Department of Cell Biology, The Scripps Research Institute, La Jolla, CA, USA
Synaptic dynamics and reorganization are fundamental features of synaptic plasticity both during synaptic circuit development and in the mature CNS underlying learning, memory, and experience-dependent circuit rearrangements. Combining in vivo time-lapse fluorescence imaging and retrospective electron microscopic analysis provides a powerful technique to decipher the rules governing dynamics of neuronal structure and synaptic connections. Here we have generated a membrane-targeted horseradish peroxidase (mHRP) that allows identification of transfected cells without obscuring the intracellular ultrastructure or organelles and in particular allows identification of synaptic sites using electron microscopy. The expression of mHRP does not affect dendritic arbor growth or dynamics of transfected neurons. Co-expression of EGFP and mHRP was used to study neuronal morphology at both the light and electron microscopic levels. mHRP expression greatly facilitates 3D reconstruction based on serial EM sections. We expect this reagent will be valuable for studying the mechanisms that guide construction of neuronal networks.
An essential open question in neuroscience concerns the identification of synaptic connections within the intact brain and the determination of the plasticity of network connectivity. A significant effort is underway to identify the entire map of network connections, the ‘connectome’ (Lichtman et al., 2008
) and to reconstruct microcircuits. Much of this work is based on the idea that all cellular profiles within a block of tissue can be identified and reconstructed with sufficient confidence and ultrastructural resolution to identify connections between neurons. The transmission electron microscope (TEM) has sufficient resolution to identify synapses, and 3D reconstructions from serial TEM sections are often used to study changes in synaptic organization and plasticity in the adult or developing CNS (Fiala et al., 1998
, 2002
; Knott et al., 2002
). Nevertheless, complete reconstruction of single neurons and identification of synaptic partners of reconstructed neurons is still very challenging due to technical issues such as relatively large sample volumes, collection of a complete series of serial thin sections, identification of specific neuronal profiles in the complete series of sections, and alignment of sections.
Identification of sites of structural plasticity and mechanisms underlying plasticity has greatly benefited from in vivo time-lapse imaging. Combining in vivo time-lapse imaging and retrospective serial section electron microscopy to identify the ultrastructural features of stable and dynamic synaptic connections is a powerful strategy to determine whether changes in dendritic and axonal arbor structure correlates with bona fide changes in synaptic connectivity (Ahmari et al., 2000
; Jontes et al., 2000
; Trachtenberg et al., 2002
; Holtmaat et al., 2005
, 2006
; De Paola et al., 2006
; Toni et al., 2007
). The current methods for these experiments typically use GFP-based time-lapse imaging followed by EGFP immunoreactivity to identify portions of the imaged neuron in the EM material (Knott et al., 2009
). These methods can obscure the ultrastructural features of the EGFP-expressing neuron and thereby limit the ability to determine the morphological properties of stable and dynamic neuronal structures.
Horseradish peroxidase (HRP) has been used extensively to label cells and allows electron microscopic analysis (Udin and Fisher, 1983
; Hamos et al., 1985
; Anderson et al., 1992
; Campbell and Shatz, 1992
). Expression of HRP from transfected cDNA has been used to study protein transport in the Golgi apparatus by adding a signal sequence and/or an endoplasmic reticulum (ER) retention sequence to target HRP to the secretory pathway (Connolly et al., 1994
). Recent work has demonstrated that expression and visualization of HRP in ER or cellular membrane preserves essential synaptic ultrastructure when expressed in neurons (Watts et al., 2004
; Schikorski et al., 2007
). Similarly, expression and detection of a fusion protein of HRP with the Drosophila wingless protein allowed ultrastructural analysis of wingless protein trafficking in well-preserved tissue (Dubois et al., 2001
), as opposed to using immunolabeling EM which often requires harsh treatment with detergents that destroys tissue ultrastructure. We have generated a membrane targeted HRP (mHRP) construct which, when co-expressed with EGFP, labels the full extent of the neuronal structure, is expressed uniformly throughout the plasma membrane, and does not affect the development of dendritic arbors. We demonstrate that co-expression of EGFP and mHRP is ideal for combining in vivo time-lapse imaging with retrospective serial section EM of the transfected neurons. mHRP expression greatly facilitates serial section EM reconstruction of neurons, particularly axons, which are often of such fine caliber that they are difficult to follow unambiguously in thin sections from unlabeled tissue. We expect that this reagent and the methods we describe here will be valuable in efforts to combine ultrastructural studies with live imaging of structural dynamics of cells.
Constructs
All experimental protocols were approved by the Cold Spring Harbor Laboratory Animal Care and Use Committee and complied with the guidelines established in the Public Health Service Guide for the Care and Use of Laboratory Animals.
We generated three types of expression constructs that contain HRP cDNA: constructs that express cytosolic proteins, proteins targeted to intracellular vesicles, and extracellular plasma membrane-targeted proteins. The HRP cDNA, which encodes 309 amino acids, was originally from UAS:HRP-CD2 (Dubois et al., 2001
). We subcloned HRP alone or HRP fused to EGFP into the pEGFP vector (Clonetech) containing the CMV promoter to generate constructs that express cytosolic proteins. We then added a kozak (GCCACC) sequence and signal sequence from human CD2 protein to the N terminal of HRP and an ER retention sequence lys-asp-glu-leu (KDEL) at the C terminal. This construct targets HRP to the lumen of intracellular vesicles. To co-express HRP and EGFP, the construct was subcloned into a dual promoter vector with two CMV promoters (provided by David Turner from University of Michigan Medical Center). One CMV promoter drives expression of EGFP for fluorescence imaging and screening, and another CMV promoter including the immediate early promoter/enhancer drives expression of HRP. To generate membrane-targeted HRP constructs, we added a signal sequence and a transmembrane domain before and after the cDNA for HRP. The transmembrane domain and perimembrane region has 45 amino acids as following: Ser-Val-Glu-Pro-Val-Ser-Cys-Pro-Glu-Lys-Gly-Leu-Asp-Ile-Tyr-Leu-Ile-Ile-Gly-Ile-Cys-Gly-Gly-Gly-Ser-Leu-Leu-Met-Val-Phe-Val-Ala-Leu-Leu-Val-Phe-Tyr-Ile-Thr-Lys-Arg-Lys-Lys-Gln-Arg. In addition, we fused HRP to the N terminal of beta chain of the insulin receptor from Xenopus laevis (Chiu et al., 2008
). We also fused the membrane targeted HRP to the N terminal of synaptic vesicle protein synaptophysin to test the utility of such fusion proteins in determining the distribution of different proteins. This fused HRP and synaptophysin construct targets HRP into the lumen of the synaptic vesicles.
Transfection and Two-Photon Time-Lapse Imaging
Whole tadpole brains, eyes, or single optic tectal neurons were electroporated with the constructs generated above in anesthetized stage 44–48 X. laevis tadpoles (Haas et al., 2002
; Bestman et al., 2006
). The concentration of the constructs ranges from 2 to 3 μg μl−1, as measured by Nanodrop (ThermoScientific). For in vivo imaging, tadpoles were first screened under an epifluorescent microscope 1 day after electroporation. Tadpoles with single bright EGFP-labeled neurons were imaged with a custom built two-photon microscope as previously described (Ruthazer et al., 2006
). The depth of two-photon images ranges from 100 to 300 μm beneath the skin, determined by the presence of skin pigment cells. The image stacks were collected at 1.5 μm z intervals using a 60× water immersion objective at 1–3× scan zoom depending on the size of neuronal tree. Neuronal structure was reconstructed from image stacks and analyzed for dendritic branch dynamics as described previously (Ruthazer et al., 2006
).
Fixation and Histochemistry
We optimized the fixation procedure to improve tissue preservation for electron microscopy of the young tadpole brain tissue. Tadpoles were fixed by a modified microwave fixation protocol. The fixative (3.5% paraformaldehyde, 1% glutaraldehyde and 0.02% CaCl2 mixture in 0.035 M sodium cacodylate buffer at pH7.4) with 0.01% fast green was pressure injected into the brain ventricle with a Picospritzer until the entire tectum was surrounded by fixative. The brain was exposed by cutting the skin and peeling off the dura and immersed in a Petri dish with 0.5 cm depth of the same fixative. The brain was microwaved for 8 s (750 W GE microwave) followed by 2 h incubation in fixative at room temperature. After wash with 0.1 M Tris buffer, the brains were dissected and embedded in an albumin-gelatin mixture. Tissue blocks were cut into 60 μm sections on a vibratome (Vibratome 1500; Technical Products International, St. Louis, MO, USA). HRP was enhanced using the tyramide signal amplification (TSA) biotin system (PerkinElmer, Emeryville, CA, USA). Briefly, the sections were incubated in solution containing biotinylated tyramide that can be catalyzed by HRP resulting in the deposition of biotin immediately adjacent to the HRP. In the absence of the TSA, the success rate of detecting diaminobenzidine (DAB) reaction product is low (less than 5%). The sections were then incubated in avidin conjugated with HRP followed by reaction with nickel-intensified DAB (Sigma) for 5–10 min.
Hippocampal Neuron Culture and Transfection
Hippocampal neuron cultures were prepared from embryonic day 18 rats. Dissociated cells were seeded onto coverslips coated with poly-l-lysine (0.1 M in borate buffer, pH = 8.0) in plating media (neurobasal media containing 50 U ml−1 penicillin, 50 g ml−1 streptomycin, 2 mM glutamax, 2% B-27 and 5% fetal bovine serum). After 24 h, plating media was replaced with feeding media (plating meda without FBS) and fed twice a week throughout the experiment. Cells were transfected with mHRP/GFP or pEGFP-C1 (Clonetech) at 21 days in vitro (DIV) with Lipofectamine 2000 (Invitrogen), fixed at 24 DIV with 4% paraformaldehyde with 4% sucrose. The mHRP signal was detected by the protocol described above.
Electron Microscopy
Selected vibratome sections that contained HRP-labeled neurons were post-fixed in 1% osmium tetroxide for one hour, dehydrated in an acetone series (50%, 70% acetone with 4% uranyl acetate, 90%, 100%), and infiltrated with EMbed-812 resin (Electron Microscopy Sciences, Fort Washington, PA, USA) (50% in acetone and 100%). The next day, sections were flat-embedded in 100% EMbed-812 resin between two sheets of Aclar plastic (Ladd Research, Williston, VT, USA) and incubated at 65°C overnight. Serial ultrathin sections (70 nm, silver-gray interference color) were cut using a diamond knife (Diatome, Biel, Switzerland) and collected on formvar-coated nickel slot grids. Each grid can hold one to five sections. Ultrathin sections were examined with a Joel electron microscope 1010. Photographs were taken with a SIA CCD digital camera at 10 k or 15 k magnification and sampled at 2048 × 2048 resolution. We adjusted the brightness and contrast of all electron micrographs to make the lightest part of the background in electron micrograph close to white in Photoshop (Adobe, version 7). For 3D reconstruction, we used a software developed by Fiala (2005)
. We used NIH ImageJ (Rasband, 1997–2009
) to measure the distance across the synaptic cleft.
Generation of Reagents
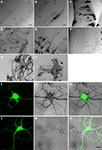
We generated 25 different constructs that contained the cDNA for HRP and subcloned them into a dual CMV promoter plasmid along with EGFP. Ten representative constructs are shown in Table 1
. After electroporation of eyes or tecta, animals were screened for EGFP expression and imaged on the two-photon microscope. After time-lapse fluorescence imaging, the HRP in the transfected cells was visualized by a DAB reaction in vibratome sections to evaluate the gross morphology of cells and the distribution of HRP (Figure 1
). Although the cytosolic HRP was likely expressed, based on the expression of the EGFP from the dual promoter construct, it did not have enzymatic activity (Figure 1
A). By adding a signal sequence at its N terminal (ssHRP, Table 1
), we observed DAB reaction products in the soma (Figure 1
B). When an ER retention sequence was added to the C terminal of HRP (ssHRPKDEL, Table 1
), the DAB reaction product labeled the whole soma and major dendritic branches (Figure 1
C), consistent with previously reported results (Schikorski et al., 2007
). When HRP was fused with single transmembrane protein CD2 or CD2 fused with EGFP, faint DAB labeling was present throughout the cell, even in tiny dendritic branches (Figure 1
D). We generated a membrane-targeted HRP construct, which we call mHRP, by adding a signal sequence and transmembrane domain from human CD2 before and after the cDNA of HRP (ssHRP-TM, Table 1
). Expression of mHRP showed an even distribution of DAB label throughout the soma and distal branches of glial cells and neurons (Figure 1
E) in the optic tectum. Fine axonal processes extending far from the transfected cell bodies were well labeled (Figure 1
F). When HRP was fused with other proteins, it was distributed in distinct locations in the neurons. For example, the HRP fused with the beta chain of the insulin receptor had a distribution in varicosities at major dendritic branches of tectal neurons (Figure 1
G), and expression of HRP fused to the N terminal of synaptophysin in retinal ganglion cells labeled axon boutons in retinotectal axon arbors (Figure 1
H). Furthermore, when we express mHRP in rat hippocampal culture neurons, we observed the DAB staining in the soma, all dendritic branches, and filopodia-like structures (Figures 1
I–N). In the following experiments, we will use mHRP to study the ultrastructure of transfected cells.
Table 1. HRP Constructs and in vivo expression.
Figure 1. HRP constructs and their expression in cells. Micrographs of vibratome sections through the optic tectum of Xenopus tadpoles in which DAB reactions are used to visualize constructs generated with cDNA from HRP and other proteins as specified: (A) cytosolic HRP. (B) EGFP/ssHRP. (C) EGFP/ssHRPKDEL. (D) EGFP/ssHRP-TM-GFP. (E,F) EGFP/ssHRP-TM. (G) EGFP/ssHRP-XIRbeta-TM. (H) ssHRP-TM-synaptophysin-EGFP. Plasmid definitions are shown in Table 1
. (I–N) Expression of mHRP in hippocampal neurons. (I–K) A hippocampal neuron, transfected with EGFP/mHRP. (I) EGFP expression. (J) The mHRP was revealed by DAB reaction. (K) Merge of EGFP and DAB signal. (L,N) A hippocampal neuron, transfected with EGFP only, showed no DAB signal. (L) EGFP expression. (M) Image following DAB reaction. (N) Merge of EGFP and DAB signal. Scale bars in (A) and (B) are 20 μm. Scale bar in (C) is 10 μm and applies to (D–H). Scale bar in (N) is 20 μm and also applies to (I–M).
mHRP does not Affect Neuronal Growth
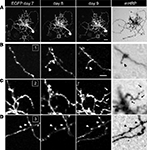
To test whether expression of the mHRP construct has detrimental effects on neuronal development, we co-expressed EGFP and mHRP in single optic tectal neurons and imaged the neurons in vivo over 9 days using two-photon microscopy. Dendritic and axonal branches were identified equally well by EGFP labeling and DAB staining (Figure 2
A). To optimize detection of mHRP, we added 0.05% tween-20 in the reaction. We find DAB labeling is evenly distributed along the membranes of dendrites and axons, including fine processes which in vivo imaging demonstrates are dynamic over a period of minutes to days (Figures 2
B–D).
Figure 2. mHRP and EGFP are detected throughout the fine structure of transfected neurons. (A) Two-photon images of a neuron expressing both EGFP and mHRP were collected once a day over 9 days. The right column shows the same mHRP-labeled neuron revealed by DAB reaction. (B–D) High magnification of insets (B–D) in (A) demonstrated that stable (B) and extended (C) dendritic branches and axonal boutons (D) can be detected equally well by both EGFP and mHRP expression. Scale bar: 20 μm in (A), 5 μm in (B), which applies to (C) and (D).
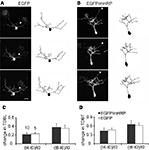
To test whether expression of EGFP and mHRP affected dendritic arbor growth rate compared to expression of EGFP alone, we transfected single neurons with co-expressed EGFP and mHRP. We collected images of neurons every 4 h over 8 h and analyzed changes in neuronal structure in the three time-lapse images (Figures 3
A,B). In our previous studies, we found that imaging tectal cells in stage 47 tadpoles three times at 4 h intervals over 8 h allowed us to detect significant increases in the growth rates of dendritic arbors, measured as total dendritic branch length (TDBL) or total dendritic branch tip numbers (TDBT; Sin et al., 2002
). Furthermore, this imaging protocol is more sensitive to changes in growth rates that may be difficult to identify in neurons that are imaged at daily intervals because the changes in arbor structure is more variable when assessed over longer intervals. We found no significant differences in the relative change in TDBL or TDBT for the two sets of neurons (Figures 3
C,D). Neurons expressing EGFP and mHRP have comparable dendrite growth rates as EGFP-expressing neurons when assayed for the relative change of TDBL [(t8-t0)/t0:0.35 ± 0.08 (EGFP) vs. 0.38 ± 0.08 (EGFP and mHRP), n = 10 and 10, Mann–Whitney test, p = 1] or TDBT [(t8-t0)/t0:0.62 ± 0.08 (EGFP) vs. 0.65 ± 0.14(EGFP and mHRP), p = 0.7, Mann–Whitney test]. Thus, expression of mHRP does not affect dendritic arbor growth and dynamics.
Figure 3. Expression of mHRP does not affect dendritic arbor growth. (A,B) Two-photon time-lapse images were collected every 4 h over 8 h 2 days after single-cell electroporation of EGFP (A) or co-expressed EGFP and mHRP (B). Drawings of the neurons from complete 3D reconstructions of two-photon imaging stacks are shown to the right of the two-photon images. The axons are marked by white arrowheads in the images. (C,D) Neurons expressing EGFP and mHRP (black bars) have same dendrite growth rates as EGFP-expressing neurons (white bars) when assayed for changes in total dendritic branch length or total dendritic branch tip number. Scale bar in (A) is 20 μm.
Ultrastructure of mHRP-Labeled Cells
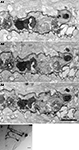
We determined the quality of labeling and the distribution of mHRP at the ultrastructural level using electron microscopy. To preserve the ultrastructural morphology, no detergents were used in any processing steps. Figure 4
A shows a light micrograph of radial glial cells in the optic tectum which span from the ventricular layer to the pial layer superficial to the tectal neuropil. The mHRP labels the cell body, major radial processes, and fine side branches of the glial cells. Serial electron microscopy (Figure 4
B) shows that mHRP is concentrated in the plasma membrane of the radial glial cells. It is intriguing to note that there are many large mHRP-labeled multivesicular organelles inside the glial processes (Figure 4
B, stars) that may be labeled by endocytosis of the plasma membrane.
Figure 4. mHRP labels the plasma membrane and endocytosed membrane of glial cells. (A1–A3) Micrographs from serial EM sections show the ultrastructure of processes from an mHRP-labeled radial glial cell. Labeled multivesicular organelles are marked as stars. (B) Vibratome section (60 μm) showing mHRP-labeled radial glial cells in the optic tectum. Scale bar: 20 μm in (A1A3), 1 μm in (B).
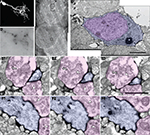
We next tested whether the combination of EGFP and mHRP labeling would allow in vivo time-lapse imaging followed by ultrastructural analysis of neurons. We transfected single neurons with co-expressed EGFP and mHRP and collected in vivo time-lapse two-photon images based on the EGFP fluorescence. After the last time point of in vivo time-lapse imaging, we fixed the animal and processed the brain to reveal the location of mHRP. mHRP labeling was present in the soma and all of the neuronal processes visualized with EGFP (Figures 5
A,B). The soma was easily identified in low magnification electron micrographs (Figure 5
C). High magnification electron micrographs showed uniform distribution of electron dense label on the plasma membrane of the transfected neuron (Figures 5
C–F). Inside the cell, the Golgi complex was labeled (Figure 5
D). Very few intracellular membrane compartments were labeled inside of dendritic branches (Figures 5
E,F). We do not observe labeling in untransfected cells (see also Figures 1 and 4
), suggesting that endogenous peroxidase activity is not significant in these tissue samples. This labeling pattern allows identification of labeled neuronal processes without obscuring intracellular structures such as mitochondria, microtubules, and vesicular structures.
Figure 5. Ultrastructure of an EGFP/mHRP labeled neuron. (A) Two-photon image of an optic tectal neuron which was labeled by single cell electroporation with co-expressed EGFP and mHRP. (B) mHRP distribution in a vibratome section through the same neuron. The distribution of mHRP was revealed by DAB reaction. (C) Low magnification image of an electron micrograph showing the location of the mHRP-labeled cell body and dendritic process in different layers within the optic tectum. The cell body layer is toward the left and the neuropil layer is to the right. (D) High magnification of an electron micrograph shows the ultrastructural morphology of the labeled cell body (blue) in (C) (large black box) and the nucleus (pink). (E1–E3) Serial electron micrographs through a labeled distal dendritic branch (blue) in (C) (small black box) show two synaptic contacts (from pink axon terminals and marker with white arrow heads). Note that the axon terminal on right which contacts the mHRP-labeled dendrite also contacts another unlabeled postsynaptic profile (white arrow). (F1–F3) Serial electron micrographs show that a labeled major dendritic branch (blue) was contacted by two presynaptic terminals (pink, white arrow heads). Scale bars are 10, 2, and 1 μm in (C,D,E). Scale bar in (E1–E3) also applies to (F1–F3).
Identification of Synapses in mHRP-Labeled Neurons
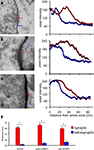
We examined the distribution of synapses in serial sections through mHRP-labeled neurons. Synapses were identified by examining at least three serial sections per candidate synapse. We used the following criteria to identify synapses: the presence of clustered synaptic vesicles opposed to the membrane of one profile in at least two serial sections and thickening of the electron dense material, including the membrane and postsynaptic density of the profile opposite to the one with clustered vesicles. Because of the concern that mHRP labeling in the dendrite membrane might obscure the postsynaptic density, we tested whether an independent measurement to identify synapses based on quantitative identification of the postsynaptic density produced a comparable distribution and number of synaptic sites. We quantified the thickness of the electron dense membrane profile opposite to the sites with obvious synaptic vesicles and neighboring sites along the plasma membrane without any synaptic vesicles. We measured the distance across the synaptic cleft starting from the active zone that has docked synaptic vesicles to the end point where pixel intensity dropped below 10% of the difference between the maximum and minimum intensity. We found that the distance from the intracellular face of the presynaptic membrane to the cytosol of the postsynaptic site was significantly greater at synaptic sites compared to extrasynaptic sites for synapses in unlabeled tissue (Figure 6
A), or tissue in which postsynaptic (Figure 6
B) or presynaptic profiles (Figure 6
C) were from mHRP-labeled neurons (Figure 6
D, n = 38, 14, and 17, p < 0.01, Mann–Whitney test). We then reviewed all the synapses in the mHRP-labeled neurons which had been identified based on the presence of clustered synaptic vesicles and tested whether they satisfied the independent criteria to identify synapses as described above. We found that the two methods identified the same number of synapses, suggesting that possible error due to oblique sectioning through mHRP-labeled membrane is not significant over the population of synapses we analyzed. This corroboration validated our criteria for synapse identification in mHRP-labeled cells.
Figure 6. Criteria to identify synaptic contacts. (A–C) Electron micrographs on the left show synaptic contact in control tissue (A), synapse with postsynaptic profiles labeled by mHRP (B), synapse with presynaptic profiles labeled by mHRP (C). Synapses are identified according to the presence of clustered synaptic vesicles opposed to the membrane of one profile in at least two serial sections. Measurements of pixel intensities taken from the locations of the red and blue lines, which mark synaptic and extrasynaptic sites (histograms on right of each image) provide an independent means to identify synaptic sites. Note that the thickness of the electron dense membrane and cleft is consistently greater at synaptic sites than at extrasynaptic sites. The average thickness at extrasynaptic sites in the pre mHRP+ samples is significantly greater than controls (p < 0.05), likely due to distribution of DAB label. White arrows in (C) mark recycled synaptic vesicles labeled by mHRP (see text for details). (D) The thickness at synaptic and extrasynaptic sites analyzed from tissues represented in panels (A–C). *p < 0.01.
We conducted an additional quantitative analysis of synapses in a 10 μm × 10 μm × 7 μm block of tissue without any mHRP labeling to address whether the presence of mHRP interferes with detection of synapses and to estimate the error in counting synapses based on criteria requiring identification of the synaptic cleft and postsynaptic density. We identified 435 potential synapses based on the presence of docked clustered synaptic vesicles opposed to one membrane. In the same data set, we identified 424 synapses that had clustered synaptic vesicles opposed to one membrane, a synaptic cleft, and a postsynaptic density. The 2% difference in synapse numbers determined by including the presence of the postsynaptic density in the criteria to identify synapses is within the error of measurement between individuals.
We found that synapses were distributed along the major dendritic shafts of neurons, which have nicely parallel microtubules (Figures 5
F1–F3), and along small dendritic branches (Figures 5
E1–E3) of mHRP-labeled developing tectal neurons. The presynaptic boutons contacting the mHRP-expressing neurons also contacted other unlabeled postsynaptic profiles (Figures 5
E1–E3). Based on the observation that all processes seen with EGFP were also labeled with mHRP (Figure 2
), the unlabeled postsynaptic profiles seen in Figure 5
E were likely from other cells not transfected with mHRP.
mHRP Facilitates 3D Reconstruction of Neurons
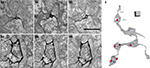
Expression of mHRP in neurons and glial could be valuable for 3D reconstruction from serial sections because processes from labeled cells can be identified relatively easily. This is a particular problem for reconstruction of axons which can have an extremely small diameter for long distances. To demonstrate the utility of the mHRP expression for neuronal reconstruction from serial EM sections, Figure 7
C shows a segment of axon branches successfully reconstructed from 262 serial sections. The mHRP was evenly distributed on very fine processes of axon branches in which the caliber of shaft was often less than 100 nm (Figures 7
A1–A3). This makes it easy to identify and trace fine axon branches. Although it appears that the DAB reaction product may diffuse within the extracellular space and lead to some ambiguity in identification of labeled processes, tracing mHRP-labeled profiles in aligned serial EM sections minimizes potential errors in reconstructing labeled cells. The thin axon branches are studded with boutons where the majority of synapses are located. Most boutons make one or multiple synaptic connections with separate postsynaptic profiles, although boutons can be occasionally seen with no apparent synaptic contacts. Reconstructions of presynaptic boutons from serial EM sections show the distribution of clustered synaptic vesicles in presynaptic terminals close to the position of postsynaptic density. Interestingly, some synaptic vesicles are labeled by mHRP. The mHRP-labeled synaptic vesicles are distributed at peripheral regions of the active zone (Figures 7
B1–B3,C), and may be labeled because of recycling of plasma membrane that contains mHRP.
Figure 7. mHRP labeling facilitates 3D reconstruction of identified axons. (A1–A3) Serial EM sections show the morphology of a fine axon shaft cut in cross section. (B1–B3) Serial EM sections show ultrastructural morphology of synaptic contacts (white arrow heads). Note that mHRP-labeled synaptic vesicles are shown in the presynaptic terminal (white arrows). (C) 3D reconstruction of a segment of an mHRP-labeled axon branch. Red dots represent the central locations of postsynaptic densities. Blue dots represented mHRP-labeled synaptic vesicles. Top arrow in C marks the location of the sections in (A) and bottom arrow marked the synaptic bouton reconstructed from sections shown in (B). Scale bar in (A) is 500 nm and also applies to (B). Scale box in (C) is 1 μm.
We have generated a membrane-targeted HRP fusion protein which, when combined with fluorescent protein labeling, allows a combination of in vivo fluorescent protein imaging and EM based ultrastructural analysis. Expression of the mHRP construct does not appear to affect the development or structural dynamics of neuronal dendrites. We demonstrate the utility of mHRP in generating 3D reconstruction of axon branches from serial EM sections. mHRP does not obscure intracellular organelles and it is therefore a valuable tool for identifying mitochondria, sites of intracellular membrane trafficking, and synaptic contacts in mHRP-labeled dendrites and axons.
Enzymatic Activity of Synthesized HRP
Here we show that synthesized cytosolic HRP proteins do not have enzymatic activity, which could be due to lack of glycosylation in the Golgi complex (Veitch, 2004
). By adding a signal sequence at the N terminal of HRP, the DAB reaction products can be detected in the soma, further suggesting that their modification in the ER and Golgi complex is necessary for enzymatic activity. This idea is further supported by the observation that adding an ER retention sequence at the C terminal allows detection of HRP activity in the cell body and major dendrites as previously reported (Connolly et al., 1994
; Schikorski et al., 2007
). By tagging HRP with a transmembrane domain from the CD2 protein, HRP can be detected in distal dendrites. The deletion of the cytosolic tail of CD2 may enhance distribution of membrane targeted HRP by elimination of potential protein–protein interactions.
Application of mHRP Constructs
mHRP expression can be combined with a variety of other labeling and imaging methods for a broad range of applications in Xenopus as well as mammalian systems. Reconstruction of neurons within the intact CNS allows the identification of synaptic connections with pre- and postsynaptic partners. Previous studies report partially- or fully-reconstructed neuronal structures by serial EM reconstruction based on neuroanatomical tracers (Hamos et al., 1985
, 1987
; Campbell and Shatz, 1992
; White et al., 2004
), photoconversion of dye (Bishop et al., 2004
), or antibody against GFP (Ahmari et al., 2000
; Jontes et al., 2000
; Trachtenberg et al., 2002
; Holtmaat et al., 2005
, 2006
; De Paola et al., 2006
; Toni et al., 2007
). mHRP expression offers several advantages over previous methods. The co-expression of mHRP with EGFP or other fluorescent reporters allows retrospective ultrastructural analysis of neurons after in vivo fluorescent protein imaging. mHRP does not disturb structural plasticity in the developing dendritic arbor, which makes it useful for studying dynamic tissues in developing or mature systems. The uniform distribution of mHRP labeling on the plasma membrane allows identification of key intracellular structures including synaptic vesicles and mitochondria. mHRP continues to label the neuronal plasma membrane for a relatively long period (>9 days). mHRP allows detection of HRP activity even after exposure to high concentrations of glutaraldehyde fixation (1% in this study), which allows better preservation of ultrastructural morphology. Finally, the DAB reaction has excellent penetration into tissue and does not require the harsh treatment with detergents used for immunodetection of anatomical tracers. These features of mHRP labeling make full 3D reconstruction of single neurons possible.
It is intriguing that the mHRP labeling is seen on synaptic vesicles present in presynaptic boutons and vesicular structures inside dendritic branches of mHRP-expressing neurons and glial cells. The intracellular organelles were likely labeled by endocytosis of the plasma membrane that contains mHRP. The presence of many mHRP-labeled multivesicular bodies in glial cells suggests a significant amount of endocytosis of plasma membranes, consistent with a high degree of receptor-mediated signaling in glia. Labeled synaptic vesicles may report vesicle recycling following synaptic transmission. The preferential location of the mHRP-labeled recycled vesicles at the periphery of the active zone is consistent with previously reported sites of vesicle endocytosis (Rizzoli and Betz, 2004
). Although further studies will be necessary to test whether membrane properties and plasticity of transfected neurons are affected by mHRP expression, the evidence of membrane cycling and normal growth rates suggest that cell physiology is not deleteriously affected by mHRP expression. In a previous report, we fused HRP to the N terminal of synaptophysin (Ruthazer et al., 2006
), which successfully labeled synaptic vesicles in axon boutons of retinal ganglion cells. When HRP was fused to the N terminal of the beta chain of insulin receptor, it has a distinctive distribution in dendritic branches. These results together with previous studies (Dubois et al., 2001
) indicate that HRP may be used as a marker for protein localization and dynamics including receptor trafficking.
In summary, expression of mHRP in plasma membranes of vertebrate neurons, glia and other cell types will be valuable for electron microscopic 3D reconstructions of cell morphology together with ultrastructural analysis of complex intracellular structures. In the CNS, mHRP expression should be particularly valuable for identification of synaptic connections to and from 3D reconstructed mHRP-labeled cells. The combination of in vivo fluorescent protein imaging and retrospective EM will help address questions relating to structural dynamics, synaptic reorganization, and membrane trafficking, as well as issues concerning network connectivity and microcircuitry of labeled neurons in the intact brain. Other potential applications, including targeted expression to specific cell types using transgenic lines and combination with postembedding immuno EM, will allow higher resolution examination of neuronal structure and network connectivity.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the NIH RO1 EY-011261 and DP10D000458 to Hollis T. Cline.
Rasband, W. S. (1997–2009). ImageJ. Bethesda, MD, National Institutes of Health. http://rsbinfonihgov/ij/
.