- 1 Feedstocks Division, Joint Bioenergy Institute, Physical Biosciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA
- 2 Department of Plant Biology and Biotechnology, University of Copenhagen, Frederiksberg, Denmark
- 3 Department of Plant and Microbial Biology, University of California, Berkeley, CA, USA
The carbohydrate active enzyme (CAZy) database is an invaluable resource for glycobiology and currently contains 45 glycosyltransferase families that are represented in plants. Glycosyltransferases (GTs) have many functions in plants, but the majority are likely to be involved in biosynthesis of polysaccharides and glycoproteins in the plant cell wall. Bioinformatic approaches and structural modeling suggest that a number of protein families in plants include GTs that have not yet been identified as such and are therefore not included in CAZy. These families include proteins with domain of unknown function (DUF) DUF23, DUF246, and DUF266. The evidence for these proteins being GTs and their possible roles in cell wall biosynthesis is discussed.
Introduction
Plant cell walls contain structural polysaccharides such as cellulose, hemicelluloses, and pectins. To assemble these polysaccharides as well as the glycan structures on glycoproteins, the plant needs extensive biosynthetic machinery, and it has been estimated that over 2000 gene products are involved in making and maintaining the wall (Carpita et al., 2001; Dhugga, 2001). The polysaccharides and other glycans are mainly synthesized by glycosyltransferases (GTs; EC 2.4.x.y). Most GTs transfer a sugar residue from an activated nucleotide sugar to a specific acceptor molecule, forming a glycosidic bond, and GTs generally display high specificity for both the sugar donor and the acceptor substrates (Breton et al., 2006). GTs are classified as “retaining” or “inverting” depending on whether glycosylation occurs with retention or inversion of stereochemistry at the anomeric carbon atom of the donor substrate. GTs have been further classified into families on the basis of amino acid sequence similarities in the carbohydrate active enzyme (CAZy) database1 (Cantarel et al., 2009). CAZy contains proteins with a demonstrated biochemical function as well as orthologous putative GTs. The CAZy database is continually increasing, adding new GTs as they are discovered, and currently contains 91 GT families numbered from GT1 to GT94 (GT36, GT46, and GT86 no longer exist). Only two folds, and variants thereof, have been observed for all structures of nucleotide–sugar-dependent GTs solved to date, termed GT-A and GT-B (Hansen et al., 2010). Whereas many GT-Bs are found to be independent of a metal ion for catalysis, most GT-A enzymes contains a conserved DxD motif that coordinates the phosphate atoms of the nucleotide donors via coordination of a divalent cation, usually Mn2+ or Mg2+ (Breton et al., 2006). Identifying the precise function of every putative plant GT represents an immense task. For example, about 1.7% of the 27,416 protein coding Arabidopsis genes are represented among 42 GT families in the database, but less than 20% of these sequences, have been annotated to date (Caffall and Mohnen, 2009; Scheller and Ulvskov, 2010) and very few of the GTs involved in cell wall biosynthesis have had their biochemical activity unambiguously demonstrated. Some of the CAZy GTs could also have lost their catalytic function and represent proteins with other functions, e.g., carbohydrate binding proteins or non-catalytic members of GT complexes as recently suggested for GAUT7 (Atmodjo et al., 2011). Cell wall biosynthesis involves two classes of GTs: The multi-membrane-spanning GTs of GT2 and GT48 and the more common type II transmembrane proteins consisting of a short cytoplasmic N-terminal tail followed by a single transmembrane helix, a stem region of variable length, and a large globular C-terminal part containing the catalytic domain. GTs that are Type II membrane proteins have been found in the endoplasmic reticulum (ER) and Golgi apparatus, while the multi-membrane-spanning members of GT2 and GT48 are associated with the Golgi apparatus or plasma membrane. The majority of GTs involved in biosynthesis of the complex cell wall polysaccharides are thought to be Golgi localized.
Given the complexity of the plant cell wall, and the fact that new GT families are regularly added to CAZy, it can be expected that some of the cell wall biosynthetic genes have yet to be identified. This could be achieved by for example forward genetics or via orthology to other newly discovered GTs followed by reverse genetics. An example of an activity with no candidates in CAZy is the β-arabinosyltransferase – a retaining GT – that transfers the innermost arabinose to hydroxyproline in extensin, and which must be present in both green algae and plants. However, analysis of retaining CAZy GTs of Chlamydomonas reinhardtii and Arabidopsis did not reveal any ortologous proteins, which did not have a putative function unrelated to extensin biosynthesis (Harholt, Paiva, Domozych and Ulvskov, unpublished). This strongly suggests that this particular β-arabinosyltransferase is in a family not yet included in CAZy. With the use of various bioinformatic strategies, Hansen et al. (2009) identified plant sequences representing a “Golgi located GT motif” in domain of unknown function (DUF) families DUF246 and DUF266, and proposed that the number of plant GT genes is underestimated. Currently, more than 3000 DUF families are represented in the protein family (Pfam) database, a large collection of protein families, grouped via sequence similarity, and hidden Markov models (HMM; Finn et al., 2010).2
The aim of this review is to briefly describe putative GTs that are not currently classified in the CAZy database. DUF246 and DUF266 had previously been identified as putative GTs (Hansen et al., 2009), while DUF23 is related to a novel GT family unknown at the time of the earlier study.
Cell Wall Associated DUF266
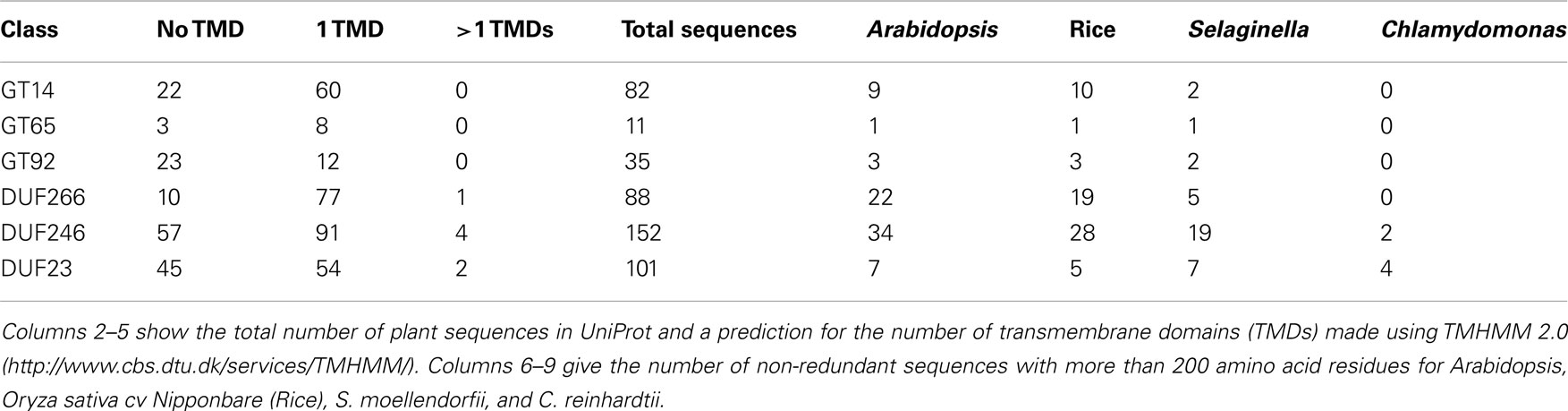
Bioinformatic studies using HMM and fold recognition identified 14 Arabidopsis genes that shared a DUF266 domain and were distantly related to GT14 (Hansen et al., 2009). These putative plant GT sequences were annotated as containing a plant specific DUF266 domain, described in the Pfam database as “likely to be glycosyltransferase related.” Fold recognition analysis and hydrophobic cluster analysis demonstrated structural similarities to the Leukocyte core-2 β1, 6N-acetylglucosaminyltransferase (C2GnT-L) family protein, which is a member of GT14 (Hansen et al., 2009). Furthermore, invariant amino acid residues were found between C2GnT-L and Arabidopsis DUF266 proteins. C2GnT-L is a Golgi localized, inverting GT with a GT-A fold (Pak et al., 2006). Quite uncommon for GTs with a GT-A fold, C2GnT-L lacks the characteristic metal ion binding DxD motif. C2GnT-L is involved in biosynthesis of mucin-type glycoproteins catalyzing the formation of the core-2 branched O-glycan (Galβ1-3[GlcNAcβ1-6]GalNac-O-Ser/Thr) from its donor and acceptor substrates, UDP-GlcNAc and the core-1 O-glycan (Galβ1-3GalNAc-O-Ser/Thr), respectively (Fukuda et al., 1996; Yeh et al., 1999; Pak et al., 2006). C2GnT-L is the only structure solved in GT14. The structure of C2GnT-L from mice was determined with β-D-galactose and/or N-acetyl-D-glucosamine as ligands (Pak et al., 2006). The putative catalytic amino acid residue (glutamic acid) of C2GnT-L is conserved in other inverting GT-A proteins and in the plant DUF266 family (see Figure 1A; Figure A4A in Appendix; Hansen et al., 2009).
Figure 1. Comparative analysis between DUF families and their related Pfam groups. (A–C) Schematic illustration of conserved motifs for (A) DUF266 and GT14, (B) DUF246 and GT65, and (C) DUF23 and GT92, based on non-redundant Arabidopsis sequences with more than 200 amino acid residues. Strickly conserved residues are in bold and residues with only two alternative amino acids are denoted by parentheses. Residues involved in substrate binding and putative catalytic residues are marked with arrowheads. GT-A structure domains (Nucleotide Binding and Acceptor, respectively) are shown as Breton et al., 2006. TMD: transmembrane domain. (D–F) Illustration of the total number of plant UniProt sequences retrieved from (D) DUF266, GT14 and the Branch domain (PF02485), (E) DUF246, GT65, and the O-fucosyltransferase domain (O-FucT; PF10250), and (F) GT92 and DUF23 (PF01697).
Concurrent with the bioinformatic identification of DUF266 as putative GTs, the rice brittle culm 10 mutant (Osbc10) was characterized. The Osbc10 mutant displayed brittleness of the plant body and morphological abnormalities including significant decrease in plant height and tiller number (Zhou et al., 2009). The corresponding protein OsBC10 was shown to be a Golgi located type II membrane protein containing a DUF266 domain. The cell wall composition showed reduced content of glucose and increased content of xylose, arabinose, and lignin, and antibody labeling identified a decrease in epitopes associated with arabinogalactan proteins (AGPs). The cellular localization, the type II membrane protein structure and the cell wall phenotypes led the authors to suggest that OsBC10 is involved in cell wall biosynthesis, although the specific enzymatic activity is still unclear. The OsBC10 protein was heterologously expressed in Chinese hamster ovary cells and demonstrated a very low in vitro activity in an assay for C2GnT-L. Thus, although OsBC10 is unlikely to have C2GnT-L activity in vivo since the core-2 branched O-glycan is almost certainly absent in plants, we conclude that the evidence for OsBC10 being a GT involved in cell wall biosynthesis is strong.
The GT14 family contains 308 sequences from viruses, bacteria, animals, and plants. It includes, beside proteins with unknown function, proteins with β-1,6-N-acetylglucosaminyl-transferase and β-xylosyltransferase activity involved in the synthesis of O-glycans in animals (Bierhuizen et al., 1993; Yeh et al., 1999; Hwang et al., 2003). A phylogenetic analysis of plant sequences from family GT14 shows that all clusters into one single subfamily (Aspeborg et al., 2005). To our knowledge, no plant GT14 gene has yet been functionally characterized. The phylogenetic relationship and sequence similarity within the PF02485 domain, between GT14 and DUF266 proteins in plants is illustrated in Figure A1 in Appendix.
Since the study of Hansen et al. (2009), the DUF266 family has been removed from the Pfam database and merged into the Branch domain (PF02485), which contains core-2 and I-branching enzymes (Bierhuizen et al., 1993; Yeh et al., 1999). The Branch domain (PF02485) contains 484 plant UniProt accessions, shared by 76 GT14, and 86 sequences of the previous DUF266 (Figure 1D). Whereas the Branch domain was found among animals, plants, vira, and bacteria, DUF266 was only found in proteins from plants and therefore likely to be involved in biological processes specific to plants.
While the majority of Arabidopsis GT14 proteins were predicted to be Golgi located, consistent with a role in cell wall biosynthesis, the predicted localization of DUF266 family proteins was more variable, with most predicted to be in Golgi and plasma membrane (Ye et al., 2011). However, as we have recently discussed, bioinformatic tools for predicting Golgi localization are highly unreliable, and a much more reliable prediction is obtained based on Pfam groupings (Oikawa et al., 2010).
Whereas the DUF266 containing OsBC10 was mainly expressed in the developing vascular bundle and sclerenchyma cells (Zhou et al., 2009), two plant GT14 members (PttGT14A and PttGT14B) were identified by expression profiling to be xylem-specific (Aspeborg et al., 2005), indicating their potential role in secondary cell wall biosynthesis. Recently, two DUF266 proteins (At1g11940 and At5g11730) were found in Golgi purified from Arabidopsis cell suspension cultures (Parsons et al., 2012) suggesting a role in primary wall biosynthesis. However, the large DUF266 family, with 22 proteins in Arabidopsis and 5 in Selaginella moellendorffii (Table 1) is likely to comprise proteins of quite different biochemical activities, and currently there is insufficient published information to make any qualified guess on what these activities might be.
DUF246 – Plant Specific Fucosyltransferases?
In the search for new GT candidates, three sequences (At1g51630, At3g02250, At5g15740) annotated DUF246 (PF03138), were identified, by the use of HMM against GT65 (Hansen et al., 2009). In TAIR3, 34 Arabidopsis sequences were found to contain a DUF246 domain, and a clear GT signature related to GT65 was demonstrated. The DUF246 motif has been found in a variety of plant species, including the evolutionary basal moss Physcomitrella patens and spikemoss S. moellendorffii (Table 1). BLAST searches with DUF246 proteins identified the single representative of Arabidopsis in GT65 (At3g05320) as the most similar protein, even though this protein is not included in DUF246 (Hansen et al., 2009).
Carbohydrate active enzyme family GT65 comprises 42 eukaryote sequences from 40 species most of which are annotated as protein-O-fucosyltransferase 1 (Pofut1), an inverting enzyme that adds fucose to serine or threonine residues in Epidermal Growth Factor-repeats (Oriol et al., 1999). The crystal structure of Pofut1 from Caenorhabditis elegans shows a manganese independent GT-B fold solved with GDP-Fucose attached in the active site (Lira-Navarrete et al., 2011). Phe261 and Phe357 bind the nucleotide sugar and mutagenesis studies showed that Arg240 is important in catalysis and binding. In C. elegans Pofut1, Asn43, and Arg240 are important catalytic residues, Asn43 being the flexible amino acid involved in fucose binding (Lira-Navarrete et al., 2011). Strict conservation of any of these amino acids could not be observed in the many plant DUF246 sequences and therefore the substrate utilized by DUF246 might be different from the GDP-Fucose utilized by Pofut1 (see Figure 1B and the alignment in Figure A4B in Appendix). However, GT65 belongs to the large fucosyltransferase superfamily, and is distantly related to families GT11, GT23, and GT68, which all represent inverting fucosyltransferases.
DUF246 is a large family, with, e.g., 34 members in Arabidopsis and 19 in S. moellendorffii (Table 1). Definitive proof that any of these proteins are in fact GTs has not been presented, but the high similarity to the other members of PF10250 makes the GT assignment very likely. The O-fucosyltransferase domain (PF10250), into which DUF246 has been merged, was found among animals, plants, vira, and bacteria, whereas DUF246 was only found in predicted protein sequences from plant species and therefore appeared to be involved in biological processes specific to plants, such as cell wall biosynthesis (Figure 1E). α-Fucosyl residues in plants are present in N-glycans, xyloglucan, AGP, and RG-II. Since fucosyltransferases involved in synthesizing xyloglucan and AGP are in GT37 (Perrin et al., 1999; Wu et al., 2010) and fucosyltransferases involved in N-glycosylation are likely in GT10 (Wilson et al., 2001), we suggest that some of the DUF246 proteins are candidates for the RG-II fucosyltransferase. However, the DUF246 family is large and contains conserved, ancient clades (see Figure A2 in Appendix) so it is unlikely that all the proteins would have the same biochemical function. Several of the DUF246 proteins are co-expressed with GTs involved in secondary cell wall biosynthesis (Oikawa et al., 2010), suggesting another role than RG-II biosynthesis. A number of other DUF246 proteins are abundant in Arabidopsis cell suspension cultures, and found in the Golgi fractions, suggesting a role in primary cell wall biosynthesis (Parsons et al., 2012).
DUF23 – A Protein Family Related to GT92
GT92 is the newest family in CAZy that contains plant members. GT92 was created based on characterization of N-glycan core α1,6-fucoside β1,4-galactosyltransferase (GALT-1) from C. elegans (Titz et al., 2009). The GT92 proteins all contain a DUF23 motif. GALT-1 encodes a manganese dependent UDP-galactose galactosyltransferase that adds β-galactose to α1,6-linked fucose at the reducing end of GalNAc in N-glycan cores. By search for homologs, the new family GT92 was created, containing eukaryotic sequences from animals and plants. However, it is interesting to note that no β1,4-galactose or α1,6-fucose has been found in plant N-glycans, to date. Three subfamilies emerged after a phylogenetic study, one including GALT-1 and homologs from various animal species, one consisting of homologs exclusively from C. elegans and Caenorhabditis briggsae and the third comprising plant proteins (Titz et al., 2009). Sequence studies showed the proteins from GT92 to share the putative metal binding DxD motif and to have a predicted type II membrane protein topology (Titz et al., 2009). Although no structure has been solved for GT92 proteins, the DxD motif suggests a GT-A fold.
Another C. elegans protein with a DUF23 motif, BAH-1 (Q9XXM0_CAEEL), is expressed in seam cells and required for microbial biofilm attachment (Drace et al., 2009). The BAH-1 protein is not included in GT92 and it is not known if it is a GT. DUF23 is assigned to clan GT-A, which contains carbohydrate interacting proteins as well as multiple nucleotide sugar-dependent GT families (e.g., GT8 and GT43)4. The DUF23 domain is found in hypothetical proteins from animals and plants, and unlike GT92, DUF23 is also found in bacteria (Drace et al., 2009; Suzuki and Yamamoto, 2010). The DUF23 embraces all the sequences of GT92 and expands beyond, comprising about three times as many plant proteins as in GT92 (Table 1; Figure 1F). All the DUF23 proteins contain two conserved cysteine residues and several charged residues, including the DxD motif, involved in substrate binding (Figure 1C; Figure A4C in Appendix; Drace et al., 2009; Suzuki and Yamamoto, 2010). Since no GT92 protein has had its structure solved it is not currently possible to determine if the essential catalytic residues are conserved between GT92 and the rest of DUF23. However, because of the similarity and the conserved DxD motif we find it highly likely that the DUF23 proteins outside GT92 are also GTs. Many of the DUF23 proteins are predicted to have a single transmembrane domain in the N-terminal part of the protein, consistent with a type II membrane protein topology.
Four of the Arabidopsis sequences containing the DUF23 motif were found to be co-expressed with GTs within CAZy either directly (GT47: At5g62220 (GT18) and GT2: CesA6) or indirectly (GT2: CesA1, CesA2, CesA3, CslC5; GT8: GATL2 and AtGolS1; and GT47: At5g62220 (GT18)) by Atted-II5 (Obayashi et al., 2011).
The putative function of the DUF23 proteins in plants is not clear. GT92 is present in basal plant species, but DUF23 proteins outside of GT92 are present even in C. reinhardtii, indicating an ancient function of these proteins (see phylogenetic analysis of plant DUF23 proteins in Figure A3 in Appendix). Given that C. reinhardtii lacks most of the polysaccharides found in plants, a conserved function of the non-GT92 members of DUF23 is more likely to be related, e.g., to glycoprotein biosynthesis than to polysaccharide biosynthesis.
Future Perspectives
For the three groups of proteins discussed here, comprising proteins currently not in CAZy, we find that there is strong evidence that they are in fact GTs. Thus, they are all good candidates for reverse genetic studies and/or heterologous expression and enzyme activity experiments. These proteins would add 64 Arabidopsis proteins to the current 463 Arabidopsis GTs in CAZy. An obvious question is how many GTs are there beyond the 529 already present or suggested GTs. In the study of Hansen et al. (2009) other families were suggested, including DUF231 and DUF248, although a clear GT signature could only be demonstrated for DUF246, DUF266, and the single protein At5g28910. DUF231 proteins have recently been shown to be involved in polysaccharide acetylation (Gille et al., 2011) and they are likely to be subunits of acetyltransferase complexes rather than GTs (Anantharaman and Aravind, 2010; Manabe et al., 2011). For DUF248 there is evidence that they may be involved in polysaccharide methylation, but no biochemical evidence has been presented (Mouille et al., 2007). However, it should be noted that GT92 was added to CAZy after the bioinformatic study of Hansen et al. (2009) but GT92 and the related DUF23 were not identified as candidates in that study, which applied stringent filters to prevent erroneous identification of too many proteins that would later turn out not to be GTs. Apparently, even though DUF23 and GT92 belong to the GT-A clan, they are sufficiently diverged from other members of the GT-A clan that they were not identified through structural modeling. It shall be interesting to see to what extent the actual structures of GT92/DUF23 proteins differ from known GT-A structures.
The fact that GT92 was missed suggests that there may well be other families of GTs in plants yet to be discovered. Some GT families were founded following the identification of novel plant GTs, e.g., GT34 and GT37, rather than by GTs from other taxonomic groups. GT37 and GT77 are families that are unique to plants. This illustrates that plants have evolved some divergent GTs that cannot easily be identified through homology with GTs from other organisms. This is not surprising given that plants have many unique properties, e.g., the complex cell wall with a structure and biological role quite different from what is found in any other organisms. Identification of such plant unique families will require isolation of mutants through forward genetics screens or indications, e.g., from gene expression and localization analyses (Manfield et al., 2004; Brown et al., 2005; Persson et al., 2005; Oikawa et al., 2010; Mutwil et al., 2011; Sharma et al., 2011). To estimate the number of such unidentified GTs that might be present is very difficult. However, an indication may be that the last plant GT that was found as an unknown protein outside CAZy was OsBC10 reported in 2009 (Zhou et al., 2009) and it belonged to DUF266, which was already predicted to be putative GTs. In general, forward screens and coexpression studies identify GTs that are already in CAZy. Therefore, apart from the three protein families discussed in this review, we think that the number of unknown plant GTs is small.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Pfam group (pfam-help@sanger.ac.uk) for retrieving the datasets of DUF266 and DUF246. This work was funded by the US Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory. Sara Fasmer Hansen was supported by a fellowship from The Carlsberg Foundation though contract 2009_01_0346 and 2010_01_0509. Jesper Harholt was funded by a Villum-Kann Rasmussen grant to the Pro-Active Plant Centre and The Danish Council for Independent Research, Technology and Production Sciences, through contract 274-09-0314.
Footnotes
References
Anantharaman, V., and Aravind, L. (2010). Novel eukaryotic enzymes modifying cell-surface biopolymers. Biol. Direct 5, 1.
Aspeborg, H., Schrader, J., Coutinho, P. M., Stam, M., Kallas, A., Djerbi, S., Nilsson, P., Denman, S., Amini, B., Sterky, F., Master, E., Sandberg, G., Mellerowicz, E., Sundberg, B., Henrissat, B., and Teeri, T. T. (2005). Carbohydrate-active enzymes involved in the secondary cell wall biogenesis in hybrid aspen. Plant Physiol. 137, 983–997.
Atmodjo, M. A., Sakuragi, Y., Zhu, X., Burrell, A. J., Mohanty, S. S., Atwood, J. A. III, Orlando, R., Scheller, H. V., and Mohnen, D. (2011). Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc. Natl. Acad. Sci. U.S.A. 108, 20225–20230.
Bierhuizen, M. F., Mattei, M. G., and Fukuda, M. (1993). Expression of the developmental I antigen by a cloned human cDNA encoding a member of a beta-1,6-N-acetylglucosaminyltransferase gene family. Genes Dev. 7, 468–478.
Breton, C., Snajdrova, L., Jeanneau, C., Koca, J., and Imberty, A. (2006). Structures and mechanisms of glycosyltransferases. Glycobiology 16, 29R–37R.
Brown, D. M., Zeef, L. A., Ellis, J., Goodacre, R., and Turner, S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295.
Caffall, K. H., and Mohnen, D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900.
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active enzymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238.
Carpita, N., Tierney, M., and Campbell, M. (2001). Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol. Biol. 47, 1–5.
Dhugga, K. S. (2001). Building the wall: genes and enzyme complexes for polysaccharide synthases. Curr. Opin. Plant Biol. 4, 488–493.
Drace, K., Mclaughlin, S., and Darby, C. (2009). Caenorhabditis elegans BAH-1 is a DUF23 protein expressed in seam cells and required for microbial biofilm binding to the cuticle. PLoS ONE 4, e6741. doi:10.1371/journal.pone.0006741
Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., Gavin, O. L., Gunasekaran, P., Ceric, G., Forslund, K., Holm, L., Sonnhammer, E. L., Eddy, S. R., and Bateman, A. (2010). The Pfam protein families database. Nucleic Acids Res. 38, D211–D222.
Fukuda, M., Bierhuizen, M. F., and Nakayama, J. (1996). Expression cloning of glycosyltransferases. Glycobiology 6, 683–689.
Gille, S., De Souza, A., Xiong, G., Benz, M., Cheng, K., Schultink, A., Reca, I. B., and Pauly, M. (2011). O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell 23, 4041–4053.
Hansen, S. F., Bettler, E., Rinnan, A., Engelsen, S. B., and Breton, C. (2010). Exploring genomes for glycosyltransferases. Mol. Biosyst. 6, 1773–1781.
Hansen, S. F., Bettler, E., Wimmerova, M., Imberty, A., Lerouxel, O., and Breton, C. (2009). Combination of several bioinformatics approaches for the identification of new putative glycosyltransferases in Arabidopsis. J. Proteome Res. 8, 743–753.
Hwang, H. Y., Olson, S. K., Brown, J. R., Esko, J. D., and Horvitz, H. R. (2003). The Caenorhabditis elegans genes sqv-2 and sqv-6, which are required for vulval morphogenesis, encode glycosaminoglycan galactosyltransferase II and xylosyltransferase. J. Biol. Chem. 278, 11735–11738.
Lira-Navarrete, E., Valero-Gonzalez, J., Villanueva, R., Martinez-Julvez, M., Tejero, T., Merino, P., Panjikar, S., and Hurtado-Guerrero, R. (2011). Structural insights into the mechanism of protein O-fucosylation. PLoS ONE 6, e25365. doi:10.1371/journal.pone.0025365
Manabe, Y., Nafisi, M., Verhertbruggen, Y., Orfila, C., Gille, S., Rautengarten, C., Cherk, C., Marcus, S. E., Somerville, S., Pauly, M., Knox, J. P., Sakuragi, Y., and Scheller, H. V. (2011). Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 155, 1068–1078.
Manfield, I. W., Orfila, C., Mccartney, L., Harholt, J., Bernal, A. J., Scheller, H. V., Gilmartin, P. M., Mikkelsen, J. D., Paul Knox, J., and Willats, W. G. (2004). Novel cell wall architecture of isoxaben-habituated Arabidopsis suspension-cultured cells: global transcript profiling and cellular analysis. Plant J. 40, 260–275.
Mouille, G., Ralet, M. C., Cavelier, C., Eland, C., Effroy, D., Hematy, K., Mccartney, L., Truong, H. N., Gaudon, V., Thibault, J. F., Marchant, A., and Hofte, H. (2007). Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 50, 605–614.
Mutwil, M., Klie, S., Tohge, T., Giorgi, F. M., Wilkins, O., Campbell, M. M., Fernie, A. R., Usadel, B., Nikoloski, Z., and Persson, S. (2011). PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23, 895–910.
Obayashi, T., Nishida, K., Kasahara, K., and Kinoshita, K. (2011). ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 52, 213–219.
Oikawa, A., Joshi, H. J., Rennie, E. A., Ebert, B., Manisseri, C., Heazlewood, J. L., and Scheller, H. V. (2010). An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS ONE 5, e15481. doi:10.1371/journal.pone.0015481
Oriol, R., Mollicone, R., Cailleau, A., Balanzino, L., and Breton, C. (1999). Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates, and bacteria. Glycobiology 9, 323–334.
Pak, J. E., Arnoux, P., Zhou, S., Sivarajah, P., Satkunarajah, M., Xing, X., and Rini, J. M. (2006). X-ray crystal structure of leukocyte type core 2 beta1,6-N-acetylglucosaminyltransferase. Evidence for a convergence of metal ion-independent glycosyltransferase mechanism. J. Biol. Chem. 281, 26693–26701.
Parsons, H. T., Christiansen, K., Knierim, B., Carroll, A., Ito, J., Batth, T. S., Smith-Moritz, A. M., Morrison, S., McInerney, P., Hadi, M. Z., Auer, M., Mukhopadhyay, A., Petzold, C. J., Scheller, H. V., Loqué, D., and Heazlewood, J. L. (2012). Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel targets involved in plantcell wall biosynthesis. Plant Physiol. (in press).
Perrin, R. M., Derocher, A. E., Bar-Peled, M., Zeng, W. Q., Norambuena, L., Orellana, A., Raikhel, N. V., and Keegstra, K. (1999). Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284, 1976–1979.
Persson, S., Wei, H. R., Milne, J., Page, G. P., and Somerville, C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102, 8633–8638.
Sharma, R., Tan, F., Jung, K. H., Sharma, M. K., Peng, Z., and Ronald, P. C. (2011). Transcriptional dynamics during cell wall removal and regeneration reveals key genes involved in cell wall development in rice. Plant Mol. Biol. 77, 391–406.
Suzuki, N., and Yamamoto, K. (2010). Molecular cloning of pigeon UDP-galactose:beta-D-galactoside alpha1,4-galactosyltransferase and UDP-galactose:beta-D-galactoside beta1,4-galactosyltransferase, two novel enzymes catalyzing the formation of Gal alpha1-4Gal beta1-4Gal beta1-4GlcNAc sequence. J. Biol. Chem. 285, 5178–5187.
Titz, A., Butschi, A., Henrissat, B., Fan, Y. Y., Hennet, T., Razzazi-Fazeli, E., Hengartner, M. O., Wilson, I. B., Kunzler, M., and Aebi, M. (2009). Molecular basis for galactosylation of core fucose residues in invertebrates: identification of Caenorhabditis elegans N-glycan core alpha1,6-fucoside beta1,4-galactosyltransferase GALT-1 as a member of a novel glycosyltransferase family. J. Biol. Chem. 284, 36223–36233.
Wilson, I. B., Rendic, D., Freilinger, A., Dumic, J., Altmann, F., Mucha, J., Muller, S., and Hauser, M. T. (2001). Cloning and expression of cDNAs encoding alpha1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim. Biophys. Acta 1527, 88–96.
Wu, Y., Williams, M., Bernard, S., Driouich, A., Showalter, A. M., and Faik, A. (2010). Functional identification of two nonredundant Arabidopsis alpha(1,2)fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285, 13638–13645.
Ye, C. Y., Li, T., Tuskan, G. A., Tschaplinski, T. J., and Yang, X. (2011). Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitis. Plant Sci. 181, 688–695.
Yeh, J. C., Ong, E., and Fukuda, M. (1999). Molecular cloning and expression of a novel beta-1, 6-N-acetylglucosaminyltransferase that forms core 2, core 4, and I branches. J. Biol. Chem. 274, 3215–3221.
Zhou, Y., Li, S., Qian, Q., Zeng, D., Zhang, M., Guo, L., Liu, X., Zhang, B., Deng, L., Luo, G., Wang, X., and Li, J. (2009). BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J. 57, 446–462.
Appendix
Figure A1. Phylogenetic trees of DUF266/GT14 containing proteins from selected plants. The tree includes sequences from arabidopsis (ARATH), rice (ORYSJ) and Selaginella moellendorffii (SELML). After manual curation of sequences the tree was generated using MUSCLE alignment, removal of gaps, phyML, and statistical analysis using SH-like approximate likelihood-ratio test (aLRT). The tree was generated using http://www.phylogeny.fr (Dereeper et al., 2008). Following sequences Q0DT08 and Q0JPU3 were omitted from the tree due to sequence mistakes, mainly intron/exon border malpredictions. Asterisks indicate the selected sequences of the multi alignment displayed in Figure A4.
Figure A2. Phylogenetic trees of DUF246/GT65 containing proteins from selected plants. The tree includes sequences from arabidopsis (ARATH), rice (ORYSJ), and S. moellendorffii (SELML). After manual curation of sequences the tree was generated using MUSCLE alignment, removal of gaps, phyML, and statistical analysis using SH-like approximate likelihood-ratio test (aLRT). The tree was generated using http://www.phylogeny.fr (Dereeper et al., 2008). Following sequences 8QTM0, D8SPE4, D8TE31, Q0DBZ9, Q53PH8, Q5N9U3, Q651P4, Q653G2, and Q7XAB1 were omitted from the tree due to sequence mistakes, mainly intron/exon border malpredictions. Asterisks indicate the selected sequences of the multi alignment displayed in Figure A4.
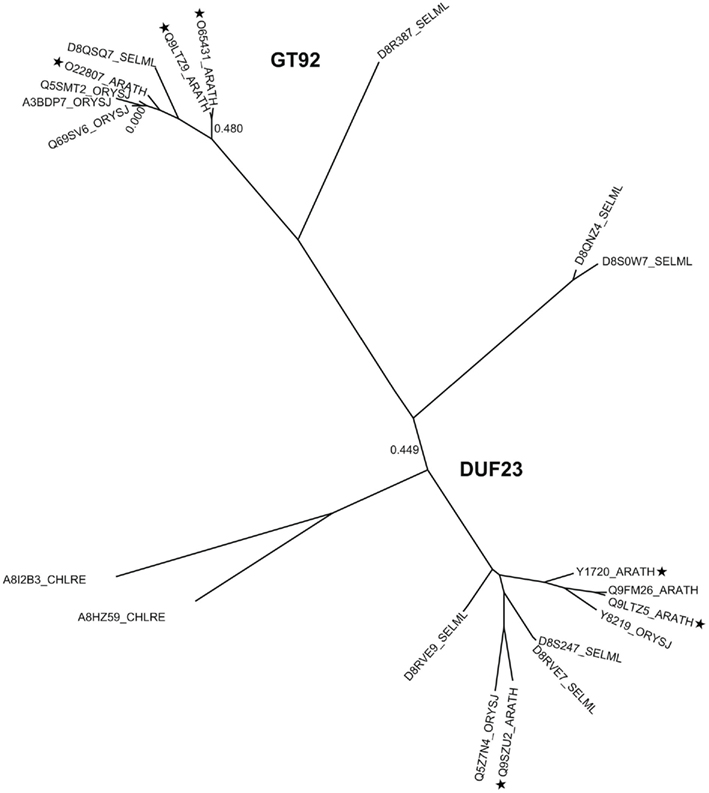
Figure A3. Phylogenetic trees of DUF23 including GT92, containing proteins from selected plants. The tree includes sequences from arabidopsis (ARATH), rice (ORYSJ), and S. moellendorffii (SELML). After manual curation of sequences the tree was generated using MUSCLE alignment, removal of gaps, phyML, and statistical analysis using SH-like approximate likelihood-ratio test (aLRT). The tree was generated using http://www.phylogeny.fr (Dereeper et al., 2008). Following sequences A8HZ53 and A8I2B1 were omitted from the tree due to sequence mistakes, mainly intron/exon border malpredictions. Asterisks indicate the selected sequences of the multi alignment displayed in Figure A4.
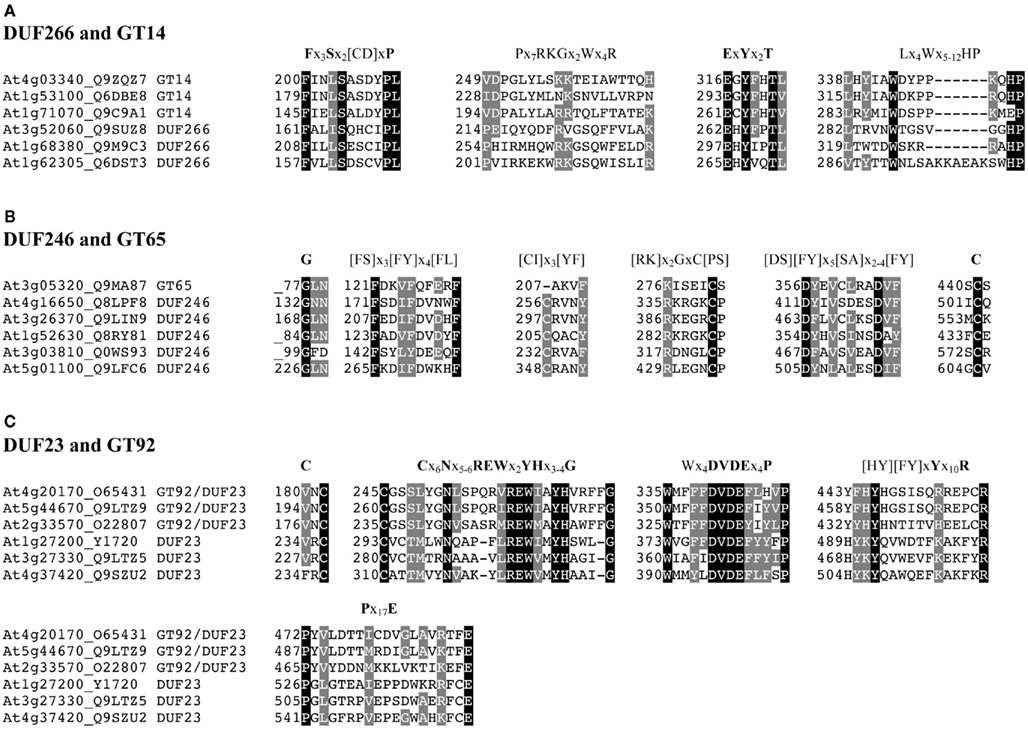
Figure A4. Multiple sequence alignment of the conserved peptide motifs identified in Arabidopsis. Representatives of protein sequences from (A) DUF266 and GT14, (B) DUF246 and GT65, and (C) DUF23 (including GT92), from different phylogenetic clades. The selected sequences are marked with asterisks in Figures A1–3. The sequence alignments were performed using ClustalW and manually refined. The invariant similar residues are indicated on a black background and other conserved positions are shaded in gray. Dashes indicate gaps. Numbers indicate the amino acid position of the first residue of each motif. Schematic illustration of conserved motifs for (A) DUF266 and GT14, (B) DUF246 and GT65, and (C) DUF23, based on non-redundant Arabidopsis sequences are shown above each alignment. Strictly conserved residues are in bold and residues with only two alternative amino acids are denoted by parentheses.
References
Keywords: cell walls, DUF23, DUF246, DUF266, glycosyltransferases
Citation: Hansen SF, Harholt J, Oikawa A and Scheller HV (2012) Plant glycosyltransferases beyond CAZy: a perspective on DUF families. Front. Plant Sci. 3:59. doi: 10.3389/fpls.2012.00059
Received: 02 February 2012; Accepted: 10 March 2012;
Published online: 28 March 2012.
Edited by:
Jose Manuel Estevez, University of Buenos Aires and CONICET, ArgentinaReviewed by:
Richard Strasser, University of Natural Resources and Life Sciences, AustriaUener Kolukisaoglu, University of Tuebingen, Germany
Copyright: © 2012 Hansen, Harholt, Oikawa and Scheller. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Henrik V. Scheller, Feedstocks Division, Joint BioEnergy Institute, Lawrence Berkeley National Laboratory, 5885 Hollis Street, Emeryville, CA 94608, USA. e-mail: hscheller@lbl.gov
 Sara Fasmer Hansen1
Sara Fasmer Hansen1