Cortical Morphogenesis during Embryonic Development Is Regulated by miR-34c and miR-204
- 1Department of Molecular Biology and Genetics, Interdisciplinary Nanoscience Center, Aarhus University, Aarhus, Denmark
- 2Department of Translational Neuroscience, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, Netherlands
- 3Neurobiology Research, Institute of Molecular Medicine, University of Southern Denmark, Odense, Denmark
- 4Laboratory for Experimental Neuropathology, Department of Pathology, Randers Hospital, Randers, Denmark
The porcine brain closely resembles the human brain in aspects such as development and morphology. Temporal miRNA profiling in the developing embryonic porcine cortex revealed a distinct set of miRNAs, including miR-34c and miR-204, which exhibited a highly specific expression profile across the time of cortical folding. These miRNAs were found to target Doublecortin (DCX), known to be involved in neuron migration during cortical folding of gyrencephalic brains. In vivo modulation of miRNA expression in mouse embryos confirmed that miR-34c and miR-204 can control neuronal migration and cortical morphogenesis, presumably by posttranscriptional regulation of DCX.
Introduction
Over the course of evolution, the mammalian brain has grown, both in volume and surface. In higher mammals, including humans, this enlargement within the confinement of the skull has been enabled by increasing the folding and convolution of the cerebral cortex (gyration) leading to increased surface of the brain. In the mammalian brain, the cerebral cortex is in charge of complex cognitive functions (Geschwind and Rakic, 2013). Normal cortical development is initiated from a functional subdivision of progenitors resulting in multiple layers (lamination), or zones, within the cortex. Neural progenitor proliferation occurs mainly in the ventricular zone of the developing cortex. Here, a high level of asymmetrical cell division of radial glial cells occurs, resulting in both new radial glial cells and more differentiated daughter cells, including neurons (Sun and Hevner, 2014). Following this step, proper lamination is primarily ensured by regulated migration of neurons and intermediate progenitors, toward the pial surface of the cortex, closest to the skull. The development ends in the cortical plate, consisting mainly of fully differentiated neurons (Geschwind and Rakic, 2013; Sun and Hevner, 2014). In gyrencephalic species, one model proposes that regional differences in proliferation and growth of intermediate progenitors cause the ridge (gyri) and groove (sulci) appearance of the folded neocortex. While the regulatory mechanisms are currently unclear, neuronal migration-dependent lamination seems to be a prerequisite for normal cortical folding. Consequently, aberrant neuronal migration is associated with cortical deformities, in the most severe form a completely smooth brain surface (lissencephaly) (Reiner, 1999). A smooth brain surface can be naturally observed in lissencephalic species such as small rodents (mice and rats) (Sun and Hevner, 2014). Mutations causing lissencephaly in gyrencephalic species are associated with brain malfunctions including mental retardation and epilepsy (des Portes et al., 1998; Shimojima et al., 2010).
The first mutation identified in humans associated with lissencephaly was in a heterotrimeric G protein, which accordingly was named lissencephaly-1 (LIS1, also known as PAFAH1B1; Reiner et al., 1993). LIS1 mutation, duplication, or knockdown has deleterious effects on gyration. Due to a role in dynein and actin polymerization, LIS1, serves an important role in brain development affecting both proliferation of neuronal precursors and impacts migration of newly formed neurons within the cortex. An alternative cause of lissencephaly, X linked-1 (LISX1), is formed by mutations in doublecortin (DCX) (des Portes et al., 1998). DCX is a microtubule-associated protein (MAP), which stabilizes microtubule protofilaments and regulates actin cytoskeleton dynamics (Tsukada et al., 2005; Slepak et al., 2012). Knockdown of DCX in utero inhibits migration of cortical neurons (Kriegstein and Noctor, 2004). Consequently, based on these lissencephaly disease models a picture emerges supporting cytoskeleton arrangement and neuronal migration as essential components of normal brain development in gyrencephalic species. Interestingly, more than 90% of children suffering from lissencephaly experience epileptic seizures (Dobyns, 2010; Herbst et al., 2016) and abnormal cortical lamination caused by perturbations in neuronal migration is the likely cause of this high seizure susceptibility (Stouffer et al., 2016). Proper lamination is a complex process highly dependent on regulation on many levels, including posttranscriptional regulation of gene expression by microRNAs (miRNAs; Sun and Hevner, 2014). The implication of miRNAs is based on the observations that (1) cortex-specific knockout of the miRNA-processing enzyme Dicer causes reduced cortex size in mice, (2) neural progenitor specific knockout of Dicer results in reduced neural progenitor pool and impaired neuronal differentiation (Saurat et al., 2013), and (3) Dicer deletion in postmitotic neurons is associated with increased neuronal cell density in the cortical plate (Davis et al., 2008). Previous studies have found that miR-22 and miR-124 affect cortical neuron migration by targeting components of the coREST/REST complex, indirectly regulating DCX expression (Volvert et al., 2014). Also miR-9 has been shown to affect migration by targeting Stathmin, a protein that increases microtubule instability (Delaloy et al., 2010) or the transcription factor, Foxp2, in conjunction with miR-132 (Clovis et al., 2012). Finally, miR-128 was shown to affect migration by regulating PHF6 (Franzoni et al., 2015). However, these miRNAs are not sharply regulated at the time of cortical folding, suggesting that, although functionally important, they are not key regulators of the process. Mir-34a, a close relative of miR-34c, was recently found to play an active role in neural cell differentiation and was shown to affect migration of neuroblasts. The authors found DCX as a target for miR-34a, which is a likely contributor to the observed role for miR-34a (Mollinari et al., 2015).
We recently profiled the expression level of miRNAs in the porcine cerebral cortex, at five different timepoints across embryonic development (manuscript in preparation). A specific subset of miRNAs exhibited a highly distinct upregulation between day 60 and 80 post gestation (time of gyration). Here we unveil a functional role for two of these miRNAs in neuronal migration, miR-34c and miR-204. Combined with our discovery that these two miRNAs show extreme expression differences across cortical folding in the pig, suggests that they are instrumental in establishing the correct cortical morphogenesis in gyrencephalic animals.
Results
Differential Expression of miRNAs in Embryonic Porcine Cortex
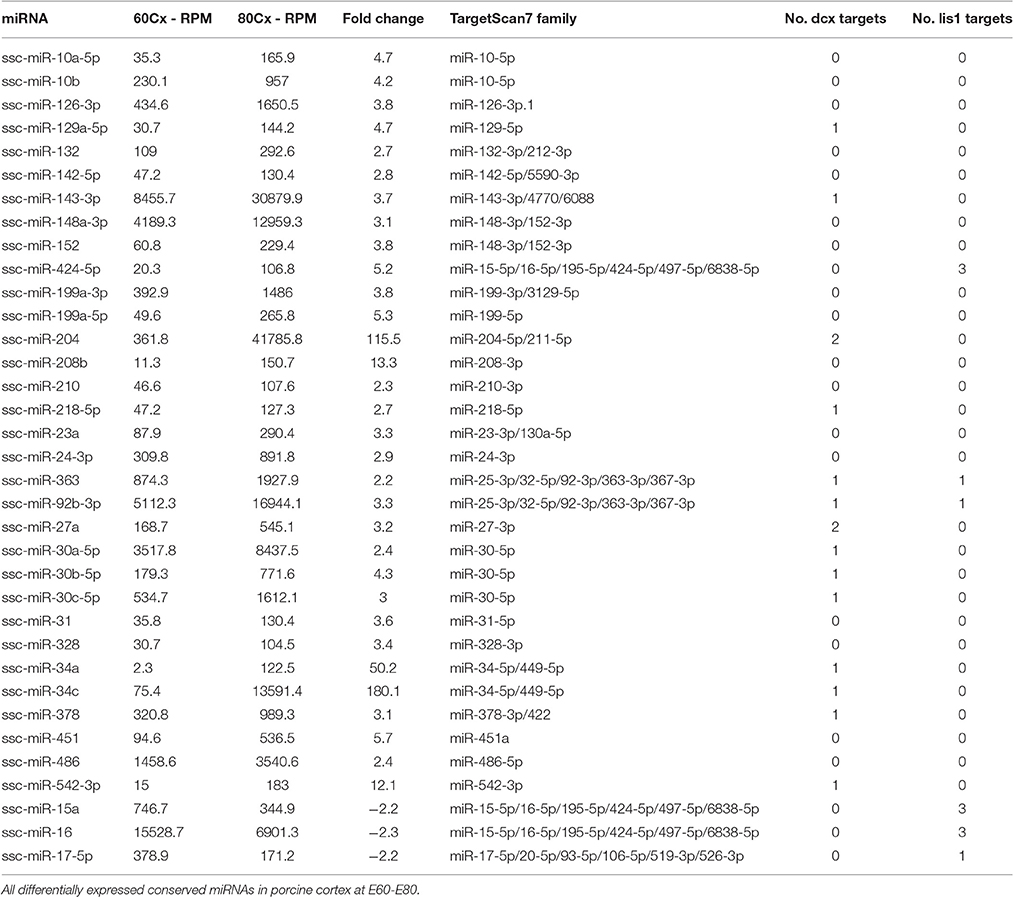
Small RNA sequencing of embryonic porcine cortex across fetal development revealed a remarkable shift in expression levels of a distinct set of miRNAs from embryonic day 60–80 (E60–E80), which is the period of cortical folding in the pig brain (manuscript in preparation). In total 49 miRNAs, expressed at least 100 reads per million (RPM), exhibited an at least two-fold change in expression in cortex from E60 to E80 (45 up- and 4 down-regulated). Restricting them to conserved miRNA families reduced the number of regulated miRNAs to 32 up- and 3 downregulated (Table 1). Conserved 3′UTR targets of this set of miRNAs were detected using TargetScan version 7 (Agarwal et al., 2015). Global miRNA targeting analysis (see Section Experimental Procedures) of the differentially expressed miRNAs revealed a list of genes predicted to experience altered miRNA-mediated control from E60 to E80 (Supplementary Table 1). Among the top 5 most significantly affected genes, DCX is of particular interest since it has been implicated in migration of cortical neurons, and hence is a prime target for miRNA-mediated control in respect to cortical morphogenesis. TargetScan predicts that 14 out of 32 (40%) upregulated miRNAs target DCX. Furthermore, none of the three downregulated miRNAs target DCX, whereas all three target LIS1 (Figures 1A,C; Table 1). In agreement with the predicted targeting of the differentially expressed miRNAs, DCX, and LIS1 are down and upregulated, respectively, upon gyration (Nielsen et al., 2010). MiR-204 and miR-34c are particularly notable upregulated miRNAs, due their almost exclusive expression in E80 cortex tissue (Figure 1B). This encouraged us to examine these miRNAs further. Detection of miR-204 and miR-34c by in situ hybridization (ISH) at E60 and E80 in the porcine cortex showed clearly increased expression at E80 for both miRNAs (Figure 2A). No expression difference was observed between sulci and gyri (Supplementary Figure 1).
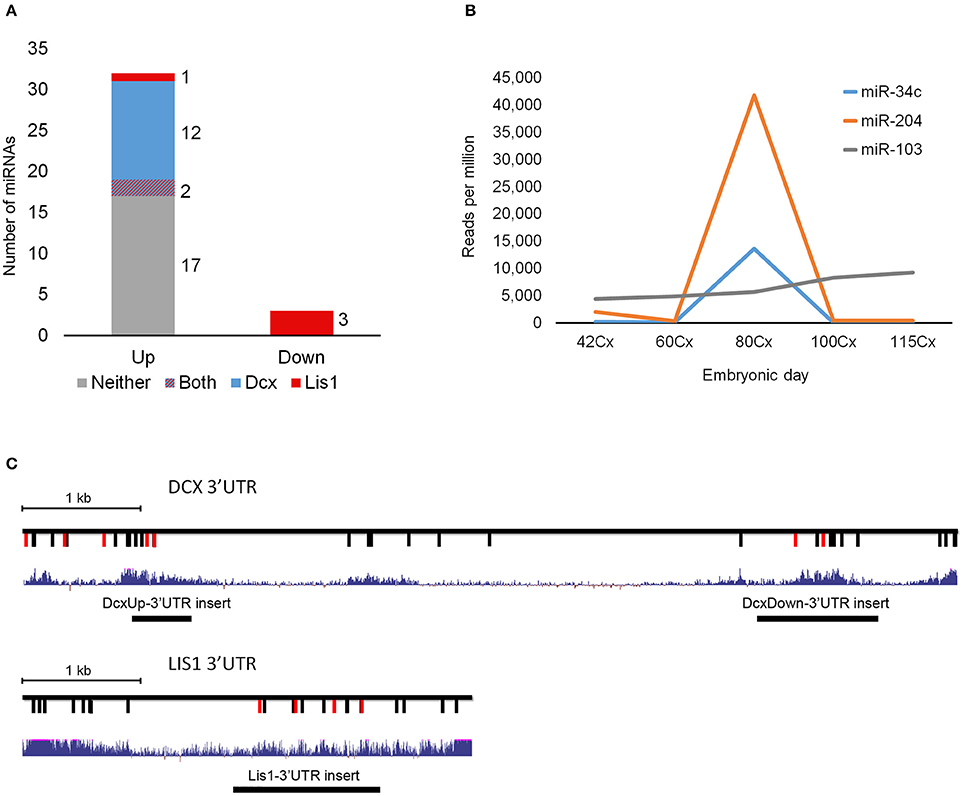
Figure 1. DCX and LIS1 targeting miRNAs. (A) The number of miRNAs up- or downregulated from E60 to E80 are shown. Color code indicates if differentially expressed miRNAs are predicted to target LIS1 (red), DCX (blue), both (red and blue), or neither (gray). (B) The RPM normalized expression of differentially regulated miR-34c and miR-204 as well as the stable miR-103. One replicate was used per timepoint. (C) Schematic illustrations of DCX and LIS1 3′UTRs with indication of conserved TargetScan7 predicted miRNA target sites. For DCX 3′UTR red vertical lines indicate target sites of upregulated miRNAs. For LIS1 3′UTR red vertical lines represent downregulated miRNA target sites. Black vertical lines indicate miRNA target sites for non-differentially expressed miRNA. Blue track shows UCSC Genome Browser's 100 vertebrates basewise conservation by PhyloP. Location of insert fragments used for luciferase reporter assay are indicated.
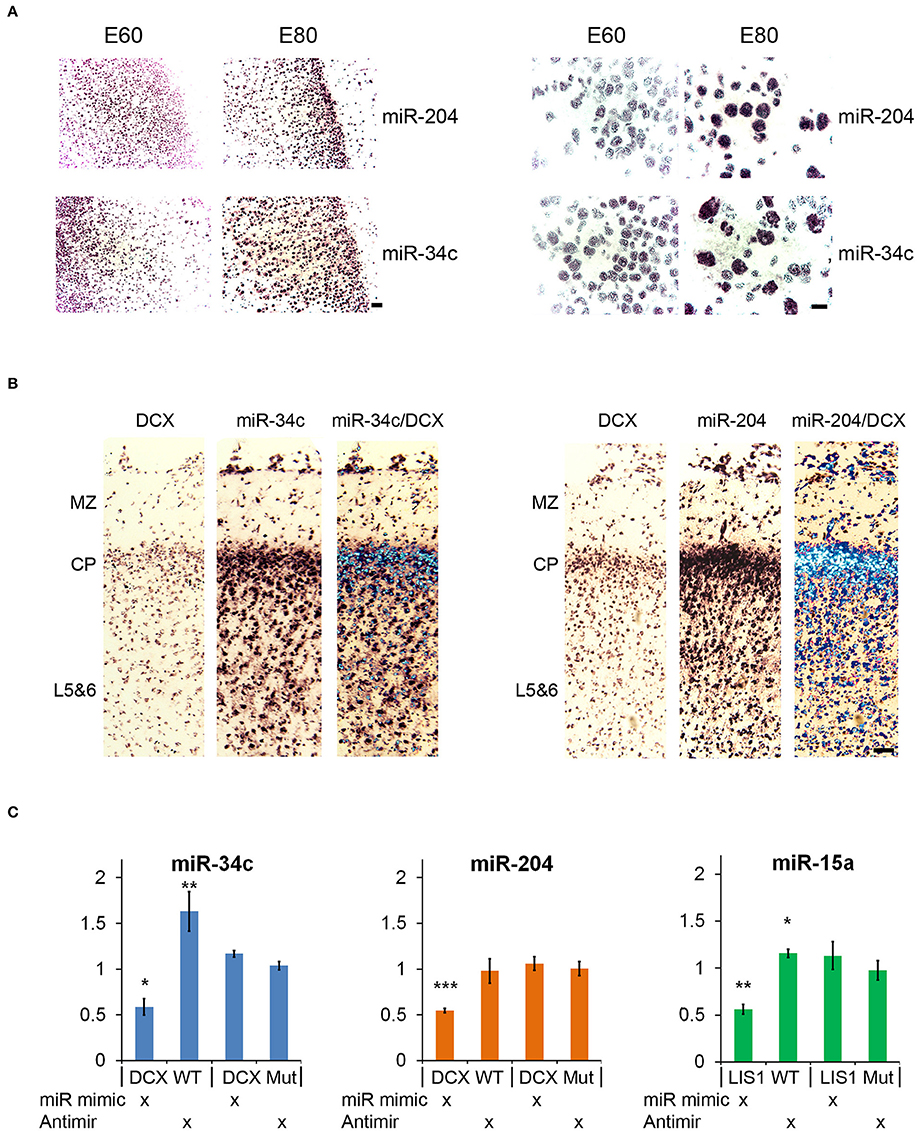
Figure 2. In situ hybridization and luciferase reporter assay. (A) In situ hybridization (ISH) images of miR-204 and miR-34c in E60 and E80 cortical porcine tissue. Upregulation from E60 to E80 is evident for both miRNAs. Scale bars: 50 mm (left panel), 10 mm (right panel). (B) Co-localization of DCX with miR-34c and miR-204 is shown by merged ISH signals as indicated. MZ, marginal zone; CP, cortical plate; L5&6, layer 5-6. Scale bar: 50 mm. (C) Luciferase 3′UTR-reporter assays. miR-34c and miR-204 mimics are shown to reduce DCX 3′UTR luciferase reporter expression, and miR-34c inhibitor increases DCX 3′UTR luciferase reporter expression. MiR-15 mimic reduces LIS1 3′UTR luciferase reporter expression, while miR-15 inhibitor slightly increases its expression. Luciferase data is normalized to relevant control (control mimic or control inhibitor), and experiments were done in biological triplicate. Data are presented as means ± standard deviation. *P < 0.05, **P < 0.01, ***P < 0.001. Independent Sample T-test.
Validation of miRNA Targeting
Both miR-34c and miR-204 are predicted by TargetScan to target DCX mRNA through 1 and 2 target sites, respectively (Table 1). This functional connection was further substantiated by co-localization of both miRNAs and DCX mRNA at most cells in the cortical plate using ISH (Figure 2B). To confirm that the predicted target sites are indeed functional targets of miR-34c and miR-204, the DCX 3′UTR was cloned downstream of a luciferase reporter and analyzed in HEK293-H cells. In accordance with being cognate targets, mimics of both miR-204 and mir-34c were able to significantly reduce luciferase expression, whereas miR-34c inhibitor increased the production from the DCX 3′UTR reporter. However, we were unable to observe any effect from miR-204 anti-miR (Figure 2C). The most prominent LIS1 targeting miRNA family contained two downregulated miRNAs, miR-15a and miR-16, with three predicted targets in the LIS1 3′UTR (Table 1). Choosing miR-15a as a representative for this miRNA family, we were able to confirm the predicted targeting, since miR-15a mimic and inhibitor reduced and increased luciferase expression from LIS1 3′UTR reporter, respectively. Mutating the predicted miR-34c, miR-204 and miR-15a target sites individually in the luciferase reporters abolished the observed effects (Figure 2C).
miR-34c and miR-204 Affect Neuronal Migration
Both miR-34c and miR-204 are conserved between mouse and pig and despite the fact that mice have a non-gyrated brain, the underlying process of neuronal migration is conserved between lissencephalic and gyrencephalic species (Kerjan and Gleeson, 2007). Hence, to explore the function of these miRNAs in vivo, miRNA mimics or inhibitors together with a GFP expressing plasmid were introduced to neuronal progenitors in the ventricular zone of the cortex by in utero electroporation (IUE) in mouse fetus at E14.5 followed by immunohistochemical characterization of the neurons that derived from these progenitors at E17.5 (van Erp et al., 2015). Analysis of migration of GFP-positive neurons in the motor cortex revealed that inhibition of miR-34c induced a robust increase in the percentage of GFP-positive cells in the cortical plate (CP; P = 0.006) and a reduction in both the intermediate zone (IZ; P = 0.007) and the subventricular zone (SVZ; P = 0.042) (Figures 3A,C). Electroporation of miR-204 inhibitor did not significantly alter the radial migration of GFP-positive cells (Figure 3C). In contrast, electroporation of miR-34c or miR-204 mimics caused cells to accumulate in the IZ (miR-34c; P = 0.001, miR-204; P = 0.004) and resulted in reduced cell numbers in the cortical plate (miR-34c; P = 0.000, miR-204 P = 0.002) compared to control mimic (Figures 3B,D). Together, these data strongly suggest that miR-34c and miR-204 regulate neuronal migration in the embryonic cortex, in line with our expression and reporter assay experiments.
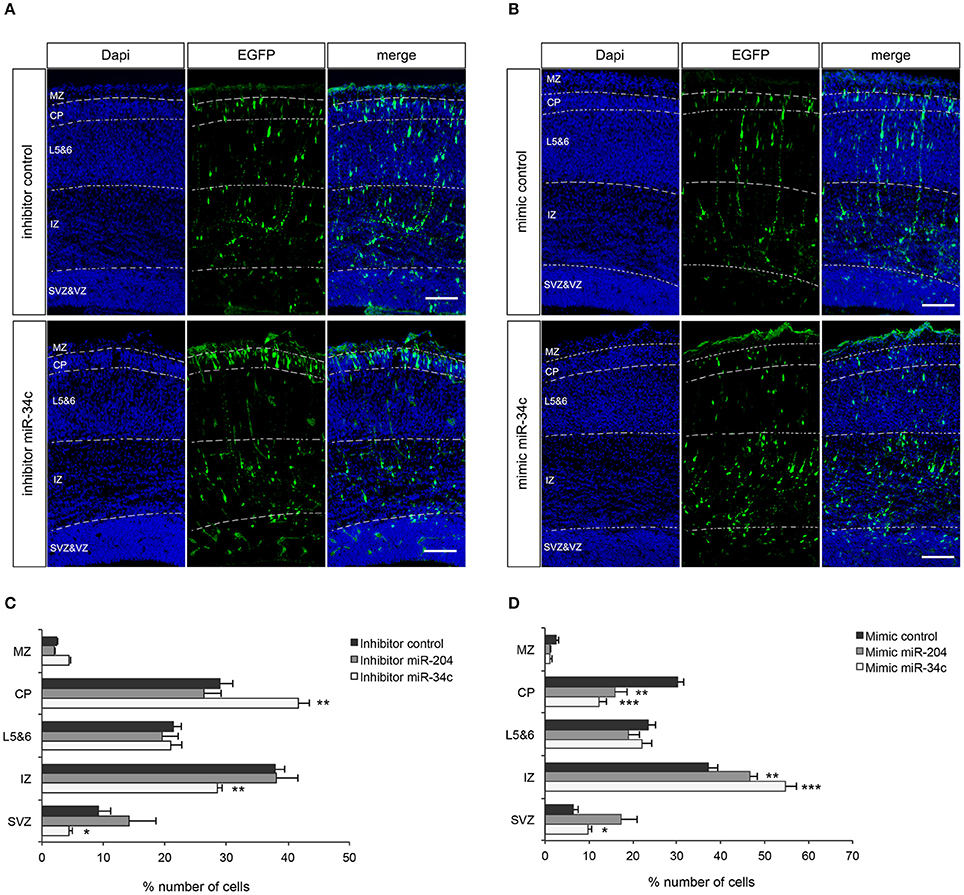
Figure 3. miR-34c and miR-204 regulate cortical neuron migration. (A,B) Coronal sections from E17.5 mouse embryos showing immunohistochemistry for GFP (green) and DAPI counterstaining (blue). Panels at the left show DAPI to indicate five different layers; MZ, marginal zone; CP, cortical plate; L5&6, layer 5-6; IZ, intermediate zone; and SVZ, subventricular zone. GFP signal indicates neurons transfected with miR-34c inhibitor (A) or mimic (B). Representative miR-204 experiment is in Supplementary Figure 2. (C,D) Quantification of GFP-positive cells (% of total) in different layers. Data are presented as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Independent Sample T-test. Number of embryos analyzed: miR-34c inhibitor: 5, miR-34c mimic: 5, miR-204 inhibitor: 6, miR-204 mimic: 7, control inhibitor: 9, control mimic: 8.
Discussion
Cortical folding is a complex process orchestrated by spatially controlled gene expression in subsections of the brain, and miRNA is likely to play key roles in the process. MiR-204 and miR-34c sequences and their predicted targets in DCX 3′UTR are conserved, implying that the effects of miR-34c and miR-204 on the migration of neurons in the mouse is most likely conserved in higher mammals, such as pigs and humans. Further support for a conserved functional relationships between DCX and miR-204/34c as well as LIS1 and miR-15a comes from the observation that the respective miRNAs can regulate luciferase expression under control of human derived DCX and LIS1 3′UTRs.
The striking finding that miR-204 and miR-34c exhibit specific expression increases in excess of 100 fold at the time of porcine gyration implies that they have key roles in the timing of the process. Notably, these two miRNAs alone change from low expression to constituting a massive 5.5% of all cortical miRNAs during gyration. miR-204 shares an identical seed sequence with mR-211 in most animals, however, this miRNA is not annotated in pig, and thus is not found in the analysis of our sequencing experiment. A recent paper reported that inactivation of two miRNA clusters, miR-34b/c and miR-449 clusters, with identical seed sequences, affected brain development, and microtubule dynamics (Wu et al., 2014). These findings are well in line with our data. The authors also find DCX to be a direct target of the miR-34-5p/449-5p family. Likewise, miR-34a has a documented role in neural cell differentiation and migration of neuroblasts, suggested to be caused by targeting of DCX (Mollinari et al., 2015). Altogether, every member of the miR-34-5p/449-5p family of miRNAs, all sharing identical seed sequences, have been found to affect brain development. Of the 6 miRNAs in this miRNA seed family (miR-34a,b,c, and miR-449a,b,c) only miR-34a and miR-34c are annotated in pig, in agreement with the detected miRNAs in our profiling experiment. Table 1 supports the shared function of miR-34a and miR-34c, given that both miRNAs show large expression increases from E60 to E80 of 50 and 180-fold, respectively. Although miR-34a and miR-34c have identical seed sequences, they have a substantial 5 nt sequence dissimilarity outside the seed, ensuring that miR-34a and miR-34c can readily be distinguished in ISH experiments via specific LNA probes.
Our finding of tightly controlled expression of miR-34c and miR-204 in brain development seems to extrapolate into neuroprotective effects in the aging animal. In a study by Liu et al. (2012), Drosophila miR-34 was found to display increased brain expression, specifically in old animals. The authors found that miR-34 over-expression ameliorated age-related neurodegeneration and increased median lifespan. This suggests that controlled upregulation of miR-34 family miRNAs is important, both in embryonic and adult animals.
Both the miR-34 family and miR-204 miRNAs are well known tumor suppressors in several cancers (He et al., 2007; Li et al., 2016) and have been linked to suppression of epithelial to mesenchymal transition (EMT; Hahn et al., 2013; Morizane et al., 2014; Li et al., 2016; Liu et al., 2016). In several cancers, including glioblastoma, metastasis is linked to the EMT process, whereby cancer cells can adopt mesenchymal traits allowing invasion into adjacent tissue (Ortensi et al., 2013). Initiation of neuronal migration is an EMT-like phenomenon, where cells lose epithelial characteristics and attain mesenchymal characteristics such as high motility (Itoh et al., 2013). Our finding, that increased miR-34c and miR-204 expression reduces neuronal migration, implies that suppression of EMT related factors could be a contributing mechanism controlling neuronal migration.
Temporal miRNA control during brain development is clearly important for animals later in life. DCX and LIS1, which our data suggest is under timing-specific miRNA-mediated control, are crucial for proper lamination of the brain. The known link between improper lamination and epilepsy (Stouffer et al., 2016) is highly relevant to our findings, since incorrect levels of both miR-34a and miR-204 have been associated with epilepsy (Hu et al., 2012; Kaalund et al., 2014; Xiang et al., 2016). Erroneous lamination in mouse brain caused by DCX knockout has been shown to result in increased seizure susceptibility and spontaneous epileptic seizures (Nosten-Bertrand et al., 2008), which the authors concluded was caused by abnormal synaptic transmission as a result of lamination defects. In humans, lissencephaly is tightly linked to epileptic seizures (Dobyns, 2010; Herbst et al., 2016), and focal cortical dysplasia (FCD), which involves some degree of disorganization of cortical lamination, is found in approximately half of the patients with medically refractory epilepsy (Wang et al., 2006). Thus, embryonic functions of miRNAs, such as miR-34c and miR-204, which affect cortical morphogenesis, are also likely to affect epilepsy susceptibility later in life.
Although we have shown that DCX is a target for both miR-34c and miR-204, and that over-expression of these miRNAs in IUE experiments significantly inhibit neuron migration in the mouse embryo, we cannot conclude that this migration effect is directly caused by inhibition of DCX. Given that single miRNAs can target multiple mRNAs, regulation of DCX may only be part of the picture. However, the strong genetic link between DCX and lissencephaly suggests that DCX may play a major role.
In summary, we show miRNA mediated control of DCX and LIS1 by miRNAs that are differentially expressed at the time of cortical folding. The repertoire of differentially expressed miRNAs at this developmental stage is likely to be crucial for timing the migration of neuronal precursors, thus affecting cortical morphogenesis. The most strongly upregulated miRNAs, miR-34c, and miR-204, were shown to alter the extent of neuronal migration in embryonic mouse brain, and their large expression increase during cortical folding in pig, suggests a pivotal role in mediating correct cortical morphogenesis of gyrencephalic animals.
Experimental Procedures
Illumina Sequencing of miRNAs
All procedures involving pigs described in present study were reviewed and approved by the Danish Experimental Animal Inspectorate (“Rådet for Dyreforsøg”), Danish Ministry of Justice. Embryonic pig brain samples were snap frozen after dissection and transferred to RNAlater-ICE (Ambion). Low molecular weight RNA (<200 nt.) was purified using the MirVana kit (Ambion). Samples were DNase treated using the TURBO DNA-free Kit (Ambion). Library preparation was performed using the TruSeq Small RNA preparation kit (Illumina) using 100 ng low molecular weight RNA. Fifty bp single end sequencing was performed at Research Centre Foulum. One replicate was sequenced per timepoint. Raw sequencing data was quality filtered and trimmed using fastX toolkit and adapter trimmed using cutadapt. Filtered sequencing reads were mapped to the mature porcine miRNAs from miRBase version 21 using Bowtie. The resultant porcine miRNA expression profile was normalized to reads per million mapping reads (RPM).
Global miRNA Targeting Analysis
Conserved predicted targets of conserved miRNAs were downloaded from TargetScan version 7 (Agarwal et al., 2015). Unique miRNA family target sites were noted for all genes. The change in targeting was defined as number of matching upregulated miRNA families subtracting the number of downregulated miRNA families from E60 to E80. We observe 26 up- and 2 downregulated unique miRNA families in cortex from E60 to E80. To test significance, we randomly picked the same number of conserved miRNA families (100,000 times), revealing whether random picking generated equal or greater changes in targeting of matching miRNA families. P-values were generated for all expressed TargetScan indexed genes by dividing the number random occurrences with equal or greater targeting than observed by the number of iterations (100,000). Only expressed genes were assayed. Expression is here defined as minimum 1 FPKM in embryonic cortex from RNA-seq data presented in (Venø et al., 2015), GEO accession: GSE71832.
In situ Hybridization
Brain slices (10 or 5 μm) from embryonic pigs (E60, E80; Venø et al., 2015) were in situ hybridized as previously described (Clausen et al., 2013). See Supplementary Table 2 for list of probes. The alkaline phosphatase-labeled probes used for miRNAs were 22 nts and 23 nts in length for miR-204 and miR-34c, respectively. The mRNA targeting alkaline phosphatase-labeled probes were 26 nts and 28 nts in length for GAPDH mRNA and DCX mRNA, respectively. Signal specificity was tested by hybridizing parallel sections with (1) 100-fold excess of unlabeled probe, (2) pre-treatment with RNase A (Pharmacia Biotech), or (3) buffer alone. All controls were devoid of signal.
The co-localization of miRNA with DCX mRNA was performed by hybridizing adjacent 5 μm thick sections with each probe, before overlaying the signals. The DCX mRNA and miRNA ISH probes were hybridized and developed for the same amount of time when performing co-localization experiments.
Toluidine Blue Staining
One series of brain sections from embryonic pigs (E60, E80) were stained with Toluidine Blue (Merck, Germany), dehydrated in graded series of alcohol (96–99% ethanol), cleared in xylene and cover-slipped in Depex (BDH Gurr, UK) as described (Clausen et al., 2006).
Luciferase Reporter Assay
Fragments of DCX or LIS1 3'UTRs were amplified by PCR using cDNA from HEK293-H. See Supplementary Table 2 for list of primers. The fragments were cloned into the psicheck2 vector and used for luciferase assay. Luciferase assay was performed by co-transfection of HEK293-H cells with miRNA mimic or inhibitor and psicheck2 reporter by standard calcium phosphate transfection. Three psicheck2 vectors with inserted fragment were made from WT sequences originating from:
DcxUp-3′UTR_WT: chrX:110543495-110543999 505 bp (The miR-34c target site).
DcxDown-3′UTR_WT: chrX:110537676-110538698 1023 bp (Both miR-204 target sites).
Lis1-3′UTR_WT: chr17:2586883-2588130 1248 bp (All three miR-15a target sites).
Figure 1C shows the location from which the inserted fragments originate. Mutated versions of the three WT psicheck2 vectors were generated, carrying specific mutations in the miRNA target sites under investigation by use of Site-directed, Ligase-Independent Mutagenesis.
Cell lysates were harvested after 24 h and used for luciferase assay using the Dual-Luciferase® Reporter Assay System (promega) according to manufacture's recommendations.
In utero Electroporation
All mouse use and care was carried out in accordance with institutional guidelines, Dutch law (Wet op de Dierproeven, 1996) and European regulation (Guideline 86/609/EEC), and with approval of the ethical animal experimentation committee (DEC). IUE was used to introduce miRNA mimics or inhibitors into the embryonic mouse cortex as described previously (van Erp et al., 2015). To visualize electroporated neurons, an EGFP expression vector was co-transfected with the mimics or inhibitors (pC1-EGFP and pCAG-EGFP both from Addgene). Electroporation was carried out at E14.5 according to an adapted protocol of (dal Maschio et al., 2012). Briefly, pregnant female mice were anesthetized with isofluorane (induction 5%; surgery 2,5%), their uterine horns were exposed, and 1.7 μl saline/Fast Green (0.3 mg/ml) mix solution containing 30 pmol RNA construct and 1.5 μg EGFP was microinjected into one of the lateral ventricles of each embryo. Embryos were electroporated with 5 impulses (30 V, 50 ms length and interval of 950 ms) using a tripolar electrode. At E17.5, embryonic brains were fixed overnight in 4% paraformaldehyde/PBS and subsequently cryoprotected in a 30% sucrose/PBS solution. Brains were sectioned at 20 μm (coronal) using a cryostat. Sections were immunostained using a rabbit anti-GFP antibody (1:1000, Thermo Fisher Scientific, Rockford, USA) overnight at 4°C (Schmidt et al., 2014). Sections were counterstained with DAPI and fluorescent staining was visualized on a Zeiss Axioskop A1 epifluorescent microscope. Neuronal migration was quantified as described previously (van Erp et al., 2015). In brief, the automated cell counting tool of ImageJ was used to determine the number of GFP-positive neurons in five different regions (marginal zone, cortical plate, layer 5–6, intermediate zone and SVZ). An independent sample T-test (SPSS software) was used to determine statistical significance.
Author Contributions
MTV conceived of the project. MTV and STV drafted the manuscript. IEH supplied embryonic porcine samples. MTV analyzed sequencing data and performed miRNA target analysis. STV performed luciferase assays. BHC and BF performed in situ hybridization. KR, JVA, and RJP performed and analyzed IUE. JK supervised the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Lundbeck Foundation and the center for integrative sequencing at Aarhus University (iSEQ) [MTV]. Netherlands Organization for Scientific Research [VICI], Epilepsiefonds (12-08, 15-05) [RJP]. The Lundbeck foundation [BC] and the Novo Nordisk foundation [BF].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fnmol.2017.00031/full#supplementary-material
References
Agarwal, V., Bell, G. W., Nam, J.-W., and Bartel, D. P. (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. doi: 10.7554/eLife.05005
Clausen, B., Fenger, C., and Finsen, B. (2013). In situ hybridization of cytokine mRNA using alkaline phosphatase-labelled oligodeoxynucleotide probes. Methods Mol. Biol. 1041, 83–91. doi: 10.1007/978-1-62703-520-0_10
Clausen, B. H., Lambertsen, K. L., and Finsen, B. (2006). Glyceraldehyde-3-phosphate dehydrogenase versus toluidine blue as a marker for infarct volume estimation following permanent middle cerebral artery occlusion in mice. Exp. Brain Res. 175, 60–67. doi: 10.1007/s00221-006-0526-3
Clovis, Y. M., Enard, W., Marinaro, F., Huttner, W. B., and De Pietri Tonelli, D. (2012). Convergent repression of Foxp2 3′UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development 139, 3332–3342. doi: 10.1242/dev.078063
dal Maschio, M., Ghezzi, D., Bony, G., Alabastri, A., Deidda, G., Brondi, M., et al. (2012). High-performance and site-directed in utero electroporation by a triple-electrode probe. Nat. Commun. 3, 960. doi: 10.1038/ncomms1961
Davis, T. H., Cuellar, T. L., Koch, S. M., Barker, A. J., Harfe, B. D., McManus, M. T., et al. (2008). Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28, 4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008
Delaloy, C., Liu, L., Lee, J. A., Su, H., Shen, F., Yang, G. Y., et al. (2010). MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6, 323–335. doi: 10.1016/j.stem.2010.02.015
des Portes, V., Pinard, J. M., Billuart, P., Vinet, M. C., Koulakoff, A., Carrié, A., et al. (1998). A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell 92, 51–61. doi: 10.1016/S0092-8674(00)80898-3
Dobyns, W. B. (2010). The clinical patterns and molecular genetics of lissencephaly and subcortical band heterotopia. Epilepsia 51(Suppl. 1), 5–9. doi: 10.1111/j.1528-1167.2009.02433.x
Franzoni, E., Booker, S. A., Parthasarathy, S., Rehfeld, F., Grosser, S., Srivatsa, S., et al. (2015). miR-128 regulates neuronal migration, outgrowth and intrinsic excitability via the intellectual disability gene Phf6. Elife 4:e04263. doi: 10.7554/eLife.04263
Geschwind, D. H., and Rakic, P. (2013). Cortical evolution: judge the brain by its cover. Neuron 80, 633–647. doi: 10.1016/j.neuron.2013.10.045
Hahn, S., Jackstadt, R., Siemens, H., Hünten, S., and Hermeking, H. (2013). SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 32, 3079–3095. doi: 10.1038/emboj.2013.236
He, X., He, L., and Hannon, G. J. (2007). The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res. 67, 11099–11101. doi: 10.1158/0008-5472.CAN-07-2672
Herbst, S. M., Proepper, C. R., Geis, T., Borggraefe, I., Hahn, A., Debus, O., et al. (2016). LIS1-associated classic lissencephaly: a retrospective, multicenter survey of the epileptogenic phenotype and response to antiepileptic drugs. Brain Dev. 38, 399–406. doi: 10.1016/j.braindev.2015.10.001
Hu, K., Xie, Y. Y., Zhang, C., Ouyang, D. S., Long, H. Y., Sun, D. N., et al. (2012). MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 13:115. doi: 10.1186/1471-2202-13-115
Itoh, Y., Moriyama, Y., Hasegawa, T., Endo, T. A., Toyoda, T., and Gotoh, Y. (2013). Scratch regulates neuronal migration onset via an epithelial-mesenchymal transition-like mechanism. Nat. Neurosci. 16, 416–425. doi: 10.1038/nn.3336
Kaalund, S. S., Venø, M. T., Bak, M., Møller, R. S., Laursen, H., Madsen, F., et al. (2014). Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-Convergence on axonal guidance. Epilepsia 55, 2017–2027. doi: 10.1111/epi.12839
Kerjan, G., and Gleeson, J. G. (2007). Genetic mechanisms underlying abnormal neuronal migration in classical lissencephaly. Trends Genet. 23, 623–630. doi: 10.1016/j.tig.2007.09.003
Kriegstein, A. R., and Noctor, S. C. (2004). Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 27, 392–399. doi: 10.1016/j.tins.2004.05.001
Li, T., Pan, H., and Li, R. (2016). The dual regulatory role of miR-204 in cancer. Tumour Biol. 37, 11667–11677. doi: 10.1007/s13277-016-5144-5
Liu, N., Landreh, M., Cao, K., Abe, M., Hendriks, G. J., Kennerdell, J. R., et al. (2012). The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482, 519–523. doi: 10.1038/nature10810
Liu, Z., Long, J., Du, R., Ge, C., Guo, K., and Xu, Y. (2016). miR-204 regulates the EMT by targeting snai1 to suppress the invasion and migration of gastric cancer. Tumour Biol. 37, 8327–8335. doi: 10.1007/s13277-015-4627-0
Mollinari, C., Racaniello, M., Berry, A., Pieri, M., de Stefano, M. C., Cardinale, A., et al. (2015). miR-34a regulates cell proliferation, morphology and function of newborn neurons resulting in improved behavioural outcomes. Cell Death Dis. 6, e1622. doi: 10.1038/cddis.2014.589
Morizane, R., Fujii, S., Monkawa, T., Hiratsuka, K., Yamaguchi, S., Homma, K., et al. (2014). miR-34c attenuates epithelial-mesenchymal transition and kidney fibrosis with ureteral obstruction. Sci. Rep. 4:4578. doi: 10.1038/srep04578
Nielsen, K. B., Kruhoffer, M., Holm, I. E., Jorgensen, A. L., and Nielsen, A. L. (2010). Identification of genes differentially expressed in the embryonic pig cerebral cortex before and after appearance of gyration. BMC Res. Notes 3:127. doi: 10.1186/1756-0500-3-127
Nosten-Bertrand, M., Kappeler, C., Dinocourt, C., Denis, C., Germain, J., Phan Dinh Tuy, F., et al. (2008). Epilepsy in Dcx knockout mice associated with discrete lamination defects and enhanced excitability in the hippocampus. PLoS ONE 3:e2473. doi: 10.1371/journal.pone.0002473
Ortensi, B., Setti, M., Osti, D., and Pelicci, G. (2013). Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res. Ther. 4, 18. doi: 10.1186/scrt166
Reiner, O. (1999). The unfolding story of two lissencephaly genes and brain development. Mol. Neurobiol. 20, 143–156. doi: 10.1007/BF02742439
Reiner, O., Carrozzo, R., Shen, Y., Wehnert, M., Faustinella, F., Dobyns, W. B., et al. (1993). Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364, 717–721. doi: 10.1038/364717a0
Saurat, N., Andersson, T., Vasistha, N. A., Molnar, Z., and Livesey, F. J. (2013). Dicer is required for neural stem cell multipotency and lineage progression during cerebral cortex development. Neural Dev. 8, 14. doi: 10.1186/1749-8104-8-14
Schmidt, E. R., Brignani, S., Adolfs, Y., Lemstra, S., Demmers, J., Vidaki, M., et al. (2014). Subdomain-mediated axon-axon signaling and chemoattraction cooperate to regulate afferent innervation of the lateral habenula. Neuron 83, 372–387. doi: 10.1016/j.neuron.2014.05.036
Shimojima, K., Sugiura, C., Takahashi, H., Ikegami, M., Takahashi, Y., Ohno, K., et al. (2010). Genomic copy number variations at 17p13.3 and epileptogenesis. Epilepsy Res. 89, 303–309. doi: 10.1016/j.eplepsyres.2010.02.002
Slepak, T. I., Salay, L. D., Lemmon, V. P., and Bixby, J. L. (2012). Dyrk kinases regulate phosphorylation of doublecortin, cytoskeletal organization, and neuronal morphology. Cytoskeleton 69, 514–527. doi: 10.1002/cm.21021
Stouffer, M. A., Golden, J. A., and Francis, F. (2016). Neuronal migration disorders: focus on the cytoskeleton and epilepsy. Neurobiol. Dis. 92, 18–45. doi: 10.1016/j.nbd.2015.08.003
Sun, T., and Hevner, R. F. (2014). Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci. 15, 217–232. doi: 10.1038/nrn3707
Tsukada, M., Prokscha, A., Ungewickell, E., and Eichele, G. (2005). Doublecortin association with actin filaments is regulated by neurabin II. J. Biol. Chem. 280, 11361–11368. doi: 10.1074/jbc.M405525200
van Erp, S., van den Heuvel, D. M., Fujita, Y., Robinson, R. A., Hellemons, A. J., Adolfs, Y., et al. (2015). Lrig2 Negatively regulates ectodomain shedding of axon guidance receptors by adam proteases. Dev. Cell 35, 537–552. doi: 10.1016/j.devcel.2015.11.008
Venø, M. T., Hansen, T. B., Venø, S. T., Clausen, B. H., Grebing, M., Finsen, B., et al. (2015). Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16:245 doi: 10.1186/s13059-015-0801-3
Volvert, M. L., Prevot, P. P., Close, P., Laguesse, S., Pirotte, S., Hemphill, J., et al. (2014). MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep. 7, 1168–1183. doi: 10.1016/j.celrep.2014.03.075
Wang, V. Y., Chang, E. F., and Barbaro, N. M. (2006). Focal cortical dysplasia: a review of pathological features, genetics, and surgical outcome. Neurosurg. Focus 20, E7. doi: 10.3171/foc.2006.20.1.8
Wu, J., Bao, J., Kim, M., Yuan, S., Tang, C., Zheng, H., et al. (2014). Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 111, E2851–E2857. doi: 10.1073/pnas.1407777111
Keywords: brain development, embryonic development, neuronal migration, microRNA, cortical morphogenesis, miR-204, miR-34c
Citation: Venø MT, Venø ST, Rehberg K, van Asperen JV, Clausen BH, Holm IE, Pasterkamp RJ, Finsen B and Kjems J (2017) Cortical Morphogenesis during Embryonic Development Is Regulated by miR-34c and miR-204. Front. Mol. Neurosci. 10:31. doi: 10.3389/fnmol.2017.00031
Received: 06 October 2016; Accepted: 26 January 2017;
Published: 09 February 2017.
Edited by:
Hermona Soreq, Hebrew University of Jerusalem, IsraelReviewed by:
Beena Pillai, Institute of Genomics and Integrative Biology (CSIR), IndiaAmit Berson, University of Pennsylvania, USA
Copyright © 2017 Venø, Venø, Rehberg, van Asperen, Clausen, Holm, Pasterkamp, Finsen and Kjems. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Morten T. Venø, mtv@mbg.au.dk
Jørgen Kjems, jk@mbg.au.dk
 Morten T. Venø1*
Morten T. Venø1*  Jessy V. van Asperen
Jessy V. van Asperen Ida E. Holm
Ida E. Holm Bente Finsen
Bente Finsen