- 1State Key Laboratory of Crop Stress, Northwest A&F University, Yangling, China
- 2College of Biology and Food Engineering, Anyang Institute of Technology, Anyang, China
- 3State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Olfaction in insects has a critical role in recognizing the host, finding food, and choosing mating partners, as well as avoiding predators. Odorant receptors (ORs), which are housed in the dendritic membrane of sensory neurons and extended into the lymph of sensilla on insect antennae, are participating in the detection of volatile compounds in insects. In the present study, we identified an OR gene, named MsepOR13, in the oriental armyworm Mythimna separata (Walker). Quantitative real-time polymerase chain reaction revealed that MsepOR13 was expressed mainly in the antennae of male and female moths. In in vitro heterologous expression experiments, MsepOR13 was widely tuned to 32 of the 67 different compounds tested. Furthermore, MsepOR13 responded to eugenol at a low concentration of 10-9 M, with an EC50 value of 3.91 × 10-6 M. The high sensitivity suggests an important role for the OR13 gene in the moth olfactory system.
Introduction
Chemoreception of odorants in the environment is critically important for the survival of insects. During evolution, insects have evolved a powerful sense of olfaction to locate hosts and mating partners, identify oviposition sites, discriminate toxic food, and escape predators (Schneider, 1969; Bruce et al., 2005; Bruyne and Baker, 2008; Hansson and Stensmyr, 2011; Gadenne et al., 2016), as they are surrounded by various chemical compounds emitted from conspecifics, predators, and host plants (Bentley and Day, 1989; Schneider, 1992; Hansson and Stensmyr, 2011). These odorants are diffused to the surface on olfactory appendages, which mainly consisting of antennae and maxillary palps (Steinbrecht, 1997), and enter the lymph through pores of the sensilla, which are hair-like structures. Odorant molecules interact with odorant-binding proteins (OBPs) in the sensilla lymph and are transferred toward the dendrites of olfactory sensory neurons (OSNs), where odorant receptors (ORs) are expressed. Activation of ORs leads to chemical information being transduced to electrical signals, which are conveyed to the antennal lobe and finally decoded by the insect brain (Vogt, 2003; Leal, 2013).
Owing to the availability of the Drosophila melanogaster genome sequence, the first insect OR was identified in D. melanogaster based on the homology of OR sequences in vertebrates and nematodes and the restricted expression of these genes in olfactory tissues (Clyne et al., 1999; Vosshall et al., 1999). Compared with G-protein coupled receptors (GPCRs), insect ORs have the opposite membrane topology, with their N-terminus inside and their C-terminus outside the cell; this is an inverse membrane topology to that found in vertebrate ORs (Buck and Axel, 1991; Benton et al., 2006; Fleischer et al., 2017; Butterwick et al., 2018). It is now generally accepted that insect ORs transduce chemical signals by forming heteromeric complexes with an OR co-receptor (Orco) that operate as non-selective cation channels (Koji et al., 2008; Wicher et al., 2008).
In recent decades, with progress in sequencing technology and bioinformatics tools, numerous ORs have been reported in many species from various insect orders, including Lepidoptera, Diptera, Hymenoptera, Coleoptera, Hemiptera, Orthoptera, and Phthiraptera. The number of OR genes varies considerably among insect species. For example, there are 65 ORs in Helicoverpa armigera (Liu et al., 2012; Zhang et al., 2015a) and 62 ORs in Mythimna separata (Du et al., 2018), based on antennal transcriptomic analysis, whereas 163 ORs have been obtained from the genome of Apis mellifera (Robertson and Wanner, 2006) and 256 ORs have been identified in the genome of Tribolium castaneum (Engsontia et al., 2008). The variation in number of ORs between insects is assumed to correlate with evolutionary adaption to certain ecological and physiological demands (Fleischer et al., 2017).
Although increasing numbers of OR genes have been identified during recent decades, the functional characterization of the encoded proteins lags significantly behind. Heterologous in vitro expression systems, such as cultured cell lines and Xenopus oocytes, and in vivo expression systems, such as the “empty neuron system” of Drosophila, have been successfully established for functional analysis of insect ORs (Dobritsa et al., 2003; Gonzalez et al., 2016; Wang et al., 2016). These systems have been applied for functional characterization of both pheromone and non-pheromone receptors in several species, including D. melanogaster (Hallem et al., 2004; Kreher et al., 2005; Hallem et al., 2006), Anopheles gambiae (Lu et al., 2007; Carey et al., 2010; Wang et al., 2010), B. mori (Sakurai et al., 2004, 2011; Nakagawa et al., 2005; Grosse-Wilde et al., 2011), Heliothis virescens (Ewald et al., 2010; Wang et al., 2011), Ostrinia nubilalis (Wanner et al., 2010; Yuji et al., 2011; Leary et al., 2012), O. furnacalis (Miura et al., 2010; Liu W. et al., 2018), Spodoptera littoralis (de Fouchier et al., 2017), Cydia pomonella (Bengtsson et al., 2014; Gonzalez et al., 2015; Cattaneo et al., 2017), H. armigera (Liu Y. et al., 2013; Cao et al., 2016; Chang et al., 2016; Di et al., 2017), H. assulta (Chang et al., 2016; Cui et al., 2018), Plutella xylostella (Sun et al., 2013; Liu Y. et al., 2018), S. exigua (Liu C. et al., 2013; Liu et al., 2014), and S. litura (Zhang et al., 2015b).
The oriental armyworm M. separata (Walker) (Lepidoptera: Noctuidae) is an economically important and common lepidopteran pest, which is widely distributed in eastern Asia and Australia, and attacks many crop plants including maize, sorghum, and rice. M. separata migrates long distances, resulting in widespread incidence, which can lead to complete crop loss (Sharma and Davies, 1983; Jiang et al., 2011). In recent years, M. separata has been observed in many regions of China and poses a severe threat to corn production. In order to control this pest, high doses of insecticides are often applied; however, this has some negative effects, including environmental pollution, insect resistance, and harm to non-target organisms (Lv et al., 2014; Duan et al., 2017). Outbreaks of M. separata represent a great challenge in crop protection worldwide (Liu et al., 2017).
Compared with the use of chemical pesticides, olfactory-baited trapping is an effective and environmentally friendly method to manage M. separata. The sex pheromone of M. separata has been used in this way (Wei, 1985; Zhu et al., 1987), but the effect was unsatisfactory for unknown reasons. Pterocarya stenoptera and Salix babylonica are also used to attract M. separata in the field (Lihuang et al., 2017), although the mechanism of attraction is unknown. In previous work, we identified the ORs in M. separata using transcriptomic analysis (Du et al., 2018), but no study on their function has been reported except for MsepOR1, responding to the major sex pheromone compound Z11-16:Ac (Mitsuno et al., 2010). In the present study, we cloned an OR, named MsepOR13, in M. separata and analyzed the expression patterns in different tissues of both sexes by quantitative real-time polymerase chain reaction (qRT-PCR). Functional analysis was completed using in vitro expression in a Xenopus oocyte system with two-electrode, voltage-clamp physiological recordings.
Materials and Methods
Insect Rearing
The M. separata colony, maintained at the laboratory of Henan Agricultural University, Zhengzhou, China, was reared on an artificial diet at 28 ± 1°C, 70% ± 5% relative humidity, and a 14 h:10 h light:dark (L:D) photoperiod. Adult male and female moths were fed with 10% sugar solution.
RNA Extraction and cDNA Synthesis
Male and female antennae, proboscises, labial palps, and legs (a mixture of female and male) of virgin male or female individuals were collected 3 days after eclosion, immediately frozen in liquid nitrogen, and stored at -70°C until RNA extraction. Total RNA of 20 adult male or female moths was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. Total RNA was dissolved in RNase-free water and gel electrophoresis was performed to assess its integrity. RNA concentration and purity were determined on a Nanodrop ND-2000 spectrophotometer (NanoDrop products, Wilmington, DE, United States).
First, total RNA was treated with DNase I (Fermentas, Glen Burnie, MD, United States) for 30 min at 37°C to remove residual gDNA. Then, 1 μg total RNA was used to synthesize single-stranded cDNA as per the First Strand cDNA Synthesis Kit (Fermentas) manufacturer’s instructions. The cDNA of antennal samples was used as a template to clone the MsepOR13 gene. The cDNA samples isolated from different female and male tissue types were used as templates for RT-qPCR.
Cloning of MsepOR13 Gene From M. separata
The sequence of MsepOR13 was identified in M. separata by transcriptomic analysis (Du et al., 2018). Specific primers were designed by Primer 5.0 (PREMIER Biosoft International, Palo Alto, CA, United States) to clone the full-length sequence of MsepOR13 (Table 1). Antennal cDNA from female and male moths was used to amplify the full-length sequence of MsepOR13 using primeSTAR HS (Premix) (TaKaRa, Dalian, China). PCR reactions of 50 μL contained 25 μL 2×primeSTAR HS (Premix), 1.5 μL sense and anti-sense primers (10 μM), 2 μL cDNA, and 20 μL double-distilled H2O. Reactions were carried out under the following conditions: 95°C for 3 min; 35 cycles of 95°C for 30 s, 57°C for 30 s, and 72°C for 1 min; and 72°C for 10 min; before being held at 16°C. PCR products were analyzed on a 1.5% agarose gel and the sequence was sub-cloned to the vector pEASY-Blunt (TransGene, Beijing, China). The sequencing was completed in Sangon Biotech, Shanghai, China.
Sequence Analysis
The amino acid sequence of MsepOR13 was determined using the ExPASy-Translate tool1. The sequence was aligned with ORs from Peridroma saucia (PsauOR, GenBank: AVF19631.1), Athetis lepigone (AlepOR19, GenBank: AOE48024.1), and Athetis dissimilis (AdisOR31, GenBank: ALM26220.1) using DNAMAN version 8 (Lynnon LLC, San Ramon, CA, United States).
Tissue Expression Profile of MsepOR13
Quantitative polymerase chain reaction was performed to determine the expression of MsepOR13. Male and female antennae, proboscises, labial palps, and legs (a mixture of female and male) were collected from 3-day-old M. separata adults after eclosion. RNA extraction and cDNA synthesis were performed following the protocol described above. MsepRPS3 was chosen as the reference gene. The primers are listed in Table 1. GoTaq qPCR Master Mix (Promega, Madison, WI, United States) was used for qPCR, and the reactions were carried out on an Applied Biosystems 7500 Fast Real-Time PCR System (ABI, Carlsbad, CA, United States). The reactions (20 μL) consisted of 10 μL GoTaq qPCR Master Mix, 0.8 μL gene primer (10 μM), 1 μL cDNA, and 7.4 μL RNase-free water. The reactions were carried out under the following conditions: 95°C for 2 min; 40 cycles of 95°C for 15 s, and 60°C for 50 s. Each qPCR reaction was performed in triplicate with three independent biological samples to check reproducibility. The melting curves were inspected to check the specificity of the primers, and the amplification efficiencies were calculated by the standard curve method. The efficiency of the primers for MsepOR13 and MsepRPS3 were 97 and 105%, respectively. MsepOR13 relative expression levels were analyzed using the relative 2-ΔΔCT quantitation method, where ΔCT = CT (MsepOR13) – CT (MsepRPS3), ΔΔCT = ΔCT (different samples) – ΔCT (legs (female and male mixture)). Statistical comparison of expression of MsepOR13 was assessed using one-way nested analysis of variance (ANOVA), followed by least-significant difference (LSD) tests.
MsepOR13 Expression in Xenopus Oocytes and Electrophysiological Recordings
The full-length MsepOR13 was first cloned into a pEASY-Blunt vector and then ligated into a pT7Ts expression vector using primers containing Apa I (GGGCCC) and Not I (GCGGCCGC) sites. The expression vector was linearized using Sma I (CCCGGG) (Fermentas, Glen Burnie, MD, United States) and the cRNA was synthesized using an mMESSAGE mMACHINE T7 kit (Ambion, Austin, TX, United States). Mature healthy Xenopus oocytes (stages V–VII) were incubated with 2 mg/mL collagenase I in pH 7.6 washing buffer consisting of 96 mM NaCl, 2 mM KCl, 5 mM MgCl2, and 5 mM HEPES at room temperature for about 1 h until almost of them were separated a signal one. Then, 27.6 ng MsepOR13 cRNA and 27.6 ng MsepOrco cRNA were microinjected together into oocytes, and the oocytes were cultured in 1× Ringer’s buffer (washing buffer supplemented with 0.8 mM CaCl2, 5% dialyzed horse serum, 50 mg/mL tetracycline, 100 mg/mL streptomycin, and 550 mg/mL sodium pyruvate) for 4–7 days. The whole-cell currents of injected oocytes were recorded with an OC-725C oocyte clamp at a holding potential of -80 mV (Warner Instruments, Hamden, CT, United States), following previously described experimental procedures (Cui et al., 2018; Liu W. et al., 2018; Liu Y. et al., 2018). Oocytes were exposed to different compounds at 10-4 M for 15 s each, in a random order, with intervals between exposures that allowed the current to return to baseline. Dose–response curves were acquired from 10-9 to 10-4 M in ascending order of concentration. All data acquisition and analysis were carried out with Digidata 1440A and Pclamp10.0 (Axon Instruments, Inc., Union City, CA, United States), and dose–response data were analyzed using GraphPad Prism 5. Statistical comparison of responses to different odors of MsepOR13 was assessed using ANOVA, followed by LSD tests.
Odorant Panel
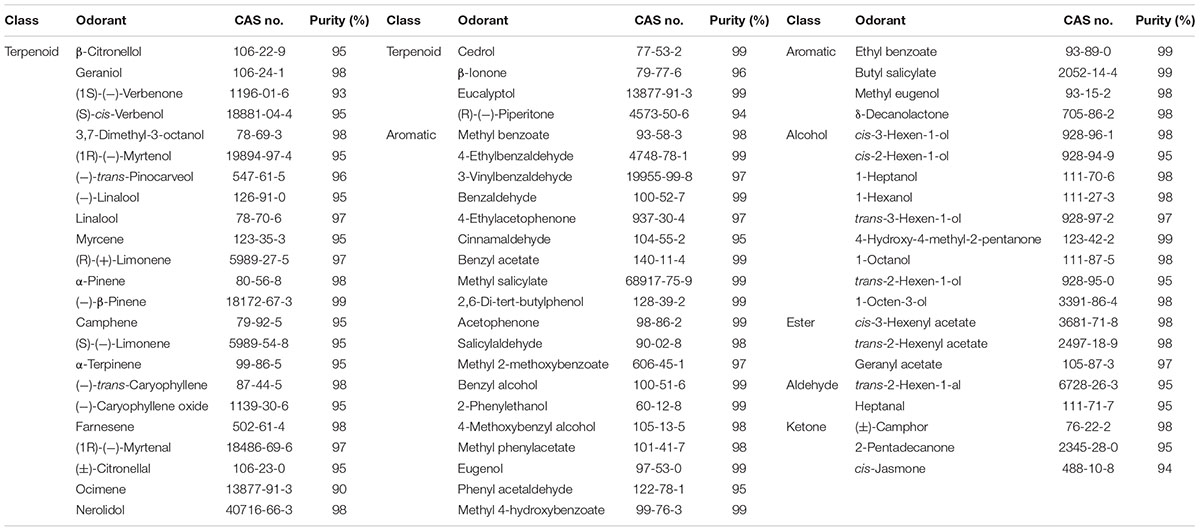
Sixty-seven plant volatile compounds purchased from Sigma-Aldrich were used in this experiment (Table 2) and were classified into six groups: terpenoid, aromatic, alcohol, ester, aldehyde, and ketone. All compounds were dissolved in dimethyl sulfoxide (DMSO) at a concentration of 1 M as stock solutions. Before the experiments, the stock solutions were diluted in 1× Ringer’s buffer to working concentrations, and 1× Ringer’s buffer containing 0.1% DMSO was used as a negative control.
Results
Gene Cloning and Sequence Analysis of MsepOR13
Based on the transcriptome of M. separata (Du et al., 2018), we obtained the full-length sequence of MsepOR13. It contained 1227 bp, encoding 408 amino acids (Figure 1) Three amino acid sequences from P. saucia (PsauOR, GenBank Accession No. AVF19631.1), A. lepigone (AlepOR19, GenBank Accession No. AOE48024.1), and A. dissimilis (AdisOR31, GenBank Accession No. ALM26220.1) were aligned with MsepOR13 (Figure 2) and found to have 84, 81, and 83% identity, respectively.

Figure 2. Alignment of the amino acid sequences of MsepOR13 to those of its homologs in other species. Amino acids identical in all sequences are marked with black shading. Numbers to the right refer to the position of the last residue in a line in each odorant receptor (OR) sequence. The horizontal lines indicate the position of predicted transmembrane domains.
Tissue Expression Profiles of MsepOR13
Quantitative polymerase chain reaction was carried out to evaluate the expression profile of MsepOR13 in different tissues of both sexes in M. separata. The results showed that MsepOR13 was mainly expressed in antennae compared with other tissues and exhibited much higher relative expression level in female antennae than male antennae (Figure 3). MsepOR13 was less expressed in proboscis and labial palp in both sexes and there was no significant difference in the expression levels of MespOR13 between leg (mixture of female and male moths) and female proboscis.
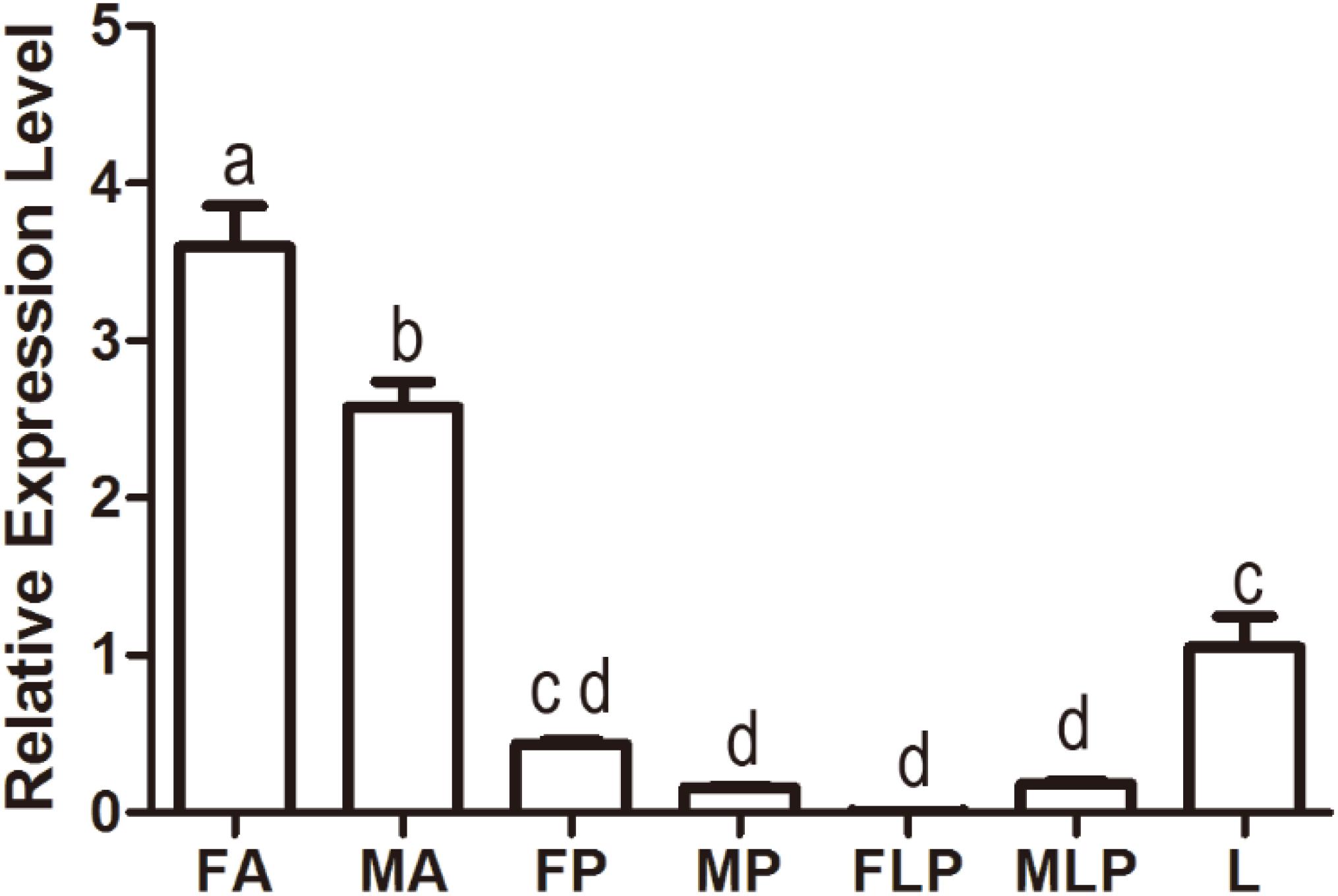
Figure 3. Tissue- and sex-specific expression of MsepOR13 in M. separata. FA, female antennae; MA, male antennae; FP, female proboscis; MP, male proboscis; FLP, female labial palp; MLP, male labial palp; L, legs (both sexes mixed). Error bars represent the standard error; those labeled with different letters are significantly different (p < 0.05, ANOVA, LSD).
Functional Characterization of MsepOR13 in the Xenopus Oocyte Expression System
The Xenopus oocyte expression system was used to identify candidate ligands for MespOR13. The cRNA of MsepOR13 and MespOrco were co-injected into Xenopus oocytes, and responses to 67 compounds were recorded using a two-electrode voltage clamp. MsepOR13 was tuned to 32 odorants from all six classes and was most sensitive to eugenol, with responses of about 3011 nA (Figures 4A,B,D). In addition, methyl eugenol and methyl phenylacetate elicited the second strongest responses, of about 1655 and 1150 nA, respectively (Figures 4A,D). Interestingly, these three main legends shared similar structure, a benzene ring (Figure 4C). The other 29 odorants elicited the same response level. Acetophenone elicited a relatively higher response (523.3 nA) and 1-hexanol elicited the lowest response with an amplitude of 60 nA (Figures 4A,D). In the dose–response study, Xenopus oocyte co-expressing MsepOR13/MsepORco responded to 10-9 M of eugenol and the peak amplitude occurred at the concentration of 10-5 M (Figure 4E). The EC50 value of eugenol was 3.91 × 10-6 M (Figure 4F).
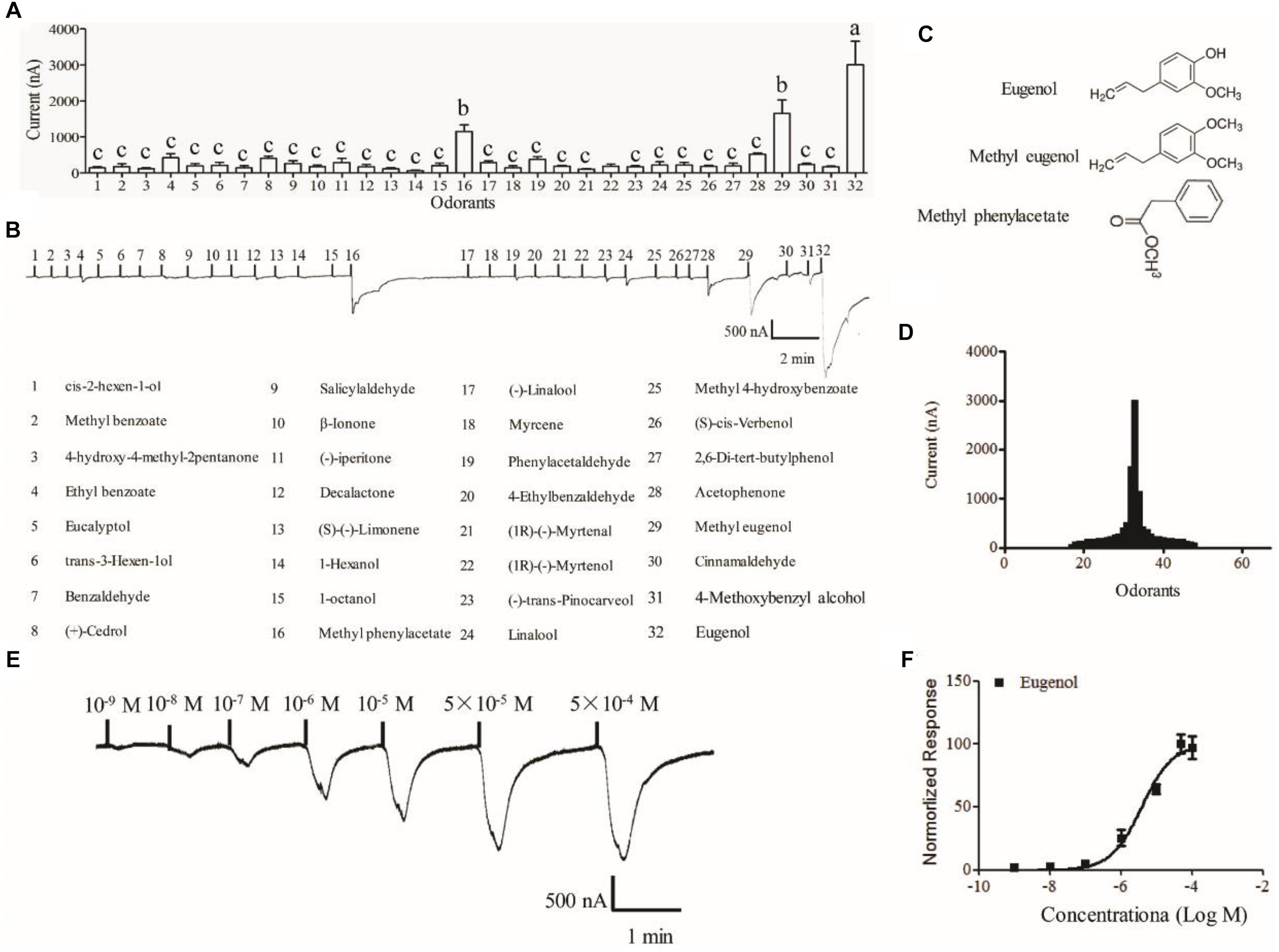
Figure 4. Functional response of Xenopus oocytes, with co-expressed MsepOR13/Orco, to volatile compounds. (A) Response profile of MsepOR13/Orco Xenopus oocytes. Error bars indicate standard error of the mean and bars labeled with different letters are significantly different (p < 0.001, ANOVA, LSD, N = 3–8); (B) responses of MsepOR13/Orco Xenopus oocytes to odorants in 10-4 M solution; (C) structure of the main ligands of MsepOR13. (D) Tuning curve for the MsepOR13 for an odorant panel comprising 67 odorants, arranged along the x-axis according to the strength of the response they elicit. The odorants that elicited the strongest responses were placed near the center of the distribution, while those that elicit the weakest responses were placed near the edges. (E) MsepOR13/Orco Xenopus oocytes stimulated with a range of eugenol concentrations. (F) Dose–response curve of MsepOR13/Orco Xenopus oocytes with eugenol. Responses were normalized by defining the maximal response as 100. The response value is given as mean ± standard error (N = 6).
Discussion
Detection of chemical odors in the environment is essential for the survival of insects. Accordingly, insects have evolved remarkable sensitive and discriminatory olfactory systems for locating hosts and food sources, identifying mating partners and oviposition sites, or escaping predators (Schneider, 1969; Hansson and Stensmyr, 2011; Gadenne et al., 2016). Previous studies have shown that ORs play an important part in the recognition of odorants and the process of chemo-electrical transduction (Leal, 2013; Wicher, 2014; Bohbot and Pitts, 2015). In this study, we cloned an OR gene, MsepOR13, in M. separata. The sequence contained 1227 bp, encoding 408 amino acids. As showed in the qPCR experiment, MsepOR13 exhibited female antennae-biased expression, which suggested that it might play a vital role in regulating female-specific behaviors, such as oviposition sites selection (Liu et al., 2015). Meanwhile, we found MsepOR13 was also expressed in legs indicating that legs might assist insects to choose suitable oviposition sites. Previous studies found that female butterflies perceive oviposition stimulant by their foreleg tarsus and further determine the suitable feeding plant for larvae in Papilio polytes (Nakayama et al., 2003). Furtherly, 4 ORs were also identified by the legs transcriptome analysis in Ectropis obliqua (Ma et al., 2016), indicating that ORs expressed in legs was a ubiquitous phenomenon. During the past decade, the sex pheromone receptors have been well-deorphanized in many Lepidoptera species. However, the identification of ligands for the non-pheromone receptor ORs has significantly lagged behind, except for a few species such as D. melanogaster (Hallem et al., 2004; Kreher et al., 2005; Hallem et al., 2006), A. gambiae (Lu et al., 2007; Carey et al., 2010; Wang et al., 2010), and S. littoralis (Montagné et al., 2012; de Fouchier et al., 2017). In this study, MsepOR13 responded to 32 odorants and only three ligands elicited relative large response; this phenomenon was also found in studies of S. littoralis (de Fouchier et al., 2017) and H. armigera (Di et al., 2017). Narrowly tuned receptors are thought to be important in the detection of odors of high biological salience. In D. melanogaster, several ORs selectively responded to odors that are necessary and sufficient for vital behaviors such as avoiding toxic microbes and choosing oviposition sites (Stensmyr et al., 2012; Dweck et al., 2013, 2015; Ronderos et al., 2014). In mosquitoes, receptors that selectively respond to human emanations play a crucial part in host recognition and blood feeding (Hughes et al., 2010; Mcbride et al., 2014). Sex pheromone perception in moths also involves such specific pathways (Miura et al., 2010; Liu Y. et al., 2018). The homolog of MsepOR13 in S. littoralis, SlitOR31 shared 80% amino acid identity with MsepOR13. But SlitOR31 was narrowly tuned to eugenol, which is different from the function of MsepOR13. The difference of their function might relate with the different environment and the selective pressures they face.
In M. separata, the three main ligands containing a benzene ring were structurally similar; a similar phenomenon has been found in functional studies of ORs in A. gambiae ((Wang et al., 2010), S. littoralis (de Fouchier et al., 2017), and H. armigera (Di et al., 2017). Among all the ligands, eugenol activated the strongest response in MsepOR13/Orco Xenopus oocytes, and could response at a 10-9 M concentration, with an EC50 value of 3.91 × 10-6 M. Actually, MspeOR13 responding to eugenol showed the similar sensitivity with the reported pheromone receptors to sex pheromones (Liu C. et al., 2013; Chang et al., 2016; Liu W. et al., 2018; Liu Y. et al., 2018), suggesting that eugenol might be important to M. separata. It has been reported that eugenol can repel the H. armigera moth (Xu, 2004), and also repel Populus yunnanensis oviposition (Ma et al., 2016). In Tribilium castaneum, eugenol has apparently repellent activity toward adults and toxic effects on both larvae and adults (Han and Huang, 2009). However, in Mamestra brassicae, eugenol was found to attract larvae and moths (Yan, 2015). The functions of eugenol with respect to M. separata require further study, especially behavioral experiments, in order to develop environmentally friendly approaches to control this economically significant insect. Based on the high sensitivity of MsepOR13 to eugenol, we predict that MsepOR13 may have an important role in the reception of eugenol in M. separata; thus, its function could be further explored using the CRISPR-Cas9 system.
Author Contributions
KZ and YF designed the experiments. HY, KL, and NL carried out the experiments. LD and SG analyzed the experimental results. JW and GW wrote the manuscript.
Funding
This work was supported by the National Public Welfare Industry (Agriculture) Scientific Research of China (201403031), the National Key Research and Development Program of China (2017YFD0201807), and the Project of Agricultural Science and Technology Innovation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Bengtsson, J. M., Gonzalez, F., Cattaneo, A. M., Montagné, N., Walker, W. B., Bengtsson, M., et al. (2014). A predicted sex pheromone receptor of codling moth Cydia pomonella detects the plant volatile pear ester. Front. Ecol. Evol. 2:33. doi: 10.3389/fevo.2014.00033
Bentley, M. D., and Day, J. F. (1989). Chemical ecology and behavioral aspects of mosquito oviposition. Annu. Rev. Entomol. 34, 401–421. doi: 10.1146/annurev.en.34.010189.002153
Benton, R., Sachse, S., Michnick, S., and Vosshall, L. (2006). Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS. Biol. 4:e20. doi: 10.1371/journal.pbio.0040020
Bohbot, J. D., and Pitts, R. J. (2015). The narrowing olfactory landscape of insect odorant receptors. Front. Ecol. Evol. 3:39. doi: 10.3389/fevo.2015.00039
Bruce, T. J., Wadhams, L. J., and Woodcock, C. M. (2005). Insect host location: a volatile situation. Trends Plant Sci. 10, 269–274. doi: 10.1016/j.tplants.2005.04.003
Bruyne, M. D., and Baker, T. C. (2008). Odor detection in insects: volatile codes. J. Chem. Ecol. 34, 882–897. doi: 10.1007/s10886-008-9485-4
Buck, L., and Axel, R. (1991). A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187. doi: 10.1016/0092-8674(91)90418-X
Butterwick, J. A., Del Marmol, J., Kim, K. H., Kahlson, M. A., Rogow, J. A., Walz, T., et al. (2018). Cryo-EM structure of the insect olfactory receptor Orco. Nature 560, 447–452. doi: 10.1038/s41586-018-0420-8
Cao, S., Liu, Y., Guo, M., and Wang, G. (2016). A conserved odorant receptor tuned to floral volatiles in three Heliothinae species. PLoS One 11:e0155029. doi: 10.1371/journal.pone.0155029
Carey, A. F., Guirong, W., Chih-Ying, S., Zwiebel, L. J., and Carlson, J. R. (2010). Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464, 66–71. doi: 10.1093/nar/gkt484
Cattaneo, A. M., Gonzalez, F., Bengtsson, J. M., Corey, E. A., Jacquinjoly, E., Montagné, N., et al. (2017). Candidate pheromone receptors of codling moth Cydia pomonella respond to pheromones and kairomones. Sci. Rep. 7:41105. doi: 10.1038/srep41105
Chang, H., Guo, M., Bing, W., Yang, L., Dong, S., and Wang, G. (2016). Sensillar expression and responses of olfactory receptors reveal different peripheral coding in two Helicoverpa species using the same pheromone components. Sci. Rep. 6:18742. doi: 10.1038/srep18742
Clyne, P. J., Warr, C. G., Freeman, M. R., Lessing, D., Kim, J., and Carlson, J. R. (1999). A novel family of divergent seven-transmembrane proteins : candidate odorant receptors in Drosophila. Neuron 22, 327–338. doi: 10.1016/S0896-6273(00)81093-4
Cui, W. C., Bing, W., Guo, M. B., Liu, Y., Jacquin-Joly, E., Yan, S. C., et al. (2018). A receptor-neuron correlate for the detection of attractive plant volatiles in Helicoverpa assulta (Lepidoptera: Noctuidae). Insect Biochem. Mol. Biol.
de Fouchier, A., Montagnã©, N., Steiner, C., Binyameen, M., Schlyter, F., Chertemps, T., et al. (2017). Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 8:15709. doi: 10.1038/ncomms15709
Di, C., Ning, C., Huang, L. Q., and Wang, C. Z. (2017). Design of larval chemical attractants based on odorant response spectra of odorant receptors in the cotton bollworm. Insect Biochem. Mol. Biol. 84, 48–62. doi: 10.1016/j.ibmb.2017.03.007
Dobritsa, A. A., van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A., and Carlson, J. R. (2003). Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37, 827–841. doi: 10.1016/S0896-6273(03)00094-1
Du, L., Zhao, X., Liang, X., Gao, X., Liu, Y., and Wang, G. (2018). Identification of candidate chemosensory genes in Mythimna separata by transcriptomic analysis. BMC Genomics 19:518. doi: 10.1186/s12864-018-4898-0
Duan, Y., Gong, Z., Wu, R., Miao, J., Jiang, Y., Li, T., et al. (2017). Transcriptome analysis of molecular mechanisms responsible for light-stress response in Mythimna separata (Walker). Sci. Rep. 7:45188. doi: 10.1038/srep45188
Dweck, H. M., Ebrahim, S. M., Farhan, A., Hansson, B., and Stensmyr, M. (2015). Olfactory proxy detection of dietary antioxidants in Drosophila. Curr. Biol. 25, 455–466. doi: 10.1016/j.cub.2014.11.062
Dweck, H. M., Ebrahim, S. M., Kromann, S., Bown, D., Hillbur, Y., Sachse, S., et al. (2013). Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 23, 2472–2480. doi: 10.1016/j.cub.2013.10.047
Engsontia, P., Sanderson, A. P., Cobb, M., Walden, K. K., Robertson, H. M., and Brown, S. (2008). The red flour beetle’s large nose: an expanded odorant receptor gene family in Tribolium castaneum. Insect Biochem. Mol. Biol. 38, 387–397. doi: 10.1016/j.ibmb.2007.10.005
Ewald, G. W., Thomas, G., Elisabeth, B., Heinz, B., and Jurgen, K. (2010). Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur. J. Neurosci. 25, 2364–2373. doi: 10.1111/j.1460-9568.2007.05512.x
Fleischer, J., Pregitzer, P., Breer, H., and Krieger, J. (2017). Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell Mol. Life. Sci. 75, 485–508. doi: 10.1007/s00018-017-2627-5
Gadenne, C., Barrozo, R. B., and Anton, S. (2016). Plasticity in insect olfaction: to smell or not to smell? Annu. Rev. Entomol. 61, 317–333. doi: 10.1146/annurev-ento-010715-023523
Gonzalez, F., Bengtsson, J. M., Walker, W. B., Sousa, M. F. R., Cattaneo, A. M., Montagné, N., et al. (2015). A conserved odorant receptor detects the same 1-Indanone analogs in a tortricid and a noctuid moth. Front. Ecol. Evol. 3:131. doi: 10.3389/fevo.2015.00131
Gonzalez, F., Witzgall, P., and Walker, W. B. (2016). Protocol for heterologous expression of insect odourant receptors in Drosophila. Front. Ecol. Evol. 4:24. doi: 10.3389/fevo.2016.00024
Grosse-Wilde, E., Kuebler, L. S., Bucks, S., Vogel, H., Wicher, D., and Hansson, B. S. (2011). Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 108, 7449–7454. doi: 10.1073/pnas.1017963108
Hallem, E., Dahanukar, A., and Carlson, J. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135. doi: 10.1016/j.inoche.2009.11.022
Hallem, E. A., Ho, M. G., and Carlson, J. R. (2004). The molecular basis of odor coding in the Drosophila antenna. Cell 117, 965–979. doi: 10.1016/j.cell.2004.05.012
Han, Q., and Huang, S. (2009). The bioactivity of eugenol against the red flour beetle Tribolium castaneum. J. Chongqing Normal Univ. 26, 16–19. doi: 10.3969/J.ISSN.1672-6693.2009.03.005
Hansson, B., and Stensmyr, M. (2011). Evolution of insect olfaction. Neuron 72, 698–711. doi: 10.1016/j.neuron.2011.11.003
Hughes, D. T., Pelletier, J., Luetje, C. W., and Leal, W. S. (2010). Odorant Receptor from the southern house mosquito narrowly tuned to the oviposition attractant skatole. J. Chem. Ecol. 36, 797–800. doi: 10.1007/s10886-010-9828-9
Jiang, X., Luo, L., Zhang, L., Sappington, T. W., and Hu, Y. (2011). Regulation of migration in Mythimna separata (Walker) in China: a review integrating environmental, physiological, hormonal, genetic, and molecular factors. Environ. Entomol. 40, 516–533. doi: 10.1603/EN10199
Koji, S., Maurizio, P., Takao, N., Tatsuro, N., Vosshall, L. B., and Kazushige, T. (2008). Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452, 1002–1006. doi: 10.1038/nature06850
Kreher, S. A., Jae Young, K., and Carlson, J. R. (2005). The molecular basis of odor coding in the Drosophila larva. Neuron 46, 445–456. doi: 10.1016/j.neuron.2005.04.007
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1146/annurev-ento-120811-153635
Leary, G. P., Allen, J. E., Bunger, P. L., Luginbill, J. B., Linn, C. E., Macallister, I. E., et al. (2012). Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. U.S.A. 109, 14081–14086. doi: 10.1073/pnas.1204661109
Lihuang, K., Zhang, Z., Kim, K., Huang, Q., and Lei, C. (2017). Antennal and behavioral responses of Mythimna separata (Walker) to three plant volatiles. Environ. Sci. Pollut. Res. Int. 24, 24953–24964. doi: 10.1007/s11356-017-0140-x
Liu, C., Liu, Y., Guo, M., Cao, D., Dong, S., and Wang, G. (2014). Narrow tuning of an odorant receptor to plant volatiles in Spodoptera exigua (Hübner). Insect Mol. Biol. 23, 487–496. doi: 10.1111/imb.12096
Liu, C., Liu, Y., Walker, W. B., Dong, S., and Wang, G. (2013). Identification and functional characterization of sex pheromone receptors in beet armyworm Spodoptera exigua (Hübner). Insect Biochem. Mol. Biol. 43, 747–754. doi: 10.1016/j.ibmb.2013.05.009
Liu, W., Jiang, X. C. W., Cao, S., Yang, B., and Wang, G. R. (2018). Functional studies of sex pheromone receptors in asian corn borer Ostrinia furnacalis. Front. Physiol. 9:591. doi: 10.3389/fphys.2018.00591
Liu, Y., Gu, S., Zhang, Y., Guo, Y., and Wang, G. (2012). Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLoS One 7:e48260. doi: 10.1371/journal.pone.0048260
Liu, Y., Liu, C., Lin, K., and Wang, G. (2013). Functional specificity of sex pheromone receptors in the cotton bollworm Helicoverpa armigera. PLoS One 8:e62094. doi: 10.1371/journal.pone.0062094
Liu, Y., Liu, Y., Jiang, X., and Wang, G. (2018). Cloning and functional characterization of three new pheromone receptors from the diamondback moth, Plutella xylostella. J. Insect. Physiol. 107, 14–22. doi: 10.1016/j.jinsphys.2018.02.005
Liu, Y., Sun, L., Cao, D., Walker, W., Zhang, Y., and Wang, G. (2015). Identification of candidate olfactory genes in Leptinotarsa decemlineata by antennal transcriptome analysis. Front. Ecol. Evol. 3:60. doi: 10.3389/fevo.2015.00060
Liu, Z., Wang, X., Lei, C., and Zhu, F. (2017). Sensory genes identification with head transcriptome of the migratory armyworm, Mythimna separata. Sci. Rep. 7:46033. doi: 10.1038/srep46033
Lu, T., Qiu, Y. T., Wang, G., Kwon, J. Y., Rutzler, M., Kwon, H. W., et al. (2007). Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Curr. Biol. 17, 1533–1544. doi: 10.1016/j.cub.2007.07.062
Lv, M., Wu, W., and Liu, H. (2014). Effects of fraxinellone on the midgut enzyme activities of the 5th instar larvae of oriental armyworm, Mythimna separata walker. Toxins 6, 2708–2718. doi: 10.3390/toxins6092708
Ma, Y., Zhang, X., Xu, Y., and Xiao, C. (2016). Effect of volatiles from Populus yunnanensis on oviposition preference of patato tuber moth, Phthorimaea operculell. Plant Prot. 42, 99–103. doi: 10.3969/j.issn.0529-1542.2016.02.017
Mcbride, C. S., Felix, B., Omondi, A. B., Spitzer, S. A., Joel, L., Rosemary, S., et al. (2014). Evolution of mosquito preference for humans linked to an odorant receptor. Nature 515, 222–227. doi: 10.1038/nature13964
Mitsuno, H., Sakurai, T. M., Yasuda, T., Kugimiya, S., Ozawa, R., Toyohara, H., et al. (2010). Identification of receptors of main sex-pheromone components of three Lepidopteran species. Eur. J. Neurosci. 28, 893–902. doi: 10.1111/j.1460-9568.2008.06429.x
Miura, N., Nakagawa, T.,and Ishikawa, Y. (2010). Broadly and narrowly tuned odorant receptors are involved in female sex pheromone reception in Ostrinia moths. Insect Biochem. Mol. Biol. 40, 64–73. doi: 10.1016/j.ibmb.2009.12.011
Montagné, N., Chertemps, T., Brigaud, I., Francois, A., Francois, M. C., de Fouchier, A., et al. (2012). Functional characterization of a sex pheromone receptor in the pest moth Spodoptera littoralis by heterologous expression in Drosophila. Eur. J. Neurosci. 36, 2588–2596. doi: 10.1111/j.1460-9568.2012.08183.x
Nakagawa, T., Sakurai, T., Nishioka, T., and Touhara, K. (2005). Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 307, 1638–1642. doi: 10.1126/science.1106267
Nakayama, T., Honda, K., Omura, H., and Hayashi, N. (2003). Oviposition stimulants for the tropical swallowtail butterfly, Papilio polytes, feeding on a rutaceous plant, Toddalia asiatica. J. Chem. Ecol. 29, 1621–1634. doi: 10.1023/a:1024274814402
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome. Res. 16, 1395–1403. doi: 10.1101/gr.5057506
Ronderos, D. S., Chun-Chieh, L., Potter, C. J., and Smith, D. P. (2014). Farnesol-detecting olfactory neurons in Drosophila. J. Neurosci. 34, 3959–3968. doi: 10.1523/jneurosci.4582-13.2014
Sakurai, T., Mitsuno, H., Haupt, S. S., Uchino, K., Yokohari, F., Nishioka, T., et al. (2011). A single sex pheromone receptor determines chemical response specificity of sexual behavior in the silkmoth Bombyx mori. PLoS Genetics 7:e1002115. doi: 10.1371/journal.pgen.1002115
Sakurai, T., Nakagawa, T. H., Mori, H., Endo, Y., Tanoue, S., Yasukochi, Y., et al. (2004). Identification and functional characterization of a sex pheromone receptor in the silkmoth Bombyx mori. Proc. Natl. Acad. Sci. U.S.A. 101, 16653–16658. doi: 10.1073/pnas.0407596101
Schneider, D. (1969). Insect olfaction: deciphering system for chemical messages. Science 163, 1031–1037. doi: 10.1126/science.163.3871.1031
Schneider, D. (1992). 100 years of pheromone research - an essay on Lepidoptera. Naturwissenschaften 79, 241–250. doi: 10.1007/BF01175388
Sharma, H. C., and Davies, J. C. (1983). The Oriental Armyworm, Mythimna Separata (Walker). Distribution, Biology and Control: a Literature Review. Miscellaneous Reports – Centre for Overseas Pest Research (UK). London: Overseas Development Administration.
Steinbrecht, R. A. (1997). Pore structures in insect olfactory sensilla: a review of data and concepts. Int. J. Insect Morphol. Embryol. 26, 229–245. doi: 10.1016/s0020-7322(97)00024-x
Stensmyr, M., Dweck, H. M., Farhan, A., Ibba, I., Strutz, A., Mukunda, L., et al. (2012). A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357. doi: 10.1016/j.cell.2012.09.046
Sun, M., Liu, Y., Walker, W. B., Liu, C., Lin, K., Gu, S., et al. (2013). Identification and characterization of pheromone receptors and interplay between receptors and pheromone binding proteins in the diamondback moth. Plutella xyllostella. PLoS One 8:e62098. doi: 10.1371/journal.pone.0062098
Vogt, R. G. (2003). Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. Insect Pheromone Biochem. Mol. Biol. 2003, 391–445. doi: 10.1016/B978-012107151-6/50016-5
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736. doi: 10.1016/S0092-8674(00)80582-6
Wang, B., Liu, Y., He, K., and Wang, G. (2016). Comparison of research methods for functional characterization of insect olfactory receptors. Sci. Rep. 6:32806. doi: 10.1038/srep32806
Wang, G., Vasquez, G. M., Schal, C., Zwiebel, L. J., and Gould, F. (2011). Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens. Insect Mol. Biol. 20, 125–133. doi: 10.1111/j.1365-2583.2010.01045.x
Wang, G. R., Carey, A. F., Carlson, J. R., and Zwiebel, L. J. (2010). Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc. Natl. Acad. Sci. U.S.A. 107, 4418–4423. doi: 10.1073/pnas.0913392107
Wanner, K. W., Nichols, A. S., Allen, J. E., Bunger, P. L., Garczynski, S. F., Linn, C. E., et al. (2010). Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One 5:e8685. doi: 10.1371/journal.pone.0008685
Wei, Z. H. (1985). Preliminary report on the sex pheromone of the armyworm Mythimna separata. Acta Entomol. Sin. 28, 348–350. doi: 10.16380/j.kcxb.1985.03.018
Wicher, D. (2014). Olfactory signaling in insects. Prog. Mol. Biol. Transl. Sci. 130, 37–54. doi: 10.1016/bs.pmbts.2014.11.002
Wicher, D., Schafer, R., Bauernfeind, R., Stensmyr, M. C., Heller, R., Heinemann, S. H., et al. (2008). Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1011. doi: 10.1038/nature06861
Xu, Y. (2004). The Test Of Components which trap Helicoverpa armigera and the design of contstitute. Master’s thesis, Henan Agricultural University, Henan.
Yan, H. (2015). Aritificial Breeding and control Technology of Mamestra Brassicae. Master’s thesis, Henan Agricultural University, Henan.
Yuji, Y., Nami, M., Ryo, N., Ken, S., and Yukio, I. (2011). Sex-linked pheromone receptor genes of the European corn borer, Ostrinia nubilalis, are in tandem arrays. PLoS One 6:e18843. doi: 10.1371/journal.pone.0018843
Zhang, J., Wang, B., Dong, S., Cao, D., Dong, J., Walker, W. B., et al. (2015a). Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS One 10:e0117054. doi: 10.1371/journal.pone.0117054
Zhang, J., Yan, S., Liu, Y., Jacquinjoly, E., Dong, S., and Wang, G. (2015b). Identification and functional characterization of sex pheromone receptors in the common cutworm (Spodoptera litura). Chem. Sens. 40, 7–16. doi: 10.1093/chemse/bju052
Keywords: Mythimna separata, odorant receptor, eugenol, Xenopus oocytes, odorant tuning
Citation: Zhang K, Feng Y, Du L, Gao S, Yan H, Li K, Liu N, Wu J and Wang G (2019) Functional Analysis of MsepOR13 in the Oriental Armyworm Mythimna separata (Walker). Front. Physiol. 10:367. doi: 10.3389/fphys.2019.00367
Received: 12 December 2018; Accepted: 18 March 2019;
Published: 09 April 2019.
Edited by:
Sylvia Anton, Institut National de la Recherche Agronomique (INRA), FranceReviewed by:
Nicolas Montagné, Sorbonne Universités, FranceLiang Sun, Tea Research Institute (CAAS), China
Copyright © 2019 Zhang, Feng, Du, Gao, Yan, Li, Liu, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junxiang Wu, junxw@nwsuaf.edu.cn Guirong Wang, grwang@ippcaas.cn
†These authors have contributed equally to this work
 Kunpeng Zhang1,2†
Kunpeng Zhang1,2† Junxiang Wu
Junxiang Wu Guirong Wang
Guirong Wang