- 1Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction, Education Ministry of China, College of Animal Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Department of Hubei Province's Engineering Research Center in Buffalo Breeding and Products, Wuhan, China
- 3Department of Biology, University of Evry Val D'essonne, Evry, France
Mature spermatozoa have highly condensed DNA that is essentially silent both transcriptionally and translationally. Therefore, post translational modifications are very important for regulating sperm motility, morphology, and for male fertility in general. Protein sumoylation was recently demonstrated in human and rodent spermatozoa, with potential consequences for sperm motility and DNA integrity. We examined the expression and localization of small ubiquitin-related modifier-1 (SUMO-1) in the sperm of water buffalo (Bubalus bubalis) using immunofluorescence analysis. We confirmed the expression of SUMO-1 in the acrosome. We further found that SUMO-1 was lost if the acrosome reaction was induced by calcium ionophore A23187. Proteins modified or conjugated by SUMO-1 in water buffalo sperm were pulled down and analyzed by mass spectrometry. Sixty proteins were identified, including proteins important for sperm morphology and motility, such as relaxin receptors and cytoskeletal proteins, including tubulin chains, actins, and dyneins. Forty-six proteins were predicted as potential sumoylation targets. The expression of SUMO-1 in the acrosome region of water buffalo sperm and the identification of potentially SUMOylated proteins important for sperm function implicates sumoylation as a crucial PTM related to sperm function.
Introduction
Spermatozoa are highly regulated processes that consist of differentiated cells based on a head, mid-piece, and tail while the mammalian sperm head is composed of a nucleus and acrosome. The acrosome is a secretory vesicle, essential for sperm-egg fusion (Ramalho-Santos et al., 2002). The acrosome contains a variety of proteins required for sperm penetration through the zona pellucida (Kim et al., 2001) and fusion with the egg (Saxena et al., 2002). Bovine sperm contain hundreds of different proteins, and most of their functions are unknown, but it is believed that they are involved in different phases of fertilization. Thus, alterations in expression levels or post-translational modifications of specific proteins could alter sperm functions, potentially optimizing semen fertility. Mature spermatozoa have highly condensed DNA, which is silent at both the transcriptional and translational level (Baker et al., 2012; Rahman et al., 2013) or poorly capable of translation (Gur and Breitbart, 2006). Therefore, post translational modifications and their ability to regulate important sperm functions, including capacitation, motility, and the acrosome reaction, are all essential for oocyte penetration and are considered as the focus of studies of individual sperm proteomes (Pixton et al., 2004; Baker et al., 2005; Aitken and Baker, 2008; Oliva et al., 2009; Brohi and Huo, 2017). The literature reveals strong relationships between proteins and post-transcriptional modifications in spermatozoa which have a positive impact on fertility of men and mice, whereas, limited studies have been reported in buffalo bulls.
Sumoylation is a reversible post-translational protein modification, involved in the modulation of many biological systems (Yeh, 2009). SUMO is a highly-conserved 11 kDa post-translational protein modifier, which has a similar structure to ubiquitin and conjugates target proteins by a similar mechanism (Marchiani et al., 2011). However, rather than targeting proteins for degradation, sumoylation induces effects, such as altering protein localization or binding partners. An isopeptide bond is created between the glycine residues at the C-terminal end of SUMO with a lysine residue ε-amino group in the target protein. This dynamic process, which is catalyzed by activation, conjugation, and ligation enzymes (Geiss-Friedlander and Melchior, 2007), is transient and reversible (Marchiani et al., 2011); De-sumoylation is catalyzed by sentrin/SUMO-specific proteases (Yeh, 2009). SUMO has been implicated in the modulation of cellular functions as diverse as replication, the repair and recombination of DNA, the transcription of RNA, trafficking between the nucleus and cytoplasm, and protein stability (Seeler and Dejean, 2003; Girdwood et al., 2004; Yurchenko et al., 2006; Geiss-Friedlander and Melchior, 2007). Sumoylated proteins have been found in both the nucleus and the cytoplasm in germ and somatic cells and the both in mitochondria and plasma membrane (Yeh, 2009; Zunino et al., 2009).
Protein sumoylation during different stages of spermatogenesis was recently demonstrated in humans (Vigodner et al., 2006, 2013; Brown et al., 2008) and rodents (Rogers et al., 2004; Vigodner and Morris, 2005; Stanton et al., 2012), with localization of SUMO1 to chromatin and other cellular domains, both in germ cells, including spermatocytes and spermatids, and in somatic cells, including Leydig, Sertoli, and peri tubular myoepithelial cells. While the role of SUMO in spermatogenesis remains undefined, it is suggested to be involved in heterochromatin stability, DNA repair mechanisms and the regulation of gene expression (Rogers et al., 2004; Vigodner et al., 2006; Stanton et al., 2012; Marchiani et al., 2014). SUMO1 expression is demonstrated in a high percentage of live, mature human sperm (Marchiani et al., 2011), However, excessive levels of sumoylation may be linked to an abnormal sperm morphology and motility (Vigodner et al., 2013; Marchiani et al., 2014) and to sperm DNA fragmentation (Marchiani et al., 2014). The localization of SUMO in sperm and its target proteins defined its promising role in different circumstances. Targets that have been identified in mature human sperm include the mitochondrial protein DRP1, the microtubule-organizing protein RanGAP1 and topoisomerase IIa, which is involved in chromatin remodeling (Marchiani et al., 2014). More studies should be conducted on the proteomic profile of sperm, particularly post-transcriptional protein modifications such as SUMOylation, which might give new insights to resolving infertility issues. The objective of this study was to evaluate SUMO1 expression and localization in ejaculated spermatozoa from Buffalo bull and identify protein targets of sumoylation in this model.
Materials and Methods
Ethics Statement (for Mice)
This current study was approved by the Research Center of Experimental Animals (Approval ID: JkXK (Hb) 200980006) of the Ethical Committee. In this present study, Kunming mice were obtained from the local Central Animal Laboratory and were kept in the experimental animal center at the University at a temperature of 22°C with access to water and food ad libitum. In this study, all of the experimental procedures were performed according the guidelines of the Committee of the Animal Research Institute of the University.
Buffalo Bull Semen
Frozen buffalo bull semen was purchased from a local Breeding Bull station of Hubei Province, Chinawere used for the present study. The Bull spermatozoa were frozen in liquid nitrogen and, for the experiments, were thawed out in a water bath at 37°C and were separated by percoll density gradient centrifugation, as described previously (Marchiani et al., 2014).
Mouse Sperm
Kunming mice sperm were obtained from the cauda epididymis by mincing the epididymis tissue in PBS. This washed mouse sperm were attached to poly-lysine coated slides for further use in experiments.
Chemicals and Solutions
The polyclonal anti-SUMO-1 antibody (catalog no: sc-9060) was purchased from Santa Cruz biotechnology, Inc. (Texas, U.S.A), the FITC-PNA staining kit was from GENMED SCIENTIFIC (Shanghai, China) and all other reagents were purchased from Sigma-Aldrich Chemical Co., USA, unless stated otherwise.
Coomassie Brilliant Blue Staining of Acrosome of Bull Sperm
Assessments of buffalo bull acrosomal status were conducted essentially as previously described (Hutt et al., 2005). Briefly, fixed spermatozoa were collected by centrifugation at 800 × g for 5 min and washed two times in 100 mM ammonium acetate, pH 9.0 (1 ml). The spermatozoa were subsequently resuspended in 0.5 ml of 100 mM ammonium acetate, pH 9.0, the sample was smeared and allowed to air-dry on glass slides. After air drying, the slides were rinsed with tap water, methanol, and water for 5 min each. The sperm were stained for 10 min at RT with Coomassie Brilliant Blue G-250 (50% methanol and 10% acetic acid). The slides were washed three times with dH2O, air-dried, and mounted in glycerol (30% in PBS) before imaging with a Nikon inverted fluorescence microscope to analyze acrosomal staining.
FITC-PNA Staining of Acrosome of Bull Sperm
The sperm acrosome and nuclei were assessed using FITC-PNA and 4, 6-diamino-2-phenylindole (DAPI) staining, respectively. The sperm acrosome status was assessed by applying FITC-PNA staining kit (GENMED SCIENTIFIC, Shanghai, China), which is specific for sperm acrosome staining. Briefly, after incubation at 37°C for 30 min, the bull spermatozoa specimen was centrifuged for 5 min at 600 × g, the sample was collected and resuspended in a preservative solution at a final concentration of 2 × 107 sperms cell/ml. A 20 μl sperm sample was smeared over a microscope glass slide at room temperature and was fixed using an ice cold fixing solution for 60 s. After drying, 200 μl of FITC-PNA stain (25 mg/ml) were spread gently over the smear and the slide was incubated in a dark chamber for 20 min at room temperature. The prepared glass slide was further washed with 200 μl PBS or cleaning solution to displace surplus stain. The sperm acrosome status was assessed and photographs were taken using an epifluorescence microscope (Olympus) at wavelengths of 480 and 530 nm.
Immunofluorescence Staining of SUMO-1 in Bull Sperm
Sperm cells, recovered from the Percoll gradient, were washed in PBS. The washed buffalo bull sperm were attached to slides and fixed in 4% formaldehyde (Beyotimes, China) for 20 min at RT and the slides were rinsed three times in PBS. The fixed sperm cells were treated for 10 min with 0.3% Triton x100, and the slides were pre-blocked for 60 min with 5% BSA placed at 4°C. The sperm cells were washed with PBS and were incubated with anti SUMO-1 antibody (1:250 in PBS/1% BSA) overnight at 4°C in a dark chamber. The cells were washed three times with PBS and were then incubated with CY-3 labeled secondary antibody (Boster Co., China) at a 1:150 dilution for 60 min at 37°C in a dark room. The cells were then washed 3 × 5 min with PBS before nuclear staining for 5 min with (DAPI, 1:5,000 in PBS) in a dark room. The specimens were washed, and the glass slides were mounted in DABCO. The images were finally analyzed with a Nikon inverted fluorescence microscope with 60 and 100x objective lenses and DAPI, FITC, and CY-3 filter settings. Minimally, 50 cells were analyzed per slide. In order to determine the co-localization of SUMO-1 with acrosome in bull sperm, after immunostaining the sperm with SUMO-1, the slides were further incubated with FITC-PNA as described above.
Protein Extraction of Buffalo Sperm and Co-immunoprecipitation
Protein extraction was performed as previously described (Sang et al., 2011; Kumar et al., 2014). Briefly, after the final wash in PBS, the sperm pellets were resuspended in 400 μl 2x Laemmli buffer [126 mM Tris/HCl, 20% glycerol, 4% SDS, 10 mM PMSF, protease inhibitor cocktail (10 mg/ml; Santa Cruz Biotechnology, CA, USA)], containing isopeptidase inhibitor NEM (Sigma), to inhibit SUMO cleavage from modified proteins. The samples were heated to 100°C (5 min), placed on ice (5 min) and centrifuged at 12,000 × g for 2 min at 4°C before collecting the supernatants. The protein concentration was determined by the BCA assay with a BSA standard (Pierce, Rockford, USA). The samples were stored at −20°C for later use.
Anti-SUMO-1 antibodies and protein A+G Agarose (Beyotimes, China) were used for the Co-Immunoprecipitation according a manufacturer's instructions. Briefly, the sperm protein extracts were incubated with anti-SUMO-1 polyclonal antibody over-night at 4°C on constant shaker. The next day, 40 μl Protein A+G Agarose were added, and the samples were incubated at 4°C with constant shaking. The resin was washed five times with ice cold PBS, and the pellets were resuspended in 2x SDS loading buffer and were heated for 5 min at 100°C before SDS-PAGE electrophoresis for western blotting or mass spectrometry.
Western Blot Analysis
After measuring the protein concentration (determined by BCA assay), the sperm lysates were mixed with 2x SDS loading buffer and were loaded over 10% polyacrylamide gels. After SDS-PAGE, the proteins were transferred to PVDF membranes (Millipore Corp., Bedford, MA). The membranes were blocked with 5% skim milk (Sigma-Aldrich), in TBST [10 mM Tris (pH 7.5), 150 mM NaCl and 0.05% Tween 20], and were then incubated with the polyclonal rabbit anti-SUMO-1 IgG (1:500) diluted in blocking buffer overnight at 4°C. Afterward the membrane was rinsed three times in TBST and was then incubated with HRP conjugated anti-rabbit IgG antibody in TBST for 60 min at room temperature and was finally rinsed three times in TBST. Detection of the bands was accomplished with a BM-enhanced chemiluminescence system (Amersham Biosciences, Piscataway, NJ).
Spermatozoa Capacitation and Acrosome Reaction In vitro
The acrosome reactions were stimulated as previously described (Kumar et al., 2014). In brief, the sperm were capacitated in vitro by thawing of frozen semen at 37°C for 40 s and centrifugation at 1,500 rpm for 25 min. The pellets were reconstituted in 2 ml of a modified Tyrode's Hepes-buffered (Sp-TALPH) washing medium and centrifuged twice, at 1,500 rpm for 10 min. After centrifugation, the pellets were washed with a modified Tyrode's bicarbonate-buffered (Sp-TALP). The pellets were re-suspended in Sp-TALP containing concentration of 25 × 106 spermatozoa in a final volume of 0.5 ml in an Eppendorf tube and capacitation was induced by the presence of 10 μg/ml heparin with an incubation for 4 h at the 38.5°C in 5% CO2. The capacitated spermatozoa were treated for 30 min with 1 mM calcium ionophore A23187 (Sigma Chemical Co.). The acrosome reaction was confirmed by FITC-PNA staining.
Mass Spectrometry Analysis of SUMO-1 Targeted Proteins in Buffalo Sperm
Protein digestion was conducted in accordance with the FASP method, essentially as earlier reported (Wisniewski et al., 2009). Briefly, the protein pellet (~30 μg) was solubilized in 30 μl SDT buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCL pH 8.0) at 90°C for 5 min. The detergent, DTT and other low molecular weight components were removed using 200 μl UA buffer (8 M Urea, 150 mM Tris Hcl pH 8.0) by repeated ultrafiltration (Microcon units, 30 kD). Then 100 μl of 0.05 M iodacetamide in UA buffer was added to block the reduced cysteine residues and the samples were incubated in the dark for 20 min. The filters were rinsed with 100 μl of UA buffer three times and then with 100 μl of 25 mM NH4HCO3 twice. Finally, the protein suspension was digested with 2 μg trypsin (Promega) in 40 μl 25 mM NH4HCO3 overnight at 37°C, and the ensuing peptides were collected. The mass spectrometry investigations were conducted in a Q Exactive MS that was linked to easy nLC (Proxeon Biosystem, now Thermo Fisher Scientific). Six microliters of each fraction was injected for the nanoLC-MS/MS investigation. A peptide admixture of 5 μg was loaded onto a C18 reverse phase column (Thermo scientific easy column, 10 cm long, 75 μm inner diameter, 3 μm resin) into buffer A (Formic acid 0.1%) and was separated by a linear gradient of buffer B (acetonitrile 80% and Formic acid 0.1%) at a 250 nl/min flow rate controlled by Inelli Flow technology over 140 min. The mass spectrometry data were procured utilizing a data dependent top10 procedure energetically selecting the most abundant precursor ions from the survey, scan (300–1800 m/z) for HCD fragmentation. The fortitude of the target value was based pAGC. The dynamical elimination period was 1 min. The investigating scans were acquired by data determination in 70,000 at m/z 200, and the resolutions for the HCD spectra were fixed in 17,500 at m/z 200. The normalized collision energy was 30 eV under a fill ratio that specializes the minimum percentage of target value probable to be attained at the maximum fill time, which was definite as 0.1%. The equipment was run in an enabled mode of peptide identification.
MS/MS spectrometry was investigated utilizing a MASCOT engine (Matrix Science, London, UK; version 2.2) with a non-redundant International Protein Index Arabidopsis sequence database v3.85, declared and released from the European Bioinformatics Institute (http://www.ebi.ac.uk/). For the identification of proteins, the following adoptions were utilized: peptide mass tolerance = 20 ppm; MS/MS tolerance = 0.1 Da; Enzyme = Trypsin; Missed cleavage = 2; fixed modification: Carbamidomethyl (C), and Variable modification: Oxidation (M). The entire described data were established on a 99% assurance for protein discovery as resolute by a false discovery rate (FDR) of ≤ 1%.
Bioinformatics Analysis
Bio-analysis was performed under Ubuntu 16.04 LTS Docker image (Hung et al., 2016) within a custom Python (Python Software Foundation, 2015) scripts using the packages biopython, pyexcel, csv, collection, panda, and matplotlib and R v3.3.1 (R Core Team, 2016) scripts within the ggplot2 and plotrix libraries. Based on the MS/MS identified proteins listed (Table S1), sequences matching with Bubalus bubalis genome (41,499, protein, assembly UMD_CASPUR_WB_2.0) were searched against the NCBI databases (September-2016, https://www.ncbi.nlm.nih.gov) (Geer et al., 2010). The Prediction of Sub Cellular Location (SCL) was performed using two packages for prediction in eukaryotic and animal proteins with single or multiple sites as follows: (I) iLoc-Euk (http://www.jci-bioinfo.cn/iLoc-Euk) (Chou et al., 2011) and (II) iLoc-Animal (http://www.jci-bioinfo.cn/iLoc-Animal) (Lin et al., 2013). The protein sequences (in fasta format) were sent via user interfaces of the dedicated package's web servers. For further understanding of the B. bubalis sperm ejaculated proteome profile, we classified proteins of interest following three strategies: (I) COG (Clusters of Orthologous Group) classification; (II) Protein function classification; and (III) GO Slim (Huntley et al., 2014) classification. The first strategy, the COG analysis, was completed with EggNOG v4.5 (a database of Orthologous Groups and functional annotation covering prokaryotic and eukaryotic species, http://eggnogdb.embl.de) (Huerta-Cepas et al., 2016). The protein sequences were submitted to the web server API. Sequences longer than 800aa were processed with HMMER v3.1b2 (A software suite widely used for analysis of homologous protein and nucleotide sequences with high speed and sensitivity, http://hmmer.org) (Jiang and Ganesan, 2016). HMMER was used for homology sequences, searching within the EggNOG mammalian database annotation (maNOG.hmm). The results were filtered based on score and E-value significance.
The second strategy was the protein function classification and Gene Ontology (GO), based. Analysis was processed by PANTHER v11 (Protein Analysis Through Evolutionary Relationships, http://pantherdb.org) (Mi et al., 2016) with a statistical overrepresentation test and default setting criteria. The third strategy consisted of GO Slim (Huntley et al., 2014) classification. The GO terms reported by PANTHER were classified by the web server tool CateGOrizer v3.218 (May06-2010, A Web-Based Program to Batch Analyze Gene Ontology Classification Categories: http://www.animalgenome.org) (Von Mering et al., 2003).
Conscious about the crucial role of protein-protein interaction (PPI) in spermatozoa process, additional information about PPIs, pathways and COG classification were collected by STRING v10. 0. (A Database of predicted functional associations between proteins, http://string-db.org) (Hu et al., 2008).
The tracking of the PTMs in the protein list previously identified by MS/MS focused on SUMOylation, which one of the most important PTMs affecting the sperm proteome of water buffalo. In order to identify SUMOylated modified protein and SUMO-1 targets, we used several tools for predicting SUMOylation sites, motifs and interactions. Thus, the protein sequences were submitted to (i) GPS-SUMO-2.0 (a tool for the prediction of sumoylation sites and SUMO-interaction motifs, http://sumosp.biocuckoo.org) (Zhao et al., 2014) and (ii) JASSA (a comprehensive tool for prediction of SUMOylation sites and SIMs, http://www.jassa.fr) (Beauclair et al., 2015). The peptide sequence visualization was performed by Seq2Logo (Thomsen and Nielsen, 2012) using the following Parameters: PSSM_LOG; Halfbits and 800 × 800.
Results
Coomassie Brilliant Blue and FITC-PNA Staining of Acrosome in Water Buffalo Sperm
In the sperm from Buffalo bull, the acrosome was clearly identified by intense blue staining with Coomassie Brilliant Blue (Figure 1A image a), and the acrosome reaction was confirmed by loss of the Coomassie staining in the acrosome region of the sperm (Figure 1A image a). The evaluation of nuclear and acrosome morphology of the frozen-thawed bull spermatozoa was assessed with DAPI and FITC-PNA staining, respectively, indicating that the acrosome was enveloped with green fluorescence of FITC in the bull spermatozoa (Figure 1A images b–d). The acrosome status of the bull spermatozoa was evaluated through FITC-PNA (Figure 1B) and was classified within four categories. In category I (Figure 1B image a), the spermatozoa showed a strong, bright fluorescence in the acrosomal cap-like structure, resulting from the PNA binding, demonstrating an intact acrosome. In category II (Figure 1B image b), the fluorescence of the acrosomal cap was disrupted or there was fluorescence in the equatorial plane. Category III (Figure 1B image c) displayed fluorescence in the post acrosomal region of the sperm, and category IV (Figure 1B image d) spermatozoa displayed no fluorescence in the acrosome area, indicating a reacted acrosome. The results demonstrated that Coomassie Brilliant Blue staining and the FITC-PNA staining of sperm could be used to evaluate the acrosome status of water buffalo sperm.
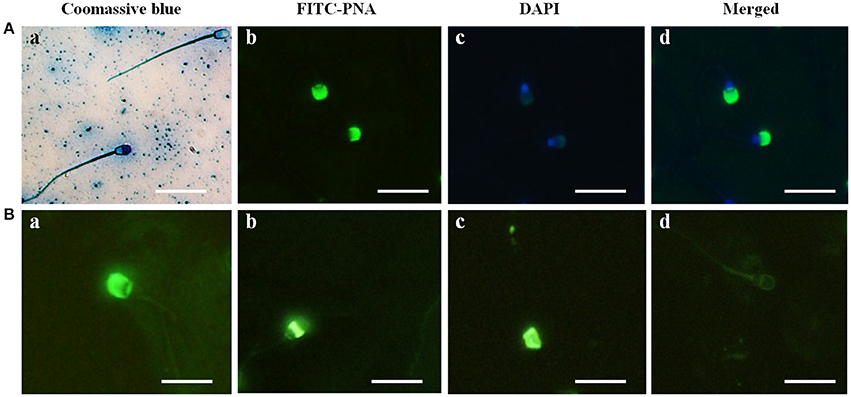
Figure 1. Acrosome identification in buffalo bull sperm and four categories of Buffalo bull sperm acrosome status. (A) (Image a) Coomassie Brilliant Blue staining of sperm with an intact and reacted acrosome, (image b) FITC-PNA stained, (image c) DAPI-stained, (image d) DAPI+FITC-PNA- stained images. (B) (Image a) intact acrosome, (image b) equatorial acrosome region, (image c) post acrosomal region, (image d) reacted acrosome. Scale bar, 20 μm; Magnification, 1,000x.
Immunocytochemical Localization of SUMO-1 in Bull Spermatozoa
The immunofluorescence staining demonstrated the mid piece, neck, head, and acrosome localization of SUMO-1 in bull spermatozoa (Figure 2A). At high magnification on the optimal immune stained samples, the SUMO-1 signal was observed in the acrosome region of sperm heads (Figure 2A). The SUMO-1 staining was not observed in the controls incubated with rabbit IgG instead of anti- SUMO-1 antibody. Additionally, signal specificity was confirmed using a secondary antibody alone under the same fluorescence intensity condition. For comparison, mouse epididymal spermatozoa were also immune-stained within anti SUMO1 antibodies. The SUMO-1 expression was similar to the bull spermatozoa (Figure 2A).
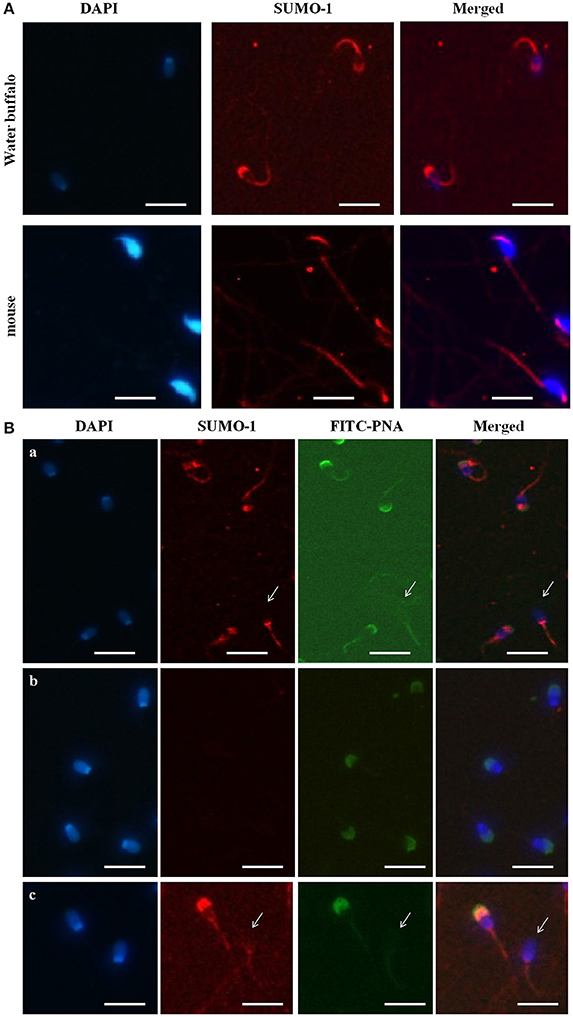
Figure 2. Localization of SUMO-1 in the acrosome of Buffalo bull and mouse spermatozoa. (A) DAPI, for nuclear staining (blue signal), SUMO-1, Anti-SUMO1 antibody followed by a CY3-conjugated secondary antibody (Red fluorescence), Merged, DAPI nuclear staining plus anti-SUMO1 antibody. (B) SUMO1 co-localization and expression in buffalo bull sperm. Collected spermatozoa were stained with SUMO1 antibody pursued by incubation with Cy-3 conjugated goat anti rabbit IgG (color red). The specimen were then restained with 4, 6-diamino-2-phenylindole (DAPI, 1:5,000 in PBS) (blue color) and FITC-PNA (flurescein isothiocyanate-conjugated peanut agglutinin) (green color). In (panel a) the superposed images shown in the spermatozoa were stained with SUMO-1 antibody followed by incubation with Cy-3 conjugated goat anti rabbit IgG. (panel b) the superposed images are shown in negative samples as control. (panel c) the images show both an intact and reacted acrosome, and the disappearance of SUMO-1 in the acrosome-reacted buffalo bull spermatozoa. Note that both SUMO-1 and FITC-PNA disappeared in acrosome region of the acrosome-reacted spermatozoa. The images were observed using a Nikon inverted fluorescence microscope. The scale bar is 100 μm.
Co-localization of SUMO-1 in Bull Spermatozoa
In order to further reveal the relationship of SUMO-1 localization with the acrosome of bull sperm, FITC-PNA staining of bull sperm was also used to co-localize the expression SUMO-1 and the acrosome in bull sperm (Figure 2B). The results demonstrated that SUMO-1 immunosignal was observed in the mid piece, neck, head, and acrosome region of the sperm head, overlapping with the PNA lectin acrosomal labeling (Figure 2B panel a). The immunosignal in the bull spermatozoa was not observed in the control sperm stained with rabbit IgG. The use of secondary antibody alone confirmed the specificity of the signal (Figure 2B panel b).
We further examined whether acrosome-localized SUMO-1 is released from buffalo bull spermatozoa during the acrosome reaction. Capacitated spermatozoa were induced to undergo acrosome reaction by Ca2+ ionophore A23187 and the acrosome status was monitored by FITC-PNA staining. The sperm showed FITC-PNA and SUMO-1 staining on an acrosomal region in spermatozoa with an intact acrosome (Figure 2B panel c, left), and this disappeared in the acrosome-reacted spermatozoa (Figure 2B panel c, right and also see Figure 2B panel a, arrow indicated sperm without an acrosome).
Expression of SUMO-1 in Capacitated and Acrosome Reacted Spermatozoa of Water Buffalo
To assess the expression profile of SUMO-1 between capacitated sperm and non-capacitated sperm in water buffalo after in vitro capacitation, protein samples extracted from the bull spermatozoa were run analyzed by western blot. After in vitro capacitation, SUMO-1-modifications were observed compared to the non-capacitated sperm (Figure 3A). After capacitation, bands at ~130, 37, and 27 kDa showed a lower intensity, a band at 30 kDa appears and bands at 85 and 50 kDa are enhanced (Figure 3A). Furthermore, after capacitation, the spermatozoa were induced to undergo the acrosome reaction by calcium ionophore A23187, and the expression of SUMO-1 in acrosome reacted sperm was also detected and was compared with the acrosome intact sperm (Figure 3B). There were subtle changes in the SUMO-1 expression profile after the acrosome reaction was induced. The acrosome reaction caused the band at 37 kDa to decrease and a smear of bands below 25 kDa disappeared (Figure 3B).
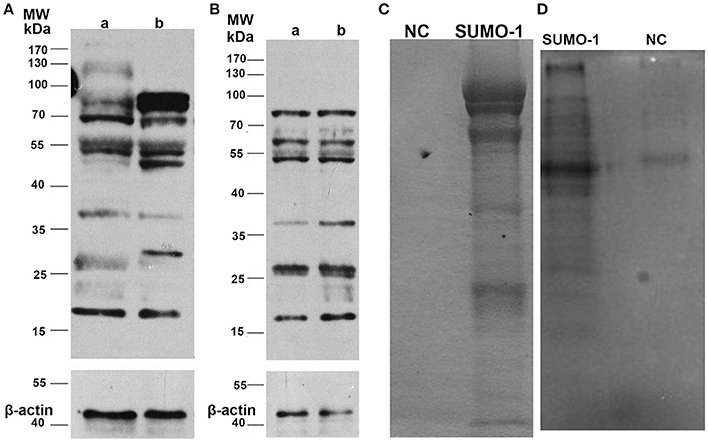
Figure 3. Expression of SUMO-1 in water buffalo spermatozoa. (A) Western blot analysis of SUMO-1 expression in water buffalo spermatozoa after in vitro capacitation, showing capacitation-induced changes in SUMO-1-modifications in sperm (lane b) compared to non-capacitated sperm (lane a). (B) Western blot analysis of acrosome reacted (lane a) vs. acrosome non-reacted (lane b) water buffalo spermatozoa. (C) Buffalo bull sperm proteins were immunoprecipatated with SUMO-1 antibody, and then run for SDS-PAGE and stained with coomassive brilliant blue. For the negative control (NC). (D) SUMO-1 conjugates were pull down from water buffalo sperm lysates by a SUMO-1 antibody and were analyzed by western blot to detect SUMO-1 expression.
SUMO-1 Conjugations and Mass Spectrometry Analysis
The SUMO-1 conjugates in water buffalo spermatozoa were first pulled down by Co-Immunoprecipitation using protein A+G agarose and the SUMO-1 antibody, and this was confirmed by Coomassive Brilliant Blue staining of the SDS-PAGE and by a Western blot analysis (Figures 3C,D). The results demonstrated that specific protein bands in the sample with SUMO-1 pull down, but not in the negative control sample (normal rabbit IgG) (Figures 3C,D), indicating that SUMO-1 specific conjugates were pulled down. These bands from the Coomassie blue stained gel (Figure 3C) were analyzed by mass spectrometry, resulting in the identification of 60 proteins that were unique to the SUMO-1 antibody sample compared to the negative control (Table S1). Ten of these proteins were identified in previous studies as being sumoylated, and these are indicated by an asterisk in Table S1.
The identified proteins have multiple functions in germ cells and sperm, including proteins associated with sperm motility, such as tubulin alpha-1C chain andalpha-1B chain, relaxin receptors RXFP4 relaxin (RLN)/insulin like family peptide receptor 4, RLN-3/INSL7 receptor 2, and LOC102283812 RLN-3 receptor 1-like, alpha and gamma actins ACTC1 and ACTG2.
Bioinformatics Findings
Database Searches
In the present work, the 60 proteins identified by MS/MS were submitted to NCBI (September-2016, https://www.ncbi.nlm.nih.gov) in order to get their correspondent (fasta) sequences. We found 110 proteins matched with B. bubalis annotated protein sequences, and 92% of them were predicted. Many isoforms (65 proteins with at least two matches) were present in the list. The lack of publication and confirmation for most of the B. bubalis sequences prompted us to keep all of isoforms for the rest of the analyses.
Sub-cellular Localization Analysis
There were eight different SCL predicted by both the iLoc-Animal (Lin et al., 2013) and iLoc-Euk (Chou et al., 2011) tools. We show that 39% of the whole cell sperm proteins are located only in the cytoplasm (Figure 4), and 31% are predicted in both the cytoplasm and nucleus. Tubulin alpha-1A chain-like [XP_006040217.1] is the only protein of the list present in the cytoskeleton in addition of his presence in the cytoplasm and nucleus. Two proteins: polyribonucleotide nucleotidyltransferase1 PNPT1 (6 isoforms) and metalloendopeptidase OMA1 (2 isoforms) known for their crucial role in the mitochondria, were predicted in both cytoplasm and mitochondrion (Table S2). We observed that cadherin EGF LAG seven-pass G-type receptor 2 [XP_006052222.1] and ADP-ribosyltransferase4 (ART4) [ABO72473.1] were located in the endoplasmic reticulum. Dombrock blood group protein (ART4) was predicted as known in humans, in the cell membrane, cytoplasm and mitochondrion also. In addition, we noted that phosphatidylinositol 3-ki0se catalytic subunit type 3-like [XP_006057019.1] is implicate in many pathways trafficking and was predicted in the cytoplasm and endosomes, which is known in humans.
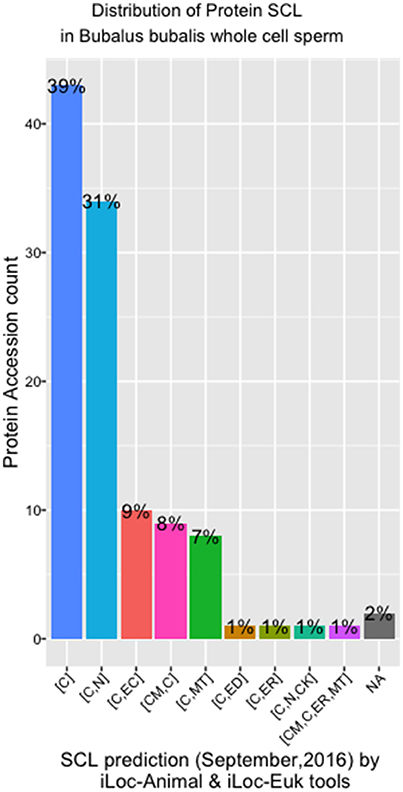
Figure 4. SCL in whole cell sperm in Bubalus bubalis. Plot describing the distribution of protein Sub-cellular localization [SCL] predicted by iLoc-Animal and iLoc-Euk tools. [C]: cytoplasm; [C,N]: cytoplasm and nculeus; [C,EC]: cytoplasm and extracellular; [CM,C]: cell membrane and cytoplasm; [C,MT]: cytoplasm and mitochondrion; [C,ED]: cytoplasm and endosome; [C,ER]: cytoplasm and endoplasmic reticulum; [C,N,CK]: cytoplasm, nucleus, cytoskeleton; [C,N,ER,MT]: cell membrane, cytoplasm, endoplasmic reticulum, and mitochondrion; [NA]: No Available results for those sequences.
Protein Classification
Those B. bubalis proteins previously identified were functionally classified through to GO and COG strategies. The EggNOG (Huerta-Cepas et al., 2016) results (270 match) were filtered based on the score and E-value criteria (Score>100, E-value <1E-180). We successfully affected 178 filtered entries in 14 COG categories (Figure 5). The COG analysis reveals that 8% of those proteins belong to (O: Post-translational modification, protein turnover, and chaperones), 23% to (Z: Cytoskeleton), 23% to (T: Signal transduction mechanisms), and 10% to (K: Transcription) COGs categories. Also, we subjected our protein list to a GO analysis (Figure 6). All of the proteins were functionally categorized in 12 groups. In our dataset, the most enriched categories (Figure 6A) were biological process (35%) followed by molecular function (19%), Cellular component (16%), and others with (3%) each. The details of GO terms of those categories are described in (Figure 6B). As shown in the figure, some important annotations were reported in Cellular component category, such as localization, locomotion, and metabolism (Figure 6C).
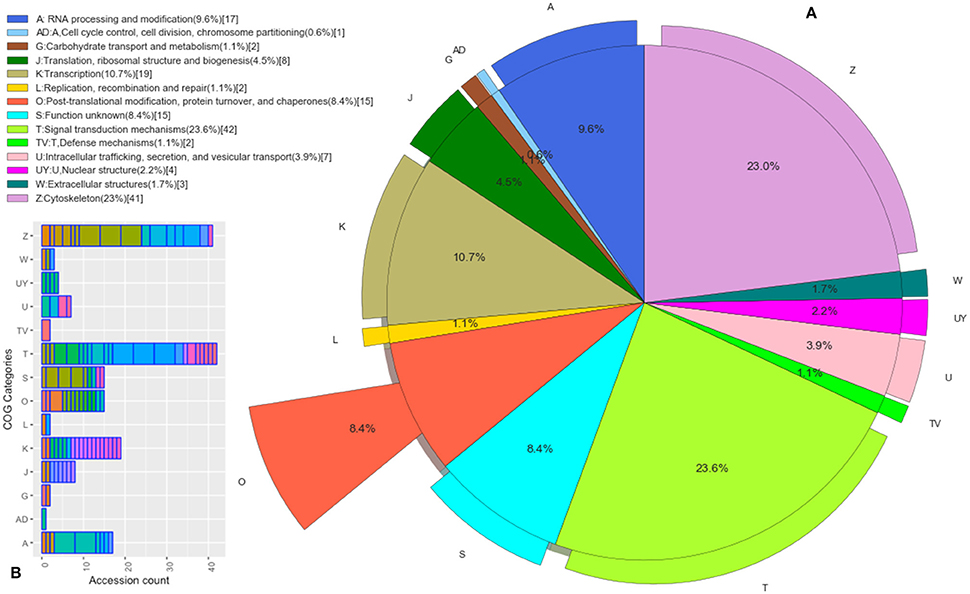
Figure 5. Functional Protein Classification (COGs). (A) Distribution of proteins count in the COGs categories. Colored boxes correspond to the unique COGs subcategories (EggNOG v4.5 terms). (B) The pie shown each COG category percentage.
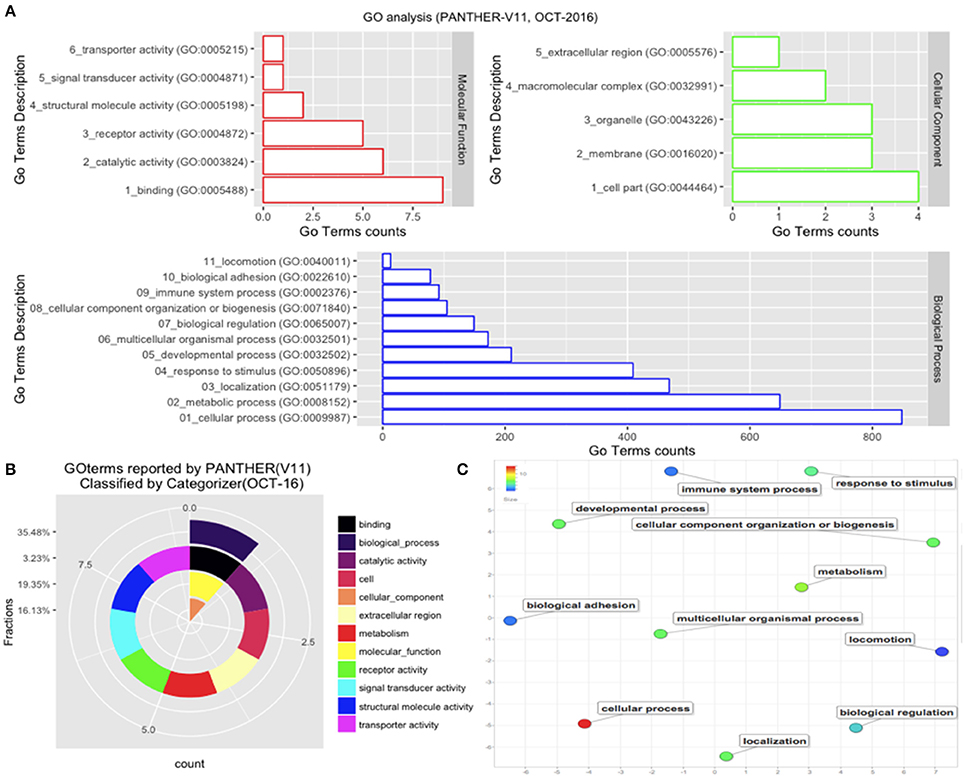
Figure 6. GO analysis. The GO analysis was performed with PANTHER (V11). (A) Details of GO terms distribution. (B) The terms were reclassified using Categorizer [http://animalgenome.org]. This represents the mapping of 22 GO terms to 127 of the “GO Slim” ancestor terms by single count. (C) Plot of GO terms details. Further analysis of GO terms was conducted by Rivgo [http://revigo.irb.hr].
PPIs and Pathways
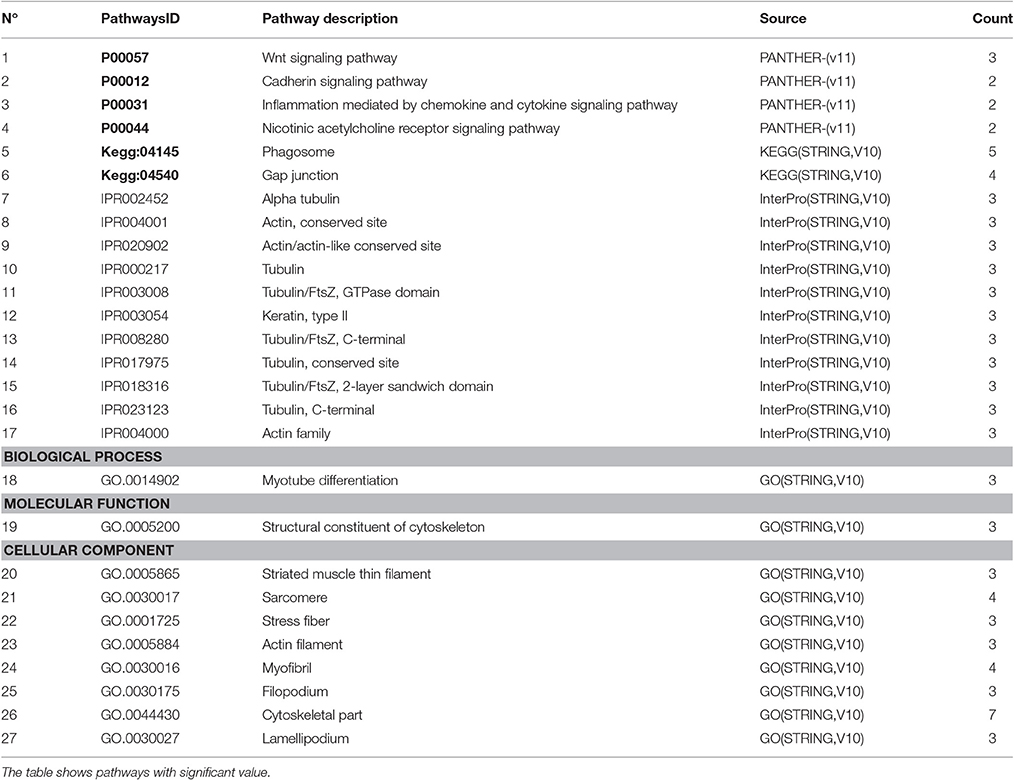
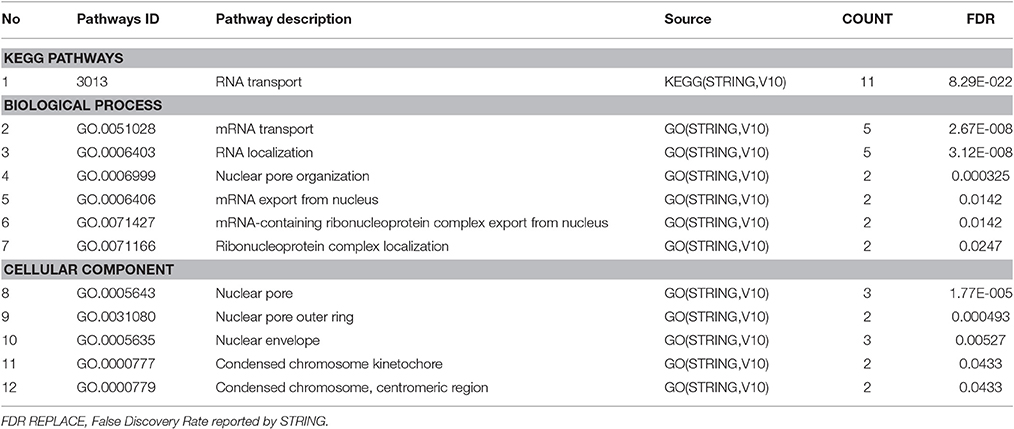
To summarize the proteomics results into a general functional model, we integrated known and predicted protein-protein interactions (PPIs) (Figure 7). The PPIs were built using STRING (Hu et al., 2008). We noted the presence of one network highlighted Tubulin-Tubulin and Actin-Tubulin interactions. In order to know more about their pathways we combined the STRING and PANTHER (Mi et al., 2016) pathway analyses. The list of relevant pathways (with at least two counts and p < 0.05) is described in Table 1. We observed several pathways confirming our previous finding. We noted that the tubulin, actin, and signaling pathways were enriched in our protein list. Those proteins are suggested to be implicated in spermatozoa acrosome assembly. This pathway analysis shows the presence of one molecular function, namely, the pathway concerning the structural constituent of the cytoskeleton. In fact, microfibrils are suggested to be the principal contractile organelles responsible for sperm motility (El Shoura, 1986).
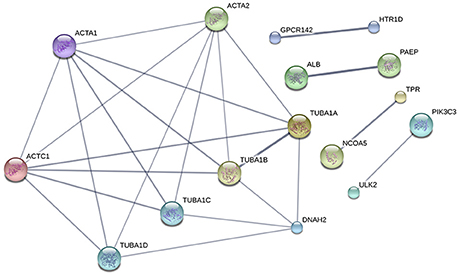
Figure 7. Protein-Protein Interaction [STRING-V10.0] (PPIs). The PPI was built using the following parameters: Bos taurus background (No match for Bubalus bubalis found, B.bubalis genome is not full and well-annotated yet). Highest confidence Score = 0.55. The dark line in the graph correspond to Highest confidence score higher than 0.7. Hide disconnected nodes in the network. PPI enrichment p-value = 0.000124, clustering coefficient: 0.902.
PTM Prediction
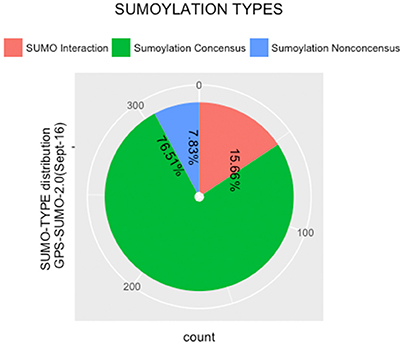
The protein sequences were scanned for potential SUMO targets. More than 400 in our identified list of proteins were reported by GPS-SUMO-v2.0 (Zhao et al., 2014) tool. We only kept 332 significant results (p < 0.05). The filtered results revealed three categories of SUMO modification (Figure 8). Only 7.8% of the proteins hold Sumoylation Non-Concensus (SNC) sites. About 15.6% of them present SUMO-Interaction (SI) sites. The majority of identified proteins (76.5%) contain Sumoylation Concensus (SC) sites. Representative peptide sequences were highlighted with Seq2Logo-v2.0 (Figure 9). Moreover, a deeper SUMO modification analysis revealed that some proteins might hold more than one SUMO modification sites type. In fact, one of them contains both SNC and SI sites, five of them contain both SNC and SC sites, eight present both SC and SI sites and three of them contain all types of SUMO modification sites (SI, SC, and SC). Twenty-two sequences hold only one type of SUMO modification site. The details are described in Table 2. In summary, we successfully predicted 46 potential SUMOylation targets between the 60 proteins identified by MS/MS. Based on 142 unique peptides (reported by the GPS-SUMO tool) analyzed, we predicted 10 Sumoylation Non-Concensus sites, 13 SUMO-interaction motifs, and 42 Sumoylation Concensus sites.
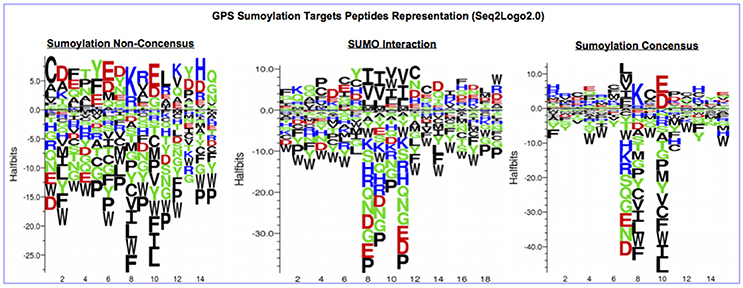
Figure 9. Seq2Logo2.0 representation of three types of SUMO modification peptides sequences reported by (GPS-SUMO2.0).
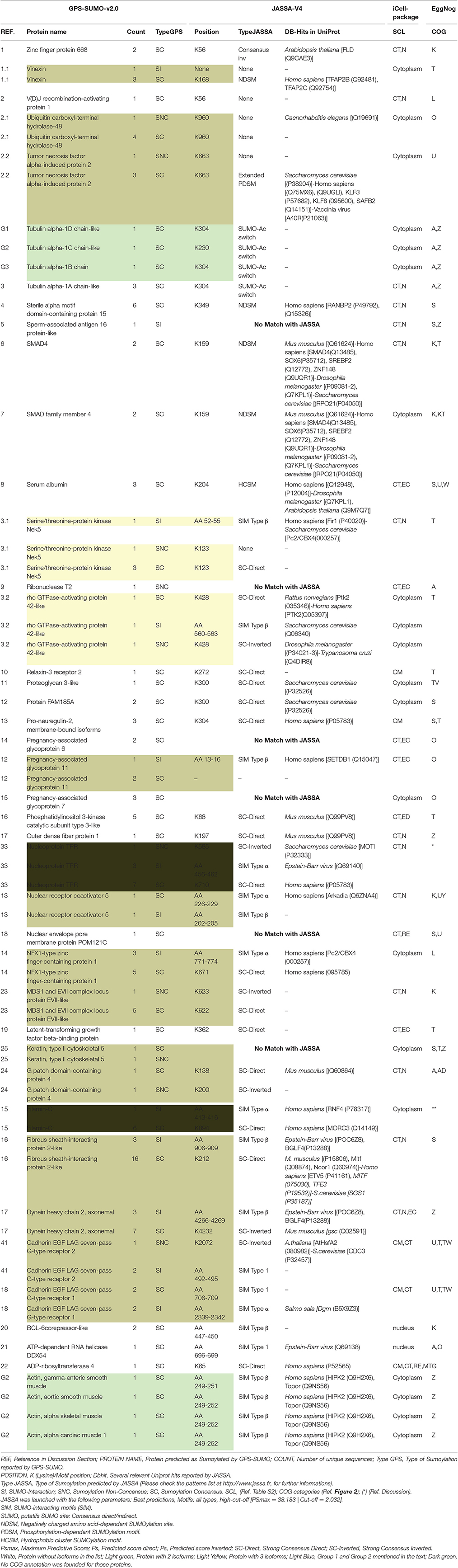
Table 2. Summary of bio-analysis findings SI, SUMO-Interaction; SNC, Sumoylation Non-Concensus; SC, Sumoylation Concensus. SCL (Ref. Table S2); COG categories (Ref. Figure 2); (*) (Ref. Discussion).
Conscious about the validation of the potential sumoylated proteins, we submitted the B. bubalis protein sequences to the JASSA tool. We used this predictor not only to confirm the sumoylated motifs and sites previously identified by GPS-SUMO-V2.0, but also to determine whether those targets were revealed in other species. Thus, as presented in Table 2, about 90% of the targets identified by GPS-SUMO-V2.0 were confirmed by JASSA. Nevertheless, this tool shows that only 52% of the targets are present in mammals, 33% in Humans and 10% in mice. We noted that 13% were predicted in Saccharomyces crvicae and 6% in Drosophila melanogaster. Furthermore, 18% of the sumoylated potential sites present SUMO-Interacting motifs type SIM Type β (Song et al., 2004) and about 6% of these show the type SUMO-Acetyl switch (Stankovic-Valentin et al., 2007).
Discussion
Sperm are essentially transcriptionally and translationally inactive. This implicates the PTM of proteins as essential in determining sperm morphology, motility, and functionality and, hence, in fundamentally influencing male fertility. To our knowledge, this is the first study of SUMO1 modification of proteins of buffalo spermatozoa. Thus, in this study, we addressed the role of sumoylation in Buffalo bull spermatozoa, an important PTM implicated in many cellular functions, such as DNA recombination, repair and replication, RNA transcription, nucleo-cytoplasmic trafficking, and protein stability. We confirmed that the expression of SUMO-1, was concentrated in the mid piece, neck, and head of the bull sperm and was lost upon induction of the sperm acrosome reaction with Ca2+ ionophore A23187. We also identified a suite of 60 bull sperm proteins that could potentially be modified by SUMO1, with implications for sperm functionality and male fertility. Importantly, 10 (17%) of these proteins were identified in other studies as sumoylated proteins, suggesting the likelihood that these are truly sumoylated proteins. This finding presents a great opportunity for future studies regarding validation of these findings and exploring the proteins in more detail.
Western blot analysis suggested that sperm capacitation induces changes in the sumoylation pattern in buffalo bull sperm. However, more sensitive techniques would be required to further dissect these differences, for example 2D gel at individual SUMO targets. This finding is in contrast to in other species, such as humans, where capacitation did not significantly alter the global level of sumoylation in sperm (Vigodner et al., 2013).
Spermatogenesis is a differentiation process with the end goal of functional sperm production, involving marked alterations in genetics, cell functions, and chromatin (Lena et al., 2016; Renata et al., 2016) Modification of sperm proteins by sumoylation, including those found in the mid piece, neck, head, and acrosomal region, therefore, could be important for many aspects of sperm function.
Sumoylation of sperm proteins at various stages of spermatogenesis was previously demonstrated in other species, including humans and rodents (Rogers et al., 2004; Vigodner and Morris, 2005; Vigodner et al., 2006, 2013; Brown et al., 2008; Marchiani et al., 2011, 2014; Stanton et al., 2012). However, the role of SUMO in spermatogenesis is as yet undefined, with involvement in processes, including heterochromatin organization, DNA repair mechanisms, and regulation of gene expression, all being suggested (Rogers et al., 2004; Vigodner et al., 2006; Stanton et al., 2012). In fact, the role of SUMO1 may be deleterious in some circumstances. While SUMO1 appears to be expressed in most mature human sperm (Marchiani et al., 2011, 2014), increased/excessive sumoylation is associated with an abnormal sperm morphology and motility (Vigodner et al., 2013; Marchiani et al., 2014) and with sperm DNA fragmentation (Marchiani et al., 2014).
In other cell types, SUMO has been shown to play a role in DNA repair. It is possible that, in sperm, SUMO may be involved an attempt to limit DNA damage or regulate mitochondrial dynamics, but in excess it may also have a deleterious effect on morphology and motility. The extent and the location of SUMO in sperm are likely to dictate the role sumoylation plays under different circumstances. Targets that have been identified in mature human sperm include the mitochondrial protein DRP1, the microtubule-organizing RanGAP1 and topoisomerase IIa, which is involved in chromatin remodeling (Marchiani et al., 2014).
We identified several proteins that are modified by SUMO1 in bull sperm. These include relaxin receptors, which have been recently associated with beneficial effects in human and boar spermatozoa motility and functionality (Ferlin et al., 2012; Feugang et al., 2015a). In boar spermatozoa, for example, relaxin and its receptors are distributed over the whole sperm length, and relaxin is particularly concentrated in the acrosome region (Feugang et al., 2015b). Our identification of the relaxin receptors RXFP4 relaxin(RLN) /INSL like family peptide 4, RLN-3/INSL7 receptor 2, and LOC102283812 RLN-3 receptor 1-like as potential SUMO1 targets in bull spermatozoa, therefore, has potential implications for sperm motility and functionality and, hence, for male fertility.
Remodeling of the cytoskeleton is also essential in sperm motility. We identified tubulin alpha-1C and alpha-1B chains, alpha and gamma actins ACTC1 and ACTG2, the axonemal protein DNAH2 dynein, and the intermediate filament components KRT4, KRT5, and KRT6B, among other cytoskeletal components, as potential SUMO1 targets in bull sperm. In un-reacted bull sperm, both actin and tubulin are shown to be localized in the equatorial segment of the acrosome actin, with actin also in the marginal acrosomal ridge of the heads of unreacted buffalo spermatozoa and tubulin around the acrosome periphery (Oikonomopoulou et al., 2009). This matches the localization we established for SUMO1 in buffalo sperm. Acrosomal expression of both actin and tubulin falls dramatically upon acrosome reaction induction (Oikonomopoulou et al., 2009), just as we identified for SUMO1 expression in buffalo sperm. The role of sumoylation in modification of cytoskeletal proteins, such as tubulin and actin, is not yet well-understood. In fact, tubulin itself is not well-recognized as a direct sumoylation target. Actually, the MAPs, including Pac1p, Bik1p, Kar9p, Tau, and Ndc80p, are more recognized targets (Alonso et al., 2015). However, we identified tubulin chains as potential direct targets of SUMO1 and suggest that this protein modification might play a role in regulating the induction of the acrosome reaction. Moreover, tubulin alpha-1C chain and ACTA1 were identified by others as sumoylated, and ACTA1 was specifically identified during mouse spermatogenesis (Becker et al., 2013; Xiao et al., 2016).
The sumoylation of actin, predominantly the nuclear form, was previously identified at position K284, and SUMO2 and SUMO3 were identified as the SUMO isoforms most likely to modify actin (Hofmann et al., 2009). However, we identified various actin types as potential SUMO1 targets in buffalo sperm. The potential effects of actin sumoylation include possible restriction of actin filament formation and the blocking of export of nuclear actin and the potential formation of alternative nuclear actin structures (Hofmann et al., 2009; Alonso et al., 2015), which again could contribute to regulating the acrosome reaction.
Sumoylation of the cytoplasmic dynein adaptor and target proteins were identified. However, no reports have yet described the direct SUMO-modulation of dynein (Alonso et al., 2015. Flagellar dynein is distinct from cytoplasmic dynein. Dynein is a molecular motor protein on the exterior doublet microtubules of sperm that generates energy for the momentum of flagella that is crucial for spermatozoa movement toward oocyte into female reproductive tract. Thus, sumoylation of the axonemal protein DNAH2 dynein has potential implications for male fertility. Meanwhile, sumoylation of the human keratin variants K8, K18, and K19 was demonstrated in hepatocytes, with implications for the organization of filaments and keratin solubility (Snider et al., 2011). More specifically, hyper sumoylation was associated with reduced keratin solubility and with chronic liver injury in both human and mouse livers. This hypersumoylation reduced the cytoprotective function of K8, K18, and K19 by retaining them in an insoluble compartment (Snider et al., 2011). Such effects on the spermatozoa keratins KRT4, KRT5, and KRT6B could therefore have a deleterious effect on sperm function.
Beyond cytoskeleton regulation, RAG1, which is essential for IgG immunoglobulin synthesis and class switching, was another potentially important SUMO1 targets identified in this study. IgG and RAG-1/RAG-2 expression was recently detected in human sperm, and an anti-IgG antibody was shown to inhibit sperm penetration of Zona-free hamster egg (Yan et al., 2016). Thus, sumoylation of RAG-1, which, to the best of our knowledge, has not previously reported, may be of importance in fertilization. In addition, SMAD4, a transcriptional regulator and key modulator of TGF-b signaling, was also identified as a SUMO1 target in buffalo sperm. The TGF-b signaling pathway was identified to be essential for maintaining adult spermatogenesis in mice (Moreno et al., 2010). The expression of this protein in the Sertoli and Leydig cells was recently identified as essential for the development of murine testes and testicular tumor suppression (Archambeault and Yao, 2014). The sumoylation of SMAD4 represses its transcriptional activity (Long et al., 2004). Thus, SMAD4 and its role in TGF-b signaling might be implicated in spermatogenesis in buffalo bull, and this pathway would be of interest to explore in the future. Other potentially important SUMO1 targets, in terms of bull fertility, include ODF1, which is involved in oocyte membrane fusion and SPAG16 (sperm associated antigen 16). ODF-1 was previously identified as a sumoylated protein human sperm (Vigodner et al., 2013). A deficiency in SPAG16 is associated with infertility in male mice due to impaired sperm motility (Zhang et al., 2006). The sumoylation of these two proteins might have affects on fertility in buffalo bulls.
It is common (but not always) that ubiquitination is associated with proteolytic pathways, and this is not the case of SUMO modification target protein, which are more implicated in PPIs, nuclear-cytoplasmic transport and the formation of specific domains (Vigodner and Morris, 2005).
To summarize our findings in Table 2, we detected 66 potential target of SUMO modification and we validated 59 of them within a second tool (JASSA). We highlighted the presence of two groups that were both predicted in the cytoplasm and present in the SC motifs. G2 is the actin protein group that is below the cytoskeleton COG category, and G1 is the tubulin protein group that is below to both the cytoskeleton and the RNA processing and modification COG categories. Furthermore, we observed an interesting motif type (SUMO-Acetyl switch) that is implicated in acetylation/deacetylation-SUMOylation switch (Stankovic-Valentin et al., 2007). This motif was detected exclusively in the G1 set of the tubulin proteins validated as SUMO targets (Table 2). Many studies (Wloga et al., 2010; Bhagwat et al., 2014) suggest that acetylated tubulin may affect sperm mobility. In addition, we noted that 18% of the predicted targets contain the SUMO-Interacting motifs type SIM Type β. This motif is suggested to be involved in the RanBP2/Nup358 and RanGAP1 interaction (Song et al., 2004). Those proteins seems to be very important for spermatozoa, since they are SUMO targets for RanGAP1 and Topoisomerase IIa in somatic and/or germ cells in mature human spermatozoa, as described in a previous study (Marchiani et al., 2014). We noted the presence of different kingdoms (plants, bacteria, virus) in the Uniport database hit match proposed by JASSA. These are known as Bovine Sperm YWHA Binding Partners (Puri et al., 2008).
PPIs between the two groups were observed (Figure 7). Furthermore, we do note two proteins without COG classification (marked with * in Table 2). The first one was TPR (XP_006060592.1), there was no match found in the EggNOG4.5 mammalian database for TPR. We reported the COG STRING results for the corresponding analysis in Figure 10C. TPR (NOG19963) seems to be connected to (COG5160: SUMO1 sentrin specific peptidase, Protease Ulp1 family which is below to [O: Post-translational modification, protein turnover, and chaperones] COG category, and COG5079: minichromosome maintenance complex component 3 associated protein and nuclear protein export factor, which is below to [DU:Cell cycle control, cell division, chromosome partitioning, Intracellular trafficking, secretion, and vesicular transport] COG category.
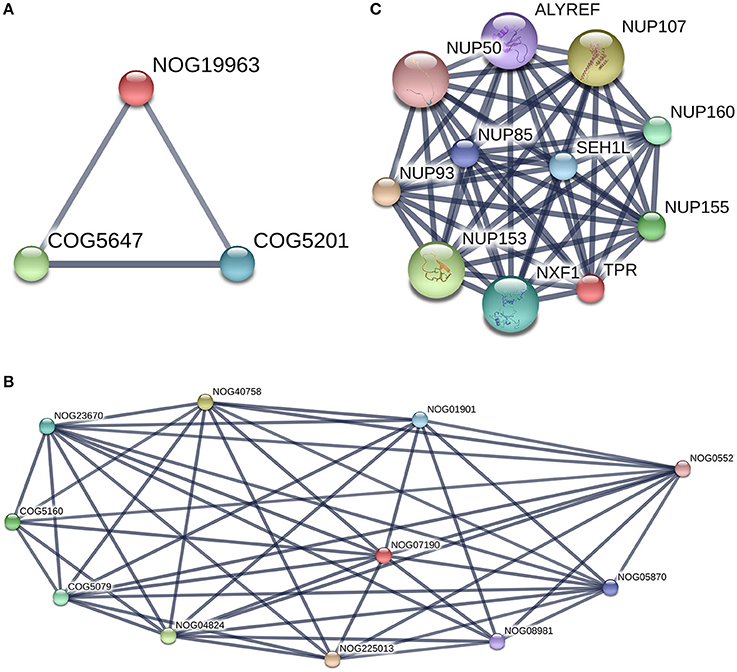
Figure 10. TPR and Filamin-C analysis with [STRING-V10.0]. Protein of interest is marked in Red in the graph. (A) TPR protein-protein interaction network. (Highest confidence Score = 0.9. Hide disconnected nodes in the network. PPI enrichment p-value = 0); (B) Filamin-C, COG analysis. This protein is marked as Non Supervised Orthologous Group (NOG19963) and is below to three edges of the graph with two COGs terms [COG5160 andCOG5079]. (C) COG analysis for TPR protein with string. TPR is marked as Non Supervised Orthologous Group (NOG07190). Two COGs terms are present in the graph [COG5646 andCOG5201].
The details of the TPR PPIs (Figure 10A) and pathways reported by STRING are described in Table 3. The second protein reported in Table 3 without a COG prediction was Filamin-C (XP_006074441.x), and it was reported (for the 7 isomers) with EggNOG as being bellow the Z (cytoskeleton category) but with a score lower than 100. With the STRING tool, we observed that Filamin-C protein formed three nodes with two other COG terms (COG5647: cullin 1, a subunit of E3 ubiquitin ligase andCOG5201: ubiquitin-dependent protein catabolic process, SCF ubiquitin ligase - SKP1 component) (Figure 10B) that were both below the (O: Post-translational modification, protein turnover, and chaperones) COG category. There was no network reported with STRING for Filamin-C, it is an uncharacterized protein in the bos genus. A better annotation of B. bubalis genome may open the way for the use of other strategies (such as methods based on homology sequences analysis) to support our findings. In addition, studies using freshly collected sperm are need to validate these findings as the freeze-thaw process is known to induce sumoylation changes.
In conclusion, we have confirmed that the expression of SUMO1 in sperm cells of buffalo bull is concentrated in the mid piece, neck, and head. We have identified a large number of SUMO1 targets which presents opportunities for future studies to contribute to a better understanding of the biological processes and pathways involved in spermatogenesis, the implications of sumoylation, including hyper sumoylation, for bull sperm function and male fertility.
Author Contributions
RB and LH conceived and designed the experiment; RB, LW, and JC performed experiments and interpreted the data; NH, ZR, CH, DW, DB, and HT contributed reagents/materials/analysis tools; RB and LH wrote and revised the paper, and all the authors approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 31171378) and the Fundamental Research Funds for the Central Universities (Program No. 2014PY045).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2017.00354/full#supplementary-material
References
Aitken, R. J., and Baker, M. A. (2008). The role of proteomics in understanding sperm cell biology. Int. J. Androl. 31, 295–302. doi: 10.1111/j.1365-2605.2007.00851.x
Alonso, A., Greenlee, M., Matts, J., Kline, J., Davis, K. J., and Miller, R. K. (2015). Emerging roles of sumoylation in the regulation of actin, microtubules, intermediate filaments, and septins. Cytoskeleton 72, 305–339. doi: 10.1002/cm.21226
Archambeault, D. R., and Yao, H. H. C. (2014). Loss of Smad4 in sertoli and leydig cells leads to testicular dysgenesis and hemorrhagic tumor formation in mice 1. Biol. Reprod. 90:62. doi: 10.1095/biolreprod.113.111393
Baker, M. A., Nixon, B., Naumovski, N., and Aitken, R. J. (2012). Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst. Biol. Reprod. Med. 58, 211–217. doi: 10.3109/19396368.2011.639844
Baker, M. A., Witherdin, R., Hetherington, L., Cunningham-Smith, K., and Aitken, R. J. (2005). Identification of post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics 5, 1003–1012. doi: 10.1002/pmic.200401100
Beauclair, G., Bridier-Nahmias, A., Zagury, J. F., Saïb, A., and Zamborlini, A. (2015). JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31, 3483–3491. doi: 10.1093/bioinformatics/btv403
Becker, J., Barysch, S. V., Karaca, S., Dittner, C., Hsiao, H. H., Diaz, M. B., et al. (2013). Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20, 525–531. doi: 10.1038/nsmb.2526
Bhagwat, S., Dalvi, V., Chandrasekhar, D., Matthew, T., Acharya, K., Gajbhiye, R., et al. (2014). Acetylated α-tubulin is reduced in individuals with poor sperm motility. Fertil. Steril. 101, 95–104. doi: 10.1016/j.fertnstert.2013.09.016
Brohi, R. D., and Huo, L. J. (2017). Post-translational modifications in spermatozoa and effects on male fertility and sperm viability. OMICS 21, 245–256. doi: 10.1089/omi.2016.0173
Brown, P. W., Hwang, K., Schlegel, P. N., and Morris, P. L. (2008). Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum. Reprod. 23, 2850–2857. doi: 10.1093/humrep/den300
Chou, K. C., Wu, Z. C., and Xiao, X. (2011). iLoc-Euk: a multi-label classifier for predicting the subcellular localization of singleplex and multiplex eukaryotic proteins. PLoS ONE 6:e18258. doi: 10.1371/journal.pone.0018258
El Shoura, S. M. (1986). Fine structure and development of the sperm of the tick Ornithodoros (Pavlovskyella) erraticus (Ixodoidea, Argasidae). J. Morphol. 190, 63–71. doi: 10.1002/jmor.1051900106
Ferlin, A., Menegazzo, M., Gianesello, L., Selice, R., and Foresta, C. (2012). Effect of relaxin on human sperm functions. J. Androl. 33, 474–482. doi: 10.2164/jandrol.110.012625
Feugang, J. M., Rodríguez-Muñoz, J. C., Dillard, D. S., Crenshaw, M. A., Willard, S. T., and Ryan, P. L. (2015a). Beneficial effects of relaxin on motility characteristics of stored boar spermatozoa. Reprod. Biol. Endocrinol. 13:24. doi: 10.1186/s12958-015-0021-4
Feugang, J. M., Greene, J. M., Sanchez-Rodríguez, H. L., Stokes, J. V., Crenshaw, M. A., Willard, S. T., et al. (2015b). Profiling of relaxin and its receptor proteins in boar reproductive tissues and spermatozoa. Reprod. Biol. Endocrinol. 13:46. doi: 10.1186/s12958-015-0043-y
Geer, L. Y., Marchler-Bauer, A., Geer, R. C., Han, L., He, J., He, S., et al. (2010). The NCBI biosystems database. Nucleic Acids Res. 38, D492–D496. doi: 10.1093/nar/gkp858
Geiss-Friedlander, R., and Melchior, F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell. Biol. 8, 947–956. doi: 10.1038/nrm2293
Girdwood, D. W., Tatham, M. H., and Hay, R. T. (2004). SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 15, 201–210. doi: 10.1016/j.semcdb.2003.12.001
Gur, Y., and Breitbart, H. (2006). Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 20, 411–416. doi: 10.1101/gad.367606
Hofmann, W. A., Arduini, A., Nicol, S. M., Camacho, C. J., Lessard, J. L., Fuller-Pace, F. V., et al. (2009). SUMOylation of nuclear actin. J. Cell Biol. 186, 193–200. doi: 10.1083/jcb.200905016
Hu, Z. L., Bao, J., and Reecy, J. M. (2008). CateGOrizer: a web-based program to batch analyze gene ontology classification categories. Online J. Bioinformatics 9, 108–112.
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Hung, L. H., Kristiyanto, D., Lee, S. B., and Yeung, K. Y. (2016). GUIdock: using Docker containers with a common graphics user interface to address the reproducibility of research. PLoS ONE 11:e0152686. doi: 10.1371/journal.pone.0152686
Huntley, R. P., Sawford, T., Martin, M. J., and O'Donovan, C. (2014). Understanding how and why the Gene Ontology and its annotations evolve: the GO within UniProt. Gigascience 3:4. doi: 10.1186/2047-217X-3-4
Hutt, D. M., Baltz, J. M., and Ngsee, J. K. (2005). Synaptotagmin, VI and VIII and syntaxin 2 are essential for the mouse sperm acrosome reaction. J. Biol. Chem. 280, 20197–20203. doi: 10.1074/jbc.M412920200
Jiang, H., and Ganesan, N. (2016). CUDAMPF: a multi-tiered parallel framework for accelerating protein sequence search in HMMER on CUDA-enabled GPU. BMC Bioinformatics 17:106. doi: 10.1186/s12859-016-0946-4
Kim, K. S., Cha, M. C., and Gerton, G. L. (2001). Mouse sperm protein sp56 is a component of the acrosomal matrix. Biol. Reprod. 64, 36–43. doi: 10.1095/biolreprod64.1.36
Kumar, R., Singh, V. K., and Atreja, S. K. (2014). Glutathione-S-transferase: role in buffalo (Bubalus bubalis) sperm capacitation and cryopreservation. Theriogenology 81, 587–598. doi: 10.1016/j.theriogenology.2013.11.012
Lena, L., Tourmente, M., and Roldan, E. R. (2016). Sexual selection of protamine 1 in mammals. Mol. Biol. Evol. 33, 174–184. doi: 10.1093/molbev/msv209
Lin, W. Z., Fang, J. A., Xiao, X., and Chou, K. C. (2013). iLoc-Animal: a multi-label learning classifier for predicting subcellular localization of animal proteins. Mol. BioSyst. 9, 634–644. doi: 10.1039/c3mb25466f
Long, J., Wang, G., He, D., and Liu, F. (2004). Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 379, 23–29. doi: 10.1042/bj20031867
Marchiani, S., Tamburrino, L., Giuliano, L., Nosi, D., Sarli, V., Gandini, L., et al. (2011). Sumo1-ylation of human spermatozoa and its relationship with semen quality. Int. J. Androl. 34, 581–593. doi: 10.1111/j.1365-2605.2010.01118.x
Marchiani, S., Tamburrino, L., Ricci, B., Nosi, D., Cambi, M., Piomboni, P., et al. (2014). SUMO1 in human sperm: new targets, role in motility and morphology and relationship with DNA damage. Reproduction 148, 453–467. doi: 10.1530/REP-14-0173
Mi, H., Poudel, S., Muruganujan, A., Casagrande, J. T., and Thomas, P. D. (2016). PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44, D336–D342. doi: 10.1093/nar/gkv1194
Moreno, S. G., Attali, M., Allemand, I., Messiaen, S., Fouchet, P., Coffigny, H., et al. (2010). TGFβ signaling in male germ cells regulates gonocyte quiescence and fertility in mice. Dev. Biol. 342, 74–84. doi: 10.1016/j.ydbio.2010.03.007
Oikonomopoulou, I., Patel, H., Watson, P. F., and Chantler, P. D. (2009). Relocation of myosin and actin, kinesin and tubulin in the acrosome reaction of bovine spermatozoa. Reprod. Fertil. Dev. 21, 364–377. doi: 10.1071/RD08166
Oliva, R., de Mateo, S., and Estanyol, J. M. (2009). Sperm cell proteomics. Proteomics 9, 1004–1017. doi: 10.1002/pmic.200800588
Python Software Foundation (2015). Python Language Reference, version 3.5.0. Available online at: https://www.python.org
Pixton, K. L., Deeks, E. D., Flesch, F. M., Moseley, F. L., Björndahl, L., Ashton, P. R., et al. (2004). Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: case report. Hum. Reprod. 19, 1438–1447. doi: 10.1093/humrep/deh224
Puri, P., Myers, K., Kline, D., and Vijayaraghavan, S. (2008). Proteomic analysis of bovine sperm YWHA binding partners identify proteins involved in signaling and metabolism. Biol. Reprod. 79, 1183–1191. doi: 10.1095/biolreprod.108.068734
Rahman, M. S., Lee, J.-S., Kwon, W.-S., and Pang, M.-G. (2013). Sperm proteomics: road to male fertility and contraception. Int. J. Endocrinol. 10, 360–986. doi: 10.1155/2013/360986
Ramalho-Santos, J., Schatten, G., and Moreno, R. D. (2002). Control of membrane fusion during spermiogenesis and the acrosome reaction. Biol. Reprod. 67, 1043–1051. doi: 10.1095/biolreprod67.4.1043
R Core Team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Renata, V. R., Federico, C., Saccone, S., Bonaccorsi, B., and Vitale, D. G. (2016). Fluorescence microscopy study on the cytoskeletal displacements during sperm differentiation in the bush-cricket Tylopsis liliifolia (Fabricius)(Orthoptera: Tettigoniidae). Microsc. Res. Tech. 79, 81–88. doi: 10.1002/jemt.22608
Rogers, R. S., Inselman, A., Handel, M. A., and Matunis, M. J. (2004). SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma 113, 233–243. doi: 10.1007/s00412-004-0311-7
Sang, L., Yang, W. C., Han, L., Liang, A. X., Hua, G. H., Xiong, J. J., et al. (2011). An immunological method to screen sex-specific proteins of bovine sperm. J. Dairy Sci. 94, 2060–2070. doi: 10.3168/jds.2010-3350
Saxena, D., Oh-Oka, T., Kadomatsu, K., Muramatsu, T., and Toshimori, K. (2002). Behaviour of a sperm surface transmembrane glycoprotein basigin during epididymal maturation and its role in fertilization in mice. Reproduction 123, 435–444. doi: 10.1530/rep.0.1230435
Seeler, J. S., and Dejean, A. (2003). Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4, 690–699. doi: 10.1038/nrm1200
Snider, N. T., Weerasinghe, S. V., I-iguez-Lluhí, J. A., Herrmann, H., and Omary, M. B. (2011). Keratin hypersumoylation alters filament dynamics and is a marker for human liver disease and keratin mutation. J. Biol. Chem. 286, 2273–2284. doi: 10.1074/jbc.M110.171314
Song, J., Durrin, L. K., Wilkinson, T. A., Krontiris, T. G., and Chen, Y. (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378. doi: 10.1073/pnas.0403498101
Stankovic-Valentin, N., Deltour, S., Seeler, J., Pinte, S., Vergoten, G., Guérardel, C., et al. (2007). An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved ψKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol. Cell. Biol. 27, 2661–2675. doi: 10.1128/MCB.01098-06
Stanton, P. G., Sluka, P., Foo, C. F., Stephens, A. N., Smith, A. I., McLachlan, R. I., et al. (2012). Proteomic changes in rat spermatogenesis in response to in vivo androgen manipulation; impact on meiotic cells. PLoS ONE 7:e41718. doi: 10.1371/journal.pone.0041718
Thomsen, M. C. F., and Nielsen, M. (2012). Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287. doi: 10.1093/nar/gks469
Vigodner, M., and Morris, P. L. (2005). Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev. Biol. 282, 480–492. doi: 10.1016/j.ydbio.2005.03.034
Vigodner, M., Ishikawa, T., Schlegel, P. N., and Morris, P. L. (2006). SUMO-1, human male germ cells development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am. J. Physiol. Endocrinol. Metab. 290, E1022–E1033. doi: 10.1152/ajpendo.00527.2005
Vigodner, M., Shrivastava, V., Gutstein, L. E., Schneider, J., Nieves, E., Goldstein, M., et al. (2013). Localization and identification of sumoylated proteins in human sperm: excessive sumoylation is a marker of defective spermatozoa. Hum. Reprod. 28, 210–223. doi: 10.1093/humrep/des317
Von Mering, C., Huynen, M., Jaeggi, D., Schmidt, S., Bork, P., and Snel, B. (2003). STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261. doi: 10.1093/nar/gkg034
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6:359. doi: 10.1038/nmeth.1322
Wloga, D., Dave, D., Meagley, J., Rogowski, K., Jerka-Dziadosz, M., and Gaertig, J. (2010). Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell 9, 184–193. doi: 10.1128/EC.00176-09
Xiao, Y., Pollack, D., Andrusier, M., Levy, A., Callaway, M., Nieves, E., et al. (2016). Identification of cell-specific targets of sumoylation during mouse spermatogenesis. Reproduction 151, 149–166. doi: 10.1530/REP-15-0239
Yan, M., Zhang, X., Pu, Q., Huang, T., Xie, Q., Wang, Y., et al. (2016). Immunoglobulin G expression in human sperm and possible functional significance. Sci. Rep. 6:20166. doi: 10.1038/srep20166
Yeh, E. T. (2009). SUMOylation and De-SUMOylation: wrestling with life's processes. J. Biol. Chem. 284, 8223–8227. doi: 10.1074/jbc.R800050200
Yurchenko, V., Xue, Z., and Sadofsky, M. J. (2006). SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell. Biol. 26, 1786–1794. doi: 10.1128/MCB.26.5.1786-1794.2006
Zhang, Z., Kostetskii, I., Tang, W., Haig-Ladewig, L., Sapiro, R., Wei, Z., et al. (2006). Deficiency of SPAG16L causes male infertility associated with impaired sperm motility. Biol. Reprod. 74, 751–759. doi: 10.1095/biolreprod.105.049254
Zhao, Q., Xie, Y., Zheng, Y., Jiang, S., Liu, W., Mu, W., et al. (2014). GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330. doi: 10.1093/nar/gku383
Keywords: Bubalus bubalis, spermatozoa, protein, post translational modification, SUMO-1
Citation: Brohi RD, Wang L, Hassine NB, Cao J, Talpur HS, Wu D, Huang C-J, Rehman Z-U, Bhattarai D and Huo L-J (2017) Expression, Localization of SUMO-1, and Analyses of Potential SUMOylated Proteins in Bubalus bubalis Spermatozoa. Front. Physiol. 8:354. doi: 10.3389/fphys.2017.00354
Received: 16 February 2017; Accepted: 15 May 2017;
Published: 13 June 2017.
Edited by:
Brett Nixon, University of Newcastle, AustraliaReviewed by:
Elisabetta Baldi, University of Florence, ItalyMargarit Vigodner, Yeshiva University, United States
Copyright © 2017 Brohi, Wang, Hassine, Cao, Talpur, Wu, Huang, Rehman, Bhattarai and Huo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Jun Huo, ljhuo@mail.hzau.edu.cn; lijunhuo@yahoo.com
 Rahim Dad Brohi
Rahim Dad Brohi Li Wang1,2
Li Wang1,2 Chun-Jie Huang
Chun-Jie Huang Li-Jun Huo
Li-Jun Huo