Dynamic nature of SecA and its associated proteins in Escherichia coli
A Corrigendum on
Dynamic nature of SecA and Its associated proteins in Escherichia coli
Adachi, S., Murakawa, Y., and Hiraga, S. (2015). Front. Microbiol. 6:75. doi: 10.3389/fmicb.2015.00075
In the original article, there was a mistake in Figure 2 as published. Panel J in the original figure was incorrectly labeled “SecA-GFP secA204.” Panel J in the corrected figure appearing below is now labeled “SecY-GFP secA204” in agreement with panels F to I presented in this section of the figure.
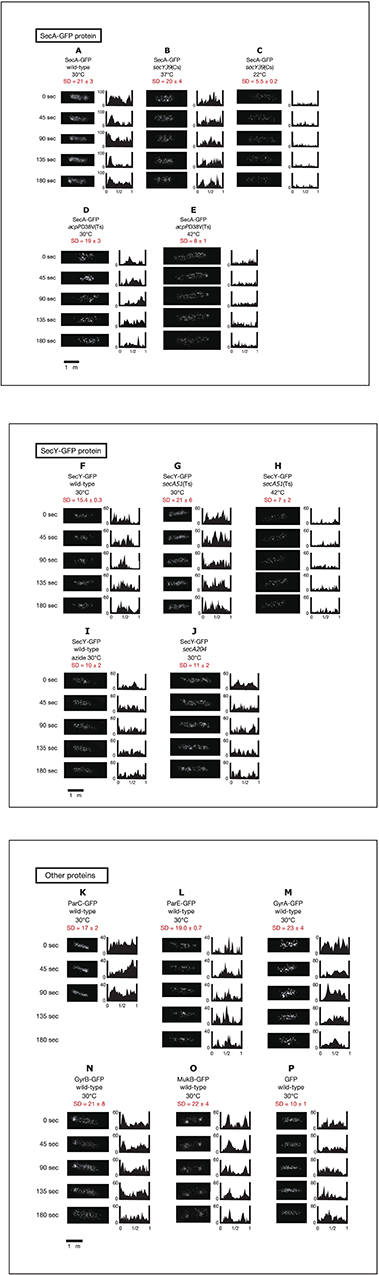
Figure 2. Time-lapse images of GFPuv4-fused proteins reveal the dynamic nature of chromosome partitioning proteins. Cells were grown in medium C and incubated with 0.1 mM IPTG for 1 h. Time-lapse images of fluorescence were taken by exposure for 100 ms. Both images and histograms of signals along long cell axes were obtained at 45-s intervals. The x-axes of histograms are subcellular positions and the y-axes of histograms are relative fluorescence units. Ninety-five Percent confidential SD (standard deviation) values of fluorescence signals along long cell axes in individual cells are also shown. (A) SecA-GFPuv4 in wild-type cells (MQ318) growing at 30°C. (B) SecA-GFPuv4 in secY39(Cs) mutant cells (MQ435) growing at the permissive temperature of 37°C. (C) SecA-GFPuv4 in secY39(Cs) mutant cells (MQ435) growing at the non-permissive temperature of 22°C. (D) SecA-GFPuv4 in acpPD38V(Ts) mutant cells (MQ456) growing at the permissive temperature of 30°C. (E) SecA-GFPuv4 in acpPD38V(Ts) mutant cells (MQ456) growing at the non-permissive temperature of 42°C. (F) SecY-GFPuv4 in wild-type cells (MQ319) growing at 30°C. (G) SecY-GFPuv4 in secA51(Ts) mutant cells (MQ748) growing at the permissive temperature of 30°C. (H) SecY-GFPuv4 in secA51(Ts) mutant cells (MQ748) growing at the non-permissive temperature of 42°C. (I) SecY-GFPuv4 in wild-type cells (MQ319) with 1 mM sodium azide growing at 30°C. The incubation time with azide was approximately 5 min. (J) SecY-GFPuv4 in secA204 azide-resistant mutant cells (MQ625) growing at 30°C (without sodium azide). (K) ParC-GFPuv4 in wild-type cells (MQ537) growing at 30°C. (L) ParE-GFPuv4 in wild-type cells (MQ539) growing at 30°C. (M) GyrA-GFPuv4 in wild-type cells (MQ323) growing at 30°C. (N) GyrB-GFPuv4 in wild-type cells (MQ324) growing at 30°C. (O) MukB-GFPuv4 in wild-type cells (MQ529) growing at 30°C. (P) GFPuv4 in wild-type cells (MQ668) growing at 30°C.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: chromosome partition, SecA, SecY, AcpP, MukB, DNA topoisomerase
Citation: Adachi S, Murakawa Y and Hiraga S (2020) Corrigendum: Dynamic nature of SecA and Its associated proteins in Escherichia coli. Front. Microbiol. 11:595994. doi: 10.3389/fmicb.2020.595994
Received: 18 August 2020; Accepted: 13 October 2020;
Published: 25 November 2020.
Edited by:
Biswarup Mukhopadhyay, Virginia Tech, United StatesReviewed by:
Anastassios Economou, KU Leuven, BelgiumCopyright © 2020 Adachi, Murakawa and Hiraga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shun Adachi, f.peregrinusns@mbox.kyoto-inet.or.jp
†Present address: Shun Adachi, Department of Microbiology, Kansai Medical University, Hirakata, Japan
Yasuhiro Murakawa, RIKEN-IFOM Joint Laboratory for Cancer Genomics, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
‡Deceased
 Shun Adachi
Shun Adachi Yasuhiro Murakawa
Yasuhiro Murakawa Sota Hiraga
Sota Hiraga