- 1Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan
- 2Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency, Saitama, Japan
Patterning of the cellulosic cell wall underlies the shape and function of plant cells. The cortical microtubule array plays a central role in the regulation of cell wall patterns. However, the regulatory mechanisms by which secondary cell wall patterns are established through cortical microtubules remain to be fully determined. Our recent study in xylem vessel cells revealed that a mutual inhibitory interaction between cortical microtubules and distinct plasma membrane domains leads to distinctive patterning in secondary cell walls. Our research revealed that the recycling of active and inactive ROP proteins by a specific GAP and GEF pair establishes distinct de novo plasma membrane domains. Active ROP recruits a plant-specific microtubule-associated protein, MIDD1, which mediates the mutual interaction between cortical microtubules and plasma membrane domains. In this mini review, we summarize recent research regarding secondary wall patterning, with a focus on the emerging interplay between plasma membrane domains and cortical microtubules through MIDD1 and ROP.
Introduction
The cellulosic cell wall plays a central role in shaping cells and determining cell function in plants. For example, physically rigid cellulose microfibrils restrict the direction of cell expansion and lead to the distinct shape of plant cells. Cortical microtubules play a central role in regulating the development of cell wall structures by controlling how the cellulose synthase complex is targeted to the plasma membrane (Paredez et al., 2006; Crowell et al., 2009; Gutierrez et al., 2009). Although cortical microtubules are closely and tightly anchored to the plasma membrane, they exhibit dynamic behaviors, such as growth, shrinkage, and branching (Shaw et al., 2003; Murata et al., 2005; Chan et al., 2009; Nakamura et al., 2010) and exhibit bundling (Dixit and Cyr, 2004), which leads to the self-organization of parallel microtubule arrays (Wasteneys and Ambrose, 2009). Several microtubule-associated proteins (MAPs) have been discovered that regulate the global dynamics of cortical microtubules in the cell. However, the molecular mechanisms underlying local regulation of cortical microtubule dynamics, for formation of structures such as patterned secondary cell walls in xylem vessels, are not well understood.
Xylem vessels are water-conductive tubes composed of dead cells called tracheary elements. Differentiating xylem cells deposit rigid water-impermeable secondary cell walls to avoid collapse of the vessel due to the negative pressure exerted as a consequence of water transport. In xylem vessels, secondary cell walls are deposited in various patterns, such as annular, spiral, reticulate, and pitted. Usually, protoxylems develop annular and spiral secondary walls while metaxylems exhibit reticulate and pitted formations. Perforations in the end walls of xylem cells and secondary wall pits in the side walls contribute to effective longitudinal and lateral water transport, respectively. Thus, the patterning of secondary cell walls is tightly coupled to function in xylem vessels (Esau, 1977).
The involvement of cortical microtubules in the regulation of secondary cell wall deposition during xylem vessel differentiation has been revealed through a number of studies. Dense cortical microtubule bundles are found beneath secondary wall thickenings and, if these cortical microtubules are pharmacologically depolymerized, the secondary cell wall patterns become severely disorganized (Hepler, 1981; Gunning and Hardham, 1982). Direct visualization of the cellulose synthase complex confirmed that patterned cortical microtubules regulate the localization of the cellulose synthase complex in xylem vessels (Wightman and Turner, 2008). Studies using isolated zinnia mesophyll cells revealed that a dramatic rearrangement of cortical microtubules leads to distinct patterns of secondary cell walls during tracheary element differentiation (Falconer and Seagull, 1985, 1986, 1988; Kobayashi et al., 1987, 1988; Fukuda and Kobayashi, 1989). Recent live cell imaging of differentiating xylem cells using Arabidopsis cell cultures further revealed that bundling and disassembly of cortical microtubules gives rise to distinct patterns of secondary cell walls (Oda et al., 2005, 2010; Oda and Hasezawa, 2006; Pesquet et al., 2010). These studies also revealed that several MAPs regulate the dynamic behavior of cortical microtubules during xylem differentiation (Oda et al., 2010; Pesquet et al., 2010; Oda and Fukuda, 2012b, 2013b). In addition, a recent study revealed that ROP GTPases generate plasma membrane domains that play critical roles in the spatial regulation of cortical microtubule dynamics, and that mutual inhibitory interactions between the plasma membrane domains and cortical microtubules establishes distinct patterns of secondary cell walls (Oda and Fukuda, 2012a). In this review, we focus on the emerging role of the dynamic interplay between plasma membrane domains and cortical microtubules in secondary cell wall patterning.
Microtubule-Depolymerizing Plasma Membrane Domain Leads Secondary Wall Pit
We previously established an Arabidopsis cultured cell line for in vitro xylem differentiation (Oda et al., 2010). In this system, VND6, a master transcription factor that prompts metaxylem vessel differentiation (Kubo et al., 2005), was introduced under the control of an estradiol inducible promoter (Zuo et al., 2000), allowing synchronous induction of metaxylem vessel differentiation at high frequency. Using this system, we found that there are several cellular regions in which cortical microtubules are unstable. Cortical microtubules are eventually depleted in these regions, resulting in the formation of secondary wall pits. This suggests that local depolymerization of cortical microtubules is the key event for formation of secondary wall pits (Oda et al., 2010). Cortical microtubules are closely anchored to the plasma membrane (Hardham and Gunning, 1978; Seagull and Heath, 1980; Lancelle et al., 1986; Giddings and Staehelin, 1988), and specific regulators of microtubule dynamics are thus expected to be locally present at the plasma membrane, mediating the formation of microtubule-depolymerizing plasma membrane domains.
MIDD1 is Localized to Microtubule-Depleting Plasma Membrane Domains
Microtubule-associated protein MIDD1 (microtubule depletion domain 1) is preferentially associated with depolymerizing cortical microtubules in the future pit region of secondary walls in xylem cells. Knockdown of MIDD1 inhibits the local disassembly of cortical microtubules, resulting in the loss of secondary wall pits. Conversely, overexpression of MIDD1 in non-xylem cells reduces cortical microtubule density. MIDD1 is composed of two coiled-coil domains: the N-terminal domain, which binds directly to microtubules, and the C-terminal domain, which is associated with specific plasma membrane domains. MIDD1 is thus thought to promote depolymerization of cortical microtubules in the plasma membrane domains, leading to the formation of secondary wall pits (Oda et al., 2010).
MIDD1 Recruits Atkinesin-13A to Locally Depolymerize Cortical Microtubules
Microtubule depletion domain 1 does not exhibit in vitro microtubule depolymerization activity. MIDD1 is therefore likely to interact with other proteins to induce microtubule disassembly. Recently, AtKinsein-13A was found to interact with MIDD1 in yeast (Mucha et al., 2010). AtKinesin-13A belongs to the kinesin-13 family (Lu et al., 2005), whose animal members have microtubule depolymerization activity (Desai et al., 1999). As expected, AtKinesin-13A exhibits microtubule depolymerization activity in vitro (Oda and Fukuda, 2013a). Knockdown of AtKinesin-13A results in smaller secondary wall pits, while overexpression of AtKinesin-13A enlarges secondary wall pits. AtKinesin-13A co-localizes and interacts with MIDD1 in secondary wall pits and promotes depolymerization of cortical microtubules. In the absence of MIDD1, however, AtKinsein-13A neither localizes to nor affects cortical microtubules (Oda and Fukuda, 2013a). Therefore, it is likely that MIDD1 is a scaffold protein that links the plasma membrane domains to AtKinesin-13A and therefore facilitates specific localized cortical microtubule depolymerization (Figure 1A).
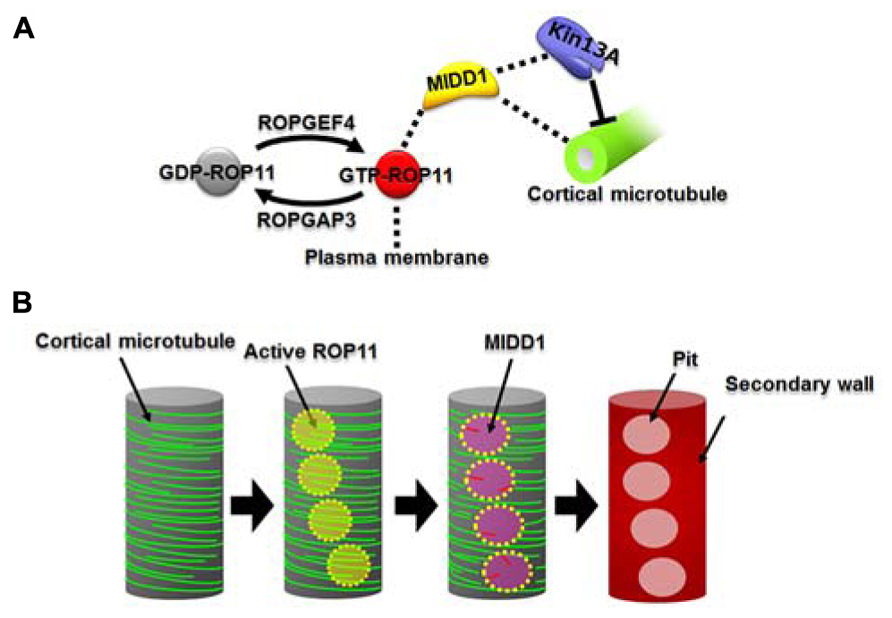
FIGURE 1. The ROP11-MIDD1 pathway produces the microtubule-depolymerizing domain. (A) MIDD1 links plasma membrane, cortical microtubules, and AtKinesin-13A. ROP11 is locally activated by ROPGEF4 and ROPGAP3. The activated ROP11 (GTP-ROP11) recruits MIDD1 to the plasma membrane, which in turn binds cortical microtubules. MIDD1 subsequently recruits AtKinsein-13A (Kin13A), which depolymerizes the cortical microtubule. (B) Locally activated ROP11 recruits MIDD1 to the plasma membrane domains. MIDD1 subsequently induces microtubule disassembly by recruiting AtKinesin-13A, resulting in the formation of secondary wall pits.
ROP GTPase Regulates Cortical Microtubule Dynamics Through the MIDD1-AtKinesin-13A Complex
Microtubule depletion domain 1 belongs to the RIP/ICR family, members of which interact with the active forms of ROP GTPases via their conserved C-terminal motif (Lavy et al., 2007; Li et al., 2008). ROP11 distributes broadly at the plasma membrane and, together with MIDD1, accumulates specifically on cortical microtubules in the secondary wall pits (Oda and Fukuda, 2012a). Further cellular analysis revealed that the active form of ROP11 recruits MIDD1 to the plasma membrane. Introduction of a constitutively activated mutant ROP11 strongly inhibits formation of secondary wall pits in metaxylem vessels by affecting pit-specific localization of AtKinesein-13A (Oda and Fukuda, 2012a, 2013a). These results suggest that locally activated ROP11 is essential for formation of secondary wall pits through the recruitment of the MIDD1-AtKinesin-13A complex (Figure 1B).
ROP GTPases Generate Microtubule-Depolymerizing Plasma Membrane Domains
ROP11 is specifically active at the secondary wall pits, posing the question of how regulation of this local activation is achieved. ROP GTPases are activated by plant-specific guanine nucleotide exchange factors (ROPGEFs) and are inactivated by ROP GTPase-activating proteins (ROPGAPs; Wu et al., 2000; Berken et al., 2005; Gu et al., 2006). ROPGAP3 and ROPGEF4 mediate the local activation and inactivation, respectively, of ROP11 in differentiating xylem cells (Oda and Fukuda, 2012a). Both ROPGEF4 and ROPGAP3 localize to plasma membrane domains in secondary wall pits. Interestingly, ROPGEF4 is concentrated at the center of the domains, while ROPGAP3 localizes much more broadly in the secondary wall pits. Knockout or knockdown of ROPGEF4 causes reduced density of secondary wall pits in metaxylem vessels, suggesting that ROPGEF4 promotes formation of secondary wall pits through local activation of ROP11 (Figure 1A; Oda and Fukuda, 2012a). Co-expression of ROP11, ROPGEF4, and ROPGAP3 in non-xylem cells causes evenly spaced patches of ROPGEF4 at the plasma membrane, which in turn activates ROP11 around the patches. This locally activated ROP11 recruits MIDD1 and finally causes local disassembly of cortical microtubules (Oda and Fukuda, 2012a).
The next question that arises is how such spontaneously activated ROP11 domains might be formed. Loss of either ROPGEF4 or ROPGAP3 abolishes the activated ROP11 domains. Furthermore, constitutively active or negative mutants of ROP11 cannot mediate this event (Oda and Fukuda, 2012a). Therefore, GTP-GDP cycling of ROP11 by ROPGEF4 and ROPGAP3 is essential for the self-organization of the activated ROP11 domains. Neither microtubules nor actin microfilaments are required for domain formation. A possible mechanism to explain this spontaneous formation of active ROP11 patches is positive feedback from active ROP11 via ROPGEF4 as some receptor-like kinases are known to interact with ROPGEFs (Zhang and McCormick, 2007; Duan et al., 2010; Chang et al., 2012; Akamatsu et al., 2013). For example, AtPRK2, a receptor-like kinase, which functions in polarized growth of pollen tubes, interacts with both ROPGEF1 and ROP GTPases, and phosphorylates ROPGEF1 to activate ROP GTPase (Chang et al., 2012). Similarly, an unknown receptor-like kinase(s) might interact with ROPGEF4 and ROP11 to initiate positive feedback and consequently form activated ROP11 patches. A negative feedback mechanism between ROPGAP3 and ROPGEF4 via ROP11 may also contribute to the self-organization of activated ROP11 patches.
Cortical Microtubules Act as a Fence Inhibiting the Movement of the ROP-MIDD1 Complex
Another important finding with respect to the formation of localized secondary wall pits is that cortical microtubules restrict the localization of active ROP11. Treatment with taxol, which stabilizes microtubules, elongates active ROP11 domains, suggesting that cortical microtubules can affect the localization of active ROP11 (Oda and Fukuda, 2012a). Further reconstruction experiments in non-xylem cells revealed that active ROP11 domains are enclosed by cortical microtubules to form polygonal structures. However, disruption of cortical microtubules by chemical treatment with oryzalin or co-expression of AtKinesin-13A resulted in round active ROP11 domains. These results strongly suggest that cortical microtubules act as a fence preventing outward diffusion of active ROP11 (Oda and Fukuda, 2012a).
How does MIDD1 contribute to this phenomenon? In non-xylem cells, cortical microtubules restricted active ROP11-MIDD1 domains when truncated MIDD1, which lacks its microtubule-binding domain, was introduced with ROP11, ROPGEF4, and ROPGAP3 (Figure 2B). In the absence of MIDD1, however, cortical microtubules did not affect the localization of ROP11 (Figure 2A). In the absence of ROP11, truncated MIDD1 was distributed broadly in the cytoplasm and its distribution was unaffected by cortical microtubules (Oda and Fukuda, 2012a). By contrast, cortical microtubules could eliminate truncated MIDD1 that was artificially anchored to the plasma membrane by fusion with a membrane-binding domain, even in the absence of ROP11 (Figure 2C). These observations strongly suggest that MIDD1 mediates the restriction of active ROP11 by cortical microtubules (Oda and Fukuda, 2012a). The precise mechanism by which cortical microtubules restrict the ROP11-MIDD1 complex at the plasma membrane remains to be determined. Cortical microtubules are closely anchored to the plasma membrane by unknown MAPs, and it is thus likely that the active ROP11 protein complex, which includes MIDD1 and AtKinesin-13A, is restricted in its ability to diffuse through the space between the cortical microtubules and the plasma membrane. Consistent with this hypothesis is the elimination of an Arabidopsis formin, AtFH1, which has a large cytoplasmic domain, from the plasma membrane by cortical microtubules (Martiniere et al., 2011). Similarly, cortical actin microfilaments limit the diffusion of plasma membrane-anchored proteins in animal cells (Kusumi et al., 2005). Instead of actin filaments, plasma-membrane-associated cortical microtubules are used in plants as membrane fences to restrict localization of plasma membrane-anchored proteins.
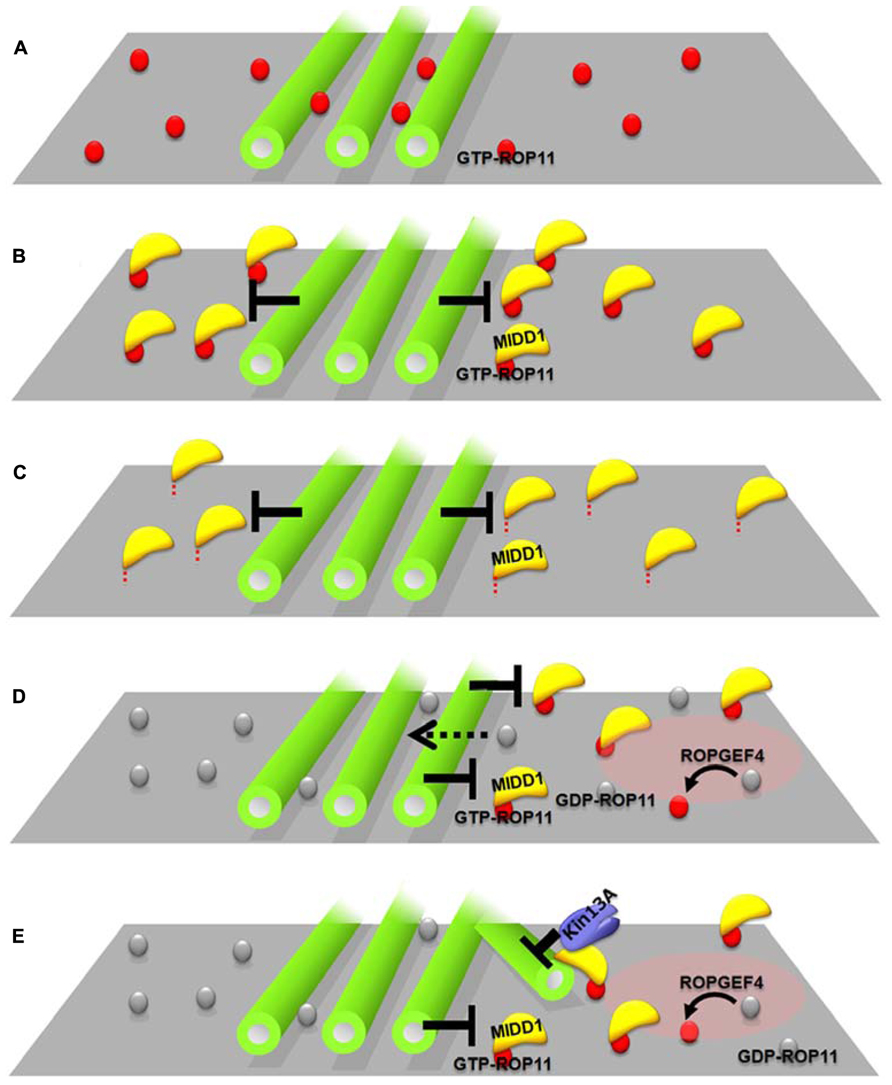
FIGURE 2. Interplay between cortical microtubules and plasma membrane domains via the ROP11-MIDD1 complex. (A–C) MIDD1 mediates elimination of active ROP11 by cortical microtubules. (A) Cortical microtubules do not affect the localization of active (GTP-) ROP11. (B) Cortical microtubules eliminate active ROP11-truncated MIDD1 complex from the plasma membrane (truncated mutant lacking microtubule-binding domain). (C) Cortical microtubules eliminate the membrane–anchored truncated MIDD1. (D,E) Interplay between cortical microtubules and plasma membrane domains in xylem cells. (D) Cortical microtubules restrict the plasma membrane domain of active ROP11 generated by ROPGEF4. Inactive ROP11 is not affected by cortical microtubules, because there is no interaction with MIDD1. (E) Active ROP11-MIDD1-AtKinesin-13A complex associates with the microtubule growing into the domain and induces depolymerization.
Interplay between Microtubules and Plasma Membrane Domains May Establish Patterns
Evenly distributed active ROP11 domains are spontaneously generated by ROPGEF4, ROPGAP3, and ROP11, and recruit the MIDD1-AtKinesin-13A complex to the domains, which in turn depolymerizes cortical microtubules at the domains. The surrounding cortical microtubules limit the localization of ROP11-MIDD1 complex to the domains, probably by inhibiting the outward lateral diffusion of the ROP11-MIDD1 complex (Figure 2D). These two interactions cause a mutual inhibition between cortical microtubules and plasma membrane domains underlain by active ROP11, resulting in spatial restriction of cortical microtubules and the plasma membrane domains (Figure 2E). Variability in the balance between these two pathways may produce a range of secondary wall pit shapes. For example, if the restriction of plasma membrane domains by cortical microtubules is dominant, reticulate secondary walls with oval pits will be formed. Conversely, if the microtubule depolymerization activity of the plasma membrane domain becomes dominant, pitted secondary walls with round pits will be formed. Therefore, this interplay between the plasma membrane domains and cortical microtubules may be a key determining factor leading to different secondary wall patterns.
Similar interplay between plasma membrane domains and cortical microtubules is reported in leaf epidermal morphogenesis. In leaf epidermis, pavement cells grow to form an interdigitating structure where lobes and indentations develop side-by-side. ROP6 and its effector RIC1 recruit microtubule-severing katanin protein to facilitate the ordering of a parallel cortical microtubule array that restricts cell growth in indentations, while ROP2/4 and their effector RIC4 enhance cortical actin microfilaments to promote local growth of lobes (Fu et al., 2005, 2009; Lin et al., 2013). The cortical actin microfilaments inhibit endocytosis of an auxin efflux carrier PIN1, promoting accumulation of PIN1 protein at the lobe forming region, which in turn promotes accumulation of auxin, which then activates the ROP6-RIC1 pathway in its neighbor cell via auxin binding protein ABP1 (Xu et al., 2010; Nagawa et al., 2012). These two signaling pathways appear to spatially restrict one another: the ROP2/4-RIC4 pathway eliminates RIC1 from the lobe forming regions and inhibits parallel ordering of cortical microtubules, while the ROP6-RIC1 pathway inhibits the ROP2/4-RIC4 pathway, preventing co-existence of these two pathways in the same area. Although the precise mechanism by which these two pathways spatially restrict each other is still unclear, one possibility is that the parallel cortical microtubules in the indentation inhibit lateral diffusion of ROP2/4-RIC4 complex on the plasma membrane, in a similar manner to the restriction of the ROP11-MIDD1 complex by cortical microtubules. It is also possible that the ROP-MIDD-AtKinesin-13 pathway contributes to the elimination of cortical microtubules from lobes and that the dynamic interplay between cortical microtubules and plasma membrane domains of ROP GTPase might be widely utilized in plant cell morphogenesis.
Concluding Remarks
Recent studies on xylem vessel differentiation and leaf epidermis morphogenesis revealed dynamic interplay between cortical microtubules and plasma membrane domains. ROP GTPases play critical roles in generating de novo plasma membrane domains and in regulating the function of the domains by recruiting different effector proteins and MAPs. In xylem cells, MIDD1 plays a central role in this interplay. Considering the diversity of the ROP GTPases and MIDD/RIP/ICR family members, the ROP-MIDD/RIP/ICR pathway may be involved in various cellular events by mediating interplay between plasma membrane domains and cortical microtubules. Recent proteomic approaches suggest that MIDD1/RIP/ICR members are also MAPs (Hamada et al., 2013). Further analysis of the pathways will provide novel insights regarding the molecular mechanisms underlying local control of plasma membrane domains coupled to microtubule dynamics in various plant cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan to Yoshihisa Oda (25114507); from the Japan Society for the Promotion of Science to Hiroo Fukuda (23227001) and Yoshihisa Oda (25440128); from the NC-CARP project to Hiroo Fukuda; and from the Japan Science and Technology Agency (JST), Precursory Research for Embryonic Science and Technology (PRESTO) to Yoshihisa Oda (20103).
References
Akamatsu, A., Wong, H. L., Fujiwara, M., Okuda, J., Nishide, K., Uno, K., et al. (2013). An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13, 465–476. doi: 10.1016/j.chom.2013.03.007
Berken, A., Thomas, C., and Wittinghofer, A. (2005). A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436, 1176–1180. doi: 10.1038/nature03883
Chan, J., Sambade, A., Calder, G., and Lloyd, C. (2009). Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21, 2298–2306. doi: 10.1105/tpc.109.069716
Chang, F., Gu, Y., Ma, H., and Yang, Z. (2012). AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol. Plant doi: 10.1093/mp/sss103
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y. D., Schumacher, K., et al. (2009). Pausing of golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154. doi: 10.1105/tpc.108.065334
Desai, A., Verma, S., Mitchison, T. J., and Walczak, C. E. (1999). Kin I kinesins are microtubule-destabilizing enzymes. Cell 96, 69–78. doi: 10.1016/S0092-8674(00)80960-5
Dixit, R., and Cyr, R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16, 3274–3284. doi: 10.1105/tpc.104.026930
Duan, Q., Kita, D., Li, C., Cheung, A. Y., and Wu, H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U.S.A. 107, 17821–17826. doi: 10.1073/pnas.1005366107
Falconer, M. M., and Seagull, R. W. (1985). Xylogenesis in tissue-culture – taxol effects on microtubule reorientation and lateral association in differentiating cells. Protoplasma 128, 157–166. doi: 10.1007/Bf01276337
Falconer, M. M., and Seagull, R. W. (1986). Xylogenesis in tissue-culture.2. Microtubules, cell-shape and secondary wall patterns. Protoplasma 133, 140–148. doi: 10.1007/Bf01304629
Falconer, M. M., and Seagull, R. W. (1988). Xylogenesis in tissue-culture.3. Continuing wall deposition during tracheary element development. Protoplasma 144, 10–16. doi: 10.1007/Bf01320275
Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G., and Yang, Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120, 687–700. doi: 10.1016/j.cell.2004.12.026
Fu, Y., Xu, T., Zhu, L., Wen, M., and Yang, Z. (2009). A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol. 19, 1827–1832. doi: 10.1016/j.cub.2009.08.052
Fukuda, H., and Kobayashi, H. (1989). Dynamic organization of the cytoskeleton during tracheary-element differentiation. Dev. Growth Differ. 31, 9–16. doi: 10.1111/j.1440-169X.1989.00009.x
Giddings, T. H. Jr., and Staehelin, L. A. (1988). Spatial relationship between microtubules and plasma-membrane rosettes during the deposition of primary wall microfibrils in Closterium sp. Planta 173, 22–30. doi: 10.1007/BF00394482
Gu, Y., Li, S., Lord, E. M., and Yang, Z. (2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18, 366–381. doi: 10.1105/tpc.105.036434
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. doi: 10.1038/ncb1886
Hamada, T., Tekeuchi, N. N., Kato, T., Fujiwara, M., Sonobe, S., Fukao, Y., et al. (2013). Purification and characterization of novel microtubule-associated proteins from Arabidopsis cell suspension cultures. Plant Physiol. doi: 10.1104/pp.113.225607 [Epub ahead of print].
Hardham, A. R., and Gunning, B. E. S. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77, 14–34.
Hepler, P. K. (1981). Morphogenesis of tracheary elements and guard cells. Cell Biol. Monogr. 8, 327–347. doi: 10.1007/978-3-7091-8602-2_12
Kobayashi, H., Fukuda, H., and Shibaoka, H. (1987). Reorganization of actin-filaments associated with the differentiation of tracheary elements in zinnia mesophyll-cells. Protoplasma 138, 69–71. doi: 10.1007/BF01281188
Kobayashi, H., Fukuda, H., and Shibaoka, H. (1988). Interrelation between the spatial disposition of actin-filaments and microtubules during the differentiation of tracheary elements in cultured zinnia cells. Protoplasma 143, 29–37. doi: 10.1007/BF01282956
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Kusumi, A., Nakada, C., Ritchie, K., Murase, K., Suzuki, K., Murakoshi, H., et al. (2005). Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34, 351–378. doi: 10.1146/annurev.biophys.34.040204.144637
Lancelle, S. A., Callaham, D. A., and Hepler, P. K. (1986). A method for rapid freeze fixation of plant cells. Protoplasma 13. doi: 10.1007/BF01285037
Lavy, M., Bloch, D., Hazak, O., Gutman, I., Poraty, L., Sorek, N., et al. (2007). A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17, 947–952. doi: 10.1016/j.cub.2007.04.038
Li, S., Gu, Y., Yan, A., Lord, E., and Yang, Z. B. (2008). RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol. Plant 1, 1021–1035. doi: 10.1093/mp/ssn051
Lin, D., Cao, L., Zhou, Z., Zhu, L., Ehrhardt, D., Yang, Z., et al. (2013). Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr. Biol. 23, 290–297. doi: 10.1016/j.cub.2013.01.022
Lu, L., Lee, Y. R., Pan, R., Maloof, J. N., and Liu, B. (2005). An internal motor kinesin is associated with the Golgi apparatus and plays a role in trichome morphogenesis in Arabidopsis. Mol. Biol. Cell 16, 811–823. doi: 10.1091/mbc.E04-05-0400
Martiniere, A., Gayral, P., Hawes, C., and Runions, J. (2011). Building bridges: formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J. 66, 354–365. doi: 10.1111/j.1365-313X.2011.04497.x
Mucha, E., Hoefle, C., Huckelhoven, R., and Berken, A. (2010). RIP3 and AtKinesin-13A – a novel interaction linking Rho proteins of plants to microtubules. Eur. J. Cell Biol. 89, 906–916. doi: 10.1016/j.ejcb.2010.08.003
Murata, T., Sonobe, S., Baskin, T. I., Hyodo, S., Hasezawa, S., Nagata, T., et al. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7, 961–968. doi: 10.1038/ncb1306
Nagawa, S., Xu, T., Lin, D., Dhonukshe, P., Zhang, X., Friml, J., et al. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10:e1001299. doi: 10.1371/journal.pbio.1001299
Nakamura, M., Ehrhardt, D. W., and Hashimoto, T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12, 1064–1070. doi: 10.1038/ncb2110
Oda, Y., and Fukuda, H. (2012a). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337, 1333–1336. doi: 10.1126/science.1222597
Oda, Y., and Fukuda, H. (2012b). Secondary cell wall patterning during xylem differentiation. Curr. Opin. Plant Biol. 15, 38–44. doi: 10.1016/j.pbi.2011.10.005
Oda, Y., and Fukuda, H. (2013a). Rho of plant GTPase signaling regulates the behavior of Arabidopsis Kinesin-13A to establish secondary cell wall patterns. Plant Cell doi: org/10.1105/tpc.113.117853 [Epub ahead of print].
Oda, Y., and Fukuda, H. (2013b). Spatial organization of xylem cell walls by ROP GTPases and microtubule-associated proteins. Curr. Opin. Plant Biol. doi: 10.1016/j.pbi.2013.10.010. [Epub ahead of print].
Oda, Y., and Hasezawa, S. (2006). Cytoskeletal organization during xylem cell differentiation. J. Plant Res. 119, 167–177. doi: 10.1007/s10265-006-0260-8
Oda, Y., Iida, Y., Kondo, Y., and Fukuda, H. (2010). Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr. Biol. 20, 1197–1202. doi: 10.1016/j.cub.2010.05.038
Oda, Y., Mimura, T., and Hasezawa, S. (2005). Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137, 1027–1036. doi: 10.1104/pp.104.052613
Paredez, A. R., Somerville, C. R., and Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Pesquet, E., Korolev, A. V., Calder, G., and Lloyd, C. W. (2010). The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr. Biol. 20, 744–749. doi: 10.1016/j.cub.2010.02.057
Seagull, R. W., and Heath, I. B. (1980). The organization of cortical microtubule arrays in the radish root hair. Protoplasma 103, 205–229. doi: 10.1007/BF01276268
Shaw, S. L., Kamyar, R., and Ehrhardt, D. W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. doi: 10.1126/science.1083529
Wasteneys, G. O., and Ambrose, J. C. (2009). Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 19, 62–71. doi: 10.1016/j.tcb.2008.11.004
Wightman, R., and Turner, S. R. (2008). The roles of the cytoskeleton during cellulose deposition at the secondary cell wall. Plant J. 54, 794–805. doi: 10.1111/j.1365-313X.2008.03444.x
Wu, G., Li, H., and Yang, Z. (2000). Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation. Plant Physiol. 124, 1625–1636. doi: 10.1104/pp.124.4.1625
Xu, T., Wen, M., Nagawa, S., Fu, Y., Chen, J. G., Wu, M. J., et al. (2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110. doi: 10.1016/j.cell.2010.09.003
Zhang, Y., and McCormick, S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 18830–18835. doi: 10.1073/pnas.0705874104
Keywords: secondary cell wall, ROP GTPase, MIDD1, cortical microtubule, xylem
Citation: Oda Y and Fukuda H (2013) The dynamic interplay of plasma membrane domains and cortical microtubules in secondary cell wall patterning. Front. Plant Sci. 4:511. doi: 10.3389/fpls.2013.00511
Received: 05 November 2013; Accepted: 28 November 2013;
Published online: 17 December 2013.
Edited by:
Christopher J. Staiger, Purdue University, USAReviewed by:
Edouard Pesquet, Umea Plant Science Centre, SwedenYing Fu, China Agricultural University, China
Copyright © 2013 Oda and Fukuda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshihisa Oda and Hiroo Fukuda, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan e-mail: oda@biol.s.u-tokyo.ac.jp; fukuda@biol.s.u-tokyo.ac.jp
 Yoshihisa Oda
Yoshihisa Oda Hiroo Fukuda
Hiroo Fukuda