- 1National Veterinary Services Laboratories, APHIS, United States Department of Agriculture, Ames, IA, United States
- 2Infectious Bacterial Diseases Research Unit, ARS, United States Department of Agriculture, Ames, IA, United States
- 3WOAH Reference Laboratory for Leptospirosis, Department of Medical Microbiology and Infection Prevention, Expertise Center for Reference and Research on Leptospirosis, Amsterdam University Medical Center, Amsterdam, Netherlands
The genus Leptospira is a diverse and unique group of bacteria comprising multiple saprophytic and pathogenic species, which survive and persist in suitable moist environments. Pathogenic species cause human and animal leptospirosis, a global and neglected zoonotic disease. Disease transmission occurs by exposure to contaminated water and moist soil environments or by contact with domestic animals and wildlife acting as reservoir hosts that shed Leptospira via urine. Here, we describe the unexpected diversity of saprophytic and pathogenic species of Leptospira isolated from water in the Midwestern United States. Samples were collected by volunteers in 11 counties in Iowa from water sources, including puddles, sewage, creeks, ponds, lakes, and rivers, during the summer of 2021. One hundred and five water samples were tested by culture for the presence of saprophytic and pathogenic species and by lipL32 qPCR specific for the detection of pathogens; 82 (78.1%) were culture positive and five (4.8%) were positive by lipL32 qPCR. Whole genome sequencing of isolates cultured from water samples identified 10 species of saprophytes, namely L. montravelensis, L. kemamanensis, L. bandrabouensis, L. bourretii, L. bouyouniensis, L. chreensis, L. ellinghausenii, L. terpstrae, L. yanagawae, and L. abararensis, as well as three novel saprophytic species. Whole genome sequencing also identified two novel pathogenic species. The remaining cultures comprised mixed populations of saprophytic species and six comprised a mixture of saprophytic and pathogenic species. One of these mixed cultures was enriched to select for a clonal isolate of pathogenic Leptospira, strain WS101.C1, which was classified as L. interrogans serogroup Djasiman serovar Djasiman. Cumulatively, 9.5% (10/105) of water samples were positive for pathogenic Leptospira. This study emphasizes the diversity of Leptospira present in water sources in the Midwestern United States and provides unique opportunities to explore the geographic diversity and evolution of this genus. The identification of known and novel pathogenic species circulating in local water sources highlights their potential usefulness as diagnostic antigens, as well as the role of water in the transmission of infection to human and animal populations. Integrating knowledge on human, animal, and environmental health is essential to control and predict risk for zoonoses.
1 Introduction
Leptospirosis is a neglected zoonotic disease with a worldwide distribution caused by pathogenic leptospires that can survive and persist in suitable moist environments (Bharti et al., 2003; Thibeaux et al., 2017; Casanovas-Massana et al., 2018; Stone et al., 2022). The genus Leptospira is diverse and divided into 72 species (Vincent et al., 2019; Casanovas-Massana et al., 2020; Grillová et al., 2021; Korba et al., 2021; Fernandes et al., 2022; Dos Santos et al., 2023). The phylogenomic analysis separates the genus into two clades: the S clade contains saprophytes isolated from the environment and not responsible for infections, compared to the P clade, which contains pathogens/intermediates responsible for infections in humans and animals. Each S and P clade is further subdivided into two subclades: S1 and S2 contain 26 and 5 species, respectively, while P1 and P2 contain 20 and 21 species, respectively.
The epidemiology of a zoonotic disease such as leptospirosis is complex and makes it a paradigm of One Health (Bharti et al., 2003; Putz and Nally, 2020; Sykes et al., 2022). Pathogenic Leptospira colonizes the proximal renal tubules of domestic animals and wildlife, which can act as reservoir hosts of infection and excrete Leptospira via urine into the environment. Transmission to humans and animals occurs by direct contact with reservoir hosts or indirect exposure to environmental water and moist soil contaminated with pathogenic Leptospira (Faine et al., 1999). Leptospires can also colonize the genital tract of domestic livestock and are shed via semen or uterine discharges (Ellis, 2015). Leptospira can be maintained in moist soil and water for weeks to months (Saito et al., 2013; Bierque et al., 2020; Stone et al., 2022).
Human leptospirosis ranges in severity from a mild, self-limited febrile illness to a fulminant, life-threatening disease (Haake and Levett, 2015). In the United States, leptospirosis is rarely recognized, and most cases are associated with occupational exposure to infected animals (Heath et al., 1965), recreational exposures (Schiemann, 1973; Sasaki et al., 1993; Katz et al., 1997; Lee et al., 2002), and individuals living in economically disadvantaged inner-city environments (Vinetz et al., 1996; Viotti et al., 2020; McNeilly et al., 2023). In the state of Iowa, outbreaks of human leptospirosis were associated with farm creeks, wildlife, and domestic livestock (Diesch and McCulloch, 1966; Diesch et al., 1970). Serogroup Pomona was isolated from humans with bovine contacts, while serogroup Icterohaemorrhagiae was isolated from a patient exposed to mud polluted with rat feces (Tjalma and Galton, 1965). The few cases where pathogenic leptospires have been isolated were from surface waters of recreational areas (Diesch and McCulloch, 1966; Braun and McCULLOCH, 1968; Crawford et al., 1969), wildlife (Diesch et al., 1970; Smith et al., 1992), and cattle and swine (Ellis and Thiermann, 1986; Bolin and Cassells, 1990; Bolin et al., 1991; Nally et al., 2018).
Integrating the knowledge of human, animal, and environmental health is essential to control and predict zoonotic diseases. While investigations on animal and human interfaces are increasing, greater incorporation of environmental and ecosystem components has been highlighted as a missing link in the One Health approach (Nally et al., 2016; Warnasekara et al., 2022). The importance and role of water in the transmission of leptospirosis is well-established (Monahan et al., 2009; Barragan et al., 2011; Bierque et al., 2020; Stone et al., 2022). Here, we describe saprophytic and pathogenic species of Leptospira isolated from water sources in the Midwestern United States, including the presence of novel species.
2 Materials and methods
2.1 Study site and sampling
This study was conducted in Iowa, a midwestern U.S. state. Water samples were collected from 11 counties in central Iowa (Polk, Story, Boone, Appanoose, Warren, Marshall, Webster, Audubon, Dallas, Hardin, and Hamilton) from puddles, sewage, creeks, ponds, lakes, and rivers during the summer months of 2021. Water samples were collected by volunteers who were provided with a kit containing a 50-mL conical tube, a 1-mL syringe, and two 15-mL conical tubes containing either 5 mL of HAN media (Hornsby et al., 2020) or 5 mL of HAN media, which contained a combination of antimicrobial agents, including sulfamethoxazole (40 μg/mL), trimethoprim (20 μg/mL), amphotericin B (5 μg/mL), fosfomycin (400 μg/mL), and 5-fluorouracil (100 μg/mL) (STAFF) (Chakraborty et al., 2011), for each water sample collected. Volunteers were asked to collect 40 mL of surface water into a sterile 50-mL conical tube. A 250-μL aliquot of each water sample was then transferred, using the 1-ml syringe, into each 15-mL tube containing media. Samples were labeled with GPS coordinates of the sample location and a description of the water sample type. Water sources were selected by volunteers, which included areas with a wildlife presence, domestic animal presence, and recreational areas. A total of 105 water samples were collected (Supplementary Table S1). Representative photographs of sample sites taken by volunteers are provided (Figure 1). The samples were transported at ambient temperature to the National Veterinary Services Laboratories, APHIS, Ames, IA, within 24 h of collection and were processed for culture and lipL32 rt-PCR.

Figure 1. Representative photographs of water sample sites. WS designation refers to those listed in Supplementary Table S1.
2.2 DNA extraction and lipL32 qPCR
In total, 40 mL of each water sample was centrifuged at 15,000 × g for 20 min at 4°C (Riediger et al., 2016). Pellets were resuspended in 500 μL PBS, and DNA was extracted using an automated Maxwell RSC cell DNA purification kit (Promega Corporation, Madison, WI) as previously described (Casanovas-Massana et al., 2018). DNA was tested in triplicate by lipL32 qPCR as previously described (Hamond et al., 2022). In brief, the lipL32 gene was amplified and detected using the following primers and probe: LipL32-47Fd (5′-GCATTACMGCTTGTGGTG-3′) and LipL32-301Rd (5′-CCGATTTCGCCWGTTGG-3′), and LipL32-189P (6-carboxyfluorescein [FAM]-5′-AAAGCCAGGACAAGCGCCG-3′-black hole quencher 1 [BHQ1]), using PerfeCTa qPCR ToughMix®, Low ROX™ (Quanta Biosciences, Gaithersburg, MD, United States). As a control for PCR inhibitors, the TaqMan® Exogenous Internal Positive Control (Applied Biosystems) was added to the master mix to confirm DNA amplification and detect the presence of amplification inhibitory substances in each sample. The sample was considered positive when duplicates or triplicates were positive with Ct values <40 (Stoddard et al., 2009; Galloway and Hoffmaster, 2015).
2.3 Culture
A 250-μL aliquot of each water sample was inoculated at the time of collection into 5 mL of HAN liquid media (Hornsby et al., 2020) or 5 mL of HAN liquid media containing STAFF (Chakraborty et al., 2011). In the laboratory, 500 μL of the inoculated media containing STAFF was then used to inoculate 2 × 5 mL of HAN media containing STAFF and incubated at 29°C and 37°C. Cultures were examined daily by darkfield microscopy for up to 6 months. Media without STAFF resulted in heavy growth of contaminants.
2.4 Genome sequencing and analysis
DNA was extracted from separate 5 mL cultures obtained from recovered isolates using the Maxwell RSC Purefood Purification Pathogen kit (Promega Corporation, Madison, WI), following the manufacturer’s instructions. The concentration of genomic DNA was determined using Qubit® (Qubit dsDNA Broad Range Assay, Qubit 3.0 fluorometer, Invitrogen, Carlsbad, CA, United States). Illumina whole-genome sequence was obtained per the manufacturer’s instructions by employing the Illumina Nextera XT DNA Library Preparation Kit and the MiSeq Desktop Sequencer (2×250 v2 paired-end chemistry, Illumina, San Diego, CA), as previously described (Hamond et al., 2022).
Genome analysis after assembly, including scaffold length and number, N50, and top BLAST identifications, was used to determine whether each assembled genome was derived from a single species’ DNA. Illumina whole genome sequencing (WGS) reads were analyzed with Kraken (Wood and Salzberg, 2014) and visually displayed with Krona (Ondov et al., 2011). This analysis assisted in evaluating genomic DNA fidelity to a single isolate and species identification. Paired-end reads were assembled with SPAdes, which was run with the careful option enabled (Bankevich et al., 2012), and assemblies were checked for quality by comparing the expected genome size with the total assembly size and verifying scaffolds as Leptospira by BLAST (Altschul et al., 1990).
Phylogenetic trees were built from assembled genomes with Mashtree (Katz et al., 2019) and Leptospira species in the National Center for Biotechnology Information (NCBI) RefSeq (O'Leary et al., 2016), which are labeled with the NCBI accession number. Samples were subsets on Mashtree groupings based on three main branches. Using subsets of assembled and NCBI RefSeq, three genome trees were built using kSNP (Gardner et al., 2015). TipAlleleCounts-ML tree output from kSNP showed approximate SNP counts between isolates and those isolates most similar to references. All data from isolates of Leptospira detected in this study are labeled according to Supplementary Table S1. Sequencing data for all samples were deposited in the NCBI GenBank database, BioProject number PRJNA944241, Supplementary Table S2.
Bracken (Bayesian Re-estimation of Abundance with Kraken) was used as a statistical method for computing the abundance of species in DNA sequences from a metagenomics sample1 (Lu et al., 2017).
For all cultures, including cultures with mixed species of Leptospira, the DNA sequence for the ppk gene was informatically extracted from each genome assembly using Geneious Prime 2022.2.2. The ppk gene was also extracted from the WGS of the reference strains obtained from NCBI using the same method. A phylogenetic tree was made for the ppk gene using the Geneious Tree Builder Tamura-Nei model and the Neighbor-Joining method. The reference strains are labeled with the NCBI accession number followed by the species and strain. In cultures with mixed species, the ppk sequences extracted from WGS assemblies include a label of 1, 2, or 3 to differentiate the separately assembled ppk genes that were derived from the mixed population.
A delineation of the species based on the genome sequences was performed by Average Nucleotide Identity (ANI) using EZBioCloud (Yoon et al., 2017). An OrthoANI result greater than or equal to 95% was considered the same species. The reference genomes for each species group were the same as those used in the ppk analysis.
2.5 Sequencing of lipL32 and lfb1
The lipL32 gene was amplified for sequencing using the same primers as the qPCR. PCR products were then purified and labeled using the Big Dye Terminator v3.1 cycle sequencing reagent (Applied Biosystems, Foster City, California, United States). The lfb1 gene was detected with primers LFB1-F (5′-CATTCATGTTTCGAATCATTTCAAA-3′) and LFB1-R (5′-GGCCCAAGTTCCTTCTAAAAG-3′) (Merien et al., 2005). Sequencing was performed using the ABI 3130XL Genetic Analyzer. Sequence data were analyzed with DNAStar’s Lasergene sequence analysis software. Consensus sequences were compared with available sequences in the GenBank database using BLAST.
2.6 Serotyping of Leptospira cultured from water sources
Leptospira was serotyped by the microscopic agglutination test (MAT) using a panel of polyclonal rabbit reference antisera representing 14 serogroups: Australis, Autumnalis, Ballum, Bataviae, Canicola, Djasiman, Grippotyphosa, Hebdomadis, Icterohaemorrhagiae, Mini, Pomona, Pyrogenes, Sejröe, Tarassovi, and (Serogroup not yet determined) serovar Room22 as previously described (Supplementary Table S3). The isolate WS101.C1 was further typed to the serovar level by the World Organisation for Animal Health (WOAH) Reference Laboratory for Leptospirosis by performing MAT with a panel of reference monoclonal antibodies that characteristically agglutinate serovars from the relevant serogroups as previously described (Terpstra et al., 1985; Hartskeerl et al., 2006; Goris and Hartskeerl, 2014).
2.7 Approach to enrich for clonal isolates of Leptospira
Sample WS101 was determined by WGS to contain both saprophytic and pathogenic species of Leptospira. To separate the pathogen and obtain a clonal isolate, the culture was grown in liquid HAN at 37°C in 5% CO2 (Hornsby et al., 2020) and enriched using polyclonal rabbit reference antisera for serogroup Djasiman before plating for screening of clonal isolates on agar plates, and as previously described (Hamond et al., 2023). To identify clonal isolates, DNA was extracted from a 5-mL culture using the Maxwell RSC Purefood Purification Pathogen kit (Promega Corporation, Madison, WI), following instructions from the manufacturer. PCR for rrs was performed as previously described (Mérien et al., 1992). PCR amplicons were sequenced, and consensus sequence data were generated and analyzed as previously described (Hamond et al., 2022).
2.8 Evaluation of virulence
All animal experimentation was conducted in accordance with protocols as reviewed and approved by the Animal Care & Use Committee at the National Animal Disease Center (ARS-2021-938) and as approved by USDA institutional guidelines. Three water samples in which pathogenic leptospires were detected [WS58 (L. sp. nov Patho P2), WS92 (L. sp. nov Patho P1), and WS101.C1 (L. interrogans serogroup Djasiman)] were used to inoculate liquid HAN medium at 29°C. The virulence of the pathogens cultured in the aforementioned manner was evaluated by intraperitoneal injection of 108 Leptospira in 1 mL into groups (n = 4 per group) of female golden Syrian hamsters (Mesocricetus auratus). A negative control group received HAN media alone. At 3 weeks post-inoculation, kidney tissues were harvested for Leptospira culture and lipL32 qPCR, as previously described (Wunder Jr et al., 2016; Fernandes et al., 2022). DNA from L. interrogans serovar Canicola strain H. Utrecht IV was used for a standard curve. Sera were collected for MAT, which was performed according to World Organization for Animal Health (WOAH) guidelines (Cole et al., 1973), using a panel of 17 antigens representative of 14 serogroups, serovar Room22 (Supplementary Table S4), and strains WS58, WS92, and WS101.C1. A titer was considered positive at ≥ 1:100.
2.9 Spatial data
Geographic Information System (GIS) mapping was prepared, and its accuracy was assessed using Google Earth software.
3 Results
3.1 lipL32 qPCR
One hundred and five water samples were collected from water sources in 11 counties in the state of Iowa, including recreational areas and those with a wildlife and domestic animal presence (Figure 1). Of these, five (4.8%) samples (WS40, WS57, WS84, WS91, and WS101) collected from five different counties (Appanoose, Webster, Polk, Boone, and Hamilton) were positive by lipL32 qPCR (Supplementary Table S1; Figure 2). Positive Ct values ranged from 24 to 37 (mean = 33.6 ± 5.5) (Supplementary Table S1).
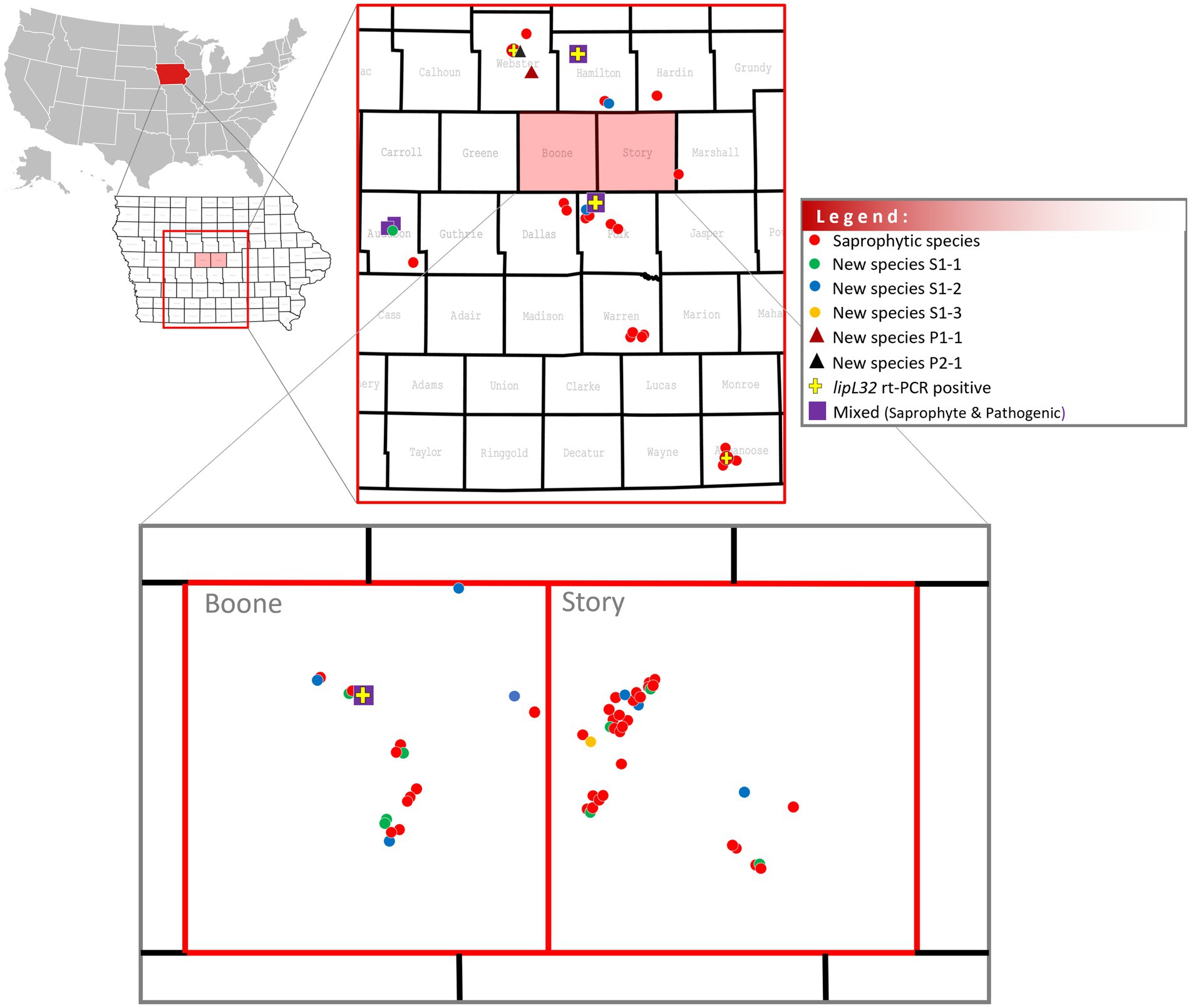
Figure 2. Distribution of saprophyte and pathogenic Leptospira in water samples in central Iowa, U.S.
3.2 Culture
Positive cultures for spirochetes were detected in 82/105 (78.1%) water samples and in all counties (Supplementary Table S1; Figure 2). Leptospira-positive cultures free of other microbial organisms were only detected in HAN media containing STAFF selective agents when incubated at 29°C.
3.3 Genotyping of cultures
Genome sequencing of leptospires cultured from water samples identified 10 different species of saprophytic Leptospira belonging to subclade S1, including three (WS1, WS43, and WS49) speciated as L. montravelensis, two (WS2 and WS57) as L. kemamanensis, seven (WS15, WS26, WS33, WS48, WS82, WS89, and WS103) as L. bandrabouensis, four (WS5, WS37, WS41, and WS75) as L. bourretii, five (WS31, WS59, WS62, WS77, and WS80) as L. bouyouniensis, two (WS34 and WS56) as L. chreensis, two (WS66 and WS70) as L. ellinghausenii, seven (WS40, WS44, WS45, WS46, WS74, WS86, and WS98) as L. terpstrae, WS63 as L. yanagawae, and WS24 as L. abararensis (Supplementary Table S1; Figure 3).
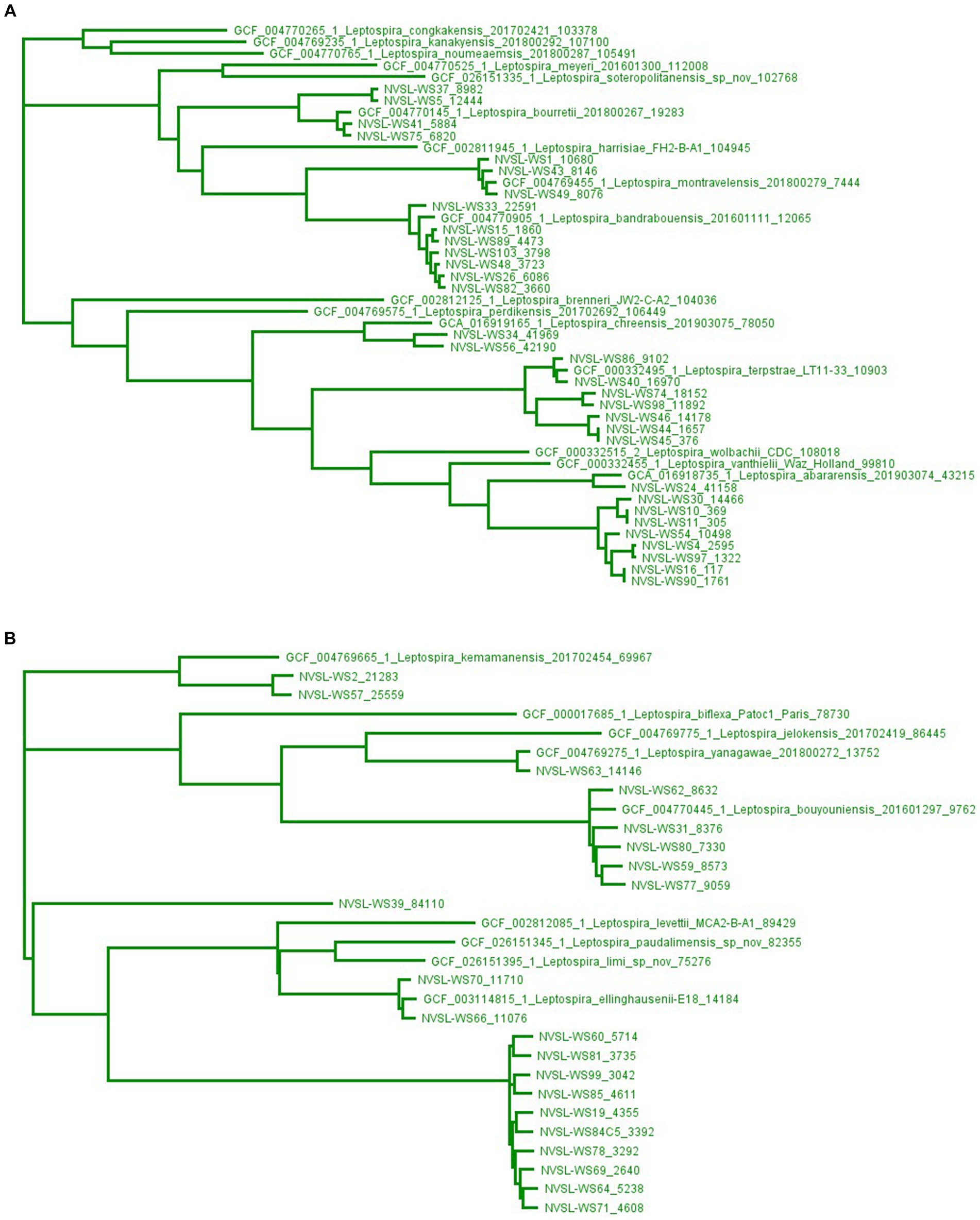
Figure 3. Maximum-likelihood phylogenetic tree created using kSNP3.0, a reference-free phylogenetic analysis tool, of isolates identified as saprophytes, subclade S1, in water samples by whole genome sequencing. The high diversity of genomes in subclade S1 by kSNP3.0 results in two groupings shown separately in Panels A,B, and as illustrated in Supplementary Figure S7. Genome sequences from GenBank are preceded by an accession number, while genome sequences from water sources (WS) are preceded by NVSL. The end number is the estimated number of SNPs to the nearest neighbor.
Analysis of genomic sequencing reads and assemblies identified mixed populations of saprophytic species in 23 cultures, including WS6, WS7, WS8, WS9, WS12, WS13, WS14, WS17, WS20, WS35, WS36, WS38, WS42, WS47, WS50, WS51, WS52, WS53, WS55, WS68, WS72, WS73, and WS76 (Supplementary Table S1; Supplementary Figure S1).
Analysis of genomic sequencing reads and assemblies also identified mixed populations of both saprophytic and pathogenic species in six cultures, including WS73, WS84, WS91, WS95, WS96, and WS101 (Supplementary Table S1; Supplementary Figure S2). The genome sequence from sample WS73 comprised 4% P1 and 83% S1, WS84 comprised 79.5% P2 and 13.3% of S1, WS91 comprised 22.5% P1 and 63.1% S1, WS95 comprised 6.5% P1 and 90.6% S1, WS96 comprised 7% P1 and 90.2% S1, and WS101 comprised 27.6% P1 and 67.2% S1 (Supplementary Figure S2). Given that water samples WS73, WS95, and WS96 had originally tested lipL32 qPCR negative, but that genomic analyses indicated the presence of DNA derived from pathogenic species, the gene sequence for lipL32 was extracted from each of these sequences to detect matching lipL32 primers and probes. Results show that the lipL32 qPCR assay has two forward primer mismatches and a missing probe sequence in the WS73 lipL32, while the sequence matching the reverse primer is missing in the lipL32 sequence from WS95 (Supplementary Figure S3). Given that water samples WS40 and WS57 had originally tested lipL32 qPCR positive, but analyses of genome sequence failed to identify pathogenic species, PCR for detection of lipL32 and lfb1 was performed, and amplicons were sequenced (Supplementary Figure S4). The results confirmed the presence of lipL32 derived from pathogenic Leptospira in samples WS40 and WS57 (Supplementary Figure S5). Sequencing of amplicons from lfb1 was not successful.
Analysis of genome sequences also identified cultures comprising new species of Leptospira, as determined by comparison of average nucleotide identity (ANI) < 95%. A new saprophytic species of Leptospira designated L. sp. nov Sapro S1.1 was assigned for strains WS4, WS10, WS11, WS16, WS30, WS54, WS90, and WS97 isolated in Boone, Story, and Audubon Counties (Supplementary Table S1; Figure 4). A new saprophytic species of Leptospira designated L. sp. nov Sapro S1.2 was assigned for strains WS19, WS60, WS64, WS69, WS71, WS78, WS81, WS85, and WS99 isolated in Boone, Polk, Hamilton, and Story Counties (Supplementary Table S1; Figure 4). A new saprophytic species of Leptospira designated L. sp. nov Sapro S1.3 was assigned for strain WS39 from Story County (Supplementary Table S1; Figure 4). A new pathogenic species of Leptospira designated L. sp. nov Patho 1 was assigned for strain WS92 (Supplementary Table S1; Figures 4, 5), and a new pathogenic species of Leptospira designated L. sp. nov Patho 2 was assigned for strain WS58 (Supplementary Table S1; Figures 4, 6), both of which were isolated in Webster County (Supplementary Table S1). Sample WS92 L. sp. nov Patho 1 originally tested lipL32 qPCR negative (Supplementary Table S1) and was still negative after additional PCRs for lipL32 and lfb1 (Supplementary Figure S4). Primers used in our PCR assays did not match or amplify sequences for lipL32 or lfb1 in WS92 (Supplementary Figure S4). Phylogeny of the WGS from all non-mixed cultures is provided in Supplementary Figure S6.

Figure 4. Average Nucleotide Identity (ANI) values to delineate the identification of new species. An OrthoANI result greater than or equal to 95% was considered the same species.

Figure 5. Maximum-likelihood phylogenetic tree created using kSNP3.0, a reference-free phylogenetic analysis tool, of isolates identified as pathogens, subclade P1, in water samples by whole genome sequencing. Genome sequences from GenBank are preceded by an accession number, while genome sequences from water sources (WS) are preceded by NVSL. The end number is the estimated number of SNPs to the nearest neighbor.

Figure 6. Maximum-likelihood phylogenetic tree created using kSNP3.0, a reference-free phylogenetic analysis tool, of isolates identified as pathogens, subclade P2, in water samples by whole genome sequencing. Genome sequences from GenBank are preceded by an accession number, while genome sequences from water sources (WS) are preceded by NVSL. The end number is the estimated number of SNPs to the nearest neighbor.
3.4 Serotyping of cultures with reference antisera
Serotyping of cultures was performed by MAT using a panel of 17 anti-Leptospira rabbit reference sera (Supplementary Table S3). No reactivity was detected for strains WS24 (L. abararensis); strains WS15, WS26, WS33, WS48, WS82, WS89, and WS103 (L. bandrabouensis); strains WS5, WS37, WS41 and WS75 (L. bourreti); strains WS31, WS59, WS62, WS77, and WS80 (L. bouyouniensis); strains WS34 and WS56 (L. chreensis); strain WS66 (L. ellinghausenii); strain WS2 (L. kemamanensis); strains WS1, WS43, and WS49 (L. montravelensis); and strains WS40, WS44, WS45, WS46, WS74, and WS98 (L. terpstrae) (Supplementary Table S1).
Low reactivity was detected for strain WS70 (L. ellinghausenii) with serogroups Hebdomadis (1:200), Autumnalis (1:100), Bataviae (1:100), Icterohaemorrhagiae (1:100), Tarassovi (1:100), Szwajizak (1:100), and Room22 (1:100); for strain WS81 (L. sp. nov Sapro S1.2) and WS57 (L. kemamanensis) to serogroup Pyrogenes (1:100); for strain WS86 (L. terpstrae) to serovar Room22 (1:200); for strain WS63 (L. yanagawae) to serogroup Pyrogenes (1:200); and for strain WS19 (L. sp. nov Sapro S1.2) to Djasiman (1:200) (Supplementary Table S1).
No reactivity was detected for strain WS58 [L. sp. Nov Patho P2] or strain WS92 [L. sp. Nov Patho P1].
3.5 Isolation and classification of a clonal isolate of pathogenic Leptospira from mixed cultures
Culture from water sample WS101 had a positive MAT of 1:51,200 when tested against reference antisera specific for serogroup Djasiman. Given that genome analysis for strain WS101 (Supplementary Table S1) suggested a population of mixed species and a relatively low Ct value of 24 for lipL32, suggesting a high number of pathogens, additional enrichment for serogroup Djasiman with reference antisera followed by plating on agar plates was performed as previously described (Hamond et al., 2023). Of the 20 colonies (WS101.C1 to WS101C.20) selected from agar plates, all were positive by PCR for rrs, and sequencing of amplicons identified L. interrogans. Complete genome sequencing of clonal isolate WS101.C1 indicated a lower number of scaffolds (n = 239 and genome size of 4,614,224 bp) than that for the mixed culture WS101 (number of scaffolds = 2,986 and genome size of 11,472,664 bp) that was identified as belonging to L. interrogans. Serotyping of strain WS101.C1 had a titer of 1:51,200 when tested with reference antisera for serogroup Djasiman. Serotyping with panels of reference monoclonal antibodies for serogroup Djasiman identified strain WS101.C1 as belonging to serovar Djasiman. Strain WS101.C1 is classified as L. interrogans serogroup Djasiman serovar Djasiman.
Attempts to isolate a pathogenic species from mixed culture WS84 were not successful, though a clonal isolate of saprophytic species L. sp. nov Sapro S1.2 was recovered and designated WS84.C5 (Figure 3; Supplementary Table S2).
3.6 Evaluation of virulence
Experimentally inoculated hamsters did not show any clinical signs of infection or weight loss after intraperitoneal inoculation with any of the three pathogenic isolates. At 3 weeks post-infection, all kidney samples from each group were negative by culture and lipL32 qPCR. MAT reactivity was detected in hamsters inoculated with WS58 [L. sp. nov Patho P2] when tested against the homologous isolate WS58 [L. sp. nov Patho P2] at 1:3200. Similarly, MAT reactivity was detected in hamsters inoculated with WS92 [L. sp. nov Patho P1] when tested against the homologous isolate WS92 [L. sp. nov Patho P1] at 1:1600. Hamsters inoculated with L. interrogans serogroup Djasiman strain WS101.C1 had a positive titer to WS101.C1 at 1:6400 and to reference strain L. interrogans serogroup Djasiman serovar Djasiman strain at 1:6400. Hamsters inoculated with strains WS58 [L. sp. nov Patho P2] or WS92 [L. sp. nov Patho P1] were seronegative for other serogroups tested (Supplementary Table S4).
4 Discussion
The advent of appropriate combinations of selective antimicrobials has facilitated a rapid and significant increase in the ability to culture saprophytic and pathogenic species of the genus Leptospira from environmental sources (Chakraborty et al., 2011). The recovery and characterization of isolates of Leptospira from water and soil provide unique opportunities to explore the geographic diversity and evolution of this genus and further explore the role of the environment in the maintenance and transmission of pathogenic species associated with human and animal leptospirosis (Bierque et al., 2020; Fernandes et al., 2022). In this study, water samples collected by volunteers in the state of Iowa in the summer of 2021 were assessed for the presence of Leptospira by culture and PCR to determine which species could be identified.
Over 78% (82/105) of collected water samples were culture-positive for Leptospira, as confirmed by WGS. This emphasizes the widespread distribution of Leptospira in local water sources (Supplementary Table S1; Figure 2). Known saprophytic species were cultured directly from 34/82 culture-positive water samples and included L. terpstrae and L. yanagawae, which were originally isolated in China and Brazil, respectively (Smythe et al., 2013), as well as the more recently described L. montravelensis, L. kemamanensis, L. bandrabouensis, L. bourretii, L. bouyouniensis, L. chreensis, L. ellinghausenii, and L. abararensis isolated in countries including New Caledonia, Malaysia, the island of Mayotte in the Indian Ocean, Algeria, and Japan (Vincent et al., 2019). In addition, three new saprophytic species were identified, as defined by an ANI <95%, by direct culture from 18/82 culture-positive water samples; L. sp. nov Sapro S1.1 was cultured directly from eight samples, L. sp. nov Sapro S1.2 was cultured directly from nine samples and a single sample identified as L. sp. nov Sapro S1.3. Phenotype, morphology, and a complete closed genome for reference strains in each of these new saprophytic species are in progress.
WGS analyses identified mixed species of saprophytes in 23/82 culture-positive samples, while mixed species of pathogens and saprophytes were identified in 6/82 culture-positive samples. Since 16S rRNA is insufficient to robustly distinguish Leptospira species, the ppk gene encoding a polyphosphate kinase was used to identify species in mixed cultures (Vincent et al., 2019). Gene sequences for ppk were extracted from WGS assemblies and clades assigned (Supplementary Table S1). Of note, phylogenies based on ppk assigned WS75 and WS41 as more closely related to L. mtsangambouensis (Supplementary Figure S1) compared to phylogenies based on WGS, which assigned these samples to L. bourretii (Supplementary Figure S6), further highlighting the diversity of this genus and the need to employ multiple genes to accurately assign species.
Of the 105 water samples tested by PCR for the presence of lipL32, a Leptospira pathogen-specific gene, to identify water samples containing pathogenic species of the P1 clade of Leptospira, five (4.8%) were positive. Sample WS101 was the only water sample that was positive for pathogenic species by both PCR and WGS analyses of the culture isolate, in which 27.6% of WGS reads were assigned as L. interrogans (Supplementary Figure S2). The presence of pathogenic Leptospira in culture-positive sample WS101 was also indicated by a positive MAT titer for the presence of serogroup Djasiman. The selection of a clonal isolate derived from this mixed culture allowed for the identification of a single species as L. interrogans serogroup Djasiman serovar Djasiman strain WS101.C1 (raw data deposited under accession number SRR25177450 in GenBank). Given that samples WS40, WS57, WS84, and WS91 were PCR positive for lipL32 but culture negative for pathogens by WGS, PCR for lipL32 was repeated and amplicons were sequenced (Supplementary Figures S4, S5). The results confirmed the presence of pathogenic leptospires when these water samples were collected and suggested that the growth of the more fastidious pathogens was outcompeted by faster-growing saprophytes before WGS of culture. Conversely, six water samples were culture-positive for pathogens by WGS but negative by PCR. Inspection of lipL32 sequences extracted from WGS assemblies of cultures positive for pathogens but PCR negative for lipL32 samples confirmed that WS73 lacked a matching probe sequence, WS92 lacked a matching forward and reverse primers sequence, WS95 lacked a matching reverse primer sequence, and both WS58 and WS84 belong to clade P2 (Supplementary Figure S3). A repeat PCR of WS96 remained negative for lipL32, despite matching probe and primers, but was positive for lfb1. The gene encoding LipL32 is present in all pathogenic species of clades P1 and P2 and is the most used target for molecular detection of pathogenic Leptospira. The extensive gene sequence polymorphisms (Stone et al., 2022) highlight the need to update primers and probe for lipL32 to account for the diversity of this genus and the detection of both clades P1 and P2.
WGS analyses of isolates WS92 and WS58 identified new pathogenic species belonging to clade P1 and P2, respectively. Additional analyses of genome sequence for WS58 using the JSpeciesWS GenomesDB database (Richter et al., 2015) determined that WS58 was highly similar to unclassified Leptospira sp. B5-022 (GenBank Accession ANIJ00000000), an isolate cultured from a rat in Denmark (data not shown): though this genome submission annotates Leptospira sp. B5-022 as L. hovindhougenii, this species has not yet been formally described or recognized. Notably, an isolate recovered from a dog in Italy also had a similar genome (Piredda et al., 2021). No evidence of infection was observed in the hamster model of leptospirosis after inoculation with strain WS92, WS98, or L. interrogans serovar Djasiman strain WS101.C1. All hamsters seroconverted, though seroconversion in hamsters inoculated with WS92 or WS58 was limited to agglutination with the challenge isolate, suggesting these strains belong to novel serogroups. As expected, hamsters inoculated with strain WS101.C1 had a positive MAT titer to the serovar Djasiman reference strain and to strain WS101.C1. Serotyping of new pathogenic species is essential to determine if they are represented in MAT diagnostic panels and to limit false negative reactions. Serogroup Djasiman is not routinely included in MAT diagnostic panels, but testing of equine samples has confirmed exposure to this serogroup (Hamond, LeCount, and Anderson, unpublished data). It remains to be determined if exposure of horses, or other domestic livestock, to serogroup Djasiman is facilitated via contaminated waterways. Phenotype, morphology, and a complete closed genome for reference strains in each new pathogenic species are in progress.
The recovery of leptospires from the environment is challenging (Chakraborty et al., 2011; Thibeaux et al., 2018) and more so for pathogenic leptospires with relatively long generation times (Picardeau, 2015). One major obstacle to recovering leptospires without contamination is the ability to select a single species from a complex polymicrobial sample. In contrast to other growth media used at 28–30°C, HAN media functions at both 28–30°C and 37°C (Picardeau, 2015; Hornsby et al., 2020). However, in this study, HAN media was only successful at 29°C, as even with the inclusion of STAFF, unacceptable levels of contaminating bacteria were cultured in environmental samples incubated at 37°C (data not shown). Other approaches have used agar plates to culture environmental samples, but our study prioritized the use of liquid media so volunteers could inoculate immediately at the time of collection. Alternative approaches could use qPCR or fluorescent antibody testing (FAT) to screen early stages of positive cultures to identify those containing pathogens, which in turn could be selected for the enrichment of clonal pathogens on agar plates. Screening of positive cultures from environmental samples by MAT also serves to identify those containing pathogenic serogroups. Such an approach is exemplified in this study by the high titer of sample WS101 for serogroup Djasiman (Supplementary Table S1).
Leptospirosis is a zoonotic, neglected, and emerging disease in the United States. Infection is associated with recreational exposures, occupational exposures to infected animals, and individuals living in economically disadvantaged urban inner-city environments (Heath Jr. et al., 1965; Sasaki et al., 1993; Vinetz et al., 1996; Katz et al., 1997; Lee et al., 2002; Haake and Levett, 2015; McNeilly et al., 2023). In Iowa, an outbreak of human leptospirosis was associated with exposure to animals and the environment (Schiemann, 1973; Lee et al., 2002). Environmental freshwater facilitates the transmission of leptospirosis in human and animal populations (Saito et al., 2013; Thibeaux et al., 2017). In this study, sample collection was limited to summer months associated with increased outdoor activities and water locales chosen by volunteers. Five samples (WS40, WS57, WS84, WS91, and WS101) were positive by qPCR, six samples (WS73, WS84, WS91, WS95, WS96, and WS101) were positive for known pathogenic species by WGS of cultures, and two (WS58 and WS92) were positive for new pathogenic species by WGS of cultures. The identification of 9.5% (10/105) of water samples positive for pathogenic Leptospira highlights the prevalence and diversity of Leptospira in water sources. This finding also highlights the need for more comprehensive investigations into the role of moist environments in maintaining pathogenic Leptospira and associated risks to animal and public health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Animal Care & Use Committee at the National Animal Disease Center (ARS-2021-938) and as approved by USDA institutional guidelines. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. KL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – review & editing. TA: Formal analysis, Investigation, Methodology, Writing – review & editing. EP: Formal analysis, Investigation, Methodology, Writing – review & editing. TS: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – review & editing. JH: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. PC: Investigation, Methodology, Writing – review & editing. HL: Formal analysis, Investigation, Methodology, Writing – review & editing. DB: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – review & editing. LS: Conceptualization, Investigation, Methodology, Project administration, Writing – review & editing. JN: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors thank Andréa Haluch, Kari Toot, Keira Stuart, Dr. Jack Schlater, Dr. Kristina Lantz, and Dr. Kimberly Lehman (NVSL, USDA) for collecting water samples; Mycobacteria & Brucella Section from the NVSL for their technical assistance in genome sequencing. This research was supported in part by an appointment to the Animal and Plant Health Inspection Service (APHIS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by ORAU under DOE contract number DE-SC0014664. All opinions expressed in this paper are the authors and do not necessarily reflect the policies and views of USDA, DOE, or ORAU/ORISE. USDA is an equal opportunity provider and employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2024.1278088/full#supplementary-material
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barragan, V. A., Mejia, M. E., Trávez, A., Zapata, S., Hartskeerl, R. A., Haake, D. A., et al. (2011). Interactions of Leptospira with environmental bacteria from surface water. Curr. Microbiol. 62, 1802–1806. doi: 10.1007/s00284-011-9931-3
Bharti, A. R., Nally, J. E., Ricaldi, J. N., Matthias, M. A., Diaz, M. M., Lovett, M. A., et al. (2003). Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3, 757–771. doi: 10.1016/S1473-3099(03)00830-2
Bierque, E., Thibeaux, R., Girault, D., Soupé-Gilbert, M.-E., and Goarant, C. (2020). A systematic review of Leptospira in water and soil environments. PLoS One 15:e0227055. doi: 10.1371/journal.pone.0227055
Bolin, C. A., and Cassells, J. A. (1990). Isolation of Leptospira interrogans serovar Bratislava from stillborn and weak pigs in Iowa. J. Am. Vet. Med. Assoc. 196, 1601–1604. doi: 10.2460/javma.1990.196.10.1601
Bolin, C. A., Cassells, J. A., Hill, H. T., Frantz, J. C., and Nielsen, J. N. (1991). Reproductive failure associated with Leptospira interrogans serovar Bratislava infection of swine. J. Vet. Diagn. Invest. 3, 152–154. doi: 10.1177/104063879100300209
Braun, J. L., and McCULLOCH, W. F. (1968). Use of 8-azaguanine to differentiate leptospires isolated from Iowa surface waters. Appl. Microbiol. 16, 174–175. doi: 10.1128/am.16.1.174-175.1968
Casanovas-Massana, A., Hamond, C., Santos, L. A., de Oliveira, D., Hacker, K. P., Balassiano, I., et al. (2020). Leptospira yasudae sp. nov. and Leptospira stimsonii sp. nov., two new species of the pathogenic group isolated from environmental sources. Int. J. Syst. Evol. Microbiol. 70, 1450–1456. doi: 10.1099/ijsem.0.003480
Casanovas-Massana, A., Pedra, G. G., Wunder, E. A., Diggle, P. J., Begon, M., and Ko, A. I. (2018). Quantification of Leptospira interrogans survival in soil and water microcosms. Appl. Environ. Microbiol. 84:e00507-18. doi: 10.1128/AEM.00507-18
Chakraborty, A., Miyahara, S., Villanueva, S. Y., Saito, M., Gloriani, N. G., and Si, Y. (2011). A novel combination of selective agents for isolation of Leptospira species. Microbiol. Immunol. 55, 494–501. doi: 10.1111/j.1348-0421.2011.00347.x
Cole, J. R., Sulzer, C. R., and Pursell, A. R. (1973). Improved microtechnique for the leptospiral microscopic agglutination test. Appl. Microbiol. 25, 976–980. doi: 10.1128/am.25.6.976-980.1973
Crawford, R. P., Braun, J. L., McCULLOCH, W. F., and Diesch, S. L. (1969). Characterization of leptospires isolated from surface waters in Iowa. Bull. Wildl. Dis. 5, 157–165. doi: 10.7589/0090-3558-5.3.157
Diesch, S. L., and McCulloch, W. F. (1966). Isolation of pathogenic leptospires from waters used for recreation. Public Health Rep. 81:299. doi: 10.2307/4592702
Diesch, S. L., Mcculloch, W. F., Braun, J. L., and Davis, J. R. (1970). Detection and ecology of leptospirosis in Iowa wildlife. J. Wildl. Dis. 6, 275–288. doi: 10.7589/0090-3558-6.4.275
Dos Santos, R. P., Carvalho, N. B., Aburjaile, F., Sousa, T., Veríssimo, G., Gomes, T., et al. (2023). Environmental biofilms from an Urban Community in Salvador, Brazil, shelter previously uncharacterized saprophytic Leptospira. Microb. Ecol. 86, 2488–2501. doi: 10.1007/s00248-023-02253-3
Ellis, W. A. (2015). Animal leptospirosis. Curr. Top. Microbiol. Immunol. 387, 99–137. doi: 10.1007/978-3-662-45059-8_6
Ellis, W., and Thiermann, A. (1986). Isolation of Leptospira interrogans serovar Bratislava from sows in Iowa. Am. J. Vet. Res. 47, 1458–1460.
Faine, S., Adler, B., Bolin, C., and Perolat, P.. (1999). Leptospira and Leptospirosis, 2nd MediSci, Melbourne, Australia.
Fernandes, L. G., Putz, E. J., Stasko, J., Lippolis, J. D., Nascimento, A. L., and Nally, J. E. (2022). Evaluation of LipL32 and LigA/LigB knockdown mutants in Leptospira interrogans Serovar Copenhageni: impacts to proteome and virulence. Front. Microbiol. 12:799012. doi: 10.3389/fmicb.2021.799012
Fernandes, L. G. V., Stone, N. E., Roe, C. C., Goris, M. G. A., van der Linden, H., Sahl, J. W., et al. (2022). Leptospira sanjuanensis sp. nov., a pathogenic species of the genus Leptospira isolated from soil in Puerto Rico. Int. J. Syst. Evol. Microbiol. 72:5560. doi: 10.1099/ijsem.0.005560
Galloway, R. L., and Hoffmaster, A. R. (2015). Optimization of LipL32 PCR assay for increased sensitivity in diagnosing leptospirosis. Diagn. Microbiol. Infect. Dis. 82, 199–200. doi: 10.1016/j.diagmicrobio.2015.03.024
Gardner, S. N., Slezak, T., and Hall, B. G. (2015). kSNP3. 0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Goris, M. G., and Hartskeerl, R. A. (2014). Leptospirosis serodiagnosis by the microscopic agglutination test. Curr. Protoc. Microbiol. 32:Unit 12E.5. doi: 10.1002/9780471729259.mc12e05s32
Grillová, L., Robinson, M. T., Chanthongthip, A., Vincent, A. T., Nieves, C., Oppelt, J., et al. (2021). Genetic diversity of Leptospira isolates in Lao PDR and genome analysis of an outbreak strain. PLoS Negl. Trop. Dis. 15:e0010076. doi: 10.1371/journal.pntd.0010076
Haake, D. A., and Levett, P. N. (2015). Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 387, 65–97. doi: 10.1007/978-3-662-45059-8_5
Hamond, C., Browne, A. S., de Wilde, L. H., Hornsby, R. L., LeCount, K., Anderson, T., et al. (2022). Assessing rodents as carriers of pathogenic Leptospira species in the U.S. Virgin Islands and their risk to animal and public health. Sci. Rep. 12:1132. doi: 10.1038/s41598-022-04846-3
Hamond, C., Dirsmith, K. L., LeCount, K., Soltero, F. V., Rivera-Garcia, S., Camp, P., et al. (2022). Leptospira borgpetersenii serovar Hardjo and Leptospira santarosai serogroup Pyrogenes isolated from bovine dairy herds in Puerto Rico. Front. Vet. Sci. 9:1025282. doi: 10.3389/fvets.2022.1025282
Hamond, C., LeCount, K., Browne, A. S., Anderson, T., Stuber, T., Hicks, J., et al. (2023). Concurrent colonization of rodent kidneys with multiple species and serogroups of pathogenic Leptospira. Appl. Environ. Microbiol. 89:e0120423. doi: 10.1128/aem.01204-23
Hartskeerl, R., Smits, H., Korver, H., Goris, M., and Terpstra, W.. (2006). Manual international course on laboratory methods for the diagnosis of leptospirosis. KIT, Amsterdam, The Netherlands.
Heath, C. W., Alexander, A. D., and Galton, M. M. (1965). Leptospirosis in the United States. N. Engl. J. Med. 273, 857–64 contd. doi: 10.1056/NEJM196510142731606
Hornsby, R. L., Alt, D. P., and Nally, J. E. (2020). Isolation and propagation of leptospires at 37 degrees C directly from the mammalian host. Sci. Rep. 10:9620. doi: 10.1038/s41598-020-66526-4
Katz, L. S., Griswold, T., Morrison, S. S., Caravas, J. A., Zhang, S., den Bakker, H. C., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4:1762. doi: 10.21105/joss.01762
Katz, A. R., Sasaki, D. M., Mumm, A. H., Escamilla, J., Middleton, C. R., and Romero, S. E. (1997). Leptospirosis on Oahu: an outbreak among military personnel associated with recreational exposure. Mil. Med. 162, 101–104. doi: 10.1093/milmed/162.2.101
Korba, A. A., Lounici, H., Kainiu, M., Vincent, A. T., Mariet, J. F., Veyrier, F. J., et al. (2021). Leptospira ainlahdjerensis sp. nov., Leptospira ainazelensis sp. nov., Leptospira abararensis sp. nov. and Leptospira chreensis sp. nov., four new species isolated from water sources in Algeria. Int. J. Syst. Evol. Microbiol. 71:5148. doi: 10.1099/ijsem.0.005148
Lee, S. H., Levy, D. A., Craun, G. F., Beach, M. J., and Calderon, R. L. (2002). Surveillance for waterborne-disease outbreaks--United States, 1999-2000. MMWR Surveill. Summ. 51, 1–47. Available at: https://digitalcommons.unmc.edu/coph_epidem_articles/95
Lu, J., Breitwieser, F., Thielen, P., and Salzberg, S. (2017). Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3:e104. doi: 10.7717/peerj-cs.104
McNeilly, B. P., Williams, D. M., Bontempo, L. J., and Gatz, J. D. (2023). 44-year-old man with hemoptysis and hypoxemic respiratory failure: a case report. Clin. Pract. Cases Emerg. Med. 7, 54–59. doi: 10.5811/cpcem.1418
Mérien, F., Amouriaux, P., Perolat, P., Baranton, G., and Saint, G. I. (1992). Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J. Clin. Microbiol. 30, 2219–2224. doi: 10.1128/jcm.30.9.2219-2224.1992
Merien, F., Portnoi, D., Bourhy, P., Charavay, F., Berlioz-Arthaud, A., and Baranton, G. (2005). A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol. Lett. 249, 139–147. doi: 10.1016/j.femsle.2005.06.011
Monahan, A., Miller, I., and Nally, J. (2009). Leptospirosis: risks during recreational activities. J. Appl. Microbiol. 107, 707–716. doi: 10.1111/j.1365-2672.2009.04220.x
Nally, J. E., Arent, Z., Bayles, D. O., Hornsby, R. L., Gilmore, C., Regan, S., et al. (2016). Emerging infectious disease implications of invasive mammalian species: the greater White-toothed shrew (Crocidura russula) is associated with a novel Serovar of pathogenic Leptospira in Ireland. PLoS Negl. Trop. Dis. 10:e0005174. doi: 10.1371/journal.pntd.0005174
Nally, J. E., Hornsby, R. L., Alt, D. P., Bayles, D., Wilson-Welder, J. H., Palmquist, D. E., et al. (2018). Isolation and characterization of pathogenic leptospires associated with cattle. Vet. Microbiol. 218, 25–30. doi: 10.1016/j.vetmic.2018.03.023
O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Ondov, B. D., Bergman, N. H., and Phillippy, A. M. (2011). Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12, 1–10. doi: 10.1186/1471-2105-12-385
Picardeau, M. (2015). Leptospira. Bergey's manual of systematics of Archaea and Bacteria, John Wiley & Sons, Inc., in association with Bergey’s Manual Trust. 1–18.
Piredda, I., Bertoldi, L., Benvenuto, G., Palmas, B., Pedditzi, A., Pintore, P., et al. (2021). First isolation and molecular typing of pathogenic and intermediate Leptospira species from urine of symptomatic dogs. Vet. Sci. 8:304. doi: 10.3390/vetsci8120304
Putz, E. J., and Nally, J. E. (2020). Investigating the immunological and biological equilibrium of reservoir hosts and pathogenic Leptospira: balancing the solution to an acute problem? Front. Microbiol. 11:2005. doi: 10.3389/fmicb.2020.02005
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2015). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Riediger, I. N., Hoffmaster, A. R., Casanovas-Massana, A., Biondo, A. W., Ko, A. I., and Stoddard, R. A. (2016). An optimized method for quantification of pathogenic Leptospira in environmental water samples. PLoS One 11:e0160523. doi: 10.1371/journal.pone.0160523
Saito, M., Villanueva, S. Y., Chakraborty, A., Miyahara, S., Segawa, T., Asoh, T., et al. (2013). Comparative analysis of Leptospira strains isolated from environmental soil and water in the Philippines and Japan. Appl. Environ. Microbiol. 79, 601–609. doi: 10.1128/AEM.02728-12
Sasaki, D. M., Pang, L., Minette, H. P., Wakida, C. K., Fujimoto, W. J., Manea, S. J., et al. (1993). Active surveillance and risk factors for leptospirosis in Hawaii. Am. J. Trop. Med. Hyg. 48, 35–43. doi: 10.4269/ajtmh.1993.48.35
Schiemann, D. (1973). Leptospirosis may result from swimming in streams and lakes. J. Environ. Health 36, 70–73.
Smith, K. E., Zimmerman, J. J., Bolin, C. A., Beran, G. W., and Hill, H. T. (1992). A survey of house mice from Iowa swine farms for infection with Leptospira interrogans serovar Bratislava. Can. Vet. J. 33, 742–744.
Smythe, L., Adler, B., Hartskeerl, R., Galloway, R., Turenne, C., and Levett, P. (2013). Classification of Leptospira genomospecies 1, 3, 4 and 5 as Leptospira alstonii sp. nov., Leptospira vanthielii sp. nov., Leptospira terpstrae sp. nov. and Leptospira yanagawae sp. nov., respectively. Int. J. Syst. Evol. Microbiol. 63, 1859–1862. doi: 10.1099/ijs.0.047324-0
Stoddard, R. A., Gee, J. E., Wilkins, P. P., McCaustland, K., and Hoffmaster, A. R. (2009). Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 64, 247–255. doi: 10.1016/j.diagmicrobio.2009.03.014
Stone, N. E., Hall, C. M., Ortiz, M., Hutton, S. M., Santana-Propper, E., Celona, K. R., et al. (2022). Diverse lineages of pathogenic Leptospira species are widespread in the environment in Puerto Rico, USA. PLoS Negl. Trop. Dis. 16:e0009959. doi: 10.1371/journal.pntd.0009959
Sykes, J. E., Haake, D. A., Gamage, C. D., Mills, W. Z., and Nally, J. E. (2022). A global one health perspective on leptospirosis in humans and animals. J. Am. Vet. Med. Assoc. 260, 1589–1596. doi: 10.2460/javma.22.06.0258:1-8
Terpstra, W. J., Korver, H., Van Leeuwen, J., Klatser, P. R., and Kolk, A. H. (1985). The classification of Sejroe group serovars of Leptospira interrogans with monoclonal antibodies. Zentralbl. Bakteriol. Mikrobiol. Hyg. Series A 259, 498–506. doi: 10.1016/S0176-6724(85)80082-1
Thibeaux, R., Geroult, S., Benezech, C., Chabaud, S., Soupé-Gilbert, M.-E., Girault, D., et al. (2017). Seeking the environmental source of leptospirosis reveals durable bacterial viability in river soils. PLoS Negl. Trop. Dis. 11:e0005414. doi: 10.1371/journal.pntd.0005414
Thibeaux, R., Girault, D., Bierque, E., Soupé-Gilbert, M.-E., Rettinger, A., Douyère, A., et al. (2018). Biodiversity of environmental Leptospira: improving identification and revisiting the diagnosis. Front. Microbiol. 9:816. doi: 10.3389/fmicb.2018.00816
Tjalma, R., and Galton, M. (1965). Human leptospirosis in Iowa. Am. J. Trop. Med. Hyg. 14, 387–396. doi: 10.4269/ajtmh.1965.14.387
Vincent, A. T., Schiettekatte, O., Goarant, C., Neela, V. K., Bernet, E., Thibeaux, R., et al. (2019). Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 13:e0007270. doi: 10.1371/journal.pntd.0007270
Vinetz, J. M., Glass, G. E., Flexner, C. E., Mueller, P., and Kaslow, D. C. (1996). Sporadic urban leptospirosis. Ann. Intern. Med. 125, 794–798. doi: 10.7326/0003-4819-125-10-199611150-00002
Viotti, J. B., Chan, J. C., Rivera, C., and Tuda, C. (2020). Sporadic leptospirosis case in Florida presenting as Weils disease. IDCases 19:e00686. doi: 10.1016/j.idcr.2019.e00686
Warnasekara, J., Srimantha, S., Senavirathna, I., Kappagoda, C., Farika, N., Nawinna, A., et al. (2022). The variable presence of Leptospira in the environment; an epidemiological explanation based on serial analysis of water samples. PLoS One 17:e0263719. doi: 10.1371/journal.pone.0263719
Wood, D. E., and Salzberg, S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, 1–12. doi: 10.1186/gb-2014-15-3-r46
Wunder, E. A. Jr., Figueira, C. P., Santos, G. R., Lourdault, K., Matthias, M. A., Vinetz, J. M., et al. (2016). Real-time PCR reveals rapid dissemination of Leptospira interrogans after intraperitoneal and conjunctival inoculation of hamsters. Infect. Immun. 84, 2105–2115. doi: 10.1128/IAI.00094-16
Keywords: Leptospira, leptospirosis, water, environment, one health
Citation: Hamond C, LeCount K, Anderson T, Putz EJ, Stuber T, Hicks J, Camp P, van der Linden H, Bayles DO, Schlater LK and Nally JE (2024) Isolation and characterization of saprophytic and pathogenic strains of Leptospira from water sources in the Midwestern United States. Front. Water. 6:1278088. doi: 10.3389/frwa.2024.1278088
Edited by:
Kovashnee Naidoo, National Health Laboratory Service (NHLS), South AfricaReviewed by:
Claudia Munoz-Zanzi, University of Minnesota, United StatesJackie Benschop, Massey University, New Zealand
Copyright © 2024 Hamond, LeCount, Anderson, Putz, Stuber, Hicks, Camp, van der Linden, Bayles, Schlater and Nally. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Camila Hamond, camila.hamond@usda.gov
 Camila Hamond
Camila Hamond Karen LeCount
Karen LeCount Tammy Anderson1
Tammy Anderson1 Ellie J. Putz
Ellie J. Putz Jessica Hicks
Jessica Hicks Patrick Camp
Patrick Camp Hans van der Linden
Hans van der Linden Darrell O. Bayles
Darrell O. Bayles Linda K. Schlater
Linda K. Schlater Jarlath E. Nally
Jarlath E. Nally