- 1Institute of Eastern-Himalaya Biodiversity Research, Dali University, Dali, Yunnan, China
- 2Erhai Watershed Ecological Environment Quality Testing Engineering Research Center of Yunnan Provincial Universities, Erhai Research Institute, West Yunnan University of Applied Sciences, Dali, Yunnan, China
- 3Colledge of Life Science, China West Normal University, Nanchong, Sichuan, China
- 4The Provincial Innovation Team of Biodiversity Conservation and Utility of the Three Parallel Rivers Region from Dali University, Dali, Yunnan, China
To cope with the damage from oxidative stress caused by hypoxia, mammals have evolved a series of physiological and biochemical traits, including antioxidant ability. Although numerous research studies about the mechanisms of hypoxia evolution have been reported, the molecular mechanisms of antioxidase-related genes in mammals living in different environments are yet to be completely understood. In this study, we constructed a dataset comprising 7 antioxidase-related genes (CAT, SOD1, SOD2, SOD3, GPX1, GPX2, and GPX3) from 43 mammalian species to implement evolutionary analysis. The results showed that six genes (CAT, SOD1, SOD2, SOD3, GPX1, and GPX3) have undergone divergent evolution based on the free-ratio (M1) model. Furthermore, multi-ratio model analyses uncovered the divergent evolution between hypoxic and non-hypoxic lineages, as well as various hypoxic lineages. In addition, the branch-site model identified 9 positively selected branches in 6 genes (CAT, SOD1, SOD2, SOD3, GPX2, and GPX3) that contained 35 positively selected sites, among which 31 positively selected sites were identified in hypoxia-tolerant branches, accounting for 89% of the total number of positively selected sites. Interestingly, 65 parallel/convergent sites were identified in the 7 genes. In summary, antioxidase-related genes are subjected to different selective pressures among hypoxia-tolerant species living in different habitats. This study provides a valuable insight into the molecular evolution of antioxidase-related genes in hypoxia evolution in mammals.
1 Introduction
Adaptation to extreme environments is an important research field in the molecular ecology, physiological ecology, and evolutionary biology of animals. Organisms usually face environmental challenges, such as low oxygen, aridity, high ultraviolet radiation, low pressure, and extreme temperatures, of which hypoxia-tolerant species adapt to poor oxygen (O2) levels in aquatic, terrestrial highland, or cave environments (Nathaniel et al., 2015). Hypoxia tolerance is the adaptive ability of organisms to cope with acute and chronic hypoxia with the production of reactive oxygen species (ROS) (Ramirez et al., 2007). ROS play key roles in the body, such as signaling between cells, immunity, and participation in normal physiological reactions (Nose, 2000; Dröge, 2002), but excessive ROS produce certain oxidative damage pressure on the body (Kaelin, 2005). Consequently, numerous species have evolved various antioxidant mechanisms, including non-enzymatic and endogenous antioxidants, to adapt to peroxidative damage in hypoxic environments (Tian et al., 2021a). Among these mechanisms, the antioxidant system is crucial for counteracting the effects of hypoxic environments in hypoxia-tolerant animals. It is primarily composed of superoxide dismutases (SODs), catalase (CAT), glutathione peroxidases (GPXs), glutathione S-transferases (GSTs), and other constituents (Dröge, 2002; Wilhelm Filho et al., 2002; Birben et al., 2012).
Mammals are a highly diverse group distributed across different habitats worldwide. Many lineages inhabit hypoxic environments, including aquatic and terrestrial. Aquatic hypoxia-tolerant species can be categorized as semi-aquatic (e.g., pinnipeds) or fully aquatic (e.g., cetaceans), and terrestrial hypoxia-tolerant species primarily inhabit caves (e.g., mole rats) and high-altitude regions (e.g., yak). Previous studies showed that the activity of antioxidases is significantly higher in fully aquatic cetaceans than in terrestrial mammals (Dröge, 2002; Birben et al., 2012). In addition, semi-aquatic elephant seals (Mirounga leonina) also exhibited higher antioxidant enzyme activities than terrestrial mammals (Wilhelm Filho et al., 2002). In some terrestrial hypoxia-tolerant species, the antioxidase activity has also shown a similar trend, such as the activity of CAT and SOD being significantly higher in the heart and liver of Tibetan pigs than in DLY pigs (the Duroc × (Landrace × Yorkshire) hybrid pig) (Hu et al., 2022), as well as upward levels of Cu/Zn-SOD, Mn-SOD, and CAT in naked mole rats (Heterocephalus glaber) (Pamenter, 2022).
Researchers have uncovered some related genes or gene families that play a role in the adaptive evolutionary mechanisms of hypoxia-tolerant mammals. Tian et al. (2021b) found that the SOD gene family has undergone adaptive evolution in cetaceans. Additionally, gene duplication of GPXs occurred, and peroxidase gene families (PRDX1 and PRDX3) expanded in the cetacean lineage (Yim et al., 2014; Zhou et al., 2018; Tian et al., 2021a). In addition, the genomes of the Tibetan antelope (Pantholops hodgsonii) showed that 247 positively selected genes are related to hypoxia tolerance, and some of them are connected with antioxidant pathways (Ge et al., 2013). Moreover, marine mammals and highland animals have evolved a convergent capacity to modulate their metabolic levels to adapt to hypoxic environments by regulating the expression of genes associated with hypoxia adaptation (Bigham et al., 2013). Generally, several studies have extensively examined the antioxidant evolutionary mechanisms among hypoxia-tolerant species. However, we need further research to understand the evolutionary pattern of different hypoxia-tolerant mammalian lineages, such as divergent evolution.
To provide further molecular evidence for the adaptive evolution of antioxidase systems in hypoxia-tolerant mammals, seven antioxidase-related genes (CAT, SOD1, SOD2, SOD3, GPX1, GPX2, and GPX3) of mammals in different habitats were used to perform an in silico analysis to solve the following scientific issues: 1) whether the antioxidase-related genes have different evolutionary patterns and 2) whether convergent evolution of antioxidase-related genes occurs in hypoxia-tolerant mammals living in diverse environments. Our study provides insights into the genetic mechanisms underlying hypoxia tolerance adaptations in mammals.
2 Materials and methods
2.1 Sequence acquisition
A total of 43 mammalian species (including hypoxia-tolerant and non-hypoxia-tolerant mammals) were selected, which covered major groups, such as Cetartiodactyla, Carnivora, Primates, and Rodentia. The gene sequences of most species were obtained from the NCBI (https://www.ncbi.nlm.nih.gov/), and the sequences of some species were obtained through the NCBI genome BLAST (genome information given in Supplementary Table S1); the sequence of the bowhead whale (Balaena mysticetus) was obtained from the website http://www.bowhead-whale.org/. The coding sequences were aligned by using webPRANK (https://www.ebi.ac.uk/goldman-srv/webprank/) (Löytynoja and Goldman, 2010). We corrected the multiple sequence alignment (MSA) in MEGA 7 by eyes (Kumar et al., 2016).
2.2 Selection pressure analysis
Selection pressure analysis is mainly based on the ratio ω of nonsynonymous (dN) to synonymous substitution (dS) of coding sequences, which is used as the main basis for detecting natural selection: ω > 1, = 1, or <1 indicates that the gene is subjected to positive, neutral, or purifying selection, respectively. The ω value was evaluated using the CodeML program in the PAML 4.9 package (Yang, 2007). The likelihood ratio test statistic (2ΔL), which approximates a chi-square (χ2) distribution, was used to compare nested likelihood models. Positively selected signals were identified using BEB analysis with posterior probabilities (PPs) of ≥0.8 (Yang et al., 2005). We used the TimeTree database (http://www.timetree.org/) (Kumar et al., 2017) and a previous study (Murphy et al., 2004) to obtain the mammalian phylogenetic tree for subsequent analysis (Supplementary Figure S1).
2.2.1 Detection of positively selected sites
We performed site modeling (M8 and M8a) using the CodeML program in the PAML package to detect sites under positive selection (Swanson et al., 2003; Yang, 2007). To detect whether the positively selected sites were restricted to specific branching lineages, the strict branch-site model was used to detect the positive selection of specific sites that may affect specific branches in the datasets (Zhang et al., 2005).
2.2.2 Branch model analysis
To understand whether the adaptive evolution of antioxidase-related genes occurs in different branches, the free-ratio (M1) model was used to examine the evolutionary rate of each branch and compared with the one-ratio model (M0), which allows only a single ω ratio for all branches.
To analyze these differences, branch models were employed using the “two-ratio (2ω),” “three-ratio (3ω),” and “five-ratio (5ω)” models, which were implemented in CodeML (Yang, 1998) (Supplementary Figure S1A–C). First, to test whether different selective pressures act on the hypoxia-tolerant and non-hypoxia-tolerant mammalian lineages, the one-ratio model that enforces the same ω ratio for all lineages was compared with the 2ω model that allows one ω ratio for all hypoxia-tolerant mammal branches and another for all remaining branches (non-hypoxia-tolerant species) (Supplementary Figure S1A). Second, to explore the rate variation between terrestrial and aquatic hypoxia-tolerant species, we used the 3ω model, which assumes independent ω values for terrestrial and aquatic hypoxia-tolerant species and the remaining lineages (Supplementary Figure S1B). Finally, the 5ω model was used to detect the differences in evolutionary rates among high-altitude, cave, fully aquatic, and semi-aquatic hypoxia-tolerant species, which was compared with the 3ω model (Supplementary Figure S1C).
2.3 Parallel/convergent evolution analysis
To investigate whether convergent adaptation was undergone in antioxidase genes among related hypoxic branches, a parallel/convergent amino acid substitution analysis was performed. First, the ancestral amino acid sequences in each gene dataset were reconstructed using the M0 model approach implemented in the CodeML program of the PAML package (Yang, 2007). We then identified parallel/convergent amino acid replacement sites between branches with convergent hypoxia tolerance (branches a–o, Figure 1). Subsequently, CONVERGE 2 was used to test whether the parallel/convergent substitutions observed in the focal branches were fixed randomly or through natural selection (Zhang and Kumar, 1997). A statistical test was conducted to compare the observed number of parallel/convergent amino acid substitutions against the expected value. When p < 0.05, it meansthe convergent/parallel substitution have been driven by natural selection, rather than random mutation.
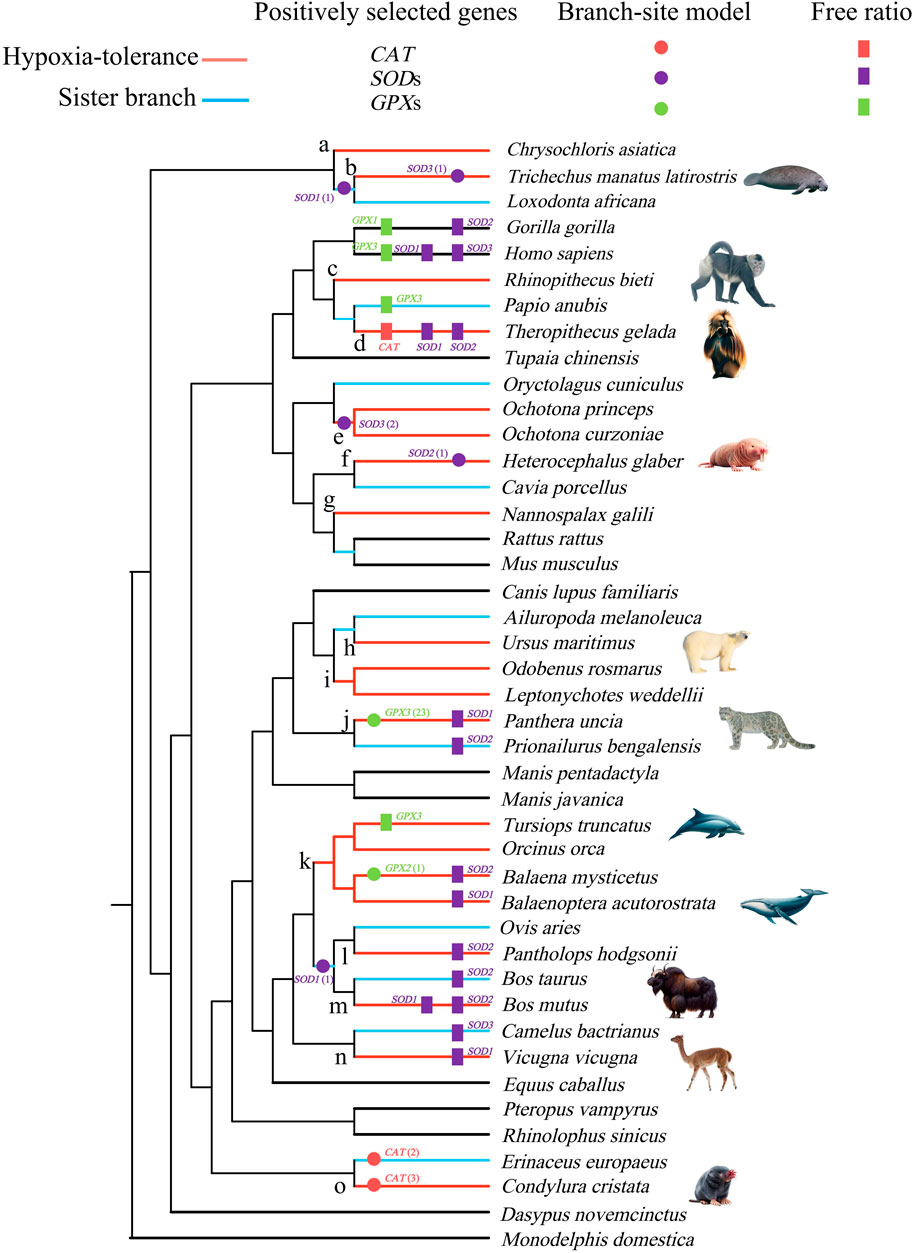
Figure 1. Positively selected lineages of catalase (CAT), superoxide dismutase (SOD), and glutathione peroxidase (GPX) genes identified by the free-ratio (rectangle) and branch-site model (circle). Branches a–o in the tree were used for the detection of convergent/parallel amino acid substitutions. The numbers in brackets behind the circle label in the tree represent the number of positively selected sites. All images are obtained from the Animal Diversity website (https://animaldiversity.org/).
2.4 Protein three-dimensional structure analysis
To gain an insight into the functional implications of the detected positively selected sites, as well as the parallel/convergent amino acid sites, we mapped these sites to the three-dimensional (3D) structures of each related protein downloaded from the AlphaFold website (https://alphafold.com/) using PyMOL (DeLano, 2002), while the GPX protein structures were predicted using the I-TASSER website (https://zhanggroup.org/I-TASSER/) (Zhang, 2008). Human CAT, SODs, and GPXs were regarded as query for the analysis. Functional information about the sites and domains of each candidate protein was obtained from the UniProt website (http://www.uniprot.org/) (UniProt Consortium, 2019) and InterPro (https://www.ebi.ac.uk/interpro/) (Paysan-Lafosse et al., 2023).
3 Results
3.1 Selection pressure for antioxidase genes of hypoxia-tolerant mammals
The site model results showed that M8 was significantly better than the neutral model M8a only in the SOD1 gene, and 7 positively selected sites were detected (12, 40, 42, 46, 47, 53, and 64) (PP > 0.80) (Supplementary Table S2).
The M0 model showed that the ω values of CAT, SOD1, SOD2, SOD3, GPX1, GPX2, and GPX3 genes were 0.124, 0.261, 0.115, 0.129, 0.073, 0.062, and 0.132, respectively (Supplementary Table S3). In the free-ratio model, six genes (CAT, SOD1, SOD2, SOD3, GPX1, and GPX3) were significantly better than that in the M0 model. A total of 20 branches were detected whose ω values were significantly greater than 1, 11 of which were branches of hypoxia-tolerant species. Most positive signals were distributed in the SOD1 and SOD2 genes (Figure 1, Supplementary Table S4).
To evaluate whether positive selection acts on specific sites in hypoxia-tolerant species (branches) and their sister branches, the much stricter branch-site model was used. The results showed that 9 positively selected branches contained 35 positively selected sites in 6 genes (CAT, SOD1, SOD2, SOD3, GPX2, and GPX3). Of the 9 branches, 6 were hypoxia-tolerant, containing 31 positively selected sites that accounted for 89% of the total (Figure 1, Supplementary Table S5). CAT exhibits positive selection along the star-nosed mole (Condylura cristata) and hedgehog (Erinaceus europaeus), with three and two positively selected sites, respectively. SOD1 exhibited positive selection in the Florida manatee (Trichechus manatus latirostris) and African elephant (Loxodonta africana) ancestors and the lineages leading to the ancestors of sheep and cattle, with one positively selected site. Moreover, evidence for positive selection was also identified in SOD2, along with the naked mole rat (H. glaber), which contained only one positively selected site. SOD3 best fits the alternative model, along with the Florida manatee branch and the last common ancestor branch of Ochotona curzoniae and Ochotona princeps, accounting for one and two positively selected sites, respectively. GPX2 exhibited positive selection in the bowhead whale, with one positively selected site. However, GPX3 had 23 positively selected sites in the snow leopard (Panthera uncia) (PP > 0.8) (Figure 1, Supplementary Table S5).
3.2 Multiple ratio analyses of different hypoxia lineages
The 2ω model was used to explore whether there is evolutionary discrepancy between hypoxia-tolerant and non-hypoxia-tolerant lineages, and the results showed that the models exhibited significance in the CAT, SOD1, and GPX2 genes. The 3ω model was used to verify the evolutionary differences in hypoxia tolerance of related genes between terrestrial and aquatic species, and the results showed significance in SOD2 and GPX3 genes. In addition, the ω ratio for aquatic hypoxia-tolerant mammals was much higher than that of terrestrial environment hypoxia-tolerant mammals. Based on the 5ω model, only SOD1 exhibited model significance among various hypoxic environments (Table 1).
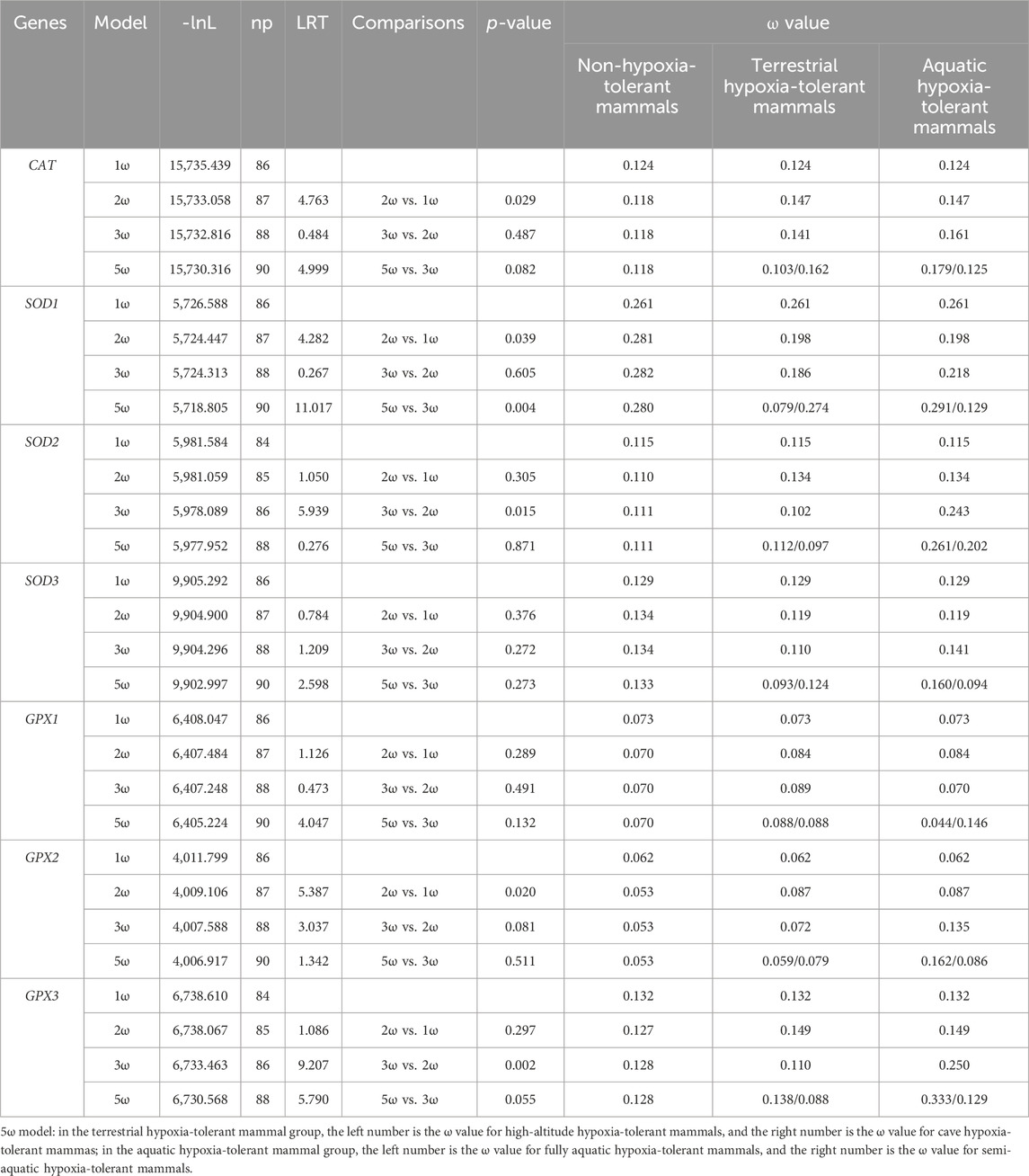
Table 1. Log-likelihood and omega values estimated under different branch models according to seven genes.
3.3 Convergent evolutionary mechanisms of hypoxia-tolerant lineages
A total of 64 parallel amino acid substitution sites and 1 convergent amino acid substitution site were detected in the hypoxia-tolerant branches of the 7 genes (Table 2). The CONVERGE 2 analysis showed p < 0.05. In addition, most of the parallel amino acid substitution sites were distributed in the branches, leading to tolerance to terrestrial hypoxia. In the high-altitude hypoxia-tolerant branches (branches d, e, j, l, and m), we detected 6 parallel amino acid substitution sites in three genes (SOD3, GPX1, and GPX3); 17 parallel amino acid substitution sites were detected in cave branches (a, g, f, and o) among 7 genes (CAT, SOD1, SOD2, SOD3, GPX1, GPX2, and GPX3). At high altitudes (branches c, d, e, and j) and in cave species (branches a, f, g, and o), 17 parallel amino acid substitution sites were detected in 6 genes (CAT, SOD1, SOD2, SOD3, GPX1, and GPX3). Only one convergent amino acid substitution site was detected in the SOD1 gene between the high-altitude and cave branches (branch a vs. e). Moreover, between fully aquatic mammals (branch b) and high-altitude species (branch e), we identified four parallel amino acid substitution sites in three genes (CAT, SOD3, and GPX1). In the high-altitude and semi-aquatic species, four parallel amino acid substitution sites were detected in four genes (SOD1, SOD3, GPX1, and GPX2). Among the cave species (branches a, f, g, and o) and fully aquatic mammals (branches b and k), nine parallel amino acid sites were detected in five genes (CAT, SOD1, SOD3, GPX1, and GPX3). In the semi-aquatic (branch i) and cave species (branches a, g, and o), four parallel amino acid substitution sites were detected in three genes (CAT, SOD3, and GPX1). Three parallel amino acid substitution sites in the semi-aquatic (branch i) and fully aquatic species (branches b and k) were identified in three genes (SOD3, GPX1, and GPX3) (Figure 1; Table 2).
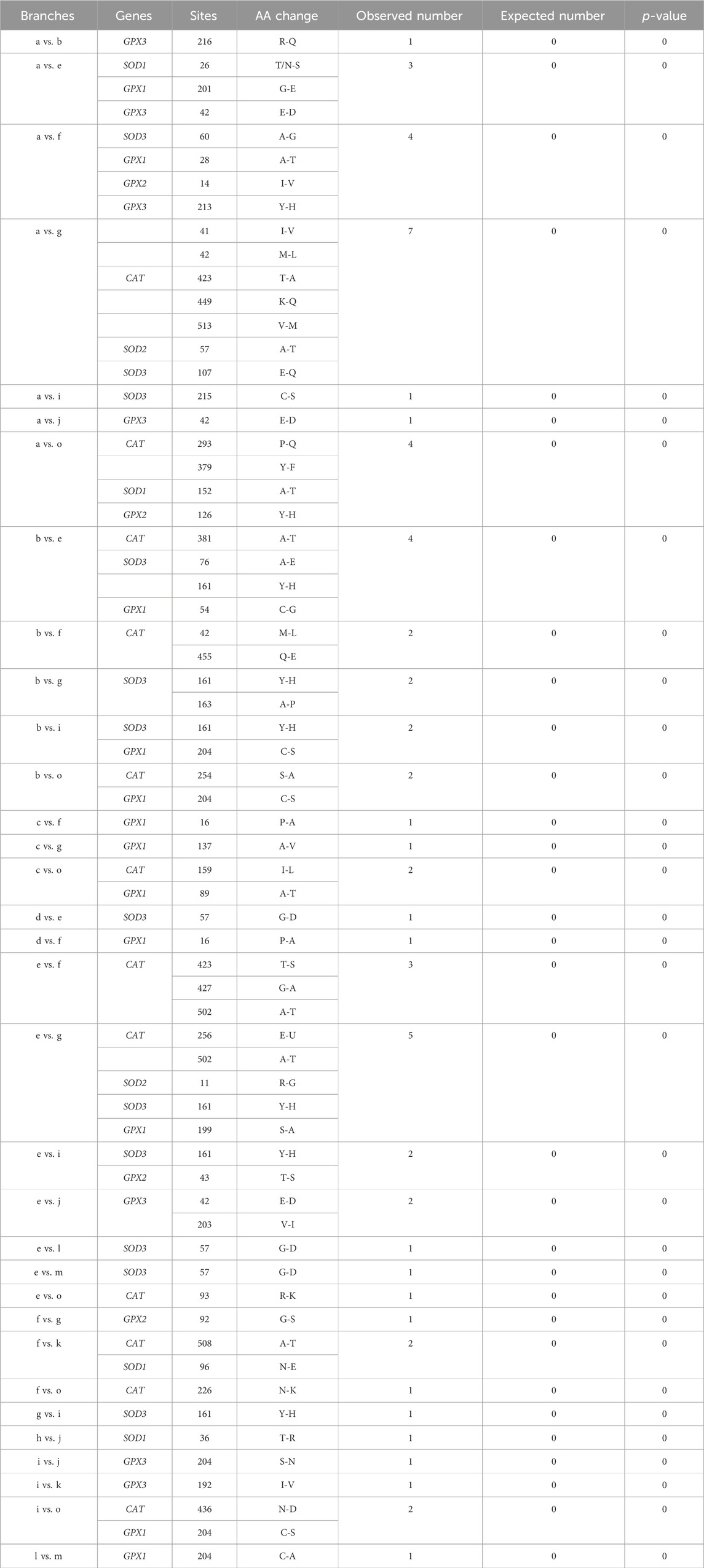
Table 2. 65 parallel/convergent amino acid substitutions sites in 7 genes in hypoxia-tolerant lineages.
3.4 3D structure analysis of antioxidase-related proteins
To gain an insight into the functional significance of the positively selected sites identified by the branch-site model and the parallel/convergent amino acid sites, we mapped these sites onto 3D structures (Figure 2, Supplementary Figure S2). In total, 21/35 positively selected sites and 19/65 convergent/parallel amino acid sites were located on important functional domains. For example, in CAT, we found that the positively selected site 444 was located next to the NADPH-binding site (445, 446). The convergent/parallel amino acid site 42 was located at the beta-strand position, site 93 was located at the turn position, and site 513 was located close to the modified residue site (511, 515). In SOD1, we found that the positively selected site 23 was located on many functionally important domains, such as the beta-strand and disulfide bond, close to the phosphoserine modification sites. Site 76 was located at the disulfide bond position. The convergent/parallel amino acid site 26 was simultaneously located at the beta-strand positions and close to the phosphoserine modification site. In SOD2, we found that the positively selected site 52 was simultaneously located on the helix, Mn/Fe-SOD-N-terminal, and next to the binding site (50) and divalent metal cation (50) positions. The convergent/parallel amino acid site 11 was located on the transport peptide and next to the natural variation site (10), whereas site 57 was located next to the modified residue site (58). In SOD3, we found that the positively selected site 87 was located on the SOD-Cu/Zn-binding domain, and site 185 was a Cu2+-binding site and was located next to the active site (181). These two sites and site 191 were located at the disulfide bond positions. The convergent/parallel amino acid site 107 was located close to the glycosylation modification site and located at the disulfide bond position, and sites 161 and 163 were located at the disulfide bond location. Sites 107, 161, and 163 were located on the beta-strand position. In GPX1, we found that the convergent/parallel amino acid site 54 was located at the helical position and located next to the active site (49) and the catalytic residue site (49); site 88 was located next to the modified residue site (89) and the dimer interface site (89); and site 201 was located at the modified residue site. In GPX2, we found that the convergent/parallel amino acid site 43 was located on the glutathione peroxidase active site, and site 14 was located at the beta-strand position. Additionally, most of the positively selected sites (12/23) in the GPX3 are located on crucial domains; most are located on the beta-strand and helix positions, such as sites 72, 132, 154, and 167. In addition, we found that the positively selected sites were 71, 72, 74, and 105, and these sites were close to the active sites, dimer interfaces, and catalytic residue sites. The convergent/parallel amino acid sites and residues 42 and 204 of GPX3 were all in the helix position (Figure 2; Supplementary Figure S2; Supplementary Tables S6 and S7).
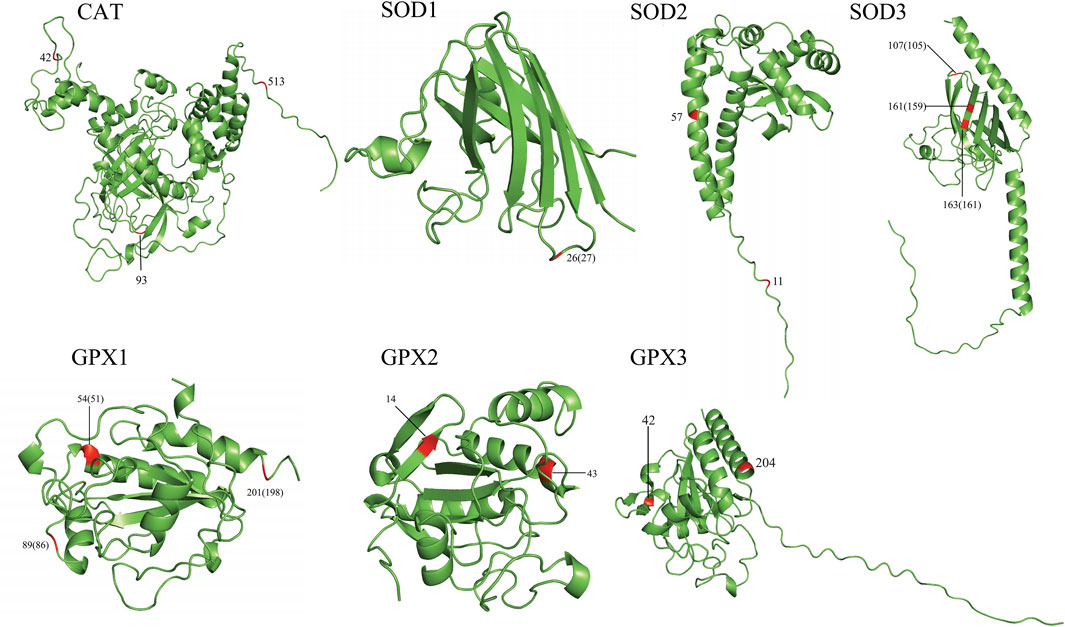
Figure 2. Annotation of the parallel/convergent amino acid sites of the seven genes on the 3D protein structure. The first number is for the order of analysis sequences used in this study. The numbers in brackets are for the true order of query sequences.
4 Discussion
4.1 Evolutionary patterns of antioxidase-related genes in hypoxia-tolerant lineages
Hypoxia-tolerant mammals have evolved adaptive mechanisms, including oxygen binding, storage and transportation capacities (Ramirez et al., 2007; Michael Panneton, 2013), energy metabolism (Tian et al., 2017), and other related tissue modifications, physiological and biochemical adaptations (Avivi et al., 2010; Helbo and Fago, 2012), and associated molecular mechanisms (Tian et al., 2019; Tian et al., 2021b) to effectively cope with hypoxic environments. As an important defense barrier, the antioxidant enzyme system plays a key role in resisting oxidative damage caused by low oxygen levels. Higher SOD activity has been observed in diving species (Miller, 2012), and a significantly higher evolutionary rate of SOD was also identified in cetaceans (Tian et al., 2021a). In addition, positive selection affects several GST genes in divergent taxonomic groups, which is indicative of pervasive adaptive evolution (Tian et al., 2019). In this study, our analyses showed that antioxidase-related genes have been subjected to adaptive evolution in hypoxia-tolerant mammals. The M0 model showed that the ω values of seven genes were significantly less than 1 (ωmin = 0.062 for GPX2, ωmax = 0.261 for SOD1), indicating that purifying selection played an important role in maintaining their function of antioxidase-related genes. Based on the M1 model, six genes exhibited significant differences (Supplementary Table S4), suggesting the divergent evolutionary rates of antioxidase-related genes across all lineages in response to diverse environments. Furthermore, the branch-site model test revealed the number of positively selected sites in antioxidase genes among the hypoxia-tolerant mammals or branches. Except for the GPX1 gene, branches subjected to positive selection were detected in six other genes, in which the number of positively selected sites accounted for 89% of the hypoxia-tolerant branches/species (Figure 1, Supplementary Table S5). Our results suggested that the core mechanisms of the adaptive evolution of antioxidase-related genes might have different patterns among different hypoxia-tolerant mammals. Antioxidase-related genes have evolved during adaptation to different hypoxic environments to facilitate their effective defense against ROS.
Mammals inhabiting diverse hypoxic environments may evolve into distinct selection pressures, even in the same lineage. For instance, cetaceans that have adapted to different diving depths have demonstrated different rates of evolution for HBA, HBB, and other associated genes (Tian et al., 2016). Hypoxia-tolerant animals exhibit remarkable adaptations in antioxidant mechanisms. For example, many marine mammals (e.g., cetaceans, pinnipeds, and manatees) can tolerate long-term hypoxia, and their recycling process of regaining oxygen after diving leads to increased ROS levels (Ramirez et al., 2007; Hermes-Lima et al., 2015). A similar tendency was detected among the antioxidase-related genes in this study. The evolutionary rates of CAT, SOD1, and GPX2 genes in hypoxia-tolerant species were significantly higher than those in non-hypoxia-tolerant species under the 2ω model, which indicated that antioxidant-related genes have gone through accelerated evolution in adapting to the hypoxic environments. Moreover, we hypothesized that physiological or genetic differences exist in the antioxidant mechanisms between terrestrial hypoxia-tolerant and aquatic hypoxia-tolerant species due to environmental heterogeneity. The 3ω model showed that SOD2 and GPX3 evolved at significantly lower rates in terrestrial hypoxic environments (plateau and caves) than in aquatic environments, which suggested that the environmental differences between terrestrial and aquatic would lead to different evolutionary patterns. Meanwhile, the higher evolutionary rates in aquatic lineages might be due to the complex aquatic environments. These results implied that the evolution of related genes would evolve to adapt to different microenvironments. Thus, we hypothesized that evolutionary patterns may have evolved in antioxidation-related genes among these typical hypoxic habits. Our speculation was confirmed in the SOD1 gene based on the 5ω model, but the specific mechanism needs to be further investigated. SOD1 is essential for the regulation and protection of cellular oxidative stress by promoting NADPH via the GAPDH/SOD1 signaling axis, its main element for the elimination of ROS (Iuchi et al., 2007; Inoue et al., 2010; Montllor-Albalate et al., 2022). In this study, the evolutionary rates of SOD1 genes vary significantly in different environments, which might be related to the different adaptive abilities of mammals to respond to oxidative stress in different hypoxic environments. In summary, the evolutionary rate of antioxidase-related genes in hypoxia-tolerant mammals varies in different habitats.
4.2 Parallel/convergent evolution of hypoxia-tolerant lineages
Hypoxia tolerance is a classic example of convergent evolution. Although hypoxia-tolerant species inhabit different environments and have different modes of adaptation to hypoxia, inherent similarities in hypoxia tolerance may drive recurrent evolution at the sequence level, which has manifested as convergent or parallel amino acid changes (Li et al., 2010). For example, echolocation in toothed whales, insect-eating bats, and pigtail mice (Typhlomys cinereus) (Shen et al., 2012; Parker et al., 2013; He et al., 2021) and convergent or parallel amino acid site substitutions have been detected in different hypoxic environments (Tian et al., 2017). In this study, genes with convergent evolution were also detected on the branches leading to hypoxia tolerance. For example, two parallel amino acid substitution sites were identified between the ancestral branches of the plateau pika and blind mole rat (Nannospalax galili) in CAT (branch e vs. g, Figure 1), and one parallel amino acid substitution site was identified between the naked mole rat (branch f) and cetacean (branch k) (Figure 1). Notably, 21 parallel amino acid substitution sites were detected in the CAT gene, accounting for 32% of the total parallel/convergent sites. Efficient scavenging of hydrogen peroxide (H2O2) is an important measure for mammals to cope with hypoxic environments during hypoxic adaptation (Baker et al., 2023), and CAT is the most important enzyme for organisms to catabolize hydrogen peroxide to enable the scavenging of oxygen radicals (Scibior and Czeczot, 2006; Kirkman and Gaetani, 2007; Ighodaro and Akinloye, 2018). Accordingly, hypoxia-tolerant species may strongly converge through hydrogen peroxide scavenging. Moreover, parallel/convergent substitutions in GPX genes were detected in almost all hypoxia-tolerant branches. GPXs are crucial in inhibiting lipid peroxidation processes (Ighodaro and Akinloye, 2018), suggesting a general convergence of hypoxia-tolerant species in inhibiting oxidative damage to lipids.
In particular, some functional sites also have evolved into convergence. For example, the convergent/parallel amino acid site 26 of SOD1 is located close to the phosphoserine residue modification site. Studies have shown that phosphoserine residues can affect secondary protein structures and alter their biological activities and catalytic functions (Shi, 2009). Site 107 of SOD3 is located close to the glycosylation modification site that may play an important role in the modification of translational and post-translational proteins (Staudacher et al., 2022), suggesting that site 107 may alter the antioxidant activity of SOD3. Studies have shown that all SODs require a catalytic metal (Cu or Mn) for their activation (Fukai and Ushio-Fukai, 2011; Skopp et al., 2019). In summary, hypoxia-tolerant species have evolved convergent antioxidant mechanisms to adapt to their respective hypoxic environments due to convergent adaptation.
4.3 Functional analysis of positively selected sites
Positively selected amino acid sites are labeled on the 3D structure of proteins. Most of these sites are located at or next to the important functional sites and domains, which might affect the structure and function of proteins, reflecting their key role in the adaptation to hypoxia. For example, the positively selected site detected by the branch-site model is in CAT, site 444 is located next to the NADPH-binding sites (445 and 446), and studies have shown that NADPH can effectively protect catalase against H2O2 (Kirkman et al., 1999). The result suggested that site 444 may play an important role in the antioxidant activity of CAT. In addition, site 52 of SOD2 is located next to the Mn/Fe-SOD-N-terminal and close to the metal cation (50), site 87 of SOD3 is located on the SOD-Cu/Zn-binding domain, and site 185 is located on the Cu2+-binding site, suggesting that these sites play an important role in the catalytic reaction of antioxidase. We also observed that some sites were located on or next to the active sites or dimer interface residues (associated with ligand binding and dimerization) (Epp et al., 1983; Maiorino et al., 1998), such as site 23 of SOD1, sites 87 and 185 of SOD3, site 89 of GPX1, site 43 of GPX2, and sites 72 and 74 of GPX3. This may alter the binding ability or catalytic activity of SOD and GPX to the substrate.
In summary, these findings must be validated by functional studies in future research studies. Therefore, it is necessary to conduct more studies on hypoxia-tolerant species and antioxidase-related genes to better understand the adaptive mechanisms of hypoxia-tolerant mammals.
5 Conclusion
Hypoxia tolerance is a complex and interesting evolutionary adaptation, and its study at the molecular level is conducive to revealing the effects of extreme hypoxic environments. Our results indicate that antioxidase-related genes have undergone different evolutionary trajectories in hypoxia- and non-hypoxia-tolerant mammals, as well as in various hypoxic environments among hypoxia-tolerant mammals. Multiple convergent/parallel amino acid substitution sites have been detected among hypoxia-tolerant mammalian lineages, and these sites were located on or next to the functional site, providing molecular evidence for the convergent mechanism of hypoxia adaptation in mammals. Our results further revealed the molecular mechanism of antioxidase-related genes involved in the adaption to hypoxia in hypoxia-tolerant mammals.
Author contributions
Q-PW: conceptualization, data curation, formal analysis, investigation, methodology, validation, and writing–original draft. C-YL: data curation, formal analysis, validation, and writing–original draft. X-HX: data curation, resources, software, validation, and writing–original draft. W-XH: visualization and writing–review and editing. Y-LG: supervision and writing–review and editing. Y-JG: supervision and writing–review and editing. YM: conceptualization, project administration, supervision, and writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by grants from the National Natural Science Foundation of China (NSFC) (No. 32360119), the start-up projects on high-level talent introduction of Dali University to YM (No. KY1916101940), the Erhai Watershed Ecological Environment Quality Testing Engineering Research Center of Yunnan provincial universities to W-XH and YM (No. DXDGCZX03), and the Yunnan Students’ Platform for Innovation and Entrepreneurship Training Program to Q-PW (No. S202210679061).
Acknowledgments
The authors thank the reviewers and the editor for their valuable comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1315677/full#supplementary-material
References
Avivi, A., Gerlach, F., Joel, A., Reuss, S., Burmester, T., Nevo, E., et al. (2010). Neuroglobin, cytoglobin, and myoglobin contribute to hypoxia adaptation of the subterranean mole rat Spalax. Proc. Natl. Acad. Sci. U.S.A. 107, 21570–21575. doi:10.1073/pnas.1015379107
Baker, A., Lin, C. C., Lett, C., Karpinska, B., Wright, M. H., and Foyer, C. H. (2023). Catalase: a critical node in the regulation of cell fate. Free Radic. Biol. Med. 199, 56–66. doi:10.1016/j.freeradbiomed.2023.02.009
Bigham, A. W., Wilson, M. J., Julian, C. G., Kiyamu, M., Vargas, E., Leon-Velarde, F., et al. (2013). Andean and Tibetan patterns of adaptation to high altitude. Am. J. Hum. Biol. 25, 190–197. doi:10.1002/ajhb.22358
Birben, E., Sahiner, U. M., Sackesen, C., Erzurum, S., and Kalayci, O. (2012). Oxidative stress and antioxidant defense. World Allergy Organ. J. 5, 9–19. doi:10.1097/WOX.0b013e3182439613
Delano, W. L. (2002). The pymol molecular graphics system. Proteins 30, 442–454. Avaliable at: http://www.pymol.org/.
Dröge, W. (2002). Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95. doi:10.1152/physrev.00018.2001
Epp, O., Ladenstein, R., and Wendel, A. (1983). The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur. J. Biochem. 133, 51–69. doi:10.1111/j.1432-1033.1983.tb07429.x
Fukai, T., and Ushio-Fukai, M. (2011). Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal 15, 1583–1606. doi:10.1089/ars.2011.3999
Ge, R. L., Cai, Q., Shen, Y. Y., San, A., Ma, L., Zhang, Y., et al. (2013). Draft genome sequence of the Tibetan antelope. Nat. Commun. 4, 1858. doi:10.1038/ncomms2860
He, K., Liu, Q., Xu, D. M., Qi, F. Y., Bai, J., He, S. W., et al. (2021). Echolocation in soft-furred tree mice. Science 372, eaay1513. doi:10.1126/science.aay1513
Helbo, S., and Fago, A. (2012). Functional properties of myoglobins from five whale species with different diving capacities. J. Exp. Biol. 215, 3403–3410. doi:10.1242/jeb.073726
Hermes-Lima, M., Moreira, D. C., Rivera-Ingraham, G. A., Giraud-Billoud, M., Genaro-Mattos, T. C., and Campos, É. G. (2015). Preparation for oxidative stress under hypoxia and metabolic depression: revisiting the proposal two decades later. Free Radic. Biol. Med. 89, 1122–1143. doi:10.1016/j.freeradbiomed.2015.07.156
Hu, H., Li, Y., Yang, Y., Xu, K., Yang, L., Qiao, S., et al. (2022). Effect of a plateau environment on the oxidation state of the heart and liver through AMPK/p38 MAPK/Nrf2-ARE signaling pathways in Tibetan and DLY pigs. Animals 12, 1219. doi:10.3390/ani12091219
Ighodaro, O. M., and Akinloye, O. A. (2018). First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 54, 287–293. doi:10.1016/j.ajme.2017.09.001
Inoue, E., Tano, K., Yoshii, H., Nakamura, J., Tada, S., Watanabe, M., et al. (2010). SOD1 is essential for the viability of DT40 cells and nuclear SOD1 functions as a guardian of genomic DNA. J. Nucleic Acids 2010, 795946. doi:10.4061/2010/795946
Iuchi, Y., Okada, F., Onuma, K., Onoda, T., Asao, H., Kobayashi, M., et al. (2007). Elevated oxidative stress in erythrocytes due to a SOD1 deficiency causes anaemia and triggers autoantibody production. Biochem. J. 402, 219–227. doi:10.1042/BJ20061386
Kaelin, W. G. (2005). ROS: really involved in oxygen sensing. Cell Metab. 1, 357–358. doi:10.1016/j.cmet.2005.05.006
Kirkman, H. N., and Gaetani, G. F. (2007). Mammalian catalase: a venerable enzyme with new mysteries. Trends Biochem. Sci. 32, 44–50. doi:10.1016/j.tibs.2006.11.003
Kirkman, H. N., Rolfo, M., Ferraris, A. M., and Gaetani, G. F. (1999). Mechanisms of protection of catalase by NADPH: kinetics and stoichiometry. J. Biol. Chem. 274, 13908–13914. doi:10.1074/jbc.274.20.13908
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi:10.1093/molbev/msx116
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi:10.1093/molbev/msw054
Li, Y., Liu, Z., Shi, P., and Zhang, J. (2010). The hearing gene Prestin unites echolocating bats and whales. Curr. Biol. 20, R55–R56. doi:10.1016/j.cub.2009.11.042
Löytynoja, A., and Goldman, N. (2010). webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinf 11, 579–587. doi:10.1186/1471-2105-11-579
Maiorino, M., Aumann, K. D., Brigelius-Flohe, R., Doria, D., van den Heuvel, J., McCarthy, J., et al. (1998). Probing the presumed catalytic triad of a selenium-containing peroxidase by mutational analysis. Z Ernahrungswiss 37, 118–121.
Michael Panneton, W. (2013). The mammalian diving response: an enigmatic reflex to preserve life? Physiology 28, 284–297. doi:10.1152/physiol.00020.2013
Miller, A. F. (2012). Superoxide dismutases: ancient enzymes and new insights. FEBS Lett. 586, 585–595. doi:10.1016/j.febslet.2011.10.048
Montllor-Albalate, C., Kim, H., Thompson, A. E., Jonke, A. P., Torres, M. P., and Reddi, A. R. (2022). Sod1 integrates oxygen availability to redox regulate NADPH production and the thiol redoxome. Proc. Natl. Acad. Sci. U. S. A. 119, e2023328119. doi:10.1073/pnas.2023328119
Murphy, W. J., Pevzner, P. A., and O'Brien, S. J. (2004). Mammalian phylogenomics comes of age. Trends Genet. 20, 631–639. doi:10.1016/j.tig.2004.09.005
Nathaniel, T. I., Williams-Hernandez, A., Hunter, A. L., Liddy, C., Peffley, D. M., Umesiri, F. E., et al. (2015). Tissue hypoxia during ischemic stroke: adaptive clues from hypoxia-tolerant animal models. Brain Res. Bull. 114, 1–12. doi:10.1016/j.brainresbull.2015.02.006
Nose, K. (2000). Role of reactive oxygen species in the regulation of physiological functions. Biol. Pharm. Bull. 23, 897–903. doi:10.1248/bpb.23.897
Pamenter, M. E. (2022). Adaptations to a hypoxic lifestyle in naked mole-rats. J. Exp. Biol. 225, jeb196725. doi:10.1242/jeb.196725
Parker, J., Tsagkogeorga, G., Cotton, J. A., Liu, Y., Provero, P., Stupka, E., et al. (2013). Genome-wide signatures of convergent evolution in echolocating mammals. Nature 502, 228–231. doi:10.1038/nature12511
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51, D418–D427. doi:10.1093/nar/gkac993
Ramirez, J. M., Folkow, L. P., and Blix, A. S. (2007). Hypoxia tolerance in mammals and birds: from the wilderness to the clinic. Annu. Rev. Physiol. 69, 113–143. doi:10.1146/annurev.physiol.69.031905.163111
Scibior, D., and Czeczot, H. (2006). Catalase: structure, properties, functions. Postepy Hig. Med. Dosw 60, 170–180.
Shen, Y. Y., Liang, L., Li, G. S., Murphy, R. W., and Zhang, Y. P. (2012). Parallel evolution of auditory genes for echolocation in bats and toothed whales. PLoS Genet. 8, e1002788. doi:10.1371/journal.pgen.1002788
Shi, Y. (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. doi:10.1016/j.cell.2009.10.006
Skopp, A., Boyd, S. D., Ullrich, M. S., Liu, L., and Winkler, D. D. (2019). Copper-zinc superoxide dismutase (Sod1) activation terminates interaction between its copper chaperone (Ccs) and the cytosolic metal-binding domain of the copper importer Ctr1. Biometals 32, 695–705. doi:10.1007/s10534-019-00206-3
Staudacher, E., Van Damme, E. J. M., and Smagghe, G. (2022). Glycosylation-the most diverse post-translational modification. Biomolecules 12, 1313. doi:10.3390/biom12091313
Swanson, W. J., Nielsen, R., and Yang, Q. (2003). Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20, 18–20. doi:10.1093/oxfordjournals.molbev.a004233
Tian, R., Geng, Y., Yang, Y., Seim, I., and Yang, G. (2021b). Oxidative stress drives divergent evolution of the glutathione peroxidase (GPX) gene family in mammals. Integr. Zool. 16, 696–711. doi:10.1111/1749-4877.12521
Tian, R., Geng, Y. P., Guo, H., Yang, C., Seim, I., and Yang, G. (2021a). Comparative analysis of the superoxide dismutase (SOD) gene family in Cetartiodactyla. J. Evol. Biol. 34, 1046–1060. doi:10.1111/jeb.13792
Tian, R., Seim, I., Ren, W., Xu, S., and Yang, G. (2019). Contraction of the ROS scavenging enzyme glutathione S-transferase gene family in cetaceans. G3 Genes, Genomes, Genet. 9, 2303–2315. doi:10.1534/g3.119.400224
Tian, R., Wang, Z., Niu, X., Zhou, K., Xu, S., and Yang, G. (2016). Evolutionary genetics of hypoxia tolerance in cetaceans during diving. Genome Biol. Evol. 8, 827–839. doi:10.1093/gbe/evw037
Tian, R., Yin, D., Liu, Y., Seim, I., Xu, S., and Yang, G. (2017). Adaptive evolution of energy metabolism-related genes in hypoxia-tolerant mammals. Front. Genet. 8, 205. doi:10.3389/fgene.2017.00205
UniProt Consortium (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi:10.1093/nar/gky1049
Wilhelm Filho, D., Sell, F., Ribeiro, L., Ghislandi, M., Carrasquedo, F., Fraga, C. G., et al. (2002). Comparison between the antioxidant status of terrestrial and diving mammals. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 133, 885–892. doi:10.1016/S1095-6433(02)00253-2
Yang, Z. (1998). Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15, 568–573. doi:10.1093/oxfordjournals.molbev.a025957
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi:10.1093/molbev/msm088
Yang, Z., Wong, W. S., and Nielsen, R. (2005). Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi:10.1093/molbev/msi097
Yim, H. S., Cho, Y. S., Guang, X., Kang, S. G., Jeong, J. Y., Cha, S. S., et al. (2014). Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 46, 88–92. doi:10.1038/ng.2835
Zhang, J., and Kumar, S. (1997). Detection of convergent and parallel evolution at the amino acid sequence level. Mol. Biol. Evol. 14, 527–536. doi:10.1093/oxfordjournals.molbev.a025789
Zhang, J., Nielsen, R., and Yang, Z. (2005). Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479. doi:10.1093/molbev/msi237
Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinforma. 9, 40. doi:10.1186/1471-2105-9-40
Keywords: hypoxia tolerance, antioxidase-related genes, adaptive evolution, convergent/parallel, mammals
Citation: Wang Q-P, Luo C-Y, Xu X-H, Hu W-X, Gai Y-L, Gong Y-J and Mu Y (2024) Adaptive evolution of antioxidase-related genes in hypoxia-tolerant mammals. Front. Genet. 15:1315677. doi: 10.3389/fgene.2024.1315677
Received: 10 October 2023; Accepted: 20 March 2024;
Published: 25 April 2024.
Edited by:
Vindhya Mohindra, National Bureau of Fish Genetic Resources (ICAR), IndiaCopyright © 2024 Wang, Luo, Xu, Hu, Gai, Gong and Mu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Lin Gai, yulingai0818@163.com; You-Jing Gong, gongyj@eastern-himalaya.cn; Yuan Mu, muy@eastern-himalaya.cn
 Qiu-Ping Wang1
Qiu-Ping Wang1 Chao-Yang Luo
Chao-Yang Luo Yu-Lin Gai
Yu-Lin Gai Yuan Mu
Yuan Mu