- Department of Pediatrics, Neonatal Intensive Care, Vito Fazzi Regional Hospital, Lecce, Italy
About two decades ago, cloning of the autoimmune regulator (AIRE) gene materialized one of the most important actors on the scene of self-tolerance. Thymic transcription of genes encoding tissue-specific antigens (ts-ags) is activated by AIRE protein and embodies the essence of thymic self-representation. Pathogenic AIRE variants cause the autoimmune polyglandular syndrome type 1, which is a rare and complex disease that is gaining attention in research on autoimmunity. The animal models of disease, although not identically reproducing the human picture, supply fundamental information on mechanisms and extent of AIRE action: thanks to its multidomain structure, AIRE localizes to chromatin enclosing the target genes, binds to histones, and offers an anchorage to multimolecular complexes involved in initiation and post-initiation events of gene transcription. In addition, AIRE enhances mRNA diversity by favoring alternative mRNA splicing. Once synthesized, ts-ags are presented to, and cause deletion of the self-reactive thymocyte clones. However, AIRE function is not restricted to the activation of gene transcription. AIRE would control presentation and transfer of self-antigens for thymic cellular interplay: such mechanism is aimed at increasing the likelihood of engagement of the thymocytes that carry the corresponding T-cell receptors. Another fundamental role of AIRE in promoting self-tolerance is related to the development of thymocyte anergy, as thymic self-representation shapes at the same time the repertoire of regulatory T cells. Finally, AIRE seems to replicate its action in the secondary lymphoid organs, albeit the cell lineage detaining such property has not been fully characterized. Delineation of AIRE functions adds interesting data to the knowledge of the mechanisms of self-tolerance and introduces exciting perspectives of therapeutic interventions against the related diseases.
Introduction
Surface receptors, enzymes, hormones, structural proteins, and other molecules act as self-antigens (self-ags) and are susceptible to autoimmune targeting in adverse circumstances. In a significant number of cases, these substances are restricted to specific tissues and for this reason are named tissue-specific antigens (ts-ags). The notion that ts-ag-encoding genes are transcribed and translated into their respective proteins within the thymus, the so-called promiscuous gene expression (PGE), dates back to the eighties, when neurohypophyseal hormones, insulin-like growth factors, and other ts-ags were found in the human and animal gland (1–4).
Later, a quantitative correlation between PGE and negative selection was established: in 1997, two research groups assayed human insulin gene (INS) expression in thymi of aborted fetuses and children dead at various ages. The researchers found that the allele classes of the variable number of tandem repeats (VNTRs) upstream of INS promoter, the so-called type-1 diabetes (T1D) susceptibility locus 2, affected INS transcription, and suggested that higher amounts of thymic insulin could promote a more effective purge of the related self-reactive thymocyte clone (5–7). Similar studies supplied valuable data on thymic PGE and led to identify several markers of autoimmunity, but did not realize the extent of the phenomenon (8–10).
In 1998, Sospedra et al. stated that the human thymus contains self-ags belonging to three classes: those synthesized in peripheral tissues and circulating at high, moderate, or low concentration; those synthesized in peripheral tissues and ordinarily undetectable in the circulation; finally, secluded self-ags, such as the retinal-S antigen and the myelin basic protein (11). Noticeably, PGE amount showed marked inter-individual variability, as confirmed by later studies (12, 13).
In 2001, Derbinski et al. assayed the expression of a large set of ts-ag-encoding genes in murine thymic stromal cells: cortical and medullary thymic epithelial cells (cTECs and mTECs, respectively), dendritic cells (DCs), and macrophages. All gene transcripts were found in mTECs, and around 50% of them were restricted to this cell sublineage (14). Detection of mRNAs from five selected genes was first obtained in 15-embryonic-day (15E) embryos and persisted into late adulthood. PGE was enhanced in UEA1hi mTECs (UEA1 stays for Ulex europaeus agglutinin 1). UEA1 labeling, in turn, was related to the co-stimulatory cluster of differentiation CD80, and, to a lesser degree, to class-II major histocompatibility complex (MHCII) antigens. Importantly, the expression of the autoimmune regulator (Aire) gene, which encodes the homonymous transcription factor, exhibited close distribution and timing (14), so that the study prompted the scientific community to inquire into the role of Aire in thymic self-representation and tolerance.
The present review is devoted to the fundamental aspects of Aire action and adverse consequences caused by its deficiency. Unless referring to the human counterparts (AIRE gene and AIRE protein), author will cite ordinarily murine gene (Aire) and protein (Aire), as the main body of scientific studies on this topic has been carried out on the animal models of disease. With regard to PGE, which is only in part dependent on Aire, author refers the kind readers to excellent reviews that delineate its extent and principles (15–17).
Ontogenesis of TECs
Generation of Mature TECs
In the murine thymus, Aire mRNA and Aire are traceable since 14E–15E (14, 18–20). Interestingly, in one of these studies the authors were able to detect Aire transcripts on a first-strand cDNA panel from 11E embryos (19). In this sense, a Chinese research group found that Aire is expressed in undifferentiated embryonic stem cells (ESCs), where it is co-stained with the stage-specific embryonic antigen 1, and that such expression attenuates upon ESC differentiation (21, 22). In ESCs, Aire associates with the spindle apparatus and plays a critical role in mitotic events (23). Hidaka et al. reported similar findings in embryoid bodies (24).
Many efforts have been produced to identify the thymic epithelial progenitor cells (TEPCs) from which Aire+ mTECs descend. Transplantation of endodermal cells of the third pharyngeal pouch from avian inter-species chimeras (25) and ectodermal-cell tracking in murine embryos (26) show that both cTECs and mTECs come from the endoderm, so that it is widely accepted that TEPCs are bipotent (27–31). In the simplest model of cTEC/mTEC commitment, TEPCs give rise simultaneously to sublineage-restricted elements. However, various research groups, on the basis of cTEC differentiation stages (32), have demonstrated that Aire+ mTECs derive from TEPCs exposing cTEC-associated markers, such as CD205, the thymoproteasome subunit β5t and the atypical CC-chemokines receptor (CCR)L1, and that such lineage persists in the postnatal thymus (33–36). Also interleukin (Il)7, which is required for T-cell development, is released by cTECs, and Il7hi cTECs can generate CD80+ mTECs through Il7–CD80lo elements (37). From this perspective, it has been possible to elaborate a model of cTEC/mTEC commitment in which mTEC sublineage diverges from a defaulted program of cTEC differentiation (38), as shown in Figure 1. Interestingly, in early organogenesis, the tight-junction claudins 3 and 4 mark the future Aire+ mTECs at the apex of the primordial endodermal layer (39). In the last few years, the researchers have focused their attention on TEPC characterization in the thymus of adult (at least 4-week-old) mice, applying different experimental settings and marker panels (40–45). Once again, markers of predetermined commitment to Aire+ mTECs have been identified (46, 47).
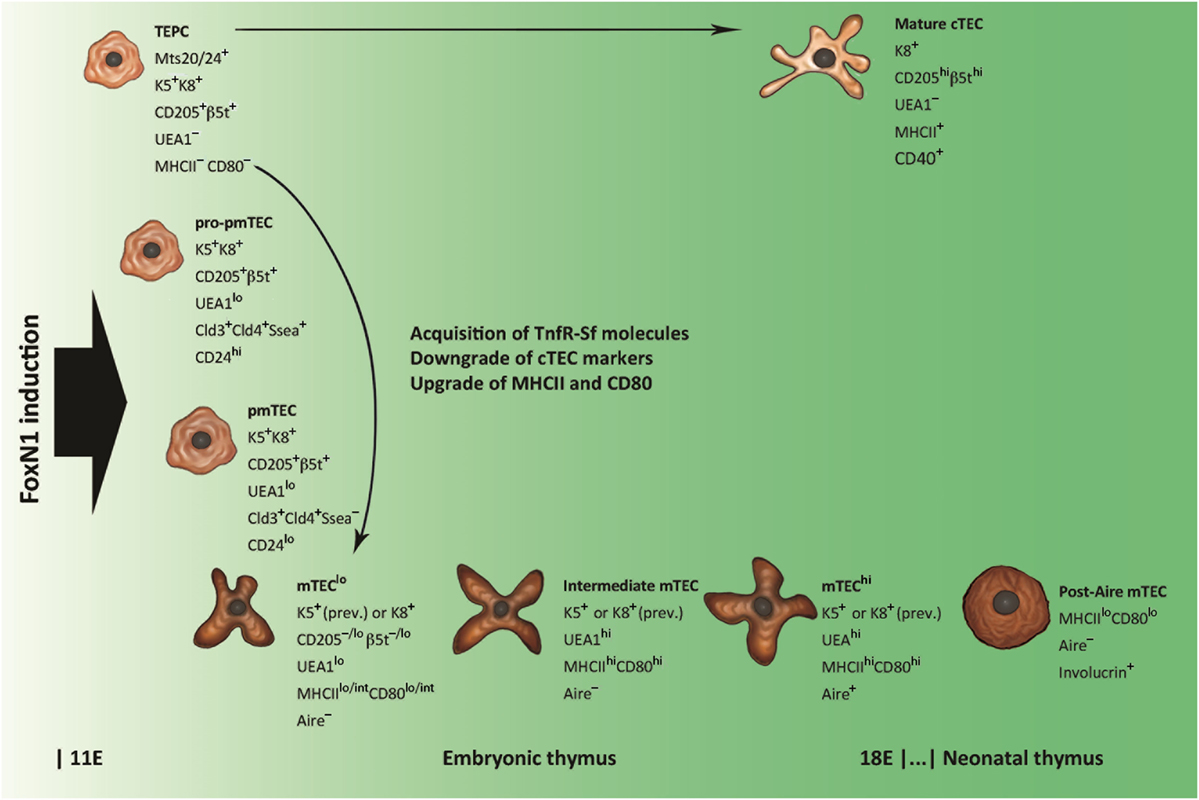
Figure 1. Schematic representation of thymic epithelial cell (TEC) differentiation. Thymic epithelial progenitor cell (TEPC) is tagged by mouse thymic stroma antibodies 20/24 (Mts 20/24), synthesizes intracellular keratins (Ks) 5 and 8 (K5 and K8, respectively), and exhibits surface markers associated with mature cortical TEC (cTEC), such as the cluster of differentiation CD205 and the thymoproteasome subunit β5t. Commitment to medullary TEC (mTEC) sublineage is restricted to claudine (Cld)-exposing elements, which, through intermediate stages of mTEC pro-precursor and precursor (pro-pmTEC and pmTEC, respectively), generate the immature mTEC (mTEClo). mTEClo differentiation into mature mTEC (mTEChi) is accompanied by enhancement of Ulex europaeus agglutinin 1 (UEA1) labeling and further upgrading of class-II major histocompatibility complex (MHCII) antigens and CD80. Lymphostromal interaction (thymic “crosstalk”) drives the emergence of pro-pmTECs by induction of molecules of the tumor necrosis factor-receptor super-family (TnfR-Sf), such as the lymphotoxin-β receptor (LtβR) and the receptor activator of nuclear factor Nf-κB (Rank). The transition from pro-pmTECs to pmTECs is characterized by loss of the stage-specific embryonic antigen 1 (Ssea) and results in a Rankhi condition. Loss of Aire expression and acquisition of keratinocyte markers typify a subset of post-Aire mTECs that emerge in the postnatal thymus.
Finally, immature cTECs and mTECs deal with the differentiation program leading to full maturity. All TECs expose the epithelial-cell adhesion molecule (EpCAM), but, while mature cTECs have a rather homogeneous phenotype, two distinct mTEC subsets exist: UEA1hi and UEA1lo mTECs, also called mTECshi and mTECslo, respectively (14, 32, 48–56). Distribution of keratins (Ks) into these subsets is not selective (48, 49, 55, 56); conversely, MHCII antigens and CD80 associate preferentially with the former (14, 50, 51, 54, 56). The expression of Aire and most ts-ag-encoding genes, in turn, is restricted to the mature, MHCIIhi or CD80hi, mTECs (14, 53, 56). These subsets represent about the same elements, which derive from their immature MHCIIloCD80lo precursors (57–59). Proliferation markers and the pattern of regeneration after pharmacological ablation indicate that a stock of MHCIIhiCD80hiAire− mTECs at an intermediate stage of differentiation exists (58–60), while predisposition to apoptosis suggests that Aire typifies terminally differentiated mTECs (57–61). Not in opposition to this evidence, later observations delineate a post-Aire mTEC stage, characterized by loss of Aire expression, suppression of PGE, reversion to MHCIIloCD80lo condition, and synthesis of keratinocyte proteins, such as desmogleins and involucrin, a soluble precursor of the envelope of the epidermal stratum corneum (62–64).
Transcriptional Regulation and Thymic “Crosstalk”
Although not fully known, there is a strict regulation of TEC ontogenesis. The thymic compartmentalization requires the transcription factor forkhead-box (Fox)N1, which is encoded at the “nude” locus: although referred to as athymic, the nude mice display an organ rudiment that includes TECs at an early stage of differentiation and is devoid of lymphoid progenitors (65). More recently, Nowell et al. have demonstrated that FoxN1, although dispensable for sublineage commitment, drives cTECs and mTECs along the program of differentiation (66). In the murine thymus, loss or downregulation of FoxN1 expression subverts the organ morphology mimicking a precocious senescence (67, 68), while FoxN1 upregulation reactivates TEPCs and reverses organ aging (69–71). These observations suggest that the thymic microenvironment reacts to FoxN1 in a dosage-sensitive manner and that FoxN1 expression is regulated in accordance with age (72).
While cTEC differentiation is induced by thymocytes at an early stage of maturation, mTEC differentiation is dependent on their full maturation and relocation. This lymphostromal interaction, the so-called thymic “crosstalk,” is achieved through two pathways enabling the nuclear factors Nf-κB (73–75). Both pathways are triggered by intercellular signals between the tumor necrosis factor (Tnf) and tumor necrosis factor-receptor (TnfR) super-families (Tnf-Sf and TnfR-Sf, respectively). TnfR-Sf members exposed on mTEC surface are the lymphotoxin-β receptor (LtβR), the receptor activator of nuclear factor Nf-κB (Rank), and CD40. LtβR and CD40 are also exposed on cTEC surface (32). There is a bulk of experimental data from the studies on the role and consequences of loss and gain of function of these molecules in the embryonic and postnatal/adult thymus (76–114). The cited studies are those that, on a targeted basis, have evaluated the impact of these changes on the generation and differentiation of Aire+ mTECs.
Unsurprisingly, differences in murine strains employed, experimental settings, and timing of observation have produced contrasting results in a number of cases. However, recent studies have set out some basic principles, highlighting that LtβR and Rank cooperate in the embryonic thymus to switch TEPCs to mTEC sublineage, while in the following step mTEC precursors become Rankhi (92, 95, 106). The release of the respective ligands is provided by T-cell subsets, such as lymphoid-tissue inducer cells and dendritic epidermal T cells, generated prior to the conventional αβ-thymocytes (98, 100, 102). Post-Aire mTEC differentiation and crosstalk of the postnatal thymus require inputs different from those acting in the embryonic period (91, 100, 101, 103). Presumably, thymic B cells and DCs participate in these processes (115, 116), while, if crosstalk is suppressed, coarse medullary cysts form, which are circumscribed by polarized mTECs (117). A careful dissection of the matter goes beyond the author scope, but essential aspects are reported in Table S1 in Supplementary Material. In addition, author refers the kind readers to excellent reviews that have thoroughly analyzed crosstalk dynamics, and the role and essentiality of each molecule involved (118–121).
Several other factors may exert inducing or inhibiting influence over mTEC development: of particular importance are the fibroblast growth factors (Fgfs), mainly Fgf7 (or keratinocyte growth factor), which is required for TEC differentiation in thymic organogenesis and regeneration (122). In murine models of graft-versus-host disease, administration of Fgf7 has proven to be decisive in the enrichment and maintenance of Aire+ mTECs able to promote T-cell reconstitution and avoid self-tolerance breaking (123–127).
Interestingly, mTEC differentiation may be reproduced in vitro by three-dimensional organotypic co-cultures engineered for dermal equivalent and based on the close relationship between skin and thymic stroma (128).
AIRE Gene and the Related Protein
Human AIRE is encoded by a gene located in the region 22q.3 of chromosome 21 (129, 130). Pathogenic AIRE variants cause the autoimmune polyglandular syndrome type 1 (APS1), characterized by chronic surface candidiasis and various autoimmune diseases involving mainly the endocrine glands (131, 132).
Murine Aire maps to chromosome 10 in a region syntenic to human 21q22 (18–20). Similarly to the human gene, Aire expression is restricted to a few cells of the thymic medulla, represented by a significant percentage of mTECshi, and, to a lesser degree, by DCs (76, 133). Presumably, Aire is synthesized and acts also in the secondary lymphoid organs, while, as reviewed elsewhere, Aire expression in other systems and cell lineages is uncertain and of doubtful meaning (134, 135).
Biophysical and Biochemical Properties
Analysis of its multidomain structure reveals that human AIRE belongs to the group of proteins able to bind to chromatin and regulate the process of gene transcription (136, 137). Starting from the N-terminus, AIRE comprises (Figure 2) a caspase-activation and recruitment domain (CARD), a nuclear localization signal (NLS), a SAND domain, and two plant-homeodomain (PHD) fingers (138). At subcellular level, AIRE localizes into small speckles uniformly distributed in the nucleoplasm and resembling the promyelocytic-leukemia nuclear bodies (NBs). In addition, it is visualized in the cytoplasm of a variable number of cells, where it forms a scaffold-like meshwork reminiscent of the intermediate filaments or microtubules (139–141). As observed in cultures of human mTECs and AIRE-transfected cell lines, AIRE has a subcellular organization following spatio-temporal cycles, and associates with the nuclear matrix (142, 143).
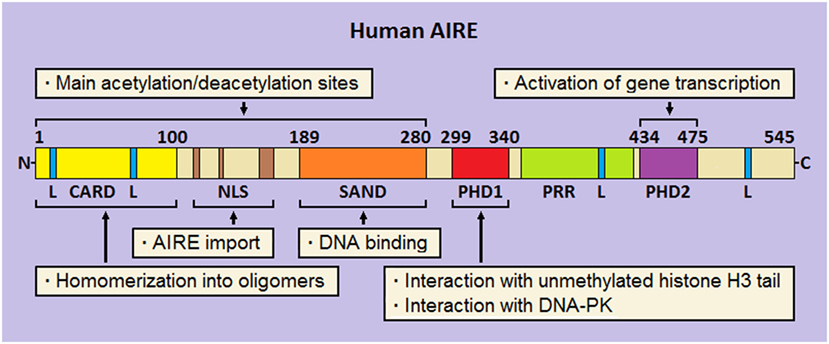
Figure 2. Schematic representation of human autoimmune regulator (AIRE). At the N-terminus, the caspase-activation and recruitment domain (CARD) and nuclear localization signal (NLS) are flanked by the SAND domain. Moving to the C-terminus, two plant-homeodomains (PHD1 and PHD2, respectively) fingers are separated by a proline-rich region (PRR). Four LxxLL (L stays for leucine) motifs are enclosed in the amino-acid chain. Preeminent domain-related properties are reported.
Homomerization into oligomers (dimers and tetramers) is an important biophysical property of AIRE, which allows binding to specific oligonucleotide motifs (144, 145). Suggestively, construction of a library of thymic consensus sequences highlighted that the promoters of several genes, among which those encoding ts-ags targeted by autoimmunity in Aire-deficient (Aire−/−) mice, enclose such motifs, albeit this mechanism represents a non-specific way of action of the protein (146).
Ability to homomerize is attributed to the AIRE N-terminus, already named homogeneously staining region (aa 1–100) by analogy with the speckled-protein SP100 (147). Two research groups demonstrated that pathogenic AIRE variants and deletion constructs involving this domain prevent oligomer formation and are unable to activate gene transcription (148, 149). Later, Ferguson et al. individuated in AIRE N-terminus a CARD (150), which is typical of pro-apoptotic proteins (151). Beside CARD, a bi- or tri-partite NLS guarantees AIRE shuttle into and out of the nucleus (152, 153).
In the middle of the amino-acid chain, the SAND domain (aa 180–280) encloses a basic amino-acid module that mediates AIRE binding to the phosphate groups of DNA (154), albeit SAND actual role is probably that of offering an anchorage to heterologous proteins (155). Importantly, CARD, NLS, and SAND domain hold most AIRE lysine residues, which are sites of acetylation (Ac) (145, 152, 153): this is a key point for proper protein localization and participation in multimolecular complexes.
At the C-terminus, AIRE is completed by two PHD fingers, named PHD1 (aa 299–340) and PHD2 (aa 434–475), which are separated by a proline-rich region. PHD fingers are cysteine-rich domains characterized by a four-cysteine, one-histidine, three-cysteine motif, which coordinates two zinc ions (156). In general, PHDs “read” the chromatin marks, mainly the degree of methylation at the tail of histone H3: importantly, AIRE PHD1 belongs to the PHD subfamily that recognizes unmethylation of H3 tail as a distinct epigenetic mark (157–159). At molecular level, opposite charges on the reciprocal surfaces facilitate the electrostatic interaction between PHD1 and H3 (160), while the methylation of some H3 amino-acid residues, mainly Arg2 and Lys4, dissociates them (161, 162). Despite a structural resemblance with PHD1, PHD2 displays a positively charged surface that makes it unsuitable to interact with histones (160). Nonetheless, its structural integrity is crucial for the activation of gene transcription, as confirmed by inherent AIRE variants (163) and deletion of the murine homolog (164). Actually, even the thirty amino acids positioned at the end of AIRE C-terminus act as an autonomous domain (165).
Finally, it should be remembered that AIRE encloses four LxxLL (L stays for leucine) motifs typical of proteins that bind to nuclear receptors and affect, as either co-activators or co-repressors, the transcriptional events (166). Interestingly, the fourth LxxLL motif lies in the C-terminus and is critical for AIRE properties (165).
Molecular Mechanisms of Action
It is now clear that Aire does not act as a conventional transcription factor by binding to consensus sequences of the target gene promoters. Rather, the protein seems to participate in coordinated events performed by multimolecular complexes (Figure 3). Several studies have been produced to elucidate Aire’s partnerships and their functional relevance. An acceleration in this field has come from the study of Abramson et al., who used AIRE-targeted co-immunoprecipitation, mass spectrometry, and RNAi-mediated mRNA knockdown to identify the pool of associated proteins (167).
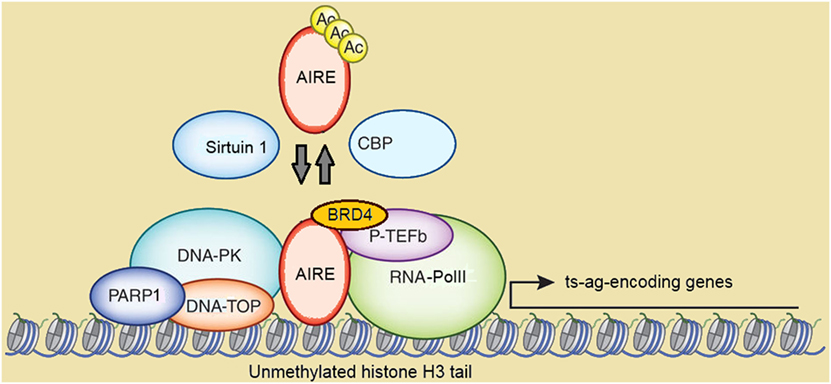
Figure 3. Schematic representation of autoimmune regulator (AIRE)-containing multimolecular complexes involved in initiation and post-initiation events of gene transcription. Abbreviations: Ac, acetylation; CBP, CREB-binding protein; DNA-PK, DNA-activated protein kinase; DNA-TOP, DNA-topoisomerase; PARP1, poly-(ADP-ribose) polymerase 1; BRD4, bromodomain-containing domain 4; P-TEFb, positive transcription elongation factor b; RNA-PolII, RNA-polymerase II; ts-ag, tissue-specific antigen. Reprinted (with changes) with the permission from Macmillan Publishers Ltd.: Peterson (171). Copyright 2015.
CREB-binding protein (CBP), which localizes in the NBs and is a co-activator of several transcription factors, was the first AIRE partner to be identified (148, 168). Following studies suggested that Ac by CBP stabilizes the subcellular distribution of AIRE, albeit data on targeted lysine residues and functional consequences were conflicting (169, 170). In a more recent study on murine mTECs, mapping of Aire lysine residues acetylated by CBP has been validated by bioinformatics-based candidate prediction. In this context, it has been highlighted that the group-III histone-deacetylase Sirtuin 1 preserves Aire-dependent PGE by deacetylation of such residues (171, 172).
Positive transcription elongation factor b (P-TEFb) and DNA-activated protein kinase (DNA-PK) are other AIRE partners (173–175). DNA-PK phosphorylates AIRE, at least in vitro, at Thr68 and Ser156 (174). Above all, DNA-PK belongs, together with other molecules co-immunoprecipitating with AIRE, to the multimolecular complex involved in DNA break and repair by non-homologous end joining (175). Among these molecules, a strong AIRE partner, as evidenced in proteomic assays, is the DNA-topoisomerase (DNA-TOP)IIα (167). DNA-TOPs are isomerase enzymes that operate on DNA topology and remove positive and negative DNA supercoils by generating transient DNA breaks: this causes local chromatin relaxation and facilitates the initiation and post-initiation events of gene transcription (176). DNA-TOPIIα performs double-stranded DNA breaks and attracts DNA-PK and poly-(ADP-ribose) polymerase 1 (PARP1). Recently, Bansal et al. have demonstrated that murine Aire and the above partners localize to long stretches of chromatin known as super-enhancers, which serve as depots of cell-specific multimolecular complexes involved in transcriptional events, and enclose the transcription start sites of most Aire-dependent genes. In the same study, the authors have indicated DNA-TopI, which introduces single-stranded DNA breaks, as a preeminent Aire partner upstream of DNA-TopIIα and DNA-TopIIβ (177). In another recent study, Guha et al. have clarified the details of the interaction between AIRE and DNA-TOPs: AIRE would exert a camptothecin- and etoposide-like function able to inhibit type-I and type-II DNA-TOP re-ligation activity. This is followed by chromatin changes attributable to DNA-PK and PARP1, and activates the transcription of low-expressed genes (178). Recently, a clinical picture resembling APS1 has been reported in two patients with pathogenic variants of the gene encoding the DNA-PK catalytic subunit. Unsurprisingly, PGE was impaired in patients’ fibroblasts transfected with AIRE (179).
Also the homeodomain-interacting protein kinase 2 (HIPK2), another serine-threonine protein kinase localized in the NBs, phosphorylates AIRE (and CBP) and exerts a repressive influence over the related properties. Interestingly, Hipk2-deficient (Hipk2−/−) mice undergo a PGE downgrade that mostly involves Aire-independent genes expressed in mTECslo, suggesting that Hipk2 operates on hypothetical transcription factors other than Aire (180).
The interaction with P-TEFb seals AIRE participation in the post-initiation events of gene transcription (173). In eukaryotic cells, gene transcription is abortive if P-Tefb does not enable elongation and pre-mRNA splicing into mature mRNA by phosphorylation and release of stalled RNA-polymerase II. As observed in human and murine cell lines, AIRE recruits P-TEFb at the transcription start sites of the target genes and enables the above sequence (181, 182). Moreover, Yoshida et al. found that the bromodomain-containing protein 4 (Brd4) forms a bridge between murine Aire and P-Tefb, and that balanced phosphorylation and Ac of Aire CARD are necessary to keep such interaction (183). Finally, interaction with the human heterogeneous nuclear ribonucleoprotein L suggests that AIRE enhances mRNA diversity by favoring alternative mRNA splicing (184), as confirmed in murine mTECs (185, 186).
Although the studies so far examined have provided a formidable contribution to the knowledge of the molecular mechanisms of Aire action, how the protein recognizes the target genes remains to be fully explained. PHD1 disruption abrogates the transcription of a part of human AIRE-dependent genes (159), while a histone H3-specific demethylase does not enlarge their number (187), so that the hypothesis that promoters of AIRE-dependent and AIRE-independent genes merely differ in chromatin marks is unsatisfying (188). A complementary mechanism may be the interaction between AIRE and the complex formed by activating-transcription-factor-7-interacting protein (ATF7IP) and methyl-CpG-binding-domain protein 1 (MBD1) (189). ATF7IP can act as either co-activator or co-repressor of gene transcription depending upon its partners, while MBD1 belongs to a family of nuclear proteins able to bind to methylated CpG dinucleotides, which characterize the promoter region of silent or low-expressed genes. Thus, coopting such repressive complex would recruit AIRE to the target genes, but the details of the interaction need further explanation. In another study, murine chromatin enclosing Aire-dependent genes exhibited marks of polycomb silencing, such as histone H3 hypomethylation at Lys4 and trimethylation at Lys27. Although Aire partnership with chromodomain-helicase-DNA members, which bind to these amino-acid residues, is controversial (163, 167), it has been suggested that such putative interaction would drive Aire to the target genes and activate gene transcription by overriding a repressive chromatin state (190).
Interaction with miRNAs
In the last few years, some research groups have put forward the hypothesis that Aire would be involved in post-transcriptional gene control by interaction with miRNAs, small (21–25 nucleotides in length) double-stranded non-protein-encoding RNAs, which join in silencing complexes able to cause translational block and mRNA degradation (191). TEC-restricted deletion of murine genes encoding molecules that participate in miRNA pathway makes the thymic environment unable to sustain thymocytes maturation and reach a proper PGE, with more or less obvious Aire dysregulation (192, 193). Observation of miRNA pattern changes throughout mTEC differentiation has led to identify a miRNA subset that affects specifically Aire mRNA translation (194–196). Conversely, Aire itself seems to condition amount and composition of miRNAs by modulating their transcription (197). Moreover, Aire would induce in genes involved in PGE a sort of refractoriness to the interaction with miRNAs, while in Aire deficiency a large number of miRNAs would achieve the target (198–200).
Mechanisms of Action in Central Tolerance
Once dissected the molecular mechanisms of Aire action, it is now appropriate to analyze in an orderly fashion the biological effects of such events. As expected, most information comes from studies on the animal models of disease. First Aire−/− mice were engineered in 2002: the animals exhibited lymphocyte infiltration invading or surrounding specific structures of various organs (for example, the portal spaces of the liver, or the gastric parietal cells, or the outer layer of the retina), paralleled by circulating antibodies to self-ags with a similar, although not exactly corresponding, pattern (201, 202). When bone marrow from either wild-type (Aire-sufficient, Aire+/+) or Aire−/− mice was transplanted into two mirror groups of lethally irradiated mice, organ infiltration and humoral autoimmunity were found only in Aire−/− recipients, independently from the donor condition (Figure 4). Obviously, PGE was impaired in Aire−/− thymi (202).
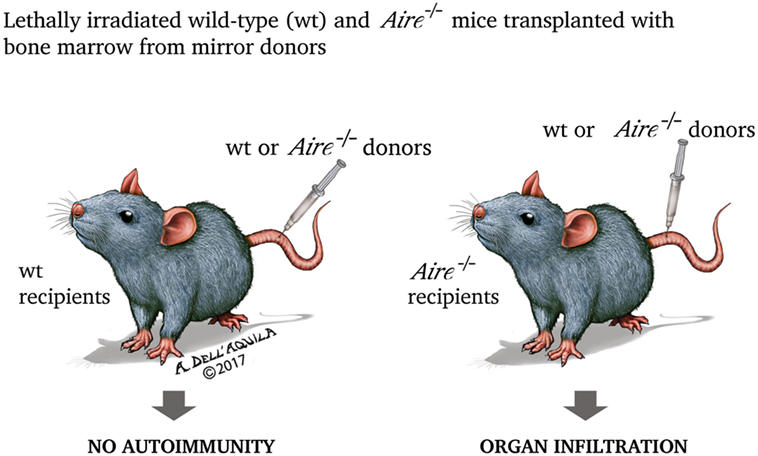
Figure 4. In the experiment by Anderson et al. (202), lethally irradiated (thus retaining only the radio-resistant stromal cells) wild-type and Aire−/− mice were transplanted with bone marrow from mirror donors. Only Aire−/− recipients, independently from the donor condition, exhibited organ damage, underlining that the property of preventing the autoimmune process is inherent to wild-type (Aire-sufficient) thymic stroma.
These principles were applied in a following study. Mice carrying T-cell receptor (TCR) loci immunized to the hen egg lysozyme were crossed with mice in which thymic expression of the related transgene was driven by the rat Ins promoter. The comparison between Aire+/+ and Aire−/− double-transgenic mice revealed that the former had a small number of TCR-specific thymocytes, which exhibited anergy markers, while failure of negative selection in Aire−/− mice caused spreading of the self-reactive T cells (203).
In the following subheadings, author will address the various modalities of Aire intervention in central tolerance: activation of PGE, presentation and transfer of self-ags, promotion of anergy by diversion to regulatory T (Treg) cells, and a putative influence over thymic cellularity.
Activation of PGE
Actually, PGE is not restricted to mTECs, but ts-ag-encoding genes expressed in cTECs are mostly lymphocyte-specific and are due to contamination by thymocytes complexed to thymic nurse cells, while those expressed in DCs and macrophages are mostly related to bone marrow-derived cell lineages (14). Recent studies indicate that Aire modulates, by induction of chemokine signals, cTEC gene transcription, and at the same time slows down cTEC metabolism and differentiation (204, 205). By contrast, Aire initiates PGE in mTECs (14, 202), as confirmed in fetal thymic organ cultures and cultures from adult thymi (206, 207). The process involves hundreds of genes whose expression overrides the ordinary sex-, tissue-, and differentiation-dependent regulation (14, 202, 208). However, Aire activates the transcription of a part of these genes, as demonstrated in Aire−/− mice (209, 210). Moreover, Aire-dependent and Aire-independent genes participating in PGE co-localize in chromosomal clusters (208–210): as seen, this phenomenon is due to the localization of Aire-containing multimolecular complexes in chromatin stretches enclosing the transcription start sites of the ts-ag-encoding genes (177).
Interesting data are available when, taking into consideration a set of functionally connected genes, thymic PGE is compared with the corresponding expression in the relevant peripheral tissue. For example, while murine casein genes (clustered on chromosome 5) are co-expressed in about 90% of mammary-gland cells of young female mice, the expression of the same genes in CD80hi mTECs exhibits a prevalence between 2 and 15%. The rate of mRNA translation into the respective proteins is even lower, so that each ts-ag is traceable in about 1–3% of mTECs (211). With regard to allele pairs, many mTECs use one chromosomal locus, with no obvious imprinting (212). At the same time, genes ordinarily imprinted in the peripheral tissues, such as the Aire-dependent gene encoding the insulin-like growth factor 2, may be expressed biallelically in mTECs (209). Another proof that gene transcription activated by Aire is regulated differently from the peripheral tissues is given by the observation that a selective deficiency in the pancreatic-duodenal homeobox 1, a master transcription factor encoded by an Aire-dependent gene, does not impair the thymic transcription of other Aire-dependent, pancreatic-islet-related genes (213).
Initially, although it was observed that clustered ts-ag-encoding genes have a higher chance of sharing the same fate, no clear pattern of co-expression emerged (211, 212). Recent studies have changed this perspective. Actually, single human mTECs shift through distinct pools of ts-ag-encoding genes. In this sense, some co-expression pools of overlapping and complementary gene sets have been individuated, which encompass intra- and inter-chromosomal distribution and align along a co-linear program of differentiation (214). Analogously, clustered Aire-dependent genes are expressed stochastically in small groups of murine mTECshi, with a significant degree of diversity between individuals (215, 216).
Other important observations may be added: first, Aire favors alternative mRNA splicing, which represents a broadening of thymic self-representation (184–186). Second, the pool of genes regulated by Aire is conditioned by the cellular environment, as demonstrated both in physiologic conditions, by comparing mTECs with the germ cells of the testis, where PGE addresses pulsed waves of scheduled apoptosis (217), and in experimental setting, by transfecting pancreatic-islet β-cells with Aire (218). Finally, the dichotomy between Aire-dependent and Aire-independent genes represents perhaps an improper simplification: it is possible that genes belonging to both categories are connected into transcriptional networks that recognize a hierarchy. This could explain how Aire regulates indirectly some genes, albeit an interaction with other transcription factors cannot be excluded (219, 220).
Self-ag Presentation and Transfer
Kuroda et al. found that Aire−/− mice display Sjögren’s syndrome-like disease of the exocrine glands, and this was associated with autoimmunity to the ubiquitous protein α-fodrin. Surprisingly, the expression of the encoding gene was not impaired by Aire deficiency, and the authors hypothesized that the autoimmune process was due to suboptimal antigen presentation and transfer (221). Initially, the features and timing of self-ag presentation by mTECs and thymic DCs were evaluated without taking into account Aire role (222, 223). Later, Hubert et al. found that some self-ags need to be transferred to the thymic DCs to be presented to the thymocytes, and that Aire is able to address this interplay (224). In another study, lethally irradiated mice transplanted with bone marrow deficient in the gene encoding the MHCII-transactivator—hereby forced to use only antigen-presenting cells (APCs) of epithelial lineage—had a higher frequency of T-cell clones with self-reactivity to an epitope of the interphotoreceptor retinoid-binding protein (Irbp), which is encoded by an Aire-dependent gene (225). More recently, it has been demonstrated that Aire+ mTECs release vesicles of endocytic origin called exosomes, which carry a high number of self-ags (226).
By contrast, another research group has provided evidence that mTECshi, through the process of macroautophagy, induce autonomously a proper thymocyte response (227). Interestingly, both Aire+ mTECs and DCs, when co-cultured with fresh thymocytes, act as APCs and re-propose in vitro the process of negative selection (228, 229).
At this point, it seems to be correct to state that self-ag presentation by mTECs and thymic DCs runs in parallel, but preeminence and degree of redundancy of the two sources remain to be deciphered (230). In a very recent study, Mouri et al. employing transgenic mice in which ovalbumin expression has been driven by either Aire or rat Ins promoter, delineate a division of labor between mTECs and thymic DCs, which configures uneven dependency on Aire and different outcomes in central tolerance (that is, negative selection versus Treg-cell generation) (231). To complicate matters, other recent data suggest that even thymic B cells display Aire expression and participate in self-ag presentation (232).
Generation of Treg Cells
As touched upon previously, thymus role in promoting self-tolerance relies not only on the process of negative selection, but also on the generation of Treg cells able to prevent and control the autoimmune process. Treg cells have a CD4+CD25+ phenotype and require FoxP3 to differentiate: initial studies suggested that Aire−/− mice have a normal number of circulating CD4+CD25+ cells (201–203), which, however, do not consist solely of the Treg-cell subset. Later, Anderson et al. (Figure 5) observed that nude mice co-engrafted with 2′-deoxyguanosine-resistant thymic stroma from wild-type and Aire−/− mice, or, in an alternative approach, recombinase-activating gene-1-deficient (Rag1−/−) mice treated with co-transfer of splenocytes from the above donors, exhibit organ infiltrates undistinguishable from those found in the animals engrafted with a single Aire−/− thymus or receiving splenocytes from Aire−/− mice only. Should Treg-cell impairment play a role in the adverse events deriving from Aire deficiency, generation of Treg cells in the co-engrafted wild-type thymus (or their presence among the co-transferred wild-type splenocytes) would prevent the autoimmune process. However, avoiding organ damage by introduction of an excess of thymic stroma (or splenocytes) from wild-type animals left reasonable doubts on the earlier conclusions (233). Similar data were obtained by Kuroda et al., albeit in this case co-engrafted thymi were employed at a fixed ratio (221). In a further study, Aire sufficiency did not enlarge, compared with a condition of Aire deficiency, Treg-cell TCR specificities on a background of TCR oligoclonality (234), but such experimental design (i.e., the utilization of transgenic mice with a restricted TCR repertoire) was questionable in itself.
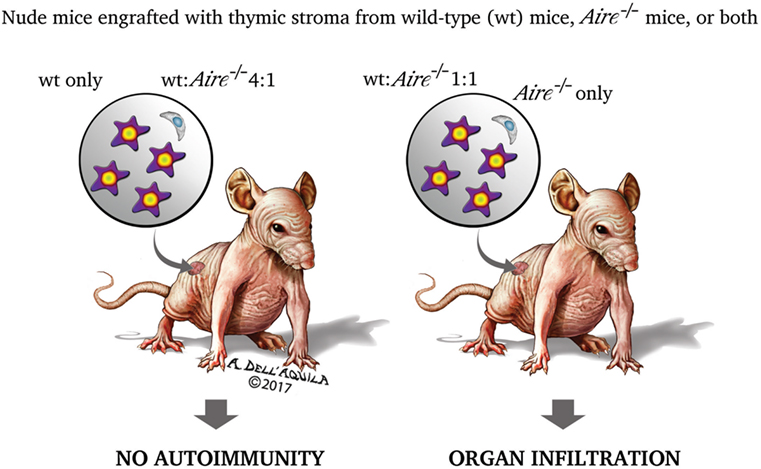
Figure 5. Two research group found that the organ damage of nude mice co-engrafted with 2′-deoxyguanosine-resistant thymic stroma from wild-type and Aire−/− donors matched that of the animals engrafted with a single Aire−/− stroma (221, 233). This led the authors to speculate that impairment of regulatory T cells (Treg cells) has no role in the autoimmune process caused by Aire deficiency. If it had, Treg cells generated in the wild-type stroma would prevent the onset of the disease. However, mice co-engrafted with an excess (4:1) of wild-type stroma had no organ damage, raising reasonable doubts on the earlier conclusions. The experiment was replicated with splenocytes co-transferred into recombinase-activating gene-1-deficient (Rag1−/−) mice, and yielded similar results.
On the other hand, the hypothesis of an Aire role in generating Treg cells moves from the observation that Aire deficiency exacerbates the organ damage in FoxP3-deficient (FoxP3−/−) mice (235). In this sense, a series of studies prove that self-ag presentation by Aire+ mTECs shapes the Treg-cell repertoire (227, 236–238). Critical factors for this process, whose efficiency correlates inversely with Treg-cell differentiation, are optimal affinity/avidity in TCR engagement and proper cytokine availability (237). Other observations propose a reissue of the relationship between Aire and conventional (effector) T cells: first, Aire+ mTECs act autonomously as APCs (227, 238), but cooperation with thymic DCs may be required for some self-ags (239, 240). Second, in the perinatal age Aire promotes the generation of a distinct compartment of Treg cells that persists into adulthood (241).
Some studies suggest that Aire promotes Treg-cell enrichment in the secondary lymphoid organs: Rag1−/− Aire+/+ recipients of T cells from Aire−/− mice show hyperproliferation of the FoxP3+ subset able to prevent overt autoimmunity (242). Coherently, consequences of Aire deficiency are made critical by constitutional defects or derailment of the mechanisms enabling Treg-cell action in the periphery (243, 244).
Unfortunately, given that Aire promotes central tolerance also to cancer-associated self-ags, generation of Treg cells with the related TCR specificities is a way to exert such unfavorable action (245–248). Finally, it is important to remember that, beside FoxP3+ major population, minor subsets of Treg cells exist: one of these, represented by CD8+CD28lo T cells, fails to control the onset of experimental colitis in Aire−/− mice (249).
Control of Thymic Cellularity
A putative role attributed to Aire is that of controlling mTEC molecular mediators that regulate thymic cellularity and dynamics (Figure 6). In this context, mTECs do not act as APCs, and their non-TCR-mediated influence relies on the production and release of cytokines.
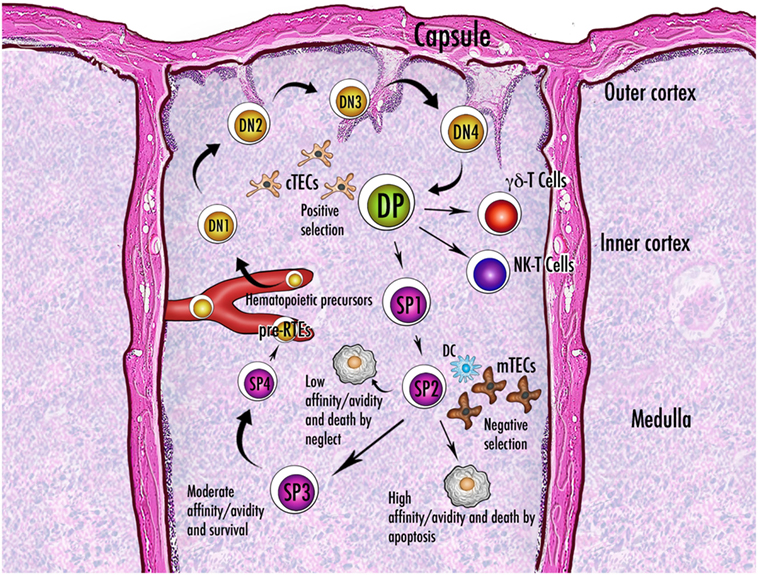
Figure 6. Schematic representation of thymocyte maturation and the processes of positive and negative selection. Thymocytes originate from bone marrow-derived pluripotent precursors, which in the most immature form lack the clusters of differentiation CD4 and CD8 and are referred to as double-negative (DN) cells. There are four stages (DN1–DN4) of DN condition, during which thymocytes move from the cortico-medullary junction to the subcapsular zone of the gland. The irreversible commitment to T-cell lineage intervenes in the passage from DN1 to DN2 condition, when expression of the recombinase-activating genes starts: only cells that succeed in rearrangement of the gene encoding the T-cell receptor (TCR) β-chain are selected for further maturation. After a brief DN4 stage, survived thymocytes become double-positive (DP) by acquisition of CD4 and CD8 and are allowed to rearrange the gene encoding the TCR α-chain. Now they deal with the processes of positive and negative selection and their fate is dictated by the interaction with the thymic stroma. Once positively selected to generate single-positive (SP) cells, the thymocytes reverse their direction and enter the medulla. While a large percentage of thymocytes do not run into an antigen fitting their TCR and die by neglect, the fate of the remainders depends on the degree of affinity with the cognate antigen. Generally, a strong affinity induces apoptosis and ensuing clonal deletion. An intermediate affinity may induce diversion to an anergic state, such as that of the regulatory T cells. In the theory of avidity, the amount of antigen determines the outcome of the interaction. There are three or four stages of maturation of the SP thymocytes, which finally reach the perivascular space as pre-recent thymic emigrants (pre-RTEs). Abbreviations: NK, natural killer; cTECs, mTECs, thymic epithelial cells (cortical, medullary); DC, dendritic cell.
The most important event is the cortex-to-medulla migration of the positively selected αβ-thymocytes. This non-inertial movement is elicited by various CC-chemokine ligands (CCLs) through their respective CCRs: CCL5, CCL17, and CCL22 interact with CCR4, while CCL19 and CCL21 interact with CCR7 (250). A lack of cytokine signal does not prevent thymocyte accumulation in the cortex and outflow from the thymus, but the process of negative selection is compromised (251). Conversely, after relocation, surviving single-positive thymocytes complete their maturation in three or four stages and enter the bloodstream as “recent thymic emigrants” (RTEs) (252). As reviewed by Cowan et al., intrathymic thymocyte migration is indispensable for the emergence of Treg-cell precursors, and involves at the same time thymocyte sublineages deputed to innate immunity, such as γδ-thymocytes and invariant natural-killer (iNK)-T cells (253).
Aire intervention in these events remains unclear. Laan et al. found that, in the murine thymus, Aire deficiency impairs the expression of the genes encoding CCR4 and CCR7 ligands, albeit the cellular source of the latter would not coincide with Aire+ mTECs (254). Other research groups found that LtβR signaling directs chemokine release by mTECs (81, 82, 85). Later, Lkhagvasuren et al. clarified that CCL21+ mTECs represent a distinct, LtβR-driven mTEC subset that emerges after the perinatal period and mostly segregates from Aire+ elements (93). A Chinese research group has dedicated a series of studies to the intra-medullary maturation of the CD4+ thymocytes, highlighting that a perinatal reduced outflow of RTEs, which play an important role in the establishment and maintenance of peripheral tolerance, deteriorates the detrimental effects of Aire deficiency (255–258). Further studies have been dedicated, with not univocal results, to Aire role in the generation, intrathymic migration and maturation of γδ-thymocytes, iNK-T cells, and DCs (259–263). Interestingly, recent studies suggest that Aire intervenes in regulating generation and function of Il17-releasing invariant and adaptive T cells, which have been linked to the early stage of the autoimmune processes (264, 265).
Organ Targeting in AIRE Deficiency
Experimentally, thymic deletion of a ts-ag-encoding gene leads ineluctably to the onset of the related autoimmune disease. Given that two murine Ins genes (Ins1 and Ins2) exist, only the latter being Aire-dependent (14, 202), Fan et al. used Cre-Lox recombination technology to restrict Ins2 deletion to Aire-expressing cells. Therefore, diabetes developed within 3 weeks. Murine strain was autoimmune-resistant and the animals displayed unimpaired tolerance to other self-ags. Importantly, the authors employed Ins1−/− mice to eliminate the interference of an Aire-independent factor, whose strength in the mechanisms of self-tolerance is undetermined (266). In the preceding study of DeVoss et al., nude mice engrafted with thymic stroma in which the Irbp gene was deleted, exhibited eye disease (267).
In contrast, Aire−/− condition causes a dysregulation, mostly a downgrade of expression, of the entire pool of Aire-dependent genes. In this chaotic perturbation of thymic PGE, the pathological consequences are determined by factors acting at various levels.
Species Specificity and Genetic Background
First, species-specific peculiarities cause remarkable differences between human APS1 and the phenotype of Aire−/− mice: in other words, the animal models of disease exhibit pathological findings not comparable with those of the APS1 patients (268–270). Nevertheless, studies on Aire−/− mice have made it possible to identify, with proven or potential connection to the human field, several targets of autoimmunity (271–278).
Moving to the intra-species level, the genetic background, more than Aire genotype, influences severity of disease and set of organs damaged in each individual, albeit in APS1 patients this is observable with some difficulty. To give a few examples of the link between geo-ethnic patient origin and clinical picture, Finnish APS1 patients have an increased prevalence of T1D (131), while autoimmune thyroiditis is common among those from Southern Italy (279). Again, chronic candidiasis is observed rarely in Iranian-Jewish APS1 patients, who generally exhibit a milder phenotype (280). It is not surprising that MHCII alleles are relevant to these differences (281).
Of course, the availability of highly inbred animal lines gives greater visibility to the phenomenon: murine autoimmune-prone strains, such as non-obese diabetic (NOD) and SJL mice, show a consistent and specific pattern of organ infiltration and self-reactivity. A relatively autoimmune-resistant strain, BALB mouse, has an intermediate prevalence of organ damage, which preferentially involves stomach and genital apparatus. Finally, an autoimmune-resistant strain, C57BL/6 (B6) mouse, shows a few components of the disease, with elective targeting to retina and prostate (221, 282). Susceptible alleles of the modifier loci—once again with privileged reference to MHCII ones—are necessary, but not always sufficient, to elicit organ damage: for example, the H2-Aβg7 allele was required to induce autoimmune pancreatitis in NOD Aire−/− mice, but was not sufficient when transferred to a B6 background (282). A related, unexpected phenomenon is the intra-organ targeting switch: typically, in NOD Aire−/− mice the autoimmune pancreatitis hits the exocrine part of the gland, and the release of autoantibodies to an acinar-cell self-ag complements the process (283).
Studies on murine chimeras add interesting data: engrafting 2′-deoxyguanosine-resistant thymic stroma from BALB or B6 Aire−/− mice into mirror nude animals, Han observed that Aire deficiency simply enhanced the restricted predisposition to autoimmunity of the recipients, independently from the genetic background of the donors. By contrast, when thymic stroma from NOD Aire−/− mice was engrafted, it dictated the spectrum of organ damage, indicating that Aire deficiency impinges on a constitutional derailment of PGE (284).
Gene Expression Variability
Not only the genetic background with which Aire deficiency overlaps, but also factors intrinsically related to Aire expression should be taken into account. Various studies demonstrate that the amount of mRNAs transcribed from murine Aire-dependent genes correlates with the level and timing of Aire mRNA (285–287), even in single cells (190). Age is an important factor able to modulate Aire expression: MHCII+ mTECs increase dramatically after birth and peak at 4 weeks of age (57). It is probable that the perinatal lymphopenia and ensuing lymphopenia-induced proliferation of Aire−/− mice are related to the infringement of the above trend and contribute to their pathological findings (288), which are reminiscent of the 3-day-thymectomized mice described by Miller (289). At the opposite, thymic involution, as depicted in 12-month-old mice, is featured by a fall in mTEC/cTEC and MHCIIhi/MHCIIlo ratios (57), and is caused by programmed aging of the primary lymphoid organs (290). The efficiency of the process of negative selection in the embryonic and neonatal thymus is confirmed by the study of Guerau-de-Arellano et al., who used a doxycycline-regulated transgene to control Aire expression, and found that self-tolerance established in the perinatal age is longstanding. The autoimmunity triggered by Aire deficiency was attenuated by transfer of previously tolerized T cells. Not surprisingly, lethal irradiation during Aire turn-off recreated the disease in adult mice (291). As cited earlier, a recent study highlights that Aire influences also the perinatal generation of Treg cells (241).
Sexual hormones seem to modulate central tolerance, explaining gender differences in susceptibility to autoimmunity. While castration prevents the decrease in thymic PGE observed in adult mice of either sex (292), androgens enhance Aire transcription and estrogens induce opposite changes acting at an epigenetic level (293, 294).
These physiologic variables score life periods at species level, but what can we say about inter-individual differences? Reappraising previous studies (11–13), Taubert et al. reiterated that human mTECs present a strong inter-individual disparity in AIRE expression and PGE. However, while mRNA from AIRE-independent genes displays restricted fluctuations uncorrelated with AIRE mRNA, variability in the transcription of the AIRE-dependent genes is wider and follows an obvious AIRE-related trend (295). Given that Liston et al., employing mice in which one Aire allele was deleted, found that PGE affects in quantitative terms the magnitude of self-reactive T cells escaping negative selection (296), it has been hypothesized that conditions of partial AIRE deficiency may represent a risk for non-syndromic autoimmunity when acting in synergy with other susceptibility factors. However, various research groups did not find an increased prevalence of AIRE variants among patients with sporadic, especially endocrine, autoimmune diseases (297–303). Instead, various patient reports suggest that some AIRE variants encode mutated chains that co-localize with the wild-type protein and undermine the activity of the oligomeric structure in a dominant manner (304–307). Reporter gene assays, in vitro structure modeling and homologous murine constructs validate such hypothesis (305–309). The resulting clinical picture is characterized by late-onset autoimmunity, milder phenotype than APS1, and incomplete penetrance (304–307).
The animal models of disease add valuable data that, once again, stress the importance of the genetic background: mTECs of murine autoimmune-prone strains display lower amounts of mRNAs from Aire and selected ts-ag-encoding genes (207, 310), and such dysregulation becomes more obvious in the stages preceding the overt disease (311). Based on the study of Venanzi et al., the difference relies on the strength of responsiveness to Aire and is no longer apparent when Aire−/− strains are compared. In the same study, the authors demonstrated that, similarly to the human thymus, there are marked inter-individual differences in the thymic expression of most ts-ag-encoding genes, even between mice homogeneously fed and housed: once again, the coefficient of variation is higher for the Aire-dependent genes and drops when the residual expression is assayed in Aire−/− thymi (312). According to the authors’ comment, this diversity may be beneficial in preventing uniform holes in central tolerance, but at the price of an unpredictable individual predisposition to autoimmunity.
AIRE and T1D: Special Case or Paradigm?
In author opinion, the relationship between APS1 and T1D resumes most principles regarding AIRE function. Insulin is a self-ag commonly targeted in T1D and encoded by an AIRE-dependent gene: this dependence was inferred from gene expression pattern in murine (14, 202) and human mTECs (208). Later, two research groups observed that, although class-III VNTR alleles induce a higher level of INS expression compared to class-I alleles, the thymic amount of insulin varies widely among individuals carrying the same VNTR haplotype and correlates better with AIRE expression (295, 313). Another research group demonstrated that AIRE is able to bind to class-I and class-III VNTRs, and that the complexes modulate INS expression (314).
At this point, one would expect that AIRE deficiency lead invariably to overt pancreatic-islet β-cell autoimmunity. Actually, T1D affects a minority of APS1 patients (131, 132, 315), so that it can be assumed that one or more additional factors modulate the related risk. Although initially no or weak influence was attributed to MHCII alleles (316, 317), following studies modified this perspective: Gylling et al. found that DQB1*0602 plays a protective role in the development of APS1-associated T1D (318). Similarly, Halonen et al. showed a negative correlation with DRB1*15-DQB1*0602 (281). Two later studies on APS1 patients drew once again attention to the risk conferred by the T1D susceptibility locus 2, but both research groups genotyped MHCII in a limited percentage of the sample and omitted to include these data in multivariate statistics (319, 320).
Therefore, we can conclude that AIRE exerts a chief role in the hierarchical regulation of thymic INS expression, but, in a condition of AIRE deficiency, unfavorable classes of VNTR alleles are needed to reduce INS transcription below a critical threshold and determine a failure in the process of negative selection. Other genetic variables, such as MHCII haplotype, may further stratify the risk by shaping organ susceptibility to autoimmunity.
AIRE: Not only “Central”
It is known that ts-ag-encoding genes are expressed also in the secondary lymphoid organs. Nonetheless, cell lineages holding this property and the related Aire role remain unresolved issues. According to two studies, main source of extra-thymic PGE would be Aire+ epithelial cells located within the lymph-nodal and splenic stroma. In an experimental setting, these cells were fostered to express an ovalbumin transgene driven by the promoter of the Aire-dependent gene encoding the intestinal fatty-acid-binding protein (321). In the other setting, Aire promoter itself drove the expression of the gene encoding the pancreatic-islet β-cell-specific glucose-6-phosphatase-related protein, a self-ag routinely undetectable in the thymus (322). Both cell types induced deletion of the TCR-specific CD8+ T-cell clones, even when the latter were transferred into lethally irradiated mice transplanted with bone marrow from β2-microglobulin-deficient donors. In this way, reconstituting DCs were unable to act as APCs (321, 322). Later, one of these research groups revised the phenotype of the extra-thymic Aire+ cells, identifying them in unconventional CD45loEpcam+MHCIIhiCD80lo bone marrow-derived APCs (323).
Reappraising their previous study (266), Grupillo et al. deleted Ins2 in CD11c-expressing cells of Ins1−/− mice. The splenic source of Aire expression and PGE was attributed to CD11cintMHCII+B220+ plasmacytoid DCs. Treg-cell number was unaffected, so that self-tolerance was necessarily deletional. An interesting aspect of this study was that only B6 mice crossed to adopt MHCII alleles typical of NOD mouse exhibited some degree of pancreatic-islet β-cell damage (324). Jointly, the studies of this research group (266, 324) suggest that thymic PGE plays a chief role in self-tolerance, and that thymic gene deletion causes inevitably the related autoimmune disease. By contrast, the same event in the secondary lymphoid organs leads to adverse consequences only on an autoimmune-prone genetic background.
Other researchers did not find a relationship between Aire and extra-thymic PGE (325, 326). In further studies, Aire was localized in uncharacterized cells of the secondary lymphoid organs (327), or even in the stroma of non-lymphoid organs where immune tolerance is strictly needed, such as the decidua basalis at the embryo implantation site (328).
Another Theory of Action
The mTEC developmental theory (329), which configures an alternative hypothesis on Aire function, moves from the observation that the pharyngeal arches can generate many types of tissue, and that distinct foci of mTECs are arranged in organoids resembling typical epithelial formations. Such organization suggests that the thymic medulla forms some sort of mosaic, whose pieces follow different programs of differentiation, and that mTECs with the largest PGE are intermediate elements that progressively restrict the pool of ts-ag-encoding genes expressed (330–334). Other evidences have been called into question, such as the small percentage of mTECs in which each ts-ag is detectable (335); the dependency on Aire of differentiation-associated genes and genes encoding master transcription factors (21–23, 336, 337); and the ultrastructure of Aire−/− thymus, in which expansion of the K8+ subset indicates mTEC inability to differentiate into distinct epithelial lineages (338, 339). Finally, the detection of a post-Aire mTEC stage (62) and the lack, once again in the murine Aire−/− thymus, of hyalinized structures equivalent to the human Hassall’s corpuscles (340), would provide the conclusive proof that Aire controls mTEC differentiation, and that its scheduled disappearance is a condition for the proper implementation of the latter (341–343). As discussed above, the same arguments have been used to build and support the well-defined theory that places Aire onto the high point of mTEC differentiation.
Is Aire expression the end-stage of an invariant differentiation program, albeit with a stochastic pattern of PGE, or does it enable, before to be lost, multiple and predetermined programs of mTEC differentiation (344, 345)? There is still no definite answer to this question, but, in author opinion, Aire mandate remains unchanged: to accomplish the largest PGE for self-tolerance induction.
Perspectives and Conclusion
To sum up, Aire activates the transcription of a large pool of ts-ag-encoding genes in mTECs. In Aire deficiency, missed self-ag presentation to the thymocytes determines a failure in the process of negative selection and the subsequent spreading of self-reactive T cells. The absolute heritable profile of the human related disease, APS1, suggests exciting implications on the topic of gene therapies.
A first approach is aimed at obtaining a functional thymus from ESCs. Following two studies in which murine ESCs were induced to generate TEPCs able to self-renew and foster thymocyte maturation (346, 347), two research groups replicated such results with human ESCs (348, 349). In one of these studies, TEPC re-aggregation with embryonic fibroblasts and following engraftment into nude mice led to mTEC differentiation and AIRE expression, albeit T-cell outflow from the thymus was short-lived (349). Similar results were achieved with human and murine induced pluripotent stem cells (350, 351).
Aire expression can be manipulated by immunologic (78, 79, 240), virus-based (285, 352), physical (286, 287), and chemical (353) methods. Nonetheless, enhancing Aire expression may impair unexpected forms of immune defense and get unwelcome surprises. As seen, genes encoding some cancer-associated ts-ags are Aire-dependent (245–248), so that it is not surprising that Aire−/− mice are able to provide a stronger immune response after melanoma challenge (354, 355).
Translating these data to hypothetical therapies of human autoimmune diseases, the cited studies suggest that, while restoring AIRE expression is the goal of gene therapy in APS1 patients, ideal profile of a tailored, AIRE-based treatment should be restricted to selected cell lineages or single AIRE-dependent genes, to avoid the pitfalls of a generalized PGE distortion.
Of course, just an increasing knowledge of PGE and the related Aire role will help to refine any strategy aiming at restoring, promoting, or strengthening the mechanisms of central and peripheral self-tolerance. Finally, author refers the kind readers to excellent preceding reviews, which recapitulate the course of discoveries over Aire, and mark chronologically doubts and insights into its function (356–380).
Author Contributions
RP is the only contributor to the Review.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author would like to thank the reviewers for their helpful and constructive comments that greatly contributed to improving this paper and the editor for his generous support during the review process. The author would also like to thank Dr. Antonio Dell’Aquila for the production of Figures 1 and 4–6.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fimmu.2018.00098/full#supplementary-material.
Abbreviations
Ac, acetylation; AIRE/Aire, autoimmune regulator; APC, antigen-presenting cell; APS1, autoimmune polyglandular syndrome type 1; ATF7IP, activating-transcription-factor-7-interacting protein; Brd4, bromodomain-containing domain 4; CARD, caspase-activation and recruitment domain; CBP, CREB-binding protein; CCL, CC-chemokine ligand; CCR, CC-chemokine receptor; DC, dendritic cell; DNA-PK, DNA-activated protein kinase; DNA-TOP/DNA-Top, DNA topoisomerase; E, embryonic day; EpCAM, epithelial-cell adhesion molecule; ESC, embryonic stem cell; Fgf, fibroblast growth factor; Fox, forkhead-box; HIPK2/Hipk2, homeodomain-interacting protein kinase 2; Il, interleukin; iNK, invariant natural killer; INS/Ins, insulin gene; Irbp, interphotoreceptor retinoid-binding protein; K, keratin; LtβR, lymphotoxin-β receptor; MBD1, methyl-CpG-binding-domain protein 1; MHC, major histocompatibility complex; NB, nuclear body; NLS, nuclear localization signal; NOD, non-obese diabetic; PARP1, poly-(ADP-ribose) polymerase 1; PGE, promiscuous gene expression; PHD, plant-homeodomain; P-TEFb/P-Tefb, positive transcription elongation factor b; Rag, recombinase-activating gene; Rank, receptor activator of nuclear factor Nf-κB; RTE, recent thymic emigrant; self-ag, self-antigen; Sf, super-family; T1D, type-1 diabetes; TCR, T-cell receptor; TEC (cTEC, mTEC), thymic epithelial cell (cortical, medullary); TEPC, thymic epithelial progenitor cell; Tnf, tumor necrosis factor; TnfR, tumor necrosis factor-receptor; Treg cell, regulatory T cell; ts-ag, tissue-specific antigen; UEA1, Ulex europaeus agglutinin 1; VNTR, variable number of tandem repeat.
References
1. Geenen V, Legros J-J, Franchimont P, Baudrihaye M, Defresne M-P, Boniver J. The neuroendocrine thymus: coexistence of oxytocin and neurophysin in the human thymus. Science (1986) 232(4749):508–11. doi:10.1126/science.3961493
2. Han VKM, D’Ercole AJ, Lund PK. Cellular localization of somatomedin (insulin-like growth factor) messenger RNA in the human fetus. Science (1987) 236(4798):193–7. doi:10.1126/science.3563497
3. Kirchner T, Hoppe F, Müller-Hermelink HK, Schalke B, Tzartos S. Acetylcholine receptor epitopes on epithelial cells of thymoma in myasthenia gravis. Lancet (1987) 329(8526):218. doi:10.1016/S0140-6736(87)90032-8
4. Fuller PJ, Verity K. Somatostatin gene expression in the thymus gland. J Immunol (1989) 143(3):1015–7.
5. Vafiadis P, Bennett ST, Colle E, Grabs R, Goodyer CG, Polychronakos C. Imprinted and genotype-specific expression of genes at the IDDM2 locus in pancreas and leucocytes. J Autoimmun (1996) 9(3):397–403. doi:10.1006/jaut.1996.0054
6. Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet (1997) 15(3):289–92. doi:10.1038/ng0397-289
7. Pugliese A, Zeller M, Fernandez A Jr, Zalcberg LJ, Bartlett RJ, Ricordi C, et al. The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet (1997) 15(3):293–7. doi:10.1038/ng0397-293
8. Heath V, Mason D, Ramirez F, Seddon B. Homeostatic mechanisms in the control of autoimmunity. Semin Immunol (1997) 9(6):375–80. doi:10.1006/smim.1997.0095
9. Hanahan D. Peripheral-antigen-expressing cells in thymic medulla: factors in self-tolerance and autoimmunity. Curr Opin Immunol (1998) 10(6):656–62. doi:10.1016/S0952-7915(98)80085-X
10. Werdelin O, Cordes U, Jensen T. Aberrant expression of tissue-specific proteins in the thymus: a hypothesis for the development of central tolerance. Scand J Immunol (1998) 47(2):95–100. doi:10.1046/j.1365-3083.1998.00280.x
11. Sospedra M, Ferrer-Francesch X, Domínguez O, Juan M, Foz-Sala M, Pujol-Borrell R. Transcription of a broad range of self-antigens in human thymus suggests a role for central mechanisms in tolerance toward peripheral antigens. J Immunol (1998) 161(11):5918–29.
12. Bruno R, Sabater L, Sospedra M, Ferrer-Francesch X, Escudero D, Martínez-Cáceres E, et al. Multiple sclerosis candidate autoantigens except myelin oligodendrocyte glycoprotein are transcribed in human thymus. Eur J Immunol (2002) 32(10):2737–47. doi:10.1002/1521-4141(2002010)32:10<2737::AID-IMMU2737>3.0.CO;2-0
13. Takase H, Yu C-R, Mahdi RM, Douek DC, DiRusso GB, Midgley FM, et al. Thymic expression of peripheral tissue antigens in humans: a remarkable variability among individuals. Int Immunol (2005) 17(8):1131–40. doi:10.1093/intimm/dxh275
14. Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol (2001) 2(11):1032–9. doi:10.1038/ni723
15. Kyewski B, Derbinski J, Gotter J, Klein L. Promiscuous gene expression and central T-cell tolerance: more than meets the eye. Trends Immunol (2002) 23(7):364–71. doi:10.1016/S1471-4906(02)02248-2
16. Kyewski B, Derbinski J. Self-representation in the thymus: an extended view. Nat Rev Immunol (2004) 4(9):688–98. doi:10.1038/nri1436
17. Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol (2006) 24:571–606. doi:10.1146/annurev.immunol.23.021704.115601
18. Mittaz L, Rossier C, Heino M, Peterson P, Krohn KJE, Gos A, et al. Isolations and characterization of the mouse Aire gene. Biochem Biophys Res Commun (1999) 255(2):483–90. doi:10.1006/bbrc.1999.0223
19. Blechschmidt K, Schweiger M, Wertz K, Poulson R, Christensen H-M, Rosenthal A, et al. The mouse Aire gene: comparative genomic sequencing, gene organization, and expression. Genome Res (1999) 9(2):158–66. doi:10.1101/gr.9.2.158
20. Wang C-Y, Shi J-D, Davoodi-Semiromi A, She J-X. Cloning of Aire, the mouse homologue of the autoimmune regulator (AIRE) gene responsible for autoimmune polyglandular syndrome type 1 (APS1). Genomics (1999) 55(3):322–6. doi:10.1006/geno.1998.5656
21. Gu B, Zhang J, Chen Q, Tao B, Wang W, Zhou Y, et al. Aire regulates the expression of differentiation-associated genes and self-renewal of embryonic stem cells. Biochem Biophys Res Commun (2010) 394(2):418–23. doi:10.1016/j.bbrc.2010.03.042
22. Gu B, Zhang J, Wang S, Song X, Xu C, Chen L, et al. Aire promotes the self-renewal of embryonic stem cells through Lin28. Stem Cells Dev (2012) 21(15):2878–90. doi:10.1089/scd.2012.0097
23. Gu B, Lambert J-P, Cockburn K, Gingras A-C, Rossant J. AIRE is a critical spindle-associated protein in embryonic stem cells. eLife (2017) 6:e28131. doi:10.7554/eLife.28131
24. Hidaka K, Nitta T, Sugawa R, Shirai M, Schwartz RJ, Amagai T, et al. Differentiation of pharyngeal endoderm from mouse embryonic stem cells. Stem Cells Dev (2010) 19(11):1735–43. doi:10.1089/scd.2009.0466
25. Le Douarin NM, Jotereau FV. Tracing of cells of the avian thymus through embryonic life in interspecific chimeras. J Exp Med (1975) 142(1):17–40. doi:10.1084/jem.142.1.17
26. Gordon J, Wilson VA, Blair NF, Sheridan J, Farley A, Wilson L, et al. Functional evidence for a single endodermal origin for the thymic epithelium. Nat Immunol (2004) 5(5):546–53. doi:10.1038/ni1064
27. Bennett AR, Farley A, Blair NF, Gordon J, Sharp L, Blackburn CC. Identification and characterization of thymic epithelial progenitor cells. Immunity (2002) 16(6):803–14. doi:10.1016/S1074-7613(02)00321-7
28. Gill J, Malin M, Holländer GA, Boyd R. Generation of a complete thymic microenvironment by MTS24+ epithelial cells. Nat Immunol (2002) 3(7):635–42. doi:10.1038/ni812
29. Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature (2006) 441(7096):988–91. doi:10.1038/nature04813
30. Bleul CC, Corbeaux T, Reuter A, Fisch P, Mönting JS, Boehm T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature (2006) 441(7096):992–6. doi:10.1038/nature04850
31. Rossi SW, Chidgey AP, Parnell SM, Jenkinson WE, Scott HS, Boyd RL, et al. Redefining epithelial progenitor potential in the developing thymus. Eur J Immunol (2007) 37(9):2411–8. doi:10.1002/eji.200737275
32. Shakib S, Desanti GE, Jenkinson WE, Parnell SM, Jenkinson EJ, Anderson G. Checkpoints in the development of thymic cortical epithelial cells. J Immunol (2009) 182(1):130–7. doi:10.4049/jimmunol.182.1.130
33. Baik S, Jenkinson EJ, Lane PJL, Anderson G, Jenkinson WE. Generation of both cortical and Aire+ medullary thymic epithelial compartments from CD205+ progenitors. Eur J Immunol (2013) 43(3):589–94. doi:10.1002/eji.201243209
34. Ohigashi I, Zuklys S, Sakata M, Mayer CE, Zhanybekova S, Murata S, et al. Aire-expressing thymic medullary epithelial cells originate from β5t-expressing progenitor cells. Proc Natl Acad Sci U S A (2013) 110(24):9885–90. doi:10.1073/pnas.1301799110
35. Mayer CE, Žuklys S, Zhanybekova S, Ohigashi I, Teh H-Y, Sansom SN, et al. Dynamic spatio-temporal contribution of single β5t+ cortical epithelial precursors to the thymus medulla. Eur J Immunol (2016) 46(4):846–56. doi:10.1002/eji.201545995
36. Ribeiro AR, Meireles C, Rodrigues PM, Alves NL. Intermediate expression of CCRL1 reveals novel subpopulations of medullary thymic epithelial cells that emerge in the postnatal thymus. Eur J Immunol (2014) 44(10):2918–24. doi:10.1002/eji.201444585
37. Ribeiro AR, Rodrigues PM, Meireles C, Di Santo JP, Alves NL. Thymocyte selection regulates the homeostasis of IL-7–expressing thymic cortical epithelial cells in vivo. J Immunol (2013) 191(3):1200–9. doi:10.4049/jimmunol.1203042
38. Alves NL, Takahama Y, Ohigashi I, Ribeiro AR, Baik S, Anderson G, et al. Serial progression of cortical and medullary thymic epithelial microenvironments. Eur J Immunol (2014) 44(1):16–22. doi:10.1002/eji.201344110
39. Hamazaki Y, Fujita H, Kobayashi T, Choi Y, Scott HS, Matsumoto M, et al. Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol (2007) 8(3):304–11. doi:10.1038/ni1438
40. Osada M, Singh VJ, Wu K, Sant’Angelo DB, Pezzano M. Label retention identifies a multipotent mesenchymal stem cell-like population in the postnatal thymus. PLoS One (2013) 8(12):e83024. doi:10.1371/journal.pone.0083024
41. Wong K, Lister NL, Barsanti M, Lim JMC, Hammett MV, Khong DM, et al. Multilineage potential and self-renewal define an epithelial progenitor cell population in the adult thymus. Cell Rep (2014) 8(4):1198–209. doi:10.1016/j.celrep.2014.07.029
42. Ucar A, Ucar O, Klug P, Matt S, Brunk F, Hofmann TG, et al. Adult thymus contains FoxN1ˉ epithelial stem cells that are bipotent for medullary and cortical thymic epithelial lineages. Immunity (2014) 41(2):257–69. doi:10.1016/j.immuni.2014.07.005
43. Ucar O, Li K, Dvornikov D, Kreutz C, Timmer J, Matt S, et al. A thymic epithelial stem cell pool persists throughout ontogeny and is modulated by TGF-β. Cell Rep (2016) 17(2):448–57. doi:10.1016/j.celrep.2016.09.027
44. Ulyanchenko S, O’Neill KE, Medley T, Farley AM, Vaidya HJ, Cook AM, et al. Identification of a bipotent epithelial progenitor population in the adult thymus. Cell Rep (2016) 14(12):2819–32. doi:10.1016/j.celrep.2016.02.080
45. Ohigashi I, Zuklys S, Sakata M, Mayer CE, Hamazaki Y, Minato N, et al. Adult thymic medullary epithelium is maintained and regenerated by lineage-restricted cells rather than bipotent progenitors. Cell Rep (2015) 13(7):1432–43. doi:10.1016/j.celrep.2015.10.012
46. Sekai M, Hamazaki Y, Minato N. Medullary thymic epithelial stem cells maintain a functional thymus to ensure lifelong central T cell tolerance. Immunity (2014) 41(5):753–61. doi:10.1016/j.immuni.2014.11.008
47. Onder L, Nindl V, Scandella E, Chai Q, Cheng H-W, Caviezel-Firner S, et al. Alternative NF-κB signaling regulates mTEC differentiation from podoplanin-expressing precursors in the cortico-medullary junction. Eur J Immunol (2015) 45(8):2218–31. doi:10.1002/eji.201545677
48. Klug DB, Carter C, Crouch E, Roop D, Conti CJ, Richie ER. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc Natl Acad Sci U S A (1998) 95(20):11822–7. doi:10.1073/pnas.95.20.11822
49. Klug DB, Carter C, Gimenez-Conti IB, Richie ER. Thymocyte-independent and thymocyte-dependent phases of epithelial patterning in the fetal thymus. J Immunol (2002) 169(6):2842–5. doi:10.4049/jimmunol.169.6.2842
50. Farr AG, Anderson SK. Epithelial heterogeneity in the murine thymus: fucose-specific lectins bind medullary epithelial cells. J Immunol (1985) 134(5):2971–7.
51. Surh CD, Gao E-K, Kosaka H, Lo D, Ahn C, Murphy DB, et al. Two subsets of epithelial cells in the thymic medulla. J Exp Med (1992) 176(2):495–505. doi:10.1084/jem.176.2.495
52. Gray DHD, Chidgey AP, Boyd RL. Analysis of thymic stromal cell populations using flow cytometry. J Immunol Methods (2002) 260(1–2):15–28. doi:10.1016/S0022-1759(01)00493-8
53. Gray DHD, Fletcher AL, Hammett M, Seach N, Ueno T, Young LF, et al. Unbiased analysis, enrichment and purification of thymic stromal cells. J Immunol Methods (2008) 329(1–2):56–66. doi:10.1016/j.jim.2007.09.010
54. McLelland BT, Gravano D, Castillo J, Montoy S, Manilay JO. Enhanced isolation of adult thymic epithelial cell subsets for multiparameter flow cytometry and gene expression analysis. J Immunol Methods (2011) 367(1–2):85–94. doi:10.1016/j.jim.2011.02.008
55. Seach N, Wong K, Hammett M, Boyd RL, Chidgey AP. Purified enzymes improve isolation and characterization of the adult thymic epithelium. J Immunol Methods (2012) 385(1–2):23–34. doi:10.1016/j.jim.2012.07.023
56. Sawanobori Y, Ueta H, Dijkstra CD, Park CG, Satou M, Kitazawa Y, et al. Three distinct subsets of thymic epithelial cells in rats and mice defined by novel antibodies. PLoS One (2014) 9(10):e109995. doi:10.1371/journal.pone.0109995
57. Gray DHD, Seach N, Ueno T, Milton MK, Liston A, Lew AM, et al. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood (2006) 108(12):3777–85. doi:10.1182/blood-2006-02-004531
58. Gäbler J, Arnold J, Kyewski B. Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur J Immunol (2007) 37(12):3363–72. doi:10.1002/eji.200737131
59. Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med (2007) 204(11):2521–8. doi:10.1084/jem.20070795
60. Fletcher AL, Lowen TE, Sakkal S, Reiseger JJ, Hammett MV, Seach N, et al. Ablation and regeneration of tolerance-inducing medullary thymic epithelial cells after cyclosporine, cyclophosphamide, and dexamethasone treatment. J Immunol (2009) 183(2):823–31. doi:10.4049/jimmunol.0900225
61. Colomé N, Collado J, Bech-Serra JJ, Liiv I, Antón LC, Peterson P, et al. Increased apoptosis after autoimmune regulator expression in epithelial cells revealed by a combined quantitative proteomics approach. J Proteome Res (2010) 9(5):2600–9. doi:10.1021/pr100044d
62. Nishikawa Y, Hirota F, Yano M, Kitajima H, Miyazaki J, Kawamoto H, et al. Biphasic Aire expression in early embryos and in medullary thymic epithelial cells before end-stage terminal differentiation. J Exp Med (2010) 207(5):963–71. doi:10.1084/jem.20092144
63. Wang X, Laan M, Bichele R, Kisand K, Scott HS, Peterson P. Post-Aire maturation of thymic medullary epithelial cells involves selective expression of keratinocyte-specific autoantigens. Front Immunol (2012) 3:e19. doi:10.3389/fimmu.2012.00019
64. Metzger TC, Khan IS, Gardner JM, Mouchess ML, Johannes KP, Krawisz AK, et al. Lineage tracing and cell ablation identify a post-Aire-expressing thymic epithelial cell population. Cell Rep (2013) 5(1):166–79. doi:10.1016/j.celrep.2013.08.038
65. Nehls M, Kyewski B, Messerle M, Waldschütz R, Schüddekopf K, Smith AJ, et al. Two genetically separable steps in the differentiation of thymic epithelium. Science (1996) 272(5263):886–9. doi:10.1126/science.272.5263.886
66. Nowell CS, Bredenkamp N, Tetélin S, Jin X, Tischner C, Vaidya H, et al. Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence. PLoS Genet (2011) 7(11):e1002348. doi:10.1371/journal.pgen.1002348
67. Guo J, Rahman M, Cheng L, Zhang S, Tvinnereim A, Su D-M. Morphogenesis and maintenance of the 3D thymic medulla and prevention of nude skin phenotype require FoxN1 in pre- and post-natal K14 epithelium. J Mol Med (Berl) (2011) 89(3):263–77. doi:10.1007/s00109-010-0700-8
68. Xia J, Wang H, Guo J, Zhang Z, Coder B, Su D-M. Age-related disruption of steady-state thymic medulla provokes autoimmune phenotype via perturbing negative selection. Aging Dis (2012) 3(3):248–59.
69. Garfin PM, Min D, Bryson JL, Serwold T, Edris B, Blackburn CC, et al. Inactivation of the RB family prevents thymus involution and promotes thymic function by direct control of Foxn1 expression. J Exp Med (2013) 210(6):1087–97. doi:10.1084/jem.20121716
70. Bredenkamp N, Nowell CS, Blackburn CC. Regeneration of the aged thymus by a single transcription factor. Development (2014) 141(8):1627–37. doi:10.1242/dev.103614
71. Jin X, Nowell CS, Ulyanchenko S, Stenhouse FH, Blackburn CC. Long-term persistence of functional thymic epithelial progenitor cells in vivo under conditions of low FOXN1 expression. PLoS One (2014) 9(12):e114842. doi:10.1371/journal.pone.0114842
72. O’Neill KE, Bredenkamp N, Tischner C, Vaidya HJ, Stenhouse FH, Peddie CD, et al. Foxn1 is dynamically regulated in thymic epithelial cells during embryogenesis and at the onset of thymic involution. PLoS One (2016) 11(3):e151666. doi:10.1371/journal.pone.0151666
73. van Ewijk W, Shores EW, Singer A. Crosstalk in the mouse thymus. Immunol Today (1994) 15(5):214–7. doi:10.1016/0167-5699(94)90246-1
74. Holländer GA, Wang B, Nichogiannopoulou A, Platenburg PP, van Ewijk W, Burakoff SJ, et al. Developmental control point in induction of thymic cortex regulated by a subpopulation of prothymocytes. Nature (1995) 373(6512):350–3. doi:10.1038/373350a0
75. van Ewijk W, Holländer G, Terhorst C, Wang B. Stepwise development of thymic microenvironments in vivo is regulated by thymocyte subsets. Development (2000) 127(8):1583–91.
76. Heino M, Peterson P, Sillanpää N, Guérin S, Wu L, Anderson G, et al. RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur J Immunol (2000) 30(7):1884–93. doi:10.1002/1521-4141(200007)30:7<1884:AID-IMMU1884>3.0.CO;2-P
77. Zuklys S, Balciunaite G, Agarwal A, Fasler-Kan E, Palmer E, Holländer GA. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). J Immunol (2000) 165(4):1976–83. doi:10.4049/jimmunol.165.4.1976
78. Chin RK, Lo JC, Sim O, Blink SE, Christiansen PA, Peterson P, et al. Lymphotoxin pathway directs thymic Aire expression. Nat Immunol (2003) 4(11):1121–7. doi:10.1038/ni982
79. Chin RK, Zhu M, Christiansen PA, Liu W, Ware C, Peltonen L, et al. Lymphotoxin pathway-directed, autoimmune regulator-independent central tolerance to arthritogenic collagen. J Immunol (2006) 177(1):290–7. doi:10.4049/jimmunol.177.1.290
80. Zhu M, Chin RK, Christiansen PA, Lo JC, Liu X, Ware C, et al. NF-κB2 is required for the establishment of central tolerance through an Aire-dependent pathway. J Clin Invest (2006) 116(11):2964–71. doi:10.1172/JCI28326
81. Zhu M, Chin RK, Tumanov AV, Liu X, Fu Y-X. Lymphotoxin β receptor is required for the migration and selection of autoreactive T cells in thymic medulla. J Immunol (2007) 179(12):8069–75. doi:10.4049/jimmunol.179.12.8069
82. Boehm T, Scheu S, Pfeffer K, Bleul CC. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTβR. J Exp Med (2003) 198(5):757–69. doi:10.1084/jem.20030794
83. Martins V, Boehm T, Bleul CC. Ltβr signaling does not regulate Aire-dependent transcripts in medullary thymic epithelial cells. J Immunol (2008) 181(1):400–7. doi:10.4049/jimmunol.181.1.400
84. Venanzi ES, Gray DHD, Benoist C, Mathis D. Lymphotoxin pathway and Aire influences on thymic medullary epithelial cells are unconnected. J Immunol (2007) 179(9):5693–700. doi:10.4049/jimmunol.179.9.5693
85. Seach N, Ueno T, Fletcher AL, Lowen T, Mattesich M, Engwerda CR, et al. The lymphotoxin pathway regulates Aire-independent expression of ectopic genes and chemokines in thymic stromal cells. J Immunol (2008) 180(8):5384–92. doi:10.4049/jimmunol.180.8.5384
86. Kajiura F, Sun S, Nomura T, Izumi K, Ueno T, Bando Y, et al. NF-κB-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J Immunol (2004) 172(4):2067–75. doi:10.4049/jimmunol.172.4.2067
87. Akiyama T, Maeda S, Yamane S, Ogino K, Kasai M, Kajiura F, et al. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science (2005) 308(5719):248–51. doi:10.1126/science.1105677
88. Kinoshita D, Hirota F, Kaisho T, Kasai M, Izumi K, Bando Y, et al. Essential role of IκB kinase α in thymic organogenesis required for the establishment of self-tolerance. J Immunol (2006) 176(7):3995–4002. doi:10.4049/jimmunol.176.7.3995
89. Zhang B, Wang Z, Ding J, Peterson P, Gunning WT, Ding H-F. NF-κB2 is required for the control of autoimmunity by regulating the development of medullary thymic epithelial cells. J Biol Chem (2006) 281(50):38617–24. doi:10.1074/jbc.M606705200
90. Lomada D, Liu B, Coghlan L, Hu Y, Richie ER. Thymus medulla formation and central tolerance are restored in IKKα–/– mice that express an IKKα transgene in keratin 5+ thymic epithelial cells. J Immunol (2007) 178(2):829–37. doi:10.4049/jimmunol.178.2.829
91. Akiyama T, Shimo Y, Yanai H, Qin J, Ohshima D, Maruyama Y, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity (2008) 29(3):423–37. doi:10.1016/j.immuni.2008.06.015
92. Mouri Y, Yano M, Shinzawa M, Shimo Y, Hirota F, Nishikawa Y, et al. Lymphotoxin signal promotes thymic organogenesis by eliciting RANK expression in the embryonic thymic stroma. J Immunol (2011) 186(9):5047–57. doi:10.4049/jimmunol.1003533
93. Lkhagvasuren E, Sakata M, Ohigashi I, Takahama Y. Lymphotoxin β receptor regulates the development of CCL21-expressing subset of postnatal medullary thymic epithelial cells. J Immunol (2013) 190(10):5110–7. doi:10.4049/jimmunol.1203203
94. Akiyama N, Shinzawa M, Miyauchi M, Yanai H, Tateishi R, Shimo Y, et al. Limitation of immune tolerance-inducing thymic epithelial cell development by Spi-B-mediated negative feedback regulation. J Exp Med (2014) 211(12):2425–38. doi:10.1084/jem.20141207
95. Akiyama N, Takizawa N, Miyauchi M, Yanai H, Tateishi R, Shinzawa M, et al. Identification of embryonic precursor cells that differentiate into thymic epithelial cells expressing autoimmune regulator. J Exp Med (2016) 213(8):1441–58. doi:10.1084/jem.20151780
96. Bonito AJ, Aloman C, Fiel MI, Danzl NM, Cha S, Weinstein EG, et al. Medullary thymic epithelial cell depletion leads to autoimmune hepatitis. J Clin Invest (2013) 123(8):3510–24. doi:10.1172/JCI65414
97. Danzl NM, Jeong S, Choi Y, Alexandropoulos K. Identification of novel thymic epithelial cell subsets whose differentiation is regulated by RANKL and Traf6. PLoS One (2014) 9(1):e86129. doi:10.1371/journal.pone.0086129
98. Rossi SW, Kim M-Y, Leibbrandt A, Parnell SM, Jenkinson WE, Glanville SH, et al. RANK signals from CD4+3– inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med (2007) 204(6):1267–72. doi:10.1084/jem.20062497
99. Heikenwalder M, Prinz M, Zeller N, Lang KS, Junt T, Rossi S, et al. Overexpression of lymphotoxin in T cells induces fulminant thymic involution. Am J Pathol (2008) 172(6):1555–70. doi:10.2353/ajpath.2008.070572
100. White AJ, Withers DR, Parnell SM, Scott HS, Finke D, Lane PJL, et al. Sequential phases in the development of Aire-expressing medullary thymic epithelial cells involve distinct cellular input. Eur J Immunol (2008) 38(4):942–7. doi:10.1002/eji.200738052
101. White AJ, Nakamura K, Jenkinson WE, Saini M, Sinclair C, Seddon B, et al. Lymphotoxin signals from positively selected thymocytes regulate the terminal differentiation of medullary thymic epithelial cells. J Immunol (2010) 185(8):4769–76. doi:10.4049/jimmunol.1002151
102. Roberts NA, White AJ, Jenkinson WE, Turchinovich G, Nakamura K, Withers DR, et al. Rank signaling links the development of invariant γδ T cell progenitors and Aire+ medullary epithelium. Immunity (2012) 36(3):427–37. doi:10.1016/j.immuni.2012.01.016
103. Desanti GE, Cowan JE, Baik S, Parnell SM, White AJ, Penninger JM, et al. Developmentally regulated availability of RANKL and CD40 ligand reveals distinct mechanisms of fetal and adult cross-talk in the thymus medulla. J Immunol (2012) 189(12):5519–26. doi:10.4049/jimmunol.1201815
104. White AJ, Jenkinson WE, Cowan JE, Parnell SM, Bacon A, Jones ND, et al. An essential role for medullary thymic epithelial cells during the intrathymic development of invariant NKT cells. J Immunol (2014) 192(6):2659–66. doi:10.4049/jimmunol.1303057
105. McCarthy NI, Cowan JE, Nakamura K, Bacon A, Baik S, White AJ, et al. Osteoprotegerin-mediated homeostasis of Rank+ thymic epithelial cells does not limit Foxp3+ regulatory T cell development. J Immunol (2015) 195(6):2675–82. doi:10.4049/jimmunol.1501226
106. Baik S, Sekai M, Hamazaki Y, Jenkinson WE, Anderson G. Relb acts downstream of medullary thymic epithelial stem cells and is essential for the emergence of RANK+ medullary epithelial progenitors. Eur J Immunol (2016) 46(4):857–62. doi:10.1002/eji.201546253
107. Cosway EJ, Lucas B, James KD, Parnell SM, Carvalho-Gaspar M, White AJ, et al. Redefining thymus medulla specialization for central tolerance. J Exp Med (2017) 214(11):3183–95. doi:10.1084/jem.20171000
108. Hikosaka Y, Nitta T, Ohigashi I, Yano K, Ishimaru N, Hayashi Y, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity (2008) 29(3):438–50. doi:10.1016/j.immuni.2008.06.018
109. Irla M, Hugues S, Gill J, Nitta T, Hikosaka Y, Williams IR, et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity (2008) 29(3):451–63. doi:10.1016/j.immuni.2008.08.007
110. Irla M, Guerri L, Guenot J, Sergé A, Lantz O, Liston A, et al. Antigen recognition by autoreactive CD4+ thymocytes drives homeostasis of the thymic medulla. PLoS One (2012) 7(12):e52591. doi:10.1371/journal.pone.0052591
111. Irla M, Guenot J, Sealy G, Reith W, Imhof BA, Sergé A. Three-dimensional visualization of the mouse thymus organization in health and immunodeficiency. J Immunol (2013) 190(2):586–96. doi:10.4049/jimmunol.1200119
112. Jenkinson SR, Williams JA, Jeon H, Zhang J, Nitta T, Ohigashi I, et al. TRAF3 enforces the requirement for T cell cross-talk in thymic medullary epithelial development. Proc Natl Acad Sci U S A (2013) 110(52):21107–12. doi:10.1073/pnas.1314859111
113. Williams JA, Zhang J, Jeon H, Nitta T, Ohigashi I, Klug D, et al. Thymic medullary epithelium and thymocyte self-tolerance require cooperation between CD28–CD80/86 and CD40–CD40L costimulatory pathways. J Immunol (2014) 192(2):630–40. doi:10.4049/jimmunol.1302550
114. Riemann M, Andreas N, Fedoseeva M, Meier E, Weih D, Freytag H, et al. Central immune tolerance depends on crosstalk between the classical and alternative NF-κB pathways in medullary thymic epithelial cells. J Autoimmun (2017) 81:56–67. doi:10.1016/j.jaut.2017.03.007
115. Akirav EM, Xu Y, Ruddle NH. Resident B cells regulate thymic expression of myelin oligodendrocyte glycoprotein. J Neuroimmunol (2011) 235(1–2):33–9. doi:10.1016/j.jneuroim.2011.03.013
116. Mouri Y, Nishijima H, Kawano H, Hirota F, Sakaguchi N, Morimoto J, et al. NF-κB-inducing kinase in thymic stroma establishes central tolerance by orchestrating cross-talk with not only thymocytes but also dendritic cells. J Immunol (2014) 193(9):4356–67. doi:10.4049/jimmunol.1400389
117. Vroegindeweij E, Crobach S, Itoi M, Satoh R, Zuklys S, Happe C, et al. Thymic cysts originate from Foxn1 positive thymic medullary epithelium. Mol Immunol (2010) 47(5):1106–13. doi:10.1016/j.molimm.2009.10.034
118. Irla M, Hollander G, Reith W. Control of central self-tolerance induction by autoreactive CD4+ thymocytes. Trends Immunol (2010) 31(2):71–9. doi:10.1016/j.it.2009.11.002
119. Nitta T, Ohigashi I, Nakagawa Y, Takahama Y. Cytokine crosstalk for thymic medulla formation. Curr Opin Immunol (2011) 23(2):190–7. doi:10.1016/j.coi.2010.12.002
120. Akiyama T, Shinzawa M, Akiyama N. TNF receptor family signaling in the development and functions of medullary thymic epithelial cells. Front Immunol (2012) 3:e278. doi:10.3389/fimmu.2012.00278
121. Lopes N, Sergé A, Ferrier P, Irla M. Thymic crosstalk coordinates medulla organization and T-cell tolerance induction. Front Immunol (2015) 6:e365. doi:10.3389/fimmu.2015.00365
122. Erickson M, Morkowski S, Lehar S, Gillard G, Beers C, Dooley J, et al. Regulation of thymic epithelium by keratinocyte growth factor. Blood (2002) 100(9):3269–78. doi:10.1182/blood-2002-04-1036
123. Rossi S, Blazar BR, Farrell CL, Danilenko DM, Lacey DL, Weinberg KI, et al. Keratinocyte growth factor preserves normal thymopoiesis and thymic microenvironment during experimental graft-versus-host disease. Blood (2002) 100(2):682–91. doi:10.1182/blood.V100.2.682
124. Kelly RM, Highfill SL, Panoskaltsis-Mortari A, Taylor PA, Boyd RL, Holländer GA, et al. Keratinocyte growth factor and androgen blockade work in concert to protect against conditioning regimen-induced thymic epithelial damage and enhance T-cell reconstitution after murine bone marrow transplantation. Blood (2008) 111(12):5734–44. doi:10.1182/blood-2008-01-136531
125. Kelly RM, Goren EM, Taylor PA, Mueller SN, Stefanski HE, Osborn MJ, et al. Short-term inhibition of p53 combined with keratinocyte growth factor improves thymic epithelial cell recovery and enhances T-cell reconstitution after murine bone marrow transplantation. Blood (2010) 115(5):1088–97. doi:10.1182/blood-2009-05-223198
126. Dertschnig S, Nusspaumer G, Ivanek R, Hauri-Hohl MM, Holländer GA, Krenger W. Epithelial cytoprotection sustains ectopic expression of tissue-restricted antigens in the thymus during murine acute GVHD. Blood (2013) 122(5):837–41. doi:10.1182/blood-2012-12-474759
127. Dertschnig S, Hauri-Hohl MM, Vollmer M, Holländer GA, Krenger W. Impaired thymic expression of tissue-restricted antigens licenses the de novo generation of autoreactive CD4+ T cells in acute GVHD. Blood (2015) 125(17):2720–3. doi:10.1182/blood-2014-08-597245
128. Pinto S, Schmidt K, Egle S, Stark H-J, Boukamp P, Kyewski B. An organotypic coculture model supporting proliferation and differentiation of medullary thymic epithelial cells and promiscuous gene expression. J Immunol (2013) 190(3):1085–93. doi:10.4049/jimmunol.1201843
129. Nagamine K, Peterson P, Scott HS, Kudoh J, Minoshima S, Heino M, et al. Positional cloning of the APECED gene. Nat Genet (1997) 17(4):393–8. doi:10.1038/ng1297-393
130. Finnish-German APECED Consortium. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet (1997) 17(4):399–403. doi:10.1038/ng1297-399
131. Ahonen P, Myllärniemi S, Sipilä I, Perheentupa J. Clinical variation of autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) in a series of 68 patients. New Engl J Med (1990) 322(26):1829–36. doi:10.1056/NEJM199006283222601
132. Betterle C, Greggio NA, Volpato M. Autoimmune polyglandular syndrome type 1. J Clin Endocrinol Metab (1998) 83(4):1049–55. doi:10.1210/jcem.83.4.4682
133. Heino M, Peterson P, Kudoh J, Nagamine K, Lagerstedt A, Ovod D, et al. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun (1999) 257(3):821–5. doi:10.1006/bbrc.1999.0308
134. Eldershaw SA, Sansom DM, Narendran P. Expression and function of the autoimmune regulator (Aire) gene in non-thymic tissue. Clin Exp Immunol (2011) 163(3):296–308. doi:10.1111/j.1365-2249.2010.04316.x
135. Perniola R. Expression of the autoimmune regulator gene and its relevance to the mechanisms of central and peripheral tolerance. Clin Dev Immunol (2012) 2012:e207403. doi:10.1155/2012/207403
136. Björses P, Aaltonen J, Horelli-Kuitunen N, Yaspo M-L, Peltonen L. Gene defect behind APECED: a new clue to autoimmunity. Hum Mol Genet (1998) 7(10):1547–53. doi:10.1093/hmg/7.10.1547
137. Peterson P, Nagamine K, Scott H, Heino M, Kudoh J, Shimizu N, et al. APECED: a monogenic autoimmune disease providing new clues to self-tolerance. Immunol Today (1998) 19(9):384–6. doi:10.1016/S0167-5699(98)01293-6
138. Perniola R, Musco G. The biophysical and biochemical properties of the autoimmune regulator (AIRE) protein. Biochim Biophys Acta (2014) 1842(2):326–37. doi:10.1016/j.bbadis.2013.11.020
139. Björses P, Pelto-Huikko M, Kaukonen J, Aaltonen J, Peltonen L, Ulmanen I. Localization of the APECED protein in distinct nuclear structures. Hum Mol Genet (1999) 8(2):259–66. doi:10.1093/hmg/8.2.259
140. Rinderle C, Christensen H-M, Schweiger S, Lehrach H, Yaspo M-L. AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered sub-cellular distribution of mutants lacking the PHD zinc fingers. Hum Mol Genet (1999) 8(2):277–90. doi:10.1093/hmg/8.2.277
141. Pitkänen J, Vähämurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. Characterization of nuclear targeting and trascriptional activation domain. J Biol Chem (2001) 276(22):19597–602. doi:10.1074/jbc.M008322200
142. Akiyoshi H, Hatakeyama S, Pitkänen J, Mouri Y, Doucas V, Kudoh J, et al. Subcellular expression of autoimmune regulator is organized in a spatiotemporal manner. J Biol Chem (2004) 279(32):33984–91. doi:10.1074/jbc.M400702200
143. Tao Y, Kupfer R, Stewart BJ, Williams-Skipp C, Crowell CK, Patel DD, et al. AIRE recruits multiple transcriptional components to specific genomic regions through tethering to nuclear matrix. Mol Immunol (2006) 43(4):335–45. doi:10.1016/j.molimm.2005.02.018
144. Kumar PG, Laloraya M, Wang C-Y, Ruan Q-G, Davoodi-Semiromi A, Kao K-J, et al. The autoimmune regulator (AIRE) is a DNA-binding protein. J Biol Chem (2001) 276(44):41357–64. doi:10.1074/jbc.M104898200
145. Purohit S, Kumar PG, Laloraya M, She J-X. Mapping DNA-binding domains of the autoimmune regulator protein. Biochem Biophys Res Commun (2005) 327(3):939–44. doi:10.1016/j.bbrc.2004.12.093
146. Ruan Q-G, Tung K, Eisenman D, Setiady Y, Eckenrode S, Yi B, et al. The autoimmune regulator directly controls the expression of genes critical for thymic epithelial function. J Immunol (2007) 178(11):7173–89. doi:10.4049/jimmunol.178.11.7173
147. Sternsdorf T, Jensen K, Reich B, Will H. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J Biol Chem (1999) 274(18):12555–66. doi:10.1074/jbc.274.18.12555
148. Pitkänen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem (2000) 275(22):16802–9. doi:10.1074/jbc.M908944199
149. Ramsey C, Bukrinsky A, Peltonen L. Systematic mutagenesis of the functional domains of AIRE reveals their role in intracellular targeting. Hum Mol Genet (2002) 11(26):3299–308. doi:10.1093/hmg/11.26.3299
150. Ferguson BJ, Alexander C, Rossi SW, Liiv I, Rebane A, Worth CL, et al. AIRE’s CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J Biol Chem (2008) 283(3):1723–31. doi:10.1074/jbc.M707211200
151. Hofmann K, Bucher P, Tschopp J. The CARD domain: a new apoptotic signalling motif. Trends Biochem Sci (1997) 22(5):155–6. doi:10.1016/S0968-0004(97)01043-8
152. Ilmarinen T, Melén K, Kangas H, Julkunen I, Ulmanen I, Eskelin P. The monopartite nuclear localization signal of autoimmune regulator mediates its nuclear import and interaction with multiple importin α molecules. FEBS J (2006) 273(2):315–24. doi:10.1111/j.1742-4658.2005.05065.x
153. Saltis M, Criscitiello MF, Ohta Y, Keefe M, Trede NS, Goitsuka R, et al. Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: a comparative analysis. Immunogenetics (2008) 60(2):105–14. doi:10.1007/s00251-007-0268-9
154. Gibson TJ, Ramu C, Gemünd C, Aasland R. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the SAND domain and is probably a transcription factor. Trends Biochem Sci (1998) 23(7):242–4. doi:10.1016/S0968-0004(98)01231-6
155. Carles CC, Fletcher JC. Missing links between histones and RNA Pol II arising from SAND? Epigenetics (2010) 5(5):381–5. doi:10.4161/epi.5.5.11956
156. Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci (1995) 20(2):56–9. doi:10.1016/S0968-0004(00)88957-4
157. Org T, Chignola F, Hetényi C, Gaetani M, Rebane A, Liiv I, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep (2008) 9(4):370–6. doi:10.1038/sj.embor.2008.11
158. Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, et al. Aire empolys a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A (2008) 105(41):15878–83. doi:10.1073/pnas.0808470105
159. Org T, Rebane A, Kisand K, Laan M, Haljasorg U, Andreson R, et al. AIRE activated tissue specific genes have histone modifications associated with inactive chromatin. Hum Mol Genet (2009) 18(24):4699–710. doi:10.1093/hmg/ddp433
160. Musco G, Peterson P. PHD finger of autoimmune regulator. An epigenetic link between the histone modifications and tissue-specific antigen expression in thymus. Epigenetics (2008) 3(6):310–4. doi:10.4161/epi.3.6.7182
161. Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, et al. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res (2009) 37(9):2951–61. doi:10.1093/nar/gkp166
162. Chakravarty S, Zeng L, Zhou M-M. Structure and site-specific recognition of histone H3 by the PHD finger of human autoimmune regulator. Structure (2009) 17(5):670–9. doi:10.1016/j.str.2009.02.017
163. Gaetani M, Matafora V, Saare M, Spiliotopoulos D, Mollica L, Quilici G, et al. AIRE-PHD fingers are structural hubs to maintain the integrity of chromatin-associated interactome. Nucleic Acids Res (2012) 40(22):11756–68. doi:10.1093/nar/gks933
164. Yang S, Bansal K, Lopes J, Benoist C, Mathis D. Aire’s plant homeodomain(PHD)-2 is critical for induction of immunological tolerance. Proc Natl Acad Sci U S A (2013) 110(5):1833–8. doi:10.1073/pnas.1222023110
165. Meloni A, Incani F, Corda D, Cao A, Rosatelli MC. Role of PHD fingers and COOH-terminal 30 amino acids in AIRE transactivation activity. Mol Immunol (2008) 45(3):805–9. doi:10.1016/j.molimm.2007.06.156
166. Plevin MJ, Mills MM, Ikura M. The LxxLL motif: a multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci (2005) 30(2):66–9. doi:10.1016/j.tibs.2004.12.001
167. Abramson J, Giraud M, Benoist C, Mathis D. Aire’s partners in the molecular control of immunological tolerance. Cell (2010) 140(1):123–35. doi:10.1016/j.cell.2009.12.030
168. Pitkänen J, Rebane A, Rowell J, Murumägi A, Ströbel P, Möll K, et al. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem Biophys Res Commun (2005) 333(3):944–53. doi:10.1016/j.bbrc.2005.05.187
169. Saare M, Rebane A, Rajashekar B, Vilo J, Peterson P. Autoimmune regulator is acetylated by transcription coactivator CBP/p300. Exp Cell Res (2012) 318(14):1767–78. doi:10.1016/j.yexcr.2012.04.013
170. Incani F, Serra ML, Meloni A, Cossu C, Saba L, Cabras T, et al. AIRE acetylation and deacetylation: effect on protein stability and transactivation activity. J Biomed Sci (2014) 21:e85. doi:10.1186/s12929-014-0085-z
172. Chuprin A, Avin A, Goldfarb Y, Herzig Y, Levi B, Jacob A, et al. The deacetylase Sirt1 is an essential regulator of Aire-mediated induction of central immunological tolerance. Nat Immunol (2015) 16(7):737–45. doi:10.1038/ni.3194
173. Oven I, Brdičková N, Kohoutec J, Vaupotič T, Narat M, Peterlin BM. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol (2007) 27(24):8815–23. doi:10.1128/MCB.01085-07
174. Liiv I, Rebane A, Org T, Saare M, Maslovskaja J, Kisand K, et al. DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim Biophys Acta (2008) 1783(1):74–83. doi:10.1016/j.bbamcr.2007.09.003
175. Žumer K, Low AK, Jiang K, Saksela K, Peterlin M. Unmodified histone H3K4 and DNA-dependent protein kinase recruit autoimmune regulator to target genes. Mol Cell Biol (2012) 32(8):1354–62. doi:10.1128/MCB.06359-11
176. Pommier Y, Sun Y, Huang SN, Nitiss JL. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol (2016) 17(11):703–21. doi:10.1038/nrm.2016.111
177. Bansal K, Yoshida H, Benoist C, Mathis D. The transcriptional regulator Aire binds to and activates super-enhancers. Nat Immunol (2017) 18(3):263–73. doi:10.1038/ni.3675
178. Guha M, Saare M, Maslovskaja J, Kisand K, Liiv I, Haljasorg U, et al. DNA breaks and chromatin structural changes enhance the transcription of autoimmune regulator target genes. J Biol Chem (2017) 292(16):6542–54. doi:10.1074/jbc.M116.764704
179. Mathieu A-L, Verronese E, Rice GI, Fouyssac F, Bertrand Y, Picard C, et al. PRKDC mutations associated with immunodeficiency, granuloma, and autoimmune regulator-dependent autoimmunity. J Allergy Clin Immunol (2015) 135(6):1578–88. doi:10.1016/j.jaci.2015.01.040
180. Rattay K, Claude J, Rezavandy E, Matt S, Hofmann TG, Kyewski B, et al. Homeodomain-interacting protein kinase 2, a novel autoimmune regulator interaction partner, modulates promiscuous gene expression in medullary thymic epithelial cells. J Immunol (2015) 194(3):921–8. doi:10.4049/jimmunol.1402694
181. Žumer K, Plemenitaš A, Saksela K, Peterlin BM. Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription. Nucleic Acids Res (2011) 39(18):7908–19. doi:10.1093/nar/gkr527
182. Giraud M, Yoshida H, Abramson J, Rahl PB, Young RA, Mathis D, et al. Aire unleashes stalled mRNA polymerase to induce ectopic gene expression in thymic epithelial cells. Proc Natl Acad Sci U S A (2012) 109(2):535–40. doi:10.1073/pnas.1119351109
183. Yoshida H, Bansal K, Schaefer U, Chapman T, Rioja I, Proekt I, et al. Brd4 bridges the transcriptional regulators, Aire and P-TEFb, to promote elongation of peripheral-tissue antigen transcripts in thymic stromal cells. Proc Natl Acad Sci U S A (2015) 112(32):E4448–57. doi:10.1073/pnas.1512081112
184. Giraud M, Jmari N, Du L, Carallis F, Nieland TJF, Perez-Campo FM, et al. An RNAi screen for Aire cofactors reveals a role for Hnrnpl in polymerase release and Aire-activated ectopic transcription. Proc Natl Acad Sci U S A (2014) 111(4):1491–6. doi:10.1073/pnas.1323535111
185. Keane P, Ceredig R, Seoighe C. Promiscuous mRNA splicing under the control of AIRE in medullary thymic epithelial cells. Bioinformatics (2015) 31(7):986–90. doi:10.1093/bioinformatics/btu785
186. Danan-Gotthold M, Guyon C, Giraud M, Levanon EY, Abramson J. Extensive RNA editing and splicing increase immune self-representation diversity in medullary thymic epithelial cells. Genome Biol (2016) 17(1):e219. doi:10.1186/s13059-016-1079-9
187. Koh AS, Kingston RE, Benoist C, Mathis D. Global relevance of Aire binding to hypomethylated lysine-4 of histone-3. Proc Natl Acad Sci U S A (2010) 107(29):13016–21. doi:10.1073/pnas.1004436107
188. Tykocinski L-O, Sinemus A, Rezavandy E, Weiland Y, Baddeley D, Cremer C, et al. Epigenetic regulation of promiscuous gene expression in thymic medullary epithelial cells. Proc Natl Acad Sci U S A (2010) 107(45):19426–31. doi:10.1073/pnas.1009265107
189. Waterfield M, Khan IS, Cortez JT, Fan U, Metzger T, Greer A, et al. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat Immunol (2014) 15(3):258–65. doi:10.1038/ni.2820
190. Sansom SN, Shikama-Dorn N, Zhanybekova S, Nusspaumer G, Macaulay IC, Deadman ME, et al. Population and single-cell genomics reveal the Aire dependency, relief from Polycomb silencing, and distribution of self-antigen expression in thymic epithelia. Genome Res (2014) 24(12):1918–31. doi:10.1101/gr.171645.113
191. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell (2004) 116(2):281–97. doi:10.1016/S0092-8674(04)00045-5
192. Zuklys S, Mayer CE, Zhanybekova S, Stefanski HE, Nusspaumer G, Gill J, et al. MicroRNAs control the maintenance of thymic epithelia and their competence for T lineage commitment and thymocyte selection. J Immunol (2012) 189(8):3894–904. doi:10.4049/jimmunol.1200783
193. Khan IS, Taniguchi RT, Fasano KJ, Anderson MS, Jeker LT. Canonical microRNAs in thymic epithelial cells promote central tolerance. Eur J Immunol (2014) 44(5):1313–9. doi:10.1002/eji.201344079
194. Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via the miR-29a mediated suppression of the IFN-α receptor. Nat Immunol (2012) 13(2):181–7. doi:10.1038/ni.2193
195. Ucar O, Tykocinski L-O, Dooley J, Liston A, Kyewski B. An evolutionarily conserved mutual interdependence between Aire and microRNAs in promiscuous gene expression. Eur J Immunol (2013) 43(7):1769–78. doi:10.1002/eji.201343343
196. Ucar O, Rattay K. Promiscuous gene expression in the thymus: a matter of epigenetics, miRNA, and more? Front Immunol (2015) 6:e93. doi:10.3389/fimmu.2015.00093
197. Macedo C, Evangelista AF, Marques MM, Octacílio-Silva S, Donadi EA, Sakamoto-Hojo ET, et al. Autoimmune regulator (Aire) controls the expression of microRNAs in medullary thymic epithelial cells. Immunobiology (2013) 218(4):554–60. doi:10.1016/j.imbio.2012.06.013
198. Passos GA, Mendes-da-Cruz DA, Oliveira EH. The thymic orchestration involving Aire, miRNAs, and cell-cell interactions during the induction of central tolerance. Front Immunol (2015) 6:e352. doi:10.3389/fimmu.2015.00352
199. Macedo C, Oliveira EH, Almeida RS, Donate PB, Fornari TA, Pezzi N, et al. Aire-dependent peripheral tissue antigen mRNAs in mTEC cells feature networking refractoriness to microRNA interaction. Immunobiology (2015) 220(1):93–102. doi:10.1016/j.imbio.2014.08.015
200. Oliveira EH, Macedo C, Collares CV, Freitas AC, Donate PB, Sakamoto-Hojo ET, et al. Aire downregulation is associated with changes in the posttranscriptional control of peripheral tissue antigens in medullary thymic epithelial cells. Front Immunol (2016) 7:e526. doi:10.3389/fimmu.2016.00526
201. Ramsey C, Winqvist O, Puhakka L, Halonen M, Moro A, Kämpe O, et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet (2002) 11(4):397–409. doi:10.1093/hmg/11.4.397
202. Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the Aire protein. Science (2002) 298(5597):1395–401. doi:10.1126/science.1075958
203. Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol (2003) 4(4):350–4. doi:10.1038/ni906
204. St-Pierre C, Brochu S, Vanegas JR, Dumont-Lagacé M, Lemieux S, Perreault C. Transcriptome sequencing of neonatal thymic epithelial cells. Sci Rep (2013) 3:e01860. doi:10.1038/srep01860
205. St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential features of AIRE-induced and AIRE-independent promiscuous gene expression in thymic epithelial cells. J Immunol (2015) 195(2):498–506. doi:10.4049/jimmunol.1500558
206. Cardoso RS, Magalhães DAR, Baião AMT, Junta CM, Macedo C, Marques MMC, et al. Onset of promiscuous gene expression in murine fetal thymus organ culture. Immunology (2006) 119(3):369–75. doi:10.1111/j.1365-2567.2006.02441.x
207. Chen J, Yang W, Yu C, Li Y. Autoimmune regulator initiates the expression of promiscuous genes in thymic epithelial cells. Immunol Invest (2008) 37(3):203–14. doi:10.1080/08820130801967841
208. Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med (2004) 199(2):155–66. doi:10.1084/jem.20031677
209. Derbinski J, Gäbler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med (2005) 202(1):33–45. doi:10.1084/jem.20050471
210. Johnnidis JB, Venanzi ES, Taxman DJ, Ting JP-Y, Benoist CO, Mathis DJ. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci U S A (2005) 102(20):7233–8. doi:10.1073/pnas.0502670102
211. Derbinski J, Pinto S, Rösch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci U S A (2008) 105(2):657–62. doi:10.1073/pnas.0707486105
212. Villaseñor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci U S A (2008) 105(41):15854–9. doi:10.1073/pnas.0808069105
213. Danso-Abeam D, Staats KA, Franckaert D, Van Den Bosch L, Liston A, Gray DHD, et al. Aire mediates thymic expression and tolerance of pancreatic antigens via an unconventional transcriptional mechanism. Eur J Immunol (2013) 43(1):75–84. doi:10.1002/eji.201242761
214. Pinto S, Michel C, Schmidt-Glenewinkel H, Harder N, Rohr K, Wild S, et al. Overlapping gene coexpression patterns in human medullary thymic epithelial cells generate self-antigen diversity. Proc Natl Acad Sci U S A (2013) 110(37):E3497–505. doi:10.1073/pnas.1308311110
215. Brennecke P, Reyes A, Pinto S, Rattay K, Nguyen M, Küchler R, et al. Single-cell transcriptome analysis reveals coordinated ectopic gene-expression patterns in medullary thymic epithelial cells. Nat Immunol (2015) 16(9):933–41. doi:10.1038/ni.3246
216. Meredith M, Zemmour D, Mathis D, Benoist C. Aire controls gene expression in the thymic epithelium with ordered stochasticity. Nat Immunol (2015) 16(9):942–9. doi:10.1038/ni.3247
217. Schaller CE, Wang CL, Beck-Engeser G, Goss L, Scott HS, Anderson MS, et al. Expression of Aire and early wave of apoptosis in spermatogenesis. J Immunol (2008) 180(3):1338–43. doi:10.4049/jimmunol.180.3.1338
218. Guerau-de-Arellano M, Mathis D, Benoist C. Trascriptional impact of Aire varies with cell type. Proc Natl Acad Sci U S A (2008) 105(37):14011–6. doi:10.1073/pnas.0806616105
219. Macedo C, Evangelista AF, Magalhães DA, Fornari TA, Linhares LL, Junta CM, et al. Evidence for a network transcriptional control of promiscuous gene expression in medullary thymic epithelial cells. Mol Immunol (2009) 46(16):3240–4. doi:10.1016/j.molimm.2009.08.002
220. Donate PB, Fornari TA, Junta CM, Magalhães DA, Macedo C, Cunha TM, et al. Collagen induced arthritis (CIA) in mice features regulatory transcriptional network connecting major histocompatibility complex (MHC H2) with autoantigen genes in the thymus. Immunobiology (2011) 216(5):591–603. doi:10.1016/j.imbio.2010.09.007
221. Kuroda N, Mitani T, Takeda N, Ishimaru N, Arakaki R, Hayashi Y, et al. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J Immunol (2005) 174(4):1862–70. doi:10.4049/jimmunol.174.4.1862
222. Gallegos AM, Bevan MJ. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J Exp Med (2004) 200(8):1039–49. doi:10.1084/jem.20041457
223. Koble C, Kyewski B. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J Exp Med (2009) 206(7):1505–13. doi:10.1084/jem.20082449
224. Hubert F-X, Kinkel SA, Davey GM, Phipson B, Mueller SN, Liston A, et al. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood (2011) 118(9):2462–72. doi:10.1182/blood-2010-06-286393
225. Taniguchi RT, DeVoss JJ, Moon JJ, Sidney J, Sette A, Jenkins MK, et al. Detection of an autoreactive T-cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)-mediated selection. Proc Natl Acad Sci U S A (2012) 109(20):7847–52. doi:10.1073/pnas.1120607109
226. Skogberg G, Lundberg V, Berglund M, Gudmundsdottir J, Telemo E, Lindgren S, et al. Human thymic epithelial primary cells produce exosomes carrying tissue-restricted antigens. Immunol Cell Biol (2015) 93(8):727–34. doi:10.1038/icb.2015.33
227. Aichinger M, Wu C, Nedjic J, Klein L. Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance. J Exp Med (2013) 210(2):287–300. doi:10.1084/jem.20122149
228. Yamaguchi Y, Takayanagi A, Chen J, Sakai K, Kudoh J, Shimizu N. Mouse thymic epithelial cell lines expressing “Aire” and peripheral tissue-specific antigens reproduce in vitro negative selection of T cells. Exp Cell Res (2011) 317(14):2019–30. doi:10.1016/j.yexcr.2011.05.002
229. Yamaguchi Y, Kudoh J, Yoshida T, Shimizu N. In vitro co-culture systems for studying molecular basis of cellular interaction between Aire-expressing medullary thymic epithelial cells and fresh thymocytes. Biol Open (2014) 3(11):1071–82. doi:10.1242/bio.201410173
230. Perry JS, Hsieh CS. Development of T-cell tolerance utilizes both cell-autonomous and cooperative presentation of self-antigen. Immunol Rev (2016) 271(1):141–55. doi:10.1111/imr.12403
231. Mouri Y, Ueda Y, Yamano T, Matsumoto M, Tsuneyama K, Kinashi T, et al. Mode of tolerance induction and requirement for Aire are governed by the cell types that express self-antigen and those that present antigen. J Immunol (2017) 199(12):3959–71. doi:10.4049/jimmunol.1700892
232. Yamano T, Nedjic J, Hinterberger M, Steinert M, Koser S, Pinto S, et al. Thymic B cells are licensed to present self antigens for central T cell tolerance induction. Immunity (2015) 42(6):1048–61. doi:10.1016/j.immuni.2015.05.013
233. Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity (2005) 23(2):227–39. doi:10.1016/j.immuni.2005.07.005
234. Daniely D, Kern J, Cebula A, Ignatowicz L. Diversity of TCRs on natural Foxp3+ T cells in mice lacking Aire expression. J Immunol (2010) 184(12):6865–73. doi:10.4049/jimmunol.0903609
235. Chen Z, Benoist C, Mathis D. How defects in central tolerance impinge on a deficiency in regulatory T cells. Proc Natl Acad Sci U S A (2005) 102(41):14735–40. doi:10.1073/pnas.0507014102
236. Aschenbrenner K, D’Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol (2007) 8(4):351–8. doi:10.1038/ni1444
237. Wirnsberger G, Mair F, Klein L. Regulatory T cell differentiation of thymocytes does not require a dedicated antigen-presenting cell but is under T cell-intrinsic developmental control. Proc Natl Acad Sci U S A (2009) 106(25):10278–83. doi:10.1073/pnas.0901877106
238. Hinterberger M, Aichinger M, Prazeres da Costa O, Voehringer D, Hoffmann R, Klein L. Autonomous role of medullary thymic epithelial cells in central CD4+ T cell tolerance. Nat Immunol (2010) 11(6):512–20. doi:10.1038/ni.1874
239. Perry JSA, Lio C-WJ, Kau AL, Nutsch K, Yang Z, Gordon JI, et al. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity (2014) 41(3):414–26. doi:10.1016/j.immuni.2014.08.007
240. Lin J, Yang L, Silva HM, Trzeciak A, Choi Y, Schwab SR, et al. Increased generation of Foxp3+ regulatory T cells by manipulating antigen presentation in the thymus. Nat Commun (2016) 7:e10562. doi:10.1038/ncomms10562
241. Yang S, Fujicado N, Kolodin D, Benoist C, Mathis D. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science (2015) 348(6234):589–94. doi:10.1126/science.aaa7017
242. Kekäläinen E, Lehto M-K, Smeds E, Miettinen A, Meri S, Jarva H, et al. Defective central tolerance in Aire-deficient mice is not sufficient to induce symptomatic autoimmunity during lymphopenia-induced T cell proliferation. Scand J Immunol (2011) 74(1):71–9. doi:10.1111/j.1365-3083.2011.02543.x
243. Teh CE, Daley SR, Enders A, Goodnow CC. T-cell regulation by casitas B-lineage lymphoma (Cblb) is a critical failsafe against autoimmune disease due to autoimmune regulator (Aire) deficiency. Proc Natl Acad Sci U S A (2010) 107(33):14709–14. doi:10.1073/pnas.1009209107
244. Aricha R, Feferman T, Scott HS, Souroujon MC, Berrih-Aknin S, Fuchs S. The susceptibility of Aire−/− mice to experimental myasthenia gravis involves alterations in regulatory T cells. J Autoimmun (2011) 36(1):16–24. doi:10.1016/j.jaut.2010.09.007
245. Malchow S, Leventhal DS, Savage PA. Organ-specific regulatory T cells of thymic origin are expanded in murine prostate tumors. Oncoimmunology (2013) 2(7):e24898. doi:10.4161/onci.24898
246. Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science (2013) 339(6124):1219–24. doi:10.1126/science.1233913
247. Leventhal DS, Gilmore DC, Berger JM, Nishi S, Lee V, Malchow S, et al. Dendritic cells coordinate the development and homeostasis of organ-specific regulatory T cells. Immunity (2016) 44(4):847–59. doi:10.1016/j.immuni.2016.01.025
248. Malchow S, Leventhal DS, Lee V, Nishi S, Socci ND, Savage PA. Aire enforces immune tolerance by directing autoreactive T cells into the regulatory T cell lineage. Immunity (2016) 44(5):1102–13. doi:10.1016/j.immuni.2016.02.009
249. Pomié C, Vicente R, Vuddamalay Y, Lundgren BA, van der Hoek M, Enault G, et al. Autoimmune regulator (AIRE)-deficient CD8+CD28low regulatory T lymphocytes fail to control experimental colitis. Proc Natl Acad Sci U S A (2011) 108(30):12437–42. doi:10.1073/pnas.1107136108
250. Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol (2006) 6(2):127–35. doi:10.1038/nri1781
251. Kurobe H, Liu C, Ueno T, Saito F, Ohigashi I, Seach N, et al. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity (2006) 24(2):165–77. doi:10.1016/j.immuni.2005.12.011
252. Xu X, Zhang S, Li P, Lu J, Xuan Q, Ge Q. Maturation and emigration of single-positive thymocytes. Clin Dev Immunol (2013) 2013:e282870. doi:10.1155/2013/282870
253. Cowan JE, Jenkinson WE, Anderson G. Thymus medulla fosters generation of natural Treg cells, invariant γδ T cells, and invariant NKT cells: what we learn from intrathymic migration. Eur J Immunol (2015) 45(3):652–60. doi:10.1002/eji.201445108
254. Laan M, Kisand K, Kont V, Möll K, Tserel L, Scott HS, et al. Autoimmune regulator deficiency results in decreased expression of CCR4 and CCR7 ligands and in delayed migration of CD4+ thymocytes. J Immunol (2009) 183(12):7682–91. doi:10.4049/jimmunol.0804133
255. Li J, Li Y, Yao J-Y, Jin R, Zhu M-Z, Qian X-P, et al. Developmental pathway of CD4+CD8– medullary thymocytes during mouse ontogeny and its defect in Aire–/– mice. Proc Natl Acad Sci U S A (2007) 104(46):18175–80. doi:10.1073/pnas.0708884104
256. Jin R, Teng F, Xu X, Yao Y, Zhang S, Sun X, et al. Redox balance of mouse medullary CD4 single-positive thymocytes. Immunol Cell Biol (2013) 91(10):634–41. doi:10.1038/icb.2013.57
257. Dong J, Chen Y, Xu X, Jin R, Teng F, Yan F, et al. Homeostatic properties and phenotypic maturation of murine CD4+ pre-thymic emigrants in the thymus. PLoS One (2013) 8(2):e56378. doi:10.1371/journal.pone.0056378
258. Jin R, Aili A, Wang Y, Wu J, Sun X, Zhang Y, et al. Critical role of SP thymocyte motility in regulation of thymic output in neonatal Aire–/– mice. Oncotarget (2017) 8(1):83–94. doi:10.18632/oncotarget.13909
259. Tuovinen H, Pöntynen N, Gylling M, Kekäläinen E, Perheentupa J, Miettinen A, et al. γδ T cells develop independently of Aire. Cell Immunol (2009) 257(1–2):5–12. doi:10.1016/j.cellimm.2009.01.012
260. Mi Q-S, Deng Z-B, Joshi SK, Wang Z-Z, Zhou L, Eckenrode S, et al. The autoimmune regulator (Aire) controls iNKT cell development and maturation. Nat Med (2006) 12(6):624–6. doi:10.1038/nm1424
261. Pitt LA, Hubert F-X, Scott HS, Godfrey DI, Berzins SP. NKT cell development in the absence of the autoimmune regulator gene (Aire). Eur J Immunol (2008) 38(10):2689–96. doi:10.1002/eji.200838553
262. Lindh E, Rosmaraki E, Berg L, Brauner H, Karlsson MCI, Peltonen L, et al. AIRE deficiency leads to impaired iNKT cell development. J Autoimmun (2010) 34(1):66–72. doi:10.1016/j.jaut.2009.07.002
263. Lei Y, Ripen AM, Ishimaru N, Ohigashi I, Nagasawa T, Jeker LT, et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med (2011) 208(2):383–94. doi:10.1084/jem.20102327
264. Jenkinson WE, McCarthy NI, Dutton EE, Cowan JE, Parnell SM, White AJ, et al. Natural Th17 cells are critically regulated by functional medullary thymic microenvironments. J Autoimmun (2015) 63:13–22. doi:10.1016/j.jaut.2015.06.008
265. Fujicado N, Mann AO, Bansal K, Romito KR, Ferre EMN, Rosenzweig SD, et al. Aire inhibits the generation of a perinatal population of interleukin-17A-producing γδ T cells to promote immunologic tolerance. Immunity (2016) 45(5):999–1012. doi:10.1016/j.immuni.2016.10.023
266. Fan Y, Rudert WA, Grupillo M, He J, Sisino G, Trucco M. Thymus-specific deletion of insulin induces autoimmune diabetes. EMBO J (2009) 28(18):2812–24. doi:10.1038/emboj.2009.212
267. DeVoss J, Hou Y, Johannes K, Lu W, Liou GI, Rinn J, et al. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med (2006) 203(12):2727–35. doi:10.1084/jem.20061864
268. Pöntynen N, Miettinen A, Arstila TP, Kämpe O, Alimohammadi M, Vaarala O, et al. Aire deficient mice do not develop the same profile of tissue-specific autoantibodies in APECED patients. J Autoimmun (2006) 27(2):96–104. doi:10.1016/j.jaut.2006.06.001
269. Kekäläinen E, Miettinen A, Arstila TP. Does the deficiency of Aire in mice really resemble human APECED? Nat Rev Immunol (2007) 7(10):1. doi:10.1038/nri2136-c1
270. Hubert F-X, Kinkel SA, Crewther PE, Cannon PZF, Webster KE, Link M, et al. Aire-deficient C57BL/6 mice mimicking the common human 13-base pair deletion mutant present with only a mild autoimmune phenotype. J Immunol (2009) 182(6):3902–18. doi:10.4049/jimmunol.0802124
271. Gavanescu I, Kessler B, Ploegh H, Benoist C, Mathis D. Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc Natl Acad Sci U S A (2007) 104(11):4583–7. doi:10.1073/pnas.0700259104
272. Hou Y, DeVoss J, Dao V, Kwek S, Simko JP, McNeel DG, et al. An aberrant prostate antigen-specific immune response causes prostatitis in mice and is associated with chronic prostatitis in humans. J Clin Invest (2009) 119(7):2031–41. doi:10.1172/JCI38332
273. Misharin AV, Nagayama Y, Aliesky HA, Rapoport B, McLachlan SM. Studies in mice deficient for the autoimmune regulator (Aire) and transgenic for the thyrotropin receptor reveal a role for Aire in tolerance for thyroid autoantigens. Endocrinology (2009) 150(6):2948–56. doi:10.1210/en.2008-1690
274. Shum AK, DeVoss J, Tan CL, Hou Y, Johannes K, O’Gorman CS, et al. Identification of an autoantigen demonstrates a link between interstitial lung disease and a defect in central tolerance. Sci Transl Med (2009) 1(9):e9ra20. doi:10.1126/scitranslmed.3000284
275. Shum AK, Alimohammadi M, Tan CL, Cheng MH, Metzger TC, Law CS, et al. BPIFB1 is a lung-specific autoantigen associated with interstitial lung disease. Sci Transl Med (2013) 5(206):e206ra139. doi:10.1126/scitranslmed.3006998
276. DeVoss JJ, LeClair NP, Hou Y, Grewal NK, Johannes KP, Lu W, et al. An autoimmune response to odorant binding protein 1a is associated with dry eye in the Aire-deficient mouse. J Immunol (2010) 184(8):4236–46. doi:10.4049/jimmunol.0902434
277. Su MA, Davini D, Cheng P, Giang K, Fan U, DeVoss JJ, et al. Defective autoimmune regulator-dependent central tolerance to myelin protein zero is linked to autoimmune peripheral neuropathy. J Immunol (2012) 188(10):4906–12. doi:10.4049/jimmunol.1200493
278. Kurisaki H, Nagao Y, Nagafuchi S, Mitsuyama M. Autoimmune gastro-pancreatitis with anti-protein disulfide isomerase-associated 2 autoantibody in Aire-deficient BALB/cAnN mice. PLoS One (2013) 8(8):e73862. doi:10.1371/journal.pone.0073862
279. Perniola R, Filograna O, Greco G, Pellegrino V. High prevalence of thyroid autoimmunity in Apulian patients with autoimmune polyglandular syndrome type 1. Thyroid (2008) 18(9):1027–9. doi:10.1089/thy.2008.0027
280. Zlotogora J, Shapiro MS. Polyglandular autoimmune syndrome type I among Iranian Jews. J Med Genet (1992) 29(11):824–6. doi:10.1136/jmg.29.11.824
281. Halonen M, Eskelin P, Myhre A-G, Perheentupa J, Husebye ES, Kämpe O, et al. AIRE mutations and human leukocyte antigen genotypes as determinants of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy phenotype. J Clin Endocrinol Metab (2002) 87(6):2568–74. doi:10.1210/jcem.87.6.8564
282. Jiang W, Anderson MA, Bronson R, Mathis D, Benoist C. Modifier loci condition autoimmunity provoked by Aire deficiency. J Exp Med (2005) 202(6):805–15. doi:10.1084/jem.20050693
283. Niki S, Oshikawa K, Mouri Y, Hirota F, Matsushima A, Yano M, et al. Alteration of intra-pancreatic target-organ specificity by abrogation of Aire in NOD mice. J Clin Invest (2006) 116(5):1292–301. doi:10.1172/JCI26971
284. Han H. Target-organ specificity of autoimmunity is modified by thymic stroma and bone marrow-derived cells. J Med Invest (2007) 54(1–2):54–64. doi:10.2152/jmi.54.54
285. Kont V, Laan M, Kisand K, Merits A, Scott HS, Peterson P. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol (2008) 45(1):25–33. doi:10.1016/j.molimm.2007.05.014
286. Oliveira EH, Macedo C, Donate PB, Almeida RS, Pezzi N, Nguyen C, et al. Expression profile of peripheral tissue antigen genes in medullary thymic epithelial cells (mTECs) is dependent on mRNA levels of autoimmune regulator (Aire). Immunobiology (2013) 218(1):96–104. doi:10.1016/j.imbio.2012.02.005
287. Pezzi N, Assis AF, Cotrim-Sousa LC, Lopes GS, Mosella MS, Lima DS, et al. Aire knockdown in medullary thymic epithelial cells affects Aire protein, deregulates cell adhesion genes and decreases thymocyte interaction. Mol Immunol (2016) 77:157–73. doi:10.1016/j.molimm.2016.08.003
288. Kisand K, Peterson P, Laan M. Lymphopenia-induced proliferation in Aire-deficient mice helps to explain their autoimmunity and differences from human patients. Front Immunol (2014) 5:e51. doi:10.3389/fimmu.2014.00051
289. Miller JFAP. Effect of neonatal thymectomy on the immunological responsiveness of the mouse. Proc R Soc Lond B Biol Sci (1962) 156(964):415–28. doi:10.1098/rspb.1962.0048
290. Palmer DB. The effect of age on thymic function. Front Immunol (2013) 4:e316. doi:10.3389/fimmu.2013.00316
291. Guerau-de-Arellano M, Martinic M, Benoist C, Mathis D. Neonatal tolerance revisited: a perinatal window for Aire control of autoimmunity. J Exp Med (2009) 206(6):1245–52. doi:10.1084/jem.20090300
292. Dumont-Lagacé M, St-Pierre C, Perreault C. Sex hormones have pervasive effects on thymic epithelial cells. Sci Rep (2015) 5:e12895. doi:10.1038/srep12895
293. Dragin N, Bismuth J, Cizeron-Clairac G, Biferi MG, Berthault C, Serraf A, et al. Estrogen-mediated downregulation of AIRE influences sexual dimorphism in autoimmune diseases. J Clin Invest (2016) 126(4):1525–37. doi:10.1172/JCI81894
294. Zhu M-L, Bakhru P, Conley B, Nelson JS, Free M, Martin A, et al. Sex bias in CNS autoimmune disease mediated by androgen control of autoimmune regulator. Nat Commun (2016) 7:e11350. doi:10.1038/ncomms11350
295. Taubert R, Schwendemann J, Kyewski B. Highly variable expression of tissue-restricted self-antigens in human thymus: implications for self-tolerance and autoimmunity. Eur J Immunol (2007) 37(3):838–48. doi:10.1002/eji.200636962
296. Liston A, Gray DHD, Lesage S, Fletcher AL, Wilson J, Webster KE, et al. Gene dosage-limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med (2004) 200(8):1015–26. doi:10.1084/jem.20040581
297. Vaidya B, Imrie H, Geatch DR, Perros P, Ball SG, Baylis PH, et al. Association analysis of the cytotoxic T lymphocyte antigen-4 (CTLA-4) and autoimmune regulator-1 (AIRE-1) genes in sporadic autoimmune Addison’s disease. J Clin Endocrinol Metab (2000) 85(2):688–91. doi:10.1210/jcem.85.2.6369
298. Nithiyananthan R, Heward JM, Allahabadia A, Barnett AH, Franklyn JA, Gough SCL. A heterozygous deletion of the autoimmune regulator (AIRE1) gene, autoimmune thyroid disease, and type 1 diabetes: no evidence for association. J Clin Endocrinol Metab (2000) 85(3):1320–2. doi:10.1210/jcem.85.3.6465
299. Meyer G, Donner H, Herwig J, Böhles H, Usadel KH, Badenhoop K. Screening for an AIRE-1 mutation in patients with Addison’s disease, type 1 diabetes, Graves’ disease and Hashimoto’s thyroiditis as well as in APECED syndrome. Clin Endocrinol (Oxf) (2001) 54(3):335–8. doi:10.1046/j.1365-2265.2001.01230.x
300. Bøe AS, Knappskog PM, Myhre AG, Sørheim JI, Husebye ES. Mutational analysis of the autoimmune regulator (AIRE) gene in sporadic autoimmune Addison’s disease can reveal patients with unidentified autoimmune polyendocrine syndrome type I. Eur J Endocrinol (2002) 146(4):519–22. doi:10.1530/eje.0.1460519
301. Török H-P, Tonenchi L, Glas J, Schiemann U, Folwaczny C. No significant association between mutations in exons 6 and 8 of the autoimmune regulator (AIRE) gene and inflammatory bowel disease. Eur J Immunogenet (2004) 31(2):83–6. doi:10.1111/j.1365-2370.2004.00449.x
302. Goswami R, Gupta N, Ray D, Rani R, Tomar N, Sarin R, et al. Polymorphisms at +49A/G and CT60 sites in the 3′ UTR of the CTLA-4 gene and APECED-related AIRE gene mutations analysis in sporadic idiopathic hypoparathyroidism. Int J Immunogenet (2005) 32(6):393–400. doi:10.1111/j.1744-313X.2005.00545.x
303. Palma A, Gianchecchi E, Palombi M, Luciano R, Di Carlo P, Crinò A, et al. Analysis of the autoimmune regulator gene in patients with autoimmune non-APECED polyendocrinopathies. Genomics (2013) 102(3):163–8. doi:10.1016/j.ygeno.2013.04.016
304. Cetani F, Barbesino G, Borsari S, Pardi E, Cianferotti L, Pinchera A, et al. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J Clin Endocrinol Metab (2001) 86(10):4747–52. doi:10.1210/jcem.86.10.7884
305. Bellacchio E, Palma A, Corrente C, Di Girolamo F, Kemp EH, Di Matteo G, et al. The possible implication of the S250C variant of the autoimmune regulator protein in a patient with autoimmunity and immunodeficiency: in silico analysis suggests a molecular pathogenic mechanism for the variant. Gene (2014) 549(2):286–94. doi:10.1016/j.gene.2014.07.064
306. Oftedal BE, Hellesen A, Erichsen MM, Bratland E, Vardi A, Perheentupa J, et al. Dominant mutations in the autoimmune regulator AIRE are associated with common organ-specific autoimmune diseases. Immunity (2015) 42(6):1185–96. doi:10.1016/j.immuni.2015.04.021
307. Abbott JK, Huoh Y-S, Reynolds PR, Yu L, Rewers M, Reddy M, et al. Dominant-negative loss of function arises from a second, more frequent variant within the SAND domain of autoimmune regulator (AIRE). J Autoimmun (2018) In press. doi:10.1016/j.jaut.2017.10.010
308. Ilmarinen T, Eskelin P, Halonen M, Rüppell T, Kilpikari R, Duran Torres G, et al. Functional analysis of SAND mutations in AIRE supports dominant inheritance of the G228W mutation. Hum Mutat (2005) 26(4):322–31. doi:10.1002/humu.20224
309. Su MA, Giang K, Žumer K, Jiang H, Oven I, Rinn JL, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J Clin Invest (2008) 118(5):1712–26. doi:10.1172/JCI34523
310. Fletcher AL, Seach N, Reiseger JJ, Lowen TE, Hammett MV, Scott HS, et al. Reduced thymic Aire expression and abnormal NF-κB2 signaling in a model of systemic autoimmunity. J Immunol (2009) 182(5):2690–9. doi:10.4049/jimmunol.0801752
311. Fornari TA, Donate PB, Macedo C, Marques MMC, Magalhães DA, Passos GAS. Age-related deregulation of Aire and peripheral tissue antigen genes in the thymic stroma of non-obese diabetic (NOD) mice is associated with autoimmune type 1 diabetes mellitus (DM-1). Mol Cell Biochem (2010) 342(1–2):21–8. doi:10.1007/s11010-010-0464-z
312. Venanzi ES, Melamed R, Mathis D, Benoist C. The variable immunological self: genetic variation and nongenetic noise in Aire-regulated transcription. Proc Natl Acad Sci U S A (2008) 105(41):15860–5. doi:10.1073/pnas.0808070105
313. Sabater L, Ferrer-Francesch X, Sospedra M, Caro P, Juan M, Pujol-Borrell R. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J Autoimmun (2005) 25(4):312–8. doi:10.1016/j.jaut.2005.08.006
314. Cai CQ, Zhang T, Breslin MB, Giraud M, Lau MS. Both polymorphic variable number of tandem repeats and autoimmune regulator modulate differential expression of insulin in human thymic epithelial cells. Diabetes (2011) 60(1):336–44. doi:10.2337/db10-0255
315. Fierabracci A. Type 1 diabetes in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome (APECED): a “rare” manifestation in a “rare” disease. Int J Mol Sci (2016) 17(7):e1106. doi:10.3390/ijms17071106
316. Tuomi T, Björses P, Falorni A, Partanen J, Perheentupa J, Lernmark Å, et al. Antibodies to glutamic acid decarboxylase and insulin-dependent diabetes in patients with autoimmune polyendocrine syndrome type I. J Clin Endocrinol Metab (1996) 81(4):1488–94. doi:10.1210/jcem.81.4.8636356
317. Huang W, Connor E, Dela Rosa T, Muir A, Schatz D, Silverstein J, et al. Although DR3-DQB1*0201 may be associated with multiple component diseases of the autoimmune polyglandular syndromes, the human leukocyte antigen DR4-DQB1*0302 haplotype is implicated only in β-cell autoimmunity. J Clin Endocrinol Metab (1996) 81(7):2559–63. doi:10.1210/jcem.81.7.8675578
318. Gylling M, Tuomi T, Björses P, Kontiainen S, Partanen J, Christie MR, et al. β-Cell autoantibodies, human leukocyte antigen II alleles, and type 1 diabetes in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab (2000) 85(12):4434–40. doi:10.1210/jcem.85.12.7120
319. Adamson KA, Cheetham TD, Kendall-Taylor P, Seckl JR, Pearce SHS. The role of the IDDM2 locus in the susceptibility of UK APS1 subjects to type 1 diabetes mellitus. Int J Immunogenet (2007) 34(1):17–21. doi:10.1111/j.1744-313X.2006.00643.x
320. Paquette J, Varin DSE, Hamelin CE, Hallgren Å, Kämpe O, Carel J-C, et al. Risk of autoimmune diabetes in APECED: association with short alleles of the 5′ insulin VNTR. Genes Immun (2010) 11(7):590–7. doi:10.1038/gene.2010.33
321. Lee J-W, Epardaud M, Sun J, Becker JE, Cheng AC, Yonekura A, et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol (2007) 8(2):181–90. doi:10.1038/ni1427
322. Gardner JM, DeVoss JJ, Friedman RS, Wong DJ, Tan YX, Zhou X, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science (2008) 321(5890):843–7. doi:10.1126/science.1159407
323. Gardner JM, Metzger TC, McMahon EJ, Au-Yeung BB, Krawisz AK, Lu W, et al. Extrathymic Aire-expressing cells are a distinct bone marrow-derived population that induce functional inactivation of CD4+ T cells. Immunity (2013) 39(3):560–72. doi:10.1016/j.immuni.2013.08.005
324. Grupillo M, Gualtierotti G, He J, Sisino G, Bottino R, Rudert WA, et al. Essential roles of insulin expression in Aire+ tolerogenic dendritic cells in maintaining peripheral self-tolerance of islet β-cells. Cell Immunol (2012) 273(2):115–23. doi:10.1016/j.cellimm.2011.12.010
325. Zheng X, Yin L, Liu Y, Zheng P. Expression of tissue-specific autoantigens in the hematopoietic cells leads to activation-induced cell death of autoreactive T cells in the secondary lymphoid organs. Eur J Immunol (2004) 34(11):3126–34. doi:10.1002/eji.200425177
326. Cohen JN, Guidi CJ, Tewalt EF, Qiao H, Rouhani SJ, Ruddell A, et al. Lymph node-resident lymphatic endothelial cells mediate peripheral tolerance via Aire-independent direct antigen presentation. J Exp Med (2010) 207(4):681–8. doi:10.1084/jem.20092465
327. Fletcher AL, Lukacs-Kornek V, Reynoso ED, Pinner SE, Bellemare-Pelletier A, Curry MS, et al. Lymph node fibroblastic reticular cells directly present peripheral tissue antigen under steady-state and inflammatory conditions. J Exp Med (2010) 207(4):689–97. doi:10.1084/jem.20092642
328. Zhang J, Chen Z, Fritz JH, Rochman Y, Leonard WJ, Plumb AW, et al. Unusual timing of CD127 expression by mouse uterine natural killer cells. J Leukoc Biol (2012) 91(3):417–26. doi:10.1189/jlb.1011501
329. Matsumoto M. Contrasting models for the roles of Aire in the differentiation program of epithelial cells in the thymic medulla. Eur J Immunol (2011) 41(1):12–7. doi:10.1002/eji.201041024
330. Farr AG, Rudensky A. Medullary thymic epithelium: a mosaic of epithelial “self”? J Exp Med (1998) 188(1):1–4. doi:10.1084/jem.188.1.1
331. Farr AG, Dooley JL, Erickson M. Organization of thymic medullary epithelial heterogeneity: implications for mechanisms of epithelial differentiation. Immunol Rev (2002) 189:20–7. doi:10.1034/j.1600-065X.2002.18903.x
332. Dooley J, Erickson M, Roelink H, Farr AG. Nude thymic rudiment lacking functional foxn1 resembles respiratory epithelium. Dev Dyn (2005) 233(4):1605–12. doi:10.1002/dvdy.20495
333. Dooley J, Erickson M, Farr AG. An organized medullary epithelial structure in the normal thymus expresses molecules of respiratory epithelium and resembles the epithelial thymic rudiment of nude mice. J Immunol (2005) 175(7):4331–7. doi:10.4049/jimmunol.175.7.4331
334. Dooley J, Erickson M, Farr AG. Lessons from thymic epithelial heterogeneity: FoxN1 and tissue-restricted gene expression by extrathymic, endodermally derived epithelium. J Immunol (2009) 183(8):5042–9. doi:10.4049/jimmunol.0901371
335. Gillard GO, Farr AG. Features of medullary thymic epithelium implicate postnatal development in maintaining epithelial heterogeneity and tissue-restricted antigen expression. J Immunol (2006) 176(10):5815–24. doi:10.4049/jimmunol.176.10.5815
336. Gillard GO, Dooley J, Erickson M, Peltonen L, Farr AG. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J Immunol (2007) 178(5):3007–15. doi:10.4049/jimmunol.178.5.3007
337. Dooley J, Erickson M, Farr AG. Alterations of the medullary epithelial compartment in the Aire-deficient thymus: implications for programs of thymic epithelial differentiation. J Immunol (2008) 181(8):5225–32. doi:10.4049/jimmunol.181.8.5225
338. Milićević NM, Milićević Ž, Miljković MD, Labudović-Borović M, Laan M, Peterson P, et al. Metallophilic macrophages are fully developed in the thymus of autoimune regulator (Aire)-deficient mice. Histochem Cell Biol (2009) 131(5):643–9. doi:10.1007/s00418-008-0553-1
339. Milićević Ž, Milićević NM, Laan M, Peterson P, Kisand K, Scott HS, et al. Ultrastructure of medullary thymic epithelial cells of autoimmune regulator (Aire)-deficient mice. Immunol Cell Biol (2010) 88(1):50–6. doi:10.1038/icb.2009.55
340. Yano M, Kuroda N, Han H, Meguro-Horike M, Nishikawa Y, Kiyonari H, et al. Aire controls the differentiation program of thymic epithelial cells in the medulla for the establishment of self-tolerance. J Exp Med (2008) 205(12):2827–38. doi:10.1084/jem.20080046
341. Nishikawa Y, Nishijima H, Matsumoto M, Morimoto J, Hirota F, Takahashi S, et al. Temporal lineage tracing of Aire-expressing cells reveals a requirement for Aire in their maturation program. J Immunol (2014) 192(6):2585–92. doi:10.4049/jimmunol.1302786
342. Nishijima H, Kitano S, Miyachi H, Morimoto J, Kawano H, Hirota F, et al. Ectopic Aire expression in the thymic cortex reveals inherent properties of Aire as a tolerogenic factor within the medulla. J Immunol (2015) 195(10):4641–9. doi:10.4049/jimmunol.1501026
343. Kawano H, Nishijima H, Morimoto J, Hirota F, Morita R, Mouri Y, et al. Aire expression is inherent to most medullary thymic epithelial cells during their differentiation program. J Immunol (2015) 195(11):5149–58. doi:10.4049/jimmunol.1501000
344. Danso-Abeam D, Humblet-Baron S, Dooley J, Liston A. Models of Aire-dependent gene regulation for thymic negative selection. Front Immunol (2011) 2:e14. doi:10.3389/fimmu.2011.00014
345. Matsumoto M, Nishikawa Y, Nishijima H, Morimoto J, Matsumoto M, Mouri Y. Which model better fits the role of Aire in the establishment of self-tolerance: the transcription model or the maturation model? Front Immunol (2013) 4:e210. doi:10.3389/fimmu.2013.00210
346. Lai L, Jin J. Generation of thymic epithelial cell progenitors by mouse embryonic stem cells. Stem Cells (2009) 27(12):3012–20. doi:10.1002/stem.238
347. Lai L, Cui C, Jin J, Hao Z, Zheng Q, Ying M, et al. Mouse embryonic stem cell-derived thymic epithelial cell progenitors enhance T-cell reconstitution after allogeneic bone marrow transplantation. Blood (2011) 118(12):3410–8. doi:10.1182/blood-2011-03-340794
348. Parent AV, Russ HA, Khan IS, LaFlam TN, Metzger TC, Anderson MS, et al. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell (2013) 13(2):219–29. doi:10.1016/j.stem.2013.04.004
349. Sun X, Xu J, Lu H, Liu W, Miao Z, Sui X, et al. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell (2013) 13(2):230–6. doi:10.1016/j.stem.2013.06.014
350. Inami Y, Yoshikai T, Ito S, Nishio N, Suzuki H, Sakurai H, et al. Differentiation of induced pluripotent stem cells to thymic epithelial cells by phenotype. Immunol Cell Biol (2011) 89(2):314–21. doi:10.1038/icb.2010.96
351. Soh C-L, Giudice A, Jenny RA, Elliott DA, Hatzistavrou T, Micallef SJ, et al. FOXNIGFP/w reporter hESCs enable identification of integrin-β4, HLA-DR, and EpCAM as markers of human PSC-derived FOXNI+ thymic epithelial progenitors. Stem Cell Rep (2014) 2(6):925–37. doi:10.1016/j.stemcr.2014.04.009
352. Ko H-J, Kinkel SA, Hubert F-X, Nasa Z, Chan J, Siatskas C, et al. Transplantation of autoimmune regulator-encoding bone marrow cells delays the onset of experimental autoimmune encephalomyelitis. Eur J Immunol (2010) 40(12):3499–509. doi:10.1002/eji.201040679
353. Yang X, Levi D, Ounissi-Benkalha H, Yu X, Qu H, Polychronakos C, et al. Screening for novel lead compounds increasing insulin expression in medullary thymic epithelial cells. Eur J Pharmacol (2012) 688(1–3):84–9. doi:10.1016/j.ejphar.2012.03.047
354. Träger U, Sierro S, Djordjevic G, Bouzo B, Khandwala S, Meloni A, et al. The immune response to melanoma is limited by thymic selection of self-antigens. PLoS One (2012) 7(4):e35005. doi:10.1371/journal.pone.0035005
355. Bakhru P, Zhu M-L, Wang H-H, Hong LK, Khan I, Mouchess M, et al. Combination central tolerance and peripheral checkpoint blockade unleashes antimelanoma immunity. JCI Insight (2017) 2(18):e93265. doi:10.1172/jci.insight.93265
356. Peterson P, Pitkänen J, Sillanpää N, Krohn K. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED): a model disease to study molecular aspects of endocrine autoimmunity. Clin Exp Immunol (2004) 135(3):348–57. doi:10.1111/j.1365-2249.2004.02384.x
357. Su MA, Anderson MS. Aire: an update. Curr Opin Immunol (2004) 16(6):746–52. doi:10.1016/j.coi.2004.09.009
358. Pereira LE, Bostik P, Ansari AA. The development of mouse APECED models provides new insight into the role of AIRE in immune regulation. Clin Dev Immunol (2005) 12(3):211–6. doi:10.1080/17402520500212589
359. Villaseñor J, Benoist C, Mathis D. AIRE and APECED: molecular insights into an autoimmune disease. Immunol Rev (2005) 204:156–64. doi:10.1111/j.0105-2896.2005.00246.x
360. Peterson P, Peltonen L. Autoimmune polyendocrinopathy syndrome type 1 (APS1) and AIRE gene: new views on molecular basis of autoimmunity. J Autoimmun (2005) 25(S):49–55. doi:10.1016/j.jaut.2005.09.022
361. Rizzi M, Ferrera F, Filaci G, Indiveri F. Disruption of immunological tolerance: role of AIRE gene in autoimmunity. Autoimmun Rev (2006) 5(2):145–7. doi:10.1016/j.autrev.2005.09.001
362. Liston A. There and back again: autoimmune polyendocrinopathy syndrome type I and the Aire knockout mouse. Drug Discov Today Dis Models (2006) 3(1):33–40. doi:10.1016/j.ddmod.2006.03.006
363. Sigal LH. Protecting against autoimmunity: tolerance and aire, the immunologic shadow, and other mechanisms of negative selection in the thymus. J Clin Rheumatol (2006) 12(1):44–6. doi:10.1097/01.rhu.0000200338.09858.16
364. DeVoss JJ, Anderson MS. Lessons on immune tolerance from the monogenic disease APS1. Curr Opin Genet Dev (2007) 17(3):193–200. doi:10.1016/j.gde.2007.04.001
366. Cheng MH, Shum AK, Anderson MS. What’s new in the Aire? Trends Immunol (2007) 28(7):321–7. doi:10.1016/j.it.2007.05.004
367. Peterson P, Org T, Rebane A. Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat Rev Immunol (2008) 8(12):948–57. doi:10.1038/nri2450
368. Ferguson BJ, Cooke A, Peterson P, Rich T. Death in the AIRE. Trends Immunol (2008) 29(7):306–12. doi:10.1016/j.it.2008.03.004
369. Mathis D, Benoist C. Aire. Annu Rev Immunol (2009) 27:287–312. doi:10.1146/annurev.immunol.25.022106.141532
370. Shikama N, Nusspaumer G, Holländer GA. Clearing the AIRE: on the pathophysiological basis of the autoimmune polyendocrinopathy syndrome type-1. Endocrinol Metab Clin North Am (2009) 38(2):273–88. doi:10.1016/j.ecl.2009.01.011
371. Cohn M. Why Aire? Compensating for late bloomers. Eur J Immunol (2009) 39(11):2969–72. doi:10.1002/eji.200939628
372. Fierabracci A. Recent insights into the role of molecular mechanisms of the autoimmune regulator (AIRE) gene in autoimmunity. Autoimmun Rev (2011) 10(3):137–43. doi:10.1016/j.autrev.2010.08.019
373. Taniguchi RT, Anderson MS. The role of Aire in clonal selection. Immunol Cell Biol (2011) 89(1):40–4. doi:10.1038/icb.2010.132
374. Metzger TC, Anderson MS. Control of central and peripheral tolerance by Aire. Immunol Rev (2011) 241(1):89–103. doi:10.1111/j.1600-065X.2011.01008.x
375. Akirav EM, Ruddle NH, Herold KC. The role of AIRE in human autoimmune disease. Nat Rev Endocrinol (2011) 7(1):25–33. doi:10.1038/nrendo.2010.200
376. Laan M, Peterson P. The many faces of Aire in central tolerance. Front Immunol (2013) 4:e326. doi:10.3389/fimmu.2013.00326
377. Chan AY, Anderson MS. Central tolerance to self revealed by the autoimmune regulator. Ann N Y Acad Sci (2015) 1356:80–9. doi:10.1111/nyas.12960
378. Abramson J, Husebye ES. Autoimmune regulator and self-tolerance – molecular and clinical aspects. Immunol Rev (2016) 271(1):127–40. doi:10.1111/imr.12419
379. Anderson MS, Su MA. AIRE expands: new roles in immune tolerance and beyond. Nat Rev Immunol (2016) 16(4):247–58. doi:10.1038/nri.2016.9
Keywords: animal disease models, autoimmune polyendocrinopathies, immune tolerance, thymus gland, transcription factors, type-1 diabetes mellitus
Citation: Perniola R (2018) Twenty Years of AIRE. Front. Immunol. 9:98. doi: 10.3389/fimmu.2018.00098
Received: 20 July 2017; Accepted: 12 January 2018;
Published: 12 February 2018
Edited by:
Adrian Liston, Flanders Institute for Biotechnology, BelgiumReviewed by:
Jakub Abramson, Weizmann Institute of Science, IsraelMitsuru Matsumoto, Tokushima University, Japan
Copyright: © 2018 Perniola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Perniola, cnBlcm5pb2xhQGhvdG1haWwuY29t
 Roberto Perniola
Roberto Perniola