- 1Department of Electrical and Computer Engineering, University of Washington, Seattle, WA, United States
- 2Department of Applied Mathematics, University of Washington, Seattle, WA, United States
Introduction: Quantifying natural behavior from video recordings is a key component in ethological studies. Markerless pose estimation methods have provided an important step toward that goal by automatically inferring kinematic body keypoints. Such methodologies warrant efficient organization and interpretation of keypoints sequences into behavioral categories. Existing approaches for behavioral interpretation often overlook the importance of representative samples in learning behavioral classifiers. Consequently, they either require extensive human annotations to train a classifier or rely on a limited set of annotations, resulting in suboptimal performance.
Methods: In this work, we introduce a general toolset which reduces the required human annotations and is applicable to various animal species. In particular, we introduce OpenLabCluster, which clusters temporal keypoint segments into clusters in the latent space, and then employ an Active Learning (AL) approach that refines the clusters and classifies them into behavioral states. The AL approach selects representative examples of segments to be annotated such that the annotation informs clustering and classification of all temporal segments. With these methodologies, OpenLabCluster contributes to faster and more accurate organization of behavioral segments with only a sparse number of them being annotated.
Results: We demonstrate OpenLabCluster performance on four different datasets, which include different animal species exhibiting natural behaviors, and show that it boosts clustering and classification compared to existing methods, even when all segments have been annotated.
Discussion: OpenLabCluster has been developed as an open-source interactive graphic interface which includes all necessary functions to perform clustering and classification, informs the scientist of the outcomes in each step, and incorporates the choices made by the scientist in further steps.
1 Introduction
Analysis and interpretation of animal behavior are essential for a multitude of biological investigations. Behavioral studies extend from ethological studies to behavioral essays as a means to investigate biological mechanisms (Sturm et al., 2019; Monosov et al., 2024; Marques et al., 2018; Johnson et al., 2020; Berman et al., 2016; Jazayeri and Afraz, 2017; Datta et al., 2019; Weber et al., 2022; McCullough and Goodhill, 2021; Pereira et al., 2019). In these studies, methodologies facilitating robust, uninterrupted, and high-resolution observations are key. Indeed, researchers have been recording animal behaviors for decades with various modalities, such as video, sound, placement of physical markers, and more (Taiwanica, 2000; Morrow-Tesch et al., 1998; Han et al., 2011; Lynch et al., 2013; Nakamura et al., 2016; Buccino et al., 2018; Bain et al., 2021). Recent enhancements in recording technologies have extended the ability for the deployment of recording devices in various environments and for extended periods of time. The enhancement in the ability to perform longer observations and in the number of modalities brings forward the need to organize, interpret, and associate recordings with identified repertoires of behaviors, i.e., perform classification of the recordings into behavioral states. Performing these operations manually would typically consume a significant amount of time and would require expertise. For many recordings, manual behavior classification becomes an unattainable task. Therefore, it is critical to develop methodologies to accelerate the classification of behavioral states and require as little involvement from the empiricist as possible (Anderson and Perona, 2014; Dell et al., 2014; Krause et al., 2013).
Early efforts in automatic behavior classification focused on raw video analysis using machine learning techniques such as convolutional neural networks (CNNs) (van Dam et al., 2020; Jia et al., 2022; Batty et al., 2019; Brattoli et al., 2021), recurrent neural networks (RNNs) (Stern et al., 2015; Murari et al., 2019), temporal Gaussian mixture models Bohnslav et al. (2021), and temporal CNNs (Marks et al., 2022). While effective in specific scenarios, video-based methods often incorporate extraneous background information and noise (e.g., camera artifacts), which can undermine reliability and require considerable computational resources due to the high-dimensional nature of video data (Marks et al., 2022). In contrast, approaches that concentrate on movement by utilizing body keypoints or kinematics-extracted from video frames-can circumvent these limitations (Xu et al., 2017; Insafutdinov et al., 2017, 2016; Sturman et al., 2020; Isik and Unal, 2023).
Markerless pose estimation techniques, such as OpenPose, DeepLabCut, Anipose, and others (Mathis et al., 2018; Deeplabcut, 2018; Cao et al., 2019; Nath et al., 2019; Lauer et al., 2022; Karashchuk et al., 2021), enable accurate keypoint detection without the need for physical markers. Furthermore, numerous related tools and approaches have also been further introduced to advance animal pose estimation (Pereira et al., 2019; Wu et al., 2020; Bala et al., 2020; Pereira et al., 2022; Zhang et al., 2021; Usman et al., 2022; Ye et al., 2024). Once body keypoints are estimated, behavioral segmentation can be achieved using unsupervised clustering methods-such as HBDSCAN (Ester et al., 1996; Hsu and Yttri, 2021), hierarchical clustering (Huang et al., 2021), and the Watershed algorithm (Meyer, 1994; Berman et al., 2014; Dunn et al., 2021)-which group similar postural states and differentiate distinct behaviors (Marques et al., 2018; Hsu and Yttri, 2021). Dimensionality reduction techniques, including principal component analysis (PCA) and uniform manifold approximation and projection (UMAP), further enhance the representation of body keypoints for effective clustering (McInnes et al., 2018; Huang et al., 2021; Kwon et al., 2024; Hsu and Yttri, 2021; Wiltschko et al., 2020).
Recent deep learning methods have advanced latent keypoint representation learning through task-specific optimization, as demonstrated by TREBA (Sun et al., 2021) and its automated extension, AutoSWAP (Tseng et al., 2022). Contrastive learning approaches have also been proposed to refine the latent space by drawing together similar behavioral samples and separating dissimilar ones (Zhou et al., 2023; Schneider et al., 2023). One of the challenges in such approaches is the selection of appropriate positive and negative samples which remains challenging without human guidance. General methods such as Predict&Cluster (Su et al., 2020) and VAME (Luxem et al., 2022) address these challenges by focusing on sequence reconstruction and future prediction, enabling the unsupervised clustering of behavioral patterns (Su et al., 2020; Luxem et al., 2022; Weinreb et al., 2024).
While unsupervised clustering can identify similar behavioral patterns (Su et al., 2020; Luxem et al., 2022; Weinreb et al., 2024), it may not effectively identify behaviors of specific interest. Supervised classification approaches address this limitation by mapping behavioral segments to behavioral categories of interest, under the guidance of annotated training data (Xu et al., 2017; Sturman et al., 2020; Segalin et al., 2021). The classification accuracy critically depends on both the choice of classifier and the quality and quantity of annotations. Early success in behavioral classification was achieved using classical machine learning approaches (Dankert et al., 2009; Jhuang et al., 2010; Segalin et al., 2021; Burgos-Artizzu et al., 2012; Hong et al., 2015; De Chaumont et al., 2019; Goodwin et al., 2024; Hsu and Yttri, 2021). These have recently been supplemented by deep learning approaches (Rousseau et al., 2000; Sakata, 2023; Zhou et al., 2023; Ye et al., 2024; Sturman et al., 2020). Nevertheless, manual annotation remains labor-intensive and subject to inter-annotator variability.
To address the need for manual annotation, methods such as SaLSa-which assigns uniform labels to pre-computed unsupervised clusters-and JAABA-which provides an interactive framework for correcting misclassifications-have been developed (Sakata, 2023; Kabra et al., 2013). Active learning (AL) techniques further streamline the process by automatically selecting samples for annotation, balancing annotation effort with classification accuracy (Cohn et al., 1996; Settles, 2009, 2012; Li and Shlizerman, 2020b; Tillmann et al., 2024). In particular, for behavior recognition, A-SOiD Tillmann et al. (2024) employs AL to prioritize samples with high prediction uncertainty for animal behavior recognition; however, uncertainty-based selection may inadvertently target redundant samples. Integrating clustering information with classifier uncertainty could improve the efficiency of sample selection.
In this study, we extend previous methods by jointly learning representations for AL and classifier training using pose estimated from video recordings, e.g., keypoints estimated using DeepLabCut (Mathis et al., 2018; Nath et al., 2019; Deeplabcut, 2018). In particular, we introduce the OpenLabCluster toolset, an AL based semi-supervised behavior classification platform embedded in a graphic interface for animal behavior classification from body keypoints. The system implements and allows the use of multiple semi-supervised AL methods. AL is performed in an iterative way, where, in each iteration, an automatic selection of a subset of candidate segments is chosen for annotation, which in turn enhances the accuracy of clustering and classification. OpenLabCluster is composed of two components illustrated in Figure 1: (1) Unsupervised deep encoder-decoder clustering of behavior representation, Cluster Maps, which depicts the representations as points and show their groupings, followed by (2) Iterative automatic selection of representations for annotation and subsequent generation of Behavior Classification Maps. In each iteration, each point in the Cluster Map is re-positioned and associated with a behavioral class (colored with a color that corresponds to a particular class). These operations are performed through the training of a clustering encoder-decoder [component (1)] along with a deep classifier [component (2)]. OpenLabCluster implements these methodologies as an open-source graphical user interface (GUI) to empower scientists with little or no deep-learning expertise to perform animal behavior classification. In addition, OpenLabCluster includes advanced options for experts.
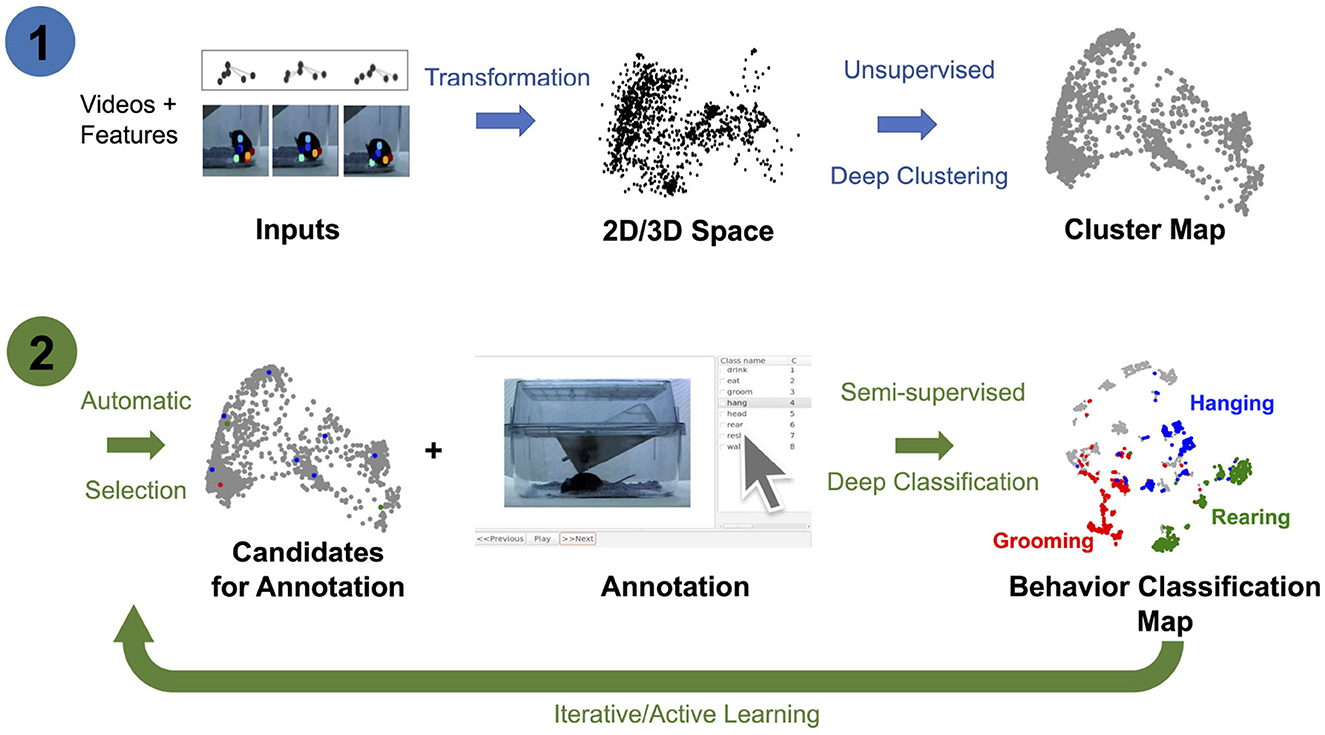
Figure 1. OpenLabCluster overview. (1) Clustering: Input of body keypoints segments is mapped to low dimensional space. Unsupervised encoder–decoder maps them to a Cluster Map. (2) Classification: AL algorithms automatically select candidates for annotation after which Cluster Map is reshaped into the Behavior Classification Map where each point is associated with a behavioral state [Grooming (red), Hanging (blue), Rearing (blue)]. Mouse images are reproduced from frames of videos provided in the dataset of Jhuang et al. (2010).
2 Results
2.1 Datasets
Behavioral states and their dynamics vary from species to species and from recordings to recordings. We use four different datasets to demonstrate OpenLabCluster applicability to various settings. The datasets include videos of behaviors of four different animal species [Mouse (Jhuang et al., 2010), Zebrafish (Marques et al., 2018), C. elegans (Yemini et al., 2013), Monkey (Bala et al., 2020)] with three types of motion features (body keypoints, kinematics, segments), as depicted in Figure 2. Two of the datasets include apriori annotated behavioral states (ground truth) (Mouse, C. elegans), while the Zebrafish dataset includes ground truth a priori predicted by another method, and the Monkey dataset does not include ground truth annotations. Three of the datasets have been temporally segmented into single-action clips (Mouse, Zebrafish, C. elegans), i.e., temporal segments, while the Monkey dataset is a continuous recording that requires segmentation into clips. We describe further details about each dataset below.
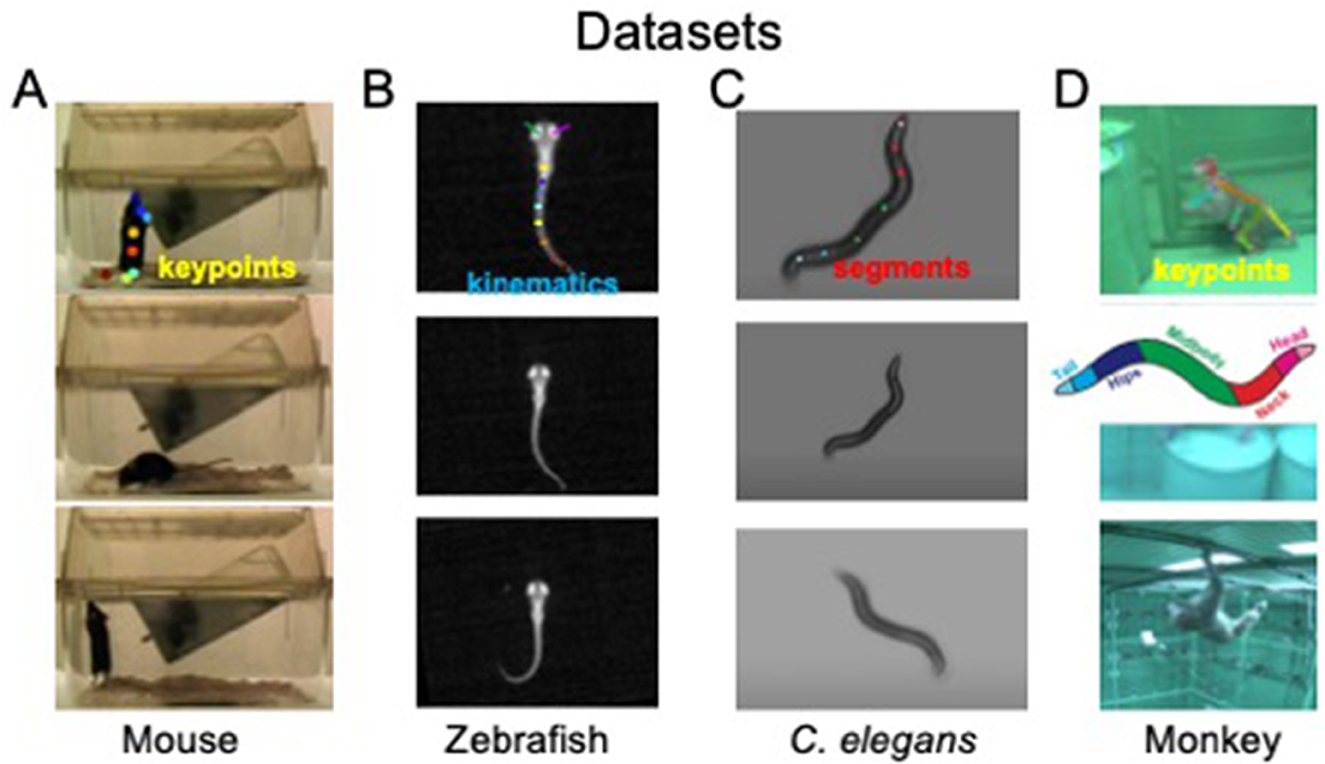
Figure 2. Visualization of four animal behavior datasets. (A) Home-Cage mouse dataset (mouse); (B) C. elegans movement dataset (C. elegans); (C) zebrafish free swimming dataset (Zebrafish); (D) OpenMonkeyStudio Macaque behaviors dataset (Monkey). The top row shows positions of extracted keypoints for each dataset. Images sources: (A) Images are reproduced from frames of videos in the dataset of Jhuang et al. (2010). (B) Images are reproduced from dataset of Marques et al. (2018). (C) Images are reproduced from datasets provided by Yemini et al. (2013). (D) Images are reproduced from Figure 8 and images of Bala et al. (2020).
2.1.1 Home-cage mouse
The dataset includes video segments of 8 identified behavioral states (Jhuang et al., 2010). The Home-Cage Mouse dataset is selected considering its clearly segmented videos, each with well-defined behavioral categories. In particular, it contains videos recorded by front cage cameras when the mouse is moving freely and exhibits natural behaviors, such as drinking, eating, grooming, hanging, micromovement, rearing, walking, and resting. Since keypoints have not been provided in this dataset, we use DeepLabcut (Mathis et al., 2018; Nath et al., 2019; Deeplabcut, 2018) to automatically mark and track eight body joint keypoints (snout, left-forelimb, right-forelimb, left-hindlimb, right-hindlimb, fore-body, hind-body, and tail) in all recorded videos frames. An example of estimated keypoints overlaid on top of the corresponding video frame from a side view is shown in Figure 2A (top). To reduce the noise that could be induced by the pose estimation procedure, we only use the segments for which DeepLabCut estimation confidence is high enough. We use 8 sessions for training the models of clustering and classification (2856 segments) and test classification accuracy on 4 other sessions (393 segments).
2.1.2 Zebrafish
The dataset includes video footage of zebrafish movements and was utilized in Marques et al. (2018) for unsupervised behavior clustering using 101 precomputed kinematic features, a procedure that identified 13 clusters which were manually related to 13 behavior prototypes (see Appendix 1.3). In the application of OpenLabCluster to this dataset, we utilize only a small subset of these features (16 features) and examine whether OpenLabCluster is able to generate classes aligned with the unsupervised clustering results obtained on full 101 features (as the ground truth). We use 5,294 segments for training and 2,781 segments for testing.
2.1.3 C. elegans
The dataset is recorded with Worm Tracker 2.0 when the worm is freely moving. The body contour is identified automatically using contrast to background from which kinematic features are calculated and constitute 98 features that correspond to body segments from head to tail in 2D coordinates, see Yemini et al. (2013) and Figure 2C. Behavioral states are divided into three classes: moving forward, moving backward, and staying stationary. We use ten sessions (a subset) to investigate the application of OpenLabCluster to this dataset, where the first 7 sessions (543 segments) are used for training and the remaining 3 sessions (196 segments) are used for testing.
2.1.4 Monkey
This dataset is from OpenMonkeyStudio repository (Bala et al., 2020) and captures freely moving macaques in a large unconstrained environment using 64 cameras encircling an open enclosure. 3D keypoints positions are reconstructed from 2D images by applying deep neural network reconstruction algorithms on the multi-view images. Among the movements, 6 behavioral classes have been identified. In contrast to other datasets, this dataset consists of continuous recordings without segmentation into action clips. We thereby segment the videos by clipping them into fixed duration clips (10 frames with 30 fps rate) which results in 919 segments, where each segment is ≈ 0.33 s long. OpenLabCluster receives the 3D body key points of each segment as inputs. Notably, a more advanced technology could be implemented to segment the videos as described in Sarfraz et al. (2021). Here, we focused on examining the ability of OpenLabCluster to work with segments that have not been pre-analyzed and thus used the simplest and most direct segmentation method.
2.2 Evaluation metrics
We evaluate the accuracy of OpenLabCluster by computing the percentage of temporal segments in the test set that OpenLabCluster correctly associated with the states given as ground truth, such that 100% accuracy will indicate that OpenLabCluster correctly classified all temporal segments in the test set. Since OpenLabCluster implements a semi-supervised approach to minimize the number of annotations for segments, we compute the accuracy given annotation budgets of overall 5%, 10%, and 20% labels to be used over the possible iterations in conjunction with AL. In particular, we test the accuracy when the Top, CS, and MI AL methods implemented in OpenLabCluster are used for the selection of temporal segments to annotate. Method details are provided in Section 4 and Appendix 1.3.
2.3 Benchmark comparison
We evaluated OpenLabCluster against established animal behavior classification approaches either with or without AL methods. For non-AL approaches, we compared against K-Nearest Neighbor (KNN) (Cover and Hart, 1967), Support Vector Machine (SVM) (Jhuang et al., 2010), SimBA's Random Forest Classifier (RFC) (Goodwin et al., 2024), and VAME with an additional classifier (VAME+C) (Luxem et al., 2022). With respect to AL, we compared our method to A-SOiD (Tillmann et al., 2024), which employs RFC with selective sampling. Furthermore, we conducted ablation studies by evaluating OpenLabCluster with decoder removed and explored alternative architectures by integrating VAME's encoder-decoder (OpenLabCluster-V) with various AL strategies (CS, TOP, and MI). Detailed experimental settings and additional results are provided in the Benchmark Details Section 1.1.
2.4 Outcomes
The results of evaluation consisting of 5 runs are shown in Tables 1, 2 and further analysis in Figures 3, 4. We summarize the main outcomes of the evaluation and their interpretation below.
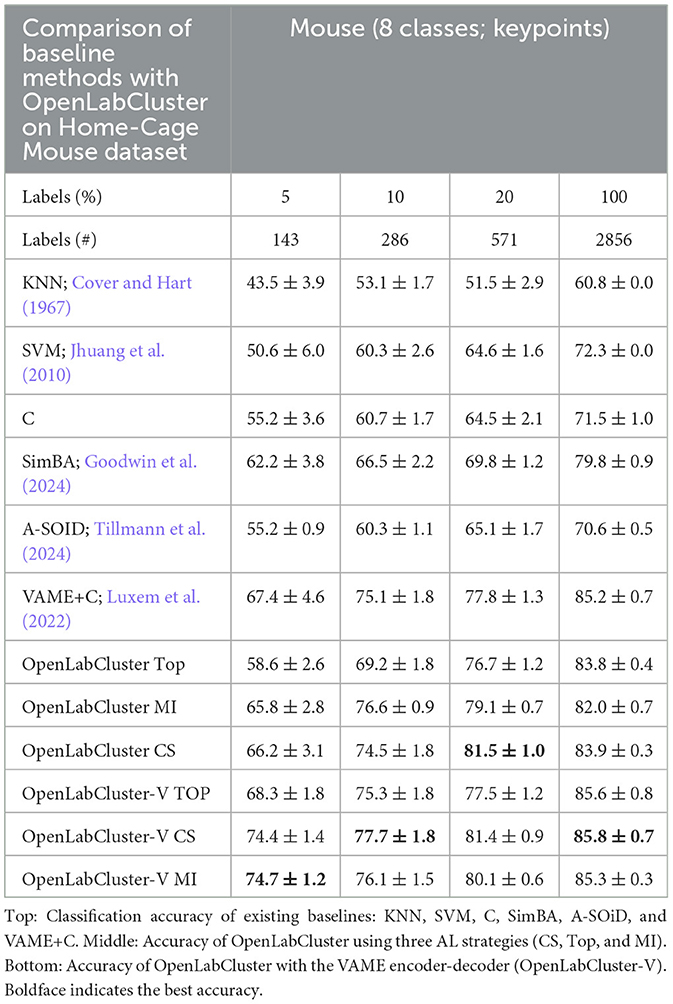
Table 1. Classification accuracy of Home-Cage Mouse behaviors for increasing number of annotated segment (reported as percentage (%)).
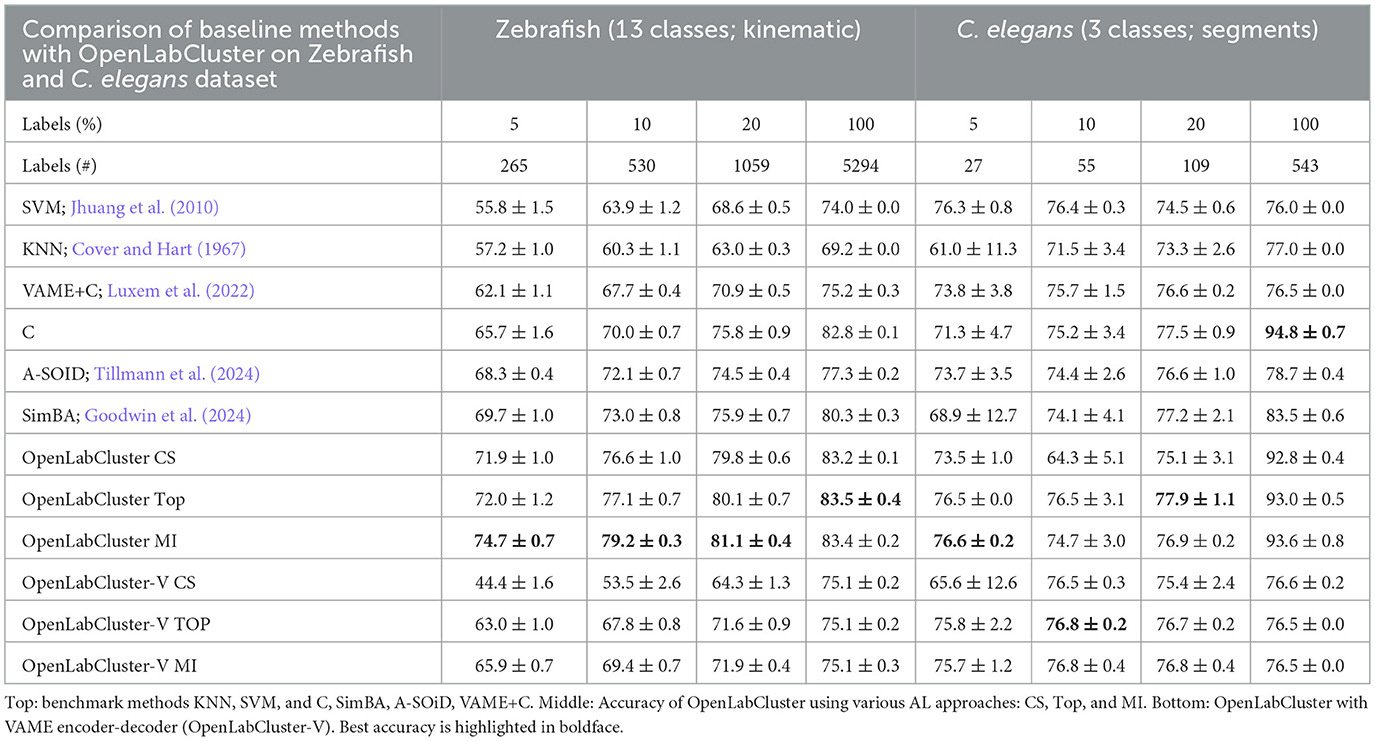
Table 2. Classification accuracy of Zebrafish and C. elegans behaviors for increasing the number of annotated segments [reported as percentage (%)].
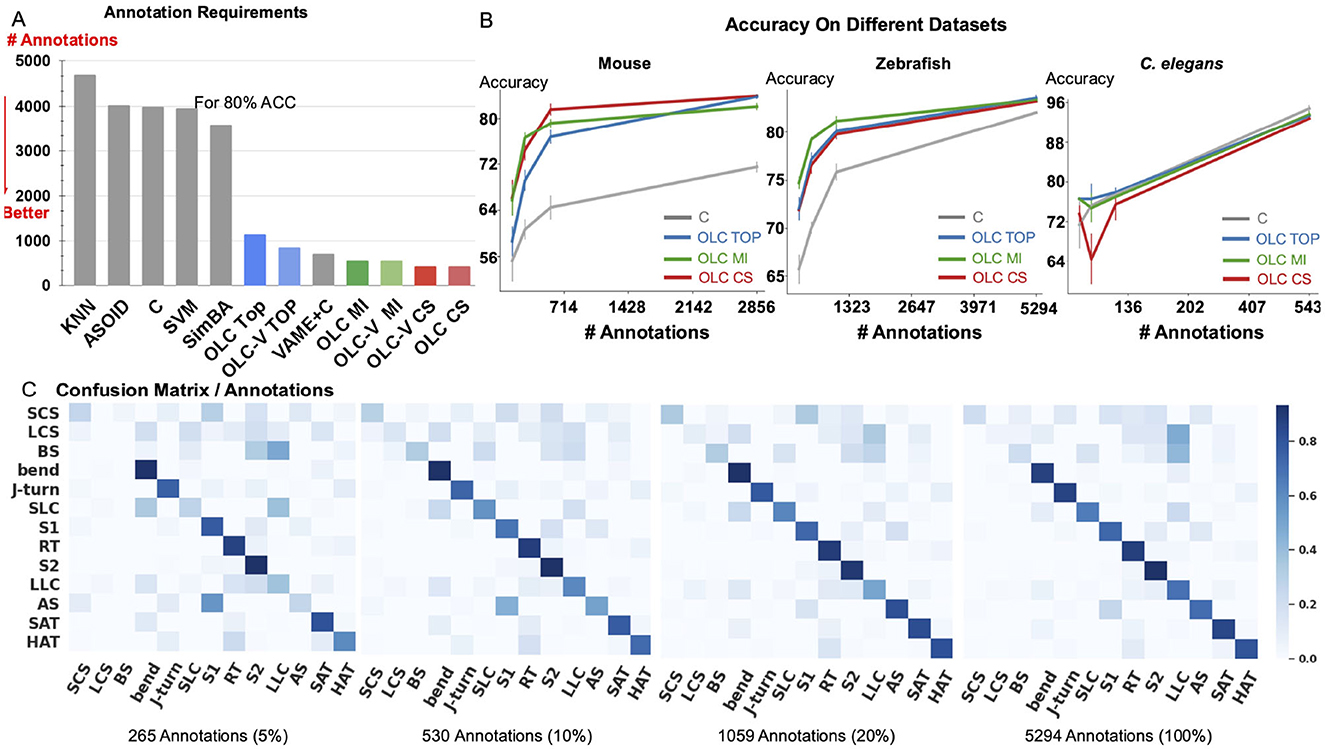
Figure 3. Relation between accuracy and annotation. (A) The amount of annotations required to achieve 80% accuracy for classification of Home-Cage Mouse behaviors. Computed for benchmark methods (KNN, SVM, and C, SimBA, A-SOiD, VAME+C), and variants of OpenLabCluster with three AL methods (Top, MI, CS). (B) Prediction accuracy with increasing annotation budget on three datasets of Mouse, C. elegans and Zebrafish. (C) Confusion matrix for zebrafish dataset for increasing annotation budget (5%, 10%, 20%, 100%).
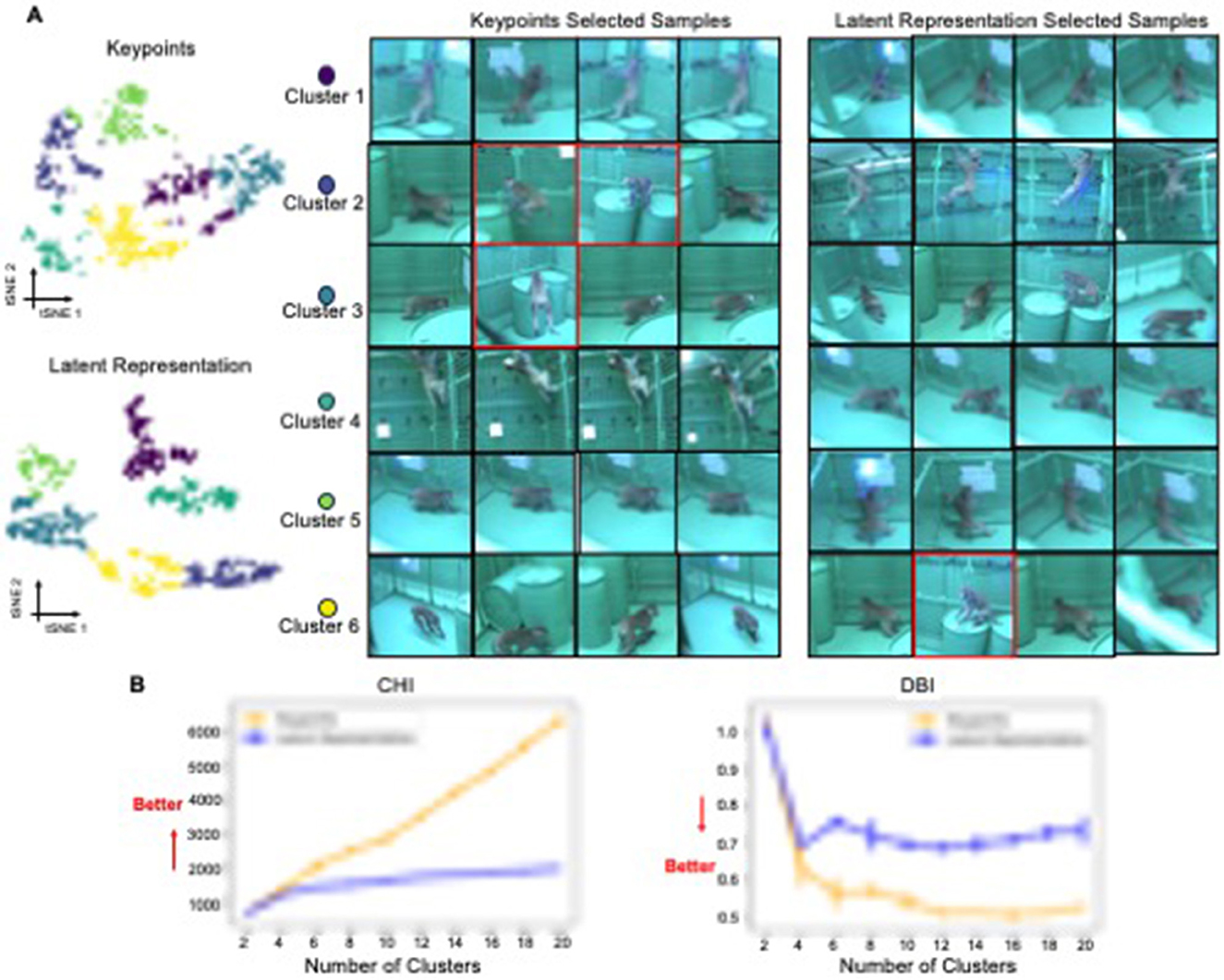
Figure 4. 2D tSNE projection of behavioral segments. (A) 2D t-SNE projections comparing representations derived from keypoints vs. a latent representation. The six K-means clusters are color-coded. For per cluster per representation, the four samples with the shortest distance to each cluster's centroid are shown as examples. The images are sourced from the dataset of Bala et al. (2020). (B) Plots of the Calinski-Harabasz (CHI) and Davies-Bouldin (DBI) scores, which assess clustering quality, for each projection method across a range of cluster numbers.
2.4.1 Accuracy of classification
We observe that the accuracy of classification of OpenLabCluster across datasets is consistently higher than standard supervised classification methods (e.g., C, SVM, and SimBA) for almost any budget of annotation. Specifically, for the Home-Cage Mouse Behavior dataset (Table 1), OpenLabCluster achieves the accuracy of 66.2% when just 143 (5% of 2,856) segments have been annotated. Accuracy improves along with the increase in the number of annotated segments, i.e., accuracy is 76.6% when 10% of segments are annotated, and 81.5% when 20% of segments are annotated. Compared to C-the encoder-only version of OpenLabCluster-OpenLabCluster achieves an average accuracy increase of approximately 12%. This improvement underscores the importance of both the encoder-decoder structure and active sample selection. Meanwhile, VAME+C, which incorporates an encoder-decoder and a classifier, shows promise by reaching an accuracy of 67.4% with only 5% annotated samples. However, it remains less optimal than OpenLabCluster-V, which attains 74.7% under the same annotation budget. These results further illustrate the effectiveness of AL for accurate behavior classification with limited labels. It should be noted that although A-SOiD also integrates active learning for sample selection, its classifier design and exclusive reliance on an uncertainty-based selection method appears to limit its performance relative to OpenLabCluster variants. Among the AL strategies TOP, CS, and MI, both CS and MI outperform TOP on the Home-Cage Mouse dataset. Notably, while AL is expected to be especially effective in sparse annotation scenarios when all segments are annotated (fully supervised scenario), the accuracy of the OpenLabCluster MI approach exceeds supervised classification approaches (C) by 12.4% (rightmost column in Table 1). This reflects the effectiveness of the targeted selection of candidates for annotation and the use of clustering latent representation to enhance the overall organization of the segments.
For Zebrafish and C. elegans datasets, OpenLabCluster consistently achieves higher accuracy, except when 100% of the C. elegans dataset is annotated, demonstrating its generalizability across various animal behavior datasets. When compared to its encoder-only variant (C), OpenLabCluster exhibits an accuracy improvement of approximately 7.8% on the zebrafish dataset and approximately 2.2% on the C. elegans dataset. These gains are less pronounced than those observed on the Home-Cage Mouse dataset. These could be associated with not having manually identified ground truth behavior states for Zebrafish and having only three classes for the C. elegans dataset which is a simpler semantic task that does not challenge classifiers. We can indeed observe that when all annotations are considered ins C. elegans dataset, all approaches perform well (above 92%) and a standard classifier achieves the best accuracy. In contrast to the results observed with the Home-Cage Mouse dataset, we find that on the Zebrafish dataset, OpenLabCluster-V underperforms OpenLabCluster and exhibits comparable performance on C. elegans. Moreover, the CS strategy appears unsuitable for OpenLabCluster-V in the Zebrafish setting, resulting in diminished outcomes.
2.4.2 The amount of required annotations
Since accuracy varies across datasets and depends on the number of classes and other aspects, we examine the relationship between accuracy and the number of required annotations. In Figure 3A, we compute the necessary number of annotations required to achieve 80% of classification accuracy with benchmark methods, OpenLabCluster and OpenLabCluster-V on the Home-Cage Mouse dataset. We observe that AL methods require only 15–20% of the annotated segments to achieve 80% of classification accuracy, whereas benchmark methods (KNN, SVM, C, A-SOiD, SimBA) require roughly nine times as many annotations as the optimal AL approaches. Among the AL methods, the MI and CS embedded variants of OpenLabCluster and OpenLabCluster-V achieve 80% accuracy with approximately 400 annotated samples. The variant with the TOP selection method turns out to be slightly less effective, requiring approximately 700 annotations.
We further visualize the effectiveness of pertaining (C vs. OpenLabCluster) under varying annotation budgets in Figure 3B. We observe that for most cases, OpenLabCluster methods lead to higher accuracy for a given number of annotations than the counterpart, encoder-only classifier. The curves indicating the accuracy of various OpenLabCluster AL methods (red, green, blue) have a clear gap between them and C curve (darkgray), especially in the mid-range of the number of annotations. However, the performance of OpenLabCluster and C is comparable on C. elegans dataset, likely due to the dataset's relative simplicity. In Figure 3C, we further examine class-wise confusion matrices for the Zebrafish dataset on 4 annotation budgets (5%, 10%, 20%, 100%). From visual inspection, it appears that the matrix that corresponds to 20% annotations is close to the matrix that corresponds to 100% annotations. This proximity suggests that the annotation of the full dataset might be redundant. Indeed, further inspection of Figure 3C indicates that samples annotated as LCS and BS classes (y-axis) by the unsupervised learning method are likely to be predicted as the LLC (x-axis) by OpenLabCluster. One possibility for the discrepancy could be annotation errors of the prior clustering method, which are taken as the ground truth. Re-examination of the dynamics of some of the features (e.g., tail angle) further supports this hypothesis and demonstrates that the methods in OpenLabCluster can potentially identify the outlier segments whose annotation settles the organization of the Behavior Classification Map (for more details see in Appendix 1.3).
2.4.3 Organization of the latent representation
Our results indicate that the Latent Representation captured by the OpenLabCluster encoder-decoder and the classifier are able to better organize behavioral segments in comparison with direct embeddings of body keypoints. We quantitatively investigate such an organization with the Monkey dataset, for which ground truth annotations and segmentation are unavailable. Specifically, we obtain the Cluster Map of the segments with OpenLabCluster through the unsupervised training stage, projecting keypoints into the latent representation space. We then depict the 2D tSNE projection of the Latent Representation and compare it with the 2D tSNE projection of body keypoints in Figure 4A. The color in both plots indicates Kmeans Clusters. We set the number clusters as 6 which is defined by the OpenMonkeyStudio dataset (Bala et al., 2020). Indeed, it can be observed that within the Cluster Map, segments are grouped into more distinct and enhanced clusters. To measure the clustering properties of each embedding, we apply clustering metrics of Calinski-Harabasz (CHI) (Caliński and Harabasz, 1974) and Davies-Bouldin (DBI) (Davies and Bouldin, 1979). CHI measures the ratio of inter- and intra-cluster dispersion, with larger values indicating better clustering. DBI measures the ratio of inter-cluster distance to intra-cluster distance, with lower values indicating better clustering. CHI and DBI are shown in the bottom of Figure 4 considering the various number of clusters (from 2 to 20 with interval 2). The comparison shows that the CHI index is higher for the Latent representation than the embedding of the keypoints regardless of the number of clusters being considered and is monotonically increasing with the number of clusters. The DBI index for the Cluster Map is lower than the index of the embedding of the keypoints and illustrating the DBI index decreasing with increasing number of clusters. This is consistent with the expectation that clustering quality will be consistent with the number of behavioral types.
3 Discussion
In this study, we introduce OpenLabCluster, a novel toolset for quantitative studies of animal behavior from video recordings in terms of automatic grouping and depiction of behavioral segments into clusters and their association with behavioral classes. OpenLabCluster works with body keypoints which describe the pose of the animal in each frame and across frames reflecting the kinematic information of the movement that is being exhibited in the segment. The advancement and the availability of automatic tools for markerless pose estimation in recent years allows the employment of such tools in conjunction with OpenLabCluster for performing almost automatic organization and interpretation of a variety of ethological experiments.
The efficacy of OpenLabCluster is attributed to two major components: (i) Unsupervised pre-training process which groups segments with similar movement patterns and disperses segments with dissimilar movement patterns (Clustering); (ii) Automatic selection of samples of segments for association with behavioral classes (AL) through which all segments class labels are associated (classification) and the clustering representation is being refined.
We evaluate OpenLabCluster performance on various datasets of recorded animal species freely behaving, such as Home-Cage Mouse, Zebrafish, C. elegans, and Monkey datasets. For the datasets for which ground-truth labels have been annotated, we show that OpenLabCluster classification accuracy exceeds the accuracy of a direct deep classifier for most annotation budget even when all segments in the training set have been annotated. The underlying reason for the efficacy of OpenLabCluster is the unsupervised pre-training stage of the encoder-decoder which establishes similarities and clusters segments with the Latent Representation of the encoder-decoder. This unsupervised pretraining through encoder-decoder modeling effectively mitigates noise from body orientation and inaccuracies in keypoint estimation to provide a summarized representation. Such a representation turns out to be useful in informing which segments could add semantic meaning of the groupings and refine the representation further.
In practice, we observe that even a sparse annotation of a few segments (5%–20% of the training set) chosen with appropriate AL methods would boost clustering and classification significantly. Classification accuracy continues to improve when more annotations are performed; however, we also observe that the increase in accuracy is primarily in the initial annotation steps, which demonstrates the importance of employing clustering in conjunction with AL selection in these critical steps. Indeed, our results demonstrate that among different AL approaches, more direct approaches such as Top are not as effective as others considering the need to include more metrics quantifying uncertainty and similarity of the segments.
As we describe in the Methodology section, OpenLabCluster includes advanced techniques of unsupervised and semi-supervised neural network training through AL (Appendix 1.1). Inspired by the DeepLabCut project (Mathis et al., 2018; Nath et al., 2019; Deeplabcut, 2018), we implement these techniques jointly with a graphic user interface to enable scientists to use the methodology to analyze various ethological experiments with no deep learning technical expertise. In addition, OpenLabCluster is an open-source project and is designed such that further methodologies and extensions would be seamlessly integrated into the project by developers. Beyond ease of use, the graphic interface is an essential part of OpenLabCluster functionality, since it visually informs the scientists of the outcomes in each iteration step. This provides the possibility to inspect the outcomes and assist with additional information "on-the-go". Specifically, OpenLabCluster allows for pointing at points (segments) in the maps, inspecting their associated videos, adding or excluding segments to be annotated, working with different low-dimensional embeddings (2D or 3D), switching between AL methods, annotating the segments within the same interface, and more.
4 Materials and methods
Existing approaches for behavior classification from keypoints are supervised and require annotation of extensive datasets before training (Xu et al., 2017; Sturman et al., 2020). The requirement limits the generalization of classification from one subject to another, from animal to animal, from a set of keypoints to another, and from one set of behaviors to another due to the need for re-annotation when such variations are introduced.
In contrast, grouping behavioral segments into similarity groups (clustering) typically it does not require annotation and could be done by finding an alternative representation of behavioral segments reflecting the differences and the similarities among segments. Both classical and deep-learning approaches address such groupings (Marques et al., 2018; Hsu and Yttri, 2021). Notably, clustering is a ‘weaker' task than classification since it does not provide the semantic association of groups with behavioral classes; however, it could be used as a preliminary stage for classification. If leveraged effectively, as a preliminary stage, clustering can direct annotation to minimize the number of segments that need to be annotated and at the same time to boost classification accuracy.
OpenLabCluster, that is primarily based on this concept, first infers a Cluster Map and then leverages it for automatic selection of sparse segments for annotation (AL) that will both inform behavior classification and enhance clustering. It iteratively converges to a detailed Behavior Classification Map where segments are grouped into similarity classes and each class is homogeneously representing a behavioral state. Below we describe the components.
4.1 Clustering
The inputs into OpenLabCluster denoted as are sets of keypoint coordinates (2D or 3D) or kinematics features for each time segment along with the video footage (image frames that correspond to these keypoints). Effectively, each input segment of keypoints is a matrix with the row dimension indicating the keypoints coordinate, e.g., the first row will indicate the x-coordinate of the first keypoint and the second row will indicate the y-coordinate of the first keypoint and so on.
The first stage of OpenLabCluster is to employ a recurrent neural network (RNN) encoder-decoder architecture that will learn a Latent Representation for the segments as shown in Figure 5. The encoder is composed of m bi-directional gated recurrent units (bi-GRU) (Cho et al., 2014) sequentially encoding time-series input into a Latent Representation (latent vector in ℝm space). Thus, each segment is represented as a point in the Latent Representation ℝm space. The decoder is composed of uni-directional GRUs that receive as input the latent vector and decode (reconstruct) the same keypoints from the latent vector. Training optimizes encoder-decoder connectivity weights such that the reconstruction loss, the distance between the segment keypoints reconstructed by the decoder and the input segment, is minimized, see the Appendix for further details (Section 1.1). This process reshapes the latent vector points in the Latent Representation space to better represent the segments similarities and distinctions.
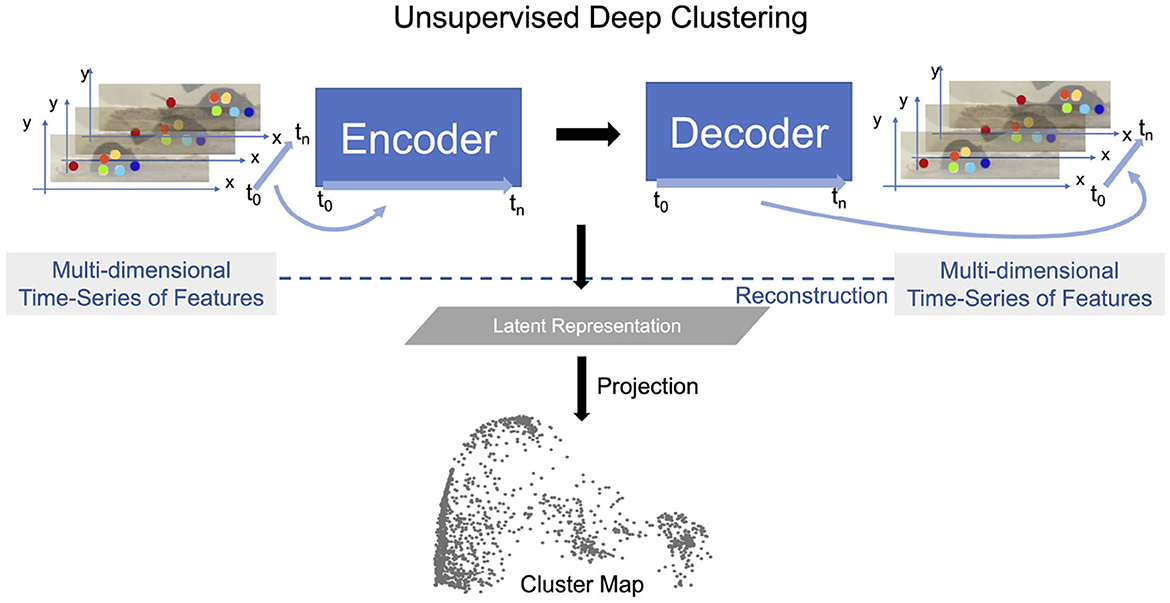
Figure 5. Latent Representation is learned by performing the reconstruction task using an encoder-decoder structure. Latent vectors (last state of the encoder) are projected onto low dimensional space with various dimension reduction techniques to visualize the Latent Representation which constitute the Cluster Map. Mouse images are reproduced from frames of videos in the dataset of Jhuang et al. (2010).
To visualize the relative locations of segments in the Latent Representation, OpenLabCluster implements various dimension reductions (from ℝm → ℝ2 or ℝm → ℝ3), such as PCA, tSNE, and UMAP, to obtain Cluster Maps, see Figure 5-bottom. Thus, each point in the Cluster Map is a reduced-dimensional Latent Representation of an input segment. From inspection of the Cluster Map on multiple examples and benchmarks, it can be observed that the Latent Representation clusters segments that represent similar movements into the same clusters, to a certain extent, typically more effectively than an application of dimension reduction techniques directly to the keypoints segments (Su et al., 2020; Su and Shlizerman, 2020).
4.2 Classification
To classify behavioral segments that have been clustered, we append a classifier, a fully connected network, to the encoder. The training of the classifier is based on segments that have been annotated and minimizes the error between the predicted behavioral states and the behavioral states given by the annotation (cross-entropy loss). When the annotated segments well represent the states and the clusters, the learned knowledge is transferable to other unlabeled segments. AL methods such as Cluster Center (Top), Core-Set (CS), and Marginal Index (MI) aim to select such representative segments by analyzing the Latent Representation. Top selects representative segments which are located at the centers of the clusters [obtained by Kmeans (Li and Shlizerman, 2020a)] in the Latent Representation space. This approach is effective at the initial stage. CS selects samples that cover the remaining samples with minimal distance (Sener and Savarese, 2018). MI is an uncertainty-based selection method, selecting samples that the network is most uncertain about. See Supplementary materials 1.1 for further details regarding these methods. Once segments for annotation are chosen by the AL method, OpenLabCluster highlights the points in the Cluster Map that represent them and their associated video segments, such that they can be annotated within the graphic interface of OpenLabCluster (choosing the most related behavioral class). When the annotations are set, the full network of encoder-decoder with appended classifier is re-trained to perform classification and predict the labels of all segments. The outcome of this process is the Behavior Classification Map which depicts both the points representing segments in clusters and associated states labels with each point (color) as illustrated in Figure 6. In this process, each time that a new set of samples is selected for annotation, the parameters of the encoder-decoder and the classifier are being tuned to generate more distinctive clusters and more accurate behavioral states classification. The process of annotation and tuning is repeated, typically until the number of annotations reaches the maximum amount of the annotation budget, or when clustering and classification appear to converge to a steady state.
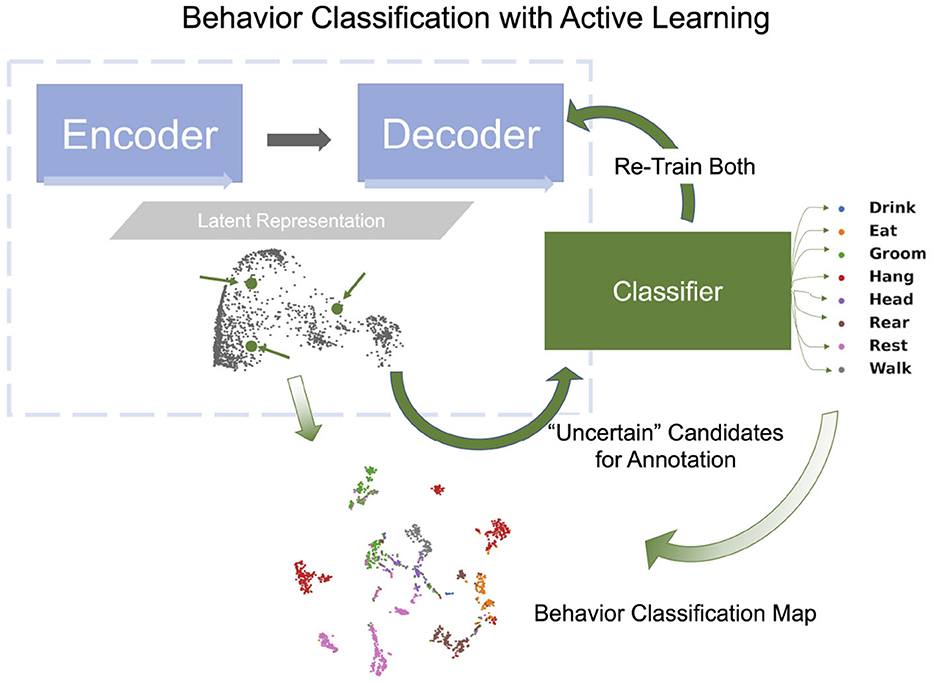
Figure 6. Behavior classification map is generated by a fully connected classifier network (green rectangle) which receives the latent vector transformed by the encoder-decoder as input and classifies them into behavior classes (example shown: 8 classes in Home-Cage Mouse Behavior dataset). Behavioral Classification Map is generated from the Cluster Map and indicates the predicted classes of all segments.
4.3 Implementation details
OpenLabCluster code (OpenLabCLuster, 2022) was developed in University of Washington UW NeuroAI Lab by Jingyuan Li and Moishe Keselman. OpenLabCluster interface is inspired by Deeplabcut (2018), which code is used as a backbone for user interface panels, interaction with the back-end, logging, and visualization. OpenLabCluster also uses Google Active Learning Playground code (Google Active Learning Playground Github Repository, 2017) for the implementation of the K-center selection method in the Core-Set AL option. For specific usage, please see the third_party folder within the OpenLabCluster code repository (OpenLabCLuster, 2022).
OpenLabCluster is available as a GitHub Repository (OpenLabCluster) https://github.com/shlizee/OpenLabCluster and also can be installed with Package Installer for Python (PIP) pip install openlabcluster Jingyuan Li (2022). The repository includes a manual, instructions, and examples.
4.4 Benchmark details
As described earlier, OpenLabCluster summarizes keypoints or kinematic features of a temporal segment into a latent representation and then classifies the behavior using this summarized representation. This approach captures the intrinsic dynamics of short behavior prototypes, in contrast to benchmark methods that compute movement features at each timestep via predefined protocols (Segalin et al., 2021; Tillmann et al., 2024) and classify behavior on a per-timestep basis. To ensure fair comparison, we concatenated the frame-wise features within each segment and applied each frame-wise classification method to the resulting representation. Specifically, for KNN (Cover and Hart, 1967), SVM (Jhuang et al., 2010), and A-SOiD (Tillmann et al., 2024), we concatenated the frame-wise features of each action segment and then employed the classifier proposed by each method for behavior recognition. For SimBA (Goodwin et al., 2024), movement features were extracted from each frame and integrated with pose-based features. The final representation was formed by concatenating these integrated features across all timesteps within the segment. VAME (Luxem et al., 2022) closely resembles OpenLabCluster by learning a unified representation for entire sequences. In VAME+C, we pre-trained VAME, appended a classifier to its latent feature-which encodes the temporal segment's dynamics-and fine-tuned the model using a classification loss. For the Home-Cage Mouse, Zebrafish, and C. elegans datasets, sequences are pre-segmented so that each segment contains a single behavioral prototype. For the OpenStudio Monkey dataset, which is continuously recorded, we divided the videos into fixed temporal windows. More advanced approaches, such as change-point detection algorithms (Edelhoff et al., 2016; Etemad et al., 2021), could also be employed for video segmentation.
Data availability statement
Publicly available datasets were analyzed in this study. These datasets can be found at: HomeCage Mouse (https://dspace.mit.edu/handle/1721.1/49527), OpenMonkeyStudio (https://github.com/OpenMonkeyStudio/OMS_Data), Zebrafish (https://data.mendeley.com/datasets/r9vn7x287r/1), C. elegans (http://wormbehavior.mrc-lmb.cam.ac.uk/).
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the study used previously published datasets.
Author contributions
JL: Formal analysis, Writing – original draft, Data curation, Visualization, Methodology, Conceptualization, Validation, Investigation, Software, Writing – review & editing. MK: Software, Investigation, Writing – review & editing, Formal analysis, Methodology. ES: Writing – review & editing, Project administration, Supervision, Investigation, Visualization, Conceptualization, Funding acquisition, Resources.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The authors acknowledge the partial support of HDR Institute: Accelerated AI Algorithms for Data-Driven Discovery (A3D3) National Science Foundation grant PHY-2117997 (JL, ES) and EFRI-BRAID-2223495 (ES). The authors also acknowledge the partial support by the Departments of Electrical Computer Engineering (JL, ES), Applied Mathematics (ES).
Acknowledgments
Authors are thankful to the Center of Computational Neuroscience and the eScience Center at the University of Washington.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnsys.2025.1630654/full#supplementary-material
References
Anderson, D. J., and Perona, P. (2014). Toward a science of computational ethology. Neuron 84, 18–31. doi: 10.1016/j.neuron.2014.09.005
Bain, M., Nagrani, A., Schofield, D., Berdugo, S., Bessa, J., Owen, J., et al. (2021). Automated audiovisual behavior recognition in wild primates. Sci. Adv. 7:eabi4883. doi: 10.1126/sciadv.abi4883
Bala, P. C., Eisenreich, B. R., Yoo, S. B. M., Hayden, B. Y., Park, H. S., and Zimmermann, J. (2020). Automated markerless pose estimation in freely moving macaques with openmonkeystudio. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-18441-5
Batty, E., Whiteway, M., Saxena, S., Biderman, D., Abe, T., Musall, S., et al. (2019). “BehaveNet: nonlinear embedding and bayesian neural decoding of behavioral videos,” in Advances in Neural Information Processing Systems, 32.
Berman, G. J., Bialek, W., and Shaevitz, J. W. (2016). Predictability and hierarchy in drosophila behavior. Proc. Nat. Acad. Sci. 113, 11943–11948. doi: 10.1073/pnas.1607601113
Berman, G. J., Choi, D. M., Bialek, W., and Shaevitz, J. W. (2014). Mapping the stereotyped behaviour of freely moving fruit flies. J. Royal Soc. Interf. 11:20140672. doi: 10.1098/rsif.2014.0672
Bohnslav, J. P., Wimalasena, N. K., Clausing, K. J., Dai, Y. Y., Yarmolinsky, D. A., Cruz, T., et al. (2021). Deepethogram, a machine learning pipeline for supervised behavior classification from raw pixels. Elife 10:e63377. doi: 10.7554/eLife.63377
Brattoli, B., Büchler, U., Dorkenwald, M., Reiser, P., Filli, L., Helmchen, F., et al. (2021). Unsupervised behaviour analysis and magnification (ubam) using deep learning. Nat. Mach. Intellig. 3, 495–506. doi: 10.1038/s42256-021-00326-x
Buccino, A. P., Lepperód, M. E., Dragly, S.-A., Häfliger, P., Fyhn, M., and Hafting, T. (2018). Open source modules for tracking animal behavior and closed-loop stimulation based on open ephys and bonsai. J. Neural Eng. 15:055002. doi: 10.1088/1741-2552/aacf45
Burgos-Artizzu, X. P., Dollár, P., Lin, D., Anderson, D. J., and Perona, P. (2012). “Social behavior recognition in continuous video,” in 2012 IEEE Conference on Computer Vision and Pattern Recognition (Providence, RI: IEEE), 1322–1329.
Caliński, T., and Harabasz, J. (1974). A dendrite method for cluster analysis. Commun. Statist.-Theory Methods 3, 1–27. doi: 10.1080/03610927408827101
Cao, Z., Hidalgo, G., Simon, T., Wei, S.-E., and Sheikh, Y. (2019). Openpose: realtime multi-person 2d pose estimation using part affinity fields. IEEE Trans. Pattern Anal. Mach. Intell. 43, 172–186. doi: 10.1109/TPAMI.2019.2929257
Cho, K., Van Merriënboer, B., Bahdanau, D., and Bengio, Y. (2014). On the properties of neural machine translation: encoder-decoder approaches. arXiv [preprint] arXiv:1409.1259. doi: 10.3115/v1/W14-4012
Cohn, D. A., Ghahramani, Z., and Jordan, M. I. (1996). Active learning with statistical models. J. Artif. Intellig. Res. 4, 129–145. doi: 10.1613/jair.295
Cover, T., and Hart, P. (1967). Nearest neighbor pattern classification. IEEE Trans. Inform. Theory 13, 21–27. doi: 10.1109/TIT.1967.1053964
Dankert, H., Wang, L., Hoopfer, E. D., Anderson, D. J., and Perona, P. (2009). Automated monitoring and analysis of social behavior in drosophila. Nat. Methods 6, 297–303. doi: 10.1038/nmeth.1310
Datta, S. R., Anderson, D. J., Branson, K., Perona, P., and Leifer, A. (2019). Computational neuroethology: a call to action. Neuron 104, 11–24. doi: 10.1016/j.neuron.2019.09.038
Davies, D. L., and Bouldin, D. W. (1979). A cluster separation measure. IEEE Trans. Pattern Analy. Mach. Intellig. 2, 224–227. doi: 10.1109/TPAMI.1979.4766909
De Chaumont, F., Ey, E., Torquet, N., Lagache, T., Dallongeville, S., Imbert, A., et al. (2019). Real-time analysis of the behaviour of groups of mice via a depth-sensing camera and machine learning. Nature Biomed. Eng. 3, 930–942. doi: 10.1038/s41551-019-0396-1
DeepLabCut (2018). A Software Package for Animal Pose Estimation Github Repository. Available online at: https://github.com/DeepLabCut/DeepLabCut
Dell, A. I., Bender, J. A., Branson, K., Couzin, I. D., de Polavieja, G. G., Noldus, L. P., et al. (2014). Automated image-based tracking and its application in ecology. Trends Ecol. Evol. 29, 417–428. doi: 10.1016/j.tree.2014.05.004
Dunn, T. W., Marshall, J. D., Severson, K. S., Aldarondo, D. E., Hildebrand, D. G., Chettih, S. N., et al. (2021). Geometric deep learning enables 3d kinematic profiling across species and environments. Nat. Methods 18, 564–573. doi: 10.1038/s41592-021-01106-6
Edelhoff, H., Signer, J., and Balkenhol, N. (2016). Path segmentation for beginners: an overview of current methods for detecting changes in animal movement patterns. Movem. Ecol. 4, 1–21. doi: 10.1186/s40462-016-0086-5
Ester, M., Kriegel, H.-P., Sander, J., and Xu, X. (1996). A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases With Noise, 226–231.
Etemad, M., Soares, A., Etemad, E., Rose, J., Torgo, L., and Matwin, S. (2021). SWS: an unsupervised trajectory segmentation algorithm based on change detection with interpolation kernels. Geoinformatica 25, 269–289. doi: 10.1007/s10707-020-00408-9
Goodwin, N. L., Choong, J. J., Hwang, S., Pitts, K., Bloom, L., Islam, A., et al. (2024). Simple behavioral analysis (SIMBA) as a platform for explainable machine learning in behavioral neuroscience. Nat. Neurosci. 27, 1411–1424. doi: 10.1038/s41593-024-01649-9
Google Active Learning Playground Github Repository (2017). Available online at: https://github.com/google/active-learning
Hong, W., Kennedy, A., Burgos-Artizzu, X. P., Zelikowsky, M., Navonne, S. G., Perona, P., et al. (2015). Automated measurement of mouse social behaviors using depth sensing, video tracking, and machine learning. Proc. Nat. Acad. Sci. 112, E5351–E5360. doi: 10.1073/pnas.1515982112
Hsu, A. I., and Yttri, E. A. (2021). B-SOiD, an open-source unsupervised algorithm for identification and fast prediction of behaviors. Nat. Commun. 12:5188. doi: 10.1038/s41467-021-25420-x
Huang, K., Han, Y., Chen, K., Pan, H., Zhao, G., Yi, W., et al. (2021). A hierarchical 3D-motion learning framework for animal spontaneous behavior mapping. Nat. Commun. 12:2784. doi: 10.1038/s41467-021-22970-y
Insafutdinov, E., Andriluka, M., Pishchulin, L., Tang, S., Levinkov, E., Andres, B., et al. (2017). “Arttrack: Articulated multi-person tracking in the wild,” in CVPR'17.
Insafutdinov, E., Pishchulin, L., Andres, B., Andriluka, M., and Schieke, B. (2016). “Deepercut: a deeper, stronger, and faster multi-person pose estimation model,” in European Conference on Computer Vision (ECCV) (Springer), 34–50.
Isik, S., and Unal, G. (2023). Open-source software for automated rodent behavioral analysis. Front. Neurosci. 17:1149027. doi: 10.3389/fnins.2023.1149027
Jazayeri, M., and Afraz, A. (2017). Navigating the neural space in search of the neural code. Neuron 93, 1003–1014. doi: 10.1016/j.neuron.2017.02.019
Jhuang, H., Garrote, E., Yu, X., Khilnani, V., Poggio, T., Steele, A. D., et al. (2010). Automated home-cage behavioural phenotyping of mice. Nat. Commun. 1, 1–10. doi: 10.1038/ncomms1064
Jia, Y., Li, S., Guo, X., Lei, B., Hu, J., Xu, X.-H., et al. (2022). Selfee, self-supervised features extraction of animal behaviors. Elife 11:e76218. doi: 10.7554/eLife.76218
Johnson, R. E., Linderman, S., Panier, T., Wee, C. L., Song, E., Herrera, K. J., et al. (2020). Probabilistic models of larval zebrafish behavior reveal structure on many scales. Curr. Biol. 30, 70–82. doi: 10.1016/j.cub.2019.11.026
Kabra, M., Robie, A. A., Rivera-Alba, M., Branson, S., and Branson, K. (2013). Jaaba: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67. doi: 10.1038/nmeth.2281
Karashchuk, P., Rupp, K. L., Dickinson, E. S., Walling-Bell, S., Sanders, E., Azim, E., et al. (2021). Anipose: a toolkit for robust markerless 3D pose estimation. Cell Rep. 36:109730. doi: 10.1016/j.celrep.2021.109730
Krause, J., Krause, S., Arlinghaus, R., Psorakis, I., Roberts, S., and Rutz, C. (2013). Reality mining of animal social systems. Trends Ecol. Evol. 28, 541–551. doi: 10.1016/j.tree.2013.06.002
Kwon, J., Kim, S., Kim, D.-K., Joo, J., Kim, S., Cha, M., et al. (2024). Subtle: An unsupervised platform with temporal link embedding that maps animal behavior. Int. J. Comput. Vis. 132, 4589–4615. doi: 10.1007/s11263-024-02072-0
Lauer, J., Zhou, M., Ye, S., Menegas, W., Schneider, S., Nath, T., et al. (2022). Multi-animal pose estimation, identification and tracking with deeplabcut. Nat. Methods 19, 496–504. doi: 10.1038/s41592-022-01443-0
Li, J., and Shlizerman, E. (2020a). Iterate &cluster: Iterative semi-supervised action recognition. arXiv [preprint] arXiv:2006.06911. doi: 10.48550/arXiv.2006.06911
Li, J., and Shlizerman, E. (2020b). Sparse semi-supervised action recognition with active learning. arXiv [preprint] arXiv:2012.01740. doi: 10.48550/arXiv.2012.01740
Luxem, K., Mocellin, P., Fuhrmann, F., Kürsch, J., Miller, S. R., Palop, J. J., et al. (2022). Identifying behavioral structure from deep variational embeddings of animal motion. Commun. Biol. 5:1267. doi: 10.1038/s42003-022-04080-7
Lynch, E., Angeloni, L., Fristrup, K., Joyce, D., and Wittemyer, G. (2013). The use of on-animal acoustical recording devices for studying animal behavior. Ecol. Evol. 3, 2030–2037. doi: 10.1002/ece3.608
Marks, M., Jin, Q., Sturman, O., von Ziegler, L., Kollmorgen, S., von der Behrens, W., et al. (2022). Deep-learning-based identification, tracking, pose estimation and behaviour classification of interacting primates and mice in complex environments. Nat. Mach. Intellig. 4, 331–340. doi: 10.1038/s42256-022-00477-5
Marques, J. C., Lackner, S., Félix, R., and Orger, M. B. (2018). Structure of the zebrafish locomotor repertoire revealed with unsupervised behavioral clustering. Curr. Biol. 28, 181–195. doi: 10.1016/j.cub.2017.12.002
Mathis, A., Mamidanna, P., Cury, K. M., Abe, T., Murthy, V. N., Mathis, M. W., et al. (2018). Deeplabcut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 21:1281. doi: 10.1038/s41593-018-0209-y
McCullough, M. H., and Goodhill, G. J. (2021). Unsupervised quantification of naturalistic animal behaviors for gaining insight into the brain. Curr. Opin. Neurobiol. 70, 89–100. doi: 10.1016/j.conb.2021.07.014
McInnes, L., Healy, J., Saul, N., and Groberger, L. (2018). Umap: Uniform manifold approximation and projection. J. Open Source Softw. 3:861. doi: 10.21105/joss.00861
Meyer, F. (1994). Topographic distance and watershed lines. Signal Proc. 38, 113–125. doi: 10.1016/0165-1684(94)90060-4
Monosov, I. E., Zimmermann, J., Frank, M. J., Mathis, M. W., and Baker, J. T. (2024). Ethological computational psychiatry: challenges and opportunities. Curr. Opin. Neurobiol. 86:102881. doi: 10.1016/j.conb.2024.102881
Morrow-Tesch, J., Dailey, J., and Jiang, H. (1998). A video data base system for studying animal behavior. J. Anim. Sci. 76, 2605–2608. doi: 10.2527/1998.76102605x
Murari, K. (2019). “Recurrent 3D convolutional network for rodent behavior recognition,” in ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Brighton: IEEE), 1174–1178.
Nakamura, T., Matsumoto, J., Nishimaru, H., Bretas, R. V., Takamura, Y., Hori, E., et al. (2016). A markerless 3D computerized motion capture system incorporating a skeleton model for monkeys. PLoS ONE 11:e0166154. doi: 10.1371/journal.pone.0166154
Nath, T., Mathis, A., Chen, A. C., Patel, A., Bethge, M., and Mathis, M. W. (2019). Using deeplabcut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 14, 2152–2176. doi: 10.1038/s41596-019-0176-0
OpenLabCLuster (2022) Github Repository. Available online at: https://github.com/shlizee/OpenLabCluster
Pereira, T. D., Aldarondo, D. E., Willmore, L., Kislin, M., Wang, S. S.-H., Murthy, M., et al. (2019). Fast animal pose estimation using deep neural networks. Nat. Methods 16, 117–125. doi: 10.1038/s41592-018-0234-5
Pereira, T. D., Tabris, N., Matsliah, A., Turner, D. M., Li, J., Ravindranath, S., et al. (2022). Sleap: A deep learning system for multi-animal pose tracking. Nat. Methods 19, 486–495. doi: 10.1038/s41592-022-01426-1
Rousseau, J., Van Lochem, P., Gispen, W., and Spruijt, B. (2000). Classification of rat behavior with an image-processing method and a neural network. Behav. Res. Methods Instrum. Comp. 32, 63–71. doi: 10.3758/BF03200789
Sakata, S. (2023). Salsa: a combinatory approach of semi-automatic labeling and long short-term memory to classify behavioral syllables. Eneuro 10:ENEURO.0201-23.2023. doi: 10.1523/ENEURO.0201-23.2023
Sarfraz, S., Murray, N., Sharma, V., Diba, A., Van Gool, L., and Stiefelhagen, R. (2021). “Temporally-weighted hierarchical clustering for unsupervised action segmentation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Nashville, TN: IEEE), 11225–11234.
Schneider, S., Lee, J. H., and Mathis, M. W. (2023). Learnable latent embeddings for joint behavioural and neural analysis. Nature 617, 360–368. doi: 10.1038/s41586-023-06031-6
Segalin, C., Williams, J., Karigo, T., Hui, M., Zelikowsky, M., Sun, J. J., et al. (2021). The mouse action recognition system (MARS) software pipeline for automated analysis of social behaviors in mice. Elife 10:e63720. doi: 10.7554/eLife.63720
Sener, O., and Savarese, S. (2018). “Active learning for convolutional neural networks: A core-set approach,” in International Conference on Learning Representations.
Settles, B. (2009). “Active learning literature survey,” in Technical report, University of Wisconsin-Madison Department of Computer Sciences.
Settles, B. (2012). Active learning. Synthesis Lect Artif Intellig Mach Learn. 6, 1–114. doi: 10.1007/978-3-031-01560-1
Stern, U., He, R., and Yang, C.-H. (2015). Analyzing animal behavior via classifying each video frame using convolutional neural networks. Sci. Rep. 5, 1–13. doi: 10.1038/srep14351
Sturm, V., Efrosinin, D., Efrosinina, N., Roland, L., Iwersen, M., Drillich, M., et al. (2019). A chaos theoretic approach to animal activity recognition. J. Mathem. Sci. 237, 730–743. doi: 10.1007/s10958-019-04199-9
Sturman, O., von Ziegler, L., Schläppi, C., Akyol, F., Privitera, M., Slominski, D., et al. (2020). Deep learning-based behavioral analysis reaches human accuracy and is capable of outperforming commercial solutions. Neuropsychopharmacology 45, 1942–1952. doi: 10.1038/s41386-020-0776-y
Su, K., Liu, X., and Shlizerman, E. (2020). “Predict and cluster: Unsupervised skeleton based action recognition,” in IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR) (Seattle, WA: IEEE).
Su, K., and Shlizerman, E. (2020). Clustering and recognition of spatiotemporal features through interpretable embedding of sequence to sequence recurrent neural networks. Front. Artif. Intellig. 3:70. doi: 10.3389/frai.2020.00070
Sun, J. J., Kennedy, A., Zhan, E., Anderson, D. J., Yue, Y., and Perona, P. (2021). “Task programming: Learning data efficient behavior representations,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (Nashville, TN: IEEE), 2876–2885.
Taiwanica, Z. (2000). Ethom: event-recording computer software for the study of animal behavior. Acta Zool. Taiwanica 11, 47–61.
Tillmann, J. F., Hsu, A. I., Schwarz, M. K., and Yttri, E. A. (2024). A-soid, an active-learning platform for expert-guided, data-efficient discovery of behavior. Nat. Methods 21, 703–711. doi: 10.1038/s41592-024-02200-1
Tseng, A., Sun, J. J., and Yue, Y. (2022). “Automatic synthesis of diverse weak supervision sources for behavior analysis,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (New Orleans, LA: IEEE), 2211–2220.
Usman, B., Tagliasacchi, A., Saenko, K., and Sud, A. (2022). “Metapose: Fast 3D pose from multiple views without 3D supervision,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (New Orleans, LA: IEEE), 6759–6770.
van Dam, E. A., Noldus, L. P., and van Gerven, M. A. (2020). Deep learning improves automated rodent behavior recognition within a specific experimental setup. J. Neurosci. Methods 332:108536. doi: 10.1016/j.jneumeth.2019.108536
Weber, R. Z., Mulders, G., Kaiser, J., Tackenberg, C., and Rust, R. (2022). Deep learning-based behavioral profiling of rodent stroke recovery. BMC Biol. 20:232. doi: 10.1186/s12915-022-01434-9
Weinreb, C., Pearl, J. E., Lin, S., Osman, M. A. M., Zhang, L., Annapragada, S., et al. (2024). Keypoint-moseq: parsing behavior by linking point tracking to pose dynamics. Nat. Methods 21, 1329–1339. doi: 10.1038/s41592-024-02318-2
Wiltschko, A. B., Tsukahara, T., Zeine, A., Anyoha, R., Gillis, W. F., Markowitz, J. E., et al. (2020). Revealing the structure of pharmacobehavioral space through motion sequencing. Nat. Neurosci. 23, 1433–1443. doi: 10.1038/s41593-020-00706-3
Wu, A., Buchanan, E. K., Whiteway, M., Schartner, M., Meijer, G., Noel, J.-P., et al. (2020). Deep graph pose: a semi-supervised deep graphical model for improved animal pose tracking. Adv. Neural Inf. Process. Syst. 33, 6040–6052. doi: 10.1101/2020.08.20.259705
Xu, C., Govindarajan, L. N., Zhang, Y., and Cheng, L. (2017). Lie-x: Depth image based articulated object pose estimation, tracking, and action recognition on lie groups. Int. J. Comput. Vis. 123, 454–478. doi: 10.1007/s11263-017-0998-6
Ye, S., Filippova, A., Lauer, J., Schneider, S., Vidal, M., Qiu, T., et al. (2024). Superanimal pretrained pose estimation models for behavioral analysis. Nat. Commun. 15:5165. doi: 10.1038/s41467-024-48792-2
Yemini, E., Jucikas, T., Grundy, L. J., Brown, A. E., and Schafer, W. R. (2013). A database of caenorhabditis elegans behavioral phenotypes. Nat. Methods 10, 877–879. doi: 10.1038/nmeth.2560
Zhang, L., Dunn, T., Marshall, J., Olveczky, B., and Linderman, S. (2021). “Animal pose estimation from video data with a hierarchical von mises-fisher-gaussian model,” in International Conference on Artificial Intelligence and Statistics (New York: PMLR), 2800–2808.
Zhou, T., Cheah, C. C. H., Chin, E. W. M., Chen, J., Farm, H. J., Goh, E. L. K., et al. (2023). Contrastivepose: a contrastive learning approach for self-supervised feature engineering for pose estimation and behavorial classification of interacting animals. Comput. Biol. Med. 165:107416. doi: 10.1016/j.compbiomed.2023.107416
Keywords: graphic user interface (GUI) for behavior recognition, animal behavior analysis, active learning, semi-supervised learning, efficient behavior recognition
Citation: Li J, Keselman M and Shlizerman E (2025) OpenLabCluster: active learning based clustering and classification of animal behaviors based on kinematic body keypoints. Front. Syst. Neurosci. 19:1630654. doi: 10.3389/fnsys.2025.1630654
Received: 18 May 2025; Accepted: 14 August 2025;
Published: 31 October 2025.
Edited by:
Konstantinos Slavakis, Institute of Science Tokyo, JapanReviewed by:
Shuzo Sakata, University of Strathclyde, United KingdomEric Allen Yttri, Carnegie Mellon University, United States
Copyright © 2025 Li, Keselman and Shlizerman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingyuan Li, amluZ3lsaTZAdXcuZWR1
 Jingyuan Li
Jingyuan Li Moishe Keselman
Moishe Keselman Eli Shlizerman
Eli Shlizerman