- 1Department of Genetics, Faculty of Science, Shahid Chamran University of Ahvaz, Ahvaz, Iran
- 2Department of Medical Genetics, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
The majority of the Human genome is transcribed into a large number of non-coding RNAs (ncRNAs) which have different roles in the cell (Haemmerle and Gutschner, 2015). Long non-coding RNAs (LncRNAs) constitute a heterogeneous group of the ncRNAs that are longer than 200 nt. They are usually capped and poly-adenylated like mRNAs (Esteller, 2011; Fritah et al., 2014). Accumulating evidence show that the lncRNAs play an important role in cancer progression. So, these molecules have been considered as potential biomarker and therapeutic targets (Hajjari and Khoshnevisan, 2013; Hajjari et al., 2013b; Huarte, 2015).
One of the well-known lncRNAs is HOTAIR (HOX transcript antisense RNA) which is known to effect on the chromatin structure (Cao, 2014; Huang et al., 2014). This trans-acting lncRNA has different target loci including tumor suppressor genes. It recruits the PRC2 and LSD1 complexes in order to repress the transcription of target genes (Hajjari and Salavaty, 2015).
Owing to this regulatory mechanism, lots of the studies reported the role of HOTAIR in progression of different cancers such as breast, colon, and gastric cancer (Reviewed in Hajjari et al., 2013a, 2014; Hajjari and Salavaty, 2015). Gupta et al. found the dysregulation of this long transcript in breast tumors (Gupta et al., 2010). Then, other studies showed the importance of the HOTAIR in poor prognosis, metastasis, invasion, and short overall survival of breast cancer. Alves et al. indicated the potential role of HOTAIR in EMT progression and cancer stem cell features (Pádua Alves et al., 2013). Also, Gökmen-Polar et al. showed the HOTAIR as a marker for lymphatic metastases in ER-negative patients (Gökmen-Polar et al., 2015). However, the results are in contrast to the results of Sorensen et al but are similar to those reported by Lu et al. (Sørensen et al., 2013; Liu et al., 2014).
There are just a few studies with limited number of samples indicating the differentiated expression of HOTAIR in breast tumors compared to normal tissues. We believe that the differential expression of HOTAIR may indicate the potential role of this lncRNA in cancer initiation and progression. This study was aimed to highlight the potential role of HOTAIR in breast cancer. In this study, we analyzed the HOTAIR expression in breast invasive carcinoma tissues derived from TCGA (The Cancer Genome Atlas) which applies RNA sequencing of large cohorts. We also validated our data in different GEO dataset. The results can highlight the role of HOTAIR in breast invasive cancer and provide the viewpoint for further analyses of HOTAIR in breast cancer progression.
For this study, The Cancer Genome browser database (https://genome-cancer.ucsc.edu/), which uses the TCGA data, was used to analyze the association between HOTAIR expression level and features of breast invasive carcinoma. “BRCA gene expression (Illumina Hiseq Percentile)” data was selected for our analysis. The expression of HOTAIR was compared between the tumor (n = 1,066) and normal samples (n = 133) of TCGA tissues. Besides, the association between HOTAIR expression level of breast tissues and ER/PR/HER2 status of tumor tissues was also checked.
For validation study, the expression of HOTAIR gene was analyzed between breast tumor and normal tissues in different GEO datasets and profiles including GSE58135, GDS2618, GDS3853, GSE69240, GSE48408 through NCBI as well as Nexus expression database (http://syslab4.nchu.edu.tw/).
Normalized expression (Z score) derived from TCGA tissues of cancer genome browser were compared between groups of study. The statistical analysis was done by t-test through R-software integrated in cancer genome browser. The P-value <0.05 were considered as significant P-value. Expression analysis between breast invasive tumor and normal tissues showed that the HOTAIR is significantly overexpressed in breast tumors compared to normal tissues (Figure 1). Categorizing the samples based on the HER2/PR/ER status demonstrated that HOTAIR is significantly over expressed in HER2 positive samples compared to negative ones. Additionally, in comparison with ER and PR positive tumors, HOTAIR is up-regulated in ER and PR negative tumors. It was found that the expression of HOTAIR is down-regulated in LuminalA, LuminalB, and normal like breast tumors subtypes (Data not shown).
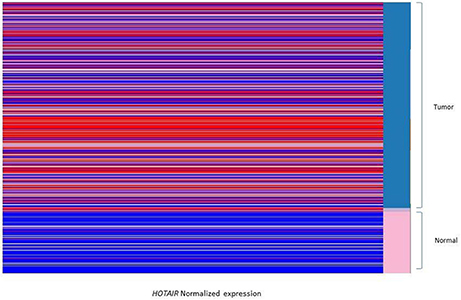
Figure 1. Heatmap displays the comparison between the HOTAIR expression of breast tumors and solid normal tissues. The data heatmap displays the normalized gene expression in red (Z > 0) and blue color (Z < 0). The HOTAIR is overexpressed in breast tumor tissues (A part of the tissues are shown as blue subgroup in right column) compared to normal tissues (shown as pink subgroup in right column). Data is derived from Cancer genome browser database.
Different GEO dataset and profiles were analyzed in order to compare the HOTAIR expression level between tumor and normal samples. The expression level between tumor and normal tissues were compared by t-test through GraphPad software. The data showed that the HOTAIR is significantly up-regulated in cancer cell lines and tumor tissues compared to normal breast samples (Table 1). To our knowledge, this is the first integrative study highlighting the over expression of HOTAIR in breast invasive carcinoma in a large cohort and different data sets. Since the breast cancer is a heterogeneous disease, the predictive power of current biomarkers is sometimes limited. So, there is a need to identify additional prognostic and predictive molecular biomarkers. The aim of this study was to examine the significance of the HOTAIR gene expression in breast cancer. Given the importance of HOTAIR in breast cancer, it promises as a potential biomarker and therapeutic target. However, because of the follow up limitations of TCGA cohort, further studies are necessary to reveal the role of HOTAIR in breast cancer initiation/progression in different cohorts with well annotation for tumor histology, and survival data.
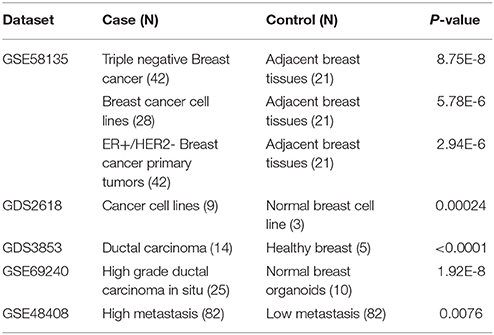
Table 1. The Up-regulation of HOTAIR in breast cancer samples (case) compared to normal samples (controls) in different GEO datasets.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Shahid Chamran University of Ahvaz for supporting this study.
References
Cao, J. (2014). The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online 16:42. doi: 10.1186/1480-9222-16-11
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12:861. doi: 10.1038/nrg3074
Fritah, S., Niclou, S. P., and Azuaje, F. (2014). Databases for lncRNAs: a comparative evaluation of emerging tools. RNA 20, 1655–1665. doi: 10.1261/rna.044040.113
Gökmen-Polar, Y., Vladislav, I. T., Neelamraju, Y., Janga, S. C., and Badve, S. (2015). Prognostic impact of HOTAIR expression is restricted to ER-negative breast cancers. Sci. Rep. 5:8765. doi: 10.1038/srep08765
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071. doi: 10.1038/nature08975
Haemmerle, M., and Gutschner, T. (2015). Long non-coding RNAs in cancer and development: where do we go from here? Int. J. Mol. Sci. 16, 1395–1405. doi: 10.3390/ijms16011395
Hajjari, M., and Salavaty, A. (2015). HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 12:1. doi: 10.7497/j.issn.2095-3941.2015.0006
Hajjari, M., and Khoshnevisan, A. (2013). Potential long non-coding RNAs to be considered as biomarkers or therapeutic targets in gastric cancer. Front. Genet. 4:210. doi: 10.3389/fgene.2013.00210
Hajjari, M., Behmanesh, M., Sadeghizadeh, M., and Zeinoddini, M. (2013a). Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med. Oncol. 30:670. doi: 10.1007/s12032-013-0670-0
Hajjari, M., Khoshnevisan, A., and Shin, Y. K. (2013b). Long non-coding RNAs in hematologic malignancies: road to translational research. Front. Genet. 4:250. doi: 10.3389/fgene.2013.00250
Hajjari, M., Khoshnevisan, A., and Shin, Y. K. (2014). Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumor Biol. 35, 10645–10663. doi: 10.1007/s13277-014-2636-z
Huang, J., Zhou, N., Watabe, K., Lu, Z., Wu, F., Xu, M., et al. (2014). Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 5:e1008. doi: 10.1038/cddis.2013.541
Liu, X. H., Sun, M., Nie, F. Q., Ge, Y. B., Zhang, E. B., Yin, D. D., et al. (2014). Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 13:92. doi: 10.1186/1476-4598-13-92
Pádua Alves, C., Fonseca, A. S., Muys, B. R., de Barros e Lima Bueno, R., Bürger, M. C., Souza, J. E., et al. (2013). Brief report: the lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 31, 2827–2832. doi: 10.1002/stem.1547
Keywords: HOTAIR, long non-coding RNA, gene expression, cancer, breast invasive carcinoma
Citation: Avazpour N, Hajjari M and Tahmasebi Birgani M (2017) HOTAIR: A Promising Long Non-coding RNA with Potential Role in Breast Invasive Carcinoma. Front. Genet. 8:170. doi: 10.3389/fgene.2017.00170
Received: 19 June 2017; Accepted: 19 October 2017;
Published: 21 November 2017.
Edited by:
Sanjeev Kumar Srivastava, Mitchell Cancer Institute, United StatesReviewed by:
Shubhanchi Nigam, University of Pittsburgh, United StatesLihua Li, University of Minnesota, United States
Vijay Kumar Prajapati, Central University of Rajasthan, India
Copyright © 2017 Avazpour, Hajjari and Tahmasebi Birgani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammadreza Hajjari, mohamad.hajari@gmail.com; m-hajari@scu.ac.ir
 Niloofar Avazpour
Niloofar Avazpour Mohammadreza Hajjari
Mohammadreza Hajjari Maryam Tahmasebi Birgani
Maryam Tahmasebi Birgani