- 1College of Biotechnology, Jiangsu University of Science and Technology, Zhenjiang, Jiangsu, China
- 2Key Sericultural Laboratory of Shaanxi, Ankang University, Ankang, Shaanxi, China
- 3Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang, Jiangsu, China
The genetic male sterile line (GMS) of the silkworm Bombyx mori is a recessive mutant that is naturally mutated from the wild-type 898WB strain. One of the major characteristics of the GMS mutant is its small larvae. Through positional cloning, candidate genes for the GMS mutant were located in a region approximately 800.5 kb long on the 24th linkage group of the silkworm. One of the genes was Bombyx mori CCAAT/enhancer-binding protein zeta (BmC/EBPZ), which is a member of the basic region-leucine zipper transcription factor family. Compared with the wild-type 898WB strain, the GMS mutant features a 9 bp insertion in the 3′end of open reading frame sequence of BmC/EBPZ gene. Moreover, the high expression level of the BmC/EBPZ gene in the testis suggests that the gene is involved in the regulation of reproduction-related genes. Using the CRISPR/Cas9-mediated knockout system, we found that the BmC/EBPZ knockout strains had the same phenotypes as the GMS mutant, that is, the larvae were small. However, the larvae of BmC/EBPZ knockout strains died during the development of the third instar. Therefore, the BmC/EBPZ gene was identified as the major gene responsible for GMS mutation.
Introduction
CCAAT/enhancer-binding proteins (C/EBPs) belong to the basic region-leucine zipper (b-ZIP) protein family (Ramji and Foka, 2002). C/EBPs were discovered as heat-stable proteins in rat liver nuclei (Johnson and McKnight, 1989). To date, six members of the C/EBP family have been identified: C/EBP α, C/EBP β, C/EBP γ, C/EBP δ, C/EBP ε and C/EBP ζ (Ramji and Foka, 2002). C/EBPs typically has three structural components: a C-terminal leucine zipper domain, DNA-binding domain, and N-terminal transcriptional activation domain (Tenen, 2003). C/EBP factors are mainly involved in cell growth and differentiation, immune response, inflammatory response, energy metabolism, tumorigenesis, and apoptosis and regulate the transcription of target genes (Nerlov, 2007; Jin et al., 2011). C/EBP α is the first described C/EBP protein and plays an important role in many cellular responses and adipocyte and hepatocyte differentiation (Lefterova et al., 2008; Li et al., 2010; Pang et al., 2021; Chowdhury et al., 2023). The first discovered C/EBP β was NF-IL6, which plays an important role in many cellular processes, such as cell differentiation, tumorigenesis, apoptosis, energy metabolism, and reproduction (Akira et al., 1990; Ghani et al., 2022). C/EBPs contain an N-terminal transcriptional activation domain, except C/EBP γ. Mice deficient in the C/EBP γ gene have high post-birth mortality rates (Lekstrom-Himes and Xanthopoulos, 1998; Kaisho et al., 1999; Renfro et al., 2022). The expression level of C/EBP δ in human and adult mouse tissues is extremely low or even undetectable. C/EBP δ not only can promote inflammatory signals but also can inhibit pro-inflammatory pathways. However, despite that it can reduce the incidence of tumors, it promotes tumor metastasis (Balamurugan and Sterneck, 2013). C/EBP ε plays a vital role in terminal neutrophil differentiation. Mice deficient in C/EBP ε exhibit abnormal terminal granulocytic differentiation and typically succumb to bacterial infections within 5 months. (Tavor et al., 2002). A C/EBP ζ named DNA damage-inducible transcript 3 is associated with the development of leukemia, melanoma, and myxoid liposarcoma (Wang et al., 2011).
C/EBPs are present in various species, including mice, humans, chickens, Xenopus laevis, Rana catesbeiana, and fish (Johnson and McKnight, 1989; Akira et al., 1990; Roman et al., 1990; Cao et al., 1991; Ron and Habener, 1992; Hartl et al., 2023). The C/EBP DmC/EBP was first reported in insects (Drosophila) in 1992 (Rørth and Montell, 1992). It plays an important role in embryonic development. C/EBP factors regulate the expression of ferritin heavy-chain genes in Aedes aegypti and sterol carrier genes in Spodoptera litura (Pham et al., 2005; Liang et al., 2015). In 2005, the C/EBP BmC/EBP was discovered in silkworm (Sourmeli et al., 2005). It was the first C/EBP discovered in lepidopterans. In silkworm, the BmC/EBPg gene regulates histone acetylation and the expression of chorion gene (Lyu et al., 2020).
The genetic male sterile silkworm line (GMS) is naturally mutated from the wild-type 898WB strain. One of the major characteristics of the GMS mutant is that the larvae are small (Supplementary Figure S1). The wild-type 898WB were reared at normal temperature and humidity standards, and GMS mutants appeared in eight moth areas when the larvae developed to the 2nd instar. The silkworm larvae in eight moth areas were observed, and the separation ratio between the normal and mutant individuals was approximately 3:1. The results showed that the GMS mutant trait was heritable and controlled by a recessive single gene. RNA-Seq results have identified many differentially expressed genes between the GMS mutant and wild-type 898WB, such as immune-related genes, serine or threonine protein kinase-encoding genes, and heat shock protein genes (Mei et al., 2022). In the present study, positional cloning and gene sequencing showed that the BmC/EBPZ gene is the major gene responsible for GMS mutation. We further verified the role of the gene in the growth and development of silkworm by using the CRISPR/Cas9-mediated knockout system. Further analysis of the important role of BmC/EBPZ gene in the silkworm can help to provide potential targets for pest control.
Materials and methods
Silkworm strains
The GMS mutant, wild-type 898WB, Nistari, and Dazao (P50) silkworm strains were maintained in the key laboratory of Sericultural Biology (Sericultural Research Institute of the Chinese Academy of Agricultural Science, Jiangsu, China). The larvae were fed with mulberry leaves and reared at 25°C ± 1°C and 65% ± 5% relative humidity.
Materials used for positional cloning
The silkworm GMS mutant (gms/gms) mated with the p50 strain (+gms/+gms) to generate F1 offspring. Then, the female F1 and male F1 offspring were backcrossed with the GMS mutant to produce BC1F (♀F1×♂GMS) and BC1M (♀GMS×♂F1) populations, respectively. The GMS mutant, F1 offspring, and p50 can be used to screen polymorphic genetic markers. No genetic exchange was observed during meiosis in the female silkworms. The BC1F populations (10 wild-type strains and 10 mutant strains) were used for screening the linkage group, and the BC1M populations (252 mutant strains) were used for constructing the molecular marker linkage map of the mutant gene.
DNA, RNA extraction, and qRT–PCR
The heads of fifth instar larvae were used in genomic DNA extraction. First, a DNA extraction buffer (50 mM Tris-cl [pH 8.0], 10 mM EDTA [pH 8.0], 50 mM NaCl, 0.1% SDS, 20 mM proteinase K) was mixed with the tissues. DNA was then purified with phenol and chloroform and washed with 75% ethanol. Then, genomic DNA was precipitated with anhydrous ethanol. Finally, the DNA was dissolved in ultrapure water and diluted to 50 ng/μL. Total RNA extraction was conducted according to our previously described method (Mei et al., 2022).
Differences in gene expression levels between the GMS mutant and wild-type 898WB were determined through qRT–PCR. The total volume of the qRT–PCR reaction was 20 μL, composed of 1 μL of primer (10 μM), 1 μL of EvaGreen (Biotum, United States), 1 μL of total RNA (50 ng/μL), 10 μL of 2× one-step RT–PCR Mix (XT Biotech, China), and 7 μL of ultrapure water. The silkworm housekeeping gene (BmGAPDH) was used to normalize the qRT-PCR results. A QuantStudio 3 system (Thermo Fisher, United States) was used for qRT–PCR. The PCR amplification program was as follows: 50°C for 30 min; 95°C for 1 min; and 45 cycles of 95°C for 15 s and 58°C for 30 s. The qRT-PCR reaction was repeated three times for each sample, and gene expression levels were analyzed by the 2−ΔΔCT method. The significance of difference was analyzed by two-tailed t-test (*p < 0.05, **p < 0.01, ***p < 0.001).
Preparation of the genetic linkage map
The simple sequence repeat (SSR) genetic markers were used for screening the linkage group and constructing a genetic linkage map. The genome sequences of the 28 linkage groups of silkworms were downloaded from SilkDB 3.0 (https://silkdb.bioinfotoolkits.net/). The primers were designed using the software Primer Premier 5.0 and synthesized by BGI (Beijing, China). A mutant gene was considered to be on the same chromosome with the SSR marker when the DNA typing of wild-type individuals in the BC1F populations were the same as that of the F1 offspring or GMS mutant parental strain and the DNA typing of mutant individuals was the same as that of the GMS mutant parental strain. After the linkage group was confirmed, other polymorphic SSR molecular markers on the linkage group were used in the construction of a linkage map, and the location of a candidate gene was finally confirmed according to the number of individuals that showed genetic exchange.
Candidate genes analysis
On the basis of the results of fine mapping, the silkworm database SilkDB 3.0 (https://silkdb.bioinfotoolkits.net/) and KAIKObase (https://kaikobase.dna.affrc.go.jp/) were used in the screening of genes located in the linkage group. The open reading frame (ORF) sequences of the candidate genes were downloaded. The structures and expression levels of the candidate genes were analyzed on the basis of the results of RNA-Seq (Mei et al., 2022) and qRT–PCR. Differences in candidate genes between the GMS mutant and wild-type 898WB were identified to confirm the gene that was mainly responsible for GMS mutation. The SMART database (http://smart.embl-heidelberg.de/) was used to predict the domain of the candidate gene, and Mega7.0 was used to construct the phylogenetic tree by Neighbor-Joining method, and other parameters were the default values of the software.
CRISPR/Cas9-mediated knockout
According to the 5ʹ-GGNN18NGG-3ʹ design rule of CRISPR/Cas9 target, two single-guide RNA (sgRNA) sites in the ORF region of BmC/EBPZ gene were designed by the online software CRISPR direct (http://crispr.dbcls.jp/). The DNA sequence containing the two targets of sgRNA1 and sgRNA2 of the BmC/EBPZ gene was amplified. We ensured whether the two sgRNA sites of the genomic DNA of the Nistari strain had mutations by cloning and sequencing to prevent CRISPR/Cas9 off-target. Two pairs of primers containing sgRNA sites were used to amplify the plasmids constructed in our laboratory, and then the sgRNA sequence was homologously recombined with the initial plasmid (pXL [IE1-DsRed-U6]) to produce transgenic plasmids containing sgRNA sites (pXL [IE1-DsRed-U6-sgRNA]). The sgRNA transgenic plasmid contains a DsRed gene that encodes a red fluorescent protein and two sgRNA sites. The DsRed gene was initiated by, I.E.,1, and the sgRNA was initiated by U6 promoter. The primers were designed according to the recombinant plasmid, and plasmid integrity was verified by cloning and sequencing.
Nistari fertilized eggs were prepared, and then the sgRNA transgenic plasmid was mixed with the PiggyBac helper plasmid and microinjected into the prepared silkworm eggs. The eggs were regarded as the G0 generation, and then the adults of the G0 generation self-mated. The sgRNA-positive silkworms that expressed red fluorescent proteins were screened under a fluorescence microscope (Nikon AZ100, Japan), and the G1-positive individuals were mated with the Cas9 transgenic silkworms expressing green fluorescent protein. Finally, double-positive silkworms that expressed both fluorescence proteins were screened in the G2 generation, which were the knockout strains.
Three mutant individuals were randomly selected and genomic DNA was extracted. PCR amplification was performed using cloned primers containing two sgRNA sites, and the amplified sequence was cloned into pMD18-T vector (TaKaRa, China) for sequencing verification. Finally, it was determined whether the target gene was successfully edited by the CRISPR/Cas9 system.
Results
Candidate genes of the GMS mutant were located on the 24th linkage group
Genomic DNA was extracted from the GMS mutant, P50, and BC1F individuals, and the extracted DNA was amplified by SSR primers for the screening of polymorphic molecular markers. The genomic DNA of 10 mutant and 10 wild-type individuals in the same batch of BC1F populations were extracted. The SSR primers with polymorphisms were used for PCR amplification and verification of the linkage relationship between the SSR polymorphic markers and GMS mutant genes. When PCR amplification was performed with polymorphic marker primer chr24-71-7 on the 24th linkage group of silkworms, the results showed that the DNA typing of mutant individuals in the BC1F populations were the same as that of the GMS mutant parental strain, and the DNA typing of wild-type individuals was the same as that of F1 offspring or GMS mutant parental strain (Figure 1A). The result indicated that the GMS mutant gene was located on the 24th linkage group of silkworms. The sequences of the primer chr24-71-7 are listed in Supplementary Table S1.
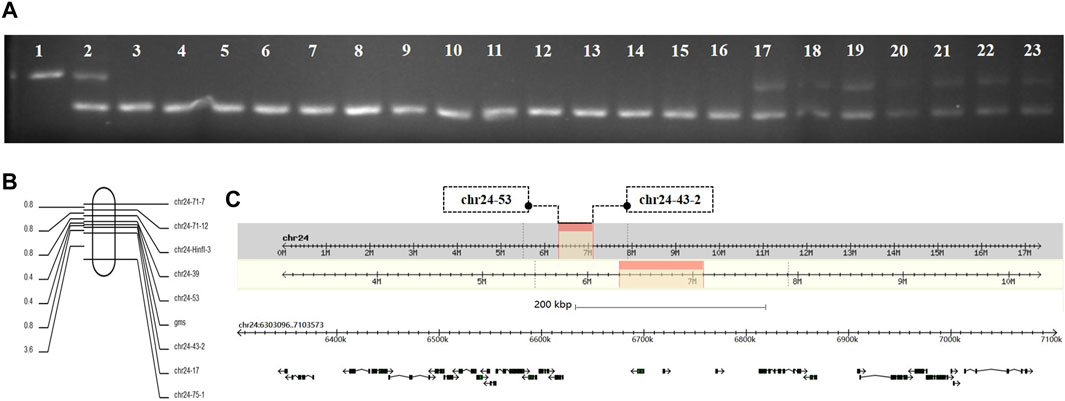
FIGURE 1. Genetic linkage group analysis of GMS candidate genes. (A) Lanes 1, 2, and 3 are the parent strain P50, BC1F offspring, and GMS mutant parent, respectively. Lanes 4–13 are BC1F populations with mutant phenotypes. Lanes 14–23 are BC1F populations with wild-type phenotypes. (B) Genetic linkage map between the GMS candidate gene and polymorphic molecular markers. (C) The physical map of the GMS candidate gene located on the 24th linkage group. The black box with arrows represents the 27 candidate genes.
Construction of the SSR molecular marker genetic linkage map
To determine the accurate location of the GMS mutant gene on the 24th linkage group, we used 252 BC1M mutants for genotype analysis. The genomic DNA of each mutant was used as a template, and primers for PCR amplification was performed using all the SSR polymorphic markers on the 24th linkage group (Supplementary Table S1). The DNA typing of mutants in the BC1M populations should be the same as that of the GMS mutant parental strain. When the GMS mutant gene of the F1 male parent showed genetic exchange with the SSR marker, the DNA typing of mutant individuals in the BC1M populations was be the same as that of the F1 offspring. According to the results of genotype analysis, a genetic linkage map of the molecular markers was drawn by Mapdraw software (Figure 1B). The mutant gene was located between polymorphic markers chr24–53 and chr24-43-2. The genetic distance between the two markers was 7.54 cM, and the physical distance was 800.5 kb (Figure 1C).
BmC/EBPZ gene was identified as the major gene responsible for GMS mutation
Fine mapping results showed that the mutant gene was located between two polymorphic markers chr24–53 and chr24-43-2 on the 24th linkage group, and this specific region contained 27 annotated genes (Supplementary Table S2). According to our transcriptome sequencing results (Mei et al., 2022), the expression levels of and structural variations in the candidate genes were analyzed. Then, on the basis of the RNA-Seq and Sanger sequencing results, a 9 bp-long sequence was inserted into the 3′end of the ORF sequence of the BmC/EBPZ gene in the GMS mutant (Figure 2A). The qRT–PCR results showed that the BmC/EBPZ gene was differentially expressed between the GMS mutant and wild-type 898WB (Figure 2B). The relative expression levels of the BmC/EBPZ gene in the different tissues of the wild-type strain (P50) were analyzed. The results showed that the expression level of BmC/EBPZ gene was higher in the testis, trachea, malpighian tubule, ovary, lower epidermis, fat body, ventral nerve cord, and brain, but nearly no expression was observed in the midgut (Figure 2C). The above data indicated that the BmC/EBPZ gene was the major gene mainly responsible for GMS mutation. The qRT–PCR primers are listed in Supplementary Table S3.
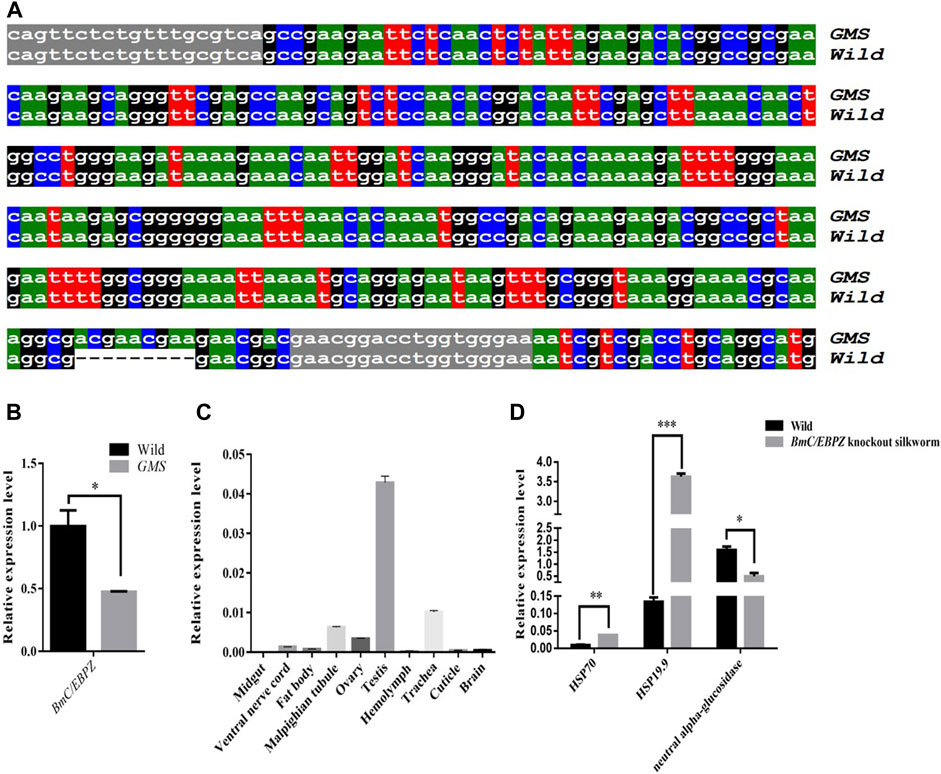
FIGURE 2. Sequencing results and relative expression analysis of the BmC/EBPZ gene. (A) Sequence alignment of the partial ORF of BmC/EBPZ gene between the GMS mutant and wild-type 898WB. Gray marker sequences indicate the position of the cloning primer. “---” indicates that the sequence deletion. (B) The relative expression levels of the BmC/EBPZ gene between the GMS mutant and wild-type 898WB. (C) The relative expression levels of the BmC/EBPZ gene in different tissues of P50. (D) The relative expression levels of HSP70, HSP19.9 and neutral alpha-glucosidase between the BmC/EBPZ gene knockout strain and wild-type 898WB.
CRISPR/Cas9-mediated knockout of the BmC/EBPZ gene
To verify whether the BmC/EBPZ gene was the major gene responsible for GMS mutation, we knocked out the expression of the BmC/EBPZ gene in the Nistari strain with the CRISPR/Cas9 system. We designed sgRNA 1 and sgRNA 2 on exons 8 and 10 in the BmC/EBPZ gene, respectively (Figure 3A). Two pairs of primers, each with two sgRNA site sequences, were synthesized, and the primers were named sgRNA1F, sgRNA1R, sgRNA2F, and sgRNA2R (Supplementary Table S4). We used the plasmids constructed in the laboratory and the above two pairs of primers for PCR amplification and recovered the amplified fragments. We used two amplified fragments and the initial plasmid to obtain a transgenic plasmid (pXL [IE1-DsRed-U6-sgRNA]) containing two sgRNA sites through homologous recombination (Supplementary Figure S2). The schematic diagram of how to connect two sgRNAs in one plasmid is shown in Supplementary Figure S3. The sequencing result of the recombinant plasmid is shown in Figure 3B. The primers were named V-F and V-R (Supplementary Table S4). We prepared a mixed injection containing recombinant plasmid (400 ng/μL) and PiggyBac helper plasmid (200 ng/μL), and injected 300 eggs. A total of 300 Nistari silkworm eggs were injected, 51% of the eggs successfully hatched into ant-silkworms, and 80% of the ant-silkworms survived to adults. The proportion of red fluorescent transgenic silkworms was 20%. The sgRNA transgenic silkworms expressing the red fluorescent protein was mated with the Cas9 transgenic silkworm expressing the green fluorescent protein to produce the BmC/EBPZ knockout strains. We obtained 11 moth batches of eggs that hatched into G2 generation mutants. Two moth batches of eggs were unfertilized, and nine moth batches of eggs were fertilized. The proportion of the fertilized eggs expressing double fluorescence was 25%. Compared with the control silkworms, all the knockout strains had small larvae, and the larvae died before the third instar. The larvae in the knockout strains varied in body size (Figure 3C). To confirm whether the above phenotypes were caused by the knockout of BmC/EBPZ gene, we extracted the genomic DNA of the BmC/EBPZ knockout strains and verified it by sequencing. Compared with the control group, the BmC/EBPZ knockout strains had different degrees of variation in genomic DNA (Figure 3D). This result confirmed that the BmC/EBPZ gene had been successfully knocked out in the BmC/EBPZ knockout strains. We performed qRT-PCR analysis on the three genes of HSP70, HSP19.9 and neutral alpha-glucosidase. The results showed that the above three genes were differentially expressed between the BmC/EBPZ gene knockout strain and the wild-type 898WB. In the BmC/EBPZ gene knockout strain, the expression levels of HSP70 and HSP19.9 genes were upregulated, while the expression levels of neutral alpha-glucosidase gene were downregulated (Figure 2D).
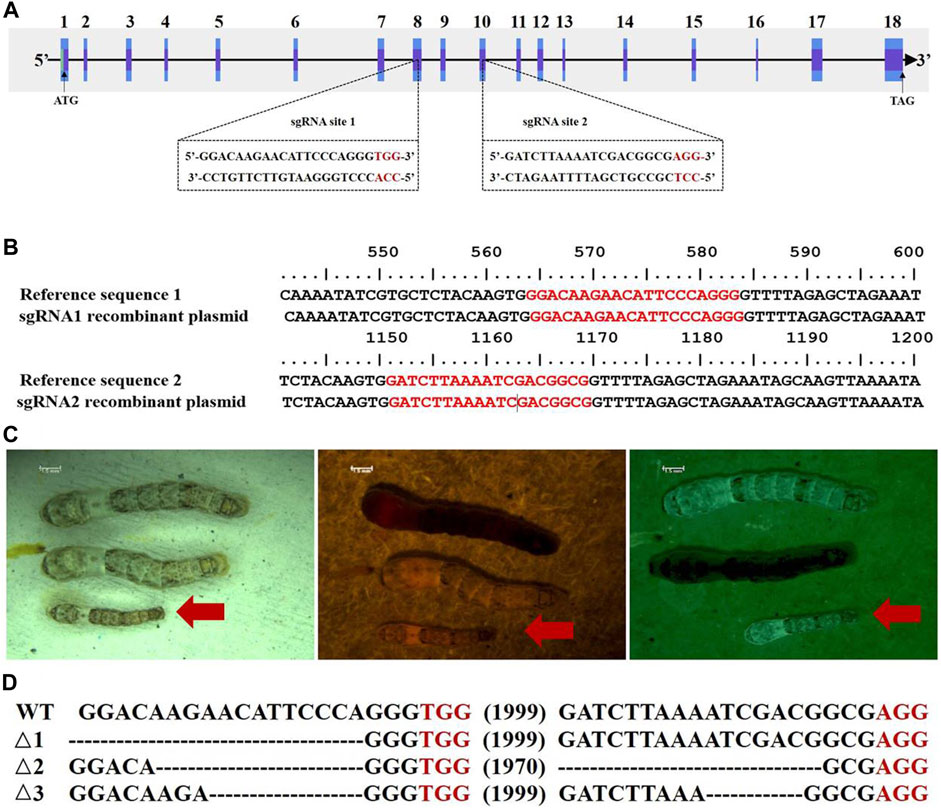
FIGURE 3. CRISPR/Cas9-mediated knockout of the BmC/EBPZ gene. (A) Schematic diagram of the sgRNA-target site in the BmC/EBPZ gene. The boxes represent exons, the black sequences represent sgRNA sequences, and the red sequences represent the protospacer adjacent motif sequences. (B) Sequencing results of sgRNA recombinant plasmid. The red sequences represent two sgRNA sequences. (C) Phenotype of the BmC/EBPZ knockout strains. Larvae under natural light (left), larvae under red light (middle) and larvae under green light (right). Larvae that can express red and green fluorescence proteins are the BmC/EBPZ knockout strains (red arrows). (D) Various deletion genotypes in the BmC/EBPZ knockout strains. WT represents wild-type 898WB. Δ1, Δ2, and Δ3 represent the three different knockout strains.
Structure and phylogenetic analysis of the BmC/EBPZ gene
The GMS mutant was caused by the 9 bp insertion in the BmC/EBPZ gene. To predict the function of the BmC/EBPZ gene in silkworms, we analyzed the protein domain with the SMART database. The BmC/EBPZ gene contained a CBF domain, which was present in the C/EBPs (Figure 4A). The two sgRNA sites were within the CBF domain of the BmC/EBPZ gene. The phylogenetic tree analysis of amino acid sequences from different species showed that the evolutionary relationship between silkworm and lepidoptera pests were close (Figure 4B).
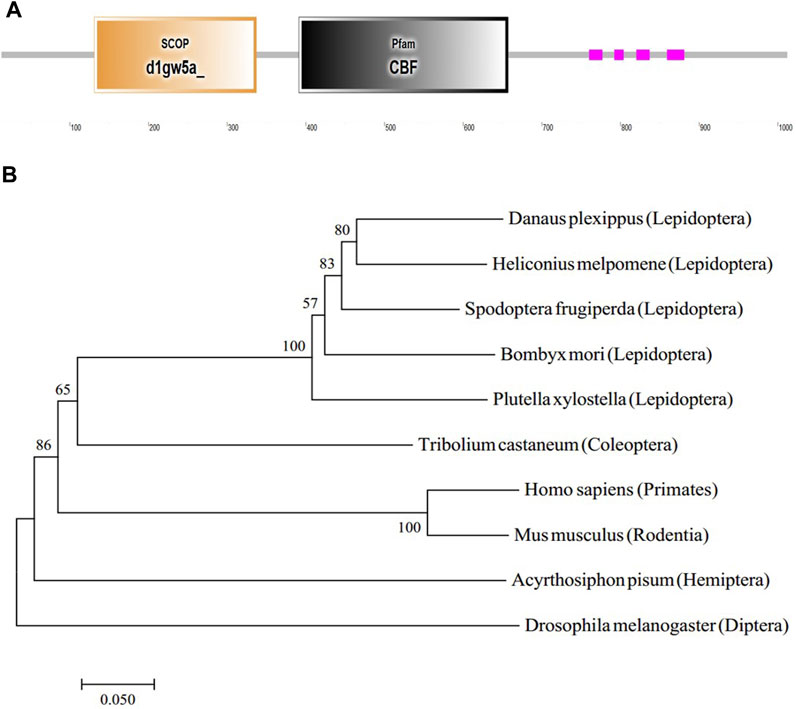
FIGURE 4. Structure and phylogenetic relationship of the BmC/EBPZ gene. (A) The predicted gene structure of the BmC/EBPZ gene. (B) Phylogenetic tree of the BmC/EBPZ gene in silkworms and other species.
Discussion
The GMS mutation is spontaneous and occurs during mass rearing for silk production. We explored the molecular mechanism underlying this mutation by RNA-Seq (Mei et al., 2022). By conducting positional cloning and using a CRISPR/Cas9 system, we found that the BmC/EBPZ gene had a relatively high expression level in the testis. The deletion of the gene affected the growth and development of the silkworms. These results suggested that the BmC/EBPZ gene is the major gene responsible for GMS mutation.
C/EBPZ, the human homolog, belongs to the C/EBP family. In acute myeloid leukemia, C/EBPZ binds to the transcriptional initiation site of the METTL3 gene and may play an important role in inflammatory response and cell differentiation (Liu et al., 2020). The genome sequences of patients with acute myeloid leukemia and gastric cancer were studied. Some of the patients carried C/EBPZ gene mutations, suggesting that the C/EBPZ gene can be used as a diagnosis or prognosis biomarker for the two types of cancer (Herold et al., 2014; Kori and Gov, 2022). Studies on the other members of the C/EBP family, such as C/EBP β and C/EBP δ, have shown that these transcription factors regulate the expression levels of genes involved in immune and inflammatory responses (Pérez-Pulido et al., 2021; Spek et al., 2021). The tissue-specific expression element C/EBP is located in the promoter of the NF-kb sequence and plays an important role in insects’ innate immunity (Liu et al., 2017). Moreover, C/EBP plays a key regulatory role in insect growth and development by enhancing histone acetylation. In silkworms, 20-hydroxyecdysone enhances the expression of the BmC/EBP gene through its receptor, thereby activating the expression of BmCBP gene, which then enhances histone H3K27 acetylation (Lyu et al., 2020).
KEGG pathway analysis of differential genes between the GMS mutant and 898WB showed that the significantly enriched pathways were metabolic pathways and ECM-receptor interaction (Mei et al., 2022). C/EBP-2, a homolog of C/EBPs, affects fat storage in C. elegans by regulating the expression of key enzymes in the energy metabolism pathway, and C/EBP-2 deficiency has been observed in Caenorhabditis elegans with a low degree of lipid accumulation (Xu et al., 2015). Lipogenesis is regulated by central cascade transcription factors, including CEBPs (Xiong et al., 2021). A transcriptome data analysis of glioma patients showed that the ECM–receptor interaction is the most significant pathway in the KEGG pathway and the number of genes that directly interacted with C/EBP in the differential co-regulation network is extremely large, indicating that CEBP plays an essential role in glioma (Aouacheria et al., 2006). C/EBPZ containing a CBF domain is essential for growth and 60S ribosomal subunit biogenesis (Edskes et al., 1998). Other proteins containing this domain stimulate transcription from the HSP70 promoter (Edskes et al., 1998). C/EBPZ can regulate cell growth and differentiation and is highly tissue specific (O'Rourke et al., 1997). In this study, we successfully constructed the BmC/EBPZ gene knockout strain. After the BmC/EBPZ gene was knocked out, we found that the expression levels of heat shock signal transduction pathway related genes HSP70 and HSP19.9 were significantly upregulated, while the expression level of glycogen metabolism pathway related gene neutral alpha-glucosidase was significantly downregulated. The results suggest that the BmC/EBPZ gene may be involved in the regulation of heat shock signal transduction pathway and glycogen metabolism pathway.
The two sgRNA sites designed in this study were found within the CBF domain of the BmC/EBPZ gene and resulted in various degrees of deletion in the BmC/EBPZ gene of the BmC/EBPZ knockout strains, leading to the loss of function of the BmC/EBPZ gene. As mentioned above, the BmC/EBPZ gene was essential for the growth and development of organisms. The deletion of the coding sequence of the BmC/EBPZ gene should be the main factor of small larvae and even lethal of the knocked out strains. However, a 9 bp insertion was found in the coding sequence of the 17th exon (non-CBF domain) of the BmC/EBPZ gene in the GMS mutant. Compared with the gene deletion of the BmC/EBPZ knockout strains, the insertion of the short fragment in the nonfunctional domain should have less functional impact on the BmC/EBPZ gene. Therefore, the GMS mutant showed small larvae, whereas the BmC/EBPZ knockout strains were lethal in addition to small larvae. These results demonstrated that the BmC/EBPZ gene regulated silkworm growth.
We also studied the relative expression level of the BmC/EBPZ gene in different tissues. The gene was highly expressed in the testis and ovary. This results was consistent with the results of (Yokoi et al., 2021). The RT-qPCR results suggested that the BmC/EBPZ gene is involved in the regulation of reproduction-related genes. The phylogenetic tree of the C/EBPZ gene of different species showed that the silkworms have a close evolutionary relationship with other Lepidoptera insects such as plutella xylostella and spodoptera frugiperda. Therefore, the study of the BmC/EBPZ gene of silkworms can facilitates the development of novel technologies for controlling Lepidoptera pests.
Data availability statement
The coding sequence of BmC/EBPZ gene of the GMS mutant has been submitted to GenBank with the accession number, OR751402.1(https://www.ncbi.nlm.nih.gov/genbank/).
Author contributions
XM: Conceptualization, Investigation, Methodology, Writing–original draft. TH: Conceptualization, Methodology, Software, Writing–review and editing. AC: Methodology, Writing–review and editing. WL: Formal Analysis, Writing–review and editing. LJ: Formal Analysis, Writing–review and editing. SZ: Formal Analysis, Writing–review and editing. DS: Investigation, Writing–review and editing. PQ: Formal Analysis, Writing–review and editing. QZ: Conceptualization, Methodology, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (No. 31972616, QZ), and the Graduate Student Scientific Research Innovation Projects of Jiangsu Province (grant number KYCX20_3104, XM) and the Key R&D Program of Jiangsu Province (Modern Agriculture) (No. BE2021346, QZ).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2024.1298869/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Comparison of body size between GMS mutant and wild-type 898WB.
SUPPLEMENTARY FIGURE 2 | Structure of the transgenic plasmid (pXL [IE1-DsRed-U6-sgRNA]).
SUPPLEMENTARY FIGURE 3 | The schematic diagram of how to connect two sgRNAs in one plasmid.
References
Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., et al. (1990). A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9, 1897–1906. doi:10.1002/j.1460-2075.1990.tb08316.x
Aouacheria A., Navratil V., Barthelaix A., Mouchiroud D., Gautier C. (2006). Bioinformatic screening of human ESTs for differentially expressed genes in normal and tumor tissues. BMC genomics 7, 94–11. doi:10.1186/1471-2164-7-94
Balamurugan K., Sterneck E. (2013). The many faces of C/EBPδ and their relevance for inflammation and cancer. Int. J. Biol. Sci. 9, 917–933. doi:10.7150/ijbs.7224
Cao Z., Umek R. M., Mcknight S. L. (1991). Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes and Dev. 5, 1538–1552. doi:10.1101/gad.5.9.1538
Chowdhury S., Singh A. K., Srivastava S., Upadhyay V., Sethi A., Siddiqui S., et al. (2023). AIP4 regulates adipocyte differentiation by targeting C/EBPα for ubiquitin-mediated proteasomal degradation. J. Cell. Biochem. 124, 961–973. doi:10.1002/jcb.30421
Edskes H. K., Ohtake Y., Wickner R. B. (1998). Mak21p of Saccharomyces cerevisiae, a homolog of human CAATT-binding protein, is essential for 60 S ribosomal subunit biogenesis. J. Biol. Chem. 273, 28912–28920. doi:10.1074/jbc.273.44.28912
Ghani L. A., Yusenko M. V., Frank D., Moorthy R., Widen J. C., DöRNER W., et al. (2022). A synthetic covalent ligand of the C/EBPβ transactivation domain inhibits acute myeloid leukemia cells. Cancer Lett. 530, 170–180. doi:10.1016/j.canlet.2022.01.024
Hartl L., Roelofs J. J., Dijk F., Bijlsma M. F., Duitman J., Spek C. A. (2023). C/EBP-Family redundancy determines patient survival and lymph node involvement in PDAC. Int. J. Mol. Sci. 24, 1537. doi:10.3390/ijms24021537
Herold T., Metzeler K. H., Vosberg S., Hartmann L., RöLLIG C., StöLZEL F., et al. (2014). Isolated trisomy 13 defines a homogeneous AML subgroup with high frequency of mutations in spliceosome genes and poor prognosis. Blood, J. Am. Soc. Hematol. 124, 1304–1311. doi:10.1182/blood-2013-12-540716
Jin F., Li Y., Ren B., Natarajan R. (2011). PU.1 and C/EBP(alpha) synergistically program distinct response to NF-kappaB activation through establishing monocyte specific enhancers. Proc. Natl. Acad. Sci. 108, 5290–5295. doi:10.1073/pnas.1017214108
Johnson P. F., Mcknight S. L. (1989). Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 58, 799–839. doi:10.1146/annurev.bi.58.070189.004055
Kaisho T., Tsutsui H., Tanaka T., Tsujimura T., Takeda K., Kawai T., et al. (1999). Impairment of natural killer cytotoxic activity and interferon γ production in CCAAT/enhancer binding protein γ–deficient mice. J. Exp. Med. 190, 1573–1582. doi:10.1084/jem.190.11.1573
Kori M., Gov E. (2022). Bioinformatics prediction and machine learning on gene expression data identifies novel gene candidates in gastric cancer. Genes 13, 2233. doi:10.3390/genes13122233
Lefterova M. I., Zhang Y., Steger D. J., Schupp M., Schug J., Cristancho A., et al. (2008). PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes and Dev. 22, 2941–2952. doi:10.1101/gad.1709008
Lekstrom-Himes J., Xanthopoulos K. G. (1998). Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273, 28545–28548. doi:10.1074/jbc.273.44.28545
Li H.-X., Xiao L., Wang C., Gao J.-L., Zhai Y.-G. (2010). Review: epigenetic regulation of adipocyte differentiation and adipogenesis. J. Zhejiang Univ. Sci. B 11, 784–791. doi:10.1631/jzus.B0900401
Liang L. N., Zhang L. L., Zeng B. J., Zheng S. C., Feng Q. L. (2015). Transcription factor CAAT/enhancer-binding protein is involved in regulation of expression of sterol carrier protein x in Spodoptera litura. Insect Mol. Biol. 24, 551–560. doi:10.1111/imb.12182
Liu S., Zhuo L., Wang J., Zhang Q., Li Q., Li G., et al. (2020). METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 10, 1631–1646.
Liu X., Li J., Wang Q., Xia H., Chen K. (2017). Functional analysis of hemolin gene from silkworm, Bombyx mori-immune and development. Invertebr. Surviv. J. 14, 330–339. doi:10.25431/1824-307X/isj.v14i1.330-339
Lyu H., Xu G., Chen P., Song Q., Feng Q., Yi Y., et al. (2020). 20-Hydroxyecdysone receptor-activated Bombyx mori CCAAT/enhancer-binding protein gamma regulates the expression of BmCBP and subsequent histone H3 lysine 27 acetylation in Bo. mori. Insect Mol. Biol. 29, 256–270. doi:10.1111/imb.12630
Mei X., Gao M., Huang T., Shen D., Xia D., Qiu Z., et al. (2022). Comparative analysis of testis transcriptome between a genetic male sterile line (GMS) and its wild-type 898WB in silkworm, Bombyx mori. Comp. Biochem. Physiology Part D Genomics Proteomics 42, 100961. doi:10.1016/j.cbd.2022.100961
Nerlov C. (2007). The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 17, 318–324. doi:10.1016/j.tcb.2007.07.004
O'Rourke J., Yuan R., Dewille J. (1997). CCAAT/enhancer-binding protein-δ (C/EBP-δ) is induced in growth-arrested mouse mammary epithelial cells. J. Biol. Chem. 272, 6291–6296. doi:10.1074/jbc.272.10.6291
Pang B., Zhang J., Zhang X., Yuan J., Shi Y., Qiao L. (2021). Inhibition of lipogenesis and induction of apoptosis by valproic acid in prostate cancer cells via the C/EBPα/SREBP-1 pathway. Acta Biochimica Biophysica Sinica 53, 354–364. doi:10.1093/abbs/gmab002
PéREZ-Pulido A. J., Asencio-CortéS G., Brokate-Llanos A. M., Brea-Calvo G., RodríGUEZ-GriñOLO R., GarzóN A., et al. (2021). Serial co-expression analysis of host factors from SARS-CoV viruses highly converges with former high-throughput screenings and proposes key regulators. Briefings Bioinforma. 22, 1038–1052. doi:10.1093/bib/bbaa419
Pham D. D., Douglass P., Chavez C., Shaffer J. (2005). Regulation of the ferritin heavy-chain homologue gene in the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 14, 223–236. doi:10.1111/j.1365-2583.2004.00550.x
Ramji D. P., Foka P. (2002). CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575. doi:10.1042/BJ20020508
Renfro Z., White B. E., Stephens K. E. (2022). CCAAT enhancer binding protein gamma (C/EBP-γ): an understudied transcription factor. Adv. Biol. Regul. 84, 100861. doi:10.1016/j.jbior.2022.100861
Roman C., Platero J. S., Shuman J., Calame K. (1990). Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes and Dev. 4, 1404–1415. doi:10.1101/gad.4.8.1404
Ron D., Habener J. F. (1992). CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes and Dev. 6, 439–453. doi:10.1101/gad.6.3.439
RøRTH P., Montell D. J. (1992). Drosophila C/EBP: a tissue-specific DNA-binding protein required for embryonic development. Genes and Dev. 6, 2299–2311. doi:10.1101/gad.6.12a.2299
Sourmeli S., Papantonis A., Lecanidou R. (2005). A novel role for the Bombyx Slbo homologue, BmC/EBP, in insect choriogenesis. Biochem. biophysical Res. Commun. 337, 713–719. doi:10.1016/j.bbrc.2005.09.103
Spek C. A., Aberson H. L., Butler J. M., De Vos A. F., Duitman J. (2021). CEBPD potentiates the macrophage inflammatory response but CEBPD knock-out macrophages fail to identify CEBPD-dependent pro-inflammatory transcriptional programs. Cells 10, 2233. doi:10.3390/cells10092233
Tavor S., Vuong P. T., Park D. J., Gombart A. F., Cohen A. H., Koeffler H. P. (2002). Macrophage functional maturation and cytokine production are impaired in C/EBPε-deficient mice. Blood, J. Am. Soc. Hematol. 99, 1794–1801. doi:10.1182/blood.v99.5.1794
Tenen D. G. (2003). Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. cancer 3, 89–101. doi:10.1038/nrc989
Wang L., Li H., Wang J., Gao W., Lin Y., Jin W., et al. (2011). C/EBP ζ targets to neutrophil gelatinase-associated lipocalin (NGAL) as a repressor for metastasis of MDA-MB-231 cells. Biochimica Biophysica Acta (BBA)-Molecular Cell Res. 1813, 1803–1813. doi:10.1016/j.bbamcr.2011.06.010
Xiong Y., Wang Y., Xu Q., Li A., Yue Y., Ma Y., et al. (2021). LKB1 regulates goat intramuscular Adipogenesis through focal adhesion pathway. Front. Physiology 12, 755598. doi:10.3389/fphys.2021.755598
Xu X.-Y., Hu J.-P., Wu M.-M., Wang L.-S., Fang N.-Y. (2015). CCAAT/enhancer-binding protein CEBP-2 controls fat consumption and fatty acid desaturation in Caenorhabditis elegans. Biochem. biophysical Res. Commun. 468, 312–318. doi:10.1016/j.bbrc.2015.10.106
Keywords: Bombyx mori, BmC/EBPZ, GMS mutant, small larvae, positional cloning, CRISPR/Cas9
Citation: Mei X, Huang T, Chen A, Liu W, Jiang L, Zhong S, Shen D, Qiao P and Zhao Q (2024) BmC/EBPZ gene is essential for the larval growth and development of silkworm, Bombyx mori. Front. Physiol. 15:1298869. doi: 10.3389/fphys.2024.1298869
Received: 22 September 2023; Accepted: 19 February 2024;
Published: 07 March 2024.
Edited by:
Muhammad Nadeem Abbas, Southwest University, ChinaReviewed by:
Kui Zhang, The University of Chicago, United StatesZhengying You, Zhejiang Sci-Tech University, China
Hui Xiao, Shaanxi Normal University, China
Copyright © 2024 Mei, Huang, Chen, Liu, Jiang, Zhong, Shen, Qiao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiaoling Zhao, qlzhao302@126.com
†These authors have contributed equally to this work and share first authorship
 Xinglin Mei
Xinglin Mei Tianchen Huang
Tianchen Huang Anli Chen
Anli Chen Weibin Liu1
Weibin Liu1