- 1 Arnold Arboretum of Harvard University, Boston, MA, USA
- 2 USDA Forest Service Pacific Southwest Research Station, Davis, CA, USA
- 3 Department of Plant Biology, University of California Davis, Davis, CA, USA
Living botanical collections include germplasm repositories, long-term experimental plantings, and botanical gardens. We present here a series of vignettes to illustrate the central role that living collections have played in plant biology research, including evo-devo research. Looking toward the future, living collections will become increasingly important in support of future evo-devo research. The driving force behind this trend is nucleic acid sequencing technologies, which are rapidly becoming more powerful and cost-effective, and which can be applied to virtually any species. This allows for more extensive sampling, including non-model organisms with unique biological features and plants from diverse phylogenetic positions. Importantly, a major challenge for sequencing-based evo-devo research is to identify, access, and propagate appropriate plant materials. We use a vignette of the ongoing 1,000 Transcriptomes project as an example of the challenges faced by such projects. We conclude by identifying some of the pinch points likely to be encountered by future evo-devo researchers, and how living collections can help address them.
Introduction: Living Botanical Collections in Historical Context
Living collections are curated for various purposes including scientific education and research. For plants, living collections include germplasm repositories – such as the world’s largest, the National Plant Germplasm System (NPGS) of USDA – that preserve plant genetic resources for research and conservation. There are also experimental research plots, seed banks, conservatories, and germplasm repositories for research that are associated with universities and research institutions. Lastly, there are the public facing botanical gardens – some 3,000 worldwide according to Botanic Gardens Conservation International. Although their raison d’être is assumed by some to be display and education, their living collections also serve vital research roles.
The earliest gardens (e.g., Padua, founded in 1545) served the apothecary and physician, but during the subsequent age of exploration, these collections grew to become living encyclopedias ripe for comparative and descriptive work. With botany and taxonomy as driving forces, scholars used these cultivated plants to describe, name, and place these species into ordered groups. Over time, some gardens diversified, while others specialized. For instance, the Palm House at the Royal Botanic Gardens, Kew, opened in 1848 and provided this institution of diverse yet temperate plants the opportunity to cultivate and study a broader range of tropical plants collected by explorer scientists. While comprehensive or general collections such as Kew and the Missouri Botanical Garden thrived and multiplied, others focused their collections. For example, in 1872, the Arnold Arboretum of Harvard University became the first public botanical garden in the US to specialize in temperate woody plants. Other institutions defined the scope of their collections geographically, such as Rancho Santa Ana Botanic Garden, founded in 1927, which has botanical collections which represent the flora of California. The resulting impressive array of botanical resources began and continue to serve scientific disciplines beyond basic taxonomy and botany, including horticulture, plant breeding, conservation, and ecology.
Living Collections Facilitate Plant Biology Research
Living collections have played diverse yet crucial roles in plant biology research. In general, identifying, acquiring, propagating, and growing appropriate plant materials are fundamental needs of almost all plant biology, and living collections are well positioned to meet these needs. This is especially true for evolution of development (evo-devo) research, which seeks to understand both the evolutionary history and molecular mechanisms underlying development and biological processes. Evo-devo research is defined by comparative approaches that require the ability to sample different ecotypes or species of interests.
The vignettes presented below illustrate the importance of living collections in plant biology research, ranging from applied research for plant breeding to evo-devo research – all of which hinged on the use of living collections. The need for access to plant materials is quickly becoming more acute as genomics-based evo-devo studies expand to encompass broader taxonomic groups and larger numbers of species. The final vignette of the 1,000 Transcriptomes Project (1KP) is used to illustrate some of the challenges and opportunities for living collections in a new age of genomics-based evo-devo.
Mr. Eddy’s Tree Breeding Station and Adaptive Traits of Forest Trees
James G. Eddy was a lumberman from the Pacific Northwest who made a fortune from forest industry in the late 1800s. Realizing that forests were being cut at a rate far exceeding replacement, Eddy proposed that forest trees could be bred to produce superior performing stocks to make up the difference. In 1925, he established a tree breeding station located at Placerville, California. Lloyd Austin was hired as the first director of the Eddy Tree Breeding Station (now known as the Institute of Forest Genetics), and quickly set about establishing an extensive arboretum of conifers.
Around the same time, researchers Clausen, Keck, and Hiesey of Stanford established that herbaceous plants showed heritable traits associated with their local environments and elevations in the east to west environmental clines of California (Clausen et al., 1941). In general, seed collected and grown from plants at their native elevations performed better than seed collected from plants of the same species at higher or lower elevations. Realizing that similar effects could have significant impacts on reforestation efforts, Austin established an elevational gradient experiment for Pinus ponderosa and P. jeffreyi. Seed were collected from trees at different elevations, and then planted in three common planting sites ranging from low to high elevation. The significance of this experiment is presaged by correspondence from Hiesey to Austin after his visit to the Institute in 1938, in which he writes “The practical application to plant breeding are obvious, and for this reason I think your plan of starting a series of transect gardens is not only basically sound, but indispensable to realizing your objective to produce the best possible races for each climatic region.”
The long-term survival and performance of the Eddy Arboretum trees was only revealed after many decades, however, and the ranking of performance for families from different elevations changed during the course of long-term growth of the plantations (Namkoong and Conkle, 1976). Only after exposure to the full range of environmental challenges (e.g., harsh winters) over many years was it revealed that higher elevation seed sources ultimately performed better at high elevation sites than did lower elevation seed sources (Conkle, 1973). This research fully established the hereditary nature of adaptation for tree species, and was the basis of “seed zones” for reforestation.
Domestication, Determinacy, and the Phaseolus World Collection
Understanding the process of domestication provides important insight into the origins of crops and gives information about wild relatives vital to plant breeding efforts. While wild relatives can be important sources of unique alleles conferring disease resistance and other useful traits for agriculture, understanding the processes of domestication can also provide insight into the evolution of developmental traits as driven by human selection. One such example is given by Phaseolus, the genus containing the common bean (P. vulgaris).
One immediate challenge of studying domestication syndromes is acquiring germplasm representative of both domesticated varieties as well as wild relatives. Paul Gepts and colleagues wanted to understand the process by which domestication occurred independently in two geographic regions, giving rise to a Mesoamerican gene pool and an Andean gene pool within modern P. vulgaris varieties. Crucial to this research was the Phaseolus World Collection at CIAT, Cali, Colombia, as well as the collection maintained at the NPGS station in Pullman, Washington. Phylogenetic analysis of samples representing 100 wild and 249 domesticated cultivars revealed nine populations, four each of Andean and Mesoamerican origins, and one consisting of the likely wild ancestor of P. vulgaris (Kwak and Gepts, 2009; Kwak et al., 2009).
Related research investigated the genetic basis underlying the two major traits selected in Phaseolus domestication: photoperiod insensitivity and determinate growth habit. Plants with these traits can be grown at varied latitudes, and produce more rapid and synchronous flowering and fruiting. A cross between parents of Andean and Mesoamerican origins yielded a genetic mapping population. Using this and an assay of variation for candidate genes affecting flowering time in Arabidopsis revealed that homologs of Terminal Flower 1 (TFL1) mapped to quantitative trait loci affecting determinacy (Kwak et al., 2008). This research shows the potential power of comparative approaches, and holds great promise for plant breeding applications.
A Hand-Drawn Picture is Worth a 1000 Words: The Eupomatiaceae of Zurich
It is hard to overemphasize the role of botanical gardens in providing researchers the ability to closely observe the growth and development of their subjects – to have “a feeling for the organism,” as Barbara McClintock put it (Keller, 1983). Throughout his career, Peter Endress’ access to living collections at the Botanic Garden of the University of Zurich has been essential to his study of floral development. In fact, his early impressions of Eupomatiaceae can be traced to his acute observations – including detailed hand drawings (Figure 1) – of flowering material in the 1970s. In an essay (Endress, 2008), he emphasized “the great heuristic value of drawings,” noting that “the process of hand drawing can provide valuable insights into patterns or processes of nature, which can scarcely be achieved by merely looking at and analyzing a picture taken by a camera or the SEM.” Subsequent studies (Endress, 2003; Kim et al., 2005) revealed and confirmed the nature of the unusual cap, or calyptra, which covers the floral organs during their development: it is a bract, not a modification of the perianth.
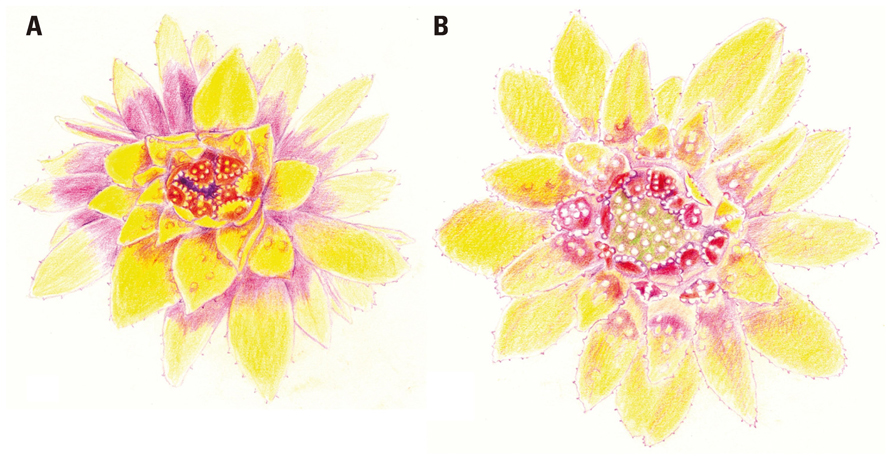
FIGURE 1. An opening flower of Eupomatia bennettii F. Muell., drawn by Peter Endress on 5 February 1979 at 09.00 h (A) and 14.00 h (B). The inner staminodes are shown bearing secretory warts during the male phase of anthesis (A), while the exposed gynoecium is seen in the female phase (B). Reproduced from Endress (2008), by permission of Oxford University Press.
Charlie Rick’s Tomatoes and the Regulation of Leaf Complexity
Charlie Rick devoted most of his long career as a botanist to the study of genetic variation in tomato (Solanum spp.), using pioneering cytological and genetic approaches. During the course of his research, he made multiple collecting expeditions to South America and the Galapagos Islands to sample the diversity found in wild species. Today, the C. M. Rick Tomato Genetics Resource Center at the University of California Davis has over 3,600 active accessions available for plant breeding and research.
Neelima Sinha has benefitted from Charlie Rick’s collections in the course of her research of leaf complexity. Neelima first met Charlie during her graduate studies at UC Berkeley. Interested in the mouse ears (Me) mutation that causes changes in the degree of leaf compoundedness, Neelima contacted Charlie, who not only granted her request for heterozygous Me seed for genetic mapping, but also included additional alleles and insight into Me phenotypes in other genetic backgrounds (N. Sinha, personal communication). Years later, Neelima acquired seed of Solanum cheesmaniae and S. galapagense through the Resource Center. Darwin first collected seed from these species, which were later characterized by J. G. Hooker for their differences in leaf compoundedness (Hooker, 1847). Neelima used them to map the causative locus to a gene, PETEROSELINUM (PTS), which encodes a novel KNOX transcription factor that lacks the homeodomain required for KNOX protein function (Kimura et al., 2008). Because KNOX proteins act as heterodimers, the PTS-encoded protein competes with legitimate KNOX proteins, which normally increase leaf compoundeness. PTS thus represents a novel mechanism by which leaf morphology is regulated through a naturally occurring mutation.
The 1,000 Transcriptomes Project and the Origin of Samples
Recent advances in sequencing technologies have greatly expanded the scope of evo-devo research (Rokas and Abbot, 2009). A bold example is the 1KP Project, a multi-institutional effort that is surveying gene expression in leaves (and for some species flowers or other tissues) from 1,000 green plant species1. In many regards, this project represents a paradigm shift in plant biology and in particular plant evo-devo research. Until recently, large sequencing projects were restricted to a modest number of well-developed model species. The rapidly decreasing cost and increasing output from “next generation” sequencing (Mardis, 2008; Schuster, 2008) now allows a different strategy, in which diverse species can be selected for study based on phylogenetic position and biological traits of interest. Sequencing the transcriptomes (as opposed to the genomes) of plants has a number of advantages (Wang et al., 2009). Transcriptomes are less complex and can be assembled de novo directly from sequence reads; they provide information about gene structure and alternative splicing; and they provide information about gene expression in the tissues sampled. Some of the broad objectives of 1KP include determining the relationship of gene expression in gametophyte-dominant versus sporophyte-dominant plants, identifying ancient polyploidization events in angiosperms, and testing for correlation between polyploidization and species richness. In addition, each transcriptome provides resources for the sequenced species, and also provides opportunities for comparative analyses across tissue types and taxa.
A number of practical issues associated with 1KP will be common to future genomic-based evo-devo studies. Obtaining appropriate plant materials from a diverse array of plant species is no small task. Some specimens were collected in the wild or from opportunistic locations including backyards – the accession for Oenothera grandiflora includes the GPS coordinates of the plant sampled and annotation to “ask the nice lady for permission to collect” at a home in Bigbee, Alabama. Field-based collections were made more challenging still because flash freezing of tissue samples using liquid nitrogen was found necessary to reliably preserve tissues for RNA isolation from the range of species sampled (D. Soltis, personal communication). Specimens from botanical gardens or other living collections are much preferred not just for ease of access, but because they have known provenance, additional collections can be made in the future, and the plants can be observed, propagated, or potentially experimentally manipulated. In addition, gene expression can be affected by a wide variety of factors ranging from environmental to genetic to developmental stage. To make comparisons in gene expression between, say, leaves of two species, it will often be desirable to have the plants for comparison grown under similar conditions and collected at the same time and growth stage. Living collections and botanical gardens in particular are well positioned to provide such materials, and 1KP benefitted from principal investigators and collaborators associated with institutions including the New York Botanical Garden, Kew, and the University of British Columbia Botanical Garden and Centre for Plant Research.
Living Collections Facilitate Genomics-Based Evo-Devo Research
While genomic and next generation sequencing approaches are well suited for evo-devo research, they alone are insufficient. Indeed, a major challenge of plant evo-devo research is to develop an initial understanding of the trait through direct observation, then assembling plant materials representing a range of taxonomic and phenotypic variation for the trait, and finally quantifying variation in that trait across taxa. This is a significant challenge when one considers the breadth of plant biodiversity, the near limitless traits to study, and the range of environments within which they occur. Fortunately, genomics researchers have direct access to the extensive range of living plant collections. Collectively, these living libraries possess a rich array of well-documented plant materials maintained by knowledgeable curatorial and horticultural staff, and are available for research and discovery.
Collections not only comprise wild-provenance material that may be challenging to obtain, but also artificial hybrids of known pedigree, and ornamental variants selected for their horticultural value that would only be recognized and able to thrive under contrived conditions. This latter group serves as a unique resource for evo-devo investigation, and contains a wealth of genetic diversity including chimeras and spontaneously arising mutants (Dosmann, 2006). The research potential of atypical forms or mutants growing but meters away from their wild-type cousins is compelling. And, because many identical cultivars as well as accessions of known wild provenance are replicated in collections around the world, it is possible to examine variability as a function of environment. It is striking that living collections also serve as repositories for future research, particularly in underdeveloped and unforeseen areas (Tanksley and McCouch, 1997; Dosmann, 2006; Donaldson, 2009). In the example presented above of the Eddy Arboretum, those trees have experienced much of the industrial age of global climate change and contain a record of their growth in the form of annual rings. They therefore represent a valuable resource for measuring the response of different genotypes to environmental change, which was unforeseen at the time of their planting.
Another important distinction among these collections is the fact that they can be well documented with data such as provenance, verification status, morphological measurements, previous research results, as well as voucher herbarium specimens and images. They are also curated by experts in systematics, horticulture, plant anatomy and morphology, and other disciplines. In fact, the horticultural and curatorial staff not only preserve, study, and document these collections, but also collaborate with the scientists who seek to use the plant material. Through collaboration, researchers can become aware of and access previously unknown genotypes of interest, while curators expand the use of their collections. While the one-on-one conversation between researcher and curator may be most effective, a majority of collections now have searchable inventories online, making it easy to learn what might be available. Because of their long traditions in horticulture, gardens and other repositories are well equipped to propagate (sexually and asexually) and cultivate fickle genotypes where researchers may have failed. Lastly, all gardens and arboreta (and a growing number of other repositories) have outreach missions which provide collaborating scholars the ability to tap into existing frameworks to facilitate the broader impact requirements associated with grant-funded work. Indeed, maintaining funding and resources for living collections is challenging, and demonstrating research value of collections is one aspect of demonstrating their importance.
While there is no single database encompassing all living collections, the majority of botanical gardens and other repositories maintain online, searchable inventories of their living collections on their institutional webpages. For example, at the Arnold Arboretum, searches can be conducted through a simple query2 as well as through an interactive map known as Collection Researcher3. Repositories within USDA’s NPGS can be searched at http://www.ars-grin.gov/npgs/index.html, and BGCI’s PlantSearch4 has the capability to draw upon over 500,000 records in botanic gardens across the globe. Assuming they already know what they wish to sample, scholars interested in accessing material are best served if they start online to see what taxa are available prior to contacting curatorial staff. Curators can then provide additional documentation as well as work with researchers to further brainstorm about the project, schedule collection dates, and provide other services (see Dosmann, 2006, for examples).
We see great opportunity ahead for genomics-based evo-devo research. Technological advances now provide scientists with a broader array of tools whereby they can rapidly and effectively reach out beyond model organisms, and will find valuable samples that are being grown and/or stored in plant collections. Researchers benefit by not only accessing germplasm of value, but through collaboration with staff who often possess unique expertise complementary to those in the research community. Through these joint efforts, not only can tremendous progress be made in the study of evo-devo, but living collections can successfully expand their utility to wider audiences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Peter Endress, Paul Gepts, Neelima Sinha, and Doug Soltis for sharing experiences and insights concerning the use of living collections in their research. Curatorial work by Michael Dosmann is supported by competitive grants by IMLS, and research by Andrew Groover is supported by competitive grants from USDA NIFA and DOE. This work was inspired by the National Evolutionary Synthesis Center meeting “Evolutionary Origins and Development of Woody Plants,” October 2011.
Footnotes
References
Clausen, J., Keck, D. D., and Hiesey, W. M. (1941). Regional differentiation in plant species. Am. Nat. 75, 231–250.
Conkle, M. T. (1973). Growth data for 29 years from the California elevational transect study of ponderosa pine. For. Sci. 19, 31–39.
Donaldson, J. S. (2009). Botanic gardens science for conservation and global change. Trends Plant Sci. 14, 608–613.
Dosmann, M. S. (2006). Research in the garden: averting the collections crisis. Bot. Rev. 72, 107–134.
Endress, P. K. (2003). Early floral development and nature of the calyptra in Eupomatiaceae (Magnoliales). Int. J. Plant Sci. 164, 489–503.
Hooker, J. D. (1847). An enumeration of the plants of the Galapagos archipelago; with descriptions of those which are new. Trans. Linn. Soc. Lond. 20, 163–233.
Keller, E. F. (1983). A Feeling for the Organism: The Life and Work of Barbara McClintock. NY, USA: Henry Holt and Company.
Kim, S., Koh, J., Ma, H., Hu, Y., Endress, P. K., Hauser, B. A., Buzgo, M., Soltis, P. S., and Soltis, D. E. (2005). Sequence and expression studies of a-, b-, and e-class MADS-box homologues in Eupomatia (Eupomatiaceae): support for the bracteate origin of the calyptra. Int. J. Plant Sci. 166, 185–198.
Kimura, S., Koenig, D., Kang, J., Yoong, F. Y., and Sinha, N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18, 672–677.
Kwak, M., and Gepts, P. (2009). Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris, Fabaceae). Theor. Appl. Genet. 118, 979–992.
Kwak, M., Kami, J. A., and Gepts, P. (2009). The putative Mesoamerican domestication center of is located in the Lerma santiago basin of Mexico. Crop Sci. 49, 554–563.
Kwak, M., Velasco, D., and Gepts, P. (2008). Mapping homologous sequences for determinacy and photoperiod sensitivity in common bean (Phaseolus vulgaris). J. Hered. 99, 283–291.
Mardis, E. R. (2008). The impact of next-generation sequencing technology on genetics. Trends Genet. 24, 133–141.
Namkoong, G., and Conkle, M. T. (1976). Time trends in genetic control of height growth in ponderosa pine. For. Sci. 22, 2–12.
Rokas, A., and Abbot, P. (2009). Harnessing genomics for evolutionary insights. Trends Ecol. Evol. 24, 192–200.
Schuster, S. C. (2008). Next-generation sequencing transforms today’s biology. Nat. Methods 5, 16–18.
Tanksley, S. D, and McCouch, S. R. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066.
Keywords: botanical gardens, genomics, plant developmental biology, next generation sequencing, outreach
Citation: Dosmann M and Groover A (2012) The importance of living botanical collections for plant biology and the “next generation” of evo-devo research. Front. Plant Sci. 3:137. doi: 10.3389/fpls.2012.00137
Received: 30 March 2012; Accepted: 06 June 2012;
Published online: 22 June 2012.
Edited by:
Elena M. Kramer, Harvard University, USAReviewed by:
Verónica S. Di Stilio, University of Washington, USAKentaro K. Shimizu, University of Zurich, Switzerland
Copyright: © 2012 Dosmann andGroover. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Andrew Groover, USDA Forest Service Pacific Southwest Research Station, Berkeley, CA, USA; Department of Plant Biology, University of California Davis, 1731 Research Park Dr, Davis, CA 95616, USA. e-mail: agroover@fs.fed.us
