- College of Veterinary Medicine, Northwest A&F University, Xianyang, China
Background: Pulmonary arterial hypertension (PAH) is a life-threatening chronic cardiopulmonary disease. However, there are limited studies reflecting the available biomarkers from separate gene expression profiles in PAH. This study explored two microarray datasets by an integrative analysis to estimate the molecular signatures in PAH.
Methods: Two microarray datasets (GSE53408 and GSE113439) were exploited to compare lung tissue transcriptomes of patients and controls with PAH and to estimate differentially expressed genes (DEGs). According to common DEGs of datasets, gene and protein overrepresentation analyses, protein–protein interactions (PPIs), DEG–transcription factor (TF) interactions, DEG–microRNA (miRNA) interactions, drug–target protein interactions, and protein subcellular localizations were conducted in this study.
Results: We obtained 38 common DEGs for these two datasets. Integration of the genome transcriptome datasets with biomolecular interactions revealed hub genes (HSP90AA1, ANGPT2, HSPD1, HSPH1, TTN, SPP1, SMC4, EEA1, and DKC1), TFs (FOXC1, FOXL1, GATA2, YY1, and SRF), and miRNAs (hsa-mir-17-5p, hsa-mir-26b-5p, hsa-mir-122-5p, hsa-mir-20a-5p, and hsa-mir-106b-5p). Protein–drug interactions indicated that two compounds, namely, nedocromil and SNX-5422, affect the identification of PAH candidate biomolecules. Moreover, the molecular signatures were mostly localized in the extracellular and nuclear areas.
Conclusions: In conclusion, several lung tissue-derived molecular signatures, highlighted in this study, might serve as novel evidence for elucidating the essential mechanisms of PAH. The potential drugs associated with these molecules could thus contribute to the development of diagnostic and therapeutic strategies to ameliorate PAH.
Introduction
Pulmonary arterial hypertension (PAH) is a rare vascular disease with an annual incidence of two cases per million (Peacock et al., 2007). PAH is defined by a mean pulmonary arterial pressure (mPAP) > 20 mmHg at rest, a pulmonary artery wedge pressure (PAWP) ≤ 15 mmHg, and a pulmonary vascular resistance (PVR) ≥ 3.0 Wood units (Gouyou et al., 2021). As an obliterative vasculopathy, PAH is characterized by high pulmonary arterial pressure, resulting in right ventricular failure and even death (Boucly et al., 2017; Tang et al., 2018). Over the past decades, the progression of effective medical treatments and the application of combined therapy have significantly improved the prognosis of patients with PAH (Sitbon et al., 2014, 2016; Galie et al., 2015). Although current diagnosis and therapy strategies have effectively ameliorated the abnormal hemodynamics and severe pulmonary vascular remodeling of PAH, and efficaciously alleviated the clinical symptoms in patients with PAH, there are still a number of patients suffering from persistent symptoms and even right heart failure (Van De Veerdonk et al., 2011, 2015). Thus, investigating PAH biomarkers is essential not just for a better understanding of PAH development but also as a key step to establish promising novel treatment strategies (Kerkhof et al., 2019).
The correlation between PAH and poor cardiorespiratory outcomes justifies the necessity of early diagnosis and treatment of this disease (Farhadi et al., 2017). The wide application of cardiac catheterization and echocardiography in clinical practice, evaluation of pulmonary artery pressure and right heart hemodynamic as well as high-resolution image data are no longer difficult to access, which significantly improve the differential diagnosis and assessment of patients with PAH (Claessen and La Gerche, 2017; Farhadi et al., 2017). Indeed, there is tremendous progress in understanding the essential pathophysiology of PAH, including the prognostic biomarkers as well as many promising therapy options (Ma et al., 2020). However, there are limited studies providing PAH-associated gene expression profiles, and a number of studies implying that there may be an extraordinary significance of investigating this type of clinical molecular biomarkers and elucidating the fundamental mechanisms involved in PAH. This may be useful in developing a new scientific-based diagnosis modality and performing targeted therapy in patients with PAH more precisely (Farhadi et al., 2017; Sullivan and Kass, 2019; Ma et al., 2020; Maremanda et al., 2020).
In recent years, bioinformatics analysis has been widely employed to investigate the microarray data to estimate differentially expressed genes (DEGs) and adopt various analyses (Kanwar, 2020). However, the limited sample size or high false-positive rate of a single microarray analysis might hinder the derivation of reliable conclusions. The present study retrieved two separate microarray datasets from gene expression omnibus (GEO) for further analyses. Common DEGs between patients with PAH and controls of these two datasets were screened to identify significant biomarkers. Potential differentially expressed genes and hub genes participating in PAH were estimated via Gene Ontology (GO) annotations, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, protein–protein interaction (PPI) network investigations, and protein subcellular localization. Eventually, a total of 38 DEGs and 9 hub genes was chosen as prospective diagnostic candidates and targeted biomarkers for PAH.
Materials and Methods
Identification of the Differentially Expressed Genes of Microarray Datasets From Lung Tissue With Patients With PAH
The high-throughput datasets of microarray gene expression in PAH were retrieved from the NCBI-GEO database (Barrett et al., 2013). The datasets were obtained from two separate studies on human lung tissues that were compared across normal individuals and patients with PAH on Affymetrix microarrays, which have been deposited in the NCBI-GEO database under the accession numbers GSE53408 and GSE113439. GSE53408 contains samples of 16 individuals, including 11 controls and five patients with idiopathic pulmonary arterial hypertension (IPAH), taken from a study originally published by Zhao et al. (2014). GSE113439 encompasses 26 lung tissue samples (11 controls and 15 cases), the PAH group contains six patients with IPAH, four patients with PAH secondary to the connective tissue disease (CTD), four patients with PAH secondary to the congenital heart disease (CHD), and one patient with chronic thromboembolic pulmonary hypertension (CTEPH). This dataset was originally analyzed by Mura and colleagues to explore DEGs in PAH compared to the controls (Mura et al., 2019). To verify the results, GSE15197 (13 control and 18 PAH lung tissues) and GSE117261 (25 control and 58 PAH lung tissues) were also used in our research. First, we normalized the gene expression datasets for log2 transformation. Thereafter, these datasets were analyzed in GEO2R of NCBI with Limma package in hypothesis testing, and the false discovery rate was regulated by Benjamini and Hochberg correction. As the cutoff criteria, a p < 0.01 was considered to select the significant DEGs. Jvenn was exploited to identify the common DEGs of two datasets and make the Venn plot (Bardou et al., 2014).
Gene Ontology and Gene Pathway Enrichment Analysis
As the bioinformatics resource, Enrichr was adopted to ascertain the common DEGs and identify their molecular functions, biological processes, cellular functions, and pathway annotations (Kuleshov et al., 2016). GO and the KEGG pathway database served as the annotation sources in this study. PANTHER database was employed to detect the over-representation of protein classes (Mi et al., 2013). Again, a p < 0.05 was considered significant in all enrichment analyses.
Protein–Protein Interaction Analysis
The PPI network of proteins encoded by DEGs was constructed by the STRING protein interaction database. Since the number of common DEGs (Szklarczyk et al., 2017) was low, the PPI network was developed with a medium confidence score (600). NetworkAnalyst was performed to visualize and analyze the topological network (Xia et al., 2015). The hub proteins were selected based on the topological indices, i.e., degree > 15.
Differentially Expressed Genes–Transcription Factor (TF) Interaction Analysis
To investigate the regulatory TFs that regulate the DEGs at the transcriptional level, we identified the interactions of TF-targeted genes with the JASPAR database and estimated the topological parameters using NetworkAnalyst (Xia et al., 2015; Khan et al., 2018).
Differentially Expressed Genes–miRNA Interaction Analysis
The regulatory miRNAs that control DEGs at the posttranscriptional level were evaluated by identifying the interactions of miRNA-target genes with TarBase and miRTarBase and by examining the topological parameters using the NetworkAnalyst (Sethupathy et al., 2006; Hsu et al., 2011; Xia et al., 2015).
Protein–Drug Interaction Analysis
The protein–drug interaction was estimated by the DrugBank database (version 5.1.8) via the NetworkAnalyst, which could highlight the potential drugs applied in the treatment of PAH (Wishart et al., 2018). The results suggested the interaction of HSP90AA1 (heat shock protein) with few drugs. Moreover, according to the protein–ligand docking server SwissDock, molecular docking analysis between HSP90AA1 and drugs was elucidated with the three-dimensional crystal structure (Grosdidier et al., 2011; Biasini et al., 2014). The protein–drug interaction analysis also indicated the interaction of HSP90AA1 with nedocromil and SNX-5422 (Wishart et al., 2018).
Investigation of Protein Subcellular Localization
WoLF PSORT helped to determine the subcellular localization of a set of proteins encoded by DEGs (Horton et al., 2007). The subcellular localization of proteins was predicted based on their amino acid sequences. This method made predictions in the light of the known sorting signals, amino acid content, and functional motifs, collected from UniProt (Universal Protein) and GO database.
Results
Transcriptomic Signatures of PAH
The microarray datasets retrieved from the lung tissues of patients with PAH were investigated, and 38 mutual core DEGs (SCARNA4, GALNT1, CCDC186, EEA1, SMC4, GCC2, ZNF721, EPRS, RAD50, POSTN, DNTTIP2, RAMP2, HSP90AA1, ZNF845, HSPD1, NEXN, CFH, ZNF267, HSP90AA6P, CD163, HIGD1B, PI15, SNORD20, ANGPT2, S100A3, TDO2, SOSTDC1, ANLN, TSHZ2, SLC7A11, MS4A15, HSPH1, DKC1, TTN, VSIG1, SPP1, RNU5D-1, and MMP8) were detected between the two datasets. The core DEGs, reflecting the transcriptomic signatures for PAH (Figure 1A), were classified into eight groups based on their functions and bioactivities as protein-modifying enzyme (8%), transporter (8%), scaffold/adaptor protein (4%), membrane traffic protein (4%), chaperone (4%), cell adhesion molecule (4%), transmembrane signal receptor (4%), defense/immunity protein (8%), calcium-binding protein (4%), cytoskeletal protein (8%), intercellular signal molecule (12%), gene-specific transcriptional regulator (15%), translational protein (4%), metabolite interconversion enzyme (12%), and chromatin/chromatin-binding or chromatin-regulatory protein (4%) (Figure 1B).
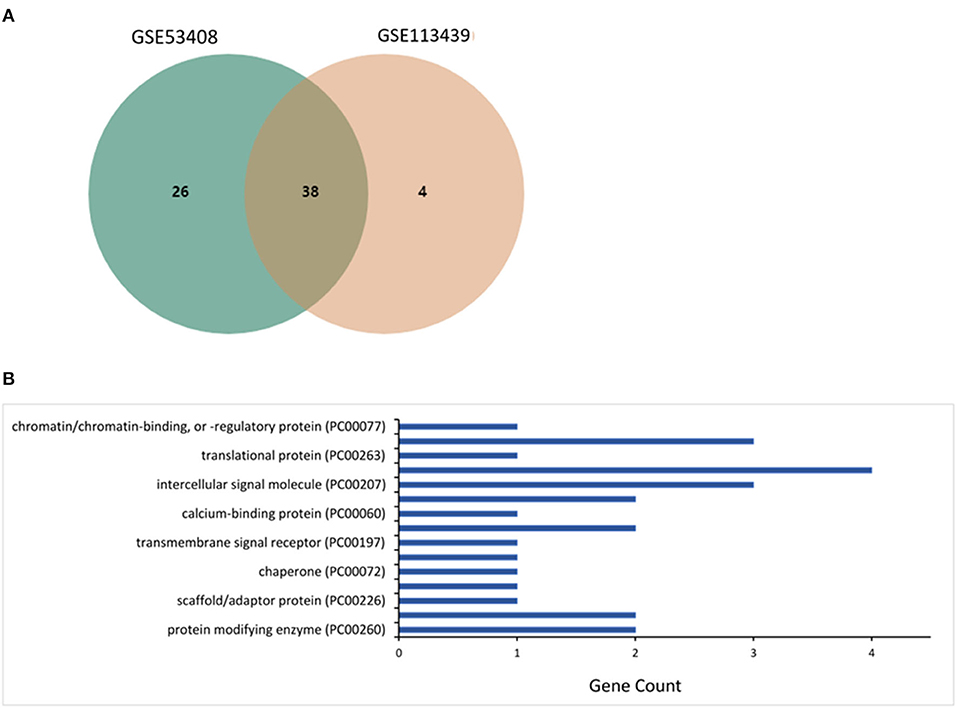
Figure 1. Investigation of differentially expressed genes (DEGs) in microarray datasets GSE53408 and GSE113439. (A) The mutual core DEGs analyzed between two datasets. (B) The over-represented protein class coded by the DEGs estimated using the PANTHER database.
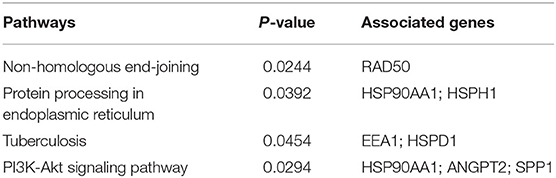
Gene-set enrichment analysis revealed the abundance of DEGs in biological processes, molecular functions, and cellular components. Table 1 summarizes the specific information. The enrichment analysis of the molecular pathway claimed that the pathways involved in non-homologous end-joining, protein processing in the endoplasmic reticulum, tuberculosis, and PI3K-Akt signaling pathway were altered (Table 2).
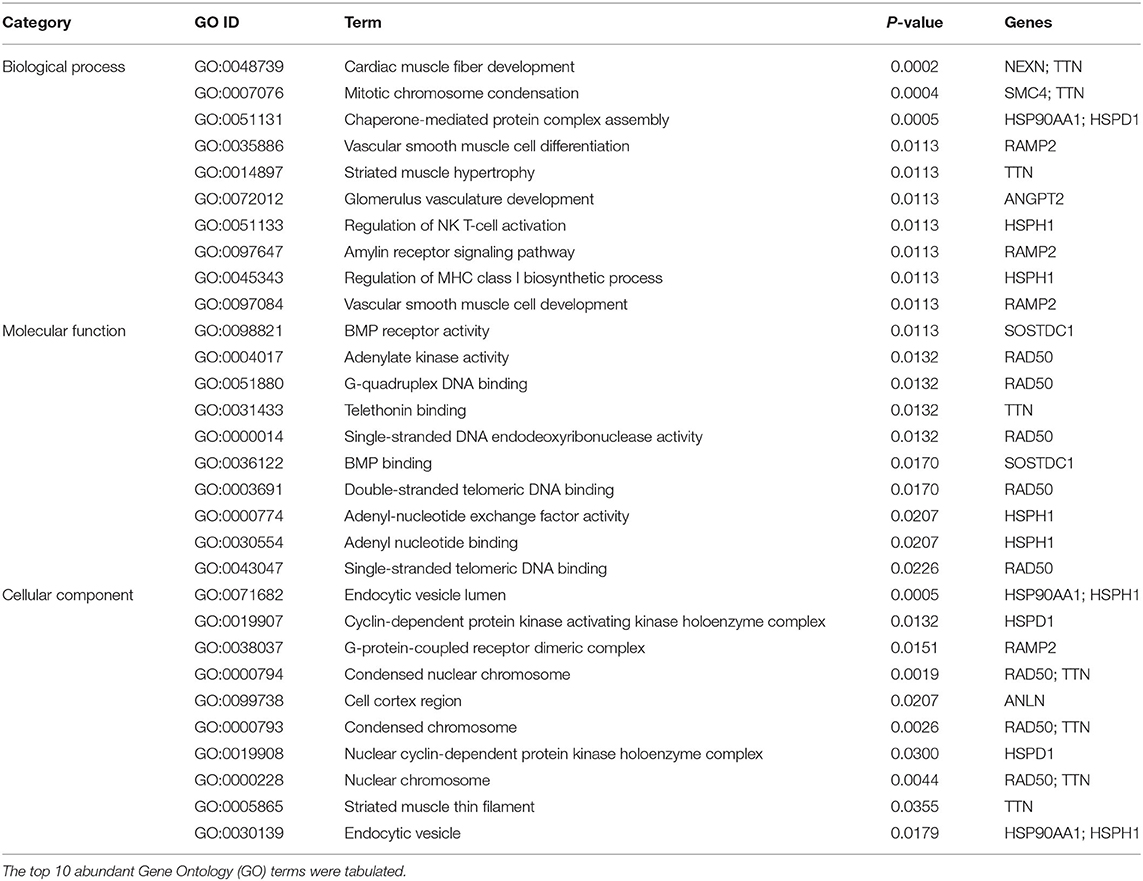
Table 1. Gene set enrichment analysis for differentially expressed genes detected from the microarray data of lung tissue with patients with pulmonary arterial hypertension (PAH).
Proteomic Signatures in PAH
To demonstrate central proteins, the protein–protein network of mutual DEGs was constructed to offer deep knowledge in the biological characterization of targeted proteins encoded by DEGs and the estimation of drug targets (Figure 2). The hub proteins responsible for transmitting signal stimulus to other proteins in networks are obtained from the topological examination of PPI networks. Based on the topological metric, the present method estimated the hub proteins, which could serve as biomarkers and drug targets in PAH. Besides, Table 3 highlights nine central hub proteins (HSP90AA1, ANGPT2, HSPD1, HSPH1, TTN, SPP1, SMC4, EEA1, and DKC1). The other two independent studies also showed that these hub proteins play an important role in PAH (Supplementary Table 1).
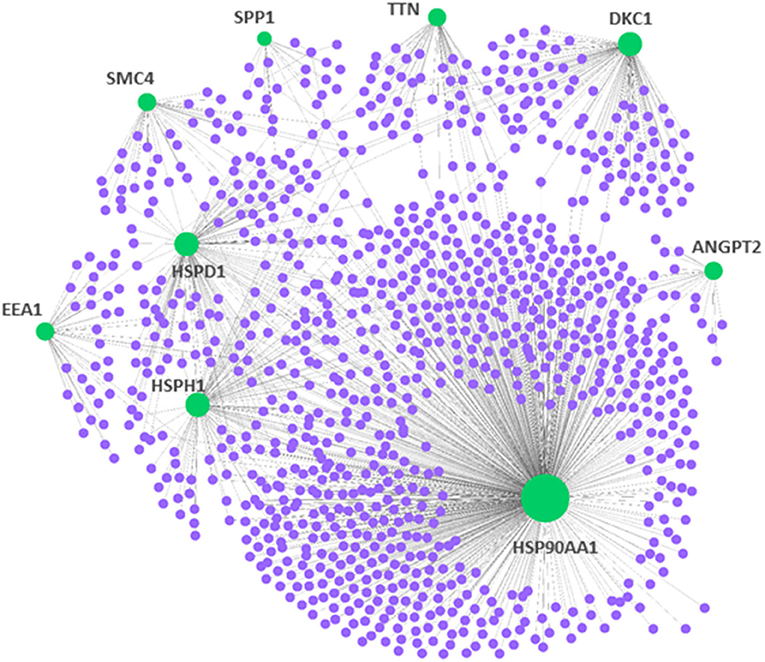
Figure 2. Protein–protein interaction network for the DEGs in pulmonary arterial hypertension (PAH). The nodes indicated the DEGs, while the edges indicated the interactions between different proteins. The medium confidence score was performed to construct the Protein–protein interaction (PPI) networks.
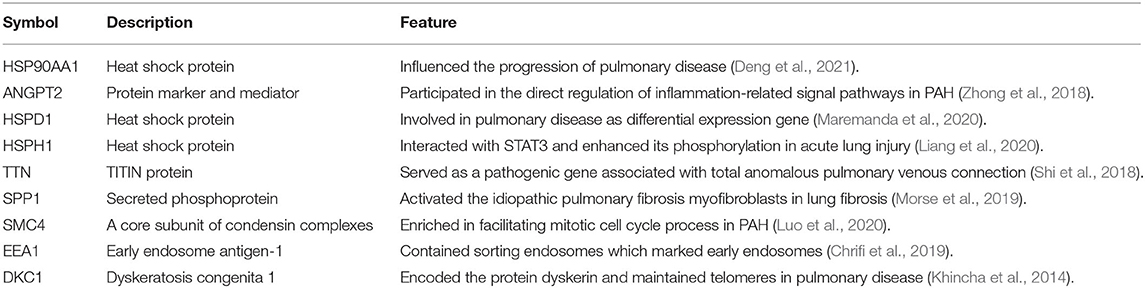
Table 3. Summary of hub proteins identified from protein–protein interactions analysis of encoded differentially expressed genes in PAH disease.
Regulatory Signatures of PAH
Exploration of the DEG–TF interaction (Figure 3) and DEG–miRNA interaction (Figure 4) with topological parameters contributed to the identification of the central regulatory biomolecules of PAH. As detailed in Table 4, the present study documented five TFs (FOXC1, FOXL1, GATA2, YY1, and SRF) and five miRNAs (hsa-mir-17-5p, hsa-mir-26b-5p, hsa-mir-122-5p, hsa-mir-20a-5p, and hsa-mir-106b-5p).
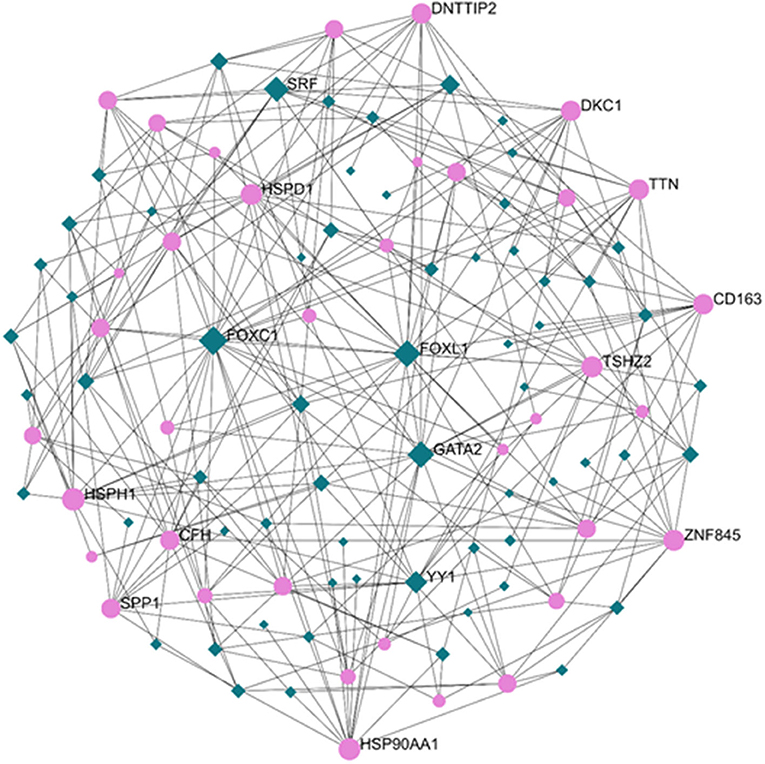
Figure 3. Regulatory networks showed the DEG–transcription factor (DEG–TF) interactions in PAH. Medium confidence score was performed to construct the regulatory networks. Green, TFs; pink, DEGs.
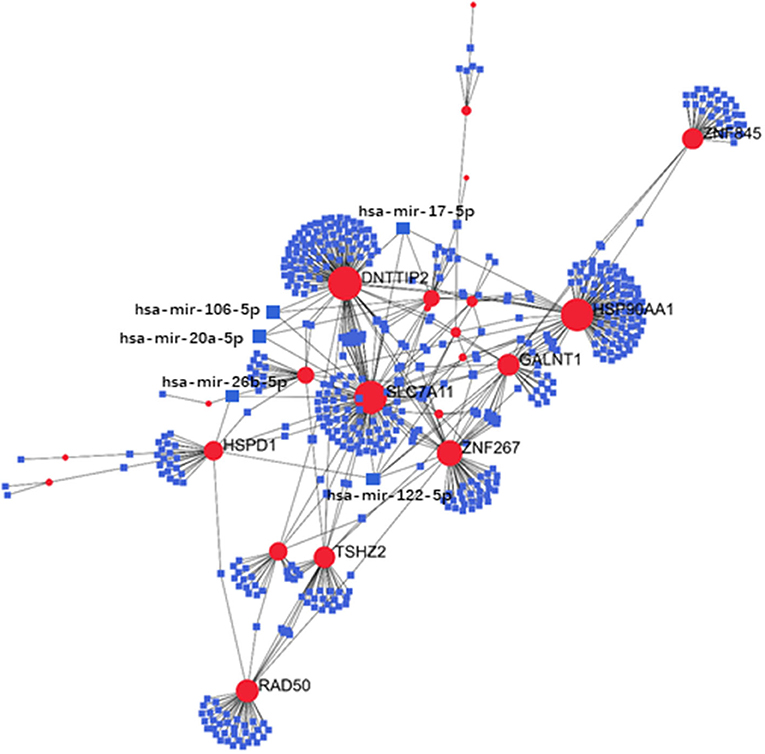
Figure 4. Regulatory networks showed the DEG–miRNA interactions in PAH. Medium confidence score was performed to construct the regulatory networks. Blue, miRNAs; red, DEGs.
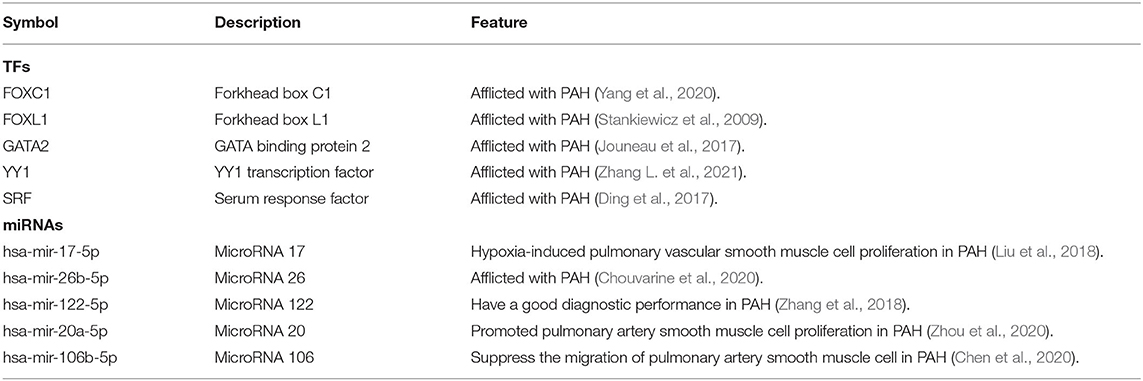
Table 4. Summary of the regulatory biomolecules [transcription factors (TFs), miRNAs] of DEGs in PAH disease identified from the interactions of DEGs with TFs and DEGs with miRNAs.
Protein–Drug Interactions
The protein–drug interaction network reported the relation of HSP90AA1 protein with the drugs nedocromil and SNX-5422 (Figure 5, Table 5). Nedocromil is a pyranoquinolone derivative that can suppress the activation of inflammatory cells, such as eosinophils, neutrophils, macrophages, mast cells, monocytes, and platelets. SNX-5422 is not only an oral agent exhibiting strong efficacy and tolerability but also a new synthetic Hsp90 inhibitor, and it was considered as a drug with breakthrough treatment and widespread applicability in a wide range of cancers. According to the statistical significance threshold of protein–drug interaction and the potential influence of the targeted protein in PAH pathogenesis, several protein–drug interactions were screened, and molecular docking simulations were carried out to determine the binding affinities of the drugs with targeted proteins (Table 5). The resultant energetic states and docking scores confirmed the thermodynamic feasibility of all these interactions.
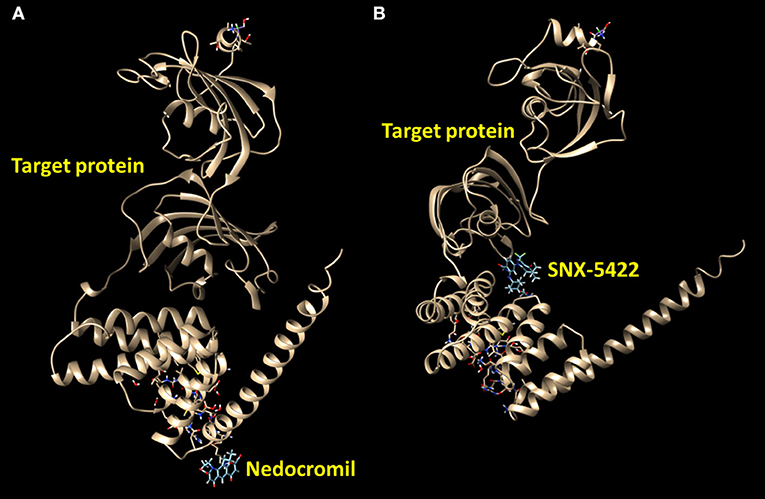
Figure 5. Predict binding modes of protein–drug interactions. Molecular graphics images were produced by the UCSF Chimera package. Binding modes were scored using their FullFitness and clustered. Clusters were then ranked based on the average FullFitness of their elements. (A) Nedocromil–protein; (B) SNX-5422–protein.
Protein Subcellular Localization
The WoLF PSORT software package can predict the subcellular localization of proteins encoded by the 38 DEGs in PAH. The proportions of these proteins in distinct subcellular compartments were computed, which indicated the localization of 89.5% of the proteins in extracellular areas, whereas the remaining 10.5% existed in the nuclear area. Notably, all hub proteins manifested the extracellular localization, including HSP90AA1, ANGPT2, HSPD1, HSPH1, TTN, SPP1, SMC4, EEA1, and DKC1.
Discussion
The diagnosis of PAH currently relies on the right heart catheterization, but the specific biomarkers for PAH diagnosis are an unmet challenge. The present research focused on the comprehensive analysis of the gene expression patterns in lung tissues of patients with PAH and exploration of the robust candidate molecular targets, which may act as potential biomarkers of PAH. Our study may, therefore, provide the relevant information regarding the progression of PAH.
With a wide application in biomedical investigation, microarray datasets have become a major resource for elucidating biomarker candidates (Budinska et al., 2013; Marisa et al., 2013). The extensive contribution of microarray gene expression profiling has also been reflected in the investigation of DEGs in various diseases, including Alzheimer's disease and breast cancer (Nami and Wang, 2018; Rahman et al., 2020). Significant alterations in the profiles of 38 genes in two separate transcriptomic datasets have been observed from the gene expression patterns in the lung tissues of patients with PAH. The enrichment analysis reported PAH-related molecular pathways in the endoplasmic reticulum and PI3K-Akt signaling pathway (Table 2) (Li et al., 2016).
Interactive analysis of PPI networks facilitates the identification of the proteins that play a key role in the pathophysiology of various diseases (Goh et al., 2007; Yang et al., 2009). The PPI analysis offers deep knowledge in the biological characterization of targeted proteins encoded by DEGs and the estimation of drug targets (Taz et al., 2021; Xu et al., 2021; Zhang T. et al., 2021). In light of DEGs, PPI networks detected a set of key hub proteins (Table 3), which may reflect the onset and development processes of vascular diseases. These hub proteins were involved in a number of biological and pathologic processes. As a hub protein, HSP90AA1 was differentially expressed in the lung tissues of patients with PAH and indulged in the inflammatory responses of the airway and the smooth muscle function of the bronchia, which reflected that HSP90AA1 may be a potential prognostic biomarker and drug target for the treatment of PAH (Deng et al., 2021). Consistent with our findings, ANGPT2 was upregulated in several inflammatory diseases and took part in the direct regulation of inflammation-related signal pathways in PAH (Zhong et al., 2018). Differential expression of HSPD1, involved in mitochondrial biogenesis, was noted in patients suffering from pulmonary diseases compared with normal controls (Maremanda et al., 2020). Interaction of HSPH1 with STAT3 may enhance its phosphorylation, which exacerbated pulmonary inflammation in acute lung injury, though there is no such report relating to PAH (Liang et al., 2020). Furthermore, TTN, a pathogenic gene in total anomalous pulmonary venous connection, appeared to play a critical role in the genetic mechanism (Shi et al., 2018). It has been suggested that TTN isoform composition was unchanged in PAH cardiomyocytes but that TTN phosphorylation was significantly decreased in patients with PAH (Rain et al., 2013). Pulmonary myofibroblasts can be activated by highly proliferative SPP1 macrophages, in turn, contributing vitally to lung fibrosis (Morse et al., 2019). Previous studies have shown that the SPP1 gene was significantly increased in PAH and played an important role by promoting pulmonary vascular smooth muscle cell (PVSMC) proliferation (Saker et al., 2016; Zeng et al., 2021). SMC4 manifested higher expression levels in patients with PAH (Luo et al., 2020). Similarly, SMC4 is a vital core subunit of condensin, which has an essential impact on mitotic chromosome condensation (Takemoto et al., 2004; Luo et al., 2020). It has been suggested that knockdown of SMC4 could inhibit Toll-like receptor-mediated production of several proinflammatory cytokines, including IL-6 and TNF-α in macrophages (Ma et al., 2020). Interaction of EEA1 with platelet-derived growth factor receptor can mark early endosomes, which are required for the progression of secondary pulmonary alveolar septa, which reflected that the internalization of TGF-R by clathrin-mediated endocytosis in EEA1-positive endosomes resulted in productive nuclear signaling via the interaction of TGF-R with Smad anchor for receptor activation (SARA) (Saker et al., 2016; Chrifi et al., 2019). DKC1 was known as a gene-encoded protein dyskerin and affected several modules of the telomere complex in pulmonary disease (Heiss et al., 1998; Khincha et al., 2014).
The interaction of DEGs with TFs and DEGs with miRNAs was also examined to highlight the transcriptional or posttranscriptional regulators associated with the mutual DEGs. Table 4 illustrates a set of regulators related to DEGs in PAH, including TFs and miRNAs. The measured transcriptional regulatory TFs (FOXC1, FOXL1, GATA2, YY1, SRF) that interacted with DEGs in PAH were in accordance with the previous observations (Stankiewicz et al., 2009; Ding et al., 2017; Yang et al., 2019, 2020; Zhang L. et al., 2021). miRNA is known as a single-stranded non-coding RNA that targets their transcripts to regulate gene expressions (Caruso et al., 2017). As potential biomarkers, miRNAs may provide breakthrough treatment strategies for the diagnosis and management of PAH (Caruso et al., 2017). Therefore, the miRNAs were explored as regulatory factors of target DEGs. The mir-17-5p identified in this study was involved in the proliferation of pulmonary vascular smooth muscle cells and could thus provide a potential novel treatment target for the control of PAH (Liu et al., 2018). In comparison with the control group, the lower concentration of miR-26b-5p in the lung tissue of patients with PAH signified its involvement in the remodeling process of PAH (Chouvarine et al., 2020). As a good biomarker for hypertension, mir-122-5p had a prominent diagnostic performance, and its dysregulation could indulge in the risk of PAH (Zhang et al., 2018). The mir-20a-5p-induced proliferation of pulmonary artery smooth muscle cells can exacerbate the development of PAH via targeting ATP-binding cassette subfamily A1 (Zhou et al., 2020). The mir-106b-5p played a key role in suppressing the migration of pulmonary artery smooth muscle cells, indicating that mir-106b-5p may act as a potential marker in PAH (Chen et al., 2020). These biomolecules may regulate target genes at transcriptional or posttranscriptional levels.
Given the importance of hub genes and their potential role in the pathogenic process in PAH, the interactions between target proteins and drugs were further studied in this research. Two drugs were spotted from the interaction network according to the DrugBank database (Table 5). A previous study has suggested that nedocromil sodium could inhibit antigen-induced shrinkage of human lung parenchymal as well as bronchial strips (Napier et al., 1990). As a therapy for non-small-cell lung cancer (NSCLC), another study also supported the evaluation of SNX-5422, especially in cases where cancer was driven by c-Met amplification and mutated epidermal growth factor receptor (EGFR) forms that were resistant to EGFR inhibitors (Rice et al., 2009). Several molecular modeling-based techniques were conducted in pharmaceutical research to evaluate complex biological systems, especially for molecular docking methods, which were broadly applied in drug designs to demonstrate the ligand conformation with binding sites of target proteins. The free energy was estimated in molecular docking methods by evaluating critical phenomena that participated in the intermolecular recognition processes in ligand–receptor binding (Chen et al., 2020). Henceforth, the binding modes of drugs with target protein HSP90AA1 were screened, and the energetically stable conformations were obtained from the existing databases (Figure 5). Moreover, the protein subcellular localizations for DEGs confirmed that extracellular and nuclear areas were the primary subcellular sites for the DEGs. This result substantiated the participation of these genes in PAH, extending from the nucleus to extracellular areas. For further drug selection to alleviate PAH, the protein subcellular localization may provide targeting sites for specific drugs. The present study documented the relationship between drugs and putative PAH molecular biomarkers; however, the consequence of molecular targets blockade was ambiguous from this study that should be taken into account for further investigation. Although the molecules are lung tissue-based and how they participate in pulmonary pathogenesis is still not known, clinical evidence has substantiated the effect of several of these identified drugs on PAH, and it may be helpful to know what are their influences on lung tissues.
Conclusion
Integrative multi-omics analysis was adopted for the evaluation of the lung tissue-based transcriptomic profiles to identify the molecular signatures at protein levels (hub proteins, TFs) and RNA levels (mRNAs, miRNAs). Based on the genome transcriptome datasets, nine hub genes (HSP90AA1, ANGPT2, HSPD1, HSPH1, TTN, SPP1, SMC4, EEA1, and DKC1) were found in this study. The significant abundance of numerous pivotal hub genes was evident in pathways that participated in endoplasmic reticulum-related protein processing and PI3K-Akt signaling pathway. We also screened TFs (FOXC1, FOXL1, GATA2, YY1, and SRF) and miRNAs (hsa-mir-17-5p, hsa-mir-26b-5p, hsa-mir-122-5p, hsa-mir-20a-5p, and hsa-mir-106b-5p) regulating the expression of DEGs in PAH. These biomolecules could be accounted as candidate system biomarkers at protein levels and RNA levels. Therefore, the potential molecular signatures obtained for PAH could be detected as transcripts in lung tissues, and these signatures warrant clinical analyses in patients with PAH to access their utility. The availability of these biomolecules in pulmonary tissues presumes the establishment of these biomarkers as a novel aspect of PAH development and progression.
Data Availability Statement
The original contributions generated for the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
DC, HZ, and WM conceived and designed the experiments. XY and TJ performed all experiments. XY, TW, and XC collected and analyzed the data. CG, HF, TJ, and FC drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31902282), Key Project of Science and Technology Coordination in Shaanxi Province (2020ZDLNY02-06) and Qinghai Province Major R&D and Transformation Project (2021-QY-211 and 2018-NK-125) and National Key Research and Development Program of China (2019YFE0119400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2021.694702/full#supplementary-material
References
Bardou, P., Mariette, J., Escudie, F., Djemiel, C., and Klopp, C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinform. 15:293. doi: 10.1186/1471-2105-15-293
Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., et al. (2013). NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41, D991–D995. doi: 10.1093/nar/gks1193
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Boucly, A., Weatherald, J., Savale, L., Jais, X., Cottin, V., Prevot, G., et al. (2017). Risk assessment, prognosis and guideline implementation in pulmonary arterial hypertension. Europ. Resp. J. 50:2017. doi: 10.1183/13993003.00889-2017
Budinska, E., Popovici, V., Tejpar, S., D'ario, G., Lapique, N., Sikora, K. O., et al. (2013). Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J. Pathol. 231, 63–76. doi: 10.1002/path.4212
Caruso, P., Dunmore, B. J., Schlosser, K., Schoors, S., Dos Santos, C., Perez-Iratxeta, C., et al. (2017). Identification of MicroRNA-124 as a major regulator of enhanced endothelial cell glycolysis in pulmonary arterial hypertension via PTBP1 (Polypyrimidine Tract Binding Protein) and pyruvate kinase M2. Circulation136:2451. doi: 10.1161/Circulationaha.117.028034
Chen, H., Ma, Q., Zhang, J., Meng, Y., Pan, L., and Tian, H. (2020). miR106b5p modulates acute pulmonary embolism via NOR1 in pulmonary artery smooth muscle cells. Int. J. Mol. Med. 45, 1525–1533. doi: 10.3892/ijmm.2020.4532
Chouvarine, P., Geldner, J., Giagnorio, R., Legchenko, E., Bertram, H., and Hansmann, G. (2020). Trans-right-ventricle and transpulmonary MicroRNA gradients in human pulmonary arterial hypertension. Pediatr. Crit. Care Med. 21, 340–349. doi: 10.1097/PCC.0000000000002207
Chrifi, I., Louzao-Martinez, L., Brandt, M. M., Van Dijk, C. G. M., Burgisser, P. E., Zhu, C., et al. (2019). CMTM4 regulates angiogenesis by promoting cell surface recycling of VE-cadherin to endothelial adherens junctions. Angiogenesi. 22, 75–93. doi: 10.1007/s10456-018-9638-1
Claessen, G., and La Gerche, A. (2017). Pulmonary vascular function during exercise: progressing toward routine clinical use. Circ. Cardiovasc. Imaging 10:326. doi: 10.1161/CIRCIMAGING.117.006326
Deng, Y. X., Zhong, J., Liu, Z. J., Wang, X. Q., and Zhang, B. (2021). Active ingredients targeting Nrf2 in the Mongolian medicine Qiwei Putao powder: systematic pharmacological prediction and validation for chronic obstructive pulmonary disease treatment. J. Ethnopharm. 265:113385. doi: 10.1016/j.jep.2020.113385
Ding, X., Zhou, S., Li, M., Cao, C., Wu, P., Sun, L., et al. (2017). Upregulation of SRF is associated with hypoxic pulmonary hypertension by promoting viability of smooth muscle cells via increasing expression of Bcl-2. J. Cell Biochem. 118, 2731–2738. doi: 10.1002/jcb.25922
Farhadi, R., Rafiei, A., Hamdamian, S., Zamani, H., and Yazdani, J. (2017). Pentraxin 3 in neonates with and without diagnosis of pulmonary hypertension. Clin. Biochem. 50, 223–227. doi: 10.1016/j.clinbiochem.2016.11.009
Galie, N., Barbera, J. A., Frost, A. E., Ghofrani, H. A., Hoeper, M. M., Mclaughlin, V. V., et al. (2015). Initial use of ambrisentan plus tadalafil in pulmonary arterial hypertension. N. Engl. J. Med. 373, 834–844. doi: 10.1056/NEJMoa1413687
Goh, K. I., Cusick, M. E., Valle, D., Childs, B., Vidal, M., and Barabasi, A. L. (2007). The human disease network. Proc. Natl. Acad. Sci. U.S.A. 104, 8685–8690. doi: 10.1073/pnas.0701361104
Gouyou, B., Grun, K., Kerschenmeyer, A., Villa, A., Matasci, M., Schrepper, A., et al. (2021). Therapeutic evaluation of antibody-based targeted delivery of interleukin 9 in experimental pulmonary hypertension. Int. J. Mol. Sci. 22:460. doi: 10.3390/ijms22073460
Grosdidier, A., Zoete, V., and Michielin, O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–277. doi: 10.1093/nar/gkr366
Heiss, N. S., Knight, S. W., Vulliamy, T. J., Klauck, S. M., Wiemann, S., Mason, P. J., et al. (1998). X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 19, 32–38. doi: 10.1038/ng0598-32
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–587. doi: 10.1093/nar/gkm259
Hsu, S. D., Lin, F. M., Wu, W. Y., Liang, C., Huang, W. C., Chan, W. L., et al. (2011). miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39, D163–169. doi: 10.1093/nar/gkq1107
Jouneau, S., Ballerie, A., Kerjouan, M., Demant, X., Blanchard, E., and Lederlin, M. (2017). Haemodynamically proven pulmonary hypertension in a patient with GATA2 deficiency-associated pulmonary alveolar proteinosis and fibrosis. Eur. Respir. J. 49:2017. doi: 10.1183/13993003.00407-2017
Kanwar, M. K. (2020). Biomarkers in pulmonary arterial hypertension: moving closer toward precision medicine? J. Heart Lung Transplant. 39, 287–288. doi: 10.1016/j.healun.2020.02.020
Kerkhof, P. L. M., Li, J. K., and Handly, N. (2019). Interpretation of a new biomarker for the right ventricle introduced to evaluate the severity of pulmonary arterial hypertension. Pulm. Circ. 9:2045894019826945. doi: 10.1177/2045894019826945
Khan, A., Fornes, O., Stigliani, A., Gheorghe, M., Castro-Mondragon, J. A., Van Der Lee, R., et al. (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266. doi: 10.1093/nar/gkx1126
Khincha, P. P., Wentzensen, I. M., Giri, N., Alter, B. P., and Savage, S. A. (2014). Response to androgen therapy in patients with dyskeratosis congenita. Br. J. Haematol. 165, 349–357. doi: 10.1111/bjh.12748
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–97. doi: 10.1093/nar/gkw377
Li, A., Zhang, S., Li, J., Liu, K., Huang, F., and Liu, B. (2016). Metformin and resveratrol inhibit Drp1-mediated mitochondrial fission and prevent ER stress-associated NLRP3 inflammasome activation in the adipose tissue of diabetic mice. Mol. Cell Endocrinol. 434, 36–47. doi: 10.1016/j.mce.2016.06.008
Liang, Y., Luo, J., Yang, N., Wang, S., Ye, M., and Pan, G. (2020). Activation of the IL-1beta/KLF2/HSPH1 pathway promotes STAT3 phosphorylation in alveolar macrophages during LPS-induced acute lung injury. Biosci. Rep. 40:72. doi: 10.1042/BSR20193572
Liu, G., Hao, P., Xu, J., Wang, L., Wang, Y., Han, R., et al. (2018). Upregulation of microRNA-17-5p contributes to hypoxia-induced proliferation in human pulmonary artery smooth muscle cells through modulation of p21 and PTEN. Respir Res. 19, 200. doi: 10.1186/s12931-018-0902-0
Luo, J., Li, H., Liu, Z., Li, C., Wang, R., Fang, J., et al. (2020). Integrative analyses of gene expression profile reveal potential crucial roles of mitotic cell cycle and microtubule cytoskeleton in pulmonary artery hypertension. BMC Med Genomics 13:86. doi: 10.1186/s12920-020-00740-x
Ma, Y., Chen, S. S., Feng, Y. Y., and Wang, H. L. (2020). Identification of novel biomarkers involved in pulmonary arterial hypertension based on multiple-microarray analysis. Biosci. Rep. 40:2346. doi: 10.1042/BSR20202346
Maremanda, K. P., Sundar, I. K., Li, D., and Rahman, I. (2020). Age-dependent assessment of genes involved in cellular senescence, telomere and mitochondrial pathways in human lung tissue of smokers, COPD and IPF: associations with SARS-CoV-2 COVID-19 ACE2-TMPRSS2-Furin-DPP4 axis. medRxiv. doi: 10.1101/2020.06.14.20129957
Marisa, L., De Reynies, A., Duval, A., Selves, J., Gaub, M. P., Vescovo, L., et al. (2013). Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 10:e1001453. doi: 10.1371/journal.pmed.1001453
Mi, H., Muruganujan, A., and Thomas, P. D. (2013). PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41(Database issue), D377–386. doi: 10.1093/nar/gks1118
Morse, C., Tabib, T., Sembrat, J., Buschur, K. L., Bittar, H. T., Valenzi, E., et al. (2019). Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 54:2018. doi: 10.1183/13993003.02441-2018
Mura, M., Cecchini, M. J., Joseph, M., and Granton, J. T. (2019). Osteopontin lung gene expression is a marker of disease severity in pulmonary arterial hypertension. Respirology 24, 1104–1110. doi: 10.1111/resp.13557
Nami, B., and Wang, Z. (2018). Genetics and Expression Profile of the Tubulin Gene Superfamily in Breast Cancer Subtypes and Its Relation to Taxane Resistance. Cancers 10:274. doi: 10.3390/cancers10080274
Napier, F. E., Shearer, M. A., and Temple, D. M. (1990). Nedocromil sodium inhibits antigen-induced contraction of human lung parenchymal and bronchial strips, and the release of sulphidopeptide-leukotriene and histamine from human lung fragments. Br. J. Pharmacol. 100, 247–250. doi: 10.1111/j.1476-5381.1990.tb15790.x
Peacock, A. J., Murphy, N. F., Mcmurray, J. J., Caballero, L., and Stewart, S. (2007). An epidemiological study of pulmonary arterial hypertension. Eur. Respir. J. 30, 104–109. doi: 10.1183/09031936.00092306
Rahman, M. R., Islam, T., Zaman, T., Shahjaman, M., Karim, M. R., Huq, F., et al. (2020). Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer's disease: insights from a systems biomedicine perspective. Genomics 112, 1290–1299. doi: 10.1016/j.ygeno.2019.07.018
Rain, S., Handoko, M. L., Trip, P., Gan, C. T., Westerhof, N., Stienen, G. J., et al. (2013). Right ventricular diastolic impairment in patients with pulmonary arterial hypertension. Circulation 128, 2016–2025, 2011–2010. doi: 10.1161/CIRCULATIONAHA.113.001873
Rice, J. W., Veal, J. M., Barabasz, A., Foley, B., Fadden, P., Scott, A., et al. (2009). Targeting of multiple signaling pathways by the Hsp90 inhibitor SNX-2112 in EGFR resistance models as a single agent or in combination with erlotinib. Oncol Res. 18, 229–242. doi: 10.3727/096504009x12596189659240
Saker, M., Lipskaia, L., Marcos, E., Abid, S., Parpaleix, A., Houssaini, A., et al. (2016). Osteopontin, a key mediator expressed by senescent pulmonary vascular cells in pulmonary hypertension. Arterioscler. Thromb. Vasc. Biol. 36, 1879–1890. doi: 10.1161/ATVBAHA.116.307839
Sethupathy, P., Corda, B., and Hatzigeorgiou, A. G. (2006). TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 12, 192–197. doi: 10.1261/rna.2239606
Shi, X., Cheng, L., Jiao, X., Chen, B., Li, Z., Liang, Y., et al. (2018). Rare copy number variants identify novel genes in sporadic total anomalous pulmonary vein connection. Front. Genet. 9:559. doi: 10.3389/fgene.2018.00559
Sitbon, O., Jais, X., Savale, L., Cottin, V., Bergot, E., Macari, E. A., et al. (2014). Upfront triple combination therapy in pulmonary arterial hypertension: a pilot study. Eur. Respir. J. 43, 1691–1697. doi: 10.1183/09031936.00116313
Sitbon, O., Sattler, C., Bertoletti, L., Savale, L., Cottin, V., Jais, X., et al. (2016). Initial dual oral combination therapy in pulmonary arterial hypertension. Eur Respir J. 47, 1727–1736. doi: 10.1183/13993003.02043-2015
Stankiewicz, P., Sen, P., Bhatt, S. S., Storer, M., Xia, Z., Bejjani, B. A., et al. (2009). Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am. J. Hum. Genet. 84, 780–791. doi: 10.1016/j.ajhg.2009.05.005
Sullivan, D. I., and Kass, D. J. (2019). Signals and signposts: biomarkers in IPF and PAH at the crossroads of clinical relevance. Respirology 24, 1044–1045. doi: 10.1111/resp.13694
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Takemoto, A., Kimura, K., Yokoyama, S., and Hanaoka, F. (2004). Cell cycle-dependent phosphorylation, nuclear localization, and activation of human condensin. J Biol Chem. 279, 4551–4559. doi: 10.1074/jbc.M310925200
Tang, H., Babicheva, A., Mcdermott, K. M., Gu, Y., Ayon, R. J., Song, S., et al. (2018). Endothelial HIF-2alpha contributes to severe pulmonary hypertension due to endothelial-to-mesenchymal transition. Am. J. Physiol. Lung Cell Mol. Physiol. 314, L256–L275. doi: 10.1152/ajplung.00096.2017
Taz, T. A., Ahmed, K., Paul, B. K., Al-Zahrani, F. A., Mahmud, S. M. H., and Moni, M. A. (2021). Identification of biomarkers and pathways for the SARS-CoV-2 infections that make complexities in pulmonary arterial hypertension patients. Brief. Bioinform. 22, 1451–1465. doi: 10.1093/bib/bbab026
Van De Veerdonk, M. C., Kind, T., Marcus, J. T., Mauritz, G. J., Heymans, M. W., Bogaard, H. J., et al. (2011). Progressive right ventricular dysfunction in patients with pulmonary arterial hypertension responding to therapy. J. Am. Coll. Cardiol. 58, 2511–2519. doi: 10.1016/j.jacc.2011.06.068
Van De Veerdonk, M. C., Marcus, J. T., Westerhof, N., De Man, F. S., Boonstra, A., Heymans, M. W., et al. (2015). Signs of right ventricular deterioration in clinically stable patients with pulmonary arterial hypertension. Chest 147, 1063–1071. doi: 10.1378/chest.14-0701
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082. doi: 10.1093/nar/gkx1037
Xia, J., Gill, E. E., and Hancock, R. E. (2015). NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 10, 823–844. doi: 10.1038/nprot.2015.052
Xu, Z., Ruan, J., Pan, L., and Chen, C. (2021). Candidate genes identified in systemic sclerosis-related pulmonary arterial hypertension were associated with immunity, inflammation, and cytokines. Cardiovasc. Ther. 2021:6651009. doi: 10.1155/2021/6651009
Yang, L., Liang, H., Meng, X., Shen, L., Guan, Z., Hei, B., et al. (2020). mmu_circ_0000790 is involved in pulmonary vascular remodeling in mice with HPH via MicroRNA-374c-Mediated FOXC1. Mol. Ther. Nucleic Acids 20, 292–307. doi: 10.1016/j.omtn.2019.12.027
Yang, L., Liang, H., Shen, L., Guan, Z., and Meng, X. (2019). LncRNA Tug1 involves in the pulmonary vascular remodeling in mice with hypoxic pulmonary hypertension via the microRNA-374c-mediated Foxc1. Life Sci. 237:116769. doi: 10.1016/j.lfs.2019.116769
Yang, X., Deignan, J. L., Qi, H., Zhu, J., Qian, S., Zhong, J., et al. (2009). Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet. 41, 415–423. doi: 10.1038/ng.325
Zeng, Y., Li, N., Zheng, Z., Chen, R., Peng, M., Liu, W., et al. (2021). Screening of hub genes associated with pulmonary arterial hypertension by integrated bioinformatic analysis. Biomed. Res. Int. 2021:6626094. doi: 10.1155/2021/6626094
Zhang, L., Zeng, X. X., Li, Y. M., Chen, S. K., Tang, L. Y., Wang, N., et al. (2021). Keratin 1 attenuates hypoxic pulmonary artery hypertension by suppressing pulmonary artery media smooth muscle expansion. Acta Physiol. 231:e13558. doi: 10.1111/apha.13558
Zhang, T., Huang, C., Luo, H., Li, J., Huang, H., Liu, X., et al. (2021). Identification of key genes and immune profile in limited cutaneous systemic sclerosis-associated pulmonary arterial hypertension by bioinformatics analysis. Life Sci. 271:119151. doi: 10.1016/j.lfs.2021.119151
Zhang, X., Wang, X., Wu, J., Peng, J., Deng, X., Shen, Y., et al. (2018). The diagnostic values of circulating miRNAs for hypertension and bioinformatics analysis. Biosci. Rep. 38:525. doi: 10.1042/BSR20180525
Zhao, Y. D., Yun, H. Z. H., Peng, J., Yin, L., Chu, L., Wu, L., et al. (2014). De novo synthesize of bile acids in pulmonary arterial hypertension lung. Metabolomics 10, 1169–1175. doi: 10.1007/s11306-014-0653-y
Zhong, Y., Catheline, D., Houeijeh, A., Sharma, D., Du, L., Besengez, C., et al. (2018). Maternal omega-3 PUFA supplementation prevents hyperoxia-induced pulmonary hypertension in the offspring. Am. J. Physiol. Lung Cell Mol. Physiol. 315, L116–L132. doi: 10.1152/ajplung.00527.2017
Keywords: systems biology, pulmonary arterial hypertension, protein-drug interaction, biomarkers, protein subcellular localization
Citation: Yao X, Jing T, Wang T, Gu C, Chen X, Chen F, Feng H, Zhao H, Chen D and Ma W (2021) Molecular Characterization and Elucidation of Pathways to Identify Novel Therapeutic Targets in Pulmonary Arterial Hypertension. Front. Physiol. 12:694702. doi: 10.3389/fphys.2021.694702
Received: 13 April 2021; Accepted: 26 May 2021;
Published: 23 July 2021.
Edited by:
Haiyang Tang, University of Arizona, United StatesReviewed by:
Yunshan Cao, Gansu Provincial Hospital, ChinaPritesh Jain, University of California, San Diego, United States
Copyright © 2021 Yao, Jing, Wang, Gu, Chen, Chen, Feng, Zhao, Chen and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiying Zhao, zhaohuiying@nwafu.edu.cn; Dekun Chen, cdk@nwafu.edu.cn; chendekun163@163.com; Wentao Ma, mawentao@nwafu.edu.cn
†These authors have contributed equally to this work
 Xiaoting Yao
Xiaoting Yao Tian Jing†
Tian Jing† Wentao Ma
Wentao Ma