- 1Anhui Province Key Laboratory of Medical Physics and Technology, Center of Medical Physics and Technology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, China
- 2Hefei Cancer Hospital, Chinese Academy of Sciences, Hefei, China
- 3Hefei Institute of Stem Cell and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Hefei, China
- 4Department of Oncology, Xinhua Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
Aurora kinase A (AURKA) is a cell cycle regulatory serine/threonine kinase that promotes cell cycle progression. It plays an important role in regulating the transition from G2 to M phase during mitosis. The association between the AURKA rs2273535 T>A polymorphism and cancer risk has been investigated, but the results remain inconsistent. To get a more accurate conclusion, we conducted a comprehensive meta-analysis of 36 case-control studies, involving 22,884 cancer cases and 30,497 healthy controls. Crude odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to determine the association of interest. Pooled analysis indicated that the AURKA rs2273535 T>A polymorphism increased the overall risk of cancer (homozygous: OR = 1.17, 95% CI = 1.04-1.33; recessive: OR = 1.15, 95% CI = 1.05-1.25; allele: OR = 1.07, 95% CI = 1.02-1.13). Stratification analysis by cancer type further showed that this polymorphism was associated with an increased breast cancer risk. This meta-analysis indicated that the AURKA rs2273535 T>A polymorphism was associated with an overall increased cancer risk, especially breast cancer. Further validation experiments are needed to strengthen our conclusion.
Introduction
Aurora kinase A (AURKA) is a cell-cycle regulatory serine/threonine kinase that promotes cell cycle progression (1). AURKA is expressed in proliferating cells, especially in the G2 and mitotic phases of the cell cycle. It has various roles in promoting cell division, including the establishment of the mitotic spindle and centrosome separation (1). The AURKA gene is located at chromosomal locus 20q13.2 according to the HUGO Gene Nomenclature Committee (HGNC), and is often amplified in cancers (2–4). AURKA amplification has been found in certain tumor types, including colorectal, leukemia, and pancreatic cancers (3, 5, 6). Overexpression of AURKA can promote cellular transformation and may potentiate the activity of other oncogenes, such as RAS, further promoting tumorigenesis (7).
AURKA rs2273535 T>A, also known as F31I or Phe31Ile, is caused by a T-to-A transversion at position 91 in the AURKA coding sequence. This single nucleotide polymorphism is located in the Aurora Box1 (aa 5-40) motif, which belongs to the NH2-terminal region of aurora-A. This motif is thought to act on ubiquitin-based proteolysis (8).
Up until now, there have been many studies that have investigated the association between the AURKA F31I polymorphism and the risk of different types of cancer in different populations. However, the results are inconsistent, most likely because the sample size is relatively small in each published study, and the impact of the polymorphism on cancer risk might be small. Therefore, we conducted a comprehensive meta-analysis by identifying as many relevant articles as possible to identify evidence for the link between the AURKA F31I polymorphism and cancer risk.
Materials and Methods
Search Strategy
Two authors (JQ and SW) conducted the search for relevant articles before December 2018, using the following terms, “AURKA or Aurora Kinase A,” “tumor or cancer or carcinoma or neoplasm,” and “polymorphism or single-nucleotide polymorphism (SNP) or variant,” in the PubMed, EMBASE, CNKI, WANFANG, and Vip databases. We also examined the references of the retrieved publications for additional eligible studies.
Eligibility Criteria
Publications retrieved from the various databases were assessed for eligibility according to the following criteria: (1) the publication was published in English or Chinese; (2) the publication evaluated the association between the AURKA gene rs2273535 polymorphism and cancer risk; (3) the publication was a case-control study; the publication with the largest number of individuals was selected if studies had duplicate subjects. In addition, publications were excluded according to the following criteria: (1) genotype data included were not sufficient to calculate an odds ratio (OR) and 95% confidence interval (CI); (2) only survival data was included; (3) the genotype frequency distribution departed from Hardy-Weinberg equilibrium (HWE) in the controls.
Data Extraction
The following information was independently extracted by two authors (SW and JQ): first author name, year of publication, cancer type, region, ethnicity, genotyping method, source of controls (hospital-based, population-based, and mixed), the genotype counts of cases, and controls for the investigated polymorphism.
Quality Assessment
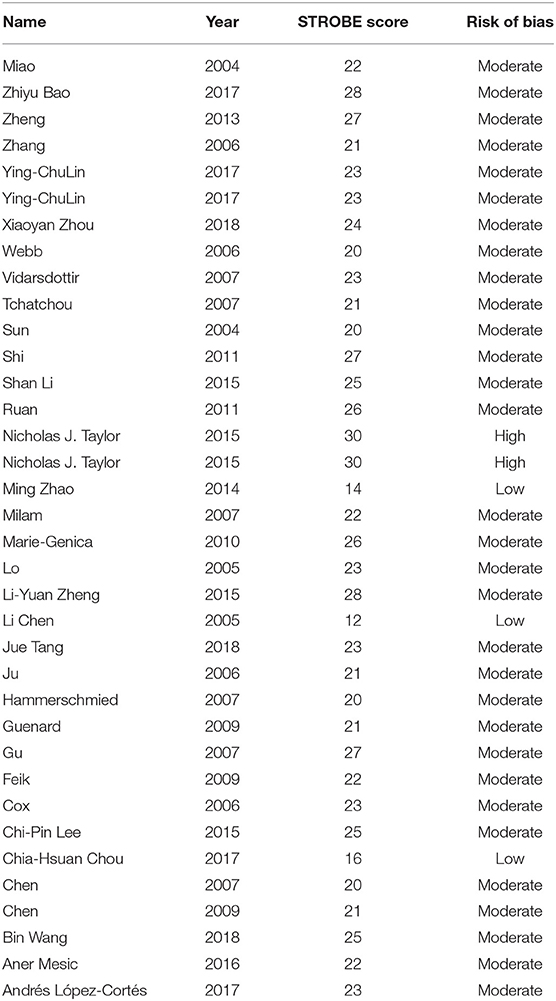
Two authors (SW and JQ) independently assessed the quality of the included studies according to the Strengthening the Reporting of Observational studies in Epidemiology (STROBE) quality scoring system (9). Forty evaluation items related to quality assessment were used in meta-analysis, with scores ranging from 0 to 40. Based on the STROBE score, the included studies were classified as low quality (0-19), moderate quality (20-29), and high quality (30-40) (10). If the studies were of low quality, moderate, or high quality, they were considered to be at high, moderate or low risk of bias, respectively. If two authors had contradictory information, a third author (MW) was consulted.
Statistical Analysis
Pooled ORs and 95% CIs were evaluated to assess the relationship between the AURKA rs2273535 T>A polymorphism and overall cancer risk under the heterozygous (AT vs. TT), homozygous (AA vs. TT), dominant (AT+AA vs. TT), recessive (AA vs. AT+TT), and allele contrast (A vs. T) models. We conducted stratification analyses by ethnicity, cancer type (“others”: one cancer type was investigated in less than 3 studies), study design (“mixed”: the source of controls contained both hospital-based and population-based subjects) and risk of bias. The Chi square-based Q-test was used to calculate the heterogeneity among studies. A random-effect model was adopted when P < 0.1 (heterogeneity). Otherwise, a fixed-effect model was adopted (11). Funnel plots were used to evaluate potential publication bias (12). All data analyses were performed using R software. All of the P values were two-tailed; P < 0.05 indicated statistical significance.
Results
Literature Search
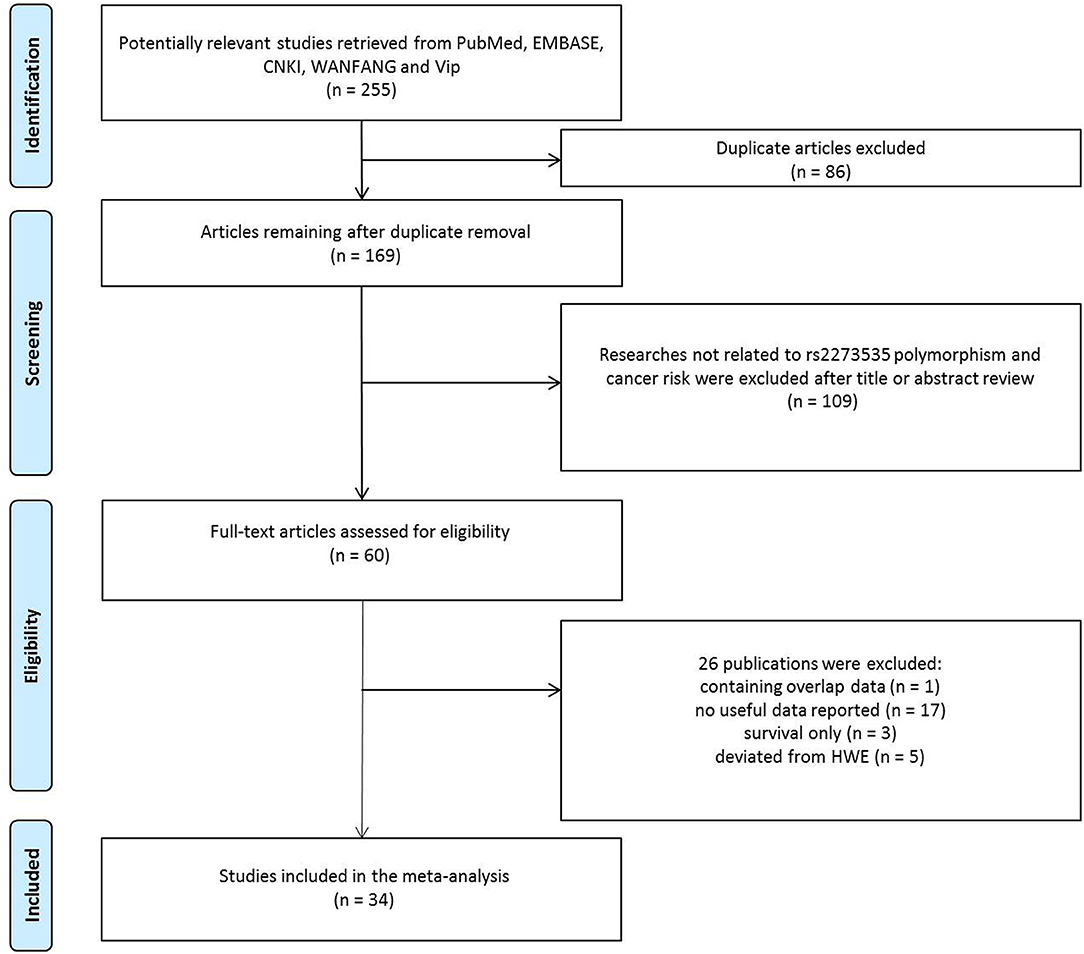
As shown in Figure 1, 255 potentially relevant studies were selected from the PubMed, CNKI, EMBASE, WANFANG, and Vip databases. We excluded 86 duplicate articles and 109 publications not investigating the association between the AURKA gene rs2273535 polymorphism and cancer risk after reviewing titles and abstracts. Then, the full texts of the remaining articles were evaluated. One publication (13) was removed for containing overlapping data. We also excluded 17 publications (14–30) because no useful data was reported to calculate ORs and 95% CIs. In addition, we eliminated 3 publications (31–33) presenting survival data only. Lastly, we excluded 5 publications (34–38) due to deviation from the HWE. Overall, 34 publications with a total of 22,884 cancer cases and 30,497 healthy controls were included in the meta-analysis.
Description and Quality of the Studies
The 34 publications actually consisted of 36 case-control studies, because 2 publications included 2 individual studies. The characteristics of these studies were shown in Supplemental Table 1. Among these publications, 12 focused on breast cancer (39–50), 5 on gastric cancer (51–55), 3 on colorectal cancer (56–58), 2 on esophageal (59, 60), 2 on liver cancer (61, 62), and 2 on oral cancer (63, 64). Moreover, there was only 1 study for each of the following cancers: lung cancer (65), neuroblastoma cancer (66), ovarian cancer (67), prostate cancer (68), renal cancer (69), urinary tract urothelial cancer (70), uterine cancer (71), bladder cancer (70), and endometrial cancer (72). As evident in Table 1, among these case-control studies, 2 of them had low risk of bias, 31 had moderate risk of bias, while 3 had high risk of bias. The shortcomings of low risk of bias research mainly focused on the lack of some descriptions in the results section (describing the numbers of individuals at each stage of the study, reasons for non-participation at each stage and a flow diagram) and methods section (describing comparability of assessment methods, address potential sources of bias, the estimation of the study size). In addition to the shortcomings involved in the low-risk bias study, the moderate-risk bias study also lacked the description in the methods section(explaining how missing data were addressed, how missing data were addressed and how quantitative variables were handled in the analyses), results section(indicating the number of participants with missing data and if relevant, considering translating estimates of relative risk into absolute risk for a meaningful time period) and discussion section (reporting category boundaries when continuous variables were categorized). High-risk bias study not only included the above-mentioned defects, but included the lack of giving the sources and methods of case ascertainment and control selection, the rationale for the choice of cases and controls and matching criteria and the number of controls, clearly defining all outcomes, exposures, predictors, potential confounders, effect modifiers and describing any sensitivity analyses in the methods section. Finally, this meta-analysis contained 19 hospital-based and 15 population-based studies.
Meta-Analysis Results
As evident in Table 2, Figures 2, 3, there is significant inter-study heterogeneity under all the genetic models; thus, we used a random-effect model. After calculating crude ORs and 95% CIs, we found that the AURKA gene rs2273535 T>A polymorphism was associated with increased overall cancer susceptibility (homozygous: OR = 1.17, 95% CI = 1.04-1.33; recessive: OR = 1.15, 95% CI = 1.05-1.25; allele: OR = 1.07, 95% CI = 1.02-1.13). Figures 2, 3 depicted forest plot of the association between the AURKA rs2273535 T>A polymorphism and overall cancer risk under the dominant and homozygous model. We performed stratification analyses by cancer type, ethnicity, study design and risk of bias. Stratification analysis further indicated that the AURKA gene rs2273535 T>A polymorphism was associated with increased risk of breast cancer (homozygous: OR = 1.28, 95% CI = 1.12-1.47; recessive: OR = 1.17, 95% CI = 1.05-1.31; allele: OR = 1.09, 95% CI = 1.02-1.17). Figures 4, 5 show stratification analysis of the association between the AURKA rs2273535 T>A polymorphism and cancer risk by cancer type under the dominant and homozygous model. We also checked the association in the Asian population. Interestingly, we only observed significant associations in Asians (recessive: OR = 1.17, 95% CI = 1.05-1.32; allele: OR = 1.09, 95% CI = 1.01-1.18). Moreover, the association remained significant in the subgroups with population-based studies (homozygous: OR = 1.25, 95% CI = 1.07-1.46; recessive: OR = 1.19, 95% CI = 1.07-1.33; allele: OR = 1.10, 95% CI = 1.03-1.19) and moderate risk of bias (homozygous: OR = 1.17, 95% CI = 1.02-1.34; recessive: OR = 1.17, 95% CI = 1.06-1.29; allele: OR = 1.08, 95% CI = 1.02-1.14). Figure 6 revealed stratification analysis of the association between the AURKA rs2273535 T>A polymorphism and cancer risk by risk of bias under the homozygous model.
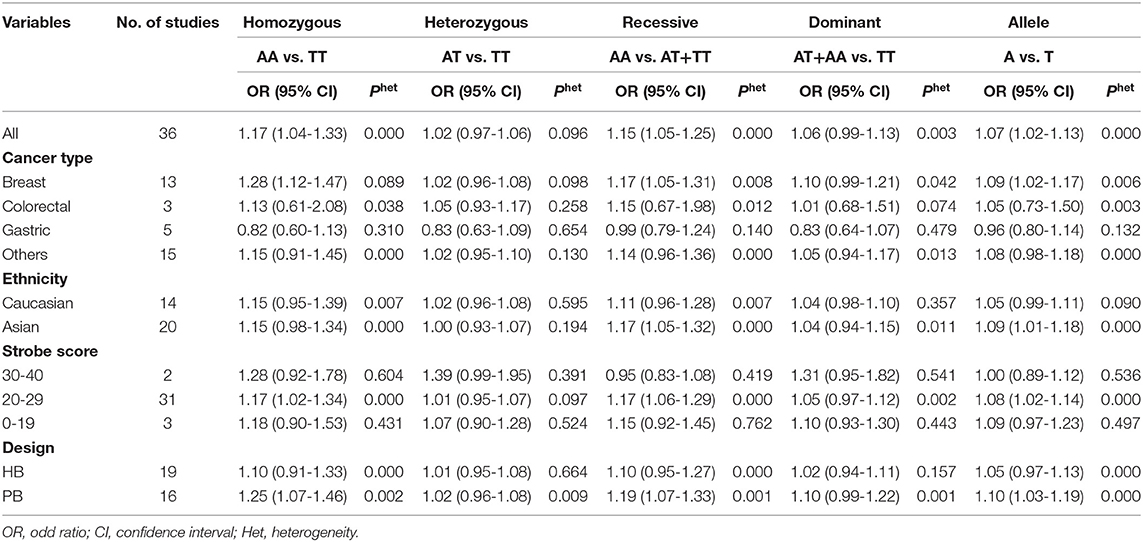
Table 2. The association between the AURKA rs2273535 T>A polymorphism and cancer risk in the meta-analysis.
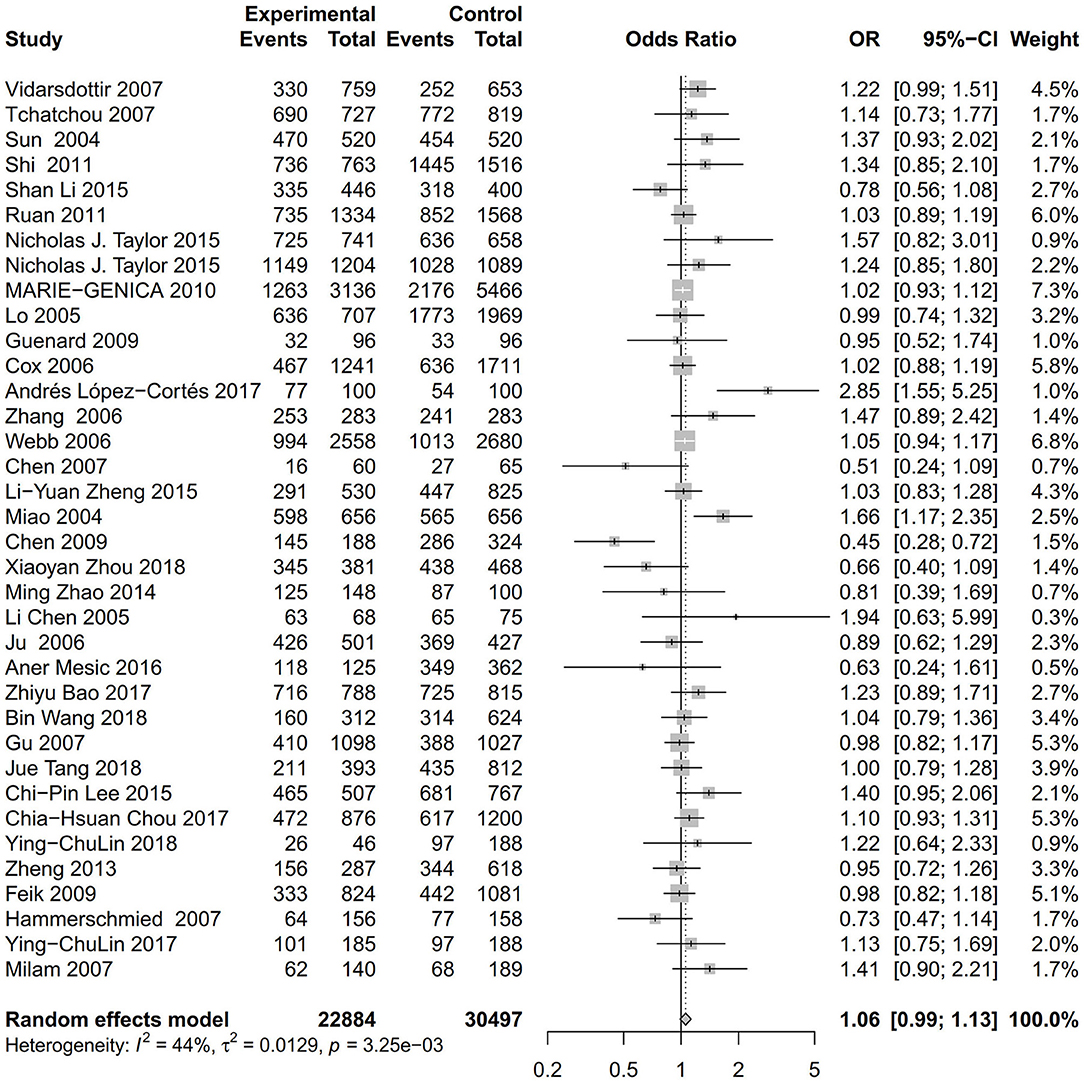
Figure 2. Forest plot of the association between the AURKA rs2273535 T>A polymorphism and overall cancer risk under the dominant model (AT+AA vs. TT).
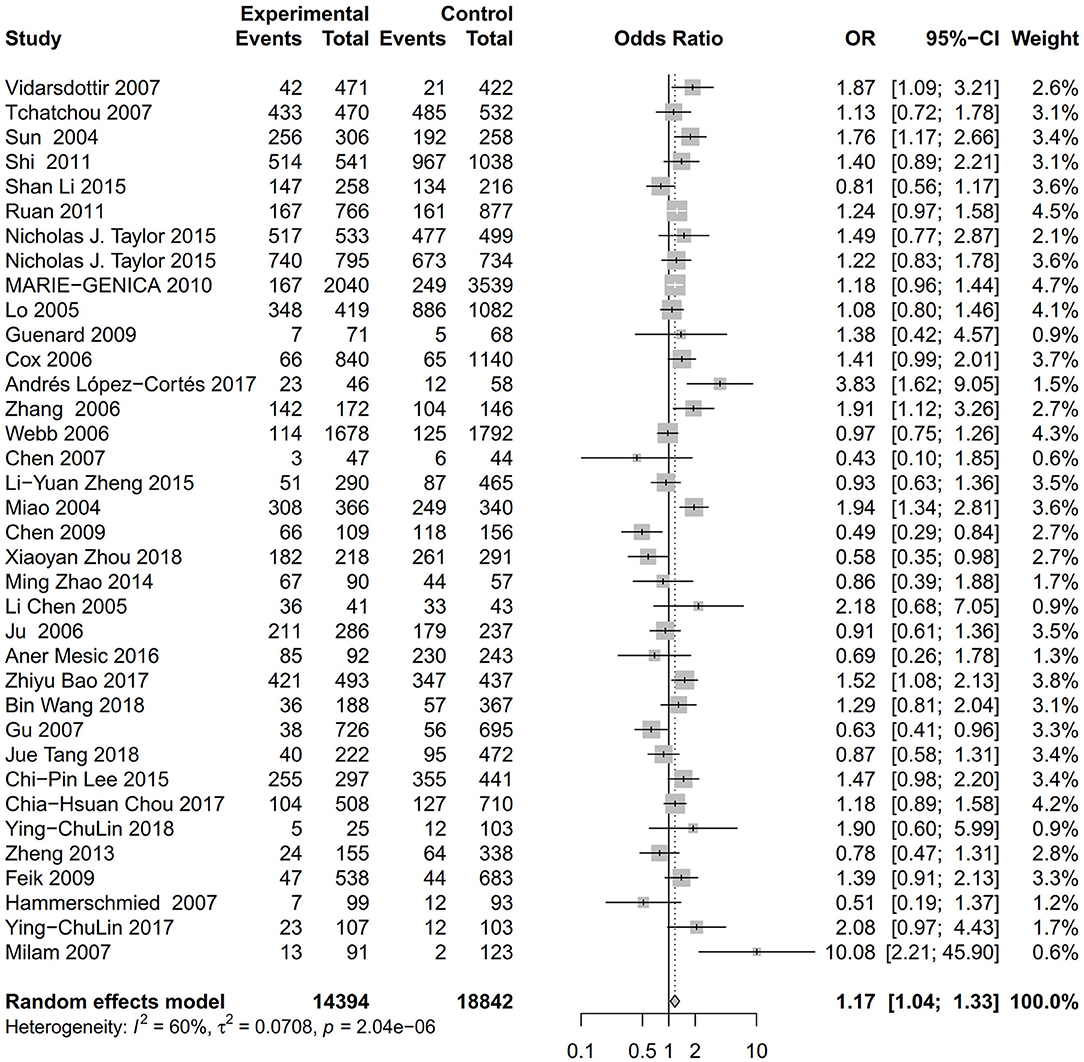
Figure 3. Forest plot of the association between the AURKA rs2273535 T>A polymorphism and overall cancer risk under the homozygous model (AA vs. TT).
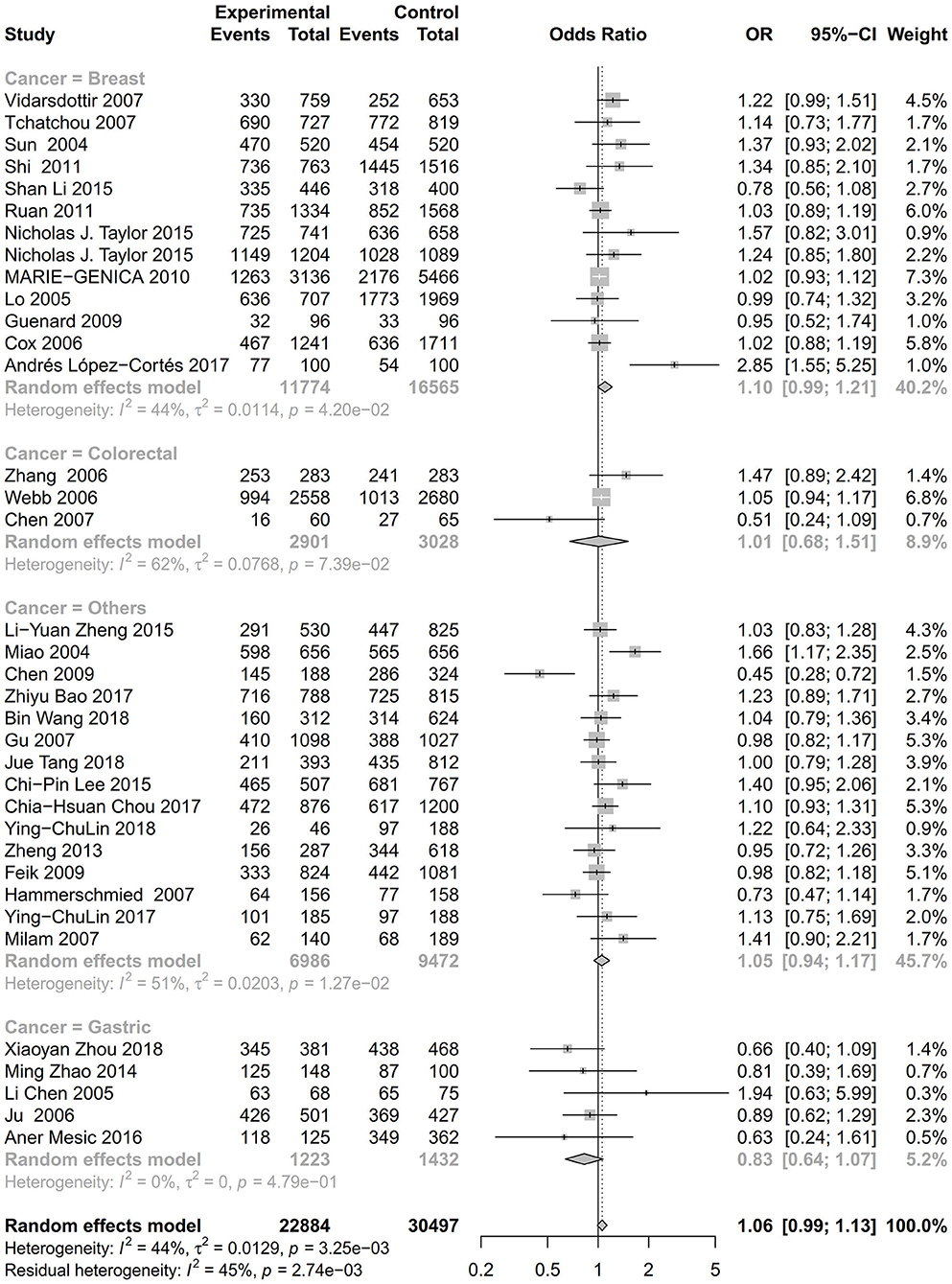
Figure 4. Stratification analysis of the association between the AURKA rs2273535 T>A polymorphism and cancer risk by cancer type under the dominant model (AT+AA vs. TT).
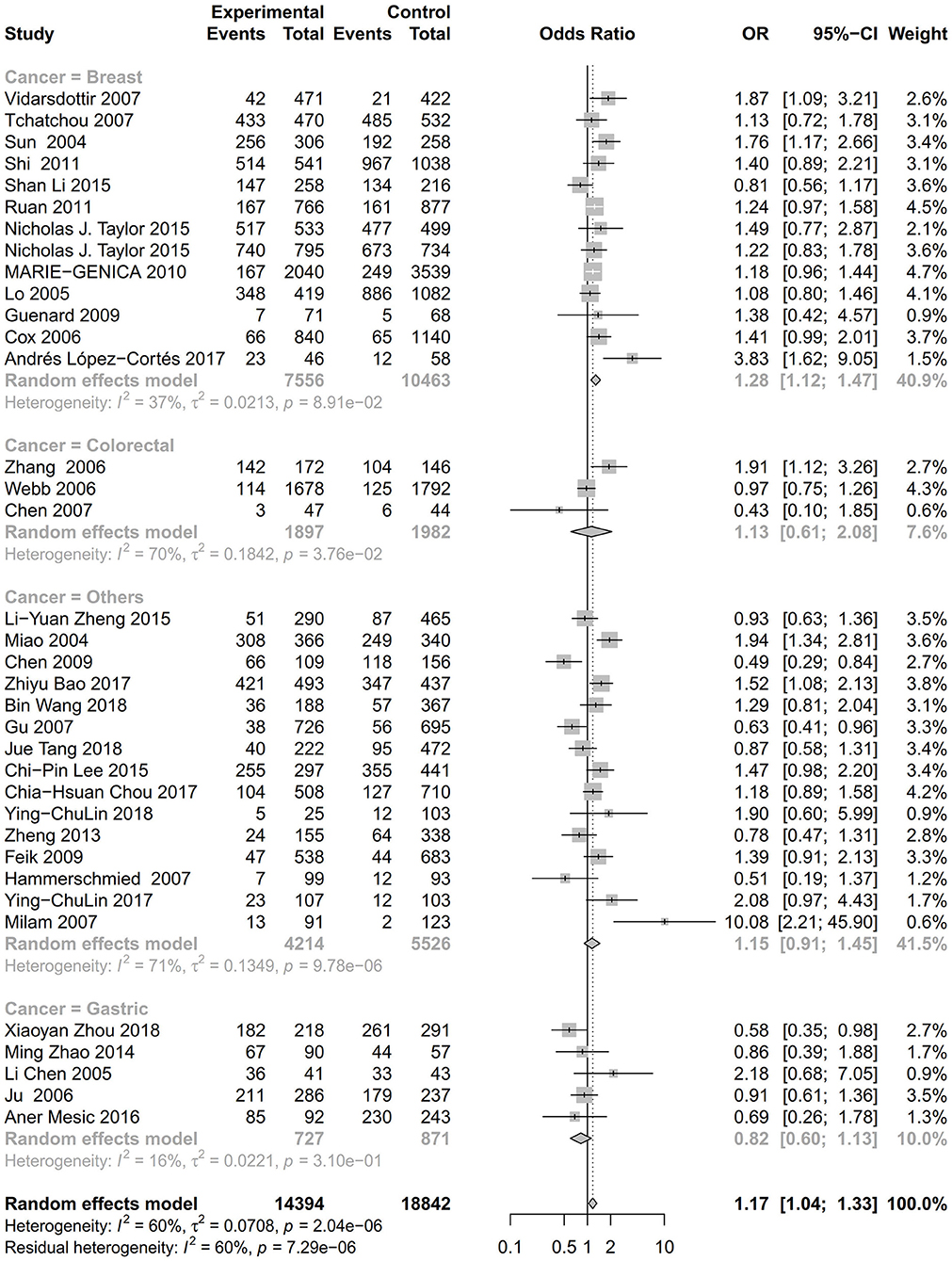
Figure 5. Stratification analysis of the association between the AURKA rs2273535 T>A polymorphism and cancer risk by cancer type under the homozygous model (AA vs. TT).
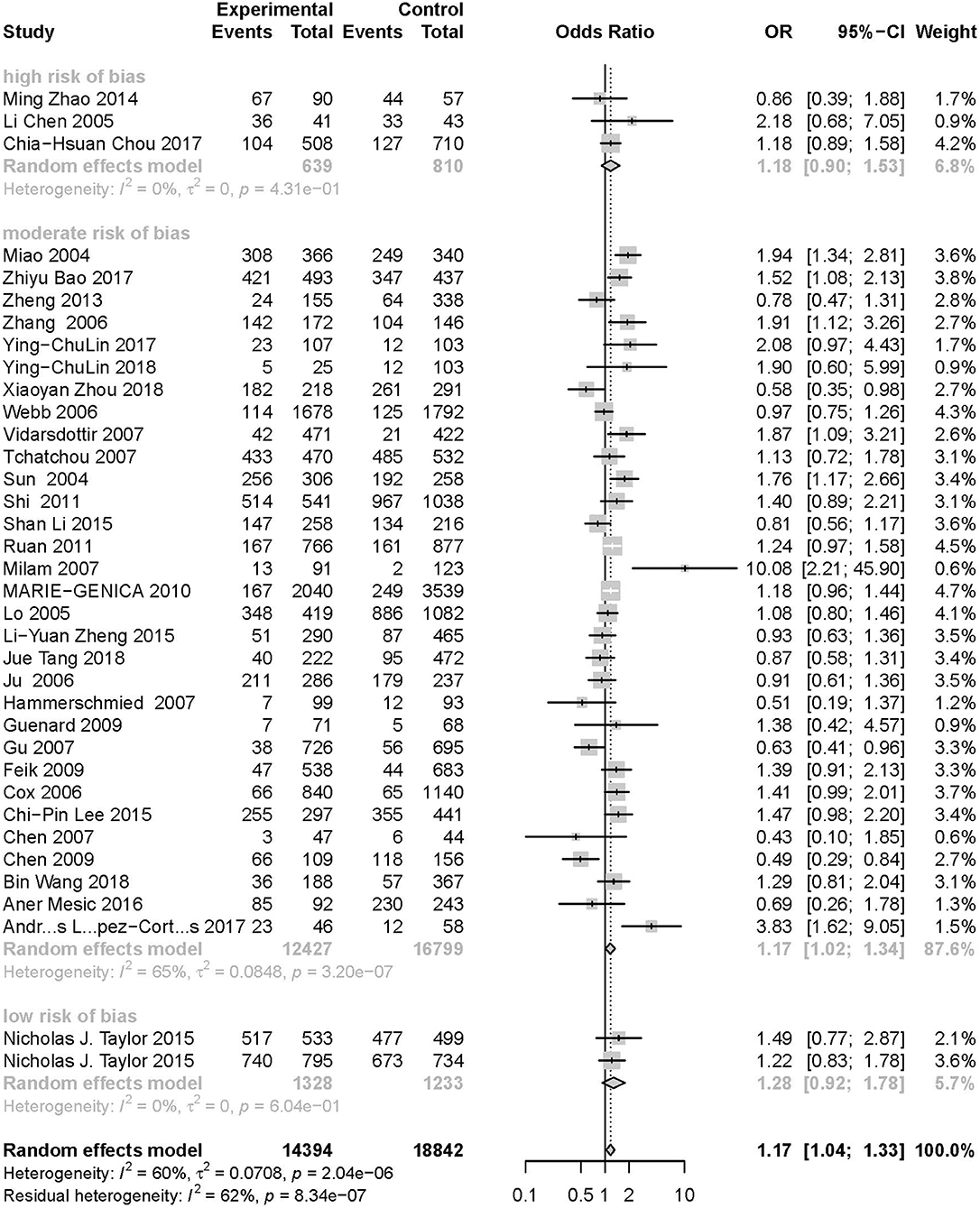
Figure 6. Stratification analysis of the association between the AURKA rs2273535 T>A polymorphism and cancer risk by risk of bias under the homozygous model (AA vs. TT).
Publication Bias
Symmetry in the funnel plots (Figures 7, 8) suggested that there was no significant publication bias in this meta-analysis (homozygous: P = 0.585; heterozygous: P = 0.939; recessive: P = 0.586; dominant: P = 0.546; allele: P = 0.657).
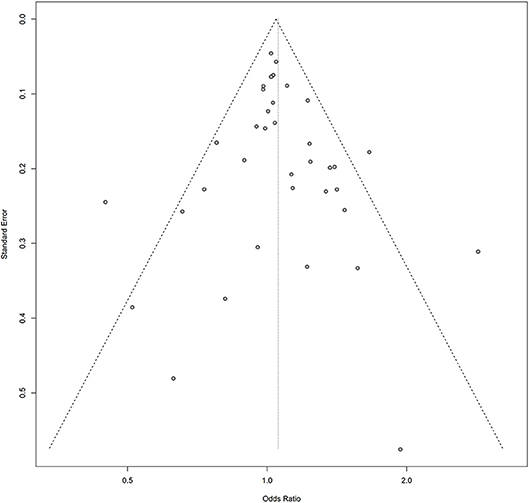
Figure 7. Funnel plot of the association between the AURKA rs2273535 T>A polymorphism and overall cancer risk under the dominant model (AT+AA vs. TT).
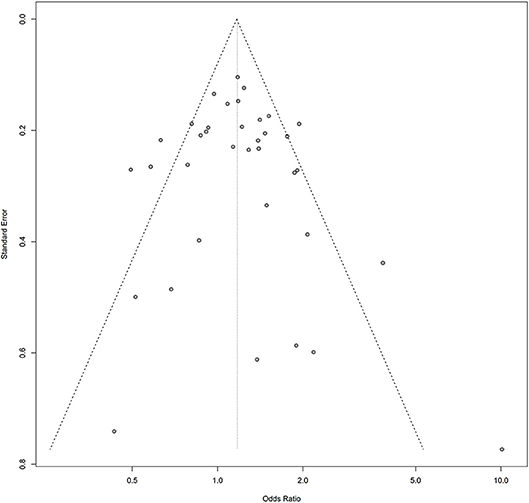
Figure 8. Funnel plot of the association between the AURKA rs2273535 T>A polymorphism and overall cancer risk under the homozygous model (AA vs. TT).
Discussion
AURKA is a key factor in regulating thetransition from G2 to M phase during mitosis. The AURKA protein includes a 129-amino acid N-terminal domain that facilitates AURKA nuclear-translocation during mitosis and a 274-amino acid C-terminal kinase catalytic domain (73). AURKA has been reportedly associated with poor prognosis in medulloblastoma and over-expression in various types of cancer (74). AURKA protein amplification and over-expression in breast and other tumors is related to centrosomal amplification, dysfunction of cytokinesis, and aneuploidy. Based on genetic mapping studies, AURKA is a potential genetic target for cancer therapy (16). The AURKA F31I polymorphism (T>A)(phenylalanine (Phe)> isoleucine (Ile)) is related to cellular transformation and distinctly enhances chromosomal instability (75). This polymorphism can also cause an obstruction in p53 binding and decreased degradation of AURKA by changing the activity of the AURKA box 1 (16). Research has shown that the stabilized over-expression of AURKA results in centrosomal amplification, abnormal cytokinesis, chromosomal instability, and the promotion of tumorigenesis. Numerous studies have been performed to explore the association between the rs2273535 polymorphism and the risk of various types of cancer. López-Cortés et al. (50) carried out a study in 2018 to investigate the role of single nucleotide polymorphism AURKA T91A (rs2273535) in a high altitude Ecuadorian Mestizo population consisting of 100 patients and 100 controls, and found a significant relationship between the rs2273535 genotype and a higher risk of breast cancer development. This association was confirmed in different types of cancer, including hepatocellular carcinoma (HCC) by Bao et al. (62) with 788 cases and 815 controls, urinary tract urothelial cancer by Lin et al. (70) with 185 cases and 188 controls, gastric cancer by Zhou et al. (51) with 381 cases and 468 controls, as well as other types of cancer. However, opposing results were also frequently reported. A case-control study containing 501 prostate cancer and 427 control subjects conducted by Feik et al. (68) revealed that the AURKA rs2273535 polymorphism was not found to be related to prostate cancer risk. Additionally, Ju et al. (54) reported that this polymorphism was not related to gastric cancer susceptibility, by studying 501 cases and 427 controls. Tang et al. (66) selected 393 cases and 812 controls, and the results indicated that none of the AURKA polymorphisms were associated with neuroblastoma susceptibility in two distinct Chinese populations. Several meta-analyses have also been conducted, and unfortunately the results were still inconclusive (76–80). In this meta-analysis, the association between the AURKA gene rs2273535 T>A polymorphism and cancer risk based on 36 eligible case-control studies, with a total of 22,884 cancer cases and 30,497 healthy controls, was estimated. Among these case-control studies, 2 of them had low risk of bias, 31 had moderate risk of bias, while 3 had high risk of bias. Most quality scores of the included studies were higher than 20 (low to moderate risk of bias). Overall, our results indicated that this polymorphism might increase the overall risk of cancer, especially breast cancer.
However, there were some limitations in this meta-analysis. First, only publications written in Chinese or English were selected. Second, the number of studies for certain cancer types was inadequate, such as colorectal cancer (<5 studies). There were 3 included studies having high risk of bias (0 ≤ STROBE score ≤ 19); further studies with low risk of bias are needed to validate the true association. In addition, other factors may also influence cancer risk, such as age and living habits. Our findings might suffer from potential confounding bias due to the lack of original data. Taken together, the results should be interpreted with caution.
To conclude, this meta-analysis suggests that the AURKA gene rs2273535 T>A polymorphism is significantly associated with an overall increased cancer risk, especially breast cancer. In the research, most of the included studies had low to moderate risk of bias. Future well-designed, large-scale studies that report upon the association of the rs2273535 polymorphism and cancer, in multiple cancer types, are required to validate the findings of this study.
Data Availability Statement
The datasets generated for this study can be found in the article/Supplementary Material.
Author Contributions
SW, MW, MZ and JN conceived and designed the study. SW, JQ, and MW conducted the literature searches, extracted the data, analyzed the data and prepared the figures and tables. SW, MW, and JQ wrote the draft of manuscript. SW, MW, and JN revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the CASHIPS Director's Fund (Grant number: YZJJ2019QN20), National Science Foundation for Young Scholars of China (Grant number: 21702207), National Natural Science Foundation of China (Grant number: 81602666) and One Hundred Talents Projects of the Chinese Academy of Sciences to JN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01040/full#supplementary-material
References
1. Carvalhal S, Ribeiro SA, Arocena M, Kasciukovic T, Temme A, Koehler K, et al. The nucleoporin ALADIN regulates Aurora A localization to ensure robust mitotic spindle formation. Mol Biol Cell. (2015) 26:3424–38. doi: 10.1091/mbc.E15-02-0113
2. Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. (1998) 20:189–93. doi: 10.1038/2496
3. Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. (1998) 17:3052–65. doi: 10.1093/emboj/17.11.3052
4. Sen S, Zhou H, Zhang RD, Yoon DS, Vakar-Lopez F, Ito S, et al. Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J Natl Cancer Inst. (2002) 94:1320–9. doi: 10.1093/jnci/94.17.1320
5. Ochi T, Fujiwara H, Suemori K, Azuma T, Yakushijin Y, Hato T, et al. Aurora-A kinase: a novel target of cellular immunotherapy for leukemia. Blood. (2009) 113:66–74. doi: 10.1182/blood-2008-06-164889
6. Li D, Zhu J, Firozi PF, Abbruzzese JL, Evans DB, Cleary K, et al. Overexpression of oncogenic STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin. Cancer Res. (2003) 9:991–7. doi: 10.1093/carcin/24.3.613
7. Tatsuka M, Sato S, Kitajima S, Suto S, Kawai H, Miyauchi M, et al. Overexpression of Aurora-A potentiates HRAS-mediated oncogenic transformation and is implicated in oral carcinogenesis. Oncogene. (2005) 24:1122–7. doi: 10.1038/sj.onc.1208293
8. Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. (2002) 16:2274–85. doi: 10.1101/gad.1007302
9. von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. (2008) 61:344–9. doi: 10.1016/j.jclinepi.2007.11.008
10. Wu S, Han Y, Hu Q, Zhang X, Cui G, Li Z, et al. Effects of common polymorphisms in the MTHFR and ACE genes on diabetic peripheral neuropathy progression: a meta-analysis. Mol Neurobiol. (2017) 54:2435–44. doi: 10.1007/s12035-016-9823-4
11. He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, et al. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. (2014) 4:6159. doi: 10.1038/srep06159
12. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. (1994) 50:1088–1101. doi: 10.2307/2533446
13. Li S. AURKA T91A Gene Polymorphism and Breast Cancer Correlation Study of Uygur and Han in Xinjiang. Wulumuqi: Xinjiang Medical University (2014).
14. Torchia EC, Chen Y, Sheng H, Katayama H, Fitzpatrick J, Brinkley WR, et al. A genetic variant of Aurora kinase A promotes genomic instability leading to highly malignant skin tumors. Cancer Res. (2009) 69:7207–15. doi: 10.1158/0008-5472.CAN-09-1059
15. Srivastava V, Sen S. Genetic variation in coding region of Aurora kinase A gene leads to cancer susceptibility. Cancer Epidemiol. (2014) 38:109–10. doi: 10.1016/j.canep.2014.03.002
16. Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. (2003) 34:403–12. doi: 10.1038/ng1220
17. Klinke OK, Mizani T, Baldwin G, Bancel B, Devouassoux-Shisheboran M, Scoazec JY, et al. KIT mutation and Loss of 14q may be sufficient for the development of clinically symptomatic very low-risk GIST. PLoS ONE. (2015) 10:e0130149. doi: 10.1371/journal.pone.0130149
18. Dillon JL, Mockus SM, Ananda G, Spotlow V, Wells WA, Tsongalis GJ, et al. Somatic gene mutation analysis of triple negative breast cancers. Breast. (2016) 29:202–207. doi: 10.1016/j.breast.2016.06.018
19. Chen GL, Hou GL, Sun F, Jiang HL, Xue JF, Li XS, et al. Upregulation of STK15 in esophageal squamous cell carcinomas in a Mongolian population. Asian Pac J Cancer Prev. (2014) 15:6021–4. doi: 10.7314/APJCP.2014.15.15.6021
20. Koleck TA, Bender CM, Clark BZ, Ryan CM, Ghotkar P, Brufsky A, et al. An exploratory study of host polymorphisms in genes that clinically characterize breast cancer tumors and pretreatment cognitive performance in breast cancer survivors. Breast Cancer (Dove Med Press). (2017) 9:95–110. doi: 10.2147/BCTT.S123785
21. Baumann A, Buchberger AMS, Piontek G, Schuttler D, Rudelius M, Reiter R, et al. The Aurora-Kinase A Phe31-Ile polymorphism as possible predictor of response to treatment in head and neck squamous cell carcinoma. Oncotarget. (2018) 9:12769–80. doi: 10.18632/oncotarget.24355
22. Liu C. The association between AURKA T91A polymorphism and breast cancer risk. Breast Cancer Res Treat. (2011) 129:281–3. doi: 10.1007/s10549-011-1497-z
23. Pickhard A, Siegl M, Baumann A, Huhn M, Wirth M, Reiter R, et al. The response of head and neck squamous cell carcinoma to cetuximab treatment depends on Aurora kinase A polymorphism. Oncotarget. (2014) 5:5428–38. doi: 10.18632/oncotarget.2117
24. Pan JY, Ajani JA, Gu J, Gong Y, Qin A, Hung M, et al. Association of Aurora-A (STK15) kinase polymorphisms with clinical outcome of esophageal cancer treated with preoperative chemoradiation. Cancer. (2012) 118:4346–53. doi: 10.1002/cncr.26581
25. Niu H, Shin H, Gao F, Zhang J, Bahamon B, Danaee H, et al. Aurora a functional single nucleotide polymorphism (SNP) correlates with clinical outcome in patients with advanced solid tumors treated with alisertib, an investigational aurora A kinase inhibitor. EBioMedicine. (2017) 25:50–7. doi: 10.1016/j.ebiom.2017.10.015
26. Chen L, Ao X, Ren Q, Wang ZN, Lu C, Xu Y, et al. Linkage disequilibrium and haplotype analysis of two single nucleotide polymorphisms in STK15 in Chinese. Acta genetica Sinica. (2005) 32:331–6.
27. Matarasso N, Bar-Shira A, Rozovski U, Rosner S, Orr-Urtreger A. Functional analysis of the Aurora Kinase A Ile31 allelic variant in human prostate. Neoplasia. (2007) 9:707–15. doi: 10.1593/neo.07322
28. Provencio M, Camps C, Cobo M, De las Penas R, Massuti B, Blanco R, et al. Prospective assessment of XRCC3, XPD and Aurora kinase A single-nucleotide polymorphisms in advanced lung cancer. Cancer Chemother. Pharmacol. (2012) 70:883–90. doi: 10.1007/s00280-012-1985-9
29. Pohl A, Azuma M, Zhang W, Yang D, Ning Y, Winder T, et al. Pharmacogenetic profiling of Aurora kinase B is associated with overall survival in metastatic colorectal cancer. Pharmacogenomics J. (2011) 11:93–9. doi: 10.1038/tpj.2010.18
30. Giacomazzi J, Aguiar E, Palmero EI, Schmidt AV, Skonieski G, Duarte Filho D, et al. Prevalence of the STK15 F31I polymorphism and its relationship with mammographic density. Braz J Med Biol Res. (2011) 44:291–6. doi: 10.1590/S0100-879X2011007500029
31. Chen J, Etzel CJ, Amos CI, Zhang Q, Viscofsky N, Lindor NM, et al. Genetic variants in the cell cycle control pathways contribute to early onset colorectal cancer in Lynch syndrome. Cancer Causes Control. (2009) 20:1769–77. doi: 10.1007/s10552-009-9416-x
32. Boonstra JJ, van Marion R, Tilanus HW, Dinjens WN. Functional polymorphisms associated with disease-free survival in resected carcinoma of the esophagus. J Gastrointest Surg. (2011) 15:48–56. doi: 10.1007/s11605-010-1358-9
33. Necchi A, Pintarelli G, Raggi D, Giannatempo P, Colombo F. Association of an aurora kinase a (AURKA) gene polymorphism with progression-free survival in patients with advanced urothelial carcinoma treated with the selective aurora kinase a inhibitor alisertib. Invest New Drugs. (2017) 35:524–8. doi: 10.1007/s10637-017-0440-5
34. Golmohammadi R, Namazi MJ, Going JJ, Derakhshan MH. A single nucleotide polymorphism in codon F31I and V57I of the AURKA gene in invasive ductal breast carcinoma in Middle East. Medicine. (2017) 96:e7933. doi: 10.1097/MD.0000000000007933
35. Couch FJ, Sinilnikova O, Vierkant RA, Pankratz VS, Fredericksen ZS, Stoppa-Lyonnet D, et al. AURKA F31I polymorphism and breast cancer risk in BRCA1 and BRCA2 mutation carriers: a consortium of investigators of modifiers of BRCA1/2 study. Cancer Epidemiol. Biomarkers Prev. (2007) 16:1416–1421. doi: 10.1158/1055-9965.EPI-07-0129
36. Wang N, Wang GY, Guo W, Dong XJ, Li Y. Study on the association between STK15 Phe31Ile polymorphisms and esophageal squamous cell carcinoma. Chin J Epidemiol. (2007) 28:394–7.
37. Chava S, Mohan V, Pasupuleti N, Latha MM, Khan IA, Upendram P, et al. Evaluation of Aurora-A gene polymorphism and esophageal cancer risk in a South Indian population. Genet Test Mol Biomarkers. (2011) 15:185–9. doi: 10.1089/gtmb.2010.0143
38. Chong ET, Goh LP, See EU, Chuah JA, Chua KH, Lee PC. Association of CYP2E1, STK15 and XRCC1 polymorphisms with risk of breast cancer in malaysian women. Asian Pac J Cancer Prev. (2016) 17:647–53. doi: 10.7314/APJCP.2016.17.2.647
39. Vidarsdottir L, Bodvarsdottir SK, Hilmarsdottir H, Tryggvadottir L, Eyfjord JE. Breast cancer risk associated with AURKA 91T –>A polymorphism in relation to BRCA mutations. Cancer Lett. (2007) 250:206–12. doi: 10.1016/j.canlet.2006.10.003
40. Tchatchou S, Wirtenberger M, Hemminki K, Sutter C, Meindl A, Wappenschmidt B, et al. Aurora kinases A and B and familial breast cancer risk. Cancer Lett. (2007) 247:266–72. doi: 10.1016/j.canlet.2006.05.002
41. Sun T, Miao X, Wang J, Tan W, Zhou Y, Yu C, et al. Functional Phe31Ile polymorphism in Aurora A and risk of breast carcinoma. Carcinogenesis. (2004) 25:2225–30. doi: 10.1093/carcin/bgh244
42. Shi H, Bevier M, Johansson R, Grzybowska E, Chen B, Eyfjord JE, et al. Single nucleotide polymorphisms in the 20q13 amplicon genes in relation to breast cancer risk and clinical outcome. Breast Cancer Res. Treat. (2011) 130:905–16. doi: 10.1007/s10549-011-1600-5
43. Li S, Li Y, Zhao ZH, Yang SE. Correlation study of AURKA T91A gene polymorphism and breast cancer susceptibility of Xinjiang Uygur and Han women. Pract J Cancer. (2015) 30: 8–11. doi: 10.3969/j.issn.1001-5930
44. Ruan Y, Song AP, Wang H, Xie YT, Han JY, Sajdik C, et al. Genetic polymorphisms in AURKA and BRCA1 are associated with breast cancer susceptibility in a Chinese Han population. J. Pathol. (2011) 225:535–43. doi: 10.1002/path.2902
45. Taylor NJ, Bensen JT, Poole C, Troester MA, Gammon MD, Luo J, et al. Genetic variation in cell cycle regulatory gene AURKA and association with intrinsic breast cancer subtype. Mol Carcinog. (2015) 54:1668–77. doi: 10.1002/mc.22238
46. MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk. Polymorphisms in the BRCA1 and ABCB1 genes modulate menopausal hormone therapy associated breast cancer risk in postmenopausal women. Breast Cancer Res Treat. (2010) 120:727–36. doi: 10.1007/s10549-009-0489-8
47. Lo YL, Yu JC, Chen ST, Yang HC, Fann CS, Mau YC, et al. Breast cancer risk associated with genotypic polymorphism of the mitosis-regulating gene Aurora-A/STK15/BTAK. Int J Cancer. (2005) 115:276–83. doi: 10.1002/ijc.20855
48. Guenard F, Labrie Y, Ouellette G, Beauparlant CJ, Durocher F. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J Hum Genet. (2009) 54:152–61. doi: 10.1038/jhg.2009.6
49. Cox DG, Hankinson SE, Hunter DJ. Polymorphisms of the AURKA (STK15/Aurora kinase) gene and breast cancer risk (United States). Cancer Causes Control. (2006) 17:81–3. doi: 10.1007/s10552-005-0429-9
50. Lopez-Cortes A, Cabrera-Andrade A, Ona-Cisneros F, Echeverria C, Rosales F, Ortiz M, et al. Breast cancer risk associated with genotype polymorphisms of the aurora kinase a gene (AURKA): a case-control study in a high altitude ecuadorian mestizo population. Pathol Oncol Res. (2018) 24:457–65. doi: 10.1007/s12253-017-0267-6
51. Zhou X, Wang P, Zhao H. The association between AURKA gene rs2273535 polymorphism and gastric cancer risk in a chinese population. Front Physiol. (2018) 9:1124. doi: 10.3389/fphys.2018.01124
52. Zhao M. The Study on the Relationship Between the Aurora-A Polymorphism and EBV-Associated Gastric Carcinoma. Qingdao: Qingdao University (2014).
53. Chen L. Genetic Polymorphisms and Disease Risks: STK15 Gene and Risk for Gastric Cancer, MMP9 Gene and Risk for Thoracic Aortic Aneurysm and Thoracic Aortic Dissection. Shenyang: China Medical University (2005).
54. Ju H, Cho H, Kim YS, Kim WH, Ihm C, Noh SM, et al. Functional polymorphism 57Val>Ile of aurora kinase A associated with increased risk of gastric cancer progression. Cancer Lett. (2006) 242:273–9. doi: 10.1016/j.canlet.2005.11.015
55. Mesic A, Rogar M, Hudler P. Association of the AURKA and AURKC gene polymorphisms with an increased risk of gastric cancer. IUBMB life. (2016) 68:634–44. doi: 10.1002/iub.1521
56. Zhang WJ, Miao XP, Sun T, Zhang XM, Qu SN, Tan W, et al. Association between genetic polymorphism in STK15 and risk of colorectal cancer in a Chinese population. Chin J Oncol. (2006) 28:43–6. doi: 10.1007/s11769-006-0026-1
57. Webb EL, Rudd MF, Houlston RS. Case-control, kin-cohort and meta-analyses provide no support for STK15 F31I as a low penetrance colorectal cancer allele. Br J Cancer. (2006) 95:1047–9. doi: 10.1038/sj.bjc.6603382
58. Chen J, Sen S, Amos CI, Wei C, Jones JS, Lynch P, et al. Association between Aurora-A kinase polymorphisms and age of onset of hereditary nonpolyposis colorectal cancer in a Caucasian population. Mol Carcinog. (2007) 46:249–56. doi: 10.1002/mc.20283
59. Chen XB, Chen GL, Liu JN, Yang JZ, Yu DK, Lin DX, et al. Genetic polymorphisms in STK15 and MMP-2 associated susceptibility to esophageal cancer in Mongolian population. Chin J Prevent Med. (2009) 43:559–64.
60. Miao X, Sun T, Wang Y, Zhang X, Tan W, Lin D. Functional STK15 Phe31Ile polymorphism is associated with the occurrence and advanced disease status of esophageal squamous cell carcinoma. Cancer Res. (2004) 64:2680–3. doi: 10.1158/0008-5472.CAN-04-0651
61. Wang B, Hsu CJ, Chou CH, Lee HL, Chiang WL, Su CM, et al. Variations in the AURKA Gene: Biomarkers for the Development and Progression of Hepatocellular Carcinoma. Int J Med Sci. (2018) 15:170–5. doi: 10.7150/ijms.22513
62. Bao Z, Lu L, Liu X, Guo B, Zhai Y, Li Y, et al. Association between the functional polymorphism Ile31Phe in the AURKA gene and susceptibility of hepatocellular carcinoma in chronic hepatitis B virus carriers. Oncotarget. (2017) 8:54904–12. doi: 10.18632/oncotarget.18613
63. Lee CP, Chiang SL, Lee CH, Tsai YS, Wang ZH, Hua CH, et al. AURKA Phe31Ile polymorphism interacted with use of alcohol, betel quid, and cigarettes at multiplicative risk of oral cancer occurrence. Clin. Oral Investig. (2015) 19:1825–32. doi: 10.1007/s00784-015-1432-5
64. Chou CH, Chou YE, Chuang CY, Yang SF, Lin CW. Combined effect of genetic polymorphisms of AURKA and environmental factors on oral cancer development in Taiwan. PLoS ONE. (2017) 12:e0171583. doi: 10.1371/journal.pone.0171583
65. Gu J, Gong Y, Huang M, Lu C, Spitz MR, Wu X. Polymorphisms of STK15 (Aurora-A) gene and lung cancer risk in Caucasians. Carcinogenesis. (2007) 28:350–5. doi: 10.1093/carcin/bgl149
66. Tang J, Qian Y, Zhu J, Zhang J, Wang FH, Zeng JH, et al. Lack of associations between AURKA gene polymorphisms and neuroblastoma susceptibility in Chinese children. Biosci Rep. (2018) 38:1–7. doi: 10.1042/BSR20180292
67. Zheng L, Song A, Ruan Y, Chen L, Liu D, Li X, et al. Genetic polymorphisms in AURKA, BRCA1, CCNE1 and CDK2 are associated with ovarian cancer susceptibility among Chinese Han women. Cancer Epidemiol. (2013) 37:639–46. doi: 10.1016/j.canep.2013.04.018
68. Feik E, Baierl A, Madersbacher S, Schatzl G, Maj-Hes A, Berges R, et al. Common genetic polymorphisms of AURKA and prostate cancer risk. Cancer Causes Control. (2009) 20:147–52. doi: 10.1007/s10552-008-9227-5
69. Hammerschmied CG, Stoehr R, Walter B, Wieland WF, Hartmann A, Blaszyk H, et al. Role of the STK15 Phe31Ile polymorphism in renal cell carcinoma. Oncol Rep. (2007) 17:3–7. doi: 10.3892/or.17.1.3
70. Lin YC, Hour TC, Tsai YC, Huang SP, Wu WJ, Chen CH, et al. Preliminary evidence of polymorphisms of cell cycle regulatory genes and their roles in urinary tract urothelial cancer susceptibility and prognosis in a Taiwan population. Urol Oncol. (2017) 35:543.e547-543.e516. doi: 10.1016/j.urolonc.2016.08.001
71. Milam MR, Gu J, Yang H, Celestino J, Wu W, Horwitz IB, et al. STK15 F31I polymorphism is associated with increased uterine cancer risk: a pilot study. Gynecol Oncol. (2007) 107:71–4. doi: 10.1016/j.ygyno.2007.05.025
72. Zheng LY, Song AP, Chen L, Liu DG, Li XH, Guo HY, et al. Association of genetic polymorphisms in AURKA, BRCA1, CCNE1 and CDK2 with the risk of endometrial carcinoma and clinicopathological parameters among Chinese Han women. Eur J Obstet Gynecol Reprod Biol. (2015) 184:65–72. doi: 10.1016/j.ejogrb.2014.11.001
73. Cheetham GM, Knegtel RM, Coll JT, Renwick SB, Swenson L, Weber P, et al. Crystal structure of aurora-2, an oncogenic serine/threonine kinase. J Biol Chem. (2002) 277:42419–22. doi: 10.1074/jbc.C200426200
74. Neben K, Korshunov A, Benner A, Wrobel G, Hahn M, Kokocinski F, et al. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. (2004) 64:3103–11. doi: 10.1158/0008-5472.CAN-03-3968
75. Chen J, Li D, Wei C, Sen S, Killary AM, Amos CI, et al. Aurora-A and p16 polymorphisms contribute to an earlier age at diagnosis of pancreatic cancer in Caucasians. Clin Cancer Res. (2007) 13:3100–4. doi: 10.1158/1078-0432.CCR-06-2319
76. Dai ZJ, Kang HF, Wang XJ, Shao YP, Lin S, Zhao Y, et al. Association between genetic polymorphisms in AURKA (rs2273535 and rs1047972) and breast cancer risk: a meta-analysis involving 37,221 subjects. Cancer Cell Int. (2014) 14:91. doi: 10.1186/s12935-014-0091-y
77. Qin K, Wu C, Wu X. Two nonsynonymous polymorphisms (F31I and V57I) of the STK15 gene and breast cancer risk: a meta-analysis based on 5966 cases and 7609 controls. J Int Med Res. (2013) 41:956–63. doi: 10.1177/0300060513490087
78. Sun H, Bai J, Chen F, Jin Y, Yu Y, Fu S. Lack of an association between AURKA T91A polymorphisms and breast cancer: a meta-analysis involving 32,141 subjects. Breast Cancer Res Treat. (2011) 125:175–9. doi: 10.1007/s10549-010-0936-6
79. Ewart-Toland A, Dai Q, Gao YT, Nagase H, Dunlop MG, Farrington SM, et al. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. (2005) 26:1368–73. doi: 10.1093/carcin/bgi085
Keywords: meta-analysis, cell cycle, AURKA F31I, tumor, cancer risk
Citation: Wang S, Qi J, Zhu M, Wang M and Nie J (2020) AURKA rs2273535 T>A Polymorphism Associated With Cancer Risk: A Systematic Review With Meta-Analysis. Front. Oncol. 10:1040. doi: 10.3389/fonc.2020.01040
Received: 17 December 2019; Accepted: 26 May 2020;
Published: 30 June 2020.
Edited by:
Massimo Broggini, Mario Negri Pharmacological Research Institute, ItalyReviewed by:
Claude Prigent, Centre National de la Recherche Scientifique (CNRS), FranceMichela Cinquini, Mario Negri Pharmacological Research Institute, Italy
Copyright © 2020 Wang, Qi, Zhu, Wang and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meiling Zhu, zhumeiling@xinhuamed.com.cn; Meng Wang, wangmeng_cssc@foxmail.com; Jinfu Nie, jeffnie@cmpt.ac.cn
†These authors have contributed equally to this work
 Shujie Wang
Shujie Wang Jian Qi1,2†
Jian Qi1,2†