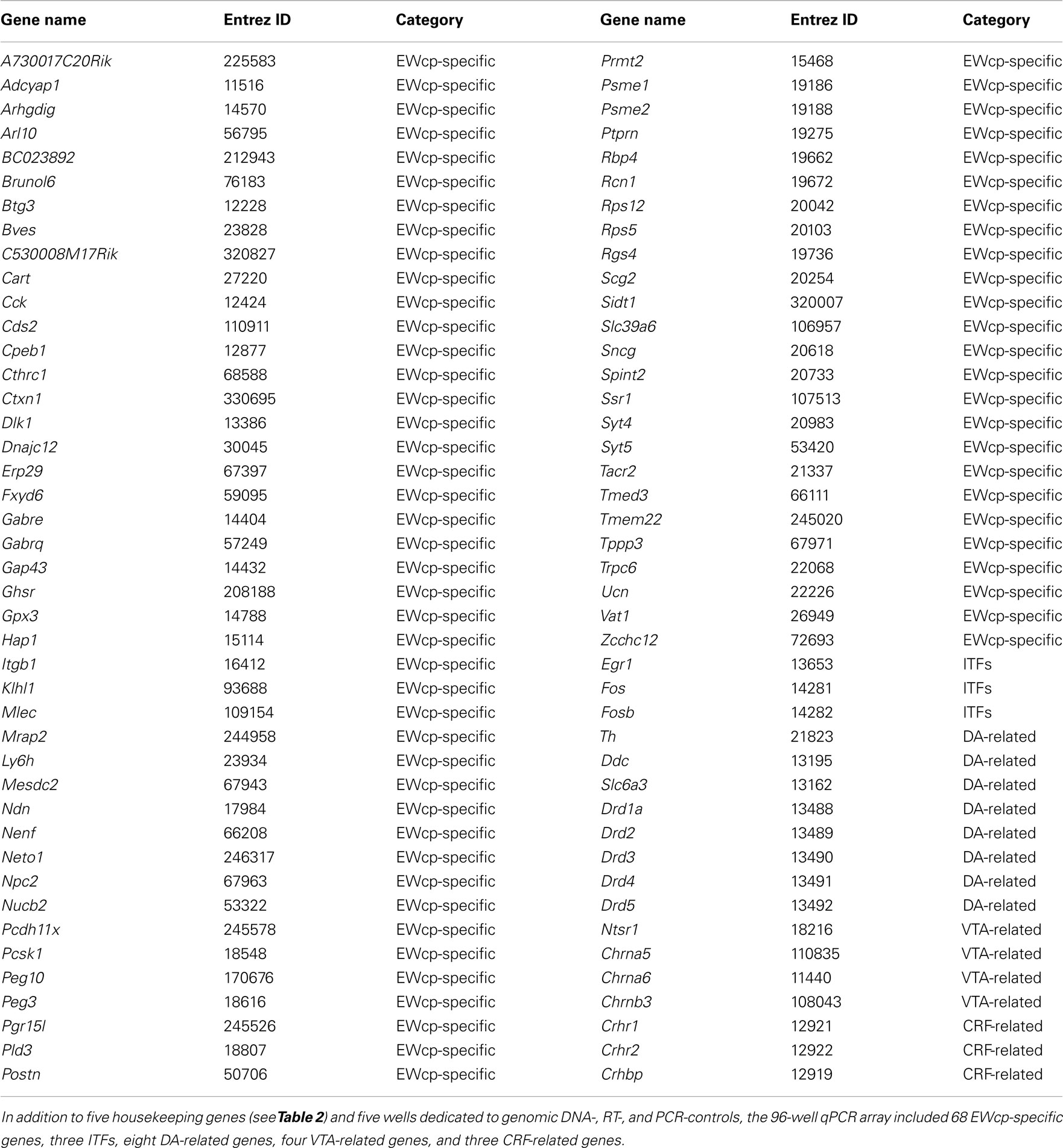
- Department of Behavioral Neuroscience, Oregon Health and Science University, Portland, OR, USA
Detailed examination of the midbrain Edinger–Westphal (EW) nucleus revealed the existence of two distinct nuclei. One population of EW preganglionic (EWpg) neurons was found to control oculomotor functions, and a separate population of EW centrally projecting (EWcp) neurons was found to contain stress- and feeding-related neuropeptides. Although it has been shown that EWcp neurons are highly responsive to drugs of abuse and behavioral stress, a genetic characterization of the EWcp was needed. To identify genetic differences in the EWcp of inbred mouse strains that differ in behaviors relevant to EWcp function, we used publicly available tools from the Allen Brain Atlas to identify 68 transcripts that were selectively expressed in the EWcp, and examined their expression within tissue punch microdissection samples containing the EWcp of adult male C57BL/6J (B6) and DBA/2J (D2) mice. Using 96-well quantitative real-time PCR (qPCR) arrays that included the EWcp-specific genes, several other genes of interest, and five housekeeping genes, we identified strain differences in expression of 11 EWcp-specific genes (BC023892, Btg3, Bves, Cart, Cck, Ghsr, Neto1, Postn, Ptprn, Rcn1, and Ucn), two immediate early genes (Egr1 and Fos), and one dopamine-related gene (Drd5). All significant expression differences were greater in B6 vs. D2 mice, and several of these were verified either at the protein level using immunohistochemistry (IHC) or in silico using microarray data sets from whole brain and other brain areas. These results demonstrate a significant advance in our understanding of the EWcp on three levels. First, we generated a list of EWcp-specific genes (most of which had not yet been reported within the EWcp in the literature) that will be informative for future studies of EWcp function. Second, due to similarity in results from qPCR and IHC, we revealed that strain differences in basal EWcp neuropeptide content are accounted for by differential transcription and number of peptidergic neurons, rather than by differential rates of peptide release. And third, our identification of differentially expressed EWcp-specific genes between B6 and D2 mice may hold powerful insight into the neurogenetic contributions of the EWcp to stress- and addiction-related behaviors.
Introduction
The Edinger–Westphal (EW) nucleus is a compact region that extends along the rostral–caudal axis of the midline within the ventromedial periaqueductal gray of the midbrain. While this region has been historically described as a cholinergic population of preganglionic neurons projecting to the ciliary ganglion to control oculomotor functions, detailed examinations have revealed that the EW is comprised of two distinct nuclei, recently designated as the EW preganglionic (EWpg) oculomotor neurons and the EW centrally projecting (EWcp) neuropeptide-containing neurons (Cavani et al., 2003; Ryabinin et al., 2005; Weitemier et al., 2005; Kozicz et al., 2011).
This updated nomenclature was required after significant confusion arose from the discoveries that a supposedly cholinergic nucleus serving ocular functions did not contain choline acetyl transferase, but was highly enriched in components of several stress- and feeding-related neuropeptide systems (Maciewicz et al., 1984; Vaughan et al., 1995; Koylu et al., 1998; Tanaka et al., 2003; Dun et al., 2005; Ryabinin et al., 2005; Weitemier et al., 2005; Zigman et al., 2006; Foo et al., 2008; Xu et al., 2011), and that this brain region was highly sensitive to alcohol self-administration (Topple et al., 1998; Bachtell et al., 1999, 2003; Ryabinin et al., 2001, 2003; Weitemier et al., 2001), as well as experimenter-administered injections of alcohol, morphine, cocaine, amphetamine, and methamphetamine (Chang et al., 1995; Ryabinin et al., 1997; Bachtell et al., 2002a; Spangler et al., 2009; Giardino et al., 2011a). Additional confusion was generated from the findings that this nucleus was sensitive to behavioral and physiological stressors (Kozicz, 2003; Gaszner et al., 2004, 2009; Korosi et al., 2005; Kozicz et al., 2008; Okere et al., 2010; Rouwette et al., 2010, 2011; Xu et al., 2010; Sterrenburg et al., 2011), as well as stimuli related to food restriction (Xu et al., 2009, 2011). Finally, the existence of two distinct nuclei was also indicated by the finding that neuropeptide-containing neurons of the EWcp projected primarily to limbic brain areas, rather than to the ciliary ganglion (Loewy and Saper, 1978; Loewy et al., 1978; Bittencourt et al., 1999; Bachtell et al., 2004; Weitemier and Ryabinin, 2005a).
Thus, the EWcp emerged as a recently identified (and therefore, poorly characterized) brain region that appeared to be especially important for regulation of responses to addictive drugs and environmental challenges (Ryabinin and Weitemier, 2006; Kozicz, 2007; Kozicz et al., 2011). In particular, our interest in this nucleus originated from several neural mapping studies in which the EWcp consistently showed a selective induction of the inducible transcription factor (ITF) c-Fos (Fos) following oral self-administration of alcohol (Topple et al., 1998; Bachtell et al., 1999, 2003; Ryabinin et al., 2001, 2003; Weitemier et al., 2001). Further experiments revealed that alcohol-induced neural activity within the EWcp occurred in 95–100% of neurons containing the neuropeptide urocortin-1 (Ucn1; Bachtell et al., 2002b; Ryabinin et al., 2003; Spangler et al., 2009). These findings suggested a potential role for EWcp–Ucn1 neurons in alcohol-related phenotypes, a hypothesis that was confirmed by studies showing that electrolytic lesions encompassing the EWcp dramatically attenuated EtOH preference in C57BL/6J (B6) mice (Bachtell et al., 2004), an effect that was later found to be dependent on the expression of Ucn1 (Giardino et al., 2011b). Also in agreement with this hypothesis were comparisons of several inbred and selectively bred rodent strains, in which higher levels of Ucn1 immunoreactivity (IR) within the EWcp were associated with a genetic predisposition toward higher alcohol consumption and greater sensitivity to some (but not all) alcohol-related phenotypes (Bachtell et al., 2002b, 2003; Kiianmaa et al., 2003; Turek et al., 2005, 2008; Ryabinin and Weitemier, 2006; Fonareva et al., 2009).
However, we were unable to determine whether these observations at the protein level resulted from differences in the magnitude of gene expression, differences in peptide release, or a difference in the total number or size of EWcp neurons. In a comparison of two inbred mouse strains that showed robust differences in alcohol-related behaviors and Ucn1-IR, our laboratory reported that neurons within the proximity of the EW in B6 mice were more numerous (and larger in size) than those of DBA/2J (D2) mice (Bachtell et al., 2002b). However, these results were obtained using a Thionin stain, which did not allow differentiation between Ucn1 neurons of the EWcp, cholinergic neurons of the EWpg, and dopaminergic (DAergic) neurons of the adjacent rostral linear nucleus of the raphe (RLi). An additional study found that B6 mice exhibited greater Ucn1-IR in individual EWcp neurons, relative to D2 mice (Weitemier and Ryabinin, 2005a). These data raised the possibility that, in addition to being driven in part by differences in the total number of neurons, the observed strain differences in EWcp–Ucn1 protein expression could also be due to differences in Ucn1 mRNA expression, or Ucn1 peptide release.
In order to determine whether differences in peptide expression could be attributed to differential expression at the mRNA level, we performed tissue punch microdissection of the EWcp region and quantified the expression of several EWcp-selective genes between B6 and D2 mice. These two well-characterized strains differ in alcohol-, stress-, and feeding-related phenotypes, which might be reflective of genetic differences within the EWcp (Ryabinin et al., 1999; Lewis et al., 2007; Yoneyama et al., 2008). Several transcripts that appeared to be selectively expressed within the EWcp were identified by resources present in the Allen Brain Atlas (ABA1; Lein et al., 2007). We extracted RNA from micropunches containing the EWcp and subjected samples to customized, 96-well plates that allowed quantitative real-time PCR (qPCR) analysis of these transcripts.
After identifying several genes that were differentially expressed within the EWcp of B6 and D2 mice by qPCR array, we used immunohistochemical (IHC) and in silico analyses to verify some of these findings. In doing so, we demonstrate that strain differences at the protein level are unlikely to be attributed to differential rates of peptide release, and are more likely due to differences in gene transcription and cell number. As such, the present findings identify several previously unexplored genes that may be integral for addiction- and stress-related behaviors regulated via the mammalian EWcp.
Materials and Methods
Animals
We studied adult (8- to 10-week-old) male C57BL/6J (B6) and DBA/2J (D2) mice that had been delivered from The Jackson Laboratory (Sacramento, CA, USA) and housed four per cage in our colony. Information from the Jax Phenome Database2 lists mean body/brain weights (in grams) for 8-week-old male B6 and D2 mice as 24.3/0.423 and 22.8/0.354, respectively. All mice received ad libitum access to food (LabDiet 5001; Richmond, IN, USA) and water, and were maintained on a 12-h light–dark schedule (lights on at 0600 hours). All experiments were performed with strict adherence to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Identification of EWcp-Specific Transcripts
We used resources available from the ABA to identify transcripts that appeared to be selectively expressed within the EWcp. The initial goal of the ABA project was to perform in situ hybridization using probes targeted against every protein-coding gene in the mouse genome in order to visualize patterns of expression within the brains of adult male B6 mice. The ABA group have systematically documented the brain expression of several 1000 mRNA transcripts, and have made this information publicly available online (Allen Brain Atlas, 2004; Lein et al., 2007).
Important for our study, the ABA has developed a “fine structure” search feature that allows searching for genes that are expressed in smaller brain structures. We began our search by browsing the expression patterns of the 50 genes identified by the fine structure feature as being located within the “Edinger–Westphal.” However, because the spatial resolution of this search feature is relatively low and does not represent the vast coronal span of the EWcp, we verified that only 27 of these 50 transcripts appeared to be selectively expressed within the EWcp. Reasons for exclusion of the other genes included either a pattern of expression that was not within the EWcp, a non-specific pattern of expression that included the EWcp as well as several other structures, or the appearance of very low expression within the EWcp.
Next, we used the “neuroblast” feature on the ABA website to find genes with similar expression patterns to those identified by the initial fine structure search. This allowed a rapid method for discovering additional genes that were also selectively expressed within EWcp. Finally, we used the AGEA gene finder, another ABA tool that finds genes within a specific brain area by allowing the user to choose any voxel in the mouse brain as a seed region and then identifying genes with expression patterns that are highly correlated with that seed space. By placing seed regions in five different voxels throughout the mouse midbrain (centered around the EWcp), we were able to identify additional genes that were specifically expressed within the EWcp, yet had not been identified by prior methods.
Thus, after beginning with 7–10 candidate transcripts that we had known were EWcp-specific (based on our prior studies and on literature searches of the EW nucleus), we were able to identify a total of 68 genes that appeared to be selectively expressed within the EWcp. It is important to note that our analysis, which relied heavily on the features included on the ABA website, was prone to false negatives (i.e., in situ probe failure). Therefore, rather than being a liberal method for assembling a list of EWcp-specific genes, this list is likely an underestimate of the number of EWcp-specific genes that are highly expressed in the adult mouse brain. The 68 identified genes were further interrogated by the qPCR array approach, as described below.
Additional Transcripts of Interest
Additional transcripts that were not selectively expressed in the EWcp were also included in the analysis, and were comprised of the following five groups: (1) three immediate early genes encoding inducible transcription factors (ITFs), included to assess differences in basal activity between strains because they are well-established markers of neuronal activity; (2) eight genes related to the dopamine (DA) system, included because the tissue punch microdissection of the EWcp may have included small quantities of DAergic neurons of the adjacent RLi, which have been shown to intermingle (but not co-localize) with the Ucn1-positive neurons of the EWcp (Bachtell et al., 2002a; Gaszner and Kozicz, 2003; Fonareva et al., 2009); (3) four genes showing robust expression in the ventral tegmental area (VTA), included because the VTA is neurochemically similar to the RLi; (4) three corticotropin-releasing factor (CRF) system genes, included because they are targets of the Ucn1 peptide, and their expression is expected within the vicinity of the EWcp if Ucn1 is released locally; and (5) five housekeeping genes, included to control for potential loading issues.
See Table 1 for a complete list of all genes of interest, and Table 2 for a complete list of all housekeeping genes.
Gene Expression Analyses
After habituation to our mouse colony, naïve mice (n = 5–7 per strain) were euthanized by CO2, and dissected brains were immediately placed inside a pre-chilled coronal brain matrix. A 1-mm-thick tissue punch containing the EWcp was isolated with a chilled 18-gage blunt needle (Figure 1), incubated in extraction buffer at 42°C for 30 min, briefly vortexed, and stored at −80°C.
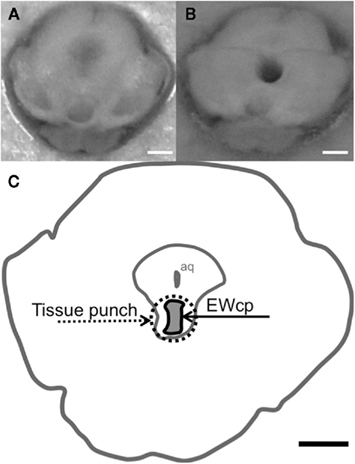
Figure 1. Tissue punch microdissection of the centrally-projecting neurons of the Edinger–Westphal nucleus. (A) Photograph demonstrating a 1-mm-thick coronal slice of the adult B6 mouse midbrain fresh after dissection, in which the EWcp is still intact and (B) photograph demonstrating a frozen slice of the midbrain in which the EWcp has been punched out with an 18-gage blunt needle and removed by making a horizontal cut with a razor blade just dorsal to the EWcp and sliding the tissue sample along the metal plate for harvesting. (C) Schematic of the location of the EWcp within a coronal slice of the midbrain (approx. −3.5 mm from bregma), indicating the area dissected by the tissue punch micropunch. Each scalebar = 1 mm.
RNA was isolated using the Arcturus PicoPure RNA Isolation Kit (Applied Biosystems) according to the kit manual, and as previously reported by our laboratory (Cservenka et al., 2010). Spectrophotometer readings were taken, and samples were diluted to match RNA concentrations. Samples were then DNase-treated at 42°C for 5 min, and upon addition of the RT cocktail, underwent first strand cDNA synthesis according to the RT2 First Strand Kit manual (Qiagen). Synthesized cDNA samples were diluted with a cocktail containing the RT2 qPCR Master Mix (Qiagen), and 25 μL of the mixture was deposited into each well of a custom-designed RT2 Profiler PCR Array for analysis by a MX3000P RT thermal cycler (Stratagene). A qPCR approach was taken instead of a microarray analysis because quantitative comparisons between the two strains could be distorted by the small amounts of tissue harvested from the EWcp (which would require high amplification of RNA). In addition, mouse microarrays are designed based on the B6 genome, yet several single nucleotide polymorphisms exist between B6 and D2 strains (Walter et al., 2009), complicating this analysis. Finally, qPCR analyses were done as biological but not technical replicates, due to the high number of housekeeping genes and additional controls already included on each array.
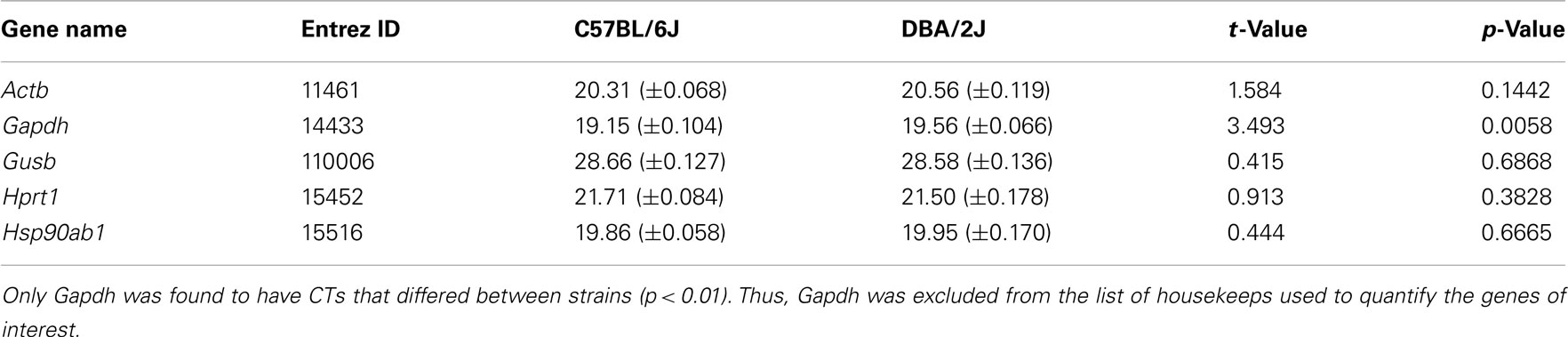
The mean cycle thresholds (CT) for the five housekeeping genes included on the qPCR array were first compared between B6 and D2 mice by t-test, and CTs of four housekeeping transcripts that did not show significant strain differences were then averaged and used to normalize the quantitative expression of all genes of interest included on the array. For each individual gene of interest, CT values were normalized by the equation 2−ΔCT, where ΔCT = the CT for the gene of interest subtracted from the mean CT value of the housekeeping genes. The mean 2−ΔCT values were compared by t-test between the two strains (significance threshold at p < 0.05), and data are presented as mean 2−ΔCT values ± SEM. Bonferroni correction for multiple comparisons was not applied, as we aimed to identify as many differentially expressed genes as possible. Such an approach relies on confirmation studies. In our case, the ABA analysis described above can serve as one such confirmation approach. Furthermore, a few of the identified genes were also analyzed at the protein level by IHC, and all identified genes were analyzed in silico using microarray data, providing additional confirmation.
In silico Analyses
Following identification of genes exhibiting strain differences in EWcp expression, we used GeneNetwork3 (GeneNetwork, 2001; Chesler et al., 2004) as an additional resource for verifying expression differences. Analysis of several microarray data sets determined whether the transcripts showing genotype-dependent expression within EWcp also differed in expression throughout whole brain, cerebellum, striatum, hippocampus, hypothalamus, neocortex, and amygdala.
For each of the identified genes, we compared the reported values for B6 and D2 mice from the following GeneNetwork data sets: UCHSC BXD Whole Brain M430 2.0 (Nov06) RMA, SJUT Cerebellum mRNA M430 (Mar05) RMA, HQF BXD Striatum ILM6.1 (Dec10v2) RankInv, Hippocampus Consortium M430v2 (June06) PDNN, INIA Hypothalamus Affy MoGene 1.0 ST (Nov10), HQF BXD Neocortex ILM6.1 (Dec10v2) RankInv Database, and INIA Amygdala Cohort Affy MoGene 1.0 ST (Mar11) RMA (Saba et al., 2006; Overall et al., 2009). Data are presented as mean ± SEM, and significance threshold was set at p < 0.05. All significant findings identified by t-test are detailed in Table 4.
Immunohistochemical Analyses
Immunohistochemistry was performed on products of three genes identified as being differentially expressed between B6 and D2 mice by the qPCR array. The selection of gene products was based on available commercial antibodies. Ucn1 and Fos were two gene products that were not included in these analyses because our previous studies had already identified differences in Ucn1-IR and Fos-IR between B6 and D2 mice (Bachtell et al., 2002b; Weitemier and Ryabinin, 2005a; Weitemier et al., 2005).
After habituation to our mouse colony, mice (n = 8 per strain) were euthanized by CO2 and underwent transcardial perfusion with 2% PFA dissolved in H2O. Brains were rapidly dissected and placed in 2% PFA for storage overnight, followed by cryoprotection in 20 and 30% sucrose dissolved in phosphate buffered saline (PBS) containing 0.1% NaN3. Coronal sections were sliced 30 μm thick on a Leica CM1850 cryostat, and slices were collected in PBS containing 0.1% NaN3.
For each gene product, six to eight slices containing the EWcp (evenly spaced along the rostral–caudal axis, from −3.2 to −3.8 mm from bregma) were chosen from each animal. Examinations of CCK- and Ptprn-IR were preceded by an antigen retrieval process. However, antigen retrieval was not necessary for examination of CART-IR, which stains heavily within mouse EWcp neurons even without this additional step (Kozicz, 2003; Cservenka et al., 2010).
For IHC procedures examining CCK and Ptprn in the EWcp, antigen retrieval consisted of rinsing the sections in PBS and then boiling the tissue in sodium citrate buffer (10 mM sodium citrate,0.05% Tween 20, pH 6.0) followed by cooling to room temperature. For all IHC procedures, slices underwent a standard DAB staining protocol identical to previous reports from our lab (Spangler et al., 2009; Giardino et al., 2011a), with the exception that primary antibodies were directed against either human CART 55-102 (H-003-60, Phoenix) human/rat/mouse CCK 26-33 (H-069-04, Phoenix), or human Ptprn (HPA-007179, Sigma-Aldrich), and were used at concentrations of 1:20,000, 1:30,000, and 1:1000, respectively. Dehydration and coverslipping methods were also identical to previous reports.
The number of CART-, CCK-, or Ptprn-positive neurons within the EWcp was counted manually using a Leica DM4000 microscope and recorded by an observer. A single value per animal was calculated by averaging the cell counts across all slices from that subject, and mean cell counts for the two strains were compared by t-test separately for each of the three gene products. One data point was excluded from the analysis of CART-IR in B6 mice, because the value was greater than 2.5 SD below the mean. No other outliers were identified. Data are presented as mean ± SEM, and significance threshold was set at p < 0.05.
Results
Analysis of Housekeeping Genes
Preliminary analysis of the five housekeeping genes included on the qPCR arrays revealed that Gapdh CT values were significantly greater in D2 vs. B6 mice (t10 = 3.49; p < 0.01; Table 2). CT values for other housekeeping genes were not different between strains (all t10 < 1.59; all p > 0.14). When Gapdh CT values were normalized to the average CT values of the remaining four housekeeps by the 2−ΔCT method, analysis revealed that Gapdh expression was greater in B6 vs. D2 mice (t10 = 2.71; p < 0.05). Therefore, all genes of interest included on the array were normalized to the average of the remaining four housekeeping genes (Actb, Gusb, Hprt1, Hsp90ab1), with Gapdh excluded.
Gene Expression Analyses
After normalization to the four remaining housekeeping genes, 14/86 genes of interest were found to differ significantly in expression between B6 and D2 strains: BC023892 (also known as Fam46a), Btg3, Bves, Cart, Cck, Ghsr, Neto1, Postn, Ptprn, Rcn1, Ucn, Egr1 (also known as zif268), Fos, and Drd5 (Figure 2). In each case, expression was greater in B6 mice, relative to D2 mice (all t10 > 2.44; all p < 0.05; Table 3).
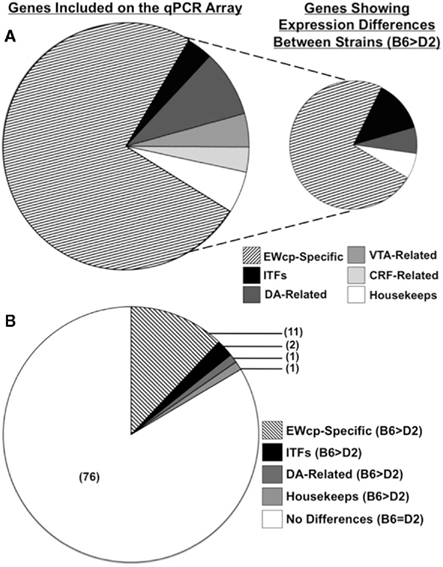
Figure 2. Genes of interest included on the qPCR array, and strain differences in EWcp gene expression identified between B6 and D2 mice. (A) Pie chart on left indicates relative proportions of gene categories included on the qPCR arrays. Pie chart on right indicates relative proportions of gene categories in which strain differences were identified. (B) Pie chart showing relative proportions of gene categories in which strain differences were identified, as compared to the proportion of genes in which no differences were identified. Note that while Ucn is both CRF-related and EWcp-specific, it was included only in the EWcp-specific category for these analyses.
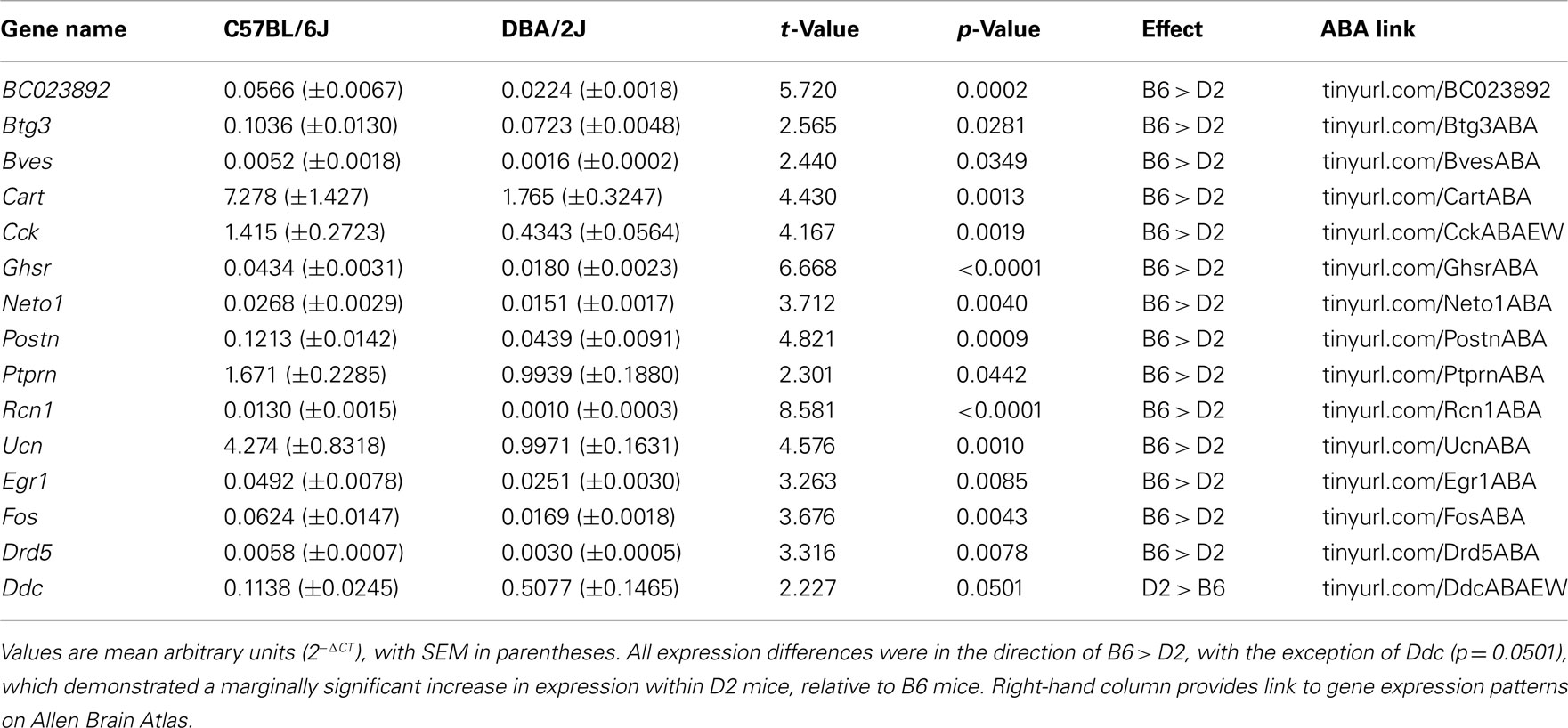
Table 3. Genes of interest showing expression differences between strains by qPCR array of EWcp micropunch.
Of these 14 genes, 11 were selectively expressed and/or enriched within the EWcp (Figures 3 and 4). Two of these genes were ITFs (Egr1 and Fos), which are not selectively expressed within EWcp, but have been known to be induced within EWcp following certain environmental stimuli (Bachtell et al., 1999; Ryabinin et al., 2001). The remaining gene, Drd5 (which encodes the dopamine receptor subtype 5) is not known to be selectively expressed within EWcp, but was included with the smaller list of DA-related genes.
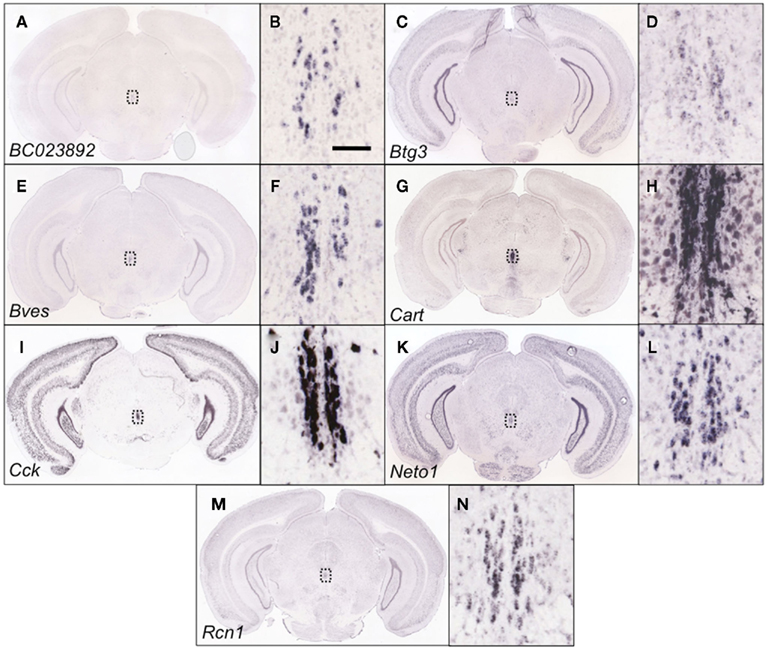
Figure 3. Genes within the EWcp-specific category that were identified as being differentially expressed between B6 and D2 mice are indeed EWcp-specific. Shown are coronal slices at approx. −3.5 mm from bregma from adult male B6 mouse brains that have undergone in situ hybridization to reveal the EWcp-specific expression of BC023892(A), Btg3 (C), Bves (E), Cart (G), Cck (I), Neto1 (K), and Rcn1 (M). The close-up images (B,D,F,H,J,L,N) show the area within the dotted line of the corresponding figure, indicating that BC023892, Btg3, Bves, Cart, Cck, Neto1, and Rcn1 demonstrate an EWcp-specific pattern of expression. Scalebar = 100 μm, and is valid for all close-up images. Images courtesy of the Allen Brain Atlas, used with permission.
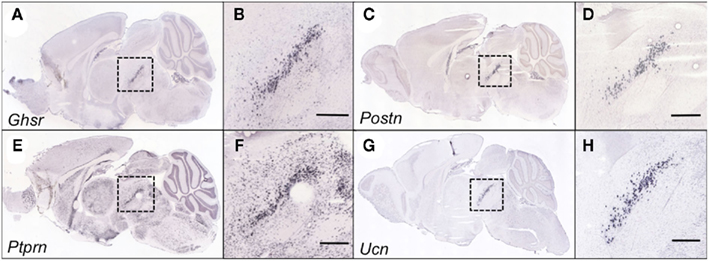
Figure 4. Genes within the EWcp-specific category that were identified as being differentially expressed between B6 and D2 mice are indeed EWcp-specific. Shown are sagittal slices at the midline from adult male B6 mouse brains that have undergone in situ hybridization to reveal the EWcp-specific expression of Ghsr(A), Postn (C), Ptprn (E), and Ucn (G). The close-up images (B,D,F,H) show the area within the dotted line, indicating that Ghsr, Postn, Ptprn, and Ucn demonstrate an EWcp-specific pattern of expression. Each scalebar = 500 μm. Images courtesy of the Allen Brain Atlas, used with permission.
In silico Analyses
Of the 14 transcripts demonstrating strain differences in EWcp expression, six of these (Btg3, Bves, Cart, Cck, Egr1, and Rcn1) were confirmed to also have significant differences in expression within whole brain and/or other brain regions (cerebellum, striatum, hippocampus, hypothalamus, neocortex, amygdala; Table 4). Consistent with qPCR array results from EWcp micropunches, all gene expression levels were greater in B6 vs. D2 mice, with the exception of Rcn1, whose genotype-dependent regulation in whole brain, cerebellum, and amygdala appeared to be opposite from that in the EWcp (Table 4).
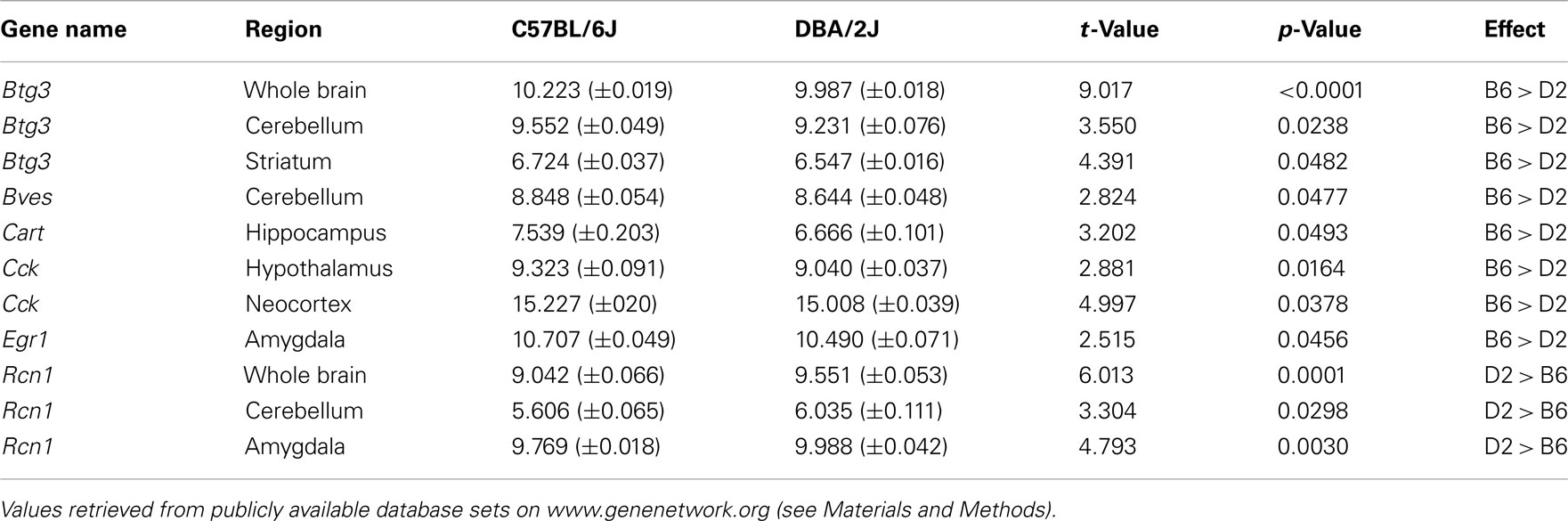
Table 4. Confirmation of qPCR array results by in silico analyses of whole brain and brain region-specific microarray data.
Immunohistochemical Analyses
In order to determine whether gene expression differences could be replicated at the protein level, we developed IHC staining protocols to visualize neurons immunoreactive for either CART, CCK, or Ptprn within the EWcp. In each case, we identified a greater number of immunoreactive neurons in B6 mice, relative to D2 mice (all t13–14 > 5.08; all p < 0.0005; Figure 5), consistent with the results from analyses of Cart, Cck, and Ptprn in the qPCR array.
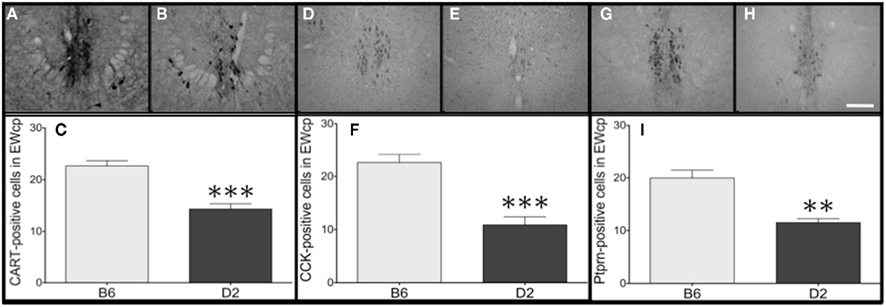
Figure 5. Gene expression differences in Cart, Cck, and Ptprn are verified at the protein level. Representative images from the EWcp (−3.4 to −3.6 mm from bregma) of B6 mice (A,D,G) and D2 mice (B,E,H). Within the EWcp, greater numbers of neurons immunoreactive for (C) CART, (F) CCK, and (I) Ptprn were found within B6 mice, relative to D2 mice. Scalebar = 500 μm, valid for all representative images. **p = 0.0002; ***p < 0.0001.
Discussion
The current study took advantage of publicly available tools in the ABA to identify several genes that were selectively expressed within the EWcp, and used tissue punch microdissection of the EWcp in combination with array expression profiling to quantify those transcripts (along with several other genes of interest) within tissue samples of the EWcp from adult male B6 and D2 mice. Our results, which expand on several previous studies that analyzed protein-level expression of ITFs and neuropeptides within the EWcp, confirm that the mRNA levels of several EWcp-specific genes and two ITFs are greater within B6 mice, relative to D2 mice. Interestingly, these findings are paralleled by differences in alcohol-related phenotypes among alcohol-preferring B6 mice and alcohol-avoiding D2 mice (Crabbe et al., 1983; Cunningham et al., 1992; Yoneyama et al., 2008).
Although prior evidence based on EWcp lesions and EWcp–Ucn1 protein expression in alcohol-preferring vs. alcohol-avoiding rodent strains suggested that EWcp–Ucn1 neurons promote alcohol drinking and food consumption (Bachtell et al., 2004; Weitemier and Ryabinin, 2005b), additional studies using intracranial injections showed that Ucn1 also decreased alcohol drinking and food consumption (Spina et al., 1996; Ryabinin et al., 2008). Thus, both a decrease in EWcp–Ucn1 tone (via. EWcp lesions) and an increase in Ucn1 tone (via. intracranial Ucn1 infusions) had similar effects on these two behaviors. One potential explanation for this apparent contradiction could be that higher Ucn1-IR within the EWcp of alcohol-preferring vs. alcohol-avoiding animals (including B6 vs. D2 mice) resulted from lower neuronal activity and less release of Ucn1 from the EWcp, rather than greater levels of Ucn1 mRNA.
Our current data provide a strong argument against a lower rate of release in B6 mice, because levels of Ucn1 mRNA were higher in the EWcp of these animals, mimicking the differences in protein expression. Thus, differences in Ucn1-IR within the EWcp of B6 vs. D2 mice are likely attributed to higher levels of Ucn1 mRNA within individual neurons (as well as a difference in the number of EWcp–Ucn1 neurons), rather than lower neural activity and lower rates of peptide release. Because EWcp–Ucn1 protein levels are reflective of EWcp–Ucn1 mRNA levels, these data support our longstanding hypothesis that greater activity of Ucn1 neurons within the EWcp is associated with a genetic predisposition toward greater alcohol intake and heightened alcohol sensitivity (Bachtell et al., 2003; Ryabinin and Weitemier, 2006). This hypothesis is also supported by our recent study in which genetic deletion of Ucn1 blunted alcohol preference and alcohol reward in mice on a B6 background (Giardino et al., 2011b).
In addition, levels of Fos and Egr1 mRNA were greater in the EWcp of B6 vs. D2 mice, arguing against the possibility that greater Ucn1-IR in B6 vs. D2 mice was due to less Ucn1 release. Although we did not directly compare baseline levels of Fos-IR in the current study, a previous experiment found that the number of Fos-IR cells was greater in B6 vs. D2 mice (Bachtell et al., 2003), consistent with our gene expression data. Since Fos and Egr1 are well-characterized markers of neural activity, this suggests that basal activity of the EWcp is higher in B6 vs. D2 mice. Given this presumed difference in neural activity, peptide release from the EWcp is likely to be higher in B6 vs. D2 mice, rather than vice versa.
An additional possibility for the seemingly contradictory relationship between Ucn1 tone and alcohol-related phenotypes was that lesions of the EWcp had the potential to eliminate DA neurons of the RLi, which intermingle with EWcp–Ucn1 neurons (Bachtell et al., 2002a; Gaszner and Kozicz, 2003; Fonareva et al., 2009). However, because there are more DA-synthesizing neurons in the RLi of alcohol-avoiding D2 mice as compared to alcohol-preferring B6 mice (D’Este et al., 2007), it remains unclear whether this neuronal population could contribute to alcohol intake and reward. Interestingly, despite this difference in the number of RLi neurons, no significant differences in transcripts characteristic of DAergic neurons were detected by qPCR.
We expanded our earlier studies examining Ucn1-IR and Fos-IR in the EWcp of B6 and D2 by detecting significantly more neurons immunoreactive for CART, CCK, and Ptprn in B6 vs. D2 mice. The protein product of Cart (cocaine- and amphetamine-regulated transcript) is a neuropeptide important for mediating drug reward and regulating food intake (Rogge et al., 2008). Our IHC analyses showed that CART has an extremely dense pattern of expression within the EWcp, a finding previously demonstrated by our lab and others across several mammalian species (Koylu et al., 1998; Kozicz, 2003; Lima et al., 2008; Cservenka et al., 2010). Here we show for the first time that EWcp–CART is differentially expressed between B6 and D2 mice at the mRNA and protein levels, suggesting that CART could be involved in similar functions as Ucn1. Since CART has been shown to co-localize with Ucn1 in EWcp (Kozicz, 2003; Cservenka et al., 2010), this result could also be due to either differences in mRNA levels per neuron and/or number of EWcp neurons between B6 and D2 mice.
The protein product of Cck (cholecystokinin) is a neuropeptide important for several functions, including regulation of food intake, anxiety-like behavior, and drug reward (Beglinger, 2002; Rotzinger and Vaccarino, 2003). Although the presence of CCK in the mammalian EWcp has been demonstrated previously (Maciewicz et al., 1984; Rattray et al., 1992), this is the first time that CCK peptide has been reported in the mouse EWcp. It is tempting to speculate that EWcp–CCK is involved in similar functions as EWcp–Ucn1 and EWcp–CART.
Although we were unable to generate a suitable IHC procedure for the protein product of growth hormone secretagogue receptor (Ghsr; the receptor for the orexigenic hormone ghrelin), previous studies from our laboratory implicate EWcp–Ghsr involvement in a mouse model of binge-like alcohol consumption (Kaur and Ryabinin, 2010), consistent with our finding of greater Ghsr mRNA expression in B6 vs. D2 mice.
Ptprn encodes protein tyrosine phosphatase, receptor type N (also known as islet antigen 2; IA-2). Other than the ABA, we are the first to report that this gene is expressed in the mammalian EWcp. The function of this gene is not well understood, despite the fact that it is a major auto-antigen in insulin-dependent diabetes mellitus and could be involved in mediating dense core vesicle release (Lu et al., 1996; Cai et al., 2004). As such, Ptprn could be involved in release of vesicles from the EWcp. This function, together with our identification of greater Ptprn expression in B6 vs. D2 mice, is an additional piece of evidence suggesting that EWcp neuronal activity is greater in B6 vs. D2 mice.
Use of in silico analyses as an additional confirmation of results from the EWcp qPCR array was largely successful, showing that at least six of the 14 identified transcripts also showed genotype-dependent expression throughout whole brain and/or cerebellum, striatum, hippocampus, hypothalamus, neocortex, and amygdala (Table 4). We speculate that although these transcripts display a typical EWcp-specific pattern within the midbrain, strain differences in expression of Btg3 and possibly Cck may generalize to several brain areas. On the other hand, the absence of consistent genotype-dependent expression of BC023892, Bves, Cart, Ghsr, Neto1, Postn, Ptprn, Ucn, Egr1, Fos, and Drd5 (and the opposite direction of difference for Rcn1) within several analyzed brain areas strengthens our conclusion that strain differences in stress-, feeding-, and addiction-related behavior may be related to expression of these genes specifically within the EWcp.
While some of these expression differences could theoretically be confirmed by Western blotting, the difficulties of dissecting relatively large quantities of EWcp from the mouse brain prevented this analysis. We anticipate that the other transcripts expressed higher in B6 vs. D2 mice also have corresponding differences in protein levels. In fact, this would be expected to be the case for nearly all EWcp-specific proteins that are co-expressed with Ucn1, because there are more Ucn1-positive neurons in B6 vs. D2 mice. Therefore, our studies are rather conservative in confirming the selectivity of gene expression within the EWcp.
It follows that greater mRNA expression within a micropunch from the EWcp region of B6 vs. D2 mice is, by itself, suggestive evidence that the gene is selectively expressed in EWcp. Thus, our finding that expression of the DA-related gene Drd5 is greater in EWcp microdissections from B6 vs. D2 mice suggests that this transcript might be expressed in EWcp neurons. The evidence for this possibility is further strengthened by the fact that other DA- and VTA-related genes were not differentially expressed between B6 vs. D2 mice, indicating a unique pattern of expression for Drd5. Drd5 is probably the least-studied DA receptor, and its potential expression and function in EWcp is an intriguing hypothesis that awaits further testing.
Our conservative use of four housekeeping genes to control for loading artifacts makes us confident in gene expression differences identified in the study. In that respect, it is interesting that we found that Gapdh was differentially expressed between B6 and D2 mice. Other studies have found that Gapdh can be regulated in the EWcp by stress (Derks et al., 2008). We would hypothesize that the observed differences in EWcp Gapdh expression are reliable, because other studies have not identified differential expression of Gapdh in whole-brain analysis of B6 and D2 mice (GeneNetwork, 2001; Shirley et al., 2004). Gapdh catalyzes an important energy-yielding step in carbohydrate metabolism, which could also serve as an indication of higher activity in the EWcp of B6 vs. D2 mice.
Taken together, we have identified at least 11 transcripts that are preferentially expressed in the EWcp, and differentially present in the EWcp of B6 vs. D2 mice. Further examination of these transcripts could shed light on the function of this recently characterized brain region, and could provide insight into the genetic underpinnings of behavioral differences between B6 and D2 mice, which serve as models of many contrasting behavioral phenotypes (including susceptibility to alcoholism, addiction, stress, and anxiety).
In broader terms, our approach illustrates how a combination of data-mining and genetic techniques can overcome the technical difficulties inherent in analyzing a distinct neuronal population. For example, the tissue punch samples that we used for our analyses contained a region larger than the EWcp itself, and the search features on the ABA provided fairly low spatial resolution. However, we were conservative in our identification of EWcp-specific genes, which led to successful utilization of the micropunch and expression profiling methods. The combination of standard gene expression analysis with a simple bioinformatics approach may prove to be a powerful technique for advancing the field of behavioral neurogenetics.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by NIH grants to Andrey E. Ryabinin (AA013738, AA016647, AA010760) and William J. Giardino (T32 DA7262-20). We thank the Allen Brain Atlas and GeneNetwork for providing data and information used in our analyses. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
References
Allen Brain Atlas. (2004). Allen Institute for Brain Science. Available at: http://www.brain-map.org
Bachtell, R. K., Tsivkovskaia, N. O., and Ryabinin, A. E. (2002a). Alcohol-induced c-Fos expression in the Edinger-Westphal nucleus: pharmacological and signal transduction mechanisms. J. Pharmacol. Exp. Ther. 302, 516–524.
Bachtell, R. K., Tsivkovskaia, N. O., and Ryabinin, A. E. (2002b). Strain differences in urocortin expression in the Edinger-Westphal nucleus and its relation to alcohol-induced hypothermia. Neuroscience 113, 421–434.
Bachtell, R. K., Wang, Y. M., Freeman, P., Risinger, F. O., and Ryabinin, A. E. (1999). Alcohol drinking produces brain region-selective changes in expression of inducible transcription factors. Brain Res. 847, 157–165.
Bachtell, R. K., Weitemier, A. Z., Galvan-Rosas, A., Tsivkovskaia, N. O., Risinger, F. O., Phillips, T. J., Grahame, N. J., and Ryabinin, A. E. (2003). The Edinger-Westphal-lateral septum urocortin pathway and its relationship to alcohol consumption. J. Neurosci. 233, 2477–2487.
Bachtell, R. K., Weitemier, A. Z., and Ryabinin, A. E. (2004). Lesions of the Edinger-Westphal nucleus in C57BL/6J mice disrupt ethanol-induced hypothermia and ethanol consumption. Eur. J. Neurosci. 20, 1613–1623.
Bittencourt, J. C., Vaughan, J., Arias, C., Rissman, R. A., Vale, W. W., and Sawchenko, P. E. (1999). Urocortin expression in rat brain: evidence against a pervasive relationship of urocortin-containing projections with targets bearing type 2 CRF receptors. J. Comp. Neurol. 415, 285–312.
Cai, T., Fukushige, T., Notkins, A. L., and Krause, M. (2004). Insulinoma-associated protein IA-2, a vesicle transmembrane protein, genetically interacts with unc-31/caps and affects neurosecretion in Caenorhabditis elegans. J. Neurosci. 24, 3115–3124.
Cavani, J. A., Reiner, A., Cuthbertson, S. L., Bittencourt, J. C., and Toledo, C. A. (2003). Evidence that urocortin is absent from neurons of the Edinger-Westphal nucleus in pigeons. Braz. J. Med. Biol. Res. 36, 1695–1700.
Chang, S. L., Patel, N. A., and Romero, A. A. (1995). Activation and desensitization of Fos immunoreactivity in the rat brain following ethanol administration. Brain Res. 679, 89–98.
Chesler, E. J., Lu, L., Wang, J., Williams, R. W., and Manly, K. F. (2004). WebQTL: rapid exploratory analysis of gene expression and genetic networks for brain and behavior. Nat. Neurosci. 7, 485–486.
Crabbe, J. C. Jr., Young, E. R., and Kosobud, A. (1983). Genetic correlations with ethanol withdrawal severity. Pharmacol. Biochem. Behav. 18(Suppl. 1), 541–547.
Cservenka, A., Spangler, E., Cote, D. M., and Ryabinin, A. E. (2010). Postnatal developmental profile of urocortin 1 and cocaine- and amphetamine-regulated transcript in the perioculomotor region of C57BL/6J mice. Brain Res. 1319, 33–43.
Cunningham, C. L., Niehus, D. R., Malott, D. H., and Prather, L. K. (1992). Genetic differences in the rewarding and activating effects of morphine and ethanol. Psychopharmacology (Berl.) 107, 385–393.
Derks, N. M., Muller, M., Gaszner, B., Tilburg-Ouwens, D. T., Roubos, E. W., and Kozicz, L. T. (2008). Housekeeping genes revisited: different expressions depending on gender, brain area and stressor. Neuroscience 156, 305–309.
D’Este, L., Casini, A., Puglisi-Allegra, S., Cabib, S., and Renda, T. G. (2007). Comparative immunohistochemical study of the dopaminergic systems in two inbred mouse strains (C57BL/6J and DBA/2J). J. Chem. Neuroanat. 33, 67–74.
Dun, S. L., Brailoiu, G. C., Mizuo, K., Yang, J., Chang, J. K., and Dun, N. J. (2005). Neuropeptide B immunoreactivity in the central nervous system of the rat. Brain Res. 1045, 157–163.
Fonareva, I., Spangler, E., Cannella, N., Sabino, V., Cottone, P., Ciccocioppo, R., Zorrilla, E. P., and Ryabinin, A. E. (2009). Increased perioculomotor urocortin 1 immunoreactivity in genetically selected alcohol preferring rats. Alcohol. Clin. Exp. Res. 33, 1956–1965.
Foo, K. S., Brismar, H., and Broberger, C. (2008). Distribution and neuropeptide coexistence of nucleobindin-2 mRNA/nesfatin-like immunoreactivity in the rat CNS. Neuroscience 156, 563–579.
Gaszner, B., Csernus, V., and Kozicz, T. (2004). Urocortinergic neurons respond in a differentiated manner to various acute stressors in the Edinger-Westphal nucleus in the rat. J. Comp. Neurol. 480, 170–179.
Gaszner, B., Jensen, K. O., Farkas, J., Reglodi, D., Csernus, V., Roubos, E. W., and Kozicz, T. (2009). Effects of maternal separation on dynamics of urocortin 1 and brain-derived neurotrophic factor in the rat non-preganglionic Edinger-Westphal nucleus. Int. J. Dev. Neurosci. 27, 439–451.
Gaszner, B., and Kozicz, T. (2003). Interaction between catecholaminergic terminals and urocortinergic neurons in the Edinger-Westphal nucleus in the rat. Brain Res. 989, 117–121.
GeneNetwork. (2001). GeneNetwork: A Tour and Tutorial. Available at: http://www.genenetwork.org
Giardino, W. J., Pastor, R., Anacker, A. M., Spangler, E., Cote, D. M., Li, J., Stenzel-Poore, M. P., Phillips, T. J., and Ryabinin, A. E. (2011a). Dissection of corticotropin-releasing factor system involvement in locomotor sensitivity to methamphetamine. Genes Brain Behav. 10, 78–89.
Giardino, W. J., Cocking, D. L., Kaur, S., Cunningham, C. L., and Ryabinin, A. E. (2011b). Urocortin-1 within the centrally-projecting Edinger-Westphal nucleus is crtical for ethanol preference. PLoS ONE 6, e26997. doi:10.1371/journal.pone.0026997
Kaur, S., and Ryabinin, A. E. (2010). Ghrelin receptor antagonism decreases alcohol consumption and activation of perioculomotor urocortin-containing neurons. Alcohol. Clin. Exp. Res. 34, 1525–1534.
Kiianmaa, K., Hyytia, P., Samson, H. H., Engel, J. A., Svensson, L., Soderpalm, B., Larsson, A., Colombo, G., Vacca, G., Finn, D. A., Bachtell, R. K., and Ryabinin, A. E. (2003). New neuronal networks involved in ethanol reinforcement. Alcohol. Clin. Exp. Res. 27, 209–219.
Korosi, A., Schotanus, S., Olivier, B., Roubos, E. W., and Kozicz, T. (2005). Chronic ether stress-induced response of urocortin 1 neurons in the Edinger-Westphal nucleus in the mouse. Brain Res. 1046, 172–179.
Koylu, E. O., Couceyro, P. R., Lambert, P. D., and Kuhar, M. J. (1998). Cocaine- and amphetamine-regulated transcript peptide immunohistochemical localization in the rat brain. J. Comp. Neurol. 391, 115–132.
Kozicz, T. (2003). Neurons colocalizing urocortin and cocaine and amphetamine-regulated transcript immunoreactivities are induced by acute lipopolysaccharide stress in the Edinger-Westphal nucleus in the rat. Neuroscience 116, 315–320.
Kozicz, T. (2007). On the role of urocortin 1 in the non-preganglionic Edinger-Westphal nucleus in stress adaptation. Gen. Comp. Endocrinol. 153, 235–240.
Kozicz, T., Bittencourt, J. C., May, P. J., Reiner, A., Gamlin, P. D., Palkovits, M., Horn, A. K., Toledo, C. A., and Ryabinin, A. E. (2011). The Edinger-Westphal nucleus: a historical, structural, and functional perspective on a dichotomous terminology. J. Comp. Neurol. 519, 1413–1434.
Kozicz, T., Bordewin, L. A., Czeh, B., Fuchs, E., and Roubos, E. W. (2008). Chronic psychosocial stress affects corticotropin-releasing factor in the paraventricular nucleus and central extended amygdala as well as urocortin 1 in the non-preganglionic Edinger-Westphal nucleus of the tree shrew. Psychoneuroendocrinology 33, 741–754.
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., Boe, A. F., Boguski, M. S., Brockway, K. S., Byrnes, E. J., Chen, L., Chen, T. M., Chin, M. C., Chong, J., Crook, B. E., Czaplinska, A., Dang, C. N., Datta, S., Dee, N. R., Desaki, A. L., Desta, T., Diep, E., Dolbeare, T. A., Donelan, M. J., Dong, H. W., Dougherty, J. G., Duncan, B. J., Ebbert, A. J., Eichele, G., Estin, L. K., Faber, C., Facer, B. A., Fields, R., Fischer, S. R., Fliss, T. P., Frensley, C., Gates, S. N., Glattfelder, K. J., Halverson, K. R., Hart, M. R., Hohmann, J. G., Howell, M. P., Jeung, D. P., Johnson, R. A., Karr, P. T., Kawal, R., Kidney, J. M., Knapik, R. H., Kuan, C. L., Lake, J. H., Laramee, A. R., Larsen, K. D., Lau, C., Lemon, T. A., Liang, A. J., Liu, Y., Luong, L. T., Michaels, J., Morgan, J. J., Morgan, R. J., Mortrud, M. T., Mosqueda, N. F., Ng, L. L., Ng, R., Orta, G. J., Overly, C. C., Pak, T. H., Parry, S. E., Pathak, S. D., Pearson, O. C., Puchalski, R. B., Riley, Z. L., Rockett, H. R., Rowland, S. A., Royall, J. J., Ruiz, M. J., Sarno, N. R., Schaffnit, K., Shapovalova, N. V., Sivisay, T., Slaughterbeck, C. R., Smith, S. C., Smith, K. A., Smith, B. I., Sodt, A. J., Stewart, N. N., Stumpf, K. R., Sunkin, S. M., Sutram, M., Tam, A., Teemer, C. D., Thaller, C., Thompson, C. L., Varnam, L. R., Visel, A., Whitlock, R. M., Wohnoutka, P. E., Wolkey, C. K., Wong, V. Y., Wood, M., Yaylaoglu, M. B., Young, R. C., Youngstrom, B. L., Yuan, X. F., Zhang, B., Zwingman, T. A., and Jones, A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176.
Lewis, S. R., Dym, C., Chai, C., Singh, A., Kest, B., and Bodnar, R. J. (2007). Genetic variance contributes to ingestive processes: a survey of eleven inbred mouse strains for fat (intralipid) intake. Physiol. Behav. 90, 82–94.
Lima, F. B., Henderson, J. A., Reddy, A. P., Tokuyama, Y., Hubert, G. W., Kuhar, M. J., and Bethea, C. L. (2008). Unique responses of midbrain CART neurons in macaques to ovarian steroids. Brain Res. 1227, 76–88.
Loewy, A. D., and Saper, C. B. (1978). Edinger-Westphal nucleus: projections to the brain stem and spinal cord in the cat. Brain Res. 150, 1–27.
Loewy, A. D., Saper, C. B., and Yamodis, N. D. (1978). Re-evaluation of the efferent projections of the Edinger-Westphal nucleus in the cat. Brain Res. 141, 153–159.
Lu, J., Li, Q., Xie, H., Chen, Z. J., Borovitskaya, A. E., Maclaren, N. K., Notkins, A. L., and Lan, M. S. (1996). Identification of a second transmembrane protein tyrosine phosphatase, IA-2beta, as an autoantigen in insulin-dependent diabetes mellitus: precursor of the 37-kDa tryptic fragment. Proc. Natl. Acad. Sci. U.S.A. 93, 2307–2311.
Maciewicz, R., Phipps, B. S., Grenier, J., and Poletti, C. E. (1984). Edinger-Westphal nucleus: cholecystokinin immunocytochemistry and projections to spinal cord and trigeminal nucleus in the cat. Brain Res. 299, 139–145.
Okere, B., Xu, L., Roubos, E. W., Sonetti, D., and Kozicz, T. (2010). Restraint stress alters the secretory activity of neurons co-expressing urocortin-1, cocaine- and amphetamine-regulated transcript peptide and nesfatin-1 in the mouse Edinger-Westphal nucleus. Brain Res. 1317, 92–99.
Overall, R. W., Kempermann, G., Peirce, J., Lu, L., Goldowitz, D., Gage, F. H., Goodwin, S., Smit, A. B., Airey, D. C., Rosen, G. D., Schalkwyk, L. C., Sutter, T. R., Nowakowski, R. S., Whatley, S., and Williams, R. W. (2009). Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front. Neurosci. 3:55. doi:10.3389/neuro.15.003.2009
Rattray, M., Savery, D., Wotherspoon, G., Priestley, J. V., and Smith, G. S. (1992). Two populations of cells that express preprocholecystokinin mRNA in ventral periaqueductal grey. Neurosci. Lett. 143, 55–59.
Rogge, G., Jones, D., Hubert, G. W., Lin, Y., and Kuhar, M. J. (2008). CART peptides: regulators of body weight, reward and other functions. Nat. Rev. Neurosci. 9, 747–758.
Rotzinger, S., and Vaccarino, F. J. (2003). Cholecystokinin receptor subtypes: role in the modulation of anxiety-related and reward-related behaviours in animal models. J. Psychiatry Neurosci. 28, 171–181.
Rouwette, T., Klemann, K., Gaszner, B., Scheffer, G. J., Roubos, E. W., Scheenen, W. J., Vissers, K., and Kozicz, T. (2011). Differential responses of corticotropin-releasing factor and urocortin 1 to acute pain stress in the rat brain. Neuroscience 183, 15–24.
Rouwette, T. P., Kozicz, T., Olde Loohuis, N. F., Gaszner, B., Vreugdenhil, E., Scheffer, G. J., Roubos, E. W., Vissers, K. C., and Scheenen, W. J. (2010). Acute pain increases phosphorylation of DCLK-long in the Edinger-Westphal nucleus but not in the hypothalamic paraventricular nucleus of the rat. J. Pain 11, 930–940.
Ryabinin, A. E., Bachtell, R. K., Freeman, P., and Risinger, F. O. (2001). ITF expression in mouse brain during acquisition of alcohol self-administration. Brain Res. 890, 192–195.
Ryabinin, A. E., Criado, J. R., Henriksen, S. J., Bloom, F. E., and Wilson, M. C. (1997). Differential sensitivity of c-Fos expression in hippocampus and other brain regions to moderate and low doses of alcohol. Mol. Psychiatry 2, 32–43.
Ryabinin, A. E., Galvan-Rosas, A., Bachtell, R. K., and Risinger, F. O. (2003). High alcohol/sucrose consumption during dark circadian phase in C57BL/6J mice: involvement of hippocampus, lateral septum and urocortin-positive cells of the Edinger-Westphal nucleus. Psychopharmacology (Berl.) 165, 296–305.
Ryabinin, A. E., Tsivkovskaia, N. O., and Ryabinin, S. A. (2005). Urocortin 1-containing neurons in the human Edinger-Westphal nucleus. Neuroscience 134, 1317–1323.
Ryabinin, A. E., Wang, Y. M., and Finn, D. A. (1999). Different levels of Fos immunoreactivity after repeated handling and injection stress in two inbred strains of mice. Pharmacol. Biochem. Behav. 63, 143–151.
Ryabinin, A. E., and Weitemier, A. Z. (2006). The urocortin 1 neurocircuit: ethanol-sensitivity and potential involvement in alcohol consumption. Brain Res. Rev. 52, 368–380.
Ryabinin, A. E., Yoneyama, N., Tanchuck, M. A., Mark, G. P., and Finn, D. A. (2008). Urocortin 1 microinjection into the mouse lateral septum regulates the acquisition and expression of alcohol consumption. Neuroscience 151, 780–790.
Saba, L., Bhave, S. V., Grahame, N., Bice, P., Lapadat, R., Belknap, J., Hoffman, P. L., and Tabakoff, B. (2006). Candidate genes and their regulatory elements: alcohol preference and tolerance. Mamm. Genome 17, 669–688.
Shirley, R. L., Walter, N. A., Reilly, M. T., Fehr, C., and Buck, K. J. (2004). Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat. Neurosci. 7, 699–700.
Spangler, E., Cote, D. M., Anacker, A. M., Mark, G. P., and Ryabinin, A. E. (2009). Differential sensitivity of the perioculomotor urocortin-containing neurons to ethanol, psychostimulants and stress in mice and rats. Neuroscience 160, 115–125.
Spina, M., Merlo-Pich, E., Chan, R. K., Basso, A. M., Rivier, J., Vale, W., and Koob, G. F. (1996). Appetite-suppressing effects of urocortin, a CRF-related neuropeptide. Science 273, 1561–1564.
Sterrenburg, L., Borch, A., Peeters, B. W., Pinter, O., Zelena, D., Roubos, E. W., and Kozicz, T. (2011). Acute ether stress differentially affects corticotropin-releasing factor and urocortin 1 in the Brattleboro rat. Brain Res. 1398, 21–29.
Tanaka, H., Yoshida, T., Miyamoto, N., Motoike, T., Kurosu, H., Shibata, K., Yamanaka, A., Williams, S. C., Richardson, J. A., Tsujino, N., Garry, M. G., Lerner, M. R., King, D. S., O’Dowd, B. F., Sakurai, T., and Yanagisawa, M. (2003). Characterization of a family of endogenous neuropeptide ligands for the G protein-coupled receptors GPR7 and GPR8. Proc. Natl. Acad. Sci. U.S.A. 100, 6251–6256.
Topple, A. N., Hunt, G. E., and McGregor, I. S. (1998). Possible neural substrates of beer-craving in rats. Neurosci. Lett. 252, 99–102.
Turek, V. F., Bennett, B., and Ryabinin, A. E. (2008). Differences in the urocortin 1 system between long-sleep and short-sleep mice. Genes Brain Behav. 7, 113–119.
Turek, V. F., Tsivkovskaia, N. O., Hyytia, P., Harding, S., Le, A. D., and Ryabinin, A. E. (2005). Urocortin 1 expression in five pairs of rat lines selectively bred for differences in alcohol drinking. Psychopharmacology (Berl.) 181, 511–517.
Vaughan, J., Donaldson, C., Bittencourt, J., Perrin, M. H., Lewis, K., Sutton, S., Chan, R., Turnbull, A. V., Lovejoy, D., Rivier, C., Sawchenko, P. E., and Vale, W. (1995). Urocortin, a mammalian neuropeptide related to fish urotensin I and to corticotropin-releasing factor. Nature 378, 287–292.
Walter, N. A., Bottomly, D., Laderas, T., Mooney, M. A., Darakjian, P., Searles, R. P., Harrington, C. A., McWeeney, S. K., Hitzemann, R., and Buck, K. J. (2009). High throughput sequencing in mice: a platform comparison identifies a preponderance of cryptic SNPs. BMC Genomics 10, 379. doi:10.1186/1471-2164-10-379
Weitemier, A. Z., and Ryabinin, A. E. (2005a). Brain region-specific regulation of urocortin 1 innervation and corticotropin-releasing factor receptor type 2 binding by ethanol exposure. Alcohol. Clin. Exp. Res. 29, 1610–1620.
Weitemier, A. Z., and Ryabinin, A. E. (2005b). Lesions of the Edinger-Westphal nucleus alter food and water consumption. Behav. Neurosci. 119, 1235–1243.
Weitemier, A. Z., Tsivkovskaia, N. O., and Ryabinin, A. E. (2005). Urocortin 1 distribution in mouse brain is strain-dependent. Neuroscience 132, 729–740.
Weitemier, A. Z., Woerner, A., Backstrom, P., Hyytia, P., and Ryabinin, A. E. (2001). Expression of c-Fos in Alko alcohol rats responding for ethanol in an operant paradigm. Alcohol. Clin. Exp. Res. 25, 704–710.
Xu, L., Bloem, B., Gaszner, B., Roubos, E. W., and Kozicz, T. (2009). Sex-specific effects of fasting on urocortin 1, cocaine- and amphetamine-regulated transcript peptide and nesfatin-1 expression in the rat Edinger-Westphal nucleus. Neuroscience 162, 1141–1149.
Xu, L., Bloem, B., Gaszner, B., Roubos, E. W., and Kozicz, T. (2010). Stress-related changes in the activity of cocaine- and amphetamine-regulated transcript and nesfatin neurons in the midbrain non-preganglionic Edinger-Westphal nucleus in the rat. Neuroscience 170, 478–488.
Xu, L., Scheenen, W. J., Leshan, R. L., Patterson, C. M., Elias, C. F., Bouwhuis, S., Roubos, E. W., Myers, M. G. Jr., and Kozicz, T. (2011). Leptin signaling modulates the activity of urocortin 1 neurons in the mouse nonpreganglionic Edinger-Westphal nucleus. Endocrinology 152, 979–988.
Yoneyama, N., Crabbe, J. C., Ford, M. M., Murillo, A., and Finn, D. A. (2008). Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol 42, 149–160.
Keywords: Edinger–Westphal, midbrain, oculomotor, neuropeptide, inducible transcription factor, immediate early gene, alcohol, urocortin
Citation: Giardino WJ, Cote DM, Li J and Ryabinin AE (2012) Characterization of genetic differences within the centrally projecting Edinger–Westphal nucleus of C57BL/6J and DBA/2J mice by expression profiling. Front. Neuroanat. 6:5. doi: 10.3389/fnana.2012.00005
Received: 25 November 2011; Paper pending published: 21 December 2011;
Accepted: 29 January 2012; Published online: 14 February 2012.
Edited by:
Li Yun-Qing, Fourth Military Medical University, ChinaReviewed by:
Richard S. Nowakowski, Florida State University, USAPaul A. Gray, Washington University, USA
Copyright: © 2012 Giardino, Cote, Li and Ryabinin. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Andrey E. Ryabinin, Department of Behavioral Neuroscience, Oregon Health and Science University, Mail Code L-470, 3181 Southwest Sam Jackson Park Road, Portland, OR 97239, USA. e-mail:cnlhYmluaW5Ab2hzdS5lZHU=
 Dawn M. Cote
Dawn M. Cote