- 1Key Laboratory of Adaptation and Evolution of Plateau Biota, Northwest Institute of Plateau Biology, Institute of Sanjiangyuan National Park, Chinese Academy of Sciences, Xining, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3School of Pharmacy, Weifang Medical University, Weifang, China
- 4Qinghai Provincial Key Laboratory of Crop Molecular Breeding, Xining, China
- 5Institute for Biology and Environmental Sciences, Carl von Ossietzky Universität Oldenburg, Oldenburg, Germany
Parnassia L., a perennial herbaceous genus in the family Celastraceae, consists of about 60 species and is mainly distributed in the Pan-Himalayan and surrounding mountainous regions. The taxonomic position and phylogenetic relationships of the genus are still controversial. Herein, we reassessed the taxonomic status of Parnassia and its intra- and inter-generic phylogeny within Celastraceae. To that end, we sequenced and assembled the whole plastid genomes and nuclear ribosomal DNA (nrDNA) of 48 species (74 individuals), including 25 species of Parnassia and 23 species from other genera of Celastraceae. We integrated high throughput sequence data with advanced statistical toolkits and performed the analyses. Our results supported the Angiosperm Phylogeny Group IV (APG IV) taxonomy which kept the genus to the family Celastraceae. Although there were topological conflicts between plastid and nrDNA phylogenetic trees, Parnassia was fully supported as a monophyletic group in all cases. We presented a first attempt to estimate the divergence of Parnassia, and molecular clock analysis indicated that the diversification occurred during the Eocene. The molecular phylogenetic results confirmed numerous taxonomic revisions, revealing that the morphological characters used in Parnassia taxonomy and systematics might have evolved multiple times. In addition, we speculated that hybridization/introgression might exist during genus evolution, which needs to be further studied. Similarly, more in-depth studies will clarify the diversification of characters and species evolution models of this genus.
Introduction
Parnassia L. (Celastraceae) comprises approximately 60 perennial herbaceous species, and is widely distributed in the Arctic and temperate regions of the Northern Hemisphere (Phillips, 1982; Hultgard, 1987; Gu and Hultgård, 2001; Simmons, 2004). Most Parnassia species are endemic to China and have limited distribution, with >30 species only restricted to the Pan-Himalayan region (Hultgard, 1987), which is considered as an important diversity center of Parnassia (Handel-Mazzetti, 1941; Phillips, 1982; Ku, 1987; Wu, 2003; Simmons, 2004). The phylogenetic position of the genus is debatable, and prior studies showed that Parnassia should be treated as a member of Saxifragaceae (Hooker and Thomson, 1858; Engler, 1930; Cronquist and Takhtadzh n, 1981; Gu and Hultgård, 2001). However, Parnassia had also been suggested being closely related to Droseraceae morphologically (especially with the characteristics gynoecium; Pace, 1912; Dahlgren, 1980), to Hypericaceae (based on androecium and Chemotaxonomy through flavonoids; Arber, 1913; Jay, 1970), to Nymphaeaceae (based on vegetative characters; Hallier, 1901), and to Ochnaceae (based on peculiar staminodes; Bennett, 1869). Many systematists (e.g., Drude, 1875; Murbeck, 1918; Sharma, 1968; Klopfer, 1972; Hultgard, 1987) even suggested that Parnassia should be recognized as a separate family or order (e.g., Takhtadzhian and Takhtajan, 1997; Wu, 2003). The taxonomic debate over the genus seems to have ended over the last decades, especially with the advancement in molecular systematics. In contrast to the long-debated morphology-based systematics, more consistent results of the phylogenetic position of Parnassia were revealed by molecular studies, which suggested that Parnassia might be more closely related to Celastraceae than other families (Savolainen et al., 1997; Soltis et al., 1997, 2000; Soltis and Soltis, 1997; Simmons et al., 2001). Other studies subsequently confirmed this result, and Parnassia has been recovered as an early derived lineage of Celastraceae (Zhang and Simmons, 2006; Simmons et al., 2012; Bacon et al., 2016; Li et al., 2019). However, all these studies were based on a limited number of species, especially for those from its center of distribution and differentiation. Similarly, the molecular DNA markers used in these studies also lacked sufficient phylogenetic signals (Fischer and Steel, 2009; Philippe et al., 2011). Therefore, it is essential to provide molecular markers with sufficient informative characters to reconstruct the phylogenetic relationship of Parnassia.
n, 1981; Gu and Hultgård, 2001). However, Parnassia had also been suggested being closely related to Droseraceae morphologically (especially with the characteristics gynoecium; Pace, 1912; Dahlgren, 1980), to Hypericaceae (based on androecium and Chemotaxonomy through flavonoids; Arber, 1913; Jay, 1970), to Nymphaeaceae (based on vegetative characters; Hallier, 1901), and to Ochnaceae (based on peculiar staminodes; Bennett, 1869). Many systematists (e.g., Drude, 1875; Murbeck, 1918; Sharma, 1968; Klopfer, 1972; Hultgard, 1987) even suggested that Parnassia should be recognized as a separate family or order (e.g., Takhtadzhian and Takhtajan, 1997; Wu, 2003). The taxonomic debate over the genus seems to have ended over the last decades, especially with the advancement in molecular systematics. In contrast to the long-debated morphology-based systematics, more consistent results of the phylogenetic position of Parnassia were revealed by molecular studies, which suggested that Parnassia might be more closely related to Celastraceae than other families (Savolainen et al., 1997; Soltis et al., 1997, 2000; Soltis and Soltis, 1997; Simmons et al., 2001). Other studies subsequently confirmed this result, and Parnassia has been recovered as an early derived lineage of Celastraceae (Zhang and Simmons, 2006; Simmons et al., 2012; Bacon et al., 2016; Li et al., 2019). However, all these studies were based on a limited number of species, especially for those from its center of distribution and differentiation. Similarly, the molecular DNA markers used in these studies also lacked sufficient phylogenetic signals (Fischer and Steel, 2009; Philippe et al., 2011). Therefore, it is essential to provide molecular markers with sufficient informative characters to reconstruct the phylogenetic relationship of Parnassia.
Besides the debatable phylogenetic position of Parnassia, infrageneric systematics of the genus were also problematic. The distribution of Parnassia species in the Pan-Himalayan region often overlaps, and considerable morphological variation may occur within the same species. Scholars often have a different understanding of taxonomic traits, which led to different opinions about the infrageneric classification of the genus, including different ranks as section and series (Drude, 1875; Franchet, 1897; Engler, 1930; Handel-Mazzetti, 1941; Ku, 1987; Wu, 2003, 2005; Shu, 2017), and a plethora of taxonomic revisions (Turner and Veldkamp, 2001; Wu et al., 2004, 2008, 2009; Shu and Zhang, 2016, 2017; Wang et al., 2018; Yu et al., 2018; Ma et al., 2020; Dai et al., 2021). However, most of these investigations were based on morphological characters and need to be validated with molecular phylogenetic studies. Molecular phylogenetics for the genus based on nuclear ITS as well plastidial trnL-F and trnT-L markers suggested that Parnassia is monophyletic and that parallel evolution of morphological traits may occur (Wu, 2005). Yu (2019) reconstructed the phylogeny of Parnassia with 19 high polymorphic fragments of the plastid genome and suggested that the complete plastid genome dataset would be more comprehensive. Different studies have revealed that phylogenetic resolution and reliability seem to be related to the taxon as well as gene sampling (Wortley et al., 2005; Philippe et al., 2011). Since prior molecular phylogeny analyses on Parnassia are inadequate, there is an urgent need to construct a stable and precise phylogenetic tree to substantiate or negate the hypotheses made about Parnassia infrageneric systematics.
With the rapid development of next-generation sequencing technology, the amount of data used in phylogenetic research increased exponentially. Large-scale genomic data, such as plastid genome sequences and nuclear ribosomal DNA (nrDNA) data, can provide more phylogenetic informative sites than those of only a few DNA fragments. Recently, the plastid genome data has been successfully applied to resolve long-standing debates in phylogenetic positions of different taxa: e.g., the phylogenetic positions of Paeoniaceae within Saxifragales (Dong et al., 2018), Selaginellaceae (Zhang et al., 2019), and sanguinolenta group (Zhang H. R. et al., 2020). In addition, the plastid genome has also been employed to resolve the phylogenetic relationships of various plant lineages (Zhai et al., 2019; Liu et al., 2020; Ma et al., 2021). However, few plastid genomes have been sequenced for Parnassia and Celastraceae so far, which hinders the process of phylogenetic study of Parnassia (e.g., Wu, 2005; Yang et al., 2012).
With the development of molecular systematics, the phylogenetic research of Parnassia has made rapid progress. However, there is still a lack of large-scale data containing substantial phylogenetic signals (e.g., the plastid genome) for inferring the phylogenetic relationship of the genus. In addition, a large number of taxonomic revisions of Parnassia in recent years need to be confirmed by more reliable molecular systematics. Therefore, the present study attempts to resolve infrageneric phylogenetic questions about Parnassia using the whole plastid genomes and nrDNA (18S-ITS1-5.8S-ITS2-26S) data. The sampled species included seven of the nine sections of Chinese Parnassia (Ku, 1995; Wu, 2003) and covered the major species of Parnassia in the Pan-Himalayan region. The aims of this study are (1) to provide a robust and highly resolved phylogenetic tree for the phylogenetic position of Parnassia; and (2) to reassess the reliability of morphological features that have been widely used for taxonomic and phylogenetic considerations.
Materials and Methods
Sampling, Genome Sequencing, and Annotation
We sampled a total of 74 individuals, including 25 species (48 individuals) of Parnassia and 23 species (24 individuals) from all other 14 genera of Celastraceae, and two outgroups (Supplementary Table 1). To show the relationships within the species, we included two or more individuals from different localities for some species. Voucher specimens for the species sampled were deposited in the Qinghai-Tibetan Plateau Museum of Biology (HNWP), Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Besides these 74 individuals, we retrieved 89 plastid genomes from the GenBank (Supplementary Table 2). In the final dataset, most orders of Superrosids were included, and two individuals of Asteraceae were selected as outgroups. We re-annotated the downloaded plastid genome sequence files using PGA (Qu et al., 2019) to ensure the accuracy of the gene coding sequences (CDSs) data obtained from GenBank in the analysis.
We extracted total genomic DNA using about 10 mg of silica-dried leaf tissue through modified CTAB protocols (Doyle and Doyle, 1987). The extracted DNAs of all the individuals were then sent to Novogene (Beijing, China). Genomic DNA library was generated using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, United States) following the manufacturer’s recommendations, and index codes were added to each sample and sequenced on an Illumina HiSeq 2500 sequencer (San Diego, CA, United States) using the paired-end option (2 × 150 bp). Sequencing quality was assessed through FastQC v. 0.11.8 (Andrews, 2010). Where necessary, we used Trimmomatic v. 0.33 (Bolger et al., 2014) to clean the sequencing data. The plastid genomes and nrDNA (18S-ITS1-5.8S-ITS2-26S) were assembled de novo using GetOrganelle (Jin et al., 2020) and then visually assembled, resulting in bandage v. 0.8.1 (Wick et al., 2015). Annotation of plastid genome was performed through the online program GeSeq1 (Tillich et al., 2017) with manual adjustment of start/stop codons and intron/exon borders in Sequin v. 15.502. The nrDNA was annotated manually with the published ribosomal data of Parnassia palustris L. (AY929353), Parnassia fimbriata K. D. Koenig (AF036496), and Trifolium repens L. (MT735335).
Sequence Alignment and Dataset Feature Evaluation
To reconstruct the phylogeny of Parnassia and its phylogenetic position among the Superrosids, we generated seven different datasets (datasets 1–7). We used 71 shared CDSs based on 88 annotated plastomes (83 plastomes from NCBI and five newly generated) to reassess the reliability of the morphological-based phylogenetic position of Parnassia (see details in Supplementary Table 3). Similarly, we used 73 shared CDSs data based on 80 annotated plastomes (six downloaded ones and the rest are newly generated) to reconstruct the infrageneric systematic of Parnassia (see details in Supplementary Table 3). To extract the shared CDSs, we used the program PhyloSuite v1.2.2 [see details in Zhang D. et al. (2020)]. We generated three different pairs of chloroplast datasets: the concatenated CDSs (datasets 1 and 4), utilizing only the first and second codon sites of the concatenated CDSs (datasets 2 and 5), and the third codon site of the concatenated CDSs (datasets 3 and 6). Additionally, we generated the nrDNA (18S-ITS1-5.8S-ITS2-26S; 5,879 bp) to explore the phylogenetic relationships of Parnassia (dataset 7). The alignment of datasets was conducted using MAFFT V.7.409 with the codon matching strategy (Katoh and Standley, 2013). To check the suitability of all the datasets, we performed the sequence substitution saturation test through DAMBE7 (Xia, 2018).
Phylogenetic Analyses
We used the maximum likelihood (ML) and Bayesian inference (BI) statistics to reconstruct the Parnassia phylogenetic tree. Before tree reconstructions, we specified the GTR + G + I model inferred by jModelTest 2.1.6 (Darriba et al., 2012) under the Akaike information criterion (AIC) (Supplementary Table 4). For the ML analysis, we used the program RAxML 8.2.12 (Stamatakis, 2014), utilizing the option of 1,000 rapid bootstrap replicates. Similarly, BI analyses were carried out by MrBayes v 3.2.7a (Ronquist et al., 2012) implemented in the CIPRES Science Gateway V 3.3 (Miller et al., 2010). Each BI analysis was conducted with two independent runs and four Monte Carlo Markov chains (MCMCs) of ten million generations, and trees were sampled every 1,000 generations. The first 25% of the sampled trees were discarded as burn-in, and the remaining were used to generate the consensus tree and calculate the Bayesian posterior probabilities (PP).
Molecular Dating Analyses and Ancestral Area Reconstruction
We used the concatenated CDSs of chloroplast (dataset 4) and estimated the divergence time of Parnassia utilizing the Bayesian statistics as implemented in BEAST v1.10.4 (Drummond et al., 2012). The parameters were set as GTR + I + Γ model and uncorrelated relaxed lognormal clock. The birth–death speciation model of the tree prior was selected according to the value of marginal likelihood, which was estimated by path sampling/stepping-stone sampling. The chain length of MCMC generations and the sampling frequency were set to 200,000,000 and 20,000, respectively. Generally, there may be many factors for the error in molecular clock analysis (e.g., taxon and gene sampling numbers, fossil calibrations strategy), which lead to bias in divergence time estimates (Sauquet et al., 2012; Bacon et al., 2016; Foster et al., 2017; Barba-Montoya et al., 2018). Due to the lack of reliable fossil evidence for the small perennial herbaceous plants of Parnassia (Coope et al., 1961), we selected four fossil records of its close relative taxa within Celastraceae as calibrations point to get a more reliable estimated divergence time (Figure 3). The strategies of divergence time estimation were as follows: Firstly, we selected the four most reliable evaluated fossils (Bacon et al., 2016) of relative taxa as fossil calibrations to avoid the generation of misleading results. Secondly, we employed a more informative dataset to reduce bias caused by a single gene (Zhai et al., 2019). We used four different times constraints based on the fossil records: (1) exponential priors (offset = 50.5, mean = 0.5) were set for the stem node of the monotypic Catha (following Poole and Wilkinson, 1999); (2) normal prior (mean = 26, St. dev. = 1) was set for the crown age of Celastrus + Tripterygium clade (following Xi et al., 2012; Bacon et al., 2016; Zhu et al., 2020); (3) exponential priors (offset = 56.5, mean = 0.833) were set for the stem node of Glyptopetalum + Euonymus (following Pitman and Rowan, 2012); and (4) secondary age was estimated from Xi et al. (2012) and Bacon et al. (2016), and a normal prior (mean = 90.6, St. dev. = 8.1) was set for the crown clade of Celastrales. We used BEAUti to generate the XML input files for BEAST, and the runs were performed on the CIPRES website (Miller et al., 2010). A total of four independent runs were followed in parallel, and the log files were combined using LogCombiner v1.10.4 (Drummond et al., 2012). We used Tracer v1.7 (Rambaut et al., 2018) to check the convergence of effective sample size (ESS). Lastly, the maximum clade credibility (MCC) tree with median heights was generated using TreeAnnotator v1.10.4 with the initial 10% trees discarded as burn-in. The final tree was visualized and edited using FigTree v.1.4.33.
To assess the possible ancestral distribution regions of Parnassia, we performed an ancestral region reconstruction analysis in RASP v4.2 (Yu et al., 2020). The distribution regions of Parnassia were defined as belonging to the Pan-Himalayan and/or non-Pan-Himalayan. According to the distribution of species, we set the parameter “Max area” to two. The biogeographic models DEC, DIVALIKE, and BAYAREALIKE with/without J-parameter were tested by the R package “BioGeoBEARS” (Matzke, 2014), and the best-fit model was selected based on the Akaike Information Criterion (AIC) values.
Results
Plastomes and Nuclear Ribosomal DNA Assemblies
We obtained a total of 778.1 Gb sequencing data for all the 74 individuals with an average of 10.51 Gb ranging from 4.7 Gb (Ribes glaciale Wallich) to 15.86 Gb (Parnassia nubicola Wallich ex Royle) (Supplementary Table 1). The sizes of complete plastome ranged from 147,646 bp (Saxifraga sinomontana J. T. Pan & Gornall) to 163,053 bp (Salacia obovatilimba S. Y. Pao), and the sizes of nrDNA (18S-ITS1-5.8S-ITS2-26S) ranged from 5,783 bp (Microtropis henryi Merrill & F. L. Freeman) to 5,879 bp (Parnassia foliosa J. D. Hooker & Thomson). All the plastid genomes obtained in this study were composed of a typical quadripartite structure, similar to the previously published genomes of Celastraceae (Supplementary Figure 1). The total number of annotated genes varied between 113 and 115, including 79–81 protein-coding genes, four rRNA genes, and 30 tRNA genes, respectively. Overall, the GC content in these species ranged from 38.10% in S. sinomontana to 36.90% in P. arborea (Supplementary Table 1).
Phylogenetic Position of Parnassia
The results about the suitability of the datasets used to investigate the phylogenetic position of Parnassia showed that the Iss values were significantly smaller than the Iss–c values (Supplementary Table 4). This suggested that these datasets had not reached substitution saturation and could be used for phylogenetic analyses (Xia et al., 2003). Though the phylogenetic trees were reconstructed with different datasets and different methods, all the results showed highly congruent topologies. Similarly, the relationships among different taxa were highly supported, suggesting that large-scale genomic data indeed significantly improved the resolution of phylogenetic trees (details see in Figure 1 and Supplementary Figures 2–5).
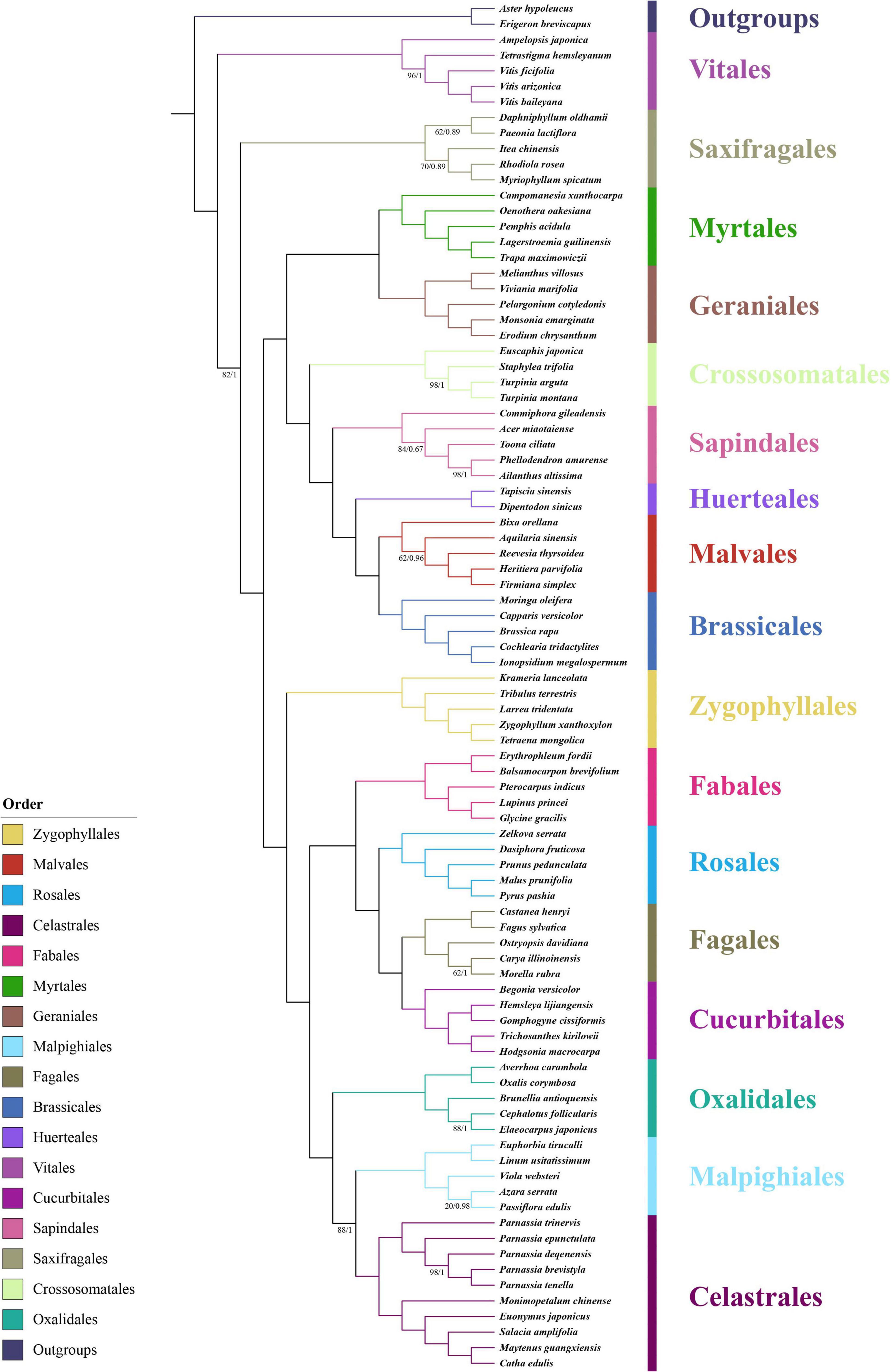
Figure 1. Phylogenetic position of the genus Parnassia in Superrosids revealed by the concatenated data sets of 71 CDSs. Numbers associated with branches are ML bootstrap values and Bayesian posterior probabilities. Nodes without numbers indicate 100% bootstrap support and 1.0 posterior probability.
Our results showed that the phylogenetic relationships among all the 17 orders of Superrosids were strongly supported, with each clade corresponding to a tribe that was defined in previous studies. Celastrales, Malpighiales, and Oxalidales formed the COM clade; Cucurbitales, Fagales, Rosales, and Fabales clustered in the Fabid clade (nitrogen-fixing clade); COM, Fabid, and Zygophyllales formed the Fabidae; while Brassicales, Malvales, Huerteales, Sapindales, Crossosomatales, Geraniales, and Myrtales grouped into the malvid clade. Similarly, the Fabidae and malvid clades were sisters to one other. All the phylogenetic trees supported the above results, except the relationship among Saxifragales, Vitales, Fabidae, and malvid. In datasets (1 and 2), the relationship was resolved as (Fabidae, malvid), Saxifragales), Vitales, while in dataset 3, the relationship was resolved as [(Fabidae, malvid), Vitales], Saxifragales (Figure 1 and Supplementary Figures 2–5). Consistent with previous studies, Parnassia was clustered with the remaining genera of Celastraceae in all trees with fully supported (bootstrap support (BS) = 100, posterior probability (PP) = 1) (Figure 1 and Supplementary Figures 2–5).
Phylogenetic Relationship of Parnassia
All the plastome datasets (datasets 4, 5, and 6) generated consistent trees with identical topologies of Parnassia, except for some uncertain clades (Figure 2 and Supplementary Figures 6–9). Phylogenetic trees showed that Parnassia was sister to the rest of Celastraceae with high support (BS = 100, PP = 1). In addition, all individuals of Parnassia were clustered together in all cases, indicating a well-supported monophyly of the genus. Parnassia sect. Nectarotrilobos Drude, the largest section in this genus, shows paraphyly, where some species of sect. Nectarotrilobos clustered with Clade I (Parnassia lutea Batalin, Parnassia nubicola Wallich ex Royle, Parnassia oreophila Hance, Parnassia trinervis Drude, Parnassia viridiflora Batalin, and Parnassia filchneri Ulbrich) and Clade II (Parnassia pusilla Wallich ex Arnott, Parnassia mysorensis F. Heyne ex Wight & Arnott, and Parnassia deqenensis T. C. Ku). However, the species of sect. Nectarotrilobos subsect. Xiphosandra (Franchet) Ku [Parnassia brevistyla (Brieger) Handel-Mazzetti, Parnassia delavayi Franchet, and Parnassia leptophylla Handel-Mazzetti] clustered together (Clade III). In addition, the species, Parnassia chinensis Franchet of sect. Nectarotrilobos was highly supported (BS = 100, PP = 1) as the sister to Parnassia longipetala Handel-Mazzetti of sect. Saxifragastrum Drude. Similarly, individuals of sect. Nectaroquinquelobos Ku (Parnassia longshengensis T. C. Ku and Parnassia gansuensis T. C. Ku) were not resolved as one clade, and so were the individuals of sect. Saxifragastrum Drude (Parnassia tenella Hooker & Thomson and P. longipetala) and sect. Fimbripetalum Drude (Parnassia foliosa Hooker & Thomson and Parnassia noemiae Franchet).
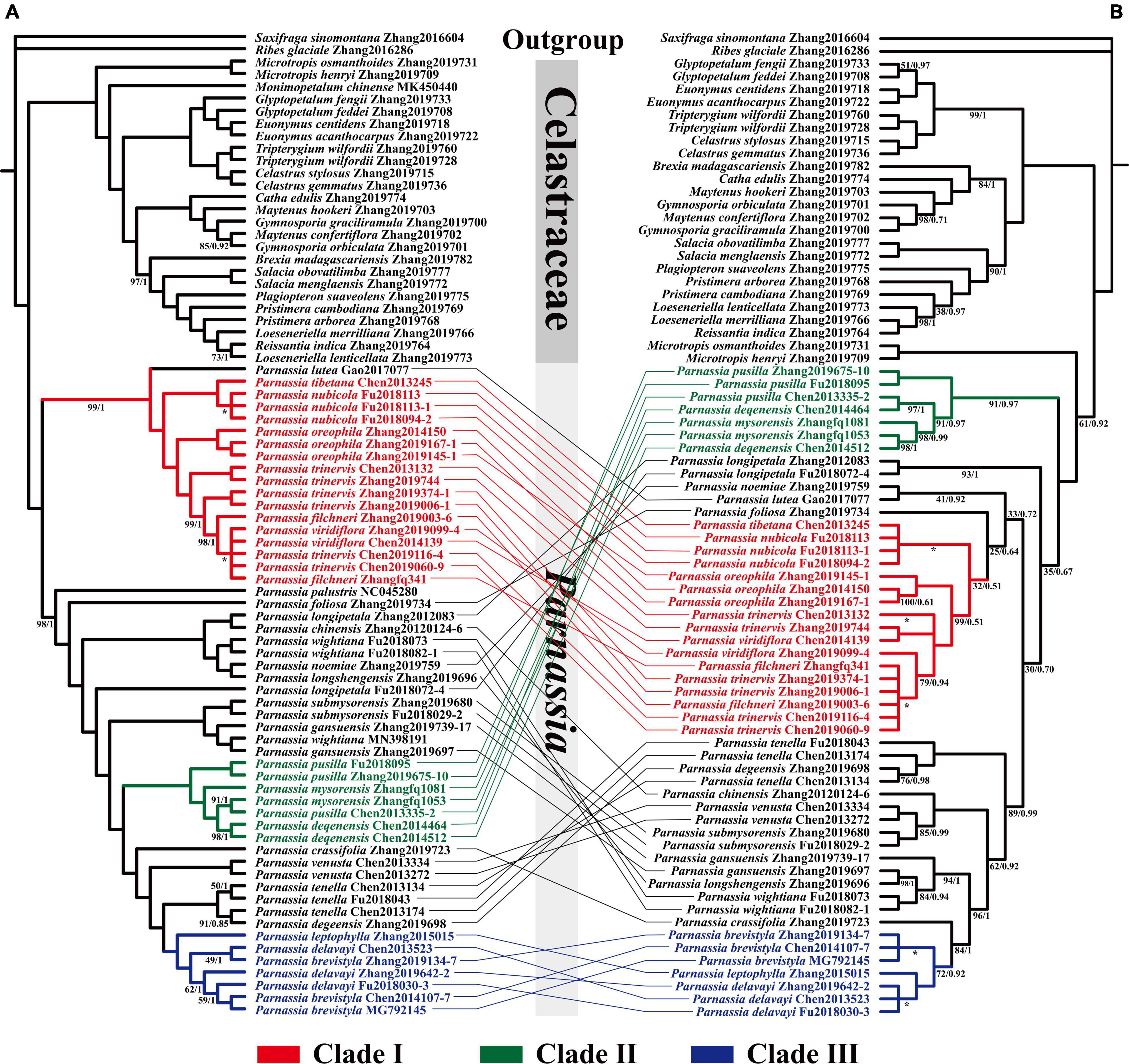
Figure 2. Phylogenetic relationship of Parnassia resolved based on 73 CDSs concatenated genes (A) and nrDNA marker (B). Numbers associated with branches are ML bootstrap values and Bayesian posterior probabilities. Nodes without numbers indicate 100% bootstrap support and 1.0 posterior probability. Nodes with * indicate that the topological structure of ML tree and BI tree is inconsistent at this branch.
The topology of BI and ML analysis, reconstructed with the nrDNA sequence data, showed the same results. However, it was incongruent with the phylogeny retrieved by plastomes datasets (Supplementary Figures 6–9). In the nrDNA phylogenetic tree, the position of Clade I and Clade II was reversed (Figure 2). Three species of sect. Nectarotrilobos (P. chinensis (Parnassia venusta Jien, Parnassia submysorensis J. T. Pan) formed a highly supported clade (BS = 100, PP = 1). Similarly, P. longshengensis, P. gansuensis, and Parnassia wightiana Wallich ex Wight & Arnott were also recovered as a clade with enough support (BS = 94, PP = 1). Besides, concordant results of both the plastome-based and the nrDNA phylogenetic trees were shown in some branches (Figure 2), i.e., individuals of Clade I, II, and III were strongly recovered with the same topology in the nrDNA tree (except for P. lutea of Clade I).
Divergence Time Estimates and Ancestral Range Inference
Bayesian relaxed molecular clock analyses suggested that the split between Parnassia and the remaining genera of Celastraceae occurred at 113.68 Mya, with the highest posterior density (HPD) intervals being 97.36–128.24 Mya (Figure 3). Within Parnassia, the first divergence occurred around 39.97 Mya (95% HPD = 28.13–52.93 Mya, Figure 3). The crown age of Clade A was inferred to be 36.18 Mya (95% HPD = 24.16–49.84 Mya, Figure 3). The mean divergence times of (P. nubicola and P. tibetana) and P. oreophila from the remaining species of Clade A were 26.37 Mya (95% HPD = 13.88–39.02 Mya) and 16.95 Mya (95% HPD = 7.20–28.83 Mya), respectively (Figure 3). The crown age of P. palustris was dated at 37.25 Mya (95% HPD = 26.57–49.67 Mya), followed by the crown age of P. foliosa at 30.30 Mya (95% HPD = 20.69–41.34 Mya) (Figure 3). The stem ages of Clade B were estimated to be 16.81 Mya (95% HPD = 10.19–23.09 Mya, Figure 3). The mean divergence times of Clade C and subsect. Xiphosandra were dated to 10.71 Mya (95% HPD = 6.10–15.78 Mya, Figure 3).
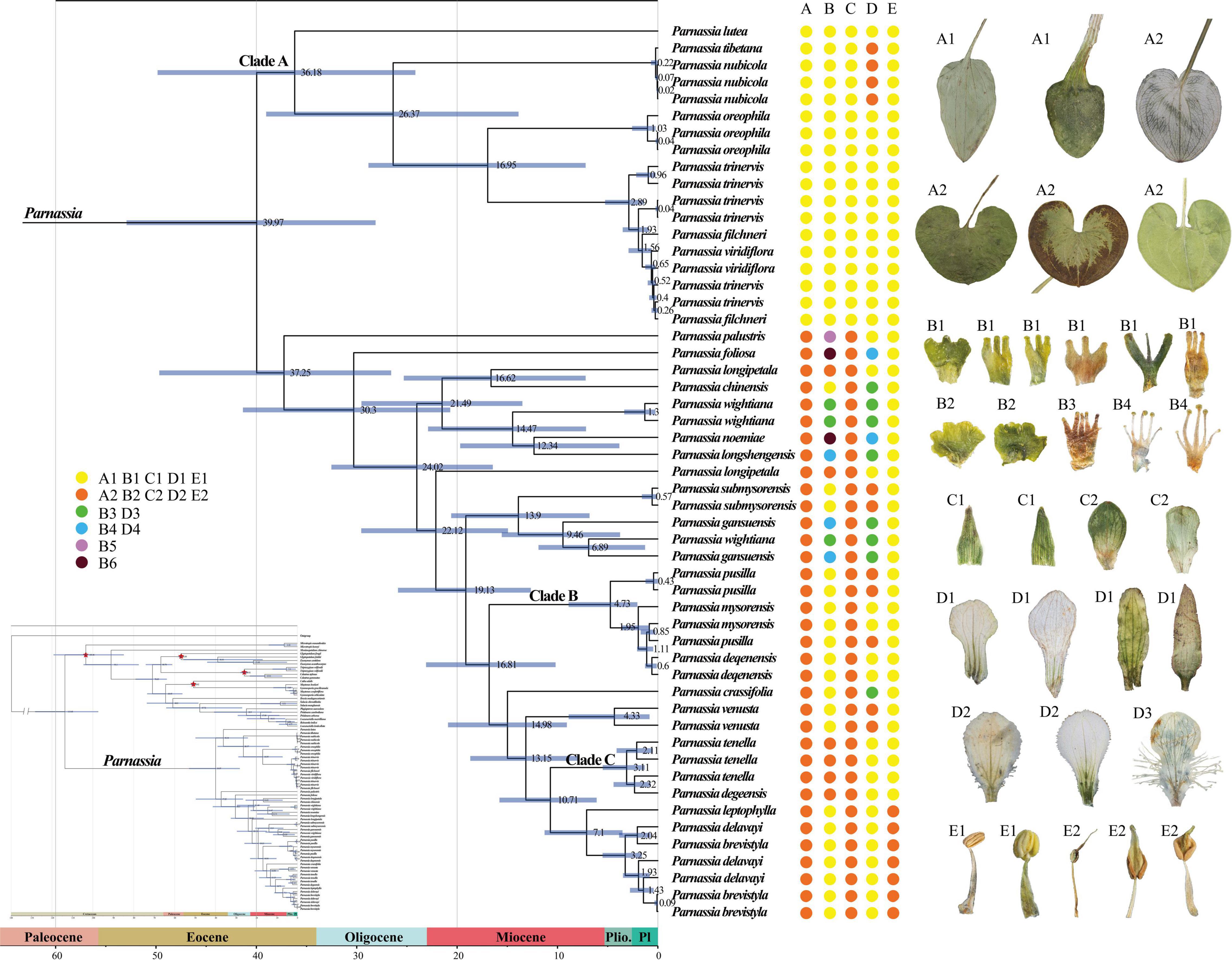
Figure 3. The dated phylogenetic tree of Parnassia is based on the dataset (4). Numbers associated with branches represent the mean estimated divergence time (Mya) and the blue bars correspond to the 95% highest posterior density (HPD) of divergence time. Red stars indicate four calibrating points. Pl, Pleistocene; Plio., Pliocene. (A–E) represent radical leaf, staminode, sepal, petal, and anther connective type, respectively. It should be noted that the characters shown in the figure only represent the characteristic types rather than the actual performance of the species. For a description of the specific feature types, please see Supplementary Table 5.
According to the AICc_wt value, the best model was BAYAREALIKE + J. The common ancestor of Parnassia probably originated in the Pan-Himalayan region (Supplementary Figure 10). The initial divergence of Parnassia took place at the Eocene and emerged as Clade A and Clade B. The diversification of Clade A was estimated to be in the non-Pan-Himalayan region, which was different from Clade B (Supplementary Figure 10).
Discussion
Recently, high throughput sequence data has been used frequently to get robust topologies and to resolve the doubtful phylogenetic positions of different taxa [details see in Dong et al. (2018); Pouchon et al. (2018), Li et al. (2019); Zhai et al. (2019), Zhang H. R. et al. (2020)]. Here, we used the plastome and nrDNA datasets to deeply reassess the phylogenetic position of Parnassia within Superrosids and provide a resolved phylogenetic tree of its species. Our results highly supported the monophyly of Parnassia; however, the infrageneric systematics of the genus showed incongruencies.
Phylogenetic Position of Parnassia in Superrosids
We reconstructed a well-resolved phylogenetic tree for 17 orders of Superrosids. The COM clade topology of all the phylogenetic trees revealed that Celastrales was sister to Malpighiales with high-support, and then this clade is sister to Oxalidales. The COM clade topology (Figure 1) obtained here was consistent with Zhang and Simmons (2006); Soltis et al. (2007), and Li et al. (2019), nevertheless, incongruent with many other previous studies (Qiu et al., 2010; Soltis et al., 2011; Ruhfel et al., 2014; Chen et al., 2016; Group et al., 2016; Sun et al., 2016). Although the sequence data used for phylogenetic analysis of COM clade has significantly increased in recent years, their systematics are still problematic or at least did not obtain well-supported topologies [details see in Li et al. (2019) and here in this study].
The phylogenetic trees based on different datasets to investigate the phylogenetic position of Saxifragales and Vitales were incongruent (Figure 1 and Supplementary Figures 2–5). The phylogenetic trees of dataset 3 recovered a similar topology as proposed by Li et al. (2019; Supplementary Figures 4, 5). However, the trees of datasets 1 and 2 showed reverse topologies of the phylogenetic position between Saxifragales and Vitales (Supplementary Figures 1–3). All the phylogenetic trees revealed that Parnassia was closely related to Celastrales (Celastraceae: Figure 1 and Supplementary Figures 2–5). Therefore, our results rejected the hypothesis based on morphology, which considered Parnassia was close to Saxifragaceae or other families (e.g., Hooker and Thomson, 1858; Hallier, 1901; Pace, 1912; Arber, 1913; Engler, 1930; Thorne, 1968; Spongberg, 1972; Bensel and Palser, 1975; Dahlgren, 1980; Cronquist and Takhtadzh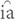 n, 1981; Ku, 1987; Gu and Hultgård, 2001). Previous studies have questioned the relationship between Parnassia and Saxifragaceae. Grund and Jensen’s (1981) comparative investigations of serological characteristics of seed proteins revealed that there was no relationship between Parnassia and Saxifragaceae. In addition, Matthews and Endress (2005) compared the floral structures of Celastrales, expounded 15 shared floral features between Parnassia and Celastraceae, and described five different floral structural characteristics that distinguished Parnassia from Saxifragaceae. Soltis et al. (1990) found that the relationship between Parnassia and Brexia was closer than Saxifragaceae by using rbcL. It was also confirmed in other later molecular phylogenetic researches (Chase et al., 1993; Morgan and Soltis, 1993; Soltis et al., 1997, 2000; Soltis and Soltis, 1997; Koontz and Soltis, 1999). Similarly, our results corroborated that Parnassia, belonging to Celastrales, was closely related to Celastraceae.
n, 1981; Ku, 1987; Gu and Hultgård, 2001). Previous studies have questioned the relationship between Parnassia and Saxifragaceae. Grund and Jensen’s (1981) comparative investigations of serological characteristics of seed proteins revealed that there was no relationship between Parnassia and Saxifragaceae. In addition, Matthews and Endress (2005) compared the floral structures of Celastrales, expounded 15 shared floral features between Parnassia and Celastraceae, and described five different floral structural characteristics that distinguished Parnassia from Saxifragaceae. Soltis et al. (1990) found that the relationship between Parnassia and Brexia was closer than Saxifragaceae by using rbcL. It was also confirmed in other later molecular phylogenetic researches (Chase et al., 1993; Morgan and Soltis, 1993; Soltis et al., 1997, 2000; Soltis and Soltis, 1997; Koontz and Soltis, 1999). Similarly, our results corroborated that Parnassia, belonging to Celastrales, was closely related to Celastraceae.
To investigate the phylogenetic position of Parnassia, we sampled species of Parnassia and 15 genera of Celastrales distributed in China and reconstructed the phylogenetic relationship. Two families were included in Celastrales: Celastraceae and Lepidobotryaceae. In previous investigations, Parnassia and Celastraceae were resolved as one clade according to the rbcL (Savolainen et al., 1997), and Lepidobotryaceae was sister to this clade (also see Zhang and Simmons, 2006). The floral morphological study revealed that Lepidobotryaceae was more distant from Parnassia and Celastraceae, and these three taxa were also supported as a group with some shared floral features (Matthews and Endress, 2005). Nevertheless, the relationship between Parnassia and Celastraceae was still controversial (details see introduction). In this study, plastome phylogenetic suggested that Parnassia was sister to Celastraceae with high support, which was consistent with the results of Simmons et al. (2012), Bacon et al. (2016), Chen et al. (2016), and Li et al. (2019). However, the nrDNA data clustered Parnassia and Microtropis together and then resolved them as sister groups to the other genera of Celastraceae (also see Simmons et al., 2012 and Sun et al., 2016). Discordance between the plastid genome and nrDNA analyses is a common phenomenon in molecular phylogenetic studies (see Lee-Yaw et al., 2019; Chen et al., 2020; Liu et al., 2020; Ma et al., 2021; Uckele et al., 2021), which was usually suggested to be a result of horizontal gene transfer (Stegemann et al., 2012), hybridization (Rieseberg et al., 1991, 1996) and incomplete lineage sorting (Degnan and Rosenberg, 2009).
In addition to the doubtful phylogenetic relationship between Parnassia and Microtropis shown in this study, the relationship among Parnassia, Perrottetia, and Mortonia is also unclear. Previous studies revealed that (Parnassia, Lepuropetalon) + (Perrottetia, Mortonia) were sister to other genera of Celastraceae, suggesting that Parnassia should be considered as an early derived lineage of Celastraceae (Simmons et al., 2001; Sun et al., 2016). Meanwhile, several studies demonstrated that Parnassia was sister to all the remaining genera of Celastraceae (Simmons et al., 2012; Bacon et al., 2016; Chen et al., 2016; Li et al., 2019). Here, it was suggested that the topology of trees based on plastome have higher resolution in comparison with the nrDNA. With the combination of the previous studies and our results of the plastome tree, we speculated that Parnassia was a basalmost taxon of Celastraceae. Nevertheless, it is still not mature enough to make clearer hypotheses before resolving the phylogenetic relationships among Parnassia, Mortonia, Perrottetia, and Microtropis.
Biogeography and Divergence Estimation of Chinese Parnassia
The divergence time in this study is congruent with the previous geographical study of Parnassia (Wu, 2005). However, due to the limited fossil calibrations points (lack of reliable fossil records of Parnassia), the results of divergence estimation in this study should be treated cautiously. Wu (2005) discussed the geographical distribution of Parnassia and supposed that its origin was in the early tertiary periods or even earlier. Bell et al. (2010) estimated the divergence times of key angiosperm lineages and showed that the crown-group age of Parnassia was estimated as 29 (17–43) Ma (exponential priors) and 34 (19–48) Ma (Lognormal priors). Here, the diversification of Parnassia was estimated to be 39.97 Mya (95% HPD = 28.13–52.93 Mya, Figure 3), which was earlier than the result of Bell et al. (2010; 34 Mya, 95% HPD = 19–48 Mya). The limited taxon coverage may reduce the accuracy of divergence time estimation [only one individual representing Parnassia in Bell et al. (2010) and Gao et al. (2017)], which may partially account for the slight inconsistency.
Most species in this study were distributed in the southeastern Qinghai-Tibetan Plateau and its surrounding mountainous regions. The ancestral area inference results indicated that the ancestral lineage of Parnassia probably originated in the Pan-Himalayan region. The north of Tibet localized uplift to the near present heights during the Eocene, while northwestern Yunnan formed its present topography during the late Eocene/early Oligocene (Spicer et al., 2021). Our results of divergence time estimation indicated that the genus Parnassia began to diverge and spread to non-Pan-Himalayan areas during the Eocene. Combining with the Qinghai-Tibetan Plateau uplift, the global climate oscillations led to a significant change in plant diversity during the Eocene–Oligocene transition period (Abels et al., 2011; Deng et al., 2020). As a result, the differentiation Parnassia species began to appear at Oligocene in the Pan-Himalayan region, and then mountain building and Asian monsoon may have combined to promote species diversification and colonization in the early to middle Miocene (Xing and Ree, 2017; Ding et al., 2020). Finally, the final uplift and climate oscillation of the Qinghai-Tibet Plateau in the late Miocene or Pliocene has jointly driven the rapid diversification of Parnassia species (Chen et al., 2012; Zhou et al., 2013). This may partially explain the high species richness of Parnassia species in the Pan-Himalayan region. Similar to previous studies in this space–time (Sun et al., 2017; Lu et al., 2018), it is probable that the large-scale expansion occurred after species diversification of Parnassia and accelerated the colonization of adjacent areas.
Infrageneric Relationships of Parnassia
We mainly focused on resolving the phylogenetic relationship of Parnassia species and advancing some supportive hypotheses about its systematics. Our results recovered a well-supported monophyletic clade of Parnassia by using both the plastome and the nrDNA datasets (Figure 2). Notably, most of the plastome-based topology branches reached a 100% support rate, which significantly improved the phylogenetic resolution (Figure 2).
The molecular systematics of Parnassia have focused on the phylogenetic position problems, and there were few studies on the phylogenetic relationship within this genus (Wu, 2005; Yang et al., 2012; Yu, 2019). In this study, more comprehensive plastome and nrDNA data were used to elaborate the phylogeny of Parnassia for the first time. Consistent with previous studies, Parnassia has been fully supported as a monophyletic group. Clade I (P. lutea, P. tibetana, P. nubicola, P. oreophila, P. trinervis, P. filchneri, and P. viridiflora) consisted of seven species of sect. Nectarotrilobos, forming a monophyletic branch (details see in Wu, 2005). Parnassia lutea, without the cauline leaves, was considered to represent the early colonizer of sect. Nectarotrilobos to high-altitude areas (Wu, 2003, 2005). Although P. lutea was located at the base of Clade I, reflecting it was an earlier element of Clade I, our results did not support its more ancient hypothesis within the genus. Ma et al. (2020) proposed that P. tibetana was synonymous to P. nubicola. Likewise, P. tibetana has been resolved as nested within the individuals of P. nubicola, and we did not find any significant molecular differences between P. tibetana and P. nubicola (Figure 2). Compared to our results of P. oreophila, Yu (2019) also demonstrated that P. oreophila, P. nubicola, P. laxmannii, P. trinervis, and P. cabulica belonged to the same clade. As inferred from the distribution range and divergence time, P. trinervis, P. filchneri, and P. viridiflora may be the species differentiated from P. oreophila in the process of colonizing to high altitude during Miocene. Hence, we herein supported the taxonomic revisions of P. tibetana and P. nubicola in Ma et al. (2020). Individuals of P. trinervis, P. filchneri, and P. viridiflora were nested within each other (Figure 2). During field investigation, we found that the distribution range of P. trinervis overlapped with P. viridiflora. Furthermore, the petal color of these two species changed from dark green to light green and white, and even different colors might occur in the same population (petal color was the key character to distinguish these two species). Therefore, we supported the recommendation that P. viridiflora should be treated as the synonym of P. trinervis by Yang et al. (2012) [also in Shu et al. (2017)]. For P. filchneri, a follow-up study is recommended to assess the relationship with P. trinervis. Previous studies suggested that P. palustris appeared earlier than other species of Parnassia and should be a relict species (Wu, 2003, 2005). Divergence time estimation supported this speculation (Figure 3), and we suggested that P. palustris was the ancestor species of Pan-Himalayan region.
Clade II consisted of three species of sect. Nectarotrilobos (P. pusilla, P. mysorensis, and P. deqenensis). For P. deqenensis, Wu (2005) suggested reducing “it” as the synonym of P. trinervis, while Shu et al. (2017) proposed reducing “it” as the synonym of P. pusilla. Our results here showed that the three species in Clade II were grouped into one branch in all trees, while individuals were nested within each other in phylogenetic trees (Figure 2). Based on the above results, we agreed with the taxonomic revision of Shu et al. (2017). Meanwhile, further studies with P. mysorensis and P. pusilla are suggested.
Clade III consisted of three species of sect. Nectarotrilobos subsect. Xiphosandra (P. brevistyla, P. delavayi, and P. leptophylla). Yu et al. (2018) found that the continuous characteristics were mistakenly used in distinguishing these three species. Hence, P. brevistyla and P. leptophylla were designated as synonyms of P. delavayi. In this study, the results of phylogenetic relationships showed that individuals of these three species were nested within each other (Figure 2), supporting the taxonomic revision of Yu et al. (2018).
There were widespread inconsistencies in the topological structure of the plastid trees and the nrDNA trees (Figure 2), such as the systemic position of P. noemiae, P. lutea, P. foliosa, P. chinensis, P. venusta, P. submysorensis, P. longshengensis, P. gansuensis, and P. wightiana were changed. The individuals of P. wightiana were resolved as polyphyletic in plastid trees (Figure 2), which could be attributed to the previously reported hybridization of this species (Yu, 2019). Furthermore, P. longipetala was not monophyletic as well in plastid trees, but clustered together in nrDNA trees, suggesting that incomplete lineage sorting, hybridization, or gene introgression events might have occurred with its related sympatric species. After comparing with many previous studies about the genus (e.g., Chen et al., 2020; Liu et al., 2020; Ma et al., 2021; Uckele et al., 2021), we speculated that widespread hybridization or introgression events might occur in Parnassia species distributed in and around the Hengduan Mountains (e.g., P. venusta, P. longshengensis, P. wightiana, P. submysorensis, and P. chinensis).
Wu (2005) and Yu (2019) explored the phylogeny of Parnassia based on morphology. However, the results showed that almost all characters (such as the shape of petal margin and the number of staminodes lobs) that have been used for taxonomy and systematic might have undergone parallel or even reverse evolution [details in Shu (2017)]. Our phylogenetic results showed that except for the characters of basal leaves and anther connective, other characteristics may not represent the evolutionary relationship of Parnassia (Figure 3). The sect. Nectarotrilobos, sect. Saxifragastrum, sect. Fimbripetalum, and sect. Nectaroquinquelobos were resolved as polyphyletic. Shu (2017) proposed reducing P. venusta and Parnassia degeensis as synonyms of Parnassia cacuminum. However, in this study, individuals of P. venusta and P. degeensis were not clustered together, which was inconsistent with traditional taxonomy revisions (Shu, 2017). The molecular phylogenetic analyses showed that a single species with continuous quantitative traits might be divided into several different species in the morphological study of Parnassia, e.g., P. brevistyla, P. delavayi, and P. leptophylla (Yu et al., 2018). In addition, different species with similar characters may be merged into one species, e.g., P. venusta, P. degeensis, and P. cacuminum (see details in Figure 3; Shu and Zhang, 2017). Characters (e.g., staminodes 3-lobed) of Parnassia might evolve at multiple times, and introgression or hybridization events might have happened in the process of evolution. This may explain why the trend of morphological character evolution of Parnassia has always been controversial in the traditional taxonomy.
Conclusion
Here we utilized the whole plastome and nrDNA (5,879 bp) markers to reconstruct the position of Parnassia within the Superrosids and phylogenetic relationships of Chinese Parnassia species. Our results corroborated that Parnassia is most closely related to Celastraceae, while follow-up studies are still needed to determine the relationship between Parnassia and several related genera. However, there was incongruence between the plastome and nuclear phylogenetic trees. In addition, we presented a first attempt to use the fossil records of Celastraceae to estimate the divergence of Chinese Parnassia, which was strongly consistent with the expected results of previous biogeographic studies. Moreover, the molecular phylogenetic results confirmed numerous taxonomic revisions and suggested that many traits have been widely used for classification, and that systematic considerations were the results of multiple evolutions. Our results will provide valuable insights into the taxonomic study of Parnassia and expand the foundation for further exploration of the evolutionary diversity of the genus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
F-QZ: conceptualization, funding acquisition, and resources. S-LC: funding acquisition, resources, and supervision. M-ZX: formal analysis, data curation, and writing – original draft. YL: resources, formal analysis, and writing – review and editing. J-YY, X-FC, and HX: resources and formal analysis. GK: formal analysis and writing – review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was partially supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0502), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA2005020405), Chinese Academy of Sciences – People’s Government of Qinghai Province on Sanjiangyuan National Park (LHZX-2021-04), Basic Research Project of Qinghai Province (2019-ZJ-7042), and Construction Project for Innovation Platform of Qinghai Province (2017-ZJ-Y14 and 2021-ZJ-Y05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Pengcheng Fu (Luoyang Normal University), Qingbo Gao (Chinese Academy of Sciences), and Liukun Jia (Chinese Academy of Sciences) for their assistance in material collection. We also thank Xiaolei Ma (Chinese Academy of Sciences) for his help with the specimen image.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.855944/full#supplementary-material
Supplementary Figure 1 | Representation of the chloroplast genome map of Parnassia trinervis. The genome was plotted using Chloroplot (https://irscope.shinyapps.io/Chloroplot/).
Supplementary Figure 2 | Phylogenetic position of Parnassia in Superrosids reconstructed with the dataset based on the first and second sites on the concatenated codons. Numbers associated with branches are ML bootstrap values. Nodes without numbers indicate 100% bootstrap support.
Supplementary Figure 3 | Phylogenetic location of Parnassia in Superrosids reconstructed with the dataset based on the first and second sites on the concatenated codons. Numbers associated with branches are Bayesian posterior probabilities. Nodes without numbers indicate posterior probability of 1.
Supplementary Figure 4 | Phylogenetic position of Parnassia in Superrosids recovered with the dataset consists of third site on the codons in concatenated. Numbers associated with branches are ML bootstrap values. Nodes without numbers indicate 100% bootstrap support.
Supplementary Figure 5 | Phylogenetic position of Parnassia in Superrosids recovered with the dataset consists of third site on the codons in concatenated. Numbers associated with branches are Bayesian posterior probabilities. Nodes without numbers indicate 1.0 posterior probability.
Supplementary Figure 6 | Phylogenetic relationship of Parnassia species resolved with the dataset of first and second sites on the concatenated codons. Numbers associated with branches are ML bootstrap values. Nodes without numbers indicate 100% bootstrap support.
Supplementary Figure 7 | Phylogenetic relationship of Parnassia species resolved with the dataset of first and second sites on the concatenated codons. Numbers associated with branches are Bayesian posterior probabilities. Nodes without numbers indicate 1.0 posterior probability.
Supplementary Figure 8 | Phylogenetic relationship of Parnassia species resolved with the dataset consists of third site on the codons in concatenated. Numbers associated with branches are ML bootstrap values. Nodes without numbers indicate 100% bootstrap support.
Supplementary Figure 9 | Phylogenetic relationship of Parnassia species resolved with the dataset consists of third site on the codons in concatenated. Numbers associated with branches are Bayesian posterior probabilities. Nodes without numbers indicate 1.0 posterior probability.
Footnotes
- ^ http://chlorobox.mpimp-golm.mpg.de/geseq.html
- ^ https://www.ncbi.nlm.nih.gov/guide/data-software
- ^ http://tree.bio.ed.ac.uk/software/figtree/
References
Abels, H. A., Dupont-Nivet, G., Xiao, G., Bosboom, R., and Krijgsman, W. (2011). Step-wise change of Asian interior climate preceding the eocene–oligocene transition (EOT). Palaeogeogr. Palaeoclimatol. Palaeoecol. 299, 399–412.
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge, MA: Babraham Institute.
Arber, A. (1913). On the structure of the androecium in Parnassia and its bearing on the affinities of the genus. Ann. Bot. 27, 491–510. doi: 10.1093/oxfordjournals.aob.a089471
Bacon, C. D., Simmons, M. P., Archer, R. H., Zhao, L. C., and Andriantiana, J. (2016). Biogeography of the Malagasy Celastraceae: multiple independent origins followed by widespread dispersal of genera from Madagascar. Mol. Phylogen. Evol. 94, 365–382. doi: 10.1016/j.ympev.2015.09.013
Barba-Montoya, J., Dos Reis, M., Schneider, H., Donoghue, P. C., and Yang, Z. (2018). Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a cretaceous terrestrial revolution. New Phytol. 218, 819–834. doi: 10.1111/nph.15011
Bell, C. D., Soltis, D. E., and Soltis, P. S. (2010). The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97, 1296–1303. doi: 10.3732/ajb.0900346
Bennett, A. W. (1869). Note on the structure and affinities of Parnassia palustris, L. Bot. J. Linn. Soc. 11, 24–31. doi: 10.1111/j.1095-8339.1869.tb00052.x
Bensel, C. R., and Palser, B. F. (1975). Floral anatomy in the Saxifragaceae sensu lato. I. Introduction, Parnassioideae and Brexioideae. Am. J. Bot. 62, 176–185. doi: 10.1002/j.1537-2197.1975.tb14050.x
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chase, M. W., Soltis, D. E., Olmstead, R. G., Morgan, D., Les, D. H., Mishler, B. D., et al. (1993). Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann. Missouri Bot. Garden 80, 528–580.
Chen, H. L., German, D. A., Al-Shehbaz, I. A., Yue, J. P., and Sun, H. (2020). Phylogeny of Euclidieae (Brassicaceae) based on plastome and nuclear ribosomal DNA data. Mol. Phylogen. Evol. 153:106940. doi: 10.1016/j.ympev.2020.106940
Chen, S., Xing, Y., Su, T., Zhou, Z., Dilcher, E. D. L., and Soltis, D. E. (2012). Phylogeographic analysis reveals significant spatial genetic structure of Incarvillea sinensis as a product of mountain building. BMC Plant Biol. 12:58. doi: 10.1186/1471-2229-12-58
Chen, Z. D., Yang, T., Lin, L., Lu, L. M., Li, H. L., Sun, M., et al. (2016). Tree of life for the genera of Chinese vascular plants. J. Syst. Evol. 54, 277–306.
Coope, G. R., Shotton, F. W., and Strachan, I. (1961). A late Pleistocene fauna and flora from Upton Warren, Worcestershire. Philos. Trans. R. Soc. Lond. B Biol. Sci. 244, 379–421.
Cronquist, A., and Takhtadzh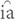 n, A. L. (1981). An Integrated System of Classification of Flowering Plants. New York, NY: Columbia University Press.
n, A. L. (1981). An Integrated System of Classification of Flowering Plants. New York, NY: Columbia University Press.
Dahlgren, R. (1980). A revised system of classification of the angiosperms. Bot. J. Linn. Soc. 80, 91–124. doi: 10.1111/j.1095-8339.1980.tb01661.x
Dai, X. S., Min, D. Z., Yang, B., Wu, D., and Li, B. (2021). Taxonomic note of Parnassia (Celastraceae) in China II: population surveys reveal that P. guilinensis is conspecific to P. xinganensis. PhytoKeys 172:67.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Degnan, J. H., and Rosenberg, N. A. (2009). Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340. doi: 10.1016/j.tree.2009.01.009
Deng, W. Y. D., Su, T., Wappler, T., Liu, J., Li, S. F., Huang, J., et al. (2020). Sharp changes in plant diversity and plant-herbivore interactions during the Eocene-Oligocene transition on the southeastern Qinghai-Tibetan Plateau. Glob. Planet. Change 194:103293.
Ding, W. N., Ree, R. H., Spicer, R. A., and Xing, Y. W. (2020). Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science 369, 578–581. doi: 10.1126/science.abb4484
Dong, W., Xu, C., Wu, P., Cheng, T., Yu, J., Zhou, S., et al. (2018). Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol. Phylogen. Evol. 126, 321–330. doi: 10.1016/j.ympev.2018.04.033
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. Bot. Soc. Am. 19, 11–15.
Drude, O. (1875). Über die Blüthengestaltung und die Verwandtschaftsverhältnisse des Genus Parnassia, nebst einer systematischen Revision seiner Arten. Linnaea 39, 239–324.
Drummond, A., Suchard, M., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Fischer, M., and Steel, M. (2009). Sequence length bounds for resolving a deep phylogenetic divergence. J. Theor. Biol. 256, 247–252. doi: 10.1016/j.jtbi.2008.09.031
Foster, C. S., Sauquet, H., Van der Merwe, M., McPherson, H., Rossetto, M., and Ho, S. Y. (2017). Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Syst. Biol. 66, 338–351. doi: 10.1093/sysbio/syw086
Gao, Q. B., Li, Y., Gengji, Z. M., Gornall, R. J., Wang, J. L., Liu, H. R., et al. (2017). Population genetic differentiation and taxonomy of three closely related species of Saxifraga (saxifragaceae) from Southern Tibet and the Hengduan Mountains. Front. Plant Sci. 8:1325. doi: 10.3389/fpls.2017.01325
Group, T. A. P., Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., Judd, W. S., et al. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20.
Grund, C., and Jensen, U. (1981). Systematic relationships of the Saxifragales revealed by serological characteristics of seed proteins. Plant Syst. Evol. 137, 1–22.
Gu, C., and Hultgård, U. M. (2001). “Parnassia,” in Flora of China, eds Z. Wu and P. H. R. Co (Beijing: Science Press), 358–379.
Hallier, H. (1901). Über die Verwandtschaftsverhältnisse der Tubifloren und Ebenalen, den die polyphyletischen Ursprung der Sympetalen und Apetalen und die Anordnung der Angiospermen üBerhaupt. Rome: FAO.
Jay, M. (1970). Quelques problèmes taxonomiques et phylogénétiques des Saxifragacees vus á la lumiére de la biochimie flavonique. Bull. Museum D’Histoire Nat. 42, 754–775.
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., Depamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 1–31. doi: 10.1186/s13059-020-02154-5
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Klopfer, K. (1972). Beiträge zur floralen Morphogenese und Histogenese der Saxifragaceae: 7. Parnassia palustris und Francoa sonchifolia. Flora 161, 320–332. doi: 10.1016/s0367-2530(17)32070-4
Koontz, J. A., and Soltis, D. E. (1999). DNA sequence data reveal polyphyly of Brexioideae (Brexiaceae; Saxifragaceae sensu lato). Plant Syst. Evol. 219, 199–208. doi: 10.1007/bf00985579
Ku, T. (1987). A revision of the genus Parnassia (Saxifragaceae) in China. Bull. Bot. Res. 7, 1–61. doi: 10.1155/2012/874256
Ku, T. C. (1995). “Parnassia,” in Flora Reipublicae Popularis Sinicae 35, eds L. T. Lu and S. M. Huang (Beijing: Science Press), 9–66.
Lee-Yaw, J. A., Grassa, C. J., Joly, S., Andrew, R. L., and Rieseberg, L. H. (2019). An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phytol. 221, 515–526. doi: 10.1111/nph.15386
Li, H. T., Yi, T. S., Gao, L. M., Ma, P. F., Zhang, T., Yang, J. B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470. doi: 10.1038/s41477-019-0421-0
Liu, B. B., Campbell, C. S., Hong, D. Y., and Wen, J. (2020). Phylogenetic relationships and chloroplast capture in the Amelanchier-Malacomeles-Peraphyllum clade (Maleae, Rosaceae): evidence from chloroplast genome and nuclear ribosomal DNA data using genome skimming. Mol. Phylogen. Evol. 147:106784. doi: 10.1016/j.ympev.2020.106784
Lu, L. M., Mao, L. F., Yang, T., Ye, J. F., Liu, B., Li, H. L., et al. (2018). Evolutionary history of the angiosperm flora of China. Nature 554, 234–238. doi: 10.1038/nature25485
Ma, Y. H., Zhou, H. L., and Shu, Y. M. (2020). Taxonomic note on Parnassia (Celastraceae): the identity of P. nubicola. PhytoKeys 154:103.
Ma, Z. Y., Nie, Z. L., Ren, C., Liu, X. Q., Zimmer, E. A., and Wen, J. (2021). Phylogenomic relationships and character evolution of the grape family (Vitaceae). Mol. Phylogen. Evol. 154:106948. doi: 10.1016/j.ympev.2020.106948
Matthews, M. L., and Endress, P. K. (2005). Comparative floral structure and systematics in Celastrales (Celastraceae, Parnassiaceae, Lepidobotryaceae). Bot. J. Linn. Soc. 149, 129–194. doi: 10.1111/j.1095-8339.2005.00445.x
Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst. Biol. 63, 951–970. doi: 10.1093/sysbio/syu056
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), (Piscataway, NJ: IEEE), 1–8.
Morgan, D. R., and Soltis, D. E. (1993). Phylogenetic relationships among members of Saxifragaceae sensu lato based on rbcL sequence data. Ann. Missouri Bot. Garden 80, 631–660.
Murbeck, S. S. (1918). Über die Organisation und verwandtschaftlichen Beziehungen der Gattung Lepuropetalon. Ark. Bot. 15, 1–12.
Philippe, H., Brinkmann, H., Lavrov, D. V., Littlewood, D. T. J., Manuel, M., Wörheide, G., et al. (2011). Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9:e1000602. doi: 10.1371/journal.pbio.1000602
Phillips, R. B. (1982). Systematics of Parnassia L.(Parnassiaceae): Generic overview and revision of North American taxa. Berkeley: University of California.
Pitman, J. K., and Rowan, E. L. (2012). Temperature and Petroleum Generation History of the Wilcox Formation, Louisiana. New York, NY: US Geological Survey.
Poole, I., and Wilkinson, H. P. (1999). A celastraceous twig from the Eocene London Clay of south-east England. Bot. J. Linn. Soc. 129, 165–176.
Pouchon, C., Fernández, A., Nassar, J. M., Boyer, F., Aubert, S., Lavergne, S., et al. (2018). Phylogenomic analysis of the explosive adaptive radiation of the Espeletia complex (Asteraceae) in the tropical Andes. Syst. Biol. 67, 1041–1060. doi: 10.1093/sysbio/syy022
Qiu, Y. L., Li, L. B., Wang, B., Xue, J. Y., Hendry, T. A., Li, R. Q., et al. (2010). Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J. Syst. Evol. 48, 391–425.
Qu, X. J., Moore, M. J., Li, D. Z., and Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 1–12. doi: 10.1186/s13007-019-0435-7
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67:901. doi: 10.1093/sysbio/syy032
Rieseberg, L. H., Beckstrom-Sternberg, S. M., Liston, A., and Arias, D. M. (1991). Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus (Asteraceae). Syst. Bot. 16, 50–76. doi: 10.2307/2418973
Rieseberg, L. H., Whitton, J., and Randal Linder, C. (1996). Molecular marker incongruence in plant hybrid zones and phylogenetic trees. Acta Bot. Neerl. 45, 243–262. doi: 10.1111/j.1438-8677.1996.tb00515.x
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ruhfel, B. R., Gitzendanner, M. A., Soltis, P. S., Soltis, D. E., and Burleigh, J. G. (2014). From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 14:23. doi: 10.1186/1471-2148-14-23
Sauquet, H., Ho, S. Y., Gandolfo, M. A., Jordan, G. J., Wilf, P., Cantrill, D. J., et al. (2012). Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales). Syst. Biol. 61, 289–313. doi: 10.1093/sysbio/syr116
Savolainen, V., Spichiger, R., and Manen, J. F. (1997). Polyphyletism of Celastrales deduced from a chloroplast noncoding DNA region. Mol. Phylogen. Evol. 7, 145–157. doi: 10.1006/mpev.1996.0380
Sharma, V. (1968). Morphology floral anatomy and embryology of parnassia nubicola wall. Phytomorphology 18, 193–213.
Shu, Y. M. (2017). Ataxonomic Revision of Parnassia L. (Celastraceae) from Pan-Himalaya. Beijing: Beijing Forestry University.
Shu, Y. M., Yu, H. Y., Mu, X. Y., and Zhang, Z. X. (2017). Checklist and typification of Parnassia (Celastraceae). Phytotaxa 309, 1–22. doi: 10.11646/phytotaxa.309.1.1
Shu, Y. M., and Zhang, Z. X. (2016). Lectotypification of Parnassia pusilla (Celastraceae). Phytotaxa 265, 98–100. doi: 10.11646/phytotaxa.265.1.13
Shu, Y. M., and Zhang, Z. X. (2017). Four new synonyms and a new combination in Parnassia (Celastraceae). PhytoKeys 77, 81–88. doi: 10.3897/phytokeys.77.11513
Simmons, M. P. (2004). “Parnassiaceae,” in Flowering Plants Dicotyledons: Celastrales, Oxalidales, Rosales, Cornales, Ericales, ed. K. Kubitzki (Berlin: Springer), 291–296. doi: 10.1007/978-3-662-07257-8_33
Simmons, M. P., McKenna, M. J., Bacon, C. D., Yakobson, K., Cappa, J. J., Archer, R. H., et al. (2012). Phylogeny of Celastraceae tribe Euonymeae inferred from morphological characters and nuclear and plastid genes. Mol. Phylogen. Evol. 62, 9–20. doi: 10.1016/j.ympev.2011.08.022
Simmons, M. P., Savolainen, V., Clevinger, C. C., Archer, R. H., and Davis, J. I. (2001). Phylogeny of the Celastraceae inferred from 26S nuclear ribosomal DNA, phytochrome B, rbcL, atpB, and morphology. Mol. Phylogen. Evol. 19, 353–366. doi: 10.1006/mpev.2001.0937
Soltis, D. E., Gitzendanner, M. A., and Soltis, P. S. (2007). A 567-taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int. J. Plant Sci. 168, 137–157. doi: 10.1086/509788
Soltis, D. E., Smith, S. A., Cellinese, N., Wurdack, K. J., Tank, D. C., Brockington, S. F., et al. (2011). Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98, 704–730. doi: 10.3732/ajb.1000404
Soltis, D. E., and Soltis, P. S. (1997). Phylogenetic relationships in Saxifragaceae sensu lato: a comparison of topologies based on 18S rDNA and rbcL sequences. Am. J. Bot. 84, 504–522. doi: 10.2307/2446027
Soltis, D. E., Soltis, P. S., Chase, M. W., Mort, M. E., Albach, D. C., Zanis, M., et al. (2000). Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 133, 381–461. doi: 10.1006/bojl.2000.0380
Soltis, D. E., Soltis, P. S., Clegg, M. T., and Durbin, M. (1990). rbcL sequence divergence and phylogenetic relationships in Saxifragaceae sensu lato. Proc. Natl. Acad. Sci. U.S.A. 87:4640. doi: 10.1073/pnas.87.12.4640
Soltis, D. E., Soltis, P. S., Nickrent, D. L., Johnson, L. A., Hahn, W. J., Hoot, S. B., et al. (1997). Angiosperm phylogeny inferred from 18S ribosomal DNA sequences. Ann. Missouri Bot. Garden 84, 1–49.
Spicer, R. A., Su, T., Valdes, P. J., Farnsworth, A., Wu, F. X., Shi, G., et al. (2021). Why ‘the uplift of the Tibetan Plateau’is a myth. Natl. Sci. Rev. 8:nwaa091. doi: 10.1093/nsr/nwaa091
Spongberg, S. A. (1972). The genera of Saxifragaceae in the southeastern United States. J. Arnold Arboretum 53, 409–498.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stegemann, S., Keuthe, M., Greiner, S., and Bock, R. (2012). Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. U.S.A. 109, 2434–2438. doi: 10.1073/pnas.1114076109
Sun, H., Zhang, J. W., Deng, T., and Boufford, D. E. (2017). Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers. 39:161. doi: 10.1016/j.pld.2017.09.004
Sun, M., Naeem, R., Su, J. X., Cao, Z. Y., Burleigh, J. G., Soltis, P. S., et al. (2016). Phylogeny of the Rosidae: a dense taxon sampling analysis. J. Syst. Evol. 54, 363–391.
Takhtadzhian, A. L., and Takhtajan, A. (1997). Diversity and Classification of Flowering Plants. New York, NY: Columbia University Press.
Thorne, R. F. (1968). Synopsis of a putatively phylogenetic classification of the flowering plants. Aliso J. Syst. Evol. Bot. 6, 57–66.
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Turner, H., and Veldkamp, J. (2001). Parnassia (Parnassiaceae) in North Sumatra. Blumea 46, 599–603.
Uckele, K. A., Adams, R. P., Schwarzbach, A. E., and Parchman, T. L. (2021). Genome-wide RAD sequencing resolves the evolutionary history of serrate leaf Juniperus and reveals discordance with chloroplast phylogeny. Mol. Phylogen. Evol. 156:107022. doi: 10.1016/j.ympev.2020.107022
Wang, Y. P., Chen, S., Shu, Y. M., Li, B., and Wu, D. (2018). Taxonomic note of Parnassia (Celastraceae) in China: the identity of P. wightiana. Phytotaxa 375, 130–134.
Wick, R. R., Schultz, M. B., Zobel, J., and Holt, K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wortley, A. H., Rudall, P. J., Harris, D. J., and Scotland, R. W. (2005). How much data are needed to resolve a difficult phylogeny? Case study in Lamiales. Syst. Biol. 54, 697–709. doi: 10.1080/10635150500221028
Wu, C. Y. (2003). The Families and Genera of Angiosperms in China: A Comprehensive Analysis. Beijing: Science Press.
Wu, D. (2005). Phylogeny and Biogeography of the genus Parnassia. Kunming: Kunming Istitute of Botany, Chinese Academy of Sciences.
Wu, D., Gao, B., Wang, H., and Li, D. (2004). Four new synonyms of four species in the Genus Parnassia (Parnassiaceae). Acta Bot. Yunnanica 26, 628–630.
Wu, D., Gao, L. M., and Moeller, M. (2009). Taxonomic notes on Parnassia section Saxifragastrum (Parnassiaceae) from China. Ann. Bot. Fenn. 46, 559–565.
Wu, D., Lu, J. M., and Wang, H. (2008). Taxonomic revision on some species of Parnassia (Parnassiaceae) from China. Plant Divers. 30:657.
Xi, Z. X., Ruhfel, B. R., Schaefer, H., Amorim, A. M., Sugumaran, M., Wurdack, K. J., et al. (2012). Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. U.S.A. 109, 17519–17524. doi: 10.1073/pnas.1205818109
Xia, X. H. (2018). DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 35, 1550–1552. doi: 10.1093/molbev/msy073
Xia, X. H., Xie, Z., Salemi, M., Chen, L., and Wang, Y. (2003). An index of substitution saturation and its application. Mol. Phylogen. Evol. 26, 1–7. doi: 10.1016/s1055-7903(02)00326-3
Xing, Y., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U.S.A. 114, E3444–E3451. doi: 10.1073/pnas.1616063114
Yang, J. B., Wang, Y. P., MÖLler, M., Gao, L. M., and Wu, D. (2012). Applying plant DNA barcodes to identify species of Parnassia (Parnassiaceae). Mol. Ecol. Resour. 12, 267–275. doi: 10.1111/j.1755-0998.2011.03095.x
Yu, H. Y. (2019). A Systematic Study on Parnassia L. (Celastraceae). Beijing: Beijing Forestry University.
Yu, H. Y., Guo, F. Y., Shu, Y. M., and Zhang, Z. X. (2018). Taxonomic note of Parnassia (Celastraceae) in China: a reassessment of Subsect. Xiphosandra. PhytoKeys 114, 43–54. doi: 10.3897/phytokeys.114.30551
Yu, Y., Blair, C., and He, X. J. (2020). RASP 4: ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 37, 604–606. doi: 10.1093/molbev/msz257
Zhai, W., Duan, X. S., Zhang, R., Guo, C. C., Li, L., Xu, G. X., et al. (2019). Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol. Phylogen. Evol. 135, 12–21. doi: 10.1016/j.ympev.2019.02.024
Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, H. R., Wei, R., Xiang, Q. P., and Zhang, X. C. (2020). Plastome-based phylogenomics resolves the placement of the sanguinolenta group in the spikemoss of lycophyte (Selaginellaceae). Mol. Phylogen. Evol. 147:106788. doi: 10.1016/j.ympev.2020.106788
Zhang, H. R., Xiang, Q. P., and Zhang, X. C. (2019). The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (Lycophyte). Genome Biol. Evol. 11, 1258–1274. doi: 10.1093/gbe/evz073
Zhang, L. B., and Simmons, M. P. (2006). Phylogeny and delimitation of the Celastrales inferred from nuclear and plastid genes. Syst. Bot. 31, 122–137. doi: 10.1600/036364406775971778
Zhou, Z., Hong, D., Niu, Y., Li, G., Nie, Z., Wen, J., et al. (2013). Phylogenetic and biogeographic analyses of the Sino-Himalayan endemic genus Cyananthus (Campanulaceae) and implications for the evolution of its sexual system. Mol. Phylogenet. Evol. 68, 482–497. doi: 10.1016/j.ympev.2013.04.027
Keywords: Parnassia, phylogeny, plastome, nuclear ribosomal DNA, divergence time, character evolution
Citation: Xia M-Z, Li Y, Zhang F-Q, Yu J-Y, Khan G, Chi X-F, Xu H and Chen S-L (2022) Reassessment of the Phylogeny and Systematics of Chinese Parnassia (Celastraceae): A Thorough Investigation Using Whole Plastomes and Nuclear Ribosomal DNA. Front. Plant Sci. 13:855944. doi: 10.3389/fpls.2022.855944
Received: 16 January 2022; Accepted: 16 February 2022;
Published: 18 March 2022.
Edited by:
Tingshuang Yi, Kunming Institute of Botany (CAS), ChinaReviewed by:
Ran Wei, Institute of Botany (CAS), ChinaBin-Bin Liu, Institute of Botany (CAS), China
Copyright © 2022 Xia, Li, Zhang, Yu, Khan, Chi, Xu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fa-Qi Zhang, fqzhang@nwipb.cas.cn; Shi-Long Chen, slchen@nwipb.cas.cn
†These authors have contributed equally to this work
 Ming-Ze Xia
Ming-Ze Xia Yan Li3†
Yan Li3† Fa-Qi Zhang
Fa-Qi Zhang Gulzar Khan
Gulzar Khan Shi-Long Chen
Shi-Long Chen