Processing of the Hepatitis E virus ORF1 nonstructural polyprotein
- 1Agharkar Research Institute, Nanobioscience Group, Pune, India
- 2Savitribai Phule Pune University, Pune, India
Hepatitis E viruses (HEV) Open Reading Frame 1 (ORF1) encodes a non-structural polyprotein. In most positive-sense RNA viruses found in animals, this non-structural polyprotein is cleaved by viral protease or host protease. However, the mechanism behind the processing of HEV polyprotein remains one of the most controversial questions in HEV biology. The role of putative HEV protease in processing is difficult to demonstrate. Recent studies have questioned the existence of HEV protease and suggested that pORF1 lacks protease activity. Conversely, studies also suggested the role of host proteases involved in the blood coagulation cascade, like thrombin, in processing the HEV pORF1 polyprotein. In summary, recent studies support the notion that pORF1 lacks protease activity and host proteases are responsible for processing pORF1. The present review compiles a thorough overview of contentious research on HEV’s papain-like cysteine protease (PCP) and highlights recent advancements in the field. We aim to discuss the challenges and opportunities in the field to focus on further research.
Introduction
Hepatitis E virus (HEV) is a single-stranded positive-sense virus belonging to the family Hepeviridae (1). The virus is further sub-classified into eight distinct genotypes (1). Among these genotypes, five (genotypes 1,2,3,4 and 5) have been identified as pathogenic to humans. HEV-genotype 1 (HEV-1) and HEV-genotype 2 (HEV-2) are responsible for human acute infections and are primarily transmitted via the fecal-oral route. Conversely, HEV-3 and HEV-4 genotypes are prevalent in developed countries and cause sporadic infections through zoonotic transmission. HEV-3 mainly circulates among wild boars, pigs, and rabbits, whereas HEV-4 is primarily found in pigs. HEV-7 has been shown to circulate in camels; however, a report also suggested possible infection in humans (2). HEV can lead to chronic infections, especially in immunocompromised individuals, with HEV-3 being responsible. Furthermore, HEV-1 infections in pregnant women can result in severe disease, characterized by mortality rates of up to 30% (3, 4).
HEV is a non-enveloped, single-stranded positive-sense RNA virus with a genome of 7.2 kb (5). The viral genomic RNA features a 7-methylguanosine RNA cap at the 5’ end and a polyadenylation tail at the 3’ end (6–8). The HEV genome contains three main open reading frames (ORFs): ORF1, ORF2, and ORF3. The genome is flanked by untranslated regions (UTRs) (9) (Figure 1). ORF1 encodes for non-structural proteins, which primarily play a role in RNA genome replication (6, 10, 11). ORF2 encodes for structural glycoprotein (12–15), while ORF3 encodes for a small multifunctional protein involved in many proviral processes (16–29). In addition, Apart from these conserved ORFs, HEV-1 possesses an additional ORF, designated as ORF4, which responds to the endoplasmic reticulum (ER) stress (30).
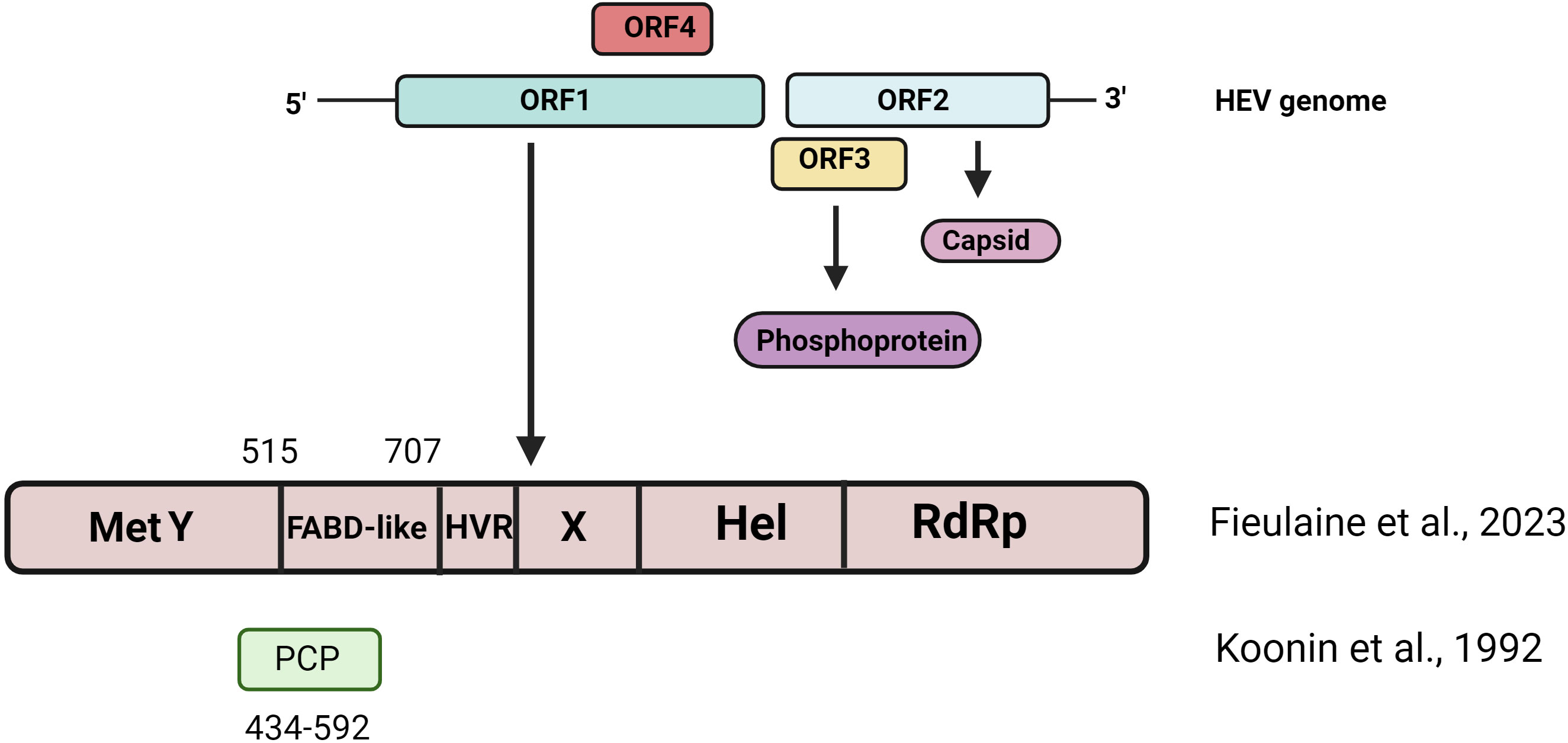
Figure 1 Schematic representation of Hepatitis E virus genome organization. HEV Genotype 1 encodes for 4 ORFs. The figure shows the domain organization originally proposed by Konnin et al., 1992 and the domain organization reported by recent computational biology studies (Fieulaine et al., 2023).
The process of HEV particle internalization into host cells remains unidentified (31). Similar to the typical positive-sense RNA viruses, translation is the initial event following HEV particle internalization. The viral genome serves as a template for translation, leading to the synthesis of a non-structural polyprotein. However, the precise mechanism of HEV pORF1 polyprotein processing and post-translational modification remains unclear. It is suggested that the viral RNA-dependent RNA polymerase (RdRp) domain is responsible for both the negative strand and sub-genomic RNA synthesis (6). Negative-sense RNA strands act as a template for synthesizing positive-sense genomic RNA, and sub-genomic RNA serves as a template for synthesizing pORF2 and pORF3 (10, 11, 32). Virus particles are believed to egress from infected cells via cellular exosomal pathway (33). HEV ORF1 encodes for a non-structural polyprotein comprising 5079 bases with a molecular mass 186-kDa (6). The pORF1 codes for non-structural functional domains, which are enzymes primarily responsible for RNA genome replication. According to bioinformatics analysis, pORF1 encompasses domains of methyl transferase, Y-domain, papain-like cysteine protease (PCP), proline-rich hinge domain, X-domain, RNA helicase, and RdRP (34). The biochemical activities of methyl transferase (35), helicase (36, 37), macro domain (38) and RdRp (39) have been experimentally demonstrated. However, activities associated with the HEV protease domain remain controversial. Despite numerous experimental studies, the primary predicted role of the protease domain in polyprotein processing remains mysterious.
Polyprotein processing in positive sense RNA viruses
In the typical life cycle of a positive-sense RNA virus, the translation of viral proteins is the first event. An intriguing feature is the compact nature of the viral genome, typically encoding just one or two ORFs (40). These ORFs encode multiple functional domains with distinct functions. Thus, proteins are translated as polyproteins and post-translationally proteolytically processed into smaller functional subunits. The strategy of polyprotein processing is a gene expression or regulation of the viral life cycle. In this type of post-translational modification, the structure, function, and biochemical activity of domains of protein subunits can be regulated. Polyprotein processing regulates the activities of different sets of enzymes with distinct activity and substrate requirements. And this regulation may be responsible for regulating viral transcription and translation. Typically, polyprotein processing is orchestrated by viral proteases’ limited and tightly regulated activity. In the case of non-structural polyproteins, they are processed into separate subunits of functional domains, including capping enzyme, helicase, protease, and RdRP (41). Polyprotein processing is well studied in various RNA virus families such as flavivirus, alphavirus, and calicivirus. However, in the case of HEV, the mechanism of processing of non-structural polyproteins remains elusive.
Alphaviruses encode for two ORFs; one encoding non-structural polyprotein is located at the 5’-end of the genome, and the ORF encoding structural genes is located at the 3’ end. The genomic RNA acts as a template for synthesizing non-structural proteins, which are processed by regulated proteolysis by viral proteases within the nsP2 domain (42). The process ensures temporal and spatial regulation of the transcription and genome regulation. During the alphavirus life cycle, sub-genomic RNA is synthesized, serving as a template for synthesizing c-terminal ORF, responsible for encoding structural proteins. The structural polyprotein undergoes proteolytic processing, combining autocatalytic activity within the capsid protein and cellular proteins. Host cell proteases, known as signalase proteins, and furin-like proteases are known to be involved in structural gene polyprotein processing (42, 43).
In the case of flaviviruses, a single long ORF produces a polyprotein, which is cleaved both co- and post-translationally into small functional proteins. The N-terminal one-fourth part of polyprotein encodes structural proteins, followed by the non-structural protein part. Polyprotein processing in flaviviruses involves host signal peptidases and the virus-encoded serine protease (44–46). Caliciviruses encode for three ORFs, and ORF1 encodes for non-structural polyproteins. The non-structural polyprotein is co-translationally processed by viral 3C-like cysteine proteinase. This proteinase activity generates non-structural proteins and capsid protein precursors (47).
These studies show the diversity of mechanisms involved in polyprotein processing, including processing by the host or virus-encoded proteases or a combination of both.
HEV protease domain and polyprotein processing
Proteases belong to a class of enzymes that catalyze the hydrolysis of peptide bonds within proteins or peptides (48, 49). Generally, proteases are classified based on catalytic residue types such as cysteine, serine, etc. (48, 49)Additionally, proteases that utilize metal ions for catalysis are known as metalloproteases. PCP represents a sub-class of cysteine proteases. These proteases contribute to diverse biological functions in all life forms, including viruses (50). HEV protease was identified as a putative papain-like protease due to its similarity with rubella virus protease in computer-assisted sequence alignments (34). In silico analysis identified various signature motifs of papain-like proteases in the HEV protease domain, such as papain-like beta-barrel fold, putative catalytic dyad (C434 and H443), putative Zinc binding domain, and disulfide bridges (51). Many researchers attempted to investigate HEV papain-like cysteine protease’s biochemical activity and role in polyprotein processing. The primary purpose of ORF1 expression studies in cell-free and prokaryotic systems was to study HEV protease’s cis- or trans-action on pORF1 polyprotein. However, none of the published studies has demonstrated HEV polyprotein processing in the cell-free system consisting of a coupled transcription-translation system with phage T7 polymerase in rabbit reticulocyte lysate (52). However, the processing status remained unclear when ORF1 was expressed in a bacterial system. In a study by Ansari et al., 2000, GST-tagged ORF1 protein was expressed in E. coli, while a prominent band of intact ORF1 protein in immunoblot analysis, smaller fragments of ORF1 were observed in the western blot, possibly due to protein degradation in E. coli (52). The challenge of expressing intact ORF1 protein in E. coli may be attributed to its larger size and codon usage pattern. A group studied pORF1 expression in insect cells using the baculovirus expression system, and ORF1 processing was studied using mass spectroscopy. This approach revealed nine ORF1 fragments, and the processing was inhibited when cells were treated with cysteine protease inhibitors (53).
Furthermore, studies have suggested atypical activities associated with the HEV protease domain. A study suggested that the HEV protease domain possesses chymotrypsin-like activity and processes the pORF1 and pORF2 (54). Another study by Karpe and Lole 2011 suggested that the HEV protease domain has deubiquitinating activity; however, its role in pORF1 polyprotein processing remained unestablished (55). This activity may contribute to regulating various signaling pathways and modulation of host immune pathways.
Many researchers have attempted to study ORF1 expression and processing in mammalian systems, falling into two categories: the plasmid-based expression of pORF1 in mammalian cells and the use of infectious replicon. The latter approach appears more relevant; however, both studies have yielded inconclusive results.
In a study, authors constructed a recombinant vaccinia virus vector expressing ORF1 and studied the expression of pORF1 in mammalian cells. In this study, no processing of ORF1 was observed; however, after extended incubation, processing of pORF1 was observed (two fragments, 107 kDa and 78 kDa) (56)]. However, when the predicted active site of protease was mutated with site-directed mutagenesis, it failed to stop the observed pORF1 processing, suggesting the possible involvement of host protease in this processing (56). On the contrary, a study that expressed genotype 1 and genotype 3 epitope-tagged pORF1 via plasmids in 293T cells did not show processing and observed an intact band of pORF1 in western blots experiments (57). Yet another study expressed pORF1 in HeLa and Huh7 cells and demonstrated localization in ER membranes. However, the pulse-chase experiment with radioactive amino acids did not reveal polyprotein processing (58). Similarly, Ansari et al. expressed pORF1 in the human hepatoma cell line HepG2, but in immune precipitation experiments, no processing was observed (52). A subgenomic replicon expressing GFP of Sar55 Genotype 1 was used in a study. Site-directed mutagenesis was employed to alter cysteine and histidine residues within the putative protease domain,. Mutant replicons showed reduced GFP expression, suggesting that the protease domain plays a vital role in HEV replication. In the same study, epitope-tagged replicons were transfected into Huh7 cells and showed the processing of pORF1 into two fragments, 35 and 78 kDa (59). Similarly, a recent study demonstrated the importance of the protease domain in HEV replication by using mutagenesis; however, this study supports the notion that pORF1 operates as a multifunctional unprocessed polyprotein (60). A functional ORF1 trans-complementation system was used to investigate HEV replication; this system showed an intact band of pORF1, suggesting the possibility that ORF1 functions as an unprocessed polyprotein (61, 62). Also, replicons expressing recombinant HEV with HA-tag were used in a study, and an intact band of pORF1 was observed (63). In contrast, the V5 epitope-tagged full-length infectious replicon system in Huh7.5 cells showed an intact band of pORF1 along with fragments of lower molecular weight, indicating the possibility of pORF1 processing (64). All these studies have yielded varied results and remain inconclusive, possibly due to factors like non-specific action of host proteases, improper protein folding, and lack of required host factor and host environment. Ideally, polyprotein processing should be studied in the context of virus replication, which may give a specific processing pattern of pORF1. However, more experiments are required to establish the processing and role of viral or host protease. These studies suggest several possibilities: 1) HEV pORF1 may not be processed at all and works are multifunctional polyprotein 2) HEV pORF1 does not have protease activity 3) Host proteases contribute to the processing, 4) Processing may be carried out in specific cellular microenvironment to a very limited extent, which is enough for virus replication.
Recently, Fieulaine et al., 2023, proposed an interesting opinion that the HEV community should stop calling the term “HEV protease” or “PCP-domain” based on computational and structural biology studies (65), while Koonin et al., 1992, suggested that HEV has a protease domain (amino acid residue 434-592); however, the confidence index for the putative PCP domain was very low compared to other HEV ORF1 domains (34). Recent studies involving X-ray crystallography and artificial intelligence (AI)-based computational studies have suggested that HEV ORF1 lacks protease activity. It is suggested that the 510-690 region is the Fatty Acid Binding-like domain (Figure 1). Recently, three independent groups used AI-based AlphaFold2 tools to predict ORF1 domains. All three studies support that HEV ORF1 lacks protease activity (60, 66, 67). It is suggested that pORF1 may not be processed, and the structural flexibility of different pORF1 domains may regulate the activities of different domains.
Role of thrombin in polyprotein processing
Recent reports suggested the possible role of coagulation proteases like thrombin and factor Xa in HEV polyprotein processing (68, 69). Thrombin, a serine protease, is composed of two polypeptide chains, namely A chain (36 amino acid residues) and B chain (259 amino acid residues) (70, 71). Thrombin plays a significant role in the blood coagulation pathway (72, 73). Its primary function is to cleave fibrinogen to form an insoluble fibrin clot (74). It can act as a procoagulant or anticoagulant (70). In normal physiological conditions, blood does not contain thrombin; however, after injury, thrombin is generated. Following vascular injury, tissue factor (TF) is exposed to blood and binds with FVIIa (75, 76). This interaction activates Factor X (FX) to form Factor Xa (FXa) (76). FXa cleaves prothrombin to form active thrombin (75, 76). Further, thrombin is a versatile enzyme and acts on various substrates. The primary function of thrombin is to convert fibrinogen to fibrin. In addition, thrombin activates platelets and various factors like FXIII, FV, FVIII, FXI, and TAFI. It also inactivates ADAMTS13 (77). In the anticoagulant phase, thrombin activates protein C and binds to antithrombin (78).
Earlier a study by Kanade et al., 2018 reported a possible role of factor Xa and thrombin in HEV polyprotein processing (68). They identified highly conserved thrombin and factor Xa sites on pORF1 (Figure 2). Subsequent experiments were performed in which the cleavage sites of thrombin and factor Xa were altered by site-directed mutagenesis on HEV sub-genomic replicons expressing the luciferase gene. These mutant replicons were transfected into permissive human hepatoma cell line Huh7-S10, and levels of luciferase were studied. Compared with the wild-type, mutant replicons significantly reduced luciferase counts. This suggests that intact thrombin and Factor Xa cleavage sites are required for HEV replication. HEV replication was significantly reduced in cells transfected with siRNA targeting thrombin and factor Xa. This observation proved that thrombin and factor Xa are essential for HEV replication. When thrombin and factor Xa inhibitors were treated on cells transfected with the capped transcript of HEV replicons, the viral replication was significantly reduced. Hence, these experiments collectively showed a significant role of thrombin and factor Xa in HEV replication (68). This study demonstrated the insights into the intracellular function of blood coagulation proteases.
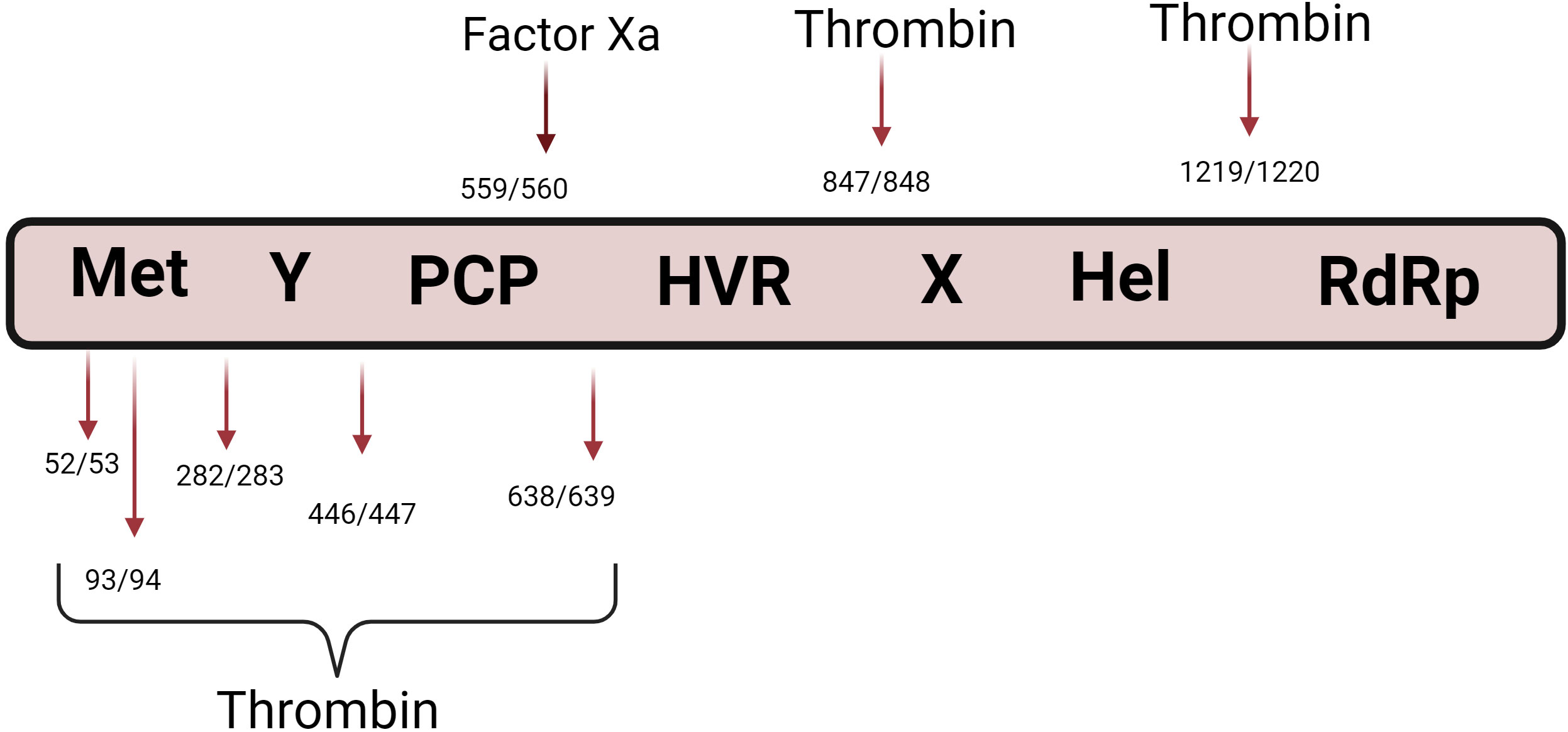
Figure 2 Role of blood coagulation proteases in HEV polyprotein processing. The figure shows the pORF1 domain organization Two independent studies by Kanade et al., 2018 and Pierce et al., 2023, reported the possible role of thrombin in HEV polyprotein processing. The figure shows the reported thrombin and Factor Xa cleavage sites on pORF1.
A recent study demonstrated seven potential thrombin cleavage sites within pORF1 (69) (Figure 2). To demonstrate the in vitro pORF1 processing, authors generated pORF1 fragments using in vitro coupled transcription and translation, and purified thrombin was added to the reactions. Authors have demonstrated the processing of pORF1 polyprotein upon the addition of thrombin. Results were further confirmed by mutagenesis of thrombin cleavage sites on pORF1 fragments. These mutant proteins were subjected to thrombin cleavage, and no processing of pORF1 fragments was observed. The authors corroborated their findings using G1 and G3 replicons expressing nano-luciferase (nLuc) instead of structural genes. In these replicons, the thrombin cleavage sites were altered by site-directed mutagenesis, and replication mutant replicons were analyzed. It was observed that the removal of thrombin cleavage significantly inhibits viral replication. Further, the authors studied the effect of serine protease inhibitors on virus replication and demonstrated that serine protease inhibitors inhibit virus replication (69). Similar to the study by Kanade et al., the authors also demonstrated the presence of intracellular thrombin (68). Using fluorescence microscopy, authors also demonstrated partial co-localization between thrombin/prothrombin and pORF1 proteins (69).
Unanswered questions and prospects
Despite many studies, polyprotein processing remains unsolved. Ideally, HEV polyprotein processing should be studied in a permissive cell culture system, and results must be confirmed by reverse genetics and the biochemical function of pORF1 domains. However, the lack of robust and accessible cell culture systems remains the most challenging in HEV replication studies. Recent structural and computational biology studies support the notion that HEV pORF1 lacks the PCP domain; instead, it has a Fatty acid binding domain (FABD)-like domain. This is an exciting development in the biology of HEV. The role of the FABD-like domain in HEV replication and pathogenesis is unknown. Conversely, recently, two studies (including ours) have supported the possible role of coagulation proteases in polyprotein processing (68, 69).
Many coagulation cascade proteins are synthesized in the liver, a primary site of HEV infection. Studies have implicated the roles of coagulation proteases like thrombin and Factor Xa in the HEV life cycle and possibly polyprotein processing. The studies conducted by Kanade et al., 2018 and Pierce et al., 2023 agree in principle (68, 69). However, further evidence is still required to prove thrombin’s direct role in pORF1 polyprotein processing. Also, whether pORF1 sub-units generated from thrombin cleavage are biochemically active or form the virus replication complex is unknown. Pierce et al., 2023 also suggested that along with thrombin, other host serine proteases like hespin in the endoplasmic reticulum may contribute to polyprotein processing. Previously, the pORF1 domain structure was studied by in silico modeling (60, 66, 79). Pierce et al., 2023 suggested that some thrombin cleavage sites were matching with some of the in silico predictions (69). Interestingly, thrombin is present across all mammals, and a broad host range of HEV supports this notion.
Interestingly, this study demonstrated the intracellular function of blood coagulation proteases. Thus, it would be interesting to study the roles of intracellular coagulation proteases. It is unlikely that the thrombin present in the Huh7 cell is generated from a classical coagulation cascade. Further studies are needed to study the synthesis and secretion of intracellular coagulation proteases in HEV-infected liver cells, animal models, and human patient samples. In summary, recent studies suggest that HEV pORF1 lacks protease activity, and possibly, host proteases may carry out processing.
Author contributions
YK: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors are thankful to the Director, Agharkar Research Institute, Pune for all the support. The figures were created using BioRender.com.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Smith DB, Izopet J, Nicot F, Simmonds P, Jameel S, Meng X-J, et al. Update: proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J Gen Virol (2020) 101:692–8. doi: 10.1099/jgv.0.001435
2. Lee G-H, Tan B-H, Chi-Yuan Teo E, Lim S-G, Dan Y-Y, Wee A, et al. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology (2016) 150:355–357.e3. doi: 10.1053/j.gastro.2015.10.048
3. Khuroo MS, Kamili S. Aetiology, clinical course and outcome of sporadic acute viral hepatitis in pregnancy*. J Viral Hepatitis (2003) 10:61–9. doi: 10.1046/j.1365-2893.2003.00398.x
4. Wu C, Wu X, Xia J. Hepatitis E virus infection during pregnancy. Virol J (2020) 17:73. doi: 10.1186/s12985-020-01343-9
5. Smith DB, Simmonds P, Jameel S, Emerson SU, Harrison TJ, Meng X-J, et al. Consensus proposals for classification of the family Hepeviridae. J Gen Virol (2014) 95:2223–32. doi: 10.1099/vir.0.068429-0
6. Wang B, Meng X-J. Structural and molecular biology of hepatitis E virus. Comput Struct Biotechnol J (2021) 19:1907–16. doi: 10.1016/j.csbj.2021.03.038
7. Emerson SU, Zhang M, Meng X-J, Nguyen H, St. Claire M, Govindarajan S, et al. Recombinant hepatitis E virus genomes infectious for primates: Importance of capping and discovery of a cis-reactive element. Proc Natl Acad Sci (2001) 98:15270–5. doi: 10.1073/pnas.251555098
8. Huang YW, Haqshenas G, Kasorndorkbua C, Halbur PG, Emerson SU, Meng XJ. Capped RNA Transcripts of Full-Length cDNA Clones of Swine Hepatitis E Virus Are Replication Competent When Transfected into Huh7 Cells and Infectious When Intrahepatically Inoculated into Pigs. J Virol (2005) 79:1552–8. doi: 10.1128/jvi.79.3.1552-1558.2005
9. Purdy MA, Harrison TJ, Jameel S, Meng X-J, Okamoto H, van der Poel WHM, et al. ICTV virus taxonomy profile: hepeviridae. J Gen Virol (2017) 98:2645–6. doi: 10.1099/jgv.0.000940
10. Kenney SP, Meng X-J. Hepatitis E virus genome structure and replication strategy. Cold Spring Harbor Perspect Med (2018) 9:a031724. doi: 10.1101/cshperspect.a031724
11. LeDesma R, Nimgaonkar I, Ploss A. Hepatitis E virus replication. Viruses (2019) 11:719. doi: 10.3390/v11080719
12. Shiota T, Li T-C, Yoshizaki S, Kato T, Wakita T, Ishii K. The hepatitis E virus capsid C-terminal region is essential for the viral life cycle: implication for viral genome encapsidation and particle stabilization. J Virol (2013) 87:6031–6. doi: 10.1128/jvi.00444-13
13. Surjit M, Jameel S, Lal SK. Cytoplasmic localization of the ORF2 protein of hepatitis E virus is dependent on its ability to undergo retrotranslocation from the endoplasmic reticulum. J Virol (2007) 81:3339–45. doi: 10.1128/jvi.02039-06
14. Montpellier C, Wychowski C, Sayed IM, Meunier J-C, Saliou J-M, Ankavay M, et al. Hepatitis E virus lifecycle and identification of 3 forms of the ORF2 capsid protein. Gastroenterology (2018) 154:211–223.e8. doi: 10.1053/j.gastro.2017.09.020
15. Yin X, Ying D, Lhomme S, Tang Z, Walker CM, Xia N, et al. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc Natl Acad Sci (2018) 115:4773–8. doi: 10.1073/pnas.1721345115
16. Zafrullah M, Ozdener MH, Panda SK, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol (1997) 71:9045–53. doi: 10.1128/jvi.71.12.9045-9053.1997
17. Kar-Roy A, Korkaya H, Oberoi R, Lal SK, Jameel S. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J Biol Chem (2004) 279:28345–57. doi: 10.1074/jbc.m400457200
18. Graff J, Nguyen H, Yu C, Elkins WR, St. Claire M, Purcell RH, et al. The open reading frame 3 gene of hepatitis E virus contains a cis -reactive element and encodes a protein required for infection of macaques. J Virol (2005) 79:6680–9. doi: 10.1128/jvi.79.11.6680-6689.2005
19. Kannan H, Fan S, Patel D, Bossis I, Zhang Y-J. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol (2009) 83:6375–82. doi: 10.1128/jvi.02571-08
20. Moin SM, Chandra V, Arya R, Jameel S. The hepatitis E virus ORF3 protein stabilizes HIF-1α and enhances HIF-1-mediated transcriptional activity through p300/CBP. Cell Microbiol (2009) 11:1409–21. doi: 10.1111/j.1462-5822.2009.01340.x
21. Moin SM, Panteva M, Jameel S. The hepatitis E virus orf3 protein protects cells from mitochondrial depolarization and death. J Biol Chem (2007) 282:21124–33. doi: 10.1074/jbc.m701696200
22. Korkaya H, Jameel S, Gupta D, Tyagi S, Kumar R, Zafrullah M, et al. The ORF3 protein of hepatitis E virus binds to src homology 3 domains and activates MAPK. J Biol Chem (2001) 276:42389–400. doi: 10.1074/jbc.m101546200
23. Chandra V, Kalia M, Hajela K, Jameel S. The ORF3 protein of hepatitis E virus delays degradation of activated growth factor receptors by interacting with CIN85 and blocking formation of the cbl-CIN85 complex. J Virol (2010) 84:3857–67. doi: 10.1128/jvi.01994-09
24. Emerson SU, Nguyen HT, Torian U, Burke D, Engle R, purcell RH. Release of genotype 1 hepatitis E virus from cultured hepatoma and polarized intestinal cells depends on open reading frame 3 protein and requires an intact PXXP motif. J Virol (2010) 84:9059–69. doi: 10.1128/jvi.00593-10
25. Nagashima S, Takahashi M, Jirintai S, Tanaka T, Nishizawa T, Yasuda J, et al. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J Gen Virol (2011) 92:2838–48. doi: 10.1099/vir.0.035378-0
26. Kenney SP, Wentworth JL, Heffron CL, Meng X-J. Replacement of the hepatitis E virus ORF3 protein PxxP motif with heterologous late domain motifs affects virus release via interaction with TSG101. Virology (2015) 486:198–208. doi: 10.1016/j.virol.2015.09.012
27. He M, Wang M, Huang Y, Peng W, Zheng Z, Xia N, et al. The ORF3 protein of genotype 1 hepatitis E virus suppresses TLR3-induced NF-κB signaling via TRADD and RIP1. Sci Rep (2016) 6:27597. doi: 10.1038/srep27597
28. Ding Q, Heller B, Capuccino JMV, Song B, Nimgaonkar I, Hrebikova G, et al. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci (2017) 114:1147–52. doi: 10.1073/pnas.1614955114
29. Gouttenoire J, Pollán A, Abrami L, Oechslin N, Mauron J, Matter M, et al. Palmitoylation mediates membrane association of hepatitis E virus ORF3 protein and is required for infectious particle secretion. PloS Pathog (2018) 14:e1007471. doi: 10.1371/journal.ppat.1007471
30. Nair VP, Anang S, Subramani C, Madhvi A, Bakshi K, Srivastava A, et al. Endoplasmic reticulum stress induced synthesis of a novel viral factor mediates efficient replication of genotype-1 hepatitis E virus. PloS Pathog (2016) 12:e1005521. doi: 10.1371/journal.ppat.1005521
31. Kalia M, Chandra V, Rahman SA, Sehgal D, Jameel S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol (2009) 83:12714–24. doi: 10.1128/jvi.00717-09
32. Williams TPE, Kasorndorkbua C, Halbur PG, Haqshenas G, Guenette DK, Toth TE, et al. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J Clin Microbiol (2001) 39:3040–6. doi: 10.1128/jcm.39.9.3040-3046.2001
33. Nagashima S, Jirintai S, Takahashi M, Kobayashi T, Tanggis, Nishizawa T, et al. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol (2014) 95:2166–75. doi: 10.1099/vir.0.066910-0
34. Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci (1992) 89:8259–63. doi: 10.1073/pnas.89.17.8259
35. Magden J, Takeda N, Li T, Auvinen P, Ahola T, Miyamura T, et al. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol (2001) 75:6249–55. doi: 10.1128/jvi.75.14.6249-6255.2001
36. Karpe YA, Lole KS. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol (2010) 84:3595–602. doi: 10.1128/jvi.02130-09
37. Karpe YA, Lole KS. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol (2010) 84:9637–41. doi: 10.1128/jvi.00492-10
38. Parvez MK. The hepatitis E virus ORF1 ‘X-domain’ residues form a putative macrodomain protein/Appr-1″-pase catalytic-site, critical for viral RNA replication. Gene (2015) 566:47–53. doi: 10.1016/j.gene.2015.04.026
39. Mahilkar S, Paingankar MS, Lole KS. Hepatitis E virus RNA-dependent RNA polymerase: RNA template specificities, recruitment and synthesis. J Gen Virol (2016) 97:2231–42. doi: 10.1099/jgv.0.000528
40. Ahlquist P, Noueiry AO, Lee W-M, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol (2003) 77:8181–6. doi: 10.1128/jvi.77.15.8181-8186.2003
41. Spall VE, Shanks M, Lomonossoff GP. Polyprotein processing as a strategy for gene expression in RNA viruses. Semin Virol (1997) 8:15–23. doi: 10.1006/smvy.1997.0102
42. Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol (2009) 4:837–56. doi: 10.2217/fmb.09.59
43. Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci (2012) 109:16534–9. doi: 10.1073/pnas.1210418109
44. van den Elsen K, Quek JP, Luo D. Molecular insights into the flavivirus replication complex. Viruses (2021) 13:956. doi: 10.3390/v13060956
45. Shiryaev SA, Strongin AY. Structural and functional parameters of the flaviviral protease: a promising antiviral drug target. Future Virol (2010) 5:593–606. doi: 10.2217/fvl.10.39
46. Fishburn AT, Pham OH, Kenaston MW, Beesabathuni NS, Shah PS. Let’s get physical: flavivirus-host protein–protein interactions in replication and pathogenesis. Front Microbiol (2022) 13:847588. doi: 10.3389/fmicb.2022.847588
47. Sosnovtseva SA, Sosnovtsev SV, Green KY. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J Virol (1999) 73:6626–33. doi: 10.1128/jvi.73.8.6626-6633.1999
48. Page MJ, Di Cera E. Evolution of peptidase diversity. J Biol Chem (2008) 283:30010–4. doi: 10.1074/jbc.m804650200
49. Rawlings ND, Bateman A. Origins of peptidases. Biochimie (2019) 166:4–18. doi: 10.1016/j.biochi.2019.07.026
50. Ozhelvaci F, Steczkiewicz K. Identification and classification of papain-like cysteine proteinases. J Biol Chem (2023) 299:104801. doi: 10.1016/j.jbc.2023.104801
51. Parvez MK, Khan AA. Molecular modeling and analysis of hepatitis E virus (HEV) papain-like cysteine protease. Virus Res (2014) 179:220–4. doi: 10.1016/j.virusres.2013.11.016
52. Ansari IH, Nanda SK, Durgapal H, Agrawal S, Mohanty SK, Gupta D, et al. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1). J Med Virol (2000) 60:275–83.
53. Sehgal D, Thomas S, Chakraborty M, Jameel S. Expression and processing of the Hepatitis E virus ORF1 nonstructural polyprotein. Virol J (2006) 3:38. doi: 10.1186/1743-422x-3-38
54. Paliwal D, Panda SK, Kapur N, Varma SPK, Durgapal H. Hepatitis E virus (HEV) protease: a chymotrypsin-like enzyme that processes both non-structural (pORF1) and capsid (pORF2) protein. J Gen Virol (2014) 95:1689–700. doi: 10.1099/vir.0.066142-0
55. Karpe YA, Lole KS. Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J Gen Virol (2011) 92:2088–92. doi: 10.1099/vir.0.033738-0
56. Ropp SL, Tam AW, Beames B, Purdy M, Frey TK. Expression of the hepatitis E virus ORF1. Arch Virol (2000) 145:1321–37. doi: 10.1007/s007050070093
57. Suppiah S, Zhou Y, Frey TK. Lack of processing of the expressed ORF1 gene product of hepatitis E virus. Virol J (2011) 8:245. doi: 10.1186/1743-422x-8-245
58. Perttilä J, Spuul P, Ahola T. Early secretory pathway localization and lack of processing for hepatitis E virus replication protein pORF1. J Gen Virol (2013) 94:807–16. doi: 10.1099/vir.0.049577-0
59. Parvez MK. Molecular characterization of hepatitis E virus ORF1 gene supports a papain-like cysteine protease (PCP)-domain activity. Virus Res (2013) 178:553–6. doi: 10.1016/j.virusres.2013.07.020
60. LeDesma R, Heller B, Biswas A, Maya S, Gili S, Higgins J, et al. Structural features stabilized by divalent cation coordination within hepatitis E virus ORF1 are critical for viral replication. eLife (2023) 12:e80529. doi: 10.7554/elife.80529
61. Ju X, Xiang G, Gong M, Yang R, Qin J, Li Y, et al. Identification of functional cis-acting RNA elements in the hepatitis E virus genome required for viral replication. PloS Pathog (2020) 16:e1008488. doi: 10.1371/journal.ppat.1008488
62. Ding Q, Nimgaonkar I, Archer NF, Bram Y, Heller B, Schwartz RE, et al. Identification of the intragenomic promoter controlling hepatitis E virus subgenomic RNA transcription. mBio (2018) 9(3):e00769–18. doi: 10.1128/mBio.00769-18
63. Szkolnicka D, Pollan A, Da Silva N, Oechslin N, Gouttenoire J, Moradpour D. Recombinant hepatitis E viruses harboring tags in the ORF1 protein. J Virol (2019) 93(19):e00459–19. doi: 10.1128/JVI.00459-1964
64. Metzger K, Bentaleb C, Hervouet K, Alexandre V, Montpellier C, Saliou J-M, et al. Processing and subcellular localization of the hepatitis E virus replicase: identification of candidate viral factories. Front Microbiol (2022) 13:828636. doi: 10.3389/fmicb.2022.828636
65. Fieulaine S, Tubiana T, Bressanelli S. Hepatitis E virus RNA replication polyprotein: taking structural biology seriously. Front Microbiol (2023) 14:1254741. doi: 10.3389/fmicb.2023.1254741
66. Fieulaine S, Tubiana T, Bressanelli S. De novo modelling of HEV replication polyprotein: Five-domain breakdown and involvement of flexibility in functional regulation. Virology (2023) 578:128–40. doi: 10.1016/j.virol.2022.12.002
67. Goulet A, Cambillau C, Roussel A, Imbert I. Structure prediction and analysis of hepatitis E virus non-structural proteins from the replication and transcription machinery by alphaFold2. Viruses (2022) 14(7):1537. doi: 10.3390/v14071537
68. Kanade GD, Pingale KD, Karpe YA. Activities of thrombin and factor xa are essential for replication of hepatitis E virus and are possibly implicated in ORF1 polyprotein processing. J Virol (2018) 92(6):e01853–17. doi: 10.1128/jvi.01853-17
69. Pierce DM, Buchanan FJT, Macrae FL, Mills JT, Cox A, Abualsaoud KM, et al. Thrombin cleavage of the hepatitis E virus polyprotein at multiple conserved locations is required for genome replication. PloS Pathog (2023) 19:e1011529. doi: 10.1371/journal.ppat.1011529
71. Papaconstantinou ME, Bah A, Di Cera E. Role of the A chain in thrombin function. Cell Mol Life Sci (2008) 65:1943–7. doi: 10.1007/s00018-008-8179-y
73. Palta S, Saroa R, Palta A. Overview of the coagulation system. Indian J Anaesthesia (2014) 58:515. doi: 10.4103/0019-5049.144643
74. Wolberg AS, Campbell RA. Thrombin generation, fibrin clot formation and hemostasis. Transfusion Apheresis Sci (2008) 38:15–23. doi: 10.1016/j.transci.2007.12.005
75. McVey JH. 3 Tissue factor pathway. Baillière’s Clin Haematology (1994) 7:469–84. doi: 10.1016/s0950-3536(05)80094-0
76. Smith SA, Travers RJ, Morrissey JH. How it all starts: Initiation of the clotting cascade. Crit Rev Biochem Mol Biol (2015) 50:326–36. doi: 10.3109/10409238.2015.1050550
77. Crawley JTB, Lam JK, Rance JB, Mollica LR, O’Donnell JS, Lane DA. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood (2005) 105:1085–93. doi: 10.1182/blood-2004-03-1101
78. Esmon CT. The normal role of Activated Protein C in maintaining homeostasis and its relevance to critical illness. Crit Care (2001) 5:S7. doi: 10.1186/cc1333
Keywords: Hepatitis E virus, viral protease, thrombin, polyprotein processing, virus replication
Citation: Karpe YA (2024) Processing of the Hepatitis E virus ORF1 nonstructural polyprotein. Front. Virol. 3:1327745. doi: 10.3389/fviro.2023.1327745
Received: 25 October 2023; Accepted: 08 December 2023;
Published: 05 January 2024.
Edited by:
Jun Arii, Kobe University, JapanReviewed by:
Xiaohui Ju, Fred Hutchinson Cancer Center, United StatesCopyright © 2024 Karpe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yogesh A. Karpe, yakarpe@aripune.org
 Yogesh A. Karpe
Yogesh A. Karpe