- Department of Plant Science, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
The cytochrome P450 (CYP) monooxygenase superfamily, belonging to heme-thiolate protein products, plays a vital role in metabolizing physiologically valuable compounds in plants. To date, CYP superfamily genes have not yet been characterized in grapevine (V. vinifera L.), and their functions remain unclear. In this study, a sum of 236 VvCYPs, divided into 46 families and clustered into nine clans, have been identified based on bioinformatics analyses in grapevine genome. The characteristics of both exon–intron organizations and motif structures further supported the close evolutionary relationships of VvCYP superfamily as well as the reliability of phylogenetic analysis. The gene number-based hierarchical cluster of CYP subfamilies of different plants demonstrated that the loss of CYP families seems to be limited to single species or single taxa. Promoter analysis elucidated various cis-regulatory elements related to phytohormone signaling, plant growth and development, as well as abiotic/biotic stress responses. The tandem duplication mainly contributed to the expansion of the VvCYP superfamily, followed by singleton duplication in grapevine. Global RNA-sequencing data of grapevine showed functional divergence of VvCYPs as diverse expression patterns of VvCYPs in various organs, tissues, and developmental phases, which were confirmed by quantitative real-time reverse transcription PCR (qRT-PCR). Taken together, our results provided valuable inventory for understanding the classification and biological functions of the VvCYPs and paved the way for further functional verification of these VvCYPs and are helpful to grapevine molecular breeding.
Introduction
The cytochrome P450 (CYP) monooxygenases constitute a huge and intricate gene superfamily of heme-thiolate proteins, which play important roles in the metabolism of various substrates, containing endogenous compounds, for example, steroids, also xenobiotic compounds including pesticides, drug, and environmental pollutants (Werck-Reichhart and Feyereisen, 2000). The CYP enzyme is probably the most catalytic biocatalyst in nature. According to its functions, CYPs can be roughly classified into two categories in plants. One has a metabolic detoxification function, such as catalyzing environmental toxins, organic dyes, and pesticides to become nontoxic substances (Robert and Douglas, 1991; Werck-Reichhart, 1995). The others participated in the biosynthesis of plant secondary substances, including terpenes, sterols, phytohormones, lignin intermediates, and so on. The CYP genes are widely found in different life forms, including multicellular eukaryotes (animals and plants), unicellular eukaryotes (fungi and protists), and prokaryotes (bacteria and archaea) (Omura, 1999; Ai et al., 2011). In 1969, Frear reported the first plant CYP gene in cotton (Gossypium hirsutum). Subsequently, many CYP genes were identified in several plant species, such as Arabidopsis thaliana (245) (Paquette et al., 2000), Medicago truncatula (151) (Li et al., 2007), Populus trichocarpa (310) (Nelson et al., 2008), Morus notabilis (174) (Ma et al., 2014), Ricinus communis (210) (Kumar et al., 2014), and Oryza sativa (326) (Wei and Chen, 2018). To date, more than 20,000 members of the CYP gene family have been characterized and named (Nelson, 2011). However, the amount of CYP genes found in each species is highly varying. Until now, more than 5,100 CYPs have been found in plants, and most of them also have been uploaded into a periodically updated Cytochrome P450 Homepage website (http://drnelson.uthsc.edu/CytochromeP450.html). The CYP genes were divided into different clans including six single-family clans (clan 51, clan 74, clan 97, clan 710, clan 711, and clan 727) and four multiple-family clans (clan 71, clan 72, clan 85, and clan 86) according to the criterion of phylogeny and homology (Nelson et al., 1996; Nelson et al., 2004).
In plants, CYPs function as a vital regulator in some growth and development processes. For example, cytochrome P450 MAX1 gene, found in A. thaliana, O. Sativa, and Solanum lycopersicum, is necessary for the biosynthesis of strigolactones, mainly involved in regulating the branch of plants (Zhang et al., 2014). The AtCYP715 is responsible for regulating the maturation of Arabidopsis flower, synchronizing volatile emanation, and petal expansion (Liu et al., 2015). CYP703, CYP704, and CYP86 are necessary for the formation of pollen exine and the synthesis of anther cutin (Shi et al., 2015). In Arabidopsis, overexpression of AtCYP78A9 results in seedless and bigger fruits (Ito and Meyerowitz, 2000). In rice, the loss-of-function of OsCYP78B5 leads to a tremendous embryo (Chen et al., 2015). The CYP members of clan 73, clan 84, clan 98 are responsible for catalyzing the biosynthetic reactions of phenylpropanoid pathway, producing a mass of phenol compound functioning as UV protectant (flavonoids), structural component (suberin and lignin), antimicrobial (isoflavonoids, lignans and coumarins), and antioxidants (polyphenols) (Mizutani and Ohta, 2010). The CYPs also participate in stress responses, for instance, salinity, drought, insect pest, oxidative stress, and chemical toxicity. The RNAi lines of OsABA8ox3 confers drought tolerance in O. sativa (Cai et al., 2015). The mutants of CYP96B4/dss1 increase the accumulation levels of abscisic acid (ABA) and ABA-metabolites responding to drought stress (Tamiru et al., 2015). Besides, OsCYP96B4 may participate in secondary cell wall formation and lipid metabolism in O. sativa (Ramamoorthy et al., 2011; Wang et al., 2016). AtCYP709B1, AtCYP709B2, and AtCYP709B3 in Arabidopsis are induced under salt stress (Mao et al., 2013). ZmCYP71C1 and ZmCYP71C3 are involved in the synthesis of DIMBOA in Zea mays, which act as natural defense against bacteria, fungi, and pest (Persans et al., 2001). Until now, only one VvCYP71BE5 has been functionally characterized (Takase et al., 2015), the majority of VvCYP superfamily members have not been investigated in grapevine.
Grapevine (Vitis spp.), with processing properties and important nutritional values, is one of the most economically important fruit crops worldwide. The availability of grapevine genome made it possible and feasible to examine and identify gene families (Boss et al., 1996; Velasco et al., 2007). Although CYP superfamily genes have important regulatory roles in plant growth and development, as well as abiotic/biotic stress, a systematic report on the genome-wide characterization of VvCYPs is still lacking in grapevine. Hence, we performed a comprehensive bioinformatics characterization and expression analysis of the VvCYP superfamily in grapevine. The present study involved the identification of putative VvCYPs via genome-wide searches, the investigation of their phylogenetic relationships, chromosomal distribution, exon–intron organizations, promoter cis-acting element analysis, motif pattern, and syntenic analysis. In addition, we also analyzed the expression profiles of the VvCYP superfamily in grapevine by RNA-seq for identifying pivotal VvCYPs that participated in grapevine growth and development and further validated selected target VvCYP genes by qRT-PCR. Additionally, some VvCYPs were strongly modulated by ABA treatment, suggesting these VvCYPs have important and diverse regulatory roles in responding to ABA. The findings of the current investigations will assist in better comprehending the classification and potential biological functions of VvCYPs and pave the way for further functional verification of these VvCYPs and are helpful to grapevine molecular breeding.
Materials and Methods
Plant Materials, Growth Conditions and ABA Treatments
The grapevine (the hybrid of V. vinifera and V. labrusca cv. ‘Jumeigui') plants, planted at the farm of Nanjing Agricultural University (Nanjing, China) under standard field conditions, were used as experimental materials. Three biological replicates, each consisting of three clusters, were sampled at each sampling date. The developmental stages were defined in terms of the criterion containing the color, size, sugar content, and softening degree of the berries (Boss et al., 1996). The berry samples were harvested at five development stages, including small green berry (SGB), big green berry (BGB), veraison berry (VB), postveraison berry (PVB), and ripening berry (RB). Additionally, initial flowering (IF), full flowering (FF), young stems (YS), roots (Ro), buds (Bu), young leaves (YL), medium leaves (ML), old leaves (OL), and tendrils (Te) were also collected. Five-year-old uniform grapevine plants were chosen for the ABA treatments. ABA treatments were performed by spraying the grape berry before veraison with a solution containing 50 ppm and 150 ppm ABA, while the control berries were only treated with distilled water. The berry was collected at veraison, postveraison, and ripening. For making one sample, nine berries were collected from three different grapevines with respect to each sampling point and treatment. All of the samples were collected in triplicate and then immediately frozen under liquid nitrogen and kept in a refrigerator at −80°C until further analysis.
Total RNA Extraction and cDNA Library Construction
The extraction of total RNA was carried out from grapevine samples by following the cetyltrimethylammonium bromide method (Jiu et al., 2015; Jiu et al., 2016). The concentration of total RNA was evaluated using NanoDrop (Thermo Fisher Scientific Inc., USA), and OD260/280 ratios were close to 2.0, and OD260/230 ratios were >2.0 for all the samples. RNA integrity was also assessed using standard denaturing agarose gel electrophoresis. The samples of total RNA were digested with DNase I (TaKaRa, Japan) against genomic DNA contamination. Then, the manufacturer's protocol was followed for the construction of cDNA libraries with 1.0 μg of total RNA samples (PrimeScriptTM First Strand cDNA synthesis kit, TaKaRa, Japan).
Mining of VvCYPs Superfamily in Grapevine
The sequence of grapevine genome was downloaded from the National Center for Biotechnology Information website (NCBI, ftp://ftp.ncbi.nlm.nih.gov/genomes/Vitis_vinifera/). To identify all VvCYP genes in the grapevine, a local BLAST search against the grapevine genome database using 245 known AtCYPs acquired by Cytochrome P450 Homepage (http://drnelson.uthsc.edu/CytochromeP450.html) was conducted, setting the threshold of the e-value as 1e-10 to confirm the detection of all potential CYP genes in the Vitis genome database (http://genomes.cribi.unipd.it/grape/index.php).
Additionally, we also used the seed file of P450 domain (PF00067) from the Pfam online software (http://pfam.xfam.org/) to acquire the Hidden Markov Model (HMM) sequences, then we performed HMM searches using HMMER3 software against the V. vinifera protein sequences. As recommended by the HMMER3 user's guide, an e-value threshold of 0.1 was performed in these searches (Finn et al., 2011). Subsequently, each candidate VvCYP was used to further confirm the Pfam database. To eliminate repetitive genes, all potential VvCYPs were aligned using the DNAMAN5.0 software and manually collated. All the nonoverlapping VvCYPs were used for further analysis.
The theoretical isoelectric points (pI) and molecular weights (MW) of all members of the grape VvCYPs were calculated using ExPASy (http://expasy.org/). The number of transmembrane domains was predicted by the TMHMM Server v. 2.0. (http://www.cbs.dtu.dk/services/TMHMM/). Additionally, the estimation of subcellular localizations of all identified VvCYPs was completed by WoLF PSORT Prediction PSORT II (http://wolfpsort.org/) and TargetP (http://www.cbs.dtu.dk/services/TargetP) software (Emanuelsson et al., 2000).
Multiple Sequence Alignments and Phylogenetic Analysis
Multiple alignments were performed on the amino acid (AA) sequences of VvCYP proteins in V. vinifera genomes utilizing the CLUSTALW software with default parameters. Subsequently, MEGA7.0 program was employed to construct a phylogenetic tree based on the alignments using the Neighbor-Joining (NJ) method and bootstrap tests were replicated 1,000 times, which were performed using the p-distance model (Ogrodzki and Forsythe, 2015).
Conserved Motifs and Exon–Intron Organization of VvCYP Genes
The AA sequences of 236 VvCYPs were analyzed using the MEME program (http://meme-suite.org/) with the following parameters (optimum width, 15–60; number of repetitions, any; and maximum number of motifs, 15) to identify the conserved motifs (Bailey and Elkan, 1994). The conserved motifs of VvCYPs were confirmed by InterPro software (http://www.ebi.ac.uk/interpro/). The gene structure display server 2.0 (GSDS, http://gsds.cbi.pku.edu.cn ) was employed to display exon–intron organizations of 236 VvCYPs (Hu et al., 2014).
Putative Promoter Cis-Acting Element Analysis
The nucleotide sequences of the VvCYP superfamily were obtained from the grapevine genome database (http://genomes.cribi.unipd.it/grape/index.php) in this study. The upstream 1,500-bp region from the start codon for all VvCYPs was regarded as the promoter sequence (Jiu et al., 2018). The putative cis-acting elements of VvCYP promoters were identified using PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, Lescot, 2002). The putative cis-acting elements involved in phytohormone responses, the regulation of plant growth and development, as well as biotic and abiotic stress responses were summarized.
Chromosomal Localization and Collinearity Analysis
A total of 236 VvCYPs were mapped to the grapevine chromosomes by analyzing their chromosomal localization. The information of chromosomal localization is available at the grapevine genome database (http://genomes.cribi.unipd.it/grape/index.php). The duplication events in the grapevine genome were acquired using the MCscanX software. For syntenic analysis, synteny blocks among the grapevine genome were obtained from the PGDD website (http://chibba.agtec.uga.edu/duplication/index/downloads) and visualized by the Circos software (http://circos.ca/) (Krzywinski et al., 2009).
Expression Analysis of VvCYPs
The expression values normalized in numerous organs and tissues were obtained from the RNA-sequencing database (Fasoli et al., 2012). The primers used for amplifying 24 VvCYP genes, designed by using Primer-BLAST online program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi? LINK_LOC = BlastHome), are listed in Table S1. Polymerase chain reaction (PCR) was used for screening all primer pairs. The expression levels were detected by qRT-PCR assay using a Bio-Rad System (Bio-Rad, CA, USA). Each PCR mixture (20 µl) consisted of 2×TB Green™ Premix Ex Taq™ II (10 µl), 1:10 diluted cDNA (1 µl), each primer (0.4 µl), and RNase-free water (8.2 µl). All reactions were run in 96-well plates, and each cDNA was analyzed in triplicate. The condition of the qRT-PCR was as follows: preincubation (95°C for 30 s) and then 40 cycles (95°C for 5 s, 60°C for 30 s). To detect the relative fold differences for each gene in each experiment, the Ct value of the genes was normalized to the Ct value for the reference genes, and the relative expression levels were calculated using the formula 2−ΔΔCT. KyActin1 was used as the internal reference control. The lowest expression levels of the samples were manually set to 1.
Subcellular Localization
The full-length cDNAs of the VvCYP710A1 and VvCYP51G1a were amplified using the primers (Table S1) and cloned into the binary vector PHB including two cauliflower mosaic virus (CaMV) 35S promoter, a translation enhancer, and a GFP fluorescent protein tag, respectively, to generate two fusion constructs (p35S-VvCYP710A1-GFP and p35S-VvCYP51G1a-GFP). After identifying two sequences, the fusion constructs and the control vector (PHB) were transformed into A. tumefaciens GV3101 strains and subsequently agroinfiltrated into the leaves of 3 to 5-week-old Nicotiana benthamiana plants. Localization of fluorescent proteins was observed 3–7 days after infiltration, the period when GFP fluorescence was optimal, by using a confocal laser scanning microscope (Zeiss LSM 780, Germany) according to the manufacturer's instructions.
Statistical Analysis
The experiment was arranged in a completely randomized design (CRD) with three replications in this study and collected data were statistically analyzed by SAS computer software (SAS Version 9.2, Institute). Analysis of variance (ANOVA) was used to determine the overall statistical significance of the data at level of P < 0.05 and data were represented as average ± STDEV (n = 3).
Results
Identification and Analysis of Full-Length VvCYPs in Grapevine
The availability of grapevine genome sequences provides the sources in the genome-wide identification of the grapevine VvCYP superfamily (Jaillon et al., 2007). After bioinformatics analyses, a total number of 236 VvCYP genes with conserved P450 domain have been identified and characterized in this study (Table S2). Here, we designated the VvCYP genes in grapevine according to the Arabidopsis CYP genes with the highest sequence similarity and following the classification of gene terminology used in the Arabidopsis. All VvCYPs were divided into 46 families and grouped into A-type (27 families) and non-A type (19 families) including 102 and 134 VvCYPs, respectively. Each of the 13 families (VvCYP701, VvCYP703, VvCYP712, VvCYP83, VvCYP98, VvCYP709, VvCYP715, VvCYP735, VvCYP702, VvCYP720, VvCYP724, VvCYP710, and VvCYP711) has only a single gene, whereas VvCYP71 with 25 members is the largest family, followed by VvCYP72 family with 21 members. The AA sequence lengths of the 236 identified VvCYP proteins ranged from 269 to 579 with an average of 484.40 AA. The molecular weights (Mw) of these VvCYP proteins ranged from 23.48 kDa (VvCYP72A10c) to 144.19 kDa (VvCYP79A2e), and the isoelectric points (pI) ranged from 4.92 (VvCYP74B2) to 5.31 (VvCYP72A10c). Other characteristics of the VvCYPs, including the chromosome location, number of transmembrane domains, gene duplication, conserved motif, and prediction of subcellular localization are also shown in Table S2.
Phylogenetic Analysis of VvCYP Genes in Grapevine
To elucidate the phylogenetic relationships among the members of the VvCYP superfamily, an unrooted tree was constructed from an alignment of their AA sequences and produced in MEGA 7.0 software by the NJ method. Based on the phylogenetic analysis, VvCYPs were grouped into nine clans (clan 51, clan 71, clan 72, clan 74, clan 85, clan 86, clan 97, clan 710, and clan 711) with well-supported bootstrap values (Figure 1). There were one family and two VvCYPs in clan 51, 19 families and 134 VvCYPs in clan 71, seven families and 32 VvCYPs in clan 72, one family and six VvCYPs in clan 74, 11 families and 34 VvCYPs in clan 85, four families and 23 VvCYPs in clan 86, one family and two VvCYPs in clan 97, one family and one VvCYP in clan 710, one family and one VvCYP in clan 711. The three largest clans (in descending order) were clan 71, clan 85 and clan 72. Furthermore, the nine clans were classified into four clusters. Clan 74 was thought to be a special one, and clan 51 and clan 85 were classified into a cluster. Clan 72, clan 86, and clan 97 were gathered in one cluster, and clan 710, clan 711, and clan 71 were closely grouped together to form another cluster.
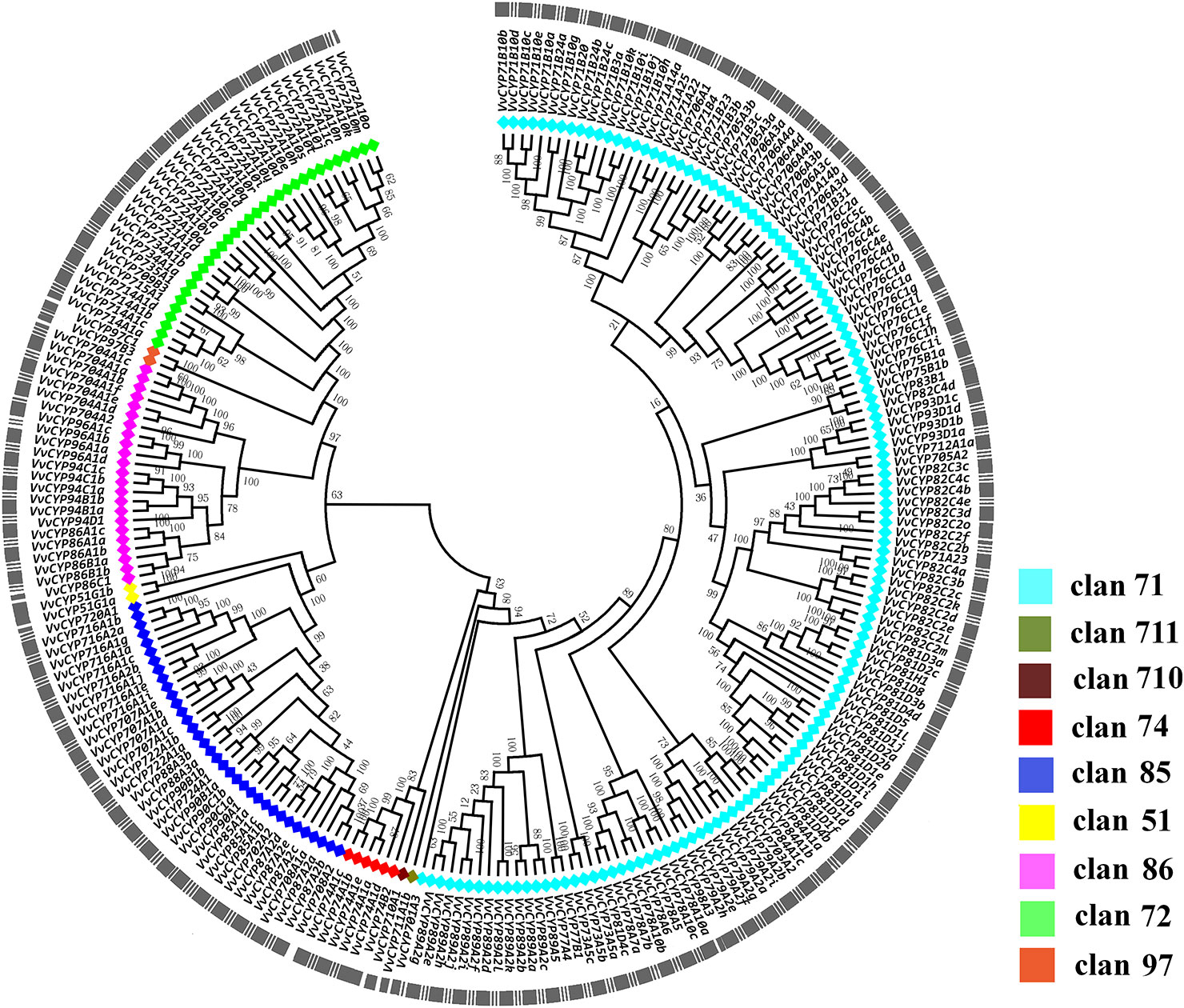
Figure 1 The phylogenetic tree of VvCYPs superfamily in grapevine. The unrooted phylogenetic tree was constructed using the full-length AA sequences of VvCYPs using MEGA7.0 by the Neighbor-Joining (NJ) method with 1000 bootstrap replicates. The entire family of VvCYPs is shown for each clan with different color: aquamarine, clan 71; olivedrab, clan 711; prunosus, clan 710; red, clan 74; royalblue, clan 85; yellow, clan 51; violet, clan 86; springgreen, clan 72; salmon, clan 97.
To further investigate the evolution and divergence of the VvCYP superfamily in grapevine, comprehensive comparisons among V. vinifera, A. thaliana, O. sativa, Populus alba, Brachypodium distachyon, Carica papaya, Salvia miltiorrhiza, Citrus clementina, Nelumbo nucifera, and Glycine max were performed. Some CYP subfamilies only present in dicots, such as CYP716, CYP720, and CYP82, whereas CYP99 and CYP723 only exist in monocots (Figure 2). Many CYP families were found in ten species, for instance, CYP88, CYP94, CYP86, CYP71, and CYP73. However, some families were only present in one of ten species, such as CYP749 in S. miltiorrhiza, CYP719 in N. nucifera. It's worth noting that all defined grapevine VvCYP subfamilies emerged after divergence of grapevine and N. nucifera. The gene number-based hierarchical cluster of the CYP subfamilies of different plants exhibited that the loss of the CYP family seems to be limited to single species or single taxa. The expansion of the CYP families can be observed in the CYP71, CYP72, CYP76, CYP81, CYP89, and CYP94 families (Figure 2). Particularly, the members of the CYP94 family are closely linked with the metabolism of aliphatic acid, and the CYP72 family with multiple functions in V. vinifera are more abundant than those in the other species.
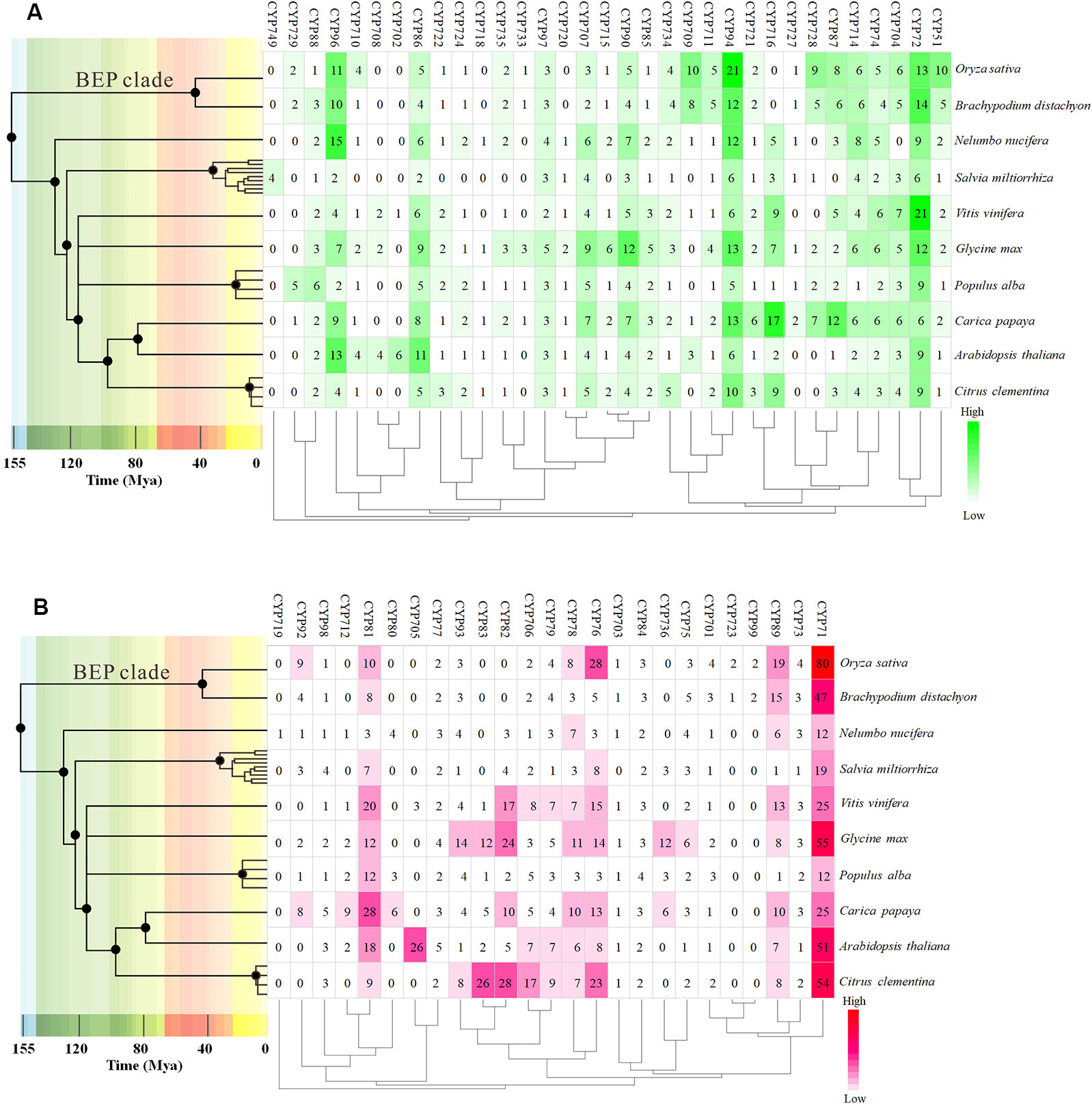
Figure 2 The distribution of CYP family sizes in whole-genome for 10 species. (A) A-type CYP families. (B) Non-A type CYP families. The time tree generated using TIMETREE web resource (http://www.timetree.org/) revealed the clock-like speciation and diversification of 10 species. The four species, Nelumbo nucifera, Salvia miltiorrhiza, Populus alba, and Carica Papaya, were instead by other species of the same genus on the time tree.
Conserved Motifs, Chromosomal Locations, Exon–Intron Organizations of VvCYP Genes
In addition to the predictions of subcellular localizations and the analysis of transmembrane domains, the evaluation of conservative motifs is one of the important means for the functional prediction of VvCYP proteins. In this study, a sum of 15 conserved motifs (motifs 1–15) were predicted using MEME online software (Figure S1). Several motifs were found in most VvCYP proteins, such as motifs 1, 2, 3, 4, and 7. However, some are only present in a specific clan, such as motifs 9 and 10 in clan 71. Additionally, clan 71 contained motifs 1–15 and motif 2 existed in all VvCYP members of clan 71, whereas clan 711 only included one member, VvCYP711A1b had motifs 1, 3, and 7 (Figure S2 and Table S3). Most of VvCYP proteins within the same clan exhibited similar motif components while a high discrepancy was observed among different clans, indicating that the VvCYP members within the same family may undertake semblable functions, and that some motifs may play a vital role in the family-specific functions. The chromosomal location of each VvCYP gene in the grapevine genome is shown in Figure 3. A total of 236 VvCYPs are unevenly distributed on 19 chromosomes. Out of the 236 VvCYPs, 33 were mapped onto chromosome (chr)18, followed by 22 on chr16, whereas only 2 VvCYPs were located on chr5. The VvCYPs with relatively high densities were detected on the bottom and middle arms of the chromosome. To better comprehend the structure of VvCYPs, the exon–intron organizations were analyzed by the alignment of cDNA sequences and corresponding genomic DNA sequences. As shown in Figure S3, 26 out of 236 VvCYPs had no intron. As Figure 4 demonstrates, grapevine VvCYP families possessed different exon numbers, ranging from one to 14. VvCYP97B3, which is a member of clan 86, contains 14 exons. Most members within the identical family showed similar exon–intron distribution characteristics in accordance with exon length and number. For example, 15 VvCYPs contain nine exons, 14 of which belong to the CYP85 family. 25 VvCYPs contain four exons, 24 of which belong to the CYP72 family. VvCYPs in the same families showed great consistence with exon length and number. Only a few of VvCYPs had no typical genetic structure. For example, VvCYP81D3b, VvCYP82C2o, VvCYP73A5a, VvCYP78A5, and VvCYP706A4d in clan 71, VvCYP72A10d, VvCYP72A10e, and VvCYP72A10q in clan 72 had a long intron and short exon organization compared with the other members within the same family. The majority of them share two (51.7%, 122/236), one (11.4%, 27/236), and five (10.6%, 25/236) exons. At the DNA level, VvCYP97B3 with 28 kb genomic sequences was the longest VvCYP gene identified in this study. The great change in structures of VvCYPs could demonstrate that the grape genome had changed significantly during its long evolutionary history. The coherence of the exon/intron organizations and motif structures further supported the close evolutionary relationships of the grapevine VvCYP superfamily as well as the reliability of the phylogenetic analysis.
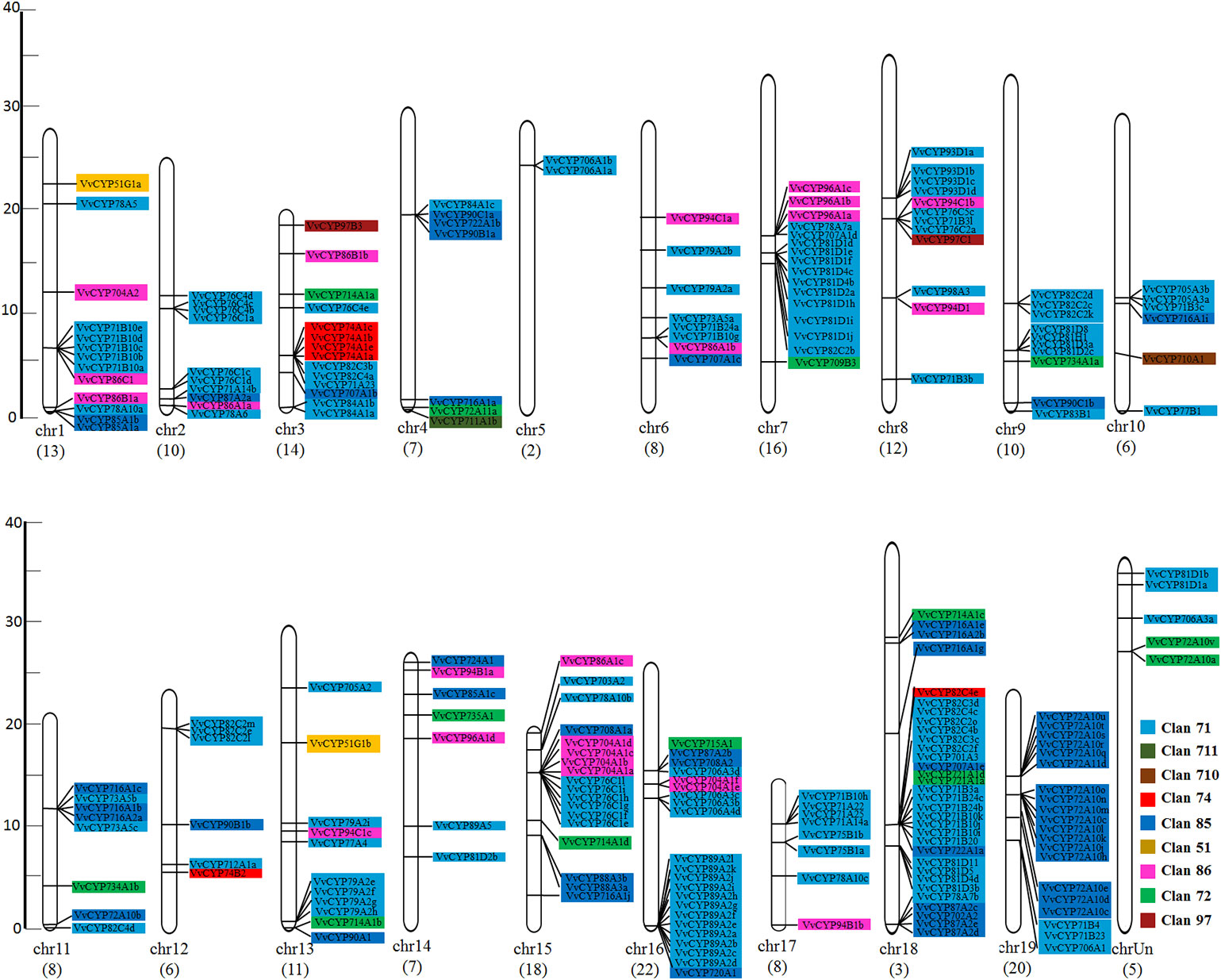
Figure 3 The chromosomal location of VvCYPs from V. vinifera. The scale represents 40 Mb chromosomal distance. The VvCYP numbers are marked on the bottom of chromosomes. The numbers on the left side of the bars designated the approximate physical position of the first exon of corresponding VvCYPs on grapevine chromosomes. The VvCYPs of each clan were shown with different background color: aquamarine, clan 71; olivedrab, clan 711; prunosus, clan 710; red, clan 74; royalblue, clan 85; yellow, clan 51; violet, clan 86; springgreen, clan 72; salmon, clan 97.
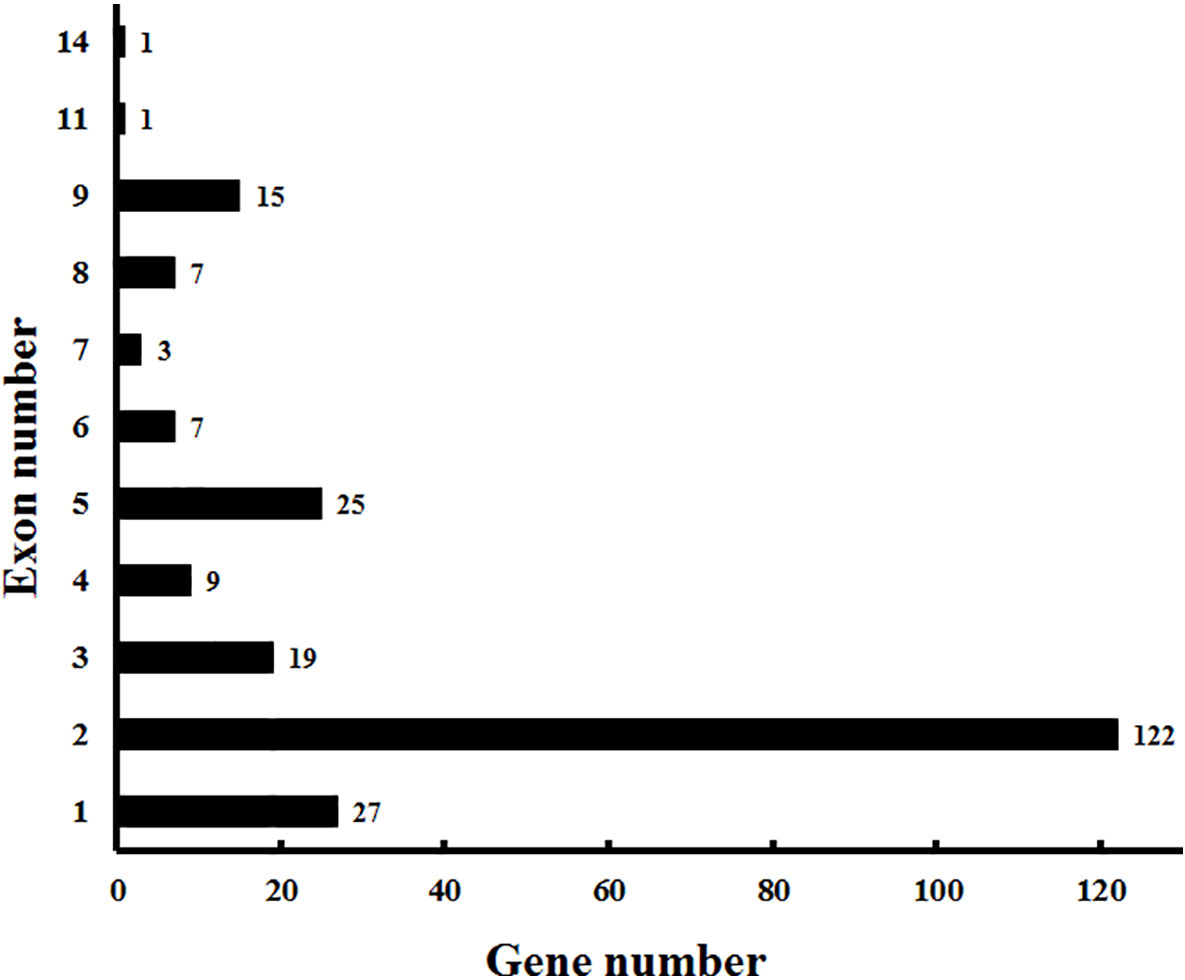
Figure 4 Exon distribution of VvCYPs in grapevine. The exon information of VvCYPs was obtained from the Vitis genome database (http://genomes.cribi.unipd.it/grape/index.php).
Gene Duplication and Syntenic Analysis of the VvCYP Genes
For the gene family expansion and evolution of novel functions, gene duplication and divergence are essential steps in the plant genome. Vitis spp. has suffered from whole-genome duplications during its generative history (Jaillon et al., 2007). Moreover, several gene duplication events including tandem duplication, WGD/segmental, and rearrangements at the chromosomal and gene levels, drive the evolution of gene families encoding proteins (Maher et al., 2006). To evaluate the effect of duplications on the VvCYP superfamily, we firstly analyzed the origins of duplicate genes for the VvCYP superfamily in the grapevine genome utilizing the MCScanX software. Each member of the VvCYPs was allocated to one of five duplication events (tandem, dispersed, singleton, WGD/segmental, and proximal). The results showed that 48.31% (114) of the VvCYPs in grapevine were duplicated from tandem event, compared with 19.92% (47) from singleton, and 16.10% (38) from proximal event, 11.02% (26) from dispersed event, and 4.66% (11) from WGD/segmental event (Table S2). The results demonstrated that tandem duplication was the major driving force for the augmentation of the VvCYP superfamily (Table S2). In addition, the chromosome distribution patterns of VvCYPs powerfully showed that tandem duplication contributed to the expansion of VvCYPs in V. vinifera. To explore further the probable evolutionary mechanisms of the VvCYP superfamily, we used a method similar to that used for the Plant Genome Duplication Database (PGDD) to identify synteny blocks across the whole grapevine genome (Figure 5). A sum of 41 segmentally duplicated gene pairs were detected in the VvCYP superfamily. As Figure 6 shows, eight segmentally duplicated gene pairs within grapevine genome included: VvCYP72A10s and VvCYP72A11c, VvCYP72A10i and VvCYP72A11d, VvCYP72A10s and VvCYP72A10i, VvCYP72A11d and VvCYP72A10j, VvCYP72A10t and VvCYP72A10k, VvCYP72A10t and VvCYP72A10n, VvCYP72A10u and VvCYP72A10q, VvCYP72A10u and VvCYP72A10t.
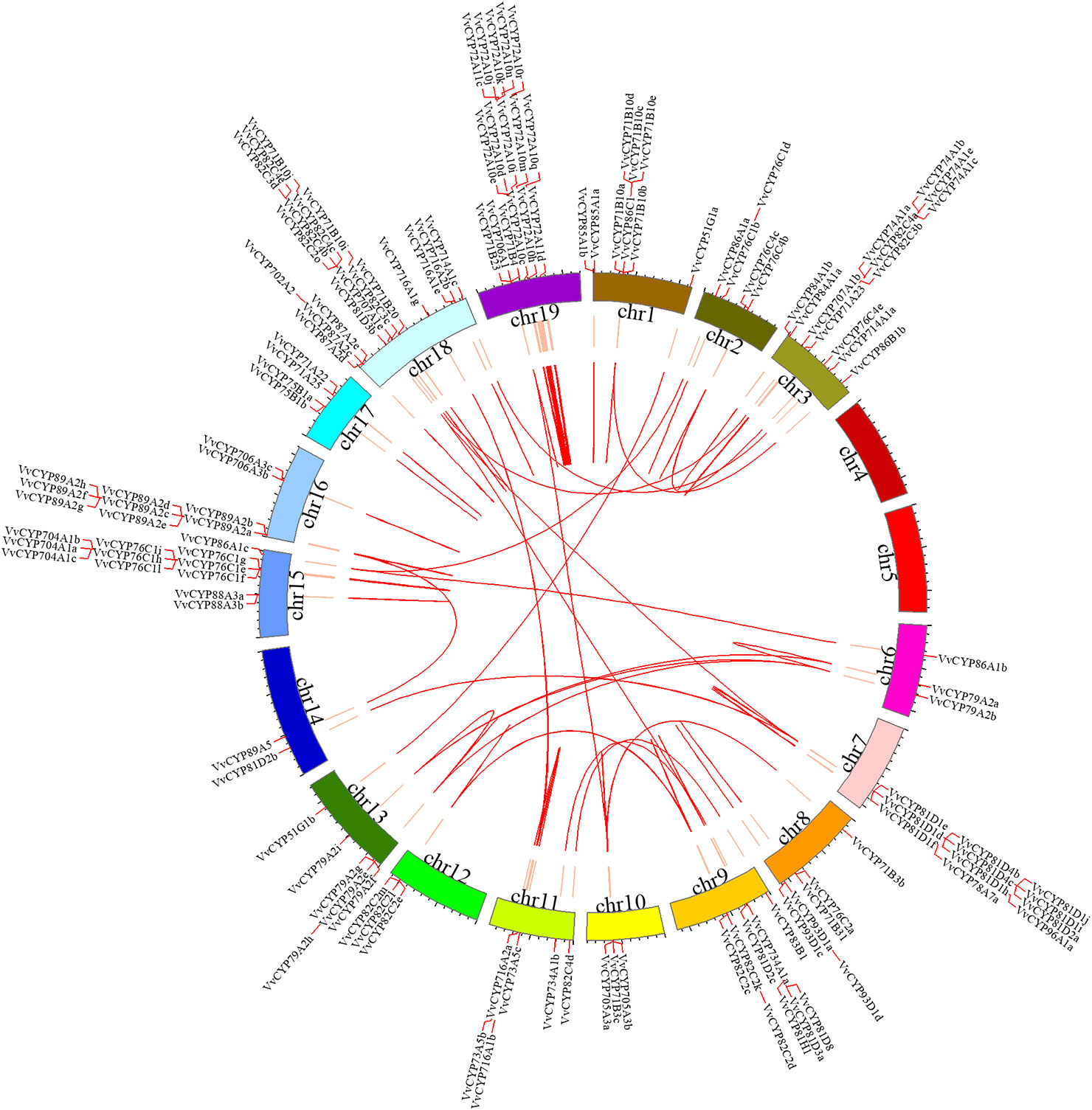
Figure 5 Syntenic block of the VvCYPs superfamily. Red lines mark the VvCYP gene positions on chromosomes. Chromosomal locations were determined according to Grape Genome Database (http://genomes.cribi.unipd.it/grape/index.php). Red curves in circle represented the syntenic relationships among VvCYP genes. The map was obtained using Circos software.
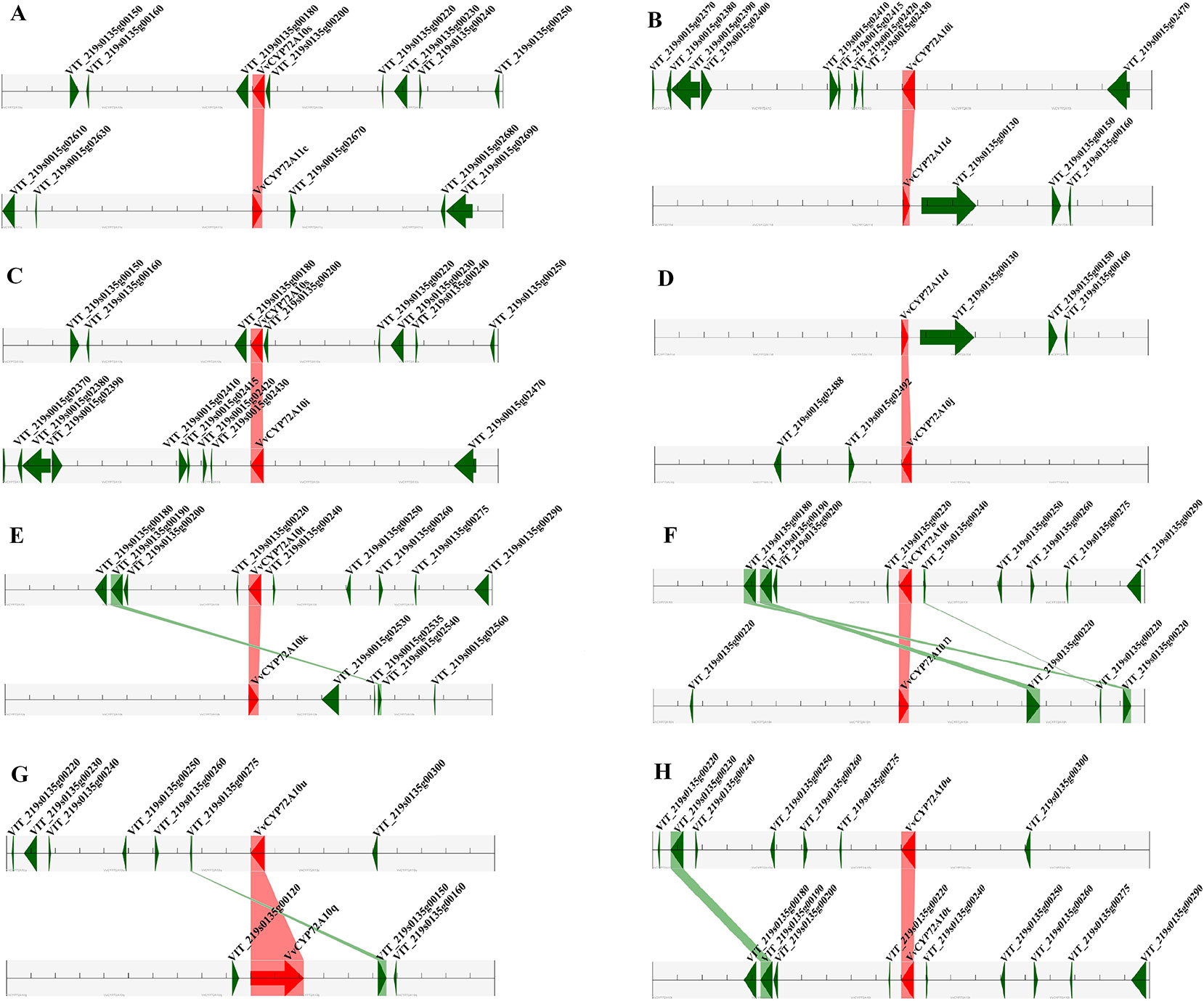
Figure 6 Segmental duplication between members of the VvCYPs subfamily. (A) VvCYP72A10s (VIT_219s0135g00190.1) and VvCYP72A11c (VIT_219s0015g02660.1), (B) VvCYP72A10i (VIT_219s0015g02440.2) and VvCYP72A11d (VIT_219s0135g00120.1), (C) VvCYP72A10s (VIT_219s0135g00190.1) and VvCYP72A10i (VIT_219s0015g02440.2), (D) VvCYP72A11d (VIT_219s0135g00120.1) and VvCYP72A10j (VIT_219s0015g02500.1), (E) VvCYP72A10t (VIT_219s0135g00230.1) and VvCYP72A10k (VIT_219s0015g02520.1), (F) VvCYP72A10t (VIT_219s0135g00230.1) and VvCYP72A10n (VIT_219s0015g02900.1), (G) VvCYP72A10u (VIT_219s0135g00290.1) and VvCYP72A10q (VIT_219s0135g00130.1), (H) VvCYP72A10u (VIT_219s0135g00290.1) and VvCYP72A10t (VIT_219s0135g00230.1). The figure shows a region of 100 kb on each side flanking the VvCYP genes. Homologous gene pairs are connected with bands. Chromosome segments are indicated by black horizontal lines, and a grey line with arrowhead represents a gene. The VvCYP genes are shown in red, and homologous genes are linked by the pink line, other genes are shown in green, and homologous genes are linked by the green line.
Identification of the Putative Cis-Acting Elements in the Promoter of VvCYP Genes
The evaluation of cis-elements in promoters is a key factor for comprehending transcriptional regulation and gene function. To recognize the putative cis-regulatory elements of VvCYPs, the promoter sequences of 236 VvCYPs were searched in the Vitis genomic database (http://genomes.cribi.unipd.it/grape/index.php). In this study, a 1,500-bp was considered as a promotor region for 221 VvCYPs, whereas, a <1,500 bp promoter sequence for the VvCYP705A2, VvCYP712A1a, VvCYP81D3b, VvCYP81D1l, VvCYP81D4d, VvCYP82C4c, VvCYP71A25, VvCYP76C1d, VvCYP76C5c, VvCYP76C2a, VvCYP82C3c, VvCYP89A2a, VvCYP706A4a, VvCYP72A10s, and VvCYP707A1d was also identified because of the existence of another gene situated at the <1,500 bp upstream (Data Sheet 1). A total of 107 putative cis-elements involved in plant hormone [e.g., auxin, ABA, MeJA, ethylene, gibberellin (GA), and salicylic acid (SA)] responses, biotic and abiotic stress responses, and plant growth and development were identified using the PlantCARE online database. As shown in Table S4, the predicted cis-elements differed among the 236 VvCYPs, and two cis-elements (TATA-box and CAAT-box) were the most abundant, which had the largest number in all 236 VvCYPs. It is quite interesting to recognize that these unique cis-elements (AuxRR-core, AACA motif and 4cl-CMA2b, etc.) were only present in one of 236 VvCYPs, implying that these gene-specific cis-elements might play a special role in regulating some biological processes. Some phytohormone-related cis-regulatory elements including the ABA response element (ABRE), the GA-responsive element (P-box), the SA-responsive element (TCA), the auxin-responsive element (TGA-element), the MeJA response element (CGTCA-motif), and the ethylene-responsive element (ERE) were found in the promoters of 144, 49, 70, 48, 115, and 172 VvCYPs, respectively. A mass of phytohormone-responsive elements were recognized in the VvCYP promoters, suggesting they could play an important role in the regulation of grape growth and development process. In several cis-elements related to plant growth and development, the zein-metabolism regulation element (O2-site), the cell growth promotion element (MYC), the circadian regulator element (circadian), the leaf development-related cis-regulatory element (HD-Zip1), the seed-specific regulation element (RY-element), the endosperm-specific expression element (GCN4_motif), the flavonoid biosynthetic regulation element (MBSI), and the meristem expression element (CAT-box) were identified in the promoters of 67, 214, 44, 19, 10, 19, 6, and 62 VvCYPs, respectively. Additionally, some stress-related cis-regulatory elements (e.g. DRE core, LTR, and WUN-motif) that are associated with drought, low-temperature, and wound responses, were found in 6 (2.54%), 97 (41.10%), and 46 (19.49%) VvCYPs, respectively.
Tissue-Specific Transcript Accumulation Patterns of VvCYP Genes in Grapevine
To gain more insight into the role that VvCYPs play in grapevine growth and development, the expression patterns of VvCYPs were analyzed in different grapevine organs and tissues based on global RNA-sequencing data (Fasoli et al., 2012). Interestingly, VvCYP74B2 and VvCYP81D5 showed consistently high expression in all 54 tissues, whereas VvCYP71A14a displayed lower expression levels in all tested tissues compared with the others (Figure 7). Additionally, many gene expression profiling exhibited that diverse temporal, spatial, and tissue-specific expression patterns of VvCYPs in all tested tissues, potentially indicating the functional divergence of VvCYPs. To further clarify possible functions of VvCYP genes in grapevine, the expression levels of selected target VvCYPs participating in grapevine growth and development were examined in different tissues including fruits, leaves, roots, buds, stems, tendrils, and flowers (Figure 8). The expression levels of various genes in different tissues were unlike, indicating that the functions of VvCYPs in different tissues were specific to the grapevine growth and development. VvCYP76C1a, VvCYP82C2b, and VvCYP93D1a, three members of clan 71, were highly expressed in tendrils, displaying that they might play a vital role in the growth and development of tendrils. Meanwhile, seven VvCYPs (VvCYP71B10a, VvCYP76C1a, VvCYP79A2a, VvCYP82C2b, VvCYP89A2a, VvCYP93D1a, and VvCYP84A1a) belonging to clan 71 are all weakly expressed in young stems. Furthermore, the expression levels of VvCYP714A1a and VvCYP72A10a, belonging to clan 72, displayed similar tendency in different tissues. In particular, VvCYP72A11a was preferentially expressed at high levels in RB, which indicated that it might make a difference in fruit maturation. VvCYP74A1a, belonging to clan 74, presented very high expression levels in RB and leaves; however, low expression levels were observed in flowers, roots, and young stems. In four members of clan 85, VvCYP707A1b and VvCYP87A2b were highly expressed in flowers; whereas VvCYP716A1a and VvCYP90A1 presented very high expression levels in ML and OL, possibly indicating their functional divergence. Additionally, several VvCYPs of clan74 displayed more extensive tissue-specific expression patterns; such as, VvCYP704A1a was hugely expressed in tendrils and buds; VvCYP94C1a displayed high relative transcript levels in YL and OL, VvCYP96A1a and VvCYP97B3 were highly expressed in ML. VvCYP710A1 showed more widespread but less tissue-specific expression patterns in different tissues, except for YS and RB. These results urged us to research the expression level of VvCYPs during different berry developmental stages.

Figure 7 Expression profiles of VvCYP genes in the V. vinifera cv “Corvina” atlas (GEO Accession: GSE36128). Data were normalized based on the mean expression value of each gene in all tissues (Fasoli et al., 2012). The mean expression values were again normalized using logarithm with the base of 2 using the Heml software. Blue and red boxes show low and high expression levels, respectively, for each gene.
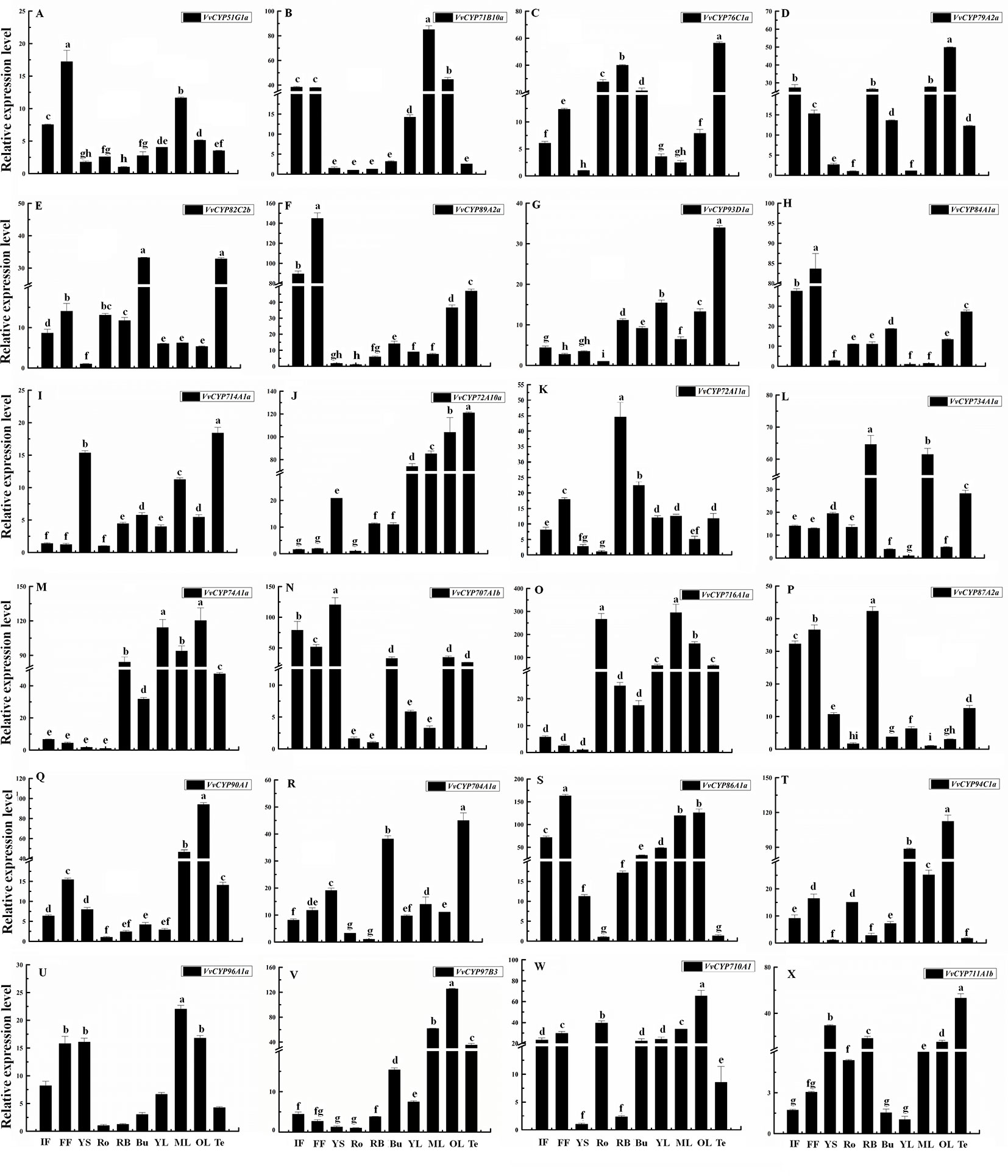
Figure 8 Relative expression levels of VvCYPs in different organs of ‘Jumeigui’ grapevine. Values were normalized against the expression data of KyActin1 and are given as means ± standard deviation among three biological replicates. Different letters indicate significant differences between genes (p< 0.05). The expression levels were calculated based on the 2−△△Ct method. Additionally, initial flowering (IF), full flowering (FF), young stems (YS), roots (Ro), buds (Bu), young leaves (YL), medium leaves (ML), old leaves (OL) and tendrils (Te). The expression levels of VvCYP51G1a (A), VvCYP71B10a (B), VvCYP76C1a (C), VvCYP79A2a (D), VvCYP82C2b (E), VvCYP89A2a (F), VvCYP93D1a (G), VvCYP84A1a (H), VvCYP714A1a (I), VvCYP72A10a (J), VvCYP72A11a (K), VvCYP734A1a (L), VvCYP74A1a (M), VvCYP707A1b (N), VvCYP716A1a (O), VvCYP87A2a (P), VvCYP90A1 (Q), VvCYP704A1a (R), VvCYP86A1a (S), VvCYP94C1a (T), VvCYP96A1a (U), VvCYP97B3 (V), VvCYP710A1 (W), and VvCYP711A1b (X) in different organs of ‘Jumeigui’ grapevine.
Transcript Accumulation Patterns of VvCYPs During Different Fruit Developmental Stages
To illuminate their functions in fruit development, qRT-PCR was employed to explore the expression levels of selected target VvCYPs belonging to different subfamilies different fruit developmental stages. As shown in Figure 9, the expression levels of six VvCYPs (VvCYP51G1a, VvCYP71B10a, VvCYP734A1a, VvCYP90A1, VvCYP94C1a, and VvCYP710A1) were down-regulated toward fruit maturity, which suggested that they might function mainly in the early period of berry development. On the contrary, the transcripts of three genes (VvCYP72A11a, VvCYP716A1a, and VvCYP87A2a) were up-regulated during the ripening process, which indicated that they might play a vital role in fruit maturation. VvCYP79A2a transcript was continuously increased from SGB to VB but declined afterward as the fruit matured. VvCYP76C1a and VvCYP86A1a transcripts showed high expression levels at SGB and RB in comparison with the other stages. Five VvCYPs (VvCYP82C2b, VvCYP714A1a, VvCYP704A1a, VvCYP96A1a, and VvCYP711A1b) were preferentially expressed at high levels in BGB, which indicated that they played a vital role in the fruit development of this stage. VvCYP89A2a, VvCYP72A10a, and VvCYP707A1b showed high expression levels at SGB and BGB in comparison with the other stages and exhibited a varying trend as they increased from SGB to BGB and decreased sharply from BGB to VB then increased progressively toward fruit maturity. VvCYP84A1a transcript showed high expression levels at SGB, BGB, and VB and then exhibited a markedly decreased trend toward fruit maturity.
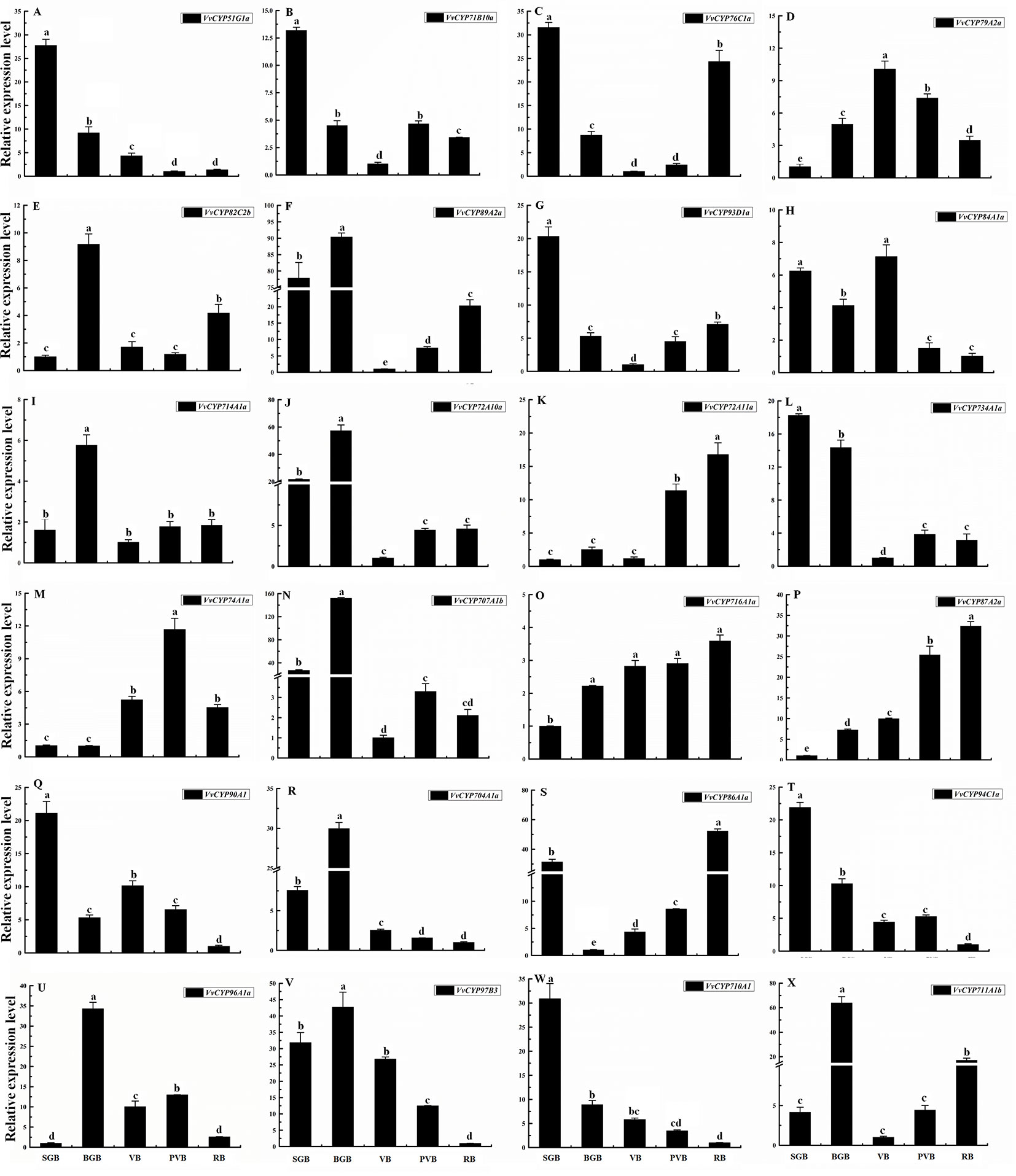
Figure 9 Relative expression levels of VvCYPs in different fruit development of ‘Jumeigui’ grapevine. Values were normalized against the expression data of KyActin1 and are given as means ± standard deviation among three biological replicates. Different letters indicate significant differences between genes (p< 0.05). The expression levels were calculated based on the 2 −△△Ct method small green berry (SGB), big green berry (BGB), veraison berry (VB), post-veraison berry (PVB) and ripening berry (RB). The expression levels of VvCYP51G1a (A), VvCYP71B10a (B), VvCYP76C1a (C), VvCYP79A2a (D), VvCYP82C2b (E), VvCYP89A2a (F), VvCYP93D1a (G), VvCYP84A1a (H), VvCYP714A1a (I), VvCYP72A10a (J), VvCYP72A11a (K), VvCYP734A1a (L), VvCYP74A1a (M), VvCYP707A1b (N), VvCYP716A1a (O), VvCYP87A2a (P), VvCYP90A1 (Q), VvCYP704A1a (R), VvCYP86A1a (S), VvCYP94C1a (T), VvCYP96A1a (U), VvCYP97B3 (V), VvCYP710A1 (W), and VvCYP711A1b (X) in different fruit development of ‘Jumeigui’ grapevine.
Transcript Analysis of VvCYP Genes in Response to ABA Treatment
Abscisic acid (ABA) have engrained roles in plant developmental processes as well as stress signaling networks (Bari and Jones, 2009). As shown in Table S4, ABA-responsive cis-element, ABRE, was observed in some VvCYPs. In order to comprehend how VvCYPs express in response to ABA treatment, qRT-PCR was employed to analyze how VvCYP transcripts respond to ABA, which has three concentration levels (0 ppm, 50 ppm, and 150 ppm). The expression levels of four VvCYPs (VvCYP82C2b, VvCYP714A1a, VvCYP86A1a, and VvCYP94C1a) were all enhanced with 50 ppm and 150 ppm ABA compared with 0 ppm ABA at VB, PVB, and RB periods, which is consistent with presence of ABA-responsive cis-element (ABRE) in the promoter of these VvCYPs, further implying ABA may play an important role in the regulation of grape berry ripening (Figure 10). On the contrary, the expression levels of four VvCYPs (VvCYP79A2a, VvCYP93D1a, VvCYP84A1a, and VvCYP72A11a) were all suppressed with 50 ppm and 150 ppm ABA compared with 0 ppm ABA at VB, PVB, and RB periods. The expression levels of two VvCYPs (VvCYP76C1a and VvCYP89A2a) were all enhanced with 50 ppm and 150 ppm ABA compared with 0 ppm ABA at VB period, whereas they were suppressed with 50 ppm and 150 ppm ABA compared with 0 ppm ABA at PVB and RB periods. The transcript accumulation of two VvCYPs (VvCYP74A1a and VvCYP707A1b) were strongly decreased with 50 ppm ABA compared with 0 ppm ABA at three periods, whereas they were suppressed with 150 ppm ABA compared with 0 ppm ABA at three periods. The expression level of VvCYP716A1a was significantly increased with 150 ppm ABA compared with 0 ppm ABA at three periods, and it was not significantly changed with 50 ppm ABA compared with 0 ppm ABA at VB and RB periods; however, it was markedly suppressed with 50 ppm ABA at PVB period. VvCYP704A1a showed a significant increase with 150 ppm ABA compared with 0 ppm and 50 ppm ABA at PVB and RB periods.
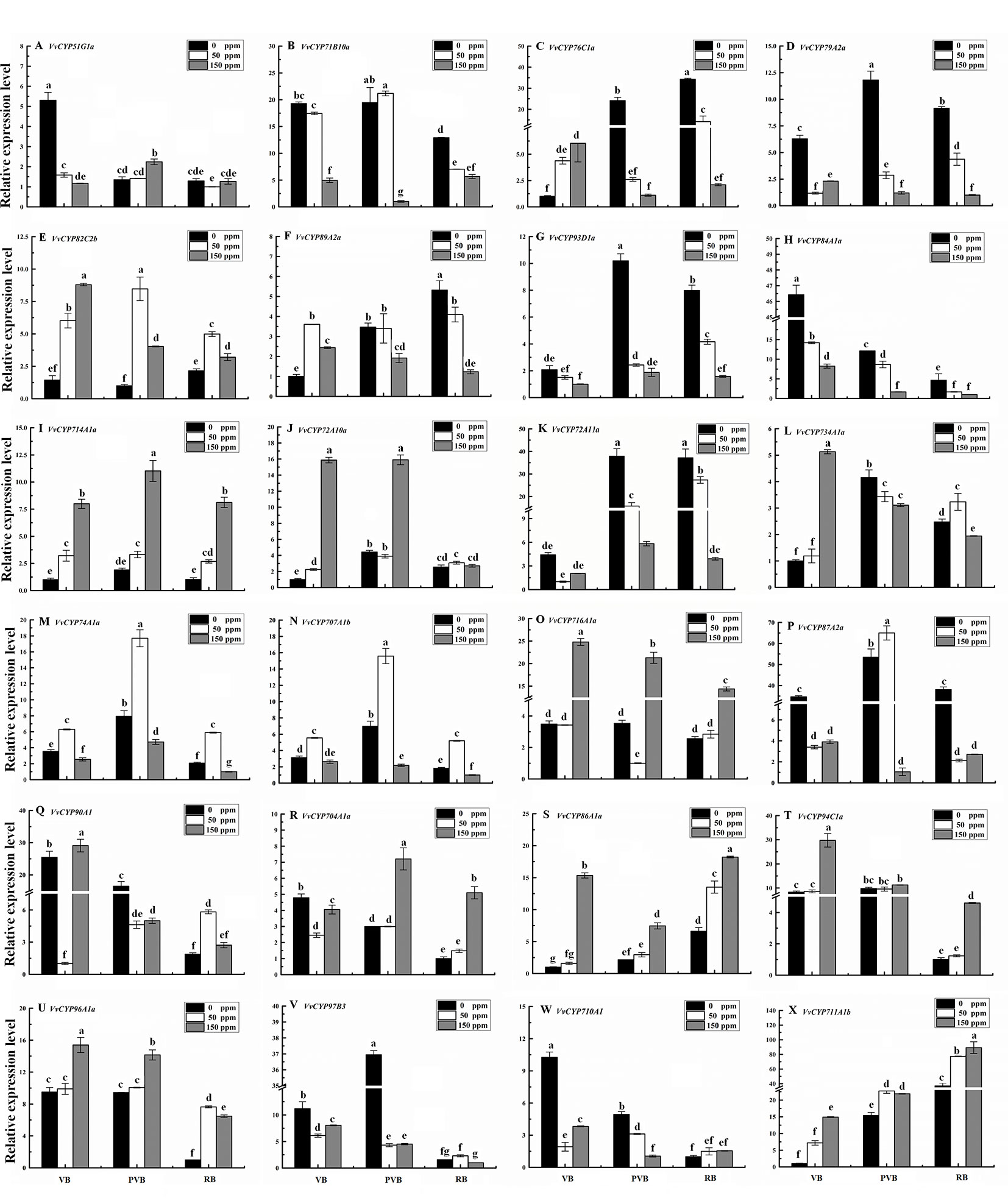
Figure 10 Relative expression levels of VvCYPs under ABA treatment. Values were normalized against the expression data of KyActin1 and are given as means ± standard deviation among three biological replicates. Different letters indicate significant differences between genes (p< 0.05). The expression levels were calculated based on the 2 −△△Ct method. The expression levels of VvCYP51G1a (A), VvCYP71B10a (B), VvCYP76C1a (C), VvCYP79A2a (D), VvCYP82C2b (E), VvCYP89A2a (F), VvCYP93D1a (G), VvCYP84A1a (H), VvCYP714A1a (I), VvCYP72A10a (J), VvCYP72A11a (K), VvCYP734A1a (L), VvCYP74A1a (M), VvCYP707A1b (N), VvCYP716A1a (O), VvCYP87A2a (P), VvCYP90A1 (Q), VvCYP704A1a (R), VvCYP86A1a (S), VvCYP94C1a (T), VvCYP96A1a (U), VvCYP97B3 (V), VvCYP710A1 (W), and VvCYP711A1b (X) under ABA treatment.
Subcellular Localizations
The subcellular localization of proteins is desirable to explore their biological functions. To investigate the function of VvCYPs, their subcellular localizations were authenticated by the fluorescent protein-tagging method. Firstly, the full-length open reading frames (ORFs) lacking the stop codon of two VvCYPs were merged to the N-terminal of the green fluorescence protein (GFP) driven by CaMV 35S promoter, generating fusion proteins p35S-VvCYP51G1a-GFP and p35S-VvCYP710A1-GFP which were agroinfiltrated into leaves of 3 to 5-week-old N. benthamiana plants. Fluorescence microscopy exhibited that the PHB-GFP was equally distributed throughout the whole cell, and the fusion proteins of p35S-VvCYP710A1-GFP and p35S-VvCYP51G1a-GFP were both expressed throughout the karyotheca and cytomembrane (Figure 11). The results demonstrated that VvCYP710A1 and VvCYP51G1a were both karyotheca- and cytomembrane-localized proteins, suggesting their functional similarity.
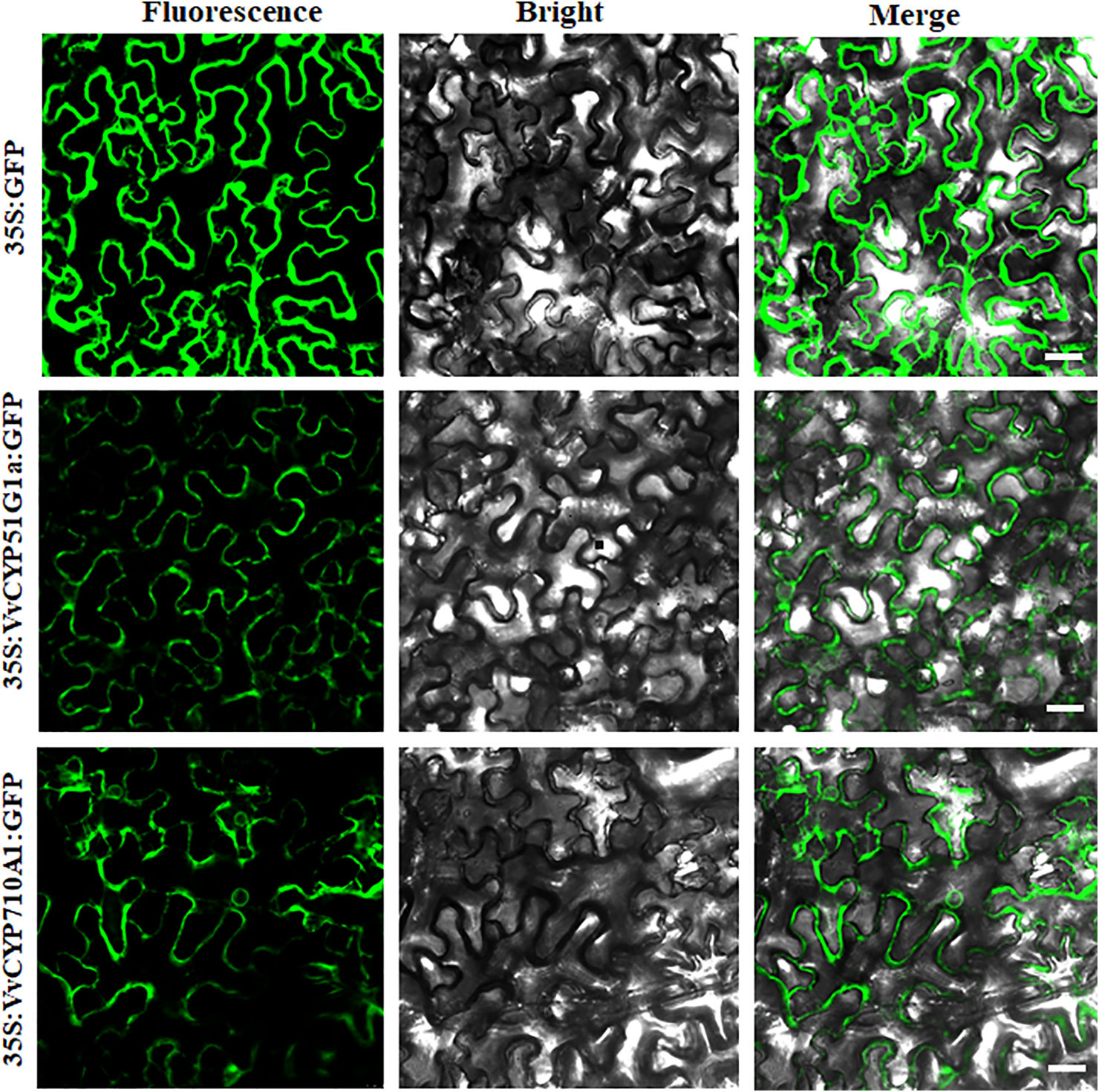
Figure 11 The leaves of 3 to 5-week-old Nicotiana benthamiana plants were transiently transformed with control, 35S-VvCYP710A1-GFP, and 35S-VvCYP51G1a-GFP. Images under the blight field (middle), fluorescence (left) and the merged images are shown on the right. Bar: 30 μm.
Discussion
The cytochrome P450 (CYP) monooxygenase superfamily, belonging to heme-thiolate protein products, plays a vital role in metabolizing physiologically important compounds in plants (Werck-Reichhart & Feyereisen, 2000). To date, many CYP genes have been reported in some studies with genome-wide characterization, such as, A. thaliana (Paquette et al., 2000), O. sativa (Wei and Chen, 2018), P. trichocarpa (Nelson et al., 2008), and M. notabilis (Ma et al., 2014). Nevertheless, no comprehensive analyses of CYP TFs in V. vinifera have been done. The present study is the first to report on the genome-wide identification of VvCYPs in grapevine, together with evolutionary relations, exon–intron organizations, conservative motifs, cis-regulatory elements, gene duplications, and syntenic analysis. Additionally, RNA-seq databases of numerous grapevine tissues were used to determine the expression pattern of all VvCYPs, and the selected pivotal genes affecting fruit development and maturity were further verified by qRT-PCR assay.
CYP Superfamily in Vitis vinifera and Their Evolution
In the present study, a sum of 236 VvCYPs were identified from the grapevine genome assigned into 46 families, and the number of VvCYP members was less than those of Arabidopsis (245), O. sativa (326), and P. trichocarpa (310), and larger than in M. notabilis (174), and C. papaya (142) (Nelson et al., 2004; Nelson et al., 2008). In contrast, 245 AtCYPs in A. thaliana were classified into 47 families, of which only CYP718 was not identified in grapevine. The OsCYPs in O. sativa were classified into 45 families, of which seven (CYP729, CYP733, CYP727, CYP728, CYP92, CYP723, and CYP99) were not found in grapevine. Eight families (CYP708, CYP702, CYP720, CYP716, CYP712, CYP705, CYP83, and CYP82) were not present in O. sativa. The quantities of CYP genes in Arabidopsis (245) and O. sativa (326) were almost 1.04 and 1.38 times more than that in V. vinifera, which were inconsistent with the coding genes in the Arabidopsis genome (35,386 genes, 135 Mb) and O. sativa genome (52,424 genes, 372 Mb) which were about 1.34 and 1.99 times more than that in grapevine (26,346 genes, 487 Mb) (Wang et al., 2012). It was observed that there were more segmental/WGD-type CYP genes in Arabidopsis and O. sativa, suggesting that the enlargement of CYP genes in Arabidopsis and O. sativa might be due to genome duplication events. Therefore, we consider that the difference of genome duplication events may be the main reason for this inconsistency. As shown in Figure 1, VvCYPs were clustered into nine clans with different functions. VvCYPs, which fell into the same clan, probably diverged from a communal ancestor.
Clan 74 which depended on neither NADPH nor molecular oxygen (O2) to catalyze interior oxygen transference have possibly arisen in early photosynthetic organisms when O2 pressure was still low on the Earth (Stumpe and Feussner, 2006). Clan 51 and clan 85 were clustered together on the phylogenetic tree, the sector may have evolved from a sterol-metabolizing CYP51 ancestor. The members of clan 85 may participate in terpenoid-related pathway, ABA catabolism, and BR biosynthesis (Vriet et al., 2013). We deduced that three clans (clan 72, clan 97, and clan 86) clustering together on the phylogenetic tree may share a common ancestor. Clan 72 is associated with cytokinin (CTK) biosynthesis and the metabolisms of isoprenoid, fatty acid, and phytohormone (GA and BR) (Takei et al., 2004; Kandel et al., 2005; Sakamoto et al., 2011; Magome et al., 2013). Some members of Clan 86 can epoxidate and hydroxylate fatty acid, fatty alcohol, or alkane and their derivatives (Li et al., 2010; Heitz et al., 2012). Clan 711 and clan 71 may have a common origin, and CYP711s play a critical role in the strigolactones (SLs) synthesis (Zhang et al., 2014). The largest clan 71, which consisted of 19 families, indicates a huge functional diversity; it participated in the metabolisms of small isoprenoid and some triterpenoid derivatives, aliphatic and aromatic amino acid derivative, fatty acid, alkaloid and hormone (Sauveplane et al., 2009; Giddings et al., 2011; Wang et al., 2012). The above results provided some powerful evidences that the duplicates diverge to obtain novel functions.
Role of Cis-Elements in the Transcriptional Regulation of VvCYP Genes
As the key unit of transcriptional regulation, cis-elements participated in the regulation of molecular networks in many biological processes (Ibraheem et al., 2010). In terms of cis-regulatory element sequences, the promoters of VvCYPs have vastly repetitive regions and some common motifs. In this study, at least 33 cis-regulatory elements (ACE, Box 4, Sp1, G-Box, MRE, etc.) were found in the promoter of VvCYPs which are required for light-driven transcriptional regulation, implying that these genes played an important role in the light-responsive process. Additionally, the occurrence of the cis-element (circadian) indicated the significance of circadian regulation in VvCYP gene expression. The results of cis-elements analyses showed the presence of unique cis-regulatory elements in the promoter of only one VvCYP gene, hence deducing the specificity of these gene expression. The unique cis-regulatory elements were recognized due to the length of their sequences (6–15 bases) but not usually easy to generate some nucleotide variabilities. Some VvCYPs (e.g. VvCYP82C2b, VvCYP714A1a, VvCYP86A1a, and VvCYP94C1a) existed in ABA-responsive element (ABRE), which were in accord with the increased expression level of these VvCYPs under the ABA treatment. In grapevine, the endosperm plays vital roles in fruit setting as well as the early stage of berry development. Promoter analysis showed that 19 of the 236 VvCYPs that harbored GCN4_motif cis-elements participated in endosperm expression. Hence, it is concluded that these VvCYPs are responsible for fruit development in grapevine.
The VvCYP Superfamily Arose Mainly Through Tandem Duplication, Accompanied by Singleton Duplication
Gene duplication is well known to be the initial dynamic force of new functions in the phylogeny of genomes and genetic systems, which is one of the main evolutionary mechanisms further leading to the divergence and speciation (Lynch, 2000; Moore and Purugganan, 2003). In the current investigations of the grapevine genome, 236 VvCYPs were further categorized into 46 families and clustered into 9 clans, mapping in 19 chromosomes or some scaffolds (Figures 1 and 3). Based on gene duplication analysis, 114 of all VvCYPs in grapevine were duplicated from tandem event, compared with 47 from the singleton event, and 38 from the proximal event, 26 from the dispersed event, and 11 from the WGD/segmental event. The results indicated that during the evolutionary process, some VvCYPs have increased rapidly, in which tandem duplication is the major mechanism for VvCYP expansion, followed by singleton duplication. However, a diverse phenomenon has been found in the OsCYP family of rice (Yin et al., 2013), where majority of the OsCYP genes arose through segmental duplication on corresponding chromosomal regions in genetic linkage maps, which is consistent with our findings in grapevine, indicating that the difference of expansion mechanism in grapevine and rice.
Potential Roles of VvCYP Genes in Vitis vinifera
Many evidences have already suggested that CYP genes are involved in various biochemical pathways and play multiple roles in the process of growth and development in plants (Werck-Reichhart and Feyereisen, 2000). We attempt to investigate the functions of VvCYPs in specific organ and development stage through expression analysis. Some VvCYPs were highly expressed in particular flower organs, such as VvCYP81D5, VvCYP89A2i, VvCYP94D1, VvCYP84A1c, VvCYP74B2, and VvCYP73A5a in stamen, VvCYP707A1b, VvCYP716A1b, VvCYP82C4d, VvCYP89A2a, and VvCYP89A2b in carpel, VvCYP96A1c, VvCYP72A1s, VvCYP71A25, VvCYP74B2, and VvCYP77A4 in petal, VvCYP89A2a, VvCYP82C2k, VvCYP82C2d, VvCYP94D1, and VvCYP71B10e in pollen (Figure 7). Multiple pieces of evidence suggested that AtCYP704B1, AtCYP86C3, and AtCYP703A2 catalyzed the hydroxylation of mid- and long-chain fatty acids during pollen exine formation (Xu et al., 2014; Yang et al., 2014; Shi et al., 2015). VvCYP704A2 shares 67.30% nucleotide sequence identity with AtCYP704B1, suggesting a close evolutionary and functional relationship between VvCYP704A2 and AtCYP704B1. VvCYP703A2 has 69.13% sequence identity with AtCYP703A2, indicating a close functional relationship that functions as the hydroxylation of fatty acid (Yang et al., 2014). VvCYP86A1a has 69.36% sequence identity with AtCYP86A8 that functions as a fatty acid ω-hydroxylase involved in cutin biosynthesis (Wellesen et al., 2001). Additionally, many VvCYP genes were highly expressed in vegetative organs, such as VvCYP51G1a, VvCYP81D2b, VvCYP89A2b, VvCYP76C1d, and VvCYP73A5a in roots, VvCYP51G1a, VvCYP74A1d, VvCYP74B2, VvCYP73A5a and VvCYP98A3 in stems, VvCYP71A25 and VvCYP74B2 in leaves, VvCYP51G1a, VvCYP77A4 and VvCYP73A5a in buds.
Leaf development and morphogenesis are regulated by phytohormones, transcriptional regulators, and mechanical properties of the tissue. To explore the VvCYP roles in the processes, qRT-PCR was used to quantify the expression levels along with the different developmental stages (young, medium, and old leaves). The transcripts of seven VvCYPs (VvCYP79A2a, VvCYP72A10a, VvCYP90A1, VvCYP86A1a, VvCYP97B3, VvCYP710A1 and VvCYP711A1b) were down-regulated toward leaf senescence, which suggested that they might function mainly in the upper period of leaf development. VvCYP87A2c has 51.96% sequence identity with OsCYP87A6 that was up-regulated in response to light and auxin signaling and in turn decreased local auxin accumulation in coleoptiles (Chaban et al., 2003). VvCYP708A2, a homolog of AtCYP708A1 which functions upstream of AtCYP705A5 and downstream of AtOSC in the biosynthesis of oxygenated triterpenoids (Field and Osbourn, 2008). VvCYP94C1a, a homolog of AtCYP94C1 that encodes a carotenoid ϵ-ring hydroxylase catalyzing the formation of lutein which modulates light energy and serves as a nonphotochemical quenching agent to deal with triplet chlorophyll (Tian et al., 2004; Ding et al., 2017), was expressed at relatively high levels. VvCYP711A1b is a homolog of OsCYP711A5 which may catalyze the biosynthesis of SLs which are carotenoid-derived plant hormones and accelerate leaves' maturity and senescence (Ueda and Kusaba, 2015). Among them, VvCYP73A5a has 74.31% sequence identity with AtCYP73A5 which catalyzes the hydroxylation of t-cinnamic acid, a key early step in the pathway (Pan et al., 2009). Additionally, VvCYP89A2i is homologous with AtCYP89A9 which is implicated in the formation of major chlorophyll catabolites during leaf senescence (Christ et al., 2013). Previous reports suggested that ABA was associated with the regulation of nonclimacteric fruit ripening (Coombe, 1992; Giovannoni, 2001; Li et al., 2011). In the present study, some VvCYP transcripts (VvCYP82C2b, VvCYP714A1a, VvCYP86A1a, VvCYP94C1a, VvCYP96A1a, and VvCYP711A1b) were increased with ABA treatment; however, VvCYP79A2a, VvCYP93D1a, VvCYP84A1a, and VvCYP72A11a had down-regulated expression after ABA treatment at different berry development stages, implying they might play distinct roles in berry development in response to ABA.
Data Availability Statement
All relevant data is contained within the article.
Author Contributions
SJ carried out the experiments, prepared the figures and wrote the manuscript. YX analyzed the data and prepared figures. JW, MA, and LW participated in the collinearity analysis and data analysis. WS and XL contributed to sample collection. IS, CM, and WX contributed with the consultation. CZ managed and designed the research and experiments. CZ and SW revised the manuscript. All authors read and approved the final manuscript.
Funding
The study was supported by grants from National Postdoctoral Program for Innovative Talents (Grant No. BX20180199), the National Key Research and Development Plan project (Grant No. 2018YFD0201305), China Postdoctoral Science Foundation (Grant No. 2018M642028).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers and editor for their careful reading and helpful comments on this manuscript. We also thank associate professor Xu Wang from Shanghai Jiao Tong University for his help in English proofreading the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.00044/full#supplementary-material
References
Ai, J., Zhu, Y., Duan, J., Yu, Q., Zhang, G., Wan, F., et al. (2011). Genome-wide analysis of cytochrome P450 monooxygenase genes in the silkworm. Bombyx mori. Gene 480, 0–50. doi: 10.1016/j.gene.2011.03.002
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Boss, P. K., Davies, C., Robinson, S. P. (1996). Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv shiraz grape berries and the implications for pathway regulation. Plant Physiol. 111, 1059–1066. doi: 10.1104/pp.111.4.1059
Cai, S., Giang, G., Ye, N., Chu, Z., Xu, X., Zhang, J., et al. (2015). A key ABA catabolic gene, OsABA8ox3, is involved in drought stress resistance in rice. PloS One 10, e0116646. doi: 10.1371/journal.pone.0116646
Chaban, C., Waller, F., Furuya, M., Nick, P. (2003). Auxin responsiveness of a novel cytochrome P450 in rice coleoptiles. Plant Physiol. 133, 2000–2009. doi: 10.1104/pp.103.022202
Chen, Y., Liu, L., Shen, Y., Liu, S., Huang, J., Long, Q., et al. (2015). Loss of function of the cytochrome P450 gene CYP78B5 causes giant embryos in rice. Plant Mol. Biol. Rep. 33, 69–83. doi: 10.1007/s11105-014-0731-3
Christ, B., Iris, S., Moser, S., Bichsel, N., Egert, A., Thomas, M., et al. (2013). Cytochrome P450 CYP89A9 is involved in the formation of major chlorophyll catabolites during leaf senescence in Arabidopsis. Plant Cell 25, 1868–1880. doi: 10.1105/tpc.113.112151
Coombe, B. G. (1992). Research on development and ripening of the grape berry. Am. J. Enol. Viticul. 43, 120–127. doi: 10.1016/S0065-2164(08)70256-9
Ding, F., Wang, M., Liu, B., Zhang, S. (2017). Exogenous melatonin mitigates photoinhibition by accelerating non-photochemical quenching in tomato seedlings exposed to moderate light during chilling. Front. Plant Sci. 8, 244. doi: 10.3389/fpls.2017.00244
Emanuelsson, O., Nielsen, H., Brunak, S., von Heijine, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Fasoli, M., Santo, S. D., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. doi: 10.1105/tpc.112.100230
Field, B., Osbourn, A. E. (2008). Metabolic diversification–independent assembly of operon-like gene clusters in different plants. Science 320, 543–547. doi: 10.1126/science.1154990
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Giddings, L. A., Liscombe, D. K., Hamilton, J. P., Childs, K. L., DellaPenna, D., Buell, C. R., et al. (2011). A stereoselective hydroxylation step of alkaloid biosynthesis by a unique cytochrome P450 in Catharanthus roseus. J. Biol. Chem. 286, 16751–16757. doi: 10.1074/jbc.M111.225383
Giovannoni, J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 725–749. doi: 10.1146/annurev.arplant.52.1.725
Heitz, T., Widemann, E., Lugan, R., Miesch, L., Ullmann, P., Desaubry, L., et al. (2012). Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287, 6296–6306. doi: 10.1074/jbc.M111.316364
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ibraheem, O., Botha, C. E., Bradley, G. (2010). In silico analysis of cis-acting regulatory elements in 5' regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 34, 268–283. doi: 10.1016/j.compbiolchem.2010.09.003
Ito, T., Meyerowitz, E. M. (2000). Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell 12, 1541–1550. doi: 10.1105/tpc.12.9.1541
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jiu, S., Zhu, X., Wang, J., Zhang, C., Fang, J. (2015). Genome-wide mapping and analysis of grapevine microRNAs and their potential target genes. Plant Genome 8, 127–132. doi: 10.3835/plantgenome2014.12.0091
Jiu, S. T., Wang, C., Zheng, T., Liu, Z., Leng, X. P., Pervaiz, T., et al. (2016). Characterization of VvPAL-like promoter from grapevine using transgenic tobacco plants. Funct. Integr. Genomic 16, 595–617. doi: 10.1007/s10142-016-0516-x
Jiu, S., Haider, M. S., Kurjogi, M. M., Zhang, K., Zhu, X., Fang., J. (2018). Genome-wide characterization and expression analysis of sugar transporter family genes in woodland strawberry. Plant Genome 11, 170103. doi: 10.3835/plantgenome2017.11.0103
Kandel, S., Morant, M., Benveniste, I., Blee, E., Werck-Reichhart, D., Pinot, F. (2005). Cloning, functional expression, and characterization of CYP709C1, the first sub-terminal hydroxylase of long chain fatty acid in plants. J. Biol. Chem. 280, 35881–35889. doi: 10.1074/jbc.M500918200
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, M. S., Babu, P. R., Rao, K. V., Reddy, V. D. (2014). Organization and classification of cytochrome p450 genes in castor (Ricinus communis L.). Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 84, 131–143. doi: 10.1007/s40011-013-0192-8
Lescot, M. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, L., Cheng, H., Gai, J., Yu, D. (2007). Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 226, 109–123. doi: 10.1007/s00425-006-0473-z
Li, H., Pinot, F., Sauveplane, V., Danièle, W., Zhang, D. (2010). Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in Rice. Plant Cell 22, 173–190. doi: 10.1105/tpc.109.070326
Li, C., Jia, H., Chai, Y., Shen, Y. (2011). Abscisic acid perception and signaling transduction in strawberry: a model for non-climacteric fruit ripening. Plant Signaling Behav. 6, 1950–1953. doi: 10.4161/psb.6.12.18024
Liu, Z. H., Boachon, B., Luqan, R., Tavares, R., Erhardt, M., Mutterer, M., et al. (2015). A conserved cytochrome P450 evolved in seed plants regulates flower maturation. Mol. Plant 8, 1751–1765. doi: 10.1016/j.molp.2015.09.002
Lynch, M. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Ma, B., Luo, Y. W., Jia, L., Qi, X. W., Zeng, Q. W., Xiang, Z. H., et al. (2014). Genome-wide identification and expression analyses of cytochrome P450 genes in mulberry (Morus notabilis). J. Integr. Plant Biol. 56, 887–901. doi: 10.1111/jipb.12141
Magome, H., Nomura, T., Hanada, A., Takeda-Kamiya, N., Ohnishi, T., Shinma, Y., et al. (2013). CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. U.S.A. 110, 1947–1952. doi: 10.1073/pnas.1215788110
Maher, C., Stein, L., Ware, D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16, 510–519. doi: 10.1101/gr.4680506
Mao, G., Timothy, S., Denyse, S., Yu, O. (2013). CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana. BMC Plant Biol. 13, 169. doi: 10.1186/1471-2229-13-169
Mizutani, M., Ohta, D. (2010). Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315. doi: 10.1146/annurev-arplant-042809-112305
Moore, R. C., Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Nelson, D. R., Koymans, L., Kamataki, T., Stegeman, J. J., Feyereisen, R., Waxman, D. J., et al. (1996). P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6, 1–42. doi: 10.1097/00008571-199602000-00002
Nelson, D. R., Schuler, M. A., Paquette, S. M., Werck-Reichhart, D., Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772. doi: 10.1104/pp.104.039826
Nelson, D. R., Ming, R., Alam, M., Schuler, M. A. (2008). Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 1, 216–235. doi: 10.1007/s12042-008-9022-1
Nelson, D. R. (2011). Progress in tracing the evolutionary paths of cytochrome P450. Biochim. Biophys. Acta 1814, 14–18. doi: 10.1016/j.bbapap.2010.08.008
Ogrodzki, P., Forsythe, S. (2015). Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genomics 16, 1–15. doi: 10.1186/s12864-015-1960-z
Omura, T. (1999). Forty years of cytochrome P450. Biochem. Biophys. Res. Commun. 266, 690–698. doi: 10.1006/bbrc.1999.1887
Pan, Y., Michael, T. P., Hudson, M. E., Kay, S. A., Chory, J., Schuler, M. A. (2009). Cytochrome P450 monooxygenases as reporters for circadian-regulated pathways. Plant Physiol. 150, 858–878. doi: 10.1104/pp.108.130757
Paquette, S. M., Bak, S., Feyereisen, R. (2000). Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 19, 307–317. doi: 10.1089/10445490050021221
Persans, M. W., Wang, J., Schuler, M. A. (2001). Characterization of maize cytochrome P450 monooxygenases induced in response to safeners and bacterial pathogens. Plant Physiol. 125, 1126–1138. doi: 10.1104/pp.125.2.1126
Ramamoorthy, R., Jiang, S. Y., Ramachandran, S. (2011). Oryza sativa cytochrome P450 family member OsCYP96B4 reduces plant height in a transcript dosage dependent manner. PloS One 6, e28069. doi: 10.1371/journal.pone.0028069
Robert, P. D., Douglas, G. L. (1991). Multiple forms of plant cytochromes P450. Plant Physiol. 96, 669–674. doi: 10.1104/pp.96.3.669
Sakamoto, T., Kawabe, A., Tokida-Segawa, A., Shimizu, B. I., Takatsuto, S., Shimada, Y., et al. (2011). Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 67, 1–12. doi: 10.1111/j.1365-313X.2011.04567.x
Sauveplane, V., Kandel, S., Kastner P, E., Ehlting., J., Compagnon, V., Werck-Reichhart, D., et al. (2009). Arabidopsis thaliana CYP77A4 is the first cytochrome P450 able to catalyze the epoxidation of free fatty acids in plants. FEBS J. 276, 719–735. doi: 10.1111/j.1742-4658.2008.06819.x
Shi, J., Cui, M., Yang, L., Kim, Y. J., Zhang, D. (2015). Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20, 741–753. doi: 10.1016/j.tplants.2015.07.010
Stumpe, M., Feussner, I. (2006). Formation of Oxylipins by CYP74 enzymes. Phytochem. Rev. 5, 347–357. doi: 10.1007/s11101-006-9038-9
Takase, H., Sasaki, K., Shinmori, H., Shinohara, A., Takata, R. (2015). Cytochrome P450 CYP71BE5 in grapevine (vitis vinifera) catalyzes the formation of the spicy aroma compound (-)-rotundone. J. Exp. Bot. 67, 787–798. doi: 10.1093/jxb/erv496
Takei, K., Yamaya, T., Sakakibara, H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of transZeatin. J. Biol. Chem. 279, 41866–41872. doi: 10.1074/jbc.M406337200
Tamiru, M., Undan, J. R., Takagi, H., Abe, A., Yoshida, K., Undan, J. Q., et al. (2015). A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in Rice (Oryza sativa L.). Plant Mol. Biol. 88, 85–99. doi: 10.1007/s11103-015-0310-5
Tian, L., Musetti, V., Kim, J., Magallanes-Lundback, M., DellaPenna, D. (2004). The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid ϵ-ring hydroxylation activity. Proc. Natl. Acad. Sci. U.S.A. 101, 402–407. doi: 10.1073/pnas.2237237100
Ueda, H., Kusaba, M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 169, 138–147. doi: 10.1104/pp.15.00325
Velasco, R., Zharkikh, A., Troggio, M., Cartwright, D. A., Cestaro, A., Pruss, D., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PloS One 2, e1326. doi: 10.1371/journal.pone.0001326
Vriet, C., Russinova, E., Reuzeau, C. (2013). From squalene to brassinolide: the steroid metabolic and signaling pathways across the plant kingdom. Mol. Plant 6, 1738–1757. doi: 10.1093/mp/sst096
Wang, Q., Hillwig, M. L., Wu, Y., Peters, R. J. (2012). CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 158, 1418–1425. doi: 10.1104/pp.111.187518
Wang, X., Cheng, Z., Zhao, Z., Gan, L., Qin, R., Zhou, K., et al. (2016). BRITTLE SHEATH1 encoding OsCYP96B4 is involved in secondary cell wall formation in rice. Plant Cell Rep. 35, 745–755. doi: 10.1007/s00299-015-1916-4
Wei, K., Chen, H. (2018). Global identification, structural analysis and expression characterization of cytochrome P450 monooxygenase superfamily in rice. BMC Genomics 19, 35. doi: 10.1186/s12864-017-4425-8
Wellesen, K., Durst, F., Pinot, F., Benveniste, I., Nettesheim, K., Wisman, E., et al. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc. Natl. Acad. Sci. USA. 98, 9694–9699. doi: 10.1073/pnas.171285998
Werck-Reichhart, D., Feyereisen, R. (2000). Cytochromes P450: a success story. Genome Biol. 1, 1–9. doi: 10.1186/gb-2000-1-6-reviews3003
Werck-Reichhart, D. (1995). Cytochromes P450 in phenylpropanoid metabolism. Drug Metab. Drug Interact. 12, 221–243. doi: 10.1515/DMDI.1995.12.3-4.221
Xu, J., Ding, Z., Vizcay-Barrena, G., Shi, J., Liang, W., Yuan, Z., et al. (2014). ABORTED MICROSPORES Acts as a master regulator of Pollen Wall formation in Arabidopsis. Plant Cell 26, 1544–1556. doi: 10.1105/tpc.114.122986
Yang, X., Wu, D., Shi, J. X., He, Y., Piont, F., Grausem, B., et al. (2014). Rice CYP703A3, a Cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 56, 979–994. doi: 10.1111/jipb.12212
Keywords: genome-wide, cytochrome P450 gene superfamily, phylogeny analysis, expression analysis, grapevine
Citation: Jiu S, Xu Y, Wang J, Wang L, Liu X, Sun W, Sabir Ia, Ma C, Xu W, Wang S, Abdullah M and Zhang C (2020) The Cytochrome P450 Monooxygenase Inventory of Grapevine (Vitis vinifera L.): Genome-Wide Identification, Evolutionary Characterization and Expression Analysis. Front. Genet. 11:44. doi: 10.3389/fgene.2020.00044
Received: 19 November 2019; Accepted: 15 January 2020;
Published: 18 February 2020.
Edited by:
Santosh Kumar Upadhyay, Panjab University, IndiaReviewed by:
Krishan M. Rai, Texas Tech University, United StatesSantosh B. Satbhai, Indian Institute of Science Education and Research Mohali, India
Copyright © 2020 Jiu, Xu, Wang, Wang, Liu, Sun, Sabir, Ma, Xu, Wang, Abdullah and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caixi Zhang, acaizh@sjtu.edu.cn
†These authors have contributed equally to this work
 Songtao Jiu
Songtao Jiu Yan Xu†
Yan Xu† Muhammad Abdullah
Muhammad Abdullah