- 1Department of Chemical and Biomolecular Engineering, Tulane University, New Orleans, LA, United States
- 2Interdisciplinary Bioinnovation PhD Program, Tulane University, New Orleans, LA, United States
The Clostridium genus is a large, diverse group consisting of Gram-positive, spore-forming, obligate anaerobic firmicutes. Among this group are historically notorious pathogens as well as several industrially relevant species with the ability to produce chemical commodities, particularly biofuels, from renewable biomass. Additionally, other species are studied for their potential use as therapeutics. Although metabolic engineering and synthetic biology have been instrumental in improving product tolerance, titer, yields, and feed stock consumption capabilities in several organisms, low transformation efficiencies and lack of synthetic biology tools and genetic parts make metabolic engineering within the Clostridium genus difficult. Progress has recently been made to overcome challenges associated with engineering various Clostridium spp. For example, developments in CRISPR tools in multiple species and strains allow greater capability to produce edits with greater precision, faster, and with higher efficiencies. In this mini-review, we will highlight these recent advances and compare them to established methods for genetic engineering in Clostridium. In addition, we discuss the current state and development of Clostridium-based promoters (constitutive and inducible) and reporters. Future progress in this area will enable more rapid development of strain engineering, which would allow for the industrial exploitation of Clostridium for several applications including bioproduction of several commodity products.
Introduction
For production of fuels and chemicals, two competing design models exist. One design paradigm aims to endow some heterologous trait (e.g., a biomass utilization or production phenotype) onto a highly editable platform organism (e.g., E. coli) with the rationale that strain engineering can be performed more quickly and in a high-throughput manner. High throughput methodologies can enable rapid construction of balanced pathways (Smanski et al., 2014), synthetically designed genetic parts (Jones et al., 2015; Rohlhill et al., 2017), and whole-genome recoding (Ostrov et al., 2016). However, importing heterologous pathways often requires significant effort to achieve the production titers attained by native producing strains. The alternative approach is to improve strains which innately have the desired trait. Such strains already contain and use necessary genes and pathways, including cofactor regeneration. Engineering strains which already have a desired phenotype can avoid potential challenges such as the metabolic burden of high expression of heterologous genes, cofactor imbalance, genetic instability of imported genes or pathways, among others (Wu et al., 2016; Czajka et al., 2017; Wang M. et al., 2017). Development of an advanced genome engineering “tool kit” in non-platform organisms brings these two models closer together.
Advancements in the synthetic biology tool kit are required to better utilize the biotechnologically important capabilities of the Clostridium genus. The Clostridium genus is home to multiple industrially relevant strains. These Gram-positive, spore-forming, obligate anaerobic firmicutes are natively capable of cellulosic and hemicellulosic biomass degradation (e.g., C. cellulolyticum) (Heinze et al., 2017), carbon fixation (e.g., C. carboxidivorans, C. ljungdahlii) (Jones et al., 2016), advanced biofuel production (e.g., C. acetobutylicum, C. beijerinckii) (Liu K. et al., 2015), platform chemical production (e.g., C. pasteurianum) (Xin et al., 2016), and acting as anti-cancer therapeutics (C. novyi-NT) (Staedtke et al., 2016). Additionally, genome editing tools are of medical interest to better understand the many pathogenic strains in the genus (e.g., C. botulinum, C. tetani, C. perfringens) (Ng et al., 2013). Clostridium genome engineering has made much progress recently in the development of synthetic biology tools, although it still lags behind workhorse organisms (e.g., E. coli). Continued progress in this genus will enable broadened engineering on new platforms. In this mini-review, we discuss recent progress in the development of synthetic biology tools for members of the Clostridium genus and compare these to the established methods. Emphasized are CRISPR-based tools for genome editing and transcriptional perturbation as well as the library of genetic parts available for use in Clostridium.
Gene Editing
ClosTron
ClosTron Technology Utilizes Group II Mobile Introns for Efficient Targeted Gene Disruption in Clostridium
Bacterial group II intron technology enables site-directed genetic disruptions based on the retrohoming of Mobile Group II introns. The mobility of Mobile Group II introns provides a convenient method of gene disruption as retrohoming is efficient and specific. The technology works by inserting an intron into chromosomal DNA through the plasmid-based monocistronic-expression of a ribonucleoprotein complex comprising RNA in a lariat configuration (acting as a ribozyme) and an intron-encoded protein (IEP). The Mobile Group II introns are minimally dependent on host factors, as the IEP (LtrA in the model system based on the Lactococcus lactis Ll.LtrB intron) performs multiple activities: maturase for facilitating RNA splicing, endonuclease for cleavage of the DNA strand opposite the RNA splice, and reverse transcriptase which uses intron RNA as template to insert DNA into the host chromosome. The host DNA repair machinery replaces intron RNA with DNA, completing the insertion. The term Targetron was first used to refer to targeted Group II introns when the L1.LtrB intron was further modified to include a retrotransposition-activated selection marker (RAM), providing a means to select for successful targeting events (Zhong et al., 2003). The RAM is inserted into the domain IV of the intron and consists a marker (often an antibiotic resistance gene) inactivated by the insertion of a Group I intron which is self-catalytically spliced out of mRNA in an orientation dependent manner. The marker gene and group I intron are oriented in the opposite directions such that it is only spliced out of the L1.LtrB mRNA, and a functional marker gene can only be expressed after successful chromosomal insertion occurs (Figure 1A).
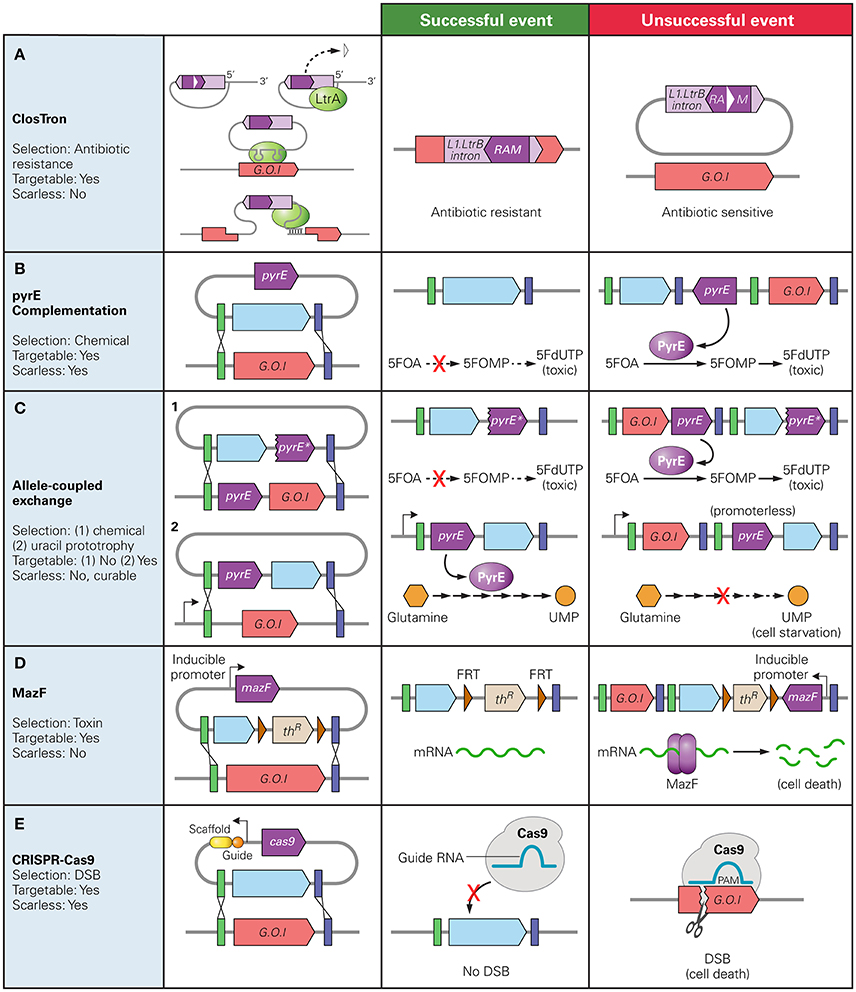
Figure 1. Counter Selection markers used in Clostridium spp. and their mechanisms of selection. The native gene of interest (G.O.I) is represented in red, desired insert in blue, and the counter selection marker gene in dark purple. The green and blue bars represent regions of homology between the chromosome and donor plasmid. (A) ClosTron: RAM disrupted by a Group I intron (white triangle) is only active after the L1.LtrB intron is inserted into the chromosome; (B) pyrE complementation: PyrE catalyzes conversion of 5-fluoroorotic acid (5FOA) to 5-fluororotidine monophosphate (5FOMP) producing toxic fluorodeoxyuridine monophosphate (FdUTP); (C) Allele-Coupled Exchange: (1) double-crossover event at the pyrE locus results in truncated version of pyrE for counter selection with same mechanism as (B), (2) successful homologous recombination inserts a promoter-less copy of the pyrE gene directly downstream a native constitutive promoter, allowing production of uracil 5′ monophosphate (UMP). Note: must be performed on pyrE deficient strain; (D) MazF protein degrades mRNA at 5′-ACA-3′ sequences; (E) Cas9: successful homologous recombination gRNA-targeted double stranded break resulting in cell death.
The specific targeting, or retrohoming, is accomplished through protein-specific DNA binding as well as programmed RNA-DNA complementarity. This complementarity over a 13-nucleotide region allows for the specification of the DNA target site by altering the intron sequence. However, the target site is limited to DNA sites compatible with the IEP. The Targetron system has been used to perform gene targeting in both Gram positive and negative bacteria (Karberg et al., 2001) including several Clostridium spp.: C. perfringens (Chen et al., 2005, 2007), C. acetobutylicum (Shao et al., 2007) and C. pasteurianum (Pyne et al., 2014). ClosTron was developed as an adaptation of Targetron technology for efficient gene targeting specifically in Clostridium species. The original ClosTron plasmid, pMTL007, tailored the commercially available E. coli Targetron vector, pACD4k-C, to include standardized genetic parts such as promoters, origins of replications and RAMs suitable for efficient gene editing in Clostridium (Heap et al., 2007). ClosTron introns are designed using a computer algorithm which identifies suitable sequences, based on a 35 bp region recognized by the IEP, to which the intron can be targeted within the desired gene (Heap et al., 2010). These introns are expressed on vectors which follow the format of the pMTL80000 modular plasmids, allowing for the availability of a range of genetic parts. Flanking the RAM with FRT sites permits reuse of the same marker after FLP-FRT recombination (Heap et al., 2010). In C. perfringens, DNA fragments of up to 1.0 kb were successfully integrated into the chromosome in addition to an ermB RAM at a frequency of 5 × 10−8 integrants per cell, a 10−4 fold decrease in integration frequency when no additional cargo is added (Heap et al., 2010).
ClosTron has been employed in targeted gene disruption across the Clostridium genus including C. acetobutylicum (Hönicke et al., 2014; Liu Z. et al., 2015; Xu M. et al., 2015), C. beijerinckii (Heap et al., 2010; Liu et al., 2016), and C. botulinum (Meaney et al., 2015, 2016), as well as the closely related species, Clostridioides difficile (Clostridium difficile, Dingle et al., 2011; Baban et al., 2013; Hensbergen et al., 2015; Lawson et al., 2016). However, gene disruption using ClosTron is limited to IEP recognition sites, the frequency of which decreases as the length of the target sequence decreases. Additionally, while a scarless deletion is often preferred for gene knockouts, ClosTron merely disrupts the gene of interest and the intron, along with its RAM, remains in the host chromosome.
Transposon-Based Random Mutagenesis
Transposon Mobile Elements Have Been Utilized in Random Mutagenesis in Clostridium
Among these, Tn916/Tn1545 family of transposons (called conjugative transposons) were among the first elements available for random gene insertions in Clostridium. These transposons consist of four functional modules responsible for conjugation, recombination, regulation and accessory functions (often antibiotic resistance). Although its expression can be plasmid-based, the conjugative ability of Tn916 (Flannagan and Clewell, 1991) has allowed its transfer from the chromosome of hosts such as E. coli and Bacillus subtilis into Clostridium species (Woolley et al., 1989). The use of Tn916-like transposons is limited by their large size, a predilection to insert in multiple copies or specific “hot spots,” and deletions at the site of insertion (Awad and Rood, 1997; Wang et al., 2000; Hussain et al., 2005). While non-conjugative transposon systems, such as Mu phage and EZ-Tn5-based transposon systems have addressed several of these concerns and have been successfully employed in Clostridium (Chen et al., 2005; Lanckriet et al., 2009; Vidal et al., 2009), they are limited by a preferential insertion into rRNA genes (Lanckriet et al., 2009; Vidal et al., 2009).
Mariner-transposable Himar1-based systems were developed for random mutagenesis in Clostridium (Cartman and Minton, 2010; Liu et al., 2013; Zhang Y. et al., 2015; Zhang et al., 2016). The Himar1 transposable element, whose sequence includes a transposase gene flanked by short inverted terminal repeat sequences (ITRs), was originally discovered in the horn fly and has been shown to insert quasi-randomly into the genomes of several bacteria species, including members of the Clostridium genus. Successful Himar1-based events have been recorded in C. acetobutylicum (Zhang Y. et al., 2015), C. perfringens (Liu et al., 2013) and C. sporogenes (Zhang J. et al., 2015). Himar1 transposase binds and cuts the element in its ITR region which, in turn, is inserted at a TA dinucleotide target site. This has been a valuable tool for mutagenesis in AT-rich Clostridium as it inserts average at one copy per cell.
Counter-Selection Markers
Counter-Selection Markers Enable the Isolation of Double Crossover Homologous Recombination Events in Clostridium spp.
Targeted gene disruption in bacteria can be achieved through plasmid-based, double crossover recombination events. The first event incorporates a vector containing a pair of homology arms flanking a cargo sequence into a target gene locus through homologous recombination (HR). In the second crossover, the specific region between the homologous sequences is deleted from the genome and is replaced by cargo DNA. Counter-selection markers are genetic elements which, when present, result in cell death; these markers are useful for selecting for chromosomal insertions which do not contain undesirable parts (e.g., the vector backbone of the recombination plasmid left over in a single-crossover event). Very few double-crossover events had been isolated within members of the Clostridium genus prior to the development of Clostridium-specific counter-selection marker systems (Awad et al., 1995; Bannam et al., 1995). Before routine usage of counter-selection markers, targeted homologous recombination in Clostridium had only been successful in a few species, the majority of which were segregationally unstable single cross-over integrations (Green and Bennett, 1996; Green et al., 1996; Nair et al., 1999; Liyanage et al., 2001; Harris et al., 2002; O'Connor et al., 2006). By contrast, the sacB gene from B. subtilis has been used to screen for double-crossover events in E. coli from the early 1990's.
One counter-selection marker method involves deactivating an easily screenable gene and then complementing the mutant strain with a heterologous version of that gene as a counter selective marker. Specifically, a disruption of pyrE, pyrF, or upp genes create uracil auxotrophic mutants which require supplementation for growth but are also resistant to the antimetabolites 5-fluoroorotic acid (5-FOA) or 5-fluorouracil (5-FU). By including a functional copy of the disrupted gene on the backbone of the donor DNA plasmid, double crossover events can be isolated as the mutants that demonstrate a resistance to 5-FOA or 5-FU (Tripathi et al., 2010; Heap et al., 2012; Croux et al., 2016) (Figure 1B). Similarly, in C. perfringens, disruption of the galKT operon produces mutants unable to produce the enzymes involved in galactose metabolism. GalK catalyzes the production of galactose-1-phosphate (Gal-1-P) from galactose, and GalT catalyzes its consumption. The accumulation of Gal-1-P is believed to inhibit cell growth by causing intracellular stress and inducing stress-responsive genes (Lee et al., 2009, 2014). By including only the galK gene and not the galT gene on the integration vector and plating mutant cells on galactose supplemented plates, unedited cells do not grow due to an accumulation of Gal-1-P while mutants that undergo a double-crossover event can be isolated (Nariya et al., 2011b).
Allelic Coupled Exchange (ACE) couples a counter selection marker gene to a desired double crossover event. This has been demonstrated in two ways. One method exploits the 5-FOA resistance conferred by a disrupted pyrE or pyrF gene. This method does not require the cells to be auxotrophic for uracil prior to recombination, nor does it rely on a heterologous version of the gene as a counter selection marker. ACE technology employs asymmetric homology arms to direct the order in which crossover events occur. The longer arm, homologous to a 1,200 bp region immediately downstream of the pyrE of pyrF directs the first crossover event in which the entire plasmid is incorporated into the genome. The second crossover event excises the plasmid backbone and is directed by the shorter arm which is homologous to a 300 bp internal region of the pyrE or pyrF gene. This second recombination replaces the wildtype pyrE gene with a truncated form thereby producing a mutant that can be screened based on 5-FOA resistance (Heap et al., 2012). Alternatively, a promoter-less heterologous pyrE gene or antibiotic marker can be inserted in the integration vector with the regions of homology such that a successful double crossover event places the silent gene directly downstream of a constitutive promoter (Heap et al., 2012). However, unlike previous methods which relied heavily on ClosTron technology to first produce auxotrophic mutants, pyrE mutants can be created utilizing ACE technology while the use of an antibiotic marker circumvents the need for a requisite mutant strain (Heap et al., 2012; Minton et al., 2016) (Figure 1C). ACE has been proven to be applicable over a range of Clostridium species, having been used for gene editing in C. acetobutylicum (Bankar et al., 2015; Ehsaan et al., 2016; Willson et al., 2016) and C. sporogenes (Heap et al., 2012; Zhang Y. et al., 2015), as well as C. difficile (Heap et al., 2012; Ng et al., 2013).
Several heterologous genes have been used for counterselection. The cytosine deaminase gene (codA) from E. coli can be used for counterselection based on the ability of the CodA protein to catalyze the conversion of 5-fluorocytosine (5-FC), an innocuous pyrimidine analog, to 5-FU (Cartman et al., 2012). codA can only be used for counterselection in strains with a functional upp gene but no native codA gene (Ehsaan et al., 2016). However, a bioinformatics survey suggests several Clostridium species contain codA homologs, restricting the applicability of codA among the genus (Al-Hinai et al., 2012).
Toxin-antitoxin systems are another useful source of counters-election markers. The E. coli-based mazF is an mRNA interferase, coded along with mazE in an operon. Under regular cell conditions, mazE binds to and inhibits mazF activity. During cellular stress, mazE is degraded, allowing mazF to bind mRNA, degrading them at 5′-ACA′3′ sequences, thereby arresting cell growth. mazF, coupled with an antibiotic resistant marker flanked by FRT sites, can be used as a counter selection marker in plasmid based homologous recombination. mazF is placed on the gene disruption plasmid under the control of an inducible lac promoter. A double crossover event can be isolated in cells able to grow on lactose-supplemented plates (Al-Hinai et al., 2012) (Figure 1D). The use of mazF requires no prior mutation for successful screening, is independent of the availability of Clostridium genetic parts, and has been shown to function across Clostridium species (Al-Hinai et al., 2012; Sandoval et al., 2015; Zhang J. et al., 2015). Flp-frt recombination has also been used to eliminate the backbone of a donor plasmid following a single-crossover event in C. acetobutylicum, allowing for the use of an antibiotic gene as a marker for the crossover event after the donor plasmid had been cured (Lee S. H. et al., 2016).
Crispr-Based Editing
CRISPR/Cas9 Allows Efficient, Marker-Less Gene Editing in Clostridium
Clustered Regularly Interspaced Palindromic Repeats (CRISPR), along with its CRISPR associated (Cas) proteins is an adaptive immunity system in prokaryotes (Barrangou et al., 2007; Wiedenheft et al., 2012). The type II CRISPR system native to the Streptococcus pyogenes bacterium was the first CRISPR system exploited for gene engineering (spyCas9) (Jinek et al., 2012). This CRISPR system consists of the single Cas9 effector protein, which can bind to and implement a double stranded break (DSB) to a targeted DNA system when co-expressed with a single guide RNA targeting a 20 bp region immediately adjacent to the protospacer adjacent to a motif (PAM). In the case of spyCas9, the PAM consensus sequence is NGG, providing many possible target sites.
The CRISPR/Cas9 system has been used as a counter-selection tool to select for homologous recombination events in several Clostridium species (Wang et al., 2015; Bruder et al., 2016; Huang et al., 2016; Nagaraju et al., 2016; Wang S. et al., 2017; Wasels et al., 2017) (Table 1). Clostridium spp. lack or have inefficient non-homologous end-joining (NHEJ) systems, so a Cas9-mediated chromosomal DSB results in cell death (Cui and Bikard, 2016; Xu et al., 2017). Thus, to select for successful homologous recombination events, one can selectively eliminate non-edited members of the population by targeting the wild type sequence (Figure 1E). Studies in E. coli have shown the DSB can enhance homologous recombination in bacteria, whereby homology directed repair (HDR) occurs after a break has been induced (Jiang et al., 2013). However, studies in different Clostridium species suggest that HDR efficiency in these species is too low to select for successful HDR events (Wang et al., 2015, 2016a; Li Q. et al., 2016). The use of CRISPR/Cas9 represents a major advancement in Clostridium gene editing in Clostridium as scarless edits are enabled.
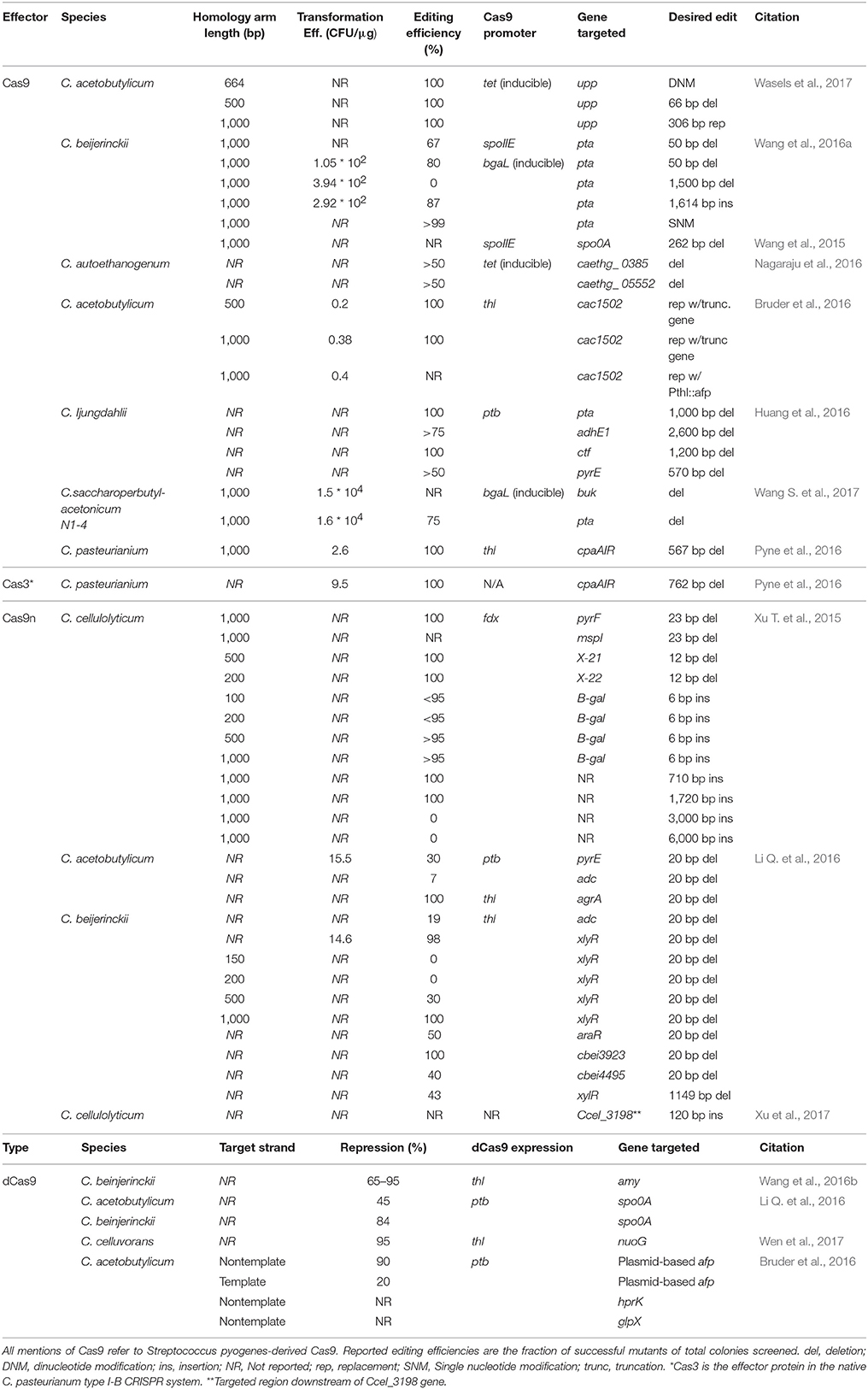
The limited number of characterized genetic parts for Clostridium poses a challenge with CRISPR/Cas9 engineering. For example, simultaneous constitutive expression of the sgRNA and Cas9 protein often resulted in few to no transformed colonies in the presence of a homologous repair donor vector, as DSBs result in cell death before recombination can occur (Bruder et al., 2016; Li Q. et al., 2016; Nagaraju et al., 2016; Wang et al., 2016a). This can be addressed by placing Cas9 expression under the control of an inducible promoter (Nagaraju et al., 2016; Wang et al., 2016a; Wang S. et al., 2017; Wasels et al., 2017). Another strategy is to use a two-plasmid system, where the donor DNA and sgRNA are introduced separately from the Cas9 gene. This method avoids the transformation of very large plasmids, which have reduced transformation efficiency, but it requires two separate transformation events (Wasels et al., 2017). Using these methods, successful recombinants were isolated at a rate up to 100% (Wasels et al., 2017) with commonly observed efficiencies of greater than 50% (Table 1).
Cas9 nickase (Cas9n) systems exploit CRISPR gene editing while circumventing the lethality associated with the co-expression of a guide RNA with Cas9. This method utilizes Cas9n, a mutated form of the Cas9 protein, with the ability to only cut one DNA strand. While simultaneous expression of Cas9 and guide RNA is fatal to cells in the absence and presence of a donor template, implementing a single nick into the genome via Cas9n allowed homologous recombination without the lethal effects of Cas9, thus permitting a mixed population of edited and unedited strains to coexist. (Xu T. et al., 2015) Therefore, several serial dilutions are required to enhance the edited population through increased likelihood of homologous recombination at the nicked site, reduced growth rate of the nicked strains, or some combination of both. CRISPRn has been used to implement gene deletions and insertions in C. acetobutylicum, C. beijerinckii and C. cellulolyticum with up to 100% efficiency (Xu T. et al., 2015; Li Q. et al., 2016; Xu et al., 2017) (Table 1).
Longer regions of homology on the donor have been shown to increase efficiency. In C. cellulolyticum, donor template arm lengths greater than 0.2 kb had an efficiency of more than 95% when compared with smaller arms which were only 55% efficient in a CRISPRn system (Xu T. et al., 2015). A similar study using Cas9 in C. acetobutylicum demonstrated an increased efficiency when homology arm lengths of 1 kb were used as opposed to 500 bp arm lengths (Bruder et al., 2016).
Application of these advanced CRISPR tools is still limited in Clostridium due to low plasmid transformation efficiencies and a lack of characterized recombineering and NHEJ tools. Recombineering, through lambda red technology, has facilitated gene engineering in E. coli via the use of linear DNA repair templates, a process that skips the cloning steps required in plasmid-based homologous recombination methods. Coupled with CRISPR, this technology enables multiplexed ssDNA recombineering events with efficiencies allowing large libraries (>105 members) to be constructed in parallel (Ronda et al., 2016; Garst et al., 2017). However, the lack of ssDNA recombineering machinery functional in Clostridium hinders the development of comparable Clostridium-based technologies. Although a RecT protein from C. perfringens demonstrated recombineering activity in C. acetobutylicum recently, the results obtained were not comparable to routine recombineering events in E. coli (Dong et al., 2014). Similarly, the expression of Ku and LigD genes from Mycobacterium tuberculosis enabled NHEJ in E. coli following the implementation of a DSB via Cas9 (Su et al., 2016). Although NHEJ related genes (ku, DNA ligase, and ligD) are found on the C. cellulolyticum, they are not highly expressed and NHEJ events have not been observed in the species after a Cas9 DSB (Xu T. et al., 2015). Heterologous expression of such genes may enable NHEJ in Clostridium.
Repurposing endogenous CRISPR systems has been proposed as an alternative to transforming and expressing CRISPR-Cas genes in Clostridium. A type I-B CRISPR system was identified in C. pasteurianum based on its genome sequence. The PAM sequence recognized by its Cas effector protein was determined via a bioinformatics survey of existing spacer sequences This system was used to target the cpaAIR gene in C. pasteurianum when transformed with a plasmid based expression copy of its CRISPR array and a homology repair template (Table 1). The donor DNA/targeting plasmid was transformed at a higher efficiency compared to a similar Cas9 based system, since the plasmid did not need to house the large CRISPR effector protein (9.5 vs. 2.6 CFU/μg) (Pyne et al., 2016). While the use of native CRISPR systems circumvents the need to express heterologous Cas effector proteins, the applicability of these systems may be limited to strains with functional CRISPR/Cas machinery and by unknown PAM sequences. Although application has not been reported in Clostridium, other CRISPR-Cas9 systems have been identified in other bacteria, including thermophilic Cas9 systems which can be utilized for gene editing in thermophiles (Mougiakos et al., 2017b) in lieu of spyCas9 as it is temperature sensitive (Mougiakos et al., 2017a).
Gene Silencing in Clostridium Can Be Achieved via a Catalytically Dead Cas9 Mutant
The catalytically dead Cas9 (dCas9) does not demonstrate endonuclease activity but retains its ability to bind to DNA at a specified target region. In a CRISPR interference (CRISPRi) system, dCas9 is targeted to a sequence by a guide RNA and binds to it, sterically hindering transcription initiation. CRISPRi has enabled simple, tunable, reversible gene knockdown at a transcriptional level. One needs only to express sgRNAs and with the aid of bioinformatics tools, can specifically implement gene expression knock-downs. This method is reversible, with no permanent change in the genome (Qi et al., 2013). Additionally, CRISPRi activity can be modulated not only through controlling the expression of dCas9 (Li X. T. et al., 2016), but also by the relative position of the dCas9 protein to the promoter and gene start site (Kim et al., 2017), allowing tight control of gene expression. CRISPRi technology has been used for gene knockdowns in C. acetobutylicum (Bruder et al., 2016; Li Q. et al., 2016), C. beijerinckii (Li Q. et al., 2016; Wang et al., 2016b) and in C. cellulovorans (Wen et al., 2017) and has been used to silence both native (Li Q. et al., 2016; Wang et al., 2016b; Wen et al., 2017) and heterologous genes (Bruder et al., 2016) (Table 1). Gene repression of up to 97% was achieved although the effectiveness of CRISPRi varies among species with similar configurations (Li Q. et al., 2016). The tunability of dCas9 has yet to be fully explored in Clostridium, as it has been in other bacterial systems (Li X. T. et al., 2016; Kim et al., 2017). In fact, one study showed activity of the knockdown target unintentionally increased over the time of fermentation (Wang et al., 2016b). This increase was attributed to variable strength of the thiolase promoter, which controlled the transcription of dCas9.
Gene expression knockdowns have also been accomplished on the translational level in Clostridium through antisense RNA (asRNA) technology. The asRNA knockdown method involves targeting an mRNA transcript using its asRNA. This method has been used to investigate the function of genes in various Clostridium species (Cooksley et al., 2010; Fagan and Fairweather, 2011; Chandrasekaran et al., 2016; Chu et al., 2016; Xu et al., 2017), several of which are essential genes, making a genetic knockout unfeasible. asRNA technology has also been used to manipulate gene expression affecting solvent titers (Tummala et al., 2003; Sillers et al., 2009). asRNAs are tunable and reversible, and have been effective in silencing, achieving as high as 90% repression levels of certain genes. However, despite their lengths (>100 bp) asRNAs have been shown to be promiscuous, binding especially to transcripts with a high homology to the target sequence (Cooksley et al., 2010). Consequently, asRNA technology requires large constructs for efficient gene repression while CRISPRi provides repression specificity, using a 20 nt sgRNA choice.
Genetic Parts
In addition to development of DNA editing tools, a “toolbox” of well-characterized biological parts including promoters, ribosomal binding sites (RBS), origin of replication (ORI), and terminators for Clostridium is in need for further advancement of metabolic engineering efforts. Expanding this toolbox would enable assemblies of individual or grouped genes to enhance productivity and yield of desired products or outcomes.
Promoters
Promoters are often the simplest way to control transcription, and promoters with various activities are valuable in any synthetic biology toolkit. However, promoters available for use in Clostridium originate from a few strains and are not always transferrable to non-native hosts. Constitutive promoters of ptb (phosphotransbutyrylase) and thl (thiolase) from C. acetobutylicum and C. pasteurianum are commonly used, however, promoter activity vary in different strains and stages of growth (Pyne et al., 2015; Lee J. et al., 2016; Yang et al., 2017). Therefore, there is a need for well-characterized promoter parts in synthetic biology.
Whole-transcriptomic sequencing offers comprehensive profiles of gene expression and transcriptome changes of an organism with high precision. Promoter motifs identified from RNA-seq analysis of E. coli and gram-positive B. subtilis are commonly used to screen for promoters in other organisms using bioinformatics tools (Borden et al., 2010). Promoter motifs are less well-established in Clostridium, thus relying on promoter motifs of B. subtilis as a reference (Paredes et al., 2004). Successful promoter prediction through bioinformatics requires more transcriptomic studies in Clostridium.
As screening for native promoters is time-consuming, mutagenesis methods such as error prone PCR, saturation mutagenesis, or site-specific mutagenesis of a known characteristic such as a hairpin are utilized to rapidly generate a library of synthetic promoters with varying strengths. A library of synthetic promoters using a web-based tool WEBLOGO was used to rapidly generate a library of promoters of various strengths by randomizing the flanking regions surrounding the -35 and -10 consensus sequences of promoter thl for C. acetobutylicum and C. ljungdahlii (Yang et al., 2017).
Several inducible promoters have been used to balance and map metabolic pathways, study function and regulation of promoters or genes, and express recombinant proteins in Clostridium: xylose-inducible promoter-repressor system from Staphylococcus xylosus (Girbal et al., 2003; Nariya et al., 2011a); lactose-inducible promoter comprising of a transcriptional regulator gene, bgaR, and a bgaL promoter from C. perfringens (Hartman et al., 2011; Al-Hinai et al., 2012; Banerjee et al., 2014); laminaribiose-inducible promoters (Mearls et al., 2015); arabinose-induced promoter (Zhang J. et al., 2015); tetracycline-inducible system (Walker and Köpke, 2015), anhydrotetracycline-inducible system (Dong et al., 2012); and radiation-induced promoter in C. acetobutylicum for cancer radiation therapy (Nuyts et al., 2001). Factors to be cognizant in using inducible promoters are its sensitivity to the inducer, as the cost of inducers may increase for large-scale production applications, and “leaky” expression in the absence of the inducer. Therefore, a quantitative measurement of transcription is necessary for characterization of promoters.
Reporters
Genetic reporters, genes which act as an observable proxy for some unobservable process, can provide measurement of gene expression and screening and characterization of promoters. An ideal reporter system has (1) high sensitivity and specificity, (2) a large dynamic range of detection, and (3) low endogenous levels of the reporter in the strain of interest. Chloramphenicol acetyltransferase (catP) has been used to select for active promoters by catalyzing the transfer reaction of an acetyl group from acetyl-CoA to the antibiotics chloramphenicol (Cm) and thiamphenicol (Bullifent et al., 1995). While Cm is effective in C. perfringens, other species such as C. acetobutylicum and C. beijerinckii NCIMB 8052 are naturally resistant to the antibiotic (Feustel et al., 2004). As the reporter mechanism is selection, it is not ideal for measurement of specific promoter activity. Interestingly, a catP-lacZ fusion reporter provides an initial screen to select for catP expressing promoters on agar with varying Cm concentrations, and subsequent colorimetric screen to measure accurate promoter activities (Yang et al., 2017).
lacZ from Thermoanaerobacterium thermosulfurigens EM1, as well as gusA (or uidA) from E. coli encoding β-galactosidase and β-glucuronidase respectively, are used to study quantitative and qualitative gene expression in Clostridium (Tummala et al., 1999; Feustel et al., 2004). A major advantage of the lacZ and gusA genes is the versatility of detection systems, depending on the substrate. lacZ reporter substrates reported in Clostridium include the substrates 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-GAL) (Feustel et al., 2004), o-nitrophenyl-β-D-galactopyranoside (ONPG) for colorimetric and spectrophotometric assays (Adcock and Saint, 2001; Tan et al., 2015), and 4-methylumbelliferyl-β-D-galactopyransoside (MUG) for fluorescent assays (Adcock and Saint, 2001). gusA reporters utilize analogous substrates to β-galactosidase assays: MUG for fluorescence, 5-bromo-4-chloro-3-indolyl-glucuronide for colorimetric, and p-nitrophenyl-β-D-glucuronide for spectrophotometric detection. Compared to lacZ, gusA is smaller and more stable with low background as a fusion reporter with the gene of interest in many organisms including Clostridium spp. (Ravagnani et al., 2000; Mani et al., 2006). Although not commonly used, other enzymatic-based reporters available include the alkaline phosphatase gene (phoZ) isolated from Enterococcus faecalis (Gibson and Caparon, 2002) and the β-1,4-endoglucanase gene (eglA) from C. saccharobutylicum (Quixley and Reid, 2000).
Bioluminescent reporters provide amplified measurements of genes expressed at low quantities. Despite the need for oxygen, luciferase reporters (luc and lucB) from North American firefly (Photinus pyralis) in Clostridium spp. can be achieved with the addition of ATP and washing cells with a neutral pH buffer (e.g., PBS) (Davis et al., 2000; Phillips-Jones, 2000; Feustel et al., 2004). In addition, luciferase fusion with secreting protein PPEP-1 in C. difficile enabled low-background luciferase expression in an anaerobic environment and luminescence measurement in an aerobic environment, albeit with a delayed signal (Oliveira Paiva et al., 2016). Likewise, bacterial luciferases (luxAB) of the lux system (luxCDABE) from Vibrio fischeri provide a luminescent output in the presence of flavin mononucleotide and long-chain aldehyde substrate in C. perfringens (Bullifent et al., 1995; Phillips-Jones, 2000). Although various strategies enable use of bioluminescent reporters in anaerobic bacteria, transcription activity is not directly correlated with the output (Iqbal et al., 2017).
Fluorescent reporters such as green fluorescent protein (GFP) and its variants allow real-time measurements of gene expression at a single cell level without the need of substrates. Although GFP variants CFP and mCherryOpt can be expressed in Clostridium, exposure to oxygen is required for the intrinsic chromophore to fluoresce (Ransom et al., 2015). FMN-based fluorescent proteins (FbFPs) including LOV (Light Oxygen Voltage) domains from plants and bacterial blue light receptors are an emerging class of fluorescent reporters for anaerobes (Drepper et al., 2007; Christie et al., 2012; Seo et al., 2018). These fluorescent reporters can be utilized with and without oxygen, light, and voltage, hence the name. They are smaller (~13 kDa) than GFP (~25 kDa) allowing relatively faster turnover rates and fusion with protein of interest without disrupting its native function. Furthermore, improved LOV (iLOV) proteins can retain 60–70% fluorescence in a broad pH range of 4–11 (Mukherjee et al., 2013). Excitation of improved LOV (iLOV) with blue-light wavelength at 450 nm emits an emission peak at 495 nm (Mukherjee et al., 2013), while GFP excitation and emission spectra are at 488 and 507 nm, respectively. iLov has been utilized as a fluorescent reporter in several Clostridium species: C. cellulolyticum (Teng et al., 2015), C. ljungdahii (Molitor et al., 2016), as well as C. difficile in which it was used for real-time measurement of protein localization and secretion via fusion to FtsZ, a cell division protein (Ransom et al., 2015; Teng et al., 2015; Buckley et al., 2016; Molitor et al., 2016). iLov expression has also been observed in C. acetobutylicum (Buckley et al., 2016).
The quantum yield (brightness) of iLOV is lower than GFP, however. Thus, there are some considerations to lower noise when using FbFPs. Flavin-based media such as reinforced clostridial medium (RCM) and PETC give high fluorescent backgrounds which can be reduced by eliminating yeast and beef extract (Molitor et al., 2016). Also, species such as C. acetabutylicum have a natural green auto-fluorescence that can increase the background signal (Buckley et al., 2016). A negative control plasmid containing a non-fluorescent reporter such as gusA can be utilized to tune excitation wavelength (i.e., 450 vs., 470 nm) to improve noise-to-signal fluorescence (Buckley et al., 2016).
Origins of Replication
As the plasmid is a fundamental synthetic biology tool, the origin of replication is fundamental to successful plasmid stability. Several origins of replication have been found to be effective in Clostridium species including: pBP1, pCB102, pCD6 and pIM13, which are included in the pMTL80000 series plasmids (Heap et al., 2009). Origins pBP1 and pCB102 are native to Clostridium (C. botulinum and C. butyricum, respectively), while pIM13 and pCD6 were isolated from B. subtilis and Clostridioides difficile, respectively. The origin-strain pair exhibit varying levels of stabilities. For example, pCB102 is stable in C. botulinum ATCC 305, but not as stable in C. acetobutylicum 824 (Minton et al., 2016).
Conditional origins are useful for when the presence of a plasmid is required only under certain conditions, such as recursive recombination editing. In C. acetobutylicum, the pAMβ1 replicon from E. faecalis, is unstable in the absence of antibiotic challenge (Lee S. H. et al., 2016). Recently, a temperature sensitive origin of replication derived from the pWV01 from Lactococcus lactis subsp. cremoris was tested in C. ljungdahlii, having the plasmid maintained at a permissive temperature of 20°C but lost by dilution at 37°C (Molitor et al., 2016).
Terminators
Terminators play an important role in gene expression stability. Often overlooked, transcriptomic studies and computational tools such as TransTerm and RNAMotif can identify and predict Clostridium terminators (Chen et al., 2011; Wang et al., 2011). The adc (acetoacetate decarboxylase) terminator from C. acetobutylicum and fdx (ferrodoxin) terminator from C. pasteurianum (Cartman and Minton, 2010; Fagan and Fairweather, 2011; Nariya et al., 2011b; Pyne et al., 2015; Zhang Y. et al., 2015) are the most commonly used. The terminator of the C. difficile 630 ferrodoxin gene, CD0164, was identified in silico and is present on the pMTL80000 plasmids routinely used in several Clostridium species (Heap et al., 2009; Ng et al., 2013). Non-native terminators, including bidirectional E. coli terminator BB1_B1010 from iGEM Parts Registry, have demonstrated stronger termination over native adc terminator (Lee J. et al., 2016).
Conclusions
Several Clostridium spp. have the potential to be platform organisms for industrial use. However, additions to the available toolkit are required for high-throughput methods of strain engineering. One consistent hurdle is low DNA transformation efficiency, commonly as low as 10 CFU/μg (Table 1). However recent efforts to improve transformation protocols (Pyne et al., 2013) and the isolation of a hyper-transformable strain have resulted in higher efficiencies (Grosse-Honebrink et al., 2017; Schwarz et al., 2017). Further directions include library-based methods requiring high transformation efficiencies which would prove to be a step change improvement over current screening method. Although several studies have focused on increasing the availability of parts such as promoters and reporters, the synthetic biology toolkit can further be expanded through the characterization of other genetic parts such as ribosomal binding sites and terminators. Improvement in these areas will accelerate progress toward a sustainable bio-based economy.
Author Contributions
RJ, NK, and NS prepared and edited the manuscript; RJ prepared figure and table.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adcock, P. W., and Saint, C. P. (2001). Rapid confirmation of Clostridium perfringens by using chromogenic and fluorogenic substrates. Appl. Environ. Microbiol. 67, 4382–4384. doi: 10.1128/AEM.67.9.4382-4384.2001
Al-Hinai, M. A., Fast, A. G., and Papoutsakis, E. T. (2012). Novel system for efficient isolation of clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA Integration. Appl. Environ. Microbiol. 78, 8112–8121. doi: 10.1128/AEM.02214-12
Awad, M. M., Bryant, A. E., Stevens, D. L., and Rood, J. I. (1995). Virulence studies on chromosomal alpha-toxin and theta-toxin mutants constructed by allelic exchange provide genetic evidence for the essential role of alpha-toxin in Clostridium perfringens-mediated gas gangrene. Mol. Microbiol. 15, 191–202. doi: 10.1111/j.1365-2958.1995.tb02234.x
Awad, M. M., and Rood, J. I. (1997). Isolation of α-toxin, θ-toxin and κ-toxin mutants of Clostridium perfringens by Tn916mutagenesis. Microb. Pathog. 22, 275–284. doi: 10.1006/mpat.1996.0115
Baban, S. T., Kuehne, S. A., Barketi-Klai, A., Cartman, S. T., Kelly, M. L., Hardie, K. R., et al. (2013). The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS ONE 8:e73026. doi: 10.1371/journal.pone.0073026
Banerjee, A., Leang, C., Ueki, T., Nevin, K. P., and Lovley, D. R. (2014). Lactose-inducible system for metabolic engineering of Clostridium ljungdahlii. Appl. Environ. Microbiol. 80, 2410–2416. doi: 10.1128/AEM.03666-13
Bankar, S. B., Jurgens, G., Survase, S. A., Ojamo, H., and Granström, T. (2015). Genetic engineering of Clostridium acetobutylicum to enhance isopropanol-butanol-ethanol production with an integrated DNA-technology approach. Renewable Energy 83(Suppl. C), 1076–1083. doi: 10.1016/j.renene.2015.05.052
Bannam, T. L., Crellin, P. K., and Rood, J. I. (1995). Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site-specific recombinase excises a circular transposon molecule. Mol. Microbiol. 16, 535–551. doi: 10.1111/j.1365-2958.1995.tb02417.x
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Borden, J. R., Jones, S. W., Indurthi, D., Chen, Y., and Papoutsakis, E. T. (2010). A genomic-library based discovery of a novel, possibly synthetic, acid-tolerance mechanism in Clostridium acetobutylicum involving non-coding RNAs and ribosomal RNA processing. Metab. Eng. 12, 268–281. doi: 10.1016/j.ymben.2009.12.004
Bruder, M. R., Pyne, M. E., Moo-Young, M., Chung, D., and Chou, C. P. (2016). Extending CRISPR-Cas9 technology from genome editing to transcriptional engineering in genus clostridium. Appl. Environ. Microbiol. 82, 6109–6117. doi: 10.1128/AEM.02128-16
Buckley, A. M., Jukes, C., Candlish, D., Irvine, J. J., Spencer, J., Fagan, R. P., et al. (2016). Lighting up Clostridium difficile: reporting gene expression using fluorescent lov domains. 6:23463. doi: 10.1038/srep23463
Bullifent, H. L., Moir, A., and Titball, R. W. (1995). The construction of a reporter system and use for the investigation of Clostridium perfringens gene expression. FEMS Microbiol. Lett. 131, 99–105. doi: 10.1111/j.1574-6968.1995.tb07761.x
Cartman, S. T., Kelly, M. L., Heeg, D., Heap, J. T., and Minton, N. P. (2012). Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl. Environ. Microbiol. 78, 4683–4690. doi: 10.1128/AEM.00249-12
Cartman, S. T., and Minton, N. P. (2010). A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl. Environ. Microbiol. 76, 1103–1109. doi: 10.1128/AEM.02525-09
Chandrasekaran, R., Kenworthy, A. K., and Lacy, D. B. (2016). Clostridium difficile Toxin A Undergoes Clathrin-Independent, PACSIN2-dependent endocytosis. PLoS Pathog. 12:30. doi: 10.1371/journal.ppat.1006070
Chen, Y., Caruso, L., McClane, B., Fisher, D., and Gupta, P. (2007). Disruption of a toxin gene by introduction of a foreign gene into the Chromosome of Clostridium perfringens using targetron induced mutagenesis. Plasmid 58, 182–189. doi: 10.1016/j.plasmid.2007.04.002
Chen, Y., Indurthi, D. C., Jones, S. W., and Papoutsakis, E. T. (2011). Small RNAs in the Genus Clostridium. MBio 2:e00340-10. doi: 10.1128/mBio.00340-10
Chen, Y., McClane, B. A., Fisher, D. J., Rood, J. I., and Gupta, P. (2005). Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71, 7542–7547. doi: 10.1128/AEM.71.11.7542-7547.2005
Christie, J. M., Hitomi, K., Arvai, A. S., Hartfield, K. A., Mettlen, M., Pratt, A. J., et al. (2012). Structural tuning of the fluorescent protein iLOV for improved photostability. J. Biol. Chem. 287, 22295–22304. doi: 10.1074/jbc.M111.318881
Chu, M., Mallozzi, M. J. G., Roxas, B. P., Bertolo, L., Monteiro, M. A., Agellon, A., et al. (2016). A Clostridium difficile cell wall glycopolymer locus influences bacterial shape, polysaccharide production and virulence. PLoS Pathog. 12:e1005946. doi: 10.1371/journal.ppat.1005946
Cooksley, C. M., Davis, I. J., Winzer, K., Chan, W. C., Peck, M. W., and Minton, N. P. (2010). Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 76, 4448–4460. doi: 10.1128/AEM.03038-09
Croux, C. N., Nguyen, N. P., Lee, J., Raynaud, C., Saint-Prix, F., Gonzalez-Pajuelo, M., et al. (2016). Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum. Biotechnol. Biofuels 9:23. doi: 10.1186/s13068-016-0432-2
Cui, L., and Bikard, D. (2016). Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 44, 4243–4251. doi: 10.1093/nar/gkw223
Czajka, J., Wang, Q., Wang, Y., and Tang, Y. J. (2017). Synthetic biology for manufacturing chemicals: constraints drive the use of non-conventional microbial platforms. Appl. Microbiol. Biotechnol. 101, 7427–7434. doi: 10.1007/s00253-017-8489-9
Davis, T. O., Henderson, I., Brehm, J. K., and Minton, N. P. (2000). Development of a transformation and gene reporter system for group II, non-proteolytic Clostridium botulinum type B strains. J. Mol. Microbiol. Biotechnol. 2, 59–69.
Dingle, T. C., Mulvey, G. L., and Armstrong, G. D. (2011). Mutagenic Analysis of the Clostridium difficile flagellar proteins, flic and flid, and their contribution to virulence in Hamsters. Infect. Immun. 79, 4061–4067. doi: 10.1128/IAI.05305-11
Dong, H., Tao, W., Gong, F., Li, Y., and Zhang, Y. (2014). A functional recT gene for recombineering of Clostridium. J. Biotechnol. 173(Suppl. C), 65–67. doi: 10.1016/j.jbiotec.2013.12.011
Dong, H., Tao, W., Zhang, Y., and Li, Y. (2012). Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: a useful tool for strain engineering. Metab. Eng. 14, 59–67. doi: 10.1016/j.ymben.2011.10.004
Drepper, T., Eggert, T., Circolone, F., Heck, A., Krausz, U. J., Guterl, K., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotech. 25, 443–445. doi: 10.1038/nbt1293
Ehsaan, M., Kuit, W., Zhang, Y., Cartman, S. T., Heap, J. T., Winzer, K., et al. (2016). Mutant generation by allelic exchange and genome resequencing of the biobutanol organism Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 9:4. doi: 10.1186/s13068-015-0410-0
Fagan, R. P., and Fairweather, N. F. (2011). Clostridium difficile has two parallel and essential sec secretion systems. J. Biol. Chem. 286, 27483–27493. doi: 10.1074/jbc.M111.263889
Feustel, L., Nakotte, S., and Durre, P. (2004). Characterization and development of two reporter gene systems for Clostridium acetobutylicum. Appl. Environ. Microbiol. 70, 798–803. doi: 10.1128/AEM.70.2.798-803.2004
Flannagan, S. E., and Clewell, D. B. (1991). Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J. Bacteriol. 173, 7136–7141. doi: 10.1128/jb.173.22.7136-7141.1991
Garst, A. D., Bassalo, M. C., Pines, G., Lynch, S. A., Halweg-Edwards, A. L., Liu, R., et al. (2017). Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55. doi: 10.1038/nbt.3718
Gibson, C. M., and Caparon, M. G. (2002). Alkaline phosphatase reporter transposon for identification of genes encoding secreted proteins in gram-positive microorganisms. Appl. Environ. Microbiol. 68, 928–932. doi: 10.1128/AEM.68.02.928-932.2002
Girbal, L., Mortier-Barrière, I., Raynaud, F., Rouanet, C., Croux, C., and Soucaille, P. (2003). Development of a sensitive gene expression reporter system and an inducible promoter-repressor system for Clostridium acetobutylicum. Appl. Environ. Microbiol. 69, 4985–4988. doi: 10.1128/AEM.69.8.4985-4988.2003
Green, E. M., and Bennett, G. N. (1996). Inactivation of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. Appl. Biochem. Biotechnol. 57, 213–221. doi: 10.1007/BF02941702
Green, E. M., Boynton, Z. L., Harris, L. M., Rudolph, F. B., Papoutsakis, E. T., and Bennett, G. N. (1996). Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142, 2079–2086. doi: 10.1099/13500872-142-8-2079
Grosse-Honebrink, A., Schwarz, K. M., Wang, H., Minton, N. P., and Zhang, Y. (2017). Improving gene transfer in Clostridium pasteurianum through the isolation of rare hypertransformable variants. Anaerobe 48(Suppl. C), 203–205. doi: 10.1016/j.anaerobe.2017.09.001
Harris, L. M., Welker, N. E., and Papoutsakis, E. T. (2002). Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184, 3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002
Hartman, A. H., Liu, H., and Melville, S. B. (2011). Construction and characterization of a lactose-inducible promoter system for controlled gene expression in Clostridium perfringens. Appl. Environ. Microbiol. 77, 471–478. doi: 10.1128/AEM.01536-10
Heap, J. T., Ehsaan, M., Cooksley, C. M., Ng, Y. K., Cartman, S. T., Winzer, K., et al. (2012). Integration of DNA into bacterial chromosomes from plasmids without a counter-selection marker. Nucleic Acids Res. 40:e59. doi: 10.1093/nar/gkr1321
Heap, J. T., Kuehne, S. A., Ehsaan, M., Cartman, S. T., Cooksley, C. M., Scott, J. C., et al. (2010). The clostron: mutagenesis in clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55. doi: 10.1016/j.mimet.2009.10.018
Heap, J. T., Pennington, O. J., Cartman, S. T., Carter, G. P., and Minton, N. P. (2007). The ClosTron: a universal gene knock-out system for the genus clostridium. J. Microbiol. Methods 70, 452–464. doi: 10.1016/j.mimet.2007.05.021
Heap, J. T., Pennington, O. J., Cartman, S. T., and Minton, N. P. (2009). A modular system for Clostridium shuttle plasmids. J. Microbiol. Methods 78, 79–85. doi: 10.1016/j.mimet.2009.05.004
Heinze, S., Mechelke, M., Kornberger, P., Liebl, W., Schwarz, W. H., and Zverlov, V. V. (2017). Identification of endoxylanase XynE from Clostridium thermocellum as the first xylanase of glycoside hydrolase family GH141. Sci. Rep. 7:11178. doi: 10.1038/s41598-017-11598-y
Hensbergen, P. J., Klychnikov, O. I., Bakker, D., Dragan, I., Kelly, M. L., Minton, N. P., et al. (2015). Clostridium difficile secreted Pro-Pro endopeptidase PPEP-1 (ZMP1/CD2830) modulates adhesion through cleavage of the collagen binding protein CD2831. FEBS Lett. 589(24, Pt B), 3952–3958. doi: 10.1016/j.febslet.2015.10.027
Hönicke, D., Lutke-Eversloh, T., Liu, Z. Y., Lehmann, D., Liebl, W., and Ehrenreich, A. (2014). Chemostat cultivation and transcriptional analyses of Clostridium acetobutylicum mutants with defects in the acid and acetone biosynthetic pathways. Appl. Microbiol. Biotechnol. 98, 9777–9794. doi: 10.1007/s00253-014-6040-9
Huang, H., Chai, C., Li, N., Rowe, P., Minton, N. P., Yang, S., et al. (2016). CRISPR/Cas9-based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth. Biol. 5, 1355–1361. doi: 10.1021/acssynbio.6b00044
Hussain, H. A., Roberts, A. P., and Mullany, P. (2005). Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J. Med. Microbiol. 54(Pt 2), 137–141. doi: 10.1099/jmm.0.45790-0
Iqbal, M., Doherty, N., Page, A. M. L., Qazi, S. N. A., Ajmera, I., Lund, P. A., et al. (2017). Reconstructing promoter activity from Lux bioluminescent reporters. PLoS Comput. Biol. 13:e1005731. doi: 10.1371/journal.pcbi.1005731
Jiang, W., Bikard, D., Cox, D., Zhang, F., and Marraffini, L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotech. 31, 233–239. doi: 10.1038/nbt.2508
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816. doi: 10.1126/science.1225829
Jones, J. A., Vernacchio, V. R., Lachance, D. M., Lebovich, M., Fu, L., Shirke, A. N., et al. (2015). ePathOptimize: a combinatorial approach for transcriptional balancing of metabolic pathways. Sci. Rep. 5:11301. doi: 10.1038/srep11301
Jones, S. W., Fast, A. G., Carlson, E. D., Wiedel, C. A., Au, J., Antoniewicz, M. R., et al. (2016). CO2 fixation by anaerobic non-photosynthetic mixotrophy for improved carbon conversion. Nat. Commun. 7:12800. doi: 10.1038/ncomms12800
Karberg, M., Guo, H., Zhong, J., Coon, R., Perutka, J., and Lambowitz, A. M. (2001). Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotech. 19, 1162–1167. doi: 10.1038/nbt1201-1162
Kim, S. K., Kim, H. W., Ahn, C. K., Park, H. E., Woo, J. D., Lee, H., et al. (2017). Efficient transcriptional gene repression by Type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth. Biol. 6, 1273–1282. doi: 10.1021/acssynbio.6b00368
Lanckriet, A., Timbermont, L., Happonen, L. J., Pajunen, M. I., Pasmans, F., Haesebrouck, F., et al. (2009). Generation of single-copy transposon insertions in Clostridium perfringens by electroporation of phage mu DNA transposition complexes. Appl. Environ. Microbiol. 75, 2638–2642. doi: 10.1128/AEM.02214-08
Lawson, P. A., Citron, D. M., Tyrrell, K. L., and Finegold, S. M. (2016). Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40, 95–99. doi: 10.1016/j.anaerobe.2016.06.008
Lee, J., Jang, S., Papoutsakis, E. T., and Lee, S. Y. (2016). Stable and enhanced gene expression in Clostridium acetobutylicum using synthetic untranslated regions with a stem-loop. J. Biotechnol. 230(Suppl. C), 40–43. doi: 10.1016/j.jbiotec.2016.05.020
Lee, S. H., Kim, H. J., Shin, Y. A., Kim, K. H., and Lee, S. J. (2016). Single crossover-mediated markerless genome engineering in Clostridium acetobutylicum. J. Microbiol. Biotechnol. 26, 725–729. doi: 10.4014/jmb.1512.12012
Lee, S. J., Trostel, A., and Adhya, S. (2014). Metabolite changes signal genetic regulatory mechanisms for robust cell behavior. MBio 5:e00972-13. doi: 10.1128/mBio.00972-13
Lee, S. J., Trostel, A., Le, P., Harinarayanan, R., Fitzgerald, P. C., and Adhya, S. (2009). Cellular stress created by intermediary metabolite imbalances. Proc. Natl. Acad. Sci. U.S.A. 106, 19515–19520. doi: 10.1073/pnas.0910586106
Li, Q., Chen, J., Minton, N. P., Zhang, Y., Wen, Z., Liu, J., et al. (2016). CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol. J. 11, 961–972. doi: 10.1002/biot.201600053
Li, X. T., Jun, Y., Erickstad, M. J., Brown, S. D., Parks, A., Court, D. L., et al. (2016). tCRISPRi: tunable and reversible, one-step control of gene expression. Sci. Rep. 6:39076. doi: 10.1038/srep39076
Liu, H., Bouillaut, L., Sonsenshein, A. L., and Melville, S. B. (2013). Use of a mariner-based transposon mutagenesis system to isolate Clostridium perfringens mutants deficient in gliding motility. J. Bacteriol. 195, 629–636. doi: 10.1128/JB.01288-12
Liu, J., Guo, T., Shen, X., Xu, J., Wang, J., Wang, Y., et al. (2016). Engineering Clostridium beijerinckii with the Cbei_4693 gene knockout for enhanced ferulic acid tolerance. J. Biotechnol. 229(Suppl. C), 53–57. doi: 10.1016/j.jbiotec.2016.04.052
Liu, K., Atiyeh, H. K., Pardo-Planas, O., Ezeji, T. C., Ujor, V., Overton, J. C., et al. (2015). Butanol production from hydrothermolysis-pretreated switchgrass: quantification of inhibitors and detoxification of hydrolyzate. Bioresour. Technol. 189, 292–301. doi: 10.1016/j.biortech.2015.04.018
Liu, Z., Qiao, K., Tian, L., Zhang, Q., Liu, Z. Y., and Li, F. L. (2015). Spontaneous large-scale autolysis in Clostridium acetobutylicum contributes to generation of more spores. Front. Microbiol. 6:950. doi: 10.3389/fmicb.2015.00950
Liyanage, H., Kashket, S., Young, M., and Kashket, E. R. (2001). Clostridium beijerinckii and Clostridium difficile detoxify methylglyoxal by a novel mechanism involving glycerol dehydrogenase. Appl. Environ. Microbiol. 67, 2004–2010. doi: 10.1128/AEM.67.5.2004-2010.2001
Mani, N., Dupuy, B., and Sonenshein, A. L. (2006). Isolation of RNA polymerase from Clostridium difficile and characterization of glutamate dehydrogenase and rRNA gene promoters in vitro and in vivo. J. Bacteriol. 188, 96–102. doi: 10.1128/JB.188.1.96-102.2006
Meaney, C. A., Cartman, S. T., McClure, P. J., and Minton, N. P. (2015). Optimal spore germination in Clostridium botulinum ATCC 3502 requires the presence of functional copies of SleB and YpeB, but not CwlJ. Anaerobe 34(Suppl. C), 86–93. doi: 10.1016/j.anaerobe.2015.04.015
Meaney, C. A., Cartman, S. T., McClure, P. J., and Minton, N. P. (2016). The role of small acid-soluble proteins (SASPs) in protection of spores of Clostridium botulinum against nitrous acid. Int. J. Food Microbiol. 216(Suppl. C), 25–30. doi: 10.1016/j.ijfoodmicro.2015.08.024
Mearls, E. B., Olson, D. G., Herring, C. D., and Lynd, L. R. (2015). Development of a regulatable plasmid-based gene expression system for Clostridium thermocellum. Appl. Microbiol. Biotechnol. 99, 7589–7599. doi: 10.1007/s00253-015-6610-5
Minton, N. P., Ehsaan, M., Humphreys, C. M., Little, G. T., Baker, J., Henstra, A. M., et al. (2016). A roadmap for gene system development in Clostridium. Anaerobe 41(Suppl. C), 104–112. doi: 10.1016/j.anaerobe.2016.05.011
Molitor, B., Kirchner, K., Henrich, A. W., Schmitz, S., and Rosenbaum, M. A. (2016). Expanding the molecular toolkit for the homoacetogen Clostridium ljungdahlii. Sci. Rep. 6:31518. doi: 10.1038/srep31518
Mougiakos, I., Bosma, E. F., Weenink, K., Vossen, E., Goijvaerts, K. J., van der Oost, J., et al. (2017a). Efficient genome editing of a facultative thermophile using mesophilic spCas9. ACS Synth. Biol. 6, 849–861. doi: 10.1021/acssynbio.6b00339
Mougiakos, I., Mohanraju, P., Bosma, E. F., Vrouwe, V., Finger Bou, M., Naduthodi, M. I. S., et al. (2017b). Characterizing a thermostable Cas9 for bacterial genome editing and silencing. Nat. Commun. 8:1647. doi: 10.1038/s41467-017-01591-4
Mukherjee, A., Walker, J., Weyant, K. B., and Schroeder, C. M. (2013). Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS ONE 8:e64753. doi: 10.1371/journal.pone.0064753
Nagaraju, S., Davies, N. K., Walker, D. J. F., Köpke, M., and Simpson, S. D. (2016). Genome editing of Clostridium autoethanogenum using CRISPR/Cas9. Biotechnol. Biofuels 9:219. doi: 10.1186/s13068-016-0638-3
Nair, R. V., Green, E. M., Watson, D. E., Bennett, G. N., and Papoutsakis, E. T. (1999). Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J. Bacteriol. 181, 319–330.
Nariya, H., Miyata, S., Kuwahara, T., and Okabe, A. (2011a). Development and characterization of a xylose-inducible gene expression system for Clostridium perfringens. Appl. Environ. Microbiol. 77, 8439–8441. doi: 10.1128/AEM.05668-11
Nariya, H., Miyata, S., Suzuki, M., Tamai, E., and Okabe, A. (2011b). Development and application of a method for counterselectable in-frame deletion in Clostridium perfringens. Appl. Environ. Microbiol. 77, 1375–1382. doi: 10.1128/AEM.01572-10
Ng, Y. K., Ehsaan, M., Philip, S., Collery, M. M., Janoir, C., Collignon, A., et al. (2013). Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE Alleles. PLoS ONE 8:e56051. doi: 10.1371/journal.pone.0056051
Nuyts, S., van Mellaert, L., Theys, J., Landuyt, W., Lambin, P., and Anne, J. (2001). The use of radiation-induced bacterial promoters in anaerobic conditions: a means to control gene expression in clostridium-mediated therapy for cancer. Radiat. Res. 155, 716–723. doi: 10.1667/0033-7587(2001)155[0716:TUORIB]2.0.CO;2
O'Connor, J. R., Lyras, D., Farrow, K. A., Adams, V., Powell, D. R., Hinds, J., et al. (2006). Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61, 1335–1351. doi: 10.1111/j.1365-2958.2006.05315.x
Oliveira Paiva, A. M., Friggen, A. H., Hossein-Javaheri, S., and Smits, W. K. (2016). The signal sequence of the abundant extracellular metalloprotease PPEP-1 can be used to secrete synthetic reporter proteins in Clostridium difficile. ACS Synth. Biol. 5, 1376–1382. doi: 10.1021/acssynbio.6b00104
Ostrov, N., Landon, M., Guell, M., Kuznetsov, G., Teramoto, J., Cervantes, N., et al. (2016). Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822. doi: 10.1126/science.aaf3639
Paredes, C. J., Rigoutsos, I., and Papoutsakis, E. T. (2004). Transcriptional organization of the Clostridium acetobutylicum genome. Nucleic Acids Res. 32, 1973–1981. doi: 10.1093/nar/gkh509
Phillips-Jones, M. K. (2000). Use of a lux reporter system for monitoring rapid changes in α-toxin gene expression in Clostridium perfringens during growth. FEMS Microbiol. Lett. 188, 29–33. doi: 10.1016/S0378-1097(00)00208-1
Pyne, M. E., Bruder, M. R., Moo-Young, M., Chung, D. A., and Chou, C. P. (2016). Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 6:25666. doi: 10.1038/srep25666
Pyne, M. E., Moo-Young, M., Chung, D. A., and Chou, C. P. (2013). Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnol. Biofuels 6:50. doi: 10.1186/1754-6834-6-50
Pyne, M. E., Moo-Young, M., Chung, D. A., and Chou, C. P. (2014). Expansion of the genetic toolkit for metabolic engineering of Clostridium pasteurianum: chromosomal gene disruption of the endogenous CpaAI restriction enzyme. Biotechnol. Biofuels 7:163. doi: 10.1186/s13068-014-0163-1
Pyne, M., Moo-Young, M., Chung, D., and Chou, C. (2015). Antisense-RNA-mediated gene downregulation in clostridium pasteurianum. Fermentation 1:113. doi: 10.3390/fermentation1010113
Qi, L. S., Matthew Larson, H., Luke Gilbert, A., Jennifer Doudna, A., Jonathan Weissman, S., Adam Arkin, P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Quixley, K. W., and Reid, S. J. (2000). Construction of a reporter gene vector for Clostridium beijerinckii using a Clostridium endoglucanase gene. J. Mol. Microbiol. Biotechnol. 2, 53–57.
Ransom, E. M., Ellermeier, C. D., and Weiss, D. S. (2015). Use of mCherry red fluorescent protein for studies of protein localization and gene expression in Clostridium difficile. Appl. Environ. Microbiol. 81, 1652–1660. doi: 10.1128/AEM.03446-14
Ravagnani, A., Jennert, K. C., Steiner, E., Grunberg, R., Jefferies, J. R., Wilkinson, S. R., et al. (2000). Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37, 1172–1185. doi: 10.1046/j.1365-2958.2000.02071.x
Rohlhill, J., Sandoval, N. R., and Papoutsakis, E. T. (2017). Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol. ACS Synth. Biol. 6, 1584–1595. doi: 10.1021/acssynbio.7b00114
Ronda, C., Pedersen, L. E., Sommer, M. O. A., and Nielsen, A. T. (2016). CRMAGE: CRISPR Optimized MAGE Recombineering. Sci. Rep. 6:19452. doi: 10.1038/srep19452
Sandoval, N. R., Venkataramanan, K. P., Groth, T. S., and Papoutsakis, E. T. (2015). Whole-genome sequence of an evolved Clostridium pasteurianum strain reveals Spo0A deficiency responsible for increased butanol production and superior growth. Biotechnol. Biofuels 8:227. doi: 10.1186/s13068-015-0408-7
Schwarz, K. M., Grosse-Honebrink, A., Derecka, K., Rotta, C., Zhang, Y., and Minton, N. P. (2017). Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum. Metab. Eng. 40(Suppl. C), 124–137. doi: 10.1016/j.ymben.2017.01.009
Seo, S. O., Lu, T., Jin, Y. S., and Blaschek, H. P. (2018). Development of an oxygen-independent flavin mononucleotide-based fluorescent reporter system in Clostridium beijerinckii and its potential applications. J. Biotechnol. 265, 119–126. doi: 10.1016/j.jbiotec.2017.11.003
Shao, L., Hu, S., Yang, Y., Gu, Y., Chen, J., Yang, Y., et al. (2007). Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 17, 963–965. doi: 10.1038/cr.2007.91
Sillers, R., Al-Hinai, M. A., and Papoutsakis, E. T. (2009). Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum Fermentations. Biotechnol. Bioeng. 102, 38–49. doi: 10.1002/bit.22058
Smanski, M. J., Bhatia, S., Zhao, D., Park, Y. L., Woodruff, L. B. A., Giannoukos, G., et al. (2014). Functional optimization of gene clusters by combinatorial design and assembly. Nat. Biotech. 32, 1241–1249. doi: 10.1038/nbt.3063
Staedtke, V., Roberts, N. J., Bai, R. Y., and Zhou, S. B. (2016). Clostridium novyi-NT in cancer therapy. Genes Dis. 3, 144–152. doi: 10.1016/j.gendis.2016.01.003
Su, T. Y., Liu, F. P., Gu, P. F., Jin, H. Y., Chang, Y. Z., Wang, Q., et al. (2016). A CRISPR-Cas9 assisted non-homologous end-joining strategy for one-step engineering of bacterial genome. Sci. Rep. 6:11. doi: 10.1038/srep37895
Tan, Y., Liu, Z. Y., Liu, Z., Zheng, H. J., and Li, F. L. (2015). Comparative transcriptome analysis between csrA-disruption Clostridium acetobutylicum and its parent strain. Mol. Biosyst. 11, 1434–1442. doi: 10.1039/C4MB00600C
Teng, L., Wang, K., Xu, J., and Xu, C. (2015). Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) as reporter for promoter screening in Clostridium cellulolyticum. J. Microbiol. Methods 119, 37–43. doi: 10.1016/j.mimet.2015.09.018
Tripathi, S. A., Olson, D. G., Argyros, D. A., Miller, B. B., Barrett, T. F., Murphy, D. M., et al. (2010). Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl. Environ. Microbiol. 76, 6591–6599. doi: 10.1128/AEM.01484-10
Tummala, S. B., Welker, N. E., and Papoutsakis, E. T. (1999). Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65, 3793–3799.
Tummala, S. B., Welker, N. E., and Papoutsakis, E. T. (2003). Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J. Bacteriol. 185, 1923–1934. doi: 10.1128/JB.185.6.1923-1934.2003
Vidal, J. E., Chen, J., Li, J., and McClane, B. A. (2009). Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS ONE 4:e6232. doi: 10.1371/journal.pone.0006232
Walker, D. J. F., and Köpke, M. (2015). Method of Producing a Recombinant Microorganism. US 14/608,132.
Wang, H., Roberts, A. P., and Mullany, P. (2000). DNA sequence of the insertional hot spot of Tn916 in the Clostridium difficile genome and discovery of a Tn916-like element in an environmental isolate integrated in the same hot spot. FEMS Microbiol. Lett. 192, 15–20. doi: 10.1111/j.1574-6968.2000.tb09352.x
Wang, M., Chen, B., Fang, Y., and Tan, T. (2017). Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 35, 1032–1039. doi: 10.1016/j.biotechadv.2017.09.008
Wang, S., Dong, S., Wang, P., Tao, Y., and Wang, Y. (2017). Genome Editing in Clostridium saccharoperbutylacetonicum N1-4 with the CRISPR-Cas9 System. Appl. Environ. Microbiol. 83:e00233-17. doi: 10.1128/AEM.00233-17
Wang, Y., Li, X., Mao, Y., and Blaschek, H. P. (2011). Single-nucleotide resolution analysis of the transcriptome structure of Clostridium beijerinckii NCIMB 8052 using RNA-Seq. BMC Genomics 12:479. doi: 10.1186/1471-2164-12-479
Wang, Y., Zhang, Z. T, Seo, S. O., Choi, K., Lu, T., Jin, Y. S., et al. (2015). Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J. Biotechnol. 200, 1–5. doi: 10.1016/j.jbiotec.2015.02.005
Wang, Y., Zhang, Z. T, Seo, S. O., Lynn, P., Lu, T., Jin, Y. S., et al. (2016a). Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable Clean mutant selection in Clostridium beijerinckii as an example. ACS Synth. Biol. 5, 721–732. doi: 10.1021/acssynbio.6b00060
Wang, Y., Zhang, Z. T., Seo, S. O., Lynn, P., Lu, T., Jin, Y. S., et al. (2016b). Gene transcription repression in Clostridium beijerinckii Using CRISPR-dCas9. Biotechnol. Bioeng. 113, 2739–2743. doi: 10.1002/bit.26020
Wasels, F., Jean-Marie, J., Collas, F., López-Contreras, A. M., and Lopes Ferreira, N. (2017). A two-plasmid inducible CRISPR/Cas9 genome editing tool for Clostridium acetobutylicum. J. Microbiol. Methods 140, 5–11. doi: 10.1016/j.mimet.2017.06.010
Wen, Z., Minton, N. P., Zhang, Y., Li, Q., Liu, J., Jiang, Y., et al. (2017). Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab. Eng. 39, 38–48. doi: 10.1016/j.ymben.2016.10.013
Wiedenheft, B., Sternberg, S. H., and Doudna, J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338. doi: 10.1038/nature10886
Willson, B. J., Kovacs, K., Wilding-Steele, T., Markus, R., Winzer, K., and Minton, N. P. (2016). Production of a functional cell wall-anchored minicellulosome by recombinant Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 9:22. doi: 10.1186/s13068-016-0526-x
Woolley, R. C., Pennock, A., Ashton, R. J., Davies, A., and Young, M. (1989). Transfer of Tn1545 and Tn916 to Clostridium acetobutylicum. Plasmid 22, 169–174. doi: 10.1016/0147-619X(89)90027-9
Wu, G., Yan, Q., Jones, J. A., Tang, Y. J., Fong, S. S., and Koffas, M. A. (2016). Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 34, 652–664. doi: 10.1016/j.tibtech.2016.02.010
Xin, B., Wang, Y., Tao, F., Li, L., Ma, C., and Xu, P. (2016). Co-utilization of glycerol and lignocellulosic hydrolysates enhances anaerobic 1,3-propanediol production by Clostridium diolis. Sci. Rep. 6:19044. doi: 10.1038/srep19044
Xu, M., Zhao, J. B., Yu, L., Tang, I. C., Xue, C., and Yang, S. T. (2015). Engineering Clostridium acetobutylicum with a histidine kinase knockout for enhanced n-butanol tolerance and production. Appl. Microbiol. Biotechnol. 99, 1011–1022. doi: 10.1007/s00253-014-6249-7
Xu, T., Li, Y., He, Z., Van Nostrand, J. D., and Zhou, J. (2017). Cas9 nickase-assisted RNA repression enables stable and efficient manipulation of essential metabolic genes in Clostridium cellulolyticum. Front. Microbiol. 8:1744. doi: 10.3389/fmicb.2017.01744
Xu, T., Li, Y., Shi, Z., Hemme, C. L., Li, Y., Nostrand, J. D. V., et al. (2015). Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 Nickase. Appl. Environ. Microbiol. 81, 4423–4431. doi: 10.1128/AEM.00873-15
Yang, G., Jia, D., Jin, L., Jiang, Y., Wang, Y., Jiang, W., et al. (2017). Rapid generation of universal synthetic promoters for controlled gene expression in both gas-fermenting and saccharolytic clostridium Species. ACS Synth. Biol. 6, 1672–1678. doi: 10.1021/acssynbio.7b00155
Zhang, J. Y., Liu, J. G., Cui, Z., and Cui, Q. (2015). A novel arabinose-inducible genetic operation system developed for Clostridium cellulolyticum. Biotechnol. Biofuels 8:36. doi: 10.1186/s13068-015-0214-2
Zhang, Y., Grosse-Honebrink, A., and Minton, N. P. (2015). A universal mariner transposon system for forward genetic studies in the genus clostridium. PLoS ONE 10:e0122411. doi: 10.1371/journal.pone.0122411
Zhang, Y., Xu, S., Chai, C., Yang, S., Jiang, W., Minton, N. P., et al. (2016). Development of an inducible transposon system for efficient random mutagenesis in Clostridium acetobutylicum. FEMS Microbiol. Lett. 363:fnw065. doi: 10.1093/femsle/fnw065
Keywords: clostridium, synthetic biology, CRISPR, metabolic engineering, biotechnology of microorganisms
Citation: Joseph RC, Kim NM and Sandoval NR (2018) Recent Developments of the Synthetic Biology Toolkit for Clostridium. Front. Microbiol. 9:154. doi: 10.3389/fmicb.2018.00154
Received: 14 October 2017; Accepted: 23 January 2018;
Published: 12 February 2018.
Edited by:
Ryan S. Senger, Virginia Tech, United StatesReviewed by:
Daniel Paredes-Sabja, Dr. Andrés Bello University, El SalvadorChristopher L. Hemme, University of Rhode Island, United States
Sang Jun Lee, Chung-Ang University, South Korea
Copyright © 2018 Joseph, Kim and Sandoval. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas R. Sandoval, bnNhbmRvdmFAdHVsYW5lLmVkdQ==
 Rochelle C. Joseph
Rochelle C. Joseph Nancy M. Kim
Nancy M. Kim Nicholas R. Sandoval
Nicholas R. Sandoval