frontline barrier in the battle against SARS-CoV-2, several studies have highlighted the importance of the analysis of samples from the airways to better characterize the direct mucosal impact of possible interventions on SARS-CoV-2 infection (11, 12).
Our results identified that RIG-I activation might be one of the pathways related to the OM-85 effect on nasopharyngeal cells during SARS-CoV-2 infection. RIG-I is activated by RNAs with 5′-(P)PP, duplex structure, and no ribose 2′-O-methylation found in several viral RNA molecules but are normally absent in the host cytosolic RNAs (13). After activation RIG-I undergoes conformational changes that allow the homotypic CARD-CARD domain to interact with MAVS. MAVS is anchored into mitochondria and induces the expression of type I interferons by signaling through TBK1 and IkB kinase ϵ, activating the transcriptional factors IRF3 and IRF7 and together with NF-κB. Antiviral responses associated with RIG-I activation are essential to control SARS-CoV-2 infection (14). The first step of replication is aborted since the interaction of RIG-I with the viral genome directly abrogates viral RNA-dependent RNA polymerase (14). Consequently, SARS-CoV-2 proteins have been described to inhibit antiviral responses by interacting with RIG-I (15, 16).
Efficient antiviral activity in the airway should point to a balanced immune response leading to viral clearance and faster patient recovery since severe viral respiratory infection is related to an aberrant immune response to uncontrolled inflammation. The role of type 1 interferon in COVID-19 severity is controversial (17). Although a large study conducted by WHO did not show the clinical benefit of subcutaneous interferon beta 1-a treatment on hospitalized COVID-19 patients (18), a phase 2 trial showed a promising result of inhaled nebulized interferon beta-1a (19). Another intervention with positive results by the inhalation route to COVID-19 patients is budesonide (20).
OM-85 can increase antiviral response inducing type 1 interferon, but is also associated with an anti-inflammatory response by increasing the levels of anti-inflammatory cytokines, such as IL-10, decreasing the levels of pro-inflammatory cytokines (IL-1β) and dampening recruitment of inflammatory cells, consequently reducing tissue damage. Other effects of OM-85 include increasing tolerogenic dendritic cells and Treg (21). These pointed out that OM-85 administration during infection boosts antiviral response, but also might control inflammatory milieu preventing tissue damage.
Our study suffers from limitations, one is associated with the number of cells obtained from the patients, limiting the analysis to a one-time point, and with one concentration of OM-85, and the second is the lack of the evaluation of OM-85 effect on virus replication on these cells. Although, previous studies have demonstrated that SARS-CoV-2 can replicate in the nasal epithelial cells (22) and after 24h of infection in cell lines (23).
In summary, we identified that OM-85 can have an antiviral effect on SARS-CoV-2-infected nasopharyngeal cells collected from COVID-19 patients. These findings highlight a potential therapeutic effect of OM-85 against viral infection on the respiratory tract, a tissue that could be directly accessed in subjects using the intranasal route. Proper human intervention studies are needed to confirm these findings so that OM-85 becomes an alternative viable therapy by intranasal route for patients with respiratory infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by CEP PUCRS CAAE no 30754220.3.0000.5336. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization, AD and RS. Formal analysis, GC, KA, JG, MD, BA. Investigation, GC, BA, LS, KA, MD and JG. Resources, PM, CB, LB. Writing—original draft preparation, GC and KA. Writing—review and editing, AD, CP, RS. Supervision, AD and RS. Project administration, AD. Funding acquisition, AD, CB, LB, PM and RS. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by FAPERGS COVID-19 20/2551-0000258-6, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001, OM-Pharma, and CNPq PQ 309400/2021-0. The funders were not involved in the study design, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
We thank Eduardo Pedraza for technical support.
Conflict of interest
Author CP is employed by the company OM-Pharma.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
The author(s) AD and CB declared that they were editorial board members of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1111619/full#supplementary-material
Supplementary Figure 1 | Characterization of cell population within the nasopharyngeal sample showing a predominance of epithelial cells by flow cytometry (A) gate strategy, (B) CD45- population (left panel), (C) cytological analysis (right panel).
Supplementary Figure 2 | Flow cytometry of the nasopharyngeal samples after treatment with OM-85 (10 µg/ml) for 24hs to analyze the expression of SARS-CoV-2 protein (A) gate strategy, (B) mean percentage of Spike protein and EpCAM positive cells from 2 patients.
References
1. Spacova I, De Boeck I, Bron PA, Delputte P, Lebeer S. Topical microbial therapeutics against respiratory viral infections. Trends Mol Med (2021) 27(6):538–53. doi: 10.1016/j.molmed.2021.03.009
2. Pasquali C, Salami O, Taneja M, Gollwitzer ES, Trompette A, Pattaroni C, et al. Enhanced mucosal antibody production and protection against respiratory infections following an orally administered bacterial extract. Front Med (Lausanne) (2014) 1:41. doi: 10.3389/fmed.2014.00041
3. Fang L, Zhou L, Tamm M, Roth M. OM-85 broncho-vaxom. Biomedicines (2021) 9(11). doi: 10.3390/biomedicines9111544
4. Antunes KH, Cassão G, Santos LD, Borges SG, Poppe J, Gonçalves JB, et al. Airway administration of bacterial lysate OM-85 protects mice against respiratory syncytial virus infection. Front Immunol (2022) 13:867022. doi: 10.3389/fimmu.2022.867022
5. Pivniouk V, Pivniouk O, DeVries A, Uhrlaub JL, Michael A, Pivniouk D, et al. The OM-85 bacterial lysate inhibits SARS-CoV-2 infection of epithelial cells by downregulating SARS-CoV-2 receptor expression. J Allergy Clin Immunol (2022) 149(3):923–33.e6. doi: 10.1016/j.jaci.2021.11.019
6. De Benedetto F, Sevieri G. Prevention of respiratory tract infections with bacterial lysate OM-85 bronchomunal in children and adults: a state of the art. Multidiscip Respir Med (2013) 8(1):33.
7. Rossi GA, Bessler W, Ballarini S, Pasquali C. Evidence that a primary anti-viral stimulation of the immune response by OM-85 reduces susceptibility to a secondary respiratory bacterial infection in mice. Ital J Pediatr (2018) 44(1):112. doi: 10.1186/s13052-018-0569-7
8. Salzmann M, Haider P, Kaun C, Brekalo M, Hartmann B, Lengheimer T, et al. Innate immune training with bacterial extracts enhances lung macrophage recruitment to protect from betacoronavirus infection. J Innate Immun (2021), 1–13.
9. Antunes KH, de Souza APD. Isolation and cell culture of human nasopharyngeal cells: A model for testing immune response and antiviral treatment. Methods Mol Biol (2022) 2511:367–73. doi: 10.1007/978-1-0716-2395-4_28
10. Antunes KH, Stein RT, FrancesChina C, da Silva EF, de Freitas DN, Silveira J, et al. Short-chain fatty acid acetate triggers antiviral response mediated by RIG-I in cells from infants with respiratory syncytial virus bronchiolitis. EBioMedicine (2022) 77:103891. doi: 10.1016/j.ebiom.2022.103891
12. Lopez J, Mommert M, Mouton W, Pizzorno A, Brengel-Pesce K, Mezidi M, et al. Early nasal type I IFN immunity against SARS-CoV-2 is compromised in patients with autoantibodies against type I IFNs. J Exp Med (2021) 218(10).
13. Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol (2020) 20(9):537–51.
14. Yamada T, Sato S, Sotoyama Y, Orba Y, Sawa H, Yamauchi H, et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat Immunol (2021) 22(7):820–8.
15. Chen X, Saccon E, Appelberg KS, Mikaeloff F, Rodriguez JE, Vinhas BS, et al. Type-I interferon signatures in SARS-CoV-2 infected Huh7 cells. Cell Death Discov (2021) 7(1):114.
16. Han L, Zhuang MW, Deng J, Zheng Y, Zhang J, Nan ML, et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J Med Virol (2021) 93(9):5376–89. doi: 10.1002/jmv.27050
17. da Silva RP, Gonçalves JIB, Zanin RF, Schuch FB, de Souza APD. Circulating type I interferon levels and COVID-19 severity: A systematic review and meta-analysis. Front Immunol (2021) 12:657363. doi: 10.3389/fimmu.2021.657363
18. Pan H, Peto R, Henao-Restrepo AM, Preziosi MP, Sathiyamoorthy V, Abdool Karim Q, et al. Repurposed antiviral drugs for covid-19 - interim WHO solidarity trial results. N Engl J Med (2021) 384(6):497–511.
19. Monk PD, Marsden RJ, Tear VJ, Brookes J, Batten TN, Mankowski M, et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med (2021) 9(2):196–206. doi: 10.1016/S2213-2600(20)30511-7
20. Yu LM, Bafadhel M, Dorward J, Hayward G, Saville BR, Gbinigie O, et al. Inhaled budesonide for COVID-19 in people at high risk of complications in the community in the UK (PRINCIPLE): a randomised, controlled, open-label, adaptive platform trial. Lancet (2021) 398(10303):843–55. doi: 10.1016/S0140-6736(21)01744-X
21. Esposito S, Cassano M, Cutrera R, Menzella F, Varricchio A, Uberti M. Expert consensus on the role of OM-85 in the management of recurrent respiratory infections: A Delphi study. Hum Vaccin Immunother (2022), 2106720. doi: 10.1080/21645515.2022.2106720
22. Ahn JH, Kim J, Hong SP, Choi SY, Yang MJ, Ju YS, et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J Clin Invest (2021) 131(13). doi: 10.1172/JCI148517
Keywords: COVID-19, SARS-CoV-2, OM-85, nasopharyngeal, RIG-I, epithelial cells
Citation: Cassão G, Antunes KH, Gonçalvez JIB, Santos LD, Abbadi BL, Bizarro CV, Machado P, Basso LA, Pasquali C, Stein RT and de Souza APD (2023) Brief research report: OM-85 reduces SARS-COV-2 viral RNA expression in nasopharyngeal cells from COVID-19 patients. Front. Virol. 3:1111619. doi: 10.3389/fviro.2023.1111619
Received: 29 November 2022; Accepted: 06 October 2023;
Published: 26 October 2023.
Edited by:
Muhammad Atif Zahoor, University Health Network (UHN), CanadaReviewed by:
Martina Kovarova, University of North Carolina at Chapel Hill, United StatesLuis Diambra, National University of La Plata, Argentina
Copyright © 2023 Cassão, Antunes, Gonçalvez, Santos, Abbadi, Bizarro, Machado, Basso, Pasquali, Stein and de Souza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Paula Duarte de Souza, ana.duarte@pucrs.br
 Gisele Cassão
Gisele Cassão Krist Helen Antunes
Krist Helen Antunes João Ismael Budelon Gonçalvez
João Ismael Budelon Gonçalvez Leonardo Duarte Santos
Leonardo Duarte Santos Bruno Lopes Abbadi
Bruno Lopes Abbadi Cristiano Valim Bizarro
Cristiano Valim Bizarro Pablo Machado
Pablo Machado Luiz Augusto Basso
Luiz Augusto Basso Christian Pasquali
Christian Pasquali Renato T. Stein
Renato T. Stein Ana Paula Duarte de Souza
Ana Paula Duarte de Souza