- 1Department of Molecular Biology and Biotechnology, Tezpur University, Tezpur, India
- 2Laboratoire des Interactions Plantes-Microorganismes, INRA, UMR 441, Castanet-Tolosan, France
- 3Laboratoire des Interactions Plantes-Microorganismes, Centre National de la Recherche Scientifique, UMR 2594, Castanet-Tolosan, France
The plant pathogen Ralstonia solanacearum has two genes encoding for the sigma factor σ54: rpoN1, located in the chromosome and rpoN2, located in a distinct “megaplasmid” replicon. In this study, individual mutants as well as a double mutant of rpoN were created in R. solanacearum strain GMI1000 in order to determine the extent of functional overlap between these two genes. By virulence assay we observed that rpoN1 is required for virulence whereas rpoN2 is not. In addition rpoN1 controls other important functions such twitching motility, natural transformation and growth on nitrate, unlike rpoN2. The rpoN1 and rpoN2 genes have different expression pattern, the expression of rpoN1 being constitutive whereas rpoN2 expression is induced in minimal medium and in the presence of plant cells. Moreover, the expression of rpoN2 is dependent upon rpoN1. Our work therefore reveals that the two rpoN genes are not functionally redundant in R. solanacearum. A list of potential σ54 targets was identified in the R. solanacearum genome and suggests that multiple traits are under the control of these regulators. Based on these findings, we provide a model describing the functional connection between RpoN1 and the PehR pathogenicity regulator and their dual role in the control of several R. solanacearum virulence determinants.
Introduction
Ralstonia solanacearum is a plant pathogenic bacterium that causes a lethal wilt disease in more than 200 plant species including many plants of economical interest such as tomato, potato, eggplant, banana, groundnut, or trees like Eucalyptus. The bacterium lives in two separate niches: in the soil as a saprophyte and inside the plant as a parasite (Genin, 2010). To coordinate the change from a saprophytic to a parasitic mode of living, an elaborate sensory network is used by the bacterium to regulate the expression of virulence and pathogenicity genes (Schell, 2000; Genin and Denny, 2012). The regulation enables growth and spread of the bacterium inside the host plant by overcoming the plant defense response. A hallmark of pathogenicity gene regulation in R. solanacearum is the interaction among different two component regulation systems (TCRSs) present in this bacterium (Schell, 2000). The TCRSs characterized in this bacterium are Phc, Prh, Peh, Sol, and Vsr systems (Yoshimochi et al., 2009; Genin and Denny, 2012). The TCRSs perceive signals from the environment and regulate expression of multiple pathogenicity factors. The interplay of the regulatory components of this intricate regulatory network is still poorly understood and the nature of several inductive environmental signals is unknown (Schell, 2000; Genin and Denny, 2012; Zuluaga et al., 2013). For example, the PehSR system has been demonstrated for its role in twitching motility, flagellar motility, production of polygalacturonases, and virulence (Allen et al., 1997; Kang et al., 2002). But the signaling molecule that activates PehS (sensor kinase) is unknown and the mechanism of PehR (response regulator) regulation of the downstream genes is yet to be demonstrated.
R. solanacearum has a bipartite genome that comprises a chromosome of 3.7 Mb and a second replicon called a megaplasmid of 2.1 Mb (Salanoubat et al., 2002). Important pathogenicity genes are distributed both on the chromosome and the megaplasmid. For example, genes encoding the type III secretion system are megaplasmid-borne whereas the phcA-phcR-S and vsrA-D, vsrB-C virulence regulatory genes are located on the chromosome. It has also been shown that paralogous gene families can be distributed on both replicons as in the case of the GALA (ripG) effector family members (Remigi et al., 2011). There is also evidence of apparent gene duplications on the two replicons as observed for some genes encoding Type III effector proteins (ripAX family), structural components such as the Flp pilus (Wairuri et al., 2012), hemagglutinins, several metabolic enzymes (including glutamine synthetase, and sarcosine oxidase), and regulatory genes. One example of the latter class is rpoN, which encodes the sigma factor σ54 (Sigma-54). Out of the two rpoN genes, the rpoN1 (RSc0408) is present in the chromosome and rpoN2 (RSp1671) is present in the megaplasmid. The role of rpoN in R. solanacearum virulence is not known.
Though rpoN gene is historically known for its role in nitrogen assimilation, the gene has also been shown to be involved in regulating other important functions in bacteria (Kohler et al., 1989). rpoN has been demonstrated to regulate multiple determinants in diverse pathogenic bacteria such as type I and type IV pili, flagellar motility, type III and type VI protein secretion systems, biosynthesis of exopolysaccharides or biofilm formation (O'Toole et al., 1997; Reitzer and Schneider, 2001; Kazmierczak et al., 2005; da Silva Neto et al., 2008; Dong and Mekalanos, 2012; Hao et al., 2013). A recent comprehensive computational study of different Sigma-54 interacting activators in bacteria indicated that Sigma-54 regulates processes that involve physical interaction of an organism with its environment like host colonization or biofilm formation (Francke et al., 2011).
In this study, we report the involvement of rpoN1 in virulence, twitching motility, natural transformation and growth on nitrate in R. solanacearum GMI1000 strain, which are functions not fulfilled by rpoN2. Our study therefore reveals that the function of the two paralogous rpoN proteins is not redundant in R. solanacearum.
Materials and Methods
Bacterial Strains and Growth Conditions
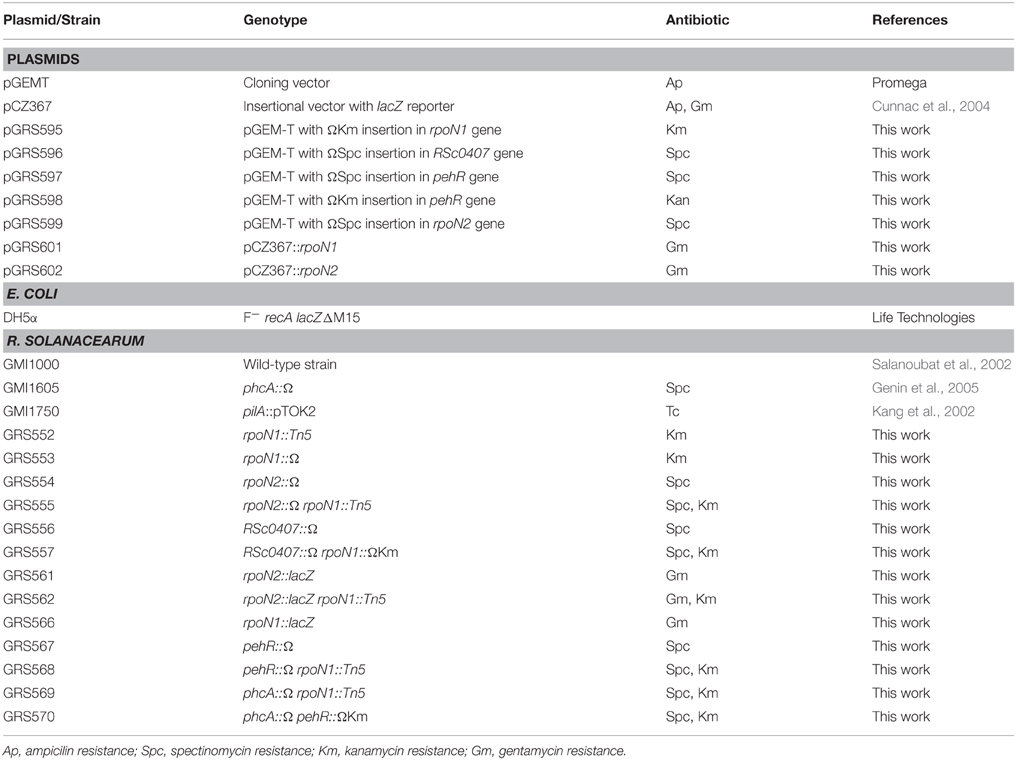
The relevant characteristics of the plasmids and bacterial strains used in this work are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani medium (Ausubel et al., 1989). R. solanacearum strains were grown in complete BG medium or in MP minimal medium supplemented with glucose (5 g L−1 at final concentration). The composition of BG medium (Plener et al., 2010) is as follows (g L−1): Bacto peptone, 10; Casamino Acids, 1; yeast extract, 1. BG medium was supplemented with glucose (5 g L−1) and triphenyltetrazolium chloride (0.05 g L−1) for agar (15 g L−1) plates. The composition of MP medium (Plener et al., 2010) is as follows (g L−1): FeSO4–7H2O, 1.25 × 10−4; (NH4)2SO4, 0.5; MgSO4–7H2O, 0.05; KH2PO4, 3.4. The pH was adjusted to 7.0 with KOH. When needed, antibiotics were added to the media at the following final concentrations (mg L−1): kanamycin, 50; spectinomycin, 40 for R. solanacearum; gentamicin, 10; tetracycline, 10; ampicillin, 100 for E. coli.
To find out if bacterial strains were capable of utilizing nitrate as nitrogen source, bacterial strains were streaked on MP minimal medium plates without (NH4)2SO4 but supplemented with KNO3 (500 μM). Difference in growth of bacterial strains having the ability to use nitrate and not having the ability to use nitrate was distinctly observed visually after 48 h of incubation.
The co-cultivation of bacteria with Arabidopsis plant cells was performed as described by Marenda et al. (1998). After growth in BG medium, bacteria were pelleted, washed and resuspended in sterile water. Hundred microliters of these suspensions, adjusted to an OD600 of 0.3, were inoculated into 10 ml of Arabidopsis cell cultures grown to a density of 40 g L−1 fresh cells. The co-cultures were maintained for 12 h at 28°C on a rotary shaker. The mixture was filtered on 20 μm pore size nylon membrane to separate bacteria and plant cells. The bacteria were then collected by centrifugation for β-galactosidase assays.
DNA Manipulations and Genetic Constructs
Standard recombinant DNA techniques were performed as described previously (Ausubel et al., 1989). Restriction enzymes, DNA ligase, and other DNA enzymes were used according to the manufacturers' recommendations. Standard PCR reactions were set up with the following reagents: 4 μL of 5X GoTaq buffer (Promega, Madison, WI), 0.6 μL DMSO (5%), 0.4 μL dNTP (10 mM), 1 μL each primer (10 μM), 0.1 μL Taq (5U/μL), template DNA 20 ng, then the volume adjusted to 20 μL with deionized water.
lacZ reporter fusions with rpoN1 and rpoN2 were created by cloning DNA fragments of these two genes into the pCZ367 integrative vector (Cunnac et al., 2004). These constructs (Table 1) were introduced by transformation in strain GMI1000 as already described (Cunnac et al., 2004).
Creation of Disruption Mutants for the rpoN1, rpoN2, pehR, and RSc0407 Genes
Disruption mutants were created with the use of the Ω interposon carrying resistance gene against either Spectinomycin or Kanamycin (Prentki and Krisch, 1984). DNA fragments encompassing the target open reading frames were PCR-amplified into the pGEMT vector. The restriction sites used for cloning the interposon and their position within the target coding sequence is shown in Supplementary Material Figure 1A and the list of primers used to amplify the gene coding sequences from the GMI1000 genome is provided in Supplementary Figure 1B. The obtained plasmids (see Table 1) were linearized and used to transform R. solanacearum as described below. Double recombination events were selected using appropriate antibiotic resistance and checked by PCR. Insertion of the Ω interposon in the rpoN1, rpoN2, pehR, RSc0887 gene sequences was confirmed by PCR amplification using one oligonucleotide specific to the interposon (5′-TGTTACCCGAGAGCTTG-3′) and a second one specific to the target gene.
The rpoN::EZ-Tn5™ strain was obtained from the R. solanacearum GMI1000 library as described by Plener et al. (2012) and available at the following web address: http://iant.toulouse.inra.fr//bacteria/annotation/site/prj/ralso/tools/mutants_db/cgi/EZLucene.cgi. Insertion and fine mapping of Tn5 in the rpoN1 gene was confirmed by the PCR using primers against the supposed flanking regions of EZ-Tn5™ insertion (Supplementary Material Figure 1A).
Procedure for R. solanacearum Transformation
The protocol used followed the method described by González et al. (2011): Ralstonia solanacearum was grown in MP medium supplemented with glycerol (20 g L−1) as sole carbon source to reach an OD600 between 0.5 and 1.0. Fifteen microliters of bacterial suspension was mixed with 3–5 μ g of linearized plasmid DNA and the mixture was deposited on a 0.45 mm cellulose nitrate filter unit placed on a BG agar medium. After 48 h of incubation at 28°C, bacteria were collected and spread on selection plates supplemented with the appropriate antibiotic.
Twitching Motility Assay
The procedure followed the one described by Liu et al. (2001). Briefly, saturated culture of bacteria was diluted in BG medium 104-fold. Ten microliters of the diluted culture was placed on BG-agar plate. The plates were kept covered, left at 28°C for 24 h growth and observed under a light microscope (Nikon Labophot) equipped with 5X objective.
Swimming Motility Assay
Swimming motility test for R. solanacearum strains was done of soft agar plates following the procedure described by Tans-Kersten et al. (2004). Soft agar BG medium plates were prepared with 0.15 and 0.2% agar. Due to the low agar concentration, the medium reaches a semi-solid state on plate after 30 min. The surface of the medium was just touched by the tip of the toothpick dipped in a saturated culture of the bacterium. Plates were incubated at 28°C. Bacteria proficient for swimming motility moves radially in all directions. A white radial zone distinctly observed can be measured. The semi-solid state of the soft-agar medium is stable for even 3 days. Motility proficient bacteria can cover the whole plate. In this study, swimming motility was estimated by the measurement of the radial zone after 24 h of incubation.
Plant Pathogenicity Assays
Virulence assays were done by soil drenching method. Inoculations were performed by watering 4-week-old tomato plants (Lycopersicum esculentum cv. Marmande) with 50 ml of a suspension containing 107 CFU ml−1. Disease development was scored daily by using a disease index scale ranging from 0 for no symptoms to 4 for completely wilted plants. Twelve plants were inoculated for each strain in at least three independent experiments. In order to analyse the data using the Kaplan–Meier representation and the non-parametric log rank test to assess the difference of the survival curves, the data was transformed as follows: all disease index lower than 2 were considered as “0” and all disease index equal of greater than 2 were considered as “1.” Statistical analysis of the results was conducted as previously described (Remigi et al., 2011). The log rank test was used to perform between-group comparisons, testing the equivalence of the Kaplan–Meier survival curves for a pair of groups. When p < 0.05 the survival curves were considered as significantly different.
β-Galactosidase Assays
β-Galactosidase assays were carried out as described by Miller (1972) with the following the modifications of Genin et al. (1992): 0.15–0.25 mL of bacterial suspension was added to Z buffer to a final volume of 0.75 mL. Cells were permeabilized with 100 μL of chloroform and 50 μL of sodium dodecyl sulfate (0.1%). The reaction was started by adding 150 μL of ONPG (4 mg/mL) and stopped by addition of 375 μL of Na2CO3 (1 M). β-Galactosidase activity was expressed in Miller units.
PatScan Analysis
Genome of strain GMI1000 was scanned for the occurrence of the following RpoN-binding site consensus TGGCAC(A, G)NNNNTTGC(A, T), with one mismatch allowed, using the PatScan tool (Dsouza et al., 1997) on the R. solanacearum GMI1000 website https://iant.toulouse.inra.fr//bacteria/annotation/cgi/ralso.cgi.
Results
Two Conserved rpoN Genes in the R. solanacearum Genome
The rpoN1 and rpoN2 coding sequences in strain GMI1000 have a moderate but significant relatedness (Blast2P: 44% identity; 62% similarity across the whole protein sequence). Both genes have a high G+C% (rpoN1: 63.80; rpoN2: 69.19) and are also similar with respect of their codon usage. BlastN analysis with individual coding sequences of the two genes revealed the presence of rpoN1 homologs in chromosomes and the presence of rpoN2 homologs in the megaplasmid in all the other tested R. solanacearum strains. Homologs of the two genes were also present in some other bacteria that were phylogenetically close to R. solanacearum (Table 2). However, Ralstonia eutropha, Cupriavidus taïwanensis, and Cupriavidus necator species have an rpoN1 homolog but no rpoN2 homolog. To get an insight into the evolutionary significance of rpoN1 and rpoN2, we compared the level of nucleotide identity for the two genes among representative strains of the R. solanacearum species complex and in some phylogenetically close bacteria. This analysis revealed that the identity level among these different strains was similar for rpoN1 and rpoN2 and followed a pattern comparable to the one of rpoD, encoding the essential sigma factor Sigma-70, which was used as control (Table 2). The nucleotide variation among the orthologous sequences in different strains is in agreement with the established species phylogeny which places phylotype I strains as more closely related to phylotype III than phylotype II or IV strains (Prior and Fegan, 2005). This observation suggests that rpoN2 arose in R. solanacearum from an ancient gene duplication or acquisition, prior to the phylotype divergence. The apparent conservation of identity for both genes also suggested that the evolutionary selection to maintain rpoN1 and rpoN2 genes has been similar in the different R. solanacearum strains.
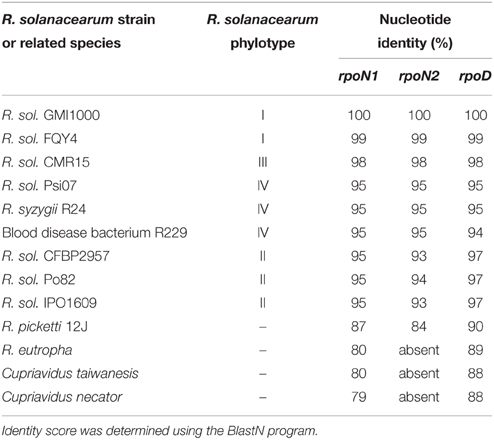
Table 2. Relatedness of the rpoN1, rpoN2, and rpoD genes with homologs within the R. solanacearum species complex and other β-Proteobacteria.
The rpoN1 Mutant is Deficient for Natural Transformation
Mutant strains for the rpoN1 and rpoN2 genes were generated. The rpoN2::Ω mutant was created by insertion of the Ω interposon as described previously (Plener et al., 2012). The rpoN1 insertion mutant was obtained from an already available EZ-Tn5™ mutant library in GMI1000 (Boucher and Genin, unpublished data). In this work we used the following mutants: an rpoN1::EZ-Tn5 mutant (Kanr) and a rpoN2::Ω Sp mutant (Spcr).
To create a double mutant, two experiments were set up (Figure 1). First, genomic DNA from rpoN2::Ω strain was used to transform rpoN1::EZ-Tn5 strain. Second, genomic DNA from rpoN1::EZ-Tn5 strain was used to transform rpoN2::Ω strain. Appropriate antibiotic medium was used to select for transformants. Surprisingly, transformants were only obtained in the second transformation experiment but not from the first transformation experiment in spite of repeating the experiment several times. As we could obtain the rpoN1 rpoN2 double mutant (rpoN1::EZ-Tn5 rpoN2::Ω; hereafter referred to rpoN1/2) in the second transformation experiment, the inability to get transformation in rpoN1::EZ-Tn5 background indicated that the GMI1000 strain with the genotype rpoN1::EZ-Tn5 was likely inefficient or strongly affected for transformation. In a separate experiment, mutation in a different gene [RSc3392::Ω ] also failed to transfer into the rpoN1::EZ-Tn5 background. This eliminated the possibility that the rpoN2 mutation was lethal in the rpoN1::EZ-Tn5 background and further supported the view that the rpoN1::EZ-Tn5 strain was inefficient for transformation. Any negative impact of EZ-Tn5 itself on transformation can be ruled out because three independent EZ-Tn5 mutants of GMI1000 strains were able to undergo transformation with rpoN2::Ω as recipient strain. Further, independent transformants could be obtained in GMI1000 using genomic DNA from the rpoN1::EZ-Tn5 strain. These new mutants were also resistant to transformation which eliminated the possible effect of any mutation at a distant locus in the earlier strain.
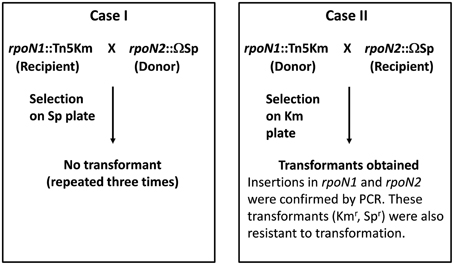
Figure 1. Genetic crosses in R. solanacearum GMI1000 to obtain an rpoN1/rpoN2 double mutant. Two transformation experiments were set up to obtain the rpoN1/rpoN2 double mutants using either the rpoN1::Tn5 or the rpoN2::Ω as receptor strains. Transformation was successful only in case II.
To eliminate the possibility of any tightly linked locus to rpoN1::EZ-Tn5 responsible for this transformation resistant phenotype, a new rpoN1 mutant was created by insertion of the Ω interposon [rpoN1::Ω (Km)] (Supplementary Material Figure 1) and this new mutant was also found to be transformation deficient. These genetic experiments confirmed that insertion mutation in rpoN1 abolished natural transformation ability in GMI1000 whereas insertion mutation in rpoN2 had no effect. As expected the rpoN1/2 double mutant was also inefficient for transformation.
A survey of the rpoN1 locus indicated that there is an ORF (RSc0407) just downstream rpoN1 and which presumably belongs to the same transcriptional unit (see https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cg). A disruption of the RSc0407 gene was created (RSc0407::Ω) and proved that this rpoN1 downstream gene had no effect on R. solanacearum transformation. Altogether, these results showed that rpoN1 is required for natural competence in R. solanacearum.
rpoN1 is Required for Twitching Motility and Growth on Nitrate
There is no previous report in any bacteria relating the role of rpoN in natural transformation. However, it has been demonstrated that type IV pili mutants of R. solanacearum are deficient for transformation as well as for twitching motility (Kang et al., 2002). In addition, there is a potential rpoN promoter sequence in the upstream region of the pilA gene in GMI1000 genome and pilA is required for the formation of type IV pili (Kang et al., 2002). It has also been reported that rpoN is involved in the formation of type IV pili and therefore twitching motility in Pseudomonas aeruginosa (Ishimoto and Lory, 1989; Semmler et al., 1999) and Neisseria elongata (Rendón et al., 2013). We thus studied twitching motility in rpoN1, rpoN2 and the rpoN1/2 mutants. We observed that the rpoN1 and rpoN1/2 mutants were twitching motility-deficient whereas the rpoN2 mutant was proficient for twitching motility as the wild type strain (Figure 2). This indicated that rpoN1 mutant is likely to be type IV pili deficient because type IV pili are required for twitching motility in R. solanacearum. The RSc0407::Ω mutation had no impact on twitching motility, thus confirming that the rpoN1 mutation had no polar effect on the downstream gene.
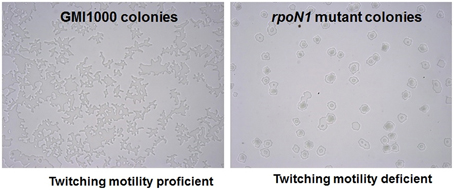
Figure 2. rpoN1 mutants are deficient in twitching motility. GMI1000 and its rpoN1 mutant derivative were checked for twitching motility under a light microscope. GMI1000 was proficient for twitching motility as evidenced by the typical finger-like projections coming out from the colony (left) whereas the rpoN1 mutant strain was formed circular shaped colonies (right).
The initial discovery of the rpoN gene in bacteria was related to its role in nitrogen assimilation (Buck et al., 2000). We studied growth of different R. solanacearum strains on 500 μM nitrate. The rpoN1 as well as rpoN1/2 mutants did not grow when nitrate was supplied as the only nitrogen source while rpoN2 and RSc0407::Ω mutants exhibited growth similar to GMI1000 in this medium (Supplementary Material Figure 2).
rpoN1 Mutants are Impaired in Virulence on Tomato Plants
To understand the role of rpoN1 and rpoN2 in R. solanacearum virulence, we independently inoculated tomato plants with rpoN1, rpoN2, rpoN1/2, RSc0407::Ω mutants and compared their virulence phenotype with the wild type GMI1000 by a soil drenching inoculation method (Figure 3). A Kaplan–Meier survival analysis shown in Figure 3A revealed that the rpoN1 and rpoN1/2 mutants were strongly reduced in pathogenicity. A log-rank test, aimed to verify the hypothesis of similarity of the survival curves, confirmed that the difference observed with the rpoN1 mutant, but not the rpoN2 mutant, was statistically significant (Figure 3B). The virulence deficiency observed in rpoN1 and as well as in rpoN1/2 double mutants could be expected because it was previously reported that twitching motility is required for full virulence of R. solanacearum (Kang et al., 2002), which is in agreement with the observed rpoN1 and rpoN1/2 twitching motility-deficient phenotype.
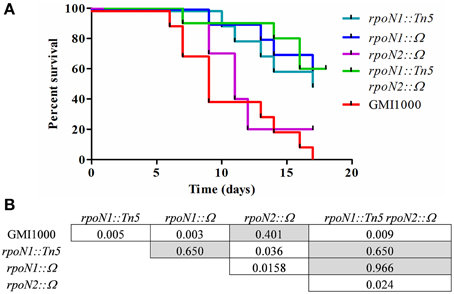
Figure 3. rpoN1 mutants are impaired in virulence. (A) Kaplan–Meier survival analysis of tomato plants inoculated with R. solanacearum GMI1000 (wild-type) and mutant derivatives. Each strain was inoculated on 12 tomato plants in three independent experiments. (B) P-values from Gehan–Breslow–Wilcoxon tests are associated with each graph. p-value of <0.05 are indicated in white boxes.
The growth proficiency of rpoN1 mutants was compared with the GMI1000 wild-type strain in complete medium. The generation time of both rpoN1 mutant strains and GMI1000 was similar (i.e., 2.0 h). On medium without antibiotic rpoN1 mutant and GMI1000 strains formed small and big colonies, respectively, which can be easily observed after 2 days of growth on solid medium (Supplementary Material Figure 3A). This small colony phenotype presumably resulted from the twitching motility-deficiency of the rpoN1 mutant and suggested that the virulence deficiency rpoN1 is not due to growth deficiency.
The rpoN1 Mutant Phenocopies the pehR Mutant for Twitching Motility
To understand any other reason apart from the twitching motility responsible for the virulence deficiency in rpoN1 mutants, we decided to compare it with the pehR mutant, since pehR is a component of the virulence regulatory network in R. solanacearum (Allen et al., 1997) which is also deficient for twitching motility (Tans-Kersten et al., 2004). We constructed a pehR disruption mutant in strain GMI1000 and compared it to the rpoN1 mutant with respect to twitching motility. This pehR mutant was also deficient for twitching motility (Supplementary Material Figure 3B). Virulence deficiency observed for the rpoN1 and pehR mutants was also found to be of similar magnitude in soil drench inoculation assay (Supplementary Material Figure 4). rpoN1 and pehR mutants were both deficient for twitching motility but the pehR mutant strain grows on nitrate unlike rpoN1.
pehR mutants are deficient for swimming motility (Allen et al., 1997). In complete medium GMI1000 exhibits no motility so we took advantage of the phcA regulatory mutant that expresses constitutive motility (Brumbley et al., 1993). To compare the role of rpoN1 and pehR in R. solanacearum swimming motility, we therefore created phcA/pehR and phcA/rpoN1 double mutants. Swimming motility assays were performed on semi-solid agar plates and motility of the strains was compared based on the measurement of the bacterial halo diameter formed after 24 h (Supplementary Material Table 1). This assay revealed that the pehR mutation, but not rpoN1, abolished the constitutive motility observed in a phcA mutant background. This phenotypic analysis of the rpoN1 and pehR mutants supported the view that both genes are jointly involved in regulating twitching motility in R. solanacearum whereas pehR appears to regulate swimming motility independently of rpoN1 and rpoN1 regulates nitrate assimilation independently of pehR.
rpoN1 Controls the Expression of rpoN2
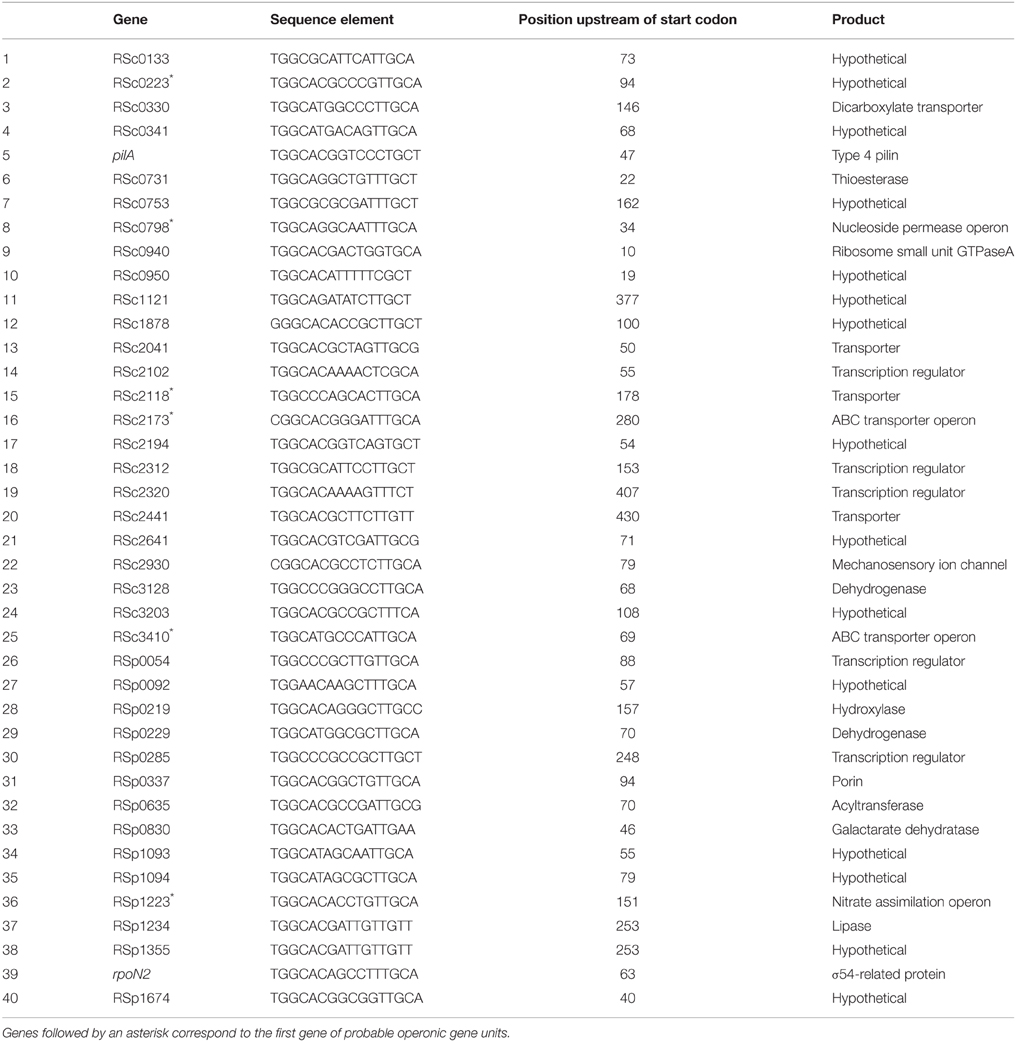
In order to identify candidate rpoN regulatory targets, the whole GMI1000 genome sequence was scanned with a consensus motif found in σ54-dependent promoters (TGGCACRNNNNTTGCW) (Barrios et al., 1999). A total 79 hits were found. Out of these 79 hits, 40 sites were considered as potential Sigma-54 target sequence because the consensus motif was located in the sense orientation in the 450 bp region immediately upstream of an annotated gene (Salanoubat et al., 2002). The list of the 40 identified genes is given in Table 3. In agreement with the former results, this list contains the pilA gene encoding the structural component of Type IV pili and the RSp1219–RSp1223 operon which was found to control nitrate assimilation in R. solanacearum (Dalsing and Allen, 2014). Among these 40 rpoN-regulated gene candidates, more than one-third encode hypothetical proteins of unknown function and six encode uncharacterized transcription regulators.
Interestingly, one of the potential σ54-dependent promoter regions was found to be upstream of rpoN2. We studied the expression of rpoN1 and rpoN2 using reporter fusions with lacZ gene (Materials and Methods). β-galactosidase expression was monitored after cultivation of bacteria in complete and minimal media, as well as in presence of Arabidopsis plant cells. Results in Figure 4 show that rpoN1 and rpoN2 display a different expression pattern: rpoN1 is significantly expressed in the three conditions tested (albeit expression appears higher in minimal medium) whereas rpoN2 is not expressed in complete medium and is specifically induced in minimal medium. In the latter condition, the expression of rpoN2 is completely abolished in an rpoN1 mutant background, thus confirming that rpoN1 regulates the expression of rpoN2 in strain GMI1000. No specific induction of rpoN1 and rpoN2 expression could be observed when bacteria were co-cultivated with plant cells.
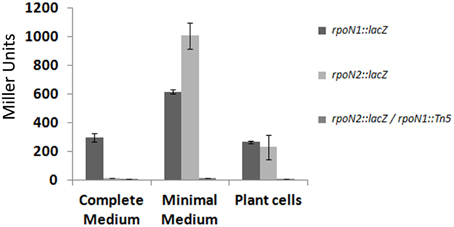
Figure 4. Expression pattern of the rpoN1 and rpoN2 genes. Expression study was carried out in complete and minimal media, and in the presence of Arabidopsis plant cells. The y-axis indicates β-galactosidase activity expressed in Miller units and the x-axis indicates the different conditions in which the expression study was carried out. Standard deviation was calculated from three different experiments performed independently.
Discussion
The role of rpoN (σ54 or also σN) was historically uncovered with the study of the regulation of nitrogen metabolism but it has been subsequently found to be involved in many other biological activities in many diverse Proteobacteria (Buck et al., 2000). Evidence of rpoN involvement in bacterial pathogenesis and virulence, mainly through the control of flagellar motility, is also documented (Kazmierczak et al., 2005). However, rpoN does not share a common role in plant pathogens: in Pseudomonas syringae, Pectobacterium carotovorum, and Erwinia amylovora rpoN is required for virulence and is involved in the regulation of the type III secretion system (Hendrickson et al., 2000a,b; Chatterjee et al., 2002; Ramos et al., 2013) but in Xanthomonas campestris pv. vesicatoria, disruption of rpoN has no impact on virulence (Horns and Bonas, 1996). A recent study revealed that in P. syringae rpoN also strongly activated the vast majority of genes involved in flagellar synthesis and motility, as well as many genes activated in planta including phytotoxins and the siderophore pyoverdin (Yu et al., 2014).
In R. solanacearum rpoN mutants still induce a hypersensitive response on tobacco (data not presented) which indicates that σ54 is not required for the functionality of the type III secretion system. However, we show in this study that rpoN1 significantly contributes to R. solanacearum pathogenesis and controls multiple traits in this bacterium. This pleiotropic phenotype was observed with two mutants carrying independent mutations in the rpoN1 gene and further analyses showed that it was independent from the rpoN1 downstream gene (RSc0407) (Supplementary Material Figure 3). Because two of the rpoN1-regulated traits (type IV pili production and nitrate assimilation) were previously found to be both required for full R. solanacearum virulence (Kang et al., 2002; Dalsing and Allen, 2014), the impaired pathogenicity phenotype of rpoN1 mutants is expectable. Altogether, these reports indicate that rpoN gene has evolved to carry out different functions in different phytopathogens, regulating virulence determinants in some species but not in others. Interestingly, deficiency for type IV pili also leads in R. solanacearum to the loss of natural transformation, a property presumably important for horizontal gene transfer and the emergence of variant strains (Coupat-Goutaland et al., 2011). To our knowledge, this is the first report of natural competence as an rpoN-regulated trait.
We found that the pehR and rpoN1 mutants are both required for the expression of type IV pili and twitching motility in R. solanacearum. Both mutants are also strongly reduced in virulence. PehR encodes a response regulator which is activated by the PehS sensor kinase (Allen et al., 1997; Tans-Kersten et al., 2004). PehS and PehR exhibit homology to the NtrB and NtrC two-component regulatory system and therefore belong to the class of bacterial enhancer binding proteins known to act as specialized activators of σ54-dependent transcription (Bush and Dixon, 2012). Based on these observations, we propose a model (Figure 5) in which PehR act as a σ54-activator protein for the control of the pilA gene required for the biogenesis of type IV pili. This hypothesis is supported by the finding that pilA contains a predicted rpoN-binding element and by a recent transcriptomic analysis of the rpoN1 mutant indicating that pilA expression is indeed rpoN1-dependent (unpublished results). Our results also indicate that PehR and RpoN control some downstream genes independently from each other: flagellar motility is a PehR-regulated trait whereas nitrate assimilation is RpoN-dependent but PehR-independent. From the inventory of putative RpoN targets listed in Table 2, it is also very likely that rpoN1 controls additional genes. For example, RSc0330, which carries an RpoN-binding element in its promoter and encodes a dicarboxylate transporter, has a homolog in P. chlororaphis which was shown to be under the control of rpoN and required for effective root colonization (Nam et al., 2006). The putative RpoN targets we identified in R. solanacearum GMI1000 comprise many genes of unknown function, transporters and transcription regulators, and their possible role in pathogenesis or plant colonization remains to be determined.
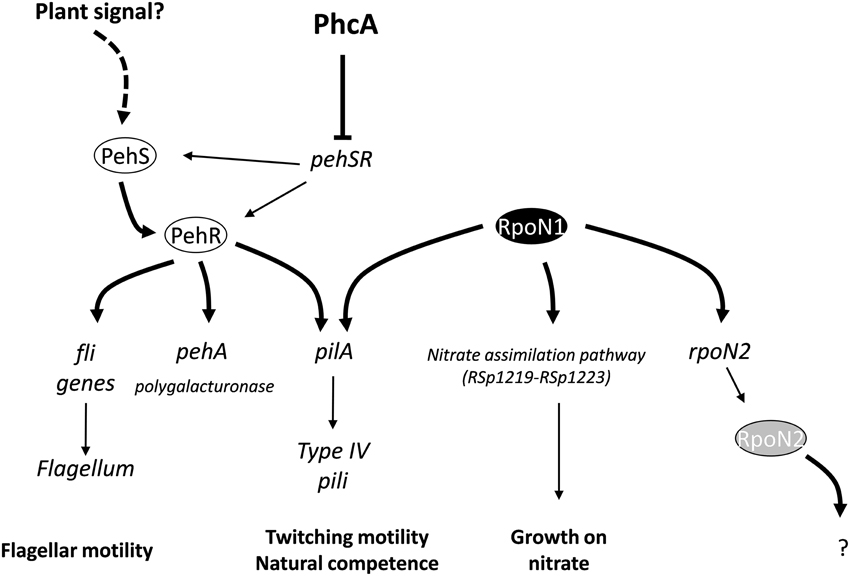
Figure 5. Model describing connection between RpoN1 and PehR and their role in the control of several R. solanacearum virulence determinants. Both rpoN1 and pehR are necessary for expression of the pilA gene which encodes the Type IV pilin, a fimbrial structure required for twitching motility and natural competence. Joint binding of PehR and RpoN1 proteins on the pilA promoter is proposed to activate the transcription of the gene. pehSR genes are under the transcriptional control of the PhcA master regulator and activity of the PehS transmembrane sensor kinase is dependent upon an unknown plant signal (Allen et al., 1997).
In most bacteria, the σ54 family contains just a single member. Relatively few organisms have two copies of rpoN, such as Bradyrhizobium japonicum, Rhizobium etli (Kullik et al., 1991; Michiels et al., 1998), some Xanthomonas oryzae pathovars (Tian et al., 2015) whereas Rhodobacter sphaeroides has up to four rpoN copies (Poggio et al., 2002). In B. japonicum, these copies are highly similar and functionally interchangeable (Michiels et al., 1998). In X. oryzae pv oryzae rpoN2 is located close to the flagellar regulon and is required for swimming motility, having therefore a distinct role from the paralogous rpoN1 gene (Tian et al., 2015).
Two copies of rpoN are generally found in β-proteobacteria having two or more replicons such as R. solanacearum, R. picketti, and many Burkholderiales. However, the second copy is absent in bacteria taxonomically related to R. solanacearum such as R. eutropha and Cupriavidus species, which are non-pathogenic. In this work we show that rpoN1 and rpoN2 are not functionally redundant in R. solanacearum since rpoN1 is required for twitching motility, natural transformation and growth on nitrate whereas rpoN2 mutant is proficient for these phenotypes. We also provide genetic evidence that expression of rpoN2 is dependent upon rpoN1 and has a distinct expression profile, being specifically induced when bacteria are grown in minimal medium or in presence of plant cells. The rpoN2 mutant is not altered in virulence and no specific phenotype could be associated to this second rpoN copy, so its role remains enigmatic. However, the broad conservation and stability of this gene in the R. solanacearum species complex, which suggest a long co-evolution of the two rpoN copies in the species, prompt us to speculate that the adaptation of the bacterium to a specific niche or environmental condition might have selected a defined regulatory role for rpoN2 during the pathogen lifecycle. Future investigations aimed to determine the specific target promoters of rpoN1 and rpoN2 at the transcriptomic level will help to define the distinct roles of these two σ54-RNA polymerases during the interaction of R. solanacearum with its host plants and its environment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Patrick Barberis for technical support. SR is thankful to DBT, Govt. of India for the long term Overseas Associateship for North East states (NE states) that helped him to come and work in France (2010–2011). SR and SG thankfully acknowledge the financial grant from CEFIPRA, New Delhi, India in the form of the Grant 4800 B1. RK thankfully acknowledges senior research fellowship from the CEFIPRA grant. The study was possible because of the study leave from Tezpur University to SR, which is thankfully acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00229/abstract
References
Allen, C., Gay, J., and Simon-Buela, L. (1997). A regulatory locus, pehSR, controls polygalacturonase production and other virulence functions in Ralstonia solanacearum. Mol. Plant Microbe Interact. 10, 1054–1064. doi: 10.1094/MPMI.1997.10.9.1054
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., et al. (1989). Current Protocols in Molecular Biology. New York, NY: Greene Publishing Associates and Wiley Interscience.
Barrios, H., Valderrama, B., and Morett, E. (1999). Compilation and analysis of σ54-dependent promoter sequences. Nucleic Acids Res. 27, 4305–4313. doi: 10.1093/nar/27.22.4305
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Brumbley, S. M., Carney, B. F., and Denny, T. P. (1993). Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator. J. Bacteriol. 175, 5477–5487.
Buck, M., Gallegos, M. T., Studholme, D. J., Guo, Y., and Gralla, J. D. (2000). The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J. Bacteriol. 182, 4129–4136. doi: 10.1128/JB.182.15.4129-4136.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Bush, M., and Dixon, R. (2012). The role of bacterial enhancer binding proteins as specialized activators of σ54-dependent transcription. Microbiol. Mol. Biol. Rev. 76, 497–529. doi: 10.1128/MMBR.00006-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Chatterjee, A., Cui, Y., and Chatterjee, A. K. (2002). Regulation of Erwinia carotovora hrpL(Ecc) (sigma-L(Ecc)), which encodes an extracytoplasmic function subfamily of sigma factor required for expression of the HRP regulon. Mol. Plant-Microbe Interact. 15, 971–980. doi: 10.1094/MPMI.2002.15.9.971
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Coupat-Goutaland, B., Bernillon, D., Guidot, A., Prior, P., Nesme, X., and Bertolla, F. (2011). Ralstonia solanacearum virulence increased following large interstrain gene transfers by natural transformation. Mol. Plant Microbe Interact. 24, 497–505. doi: 10.1094/MPMI-09-10-0197
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cunnac, S., Occhialini, A., Barberis, P., Boucher, C., and Genin, S. (2004). Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128. doi: 10.1111/j.1365-2958.2004.04118.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dalsing, B. L., and Allen, C. (2014). Nitrate assimilation contributes to Ralstonia solanacearum root attachment, stem colonization, and virulence. J. Bacteriol. 196, 949–960. doi: 10.1128/JB.01378-13
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
da Silva Neto, J. F., Koide, T., Abe, C. M., Gomes, S. L., and Marques, M. V. (2008). Role of sigma 54 in the regulation of genes involved in type I and type IV pili biogenesis in Xylella fastidiosa. Arch. Microbiol. 189, 249–261. doi: 10.1007/s00203-007-0314-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dong, G., and Mekalanos, J. J. (2012). Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res. 40, 7766–7775. doi: 10.1093/nar/gks567
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dsouza, M., Larsen, N., and Overbeek, R. (1997). Searching for patterns in genomic data. Trends Genet. 13, 497–498. doi: 10.1016/S0168-9525(97)01347-4
Francke, C., Groot Kormelink, T., Hagemeijer, Y., Overmars, L., Sluijter, V., Moezelaar, R., et al. (2011). Comparative analyses imply that the enigmatic Sigma factor 54 is a central controller of the bacterial exterior. BMC Genomics 12:385. doi: 10.1186/1471-2164-12-385
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Genin, S. (2010). Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187, 920–928. doi: 10.1111/j.1469-8137.2010.03397.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Genin, S., Brito, B., Denny, T. P., and Boucher, C. (2005). Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. doi: 10.1016/j.febslet.2005.02.058
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Genin, S., Gough, C. L., Zischek, C., and Boucher, C. A. (1992). Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol. 6, 3065–3076.
Genin, S., and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. doi: 10.1146/annurev-phyto-081211-173000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
González, A., Plener, L., Restrepo, S., Boucher, C., and Genin, S. (2011). Detection and functional characterization of a large genomic deletion resulting in decreased pathogenicity in Ralstonia solanacearum race 3 biovar 2 strains. Environ. Microbiol. 13, 3172–3185. doi: 10.1111/j.1462-2920.2011.02636.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hao, B., Mo, Z. L., Xiao, P., Pan, H. J., Lan, X., and Li, G. Y. (2013). Role of alternative sigma factor 54 (RpoN) from Vibrio anguillarum M3 in protease secretion, exopolysaccharide production, biofilm formation, and virulence. Appl. Microbiol. Biotechnol. 97, 2575–2585. doi: 10.1007/s00253-012-4372-x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hendrickson, E. L., Guevera, P., and Ausubel, F. M. (2000a). The alternative sigma factor RpoN is required for hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J. Bacteriol. 182, 3508–3516. doi: 10.1128/JB.182.12.3508-3516.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hendrickson, E. L., Guevera, P., Penaloza-Vazquez, A., Shao, J., Bender, C., and Ausubel, F. M. (2000b). Virulence of the phytopathogen Pseudomonas syringae pv. maculicola is rpoN dependent. J. Bacteriol. 82, 3498–3507. doi: 10.1128/JB.182.12.3498-3507.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Horns, T., and Bonas, U. (1996). The rpoN gene of Xanthomonas campestris pv. vesicatoria is not required for pathogenicity. Mol. Plant Microbe Interact. 9, 856–859. doi: 10.1094/MPMI-9-0856
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ishimoto, K. S., and Lory, S. (1989). Formation of pilin in Pseudomonas aeruginosa requires the alternative σ factor (RpoN) of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 86, 1954–1957. doi: 10.1073/pnas.86.6.1954
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kang, Y., Liu, H., Genin, S., Schell, M., and Denny, T. P. (2002). Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46, 427–437. doi: 10.1046/j.1365-2958.2002.03187.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kazmierczak, M. J., Wiedmann, M., and Boor, K. J. (2005). Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69, 527–543. doi: 10.1128/MMBR.69.4.527-543.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kohler, T., Haryama, S., Ramos, J.-L., and Timmis, K. N. (1989). Involvement of Pseudomonas putida RpoN σ factor in regulation of various metabolic functions. J. Bacteriol. 171, 4326–4333.
Kullik, I., Fritsche, S., Knobel, H., Sanjuan, J., Hennecke, H., and Fisher, H.-M. (1991). Bradyrhizobium japonicum has two differentially regulated functional homologs of the σ54 gene. J. Bacteriol. 173, 1125–1138.
Liu, H., Kang, Y., Genin, S., Schell, M. A., and Denny, T. P. (2001). Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147, 3215–3229.
Marenda, M., Brito, B., Callard, D., Genin, S., Barberis, P., Boucher, C., et al. (1998). PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27, 437–443. doi: 10.1046/j.1365-2958.1998.00692.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Michiels, J., Moris, M., Dombrecht, B., Verreth, C., and Vanderleyden, J. (1998). Differential regulation of Rhizobium etli rpoN2 gene expression during symbiosis and free-living growth. J. Bacteriol. 180, 3620–3628.
Miller, J. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Nam, H. S., Anderson, A. J., Yang, K. Y., Cho, B. H., and Kim, Y. C. (2006). The dctA gene of Pseudomonas chlororaphis O6 is under RpoN control and is required for effective root colonization and induction of systemic resistance. FEMS Microbiol. Lett. 256, 98–104. doi: 10.1111/j.1574-6968.2006.00092.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
O'Toole, R., Milton, D. L., Horstedt, P., and Wolf-Watz, H. (1997). RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but intraperitoneal route of inoculation. Microbiology 143, 3849–3859.
Plener, L., Boistard, P., González, A., Boucher, C., and Genin, S. (2012). Metabolic adaptation of Ralstonia solanacearum during plant infection: a methionine biosynthesis case study. PLoS ONE 7:e36877. doi: 10.1371/journal.pone.0036877
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Plener, L., Manfredi, P., Valls, M., and Genin, S. (2010). PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum. J. Bacteriol. 192, 1011–1019. doi: 10.1128/JB.01189-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Poggio, S., Osorio, A., Dreyfus, G., and Camarena, L. (2002). The four different sigma(54) factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 46, 75–85. doi: 10.1046/j.1365-2958.2002.03158.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prentki, P., and Krisch, H. M. (1984). In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29, 303–313. doi: 10.1016/0378-1119(84)90059-3
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Prior, P., and Fegan, M. (2005). “How complex is the Ralstonia solanacearum species complex,” in Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex, 1st Edn., eds C. Allen, P. Prior, and A. C. Hayward (St. Paul, MN: APS Press), 449–461.
Ramos, L. S., Lehman, B. L., Sinn, J. P., Pfeufer, E. E., Halbrendt, N. O., and McNellis, T. W. (2013). The fire blight pathogen Erwinia amylovora requires the rpoN gene for pathogenicity in apple. Mol. Plant Pathol. 14, 838–843. doi: 10.1111/mpp.12045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reitzer, L., and Schneider, B. L. (2001). Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65, 422–444. doi: 10.1128/MMBR.65.3.422-444.2001
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Remigi, P., Anisimova, M., Guidot, A., Genin, S., and Peeters, N. (2011). Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192, 976–987. doi: 10.1111/j.1469-8137.2011.03854.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rendón, M. A., Hockenberry, A. M., McManus, S. A., and So, M. (2013). Sigma factor RpoN (σ54) regulates pilE transcription in commensal Neisseria elongata. Mol. Microbiol. 90, 103–113. doi: 10.1111/mmi.12350
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. doi: 10.1038/415497a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schell, M. A. (2000). Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38, 263–292. doi: 10.1146/annurev.phyto.38.1.263
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Semmler, A. B. T., Whitchurch, C. B., and Mattick, J. S. (1999). A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145, 2863–2873.
Tans-Kersten, J., Brown, D., and Allen, C. (2004). Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant Microbe Interact. 17, 686–695. doi: 10.1094/MPMI.2004.17.6.686
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Tian, F., Yu, C., Li, H., Wu, X., Li, B., Chen, H., et al. (2015). Alternative sigma factor RpoN2 is required for flagellar motility and full virulence of Xanthomonas oryzae pv. oryzae. Microbiol Res. 170, 177–183. doi: 10.1016/j.micres.2014.07.002
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wairuri, C. K., van der Waals, J. E., van Schalkwyk, A., and Theron, J. (2012). Ralstonia solanacearum needs Flp pili for virulence on potato. Mol. Plant Microbe Interact. 25, 546–556. doi: 10.1094/MPMI-06-11-0166
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshimochi, T., Hikichi, Y., Kiba, A., and Ohnishi, K. (2009). The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signalling proteins. J. Bacteriol. 191, 3424–3428. doi: 10.1128/JB.01113-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yu, X., Lund, S. P., Greenwald, J. W., Records, A. H., Scott, R. A., Nettleton, D., et al. (2014). Transcriptional analysis of the global regulatory networks during leaf colonization. mBio 5, e01683–e01614. doi: 10.1128/mBio.01683-14
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zuluaga, A. P., Puigvert, M., and Valls, M. (2013). Novel plant inputs influencing Ralstonia solanacearum during infection. Front. Microbiol. 4:349. doi: 10.3389/fmicb.2013.00349
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: sigma 54, pathogenicity, bacterial wilt, alternative sigma factor, regulation, tomato, natural transformation, type IV pili
Citation: Ray SK, Kumar R, Peeters N, Boucher C and Genin S (2015) rpoN1, but not rpoN2, is required for twitching motility, natural competence, growth on nitrate, and virulence of Ralstonia solanacearum. Front. Microbiol. 6:229. doi: 10.3389/fmicb.2015.00229
Received: 15 December 2014; Accepted: 08 March 2015;
Published: 24 March 2015.
Edited by:
Stanton B. Gelvin, Purdue University, USACopyright © 2015 Ray, Kumar, Peeters, Boucher and Genin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephane Genin, Laboratoire des Interactions Plantes Micro-organismes, Centre national de la recherche scientifique, UMR Centre National de la Recherche Scientifique 2594/INRA 441, Chemin de Borde Rouge – Auzeville CS 52627, F-31326 Castanet-Tolosan Cedex, France stephane.genin@toulouse.inra.fr
 Suvendra K. Ray1
Suvendra K. Ray1 Nemo Peeters
Nemo Peeters Stephane Genin
Stephane Genin