- 1State Key Laboratory of Cotton Biology, The Ministry of Agriculture, Institute of Cotton Research, Chinese Academy of Agricultural Science, Anyang, China
- 2Key Laboratory of Biological and Genetic Breeding of Cotton, The Ministry of Agriculture, Institute of Cotton Research, Chinese Academy of Agricultural Science, Anyang, China
- 3Institute of Molecular Biology and Biotechnology, The University of Lahore, Lahore, Pakistan
- 4FB Genetics Four Brothers Group, Lahore, Pakistan
- 5University Institute of Physical Therapy, The University of Lahore, Lahore, Pakistan
- 6Zhejiang University, Hangzhou, China
- 7School of Biotechnology and Food Engineering, Anyang Institute of Technology, Anyang, China
- 8National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China
- 9Zhengzhou Research Base, State Key Laboratory of Cotton Biology, Zhengzhou University, Zhengzhou, China
Lack of precise information about the candidate genes involved in a complex quantitative trait is a major obstacle in the cotton fiber quality improvement, and thus, overall genetic gain in conventional phenotypic selection is low. Recent molecular interventions and advancements in genome sequencing have led to the development of high-throughput molecular markers, quantitative trait locus (QTL) fine mapping, and single nucleotide polymorphisms (SNPs). These advanced tools have resolved the existing bottlenecks in trait-specific breeding. This review demonstrates the significance of chromosomes 3, 7, 9, 11, and 12 of sub-genomes A and D carrying candidate genes for fiber quality. However, chromosome 7 carrying SNPs for stable and potent QTLs related to fiber quality provides great insights for fiber quality-targeted research. This information can be validated by marker-assisted selection (MAS) and transgene in Arabidopsis and subsequently in cotton.
Introduction
Cotton is a major cash crop cultivated worldwide (Nadeem et al., 2021). It belongs to the Hibiscus or mallow family (Malvaceae) (Wendel et al., 2009). Cotton as a natural fiber is considered a potential cash crop mainly for the textile industry. The demand of the textile industry is increasing due to the high consumption rate and considerable marketability. It is planted over 80 countries in the world. Of the 52 species of Gossypium known to date, only four species, i.e., G. hirsutum, G. barbadense, G. arboreum, and G. herbaceum are mostly cultivated around the globe (Fryxell, 1992; Kunbo et al., 2018). Gossypium hirsutum has considerable adaptability to different environments and better yield compared with the other cultivated species, covering almost 95% needs of the global population (Chen et al., 2007). Allotetraploid cotton was originated by the hybridization of extinct progenitors of G. herbaceum (A1) or G. arboreum (A2) and G. raimondii (D5) (Wendel, 1989). Upland cotton (G. hirsutum) was first domesticated about 4,000–5,000 years ago. Later on, evolutionary studies and domestications divided the upland cotton into three groups, namely, improved modern cultivars, land races, and semi-wild (Fang et al., 2017). Gossypium hirsutum belonging to semi-wild is recognized with seven races (Hutchinson, 1951). The genus Gossypium consists of 45 diploid (2n = 2× = 26) and seven tetraploid (2n = 4× = 52) species. The diploid species are clustered into eight genomic groups (A–G and K) (Wang M. et al., 2019). Interspecific hybridization generated allotetraploids in America, and subsequently, polyploidization of A and D sub-genomes took place. Approximately, 1–2 million years ago, these two sub-genomes reassembled during the geographical change. Probably, G. herbaceum (A1) and G. arboreum (A2) are the ancestral donors of genome A, while G. raimondii (D5) is the donor of genome D (Hu et al., 2019). Normally, species of A genome bring out spinnable fibers, while D genome species are not cultivated. Approximately, worldwide, more than 90% annual cotton is met by G. hirsutum (AD)1-Upland cotton (USDA, 2019b).
Generally, the fiber length and strength are principal among all other fiber quality traits including fiber uniformity and micronaire (Yang et al., 2016). Fiber quality traits are paramount for the spinning technology in the textile industry, and they influence the fiber processing and dyeing consistency (Rodgers et al., 2017). Both fiber quality and yield are complex quantitative traits. The fiber yield trait is controlled by multiple genes and is much influenced by epigenetics, whereas the fiber quality is mainly controlled by the gene additive effect and is less affected by the environment (Ming-bao et al., 2008; Zhang et al., 2017).
Although fiber development studies are being carried out for many years, the molecular bases are still hardly known (Zhang B. et al., 2013). Molecular markers are potential tools for QTL mapping and dissecting the molecular processes of fiber quality characteristics. At least, 1,075 and 1,059 QTLs from 58 to 30 research studies, respectively, have been suggested with intraspecific and interspecific crosses (Said et al., 2015). These can further be validated by gene cloning to dissect the processes of complex traits. Moreover, functional studies explain the underlying molecular processes of fiber development in upland cotton.
Genome sequencing provides recent insights into the genome of cotton and has explored significant variations among the species or cultivars. The availability of the whole-genome sequence provides comprehensive information of fiber and fiber-related characteristics. The initial research study was started with the closest diploid progenitor G. raimondii (D5) and G. arboreum (A2) (Zhang et al., 2015a) which expanded to other tetraploid species G. barbadense (AD)2 and G. hirsutum (AD)1 (Hu et al., 2019). Allotetraploid species are economically significant in natural fiber production worldwide. Gossypium hirsutum produces better fiber yield, whereas G. barbadense even under harsh environmental conditions, can produce superior quality fiber over a process of evolution. The genetic foundations of these interspecies divergences are unrevealed. The information generated on the genome assembly and sequencing enables the breeders to revamp the fiber quality and resilience to variable environments (Yang et al., 2019). Reverse genetic techniques such as gene silencing (Kuttenkeuler and Boutros, 2004), targeted gene disruption by homologous recombination (Iida and Terada, 2004), insertional mutagenesis (Stemple, 2004), and chemical mutagenesis (Gilchrist and Haughn, 2005) play a vital role in identifying the function of the gene phenotypically. The genome sequencing information can help breeders to better understand the variations in the genome structure of chromosomes over-time, producing better fiber yield and quality. Based on this genetic information, the studies can be expanded for transgenes using recent molecular approaches. This comprehensive review provides the significance of different stable QTLs and candidate genes located on different regions of the chromosomes, with a special insight of chromosome 7 in upland cotton exploring the potential for improved fiber quality and quantity.
Cotton Fiber Structure and Composition
Primarily, the cotton fiber is composed of 88.0–96.5% of cellulose (Goldwaith and Guthrie, 1954); however, a secondary cell wall is solely made of cellulose. The non-cellulosic components are located either on the cuticle or inside the lumen. They include proteins (1.0–1.9%), waxes (0.4–1.2%), pectins (0.4–1.2%), inorganics (0.7–1.6%), and other (0.5–8.0%) substances (Fan et al., 2005). The composition of cotton fiber undergoes dynamic changes throughout its development. During the early stage of 0–2 DPA, 1) xyloglucan is depleted, and the outer sheath is developed 2) an epitope (1–6)- β-D-galactan is lost carrying arabinose (Vaughn and Turley, 1999; Bowling et al., 2011), and 3) synthesis of the cotton fiber middle lamella (CFML) begins. This outer thin primary cell wall augmented with fucosylated and non-fucosylated xyloglucan and homogalacturonan helps connect adjacent fibers into tissue-like bundles, assembling into firmly packed fibers within the confined boll space (Singh et al., 2009). The inner primary wall of the cotton fiber contains ∼22% crystalline cellulose fibrils surrounded by pectin and xyloglucan (Singh et al., 2009). In the cotton fiber, the glycome profiling of the fiber shows the presence of xylan and acetylated mannan (Pattathil et al., 2010). During the transition phase of primary and secondary cell wall deposition, degradative enzymes break down the CFML to release the fibers individually—a phenomenon in which primary wall-related sugars are lessened, cell wall hydrolases are upregulated, pectin is reduced, and xyloglucan molecular mass, as secondary wall synthesis, begins (Gou et al., 2007; Singh et al., 2009).
A unique winding of cell wall layers occurs during the transition of primary and secondary walls. This unique winding is similar to the S1 layer in the wood fiber (Seagull, 1993) (Figure 1). During the same transition phase rearrangement of microtubules, a decline in the respiration rate and change in the concentration of metabolic sugars occur, whereas the cellulose synthesis rate increases to ∼35% (w/w), and the CFML undergoes degradation (Singh et al., 2009). Although, wall thickening is minimal at this stage, fiber strength increases significantly, seemingly because of differentially placed cellulose microfibrils in the layer. So the secondary wall of cotton fiber becomes 3–6 μm thick (Hinchliffe et al., 2011).
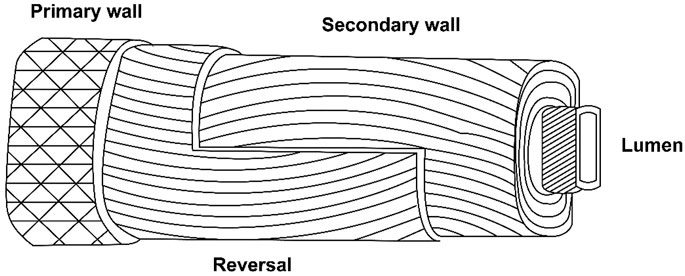
FIGURE 1. Generalized structure of the cotton fiber (M. Dochia and Z. Roskwitalski, 2012).
Cotton Fiber Developmental Biology
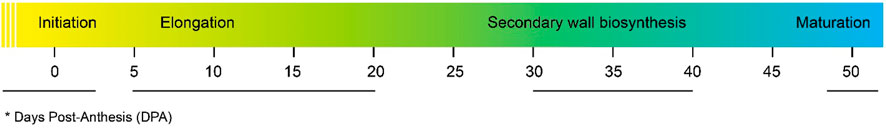
The cotton fiber is a seed-borne epidermal trichome. It is made of thickened and elongated single cells. The turgor pressure in the cell causes an expansion in the cotton fiber. (Cosgrove, 1997). Fiber development occurs 3 days before and after anthesis coupled with enlargement of epidermal cells and protrusion of trichomes. Both the lint percentage and lint index are positively associated with the density of trichome protrusion (0 DPA) and fiber initials (1 DPA) in some cultivars of G. hirsutum (Li et al., 2009). Elongation of the fiber starts within a period of 2DPA to 20DPA, followed by initiation. The twisting of elongating fibers generates several fibers (Haigler et al., 2012). Cellulose synthesis takes place at 10–16 DPA, stimulating the closing of plasmodesmata that is paramount for fiber elongation. At the cellular level, the mechanism of the GhVINI gene expression (Wang et al., 2010), expression of pectin biosynthesis genes (Pang et al., 2010), expression of expansins (Li et al., 2016a), and aquaporin proteins and their corresponding genes is stimulated, which plays a key role in development of cotton fibers (Liu et al., 2008). Additionally, during cotton fiber differentiation, DNA methylation has been reported, which is related with the modification of chromatin (Wang et al., 2016).
Fiber elongation stops during the transition of cell elongation to cell wall biosynthesis making the secondary cell wall (SCW) thickened with the upregulation of SCW genes. The rate of synthesis of cellulose, the orientation of its fibrils, and the individual length of the cellulose chain synthesized by cellulose synthase directly deals with the thickness of the SCW (Betancur et al., 2010). In sclerenchymatous cells, several flavonoids and lignin biosynthesis pathways are involved in the regulation of SCW synthesis (Zhao and Dixon, 2011) (Figure 2).
The single cell expression system provides greater insight on understanding the mechanism of fiber development (Haigler et al., 2009). During cell wall thickening, the primary cell wall (PCW) feature of Arabidopsis leaf trichomes relates to the SCW of the cotton fiber (Betancur et al., 2010).
Four stress-related genes were identified, and the expression pattern was analyzed. The phenotypic data represented that abiotic stress affects the overexpression of the cotton genes in Arabidopsis (Li et al., 2020). In another study, it was reported that the treatment of sodium chloride enhances the gene expression for ∆-pyrroline-5-carboxylase synthetase, sucrose synthase, cellulose synthase A, and sucrose phosphate synthase in leaves and fibers of the transgenic plants (Kesanakurti et al., 2010). Furthermore, the study reported that salt stress shows diverse responses over O-methyltransferase genes during fiber development and may be involved in the salt tolerance of Gossypium species (Hafeez et al., 2021). This characteristic provides a roadmap to further validate the functions of related genes.
Role of Major Genes in Fiber Quality Development Across the Whole Genome
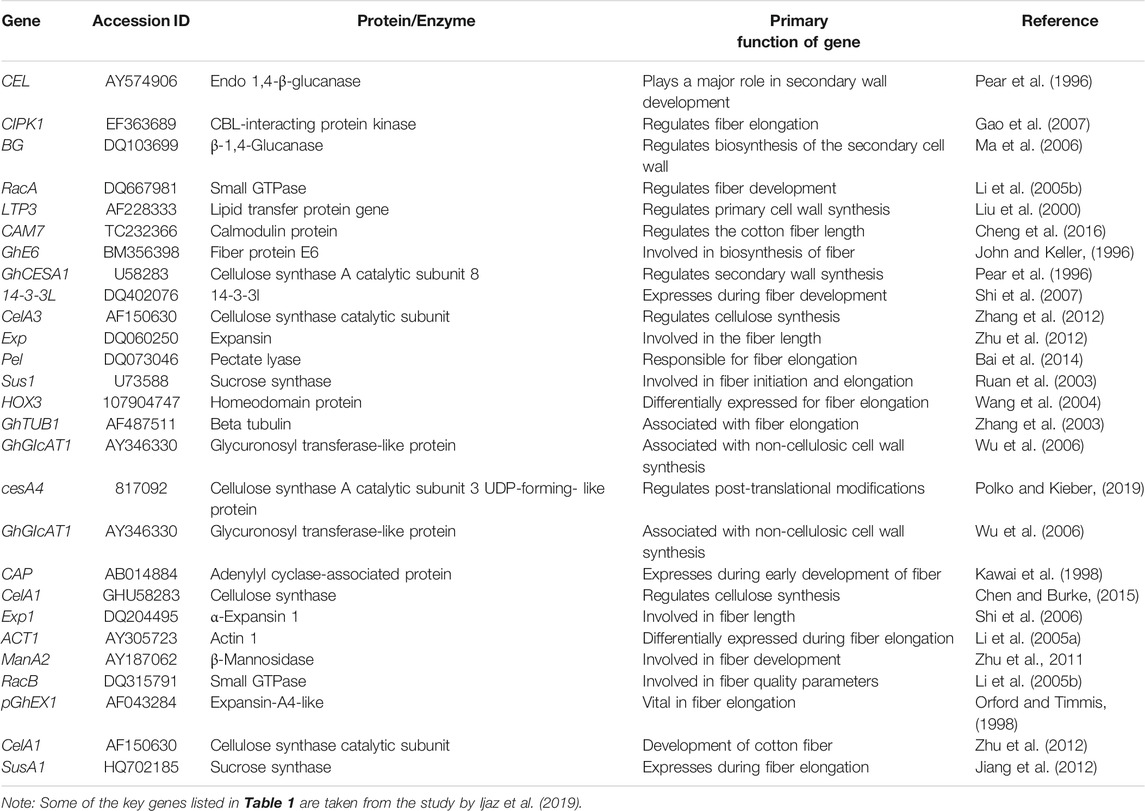
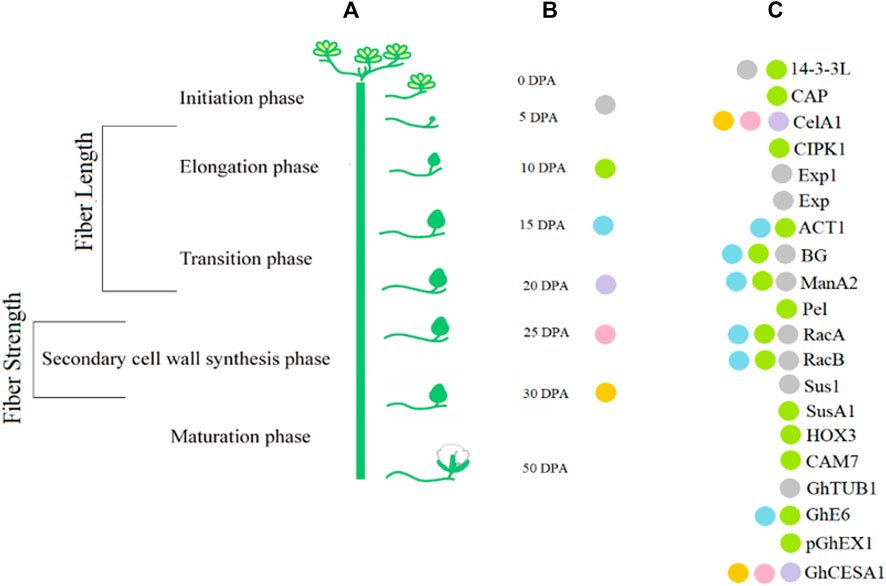
Transcript profiling is important in providing information about various phases of fiber development. Transcription factor (TF) families such as MYB, C2H2, and WRKY are involved in fiber initiation (Zhang et al., 2010). Different genes are differentially expressed at earlier stages of fiber initiation and development in different species of Gossypium (Samuel Yang et al., 2006; Zhang et al., 2010). Plant hormones also play a key role in fiber development. For example, GhPIN-mediated auxin transport is involved in the ovule-specific suppression of the GhPIN gene, which reveals its involvement in fiber initiation (Zhang J. et al., 2016). A candidate gene Pel (Accession ID: DQ073046) on chromosome 7 has been reported, which is involved in the breakdown of de-esterified pectin and in the fiber elongation process (Bai et al., 2014). Some of the potential genes with their accession numbers are listed in Table 1. A schematic diagram represents the involvement of genes associated with the fiber strength and length (Figure 3).
Molecular breeding has indicated some potential genes with a notable role in the fiber quality, yet they need to be verified through functional genomics or other advanced molecular techniques including the transgenic technology (Table 2). Two major genomic regions (MGR1 and MGR2) of upland cotton on chromosome Dt7 were reported with four potential candidate genes corresponding to the fiber length and fiber strength. It was found that MGRI regions contain three putative candidate genes qualified for the fiber length (FL), while the MGR2 region corresponding to CotAD_35088 qualified for both fiber length and fiber strength (Su et al., 2016). Similarly, a CSSL line (MBI7747 and MBI7561) was established with a cross of high-yielding CCRI45 (G. hirsutum) and high-quality fiber Hai1 (G. barbadense) species, and potential candidate genes were found (Lu et al., 2017).
During fiber development, differential expression studies help to find out fiber quality-related genes. For example, two recombinant inbred lines (RILs), differing in fiber quality, were taken from the intra-hirsutum population. The expression profiling differences and fiber quality-related genes were explored by RNA sequencing. It was noted that differentially expressed genes (DEG) 72/27, 1,137/1,584, 437/393, 1,019/184, and 2,555/1,479 were up/downregulated in lines (L1 high-fiber quality and L2 low-fiber quality) at 10, 15, 20, 25, and 30-day post-anthesis, respectively. Hence, 363 DEGs between L1 and L2 were co-localized in fiber strength QTL. Seven expression profiles were identified through short time-series expression minor (STEM) analysis. Kyoto Encyclopedia of Gene and Genomes (KEGG) and gene ontology (GO) annotations were performed to identify differences in the gene function associated with two lines L1 and L2 (Zou et al., 2019). The five modules of specific fiber development stages mainly for fiber quality were assessed by co-expression network analysis. It was noted that the relationship between hub and other genes can be revealed by the correlation network (Zou et al., 2019). Furthermore, transcriptome and QTL analysis of 780 differently expressed genes were studied (Wang et al., 2020).
Significance of Current Technologies and Cotton Fiber Development
Molecular Markers and the Linkage Map for Quantitative Trait Locus Identification
The high-density linkage map, germplasm evolution, and phylogenetic analysis are concomitant with the implication of molecular markers. These applications are highly helpful and efficient for the mapping of stable QTLs and recognition of genes related with development of fibers. Reinisch et al. (1994) developed the first genetic map using 705 RFLP markers that covered a length of 4,675 cm. Subsequently, different genetic maps were developed for fiber quality traits (Kohel et al., 2001). Molecular markers have become an integral component for the determination of fiber quality-related traits. The information can be applied across chromosomes of the Gossypium species to identify fiber quality-related QTLs and candidate genes.
Recombination inbred lines (RILs) and doubled haploids, F2, F2:3, and BC1 are primary populations (Cao et al., 2015; Islam et al., 2016a), while secondary populations are chromosome segment substitution lines (CSSLs) used in cotton (Islam et al., 2016b). The crossing of a donor and a recipient with several backcrossing with the recurrent parents develop CSSLs. The offspring and the recurrent parents have the same genetic background with a difference of single nucleotide or an Indel. CSSLs are considered ideal and efficient material for QTL analysis, fine mapping, and positioning cloning (Guo et al., 2013). The first CSSLs were developed by Esha and Zamir in 1994 and detected six QTLs in tomato related to its quality. Subsequently, these lines were successfully developed in maize, wheat, peanut, rice, soyabean, and other plants (Thomson et al., 2003; Zhou G. et al., 2015; He et al., 2015). The genetic map resulted from G. hirsutum and G. barbadense helps identify the QTLs related to fiber yield and quality. (Shi et al., 2015). It is reported that the development of CSSLs from TM-1 and Hai7124 increased the fiber quality (Wang et al., 2012). In another research, 51 QTLs were reported in 116 lineages of CSSLs using CCRI45 and Hai1 (Yang et al., 2009). The construction of chromosome segment substitution lines in Gossypium species is getting more attention to attain good fiber quality traits such as the fiber length and strength. This approach can be exploited across the economically important chromosome 7 to identify the most suitable stable QTLs and candidate genes.
Sequencing in Cotton Fiber Development Using Advanced Next Generation Tools
The development of high density and precise QTL genetic linkage maps is due to the use of molecular markers associated with the availability of genome sequences of G. raimondii, G. arboreum, G. hirsutum, and G. barbadense (Yuan et al., 2016). Subsequently, high-throughput genotyping provided a deep insight for the better understanding of the genome-wide study. For example, cotton SNP63K and cotton SNP80K enabled the researchers to better understand the genetic mapping, genome, genomic selection, and genomic diversity within Gossypium (Cai et al., 2017). Different SNP markers can be deployed on common QTLs found under diverse environments for fine mapping and candidate gene identification (Su et al., 2016; Tan et al., 2018). This could be further boosted by the advancement of next-generation sequencing which includes restriction site-associated DNA (Jia et al., 2016). For example, a candidate gene Pel (Accession ID: DQ073046) on chromosome 7 has been reported which is involved in the breakdown of de-esterified pectin and during fiber elongation (Bai et al., 2014). Hence, by the use and exploitation of high-throughput sequencing techniques, as mentioned earlier, many differentially expressed genes, playing important roles in fiber quality traits, can be discovered.
The Recent Developments in Genotyping and Quantitative Trait Locus Mapping
The rapid developments in bioinformatics and availability of the cotton genome have allowed to efficiently use SSRs and SNPs for effective genotyping and QTL mapping in cotton for both wild and domesticated species using intraspecific populations under multiple generations and diverse environments (Lacape et al., 2005; Wang et al., 2007; Zhang et al., 2009). For example, a recombinant inbred line (RIL) was developed by crossing cultivar Yumian1 and CA3084 (Ali et al., 2018). By using the SLAF-seq technology a genetic map consisting of 6,254 SNPs was developed with an average distance of 0.5 cm among the markers. A total of 95 QTLs for fiber quality were discovered, and among these, 55 were stable across multiple environments, whereas nine of the stable QTLs were found in three different environments (Ali et al., 2018). In another study, an interspecific cross between HS46 and MARCABUCAG8US-1-88 resulted in 188 F8 RILs (Li et al., 2016b). These were genotyped by the cotton SNP63K assays. During the cropping season under standard conditions, field trials were conducted at Sanya, Hainan province. Significant differences (p ≤ 0.05) were observed among the populations and environments. A linkage map consisting of 2,618 polymorphic SNP markers was developed with a density of 0.68 cM per marker. A total of 71 QTLs were found across 21 chromosomes with 16 as stable QTLs in two environments. On chromosomes 5, 9, 10, 14, 19, and 20, twelve various regions were identified. which are believed to be involved in the control of one hotspot or a group of characters (Li et al., 2016b).
Single nucleotide polymorphisms (SNPs) are significant in determining the population structure (Jain et al., 2014). The presence of SNPs in a coding region and regulatory sequence is of great importance because it may change the gene function (Hulse-Kemp et al., 2015b). Various techniques have been used for the discovery of SNPs in the cotton genome such as the single-copy sequence, BAC-end sequences, transcriptome-sequencing, reduced representation libraries (RRL) techniques, and genotyping by sequencing (GBS) (Hulse-Kemp et al., 2015a). The GBS is the most widely adopted method for the detection of SNPs in the crop due to being cost-effective and having rare chances of error (Majeed et al., 2019). However, re-sequencing is equally important for SNP studies (Bentley, 2006). SLAF-seq is also being used in many studies for uncovering the SNPs and Indel variations in the tetraploid cotton (Shen et al., 2017). Moreover, Pacific Biosciences Nanopore (Ashrafi et al., 2015) and Rosch Illumina 454 (Kumar et al., 2012) sequencing studies are meant for the determination of SNPs as long reads and as short reads, respectively, which can be performed with these techniques.
GWAS was performed on a panel of 355 accessions of upland cotton (Su et al., 2016). In this experiment, two major genomic regions such as MGR1 and MGR2 were deployed. A significant number of SNPs such as 16, 10, and 7 were determined related to fiber length, fiber strength, and uniformity on chromosome Dt7, respectively. This discovery of SNPs detected four candidate genes for the fiber length with different SNP loci (Su et al., 2016) meant for membrane protein, B3 domain-containing protein, membrane protein, and pentatricopeptide repeat-containing protein, respectively (www.cottonfgd.org). In another genome-wide association study, a total of 10,511 SNPs were distributed across 26 chromosomes of cotton, and only 46 significant SNPs relating to fiber quality were detected (Sun et al., 2017). Furthermore, these SNPs were scattered across 15 chromosomes, and 612 unique candidate genes were detected. Most of the candidate genes were found to be related to polysaccharide synthesis, signal transduction, and protein localization. In this study, on chromosomes Dt11 and At07, two haplotypes of fiber strength and length were identified. By combining transcriptome analysis and GWAS, 120 and 163 fiber quality-related genes were discovered, and out of these, 19 genes were screened as promising genes (Sun et al., 2017).
In a subsequent study, the genome-wide association revealed 209 and 139 QTNs for yield components and fiber quality traits, respectively (Liu et al., 2018). Among the total QTNs, 57 were found as stable in two environments. The candidate genes observed through 57 stable QTLs were analyzed with those related with QTN, and it was found that 35 candidate genes were common in both. However, it was also reported that four genes were pleiotropic among these common candidate genes (Liu et al., 2018). In another investigation, a GWAS-based high-density cotton SNP80K was used for the identification of genes related to fiber quality (Dong et al., 2019). Only 30 SNPs were found from 408 cotton accessions related with fiber quality traits across six environments. Seven loci were found to be the same among these SNPs with 128 candidate genes predicted in the vicinity of 1 Mb. Two major genome regions (GR1 and GR2) on chromosomes A07 and A13 associated with varied fiber qualities were found in multiple environments. Among these, 22 candidate genes were annotated using RNA-seq, and of these, 11 were expressed at different developmental stages (Dong et al., 2019).
As discussed earlier, the SNP discovery associated with fiber quality traits has opened new horizons in biotechnology. A single SNP in a coding region can alter the gene function. With the growing demand for cotton and challenges this crop cultivation, processing and marketing are facing, different protocols and assays are being extensively employed for the determination of stable QTLs, QTNs, and SNPs linked with the fiber quality over the genome of tetraploid cotton. It was experienced that like others, chromosome 7 has a fundamental role for fiber quality development. The role of SNPs for the detection of QTLs associated with the fiber quality can be seen in Table 3.
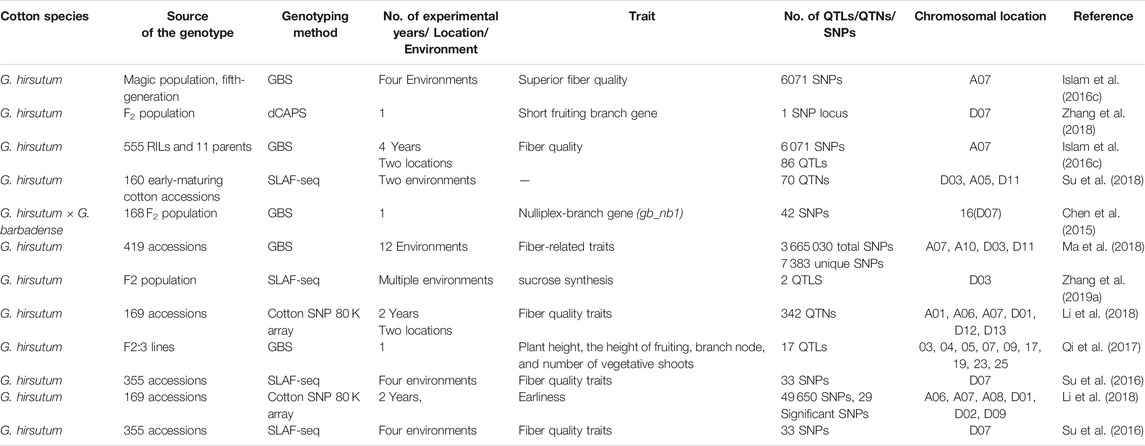
TABLE 3. Role of SNPs for the determination of QTLs of fiber quality traits across chromosome 7 in G. hirsutum
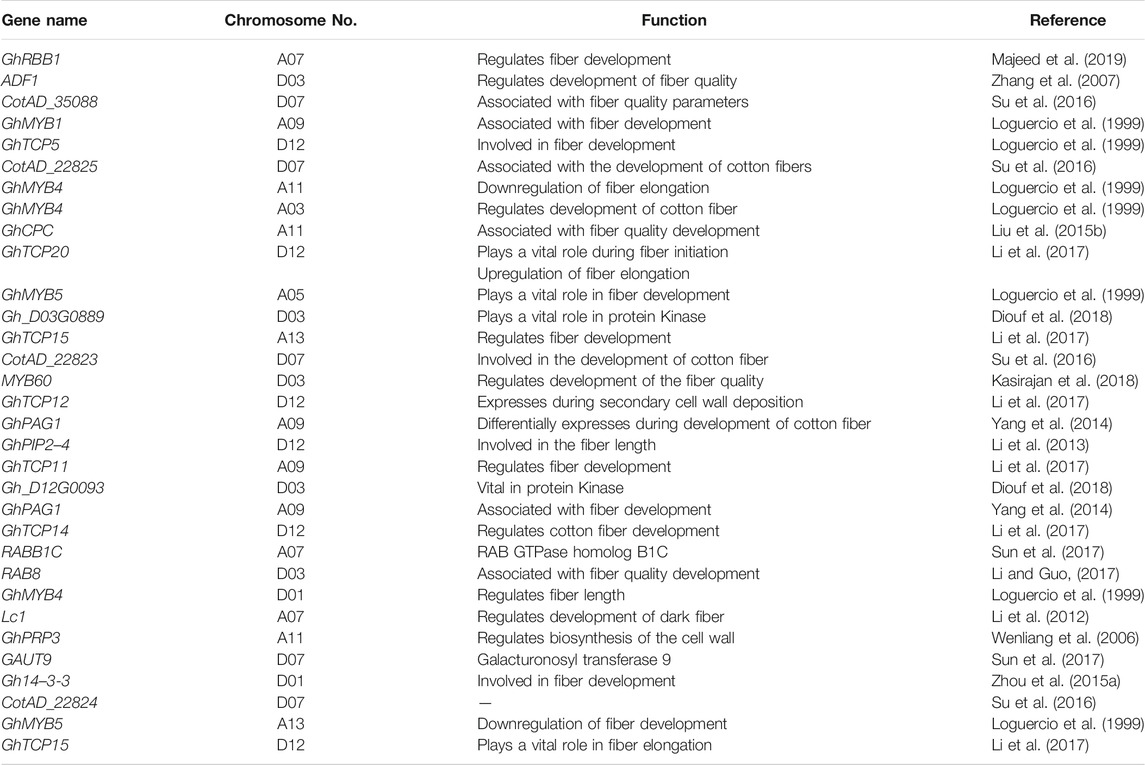
It is much evident from the aforementioned studies conducted across the whole genome of G. hirsutum L. that different sequencing techniques such as SSR, GBS, SLAF-seq, CottonSNP63K, and CottonSNP80K significantly help identify the stable QTLs and significant SNPs across different chromosomes and reveal their importance in fiber quality development. In Table 1, 2, different genes related with fiber quality development are reported across the whole genome of cotton. It is depicted from the information mentioned previously and in Tables 1,2 that chromosomes 3, 7, 9, 11, and 12 of sub-genomes A and D are harboring most of the genes associated with fiber quality development at different stages in cotton. In this study, we will continue with the importance of chromosome 7 as various studies conducted in our laboratory group have shown different stable QTLs and differentially expressed candidate genes associated with fiber quality traits. The information mentioned in Tables 3 and 4 reveals the importance of chromosome 7 containing hotspots related to cotton fiber quality.
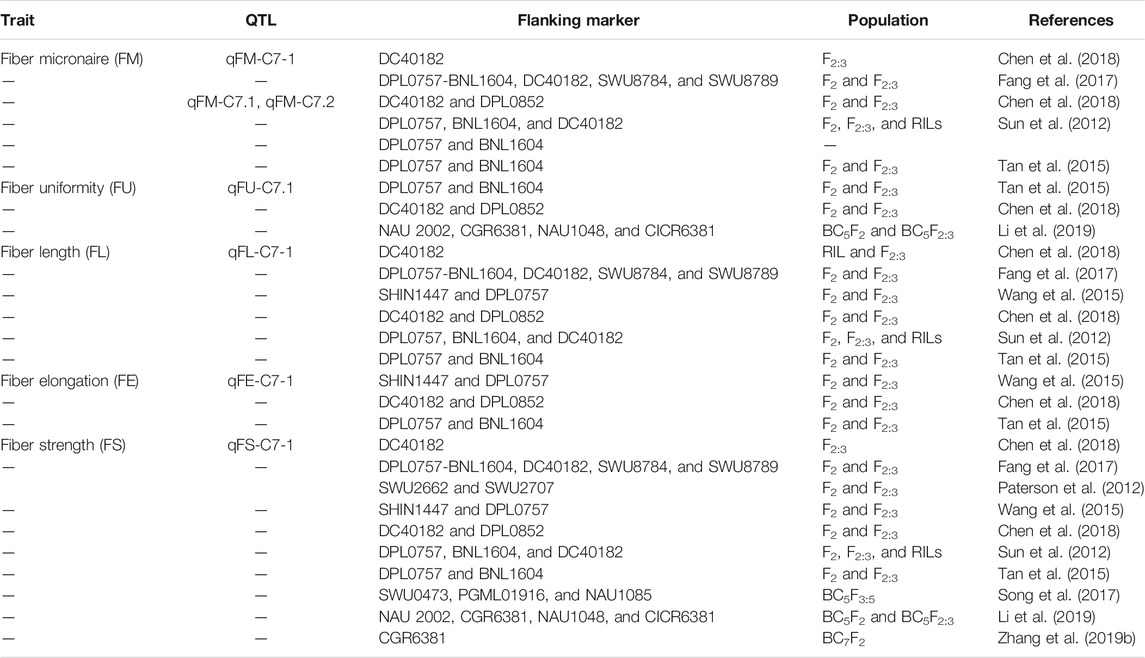
TABLE 4. Prospective QTLs of fiber development located on chromosome 7 in G. hirsutum using SSR markers.
Quantitative Trait Loci of Fiber Quality Development Across Chromosome 7
As mentioned earlier, the fiber length and fiber strength are major economic traits in cotton production. The QTL studies under different environments and across multiple generations allow scientists to identify common QTLs for marker-assisted selection (MAS). For example, by deploying the SSR markers, several QTLs were identified for fiber length, fiber strength, fiber micronaire, and fiber elongation (Tan et al., 2015). In another study, stable QTLs for fiber length and micronaire on chromosome 7 were detected, which could be further validated for fine mapping and identification of candidate genes (Ali et al., 2018). The QTL clusters of fiber quality under different environments were also detected on chromosomes c4, c7, c14, and c25 (Jamshed et al., 2016). The superior alleles of fiber quality were introgressed from G. barbadense to G. hirsutum which were found to be linked in five QTL clusters associated with potential candidate genes for fiber development (Chen et al., 2018).
Fiber quality traits are affected by the environment in which the crop is subjected to grow. However, improved lines always perform well compared with corresponding recurrent parent lines in each environment (Tan et al., 2015). As discussed previously, the fiber length and strength are economic traits, and they correlate positively with each other. Different studies have been performed in different environments for the identification of stable QTLs of fiber quality, especially FS and FL across the whole cotton genome. It has been shown by various findings that chromosome 7 contributes potentially in fiber development. For example, a major stable QTL of fiber strength qFS07.1 was identified on chromosome 7 with favorable alleles contributed by the cultivar Yumian1 in five different environments (Tan et al., 2015). A fine mapped population was produced for qFS07.1 by crossing CCRI35 and RILs with Yumian1 allele using SSR primers according to G. raimondii and G. arboreum genomes (Paterson et al., 2012; Li et al., 2014). Some extensive studies across chromosome 7 have reported some stable QTLs for fiber quality traits using different mapping techniques; they have been described as follows in Table 4.
In upland cotton (G. hirsutum), a three-parent composite population was used to identify stable QTLs of fiber quality traits on chromosome 7 between HAU1367 and HAU2282 (Zhang et al., 2012). Another study using F2 and F2:3 family lines derived from Luyuan (LY343) and Lumianyan (LMY22) was conducted, and some fiber quality-related traits were identified on chromosome 7 flanked by SSR markers (Wang et al., 2015). The upland cotton was crossed between 0 and 153 and sGK9708 for F2, F2:3 and RIL population, which allowed in identifying fiber quality traits across chromosome 7 using markers DPL0757, DC40182, and BNL 1604 (Sun et al., 2012; Fang et al., 2017). A series of improved lines (BC2F3) of super fiber quality were developed using marker-assisted selection (MAS) (Cao et al., 2015). One of the improved lines, 3,326–7, was transferred to an introgression line (ILo88-A7-3) derived from a cross of G. hirsutum acc. TM-1 and G. barbadense cv. Hai7124. It was found that this line had a consistency of producing super quality fiber properties. It was further revealed by substitution mapping of 229 BC3F2 recombinants with 207 BC3F3 and BC3F4 lines that qFLS-chr.7 and qFS-chr.7 were anchored to the same target position with an interval of 0.36-cm using markers NAU3735 and NAU845, while qFM-chr.7 was mapped with an interval of 0.44 cm using SSR markers. This positive relationship of tightly linked QTLs demonstrates their contribution to fiber quality development, especially in fiber strength and fiber length (Cao et al., 2015).
CSSLs are used as an ideal material for the identification of genetic effects. In a study, a material of CCRI45 (G. hirsutum) was crossed with Hai1 (G. barbadense) and MBI9915, and the CSSL was selected at BC4F3:5 (Li et al., 2019). A total of 2,292 SSR markers were applied which covered the whole tetraploid cotton genome. A total of 129 QTLs were detected including 103 for fiber quality and 17 for fiber yield across 17 chromosomes with a phenotypic effect of 0.85–30.35%. Out of the total, 39 were stable, 53 were common, and 76 were new with a phenotypic variation of 30.2, 41.1, and 58.9%, respectively, with 86 of favorable effects on related traits. It was also noted that more QTLs were distributed across the Dt sub-genome than the At sub-genome. Out of 25 stable QTL clusters detected across 22 chromosomes, only six introgressed segments with a special focus on Seg-A07-2 were found as important candidate chromosome regions for fiber quality (Li et al., 2019).
In another investigation, a new CSSL material, MBI9915, was obtained by crossing CCRI36 (G. hirsutum) with Hai1 (G. barbadense) (Song et al., 2017). An F2 population of 1,537 individuals was constructed by crossing MBI9915 and CCRI36, and 347 individuals from the F2 population were randomly selected for the study of the genetic effects of the introgressed chromosome segments. A total of 18 and six QTLs were detected for fiber quality and fiber yield, respectively, in two segregating populations with the cumulative phenotypic variation of 0.81–9.51% on chromosome 7. Among these QTLs, six were detected consistently, as detected in previous studies. An interaction of the fiber length with fiber strength was identified in a total of 13 pairs with each trait in two generations. The study concluded with five important chromosome segments having important effects on fiber quality and yield (Song et al., 2017).
Fiber quality and yield-associated genes were identified by constructing a high-density genetic map using SLAF-seq, while incorporating 239 RILs derived from LMY22 (high-yielding G. hirsutum L. cultivars) x LY343 (superior fiber quality germplasm with G. barbadense L. introgressions) (Wang F. et al., 2019). The resulted genetic map was spanned over 3,426.57 cm which included 3556 SLAF-seq–based SNPs and 199 SSR marker loci. Under seven different environments, 67 QTLs for fiber quality and 37 QTLs for fiber yield were detected making a total of 104 QTLs. It was noted that on chromosome 12 with 19 QTL clusters, 66 QTLs were co-located, and 24 stable QTLs were determined under more than three different environments. The role of the genomic component LY343 was also investigated on fiber quality-related traits which indicated the superior contribution of G. hirsutum races than G. barbadense (Wang F. et al., 2019).
As discussed previously, the economic value of cotton relates to the quality of fiber, and enzymes play a vital role in maintaining the fiber quality. In a study, it was reported that the laccase enzyme plays a crucial role in fiber elongation, lignification, and plant pigmentation (Balasubramanian et al., 2016). Some laccase genes have been identified genome-wide of cultivated G. hirsutum and its diploid progenitor (G. arboreum and G. raimondii) cotton species. The gene expression, enzymatic activity, and biochemical analysis have shown that the laccase enzyme is differentially expressed at different stages of fiber development, particularly at 25DPA (Days Post-Anthesis). During this study, it was noted that chromosomes 7 and 13 of G. arboreum contained the highest number of laccase genes, i.e., eight on each, indicating a significant contribution of laccase in fiber development (Balasubramanian et al., 2016).
A 63K Illumina Infinium SNP array was performed on 719 accessions of upland cotton for phenotyping and genetic variation analysis for their better understanding (Sun et al., 2017). Forty-six significant SNPs with five fiber quality traits were detected of the total 10,511 polymorphic SNPs distributed across 26 chromosomes. A total of 10,511 polymorphic SNPs were scattered across 26 chromosomes. Forty-six SNPs were found significantly associated with five fiber quality traits (Sun et al., 2017). The distribution of these significantly found SNPs across 15 chromosomes indicated their relatedness in 612 unique candidate genes with their involvement mainly in polysaccharide synthesis, signal transduction, and protein translocation. The significance of the study relates to the determination of two major haplotypes of the fiber length and strength across chromosomes Dt11 and At07, providing an insight into the genetic basis of fiber quality in G. hirsutum (Sun et al., 2017). In upland cotton, a recombinant inbred line was developed, and a consensus map was constructed using three types of markers under 17 environments Zhang et al., 2020). The study revealed some stable QTLs for fiber quality traits across chromosomes 4, 6, 13, 21, and 25 including chromosome 7. Out of the total, three stable QTLs were determined for the fiber length and strength, while two for fiber micronaire. Genomic and bioinformatics approaches are efficient to find and retrieve target QTL regions. The information on the fiber development mechanism can be deduced from the integration of transcriptome analysis and QTL mapping. Different reports and studies on fiber quality development can be further improved by making interspecific crosses and cotton genome information. Molecular markers, cotton genomes, and transcriptome information are vibrant to decipher and dissect fiber development mechanisms to cultivate superior varieties with improved fiber quality across chromosome 7 of Gossypium species.
A composite interval model (CIM) can be incorporated for QTLs or genes using molecular markers with several segregation patterns. The proposed model assists in the determination of QTLs, segregation pattern, the estimation of their position, and the interference of their linkage phase with the markers. It has been reported that a simulation study shows the significance of the model (Gazaffi et al., 2014).
Conclusion and Future Prospects
Several differentially expressed genes associated with fiber quality are reported using the marker-assisted selection strategy. Decades ago, many QTLs both in diploid and tetraploid cotton species have been reported in different populations having different characteristics from their parents. Genome sequencing provided breeders and scientists to develop high-density genetic maps based on SSR and SNPs, phenotypic and genotypic interactions, QTL identification, and fine mapping for candidate gene selection across chromosomes. The cotton genome, DNA markers, and transcriptome studies reveal the significant role of different chromosomes harboring genes associated with the fiber quality, as described in Tables. It was noticed that chromosomes 3, 7, 9, 11, and 12 associated with sub-genomes A and D contribute significantly to fiber quality. However, based on this information, our laboratory is focusing on chromosome 7 in dissecting the mechanisms of fiber quality improvement. Furthermore, RT-qPCR and molecular cloning demonstrate the differential expression and structure of candidate genes located on chromosome 7. A great insight is expected into the genetic basis of fiber-related traits such as the fiber strength and fiber length on chromosome 7 for improvement in cotton. The contribution of chromosome 7 carrying candidate genes in fiber quality development requires more attention in the future for QTL fine mapping, marker-assisted selection (MAS) breeding, and ultimately transgene in Arabidopsis and subsequently in cotton.
Author Contributions
AR and MZ wrote the initial draft of the manuscript. AA, AH, MH and XD made all necessary corrections. XG, FS, PL and YS carried out final editing of the manuscript. WG and MR proofread the manuscript. Final approval for publication was given by the group leader at the Institute of cotton research YY.
Funding
This work was funded by the National Key R&D Program of China (2017YFD0101603-11 and 2016YFD0100500), the Natural Science Foundation of China (31471538 and 31371668), the Agricultural Science and Technology Innovation Program for CAAS (CAAS-ASTIP-ICRCAAS), the National High Technology Research and Development Program of China (2012AA101108 and 2009AA101104), and the Central Level of the Scientific Research Institutes for Basic R&D Special Fund Business (1610162014008).
Conflict of Interest
AA was employed by the company FB Genetics Four Brothers Group.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ali, I., Teng, Z., Bai, Y., Yang, Q., Hao, Y., Hou, J., et al. (2018). A High Density SLAF-SNP Genetic Map and QTL Detection for Fibre Quality Traits in Gossypium Hirsutum. BMC Genomics 1, 879. doi:10.1186/s12864-018-5294-5
Ashrafi, H., Hulse Kemp, A. M., Wang, F., Yang, S. S., Guan, X., Jones, D. C., et al. (2015). Long Read Transcriptome Assembly of Cotton (Gossypium Hirsutum L.) and Intraspecific Single Nucleotide. Polymorphism Discov. 2, 1–14. doi:10.3835/plantgenome2014.10.0068
Bai, W. Q., Xiao, Y. H., Zhao, J., Song, S. Q., Hu, L., Zeng, J. Y., et al. (2014). Gibberellin Overproduction Promotes Sucrose Synthase Expression and Secondary Cell wall Deposition in Cotton Fibers. PLoS One 9, e96537. doi:10.1371/journal.pone.0096537
Balasubramanian, V. K., Rai, K. M., Thu, S. W., Hii, M. M., and Mendu, V. (2016). Genome-wide Identification of Multifunctional Laccase Gene Family in Cotton (Gossypium spp.). Expr. Biochem. Anal. during fiber Dev. Scientific Rep. 1, 1–16. doi:10.1038/srep34309
Bentley, D. R. (2006). Whole-genome Re-sequencing. Curr. Opin. Genet. Dev. 6, 545–552. doi:10.1016/j.gde.2006.10.009
Betancur, L., Singh, B., Rapp, R. A., Wendel, J. F., Marks, M. D., Roberts, A. W., et al. (2010). Phylogenetically Distinct Cellulose Synthase Genes Support Secondary wall Thickening in Arabidopsis Shoot Trichomes and Cotton Fiber. J. Integr. Plant Biol. 52, 205–220. doi:10.1111/j.1744-7909.2010.00934.x
Bowling, A. J., Vaughn, K. C., and Turley, R. B. (2011). Polysaccharide and Glycoprotein Distribution in the Epidermis of Cotton Ovules during Early Fiber Initiation and Growth. Protoplasma 248, 579–590. doi:10.1007/s00709-010-0212-y
Cai, C., Zhu, G., Zhang, T., and Guo, W. (2017). High-density 80 K SNP Array Is a Powerful Tool for Genotyping G. Hirsutum Accessions and Genome Analysis. BMC Genomics 1, 654. doi:10.1186/s12864-017-4062-2
Cao, Z., Zhu, X., Chen, H., and Zhang, T. (2015). Fine Mapping of Clustered Quantitative Trait Loci for Fiber Quality on Chromosome 7 Using a Gossypium barbadense Introgressed Line. Mol. Breed. 11, 215. doi:10.1007/s11032-015-0393-3
Chen, J., and Burke, J. J. (2015). Developing Fiber Specific Promoter-Reporter Transgenic Lines to Study the Effect of Abiotic Stresses on Fiber Development in Cotton. PloS one 6, e0129870. doi:10.1371/journal.pone.0129870
Chen, W., Yao, J., Chu, L., Yuan, Z., Li, Y., and Zhang, Y. (2015). Genetic Mapping of the Nulliplex-branch Gene (Gb_nb1) in Cotton Using Next-Generation Sequencing Theor. Appl. Genet. 3, 539–547. doi:10.1007/s00122-014-2452-2
Chen, Y., Liu, G., Ma, H., Song, Z., Zhang, C., Zhang, J., et al. (2018). Identification of Introgressed Alleles Conferring High Fiber Quality Derived from Gossypium barbadense L. In Secondary Mapping Populations of G. Hirsutum L. Front. Plant Sci., 1023. doi:10.3389/fpls.2018.01023
Chen, Z. J., Scheffler, B. E., Dennis, E., Triplett, B. A., Zhang, T., Guo, W., et al. (2007). Toward Sequencing Cotton (Gossypium) Genomes: Figure 1. Plant Physiol. 145, 1303–1310. doi:10.1104/pp.107.107672
Cheng, Y., Lu, L., Yang, Z., Wu, Z., Qin, W., Yu, D., et al. (2016). GhCaM7-like , a Calcium Sensor Gene, Influences Cotton Fiber Elongation and Biomass Production. Plant Physiol. Biochem. 109, 128–136. doi:10.1016/j.plaphy.2016.09.009
Cosgrove, D. J. (1997). Assembly and Enlargement of the Primary Cell wall in Plants. Annu. Rev. Cel Dev. Biol. 13, 171–201. doi:10.1146/annurev.cellbio.13.1.171
Zhang, C., Li, L., Liu, Q., Gu, L., Huang, J., Wei, H., et al. Identification of Loci and Candidate Genes Responsible for Fiber Length in upland Cotton (Gossypium Hirsutum L.) via Association Mapping and Linkage Analyses Front. Plant Sci. (2019a), pp. 53. doi:10.3389/fpls.2019.00053
Diouf, L., Magwanga, R. O., Gong, W., He, S., Pan, Z., Jia, Y. H., et al. (2018). QTL Mapping of Fiber Quality and Yield-Related Traits in an Intra-specific upland Cotton Using Genotype by Sequencing (GBS). Int. J. Mol. Sci. 2, 441. doi:10.3390/ijms19020441
Dochia, M., Sirghie, C., Kozłowski, R.M., and Roskwitalski, Z. 2012. Cotton fibres. In Handbook of natural fibres (pp. 11–23). Woodhead Publishing.
Dong, C., Wang, J., Yu, Y., Ju, L., Zhou, X., Ma, X., et al. (2019). Identifying Functional Genes Influencing Gossypium Hirsutum Fiber Quality. Front. Plant Sci., 1968. doi:10.3389/fpls.2018.01968
Fan, T., Sun, B., Gu, J., Zhang, D., and Lau, L. W. M. (2005). Biomorphic Al2O3 Fibers Synthesized Using Cotton as Bio-Templates. Scripta Materialia 53, 893–897. doi:10.1016/j.scriptamat.2005.06.036
Fang, L., Gong, H., Hu, Y., Liu, C., Zhou, B., Huang, T., et al. (2017). Genomic Insights into Divergence and Dual Domestication of Cultivated Allotetraploid Cottons. Genome Biol. 18, 33. doi:10.1186/s13059-017-1167-5
Fryxell, P. (1992). A Revised Taxonomic Interpretation of Gossypium L (Malvaceae) Rheedea. US Department of Agriculture, Texas, 108–116.
Gao, P., Zhao, P.-M., Wang, J., Wang, H.-Y., Wu, X.-M., and Xia, G.-X. (2007). Identification of Genes Preferentially Expressed in Cotton Fibers: A Possible Role of Calcium Signaling in Cotton Fiber Elongation. Plant Sci. 173, 61–69. doi:10.1016/j.plantsci.2007.04.008
Gazaffi, R., Margarido, G. R., Pastina, M. M., Mollinari, M., and Garcia, A. A. F. (2014). A Model for Quantitative Trait Loci Mapping, Linkage Phase, and Segregation Pattern Estimation for a Full-Sib Progeny. Tree Genet. Genomes 4, 791–801. doi:10.1007/s11295-013-0664-2
Gilchrist, E. J., and Haughn, G. W. (2005). TILLING without a Plough: a New Method with Applications for Reverse Genetics. Curr. Opin. Plant Biol. 8, 211–215. doi:10.1016/j.pbi.2005.01.004
Gou, J.-Y., Wang, L.-J., Chen, S.-P., Hu, W.-L., and Chen, X.-Y. (2007). Gene Expression and Metabolite Profiles of Cotton Fiber during Cell Elongation and Secondary Cell wall Synthesis. Cell Res 17, 422–434. doi:10.1038/sj.cr.7310150
Guo, X., Guo, Y., Ma, J., Wang, F., Sun, M., Gui, L., et al. (2013). Mapping Heterotic Loci for Yield and Agronomic Traits Using Chromosome Segment Introgression Lines in Cotton. J. Integr. Plant Biol. 8, 759–774. doi:10.1111/jipb.12054
Hafeez, A., Gě, Q., Zhāng, Q., Lǐ, J., Gōng, J., Liú, R., et al. (2021). Multi-responses of O-Methyltransferase Genes to Salt Stress and Fiber Development of Gossypium Species. BMC Plant Biol. 1, 1–17. doi:10.1186/s12870-020-02786-6
Haigler, C. H., Betancur, L., Stiff, M. R., and Tuttle, J. R. (2012). Cotton Fiber: a Powerful Single-Cell Model for Cell wall and Cellulose Research. Front. Plant Sci. 3, 104. doi:10.3389/fpls.2012.00104
Haigler, C. H., Singh, B., Wang, G., and Zhang, D. (2009). “Genomics of Cotton Fiber Secondary wall Deposition and Cellulose Biogenesis,” in Genetics and Genomics of Cotton (New York: Springer), 385–417. doi:10.1007/978-0-387-70810-2_16
He, Q., Yang, H., Xiang, S., Tian, D., Wang, W., Zhao, T., et al. (2015). Fine Mapping of the Genetic Locus L1 Conferring Black Pods Using a Chromosome Segment Substitution Line Population of Soybean. Plant Breed. 4, 437–445. doi:10.1111/pbr.12272
Hinchliffe, D. J., Meredith, W. R., Delhom, C. D., Thibodeaux, D. P., and Fang, D. D. (2011). Elevated Growing Degree Days Influence Transition Stage Timing during Cotton Fiber Development Resulting in Increased Fiber‐Bundle Strength. Crop Sci. 51, 1683–1692. doi:10.2135/cropsci2010.10.0569
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium Hirsutum Genomes Provide Insights into the Origin and Evolution of Allotetraploid Cotton. Nat. Genet. 51, 739–748. doi:10.1038/s41588-019-0371-5
Hulse-Kemp, A. M., Ashrafi, H., Stoffel, K., Zheng, X., Saski, C. A., Scheffler, B. E., et al. (2015b). BAC-end Sequence-Based SNP Mining in Allotetraploid Cotton (Gossypium) Utilizing Resequencing Data, Phylogenetic Inferences, and Perspectives for Genetic Mapping. G3: Genes, Genomes, Genet. 6, 1095–1105. doi:10.1534/g3.115.017749
Hulse-Kemp, A. M., Lemm, J., Plieske, J., Ashrafi, H., Buyyarapu, R., Fang, D. D., et al. (2015a). Development of a 63K SNP Array for Cotton and High-Density Mapping of Intraspecific and Interspecific Populations of Gossypium Spp. G3: Genes, Genomes, Genet. 6, 1187–1209. doi:10.1534/g3.115.018416
Hutchinson, J. B. (1951). Intra-specific Differentiation in Gossypium Hirsutum. Heredity 5, 161–193. doi:10.1038/hdy.1951.19
Iida, S., and Terada, R. (2004). A Tale of Two Integrations, Transgene and T-DNA: Gene Targeting by Homologous Recombination in rice. Curr. Opin. Biotechnol. 15, 132–138. doi:10.1016/j.copbio.2004.02.005
Islam, M. S., Fang, D. D., Thyssen, G. N., Delhom, C. D., Liu, Y., and Kim, H. J. (2016b). Comparative Fiber Property and Transcriptome Analyses Reveal Key Genes Potentially Related to High Fiber Strength in Cotton (Gossypium Hirsutum L.) Line MD52ne. BMC Plant Biol. 1, 36. doi:10.1186/s12870-016-0727-2
Islam, M. S., Thyssen, G. N., Jenkins, J. N., Zeng, L., Delhom, C. D., McCarty, J. C., et al. (2016c). A MAGIC Population-Based Genome-wide Association Study Reveals Functional Association of GhRBB1_A07 Gene with superior Fiber Quality in Cotton. BMC Genomics 1, 903. doi:10.1186/s12864-016-3249-2
Islam, M. S., Zeng, L., Thyssen, G. N., Delhom, C. D., Kim, H. J., Li, P., et al. (2016a). Mapping by Sequencing in Cotton (Gossypium Hirsutum) Line MD52ne Identified Candidate Genes for Fiber Strength and its Related Quality Attributes. Theor. Appl. Genet. 6, 1071–1086. doi:10.1007/s00122-016-2684-4
Jain, M., Moharana, K. C., Shankar, R., Kumari, R., and Garg, R. (2014). Genomewide Discovery of DNA Polymorphisms in rice Cultivars with Contrasting Drought and Salinity Stress Response and Their Functional Relevance. Plant Biotechnol. J. 2, 253–264. doi:10.1111/pbi.12133
Jamshed, M., Jia, F., Gong, J., Palanga, K. K., Shi, Y., et al. (2016). Identification of Stable Quantitative Trait Loci (QTLs) for Fiber Quality Traits Across Multiple Environments in Gossypium hirsutum Recombinant Inbred Line Population. BMC Genomics 17, 197. doi:10.1186/s12864-016-2560-2
Jia, X., Pang, C., Wei, H., Wang, H., Ma, Q., Yang, J., et al. (2016). High-density Linkage Map Construction and QTL Analysis for Earliness-Related Traits in Gossypium Hirsutum L. BMC Genomics 1, 909. doi:10.1186/s12864-016-3269-y
Jiang, Y., Guo, W., Zhu, H., Ruan, Y.-L., and Zhang, T. (2012). Overexpression of GhSusA1 Increases Plant Biomass and Improves Cotton Fiber Yield and Quality. Plant Biotechnol. J. 10, 301–312. doi:10.1111/j.1467-7652.2011.00662.x
John, M. E., and Keller, G. (1996). Metabolic Pathway Engineering in Cotton: Biosynthesis of Polyhydroxybutyrate in Fiber Cells. Proc. Natl. Acad. Sci. 93, 12768–12773. doi:10.1073/pnas.93.23.12768
Kasirajan, L., Hoang, N. V., Furtado, A., Botha, F. C., and Henry, R. J. (2018). Henry Transcriptome Analysis Highlights Key Differentially Expressed Genes Involved in Cellulose and Lignin Biosynthesis of Sugarcane Genotypes Varying in Fiber Content. Sci. Rep. 1, 1–16. doi:10.1038/s41598-018-30033-4
Kawai, M., Aotsuka, S., and Uchimiya, H. (1998). Isolation of a Cotton CAP Gene: a Homologue of Adenylyl Cyclase-Associated Protein Highly Expressed during Fiber Elongation. Plant Cel. Physiol. 39, 1380–1383. doi:10.1093/oxfordjournals.pcp.a029346
Kesanakurti, D., Jami, S., and Kirti, P. J. P. M. B. (2010). Constitutive Expression of Mustard Annexin, AnnBj1 Enhances Abiotic Stress Tolerance and Fiber Quality in Cotton under Stress. Plant Mol. Biol. 3, 293–308. doi:10.1007/s11103-010-9615-6
Kohel, R. J., Yu, J., Park, Y.-H., and Lazo, G. R. (2001). Mol. Mapp. characterization traits controlling fiber Qual. cotton Euphytica 2, 163–172. doi:10.1023/a:1012263413418
Kumar, S., Banks, T. W., and Cloutier, S. (2012). SNP Discovery through Next-Generation Sequencing and its Applications. Int. J. Plant Genomics 2012, 831460. doi:10.1155/2012/831460
Kunbo, W., Wendel, J. F., and Jinping, H. (2018). Designations for Individual Genomes and Chromosomes in Gossypium. J. Cotton Res. 1, 3. doi:10.1186/s42397-018-0002-1
Kuttenkeuler, D., and Boutros, M. J. (2004). Genome-wide RNAi as a Route to Gene Function in Drosophila. Brief. Funct. Genomics Proteomics 3, 168–176. doi:10.1093/bfgp/3.2.168
Lacape, J.-M., Nguyen, T.-B., Courtois, B., Belot, J.-L., Giband, M., Gourlot, J.-P., et al. (2005). QTL Analysis of Cotton Fiber Quality Using Multiple Gossypium Hirsutum× Gossypium barbadense Backcross Generations. Crop Sci. 1, 123–140.
Li, C., Dong, Y., Zhao, T., Li, L., Li, C., Yu, E., et al. (2016b). Genome-wide SNP Linkage Mapping and QTL Analysis for Fiber Quality and Yield Traits in the upland Cotton Recombinant Inbred Lines Population. Front. Plant Sci., 1356. doi:10.3389/fpls.2016.01356
Li, C., Dong, Y., Zhao, T., Li, L., Li, C., Yu, E., et al. (2016a). Genome-Wide SNP Linkage Mapping and QTL Analysis for Fiber Quality and Yield Traits in the Upland Cotton Recombinant Inbred Lines Population. Front. Plant Sci. 7, 1356. doi:10.3389/fpls.2016.01356
Li, C., Guo, W., and Zhang, T. (2009). Fiber Initiation Development in Upland Cotton (Gossypium Hirsutum L.) Cultivars Varying in Lint Percentage. Euphytica 165, 223. doi:10.1007/s10681-008-9740-3
Li, C., Wang, Y., Ai, N., Li, Y., and Song, J. (2018). A Genome Wide Association Study of Early‐maturation Traits in upland Cotton Based on the CottonSNP80K Array. J. Integr. Plant Biol. 10, 970–985. doi:10.1111/jipb.12673
Li, D.-D., Ruan, X.-M., Zhang, J., Wu, Y.-J., Wang, X.-L., and Li, X.-B. (2013). Cotton Plasma Membrane Intrinsic Protein 2s (PIP2s) Selectively Interact to Regulate Their Water Channel Activities and Are Required for Fibre Development. New Phytol. 199, 695–707. doi:10.1111/nph.12309
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome Sequence of the Cultivated Cotton Gossypium Arboreum. Nat. Genet. 6, 567–572. doi:10.1038/ng.2987
Li, F., Ning, X., Qiu, X., Su, C., Yao, J., and Tian, L. (2012). Genetic Mapping of the Dark Brown Fiber Lc1 Gene in Tetraploid Cotton. Sci. Agric. Sin 6.
Li, P., and Guo, W. (2017). Guo Genome-wide Characterization of the Rab Gene Family in Gossypium by Comparative Analysis. Bot. Stud. 1, 26. doi:10.1186/s40529-017-0181-y
Li, S., Chen, H., Hou, Z., Li, Y., Yang, C., Wang, D., et al. (2020). Screening of Abiotic Stress‐responsive Cotton Genes Using a Cotton Full‐length cDNA Overexpressing Arabidopsis Library. J. Integr. Plant Biol. 62, 998–1016. doi:10.1111/jipb.12861
Li, S. Q., Liu, A. Y., Kong, L. L., Gong, J. W., Li, J. W., Gong, W. K., et al. (2019). Shang QTL Mapping and Genetic Effect of Chromosome Segment Substitution Lines with Excellent Fiber Quality from Gossypium Hirsutum× Gossypium barbadense. Mol. Genet. Genomics 5, 1123–1136. doi:10.1007/s00438-019-01566-8
Li, W., Li, D. D., Han, L. H., Tao, M., Hu, Q. Q., Wu, W. Y., et al. (2017). Genome-wide Identification and Characterization of TCP Transcription Factor Genes in upland Cotton (Gossypium Hirsutum). Sci. Rep. 7, 10118–10214. doi:10.1038/s41598-017-10609-2
Li, X.-B., Xiao, Y.-H., Luo, M., Hou, L., Li, D.-M., Luo, X.-Y., et al. (2005b). Cloning and Expression Analysis of Two Rac Genes from Cotton (Gossypium Hirsutum L.). Acta Genetica Sinica 1, 72–78.
Li, X.-B., Fan, X.-P., Wang, X.-L., Cai, L., and Yang, W.-C. (2005a). The Cotton ACTIN1 Gene Is Functionally Expressed in Fibers and Participates in Fiber Elongation. Plant Cell 17, 859–875. doi:10.1105/tpc.104.029629
Ijaz, B., Zhao, N., Kong, J., and Hua, J. (2019). Fiber Quality Improvement in Upland Cotton (Gossypium hirsutum L.): Quantitative Trait Loci Mapping and Marker Assisted Selection Application. Front. Plant Sci. 10, 1585. doi:10.3389/fpls.2019.01585
Liu, B., Zhu, Y., and Zhang, T. (2015b). The R3-MYB Gene GhCPC Negatively Regulates Cotton Fiber Elongation. PloS one 10, e0116272. doi:10.1371/journal.pone.0116272
Liu, D., Tu, L., Wang, L., Li, Y., Zhu, L., and Zhang, X. (2008). Characterization and Expression of Plasma and Tonoplast Membrane Aquaporins in Elongating Cotton Fibers. Plant Cel Rep 27, 1385–1394. doi:10.1007/s00299-008-0545-6
Liu, H.-C., Creech, R. G., Jenkins, J. N., and Ma, D.-P. (2000). Cloning and Promoter Analysis of the Cotton Lipid Transfer Protein Gene Ltp311The Nucleotide Sequence Data Reported Will Appear in the GenBank Nucleotide Sequence Databases under the Accession Number AF228333. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1487, 106–111. doi:10.1016/s1388-1981(00)00072-x
Loguercio, L. L., Zhang, J.-Q., and Wilkins, T. A. (1999). Differential Regulation of Six Novel MYB-Domain Genes Defines Two Distinct Expression Patterns in Allotetraploid Cotton (Gossypium Hirsutum L.). Mol. Gen. Genet. 261, 660–671. doi:10.1007/s004380050009
Lu, Q., Shi, Y., Xiao, X., Li, P., Gong, J., Gong, W., et al. (2017). Transcriptome Analysis Suggests that Chromosome Introgression Fragments from Sea Island Cotton (Gossypium barbadense) Increase Fiber Strength in Upland Cotton (Gossypium Hirsutum). Genetics 7, 3469–3479. doi:10.1534/g3.117.300108
Ma, G.-J., Zhang, T.-Z., and Guo, W.-Z. (2006). Cloning and Characterization of cottonGhBGgene Encoding β-glucosidase. DNA Seq. 17, 355–362. doi:10.1080/10425170600807454
Ma, Z., He, S., Wang, X., Sun, J., Zhang, Y., Zhang, G., et al. (2018). Resequencing a Core Collection of upland Cotton Identifies Genomic Variation and Loci Influencing Fiber Quality and Yield. Nat. Genet. 6, 803–813. doi:10.1038/s41588-018-0119-7
Majeed, S., Rana, I. A., Atif, R. M., Ali, Z., Hinze, L., and Azhar, M. T. (2019). Role of SNPs in Determining QTLs for Major Traits in Cotton. J. Cotton Res. 1, 5. doi:10.1186/s42397-019-0022-5
Ming-bao, L., Xiang-mo, G., Yong-shan, Z., and Jin-bo, Y. (2008). Genetic Effect on Yield and Fiber Quality Traits of 16 Chromosome Substitution Lines in upland Cotton. Agric. Sci. China 7, 1290–1297. doi:10.1016/s1671-2927(08)60177-7
Nadeem, M., Iqbal, N., Waheed, U., Zia, Z. U., Ali, M., and Khan, Z. J. (2021). Comparative Expression Studies of Fiber Related Genes in Cotton Spp. J. Bioresour. Manag. 8, 14. doi:10.35691/JBM.1202.0173
Orford, S. J., and Timmis, J. N. (1998). Specific Expression of an Expansin Gene during Elongation of Cotton Fibres. Biochim. Biophys. Acta (Bba) - Gene Struct. Expr. 1398, 342–346. doi:10.1016/s0167-4781(98)00065-7
Pang, C.-Y., Wang, H., Pang, Y., Xu, C., Jiao, Y., Qin, Y.-M., et al. (2010). Comparative Proteomics Indicates that Biosynthesis of Pectic Precursors Is Important for Cotton Fiber and Arabidopsis Root Hair Elongation. Mol. Cell Proteomics 9, 2019–2033. doi:10.1074/mcp.m110.000349
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated Polyploidization of Gossypium Genomes and the Evolution of Spinnable Cotton Fibres. Nature 492, 423–427. doi:10.1038/nature11798
Pattathil, S., Avci, U., Baldwin, D., Swennes, A. G., McGill, J. A., Popper, Z., et al. (2010). A Comprehensive Toolkit of Plant Cell Wall Glycan-Directed Monoclonal Antibodies. Plant Physiol. 153, 514–525. doi:10.1104/pp.109.151985
Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P., and Stalker, D. M. (1996). Higher Plants Contain Homologs of the Bacterial celA Genes Encoding the Catalytic Subunit of Cellulose Synthase. Proc. Natl. Acad. Sci. 93, 12637–12642. doi:10.1073/pnas.93.22.12637
Polko, J. K., and Kieber, J. J. (2019). The Regulation of Cellulose Biosynthesis in Plants. Plant Cell 31, 282–296. doi:10.1105/tpc.18.00760
Qi, H., Wang, N., Qiao, W., Xu, Q., Zhou, H., Shi, J., et al. (2017). Construction of a High-Density Genetic Map Using Genotyping by Sequencing (GBS) for Quantitative Trait Loci (QTL) Analysis of Three Plant Morphological Traits in upland Cotton (Gossypium Hirsutum L.). Euphytica 4, 83. doi:10.1007/s10681-017-1867-7
Liu, R., Gong, J., Xiao, X., Zhang, Z., Li, J., Liu, A., et al. GWAS Analysis and QTL Identification of Fiber Quality Traits and Yield Components in upland Cotton Using Enriched High-Density SNP Markers. Front. Plant Sci. (2018), pp. 1067 doi:10.3389/fpls.2018.01067
Reinisch, A. J., Dong, J. M., Brubaker, C. L., Stelly, D. M., Wendel, J. F., and Paterson, A. H. (1994). A Detailed RFLP Map of Cotton, Gossypium hirsutum x Gossypium barbadense: Chromosome Organization and Evolution in a Disomic Polyploid Genome. Genetics 138, 829–847.
Rodgers, J., Zumba, J., and Fortier, C. (2017). Measurement Comparison of Cotton Fiber Micronaire and its Components by Portable Near Infrared Spectroscopy Instruments. Textile Res. J. 87, 57–69. doi:10.1177/0040517515622153
Ruan, Y.-L., Llewellyn, D. J., and Furbank, R. T. (2003). Suppression of Sucrose Synthase Gene Expression Represses Cotton Fiber Cell Initiation, Elongation, and Seed Development. Plant Cell 15, 952–964. doi:10.1105/tpc.010108
Said, J. I., Song, M., Wang, H., Lin, Z., Zhang, X., Fang, D. D., et al. (2015). A Comparative Meta-Analysis of QTL between Intraspecific Gossypium Hirsutum and Interspecific G. Hirsutum× G. barbadense Populations. Mol. Genet. Genomics 3, 1003–1025.
Samuel Yang, S., Cheung, F., Lee, J. J., Ha, M., Wei, N. E., Sze, S.-H., et al. (2006). Accumulation of Genome-specific Transcripts, Transcription Factors and Phytohormonal Regulators during Early Stages of Fiber Cell Development in Allotetraploid Cotton. Plant J. 47, 761–775. doi:10.1111/j.1365-313x.2006.02829.x
Seagull, R. W. (1993). Cytoskeletal Involvement in Cotton Fiber Growth and Development. Micron 24, 643–660. doi:10.1016/0968-4328(93)90042-y
Shen, C., Jin, X., Zhu, D., and Lin, Z. (2017). Uncovering SNP and Indel Variations of Tetraploid Cottons by SLAF-Seq. BMC Genomics 1, 247. doi:10.1186/s12864-017-3643-4
Shi, H., Wang, X., Li, D., Tang, W., Wang, H., Xu, W., et al. (2007). Molecular Characterization of Cotton 14-3-3L Gene Preferentially Expressed during Fiber Elongation. J. Genet. Genomics 34, 151–159. doi:10.1016/s1673-8527(07)60016-2
Shi, Y.-H., Zhu, S.-W., Mao, X.-Z., Feng, J.-X., Qin, Y.-M., Zhang, L., et al. (2006). Transcriptome Profiling, Molecular Biological, and Physiological Studies Reveal a Major Role for Ethylene in Cotton Fiber Cell Elongation. plant cell 18, 651–664. doi:10.1105/tpc.105.040303
Shi, Y., Li, W., Li, A., Ge, R., Zhang, B., Li, J., et al. (2015). Constructing a High Density Linkage Map for Gossypium Hirsutum× Gossypium barbadense and Identifying QTLs for Lint Percentage. J. Integr. Plant Biol. 5, 450–467. doi:10.1111/jipb.12288
Singh, B., Avci, U., Eichler Inwood, S. E., Grimson, M. J., Landgraf, J., Mohnen, D., et al. (2009). A Specialized Outer Layer of the Primary Cell Wall Joins Elongating Cotton Fibers into Tissue-like Bundles. Plant Physiol. 150, 684–699. doi:10.1104/pp.109.135459
Song, W., Wang, M., Su, W., Lu, Q., Xiao, X., Cai, J., et al. (2017). Genetic and Phenotypic Effects of Chromosome Segments Introgressed from Gossypium barbadense into Gossypium Hirsutum. PloS one 9, e0184882. doi:10.1371/journal.pone.0184882
Stemple, D. L. (2004). TILLING - a High-Throughput Harvest for Functional Genomics. Nat. Rev. Genet. 5, 145–150. doi:10.1038/nrg1273
Su, J., Li, L., Pang, C., Wei, H., Wang, C., Song, M., et al. (2016). Two Genomic Regions Associated with Fiber Quality Traits in Chinese upland Cotton under Apparent Breeding Selection. Sci. Rep. 6, 38496. doi:10.1038/srep38496
Su, J., Ma, Q., Li, M., Hao, F., and Wang, C. (2018). Multi-locus Genome-wide Association Studies of Fiber-Quality Related Traits in Chinese Early-Maturity upland Cotton. Front. Plant Sci., 1169. doi:10.3389/fpls.2018.01169
Sun, F.-D., Zhang, J.-H., Wang, S.-F., Gong, W.-K., Shi, Y.-Z., Liu, A.-Y., et al. (2012). QTL Mapping for Fiber Quality Traits across Multiple Generations and Environments in upland Cotton. Mol. Breed. 1, 569–582. doi:10.1007/s11032-011-9645-z
Sun, Z., Wang, X., Liu, Z., Gu, Q., Zhang, Y., Li, Z., et al. (2017). Genome-wide Association Study Discovered Genetic Variation and Candidate Genes of Fibre Quality Traits in Gossypium Hirsutum L. Plant Biotechnol. J. 15, 982–996. doi:10.1111/pbi.12693
Tan, Z., Fang, X., Tang, S., Zhang, J., Liu, D., Teng, Z., et al. (2015). Genetic Map and QTL Controlling Fiber Quality Traits in upland Cotton (Gossypium Hirsutum L.). Euphytica 3, 615–628. doi:10.1007/s10681-014-1288-9
Tan, Z., Zhang, Z., Sun, X., Li, Q., Sun, Y., Yang, P., et al. (2018). Genetic Map Construction and Fiber Quality QTL Mapping Using the CottonSNP80K Array in upland Cotton Frontiers in Plant Science. Front. Plant Sci., 225. doi:10.3389/fpls.2018.00225
Thomson, M., Tai, T., McClung, A., Lai, X., Hinga, M., Lobos, K., et al. (2003). Mapping Quantitative Trait Loci for Yield, Yield Components and Morphological Traits in an Advanced Backcross Population between Oryza Rufipogon and the Oryza Sativa Cultivar Jefferson Theor. Appl. Genet. 3, 479–493. doi:10.1007/s00122-003-1270-8
Vaughn, K. C., and Turley, R. B. (1999). The Primary walls of Cotton Fibers Contain an Ensheathing Pectin Layer. Protoplasma 209, 226–237. doi:10.1007/bf01453451
Wang, F., Zhang, J., Chen, Y., Zhang, C., Gong, J., Song, Z., et al. (2019b). Identification of Candidate Genes for Key Fibre-Related QTL S and Derivation of Favourable Alleles in Gossypium Hirsutum Recombinant Inbred Lines with G. barbadense Introgressions. Plant Biotechnol. J. 18, 707. doi:10.1111/pbi.13237
Wang, H., Huang, C., Guo, H., Li, X., Zhao, W., Dai, B., et al. (2015). QTL Mapping for Fiber and Yield Traits in upland Cotton under Multiple Environments. PLoS One 6. doi:10.1371/journal.pone.0130742
Wang, H., Huang, C., Zhao, W., Dai, B., Shen, C., Zhang, B., et al. (2016). Identification of QTL for Fiber Quality and Yield Traits Using Two Immortalized Backcross Populations in upland Cotton. PLoS One 12. doi:10.1371/journal.pone.0166970
Wang, H., Zhang, R., Shen, C., Li, X., Zhu, D., and Lin, Z. (2020). Transcriptome and QTL Analyses Reveal Candidate Genes for Fiber Quality in Upland Cotton. Crop J. 8, 98–106. doi:10.1016/j.cj.2019.05.002
Wang, L., Li, X.-R., Lian, H., Ni, D.-A., He, Y.-k., Chen, X.-Y., et al. (2010). Evidence that High Activity of Vacuolar Invertase Is Required for Cotton Fiber and Arabidopsis Root Elongation through Osmotic Dependent and Independent Pathways, Respectively. Respectively Plant Physiol. 154, 744–756. doi:10.1104/pp.110.162487
Wang, M., Tu, L., Yuan, D., Zhu, D., Shen, C., Li, J., et al. (2019a). Reference Genome Sequences of Two Cultivated Allotetraploid Cottons, Gossypium Hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi:10.1038/s41588-018-0282-x
Wang, P., Zhu, Y., Song, X., Cao, Z., Ding, Y., Liu, B., et al. (2012). Inheritance of Long Staple Fiber Quality Traits of Gossypium barbadense in G. Hirsutum Background Using CSILs Theor. Appl. Genet. 8, 1415–1428. doi:10.1007/s00122-012-1797-7
Wang, S., Wang, J.-W., Yu, N., Li, C.-H., Luo, B., Gou, J.-Y., et al. (2004). Control of Plant Trichome Development by a Cotton Fiber MYB Gene[W]. Plant Cell 16, 2323–2334. doi:10.1105/tpc.104.024844
Wendel, J. F., Brubaker, C., Alvarez, I., Cronn, R., and Stewart, J. M. (2009). “Evolution and Natural History of the Cotton Genus,” in Genetics and Genomics of Cotton (New York: Springer), 3–22. doi:10.1007/978-0-387-70810-2_1
Wendel, J. F. (1989). New World Tetraploid Cottons Contain Old World Cytoplasm. Proc. Natl. Acad. Sci. 86, 4132–4136. doi:10.1073/pnas.86.11.4132
Wenliang, X., Huang, G., Wang, X., Wang, H., and Xuebao, L. (2006). Molecular Characterization and Expression Analysis of Five Novel Genes Encoding Proline-Rich Proteins in Cotton. Gossypium hirsutum) Prog. Biochem. Biophys. 05.
Wu, Y., Machado, A. C., White, R. G., Llewellyn, D. J., and Dennis, E. S. (2006). Expression Profiling Identifies Genes Expressed Early during Lint Fibre Initiation in Cotton. Plant Cel Physiol. 47, 107–127. doi:10.1093/pcp/pci228
Yang, X., Wang, Y., Zhang, G., Wang, X., Wu, L., Ke, H., et al. (2016). Detection and Validation of One Stable Fiber Strength QTL on C9 in Tetraploid Cotton. Mol. Genet. Genomics 291, 1625–1638. doi:10.1007/s00438-016-1206-z
Yang, Z., Ge, X., Yang, Z., Qin, W., Sun, G., Wang, Z., et al. (2019). Extensive Intraspecific Gene Order and Gene Structural Variations in upland Cotton Cultivars. Nat. Commun. 10, 2989–3013. doi:10.1038/s41467-019-10820-x
Yang, Z., Li, J., Li, A., Zhang, B., Liu, G., Li, J., et al. (2009). Developing Chromosome Segment Substitution Lines (CSSLs) in Cotton (Gossypium) Using Advanced Backcross and MAS. Mol. Plant Breed 7 (2), 233–241.
Yang, Z., Zhang, C., Yang, X., Liu, K., Wu, Z., Zhang, X., et al. (2014). PAG1, a Cotton Brassinosteroid Catabolism Gene, Modulates Fiber Elongation. New Phytol. 203, 437–448. doi:10.1111/nph.12824
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2016). The Genome Sequence of Sea-Island Cotton (Gossypium Barbadense) Provides Insights into the Allopolyploidization and Development of Superior Spinnable Fibres Scientific Reports. Sci. Rep., 17662. doi:10.1038/srep17662
Zeng, L., Meredith, W. R., Gutiérrez, O. A., and Boykin, D. L. (2009). Identification of Associations between SSR Markers and Fiber Traits in an Exotic Germplasm Derived from Multiple Crosses Among Gossypium Tetraploid Species Theor. Appl. Genet. 1, 93–103. doi:10.1007/s00122-009-1020-7
Zhang, B., Yang, Y.-W., Zhang, Y., and Liu, J.-Y. (2013a). A High-Confidence Reference Dataset of Differentially Expressed Proteins in Elongating Cotton Fiber Cells. ProteomicsProteomics 13, 1159–1163. doi:10.1002/pmic.201200176
Zhang, C., Li, L., Liu, Q., Gu, L., Huang, J., Wei, H., et al. (2019b). Identification of Loci and Candidate Genes Responsible for Fiber Length in upland Cotton (Gossypium Hirsutum L.) via Association Mapping and Linkage Analyses. Front. Plant Sci., 53. doi:10.3389/fpls.2019.00053
Zhang, C., Guo, L., Wang, X., Zhang, H., Shi, H., Xu, W., et al. (2007). Genomics Mol. characterization four ADF genes differentially expressed cotton 4, 347–354. doi:10.1016/s1673-8527(07)60037-x
Zhang, F., Zuo, K., Zhang, J., Liu, X., Zhang, L., Sun, X., et al. (2010). An L1 Box Binding Protein, GbML1, Interacts with GbMYB25 to Control Cotton Fibre Development. J. Exp. Bot. 61, 3599–3613. doi:10.1093/jxb/erq173
Zhang, J., Wu, M., Yu, J., Li, X., and Pei, W. (2016a). Breeding Potential of Introgression Lines Developed from Interspecific Crossing between upland Cotton (Gossypium Hirsutum) and Gossypium barbadense: Heterosis, Combining Ability and Genetic Effects. PLoS One 1. doi:10.1371/journal.pone.0143646
Zhang, J. F., Abdelraheem, A., and Wu, J. X. (2017). Heterosis, Combining Ability and Genetic Effect, and Relationship with Genetic Distance Based on a Diallel of Hybrids from Five Diverse Gossypium barbadense Cotton Genotypes. Euphytica 213, 208. doi:10.1007/s10681-017-1997-y
Zhang, M., Zhang, Y., Huang, J. J., Zhang, X., Lee, M. K., Stelly, D. M., et al. (2012). Genome Physical Mapping of Polyploids: a BIBAC Physical Map of Cultivated Tetraploid Cotton, Gossypium Hirsutum L. PLoS One 7, e33644. doi:10.1371/journal.pone.0033644
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015a). Sequencing of Allotetraploid Cotton (Gossypium Hirsutum L. Acc. TM-1) Provides a Resource for Fiber Improvement. Nat. Biotechnol. 33, 531–537. doi:10.1038/nbt.3207
Zhang, T., Yuan, Y., Yu, J., Guo, W., and Kohel, R. J. (2003). Molecular Tagging of a Major QTL for Fiber Strength in Upland Cotton and its Marker-Assisted Selection. Theor. Appl. Genet. 106, 262–268. doi:10.1007/s00122-002-1101-3
Zhang, Y., Changhui, F., Shu, B., Xiaogang, W., Zhang, J., Songbo, X., et al. (2018). Analysis of Short Fruiting branch Gene and Marker-Assisted Selection with SNP Linked to its Trait in upland Cotton. J. Cotton Res. 1, 5. doi:10.1186/s42397-018-0001-2
Zhang, Z., Li, J., Jamshed, M., Shi, Y., Liu, A., Gong, J., et al. (2020). Genome Wide Quantitative Trait Loci Reveal the Genetic Basis of Cotton Fibre Quality and Yield Related Traits in a Gossypium Hirsutum Recombinant Inbred Line Population. Plant Biotechnol. J. 1, 239–253. doi:10.1111/pbi.13191
Zhao, Q., and Dixon, R. A. (2011). Transcriptional Networks for Lignin Biosynthesis: More Complex Than We Thought? Trends Plant Sci. 16, 227–233. doi:10.1016/j.tplants.2010.12.005
Zhou, G., Zhu, Q., Yang, G., Huang, J., Cheng, S., Yue, B., et al. (2015b). 2 Is a Pleiotropic QTL for Kernel Number Per Row, Ear Length and Ear Weight in maize (Zea mays L.). Euphytica 2, 429–436. doi:10.1007/s10681-014-1307-x
Zhou, Y., Zhang, Z.-T., Li, M., Wei, X.-Z., Li, X.-J., Li, B.-Y., et al. (2015a). Cotton (Gossypium Hirsutum) 14-3-3 Proteins Participate in Regulation of Fibre Initiation and Elongation by Modulating Brassinosteroid Signalling. Plant Biotechnol. J. 13, 269–280. doi:10.1111/pbi.12275
Zhu, H., Han, X., Lv, J., Zhao, L., Xu, X., Zhang, T., et al. (2011). Structure, Expression Differentiation and Evolution of Duplicated Fiber Developmental Genes in Gossypium barbadense and G. Hirsutum. BMC Plant Biol. 11, 40. doi:10.1186/1471-2229-11-40
Zhu, H., Lv, J., Zhao, L., Tong, X., Zhou, B., Zhang, T., et al. (2012). Molecular Evolution and Phylogenetic Analysis of Genes Related to Cotton Fibers Development from Wild and Domesticated Cotton Species in Gossypium. Mol. Phylogenet. Evol. 63, 589–597. doi:10.1016/j.ympev.2012.01.025
Zhu, H., Han, X., Lv, J., Zhao, L., Xu, X., Zhang, T., et al. (2011). Structure, Expression Differentiation and Evolution of Duplicated Fiber Developmental Genes in Gossypium barbadense and G. hirsutum. BMC Plant Biol. 1, 40.
Keywords: fiber quality, cotton fiber development, chromosome 7 significance, Gossypium species, stable QTLs, SNPs
Citation: Razzaq A, Zafar MM, Ali A, Hafeez A, Sharif F, Guan X, Deng X, Pengtao L, Shi Y, Haroon M, Gong W, Ren M and Yuan Y (2022) The Pivotal Role of Major Chromosomes of Sub-Genomes A and D in Fiber Quality Traits of Cotton. Front. Genet. 12:642595. doi: 10.3389/fgene.2021.642595
Received: 16 December 2020; Accepted: 25 October 2021;
Published: 24 March 2022.
Edited by:
Maximiller Dal-Bianco, Universidade Federal de Viçosa, BrazilReviewed by:
Waltram Ravelombola, Texas A&M University, United StatesGhulam Mustafa, University of Agriculture, Faisalabad, Pakistan
Muhammad Irfan, Forman Christian College, Pakistan
Copyright © 2022 Razzaq, Zafar, Ali, Hafeez, Sharif, Guan, Deng, Pengtao, Shi, Haroon, Gong, Ren and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdul Razzaq, biolformanite@gmail.com; Youlu Yuan , yuanyoulu@caas.cn; Maozhi Ren, renmaozhi01@caas.cn
†These authors have contributed equally to this work
 Abdul Razzaq
Abdul Razzaq Muhammad Mubashar Zafar
Muhammad Mubashar Zafar Arfan Ali4
Arfan Ali4 Abdul Hafeez
Abdul Hafeez Maozhi Ren
Maozhi Ren Youlu Yuan
Youlu Yuan